Abstract
Background:
There are currently no biomarkers for early breast cancer patient populations at risk of bone metastasis. Identification of mediators of bone metastasis could be of clinical interest.
Methods:
A de novo unbiased screening approach based on selection of highly bone metastatic breast cancer cells in vivo was used to determine copy number aberrations (CNAs) associated with bone metastasis. The CNAs associated with bone metastasis were examined in independent primary breast cancer datasets with annotated clinical follow-up. The MAF gene encoded within the CNA associated with bone metastasis was subjected to gain and loss of function validation in breast cancer cells (MCF7, T47D, ZR-75, and 4T1), its downstream mechanism validated, and tested in clinical samples. A multivariable Cox cause-specific hazard model with competing events (death) was used to test the association between 16q23 or MAF and bone metastasis. All statistical tests were two-sided.
Results:
16q23 gain CNA encoding the transcription factor MAF mediates breast cancer bone metastasis through the control of PTHrP. 16q23 gain (hazard ratio (HR) for bone metastasis = 14.5, 95% confidence interval (CI) = 6.4 to 32.9, P < .001) as well as MAF overexpression (HR for bone metastasis = 2.5, 95% CI = 1.7 to 3.8, P < .001) in primary breast tumors were specifically associated with risk of metastasis to bone but not to other organs.
Conclusions:
These results suggest that MAF is a mediator of breast cancer bone metastasis. 16q23 gain or MAF protein overexpression in tumors may help to select patients at risk of bone relapse.
Women with primary breast cancer (BC) are at risk of distant metastatic relapse many years or decades after surgery. Adjuvant (ie, postoperative) systemic treatments, such as chemotherapy, hormonal therapy (in estrogen receptor–positive tumors [ER+]) and anti-HER2 therapy (in HER2-positive tumors), may eradicate micrometastatic disease and thereby reduce the risk of metastatic relapse. However, current adjuvant treatments are toxic (particularly chemotherapy), only benefit a subset of women, and the prevention of overt metastasis is non–organ specific. BC is a highly heterogeneous disease, and there is clinical evidence of distinct patterns of disease relapse (1). In fact, the capacity of metastatic BC cells to grow in different environments may give rise to metastatic speciation (2).
The discovery in past years of mediators of organ-specific metastasis in breast and other cancers suggests that it might be possible to identify clinically actionable biomarkers that specifically predict bone metastasis risk in breast cancer. Unbiased testing for recurrent copy number aberrations (CNAs) in large datasets aimed at the identification of bone metastasis risk mediators is a feasible approach that may provide novel data and insights. CNAs have been observed in several human cancers, and these events are usually associated with the presence of mediators of malignancy and clinical outcome, for example 17q12 and the HER2 oncogene (3). However, CNAs associated with tissue-specific metastasis remained uncharacterized.
Here we followed an unbiased screening approach to test the hypothesis that BC metastasizes to the bone by selecting mediators for homing, survival, and colonization that result from genomic alterations. Using this approach, we identified that the 16q23 gain, through the v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog (MAF) gene, mediates bone metastasis in breast cancer and independently associates with risk of bone metastasis but not metastasis to other sites.
Methods
Cell Culture
The MCF7, ZR-75, and T47D human breast cancer (BC) cell lines and 4T1 murine BC cells were purchased from American Type Culture Collection (ATCC). These cell lines and their genetically modified derivatives were maintained as described previously (4). Cells were maintained in the indicated media as per ATCC guidelines and were routinely tested for well-reported breast cancer markers, including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Thus, cells were grown in DMEM or RPMI medium (Gibco), supplemented with 10% fetal bovine serum (FBS) (Biological Industries Kibbutz Beit Haemek, Israel), Glutamine 0.29mg/mL (Biological Industries Kibbutz Beit Haemek, Israel), Penicillin 100U/mL (Biological Industries) and Streptomycin 0.1mg/mL (Biological Industries Kibbutz Beit Haemek, Israel). Further details are given in the Supplementary Methods (available online).
Animal Studies and Xenografts
All animal work was approved by the institutional animal care and use committee of Institute for Research in Biomedicine (IRB) Barcelona and performed following the principles of laboratory animal care (as mandatory per European Union and Local government laws). Female BALB/c nude mice of 11 weeks of age (n = 9–37 per group) were used for all studies. Mice were anesthetized with ketamine (100mg/kg body weight) and xylazine (10mg/kg body weight), and after injection imaged mice were monitored weekly using IVIS imaging. Further details are given in the Supplementary Methods (available online).
Oligonucleotide Array Assays
RNA sample collection and generation of biotinylated complementary RNA (cRNA) probe were carried out essentially as described in the standard Affymetrix (Santa Clara, CA) GeneChip protocol. Ten micrograms of total RNA were used to prepare a cRNA probe using a Custom Superscript Kit (Invitrogen Carlsband, CA). For expression profiling, 25ng of RNA per sample was processed using isothermal amplification SPIA Biotin System (NuGEN technologies San Carlos, CA). Each sample was hybridized with an Affymetrix Human Genome U133APlus2.0 microarray at the IRB Barcelona Functional Genomics Core Facility. Further details are given in the Supplementary Methods (available online).
Analysis of Copy Number Alterations in Expression Data
The detection of CNAs by means of expression profile analysis is based on strong correlation between the genomic alterations and the aberrant gene expression in the affected genomic regions. While the detection of CNAs using gene expression analysis is possible, difficulties arise from the type of starting expression data (8). We used the function findCopyNumber from the Bioconductor phenoTest package, which implements an approach similar to the one applied by Hu et al. (5) to find regions with CNAs in the MSKCC/EMC cohort. Enrichment scores (in our case log hazard ratios) and the chromosomal positions of the scores allowed us to distinguish areas in which the enrichment was higher/lower than expected when the positions were assigned at random. Further details are given in the Supplementary Methods (available online).
Fluorescence in Situ Hybridization and Other Methods
Cells were processed as described (6). The slides were incubated with MAF probe BAC RP11-1068n20. This probe covered a 197kb segment at chr16: 79,460,645-79,657,297, a region including the full MAF gene (chr16: 79,625,745 to 79,639,622, 14kb) and excluding WWOX and the FRA16D fragile site. In parallel, a CEP16 (centromeric chr16, 16q11.2) (Abbot Chicago, IL) probe was used to score 16q23 CNAs. Findings were confirmed with an independent MAF/CEP16 probe (Inbiomotion Barcelona, Spain). DAPI counterstain was applied, and images were acquired with a Leica TCS-SP5 confocal microscope. The percentage of 16q23 CNA–positive cells was determined based on minimum of 100 counted cells for each condition. Information on protein extraction and western blots, chromatin immunoprecipitation, quantitative real-time polymerase chain reaction (qRT-PCR), histopathology and immunohistochemistry, reporter assays, osteoclast differentiation assay, and cell migration and adhesion assays is given in the Supplementary Methods (available online).
Patient Gene Expression Datasets
Information on the patients was downloaded from GEO (7). The following cohorts were used: A) MSKCC/EMC. Pooled GSE2603, GSE2034, and GSE12276; B) GSE14020. ER+ patients were selected on the basis of the bimodality of gene ESR1. C) Patient expression profiles and clinical annotations were downloaded as described in van de Vijver et al. (8). Further details are given in the Supplementary Methods (available online).
Statistical Methods
Cumulative incidence was estimated with Cox proportional hazard models and likelihood ratio tests. The assumption of proportionality was checked using the supremum test for proportional hazards assumption. This test yields a significant P value if this assumption is violated.
Of note, cumulative incidence for validation dataset II was calculated using Cox cause-specific hazard model with competing events (death). Cumulative incidence functions for recurrence were estimated. These functions estimate the actual percentage of patients who will experience the various competing events within the study cohorts as opposed to the overestimated percentages obtained with the Kaplan-Meier method based on the cause-specific hazards.
For survival analyses, a multivariable Cox proportional hazard model was fitted to test the correlation between high MAF-expressing vs the rest of the tumors and bone metastasis. Tumor size, lymph node status, tumor grade, and proliferation were used as adjustment variables. R’s function step was used to perform backward elimination by AIC. P values were obtained with Cox proportional hazards likelihood ratio tests.
The Wilcoxon rank-sum test was used to assess the agreement of immunohistochemistry (IHC) and florescence in situ hybridization (FISH) events in each sample and Pearson correlation was used to estimate the correlation of IHC and FISH events. Data were analyzed using R as well as Prism 6 (GraphPad Software Inc.). All statistical tests were two-sided.
Results
Identification of CNAs Associated With Bone Metastasis in BC
We developed an experimental xenograft mouse model based on ER+ moderately bone metastatic MCF7 BC cells to derive cells with higher propensity to metastasize to the bone (9–14). To perform in vivo selection we introduced MCF7 luciferase-expressing cells in the left ventricle of BALB/c nude mice and monitored their growth by quantitative bioluminescence imaging. After three rounds of passaging, we selected cells with a statistically higher rate of bone metastasis than parental MCF7 cells, which we termed BoM2 (Figure 1A). The tumor growth when tested by orthotopic injection at the mammary fat pad was similar between parental MCF7 and BoM2 cells, and both were dependent on estrogen gene responses (Supplementary Figure 1, A-C, available online).
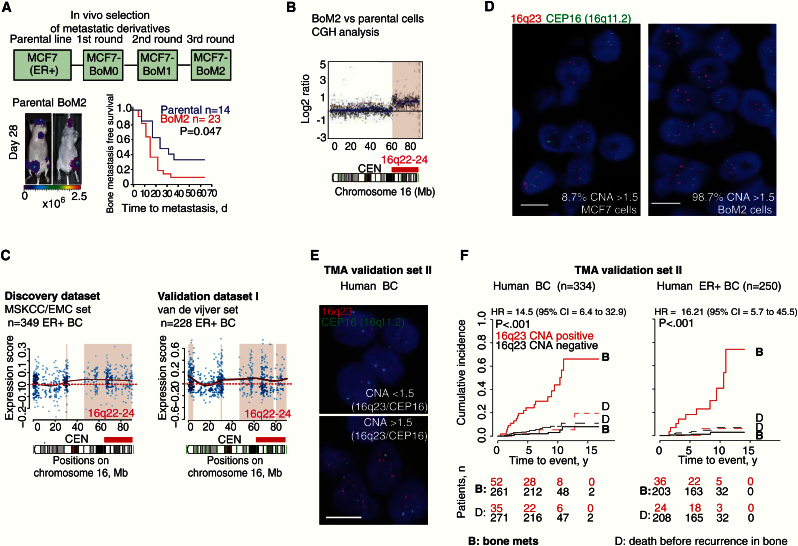
Figure 1.
16q23 gain associated with breast cancer (BC) bone metastasis. A) In vivo selection of subsequent bone metastatic derivatives from MCF7 parental cell line with representative bioluminescent images. Kaplan-Meier curve shows bone metastasis–free survival for parental or BoM2 cells injected via left ventricle. P value was obtained using two-sided log-rank test. B) Comparative genomic hybridization (CGH) analysis for Chr 16 compares BoM2 with MCF7 parental cells. Black dots and blue horizontal lines represent normalized log2 intensity ratios and segments, respectively. Red bar underlines 16q22-24 DNA genomic gain and CEN centromeric region. C) Analysis of copy number aberrations on the basis of gene expression (ACE-like Algorithm) for Chr 16. Genomic areas of statistically significant association of increased gene expression with metastasis in estrogen receptor–positive (ER+) BC tumors (discovery MSKCC/EMC dataset) and validation (van de Vijver dataset) are colored. Solid red line indicates differences in gene expression; dashed red line indicates the expected lack of variation between populations. D) Representative images of florescence in situ hybridization (FISH) analysis in BC cell lines with the indicated probes. Scale bar = 10 μm. E) Top, FISH image of patient sample without 16q23 gain (16q23/CEP16 < than 1.5 copies), and bottom, patients with 16q23 gain (16q23/CEP16> than 1.5 copies). Scale bar = 10 μm. F) Cumulative incidence plot of bone metastasis at anytime, using death before recurrence in bone as a competing event for all (left) and ER+ primary tumors (right) in Spanish dataset (n = 334, n = 250 respectively). Patients were stratified according to 16q23/CEP16 ratio per cell, as CNA-negative (<1.5) and 16q23/CEP16 CNA-positive (> or = 1.5). A minimum of 50 cells per core and three cores per tumor were scored. P values were obtained after fitting Cox proportional hazards model with competing events. BC = breast cancer; CI = confidence interval; ER = estrogen receptor; HR = hazard ratio.
We studied genomic copy number aberrations (CNAs) in BoM2 cells by comparative genomic hybridization. We uncovered substantial losses in chromosomes 6, 12, 19, 20, and 21 and one substantial gain (16q22-q24) in BoM2 compared with the parental population (Figure 1B; Supplementary Figure 1D, available online). Using an ACE-like algorithm (Analysis of CNAs by Expression data [5]), we associated variations in CNA with metastasis risk in 349 ER+ primary BC patients (discovery MSKCC/EMC BC set) (12). Only the 16q22-24 chromosomal gain showed a substantial association with metastasis (Figure 1C; Supplementary Figure 1E, available online). This association was confirmed by applying the ACE-like algorithm to an independent validation cohort I (van de Vijver BC set) (Figure 1C) (8). Patients with 16q22-24 CNA-positive ER+ BC tumors (MSKCC/EMC) had a higher cumulative rate of metastasis (hazard ratio [HR] = 1.37, 95% confidence interval [CI] = 1.01 to 1.88, P = .048) (Supplementary Figure 1F, available online). Next, we confirmed 16q23 copy gain in BoM2 cells (98.7% of cells) compared with parental MCF7 cells (8.7% of cells) by means of the ratio of a 16q23 and a CEP16 (16q11.2) centromeric FISH probe (Figure 1D). We also characterized additional breast cancer cell lines and showed that they had varying degrees of 16q23 gain (T47D 13.7% and ZR-75 47.5%) (Supplementary Figure 1G, available online).
Association of 16q23 Genomic Gain With Breast Tumors That Develop Bone Metastasis
Next, we tested 16q23 gain by FISH in paraffin-embedded samples from an independent validation set II of primary stage I-III BC specimens from patients with annotated clinical follow-up (Spanish dataset) (15). We found that 14% of the 334 primary BCs tested were classified as positive for 16q23 gain, defined by at least 1.5 copies of the 16q23 region normalized to the CEP16 centromeric probe per cell (a minimum of 50 cells per specimen were scored) (Figure 1E). 16q23 gain–positive tumors were at a high cumulative incidence rate of bone metastasis at any time (HR = 14.5, 95% I = 6.4 to 32.9, P < .001; hazard ratio was calculated considering death before recurrence in bone as a competing event) (Table 1 and Figure 1F; Supplementary Tables 1 and 2, available online). Moreover, 16q23 gain significantly associated with poor overall survival (OS) (HR for OS = 3.8, 95% CI = 2.0 to 7.4, P < .001) and not with other nonvisceral and visceral metastasis (Supplementary Figure 1, H and I, available online).
Table 1.
Cumulative incidence of recurrence in bone at any time in patients with 16q23 CNA*
| Variable | Univariate (n = 313) | Multivariable (n = 313) | ||
|---|---|---|---|---|
| HR (95% CI) |
P | HR (95% CI) |
P | |
| Menopausal status | .597 | - | ||
| Premenopausal | 1.00 (referent) | - | ||
| Postmenopausal | 0.81 (0.37 to 1.74) |
- - |
||
| Tumor size, mm | .12 | .299 | ||
| ≤20 | 1.00 (referent) | 1.00 (referent) | ||
| 21–50 | 1.86 (0.87 to 3.93) |
1.56 (0.63 to 3.81) |
||
| >50 | 1.73 (0.60 to 5.01) |
0.56 (0.12 to 2.57) |
||
| Tumor grade | .174 | .160 | ||
| I | 1.00 (referent) | 1.00 (referent) | ||
| II | 0.87 (0.41 1.83) |
2.75 (0.32 to 23.33) |
||
| III | 1.58 (0.75 to 3.33) |
1.12 (0.12 to 10.27) |
||
| Lymph nodes | .120 | .255 | ||
| None | 1.00 (referent) | 1.00 (referent) | ||
| 1–3 | 0.94 (0.41 to 2.14) |
1.20 (0.46 to 3.11) |
||
| 4–9 | 1.27 (0.38 to 4.24) |
1.79 (0.33 to 9.54) |
||
| >9 | 3.39 (1.17 to 9.81) |
4.57 (1.16 to 18.01) |
||
| Hormonal receptor status | .414 | - | ||
| Negative | 1.00 (referent) | - | ||
| Positive | 0.71 (0.32 to 1.57) |
- - |
||
| HER2 status | .580 | - | ||
| Negative | 1.00 (referent) | - | ||
| Positive | 1.30 (0.52 to 3.22) |
- - |
||
| Proliferation (Ki-67) | .010 | .263 | ||
| Low proliferation (<15%) | 1.00 (referent) | 1.00 (referent) | ||
| High proliferation (≥15%) | 2.81 (1.37 to 6.34) |
1.65 (0.68 to 3.98) |
||
| 16q23 (FISH CNA) | 1x10-11 | 3x10-10 | ||
| Below 1.5 | 1.00 (referent) | 1.00 (referent) | ||
| Equal or More 1.5 | 14.5 (6.4 to 32.9) |
18.02 (6.69 to 48.53) |
||
* Measured from date of primary tumor surgical resection. Analyzed by Cox cause-specific hazards model with competing events (death before recurrence in bone). The risk of the competing event is reported in Supplementary Table 2 (available online). CI = confidence interval; CNA = copy number aberration; HER2 = human epidermal growth factor receptor 2; HR = hazard ratio.
In this series, 16q23 gain–positive tumors were statistically significantly associated with high-grade larger tumor size and high proliferation (Ki67 > 15%), but not with age, menopausal status, ER, PR, HER2, or lymph node (LN) involvement (Supplementary Table 1, available online). In a multivariable competing risk Cox analysis, combining tumor size, grade, proliferation, LN status, and 16q23 gain, the latter retained a statistically significant hazard ratio of cumulative bone metastasis at any time of 18.02 (95% CI = 6.69 to 48.53, P < .001) (Table 1; Supplementary Table 2, available online). Thus, 16q23 gain was an independent marker of bone metastasis, including nodal status (Sensitivity [Se] = 0.71, Specificity [Sp] = 0.88 for cumulative incidence of bone metastasis ever; median follow-up is similar in both categories (Supplementary Table 1 and Supplementary Figure 1J, available online), with a 97% negative predictive value (NPV). The relationship between 16q23 gain and bone metastasis was statistically significant in ER+, triple-negative (TN), and HER2+ tumors (Figure 1F; Supplementary Figure 1K, available online).
Association of MAF Expression in the 16q23 Region With Bone Metastasis
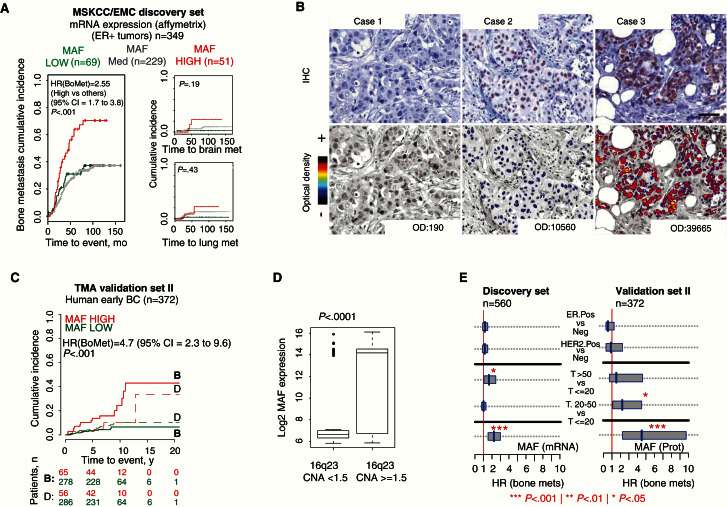
We examined possible bone mediators in the 16q23 region by identifying genes that were more than two-fold (P < .05) differentially expressed in BoM2 derivatives compared with parental MCF7 cells (Supplementary Figure 2A, available online). Among the two candidates, MAF and SLC9A5 (Supplementary Figure 2, A-D, available online), the Cox hazard ratio for the association of mRNA expression and bone metastasis showed statistical significance for MAF (P = .032) but not for the SLC9A5 (P = .95) (MSKCC/EMC BC dataset). Patients with BC with high MAF expression showed a greater cumulative incidence of metastasis to bone—but not to brain or lung—than the rest (HR for bone metastasis = 2.5, 95% CI = 1.7 to 3.8, P < .001) (Figure 2A; Supplementary Figure 2E, available online). Next, we confirmed MAF gain in 9.1% of the 1079 tumors evaluated by the The Cancer Genome Atlas BC project (16). This percentage was smaller than in the FISH validated Spanish dataset II; however, this may be related, at least in part, to the higher sensitivity of our test. MAF, a potential genetic driver of the 16q23 region, is a transcription factor of the AP-1 family that has been reported to contribute to transformation in 60% of Angioimmunoblastic T-cell lymphoma (AITL) (17,18) and 50% of multiple myeloma (MM) patients (19). However, to date, MAF has not been associated with BC cell transformation, tumor progression, or metastasis (20).
Figure 2.
MAF expression associated with bone metastasis in breast cancer (BC). A) Cumulative incidence plot of bone, brain, and lung metastasis in estrogen recptor–positive (ER+) primary BC patients (discovery MSKCC/EMC dataset). MAF expression: low (<mean - SD), medium (≥ mean - SD and ≤ mean + SD) and high (> mean + SD). P values were obtained after fitting Cox proportional hazard models and performing two-sided likelihood ratio tests. B) Representative MAF immunostainings of primary BC tissues. Case 1 represents MAF-negative tumors (optical density [OD] < 1000). Cases 2 and 3 are MAF-positive tumors (OD > 1000 and > 25000, respectively). Scale bar = 50 μm. C) Cumulative incidence plot of bone metastasis at any time, considering death as a competing event in Spanish dataset. High MAF-expressing group (red line, OD > 1000); low MAF-expressing group (green line, OD < 1000). P values were obtained after fitting Cox proportional hazards model with competing events. D) Box plot with the MAF protein log2 score (immunohistochemistry [IHC], OD values) on the vertical axis and amplified vs not-amplified categories on the horizontal axis. P value scored by two-sided Wilcoxon signed-rank test. Boxes represent interquartile range and median. Whiskers represent minimum and maximum values. E) 95% confidence intervals for the Cox proportional hazard ratios (HRs) are illustrated for the discovery and validation cohort for selected values of covariates represented. MAF is measured at the mRNA level (discovery set) and protein level (validation set). BC = breast cancer; CI = confidence interval; ER = estrogen receptor; HER2 = human epidermal growth factor receptor 2; HR = hazard ratio; OD = optical density.
To characterize the relationship between MAF and bone metastasis, we analyzed MAF protein expression by immunohistochemistry in the validation dataset II (high MAF defined as >1000 optical density [OD], cutoff determined based on the receiver operating characteristic curve) (Figure 2B; Supplementary Figure 2F, available online). High MAF protein staining associated with a high cumulative risk of metastasis to bone at any time but not to other nonvisceral and visceral sites (HR for bone metastasis = 4.68, 95% CI = 2.29 to 9.57, P < .001) (Figure 2C and Table 2; Supplementary Tables 3 and 4, available online). MAF protein expression by IHC and 16q23 gain by FISH were correlated (r = 0.52, P < .001) (Figure 2D; Supplementary Figure 2G, available online). MAF protein expression enabled better assignment of positive bone metastasis patients than MAF mRNA expression (Figure 2E). ER+ BC patients with MAF OD above 25 000 developed bone metastasis with almost complete penetrance (sensitivity = 0.36, specificity = 0.99, HR for bone met at any time = 21.3, 95% C.I = 8.3 to 54.7, P < .001) (Supplementary Figure 2H, available online). In triple-negative disease, 1000 OD cutoff–identified patients at high risk of bone metastasis (sensitivity = 0.75, specificity = 0.83; HR for bone met at any time = 10.7, 95% CI = 2.2 to 53.3, P = .001) (Supplementary Figure 2H, available online). In HER2+ patients, no statistical significance was achieved (Supplementary Figure 2I, available online). In a multivariable analysis, MAF protein expression retained its predictive value for bone metastasis independently of traditional clinico-pathological parameters (HR for bone metastasis at any time = 5.28, 95% CI = 2.5 to 11.2, P < .001) (Table 2; Supplementary Tables 3 and 4, available online).
Table 2.
Cumulative incidence of recurrence in bone at any time in patients based on MAF expression (IHC)*
| Variable | Univariate (n = 343) | Multivariable (n = 343) | ||
|---|---|---|---|---|
| HR (95% CI) |
P | HR (95% CI) |
P | |
| Menopausal status | .676 | - | ||
| Premenopausal | 1.00 (referent) | - | ||
| Postmenopausal | 0.85 (0.40 to 1.80) |
- - |
||
| Tumor size, mm | .007 | .151 | ||
| ≤20 | 1.00 (referent) | 1.00 (referent) | ||
| 21–50 | 2.13 (1.03 to 4.37) |
2.21 (0.95 to 5.13) |
||
| >50 | 1.56 (0.54 to 4.47) |
2.34 (0.65 to 8.41) |
||
| Tumor grade | .046 | .121 | ||
| I | 1.00 (referent) | 1.00 (referent) | ||
| II | 1.42 (0.69 to 2.93) |
4.27 (0.55 to 32.99) |
||
| III | 1.27 (0.61 to 2.65) |
2.37 (0.28 to 19.68) |
||
| Lymph nodes | .007 | .047 | ||
| None | 1.00 (referent) | 1.00 (referent) | ||
| 1–3 | 1.06 (0.47 to 2.39) |
1.22 (0.49 to 3.02) |
||
| 4–9 | 0.63 (0.15 to 2.65) |
0.67 (0.14 to 3.23) |
||
| >9 | 5.14 (2.20 to 11.99) |
4.04 (1.46 to 11.15) |
||
| Hormonal receptor status | .167 | - | ||
| Negative | 1.00 (referent) | - | ||
| Positive | 0.58 (0.27 to 1.22) |
- - |
||
| HER2 status | .670 | - | ||
| Negative | 1.00 (referent) | - | ||
| Positive | 0.81 (0.31 to 2.13) |
- - |
||
| Proliferation (Ki-67) | .014 | .044 | ||
| Low proliferation (<15%) | 1.00 (referent) | 1.00 (referent) | ||
| High proliferation (≥15%) | 2.52 (1.23 to 5.18) |
2.23 (1.03 to 4.84) |
||
| cMAF (IHC) | 5x10−5 | 2x10−5 | ||
| Nonoverexpression | 1.00 (referent) | 1.00 (referent) | ||
| Overexpression | 4.68 (2.29 to 9.57) |
5.28 (2.50 to 11.20) |
||
* Measured from date of primary tumor surgical resection. Analyzed by Cox cause-specific hazards model with competing events (death before recurrence in bone). The risk of competing events is reported in Supplementary Table 4 (available online). CI = confidence interval; HER2 = human epidermal growth factor receptor 2; HR = hazard ratio; IHC = immunohistochemistry.
Effect of MAF on BC Metastasis to Bone
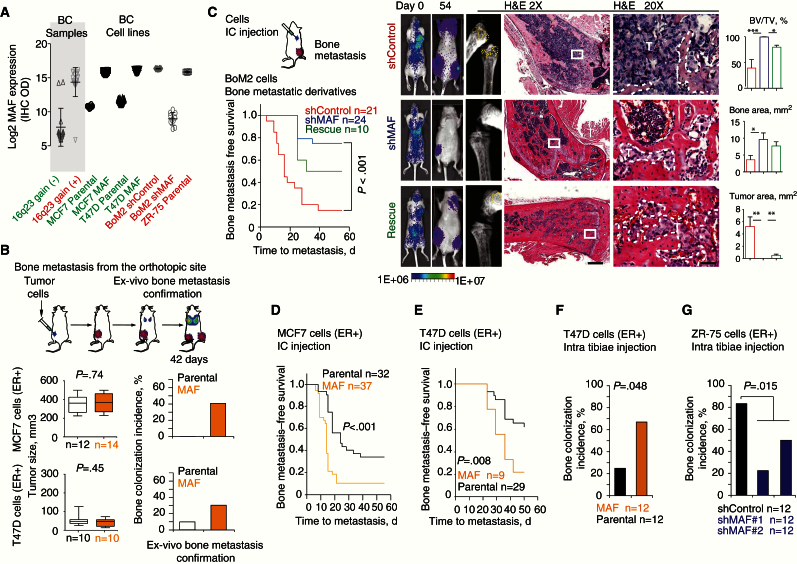
Next, we functionally validated the role of MAF in bone metastasis by MAF-stable downregulation or overexpression in a panel of BC cell lines. Interestingly, MAF protein levels in the 16q23 gain–positive BoM2, MCF7 MAF–overexpressing, and ZR-75 cells were comparable with those in 16q23 gain–positive tumors, whereas MCF7 parental, MAF-depleted BoM2, and T47D cells expressed MAF similarly to 16q23 gain–negative tumors (Figure 3A; Supplementary Figure 3, A and B, available online).
Figure 3.
MAF mediation of bone metastasis in breast cancer (BC) cells. A) MAF protein log2 score (immunohistochemistry [IHC], optical density values) on the vertical axis and various BC patient samples (n = 20) and BC cell lines (n = 10–20 sections) on the horizontal axis were plotted. Amplified (red) and not-amplified (green) categories are highlighted. B) Bone metastasis was scored from MAF-overexpressing and control parental cells implanted in the mammary fat pad. Mice bearing size-matched tumors (>300 or >100mm3 for MCF7 or T47D, respectively) at day 42 were considered when evaluating bone metastasis. Individual tumors are represented as box plots with median, interquartile range, and min and max values. Bone colonization incidence is plotted. P value scored by two-sided Wilcoxon signed-rank test. C) Schematic representation of injection via left ventricle. Kaplan-Meier curve of bone metastasis–free survival for BoM2 shControl, shMAF, and rescue cells. P value was obtained using two-sided log-rank test. Representative bioluminescent images and hematoxylin and eosin (H&E) staining at days 0 and 54 (endpoint) of bone metastasis for each group are shown. T = tumor area. 500 and 50 μm scale bars were used for middle and right panels, respectively. Histomorphometric analysis of bone metastasis lesions is depicted (BV = bone area; TV = tumor area). P values scored by two-sided Wilcoxon signed-rank test, *<.05, **<.01, ***<.001. D) Kaplan-Meier curves of bone metastasis–free survival for MCF7 parental and MAF-overexpressing cells injected via left ventricle of mice. P value was obtained using two-sided log-rank test. E) Kaplan-Meier curve of bone metastasis–free survival for T47D parental and MAF-expressing cells injected via left ventricle of mice. P value was obtained using two-sided log-rank test. F) Bone colonization incidence 14 days postintratibiae inoculation of cells in mice. (E) Two-sided Fisher test was used to score significance. G) Bone colonization incidence 28 days postintratibiae inoculation of ZR-75 control and MAF short hairpin carrying cells. Two-sided Fisher test was used to score significance. BC = breast cancer; CI = confidence interval; ER = estrogen receptor; H&E = hematoxylin and eosin; HER2 = human epidermal growth factor receptor 2; HR = hazard ratio; OD = optical density.
Orthotopically implanted mock and MAF-overexpressing MCF7 and T47D cells in mice grew similarly, indicating that MAF did not contribute to proliferation, but the MAF-expressing groups showed enhanced bone metastatic capacity (Figure 3B; Supplementary Figure 3, C-G, available online). Consistently, only 23% of the mice inoculated via the left ventricle with BoM2 MAF knockdown cells developed bone metastasis detectable by luciferase activity at day 54 postinjection, compared with 90% in shControl BoM2 cells or 50% in the shMAF BoM2 cells with rescued MAF expression (Figure 3C; Supplementary Figure 3, C and D, available online). The reduction in bone metastasis in MAF-depleted cells was accompanied by a decrease in the extent of hind limb lesions, as determined by photon flux ex vivo and histomorphometric analysis (Figure 3C; Supplementary Figure 3H, available online). In contrast, MAF overexpression (Supplementary Figure 3, C-E, available online) enhanced the capacity of MCF7 to metastasize to bone (Figure 3D; Supplementary Figure 3, I and J, available online) but did not support lung or adrenal colonization in different experimental settings (Supplementary Figure 3, K and L, available online).
Similar results were observed in T47D cells that are poorly metastatic (9,10). Inoculation of T47D MAF–overexpressing cells statistically significantly reduced bone metastasis–free survival (Figure 3E; Supplementary Figure 3, A, B, and M, available online) but did not influence metastasis rate in other sites (Supplementary Figure 3, N and O, available online). Upon in-tibiae implantation, MAF expression increased bone colonization of T47D (ER+) (Figure 3F) and 4T1 (triple-negative) cells (Supplementary Figure 3P, available online), whereas MAF depletion in ZR75 (ER+) cells impaired this process (Figure 3G). Collectively, these results confirmed MAF as a mediator of bone metastasis in the various BC cells.
MAF-Induced BC Cell–Bone Stroma Interactions
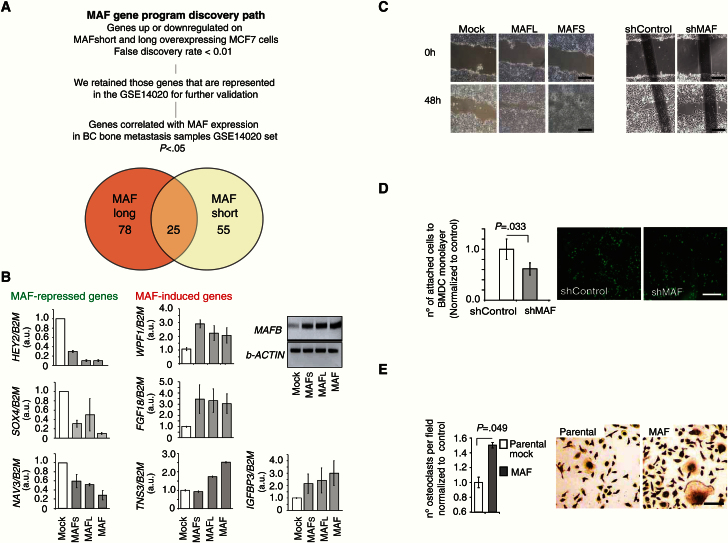
We combined two independent analyses to identify genes that were transcriptional targets of MAF-short and/or -long isoforms and whose expression in a BC metastasis sample set was significantly correlated with MAF expression. First, we identified genes significantly up or downregulated upon MAF-short and/or -long overexpression in MCF7 cells. Subsequently, we eliminated those genes in the list whose expression was not significantly correlated (P ≥ .05) with MAF expression in BC metastasis patient samples (GSE14020; liver, brain, bone, and lung BC metastasis sample dataset) (Figure 4A) (21). The resulting 148 putative MAF target genes were named MAF metastasis program (Supplementary Table 5, available online), and, of those, 25 genes were common targets of MAF-short and -long isoforms (Figure 4A; Supplementary Figure 4A, available online). The expression pattern of some of these genes in MAF-expressing cells was confirmed by qRT-PCR analysis (Figure 4B). These results suggested that MAF transcriptionally controls a collection of events that may broadly support functions required for bone metastasis, such as migration (Figure 4C; Supplementary Figure 4B), adhesion to bone marrow–derived cells (Figure 4D), and osteoclast differentiation (Figure 4E; Supplementary Figure 4C, available online).
Figure 4.
MAF regulation of expression of genes associated with bone metastasis in breast cancer (BC). A) Discovery path for the identification of genes whose expression is up- or down-regulated in MAF short- and/or long-expressing cells compared with parental Mock-MCF7 cells. Pearson correlation and fold change bigger than two and Bayesian false discovery rate below 5%. B) Expression levels of genes identified in (A) parental control, MAF-short and MAF-long isoform-overexpressing cells measured by quantitative real-time polymerase chain reaction using indicated TaqMan or SybrGreen probes. B2M or b-ACTIN were used as normalization controls. Data is mean of three experiments with SD. C) Representative images from three independent experiments show wound-closing assay at initial and end timepoints. The extent of wound closure was assessed after 48 hours. Scale bar = 200 μm. D) Representative image of attached green-labeled tumor cells to monolayer of BMSC. Data from three independent experiments is presented as average with SD. P value scored by two-sided Wilcoxon signed-rank test. Scale bar = 500 μm. E) Quantification of the number of TRAP+ differentiated multinucleated (>3 nuclei) osteoclasts per field, normalized to control; representative images are shown. Scale bar = 100 μm. Conditioned media from the indicated cellular populations were used. Data are means from three independent experiments with SD. P value scored by two-sided Wilcoxon signed-rank test.
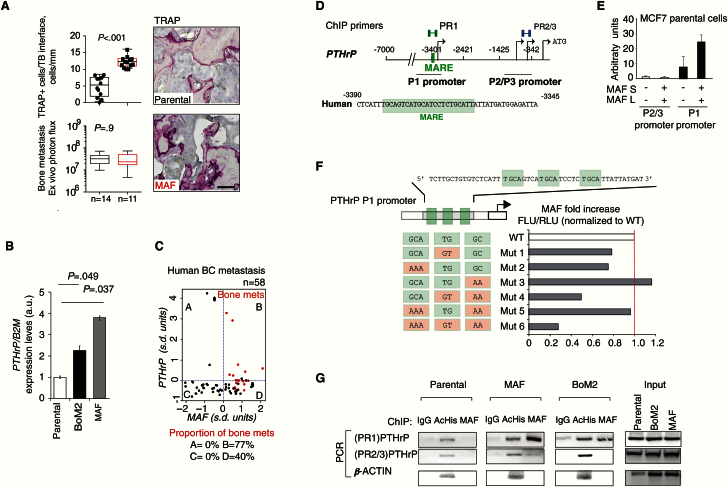
In size-matched bone metastatic lesions, MAF expression resulted in an increased number of activated tartrate-resistant acid phosphatase multinucleated osteoclasts (TRAP+ cells) along the lesion perimeter (Figure 5A). We then examined the known mediator of bone stroma–tumor interactions PTHrP (parathyroid hormone-related protein) (22) that is one of the MAF metastasis program genes (Figure 5B; Supplementary Figures 4A and 5A, available online) (23,24). Bone metastasis co-expressed high MAF and PTHrP in 77% of the specimens tested (Figure 5C). MAF-enhanced induction of osteoclast differentiation from bone marrow–derived cells in vitro was suppressed upon co-incubation with a PTHrP antagonist peptide (PTHrP-AN) (Supplementary Figure 4C, available online).
Figure 5.
MAF transcriptional control of bone-modifying cytokine PTHrP. A) Upper left panel: Quantification of TRAP+ cells per bone perimeter along the bone-tumor interface of metastasis. Data are presented as box plots with median, interquartile range, and min and max values. Lower left panel: Ex vivo quantification of the size-match bone metastasis. P value scored by two-sided Wilcoxon signed-rank test. Right panels, scale bar = 100 μm. B) PTHrP expression levels measured by quantitative real-time polymerase chain reaction in MCF7 parental, BoM2, and MAF-overexpressing cells. Data are means from three independent experiments with SD. P values scored by two-sided Wilcoxon signed-rank test. C) Dot chart of the standardized mRNA expression of MAF against the standardized mRNA expression of PTHrP in human breast cancer (BC) metastasis (GSE14020). The red dots depict bone, and black dots soft tissue metastasis. The blue dotted lines show the average MAF or PTHrP expression. Table represents the proportion of bone metastases in each quadrant above. D) Schematic representation of the PTHrP promoter regions (P1 and P2/P3) and regions amplified in ChIP assays (primer sets, PR1, and PR4). The nucleotide sequence of the human PTHrP MAF response element (MARE) is shown (green box). E) Luciferase activity of PTHrP promoter, P1, or P2/3, reporter plasmid in MCF7 cells transfected with control or MAF-short and -long isoforms expressing vectors normalized to control condition. Data are means from three independent experiments with SD. F) Luciferase activity of mutant PTHrP P1 promoter reporters in MCF7 cells normalized to wild-type. Data representative from three independent experiments are shown. G) ChIP assays of parental, MAF-expressing, and BoM2 MCF7-derivative cells performed with the indicated antibodies and polymerase chain reaction primers. PTHrP promoter proximal region (P2/P3) and the β-ACTIN promoters were used as a negative control. BC = breast cancer; PCR = polymerase chain reaction; WT = wild-type.
Previous studies showed that both MAF and PTHrP play a role in chondrocyte formation (25,26), thus suggesting a potential relationship between these genes. We addressed whether PTHrP and MAF had direct interaction in bone metastasis and/or human BC, because this possibility had not been reported to date. We confirmed that the -3401/-2421bp PTHrP P1 promoter, containing a MARE-binding region, increased activity by overexpression of MAF. No effect of MAF on other PTHrP promoter regions was seen (Figure 5, D and E). Mutations that disrupt all MARE binding sites (Mutant 6) abrogated PTHrP P1 promoter response to MAF (Figure 5F). Evidence of a direct interaction was obtained by chromatin immunoprecipitation (ChIP) analysis, with MAF binding observed at P1 but not at the P2/3 proximal promoter region (Figure 5G).
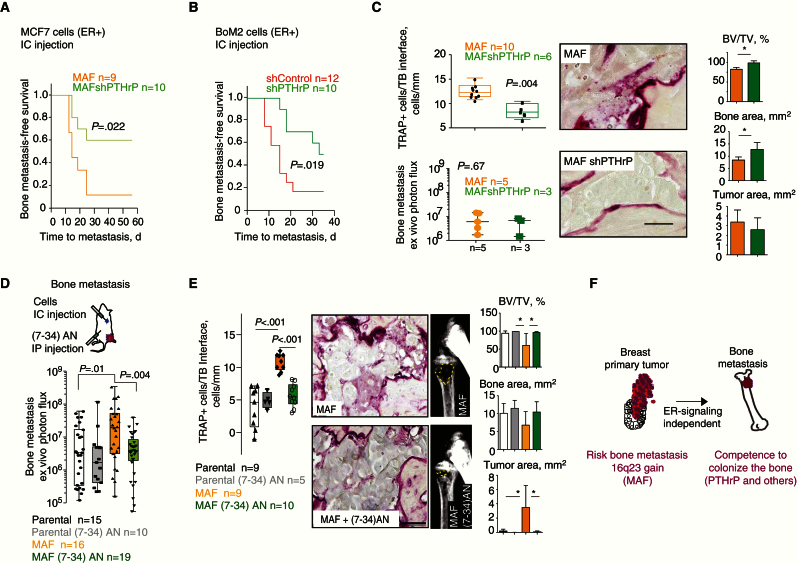
Next, we sought to validate the contribution of PTHrP to MAF-driven bone metastasis in BC cells. We reduced PTHrP function in MAF-expressing MCF7 cells by shRNA (Supplementary Figure 5B, available online) or used systemic PTHrP-AN. MAF-expressing PTHrP-depleted cells had impaired ability to form bone metastases but not metastases at other sites (Figure 6A; Supplementary Figure 5C, available online). Similarly, PTHrP downregulation blunted the capacity of BoM2 cells to colonize the bones (Figure 6B) but not other distant sites (Supplementary Figure 5D, available online). Consistent with the concept that the interaction of breast cancer cells with the niche was central to MAF-mediated bone colonization, depletion of PTHrP or treatment with PTHrP-AN in BoM2 and MCF7/MAF-overexpressing cells not only decreased the number of bone metastasis but also osteoclasts (TRAP+ cells) along the lesion perimeter (Figure 6, C-E; Supplementary Figure 5, E and F, available online). Collectively, these results support the hypothesis that MAF-mediated PTHrP expression is an important factor for the MAF-driven metastasis tumor cell–stroma interactions that foster BC bone metastasis.
Figure 6.
PTHLH contributes to MAF driven bone metastasis in breast cancer (BC). A) Kaplan-Meier curve of bone metastasis–free survival for shControl or shPTHrP MCF7 parental MAF-expressing cells injected into the left ventricle. P value was obtained using two-sided log-rank test. B) Kaplan-Meier curve of bone metastasis–free survival for shControl or shPTHrP BoM2 cells injected via left ventricle. P value was obtained using log-rank test. C) Quantification of TRAP+ cells per tumor bone (TB) perimeter along the bone-metastasis interface. Data are presented as box plots with median, interquartile range, and min and max values. Ex vivo quantification of the size-matched bone metastasis analyzed was determined. Scale bar = 20 μm. Histomorphometric analysis of bone metastasis lesions is depicted (BV = bone area; TV = tumor area). Data are means, and error bars are standard deviations. P values scored by two-sided Wilcoxon signed-rank test, *<.05. D) Schematic representation of the experiment. Quantification of ex vivo bioluminescent signal at hind limbs at day 54 with representative images for indicated experimental groups. Data are presented as box plots with median, interquartile range, and min and max values. P values scored by two-sided Wilcoxon signed-rank test. E) Quantification of TRAP+ cells per TB perimeter along the bone-metastasis interface and representative images. Data are presented as box plots with median, interquartile range, and min and max values. Scale bar = 100 μm. Histomorphometric analysis of bone metastasis lesions is depicted (BV = bone area; TV = tumor area). Data are means, and error bars are standard deviations. P values scored by two-sided Wilcoxon signed-rank test, *<.05. F) Model showing how 16q23 DNA genomic gain identifies breast primary tumors that will metastasize to the bone. ER = estrogen receptor.
Discussion
We provide novel evidence on the association of 16q23 gain to high risk of bone relapse in patients with early BC. We also show that MAF, a gene within this genomic gain, drives the molecular processes of bone colonization. Interestingly, the acquisition of high MAF expression parallels that observed in MM and AITL, where several copies of the MAF genomic region or a t(14,16) translocation are gained, leading to transformation and aggressive osteolytic bone colonization (20). These observations suggest that the biology of metastasis mechanisms to the bone may rely on a common mediator that exerts similar or different functions depending on the tumor of origin.
Our results show that MAF controls the expression of a set of genes that collectively support several steps of BC cell metastasis to bone through a series of cell-autonomous and niche-related functions (27). These observations open up the possibility of using MAF as a molecular target for the prevention or treatment of bone metastasis (28). Among the MAF target genes, we identified PTHrP. Whereas PTHrP expression in primary tumors is associated with risk of bone metastasis in T4 and positive nodal status–primary BC (29), it failed to predict bone metastasis in early stage breast tumors (30). These results suggest that PTHrP per se is not sufficient to trigger bone metastasis in early stage BC unless a higher degree of transformation (T4 and positive nodal status) is present. Consistently, our results suggest that only within the right context, for example MAF gain leading to the acquisition of various functions that support bone metastasis such as migration, adhesion, etc., does PTHrP expression become an advantage to cancer cells that colonize the bone.
This study also had some limitations. Currently there are no mouse models of breast cancer bone metastasis, making it difficult to extrapolate our experimental results into the clinical setting. In addition, in some of the models analyzed we scored for metastatic bone colonization rather than metastasis from the breast primary site to the bone. Thus, particular focus on the late steps of colonization was unavoidable. The identification of novel drivers of breast cancer bone metastasis, being tested in clinically relevant samples, allows overcoming these limitations.
Our preclinical findings, as well as results from a clinical discovery series, led us to test 16q23 gain by FISH in paraffin-embedded samples from patients with primary BC and clinical follow-up. We detected 16q23 CNA in 14% of a total of 334 primary BC specimens and showed that it had a statistically significant and independent association with the risk of bone metastasis. This observation is in agreement with the expected frequency of BC patients who develop bone metastasis (10%–20%) after a 15-year follow-up (1, 31). The 16q region contains a well-defined fragile site (FRA16) located at the FOR (WWOX) locus (32). Alterations on FRA16 are observed in various tumor types with diverse clinical outcome associations; 16q loss of heterozygosity (33), as well as 1q gain/16q loss (34), was associated with a positive outcome in luminal BC (3); 16q loss was related to prostate cancer progression (35), and 16q gain with poor outcome in MM (19). In tumor cells, fragile sites have been associated with translocations, deletions, amplifications, and integration sites (36,37), which might explain the promiscuity of this region.
In addition, we developed an immunohistochemical assay to detect MAF expression. In primary tumors, MAF protein levels measured by IHC and 16q23 gain by FISH were significantly correlated, and both were associated with a high cumulative incidence of metastasis to bone but not to other nonvisceral or visceral sites. On the basis of the mechanistic and clinical data presented above, we propose that the 16q23 gain is selectively associated with bone metastasis risk in early-stage BC and that MAF encoded within this region mediates BC metastasis to bone (Figure 6F). This novel finding may enable the identification of patients at high risk of bone metastasis in a timely fashion. Clinical trials involving thousands of patients have tested or are currently testing the capacity of bisphosphonates or denosumab to prevent bone metastasis. The results of these trials have not yet influenced routine clinical practice (38–40). In an era of personalized medicine, the incorporation of a biomarker to identify those individuals most likely to benefit from bone-targeted agents is urgently needed (41). Testing 16q23 gain and MAF protein expression in breast cancer specimens from patients included in these trials to assess their predictive value to select patients who benefit from adjuvant bone-modified agents is warranted (42).
Funding
MP and SG is supported by “La Caixa” PhD fellowship program. AAE and AB hold PhD fellowships from the Spanish Ministerio de Ciencia e Innovación (MICINN). MTS is supported by the IRB Barcelona PhD program. JU is a “Juan de la Cierva” Researcher (MICINN). FIS PI12/00680, PI12/01552, and PI12/01421 supported JA, ALl, and FR, respectively. JA, ALl, and AP were part of RD12/0036/0051, RD12/0036/0070, and RD12/0036/0042, and FR is part of biobanc RD/09/0076/00101. JA and FR are recipients of intensification grant ISCIII, 2009SGR321, XBTC, MARBiobanc, and Cellex. RRG was supported by the Institució Catalana de Recerca i Estudis Avançats. Support and structural funds were provided by the BBVA Foundation, the Generalitat de Catalunya (2014 SGR 535), and the Spanish Ministerio de Ciencia e Innovación (MICINN) (SAF2013-46196) to RRG.
Supplementary Material
We would like to thank the Functional Genomics, Microscopy, and Cytometry core facilities of Institute for Research in Biomedicine (IRB) Barcelona and the UB. We thank David L. Lacey, Angel R. Nebreda, and David Rosell for critical reading of the manuscript.
Author Contributions: MP designed and performed experiments, analyzed experimental and patient data, and participated in manuscript preparation. AAE designed and established the model system and performed microarrays and FISH. FR did the pathological analysis and contributed to the Spanish dataset. MT established the model system. EP adapted the ACE-like algorithm and provided support for the statistical studies of patient data. XGA reviewed all the statistical analyses. MM, SG, AB, and JU performed the experiments. MG performed the experiments with animal models. AR and ALL assisted with the collection of samples from Spanish patients. AP did the analysis using the The Cancer Genoma Atlas breast cancer data. RC, LN, and JJM participated in text preparation. JA assisted with the collection of samples from Spanish patients, clinico-pathological correlation, and text preparation. RRG conceived and designed the project, analyzed data, supervised the overall project, and wrote the manuscript.
RRG declares shares of Inbiomotion SL for a value of less than $10 000. Inbiomotion SL develops part of the technology described herein.
References
- 1. Kennecke H, Yerushalmi R, Woods R, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28 (20):3271–3277. [DOI] [PubMed] [Google Scholar]
- 2. Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9 (4):274–284. [DOI] [PubMed] [Google Scholar]
- 3. Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486 (7403):346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tarragona M, Pavlovic M, Arnal-Estape A, et al. Identification of NOG as a specific breast cancer bone metastasis-supporting gene. J Biol Chem. 2012;287 (25):21346–21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hu G, Chong RA, Yang Q, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15 (1):9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arnal-Estape A, Tarragona M, Morales M, et al. HER2 silences tumor suppression in breast cancer cells by switching expression of C/EBPss isoforms. Cancer Res. 2010;70 (23):9927–9936. [DOI] [PubMed] [Google Scholar]
- 7. Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: mining tens of millions of expression profiles--database and tools update. Nucleic Acids Res. 2007;35(Database issue):D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347 (25):1999–2009. [DOI] [PubMed] [Google Scholar]
- 9. Lu X, Mu E, Wei Y, et al. VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging alpha4beta1-positive osteoclast progenitors. Cancer Cell. 2011;20 (6):701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yin JJ, Mohammad KS, Kakonen SM, et al. A causal role for endothelin-1 in the pathogenesis of osteoblastic bone metastases. Proc Natl Acad Sci U S A. 2003;100 (19):10954–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3 (6):537–549. [DOI] [PubMed] [Google Scholar]
- 12. Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459 (7249):1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheung WK, Zhao M, Liu Z, et al. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23 (6):725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vanharanta S, Shu W, Brenet F, et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19 (1):50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rojo F, Garcia-Parra J, Zazo S, et al. Nuclear PARP-1 protein overexpression is associated with poor overall survival in early breast cancer. Ann Oncol. 2011;23 (5):1156–1164. [DOI] [PubMed] [Google Scholar]
- 16. CancerGenomeAtlasNetwork. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490 (7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morito N, Yoh K, Fujioka Y, et al. Overexpression of c-Maf contributes to T-cell lymphoma in both mice and human. Cancer Res. 2006;66 (2):812–819. [DOI] [PubMed] [Google Scholar]
- 18. Murakami YI, Yatabe Y, Sakaguchi T, et al. c-Maf expression in angioimmunoblastic T-cell lymphoma. Am J Surg Pathol. 2007;31 (11):1695–1702. [DOI] [PubMed] [Google Scholar]
- 19. Hurt EM, Wiestner A, Rosenwald A, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5 (2):191–199. [DOI] [PubMed] [Google Scholar]
- 20. Eychene A, Rocques N, Pouponnot C. A new MAFia in cancer. Nat Rev Cancer. 2008;8 (9):683–693. [DOI] [PubMed] [Google Scholar]
- 21. Zhang XH, Wang Q, Gerald W, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16 (1):67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guise TA, Yin JJ, Thomas RJ, et al. Parathyroid hormone-related protein (PTHrP)-(1–139) isoform is efficiently secreted in vitro and enhances breast cancer metastasis to bone in vivo. Bone. 2002;30 (5):670–676. [DOI] [PubMed] [Google Scholar]
- 23. Lacey DL, Boyle WJ, Simonet WS, et al. Bench to bedside: elucidation of the OPG-RANK-RANKL pathway and the development of denosumab. Nat Rev Drug Discov. 2012;11 (5):401–419. [DOI] [PubMed] [Google Scholar]
- 24. Guise TA, Kozlow WM, Heras-Herzig A, et al. Molecular mechanisms of breast cancer metastases to bone. Clin Breast Cancer. 2005;5(Suppl2):S46–S53. [DOI] [PubMed] [Google Scholar]
- 25. MacLean HE, Kim JI, Glimcher MJ, et al. Absence of transcription factor c-maf causes abnormal terminal differentiation of hypertrophic chondrocytes during endochondral bone development. Dev Biol. 2003;262 (1):51–63. [DOI] [PubMed] [Google Scholar]
- 26. Lanske B, Karaplis AC, Lee K, et al. PTH/PTHrP receptor in early development and Indian hedgehog-regulated bone growth. Science. 1996;273 (5275):663–666. [DOI] [PubMed] [Google Scholar]
- 27. Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8 (5):341–352. [DOI] [PubMed] [Google Scholar]
- 28. Eckhardt BL, Francis PA, Parker BS, et al. Strategies for the discovery and development of therapies for metastatic breast cancer. Nat Rev Drug Discov. 2012;11 (6):479–497. [DOI] [PubMed] [Google Scholar]
- 29. Takagaki K, Takashima T, Onoda N, et al. Parathyroid hormone-related protein expression, in combination with nodal status, predicts bone metastasis and prognosis of breast cancer patients. Exp Ther Med. 2012;3 (6):963–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Henderson MA, Danks JA, Slavin JL, et al. Parathyroid hormone-related protein localization in breast cancers predict improved prognosis. Cancer Res. 2006;66 (4):2250–2256. [DOI] [PubMed] [Google Scholar]
- 31. Yerushalmi R, Woods R, Kennecke H, et al. Patterns of relapse in breast cancer: changes over time. Breast Cancer Res Treat. 2010;120 (3):753–759. [DOI] [PubMed] [Google Scholar]
- 32. Chang NS, Doherty J, Ensign A, et al. Molecular mechanisms underlying WOX1 activation during apoptotic and stress responses. Biochem Pharmacol. 2003;66 (8):1347–1354. [DOI] [PubMed] [Google Scholar]
- 33. Hansen LL, Yilmaz M, Overgaard J, et al. Allelic loss of 16q23.2–24.2 is an independent marker of good prognosis in primary breast cancer. Cancer Res. 1998;58 (10):2166–2169. [PubMed] [Google Scholar]
- 34. Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15 (2):441–451. [DOI] [PubMed] [Google Scholar]
- 35. Harkonen P, Kyllonen AP, Nordling S, et al. Loss of heterozygosity in chromosomal region 16q24.3 associated with progression of prostate cancer. Prostate. 2005;62 (3):267–274. [DOI] [PubMed] [Google Scholar]
- 36. Myllykangas S, Himberg J, Bohling T, et al. DNA copy number amplification profiling of human neoplasms. Oncogene. 2006;25 (55):7324–7332. [DOI] [PubMed] [Google Scholar]
- 37. Coquelle A, Pipiras E, Toledo F, et al. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell. 1997;89 (2):215–225. [DOI] [PubMed] [Google Scholar]
- 38. Coleman RE, Marshall H, Cameron D, et al. Breast-cancer adjuvant therapy with zoledronic acid. N Engl J Med. 2011;365 (15):1396–1405. [DOI] [PubMed] [Google Scholar]
- 39. Gnant M, Mlineritsch B, Stoeger H, et al. Adjuvant endocrine therapy plus zoledronic acid in premenopausal women with early-stage breast cancer: 62-month follow-up from the ABCSG-12 randomised trial. Lancet Oncol. 2011;12 (7):631–641. [DOI] [PubMed] [Google Scholar]
- 40. NCT01077154. US National Library of Medicine. ClinicalTrials.gov. [Google Scholar]
- 41. Coleman RE. Bone cancer in 2011: Prevention and treatment of bone metastases. Nat Rev Clin Oncol. 2011;9 (2):76–78. [DOI] [PubMed] [Google Scholar]
- 42. Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009;101 (21):1446–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.