Abstract
Apoptosis is a fundamental process required for proper embryonic development. Various methods have been described to detect apoptosis both in vitro as well as in vivo. Activation of caspases represents the key event in the apoptotic process. To dissect the molecular events leading to caspase activation, we have been using cell-free systems that recapitulate the mitochondrial death pathway. In the cell-free apoptosis assays, we either detect caspase activation in stimulated cells by utilizing subcellular fractions or reconstitute various components in cytosol (or mitochondria) to study molecular mechanisms of caspase activation. In either case, we utilize Western blot and/or substrate cleavage to monitor caspase activation. Using in vitro reconstitution approach of caspase activation, we have discovered various factors that regulate caspase activity. Therefore, cell-free system not only is an invaluable tool to study apoptosis signaling but also provides molecular insight on caspase activation patterns and inhibitor specificities.
Keywords: Apoptosis, Apoptosome, Cytochrome c, Cell-Free Reconstitution, Substrate Cleavage, Caspase Activation, Mitochondria, Cytoplasm, Apaf-1
1. Introduction
Apoptosis plays an essential role in animal development and in maintaining the homeostasis of adult tissues (1). Deficiency in apoptosis is a hallmark of cancer and autoimmune diseases whereas excessive apoptosis is implicated in neurodegenerative diseases, strokes and cardiac diseases. The family of caspases (cysteine aspartic acid-specific protease) is the key effectors in the execution of apoptotic cell death (2). Caspases are synthesized as inactive proenzymes, which become proteolytically cleaved during apoptosis to generate active enzymes. Activated caspases then cleave cellular proteins such as poly(ADP-ribose) polymerase (PARP) to dismantle the dying cells (3). In response to stress, cells release cytochrome c from the intermembrane space of the mitochondria to the cytosol. The released cytochrome c binds to and activates the adaptor protein Apaf-1, which in turn activates the initiator procaspase-9 in the presence of ATP, leading to the formation of apoptosome and subsequent activation of “executioner” caspases such as caspase-3, 6, or 7 (4).
We have been using cell-free systems to detect apoptotic activity/caspase activation in cytosolic or mitochondrial extracts (5-10). We generally use two approaches to detect apoptosis. In the first, we isolate cytosolic or mitochondrial extracts from cells that have been treated in culture with an apoptosis-inducing agent. In the second, purified cytosolic extracts from untreated cells is used in reconstitution experiments with addition of bovine cytochrome c or recombinant active caspases. Apoptotic activity in these extracts can be examined by the measurement of enzymatic caspase activity, and/or by Western blots of proteins processed during apoptosis (i.e., caspases and their substrates). It was in 1993 when the first paper described that a cell-free system could mimic characteristic features of apoptosis in intact cells (11). Later, many other investigators have used cell-free systems successfully for dissection of biochemical mechanisms during the apoptotic process, such as the identification and characterization of the 'apoptosome', AIF (apoptosis-inducing factor), and the DNA fragmentation factor ICAD (12-14). Here we describe our protocols for the detection of caspase activation in cell-free systems (5-10).
2. Materials
2.1. Cell Culture and Subcellular Fractionation
For cell culture we used Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco Grand Island, NY) supplemented with 10% fetal bovine serum (FBS, HyClone) and 1% Penicillin and Streptomycin (see Note 1).
Staurosporine (Sigma, St. Louis, MO), dissolved in tissue-culture grade dimethyl sulfoxide (DMSO) at 1 mM, stored in aliquots at -20°C, and then added to cell-culture dishes as required.
Solution of trypsin (0.25%) and ethylenediamine tetraacetic acid (EDTA) (1 mM) from Gibco/BRL used for harvesting cells from the dishes.
1X Phosphate buffered saline (PBS): 137 mM sodium chloride (NaCl), 2.7 mM potassium chloride (KCl), 4.3 mM disodium hydrogen phosphate (Na2HPO4.7H20), 1.4 mM potassium dihydrogen phosphate (KH2PO4).
Teflon cell scrapers (Fisher Scientific).
Homogenizing (hypotonic) buffer: 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM Sodium EGTA, 1 mM DTT, 250 mM sucrose and mixture of protease inhibitors (Sigma).
Dounce homogenizer using high clearance pestle from Fischer Scientific.
TNC buffer (10 mM Tris Acetate, pH 8.0, 0.5 % NP-40, 5 mM CaCl2).
Small-volume ultracentrifugation tubes (i.e., less than 5 ml; Beckman Coulter, Inc.)
2.2. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Micro-BCA Protein Assay Kit (Pierce Biotechnology, Inc.)
For resolving gel: 1.5 M Tris-HCl, pH 8.8, 10% sodium dodecyl sulfate (SDS). Store at room temperature (see Note 2).
For stacking gel: 1.0 M Tris-HCl, pH 6.8, 10 % SDS. Store at room temperature.
Thirty percent acrylamide/bis solution (in 29:1 ratio in deionized distilled water) and N,N,N,N'-Tetramethyl-ethylenediamine, TEMED (Bio-Rad, Hercules, CA) (see Note 3).
Ammonium persulfate: prepare 10% solution in distilled water and immediately freeze in single use (200 μl) aliquots at -20°C.
Running buffer: 25 mM Tris, 250 mM glycine, 0.1% (w/v) SDS. Prepare 5X or 10 X and store at room temperature or alternatively could be purchased from BioRad.
Prestained molecular weight markers: Low-range markers (Bio-Rad).
6X SDS gel-loading buffer: 350 mM Tris-HCl, pH 6.8, 10 % (w/v) SDS, 30 % (w/v) glycerol, 9.25 % dithiothreitol (DTT), 0.02% (w/v) bromophenol blue. Make 0.5 ml aliquots and store at -80°C (See Note 4).
2.3. Western Blotting
Transfer Buffer: 24 mM Tris (do not adjust pH), 192 mM glycine, 20% (v/v) methanol.
Supported nitrocellulose membrane from BioRad, 3 MM chromatography paper from Fisher Scientific.
Tris-buffered saline with Tween 20 (TBS-T): Prepare 10X stock with 1.37 M NaCl, 200 mM Tris-HCl, pH 7.5; store at room temperature. Before using, make 1X solution in distilled water with addition of 0.1% Tween-20.
Blocking buffer: 5% (w/v) nonfat dry milk in TBS-T.
Primary and secondary antibody dilution buffer: TBS-T supplemented with 3% (w/v) nonfat dry milk.
Secondary antibody: Anti-rabbit or mouse IgG (depending on the primary antibody) conjugated to horseradish peroxidase (Amersham Biosciences).
Enhanced chemiluminescence (ECL) reagents from Amersham Biosciences.
Autoradiography X-ray film from Fisher Scientific.
2.4. Stripping and Reprobing Blots for Caspase-3 and Actin
Stripping buffer: 62.5 mM Tris-HCl, pH 6.8, 2% (w/v) SDS. Store at room temperature. Warm to working temperature of 55°C and add 100 mM β-mercaptoethanol (see Note 5).
Primary antibody: Anti-caspase-9 (Chemicon), anti-caspase-3 (Biomol), and ant-actin (ICN).
2.5. Substrate Cleavage Assay for Caspases
Caspase reaction buffer: 50 mM HEPES, pH 7.4, 100 mM NaC1, 0.1% CHAPS, 10 mM DTT, 1 mM EDTA, 10% glycerol. Always prepare fresh reaction buffer.
Ac-DEVD-AFC and Ac-LEHD-AFC (Biomol) dissolved in DMSO to the stock concentration of 10 mM and make aliquots and store at −80°C (see Note 6).
7-amino-4-trifluoromethyl-coumarin (AFC) from Sigma.
3. Methods
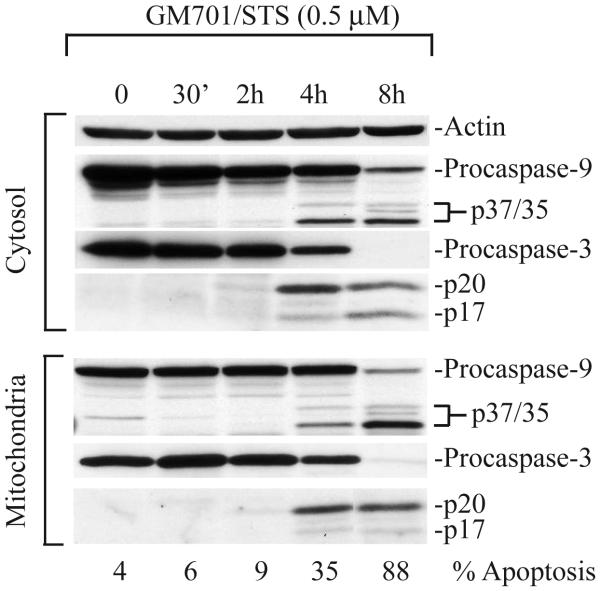
During apoptosis, procaspase-9 (~46 kD) is processed to generate the p37/p35 kD fragments. In our experiments, we have utilized an antibody that recognizes both the proform and the p37/p35 bands. As illustrated in Fig. 1, in GM701 fibroblasts treated with staurosporine (STS), the p37/p35 fragments were detected in cytosolic as well as in the mitochondrial fractions. Similarly, procaspase-3 (~32 kD) is processed to the p20/p17 bands, the latter representing catalytically active caspase-3 (5-10; Fig. 1). In such Western blotting assays, apoptosis should preferably be quantified side by side using DAPI staining to identify apoptotic nuclei (Fig. 1, bottom). This is important because cleavage of procaspase-9 does not indicate that the enzyme is active although procaspase-3 cleavage does suggest its proteolytic activation.
Figure 1.
Proteolytic processing of procaspase-9 and -3 in GM701 cells treated with STS. Thirty (cytosol) or 60 (mitochondria) μg of proteins was used in Western blotting for caspase-9, caspase-3, or actin. Modified from ref. 6.
In vitro reconstitution experiment is a relatively novel approach to mimic and study caspase activation in vivo. Using freshly purified cytosol, we could readily reconstitute caspase activation with the addition of cytochrome c alone (Fig. 2). Many other investigators have used dATP or ATP (around 1 mM) together with cytochrome c to initiate caspase processing in such reconstitution systems. We, on the other hand, have found that freshly purified cytosols contain sufficient amount of dATP or ATP (generally in mM range) to support cytochrome c-initiated caspase activation. Below we describe our general protocol for cell-free caspase activation analyzed by Western blotting and/or LEHDase/DEVDase activity assays (Fig. 2).
Figure 2.
Cytochrome c initiates caspase activation without addition of dATP or ATP. Fresh GM701 cytosol (3 μg/μl) was incubated with cytochrome c (15 μg/ml) for the time periods indicated. At the end, Western blotting was performed to detect procaspase-9 and -3 processing (A). Fifty μg of reaction mixture was also used to determine LEHDase and DEVDase activities (B). Modified from ref. 9.
3.1. Subcellular Fractionation
Treat cultured cells (e.g., GM701; ~10 million) with an apoptotic stimulus (e.g., staurosporine) or vehicle control. Harvest (using a cell scraper or trypsin/EDTA) and wash both treated and mock-treated cells twice with ice-cold 1X PBS.
Suspend washed cells in 600 μl of homogenizing (hypotonic) buffer and incubate on ice for 30 min.
Homogenize the cell suspension with a Dounce homogenizer using high clearance pestle (140 strokes) (see Note 7).
Centrifuge at 1,000 g for 5 min to remove nuclei and unbroken cells (see Note 8).
Centrifuge the resulting supernatant again at 10,000 g for 20 min at 4°C to obtain the pellet, which is enriched in mitochondria.
The resulting supernatant is further subjected to ultracentrifugation at 100,000 g for 1 hr at 4°C to obtain cytosol (or S100).
Mitochondrial fractions are washed thrice in homogenizing buffer and then solubilized in 60 μl of TNC buffer containing protease inhibitors (see note 9).
Measure protein concentrations of the prepared mitochondrial and cytosolic fractions using Micro BCA Protein Assay Kit.
3.2. Cell-free Reconstitution Experiments
Cell-free reactions are performed in homogenizing buffer in a total volume of 100 μl.
Purified cytosols (3 mg/ml) are activated by adding bovine cytochrome c (15 μg/ml; Sigma) without (d)ATP and incubated at 37°C for 150 min (see Note 10).
After incubation, samples are used for either substrate cleavage assays for caspase-9 (LEHDase) and caspase-3 (DEVDase) or procaspase cleavage by Western blotting.
3.3. Preparation of SDS-PAGE Gels
Clean the glass plates thoroughly with a rinsable detergent, rinse extensively with distilled water, and assemble according to the manufacturer’s instructions.
Depending upon the size of apparatus, prepare 10 ml reaction mix for 15% resolving gel by mixing in a 50 ml disposable plastic tube or conical flask in following order: 2.3 ml distilled water, 5.0 ml of 30% acrylamide solution, 2.5 ml of 1.5 M Tris-Cl, pH 8.8, 0.1 ml SDS, and 0.1 ml ammonium persulfate; mix and then add 4 ml of TEMED. Mix immediately and proceed to the next step. Polymerization begins as soon as TEMED is added.
Using Pasteur pipette, pour the above acrylamide solution into the gap between the glass plates. Leave one cm space below the length of the comb for stacking gel. Gel should be in vertical position and overlay a thin layer of distilled water. Leave the gel at room temperature for 30 min to polymerize.
Pour off the water and wash several times with water to remove unpolymerized acrylamide and drain all the liquid using paper towels.
Depending on the size of gel, prepare stacking gel by mixing 2.7 ml distilled water, 0.67 ml acrylamide, 0.5 ml 1.0 M Tris, pH 6.8, 40 μl of 10% SDS, 40 μl of ammonium persulfate, and then add 4 μl of TEMED. Mix immediately and, without delay, pour the stacking gel solution directly on the polymerized resolving gel. Immediately insert the comb while avoiding air bubbles, add more stacking gel to fill the spaces of the comb completely, and leave it at room temperature for 30 min to polymerize.
3.3 Preparation of Samples and Running Gels
While stacking gels is polymerizing, take 20-50 μg protein (from mitochondrial or cytosolic fraction) per lane in a total volume of 30 μl (for 18-well gel) or 40 μl (for 12-well gel). Make up the volume with 1XPBS. For Western blotting of the reconstitution experiments, 30 - 40 μl of reaction samples after incubation is used.
Add 6 μl (for 18-well gel) or 8 μl (for 12-well gel) of 6X SDS-loading buffer. Boil for 5-10 min in a heating block and centrifuge for 1 min to collect samples to the bottom of the tubes.
Once the stacking gel has set, carefully remove the comb and use a 3-ml syringe fitted with a 22-gauge needle to wash the wells with running buffer. Mount the gel in electrophoresis apparatus and add the Tris-Glycine running buffer to the upper and lower chambers of the gel unit and remove any trapped air bubbles at the bottom of the gel or in the wells.
Load 30 μl or 40 μl sample (depending on the capacity of the wells) onto 15% SDS-PAGE gels. Use one well for low-range prestained protein markers. Also load 1X SDS sampleloading buffer in any empty wells.
Attach the electrophoresis apparatus to power supply and first run at 80 Volts for 20-30 min. When the bromophenol blue dye has moved to resolving gel increase the voltage to 120 Volts and run the gel until the dye reaches the bottom. This process generally takes 2-3 hr.
3.4 Transfer of Proteins and Western Blotting
While the SDS-PAGE gel is still running, prepare transfer buffer and keep in cold room.
Soak chromatography paper and fiber pads in transfer buffer 10-20 min before start of transfer and also soak nitrocellulose membrane in transfer buffer. If PVDF membrane is to be used, soak in 100% methanol for 5- 10 min.
Once the bromophenol blue dye has reached the bottom of the gel, disconnect power supply, remove the gel from the gel holding apparatus and cut out the stacking gel. Detach the dye-containing gel at the bottom and wash for five minutes in transfer buffer on a rotating shaker.
Arrange the transfer cassettes in following order: Black side of cassette on bottom, fiber pad, single sheet of same-sized chromatography paper, gel (marker side of the gel on right), nitrocellulose membrane, one sheet of chromatography paper and then fiber pad. Close the cassette and avoid and remove air bubbles in every step.
Submerge the resulting cassette sandwich in a transfer tank that contains transfer buffer. The cassette is placed into the transfer tank such that the nitrocellulose membrane is between the gel and the anode. This orientation is very critical otherwise the proteins will be lost from the gel into the buffer rather than transferred to the nitrocellulose membrane.
Insert a small magnet and ice pack in the transfer tank and run at 100 Volts with slow stir of the magnetic stirrer for 1-2 h depending on the molecular weights of the proteins to be transferred. For proteins up to 50 kDa, a 75-minute transfer should be sufficient.
After completion of the transfer, cut the lower right-hand side of membrane before taking it out and this will become lower left-hand side to mark the transfer side (i.e., protein transfer side is up) and wash two times with 1X TBS-T for 5 min each.
Block the membrane with 5% non-fat dry milk in 1X TBS-T for 1 hr at room temperature on a rocking platform. At the end of incubation, wash the membrane one time with 1X TBS-T for 5 min.
Probe with rabbit polyclonal antibody for caspase-9 from Chemicon (Cat # AB16970) diluted (1000X) in 1X TBS-T containing 3% non-fat dry milk for 2 hr at room temperature. At the end of incubation, wash the membrane four times with 1X TBS-T for 10 min each.
Probe with secondary antibody, rabbit IgG conjugated to horseradish peroxidase, diluted (5000X) in 1X TBS-T containing 3% non-fat dry milk, for 1 hr at room temperature. After incubation, wash the membrane four times with 1X TBS-T for 10 min each.
During washing, 2 ml aliquots of ECL reagents (i.e., 2 ml of ECL A and B) are warmed separately at room temperature. Just before use mix ECL reagents in equal ratio, pour directly on the membrane and incubate for one minute and then immediately exposed to X-ray film to detect signals. ECL incubation and detection should be performed at room temperature in a dark room having safe red light (see note 11).
3.5. Reprobing the Membrane Blots for Caspase-3 and Marker Proteins
After completion of caspase-9 Western blotting and once a satisfactory exposure for the result of the processed caspase-9 has been obtained, the membrane is stripped and then reprobed one by one with antibodies that recognize the processed caspase-3 and actin, respectively, for a loading control that confirms equal recovery of the samples through the procedure. If the molecular weights of the target molecules are very different, the two (or more) antibodies can be added simultaneously for reprobing.
Stripping buffer (50 ml per blot—see Note 5) is warmed to 55°C and then β-mercaptoethanol is added. The blot is incubated for 30 min with continuous slow agitation.
Once the blot is stripped, it is extensively washed in TBS-T buffer (three times with 50 ml for each wash for 10 min), and then blocked again in blocking buffer.
The membrane is then ready to be reprobed with anti-caspase-3 (1:3000 in TBS-T) with washes, secondary antibody, and ECL detection as described above. This process is repeated for actin (1:5000) or any other molecule(s). When properly done, the stripping-reprobing process can be repeated for up to 5-8 times. Some examples are shown in Fig. 1 and Fig. 2.
3.6. LEHDase (for caspase-9) and DEVDase (for caspase-3) Activity Measurement
For caspase activity measurement, 30-50 μg of mitochondrial or cytosolic proteins is added to a reaction mixture containing 30 μM fluorogenic peptide substrates, Ac-DEVD-AFC or Ac-LEHD-AFC in a total volume of 100 μl.
Similarly, at the end of reconstitution experiments, 30-50 μg of reconstituted sample is added to the reaction mixture described above.
Production of 7-amino-4-trifluoromethyl-coumarin (AFC) is monitored in a spectrofluorimeter (Hitachi F-2000 fluorescence spectrophotometer) using excitation wavelength 400 nm and emission wavelength 505 nm (see note 12).
The fluorescent units are converted into nanomoles of AFC released per hour per milligram of protein using a standard curve. The results are generally presented as fold activation over the control (Fig. 2).
Acknowledgements
This work was supported in part by an NIH K01 award to DC (7K01CA123142) and by grants from NIH (R01-AG023374, R01-ES015888, and R21-ES015893-01A1), Department of Defense (W81XWH-07-1-0616 and PC073751), and Elsa Pardee Foundation to DGT.
Footnotes
Fetal bovine serum should be heat-inactivated in waterbath prior to use at 56°C for 30 min and make aliquots in a 50 ml disposable plastic tube.
Wear gloves and mask while handling SDS powder to prevent inhalation of the fine powder. Alternatively, premade polyacrylamide gels can be purchased from commercial sources.
Acrylamide is highly hazardous (neurotoxic) and should not be purchased in a powder form unless absolutely necessary. It is now available in premixed form from various suppliers. Always take precaution while handling unpolymerized acrylamide. TEMED should be stored at room temperature in a desiccator.
The 6X SDS-loading buffer, when stored as aliquots at −80°C, is stable for up to one year. Repeated freezing and thawing is not recommended.
2-β-mercaptoethanol is toxic and gives a very unpleasant smell in the laboratory. Use tight container and proper care while handling it.
Ac-DEVD-AFC, Ac-LEHD-AFC and AFC are light sensitive.
When homogenizing cells, take care not to overhomogenize because this will damage mitochondria and cytochrome c will leak out in control cells also. To prevent overhomogenization, monitor cells under a microscope every 50 strokes to achieve an optimal 60-80 % of cell breakage. Do not try to achieve 100% cell breakage.
Take 2 μl of supernatant and observe under a microscope. If some nuclei or unbroken cells are observed in the supernatant, recentrifuge for 5 min at 1,000 g.
Decrease or increase the amount of TNC buffer to obtain desired concentration of mitochondrial lysates.
Various investigators use 1 mM dATP or ATP to reconstitute caspase activation in cell-free system. We find that dATP or ATP is not required for cytochrome c-initiated caspase activation when fresh cytosol is used in such assays.
If signal is very weak with ECL, ECL plus could be used as alternative detection reagent.
It is very important to use proper filter for excitation (400 nm) and emission (505 nm) for caspase activity measurement.
References
- 1.Horvitz HR. Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res. 1999;59:1701S–1706S. [PubMed] [Google Scholar]
- 2.Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- 3.Thornberry NA, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 4.Wang X. The expanding role of mitochondria in apoptosis. Genes & Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- 5.Chandra D, Liu JW, Tang DG. Early mitochondrial activation and cytochrome c up-regulation during apoptosis. J. Biol. Chem. 2002;277:50842–54. doi: 10.1074/jbc.M207622200. [DOI] [PubMed] [Google Scholar]
- 6.Chandra D, Tang DG. Mitochondrially localized active caspase-9 and caspase-3 result mostly from translocation from the cytosol and partly from caspase-mediated activation in the organelle. Lack of evidence for Apaf-1-mediated procaspase-9 activation in the mitochondria. J. Biol. Chem. 2003;278:17408–20. doi: 10.1074/jbc.M300750200. [DOI] [PubMed] [Google Scholar]
- 7.Chandra D, Choy G, Deng X, Bhatia B, Daniel P, Tang DG. Association of active caspase 8 with the mitochondrial membrane during apoptosis:potential roles in cleaving BAP31 and caspase 3 and mediating mitochondrion-endoplasmic reticulum cross talk in etoposide-induced cell death. Mol.Cell. Biol. 2004;24:6592–607. doi: 10.1128/MCB.24.15.6592-6607.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandra D, Choy G, Daniel PT, Tang DG. Bax-dependent regulation of Bak by voltage-dependent anion channel 2. J. Biol. Chem. 2005;280:19051–61. doi: 10.1074/jbc.M501391200. [DOI] [PubMed] [Google Scholar]
- 9.Chandra D, Bratton SB, Person MD, Tian Y, Martin AG, Ayres M, Fearnhead HO, Gandhi V, Tang DG. Intracellular nucleotides act as critical prosurvival factors by binding to cytochrome C and inhibiting apoptosome. Cell. 2006;125:1333–46. doi: 10.1016/j.cell.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 10.Chandra D, Choy G, Tang DG. Cytosolic accumulation of HSP60 during apoptosis with or without apparent mitochondrial release: evidence that its pro-apoptotic or pro-survival functions involve differential interactions with caspase-3. J. Biol. Chem. 2007;282:31289–301. doi: 10.1074/jbc.M702777200. [DOI] [PubMed] [Google Scholar]
- 11.Lazebnik YA, Cole S, Cooke CA, Nelson WG, Earnshaw WC. Nuclear events of apoptosis in vitro in cell-free mitotic extracts: a model system for analysis of the active phase of apoptosis. J. Cell. Biol. 1993;123:7–22. doi: 10.1083/jcb.123.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–13. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 13.Susin SA, Lorenzo K, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–6. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 14.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]