Abstract
Background
Rhabdoid tumors (RTs) are aggressive tumors of early childhood that occur most often in brain (AT/RTs) or kidney (KRTs). Regardless of location, they are characterized by loss of functional SMARCB1 protein, a component of the SWI/SNF chromatin remodeling complex. The aim of this study was to determine genes and biological process dysregulated in common to both AT/RTs and KRTs.
Procedure
Gene expression for AT/RTs was compared to that of other brain tumors and normal brain using microarray data from our lab. Similar analysis was performed for KRTs and other kidney tumors and normal kidney using data from GEO. Dysregulated genes common to both analyses were analyzed for functional significance.
Results
Unsupervised hierarchical clustering of RTs identified 3 major subsets: 2 comprised of AT/RTs, and 1 of KRTs. Compared to other tumors, 1187, 663 and 539 genes were dysregulated in each subset, respectively. Only 14 dysregulated genes were common to all 3 subsets. Compared to normal tissue, 5209, 4275 and 2841 genes were dysregulated in each subset, with an overlap of 610 dysregulated genes. Among these genes, processes associated with cell proliferation, MYC activation, and epigenetic dysregulation were common to all 3 RT subsets.
Conclusions
The low overlap of dysregulated genes in AT/RTs and KRTs suggests that factors in addition to SMARCB1 loss play a role in determining subsequent gene expression. Drugs which target cell cycle or epigenetic genes may be useful in all RTs. Additionally, targeted therapies tailored to specific RT subset molecular profiles should be considered.
Keywords: atypical teratoid/rhabdoid tumor, INI-1, kidney rhabdoid tumor, microarray, SMARCB1
INTRODUCTION
Rhabdoid tumors (RTs) are aggressive tumors occurring most frequently in children under 2 years of age. They often occur in the kidneys (KRTs) or CNS (Atypical teratoid / rhabdoid tumors, AT/RTs). The molecular hallmark of RTs is loss of functional SMARCB1 protein (aka INI-1, hSNF5, BAF47), a component of the SWI/SNF chromatin remodeling complex [1, 2]. Survival is poor even with current treatments such as high-dose chemotherapy or multimodal treatment based on sarcoma regimens [3–5]. Furthermore, these therapies result in significant long-term morbidity and risk treatment-related death.
To better understand the biology driving RTs, a recent study compared gene expression of KRTs to other pediatric kidney tumors [6]. They found many down-regulated genes in KRTs were associated with epigenetic repression by polycomb repressive complexes (PRCs) in embryonic stem cells (ESC). The oncogene MYC (c-MYC), as well as many of its targets, was up-regulated.
It is unknown whether these findings in KRTs apply to RTs from other locations. One study compared gene expression in a group of 10 RTs (5 AT/RTs, 3 KRTs and 2 extra-renal RTs) to other brain tumors and concluded they were more molecularly similar to each other than to other brain tumors [7]. Another study, however, identified numerous differences in cell surface markers between an AT/RT-derived cell line compared to a soft-tissue abdominal RT cell line using flow cytometry [8]. Immunostaining of tissue microarrays from AT/RTs and non-CNS RTs showed similar protein expression for 20 ESC-related genes, but variability for genes in the p16INK4A and p14ARF pathways [9, 10].
None of these studies has focused on comprehensively addressing similarities and differences among RTs from different locations. This knowledge is particularly pertinent to the design of clinical trials involving RTs. It would also shed light on fundamental processes associated with the role of SMARCB1 loss in tumorigenesis. To address this question, we used our extensive bank of microarray data to compare the gene expression profiles of AT/RT tumor samples to those of other WHO grade IV pediatric brain tumors and normal brain. We performed a similar analysis using published microarray data for KRTs, other kidney tumors and normal kidney [6]. Finally, we analyzed the overlaps to identify common dysregulated genes and molecular processes and possible therapeutic targets among both AT/RTs and KRTs.
MATERIALS AND METHODS
Patient samples
Twenty AT/RTs, 42 WHO grade IV pediatric brain tumors (21 medulloblastomas (MEDs), 9 primitive neuroectodermal tumors (PNETs), and 12 glioblastomas (GBMs)), and 9 non-tumor normal brain samples, all collected at Children’s Hospital, Colorado, were used for this study. All AT/RTs were verified by immunohistochemistry to show loss of BAF47 (SMARCB1). Eighteen of the 20 AT/RTs have been previously published. All tumors were collected at initial surgery and snap-frozen. [11]. Sample preparation and microarray processing were performed for all samples as previously described [11]. Microarray data are available from GEO (http://www.ncbi.nlm.nih.gov/geo/), accession number GSE35493.
Microarray data for 10 KRTs, 16 kidney clear cell sarcomas, 12 cellular mesoblastic nephromas, and 15 Wilms tumors were downloaded from GEO (accession GSE11482). These samples were previously published by another lab comparing gene expression in KRTs to other pediatric kidney tumors [6]. As this dataset did not include normal kidney samples, additional data for normal kidney were selected from several studies in GEO (GSE2004, GSE6280, GSE15641). All downloaded kidney data were normalized together in the same manner as the brain samples, using gcRMA.
All kidney samples were profiled on Affymetrix U133A GeneChips (~13,000 annotated genes). However, the brain samples used Affymetrix U133 Plus2 GeneChips, a combination of U133A and U133B (~18,000 genes). Therefore, while all genes dysregulated in AT/RTs were reported, genes in common were limited to those on U133A.
Additional details of the samples used for this study are provided in Supplementary Table I.
Data Analysis
Data analysis using hierarchical clustering, NIH Database for Annotation, Visualization, and Integrated Discovery (DAVID) and Gene Set Enrichment Analysis (GSEA) bioinformatics tools has been previously described [11].
Genes that discriminated the tumor subtypes were identified using analysis of variance. Genes with p < 0.001 and fold difference ≥ 2 or ≤ 0.5 for RTs in all comparisons to each non-RT group were considered significant. At this p-value all FDRs were < 0.01.
Differential gene expression of each RT group compared to normal samples was calculated using Student’s two-sample t-tests. A False Discovery Rate (FDR) < 0.01 (associated with P-values < 0.001) with a mean fold difference ≥ 2 or ≤ 0.5 was considered significant.
Functional analysis of genes was performed with DAVID using the Protein Analysis Through Evolutionary Relationships (PANTHER) Biological Process database [12–14]. Functional enrichment was also analyzed with GSEA, using published genesets associated with developmental and cancer pathways. Activated or inhibited upstream transcriptional regulators were identified using Ingenuity (Ingenuity® Systems, www.ingenuity.com). Dysregulated genes, along with their direction of change, were compared to known up and down-regulated targets of transcriptional regulators, as published in the literature and compiled in Ingenuity’s KnowledgeBase. A z-score predicted the status of the upstream regulator (activated or inhibited). Absolute z ≥ 2 was considered significant. Ingenuity was also used to identify drugs that target particular genes, based on published literature.
RESULTS
Identification of a gene signature that distinguishes RTs from other tumors
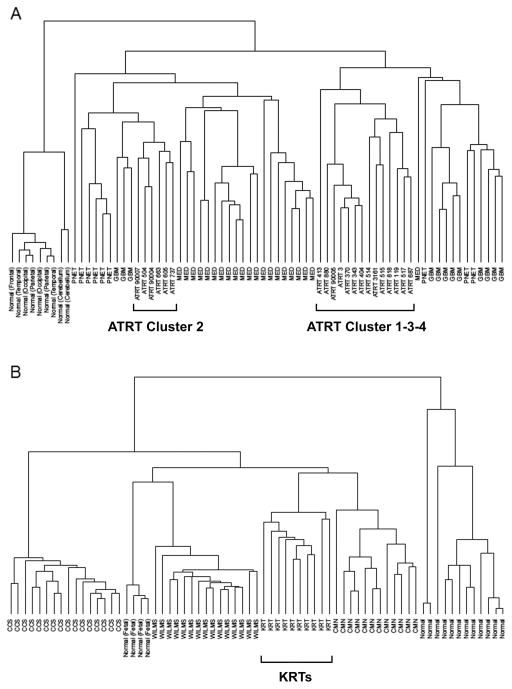
To confirm if a gene signature exists for RTs, we used unsupervised hierarchical clustering to examine whether RT gene expression profiles were distinct from other tumors and normal tissue. Because the kidney data covered only a subset of the data profiled for the brain samples (~13,000 vs. 18,000 annotated genes), we examined these datasets separately. Clustering of AT/RTs, MEDs, PNETs, GBMs, and normal brain showed that AT/RTs formed two distinct subsets (Fig. 1A). Examination of these subsets revealed that the smaller one was composed of the same samples which formed Cluster 2 in our previous analysis of gene expression profiling within AT/RTs, while the other (hereafter referred to as Cluster 1-3-4) was composed of the remaining AT/RT samples organized into the same intra-diagnostic groups previously referred to as Cluster 1, Cluster 3 and Cluster 4 [11]. Two additional new AT/RT samples grouped within Cluster 3 and Cluster 4, respectively. These results were stable across a broad range of cutoffs based on variance (from 0 to 90%). Clustering of the AT/RTs did not correspond to tumor location.
Fig 1.
Unsupervised hierarchical clustering results. A: Using 20 AT/RTs, 42 other pediatric high-grade brain tumors, and 9 pediatric non-tumor brain samples, genes with the top 25% variance, average linkage and spearman distances. B: Using 10 kidney rhabdoid tumors, 43 other kidney tumors, and 16 normal kidney samples, genes with the top 25% variance, average linkage and Spearman distances.
A similar analysis of the kidney samples, on the other hand, showed all KRTs clustered on 1 main branch separate from all other samples (Fig. 1B). There were no clear subgroups within KRTs. Again, these results were highly stable across a range of cutoffs.
The results above showed that gene expression did distinguish AT/RTs and KRTs from other tumors arising in similar tissues. Next, we used ANOVA to determine the genes that best distinguished RTs from other tumors. Using all AT/RTs, 765 genes showed gene expression distinct from other high-grade pediatric brain tumors and non-tumor samples, 351 up- and 414 down-regulated (Supplementary Table II). Because the unsupervised hierarchical clustering showed the AT/RTs separated into 2 quite separate clusters, this analysis was repeated separately for each subset. Comparing only the Cluster 1-3-4 AT/RTs to the other non-AT/RT samples, 1187 genes (525 up, 665 down) were identified (Supplementary Table III). This substantial increase in spite of the smaller sample size indicates this subset shows less heterogeneity. Performing the analysis with only Cluster 2 found 633 genes (346 up, 287 down) (Supplementary Table IV). Similar analysis found 539 genes (315 up and 225 down) distinguished KRTs from other kidney tumors (Supplementary Table V).
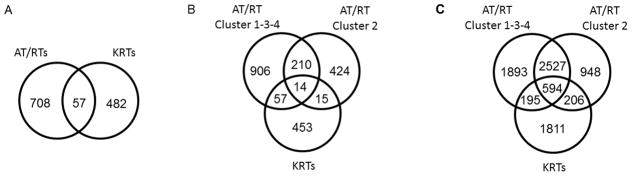
To determine genes common to all RTs distinct from other tumors, intersections of the above gene lists were taken. Using all AT/RTs as one set, 57 of the distinguishing genes were similarly dysregulated (i.e., in the same direction) in both AT/RTs and KRTs (30 up, 27 down) (Fig. 2A). If considered as 3 separate subsets (AT/RT Cluster 1-3-4, AT/RT Cluster 2, and KRTs), the 2 AT/RT subsets shared 224 genes in common (90 up, 134 down), while the intersection of all 3 subsets was only 14 (3 up, 11 down) (Fig. 2B, Table I). The KRTs shared more genes in common with AT/RT Cluster 1-3-4 than with AT/RT Cluster 2 (71 versus 29, respectively).
Fig 2. Venn diagrams showing overlaps in dysregulated genes among RT subsets.
Analysis of variance was used to determine dysregulated genes associated with each RT subset compared to other tumors or normal tissue. A: overlap of all AT/RTs (n=20) compared to other brain tumors with that of KRTs (n=10) compared to other kidney tumors. B: 3-way overlap of AT/RT Cluster 1-3-4 (n=14) compared to other brain tumors, AT/RT Cluster 2 (n=6) compared to other brain tumors, and KRTs compared to other kidney tumors. AT/RT subsets reflect hierarchical clustering results (Fig. 1); subset names based on AT/RT intra-diagnostic clustering groups previously reported [11]. C: Using same subsets as B, with comparisons to respective normal tissues rather than other tumors.
TABLE I.
Genes significantly dysregulated in all 3 RT groups (AT/RT Cluster1-3-4, AT/RT Cluster2, KRT) compared to other tumors in respective tissue.
| Gene | Direction in RTs | AT/RT 134 P-value |
AT/RT 134 FC |
AT/RT 2 P-value |
AT/RT 2 FC |
KRT P-value |
KRT FC |
|---|---|---|---|---|---|---|---|
| TRIL | Down | 2.70E-07 | 0.27 | 4.66E-05 | 0.35 | 3.15E-24 | 0.01 |
| H3F3B | Down | 7.04E-13 | 0.16 | 5.26E-07 | 0.41 | 2.63E-07 | 0.23 |
| SELENBP1 | Down | 5.45E-07 | 0.39 | 1.30E-05 | 0.30 | 3.43E-17 | 0.07 |
| SMARCB1 | Down | 6.38E-15 | 0.17 | 2.81E-08 | 0.34 | 5.67E-18 | 0.12 |
| DOCK4 | Down | 9.57E-13 | 0.13 | 4.30E-06 | 0.46 | 8.82E-17 | 0.07 |
| BIN1 | Down | 1.26E-13 | 0.15 | 5.03E-10 | 0.17 | 7.76E-09 | 0.22 |
| NRIP1 | Down | 3.86E-07 | 0.15 | 6.94E-09 | 0.16 | 2.26E-10 | 0.11 |
| LMO4 | Down | 1.68E-06 | 0.27 | 3.05E-05 | 0.34 | 1.30E-23 | 0.03 |
| PPP3CA | Down | 1.94E-13 | 0.30 | 1.92E-14 | 0.35 | 7.54E-19 | 0.04 |
| PIK3R1 | Down | 6.71E-08 | 0.43 | 3.45E-06 | 0.38 | 1.68E-06 | 0.22 |
| HECA | Down | 4.32E-06 | 0.43 | 2.40E-04 | 0.45 | 2.16E-09 | 0.22 |
| EZR | Up | 1.05E-04 | 2.1 | 2.52E-04 | 3.3 | 1.80E-07 | 3.4 |
| SLC2A3 | Up | 2.60E-11 | 3.1 | 3.58E-08 | 2.7 | 3.13E-12 | 10.9 |
| THBS3 | Up | 1.32E-09 | 7.6 | 5.77E-05 | 6.3 | 2.26E-14 | 5.3 |
FC = Fold Change
Among the 14 genes common to all 3 subsets, 5 genes have been associated with SMARCB1 loss by previous studies: DOCK4, LMO4, SLC2A3, THBS3, and SMARCB1 itself [7, 15, 16]. LMO4, SLC2A3 and THBS3 have been reported by more than one study, suggesting they are consistently associated with SMARCB1 loss.
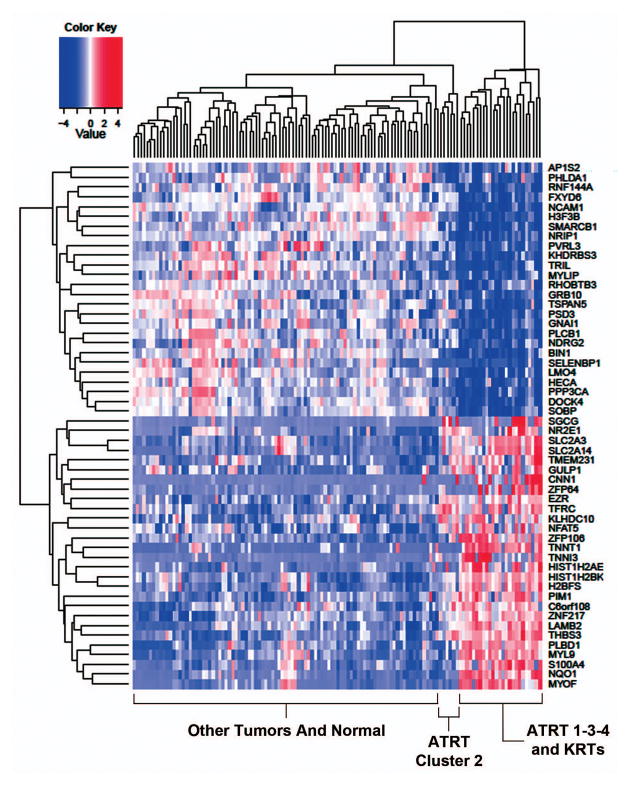
A heatmap of the genes common to AT/RTs and KRTs shows that these genes separate RTs, regardless of their location, from other tumors (Fig. 3). The KRT samples intermix with the samples from AT/RT Cluster 1-3-4, while AT/RT Cluster 2 is on a separate branch. Among the up-regulated genes, significant enrichment was seen for processes associated with mesoderm, particularly muscle contraction and development. Signal transduction was enriched among down-regulated genes.
Fig 3.
Heatmap of genes dysregulated in both AT/RTs and KRTs, compared to other tumors. The expression of dysregulated genes common to both AT/RTs (n=20) and KRTs (n=10) compared to other brain and kidney tumors, respectively, is shown. Normal samples are also included on the heatmap for comparison.
Geneset enrichment analysis was used to look for the association of RTs with pathways and processes involved in development and cancer, compared to other tumors and normal tissues. Each subset of RTs (AT/RTs Cluster 1-3-4, AT/RTs Cluster 2, and KRTs) was compared separately to their respective other tumors and normal tissue. No genesets showed statistical significance (p < .05) in all 3 comparisons. Three genesets related to gene promoter occupancy by PRCs, which results in epigenetic modifications that cause gene repression, were significantly associated with down-regulated genes in both AT/RT Cluster 1-3-4 and KRTs, but not in AT/RT Cluster 2. Hedgehog signaling was significantly up-regulated in both AT/RT subsets.
Identification of a gene signature that distinguishes RTs from normal tissue
Our analysis identified only a small set of genes that were specifically and consistently dysregulated in all RTs when compared to other tumors. This suggests that the tumorigenic property of SMARCB1 loss also involves mechanisms common to other tumors. We therefore examined genes and processes dysregulated in RTs compared to normal tissue. We performed this analysis with all 3 RT subsets, to avoid losing information specific to the 2 AT/RT groups.
This analysis identified 5209, 4275 and 2841 genes dysregulated in AT/RT Cluster 1-3-4, Cluster 2, and KRTs, respectively, compared to normal tissues (Fig. 2C). Overall, 610 genes were dysregulated in the same direction in all 3 subsets (Supplementary Table VI). Processes associated with cell proliferation, cell cycle, chromatin processing/remodeling and mRNA splicing were enriched among all RTs (Supplementary Table VII). Genesets associated with cell proliferation, embryonic stem cells, P53 signaling, and MYC targets among up-regulated genes, and with polycomb occupancy among down-regulated genes, were also enriched in all (Supplementary Table VIII).
Among the 610 dysregulated genes in common to all 3 RT subsets, Ingenuity Pathways Analysis identified 55 gene transcriptional regulators that appeared to be activated or inhibited, based on the expression levels of their downstream targets, at a statistically significant confidence level (Supplementary Table IX). Notably, inhibition of SMARCB1 was identified with this method, as was inhibition of CDKN1A and CDKN2A, and activation of CCND1, findings which have been previously reported by other studies of RTs, supporting the validity of this approach [9, 17, 18]. The most strongly activated regulator was MYC, which was consistent with results seen by GSEA, while RB1 showed the strongest predicted inhibition.
The tumor samples used in this study undoubtedly contain some percentage of cells from surrounding normal tissues. Thus, it is possible that genes identified above could be due to differing expression in contaminating normal cells, rather than to differences between the tumors themselves. To evaluate this possibility, we quantile normalized all samples together and identified 1577 genes that showed similar expression between normal brain and kidney (p > .5, absolute fold change < 1.5). If expression differences were primarily due to contaminating normal cells, then one would also expect to see little difference between AT/RTs and KRTs among these genes. However, 342 (22%) of these showed significant differences (p<.01, absolute fold change > 2) between AT/RTs and KRTs. These results suggest that differences do indeed exist between the tumors themselves.
Epigenetic effector genes are dysregulated in RTs
Epigenetic modifications affect gene expression and are orchestrated by a large number of genes with a variety of functions. The enrichment of genes associated with chromatin processing/remodeling, the down-regulation of genes associated with polycomb occupancy, and the activation or inhibition of transcription networks associated with EP400, Hdacs, KDM5B, SMARCE1 and SMARCB1 (Supplementary Table IX) reported above suggest that epigenetic dysregulation is a key element common to RTs. We therefore examined our data to identify genes responsible for epigenetic modifications, in addition to SMARCB1, that are dysregulated in RTs. A comprehensive list of genes involved with making epigenetic modifications was manually compiled from publicly available sources (286 genes, Supplementary Table X). These genes are hereafter referred to as epigenetic effectors.
Similar percentages of epigenetic effectors (~30%) were found among the significantly dysregulated genes in each of the 3 RT groups. 23 genes were dysregulated in all 3 RT subsets (Table II). The most strongly dysregulated epigenetic effectors common to all 3 were HMGA2 (93 fold up-regulated overall) and EZH2 (39 fold up-regulated).
TABLE II.
Genes responsible for epigenetic modifications (epigenetic effectors) significantly dysregulated in all 3 RT subsets (AT/RT Cluster1-3-4, AT/RT Cluster 2, KRT) compared to normal.
| Gene | Member of | Dir in RTs | AT/RT 134 P-value |
AT/RT 134 FC |
AT/RT 2 P-value |
AT/RT 2 FC |
KRT P-value |
KRT FC |
|---|---|---|---|---|---|---|---|---|
| ACTL6A | Trithorax (SWI/SNF) | Up | 1.77E-11 | 10.3 | 5.95E-08 | 10.3 | 4.25E-08 | 4.9 |
| ARID1A | Trithorax (SWI/SNF) | Up | 1.42E-07 | 2.6 | 1.94E-09 | 3.7 | 8.46E-08 | 2.8 |
| ASF1B | Up | 2.38E-06 | 10.6 | 1.01E-06 | 8.2 | 3.28E-08 | 4.9 | |
| BAZ1A | Trithorax (ISWI) | Up | 3.37E-06 | 6.0 | 3.63E-09 | 14.5 | 5.60E-05 | 3.4 |
| CBX7 | Polycomb (PRC1) | Down | 2.69E-11 | 0.02 | 1.58E-10 | 0.04 | 8.49E-07 | .10 |
| CHD1L | Trithorax | Up | 7.00E-04 | 2.1 | 1.14E-03 | 2.2 | 2.05E-12 | 4.8 |
| CHD4 | Trithorax | Up | 4.53E-04 | 2.4 | 6.52E-05 | 3.4 | 7.38E-04 | 2.9 |
| EZH2 | Polycomb (PRC2) | Up | 2.96E-10 | 30.7 | 2.05E-09 | 108.0 | 4.40E-12 | 17.3 |
| HAT1 | Up | 2.17E-04 | 2.4 | 7.83E-04 | 2.2 | 6.32E-04 | 2.0 | |
| HDAC2 | Up | 2.92E-07 | 2.3 | 3.94E-06 | 3.4 | 2.22E-06 | 3.8 | |
| HELLS | Trithorax | Up | 9.85E-05 | 5.1 | 1.55E-05 | 18.0 | 9.64E-11 | 15.2 |
| HMGA2 | Up | 1.64E-08 | 394.4 | 3.40E-04 | 70.4 | 3.49E-05 | 29.0 | |
| HMGB2 | Up | 4.37E-06 | 3.3 | 9.09E-09 | 7.0 | 1.75E-09 | 5.6 | |
| HMGN1 | Up | 2.27E-06 | 2.4 | 1.17E-06 | 3.4 | 2.66E-06 | 2.3 | |
| PCGF1 | Polycomb (PRC1) | Up | 2.62E-05 | 5.0 | 5.07E-04 | 5.3 | 1.00E-06 | 2.3 |
| PHF21A | Up | 5.06E-04 | 2.5 | 3.78E-04 | 2.6 | 1.34E-12 | 4.8 | |
| PRMT1 | Up | 7.35E-05 | 2.0 | 4.23E-08 | 2.8 | 2.06E-05 | 5.9 | |
| RBBP4 | Both (PRC2) | Up | 1.53E-04 | 2.2 | 3.59E-04 | 2.8 | 3.76E-05 | 4.2 |
| SETDB1 | Up | 3.37E-06 | 2.9 | 7.98E-06 | 4.9 | 1.10E-05 | 2.1 | |
| SMARCAL1 | Trithorax | Up | 1.16E-05 | 2.9 | 9.68E-04 | 2.6 | 1.34E-07 | 2.4 |
| SMARCC1 | Trithorax (SWI/SNF) | Up | 1.23E-08 | 4.7 | 3.02E-10 | 5.1 | 3.37E-06 | 2.7 |
| SSRP1 | Up | 1.52E-05 | 2.2 | 1.82E-07 | 3.4 | 3.23E-07 | 4.1 | |
| WHSC1 | Up | 7.21E-06 | 3.4 | 1.50E-07 | 6.0 | 3.99E-10 | 6.5 |
FC = Fold Change
Multiple genes associated with SWI/SNF or polycomb complex 1 or 2 (PRC1 or PRC2) were dysregulated. Thus, both the genes normally regulated by epigenetic modifications, as well as the epigenetic effector genes themselves, were dysregulated in RTs.
Potential therapeutic targets
The 395 genes significantly up-regulated in all 3 RT subsets versus normal were examined for potential therapeutic targets. Nineteen genes were up-regulated and targetable by clinically relevant therapeutic agents (Table III). TOP2A, targeted by etoposide, doxorubicin and others, showed the highest relative expression for RTs, with greater than 200-fold up-regulation. Gemcitabine and flavopiridol both targeted multiple genes up-regulated in RTs. Seventeen kinases that do not have clinically relevant drugs currently were up-regulated in RTs (Supplementary Table XI). Among these MELK, BUB1B, PBK, TTK, and BUB1 showed the highest up-regulation.
TABLE III.
Genes significantly upregulated in all 3 RT groups (AT/RT Cluster 1-3-4, AT/RT Cluster 2, KRT) compared to normal tissue that are targeted by therapeutic drugs.
| Gene | Average Fold Change | Drugs |
|---|---|---|
| TOP2A | 245.1 | etoposide, epirubicin, doxorubicin, daunorubicin, and others |
| RRM2 | 56.3 | gemcitabine, triapine, hydroxyurea, fludarabine phosphate |
| CDK1 | 55.8 | flavopiridol |
| TYMS | 52.7 | 5-fluorouracil, flucytosine, plevitrexed, nolatrexed, capecitabine, trifluridine, floxuridine, LY231514 |
| AURKA | 26.8 | tozasertib, MLN8054 |
| AURKB | 14.8 | tozasertib, AZD-1152, GSK1070916 |
| SMO | 13.7 | vismodegib |
| CHEK1 | 12.0 | UCN-01 |
| POLE2 | 11.9 | gemcitabine |
| CDK2 | 11.6 | flavopiridol, BMS-387032 |
| CDK4 | 8.0 | flavopiridol, PD-0332991 |
| FGFR1 | 6.6 | pazopanib |
| ABL1 | 6.4 | saracatinib, imatinib, temozolomide |
| POLD1 | 5.9 | nelarabine, MB07133, clofarabine, cytarabine, trifluridine, vidarabine, entecavir |
| TUBB6 | 5.8 | brentuximab vedotin, cabazitaxel |
| RRM1 | 4.4 | gemcitabine, clofarabine, fludarabine phosphate |
| DHFR | 3.6 | methotrexate |
| IMPDH2 | 3.5 | thioguanine, VX-944, interferon alfa-2b/ribavirin, mycophenolic acid, ribavirin |
| HDAC2 | 3.1 | tributyrin, belinostat, pyroxamide, vorinostat, romidepsin |
Several drugs were also suggested by Ingenuity upstream regulator analysis. Significant dysregulation of down-stream genes consistent with potential for treatment efficacy was seen with doxorubicin (p < 1×10−10), fulvestrant (p < 1×10−9), and tanespimycin (HSP90 inhibitor) (p < 1×10−5).
DISCUSSION
All pediatric RTs are distinguished by loss of functional SMARCB1 protein. However, only a few studies have compared RTs from different tissues to assess similarities or differences in gene or protein expression, and they have examined only a limited number of genes [7–10]. An understanding of the mechanisms underlying RTs, and the extent to which they are shared or different among RTs arising in different locations, is important given the expanding numbers of therapies that target specific biological processes or pathways. The current study examined comprehensive gene expression of AT/RTs and KRTs to determine if a common gene signature exists for RTs regardless of tumor location and if so, what this signature suggests about processes and pathways disrupted by SMARCB1 loss.
We found only a very limited number of dysregulated genes are common to both AT/RTs and KRTs when compared to other tumors from CNS and kidney, respectively. Among these, however, were confirmed several genes that have previously been associated with SMARCB1 by multiple studies - LMO4, SLC2A3, THBS3 - suggesting that there are some genes that are consistently affected by SMARCB1 expression [7, 15, 16]. The majority of dysregulated genes, however, differed substantially between AT/RTs and KRTs. These observations of multiple expression differences between RT subsets are consistent with earlier results from studies where SMARCB1 was knocked out or re-expressed in various cell types: only 7% of approximately 1500 genes associated with SMARCB1 change were reported by more than one of the studies [7, 15, 16, 19, 20].
As previously reported, KRTs showed substantial down-regulation of neural crest stem cell markers compared to other kidney tumors. [6] Many of these same markers were either not dysregulated, or showed dysregulation in the opposite direction, among the AT/RTs. Our results suggest several conclusions. First, the specific dysregulated gene expression seen upon loss of SMARCB1 appears dependent on additional factors, since a singular molecular expression pattern does not emerge across RTs from different sites. Even within AT/RTs, gene expression differences suggest 2 or more molecular groups exist, consistent with our previously reported findings on AT/RT heterogeneity[11]. One factor affecting gene expression may be the specific cellular context in which SMARCB1 loss occurs. The molecular differences seen between AT/RT and KRTs suggest that these tumors may arise from different cell types. Our data also raise the possibility that AT/RTs themselves could arise from more than one precursor cell type, similar to medulloblastomas. Our report of an AT/RT apparently evolving spontaneously from a ganglioglioma, and several reports of AT/RTs arising after treatment of gangliogliomas or pleomorphic xanthoastrocytomas, give further credence to this possibility [21–24].
The limited number of dysregulated genes common to all RTs when compared to other tumors suggests, secondly, that changes leading to tumorigenesis upon loss of SMARCB1 also involve genes and mechanisms that drive other aggressive tumors. We also compared AT/RTs and KRTs to normal brain and kidney, respectively. While substantial differences were still observed between the RT subsets, several common biological processes did emerge. In addition to the expected up-regulation of multiple processes associated with cell proliferation and cell cycle, these included up-regulation of MYC targets and of mRNA splicing, and dysregulation of p53 signaling (Supplemental Tables VII and VIII). Consistent with up-regulation of a MYC transcriptional program in RTs, both MYC and MYCN were found to be up-regulated in a newly developed AT/RT cell line and anti-MYC drug S2T1-6OTD significantly reduced cell proliferation in 2 AT/RT cell lines [25, 26].
Enrichment of several biological processes pointed to the importance of epigenetic changes in RTs compared to normal tissues. These processes include the down-regulation of genes associated with PRC occupancy (PRCs repress gene expression through histone modifications), as well as the enrichment of genes associated with chromatin processing/remodeling, and possible activation or inhibition of transcription networks associated with several epigenetic effectors. We identified 23 epigenetic effector genes dysregulated in both AT/RTs and KRTs These included multiple genes associated with SWI/SNF or PRC1/2, the most aberrant being EZH2 (38 fold up, PRC2) and CBX7 (23 fold down, PRC1). Recent studies have shown that EZH2 is also up-regulated at the protein level in RTs [10, 27]. However, similar up-regulation of EZH2 mRNA levels was seen in all of the high-grade kidney and brain tumors in this study, as in other studies [28–30]. Thus, up-regulation of EZH2, per se, does not explain the increased down regulation of genes associated with polycomb occupancy seen in RTs compared to other tumors. It has been suggested that in RTs SWI/SNF, missing the SMARCB1 component, may fail to evict PRCs from critical genes needed for differentiation, resulting in gene repression and continued self-renewal and proliferation [6, 27, 31]. The stronger down regulation of many of these genes in RTs compared to other tumors that also show up-regulated levels of EZH2 supports this hypothesis.
Results of this study suggest several possibilities for targeted therapies that might be effective for both AT/RTs and KRTs, and therefore may be of interest for future research. The high elevation of TOP2A transcription observed in our samples provides a rational basis for agents such as etoposide in therapy for RTs. Results also suggest HDAC inhibitors (HDACIs) have potential. Studies have shown that HDACIs up-regulate multiple cyclin-dependent kinase inhibitors and block proliferation in rhabdoid cells [32–35]. Recently, our lab has also shown that HDACIs sensitize rhabdoid cells to ionizing radiation [36]. AURKA, which has an inhibitor in clinical trials, showed up-regulation in all RT groups in the current study. We have previously shown inhibition of AURKA in vitro induces apoptosis in rhabdoid cells when used as a single agent, and produces a synergistic effect in combination with radiation [37]. Several up-regulated kinases without current targeted therapies were also identified as possible targets by our study, including MELK, BUB1B, and others (Supplemental Table XI).
The meta-analysis presented here has several limitations. Brain and kidney samples were processed by different laboratories, which can introduce systematic bias. Tumor samples also contained varying amounts of non-tumor cells. Approximately 1/3 of known genes could not be evaluated across both AT/RTs and KRTs, as data was only available for Affymetrix U133A genes for the kidney tumors. mRNA differences also need to be evaluated at the protein level and by functional assays. However, we view this comprehensive initial survey of a relatively large number of these uncommon tumors as a useful starting point for future studies.
In summary, this study suggests that the effect of SMARCB1 loss on gene expression results in consistent dysregulation of only a limited number of genes, and that other factors also play a role in determining the gene expression profile seen in RTs. Up-regulation of cell cycle genes, and aberrant epigenetic regulation may offer drug targets that are common to all RTs. However, the large number of genes that show dysregulation in only a subset of these tumors also provides opportunities for therapies tailored to particular molecular profiles.
Supplementary Material
Acknowledgments
The authors thank the Morgan Adams Foundation for financial support of this project.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
References
- 1.Biegel JA, Tan L, Zhang F, et al. Alterations of the hsnf5/ini1 gene in central nervous system atypical teratoid/rhabdoid tumors and renal and extrarenal rhabdoid tumors. Clin Cancer Res. 2002;8:3461–3467. [PubMed] [Google Scholar]
- 2.Judkins AR, Mauger J, Ht A, et al. Immunohistochemical analysis of hsnf5/ini1 in pediatric cns neoplasms. Am J Surg Pathol. 2004;28:644–650. doi: 10.1097/00000478-200405000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Hilden JM. Central nervous system atypical teratoid/rhabdoid tumor: Results of therapy in children enrolled in a registry. J Clin Oncol. 2004;22:2877–2884. doi: 10.1200/JCO.2004.07.073. [DOI] [PubMed] [Google Scholar]
- 4.Garre ML, Tekautz T. Role of high-dose chemotherapy (hdct) in treatment of atypical teratoid/rhabdoid tumors (at/rts) Pediatr Blood Cancer. 2010;54:647–648. doi: 10.1002/pbc.22377. [DOI] [PubMed] [Google Scholar]
- 5.Chi SN, Zimmerman MA, Yao X, et al. Intensive multimodality treatment for children with newly diagnosed cns atypical teratoid rhabdoid tumor. J Clin Oncol. 2009;27:385–389. doi: 10.1200/JCO.2008.18.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadd S, Sredni ST, Huang CC, et al. Rhabdoid tumor: Gene expression clues to pathogenesis and potential therapeutic targets. Lab Invest. 2010;90:724–738. doi: 10.1038/labinvest.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 8.Albanese P, Belin M, Delattre O. The tumour suppressor hsnf5/ini1 controls the differentiation potential of malignant rhabdoid cells. Eur J Cancer. 2006;42:2326–2334. doi: 10.1016/j.ejca.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Venneti S, Le P, Martinez D, et al. P16ink4a and p14arf tumor suppressor pathways are deregulated in malignant rhabdoid tumors. J Neuropathol Exp Neurol. 2011;70:596–609. doi: 10.1097/NEN.0b013e31822146ca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venneti S, Le P, Martinez D, et al. Malignant rhabdoid tumors express stem cell factors, which relate to the expression of ezh2 and id proteins. Am J Surg Pathol. 2011;35:1463–1472. doi: 10.1097/PAS.0b013e318224d2cd. [DOI] [PubMed] [Google Scholar]
- 11.Birks DK, Donson AM, Patel PR, et al. High expression of bmp pathway genes distinguishes a subset of atypical teratoid/rhabdoid tumors associated with shorter survival. Neuro Oncol. 2011;13:1296–1307. doi: 10.1093/neuonc/nor140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis G, Jr, Sherman BT, Hosack DA, et al. David: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Thomas PD, Kejariwal A, Campbell MJ, et al. Panther: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medjkane S, Novikov E, Versteege I, et al. The tumor suppressor hsnf5/ini1 modulates cell growth and actin cytoskeleton organization. Cancer Res. 2004;64:3406–3413. doi: 10.1158/0008-5472.CAN-03-3004. [DOI] [PubMed] [Google Scholar]
- 16.Isakoff MS, Sansam CG, Tamayo P, et al. Inactivation of the snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc Natl Acad Sci U S A. 2005;102:17745–17750. doi: 10.1073/pnas.0509014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chai J, Charboneau AL, Betz BL, et al. Loss of the hsnf5 gene concomitantly inactivates p21cip/waf1 and p16ink4a activity associated with replicative senescence in a204 rhabdoid tumor cells. Cancer Res. 2005;65:10192–10198. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- 18.Zhang ZK, Davies KP, Allen J, et al. Cell cycle arrest and repression of cyclin d1 transcription by ini1/hsnf5. Mol Cell Biol. 2002;22:5975–5988. doi: 10.1128/MCB.22.16.5975-5988.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma HI, Kao CL, Lee YY, et al. Differential expression profiling between atypical teratoid/rhabdoid and medulloblastoma tumor in vitro and in vivo using microarray analysis. Childs Nerv Syst. 2010;26:293–303. doi: 10.1007/s00381-009-1016-2. [DOI] [PubMed] [Google Scholar]
- 20.Morozov A, Lee SJ, Zhang ZK, et al. Ini1 induces interferon signaling and spindle checkpoint in rhabdoid tumors. Clin Cancer Res. 2007;13:4721–4730. doi: 10.1158/1078-0432.CCR-07-0054. [DOI] [PubMed] [Google Scholar]
- 21.Allen JC, Judkins AR, Rosenblum MK, et al. Atypical teratoid/rhabdoid tumor evolving from an optic pathway ganglioglioma: Case study. Neuro Oncol. 2006;8:79–82. doi: 10.1215/S1522851705000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chacko G, Chacko AG, Dunham CP, et al. Atypical teratoid/rhabdoid tumor arising in the setting of a pleomorphic xanthoastrocytoma. J Neurooncol. 2007;84:217–222. doi: 10.1007/s11060-007-9361-z. [DOI] [PubMed] [Google Scholar]
- 23.Dougherty MJ, Santi M, Brose MS, et al. Activating mutations in braf characterize a spectrum of pediatric low-grade gliomas. Neuro Oncol. 2010;12:621–630. doi: 10.1093/neuonc/noq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinschmidt-DeMasters BK, Birks DK, Aisner DL, et al. Atypical teratoid/rhabdoid tumor arising in a ganglioglioma: Genetic characterization. Am J Surg Pathol. 2011;35:1894–1901. doi: 10.1097/PAS.0b013e3182382a3f. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, Erdreich-Epstein A, Gonzalez-Gomez I, et al. Novel cell lines established from pediatric brain tumors. J Neurooncol. 2011 doi: 10.1007/s11060-011-0756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shalaby T, von Bueren AO, Hurlimann ML, et al. Disabling c-myc in childhood medulloblastoma and atypical teratoid/rhabdoid tumor cells by the potent g-quadruplex interactive agent s2t1-6otd. Mol Cancer Ther. 2010;9:167–179. doi: 10.1158/1535-7163.MCT-09-0586. [DOI] [PubMed] [Google Scholar]
- 27.Wilson BG, Wang X, Shen X, et al. Epigenetic antagonism between polycomb and swi/snf complexes during oncogenic transformation. Cancer Cell. 2010;18:316–328. doi: 10.1016/j.ccr.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crea F, Hurt EM, Farrar WL. Clinical significance of polycomb gene expression in brain tumors. Mol Cancer. 2010;9:265. doi: 10.1186/1476-4598-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alimova I, Venkataraman S, Harris P, et al. Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int J Cancer. 2012 doi: 10.1002/ijc.27455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagener N, Macher-Goeppinger S, Pritsch M, et al. Enhancer of zeste homolog 2 (ezh2) expression is an independent prognostic factor in renal cell carcinoma. BMC Cancer. 2010;10:524. doi: 10.1186/1471-2407-10-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kia SK, Gorski MM, Giannakopoulos S, et al. Swi/snf mediates polycomb eviction and epigenetic reprogramming of the ink4b-arf-ink4a locus. Mol Cell Biol. 2008;28:3457–3464. doi: 10.1128/MCB.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaboin J, Wild J, Hamidi H, et al. Ms-27-275, an inhibitor of histone deacetylase, has marked in vitro and in vivo antitumor activity against pediatric solid tumors. Cancer Res. 2002;62:6108–6115. [PubMed] [Google Scholar]
- 33.Graham C. Evaluation of the antitumor efficacy, pharmacokinetics, and pharmacodynamics of the histone deacetylase inhibitor depsipeptide in childhood cancer models in vivo. Clin Cancer Res. 2006;12:223–234. doi: 10.1158/1078-0432.CCR-05-1225. [DOI] [PubMed] [Google Scholar]
- 34.Furchert SE, Lanvers-Kaminsky C, Juurgens H, et al. Inhibitors of histone deacetylases as potential therapeutic tools for high-risk embryonal tumors of the nervous system of childhood. Int J Cancer. 2007;120:1787–1794. doi: 10.1002/ijc.22401. [DOI] [PubMed] [Google Scholar]
- 35.Algar EM, Muscat A, Dagar V, et al. Imprinted cdkn1c is a tumor suppressor in rhabdoid tumor and activated by restoration of smarcb1 and histone deacetylase inhibitors. PLoS ONE. 2009;4:e4482. doi: 10.1371/journal.pone.0004482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knipstein JA, Birks DK, Donson AM, et al. Histone deacetylase inhibition decreases proliferation and potentiates the effect of ionizing radiation in atypical teratoid/rhabdoid tumor cells. Neuro Oncol. 2012;14:175–183. doi: 10.1093/neuonc/nor208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venkataraman S, Alimova I, Tello T, et al. Targeting aurora kinase a enhances radiation sensitivity of atypical teratoid rhabdoid tumor cells. J Neurooncol. 2012;107:517–526. doi: 10.1007/s11060-011-0795-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.