Abstract
New epigenetic technologies may uncover etiopathogenic mechanisms of major psychosis. In this study, we applied padlock probe-based ultra-deep bisulfite sequencing for fine mapping of modified cytosines of the HLA complex group 9 (nonprotein coding) gene in the postmortem brains of individuals affected with schizophrenia or bipolar disorder and unaffected controls. Significant differences between patients and controls were detected in both CpG and CpH modifications. In addition, we identified epigenetic age effects, DNA modification differences between sense and anti-sense strands, and demonstrated how DNA modification data can be used in clustering of patient populations. Our findings revealed new epigenetic complexities but also highlighted the potential of DNA modification approaches in the search of heterogeneous causes of major psychiatric disease.
Key words: epigenetics, brain, schizophrenia, bipolar disorder, bisulfite sequencing, padlock probes
Introduction
Family, twin, and adoption studies have shown that both heritable and nonheritable risk factors contribute to psychiatric diseases.1 Along with genetic and environmental causes, a third group of etiological factors, namely acquired and inherited epigenetic misregulation, may play an etiopathogenic role in schizophrenia (SCZ) and bipolar disorder (BPD) which are together termed major psychosis.2 Epigenetics refers to the heritable and potentially reversible modifications of DNA and histone proteins that regulate gene expression and other genomic functions without changing the underlying DNA sequences.3 Epigenetic factors are vital for development, differentiation, and maintenance of tissue specific cellular phenotypes,4,5 can respond to environmental cues,6 and are subject to stochastic variation over time.7,8 Putative epigenetic misregulation can offer new insights into a number of non-Mendelian complexities of major psychosis such as fluctuating course, discordance of monozygotic twins, sex- and parent of origin-effects.9–13
A number of epigenetic studies have attempted to identify specific DNA and histone modification factors contributing to major psychosis.14–16 In addition to the new insights, DNA modification (this term includes methylated, hydroxymethylated, carboxylated, and formylated cytosines) studies have also highlighted challenges and limitations of our current approaches that need to be addressed. DNA modification maps generated using enrichment-based techniques, such as methylated DNA immunoprecipitation (MeDIP) and methylated DNA capture by affinity purification (MethylCap) have, at most, a resolution of several hundred nucleotides.17 Single cytosine resolution is feasible with bisulfite-conversion approaches, although the cost of bisulfite sequencing has traditionally limited this analysis to single genes or short genomic regions. Recent advances in next generation sequencing technologies have significantly expanded the scope of DNA modification studies. Whole genome bisulfite sequencing (WGBS) now offers comprehensive genomic coverage at a single nucleotide resolution, yielding quantitative determination of cytosine modification density, as well as strand- and allele-specific modification profiling.18–22 Yet, due to considerable cost, WGBS based studies remain limited by low read depth and/or small sample size.
Because WGBS is not yet practical for large-scale populational studies, we attempted a pilot study which contains elements of an ideal epigenomics study at a much more manageable level of cost and time. Bisulfite padlock sequencing (also titled bisulfite padlock probes23–25) offers a new approach that combines high read depth with multiplexed padlock probes to yield reliable interrogation of a large number of loci. This technique relies on sequence specific padlock probes that capture targeted DNA regions after bisulfite conversion and are subsequently amplified and sequenced (supplementary figure 1). With recent advances in bench top sequencing platforms, such as Illumina MiSeq, bisulfite padlock sequencing can now be leveraged to generate a large volume of data in a short time with limited funds. We applied this approach to HLA complex group 9 (nonprotein coding) gene (HCG9), which was shown to exhibit epigenetic differences in our microarray-based DNA modification study of major psychosis26 and was later validated by bisulfite pyrosequencing mapping.27 In this study, we investigated HCG9 cytosine modification density at CpG and CpH sites (where H is A, C or T) using ultra-deep bisulfite sequencing in a cohort of prefrontal cortex samples from patients affected with SCZ or BPD and control individuals.
Methods
Samples
We tested 22 postmortem prefrontal cortex (BA10) samples from individuals who were affected with SCZ (mean age ± SD. 58.4±14.6 years, mean postmortem interval [PMI] ± SD. 23.6±6.2 h, and male to female ratio 2.6:1); 27 samples from BPD patients (age 62.8±17.4 years, PMI 22.1±7.1 h, and male to female ratio 0.8:1); and 32 samples from controls (age 57.3±17.5 years, PMI 21.8±4.5 h, and male to female ratio 2.2:1). Detailed demographic information, including postmortem neuropathology findings, is provided in the supplementary table 1. Samples were provided by the Harvard Brain Tissue Resource Center, McLean Hospital.
Padlock Probe Design, Capture, and DNA Sequencing
Genomic DNA was extracted using standard phenol-chloroform method and subsequently bisulfite converted using the EZ-96 DNA Methylation-Lightning Kit (Zymo Research Corporation) according to manufacturer’s protocol. Padlock probes were designed with ppDesigner25 for sense and antisense strand of the full length HCG9, plus 1kb upstream and downstream of the gene. The padlock probes were then manually interrogated for presence of potentially confounding CpG dinucleotides within the last 5 nucleotides of the annealing arms. In cases when adjustments were not possible, CpG dinucleotide was replaced with a CpR, where R is a mixture of G and A, corresponding to the complementary base pair of a methylated/hydroxymethylated cytosine, or an unmodified cytosine, respectively. The annealing arms of padlock probes overlapping single nucleotide polymorphisms (SNPs) were similarly adjusted. Probes found binding to repetitive elements were removed and a total of 57 probes were then synthesized with 5’ phosphorylation modification (IDT).
400ng of sodium bisulfite treated genomic DNA was mixed with 6 µL of the padlock probe mixture containing 0.5 pM of each probe and 1× Ampligase buffer to a final volume of 25 µL. DNA was denatured at 95°C for 10min and probe hybridization was performed by gradual drop of temperature at 1°C/min to 55°C, followed by incubation at 55°C, 50°C, and 45°C for 5 h each with 1°C/min gradual drops in between. For circularization of annealed probes, 2.5 µL of HLN buffer (20 µL of 10mM dNTP, 100 µL of 10× Ampligase buffer, 500U Ampligase, 200 µL of Hemo KlenTaq [NEB, USA] in 1mL total volume) was added, and the reaction was incubated at 45°C for 5h, followed by a gradual increase (1°C/min) to 50°C, 5h at 50°C, a gradual increase to 55°C, and 5h at 55°C. This was followed by enzyme inactivation at 94°C for 2min. To digest any remaining linear DNA after circularization, 3 µL of exo mix (200U exonuclease III and 10U exonuclease I [NEB]) was added followed by incubation at 37°C for 1h and enzyme inactivation at 94°C for 2min. This was repeated 3 times.
2 µL of circularized DNA was mixed with 1 µL of 10 µM pp_Famp primer, 1 µL of 10 µM barcoded pp_Ramp primer, and 25 µL of 2X Phusion High-Fidelity PCR Master Mix (GC Buffer) in a 50 µL reaction. Thermo-cycling conditions were as follows: 98°C for 30 s; 35 cycles, each 98°C for 10 s, 58°C for 30 s, 72°C for 10 s; 72°C for 5min. The desired product size of ~300bp was confirmed on a 2% agarose gel. All PCR amplicons were pooled and purified with Ampure XP beads (Beckman Coulter) using manufacturer’s protocol. The PCR library was sequenced using MiSeq platform (Illumina). We used 60% PhiX spike-in to compensate for low base-pair diversity inherent to bisulfite conversion. Sequence information for padlock probes, PCR primers, and sequencing primers are available from the authors upon request.
Whole Genome Amplification
40ng of human genomic DNA was mixed with 2 µL of 10X phi29 reaction buffer and 2 µL of exo-resistant random primer (Thermo Scientific) in a 17 µL reaction. The mixture was incubated at 95°C for 5min and then gradually cooled to 30°C (1°C/15 s). 0.5 µL dNTP mix (10mM), 0.5 µL BSA (10mg/ml), 1 µL phi29, and 1 µL inorganic pyrophosphatase (Thermo Scientific) were added to the reaction. The mixture was then incubated for 6 h at 30°C. High molecular weight amplification products were observed on an agarose gel, and the amplicons were purified using standard ethanol precipitation.
Data Analysis
The HCG9 reference sequence was obtained from the UCSC Genome Browser (hg19). Each sequencing read was trimmed to remove low quality bases (Q < 20) and short reads (<80 bp). To ensure that mapping was independent of cytosine modification status, all “C”s in the reference and read sequence were replaced with “T”s before mapping. Mapping of sequenced reads was performed for both sense and anti-sense strands with a maximum mismatch of 2 nucleotides. After sequence alignment was complete, number of “C”s and “T”s were tallied at CpG and CpH sites for each strand within the HCG9 region. Modification status, measured as the ratio of modified cytosine (modC) to total cytosine was determined by modC/C ratio= NC/(NC + NT), where NC is the number of counts for cytosine and NT is the number of counts for thymine. modC estimated through bisulfite sequencing in this experiment refers to methylated plus hydroxymethylated cytosines.28
Data analysis was performed with R29 and SPSS30 (IBM Corp.). R Bioconductor31 packages “ShortRead”32 and “Biostrings”33 were used for input and quality assessment of sequence reads and alignment to reference genome, respectively. R package “FactomineR”34 was used for principle component analysis (PCA). Because the densities of modified cytosines were not normally distributed (Shapiro-Wilk test, P < .05), we used nonparametric tests to identify significant differences among groups. Results are presented as mean ± SEM. Cross-sample range for biological samples and technical replicates were calculated as maximum modification at a single cytosine minus minimum modification (modC/C ratio [max–min]). Nucleosome occupancy scores were generated for each nucleotide in the investigated HCG9 region using nucleosome prediction algorithms.35,36 Euclidean distances for each sample were calculated for all CpG and CpH sites combined.
Because there is genetic overlap between SCZ and BPD37,38 and familial aggregation of the 2 disorders,39 some of our analyses were performed on the combined sample.
Results
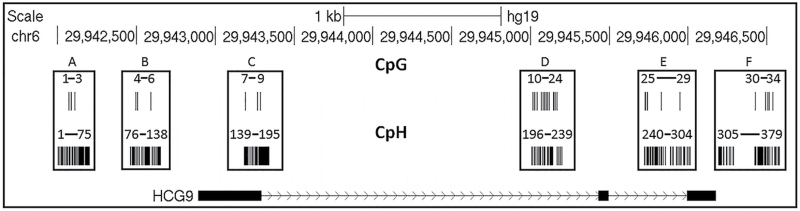
Padlock probes were designed for the total length of ~5kb, which includes the full length HCG9, plus 1kb upstream and downstream of the gene. Not all probes, however, generated sufficient number of reads, and only CpG and CpH sites with read depth ≥ 30X in at least 10 samples for both cases and controls were considered. Sites that did not meet this selection criterion were removed from further analysis. Each DNA sample was thus characterized by 96 470±8 733 (mean ± SEM) reads (supplementary figure 2). These reads covered a total of 34 of 149 possible CpG sites and 379 of 2 364 possible CpH sites. Figure 1 shows genomic distribution of investigated CpG and CpH sites. On average, each sample generated reliable DNA modification estimates for 21 CpG sites and 213 CpH sites.
Fig. 1.
The finely mapped DNA modification regions of HCG9 (UCSC Genome browser, version hg19). Black bars represent investigated CpG (n = 34) and CpH (n = 379) sites while the numbers represent their relative position. The interrogated sequence is further broken down into 6 regions (A–F).
To estimate the bisulfite conversion rate, we used DNA devoid of any modifications. This control was prepared using whole genome amplification of 2 unrelated genomic DNA samples, which were subjected to bisulfite conversion and deep sequencing using the same set of padlock probes. Cytosine conversion rate to thymine was calculated at 99.34%, indicating highly efficient bisulfite conversion. We also observed greater biological variability than technical variability in our dataset. Technical replicates of 8 samples showed higher pairwise correlation compared to biological samples (Pearson’s r = 0.82±0.06 and 0.77±0.02, respectively) and cross-sample range was significantly higher for biological samples compared to technical replicates (0.31±0.03 and 0.13±0.01, respectively, Mann-Whitney U test, P = 4.8 x 10− 5).
CpH Modification Differs in the Sense and Anti-Sense HCG9 Strands
One of the important advantages of bisulfite sequencing is the ability to investigate DNA modification profiles separately on the sense and anti-sense strands. Mean CpG modification was found to be similar for sense and anti-sense strand (modC/C ratio; 0.83±0.18 and 0.85±0.13, respectively, Wilcoxon signed ranks test, P = .6). This was also true for BPD, SCZ and control groups when analyzed separately.
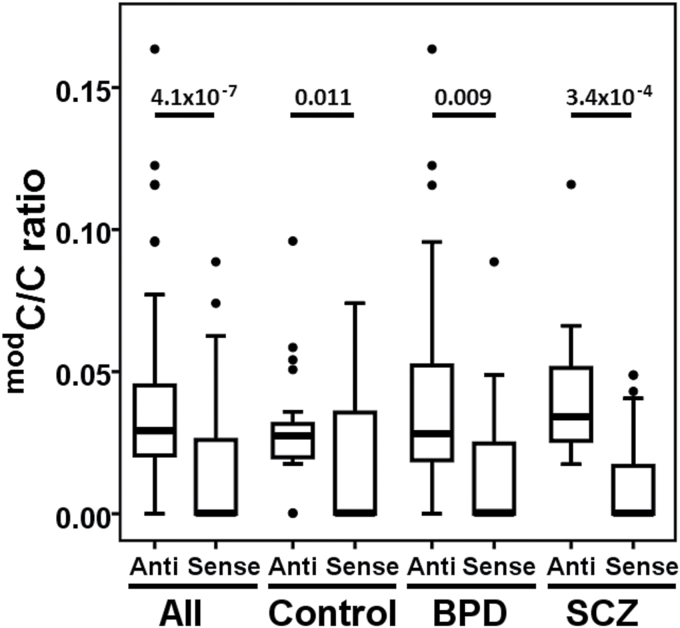
Recent publications have reported presence of CpH modification in adult brains of both humans and mice.18,19,40,41 In our dataset, we observed a mean CpH modification density of 1.69%, although 17 CpH sites far exceeded this average with modification density greater than 10%. Because CpH sites are asymmetric on the 2 DNA strands and cannot be compared individually, we analyzed modification density across all CpH sites to estimate differences between sense and anti-sense strands. We found significantly lower modification levels on the sense strand compared to the anti-sense strand (modC/C ratio; 0.01±0.002 and 0.02±0.002, respectively, Wilcoxon signed ranks test, P = 7.7 x 10− 7). A potential bias in our estimation of strand differences could be due to greater number of CpH sites on the anti-sense strand (n = 289) compared to the sense strand (n = 90). To address this potential issue, we compared CpH modification density from regions E and F (figure 1) with identical number of CpH sites (n = 42) on each strand. Consistent with the earlier observation, the sense strand exhibited significantly lower DNA modification compared to anti-sense strand (modC/C ratio; 0.014±0.003 and 0.04±0.003, respectively, P = 4.1 x 10− 7). As shown in figure 2, significantly lower modification density for sense strand vs anti-sense strand was similarly observed for control, BPD and SCZ groups (P ≤ .011).
Fig. 2.
Strand bias of CpH modification. Boxplots represent CpH modification density for sense and anti-sense strands for region E and F (chr6: 29945722–815 and 29946419–576, respectively) with equal number of CpH sites on both strands. Significant P values are provided (Wilcoxon signed ranks test) above the boxplots.
CpA Dinucleotides and Nucleosome Linker Regions are Enriched in CpH Modification
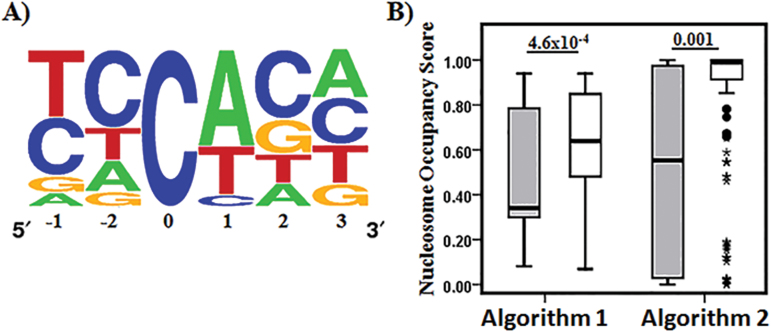
To further explore the biological roles of CpH modification we focused on modified CpH sites where ≥ 2% density of modC were present in at least 10 individuals. Consistent with previous findings,19,42 sequence logo43 generated from multiple sequence alignment of the selected CpH sites showed CpH modification occurs predominantly at CpA dinucleotides of HCG9 in brain genomic DNA (figure 3A). We determined CpA dinucleotide frequency in both strands and found them to be similar (6.8% and 6.2% for sense and anti-sense strands, respectively), despite significantly higher CpH modification in the anti-sense strand.
Fig. 3.
Comparison of modified and unmodified CpH sites. (A) CpA is overrepresented at modified CpH sites. Sequence logo43 generated from multiple sequence alignment of modified CpH sites (ie, CpH sites with ≥2% modification density in at least 10 individuals) with modified cytosine at position 0. (B) Nucleosome occupancy score determined for modified (grey bars) and unmodified (white bars) CpH sites by nucleosome occupancy prediction algorithm 135 and 2.36 Significantly lower score for modified CpH sites indicate they are present in nucleosome void regions of HCG9. P values above boxplots were obtained from Mann-Whitney U test.
Next, we compared the selected CpH sites to the remaining unmodified CpH sites for nucleosome occupancy using 2 different algorithms,35,36 which assign a score for each nucleotide ranging between 0 (linker region) and 1 (nucleosome center). Both algorithms predicted a significantly lower mean nucleosome occupancy score (Mann-Whitney U test, P < .01; figure 3B) indicating that modified CpH sites were predominantly present in linker regions.
DNA Modification Differences in Major Psychosis and Control Brains
To identify significant differences in DNA modification between cases and controls, we eliminated effects of age, sex, PMI, and neuropathological differences through linear model transformation followed by nonparametric Mann-Whitney U test. Mean CpH modification density at selected CpH sites was significantly higher in major psychosis samples compared to controls (modC/C ratio, 0.016±0.01 and −0.024±0.008, respectively, P = .006; negative mean DNA modification densities are a result of linear model correction). Similar results were also obtained when SCZ and BPD groups were analyzed separately (modC/C ratio; SCZ = 0.022±0.01 vs control = −0.015±0.008, P = .01; BPD = 0.022±0.02 vs control = −0.019±0.008; P = .02). In the analysis of individual cytosines (CpGs 1–34, figure 1), we detected significant differences in DNA modification at CpG 29, located in the second intron of HCG9, between BPD and controls (modC/C ratio; 0.10±0.01 and −0.09±0.05, respectively, Bonferroni corrected P = .004). Although BPD and controls were not matched for sex we did not find any significant association between sex and CpH or CpG 29 modification density.
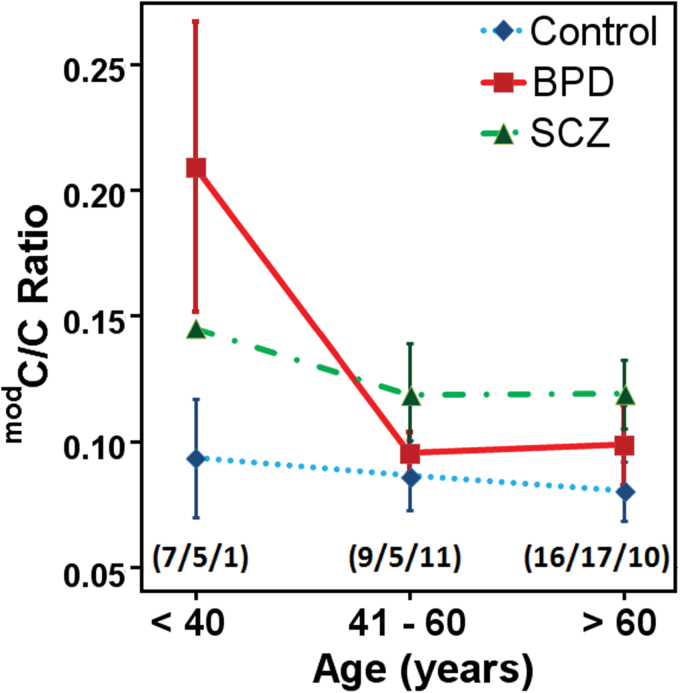
To determine how CpH modification levels change at HCG9 with age, we divided the brain samples into 3 age groups (ie, < 40 yr, 41–60 yr and > 60 yr) and compared the CpH modification density at selected CpH sites amongst the groups. For BPD patients, we found significant DNA modification differences between different age groups (Kruskal-Wallis test, P = .027) with sharp decline in CpH modification from age group < 40 years to 41–60 years (figure 4). In SCZ patients a similar decline in CpH modification for age groups < 40 years to 41–60 years was observed, however, sample numbers were too low for statistical comparison. No significant changes were observed in control samples (P = .85). Significant age induced decline of CpH modification in BPD was also observed after eliminating the effects of sex, PMI, and neuropathological differences (modC/C ratio; <40years = 0.09±0.05, 41–60 years = −0.01±0.01, >60years = −0.002±0.016, Kruskal-Wallis test, P = .04).
Fig. 4.
Age-stratified changes in HCG9 CpH modification density in the brain. Line plots represent mean CpH modification density for control, BPD, and SCZ individuals for different age groups. Numbers of samples in each age group are presented in brackets as (controls/BPD/SCZ) and error bars represent SEM. Most dramatic difference was observed in BPD age group younger than 40 years compared to the older BPD and SCZ patients as well as controls in all age categories.
Epigenetic profiles vary significantly between neuronal and glia cells42 and inter-individual variation in brain cellular composition may generate false positive epigenetic differences.44 For a subset of BPD and control brain samples (n = 34, supplementary table 1) we had access to data on the proportions of neurons and glia, which were estimated using a cell epigenotype specific model.44 DNA modification at selected CpH sites and CpG 29 were found to be significantly higher in BPD samples compared to controls after correction for neuron and glia proportions, age, sex, PMI, and neuropathological differences (modC/C ratio; CpH: BPD = 0.020±0.016, controls = −0.026±0.016, P = .03; CpG 29: BPD = 0.10±0.02, controls = −0.11±0.05, Bonferroni corrected P = .002). Similar to the age effects detected in the whole BPD sample, BPD subsample showed age-dependent differences in CpH modification density after correction for neuron and glia proportions, sex, PMI, and neuropathological differences (<40years = 0.13±0.06, 41–60 years = −0.001±0.014, >60 years = −0.02±0.006, Kruskal-Wallis test, P = .01). No significant differences were identified in control samples (P = .6).
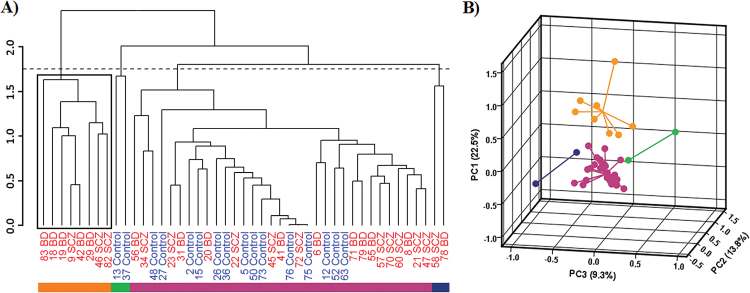
Hierarchical Clustering of HCG9 Modification in Major Psychosis
Comparison of Euclidean distances between samples revealed higher variation in DNA modification for major psychosis compared to control individuals (3.5±0.03 and 3.2±0.04, respectively; Student’s t test, P = 1.9 × 10−10). Variability in epigenetic profiles was further investigated using hierarchical clustering and PCA of HCG9. Hierarchical clustering of region A (figure 1) identified a subgroup of major psychosis samples, which clustered together and formed a separate clade from the rest of the samples (figure 5A). Figure 5B shows separation of the identified clusters in 3 dimensional space based on the first 3 principal components. As expected, we found CpH modification to be a significant driving factor behind the variance explained by the first principle component (Fisher’s combined probability test using Benjamin-Hochberg false discovery rate corrected Person’s correlation P value between PC1 and CpH modification; P = 6.5 × 10− 5). The separation of clusters was not associated with age, sex, or neuropathological changes. Lack of clinical information and relatively small brain sample size prohibited further exploration of associations between clusters and phenotypic peculiarities of major psychosis.
Fig. 5.
Inter-individual variation of modification density in major psychosis and control samples for HCG9 region A. Samples with no missing value for 3 CpG and 75 CpH sites in region A were included. (A) Dendrogram obtained from hierarchical clustering for individual samples (SCZ and BPD in red and control in blue). Bottom color bar represents clusters identified at the cut height indicated by the horizontal dashed line. A subset of major psychosis samples (black box) formed a separate cluster from the remaining samples. (B) 3 dimensional representation of first 3 principal components. Each dot represents an individual while dot colors represent the cluster to which they belong. The lines connecting each dot merge at the geometric center of each cluster. PC1, PC2 and PC3 are shown on the y-, x-, and z- axis, respectively, and variance explained by each principal component is represented in brackets.
Discussion
Our primary objective was to explore new opportunities and challenges with ultra-deep bisulfite sequencing in epigenomic studies of psychiatric diseases using the recently developed bisulfite padlock probe-based approach. This technology offers an alternative way to interrogate multiple genomic regions in large sample sets because WGBS remains cost prohibitive. We detected significant modification differences in both CpH and CpG dinucleotides of HCG9 between major psychosis and control individuals. Our study underlines the importance of mapping modified cytosines in a strand specific manner. Many psychiatric epigenetic studies have not considered the potential asymmetry of DNA modification thus far. This may result in missing both disease epimutations and information about the molecular mechanisms of epigenetic misregulation in diseased brains. Our findings of CpH modification differences in sense and antisense strands of HCG9 are consistent with earlier studies on mammalian embryonic stem cells.21,45 Similar to those studies, we detected CpH modification predominantly at CpA dinucleotides. The functional role of CpH modification is still unclear but recent studies have shown that gene body CpH modification is inversely proportional to the abundance of the associated transcripts.18,20 Age associated changes in CpH modification may shed new light in psychiatric research. Although limited to 1 gene, the sharp decline of CpH modification observed in >40 year old BPD patients is consistent with the observation that some aging BPD patients demonstrate diminished symptoms, and significant improvements in the latter parts of their lives.46,47 Identification of epigenetic markers, which exhibit significant differences in young BPD and SCZ patients but eventually return to control levels, may help to understand the origin and etiopathogenic mechanisms of major psychosis.
Our hierarchical clustering analysis, although based on a single gene and therefore of limited scope, may help divide clinically indistinguishable phenotypes of major psychosis into etiologically distinct subgroups. It has been generally accepted that SCZ (amongst many other psychiatric diseases) represents a “fuzzy cluster” of syndromes which are heterogeneous from clinical, pathological, and etiological point of view.48 Individual epigenetic profiles may, therefore, identify disease subtypes that cannot be differentiated clinically or by using other molecular approaches. To this end, our cluster and PCA based analysis of modification density at CpG and CpH sites resulted in delineation of subgroups of major psychosis samples with different HCG9 modification profiles.
Several aspects of this study can be further improved upon. First, much larger sample size of brain and other tissues would have higher power to detect statistically significant differences between affected individuals and controls. Additional clinical information, such as age of disease onset, lifetime history of illness, antipsychotic use, and use of alcohol and other recreational drugs, would reduce the chance for false associations caused by confounding factors. Secondly, efficiency of the padlock based approach can be significantly improved both in terms of the scope of the covered genome and performance quality at each padlock target. The investigated 1.4kb region of HCG9 translates to approximately one-third of the probes that were initially designed. The regions that were not interrogated could have resulted from degradation of DNA during bisulfite reaction and insufficient specificity of some probes. Furthermore, uneven read depth might result from unequal amplification efficiency of probes and possible nonideal reaction conditions caused by the replacement of the discontinued Stoffel fragment polymerase with Hemo KlenTaq polymerase. Further improvement of the technology is required to obtain even coverage across probe set. Finally, although bisulfite sequencing is a powerful approach, interrogation of CpH modification may require additional efforts. When designing PCR primers for bisulfite converted DNA it is usually assumed that cytosines within CpH dinucleotides are not modified and therefore converted to uracil. This may create bias for amplification in the presence of modified CpH sites. Similarly, the instances of probe arms overlapping with SNPs and CpG sites should be minimized to limit potential annealing biases. Therefore, for reliable estimation of modification levels, combination of different, overlapping padlock probes interrogating the same target region should be utilized.
Supplementary Material
Supplementary material is available at http://schizophreniabulletin.oxfordjournals.org.
Funding
Canadian Institutes of Health Research (77689, 119451); the US National Institute of Mental Health (MH074127, MH088413); and Brain Canada to A.P.
Supplementary Material
Acknowledgments
We would like to thank Dr Viviane Labrie for editorial help and the Harvard Brain Tissue Resource Center, McLean Hospital (USA) for providing the brain tissues. A.P. is the Tapscott Chair in Schizophrenia Studies, University of Toronto.
References
- 1.Shih RA, Belmonte PL, Zandi PP. A review of the evidence from family, twin and adoption studies for a genetic contribution to adult psychiatric disorders. Int Rev Psychiatry. 2004;16:260–283. [DOI] [PubMed]
- 2. Pidsley R, Mill J. Epigenetic studies of psychosis: current findings, methodological approaches, and implications for postmortem research. Biol Psychiatry. 2011;69:146–156. [DOI] [PubMed] [Google Scholar]
- 3. Henikoff S, Matzke MA. Exploring and explaining epigenetic effects. Trends Genet. 1997;13:293–295. [DOI] [PubMed] [Google Scholar]
- 4. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Serrano L, Vazquez BN, Tischfield J. Chromatin structure, pluripotency and differentiation. Exp Biol Med. 2013;238:259–270. [DOI] [PubMed] [Google Scholar]
- 6. Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci U S A. 2010;107(suppl 1):1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ushijima T, Watanabe N, Okochi E, Kaneda A, Sugimura T, Miyamoto K. Fidelity of the methylation pattern and its variation in the genome. Genome Res. 2003;13:868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gabory A, Attig L, Junien C. Sexual dimorphism in environmental epigenetic programming. Mol Cell Endocrinol. 2009;304:8–18. [DOI] [PubMed] [Google Scholar]
- 10. Kaminsky Z, Wang SC, Petronis A. Complex disease, gender and epigenetics. Ann Med. 2006;38:530–544. [DOI] [PubMed] [Google Scholar]
- 11. Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mill J, Dempster E, Caspi A, Williams B, Moffitt T, Craig I. Evidence for monozygotic twin (MZ) discordance in methylation level at two CpG sites in the promoter region of the catechol-O-methyltransferase (COMT) gene. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:421–425. [DOI] [PubMed] [Google Scholar]
- 13. Petronis A, Gottesman II, Kan P, et al. Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance? Schizophr Bull. 2003;29:169–178. [DOI] [PubMed] [Google Scholar]
- 14. Akbarian S. Epigenetic mechanisms in schizophrenia. Dialogues Clin Neurosci. 2014;16:405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iwamoto K, Kato T. Epigenetic profiling in schizophrenia and major mental disorders. Neuropsychobiology. 2009;60:5–11. [DOI] [PubMed] [Google Scholar]
- 16. Dempster E, Viana J, Pidsley R, Mill J. Epigenetic studies of schizophrenia: progress, predicaments, and promises for the future. Schizophr Bull. 2013;39:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Varley KE, Gertz J, Bowling KM, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo JU, Su Y, Shin JH, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci. 2014;17:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ziller MJ, Gu H, Muller F, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng J, Shoemaker R, Xie B, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Diep D, Plongthongkum N, Gore A, Fung HL, Shoemaker R, Zhang K. Library-free methylation sequencing with bisulfite padlock probes. Nat Methods. 2012;9:270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaminsky Z, Tochigi M, Jia P, et al. A multi-tissue analysis identifies HLA complex group 9 gene methylation differences in bipolar disorder. Mol Psychiatry. 2012;17:728–740. [DOI] [PubMed] [Google Scholar]
- 28. Nestor C, Ruzov A, Meehan R, Dunican D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010;48:317–319. [DOI] [PubMed] [Google Scholar]
- 29. R Core Team. A Language and Environment for Statistical Computing. [Computer Program]. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 30. IBM Corp. SPSS Statistics for Windows [Computer Program]. Version 21.0. Armonk, NY: IBM Corp; 2012. [Google Scholar]
- 31. Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morgan M, Anders S, Lawrence M, Aboyoun P, Pagès H, Gentleman R. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pages H, Aboyoun P, Gentleman R, DebRoy S. Biostrings: String Objects Representing Biological Sequences, and Matching Algorithms. R Package Version 2.36.1. [Google Scholar]
- 34. Le S, Josse J, Husson F. FactoMineR: An R Package for Multivariate Analysis. Journal of Statistical Software. 2008;25:1–18. [Google Scholar]
- 35. Kaplan N, Moore IK, Fondufe-Mittendorf Y, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xi L, Fondufe-Mittendorf Y, Xia L, Flatow J, Widom J, Wang JP. Predicting nucleosome positioning using a duration Hidden Markov Model. BMC Bioinformatics. 2010;11:1471–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Purcell SM, Wray NR, Stone JL, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rasic D, Hajek T, Alda M, Uher R. Risk of mental illness in offspring of parents with schizophrenia, bipolar disorder, and major depressive disorder: a meta-analysis of family high-risk studies. Schizophr Bull. 2014;40:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xie W, Barr CL, Kim A, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148:816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ziller MJ, Müller F, Liao J, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7:e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kozlenkov A, Roussos P, Timashpolsky A, et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic Acids Res. 2014;42:109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guo W, Chung WY, Qian M, Pellegrini M, Zhang MQ. Characterizing the strand-specific distribution of non-CpG methylation in human pluripotent cells. Nucleic Acids Res. 2014;42:3009–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Angst J, Preisig M. Course of a clinical cohort of unipolar, bipolar and schizoaffective patients. Results of a prospective study from 1959 to 1985. Schweiz Arch Neurol Psychiatr. 1995;146:5–16. [PubMed] [Google Scholar]
- 47. Tsuang MT, Woolson RF, Fleming JA. Long-term outcome of major psychoses. I. Schizophrenia and affective disorders compared with psychiatrically symptom-free surgical conditions. Arch Gen Psychiatry. 1979;36:1295–1301. [DOI] [PubMed] [Google Scholar]
- 48. Keshavan MS, Nasrallah HA, Tandon R. Schizophrenia, “Just the Facts” 6. Moving ahead with the schizophrenia concept: from the elephant to the mouse. Schizophr Res. 2011;127:3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.