Abstract
Polyglutamine-repeat disorders are part of a larger family of neurodegenerative diseases characterized by protein misfolding and aggregation. In spinal and bulbar muscular atrophy (SBMA), polyglutamine expansion within the androgen receptor (AR) causes progressive debilitating muscular atrophy and lower motor neuron loss in males. Although soluble polyglutamine-expanded aggregation species are considered toxic intermediates in the aggregation process, relatively little is known about the spectrum of structures that are formed. Here we identify novel polyglutamine-expanded AR aggregates that are SDS-soluble and bind the toxicity-predicting antibody 3B5H10. Soluble, 3B5H10-reactive aggregation species exist in low-density conformations and are larger by atomic force microscopy, suggesting that they may be less compact than later-stage, insoluble aggregates. We demonstrate disease-relevance in vivo and draw correlations with toxicity in vitro.
1. Introduction
Spinal and bulbar muscular atrophy (SBMA) is a slowly progressive X-linked disorder caused by an expanded polyglutamine tract in the androgen receptor (AR) protein. Clinically, SBMA patients display signs of lower motor neuron loss and muscle atrophy. The disease manifestations occur primarily in males, due to the dependence of the disease process on ligand binding of either testosterone or dihydrotestosterone (DHT) to the mutant AR (Chevalier-Larsen et al., 2004; Katsuno et al., 2002; Takeyama et al., 2002; Walcott and Merry, 2002). Several additional native functions of the androgen receptor are required for toxicity in model systems, including nuclear localization, an interdomain amino-/carboxyl (N-/C-) interaction, and AR acetylation (Montie et al., 2009; Montie et al., 2011; Nedelsky et al., 2010; Orr et al., 2010). A hallmark pathology of SBMA is the formation of nuclear inclusions – a feature shared with all 9 of the known neurodegenerative diseases caused by polyglutamine expansion, including Huntington’s disease (HD) (reviewed by (Orr and Zoghbi, 2007). This suggests that common features may exist between misfolded polyglutamine-expanded protein species and cellular toxicity.
We previously reported that motor neurons expressing polyglutamine-expanded AR die before detectable inclusion body formation (Heine et al., 2015), implicating a role for pre-inclusion polyglutamine-expanded AR aggregation species in toxicity. In Huntington’s disease, inclusion bodies are thought to promote survival through a process or processes that involve the sequestration of misfolded polyglutamine-expanded huntingtin (Arrasate et al., 2004; Miller et al., 2011) and the restoration of the ubiquitin-proteasome system (Mitra et al., 2009). To this end, the protein level, conformation, and stability of pre-inclusion huntingtin species are thought to predict toxicity (Miller et al., 2011; Mitra et al., 2009; Tsvetkov et al., 2014).
The mechanisms by which misfolded proteins cause toxicity remain unclear. Prevailing hypotheses include enhanced functions of the native protein (Duvick et al., 2010; Nedelsky et al., 2010), aberrant protein interactions (Ratovitski et al., 2012; Steffan et al., 2000), and perturbation of global protein folding homeostasis (Gidalevitz et al., 2006; Olzscha et al., 2011). While the role of particular subsets of aggregation species in these pathways is controversial (reviewed by (Wetzel, 2013)), research using conformation-specific antibodies supports the hypothesis that monomeric or low-order early aggregation species may promote cell death through a misfolding event that leads to a ‘toxic’ conformation (Miller et al., 2011; Nucifora et al., 2012). However, gaining an understanding of the biochemical nature of such species, as they are formed in the cellular environment, has been challenging. The ability to identify species that correlate with toxicity upon manipulation of critical post-translational modifications (reviewed by (Pennuto et al., 2009)), interdomain interactions (Orr et al., 2010), and protein-protein interactions (Lim et al., 2008), may lead to a new understanding of the nature of toxic protein aggregation species.
We recently used SDS-agarose gel electrophoresis (SDS-AGE) to demonstrate that soluble and insoluble polyglutamine-expanded AR species contain full-length androgen receptor and that the proteolytic event that produces an amino-terminal fragment occurs after the formation of nuclear inclusions (Heine et al., 2015). Here we expand our characterization of SDS-AGE-resolvable species to determine the nature of the polyglutamine-expanded AR as it forms aggregates within the context of the cellular milieu. Using a novel method to isolate soluble and insoluble protein species, we captured discrete populations of protein aggregates for further analysis. Our results identify previously uncharacterized AR aggregation species that correlate with toxicity in vitro, and which are also detected in vivo, in a transgenic mouse model of SBMA. Our results suggest a role for these structures in the disease process.
2. Results
2.1. Soluble, 3B5H10-reactive polyglutamine-expanded AR aggregates bind the toxicity-predicting antibody 3B5H10 and exist in low-density conformations
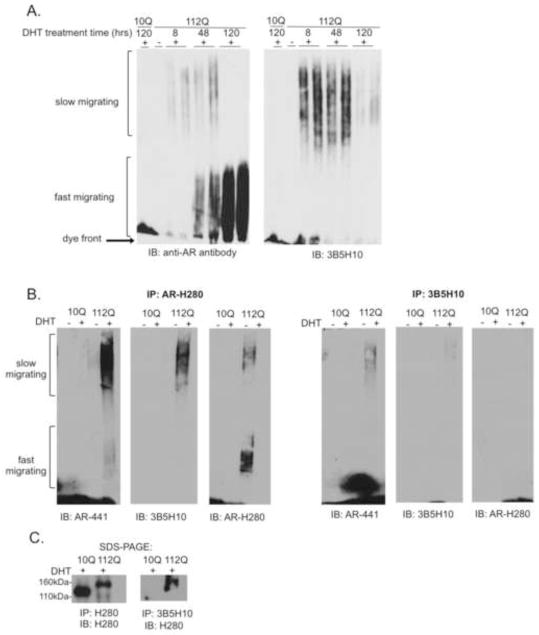
We previously demonstrated that SDS-agarose gel electrophoresis (SDS-AGE) is a sensitive and specific method for resolving both soluble and insoluble polyglutamine-expanded AR aggregation species (Heine et al., 2015). We found that soluble AR aggregation species contain full-length androgen receptor and bind the toxicity-predicting antibody 3B5H10. Although these species form early in the course of hormone-treatment, before the detection of cytological inclusions, they have a slower migration pattern by SDS-AGE when compared to insoluble AR aggregation species (Figure 1A). In contrast, the resolution of faster-migrating, 3B5H10-non-immunreactive species correlates with the detection of nuclear inclusions by fluorescence microscopy (Heine et al., 2015). This, together with their relative insolubility (Heine et al., 2015), suggests that fast-migrating species may be the biochemical correlate of, or at least associated with, cytological inclusions.
Figure 1. Soluble, 3B5H10-reactive polyglutamine-expanded AR aggregation species precede the formation of nuclear inclusions.
(A) DHT-treated PC12 cells expressing AR10Q or AR112Q were lysed in 2% SDS and resolved by SDS-agarose gel electrophoresis (SDS-AGE). Two distinct populations (indicated) of AR aggregation species were resolved in a DHT- and polyglutamine-length dependent manner. Slow-migrating species form early in the course of hormone-treatment and bind the toxicity-predicting antibody 3B5H10.
(B) PC12 cells expressing AR10Q or AR112Q were treated for 120hrs with DHT. Cells were lysed in RIPA and subject to immunoprecipitation with a conformation-specific antibody, 3B5H10, or a pan-AR antibody, AR(H280). Western analysis was performed with AR(441), which detects the full-length protein; the blots were then stripped and reprobed with 3B5H10, followed by AR(H280). To evaluate the efficiency of the IP and 10% of each immunoprecipitate was run on SDS-PAGE; levels of monomer were evaluated through western analysis with AR(H280).
Since we were able to detect soluble 3B5H10-reactive aggregation species that are resistant to lysis in 2% SDS, we wondered if the epitope on slow-migrating species that is recognized by 3B5H10 is also available under non-denaturing conditions. To test this, we lysed cells under mildly denaturing conditions and performed immunoprecipitation (IP) with 3B5H10. SDS-AGE analysis following IP with 3B5H10 revealed the recovery of slow-migrating species (Figure 1B). By comparison, both slow- and fast-migrating species were purified following IP with a non-conformational anti-AR antibody (Figure 1B). SDS-PAGE analysis confirmed that 3B5H10 binding is specific to the polyglutamine-expanded form of AR (Figure 1C). The reduced IP efficiency with 3B5H10, compared to that of the non-conformational anti-AR antibody, makes it difficult to conclude that 3B5H10 specifically binds slow-migrating species. Regardless, the IP results indicate that slow-migrating species are not merely an artifact of the denaturing lysis conditions. Collectively, these results suggest that, within cells, the polyglutamine-expanded AR adopts one or more soluble conformations that present a 3B5H10-binding epitope.
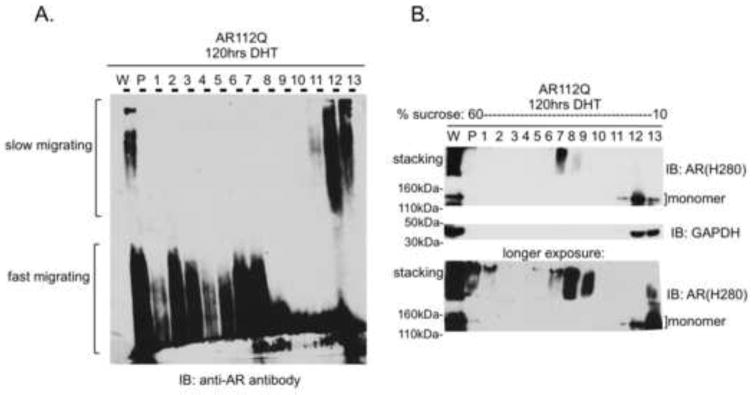
Because slow-migrating species form early in the course of hormone-treatment, before the formation of detectable nuclear inclusions, their delayed migration by SDS-AGE seemed counter-intuitive. We reasoned that slow-migrating species may represent the mutant AR in an extended conformation, while fast-migrating species may represent the mutant AR in a more compact conformation, such as that of a nuclear inclusion. The sedimentation of fast-, but not slow-, migrating AR species under conditions of ultracentrifugation (Heine et al., 2015) supports this interpretation. Moreover, this interpretation predicts that fast-migrating species are more dense than slow-migrating species. To test this idea, we used sucrose density gradient centrifugation to fractionate slow- and fast-migrating species according to their respective densities. We found that fast-migrating species fractionate in sucrose at higher densities than do slow-migrating species (Figure 2A). In addition, SDS-PAGE analysis of a sample of each density fraction revealed that the monomeric form of polyglutamine-expanded AR fractionates at the same densities as slow-migrating species (Figure 2B). Conversely, aggregation species that are retained in the stacking layer during SDS-PAGE were detected in the same higher density fractions as fast-migrating species.
Figure 2. Slow-migrating polyglutamine-expanded AR aggregates are less dense than fast-migrating polyglutamine-expanded AR aggregates.
(A) PC12 cells expressing AR10Q or AR112Q were treated with DHT and lysates were fractionated according to density on a 10–60% sucrose gradient. Slow-migrating species are retained in low-density fractions.
(B) Ten percent of each sucrose fraction was resolved by SDS-PAGE. AR112Q monomer is largely found in low-density fractions while high molecular weight species that are retained in the stacking layer of SDS-PAGE predominate in high-density fractions.
2.2. Slow-migrating AR aggregates represent novel biochemical entities
We had previously shown that AR species that are retained in the stacking layer of SDS-PAGE sediment by ultracentrifugation, suggesting that they are insoluble in solution (Heine et al., 2015). This insolubility suggests that the aggregation species that are retained in the stacking layer by SDS-PAGE are comparable to the fast-migrating species observed by SDS-AGE. Although the species that are retained in the stacking layer by SDS-PAGE have been interpreted as oligomers (Li et al., 2007), our results suggest that they may represent either higher-order AR aggregates or species that associate with and have similar biochemical characteristics as nuclear inclusions. To test this hypothesis, we next sought to evaluate the behavior of both slow- and fast-migrating species on SDS-PAGE.
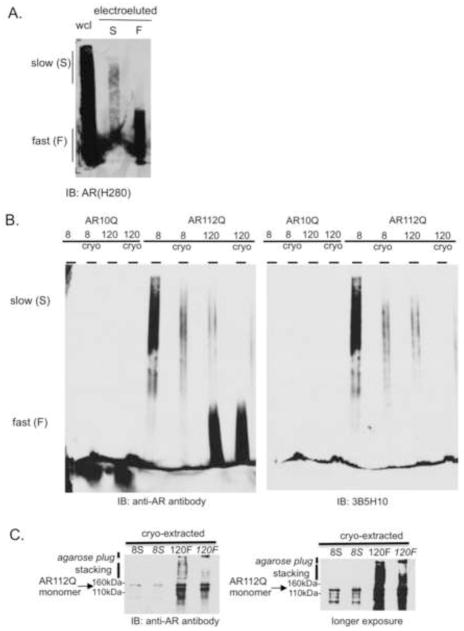
To determine if slow-migrating species enter or are retained in the stacking layer of SDS-PAGE, we developed a method to purify them from the agarose matrix in a manner that preserves their biochemical properties. The recovery of slow-migrating species by electro-elution into dialysis tubing was poor compared to the recovery of fast-migrating species (Figure 3A). We observed that slow-migrating species were retained in the agarose gel despite an additional 36 hours of electro-elution (data not shown), suggesting that they had become entrapped in the agarose matrix. To circumvent this problem, we adapted an established method, developed for the cryo-extraction of nucleic acids from agarose gels (Kurien and Scofield, 2002; Thuring et al., 1975), for the purification of protein aggregates. By snap-freezing sections of the agarose gel in liquid nitrogen, we were able to purify both slow- and fast-migrating species, respectively (Figure 3B). Importantly, this method preserved the intrinsic biochemical properties of each subset of species: cryo-extracted slow-migrating species retained their slow-migrating, 3B5H10-reactive characteristics while cryo-extracted fast-migrating species remained fast-migrating and 3B5H10-unreactive.
Figure 3. Slow-migrating polyglutamine-expanded AR species are not detected by SDS-PAGE.
(A) Poor recovery of slow-migrating species compared to fast-migrating species following electro-elution. (B) A cryo-extraction method to purify protein preserves the migration pattern of slow- and fast-migrating species. Briefly, either the top half of the gel (containing slow-migrating species) or the bottom half of the gel (containing fast-migrating species) was snap-frozen; the agarose was then pelleted and the aqueous protein extract was collected. Slow-migrating AR112Q species retain 3B5H10 immunoreactivity following cryo-extraction (right panel). cryo = cryo-extraction.
(C) Purified slow-migrating AR112Q species are not detected in the stacking layer by SDS-PAGE. Purified fast-migrating AR112Q species are detected in the stacking layer by SDS-PAGE. 8S = Slow-migrating AR112Q species purified from PC12 cell lysates treated for 8hrs with DHT. 120F = Fast-migrating AR112Q species purified from PC12 cell lysates treated for 120hrs with DHT. Italic font indicates samples run with the addition of a 0.1% SDS-1% agarose plug layered into the well of the SDS-polyacrylamide gel lane.
We next sought to determine how the purified slow- and fast-migrating species behaved in one of the most common assays for evaluating biochemical aggregation species: retention in the stacking layer of SDS-PAGE. We found that slow-migrating species migrated in identical fashion to monomeric AR by SDS-PAGE, while fast-migrating species were detected in both the stacking and resolving gels (Figure 3C). This finding suggests that slow-migrating species either consist of monomeric AR or can be dissociated under SDS-PAGE conditions to monomeric AR. To determine if any of the aggregation species were too large to enter the stacking layer of SDS-PAGE, we poured an agarose plug (0.1% SDS-1% agarose) into SDS-PAGE wells in an attempt to identify additional species within the larger pore size of the agarose gel matrix (Figure 3C). Although we were unable to detect slow-migrating species in the agarose plug, we cannot rule out the possibility that they exist in a quantity below the detection threshold. Despite this caveat, the identification of a novel slow-migrating SDS-AGE AR aggregation species that is only detectable as a monomer on SDS-PAGE suggests that this species has not been previously described.
2.3. Atomic force microscopy reveals that slow-migrating AR species are larger than fast-migrating species
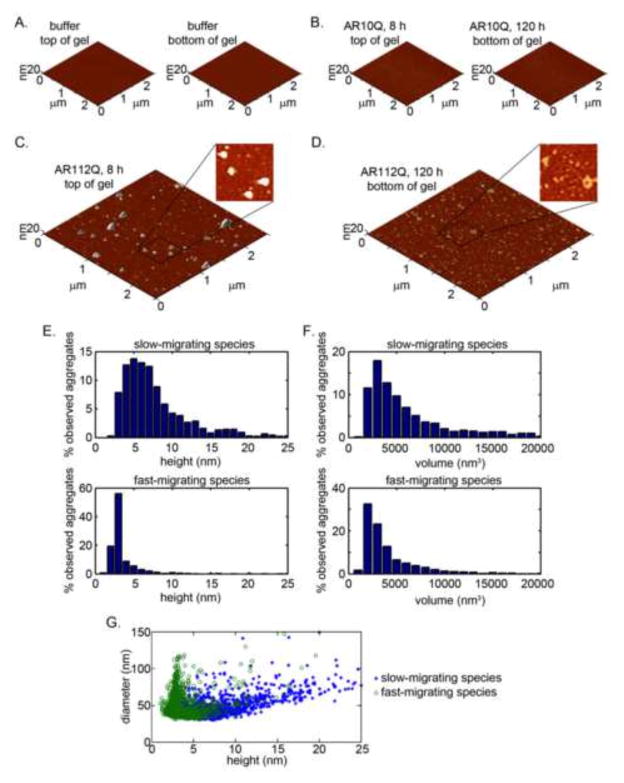
Two prior studies revealed distinct AR aggregation species by AFM (Jochum et al., 2012; Li et al., 2007). In one study (Jochum et al., 2012), polyglutamine-expanded AR formed oligomeric fibrils up to 300–600 nm in length and heights of 5–10 nm (20 nm at the termini); in this study, smaller, distinct AR aggregation species were observed even in the absence of hormone and an expanded polyglutamine tract. Another study (Li et al., 2007) observed polyglutamine-expanded AR within ordered oligomers with heights spanning 2–10 nM (Li et al., 2007). This height range is consistent with oligomers formed by polyglutamine-expanded huntingtin (Angeli et al., 2010; Legleiter et al., 2009; Legleiter et al., 2010; Nucifora et al., 2012). However, the Li et al. (Li et al., 2007) study was performed with AR aggregation species characterized by retention in the stacking layer of SDS-PAGE. We thus investigated how slow- and fast-migrating SDS-AGE species compare with the spectrum of structures previously described. To address this question, we carried out AFM on cryo-extracted samples from the top and bottom halves of SDS-AGE, which contain slow- or fast-migrating species, respectively. Very few particles were detected in the control cryo-extracted gel samples extracted after electrophoresis of either buffer alone or unexpanded AR10Q-containing protein lysates (Figure 4A–B). In contrast, we detected a large number of particles in the hormone-treated AR112Q samples (Figure 4C–D). Purified extracts of slow-migrating species contained large, amorphous aggregates with significant heterogeneity in height (Figure 4C, E). In comparison, purified extracts of fast-migrating species contained smaller aggregates that were more homogeneous in size (Figure 4D, F). The height spectrum of fast-migrating species ranged from 2–10 nM, with most particles less than 5 nM in height; this is consistent with previously described polyglutamine-expanded aggregates (Burke et al., 2013a; Legleiter et al., 2009; Legleiter et al., 2010; Miller et al., 2011). However, the range of heights detected in samples extracted from slow-migrating species extended to 25 nm, with ~20% of the observed particles exceeding 10 nM (Figure 4D). Although the samples purified from slow-migrating extracts demonstrated a correlation between height and diameter, we did not detect a similar relationship in samples purified from fast-migrating extracts, suggesting that these species represent discrete structures (Figure 4G).
Figure 4. Slow-migrating polyglutamine-expanded AR species form large, heterogeneous aggregates as detected by atomic force microscopy.
(A–B) Few particles are detected in AFM images of cryo-extracted samples from the buffer alone and from AR10Q cell lysates treated for 8hrs or 120hrs with DHT.
(C) AR112Q cells were lysed after 8hrs in DHT, at a time that precedes the formation of significant nuclear inclusions. Lysates were resolved by SDS-AGE and top of the gel, containing slow-migrating species, was subject to cryo-extraction and imaging by AFM. Large, amorphous particles were detected on the mica.
(D) AR112Q cells were lysed after 120hrs in DHT, when considerable nuclear inclusions have formed. Lysates were resolved by SDS-AGE and the bottom of the gel, containing fast-migrating species, was subject to cryo-extraction and imaging by AFM. The detected particles were smaller and amorphous.
(E–F) Quantification of numbers of particles per unit area for slow- and fast- migrating species.
(G) Correlation analysis between slow- and fast-migrating species demonstrates distinct and non-overlapping populations.
2.4. Slow-migrating AR species correlate with toxicity in vitro and in vivo
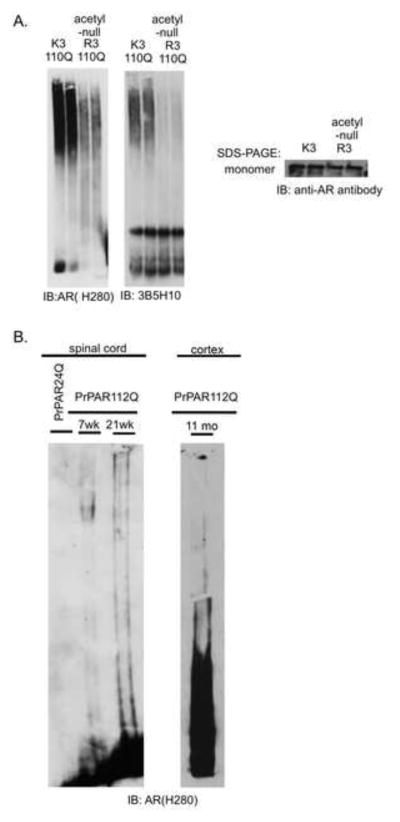
The reactivity of slow-migrating species with the toxicity-predicting antibody 3B5H10 suggested that they may correlate with toxicity. If 3B5H10-reactive slow-migrating polyglutamine-expanded AR species correlate with toxicity, we hypothesized that we would see a reduction in slow-migrating species in cell lines expressing protective mutations in an otherwise toxic polyglutamine-expanded AR. We previously showed that acetylation of the AR is necessary for toxicity and aggregation (Montie et al., 2011); therefore, we evaluated the presence of 3B5H10-immunoreactive slow-migrating species in cells expressing a mutation (lysine to arginine) in the polyglutamine-expanded AR that renders it incapable of becoming acetylated. Our results reveal that cells expressing the protective acetyl-null mutation in the polyglutamine-expanded AR form fewer slow-migrating, 3B5H10-reactive species (Figure 5A), consistent with the hypothesis that slow-migrating species are correlated with cellular toxicity.
Figure 5. Slow-migrating species correlate with toxicity in vitro and in vivo.
(A) PC12 cell lines expressing AR with protective mutations form fewer slow-migrating, AR- and 3B5H10-reactive species. K3-110Q refers to polyglutamine-expanded AR (110Q) with intact lysines at residues 630, 632 and 633. R3-110Q refers to polyglutamine-expanded AR (110Q) with mutations of lysines 630, 632 and 633 to the acetylation-null amino acid arginine (Montie et al., 2011).
(B) Spinal cord nuclear extracts (left) from male PrPAR112Q transgenic mice display slow- and fast-migrating aggregation species. Slow-migrating species are detected as early as 7 weeks, at an age that precedes the formation of large nuclear inclusions. Cortical nuclear extracts (right) reveal both slow- and fast-migrating species at 11 months of age. Spinal cord samples represent concentrated nuclear extracts from 2 mg of protein extract; cortical sample represents unconcentrated nuclear extract from 500 μg of protein extract.
We had previously shown that both slow- and fast-migrating SDS aggregation species are detected in vivo (Heine et al., 2015), and that slow-migrating species appear as early as 7.5 weeks of age in transgenic male SBMA mice, after the rise of androgen levels and just prior to the onset of motor symptoms. In contrast, fast-migrating species, while detectable at 7.5 weeks, accumulated later in the course of disease, at 21 wks of age (Figure 5B), and 11 months (Heine et al., 2015) when significant nuclear inclusions are present. These aggregation species are also seen in the cortex of transgenic mice (Figure 5B); continuing studies will evaluate the biochemical similarities and differences between aggregation species observed in distinct brain regions. Collectively, these data support the idea that slow-migrating species appear early in the disease course and correlate with toxicity, both in vitro and in vivo.
3. Discussion
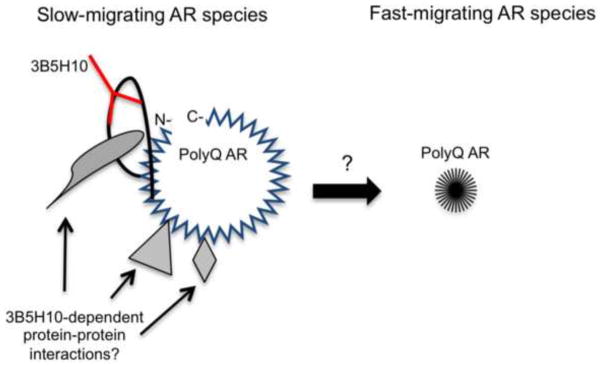
The studies described here reveal the identification what appears to be a previously uncharacterized polyglutamine-expanded AR aggregation species that is strongly correlated with toxicity. Moreover, we developed a novel protein extraction method to demonstrate that soluble, 3B5H10-reactive aggregation species exist in low-density conformations but are larger by atomic force microscopy, suggesting that they may be less compact than insoluble aggregates (Figure 6). We found that cell lines expressing an acetylation-deficient polyglutamine-expanded AR, previously shown to protect against hormone-dependent toxicity (Montie et al., 2011), contain reduced levels of the 3B5H10-reactive slow-migrating AR aggregation species. The correlation with toxicity, combined with evidence that these species exist in vivo, suggests that they may have relevance to the disease process. One caveat to these conclusions is that the cell models used here express mutant AR with a polyglutamine tract that is longer than that observed in SBMA patients. However, our preliminary studies of iPS cells derived from SBMA patients (iPS cells described in (Grunseich et al., 2014)) reveal similar fast- and slow-migrating species (data not shown). In ongoing studies, we will further characterize these species in iPS cells and other models with shorter repeat lengths.
Figure 6. Schematic of proposed aggregation pathway.
Polyglutamine-expanded AR entities that misfold early in the course of disease are distinguished by 3B5H10-immunoreactivity, lower densities, and larger sizes. Later-stage, insoluble aggregates display higher densities and smaller sizes, and are unable to bind 3B5H10. Whether slow-migrating polyglutamine-expanded AR species become fast-migrating polyglutamine-expanded AR species is under investigation.
Previous studies of polyglutamine-expanded AR aggregates have identified species with heights ranging from 2–10 nm (Jochum et al., 2012; Li et al., 2007). One study (Li et al., 2007) interpreted this height range to be consistent with multiple amino-terminal fragments of the polyglutamine-expanded AR; this conclusion was based, in part, on the assumption that protein density is consistent between aggregated forms. While this calculation is a conventional method for estimating the number of particles in an individual aggregate, our data suggest that this may not be an accurate assessment for aggregates formed by the polyglutamine-expanded AR. Moreover, the slow-migrating, low-density AR aggregation species evaluated here consist of full-length, rather than proteolyzed fragments of, AR (Heine et al., 2015). Whether the heterogeneity in densities of polyglutamine protein aggregation species is applicable to other polyglutamine-expanded diseases is further challenged by recent evidence that aggregates formed by polyglutamine-expanded atrophin-1 also display heterogeneous densities (Hinz et al., 2012). Finally, although the analyses of SDS-AGE-resolvable polyglutamine-expanded huntingtin species relied on molecular weight estimates to predict aggregate size (Legleiter et al., 2010; Miller et al., 2011), our data suggest that conformation and density are critical parameters in determining aggregate size.
Our results raise several questions with regard to the uniqueness of the protein species described here. Numerous groups have used SDS-AGE to resolve polyglutamine-expanded aggregation species (Legleiter et al., 2009; Legleiter et al., 2010; Miller et al., 2011; Nucifora et al., 2012; Sontag et al., 2012; Weiss et al., 2008), yet the presence of distinctly migrating species has not been previously reported. One possible explanation for this difference is that slow-migrating species are a unique feature of the polyglutamine-expanded AR. It is unlikely that this is due to a difference in the size of the AR protein. Data from cells expressing huntingtin with a range of polyglutamine expansion tracts demonstrate that longer polyglutamine tracts, and thus a larger protein size, seems to accelerate the migration of aggregation species by SDS-AGE (Legleiter et al., 2010). The faster migration observed with longer polyglutamine tracts is consistent with our hypothesis that more compact conformations may result in faster migration. The novel observation of slow-migrating AR species may reflect intrinsic features of specific AR functional domains. It may be that slow-migrating species have lipophilic properties, resulting from the presence of lipophilic hormone in the ligand-binding pocket, or from interactions with lipid membranes, as has been shown with other polyglutamine-expanded peptides (Burke et al., 2013b; Chaibva et al., 2014). Alternatively, the low density of slow-migrating species might arise from aberrant conformation of AR structural domains.
The transient nature of slow-migrating species (Fig 1A) suggests that this is a short-lived aggregation species that forms early and decreases in abundance with increasing hormone-treatment time. The analysis of early hormone-treatment times using this sensitive biochemical method may have facilitated the identification of these species. Whether these species are produced chronically during disease progression is under investigation.
A major unanswered question is whether slow-migrating species become fast-migrating species or whether they represent off-pathway intermediates. Studies of polyglutamine-expanded huntingtin showed that 3B5H10-reactive mutant huntingtin conformers are sequestered into inclusion bodies (Miller et al., 2011), thus protecting neurons from ‘toxic’ conformations of polyglutamine-expanded huntingtin that may participate in aberrant protein interactions or cause protein folding stress. Moreover, in mutant AR-expressing cells, diffuse nuclear 3B5H10 immunofluorescence was reduced to background levels upon the first appearance of nuclear AR puncta, the precursors to mature nuclear inclusions (Heine et al., 2015), consistent with the absence of 3B5H10 immunoreactivity in fast-migrating SDS-AGE species. Experiments to determine if 3B5H10-reactive slow-migrating species become sequestered by the formation of fast-migrating insoluble species may shed insight on the role of aggregation species in SBMA. Determining whether and how aberrant protein interactions form with 3B5H10-reactive AR species is likely to also shed valuable mechanistic insight. Further investigations are necessary to understand the fate of slow-migrating species and determine their role in toxicity.
4. Experimental Procedures
4.1. Immunoprecipitation
AR expression was induced in PC12 cells (AR112Q- and AR10Q-containing) and cells were treated with DHT (10 nM) for 120 hrs. Cells were lysed with RIPA lysis buffer (50 mM Tris-HCl, pH 8.0, 0.15 M NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, and protease inhibitors) and 500 μg of protein from each sample was incubated overnight at 4°C with or without 1.5 μg of antibody (AR(H280) or 3B5H10. Capture of the immunoprecipitated AR was carried out using Protein G Dynabeads (Invitrogen) for 30 minutes at room temperature, followed by separation from the unbound sample using a Dynal magnet. Samples were eluted from Dynabeads with 2x SDS-AGE Laemmli buffer and boiled for 5 minutes. Samples were evaluated by SDS-AGE and western analysis was performed using AR(H280) (1:1000), AR(441) (1:100), or 3B5H10 (1:10,000). Following AR(441) detection, membranes were stripped and probed for 3B5H10 and then probed for AR(H280).
4.2. SDS-agarose gel electrophoresis (SDS-AGE)
PC12 cells were lysed in 2% SDS (2% SDS, 10 mM Tris pH 8.0, 150 nM NaCl) and diluted 1:1 in non-reducing Laemmli sample buffer. Samples were boiled for 5 minutes and electrophoresed through a 0.1% SDS-1% agarose matrix as previously described.22 PC12 cell lysates (100 ug) were electrophoresed for 10–11 hrs at 4°C on a 14 cm-long x 8 cm-thick 1% agarose gel containing 0.1% SDS in 375 mM Tris-HCL, pH 8.8. Lysates were then transferred to PVDF membrane at 200 mA for 1.5 hrs using a semi-dry transfer apparatus (Owl HEP Semidry Electroblotting Systems, Thermoscientific). A 4 kg weight was placed on top of the transfer apparatus to accommodate thinning of the agarose gel during transfer. Western analysis was performed using AR(H280) (1:1000) or 3B5H10 (1:10,000).
4.3. Solubility Assay
PC12 cells were induced to express equivalent amounts of AR10Q or AR112Q and treated with 10nM DHT for 120hrs. Cells were lysed in 2% SDS (see above) and centrifuged at 314,000 x g for 30 min at 15°C using a TLA100.1 Beckman rotor. The supernatant was collected and the pellet was washed twice in 2% SDS. The supernatant, containing soluble protein, and the pellet, containing insoluble protein, were analyzed by SDS-AGE and SDS-PAGE.
4.4. Density fractionation
Cell lysates from AR112Q-expressing PC12 cells treated for 120 hrs in DHT were fractionated on a 10–60% w/v sucrose gradient (dissolved in 375 mM Tris-HCL containing 0.1% SDS). Briefly, 1.5 mg of protein was centrifuged at 35,000 rpm for 16 hrs at 15°C using a Sorval WX rotor. Fractions of equal volume were collected and concentrated using 30,000 kDa Amicon Centricon tubes (Millipore) and electrophoresed by SDS-AGE. To determine at which sucrose density AR monomers fractionate, 10% of each fraction was electrophoresed by SDS-PAGE.
4.5. Electro-elution
Lysates (200 μg) from PC12 cells induced to express AR112Q and treated for 120 hrs with DHT were electrophoresed by SDS-AGE for 10–11 hrs as described above. Vertical lanes (~1 cm wide) were cut from the agarose gel; the gel segments were cut in half 6 cm from the top of the well, with the top half containing slow-migrating species and the bottom-half containing fast-migrating species (as confirmed by parallel western analyses). The gel segments were then placed in dialysis tubing (Spectra/Por 1, MWCO 6000–8000) containing SDS-AGE running buffer. The dialysis tubes containing the agarose gel segments were then submerged in an electrophoresis chamber in a direction perpendicular to their original orientation and subjected to an electrical field of 100–120V for 12–36 hrs at 4°C. The buffer within the dialysis tubing, containing the electro-eluted protein species, was collected (~1mL total) and concentrated using 30,000 kDa Amicon Centricon filters (Millipore). The concentrated samples were then electrophoresed by SDS-AGE to evaluate recovery. To determine if protein species were left in the agarose gel, the gel segments from the dialysis tube were transferred using a semi-dry apparatus (described above) and subjected to western analysis using an anti-AR antibody (AR(H280)).
4.6. Cryo-extraction
PC12 cells were induced to express AR112Q and treated with DHT for 8 hrs or 120 hrs. To purify slow-migrating species, cells treated for 8 hrs were lysed in 2% SDS and 30–50 mg of protein was resolved by SDS-AGE. The gel was cut at a distance 6 cm from the well and the top-half (containing slow-migrating species) was placed into 6 pre-gauged 15 ml polystyrene conical tubes and snap-frozen in liquid nitrogen. To purify fast-migrating species, cells treated with DHT for 120 hrs were lysed in 2% SDS (2% SDS, 10 mM Tris pH 8.0, 150 nM NaCl) and 15–20 mg of protein was resolved by SDS-AGE. The gel was cut at a distance 7 cm from the well and the bottom-half was snap-frozen in liquid nitrogen. The 15 ml conical tubes containing segments of frozen agarose were then placed in 50 mL conical tubes as a receptacle for the aqueous extract. The agarose was then pelleted by centrifugation at 3,000 x g for 20 min at 4°C. The aqueous extract, containing the protein species, was collected and concentrated using 30,000 kDa Amicon Centricon filtration devices (Milllipore). Samples were divided into thirds and subjected to an additional snap-freeze to control for sample processing for atomic force microscopy. Every sample that was analyzed by AFM was also evaluated by SDS-AGE to ensure the nature of slow-migrating 3B5H10-reactive species and fast-migrating 3B5H10-unreactive species
4.7. Atomic force microscopy
Cryo-extracted samples were deposited on freshly cleaved mica and left on the substrate for 1 min. The mica substrate was then washed with 200 μL of ultrapure water and dried with a gentle stream of nitrogen. These samples were imaged via ex situ AFM using a Nanoscope V MultiMode scanning probe microscope (Veeco, Santa Barbara, CA) equipped with a closed-loop vertical engage J-scanner. Images were taken with diving board-shaped oxide-sharpened silicon cantilever with a nominal spring constant of 40 N/m and resonance frequency of 300 kHz. AFM images were analyzed with Matlab equipped with the image processing toolbox (Mathworks, Natick, MA) as described (Legleiter et al., 2010). For this analysis, particles were defined as features taller than 0.7 nM that contained at least 8 pixels. Volumes presented represent raw experimental values and contain some error associated with the size and shape of the imaging tip; this error is relatively uniform across all images since they were collected with the same tip.
4.8. Mouse nuclear extracts
Spinal cords were dissected from male transgenic mice expressing human AR with either 24 CAGs or 112 CAGs (Chevalier-Larsen et al., 2004) at 7 weeks or 21 weeks of age. Nuclear extractions were performed through homogenization of freshly dissected tissue in a hypotonic lysis buffer (10 mM HEPES, pH 7.9 at 4°C, 1.5 mM MgCl2, 10 mM KCL, 0.5 mM DTT). The spinal cord lysates were centrifuged at 3,000 x g for 10 min at 4°C. The pellets, containing the nuclear extract, were then resuspended in 2% SDS lysis buffer (2% SDS, 10 mM Tris pH 8.0, 150 nM NaCl). 2 mg of spinal cord lysates was concentrated and resolved by SDS-AGE. Western analysis was performed using AR(H280) (1:1000).
Highlights.
B5H10-reactive polyglutamine-expanded AR species are slow-migrating and less dense than non-3B5H10-reactive species.
Slow-migrating AR species have heterogeneous sizes by AFM.
Slow-migrating AR species correlate with toxicity in vitro.
Acknowledgments
Funding was provided by grants to DEM from the NIH (R01 NS032214 and R01 NS076919). TRB was supported in part by a Dubbs Scholar Fellowship Award from Thomas Jefferson University. We are grateful to Dr. Steve Finkbeiner for providing antibody 3B5H10 and to the members of the Merry laboratory for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angeli S, Shao J, Diamond MI. F-actin binding regions on the androgen receptor and huntingtin increase aggregation and alter aggregate characteristics. PLoS One. 2010;5:e9053. doi: 10.1371/journal.pone.0009053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Burke KA, Hensal KM, Umbaugh CS, Chaibva M, Legleiter J. Huntingtin disrupts lipid bilayers in a polyQ-length dependent manner. Biochim Biophys Acta. 2013a;1828:1953–1961. doi: 10.1016/j.bbamem.2013.04.025. [DOI] [PubMed] [Google Scholar]
- Burke KA, Kauffman KJ, Umbaugh CS, Frey SL, Legleiter J. The interaction of polygltuamine peptides with lipid membranes is regulated by flanking sequences associate with huntingtin. J Biol Chem. 2013b;287:2068–2078. doi: 10.1074/jbc.M112.446237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaibva M, Burke KA, Legleiter J. Curvature enhances binding and aggregation of huntingtin at lipid membranes. Biochemistry. 2014;53:2355–2365. doi: 10.1021/bi401619q. [DOI] [PubMed] [Google Scholar]
- Chevalier-Larsen ES, O’Brien CJ, Wang H, Jenkins SC, Holder L, Lieberman AP, Merry DE. Castration restores function and neurofilament alterations of aged symptomatic males in a transgenic mouse model of spinal and bulbar muscular atrophy. J Neurosci. 2004;24:4778–4786. doi: 10.1523/JNEUROSCI.0808-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick L, Barnes J, Ebner B, Agrawal S, Andresen M, Lim J, Giesler GJ, Zoghbi HY, Orr HT. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67:929–35. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T, Ben-Zvi A, Ho KH, Brignull HR, Morimoto RI. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;5766:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Grunseich C, Zukosky K, Kats IR, Ghosh L, Harmison GG, Bott LC, Rinaldi C, Chen KL, Chen G, Boehm M, Fischbeck KH. Stem cell-derived motor neurons from spinal and bulbar muscular atrophy patients. Neurobiol Dis. 2014;70:12–20. doi: 10.1016/j.nbd.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine EM, Berger TR, Pluciennik A, Orr CR, Zboray L, Merry DE. Proteasome-mediated proteolysis of the polyglutamine-expanded androgen receptor is a late event in spinal and bulbar muscular atrophy (SBMA) pathogenesis. J Biol Chem. 2015;290:12572–84. doi: 10.1074/jbc.M114.617894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz J, Lenhnhardt L, Zakrzewski S, Zhang G, Ignatova Z. Polyglutamine expansion alters the dynamics and molecular architecture of aggregates in dentatorubropallidoluysian atrophy. J Biol Chem. 2012;287:14993–15005. doi: 10.1074/jbc.M111.318915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum T, Ritz ME, Schuster C, Funderburk SF, Jehle K, Schmitz K, Brinkmann F, Hirtz M, Moss D, Cato ACB. Toxic and non-toxic aggregates from the SBMA and normal forms of androgen receptor have distinct oligomeric structures. Biochim Biophys Acta. 2012;1822:1070–1078. doi: 10.1016/j.bbadis.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, Sobue G. Testosterone reduction prevents phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Neuron. 2002;35:843–54. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- Kurien BT, Scofield RH. Extraction of nucleic acid fragments from gels. Anal Biochem. 2002;302:1–9. doi: 10.1006/abio.2001.5526. [DOI] [PubMed] [Google Scholar]
- Legleiter J, Lotz GP, Miller J, Ko J, Ng C, Williams GL, Finkbeiner S, Patterson PH, Muchowski PJ. Monoclonal antibodies recognize distinct conformational epitopes formed by polyglutamine in a mutant huntingtin fragment. J Biol Chem. 2009;284:21647–21658. doi: 10.1074/jbc.M109.016923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legleiter J, Mitchell E, Lotz GP, Sapp E, Ng C, DiFiglia M, Thompson LM, Muchowski PJ. Mutant huntintin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J Biol Chem. 2010;285:14777–14790. doi: 10.1074/jbc.M109.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chevalier-Larsen ES, Merry DE, Diamond MI. Soluble androgen receptor oligomers underlie pathology in a mouse model of SBMA. J Biol Chem. 2007;5:3157–3164. doi: 10.1074/jbc.M609972200. [DOI] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;7188:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, Arrasate M, Brooks E, Libeu CP, Legleiter J, Hatters D, Curtis J, Cheung K, Krishnan P, Mitra S, Widjaja K, Shaby BA, Lotz GP, Newhouse Y, Mitchell EJ, Osmand A, Gray M, Thlasiramin V, Saudou F, Segal M, Yang XW, Masliah E, Thompson LM, Muchowski PJ, Weisgraber KH, Finkbeiner S. Identifying polyglutamine protein species in situ that best predict neurodegeneration. Nat Chem Biol. 2011;7:925–934. doi: 10.1038/nchembio.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Tsvetkov AS, Finkbeiner S. Single neuron ubiquitin-proteasome dynamics accompanying inclusion body formation in Huntington Disease. J Biol Chem. 2009;284:4398–4403. doi: 10.1074/jbc.M806269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montie HL, Cho MS, Holder L, Liu Y, Tsvetkov AS, Finkbeiner S, Merry DE. Cytoplasmic retention of polyglutamine-expanded androgen receptor ameliorates disease via autophagy in a mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2009;18:1937–50. doi: 10.1093/hmg/ddp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montie HL, Pestell RG, Merry DE. SIRT1 modulates aggregation and toxicity through deacetylation of the androgen receptor in cell models of SBMA. J Neurosci. 2011;31:17425–17436. doi: 10.1523/JNEUROSCI.3958-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP. Native functions of the androgen receptor are essential to pathogenesis in a Drosophila model of spinal and bulbar muscular atrophy. Neuron. 2010;67:936–952. doi: 10.1016/j.neuron.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora LG, Burke KA, Feng X, Arbez N, Zhu SE, Miller J, Yang G, Ratovitski T, Delannoy M, Muchowski PJ, Finkbeiner S, Legleiter J, Ross CA, Poirier MA. Identification of novel potentially toxic oligomers formed in vitro from mammalian-derived expanded huntingtin exon-1 protein. J Biol Chem. 2012;287:16017–16028. doi: 10.1074/jbc.M111.252577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144:67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- Orr CR, Montie HL, Liu Y, Bolzoni E, Jenkins SC, Wilson EM, Joseph JD, McDonnell DP, Merry DE. An interdomain interaction of the androgen receptor is required for its aggregation and toxicity in spinal and bulbar muscular atrophy. J Biol Chem. 2010;285:35567–35577. doi: 10.1074/jbc.M110.146845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pennuto M, Palazzolo I, Poletti A. Post-translational modifications of expanded polyglutamine proteins: impact on neurotoxicity. Hum Mol Genet. 2009;18:R40–47. doi: 10.1093/hmg/ddn412. [DOI] [PubMed] [Google Scholar]
- Ratovitski T, Chighladze E, Arbez N, Boronina T, Herbrich S, Cole RN, Ross CA. Huntingtin protein interactions altered by polyglutamine expansion as determined by quantitative proteomic analysis. Cell Cycle. 2012;11:2006–2021. doi: 10.4161/cc.20423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag EM, Lotz GP, Yang G, Sontag CJ, Cummings BJ, Glabe C, Muchowski PJ, Thompson LM. Detection of mutant Huntingtin aggregation conformers and modulation of SDS-soluble fibrillar oligomers by small molecules. J Huntingtons Dis. 2012;1:127–140. doi: 10.3233/JHD-2012-129004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, Wanker E, Bates G, Housman D, Thompson L. The huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci (USA) 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeyama K, Ito S, Yamamoto A, Tanimoto H, Furutani T, Kanuka H, Miura M, Tabata T, Kato S. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35:855–64. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Thuring RW, Sanders JP, Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975;66:213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Tsvetkov AS, Barmada S, Ando DM, Shaby BA, Finkbeiner S. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2014;9:586–592. doi: 10.1038/nchembio.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcott JL, Merry DE. Ligand promotes intranuclear inclusions in a novel cell model of spinal and bulbar muscular atrophy. J Biol Chem. 2002;277:50855–9. doi: 10.1074/jbc.M209466200. [DOI] [PubMed] [Google Scholar]
- Weiss A, Klein C, Woodman B, Sathasivam K, Bibel M, Regulier E, Bates GP, Paganetti P. Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington’s disease. J Neurochem. 2008;104:846–848. doi: 10.1111/j.1471-4159.2007.05032.x. [DOI] [PubMed] [Google Scholar]
- Wetzel R. Physical chemistry of polyglutamine: intriguing tales of a monotonous sequence. J Mol Biol. 2013;421:466–490. doi: 10.1016/j.jmb.2012.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]