Abstract
Objective
Mutations in receptor expression enhancing protein 1 (REEP1) are associated with hereditary spastic paraplegias (HSPs). Although axonal degeneration is thought to be a predominant feature in HSP, the role of REEP1 mutations in degeneration is largely unknown. Previous studies have implicated a role for REEP1 in the ER, whereas others localized REEP1 with mitochondria. We sought to resolve the cellular localization of REEP1 and to further elucidate the pathobiology underlying REEP1 mutations in patients.
Methods
A combination of cellular imaging and biochemical approaches was used to refine the cellular localization of REEP1. Next, Reep1 mutations associated with HSP were functionally tested in neuritic growth and degeneration assays using mouse cortical culture. Finally, a novel assay was developed and used with wild type and mutant Reep1s to measure the interactions between the ER and mitochondria.
Results
We found that REEP1 is present at the ER-mitochondria interface, and it contains subdomains for mitochondrial as well as ER localization. Knockdown of Reep1 and the expression of pathological Reep1 mutations resulted in neuritic growth defects and degeneration. Finally, using our novel split-RLuc8 assay, we show REEP1 facilitates ER-mitochondria interactions, a function diminished by disease-associated mutations.
Interpretation
Our data potentially reconcile the current conflicting reports regarding REEP1 being either an ER or a mitochondrial protein. Furthermore, our results connect, for the first time, the disrupted ER-mitochondria interactions to a failure in maintaining health of long axons in HSPs. Finally, the split-RLuc8 assay offers a new tool to identify potential drugs for multiple neurodegenerative diseases with ER-mitochondria interaction defects.
Introduction
Hereditary spastic paraplegias (HSPs), also known as spastic paraplegias (SPGs), are a heterogeneous group of disorders that share clinical symptoms of progressive weakness and spasticity mainly in the lower extremities due to axonal degeneration. More than 70 distinct genetic loci have been mapped in HSP, many of which implicate common cellular processes.1–3 Among these are genes involved in membrane shaping and trafficking; mitochondrial function; myelination and lipid metabolism; protein folding and ER stress response; and axonal tract development.1,2,4
REEP1 is believed to encode an ER protein and mutations are associated with autosomal dominant HSP (SPG31) as well as distal hereditary motor neuropathy5–7. There are six members of the REEP proteins, REEP1–6, which can be grouped into two subfamilies, REEP1–4 and REEP5–6, based on structural and sequence homology. They share sequence homology with yeast Yop1 and plant HVA22 protein and all belong to the Yip (Ypt-interacting protein) family8. REEP proteins are known to function in ER membrane shaping: each contains REEP homology domain which has been shown to insert into membranes as a hairpin and actively bend membranes into curved structure. This domain also mediates physical interaction with atlastin-1, spastin, and reticulon.5,9–11 Although having a defined role in ER shaping, REEP1 has also been reported as a mitochondrial protein12; thus its intracellular localization remains controversial. Interestingly, mitochondrial dysfunctions, such as impaired fusion/fission and defective bioenergetics, have been reported in REEP1-associated HSP patients.13–15 This further suggests a mitochondrial related role for REEP1, although it is also possible that ER-localized REEP1 could affect mitochondrial functions given the tight association between the ER and mitochondria.13,16
The physical interactions between the ER and mitochondria play important roles in many aspects of cellular functions. The contact sites, known as mitochondria-associated ER membranes (MAMs), serve as channels for transporting molecules (e.g. membrane lipids, proteins, and Ca2+) between the two organelles and as hubs for mitochondrial fusion/fission and autophagosome formation.17–20 Alterations in ER-mitochondrial contact often leads to mitochondrial dysfunction and compromised ER function.1,2,4,21 Recent evidence suggests that the deregulation of ER-mitochondrial contacts and impaired Ca2+ transfer at such sites is a common pathogenic mechanism in various neurodegenerative diseases, including Parkinson’s disease, Alzheimer’s disease, and amyotrophic lateral sclerosis.5–7,22–27
In this study we have found that disease-associated Reep1 mutations lead to developmental defects and neuritic degeneration in mouse primary cortical neurons. We show that REEP1 contains subdomains for both mitochondrial and ER localization and that REEP1 is detected in MAMs. Using a novel split-RLuc8 reassembly assay, we demonstrate that REEP1 can facilitate ER-mitochondrial interactions in live cells and that this function is abrogated in the presence of disease-associated mutations. Together these data provide evidence linking altered ER-mitochondria interactions to REEP1-mediated HSP and offer new avenues for potential therapeutic intervention for patients with HSP.
Materials and Methods
DNA constructs
The full length clone of Reep1 was obtained by screening embryonic day 12.5 (E12.5) mouse cDNA library (in pBluescriptII-SK, provided by Doug Epstein), using the partial cDNA of Reep1 from a subtractive hybridization screen.8,28 For Reep1 wild type (Reep1WT) and disease mutant constructs (Reep1P19R, Reep1A20E, Reep11–115, Reep11–139, and Reep11–176), each DNA sequence was PCRed out using 5’ primers containing EcoR I site and CACCATG, and 3’ primers containing the stop codon sequence and Mlu I site and cloned into pCAG-IRES-GFP vector. For GFP-fused Reep1 constructs used to determine protein expression, PCR products encoding each disease mutant construct as well as wild type construct were cloned into pcDNA3.1/CT-GFP TOPO in frame (Life Technologies). For Reep1P19R and Reep1A20E, site directed mutagenesis (Stratagene Chameleon Kit) was used to introduce each point mutation. For Reep1 shRNA constructs, the annealed oligonucleotide was subcloned into pLL3.7 vector. The sequences used include the non-targeting negative control (5’-CAGTCGCGTTTGCGACTGG-3’; Dharmacon), target 1 (5’-GGAAGTTTGTTCATCCCAC-3’), target 2 (5’-GGGTGACCTTTACGTCTTC-3’), and target 3 (5’-GGCTAGAGAAATAGTGTTG-3’). For V5-tagged Reep1, the full length Reep1 was PCRed out and cloned into pcDNA3.1/V5-His-TOPO (Life Technologies) in frame. For GFP-tagged Reep1s used for localization study, PCR products encoding amino acid residues (a.a.) 1–201, 1–115, 116–201, and 116–157 of the mouse Reep1 were cloned into pcDNA3.1/CT-GFP TOPO in frame (Life Technologies). Untagged Reep1 constructs were generated in pcDNA3.1 TOPO with the same cloning strategy. For the split-RLuc8, Renilla luciferase 8 (RLuc8) in pBAD/Myc-His5,9–11,29 was used for the initial template. The split site of the RLuc8 was selected according to the previous reports.12,30,31 For Mito-RLuc8N (the N-terminal half of the RLuc8 targeted to the mitochondria), DNA sequences encoding a.a. 1–91 of the RLuc8 were fused to the 3’ end of the Flag tag sequence and the previously reported mitochondria-targeting sequence of the mouse AKAP1 (a.a. residues 34–63)13–15,32 in pcDNA3.1 TOPO by PCR. For RLuc8C-ER (the C-terminal half of the RLuc8 targeted to ER), DNA sequences encoding a.a. 92–311 of the RLuc8 were fused to the 5’ end of the myc tag sequence and the previously described ER localization sequence of the yeast UBC613,16,32 by PCR in pcDNA3.1 TOPO. For Reep1116–157RLuc8N used for subcellular localization study, DNA sequences encoding RLuc8N (a.a. 1–91) was fused to the 3’ end of the DNA sequences encoding REEP1116–157 by PCR in pcDNA3.1 TOPO. All constructs and mutations were confirmed by sequencing. ER-dsRed (pDsRed2-ER) and GFP-Sec61β (Plasmid #15108) were purchased from Clontech Laboratories and Addgene, respectively.
Cell culture and transfection
For immunofluorescence studies, HeLa cells were plated at a density of 1.5–2×104 cells/well in 8 well-chamber slides (Thermo Scientific). 16 hours after plating, cells were transfected via polyethylenimine (PEI, High Potency Linear, 40 kDa, Polysciences #24765)-mediated transfection with 200 ng of total DNAs per well. 24 hours after transfection, cells were fixed and used for immunofluorescence studies. For Western blot analysis, HEK293T or HeLa cells were plated at a density of 2×105 cells/well in 6 well-plates. After overnight incubation, cells were transfected via PEI-mediated transfection with 2–3 ug of total DNAs per well. For primary cortical cultures, dissociated cell cultures were generated from E15.5 CD1 mouse embryos as described with minor modifications.17–20,33 Briefly, the cerebral cortex was dissected out in Hank’s Balanced Salt Solution buffered with 0.1 M HEPES (pH 7.3) and dissociated with 0.25% trypsin-EDTA (Life Technologies) for 20 min at 37°C, followed by trituration. Dissociated cells were initially plated in neural plating medium (MEM, 10% FBS, 0.6% D-Glucose (w/v), and 10 U/mL penicillin-streptomycin) at a density of 5–8×104 cells/well on 24-well plates with coverslips (12 mm, Fisher Scientific) coated with poly-D-lysine (100 µg/mL). Five hours after plating, the medium was replaced with pre-conditioned neural maintenance medium (Neurobasal medium with B-27 supplement, Glutamax-I supplement, and 10 U/mL penicillin-streptomycin), which was prepared from 7–10 day old astroglial culture (1 day incubation). For primary cortical neuron transfections, the Ca2+-phosphate mediated method was used on 3-day old cultures as previously described with slight modification.21,34 A total of 0.5–1 µg of DNA was used for 12 mm coverslips in 24 well plates. Twenty min before the transfection, the neural maintenance medium was changed to transfection medium (same as neural maintenance medium but without penicillin-streptomycin). Four to five days after transfection, neurons were fixed with 4% paraformaldehyde and used for immunofluorescence analyses. The transfection rates among the Reep1 constructs in the primary cells were similar to each other.
RNA in situ hybridization
Whole mount (E9.5) and section (E14.5 and postnatal day 1; P1) RNA in situ hybridization using digoxigenin-labeled Reep1 riboprobes was performed as previously described.22–27,35,36
Northern Blot analysis
Adult tissue Northern blotting was performed as previously described using mouse multiple-tissue Northern blot (Clontech), ULTRAhyb (Ambion) and Reep1 RNA probes.28
Quantification of degenerating neurites and Sholl analysis
For quantification of degenerating neurites, sixteen 20× (objective) fields per sample were blindly picked and imaged using Zeiss Zen Pro software connected with Hamamatsu ORCA-Flash 4.0 camera attached to a Zeiss Observer Z1 inverted microscope. A 5×5 grid (the area of each square in the grid = 5000 µm2) was superimposed on each image with Grid plug-in for Fiji. Neurites in each square of the grid were manually scored as either whole or beaded as previously described,29,37 and values were expressed as a percentage of degenerating neurites (beaded neurites) among total transfected neurites (sum of beaded and whole neurites). Values from each square (total 16 squares) were averaged for each image, and the resulting values from each image (total 10 images) were averaged for each construct (n=160 per construct from two independent experiments). For Sholl analysis with Reep1 constructs, the entire coverslip (12 mm) was imaged using Zeiss Zen Pro software connected with Hamamatsu ORCA-Flash 4.0 camera attached to a Zeiss Observer Z1 inverted microscope (10× objective). 30 neurons from each sample (10 neurons per coverslip, three independent experiments) were randomly picked and cropped out, neurites of each neuron were traced using Simple Neurite Trace plug-in for Fiji, and automated analysis was carried out using Sholl Analysis in Fiji.30,31,38 For Sholl analysis with shRNAs, 30 to 40 neurons from each sample were randomly picked for imaging (Image-Pro software connected with Hamamatsu C5810 camera attached to a Nikon TE-300 microscope), each image was overlaid with concentric circles (25 µm intervals) generated in Photoshop, and Sholl analysis was performed manually by counting the numbers of neurites that intersected each circle.
Biochemical fractionation of the mouse brain
Embryonic mouse brains (E17.5) were fractionated into cytosol, crude mitochondria, pure mitochondria, ER, and MAM fractions as previously described.32,39,40
Western blot
HEK293 or HeLa cells were transfected with DNA constructs or shRNA constructs as indicated. At 48 or 72 hrs after transfection, the cells were lysed in lysis buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% TritonX-100) containing a protease inhibitor cocktail (Roche). 10–20 µg of total protein was loaded in each lane, separated by 10% or 4–12% SDS/PAGE and blotted onto PVDF membrane. The blots were blocked with 5% milk, followed by incubation with indicated primary antibodies including anti-V5 antibody (Invitrogen, mouse monoclonal, 1: 5000), anti-GFP antibody (Santa Cruz, rabbit polyclonal, 1:1000), anti-REEP1 antibody (Sigma-Aldrich, rabbit polyclonal, 1:1000), or anti-α tubulin antibody (Santa Cruz, mouse monoclonal, 1:1000), and incubation with HRP-conjugated goat anti-rabbit or mouse IgG secondary antibody (1:5000, Jackson Immunology). For subcellular fractionation samples, 30 µg of protein for each fraction was loaded in each lane and separated by 4–12% SDS/PAGE. The primary antibodies used for subcellular fractionation samples include anti-ATP5A (Abcam, mouse monoclonal, 1:1000), calnexin (BD Biosciences, mouse monoclonal, 1:2000), sigma-1R (ProteinTech, rabbit polyclonal, 1:2000), and REEP1 (Sigma-Aldrich, rabbit polyclonal, 1:1000) antibodies. Blots were developed using ECL kit (SuperSignal West Pico- or Femto Chemiluminescent Substrate, Thermo Scientific) and scanned using ChemiDoc™MP imaging system (Bio-Rad). Quantification of REEP1 expression was normalized to GFP expression using Image Lab software (Bio-Rad).
Immunofluorescent staining and MitoTracker treatment
For immunofluorescent staining, HeLa cells were fixed with 4% paraformaldehyde for 10 min or with ice-cold methanol for 5 min at room temperature, rinsed with PBS, and treated with 0.1% triton X-100 for 5 min. After blocking in 10% goat serum in PBS, cells were incubated with a primary antibody, including anti-REEP1 (Sigma-Aldrich, rabbit polyclonal, 1:50), calreticulin (abcam, chicken polyclonal, 1:5000), or TOM20 (Santa Cruz Biotechnology, rabbit polyclonal, 1:500) antibodies at 4°C overnight. After rinsing with PBS, cells were incubated with appropriate secondary antibodies conjugated with a fluorescent dye (Alexa-fluor 488 or Alexa-fluor 594; Invitrogen) for 1 hour at the room temperature. After rinsing with PBS, cells were incubated with DAPI (Molecular Probes) for nuclear staining, and mounted with mounting medium (Southern Biotech). For MitoTracker treatment, before the fixation step, cells were incubated with 100 nM of MitoTracker® Red CMXRos (Invitrogen) for 30 min and then incubated with new medium for 30 min to wash out unincorporated residual MitoTrackers from the cells. Images were captured with Zeiss Zen Pro software using Hamamatsu ORCA-Flash4.0 camera attached to a Zeiss Observer Z1 inverted microscope, or Zeiss LSM 710 confocal microscope.
Split-RLuc8 reassembly assay
HEK293T cells (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium (Life Technologies) with 10% fetal bovine serum (Life Technologies) at 37°C with 5% CO2. Cells (4 × 105/well) were plated in 12-well culture plate the day before transfection. Expression constructs, Mito-RLuc8N and/or RLuc8C-ER were co-transfected with the indicated constructs into HEK293T cells using PEI (Polysciences). The cells were split into 96-well plate (4 × 104/well) at 6 hr post-transfection and further incubated for 24 hrs at which time Enduren live cell substrate (Promega) was added in culture medium for 2.5–3 hrs (30 µM). The luminescence was measured by POLARstar Omega microplate reader (BMG LABTECH).
Statistical analysis
Prism 6 statistical software (GraphPad) was used to generate charts, and unpaired t-test (two-tailed) with Welch’s correction was used to analyze statistical difference. P < 0.05 was considered statistically significant.
RESULTS
Reep1 is expressed in the brain, nerve ganglia, spinal cord, and somites during development
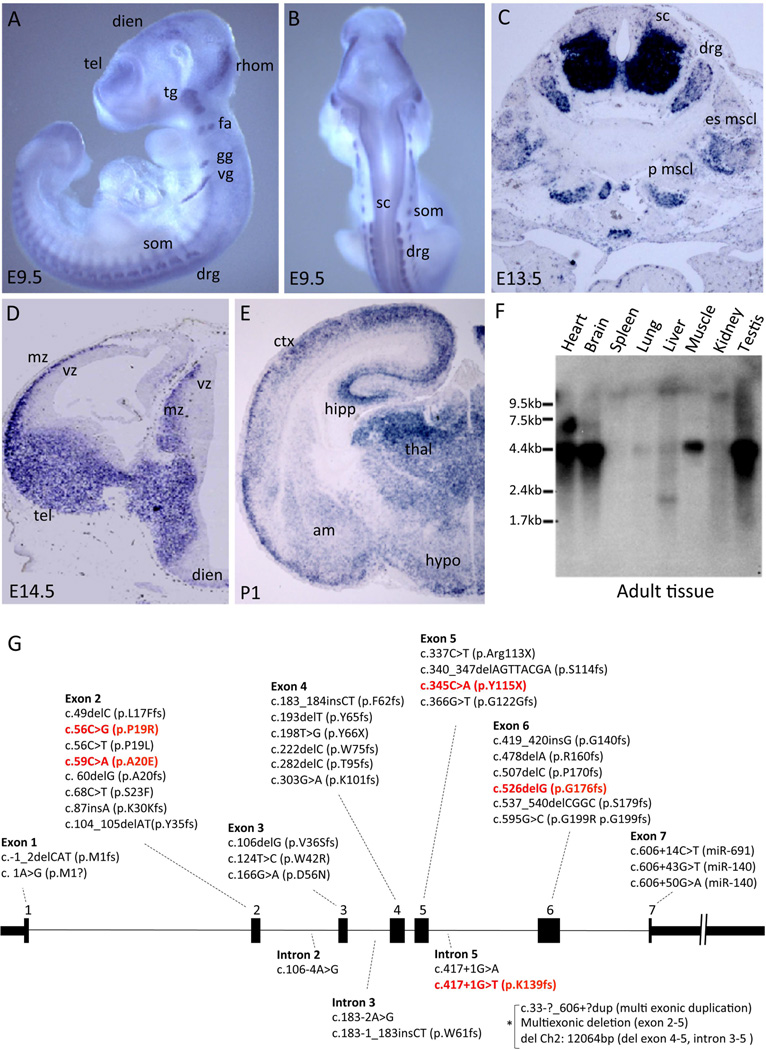
We identified Reep1 in a previous screen for novel genes involved in mouse forebrain development.28,32 To validate the screen and begin characterizing a function for REEP1, we first analyzed the expression of Reep1 by RNA in situ hybridization. On embryonic day 9.5 (E9.5), mouse Reep1 was detected in the brain, spinal cord, peripheral nerves, and somites (Fig 1A and B). Its expression in the brain was observed in the telencephalon, diencephalon, and rhombencephalon. Reep1 was also present in the spinal cord and several cranial nerve ganglia including the trigeminal, facio-acustic, glossopharyngeal, and vagal ganglia as well as the dorsal root ganglia (Fig 1A and B). Its expression in the muscle tissues, such as the erector spinae muscle (es mscl) and psoas major muscle (p mscl), were also detected as seen in E13.5 embryos (Fig 1C). By E14.5, Reep1 showed prominent expression in the mantle zone of the telencephalon and diencephalon, where the postmitotic neurons reside, with little or no expression in the proliferative ventricular zone (Fig 1D). In the forebrain of the neonate (P1), Reep1 transcripts were predominantly detected in the neocortex, hippocampus, and thalamus, while weaker labeling was found in the amygdala and hypothalamus (Fig 1E). A Northern blot analysis using adult mouse tissue showed that Reep1 is expressed in the brain as well as in the heart, skeletal muscle and testis (Fig 1F).
Figure 1. Reep1 expression in the mouse.
(A–B) Whole mount RNA in situ hybridization of Reep1 (embryonic day 9.5; E9.5) (A, side view; B, rear view). Reep1 mRNAs were detected in the brain (telencephalon=tel, diencephalon=dien, rhombencephalon=rhom), spinal cord (sc), peripheral nerve ganglia (trigeminal=tg, facio-acustic=fa, glossopharyngeal=gg, vagal=vg, dorsal root=drg), and somites (som). (C) Reep1 RNA in situ hybridization on coronal section of the mouse spinal cord (E13.5). Reep1 is detected in the ventral spinal cord (sc), dorsal root ganglia (drg), erector spinae muscle (es mscl), and psoas major muscle (p muscl) (D–E) Reep1 RNA in situ hybridization on coronal sections of the mouse forebrain (E14.5 and P1). Reep1 is expressed exclusively in the mantle zone (mz) but not in the ventricular zone (vz). Its expression was detected in the neocortex (ctx), hippocampus (hipp), thalamus (thal), amygdala (am), and hypothalamus (hyp). (F) Mouse adult tissue Northern blot analysis detected Reep1 in the brain as well as in the heart, skeletal muscle and testis. (G) Schematic of the genomic structure (black boxes are exons 1 through 7) with known HSP mutations indicated.12,13,15,39,43,49,65–68 Mutations are distributed over the entire gene (those in red indicate the five REEP1 mutations modeled in this study). * denotes duplication or deletion mutations spanning more than one exon or intron.
HSP-associated Reep1 mutations lead to neuritic degeneration in mouse cortical neurons
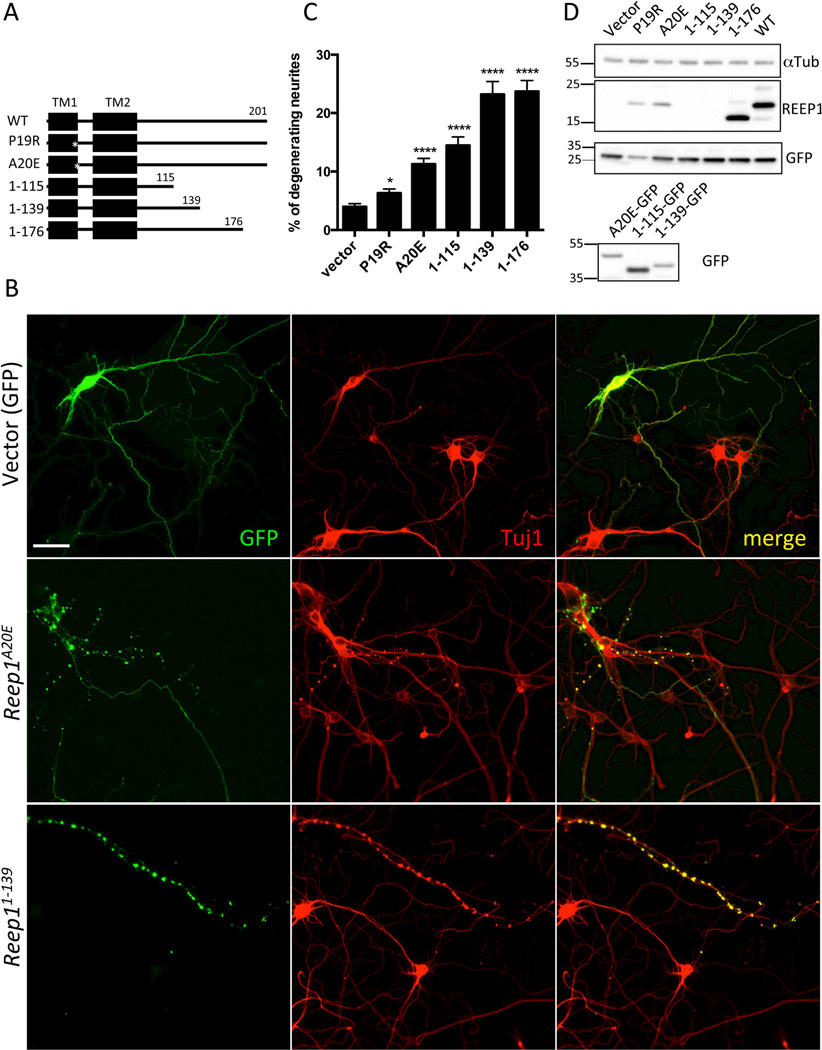
Mutations in REEP1 are known to cause hereditary spastic paraplegia with more than 40 mutations having been identified to date (Figure 1G). Axonal degeneration, predominantly in cortical motor neurons, is a common pathological feature of HSP.33,41 In order to model REEP1-associated degeneration in neurons, mouse primary cortical cultures were transfected with a construct expressing wild type mouse Reep1 or a construct expressing an HSP-associated mutant Reep1 in pCAG-IRES-GFP vector. The HSP-associated mutant Reep1 constructs included two point mutation constructs, Reep1P19R and Reep1A20E, and three truncated constructs, Reep11–115, Reep11–139, and Reep11–176, corresponding to the five human mutations, c.56C>G (p.P19R), c.59C>A (p.A20E), c.345C>A (p. Y115X), c.417+1G>T (p.K139fs), and c.526delG (p.G176fs)(Fig 2A)12,13,34,42,43. Beaded neurites, which are early signs of neurodegeneration,35–37,44 were observed in neurons transfected with mutant Reep1 constructs, as indicated by GFP (transfected cells) and Tuj1 (neuron-specific class III beta-tubulin) expression (Fig 2B). The degree of neurite degeneration was quantified as the percentage of beaded neurites among total transfected neurites (vector, 3.99 ± 0.52; Reep1P19R, 6.32 ± 0.69; Reep1A20E, 11.27 ± 0.98; Reep11–115, 14.46 ± 1.46; Reep11–139, 23.19 ± 2.20; Reep11–176, 23.69 ± 1.90; Reep1WT, 18.36 ± 2.31; values are in percentage; n=160 per construct) (Fig 2C). Reep1 mutant constructs showed a significant, though varying, increase in the percentage of degenerating neural processes when compared to a vector construct encoding GFP alone. To control for the variation in protein expression levels, we normalized the percentage of degenerating neuronal processes to the level of protein expression, which was quantified by Western blot (Fig 2D, upper panel) (Reep1P19R, 17.35 ± 1.90; Reep1A20E, 35.52 ± 3.08; Reep11–176, 14.05 ± 1.11; Reep1WT, 11.19 ± 1.40; values are in arbitrary unit). The REEP1–115 and REEP11–139 truncated mutant proteins are not recognized by REEP1 antibody due to a deletion of the epitope region, thus normalization was not performed with these constructs. To determine if these two constructs were expressed, we generated GFP fusion constructs, expressed them in cell lines (HEK293T and HeLa) and Western blotted with a GFP antibody (Fig 2D, lower panel). After normalization, the neuritic degeneration rates of the neurons expressing REEP1P19Rand REEP1A20E were significantly increased compared to that of the neurons expressing wild type REEP1 (P=0.0187 and P < 0.0001, respectively). Our data provide evidence that disease-associated Reep1 mutations result in neuritic degeneration in mouse cortical neurons, possibly modeling human mutations with a similar degeneration phenotype.
Figure 2. Overexpression of HSP-linked Reep1 mutant constructs leads to neuritic degeneration in primary cortical neurons.
(A) Diagram of Reep1 wild type and mutant constructs. Numbers above the line indicate amino acid residues. TM stands for putative transmembrane domain, and asterisks indicate the locations of the point mutations. (B) Representative images of the primary neurons transfected with the indicated Reep1 constructs or control vector. GFP staining indicates transfected cells and Tuj1 antibody was used for neuron-specific class III beta-tubulin staining. Beaded neurites indicate neuritic degeneration. Scale bar is 50 µm. (C) Quantification of the beaded neurites of the primary cortical neurons transfected with the indicated DNA constructs. Bars represent mean value ± SEM (*, P = 0.0159; ****, P < 0.0001; unpaired t-test; n = 160 per construct). (D) A representative Western blot (upper panel) shows expression levels of each Reep1 construct (in pCIG) used to transfect primary culture. As REEP1 antibody does not recognize REEP11–115 and REEP11–139, GFP-tagged Reep1 constructs were used to check their expression in Western blot (lower panel). The transfection efficiency of each construct is comparable. αTub: α-Tubulin
HSP-associated Reep1 mutations result in defective neurite growth in mouse cortical neurons
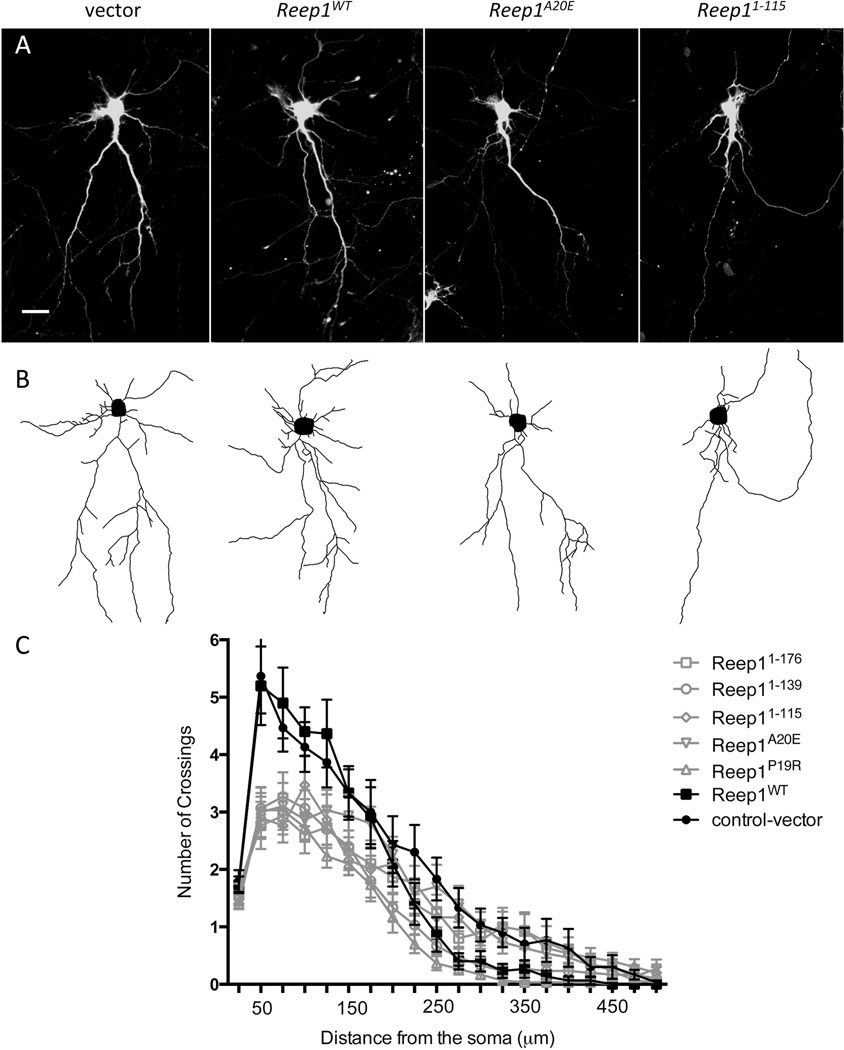
Several of the abnormalities observed in HSP patients, including the thin corpus callosum, have been proposed to be developmental in origin and potentially reflect a defect in neurite outgrowth.4,28,45,46 More recently, it has been reported that neurons derived from human pluripotent stem cells (hiPSCs) from HSP patients carrying SPASTIN (SPG4) mutations have a decreased neuritic complexity.37,47 To investigate whether Reep1 mutations associated with HSP would also result in neuritic growth abnormalities, we transfected cortical neural cultures with wild type or mutant Reep1 expression constructs. Reep1 mutant constructs led to reduction in neuritic complexity when compared to that of the wild type or vector construct; similar to what had been observed in neurons derived from SPG4 hiPSCs (Fig 3A). Individual neuronal images were used to trace neurites semi-automatically (Fig 3B) and measure the level of neuritic complexity by Sholl analysis (See Materials and Methods for detail) (Fig 3C). Neurons expressing disease-associated Reep1 mutant constructs had a significantly decreased number of crossings of the proximal neurites compared to those expressing a control vector or wild type Reep1 (Fig 3C). The effect on distal neurites was not significant, although highly variable between mutations.
Figure 3. Overexpression of HSP-linked Reep1 mutant constructs leads to defective neurite growth in primary cortical neurons.
(A) Representative images of the primary cortical neurons transfected with GFP-tagged control vector, Reep1WT, Reep1A20E or Reep11–115, and stained with GFP antibody. (B) Neurite tracings of the corresponding images in (A) using Simple Neurite Tracing plug-in for Fiji. Note some of the GFP positive neurites belonging to adjacent neurons were not traced. (C) Sholl analysis profiles of the primary cortical neurons transfected with the indicated constructs (n=30 neurons per construct, 3 independent experiments). Intersections were counted at 25 µm intervals from the soma center to a radius of 500 µm. Curves represent mean intersection values ± SEM. Scale bar in A is 50 µm.
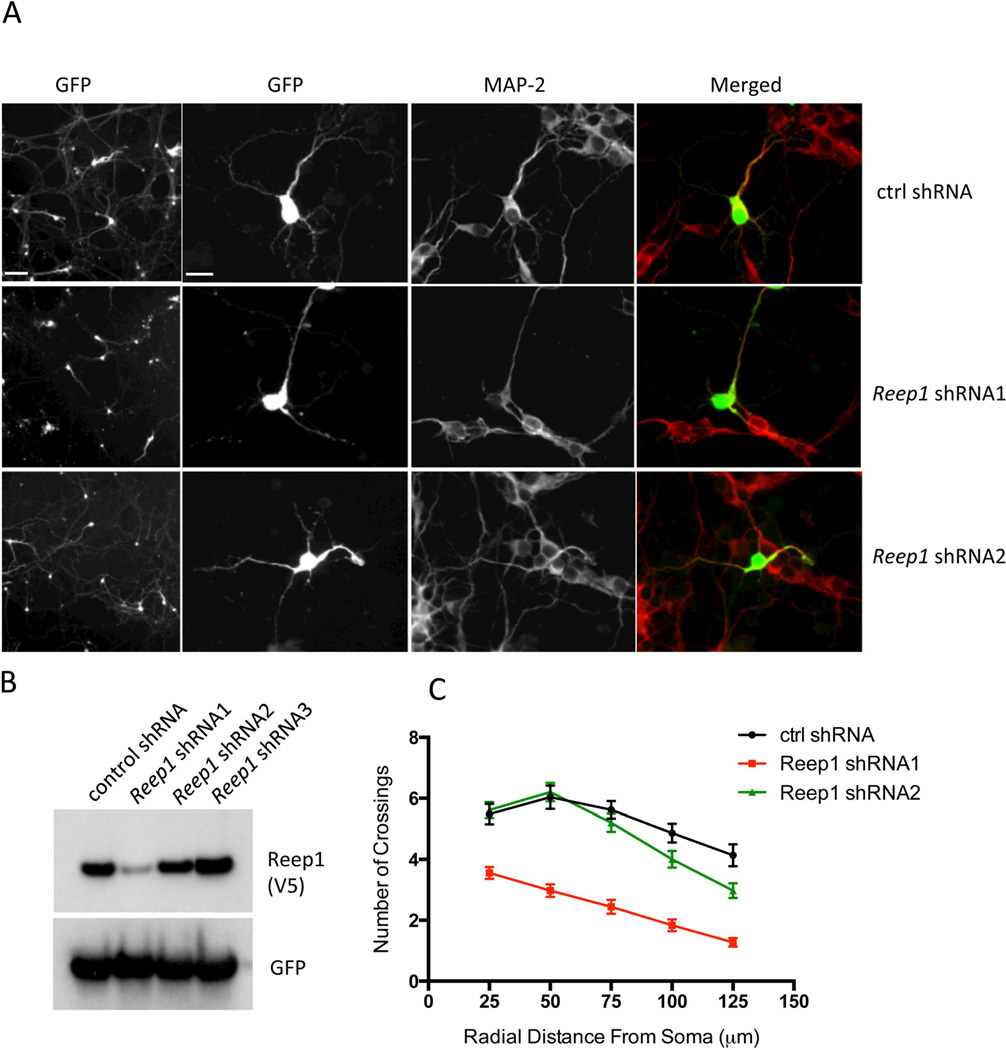
In order to exclude an overexpression artifact, we also transfected primary neurons with Reep1 specific shRNAs or control shRNA. Neurons transfected with Reep1 shRNA1, the most effective knock down construct (Fig 4B), exhibited a decrease in neuritic branch formation (Fig 4A). The Sholl analysis shows that the number of crossings in Reep1 shRNA1 expressing neurons is significantly reduced compared to the control or shRNA2 (Fig 4C). These data confirm that the endogenous REEP1 is necessary for normal neuritic branching and growth, and that neuritic defects observed with mutant REEP1 are not simply due to an overexpression artifact. These data suggest that mutations in REEP1 also impair neurite development, a finding that might explain the thin corpus callosum found in some patients.
Figure 4. The effect of endogenous REEP1 on neurite growth in primary cortical neurons.
(A) Representative images of the primary cortical neurons transfected with control shRNA or Reep1 specific shRNA1 or shRNA2 (see panel B below). Low (left panels) and high magnification (second panels from left) images of GFP fluorescence identify transfected cells. MAP-2 antibody was used for neuron-specific microtubule staining. Neurons transfected with the Reep1 shRNA1 had more simplified neurite morphology compared to those with the control shRNA. (B) Western blot to detect the level of REEP1 after co-transfecting Reep1-V5 with the indicated shRNA constructs to HEK293T cells. Reep1-specific shRNA1 was the most effective while the other two shRNAs had little or low activity. (C) Sholl analysis profiles of the primary cortical neurons transfected with the indicated shRNA constructs. Intersections were counted at 25 µm intervals from the soma center to a radius of 125 µm. Curves represent mean ± SEM (n=30 neurons). Scale bars in A are 100 and 50 µm (from the left).
REEP1 is detected in the ER-mitochondria contact sites known as mitochondria-associated ER membranes (MAMs)
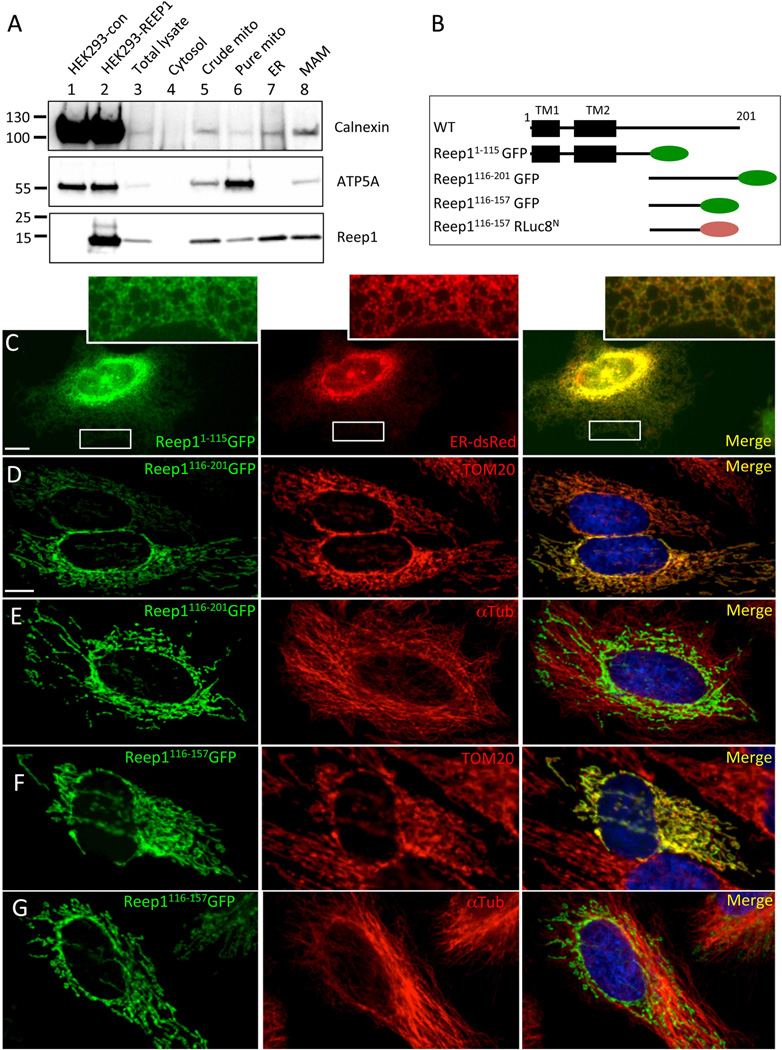
To explore how Reep1 mutations might give rise to the observed neuritic defects, we next examined the cellular function of REEP1. We first determined the subcellular localization of REEP1. It was initially reported as a mitochondrial protein, but later studies localized it in the ER.5,12,3839,40,48 The possibility of it being in the ER-mitochondria interface had been raised but never tested.13,16,28 Thus, we took a biochemical approach to determine if REEP1 is detected in the ER-mitochondria contact sites known as mitochondria-associated ER membranes (MAMs). Mouse embryonic brain extract (E17.5) was used for subcellular fractionation and assayed for the presence of REEP1 along with known markers in each fraction (Fig 5A). As expected, calnexin, an ER protein known to be enriched at the MAM,19,40,41 was present in the ER and MAM fractions but little in the pure mitochondrial fraction. Conversely, ATP5A, a mitochondrial protein, was predominantly detected in the pure mitochondrial fraction, but none in the ER. For REEP1, it was mainly detected in the ER but also significantly in the MAMs, providing evidence that it is present in the ER-mitochondria interface. Low level of REEP1 was also detected in the pure mitochondrial fraction, most likely due to small amounts of ER contamination, although we cannot rule out the possibility that some REEP1 might be present in the mitochondria.
Figure 5. REEP1 is detected in the mitochondria, ER, and ER-mitochondrial contact sites (MAM).
(A) Western blot using standard subcellular fractionation of mouse brain homogenate (E17.5) into cytosolic, crude mitochondrial, pure mitochondrial, ER, and MAM fractions (ATP-5A, mitochondrial marker; calnexin, ER and MAM marker). REEP1 was mainly detected in the ER and MAM fractions. (B) Schematic diagram of Reep1 wild type and mutant constructs. Numbers above the line indicate amino acid residues. TM stands for putative transmembrane domain and oval shapes depict GFP (green) or RLuc8N (brown). (C) HeLa cells transfected with the Reep11–115-GFP construct show co-localization with the ER (ER-dsRed), whereas the REEP1116–201-GFP (D, E) or Reep1116–157-GFP (F, G) is co-localized with mitochondria (TOM20) (D, F), but not with microtubules (αTub) (E, G). Images of each inlet in C are enlarged images of the boxed areas below. αTub: α–tubulin. Scale bars in C and D are 10 µm.
We next sought to determine whether REEP1 can be targeted to the mitochondria. We expressed a series of GFP-tagged Reep1 constructs (see schematic diagram in Fig 5B) in HeLa cells and examined their subcellular localization. Both full-length REEP1 as well as an N-terminal protein, REEP11–115, were detected in the ER as determined by overlap with ER-dsRed (Fig 5C) or calreticulin (Fig 8B); both ER markers. In contrast, the C-terminal protein, REEP1116–201, targeted to the mitochondria as demonstrated by its overlap with TOM20, a mitochondrial marker (Fig 5D). With another GFP-tagged deletion construct, Reep1116–157, we further honed down the domain required for mitochondrial localization to a.a. 116–157 (Fig 5F). Remarkably, when the amino acids 116–201 or 116–157 of REEP1 were attached to another protein sequence (Renilla luciferase 8), it was sufficient to target the heterologous protein to the mitochondria (data not shown), confirming the specificity of the mitochondrial targeting of the REEP1116–201 or REEP1116–157 independent of the tag. As the C-terminal domain of REEP1 is known to bind microtubules, we next examined whether REEP1116–201 or REEP1116–157 localization in the mitochondria is simply due to their alignment with microtubules. Co-staining with α-Tubulin ruled out this possibility as their mitochondrial expression does not overlap with α-Tubulin staining (Fig5E and 5G). Collectively, these data indicate that the N-terminal domain of the REEP1 (a.a. 1–115) is responsible for ER localization, whereas the middle domain (a.a. 116–157) is sufficient for mitochondrial localization. Together our results provide evidence of REEP1 being localized at the ER and mitochondria interface, potentially reconciling contradicting reports on REEP1 subcellular localization.5,6,12,13,42,43
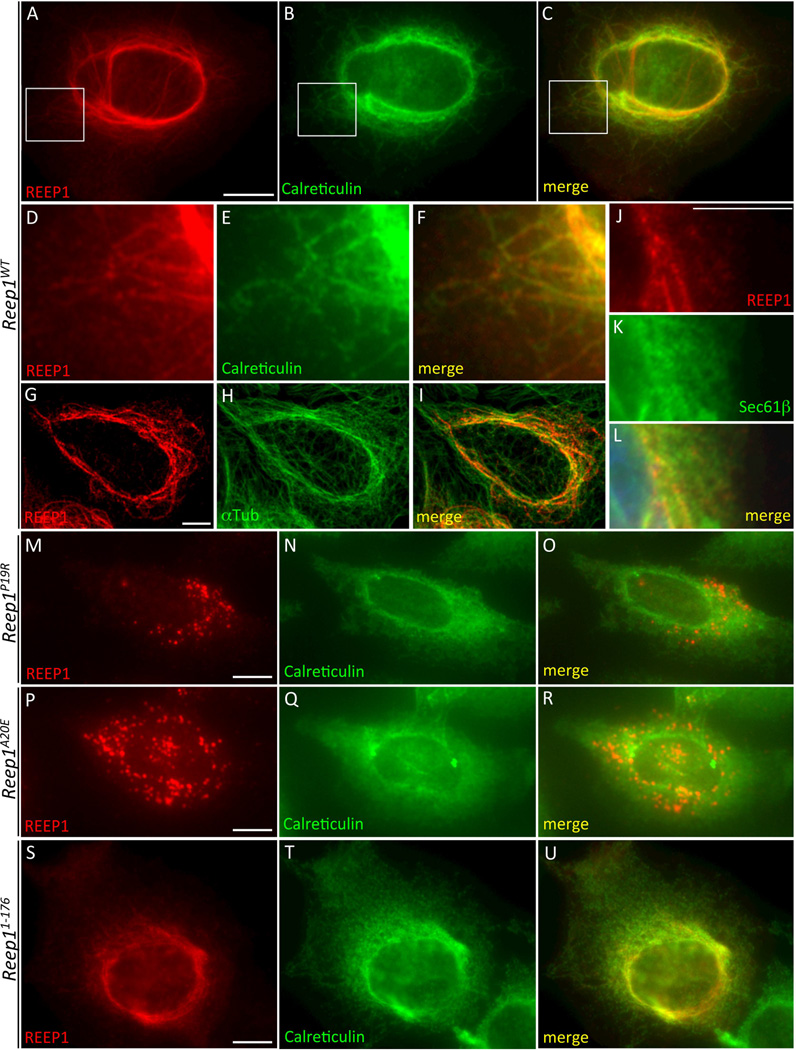
Figure 8.
REEP1 disease mutants do not shape ER into tubular pattern unlike wild type REEP1. HeLa cells expressing each indicated construct on the left were immunostained with REEP1 (A, D, G, J, M, P, S) and calreticulin (B, E, N, Q, T) or α-tubulin (H), or detected with GFP-Sec61β fluorescence (K). Wild type REEP1 (A–L) changed ER pattern into tubular whereas mutant REEP1s (M–U) did not, except REEP11–176, which showed mild changes. Scale bars are 10 µm.
REEP1, but not HSP-associated mutations, facilitates ER-mitochondrial contacts
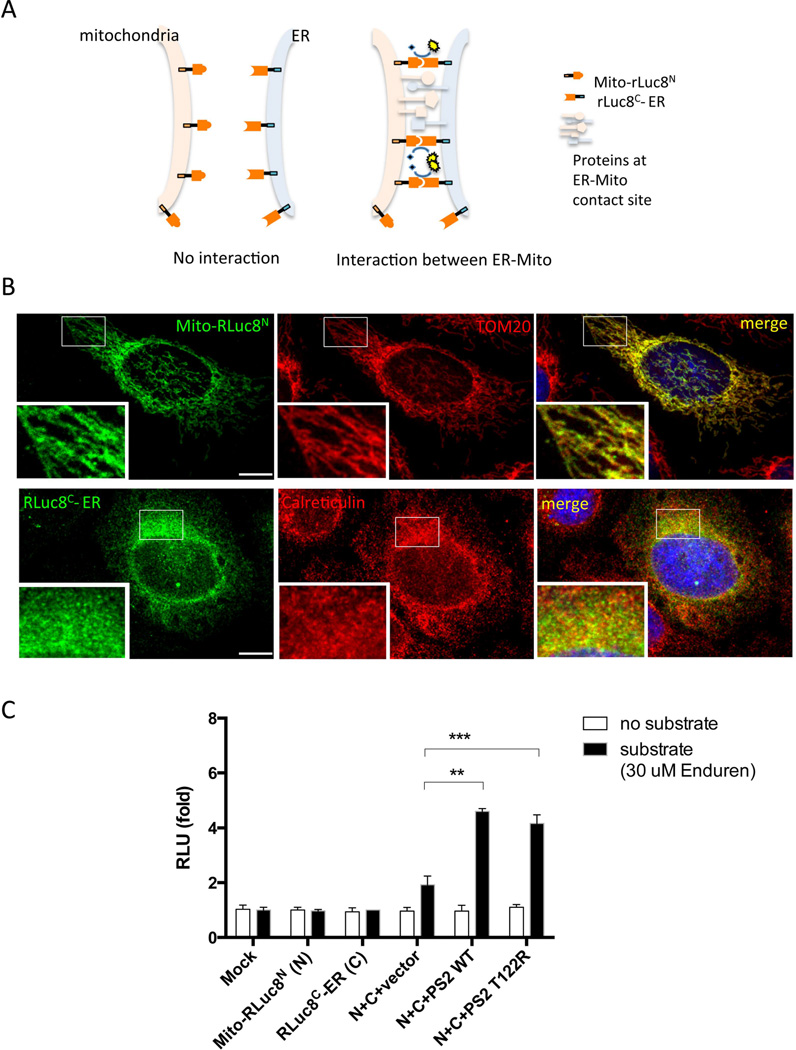
Based on our data indicating that REEP1 resides at the ER-mitochondria contact sites, we hypothesized that REEP1 might play a functional role in ER-mitochondrial contact formation. To test our hypothesis, we developed a ‘split-Renilla Luciferase 8 (RLuc8) reassembly assay’, a tool to quantitatively measure the level of ER-mitochondria interactions in live cells. RLuc8 was split into N-terminal (RLuc8N) and C-terminal (RLuc8C) halves based on the previously determined split site,30,37,44 and each half was conjugated with either a mitochondrial or an ER targeting sequence (see Materials and Methods for detail). The resulting constructs were named Mito-RLuc8N and RLuc8C-ER, respectively (Fig 6A). We verified their targeting to the mitochondria or ER in the cells by transfecting these constructs and determining their co-localization with TOM 20 or calreticulin (Fig 6B). Next, these constructs were transfected into HEK293T cells and luciferase activity measured (Fig 6C). Neither mitochondria-targeted N-terminal RLuc8 (Mito-RLuc8N), or ER-targeted C-terminal RLuc8 (RLuc8C-ER) alone produced significant luciferase activity (Fig 6C). However, when presenilin2 (PS2) wild type or T122R mutant, known to facilitate ER-mitochondria interactions,4,22,45,46 was co-transfected with Mito-RLuc8N and RLuc8C-ER, a significant increase in luciferase activity was detected when compared to that of an empty vector (Fig 6C). These results validate our split-RLuc8 assay as a method to functionally assess the interaction between the ER and mitochondria.
Figure 6. Split-RLuc8 reassembly assay can measure the level of ER-mitochondria interactions.
(A) A schematic model depicting the split-RLuc8 reassembly assay. Where no interaction between ER and mitochondria exists, Mito-RLuc8N and RLuc8C-ER fail to interact and thus do not reconstitute RLuc8 activity. In contrast, where the ER and mitochondria interact, Mito-RLuc8N moves into proximity of RLuc8C-ER reconstituting full enzymatic activity, which is detected by the luminescence conversion of substrate. (B) HeLa cells expressing Mito-RLuc8N were immunostained with Flag (tagged to Mito-RLuc8N)-antibody together with TOM20. For RLuc8C-ER, HeLa cells expressing RLuc8C-ER and ER-dsRed were detected with Myc (tagged to RLuc8C-ER)-antibody immunostaining and ER-dsRed fluorescence. (C) Split-RLuc8 reassembly assay performed in HEK293T cells transfected with the indicated constructs in the presence or absence of the substrate. Relative luminescence was measured 24 hrs after transfection (RLU stands for relative luminescence units). Abbreviations: Mito-RLuc8N, mitochondria targeting sequence-N terminal half of the Renilla luciferase 8; RLucC-ER, C terminal half of the Renilla luciferase 8-ER targeting sequence; PS2 WT, presenilin 2 wild type; PS2 T122R, presenilin 2 T122R mutant. Bars represent mean ± SD (**, P = 0.0024; ***, P = 0.0009; unpaired t-test; n=3). Scale bars in B are 10 µm.
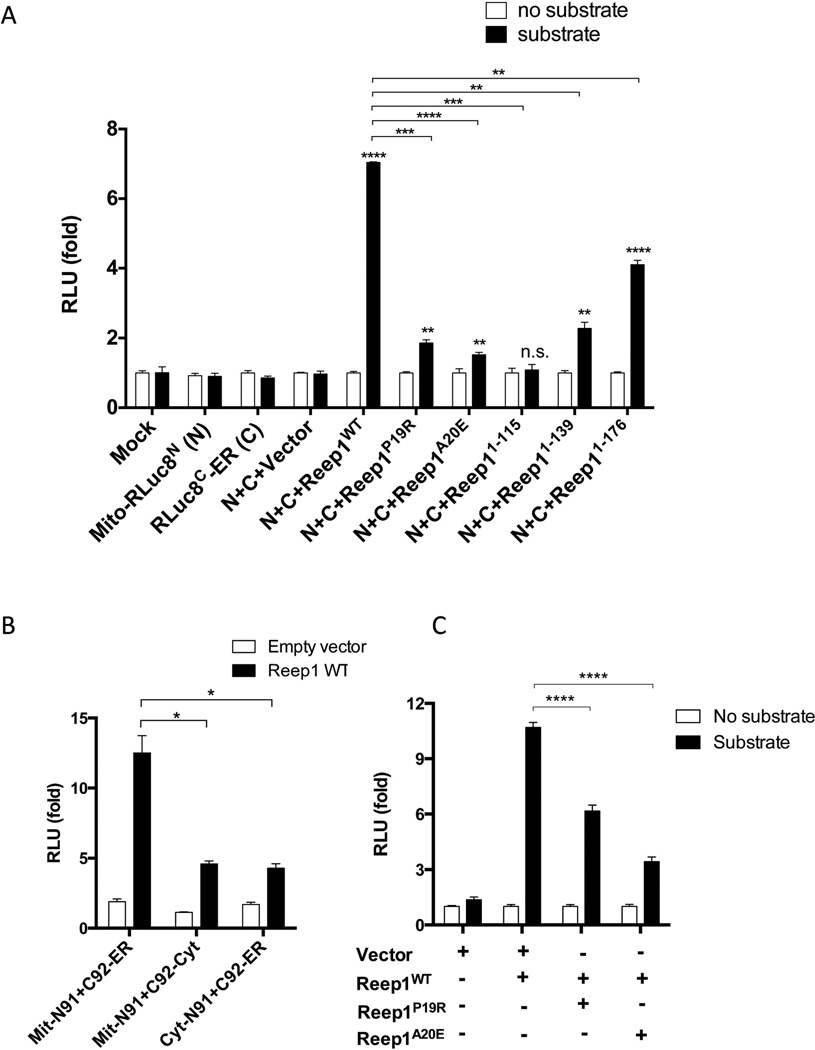
Using this assay, we next examined if co-transfection of Reep1 can increase ER-mitochondria interactions. Reep1 was co-transfected with the split-RLuc8 constructs and luciferase activity was measured (Fig 7A). Wild type Reep1 significantly increased luciferase activity when compared to the empty vector, indicating that it can indeed facilitate the interaction between the ER and mitochondria. When one of the split-RLuc8 constructs was expressed in the cytoplasm instead of being targeted to the ER- or mitochondria, the luciferase activity was minimally increased when compared to wild type (Fig 7B). This result supports that the ER and mitochondria specific localizations of the split-RLuc8 constructs are required for the restoration of RLuc8 activity. Next, our series of disease-associated Reep1 mutations, Reep1P19R, Reep1A20E, Reep11–115, Reep11–139, and Reep11–176 were used in our split-RLuc8 assay. None of these induced luciferase activity to the extent observed with wild type Reep1 (Fig 7A). Reep11–115, which only includes the ER association domain but not the mitochondrial domain, did not activate luciferase above baseline. Similarly, Reep1P19R and Reep1A20E, which possess point mutations in the putative transmembrane sequence within the ER association domain,5,12,47 also had relatively little effect. The other two truncated mutant constructs, Reep11–115 and Reep11–139, showed some activity but less than a control. Interestingly, Reep11–176 which contains both ER- and mitochondria- association domains, but not the C-terminal domain known to interact with microtubules,5,48 resulted in the most significant increase in luciferase activity, reaching more than 50% of the wild type. These results indicate that the disease-associated Reep1 mutations impair REEP1’s ability to facilitate ER-mitochondria contact. Furthermore, our data also suggest that in addition to the ER- and mitochondria association domains, the microtubule association domain is necessary for its full activity.
Figure 7. Wild type REEP1, but not REEP1s with HSP-associated mutations, facilitates ER-mitochondria contact formation.
(A) Split-RLuc8 reassembly assay performed with the indicated constructs. Wild type Reep1 shows the greatest increase in luminescence, while Reep1s with mutations either have little or no effect. Bars represent mean ± SEM. The statistical difference was analyzed using unpaired t-test between the vector versus each Reep1 construct (asterisks marked on top of each bar; ****, P < 0.0001; **, P = 0.0018; **, P = 0.0082; ns, non-significant; **, P = 0.0080; ****, P < 0.0001, from left to right in the graph; n=3), and between the wild type Reep1 and each mutant construct (asterisks marked on top of each bracket; ***, P = 0001; ****, P < 0.0001; ***, P = 0.0005; **, P = 0.0012; **, P = 0.0014, from left to right; n=3). Abbreviations: RLuc8, Renilla luciferase8; Mito-RLuc8N, mitochondria targeting sequence-N terminal half of the Renilla luciferase 8; RLucC-ER, C terminal half of the Renilla luciferase 8-ER targeting sequence; C, RLucC-ER; N, Mito-RLucN (B) Split-RLuc8 reassembly assay performed with the indicated constructs. Mitochondria- and ER- targeting is required for the full activity of REEP1 in split-RLuc8 assay. Bars represent mean ± SEM (*, P < 0.05; unpaired t-test; n=3). Abbreviations: Mit-N91, mitochondria targeted RLucN; C92-ER, ER targeted RLucC; cyt-N91, cytoplasm targeted RLucN; C92-cyto, cytoplasm targeted RLucC (C) Split-RLuc8 reassembly assay performed with co-transfection of Reep1WT with Reep1P19R or Reep1A20E. Bars represent mean ± SEM (****, P < 0.0001; unpaired t-test; n=3).
Haploinsufficiency or loss-of-function has been suggested for the underlying molecular genetic mechanism of REEP1 in SPG31.6,13,16,49 The differences in luciferase activity observed in point mutations and truncated mutations (Fig 7A), prompted us to test if Reep1 mutations also function in a dominant negative fashion. Thus, we examined if Reep1P19R or Reep1A20E inhibit the function of wild type Reep1. For this, Reep1P19R or Reep1A20E was co-transfected with the Reep1WT and the split-RLuc8 constructs, and luciferase activity was measured (Fig 7C). The addition of Reep1P19R or Reep1A20E significantly decreased the activity of the wild type Reep1, raising the possibility that these mutations can potentially function in a dominant negative fashion, at least in the context of modulating ER-mitochondria interactions. Truncated mutations, Reep11–115, Reep11–139, and Reep11–176 were also tested but the results were highly variable precluding definitive interpretation.
Finally, previous studies showed that the ER-mitochondrial contact sites have diverse structural features and in some cases the ER membrane completely wraps around the mitochondria.19,32,40,50 Thus, maintaining the ER membrane as a curved structure is crucial at these contact sites. Given that REEP1 is thought to shape ER membrane into tubular structures5,6,12, we examined whether the REEP1 mutant proteins retain this function. HeLa cells were transfected with wild type or mutant Reep1 constructs and co-immunostained with calreticulin or GFP-Sec61β marker (Fig 8). Cells transfected with the wild type Reep1 showed a tubular ER pattern as revealed by calreticulin (Fig 8A–F), GFP-Sec61β (Fig 8J–L), and α-Tubulin (Fig 8G–I) staining, consistently with previous reports.5 However, the Reep1 mutant constructs, Reep1P19R, Reep1A20E, Reep11–115, and Reep11–139 (Fig 8M–R and data not shown) did not change ER into tubular structures, although Reep11–176 had a mild effect (Fig 8S–U). These results suggest that the ER shaping function of Reep1 might be associated with ER-mitochondrial contact formation directly or indirectly, although further studies are required to confirm this observation. Taken together, our data support a model where REEP1 can facilitate physical ER-mitochondria interactions, possibly through its membrane shaping function, and this function is compromised in disease-associated Reep1 mutations.
Discussion
In this study we have found that REEP1 is present at the ER-mitochondria contact sites, where it can facilitate ER-mitochondria interactions. Furthermore, we found that HSP-associated Reep1 mutations impair these interactions, possibly explaining defects in neuritic growth and the degeneration observed in cultured cortical neurons. This novel function provides new insights into the pathogenesis of HSP that will potentially inform new therapeutic options for HSP patients with REEP1 mutations.
REEP1 subcellular localization and its function at the MAM
When REEP1 was initially identified as an HSP gene, its protein product was reported to localize in the mitochondria12,22. Later studies, however, showed that REEP1 localized to the ER5,6(ref). It is not clear whether this discrepancy is due to the differences in REEP1 antibody specificity or due to different cell types. Our data show REEP1 contains both a mitochondrial localization domain and an ER localization domain, suggesting the possibility that REEP1 associates with mitochondria as well as ER, and potentially reconciles the previous conflicting data,5,6,12 although it is not direct evidence.
Our results also suggest that REEP1 is present and functions in the MAMs. How does REEP1 facilitate the ER-mitochondria interactions? At least three possibilities exist. First, it could directly act as a tethering protein, bridging the two organelles through the formation of homodimers, heterodimers or oligomers. Supporting this model, REEP1 is able to form oligomers and is known to interact with HSP related proteins, spastin and atlastin-1, through hydrophobic domains.5,6,49 Based on our data, it is also reasonable to posit that ER- and mitochondria- localized REEP1 can form dimers with each other, thus bridging the two organelles, similar to MFN2.32,50,51
Second, REEP1 might indirectly facilitate ER-mitochondria interactions through bending ER membranes, making it topologically possible for the ER to wrap around mitochondria. Ultrastructural data indicate that ER-mitochondria contacts are diverse: in most cases ER tubules form single contacts covering approximately 10% of the mitochondrial surface, while in other cases the ER tubules cover one-half or even completely envelope the mitochondria.5,32,52,53 Thus, it is conceivable that the membrane shaping function of REEP1 could indirectly enable ER to form contacts with mitochondria.
Finally, REEP1 could function as a molecular chaperone or adaptor that stabilizes or distributes other proteins, i.e. tethering proteins, at the MAM, thus indirectly facilitating ER-mitochondria interactions. A molecular chaperone function of REEP1 has been suggested based on its sequence homology to the plant stress-induced chaperone, HVA22s.5,54 In fact, there are multiple molecular chaperones enriched at the MAM, including sigma-1R, BIP, GRP75, and HSP60; REEP1 could function as another.12,18 The three roles described here appear to be most reasonable, although other possible functions cannot be excluded, and future studies will be directed at elucidating the precise function.
Altered ER-mitochondria contacts and neurodegeneration
Physical contacts between the ER and mitochondria have been observed and functionally implicated in axons, dendrites, and neuronal soma.5,6,55–58 For example, in the synapse, ER-mitochondrial contact functions in intracellular Ca2+ signaling and for maintaining and modulating synaptic activity.5,6,12,56 More recently, disruptions in ER-mitochondrial contacts have been proposed as a pathogenic mechanism leading to several neurodegenerative diseases.5,21 Presenilins, whose mutations are associated with familial forms of Alzheimer’s disease (AD), are enriched in MAMs.51,59 The T122R mutation in presenilin 2 (PS2-T122R) increases ER-mitochondria interactions, resulting in elevated Ca2+ transfer from the ER to mitochondria and leading to chronic mitochondrial Ca2+ overload, which is believed to damage metabolic function and eventually cause neuronal death.18,22,32,52,53 In a similar fashion, α-synuclein, parkin, and DJ-1, whose mutations are responsible for familial forms of Parkinson’s disease (PD), and VAPB, whose mutations are associated with amyotropic lateral sclerosis (ALS), are also detected in the MAM fraction and their disease mutations alter ER-mitochondria interactions.25,26,54–58,60–62
The axonal injury model has been used to investigate how alterations in ER-mitochondria interactions might lead to degeneration.55,56,63,64 A mechanical axotomy disrupts Ca2+ equilibrium in the axon, activating plasma membrane channels and inducing Ca2+ release from the axonal ER. Ca2+ release can be buffered by ER-associated mitochondria. Increased contacts between them will lead to mitochondrial Ca2+ overload, which eventually triggers opening of the mitochondrial permeability transition pore (mPTP). mPTP opening leads to Ca2+ overload in the axon, increased generation of reactive oxygen species, and a drop in ATP production.21,55 As a result, the direction of the Na+/Ca2+ antiporter changes, and additional calcium enters from the extracellular space, ultimately leading to axonal degeneration.55,59,63,64 These data linking ER Ca2+ release with mPTP for axonal degeneration are consistent with those obtained in non-neuronal cells leading to cell death,22,32 and they suggest that a similar cascade might happen in neurodegenerative diseases including REEP1-associated HSPs.
The effect of disease-associated Reep1 mutations on ER-mitochondria interactions, neuritic growth and degeneration
While the disease-associated mutant proteins in AD, PD, and ALS increase ER-mitochondria contacts, the opposite is observed with REEP1 disease-associated mutations. Thus, it appears that not only an abnormal increase, but also an abnormal decrease, in ER-mitochondria communication might result in neurodegeneration. Support for this result comes from the knock down study of either of the two proteins essential for MAM tethering, sigma-1 receptor and phosphofurin acidic cluster sorting protein-2 (PACS-2), which resulted in neuronal degeneration.25,26,39,60–62 In healthy cells, the ER-mitochondria tethering ensures the propagation of inositol 1,4,5-triphosphate receptor (IP3R)-linked Ca2+ signals to the mitochondria to coordinate ATP production and to enable mitochondrial Ca2+ buffering. In the situation where the ER-mitochondria coupling is reduced, such as in cells with sigma-1 receptor or PACS-2 knocked down, or cells expressing Reep1 disease mutations, the Ca2+ signal propagation to the mitochondria is suppressed and the Ca2+-dependent control of mitochondrial metabolism likely becomes at risk. Consistent with this, cells obtained from HSP patients with REEP1 mutations showed reduced mitochondrial respiratory rate and oxygen consumption.13,14,55,63,64 In contrast, when the coupling is enhanced, as in the cells with mutations in PS2, α-synuclein, parkin and DJ-1, mitochondria are susceptible to Ca2+ overloading and may undergo mPTP permeabilization, thus committing the cells to a cell death pathway. Thus, maintaining proper ER-mitochondria interactions seems to be pivotal for maintaining normal mitochondrial function and ultimately neuronal integrity.
The most common type of REEP1 mutations reported to date is small frame shift mutations, which generate pre-mature stops.49,55 For this reason, haploinsufficiency was suggested as a major molecular genetic mechanism of SPG31. A recent Reep1 knock-out mouse study also supported loss-of-function or haploinsufficency model.6,55,63,64 Interestingly, our data with two point mutations, Reep1P19R and Reep1A20E, suggest that these non-functional mutant REEP1 proteins, which lost the ability to facilitate ER-mitochondria interactions (Fig 7A), interfere with the function of wild type REEP1, giving a dominant negative effect (Fig 7C). Without neuropathological studies of the postmortem brain or spinal cord tissue from REEP1-associated HSP patients with known genotype, it is difficult to correlate our results with human phenotype. Furthermore, the stability and proper folding of the REEP1 point mutants and truncation mutant proteins need to be assessed in future studies through purification of the REEP1 and folding analyses such as circular dichroism or dynamic light scattering.
In addition to the neuritic degeneration, our Scholl analysis indicates that Reep1 disease mutations lead to a neuritic growth defect, similar to Reep1 specific knock down. These data suggest REEP1 is also required for normal neuritic development. In agreement with our results, a recent study using neurons derived from human induced pluripotent stem cells from patients with SPG4 mutations reported a decrease in the complexity of neurites. In contrast, no developmental defects were observed in primary neuronal culture from Reep1 mutant mice.6,32 This discrepancy between the knock out mouse study and our results could reflect difference in knock down versus knock out approaches. In shRNA-mediated knock down, Reep1 is abruptly depleted, and cells may not be able to acutely adapt to this change. In contrast, mutant mice never express REEP1 and may compensate through unknown mechanisms.
In conclusion, our data provide compelling evidence that mutations in Reep1 alter ER-mitochondria interactions and that disruption in this cellular process may be a new pathogenic mechanism in REEP1-linked HSP. As the deregulation of this interaction between the two organelles is a common pathway in several neurodegenerative diseases, the split-RLuc8 reassembly assay we developed should provide a valuable tool for potential drug screening in multiple neurodegenerative diseases.
Acknowledgements
This work was supported by the NIH (NS46616) and the Ramzi Cotran Endowed Chair, Harvard Medical School. We thank Dr. Dennis Selkoe’s lab (Brigham and Women’s Hospital) for sharing their primary neuronal culture protocol, Dr. Ennio Chiocca’s lab (Brigham and Women’s Hospital) for shared resources, Dr. Matthew LaVoie’s lab (Brigham and Women’s Hospital) for reagents, Dr. Doug Epstein (Perelman School of Medicine, University of Pennsylvania) for a mouse cDNA library, Dr. Sanjiv Gambhir (Stanford School of Medicine) for Renilla luciferase 8 construct, and Dr. Paola Pizzo (University of Padova) for the presenilin 2 constructs.
Footnotes
Conflict of Interest Disclosure
The authors declare no competing financial interests.
Author Contributions
Y.L., I-T.C., G.C., and J.A.G conceived and designed the study. Y.L., I-T.C., G.C., and L.J.S., acquired the data. Y.L., I-T.C., G.C., J.A.G., conducted the data analysis. Y.L. and J.A.G. drafted the manuscript and all authors were involved in subsequent revisions. Y.L. and I-T.C. contributed equally.
References
- 1.Blackstone C. Cellular Pathways of Hereditary Spastic Paraplegia. Annu. Rev. Neurosci. 2012;35(1):25–47. doi: 10.1146/annurev-neuro-062111-150400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackstone C, O'Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat. Rev. Neurosci. 2010;12(1):31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fink JK. Hereditary spastic paraplegia: clinical principles and genetic advances. Semin Neurol. 2014;34(3):293–305. doi: 10.1055/s-0034-1386767. [DOI] [PubMed] [Google Scholar]
- 4.Fink JK. Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 2013;126(3):307–328. doi: 10.1007/s00401-013-1115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SH, Zhu P-P, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J. Clin. Invest. 2010;120(4):1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beetz C, Koch N, Khundadze M, et al. A spastic paraplegia mouse model reveals REEP1-dependent ER shaping. J. Clin. Invest. 2013;123(10):4273–4282. doi: 10.1172/JCI65665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beetz C, Pieber TR, Hertel N, et al. Exome Sequencing Identifies a REEP1 Mutation Involved in Distal Hereditary Motor Neuropathy Type V. The American Journal of Human Genetics. 2012;91(1):139–145. doi: 10.1016/j.ajhg.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Björk S, Hurt CM, Ho VK, Angelotti T. REEPs Are Membrane Shaping Adapter Proteins That Modulate Specific G Protein-Coupled Receptor Trafficking by Affecting ER Cargo Capacity. PLoS ONE. 2013;8(10):e76366. doi: 10.1371/journal.pone.0076366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hashimoto Y, Shirane M, Matsuzaki F, et al. Protrudin Regulates Endoplasmic Reticulum Morphology and Function Associated with the Pathogenesis of Hereditary Spastic Paraplegia. J Biol Chem. 2014;289(19):12946–12961. doi: 10.1074/jbc.M113.528687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montenegro G, Rebelo AP, Connell J, et al. Mutations in the ER-shaping protein reticulon 2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J. Clin. Invest. 2012;122(2):538–544. doi: 10.1172/JCI60560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shibata YY, Voss CC, Rist JMJ, et al. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283(27):18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Züchner S, Wang G, Tran-Viet K-N, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. The American Journal of Human Genetics. 2006;79(2):365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goizet C, Depienne C, Benard G, et al. REEP1 mutations in SPG31: Frequency, mutational spectrum, and potential association with mitochondrial morpho-functional dysfunction. Hum. Mutat. 2011;32(10):1118–1127. doi: 10.1002/humu.21542. [DOI] [PubMed] [Google Scholar]
- 14.McDermott CJ, Taylor RW, Hayes C, et al. Investigation of mitochondrial function in hereditary spastic paraparesis. Neuroreport. 2003;14(3):485–488. doi: 10.1097/00001756-200303030-00038. [DOI] [PubMed] [Google Scholar]
- 15.Hewamadduma C, McDermott C, Kirby J, et al. New pedigrees and novel mutation expand the phenotype of REEP1-associated hereditary spastic paraplegia (HSP) Neurogenetics. 2008;10(2):105–110. doi: 10.1007/s10048-008-0163-z. [DOI] [PubMed] [Google Scholar]
- 16.Deutch AY, Hedera P, Colbran RJ. REEPing the benefits of an animal model of hereditary spastic paraplegia. J. Clin. Invest. 2013;123(10):4134–4136. doi: 10.1172/JCI72324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowland AA, Voeltz GK. Endoplasmic reticulum–mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13(10):607–615. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi T, Rizzuto R, Hajnóczky G, Su T-P. MAM: more than just a housekeeper. Trends in Cell Biology. 2009;19(2):81–88. doi: 10.1016/j.tcb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto M, Hayashi T. New Insights into the Role of Mitochondria-Associated Endoplasmic Reticulum Membrane. Int Rev of Cell Mol Biol. 2011;292:73–117. doi: 10.1016/B978-0-12-386033-0.00002-5. [DOI] [PubMed] [Google Scholar]
- 20.Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER–mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 21.Calì T, Ottolini D, Brini M. Calcium and Endoplasmic Reticulum-Mitochondria Tethering in Neurodegeneration. DNA and Cell Biology. 2013;32(4):140–146. doi: 10.1089/dna.2013.2011. [DOI] [PubMed] [Google Scholar]
- 22.Zampese E, Fasolato C, Kipanyula MJ, et al. Presenilin 2 modulates endoplasmic reticulum (ER)–mitochondria interactions and Ca2+ cross-talk. Proc Natl Acad Sci USA. 2011;108(7):2777–2782. doi: 10.1073/pnas.1100735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Area-Gomez E, del Carmen Lara Castillo M, Tambini MD, et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. The EMBO Journal. 2012;31(21):4106–4123. doi: 10.1038/emboj.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cali T, Ottolini D, Negro A, Brini M. α-Synuclein Controls Mitochondrial Calcium Homeostasis by Enhancing Endoplasmic Reticulum-Mitochondria Interactions. J Biol Chem. 2012;287(22):17914–17929. doi: 10.1074/jbc.M111.302794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guardia-Laguarta C, Area-Gomez E, Rub C, et al. α-Synuclein Is Localized to Mitochondria-Associated ER Membranes. J Neurosci. 2013;34(1):249–259. doi: 10.1523/JNEUROSCI.2507-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ottolini D, Cali T, Negro A, Brini M. The Parkinson disease-related protein DJ-1 counteracts mitochondrial impairment induced by the tumour suppressor protein p53 by enhancing endoplasmic reticulum-mitochondria tethering. Hum Mol Genet. 2013;22(11):2152–2168. doi: 10.1093/hmg/ddt068. [DOI] [PubMed] [Google Scholar]
- 27.De Vos KJ, Morotz GM, Stoica R, et al. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum Mol Genet. 2012;21(6):1299–1311. doi: 10.1093/hmg/ddr559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho G, Lim Y, Zand D, Golden JA. Sizn1 Is a Novel Protein That Functions as a Transcriptional Coactivator of Bone Morphogenic Protein Signaling. Mol Cell Biol. 2008;28(5):1565–1572. doi: 10.1128/MCB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loening AM. Consensus guided mutagenesis of Renilla luciferase yields enhanced stability and light output. Protein Engineering Design and Selection. 2006;19(9):391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 30.Kaihara A, Kawai Y, Sato M, et al. Locating a Protein–Protein Interaction in Living Cells via Split RenillaLuciferase Complementation. Anal. Chem. 2003;75(16):4176–4181. doi: 10.1021/ac0300800. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, Choi S-Y, Frohman MA. Mitochondrion. Mitochondrion. 2010;10(5):559–566. doi: 10.1016/j.mito.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Csordas G, Renken C, Varnai P, et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006;174(7):915–921. doi: 10.1083/jcb.200604016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin M, Shepardson N, Yang T, et al. Soluble amyloid β-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc Natl Acad Sci USA. 2011;108(14):5819–5824. doi: 10.1073/pnas.1017033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1(2):695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 35.Lim Y, Golden JA. Expression pattern of cLhx2b, cZic1 and cZic3 in the developing chick diencephalon. Mech Dev. 2002;115(1–2):147–150. doi: 10.1016/s0925-4773(02)00091-6. [DOI] [PubMed] [Google Scholar]
- 36.Lim Y, Cho G, Minarcik J, Golden J. Altered BMP signaling disrupts chick diencephalic development. Mech Dev. 2005;122(4):603–620. doi: 10.1016/j.mod.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Hosie KA, King AE, Blizzard CA, et al. Chronic excitotoxin-induced axon degeneration in a compartmented neuronal culture model. ASN NEURO. 2012;4(1):47–57. doi: 10.1042/AN20110031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binley KE, Ng WS, Tribble JR, et al. Journal of Neuroscience Methods. Journal of Neuroscience Methods. 2014;225:65–70. doi: 10.1016/j.jneumeth.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Hedskog L, Pinho CM, Filadi R, et al. Modulation of the endoplasmic reticulum–mitochondria interface in Alzheimer’s disease and related models. Proc Natl Acad Sci USA. 2013;110(19):7916–7921. doi: 10.1073/pnas.1300677110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wieckowski MR, Giorgi C, Lebiedzinska M, et al. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4(11):1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 41.DeLuca GC, Ebers GC, Esiri MM. The extent of axonal loss in the long tracts in hereditary spastic paraplegia. Neuropathol Appl Neurobiol. 2004;30(6):576–584. doi: 10.1111/j.1365-2990.2004.00587.x. [DOI] [PubMed] [Google Scholar]
- 42.Beetz C, Schüle R, Deconinck T, et al. REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain. 2008;131(Pt 4):1078–1086. doi: 10.1093/brain/awn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlang KJ, Arning L, Epplen JT, Stemmler S. Autosomal dominant hereditary spastic paraplegia: Novel mutations in the REEP1 gene (SPG31) BMC Med Genet. 2008;9(1):71. doi: 10.1186/1471-2350-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasaki Y, Vohra BPS, Lund FE, Milbrandt J. Nicotinamide Mononucleotide Adenylyl Transferase-Mediated Axonal Protection Requires Enzymatic Activity But Not Increased Levels of Neuronal Nicotinamide Adenine Dinucleotide. J Neurosci. 2009;29(17):5525–5535. doi: 10.1523/JNEUROSCI.5469-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Orlacchio A, Montieri P, Babalini C, et al. Late-onset hereditary spastic paraplegia with thin corpus callosum caused by a new SPG3A mutation. J Neurol. 2011;258(7):1361–1363. doi: 10.1007/s00415-011-5934-z. [DOI] [PubMed] [Google Scholar]
- 46.França MC, D'Abreu A, Maurer-Morelli CV, et al. Prospective neuroimaging study in hereditary spastic paraplegia with thin corpus callosum. Mov Disord. 2007;22(11):1556–1562. doi: 10.1002/mds.21480. [DOI] [PubMed] [Google Scholar]
- 47.Havlicek S, Kohl Z, Mishra HK, et al. Gene dosage-dependent rescue of HSP neurite defects in SPG4 patients' neurons. Hum Mol Genet. 2014;23(10):2527–2541. doi: 10.1093/hmg/ddt644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurt CM, Björk S, Ho VK, et al. REEP1 and REEP2 proteins are preferentially expressed in neuronal and neuronal-like exocytotic tissues. Brain Research. 2014;1545(C):12–22. doi: 10.1016/j.brainres.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beetz C, Schule R, Deconinck T, et al. REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain. 2008;131(4):1078–1086. doi: 10.1093/brain/awn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedman JR, Lackner LL, West M, et al. ER Tubules Mark Sites of Mitochondrial Division. Science. 2011;334(6054):358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 52.de Meis L, Ketzer LA, da Costa RM, et al. Fusion of the Endoplasmic Reticulum and Mitochondrial Outer Membrane in Rats Brown Adipose Tissue: Activation of Thermogenesis by Ca2+ PLoS ONE. 2010;5(3):e9439. doi: 10.1371/journal.pone.0009439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai J, Kuo K-H, Leo JM, et al. Rearrangement of the close contact between the mitochondria and the sarcoplasmic reticulum in airway smooth muscle. Cell Calcium. 2005;37(4):333–340. doi: 10.1016/j.ceca.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 54.Saito H, Kubota M, Roberts RW, et al. RTP family members induce functional expression of mammalian odorant receptors. Cell. 2004;119(5):679–691. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 55.Villegas R, Martinez NW, Lillo J, et al. Calcium Release from Intra-Axonal Endoplasmic Reticulum Leads to Axon Degeneration through Mitochondrial Dysfunction. J Neurosci. 2014;34(21):7179–7189. doi: 10.1523/JNEUROSCI.4784-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mironov SL, Symonchuk N. ER vesicles and mitochondria move and communicate at synapses. J Cell Sci. 2006;119(23):4926–4934. doi: 10.1242/jcs.03254. [DOI] [PubMed] [Google Scholar]
- 57.Tsukita S, ishikawa H. Three-dimensional distribution of smooth endoplasmic reticulum in myelinated axons. Journal of Electron microscopy. 1976;25(3):141–149. [PubMed] [Google Scholar]
- 58.Ellisman MH, Porter KR. Microtrabecular structure of the axoplasmic matrix: visualization of cross-linking structures and their distribution. J. Cell Biol. 1980;87:464–479. doi: 10.1083/jcb.87.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Area-Gomez E, de Groof AJC, Boldogh I, et al. Short Communication. The American Journal of Pathology. 2010;175(5):1810–1816. doi: 10.2353/ajpath.2009.090219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim Y, Kehm VM, Lee EB, et al. α-Syn Suppression Reverses Synaptic and Memory Defects in a Mouse Model of Dementia with Lewy Bodies. J Neurosci. 2011;31(27):10076–10087. doi: 10.1523/JNEUROSCI.0618-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoica R, De Vos KJ, Paillusson SEB, et al. ER–mitochondria associations are regulated by the VAPB–PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nature Commun. 2014;5:3996. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calì T, Ottolini D, Negro A, Brini M. Biochimica et Biophysica Acta. BBA - Molecular Basis of Disease. 2013;1832(4):495–508. doi: 10.1016/j.bbadis.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 63.Barrientos SA, Martinez NW, Yoo S, et al. Axonal Degeneration Is Mediated by the Mitochondrial Permeability Transition Pore. J Neurosci. 2011;31(3):966–978. doi: 10.1523/JNEUROSCI.4065-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Court FA, Coleman MP. Mitochondria as a central sensor for axonal degenerative stimuli. Trends Neurosci. 2012;35(6):364–372. doi: 10.1016/j.tins.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 65.Liu SG, Che FY, Heng XY, et al. Clinical and genetic study of a novel mutation in the REEP1gene. Synapse. 2009;63(3):201–205. doi: 10.1002/syn.20602. [DOI] [PubMed] [Google Scholar]
- 66.Battini R, Fogli A, Borghetti D, et al. Clinical and genetic findings in a series of Italian children with pure hereditary spastic paraplegia. European Journal of Neurology. 2010;18(1):150–157. doi: 10.1111/j.1468-1331.2010.03102.x. [DOI] [PubMed] [Google Scholar]
- 67.Bot ST, Veldink JH, Vermeer S, et al. ATL1 and REEP1 mutations in hereditary and sporadic upper motor neuron syndromes. J Neurol. 2012;260(3):869–875. doi: 10.1007/s00415-012-6723-z. [DOI] [PubMed] [Google Scholar]
- 68.Ishiura H, Takahashi Y, Hayashi T, et al. Molecular epidemiology and clinical spectrum of hereditary spastic paraplegia in the Japanese population based on comprehensive mutational analyses. J Human Genetics. 2014:1–10. doi: 10.1038/jhg.2013.139. [DOI] [PubMed] [Google Scholar]