Abstract
Despite improvements in survival rates for children with cancer since the 1960s, progress for many pediatric malignancies has slowed over the past 2 decades. With the recent advances in our understanding of the genomic landscape of pediatric cancer, there is now enthusiasm for individualized cancer therapy based on genomic profiling of patients’ tumors. However, several obstacles to effective personalized cancer therapy remain. For example, relatively little data from prospective clinical trials demonstrate the selective efficacy of molecular-targeted therapeutics based on somatic mutations in the patient's tumor. In this commentary, we discuss recent advances in preclinical testing for pediatric cancer and provide recommendations for providing scientific justification and translational relevance for novel therapeutic combinations for childhood cancer. Establishing rigorous criteria for defining and validating druggable mutations will be essential for the success of ongoing and future clinical genomic trials for pediatric malignancies.
Keywords: preclinical models, pharmacokinetics, pharmacodynamics, pediatric solid tumors
Introduction
The overall survival rate of children with cancer has improved significantly over the past 50 years (1). One factor that has contributed to this success is the formation of multicenter cooperative groups. The number of pediatric patients with cancer is small, and cooperative groups provide a mechanism to coordinate clinical trial development, accrue patients, and evaluate the outcome of those trials much more quickly than would be possible at individual centers. Indeed, most children with cancer are enrolled on clinical trials, which is not the case for adults with cancer (2).
Despite these coordinated efforts, the overall survival rate for many pediatric cancers has plateaued during the past 20 years. In particular, the survival rate of children with recurrent or metastatic cancer has remained below 30% (1). This plateau comes at a time when pediatric oncologists have access to a growing number of new and diverse oncology drugs. In addition to advances in drug development, next-generation sequencing studies over the previous 5 years (3, 4) have led to the development of CLIA-certified laboratories that can provide the complete genomic profile of each patient's tumor. Thus, for the first time, pediatric oncologists can review the genomic landscape of a patient's tumor and then select appropriate molecular-targeted therapeutics to incorporate into that patient's treatment regimen. To that end, several institutions have launched clinical genomic initiatives for pediatric patients with cancer, and a national effort called “pediatric MATCH” is being developed (http://www.cancer.gov/researchandfunding/areas/clinical-trials/match#pediatric). Despite optimism about the potential benefits of individualized therapies, major barriers to bringing such therapies into the clinic remain (5, 6).
One major barrier to the successful implementation of individualized therapy is the lack of appropriate preclinical models for many pediatric cancers. In addition, even in cases where preclinical models do exist, it is not always clear how best to use those models to inform clinical decision-making. A key challenge for the pediatric research community is to develop models that closely recapitulate the genetic heterogeneity of the corresponding human cancer. In this commentary, we will focus on the use of preclinical models in basic and translational research to evaluate new therapies for children with cancer. We will also discuss the concept of druggable cancer mutations and suggest guidelines for establishing scientific justification and translational relevance of molecular-targeted therapeutics in preclinical studies.
Scientific Justification of Druggable Mutations Using Preclinical Models
Most of the clinical treatment protocols developed in the 1970s–1990s were empirically derived through trial and error because of our limited knowledge of the molecular underpinning and the complexity of the cancers they were designed to treat. As our knowledge of specific molecular targets has increased, it has become evident that targeted therapies are needed to improve the survival and decrease the toxicity of treatment for pediatric patients with cancer. An expansive body of literature demonstrates that targeted therapeutics are effective only if the pathways they target are essential in the tumor. For example, the PDGFRA antagonist crenolanib may be justified in treating pediatric patients with glioma whose tumors have PDGFRA amplifications, but it would not be warranted in children with other types of brain tumors that do not harbor that genetic lesion (NCT01229644 and NCT01393912 at clinicaltrials.gov). Also, it is now clear that intra- and inter-patient tumor heterogeneity can further confound efforts to incorporate molecular-targeted therapeutics into clinical trials (4, 7, 8).
Well-characterized animal models can be used to advance our understanding of the basic biology of diverse tumor types. In addition, they can serve as a platform for discovering tumor vulnerabilities and testing hypotheses about the genetic factors that contribute to cancer initiation and/or progression. We would also argue that preclinical models are an essential tool for providing scientific justification and translational relevance for new therapies and therapeutic combinations in pediatric cancer. In this commentary, scientific justification for a particular therapeutic approach is achieved by demonstrating that the pathway being altered by the therapy is essential for tumor survival and/or growth. Translational relevance of a therapeutic approach is achieved by demonstrating the same effect on the tumor in vivo using clinically relevant treatment regimens. It is possible to have scientifically justified therapies that lack translational relevance, and the converse is also true. Each preclinical model has strengths and weaknesses, and the challenge is selecting the appropriate model for the specific goals of the study in order to provide both scientific justification and translational relevance before moving into clinical trials.
Below, we discuss 3 of the most commonly used preclinical models of pediatric cancer: zebrafish models, genetically engineered mouse models (GEMMs), and patient-derived xenograft (PDX) models. These animal models have been used to advance basic cancer research, to identify cancer subtype–specific vulnerabilities, and to demonstrate complex genetic interactions in vivo. Although all 3 models are used in each area of research, we will highlight their unique strengths for particular applications in multidisciplinary oncology research programs.
Using Zebrafish in Basic Cancer Research
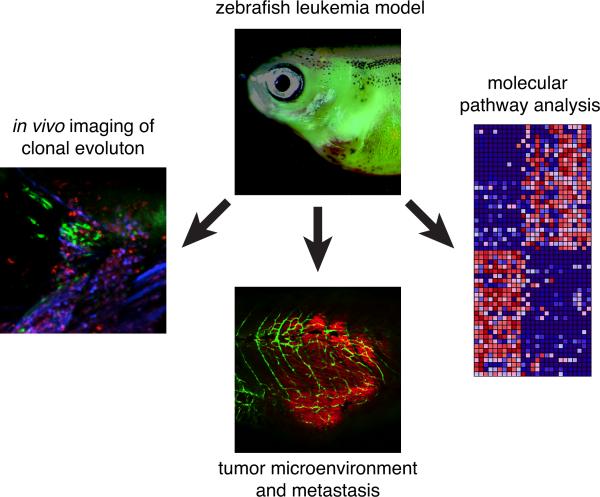
Zebrafish models are widely used in pediatric cancer research because they are genetically tractable, optically clear (i.e., they permit direct visualization of cancer processes in fluorescent transgenic fish), have a short generation time, and are more cost-effective than mice. Cancer subtype–specific mutations can be introduced into particular cell lineages at different developmental stages to model cancer initiation. The earliest examples are transgenic zebrafish models of T-cell acute lymphoblastic leukemia (T-ALL), in which MYC or NOTCH elicited transformation (9, 10). These models are histopathologically and molecularly similar to human T-ALL (11). Transgenesis has also been used to create TEL-AML–induced B cell acute lymphoblastic leukemia (12), RAS-driven embryonal rhabdomyosarcoma (13), and oncogenic ALK-mutant neuroblastoma (14). Another major advantage of zebrafish relates to visualizing cellular mechanisms of cancer progression, including neovascularization, tumor heterogeneity, clonal evolution, and invasion (Fig. 1) (15). Zebrafish are also amenable to large-scale transplantation approaches designed to observe the functional effects of clonal evolution on drug resistance in vivo (16).
Figure 1. Zebrafish in oncology research.
Diagram highlighting some of the advantages of zebrafish for oncology research including live-cell imaging of clonal heterogeneity and clonal evolution, studies on tumor cell interactions with the microenvironment, and pathway analysis for testing molecular-targeted therapeutics.
Zebrafish have been used in drug screens to identify the molecular pathways that drive cancer growth (17) and to identify new drugs that curb tumor growth in vivo (18-20). Despite the utility of these approaches, a major hurdle has been moving these findings into mammalian models because most zebrafish laboratories do not use murine cancer models (GEMMs or PDX models).
As pediatric oncologists advance novel molecular-targeted therapeutic combinations in the context of clinical genomics, it is important to consider tumor heterogeneity and clonal evolution as a result of treatment. Rare subclones within solid tumors at diagnosis can escape treatment, seed a recurrent lesion, and then continue to evolve and escape treatment during subsequent therapeutic interventions (Fig. 2) (8). Currently, there are no models of these complex processes in vivo. A combined approach that uses live imaging and genomic profiling of zebrafish could be one route to elucidating some of the underlying mechanisms of cancer recurrence in children.
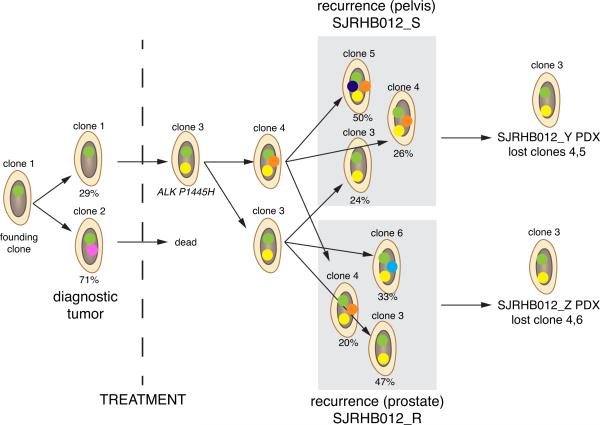
Figure 2. Clonal selection in PDX models.
The clonal evolution of a rhabdomyosarcoma is represented. Each colored dot represents a cluster of SNVs that was used to track the clonal evolution of the tumor. At diagnosis, the tumor had 2 major clones: 1of those clones (clone 2) was eliminated with treatment. Clone 3 then continued to evolve and seeded 2 different recurrent sites in the patient (prostate and pelvis). Orthotopic PDX tumors were derived from those sites, and the minor clones were lost; clone 3 remained the dominant clone in the PDX models.
Despite the advantages of zebrafish for imaging cancer processes and genetically screening chemical compounds, these models have limitations and challenges. Although some cellular pathways are evolutionarily conserved, others are not. For example, zebrafish do not have an ARF locus (21), and MYC is not a downstream target of NOTCH in zebrafish (22, 23). Another hurdle has been the lack of antibody reagents, which has hampered our ability to assess the role of immune cell subtypes in tumor progression, define critical interactions between cancer cells and the tumor microenvironment, and fully characterize tumor cell heterogeneity. Moreover, fish are ectothermic, and their unique adaptations that maintain homeostasis under variable temperature conditions may confound the interpretation of cellular processes in human cancer. Human cells have been transplanted into larval zebrafish (24); however, those studies are limited by immune rejection and the small number of cells that can be injected (10-100 cells/injection) (9, 25). Recent advances in generating immunocompromised zebrafish for cell transplantation studies may help resolve the problem of immune rejection, but have yet to be used in human xenograft studies (26). Finally, unlike mammals, fish continue to grow throughout their lifetime and exhibit remarkable mechanisms of tissue repair after injury that have not been found in mammals. Whether these unique processes affect cancer initiation or progression remains unstudied. In total, zebrafish have rapidly become a useful experimental model system in which to elucidate the fundamental principles of cancer initiation, progression, and dissemination. Zebrafish should be considered as a discovery tool that requires the extension of findings into mammalian models and patients’ tumors.
Modeling Human Cancer Mutations in Genetically Engineered Mouse Models
The mouse is the most widely used and well-validated model for cancer research. More than 20 years ago, the role of p53 as a tumor suppressor was discovered by deleting the gene in the mouse (27). Early studies also indicated that in some cases, such as that of retinoblastoma gene Rb1, GEMMs do not always recapitulate the genetics of human tumor susceptibility (28). Today, more sophisticated approaches are used that incorporate conditional deletion of tumor suppressors or activation of oncogenes in specific tissues with temporal control (29, 30). Because of these approaches, GEMMs have been widely used to identify the cells of origin for different types of cancer. TALENs and CRISPR/Cas9 have further streamlined targeted manipulation of the mouse genome (31-34). GEMMs can also be used to recapitulate the expression of fusion proteins that result from chromosomal translocations such as those found in Ewing sarcoma and rhabdomyosarcoma (35-37). Therefore, GEMMs are ideally suited for testing the contributions of individual genetic lesions to cancer initiation and/or progression and for genetically validating potential therapeutic targets. For example, by using two very similar GEMMs of retinoblastoma and targeting MDM2/MDMX with the small-molecule inhibitor nutlin-3a, it was shown that nutlin-3a specifically targets the p53 pathway (38). This study demonstrated that GEMMs provide a unique opportunity to establish scientific justification and test the specificity of molecular-targeted therapeutics for specific pediatric cancers.
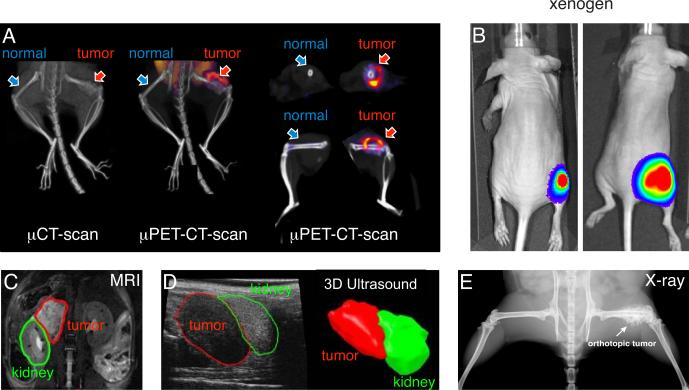
One advantage of murine models of pediatric cancer is their anatomical and physiological similarity to humans. Drug-dosing routes (e.g., oral, intravenous, infusion pumps) and schedules used to treat children with cancer can often be recapitulated in murine models, and comparable routine assays (e.g., CBC-D and blood chemistries) can be performed to monitor toxicity when it is relevant to the drug's toxicity in patients. Pharmacokinetic (PK) and pharmacodynamic (PD) studies are routinely performed in mice, and the availability of those data and methods is a major advantage when calculating the murine equivalent dose (MED) of chemotherapeutics used in humans. Diagnostic imaging modalities (e.g., X-ray, MRI, PET-CT) and surgical methods (39) that are analogous to those used in the clinic are also widely available for GEMMs (Fig. 3). Despite the challenges associated with working with a species that is 1,000 times smaller than humans, many of the procedures performed in pediatric oncology can be applied in murine preclinical models. From a basic research perspective, the major advantage of GEMMs is the ability to directly test in mammals whether a particular genetic lesion contributes to tumor initiation and progression in vivo or whether it is relevant to therapeutic response. This is important because many recurrent genetic mutations have been identified in the genomic profiling of pediatric cancers, but it is not yet clear how those mutations contribute to tumorigenesis or treatment efficacy. A further advantage of working with GEMMs is that they can recapitulate cancer in the setting of an intact immune system, something that is difficult to achieve with PDX models in immunocompromised mice.
Figure 3. Diagnostic imaging modalities used in murine models of cancer.
A) Representative PET and CT images using C11-methionine for a Ewing sarcoma orthotopic xenograft in the femur. B) Photograph of an immunocompromised mouse with an orthotopic rhabdomyosarcoma xenograft that is labeled with firefly luciferase for Xenogen imaging. C) MRI image of an orthotopic neuroblastoma xenograft (red) in the para-adrenal space adjacent to the kidney (green). D) 3D ultrasound of the same neuroblastoma xenograft shown in (C). E) X-ray image of an orthotopic Ewing sarcoma tumor showing the “hair-on-end” appearance that is typical of Ewing sarcoma.
Despite all of the advantages of murine models and GEMMs in particular, some important limitations must be considered. For example, the chromosomal structure and organization and the epigenome in mice differ from those in humans (40). Several recent studies have shown that the epigenomes of GEMMs of pediatric cancer have very little overlap with those of their human counterparts (40). Moreover, point mutations, insertions/deletions, chromosomal lesions, and aneuploidy differ across murine and human tumors (40). These differences can significantly affect basic research on cancer initiation and progression and can directly influence efforts to establish translational relevance of novel therapeutic combinations. In addition, some pediatric cancers (e.g., Ewing sarcoma) have proven to be quite difficult to model in mice, and the spectrum of cancers induced in mice by specific genetic alterations often does not recapitulate the spectrum of cancers caused by those same mutations in humans. In GEMMs in which the oncogenic driver is also the therapeutic target, it may be useful to compare drug efficacy in PDX models and/or in related GEMMs that have perturbations in the same pathway through alternative genetic lesions such as the several different GEMMs of the hedgehog subgroup of medulloblastoma (41). Despite these limitations, GEMMs are currently the most effective and efficient systems in which to study the contribution of individual genetic lesions to tumorigenesis in mammals.
Exploiting Patient-Derived Xenograft Models to Identify Tumor Vulnerabilities
Patients’ tumor samples can be directly implanted in mice to overcome some of the limitations of tumor cell lines. The derivation of severely immunocompromised mice (NOD-SCID IL2Rgamma, or NSG) mice has facilitated the generation of patient-derived xenografts (PDXs) of childhood cancer. The major advantage of PDX models of pediatric cancer is that the tumor cells potentially reflect the genomic heterogeneity found in the human disease more accurately than do other disease models. Previous work has demonstrated that PDX models recapitulate many of the features of the original tumor (8, 42-44). PDX models can increase the scientific justification and translational relevance of individualized, molecular-targeted therapy because multiple patient-derived tumors can be tested simultaneously to capture the clinical heterogeneity across patients. Adequate use of PDX models for individualized preclinical studies requires the demonstration that the tumors retain the genetic lesion targeted by the therapeutic being tested. If multiple tumors are established from different patients and only some of the tumors express the druggable mutations, then the tumors that do not express those mutations provide a useful control group for scientific justification of pathway perturbation.
Clonal heterogeneity and evolution are important factors to consider when using PDX models because we do not know whether one dominant clone grows or if the model maintains the clonal heterogeneity of the original tumor. For example, rare clones in the patient's original tumor can emerge as major clones in PDX models (45). By establishing a large collection of several independent PDX models from each patient, we may be able to study those complex cellular mechanisms in vivo. This ability is particularly relevant for modeling recurrent disease, which is a major clinical challenge (1). For example, if a PDX is derived from a patient at diagnosis and then an additional PDX is derived from a subsequent recurrent tumor, these models can be very useful to study the clonal evolution and changes in drug sensitivity that are associated with disease recurrence. Moreover, it may be possible to experimentally model disease recurrence by exposing the primary tumor's orthotopic PDX to the same treatment regimen used in patients and then determining whether the clonal evolution of that tumor in the mouse model recapitulates that seen in patients.
Three major limitations of orthotopic PDX models are relevant to basic research aimed at providing scientific justification for novel anticancer therapies. The first limitation is that the early stages of tumor initiation are usually not recapitulated in these models because the injected cells are derived from patients with advanced-stage disease and, thus, are fully transformed. Indeed, anecdotal evidence from our groups suggests that in many cases more advanced tumors are much more likely to grow as xenografts. Thus, it may be difficult to model early-stage tumors or low-risk disease with patient-derived tumors. To overcome this limitation, researchers are now deriving induced pluripotent stem cells (iPSCs) from patients that harbor genetic lesions associated with cancer predisposition and differentiating those stem cells into tissue-specific progenitors to study tumor initiation (46). It is also feasible to model tumors by introducing cancer-initiating mutations into stem cells (47).
The second limitation is that although normal tumor-associated murine cells (e.g., macrophages and vascular endothelial cells) infiltrate the orthotopic xenograft (Dyer and Murray, unpublished), whether there are species-to-species differences in those interactions that could alter tumor growth is unknown. Also, because the mice are immunocompromised, the contribution of the immune system cannot be studied in orthotopic PDX models. This is particularly important because of the recent advances in cancer immunotherapy. For example, results of a recent study using murine osteosarcoma cells in immunocompetent mice suggested that α-CTLA-4/α-PD-L1 immunotherapy can help control metastatic disease (48). However, there may be differences between mice and humans related to immunotherapy and there is currently no system to validate those findings in PDX models because the recipient mice are immunocompromised. Despite several important recent advances (49-52), we still do not have reliable biomarkers and/or prognostic factors that allow investigators to identify the pediatric cancer patients that are likely to respond to α-CTLA-4/α-PD-L1 immunotherapy. The lack of predictive biomarkers and relevant preclinical models with PDXs and human immune cells combined with the small patient population makes it difficult to provide scientific justification and translational relevance for immunomodulatory therapy for childhood cancer at this time. Recent developments in humanized mice that can support the human immune system may eventually make it possible to reconstitute a patient's immune system with their corresponding tumor to address fundamental questions in tumor immunology and immunotherapy and to provide critical preclinical data for cancer immunotherapy clinical trials (53).
The third limitation of PDX models is the clonal heterogeneity and evolution of the tumor during treatment. As mentioned above, this has not been well studied, and whether these complex cellular processes are recapitulated in orthotopic PDX models is unclear. Despite these limitations, orthotopic PDX mice are a very useful model of advanced-stage human cancer in which to establish the scientific justifications for molecular-targeted therapy.
Translational Relevance of Druggable Mutations
To move a new therapy into a clinical trial for children with cancer, one must first establish the therapy's scientific justification and translational relevance. Ideally, the careful use of preclinical models can advance our understanding of the fundamental processes of cancer initiation and progression, and those results, in turn, can identify novel therapeutics, thereby showing the scientific justification of the new therapeutic approach. However, many promising new therapies were scientifically justified but failed in clinical trials because they lacked translational relevance. In many cases, the failures were related to the therapeutic window (i.e., the difference between the minimum drug exposure required to achieve an antitumor effect and the maximum exposure tolerated by the patient). If the therapeutic window is narrow, then the drug is very difficult to administer in a clinical setting, and its side effects often limit clinical efficacy. A large therapeutic window provides more flexibility in dosing regimens and drug combinations. This is particularly important for pediatric cancer because new therapies are often used in combination with multidrug regimens that constitute the standard of care for patients at diagnosis or first recurrence. Anticipating and managing toxicity associated with a novel agent is a major challenge in pediatric oncology clinical research.
Each preclinical model has strengths and weaknesses relevant to translational research, as they do in basic research. Although immersing a developing embryo or adult zebrafish in water containing drugs is feasible, many of the newer molecular-targeted therapeutics are poorly soluble in water. Drug penetration from the water, through the fish, to the tumor site has not been studied. Therefore, it is currently not possible to relate drug concentration in water to tumor response and PK/PD in zebrafish models of pediatric cancer. Moreover, humans and mice are often dosed on schedules that have been tailored to maximize the antitumor effects and minimize the side effects. Changes in drug levels in the plasma and tumor of patients after administration are related to the drug's absorption, distribution, metabolism, and excretion (ADME). Those features are likely not recapitulated in zebrafish immersed in water that has a constant concentration of the drug. Although oral gavage and microinjection of drugs into the circulation is possible and PK/PD measurements are now being used in zebrafish, these experiments will likely be labor intensive and will require validation in GEMMs and PDX models. Similarly, long-term studies that mimic the multiple courses of therapy in clinical trials (typically 3-4 weeks per course) have yet to be described using fish. Although zebrafish are a powerful discovery tool for prioritizing drug classes and assessing combination therapies, establishing translational relevance requires further preclinical validation using GEMMs and PDX models in mice.
GEMMs and PDX models have several advantages in terms of providing translational relevance. First, the route of administration and ADME are similar in mice and humans. Some drugs (e.g., irinotecan) have properties that differ between species (54, 55), but the cross-species relevance can be directly tested and validated with careful PK analyses in both species. In the case of irinotecan, a knockout mouse was developed to correct the cross-species difference and more closely mimic irinotecan's ADME in humans (54, 55). When a drug's ADME is similar in mice and humans, clinically relevant doses and dosing schedules can be used to establish the translational relevance of new drug combinations.
GEMMs and orthotopic PDX models that harbor druggable mutations can be developed to establish scientific justification and translational relevance of particular drug/mutation pairs. It is also feasible to perform preclinical studies over extended periods of time in mice (24-30 weeks) to mimic the multiple courses of treatment given to children with cancer. As mentioned above, diagnostic imaging and laboratory assays that are relevant to pediatric oncology now have preclinical counterparts that can be used to assess treatment response in mice.
Although GEMMs and PDX models share many advantages, they also have some differences that should be considered when designing translational relevance studies. GEMMs can be generated to recapitulate tumor initiation and progression in the appropriate cellular lineage, but PDX models cannot. PDX models developed in the flank may show differences caused by the tumor microenvironment that affect PK, tumor growth, and treatment response. This issue can be partially overcome by using orthotopic PDX models (8, 56) to more accurately model the tumor/organ-site interactions, but the immune system interactions can be modeled only with humanized mice (53). This point is particularly important because the immune system plays a crucial, well-established role in tumor response, and immunomodulatory therapies such as α-GD2 antibody for neuroblastoma (57) or α-CTLA-4/α-PD-L1 for osteosarcoma (48) cannot be tested in immunocompromised PDX models. It is unclear whether other cells that contribute to the tumor microenvironment are similar or different in PDX models and GEMMs. For example, murine macrophages infiltrate tumors in GEMMs and orthotopic PDX models, but it is unclear whether they are the same subtype of macrophages (M1 or M2) and whether they function differently in GEMMs and orthotopic PDX tumors (58). Similarly, the tumor vasculature in GEMM and orthotopic PDX tumors is murine-derived, and species differences in vascular endothelial cells have not been extensively studied in this context.
Many investigators introduce potent genetic lesions in GEMMs to promote tumorigenesis even though those lesions are not present in the human tumor population. This practice makes it very difficult to interpret the response to therapy in the context of translational relevance to clinical trials. Similarly, naturally occurring secondary genetic lesions that arise in GEMMs after tumor initiation may differ across species. These lesions can include individual point mutations, larger chromosomal rearrangements, or aneuploidy (40). Moreover, given the importance of the epigenome in pediatric cancer initiation and progression, differences in the epigenomic landscape may confound efforts to provide translational relevance (40, 59).
Some of the most extensively characterized GEMMs of pediatric cancer are the retinoblastoma models. Molecular, cellular, neuroanatomical, neurochemical, genomic, and epigenomic profiling of this disease has shown that, in many ways, these GEMMs are very similar to human tumors (60). When the GEMMs of retinoblastoma were treated with the standard chemotherapy regimen (i.e., vincristine, carboplatin, and etoposide) at clinically relevant, PK-guided doses and on clinically relevant schedules, virtually all of the mice had a complete and durable response (38). In contrast, an orthotopic PDX model of advanced stage invasive retinoblastoma (Reese-Ellsworth stage V) treated with the same drugs at the same dose and schedule had no significant response (38). One interpretation of those results is that the retinoblastoma GEMMs faithfully recapitulate the human disease because the mice respond to the same drugs at the same dose and schedule as used in the clinic. However, in patients, combinations of vincristine and carboplatin or vincristine with carboplatin and etoposide are not sufficient to cure all retinoblastoma patients as in the mice (61). Moreover, the most significant benefit of chemotherapy in children with retinoblastoma is for early-stage disease (Reese-Ellsworth group I-III) (61). Therefore, the GEMMs may be a suitable model for a less-aggressive form of early-stage retinoblastoma (Reese-Ellsworth group I-III), and the PDX models may be a better model of advanced-stage disease (Reese-Ellsworth group IV, V). Thus, although the GEMMs provide genetic validation of drug targeting, they do not provide direct translational relevance for the most aggressive forms of retinoblastoma (38).
Recommendations for Using Preclinical Models to Establish Translational Relevance
Historically, preclinical models were used to provide scientific justification but not translational relevance. This distinction is important. Small pilot studies in PDX models can contribute to the scientific justification for a given targeted therapy. However, to establish translational relevance, stricter criteria should be included. A typical preclinical pediatric cancer study may involve a flank xenograft of a cell line or a PDX in immunocompromised mice. The tumor cells are often incompletely characterized, and their fundamental properties (e.g., genomic mutations, passage number, and histopathologic features) are not documented. Drugs are typically administered as a single agent, on a daily schedule, and at the murine maximum-tolerated dose, which is rarely relevant to the dose, exposure, and schedule used to treat patients. Preclinical efficacy studies are rarely blinded, and they are not benchmarked to standard-of-care treatment regimens with clinically relevant doses and schedules. The number of mice is small (10 or fewer per treatment group), and the treatments rarely extend beyond the equivalent of a single course of chemotherapy in patients (7-21 days). Progressive disease, stable disease, partial response, and complete response in PDX models are difficult to relate to clinical outcomes because caliper measurements of a flank tumor do not directly equate with a patient's response in the clinic.
St. Jude Children's Research Hospital has developed a new paradigm for establishing translational relevance in preclinical models of pediatric cancer (39). Most drugs tested in children have undergone Phase I testing in adult patients, and the paradigm is based on the assumption that a pediatric Phase I trial will be performed to determine the recommended Phase II dose and schedule. This paradigm is based on the following 10 criteria:
-
1)
Randomization: A randomization schedule should be designed by a biostatistician before the preclinical study, as is done for clinical trials. Also, interim analysis and rules for censure (e.g., mouse death caused by unrelated events) should be established before testing begins.
-
2)
Double-Blind Study Design: As with clinical trials, inadvertent bias is prevalent in preclinical testing. Efforts should be taken to design a double-blind study in which different individuals administer drugs and monitor response without any knowledge of the treatment groups (39).
-
3)
Pharmacokinetics: In addition to providing data to determine the MED of the recommended pediatric Phase II dose, PK studies should be performed to determine the penetration and retention of the drug in the tumor.
-
4)
Pharmacodynamics: To directly demonstrate that the drug penetrates tumor cells and perturbs the targeted pathway, PD analysis should be performed.
-
5)
Clinically Relevant Drug Dose: The dose and route of drug administration in preclinical models should approximate the plasma exposure measured in pediatric patients if known.
-
6)
Clinically Relevant Drug Combinations: In pediatric oncology, new drugs used in a Phase II trial are often introduced in the context of an existing treatment regimen. Therefore, clinically relevant drug combinations should be considered to complement the single-agent efficacy studies in preclinical models.
-
7)
Clinically Relevant Drug Schedule: Each course of therapy should be the same as that used in clinical trials (typically 3-4 weeks/course), and multiple courses should be incorporated in preclinical testing regimens.
-
8)
Monitoring Toxicity Relevant to Pediatric Patients: The hematologic and other organ (i.e., kidney and liver) toxicities that are anticipated for a particular drug combination should be monitored during each course of therapy, and the relation between drug exposure and toxicity should be established.
-
9)
Tumor Validation: The tumor should be extensively characterized and accurately recapitulate the histology and genomic features of patients’ tumors. Epigenomic profiling may be important for certain tumor types. If possible, the site of the tumor in the preclinical model should match that in patients to recapitulate tumor growth, spreading, and drug exposure at that organ. If subcutaneous tumors are used, follow-up studies should be attempted in an orthotopic site to confirm efficacy.
-
10)
Efficacy Measurements Relevant to Patient Outcome: Measures of progressive disease, stable disease, partial response, and complete response should be defined before the study begins and used to power the study. Typically, 10-20 mice per group are required to properly power a double-blind, placebo-controlled, randomized efficacy study in mice (39), although this may depend on the biological variability of the specific tumor model. Ideally, all data related to tumor response, histopathology, and toxicity should be made available to other investigators via a centralized database that is validated and secured (39).
In addition to these 10 criteria, specific guidelines and standard operating procedures for using preclinical models to establish translational relevance are freely available online (62). The Childhood Solid Tumor Network preclinical-testing paradigm for providing translational relevance incorporates Phase I, II, and III studies that mimic those in patients. Before starting a preclinical Phase I study, one must provide scientific justification. Also, PK analysis must be performed to establish the MED and determine the duration and concentration of drug penetration in the tumor. PD assays should be performed to link drug penetration with inhibition of the signaling pathway of interest in the tumor cells. Ideally, the preclinical tumor should be at the orthotopic site to better recapitulate the tumor microenvironment and drug distribution (39, 63).
Preclinical Phase I Study
Once the PK, PD, and MED of the drug are established, the next step is the preclinical Phase I study (39). These studies are designed to establish the tolerability of the drug combination at a particular dose and schedule that is clinically relevant for children. Toxicities should be monitored via body weight, blood counts (CBC-D), and blood chemistries. The route of administration is also important because it will affect the absorption of the drug and may influence tolerability and efficacy.
Preclinical Phase II Study
Once tolerability is established and a recommended dose is identified, a preclinical Phase II study is performed to determine whether the drug has antitumor activity in vivo (39). Preclinical Phase II studies are small (5 mice per treatment group), randomized, and placebo controlled. One treatment group receives the standard-of-care regimen to establish a baseline response. Preclinical Phase II studies are not blinded nor are they powered to provide statistical significance. The criteria for progressive disease, stable disease, partial response, and complete response are established before the study. Typically, animals receive up to 4 courses of treatment (3 weeks per course). Preclinical Phase II trials are ideally suited to refine schedules and combinations because they are not double-blinded and involve few animals. The goal is to detect any evidence of drug activity and use that data to prioritize drug combinations to move forward into preclinical Phase III studies. In addition to efficacy, other considerations, such as availability of the drug for clinical development in children, may influence the decision to move into preclinical Phase III studies. Similarly, the toxicity may be important if it overlaps with the toxicity of the standard-of-care regimen that will serve as the foundation for addition of the new agent.
Preclinical Phase III Study
Preclinical Phase III studies are double-blind, randomized, placebo-controlled trials with a sufficient number of animals (approximately 20 animals per treatment group) to provide statistical significance. As with preclinical Phase II studies, the response criteria are defined up front, and the animals receive 4 to 6 courses of therapy. For aggressive tumors, the placebo group will often progress rapidly (39); this timing is helpful for identifying effective treatments that lead to complete and durable responses (39). An appropriate standard-of-care treatment group is included. Each mouse has a unique murine medical record number, and the data are entered into a central database before the treatment groups are de-identified, helping to ensure the accuracy of the data and providing a convenient forum for sharing it.
One exciting opportunity for a preclinical Phase III study design would incorporate multiple individual PDX tumors from different patients. For each individual PDX, replicate mice would be used to establish statistical significance of the response in that particular tumor. However, to capture the clinical heterogeneity of a particular tumor, multiple PDX tumors from different individuals would be incorporated into the study. The individual mice would be randomized across the different PDX tumors and their replicates in double-blind placebo-controlled trials. This approach would also be ideal for comparing the responses of diagnostic tumors and recurrent tumors from the same individual and across different patients.
Data from preclinical studies provide translational relevance that can directly influence clinical trials. Here, we have outlined one paradigm established in the Developmental Biology and Solid Tumor Program at St. Jude Children's Research Hospital. However, alternative approaches are possible if they address the 10 key points outlined above. The distinction between providing scientific justification and establishing translational relevance of new therapeutics is essential to pediatric cancer and rare adult cancer research because the number of patients with those diseases is so small. Therefore, the most promising agents in the most appropriate dose, route, and combination must be elucidated in the laboratory before moving into the clinic. This approach is particularly important for the success of cooperative groups that invest years developing clinical trial concepts. In that setting, scientific justification and translational relevance must be established early so that trials not meeting the benchmark can be aborted efficiently.
Targeting Druggable Mutations in Pediatric Cancer
In most clinical genomic trials, mutations and their respective molecular-targeted drugs are ranked based on the data supporting their scientific justification and translational relevance. Individual protocols may have different criteria, but the concept of ranking mutations is a common theme. This approach does not take into account the sequence analysis or the possibility of tumor heterogeneity and clonal evolution. Even if we assume that every cell in a tumor has the same bona fide mutation, categorizing that mutation as druggable is still a topic of debate and an emerging area of investigation. Thus, a rigorous and robust standardized definition of druggable is crucial to help align clinical genomic trials and provide reasonable expectations for patients and their families. Even if a particular mutation is deemed to have scientific justification for being druggable, establishing its translational relevance is equally important. For many targets, several competing drugs are being developed, and the translational relevance of each drug may prove essential for successful targeting in patients.
The presence of a recurrent somatic mutation in cancer suggests that such a mutation provides a growth advantage to the tumor. If that recurrent mutation is associated with advanced-stage disease or poor outcome, then it has additional value as a biomarker. However, without careful laboratory research, such an association does not prove that a mutation is druggable. Although the mutation may be actionable (i.e., it helps identify patients whose disease should be considered high-risk), it is not necessarily druggable. First, one would need to show that the mutation is necessary and/or sufficient for tumorigenesis in vivo. This can be done using any of the 3 preclinical models described herein if their strengths and weaknesses are taken into consideration. Next, drug sensitivity associated with the particular genetic lesion should be established. The mutations that are necessary and/or sufficient for tumorigenesis far outnumber current molecular-targeted therapeutics; thus, not all mutations that induce tumors are druggable. Finally, if preclinical studies of the drug provide scientific justification, then translational relevance should be established using the 10 aforementioned criteria and preclinical Phase I, II, and III studies. If these efforts are successful, then individualized treatment regimens have a much higher probability of success. Here we provide 3 examples from solid tumor studies to highlight these concepts.
The first example is ATRX mutation in neuroblastoma. The ATRX gene encodes a protein component of the SWI/SNF complex that is involved in histone H3.3 deposition at telomeres and other regions in the genome (64, 65). ATRX-mutant tumors use a unique mechanism of telomere maintenance called alternative lengthening of telomeres (ALT) to promote cell proliferation, and ATRX mutations are associated with indolent forms of stage 4 neuroblastoma in older patients who tend to have poor rates of overall survival (66, 67). Although the association of ATRX mutations with poor outcome and the high rate of somatic mutations in older patients with stage 4 neuroblastoma meet the first criterion for druggable mutations, ATRX falls short on the other two criteria. ATRX-mutant tumors may be more sensitive to ATR inhibitors than are wild-type ATRX tumors (65), but this has not been demonstrated in vivo. Moreover, the translational relevance of ATR inhibitors for ATRX-mutant neuroblastomas has not been demonstrated.
The second example is oncogenic RAS mutation in rhabdomyosarcoma. A substantial portion of children with embryonal rhabdomyosarcoma express RAS mutations that are associated with intermediate- or high-risk disease (8). In preclinical and cell-based models, RAS mutations are necessary and sufficient for tumorigenesis (13, 17). However, individual drugs or combinations of drugs that are thought to perturb RAS signaling in cancer are not selective for RAS-mutant embryonal rhabdomyosarcoma tumors versus those that express wild-type RAS (8). Therefore, the agents have not been tested in PDX models to establish translational relevance. Although downstream modulators of RAS signaling, such as MEK and AKT/mTORC1, can be inhibited with small-molecule inhibitors, such approaches in colon and lung cancers have led to partial responses, which ultimately progress because of therapeutic resistance. Thus, although RAS is a well-established oncogenic lesion, no current molecular-targeted therapeutic has shown selective sensitivity or translational relevance. Accordingly, RAS mutations meet only 1 of 3 criteria and should not be considered druggable. If novel agents targeting RAS are developed or more effective therapies to inhibit its downstream effector pathways are tested, then translational studies in mice will be clearly warranted.
The third example is oncogenic ALK mutation in neuroblastoma. In zebrafish models and GEMMs, ALK mutations are necessary and sufficient for tumorigenesis in vivo. Moreover, small-molecule inhibitors targeting ALK have been developed, and the efficacy of those drugs in treating particular lesions has been examined (14, 68-73). Scientific justification for targeting ALK has been established in vivo by using animal models, but translational relevance has not been established. Clinical trials of ALK-mutant neuroblastoma treatment with ALK inhibitors are ongoing (74). In the Phase I study of crizotinib, only 1 of 11 patients with ALK-mutated neuroblastoma experienced an objective response. Several of those patients’ tumors had oncogenic ALK mutations that were predicted to be sensitive to crizotinib (75). In contrast, 8 of 9 patients with ALK-translocated anaplastic large cell lymphoma experienced objective responses. Taken together, these data suggest that crizotinib can penetrate the tumor and perturb ALK-mediated oncogenic signaling, but it is unclear why there were so few responses in neuroblastoma. Some details of dosing, schedule, and drug combinations can be elucidated in patients, but the effective translation of ALK inhibitors into the clinic would benefit from studies in preclinical models to establish the mutation's translational relevance. Thus, ALK is a druggable mutation that meets 2 of 3 criteria for moving into clinic trials, and with some additional translational studies, it could emerge as the standard for individualized treatment for children with neuroblastoma.
Recent studies targeting the DNA damage response in Ewing sarcoma are examples of clinical trial development with both scientific justification and translational relevance (39). Ewing sarcoma cells have defects in DNA damage repair that can be exploited by combining PARP inhibitors and DNA-damaging agents such as irinotecan or temozolomide. There are predictive biomarkers (SLNF11 and PARP1 expression) of sensitivity to these drugs, and knockdown of PARP1 protein expression led to reduced sensitivity (39). Translational relevance was established according to the 10 criteria described above and preclinical phase I,II,III testing, leading to the ongoing clinical trial with talazoparib combined with irinotecan with or without temozolomide (Clinicaltrials.gov: NCT02392793). If tolerability and efficacy can be established for these drug combinations in patients with Ewing sarcoma, then it will provide an important proof-of-principle for the strategies outlined here.
If we consider the current and planned clinical genomic trials, we have a long way to go to establish scientific justification and translational relevance of particular druggable mutations. For example, in the iCAT trial (http://meetinglibrary.asco.org/content/133535-144), which used a targeted resequencing platform, 16 of 54 patients (30%) received a therapeutic recommendation. Among those, only 1 had a tier-1 mutation, and that was an ALK-mutant neuroblastoma. All other therapeutic recommendations in iCAT had even less scientific and translational evidence. The goal of the iCAT trial was to determine the feasibility of providing therapeutic recommendations based on genomic analysis of patients’ tumors. In this regard, it was a successful trial. However, it also highlights our lack of understanding of the relationship between particular genetic lesions in cancer and response to molecular-targeted chemotherapy. On the basis of the results of the iCAT and other similar genomic studies, a combination of GEMM, PDX, and zebrafish model studies could be designed using currently available drugs to test the relevance of a specific mutation or combination of mutations.
Conclusions
The risk of making treatment recommendations that are not grounded in basic, translational, or clinical research is that the value of individualized therapy using clinical genomics will become underappreciated. We propose that the emphasis of pediatric clinical genomics should be on making recommendations that have scientific justification and translational relevance. As a research community, we should share expertise and resources to efficiently establish scientific justification and translational relevance of molecular-targeted therapeutics for the most-common mutations associated with pediatric cancer. As a result, pediatric cancer care would be substantially advanced as more informed recommendations are made in the clinic. The investment of over $1 million in the Childhood Solid Tumor Network(62) is one step in that direction.
Acknowledgements
We thank Cherise Guess for editing the manuscript.
Financial Support: This work was supported by NCI grants (CA21765 and CA168875 [MAD]; CA159859 [RWR]), NIH grants (EY014867 and EY018599 [MAD]; U54CA168512 and R01CA154923 [DML]), the American Lebanese Syrian Associated Charities [MAD], Alex's Lemonade Stand Foundation, Howard Hughes Medical Institute [MAD], the Live Like Bella Foundation [DML], and a Leadership Award from the California Institute of Regenerative Medicine (LA1-01747 [RWR].
Footnotes
Conflict of Interest: The authors have no conflict of interest to report.
References
- 1.Smith MA, Altekruse SF, Adamson PC, Reaman GH, Seibel NL. Declining childhood and adolescent cancer mortality. Cancer. 2014;120(16):2497–506. doi: 10.1002/cncr.28748. Epub 2014/05/24. doi: 10.1002/cncr.28748. PubMed PMID: 24853691; PubMed Central PMCID: PMC4136455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan R, Federico S, Dyer MA. The war on cancer: have we won the battle but lost the war? Oncotarget. 2010;1(2):77–83. doi: 10.18632/oncotarget.111. Epub 2010/09/30. PubMed PMID: 20877440; PubMed Central PMCID: PMC2945373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downing JR, Wilson RK, Zhang J, Mardis ER, Pui CH, Ding L, et al. The Pediatric Cancer Genome Project. Nature genetics. 2012;44(6):619–22. doi: 10.1038/ng.2287. Epub 2012/05/30. doi: 10.1038/ng.2287. PubMed PMID: 22641210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, Pappo A, Dyer MA. Pediatric solid tumor genomics and developmental pliancy. Oncogene. 2015 doi: 10.1038/onc.2014.474. Epub 2015/02/03. doi: 10.1038/onc.2014.474. PubMed PMID: 25639868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hobin JA, Deschamps AM, Bockman R, Cohen S, Dechow P, Eng C, et al. Engaging basic scientists in translational research: identifying opportunities, overcoming obstacles. J Transl Med. 2012;10:72. doi: 10.1186/1479-5876-10-72. Epub 2012/04/17. doi: 10.1186/1479-5876-10-72. PubMed PMID: 22500917; PubMed Central PMCID: PMC3419626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobin JA, Galbraith RA. Engaging basic scientists in translational research. Faseb J. 2012;26(6):2227–30. doi: 10.1096/fj.12-0601ufm. Epub 2012/06/02. doi: 10.1096/fj.12-0601ufm. PubMed PMID: 22653563. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Bahrami A, Pappo A, Easton J, Dalton J, Hedlund E, et al. Recurrent Somatic Structural Variations Contribute to Tumorigenesis in Pediatric Osteosarcoma. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.03.003. Epub 2014/04/08. doi: 10.1016/j.celrep.2014.03.003. PubMed PMID: 24703847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24(6):710–24. doi: 10.1016/j.ccr.2013.11.002. Epub 2013/12/18. doi: 10.1016/j.ccr.2013.11.002. PubMed PMID: 24332040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, et al. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299(5608):887–90. doi: 10.1126/science.1080280. Epub 2003/02/08. doi: 10.1126/science.1080280. PubMed PMID: 12574629. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Jette C, Kanki JP, Aster JC, Look AT, Griffin JD. NOTCH1-induced T-cell leukemia in transgenic zebrafish. Leukemia. 2007;21(3):462–71. doi: 10.1038/sj.leu.2404546. Epub 2007/01/26. doi: 10.1038/sj.leu.2404546. PubMed PMID: 17252014. [DOI] [PubMed] [Google Scholar]
- 11.Langenau DM, Zon LI. The zebrafish: a new model of T-cell and thymic development. Nat Rev Immunol. 2005;5(4):307–17. doi: 10.1038/nri1590. Epub 2005/04/02. doi: 10.1038/nri1590. PubMed PMID: 15803150. [DOI] [PubMed] [Google Scholar]
- 12.Sabaawy HE, Azuma M, Embree LJ, Tsai HJ, Starost MF, Hickstein DD. TEL-AML1 transgenic zebrafish model of precursor B cell acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103(41):15166–71. doi: 10.1073/pnas.0603349103. Epub 2006/10/04. doi: 10.1073/pnas.0603349103. PubMed PMID: 17015828; PubMed Central PMCID: PMC1622794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21(11):1382–95. doi: 10.1101/gad.1545007. Epub 2007/05/19. doi: 10.1101/gad.1545007. PubMed PMID: 17510286; PubMed Central PMCID: PMC1877750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu S, Lee JS, Guo F, Shin J, Perez-Atayde AR, Kutok JL, et al. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21(3):362–73. doi: 10.1016/j.ccr.2012.02.010. Epub 2012/03/24. doi: 10.1016/j.ccr.2012.02.010. PubMed PMID: 22439933; PubMed Central PMCID: PMC3315700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ignatius MS, Chen E, Elpek NM, Fuller AZ, Tenente IM, Clagg R, et al. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell. 2012;21(5):680–93. doi: 10.1016/j.ccr.2012.03.043. Epub 2012/05/26. doi: 10.1016/j.ccr.2012.03.043. PubMed PMID: 22624717; PubMed Central PMCID: PMC3381357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackburn JS, Liu S, Wilder JL, Dobrinski KP, Lobbardi R, Moore FE, et al. Clonal evolution enhances leukemia-propagating cell frequency in T cell acute lymphoblastic leukemia through Akt/mTORC1 pathway activation. Cancer Cell. 2014;25(3):366–78. doi: 10.1016/j.ccr.2014.01.032. Epub 2014/03/13. doi: 10.1016/j.ccr.2014.01.032. PubMed PMID: 24613413; PubMed Central PMCID: PMC3992437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le X, Pugach EK, Hettmer S, Storer NY, Liu J, Wills AA, et al. A novel chemical screening strategy in zebrafish identifies common pathways in embryogenesis and rhabdomyosarcoma development. Development. 2013;140(11):2354–64. doi: 10.1242/dev.088427. Epub 2013/04/26. doi: 10.1242/dev.088427. PubMed PMID: 23615277; PubMed Central PMCID: PMC3653557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridges S, Heaton WL, Joshi D, Choi H, Eiring A, Batchelor L, et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood. 2012;119(24):5621–31. doi: 10.1182/blood-2011-12-398818. Epub 2012/04/12. doi: 10.1182/blood-2011-12-398818. PubMed PMID: 22490804; PubMed Central PMCID: PMC3382926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez A, Pan L, Groen RW, Baleydier F, Kentsis A, Marineau J, et al. Phenothiazines induce PP2A-mediated apoptosis in T cell acute lymphoblastic leukemia. J Clin Invest. 2014;124(2):644–55. doi: 10.1172/JCI65093. Epub 2014/01/10. doi: 10.1172/JCI65093. PubMed PMID: 24401270; PubMed Central PMCID: PMC3904599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471(7339):518–22. doi: 10.1038/nature09882. Epub 2011/03/25. doi: 10.1038/nature09882. PubMed PMID: 21430780; PubMed Central PMCID: PMC3759979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez A, Feng H, Stevenson K, Neuberg DS, Calzada O, Zhou Y, et al. Loss of function tp53 mutations do not accelerate the onset of myc-induced T-cell acute lymphoblastic leukaemia in the zebrafish. Br J Haematol. 2014;166(1):84–90. doi: 10.1111/bjh.12851. Epub 2014/04/03. doi: 10.1111/bjh.12851. PubMed PMID: 24690081; PubMed Central PMCID: PMC4234197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackburn JS, Liu S, Raiser DM, Martinez SA, Feng H, Meeker ND, et al. Notch signaling expands a pre-malignant pool of T-cell acute lymphoblastic leukemia clones without affecting leukemia-propagating cell frequency. Leukemia. 2012;26(9):2069–78. doi: 10.1038/leu.2012.116. Epub 2012/04/28. doi: 10.1038/leu.2012.116. PubMed PMID: 22538478; PubMed Central PMCID: PMC3435461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herranz D, Ambesi-Impiombato A, Palomero T, Schnell SA, Belver L, Wendorff AA, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20(10):1130–7. doi: 10.1038/nm.3665. Epub 2014/09/10. doi: 10.1038/nm.3665. PubMed PMID: 25194570; PubMed Central PMCID: PMC4192073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konantz M, Balci TB, Hartwig UF, Dellaire G, Andre MC, Berman JN, et al. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci. 2012;1266:124–37. doi: 10.1111/j.1749-6632.2012.06575.x. Epub 2012/08/21. doi: 10.1111/j.1749-6632.2012.06575.x. PubMed PMID: 22901264. [DOI] [PubMed] [Google Scholar]
- 25.Smith AC, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV, et al. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010;115(16):3296–303. doi: 10.1182/blood-2009-10-246488. Epub 2010/01/09. doi: 10.1182/blood-2009-10-246488. PubMed PMID: 20056790; PubMed Central PMCID: PMC2858492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Q, Abdelfattah NS, Blackburn JS, Moore JC, Martinez SA, Moore FE, et al. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat Methods. 2014;11(8):821–4. doi: 10.1038/nmeth.3031. Epub 2014/07/22. doi: 10.1038/nmeth.3031. PubMed PMID: 25042784; PubMed Central PMCID: PMC4294527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr., Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–21. doi: 10.1038/356215a0. Epub 1992/03/29. doi: 10.1038/356215a0. PubMed PMID: 1552940. [DOI] [PubMed] [Google Scholar]
- 28.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359(6393):295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 29.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nature reviews Cancer. 2002;2(4):251–65. doi: 10.1038/nrc777. Epub 2002/05/11. doi: 10.1038/nrc777. PubMed PMID: 12001987. [DOI] [PubMed] [Google Scholar]
- 30.Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nature reviews Drug discovery. 2006;5(9):741–54. doi: 10.1038/nrd2110. Epub 2006/08/18. doi: 10.1038/nrd2110. PubMed PMID: 16915232. [DOI] [PubMed] [Google Scholar]
- 31.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi: 10.1016/j.cell.2013.04.025. Epub 2013/05/07. doi: 10.1016/j.cell.2013.04.025. PubMed PMID: 23643243; PubMed Central PMCID: PMC3969854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wefers B, Meyer M, Ortiz O, Hrabe de Angelis M, Hansen J, Wurst W, et al. Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proc Natl Acad Sci U S A. 2013;110(10):3782–7. doi: 10.1073/pnas.1218721110. Epub 2013/02/22. doi: 10.1073/pnas.1218721110. PubMed PMID: 23426636; PubMed Central PMCID: PMC3593923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–7. doi: 10.1038/nature13902. Epub 2014/10/23. doi: 10.1038/nature13902. PubMed PMID: 25337876; PubMed Central PMCID: PMC4270925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516(7531):428–31. doi: 10.1038/nature13906. Epub 2014/10/23. doi: 10.1038/nature13906. PubMed PMID: 25337879; PubMed Central PMCID: PMC4292871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haldar M, Randall RL, Capecchi MR. Synovial sarcoma: from genetics to genetic-based animal modeling. Clin Orthop Relat Res. 2008;466(9):2156–67. doi: 10.1007/s11999-008-0340-2. Epub 2008/06/20. doi: 10.1007/s11999-008-0340-2. PubMed PMID: 18563504; PubMed Central PMCID: PMC2492998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes Dev. 2004;18(21):2614–26. doi: 10.1101/gad.1244004. Epub 2004/10/19. doi: gad.1244004 [pii] 10.1101/gad.1244004. PubMed PMID: 15489287; PubMed Central PMCID: PMC525542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Straessler KM, Jones KB, Hu H, Jin H, van de Rijn M, Capecchi MR. Modeling clear cell sarcomagenesis in the mouse: cell of origin differentiation state impacts tumor characteristics. Cancer Cell. 2013;23(2):215–27. doi: 10.1016/j.ccr.2012.12.019. Epub 2013/02/16. doi: 10.1016/j.ccr.2012.12.019. PubMed PMID: 23410975; PubMed Central PMCID: PMC3640275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennan RC, Federico S, Bradley C, Zhang J, Flores-Otero J, Wilson M, et al. Targeting the p53 pathway in retinoblastoma with subconjunctival Nutlin-3a. Cancer research. 2011;71(12):4205–13. doi: 10.1158/0008-5472.CAN-11-0058. Epub 2011/04/26. doi: 10.1158/0008-5472.CAN-11-0058. PubMed PMID: 21515735; PubMed Central PMCID: PMC3116943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart E, Goshorn R, Bradley C, Griffiths LM, Benavente C, Twarog NR, et al. Targeting the DNA repair pathway in Ewing sarcoma. Cell Rep. 2014;9(3):829–41. doi: 10.1016/j.celrep.2014.09.028. Epub 2014/12/02. doi: 10.1016/j.celrep.2014.09.028. PubMed PMID: 25437539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benavente CA, McEvoy JD, Finkelstein D, Wei L, Kang G, Wang YD, et al. Cross-species genomic and epigenomic landscape of retinoblastoma. Oncotarget. 2013;4(6):844–59. doi: 10.18632/oncotarget.1051. Epub 2013/06/15. PubMed PMID: 23765217; PubMed Central PMCID: PMC3757242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng JM, Curran T. The Hedgehog's tale: developing strategies for targeting cancer. Nat Rev Cancer. 2011;11(7):493–501. doi: 10.1038/nrc3079. Epub 2011/05/27. doi: 10.1038/nrc3079. PubMed PMID: 21614026; PubMed Central PMCID: PMC3576812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao X, Liu Z, Yu L, Zhang Y, Baxter P, Voicu H, et al. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro Oncol. 2012;14(5):574–83. doi: 10.1093/neuonc/nos061. Epub 2012/03/31. doi: 10.1093/neuonc/nos061. PubMed PMID: 22459127; PubMed Central PMCID: PMC3337308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart E, Shelat AS, Bradley C, Chen X, Federico S, Thiagaragan S, et al. Development and Characterization of a Human Orthotopic Neuroblastoma Xenograft. Developmental Biology. 2015 doi: 10.1016/j.ydbio.2015.02.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9(6):338–50. doi: 10.1038/nrclinonc.2012.61. Epub 2012/04/18. doi: 10.1038/nrclinonc.2012.61. PubMed PMID: 22508028; PubMed Central PMCID: PMC3928688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518(7539):422–6. doi: 10.1038/nature13952. Epub 2014/12/04. doi: 10.1038/nature13952. PubMed PMID: 25470049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee DF, Su J, Kim HS, Chang B, Papatsenko D, Zhao R, et al. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161(2):240–54. doi: 10.1016/j.cell.2015.02.045. Epub 2015/04/11. doi: 10.1016/j.cell.2015.02.045. PubMed PMID: 25860607; PubMed Central PMCID: PMC4397979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346(6216):1529–33. doi: 10.1126/science.1253799. Epub 2014/12/20. doi: 10.1126/science.1253799. PubMed PMID: 25525250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lussier DM, Johnson JL, Hingorani P, Blattman JN. Combination immunotherapy with alpha-CTLA-4 and alpha-PD-L1 antibody blockade prevents immune escape and leads to complete control of metastatic osteosarcoma. J Immunother Cancer. 2015;3:21. doi: 10.1186/s40425-015-0067-z. Epub 2015/05/21. doi: 10.1186/s40425-015-0067-z. PubMed PMID: 25992292; PubMed Central PMCID: PMC4437699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kreiter S, Vormehr M, van de Roemer N, Diken M, Lower M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520(7549):692–6. doi: 10.1038/nature14426. Epub 2015/04/23. doi: 10.1038/nature14426. PubMed PMID: 25901682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–99. doi: 10.1056/NEJMoa1406498. Epub 2014/11/20. doi: 10.1056/NEJMoa1406498. PubMed PMID: 25409260; PubMed Central PMCID: PMC4315319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–71. doi: 10.1038/nature13954. Epub 2014/11/28. doi: 10.1038/nature13954. PubMed PMID: 25428505; PubMed Central PMCID: PMC4246418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–81. doi: 10.1038/nature13988. Epub 2014/11/28. doi: 10.1038/nature13988. PubMed PMID: 25428507; PubMed Central PMCID: PMC4279952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, et al. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol. 2014;32(4):364–72. doi: 10.1038/nbt.2858. Epub 2014/03/19. doi: 10.1038/nbt.2858. PubMed PMID: 24633240; PubMed Central PMCID: PMC4017589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morton CL, Iacono L, Hyatt JL, Taylor KR, Cheshire PJ, Houghton PJ, et al. Activation and antitumor activity of CPT-11 in plasma esterase-deficient mice. Cancer Chemother Pharmacol. 2005;56(6):629–36. doi: 10.1007/s00280-005-1027-y. Epub 2005/05/27. doi: 10.1007/s00280-005-1027-y. PubMed PMID: 15918039. [DOI] [PubMed] [Google Scholar]
- 55.Morton CL, Wierdl M, Oliver L, Ma MK, Danks MK, Stewart CF, et al. Activation of CPT-11 in mice: identification and analysis of a highly effective plasma esterase. Cancer Res. 2000;60(15):4206–10. Epub 2000/08/17. PubMed PMID: 10945631. [PubMed] [Google Scholar]
- 56.Zhang J, Benavente C, McEvoy J, Flores-Otero J, Ding L, Chen X, et al. Genetic and Epigenetic Analysis of Human Retinoblastoma Identifies SYK as a Novel Target for Therapy. Nature. 2011 in revision. [Google Scholar]
- 57.Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman SG, Chen HX, et al. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. The New England journal of medicine. 2010;363(14):1324–34. doi: 10.1056/NEJMoa0911123. Epub 2010/10/01. doi: 10.1056/NEJMoa0911123. PubMed PMID: 20879881; PubMed Central PMCID: PMC3086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bronte V, Murray PJ. Understanding local macrophage phenotypes in disease: modulating macrophage function to treat cancer. Nat Med. 2015;21(2):117–9. doi: 10.1038/nm.3794. Epub 2015/02/06. doi: 10.1038/nm.3794. PubMed PMID: 25654601. [DOI] [PubMed] [Google Scholar]
- 59.Zhang J, Benavente CA, McEvoy J, Flores-Otero J, Ding L, Chen X, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481(7381):329–34. doi: 10.1038/nature10733. Epub 2012/01/13. doi: 10.1038/nature10733. PubMed PMID: 22237022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McEvoy J, Flores-Otero J, Zhang J, Nemeth K, Brennan R, Bradley C, et al. Coexpression of normally incompatible developmental pathways in retinoblastoma genesis. Cancer Cell. 2011;20(2):260–75. doi: 10.1016/j.ccr.2011.07.005. Epub 2011/08/16. doi: 10.1016/j.ccr.2011.07.005. PubMed PMID: 21840489; PubMed Central PMCID: PMC3551581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Galindo C, Wilson MW, Haik BG, Merchant TE, Billups CA, Shah N, et al. Treatment of intraocular retinoblastoma with vincristine and carboplatin. J Clin Oncol. 2003;21(10):2019–25. doi: 10.1200/JCO.2003.09.103. Epub 2003/05/14. doi: 10.1200/JCO.2003.09.103. PubMed PMID: 12743157. [DOI] [PubMed] [Google Scholar]
- 62.Stewart E, Federico S, Karlstrom A, Shelat A, Sablauer A, Pappo A, et al. The childhood solid tumor network: A new resource for the developmental biology and oncology research Communities. Dev Biol. 2015 doi: 10.1016/j.ydbio.2015.03.001. Epub 2015/04/12. doi: 10.1016/j.ydbio.2015.02.002. PubMed PMID: 25863122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang F, Tagen M, Throm S, Mallari J, Miller L, Guy RK, et al. Whole-body physiologically based pharmacokinetic model for nutlin-3a in mice after intravenous and oral administration. Drug metabolism and disposition: the biological fate of chemicals. 2011;39(1):15–21. doi: 10.1124/dmd.110.035915. Epub 2010/10/16. doi: 10.1124/dmd.110.035915. PubMed PMID: 20947617; PubMed Central PMCID: PMC3014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voon HP, Hughes JR, Rode C, De La Rosa-Velazquez IA, Jenuwein T, Feil R, et al. ATRX Plays a Key Role in Maintaining Silencing at Interstitial Heterochromatic Loci and Imprinted Genes. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.03.036. Epub 2015/04/14. doi: 10.1016/j.celrep.2015.03.036. PubMed PMID: 25865896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flynn RL, Cox KE, Jeitany M, Wakimoto H, Bryll AR, Ganem NJ, et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science. 2015;347(6219):273–7. doi: 10.1126/science.1257216. Epub 2015/01/17. doi: 10.1126/science.1257216. PubMed PMID: 25593184; PubMed Central PMCID: PMC4358324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA : the journal of the American Medical Association. 2012;307(10):1062–71. doi: 10.1001/jama.2012.228. Epub 2012/03/15. doi: 10.1001/jama.2012.228. PubMed PMID: 22416102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13(6):397–411. doi: 10.1038/nrc3526. Epub 2013/05/25. doi: 10.1038/nrc3526. PubMed PMID: 23702928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Devoto M, Specchia C, Laudenslager M, Longo L, Hakonarson H, Maris J, et al. Genome-wide linkage analysis to identify genetic modifiers of ALK mutation penetrance in familial neuroblastoma. Human heredity. 2011;71(2):135–9. doi: 10.1159/000324843. Epub 2011/07/08. doi: 10.1159/000324843. PubMed PMID: 21734404; PubMed Central PMCID: PMC3136385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455(7215):930–5. doi: 10.1038/nature07261. Epub 2008/08/30. doi: 10.1038/nature07261. PubMed PMID: 18724359; PubMed Central PMCID: PMC2672043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berry T, Luther W, Bhatnagar N, Jamin Y, Poon E, Sanda T, et al. The ALK(F1174L) mutation potentiates the oncogenic activity of MYCN in neuroblastoma. Cancer Cell. 2012;22(1):117–30. doi: 10.1016/j.ccr.2012.06.001. Epub 2012/07/14. doi: 10.1016/j.ccr.2012.06.001. PubMed PMID: 22789543; PubMed Central PMCID: PMC3417812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carpenter EL, Mosse YP. Targeting ALK in neuroblastoma--preclinical and clinical advancements. Nat Rev Clin Oncol. 2012;9(7):391–9. doi: 10.1038/nrclinonc.2012.72. Epub 2012/05/16. doi: 10.1038/nrclinonc.2012.72. PubMed PMID: 22585002; PubMed Central PMCID: PMC3683972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heukamp LC, Thor T, Schramm A, De Preter K, Kumps C, De Wilde B, et al. Targeted expression of mutated ALK induces neuroblastoma in transgenic mice. Science translational medicine. 2012;4(141):141ra91. doi: 10.1126/scitranslmed.3003967. Epub 2012/07/06. doi: 10.1126/scitranslmed.3003967. PubMed PMID: 22764207. [DOI] [PubMed] [Google Scholar]
- 73.Schulte JH, Lindner S, Bohrer A, Maurer J, De Preter K, Lefever S, et al. MYCN and ALKF1174L are sufficient to drive neuroblastoma development from neural crest progenitor cells. Oncogene. 2012 doi: 10.1038/onc.2012.106. Epub 2012/04/10. doi: 10.1038/onc.2012.106. PubMed PMID: 22484425. [DOI] [PubMed] [Google Scholar]
- 74.Mosse YP, Lim MS, Voss SD, Wilner K, Ruffner K, Laliberte J, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: a Children's Oncology Group phase 1 consortium study. Lancet Oncol. 2013;14(6):472–80. doi: 10.1016/S1470-2045(13)70095-0. Epub 2013/04/20. doi: 10.1016/S1470-2045(13)70095-0. PubMed PMID: 23598171; PubMed Central PMCID: PMC3730818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bresler SC, Weiser DA, Huwe PJ, Park JH, Krytska K, Ryles H, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26(5):682–94. doi: 10.1016/j.ccell.2014.09.019. Epub 2014/12/18. doi: 10.1016/j.ccell.2014.09.019. PubMed PMID: 25517749; PubMed Central PMCID: PMC4269829. [DOI] [PMC free article] [PubMed] [Google Scholar]