Abstract
BACKGROUND AND PURPOSE:
Relative cerebral blood volume, as measured by T2*-weighted dynamic susceptibility-weighted contrast-enhanced MRI, represents the most robust and widely used perfusion MR imaging metric in neuro-oncology. Our aim was to determine whether differences in modeling implementation will impact the correction of leakage effects (from blood-brain barrier disruption) and the accuracy of relative CBV calculations as measured on T2*-weighted dynamic susceptibility-weighted contrast-enhanced MR imaging at 3T field strength.
MATERIALS AND METHODS:
This study included 52 patients with glioma undergoing DSC MR imaging. Thirty-six patients underwent both non-preload dose– and preload dose–corrected DSC acquisitions, with 16 patients undergoing preload dose–corrected acquisitions only. For each acquisition, we generated 2 sets of relative CBV metrics by using 2 separate, widely published, FDA-approved commercial software packages: IB Neuro and nordicICE. We calculated 4 relative CBV metrics within tumor volumes: mean relative CBV, mode relative CBV, percentage of voxels with relative CBV > 1.75, and percentage of voxels with relative CBV > 1.0 (fractional tumor burden). We determined Pearson (r) and Spearman (ρ) correlations between non-preload dose– and preload dose–corrected metrics. In a subset of patients with recurrent glioblastoma (n = 25), we determined receiver operating characteristic area under the curve for fractional tumor burden accuracy to predict the tissue diagnosis of tumor recurrence versus posttreatment effect. We also determined correlations between rCBV and microvessel area from stereotactic biopsies (n = 29) in 12 patients.
RESULTS:
With IB Neuro, relative CBV metrics correlated highly between non-preload dose– and preload dose–corrected conditions for fractional tumor burden (r = 0.96, ρ = 0.94), percentage > 1.75 (r = 0.93, ρ = 0.91), mean (r = 0.87, ρ = 0.86), and mode (r = 0.78, ρ = 0.76). These correlations dropped substantially with nordicICE. With fractional tumor burden, IB Neuro was more accurate than nordicICE in diagnosing tumor versus posttreatment effect (area under the curve = 0.85 versus 0.67) (P < .01). The highest relative CBV–microvessel area correlations required preload dose and IB Neuro (r = 0.64, ρ = 0.58, P = .001).
CONCLUSIONS:
Different implementations of perfusion MR imaging software modeling can impact the accuracy of leakage correction, relative CBV calculation, and correlations with histologic benchmarks.
Perfusion MR imaging (pMRI) has emerged as a powerful diagnostic tool in neuro-oncology. Multiple independent studies have shown how measures of microvessel volume, which are linked closely to histologic identity and malignant potential, can facilitate diagnoses that have historically eluded conventional MR imaging.1–7 For instance, the metric relative cerebral blood volume (rCBV), as measured by dynamic susceptibility-weighted contrast-enhanced pMRI, can identify high-grade components within nonenhancing glioma,6,7 distinguish tumor recurrence from posttreatment effects (ie, pseudoprogression, radiation necrosis),8–11 and predict tumoral response and patient survival after targeted therapy.12–16
Despite the potential clinical impact of pMRI, broad-scale integration has been slowed by the need to define optimal methodologic conditions to maximize rCBV accuracy. While a number of factors can affect rCBV measurements (eg, image acquisition, motion correction, signal fitting, and mathematic modeling), most methodologic studies have focused on techniques that correct for T1-weighted leakage errors from blood-brain barrier disruption and T2/T2*-weighted residual errors from contrast recirculation within tortuous microvasculature.17–24 Specifically, DSC relies on the assumptions that gadolinium-based contrast agents (GBCA) transit through tissue as a single bolus and remain within the vascular lumen. Yet, these premises are often violated in the setting of high-grade glioma, increasing the likelihood of rCBV inaccuracies.
On the basis of previous comparison studies, the administration of GBCA preload dose (PLD) and the subsequent use of software modeling (during image postprocessing) offer the most effective methods for rCBV correction.17–19 PLD, given before DSC acquisition, minimizes T1 leakage effects by presaturating tissue T1 signal and decreasing subsequent GBCA extravascular diffusion.17–22,25,26 Because of theoretic dose-dependent risks of nephrogenic systemic fibrosis, the GBCA dose is generally minimized, with most studies showing effective T1 leakage-correction with a PLD as low as 0.05–0.1 mmol/kg.19 Additionally, modeling correction has proved necessary to correct residual T1 errors and T2/T2*-weighted recirculation effects following PLD. While a number of modeling algorithms have been proposed, the method published by Boxerman et al17 remains the most highly cited and validated algorithm to date, and it is widely considered the standard for DSC-pMRI.
Generally speaking, modeling correction requires implementation of mathematic algorithms through computer software programs developed either in-house by individual academic centers or incorporated within vendor-supplied commercial packages. Vendor-supplied options offer the advantage of wide availability and ease of standardization across multiple institutions, but the methods by which the algorithms are implemented can vary by vendor. While we generally assume negligible differences in how various software programs incorporate mathematic modeling to calculate rCBV, this assumption has not been directly tested, particularly with validation against standard benchmarks such as histology.
In this study, we compared 2 commonly published, commercially available implementations of the Boxerman algorithm,17 as integrated within the IB Neuro (IBN, Version 1.1; Imaging Biometrics, Elm Grove, Wisconsin) and nordicICE (NICE, Version 2.3.13; NordicNeuroLab, Bergen, Norway) software packages.8,9,14–18,20,25–28 We present data from a cohort of 52 patients with glioma who underwent DSC-pMRI acquisition at the time of clinical MR imaging. The goals of this study are to determine the equivalency of modeling implementation and rCBV calculation across platforms and to assess whether rCBV variations, if present, will significantly impact correlations with histologic benchmarks. Our overarching goal is to provide information that will help work toward consensus and standardization of pMRI methodology.
Materials and Methods
Subjects
We searched our data base (2007–2013) for patients with histopathologically confirmed glioma who had conventional 3T MR imaging with pMRI at 2 different institutions. We included patients in whom the same examination contained 2 separate DSC-pMRI acquisitions (and separate bolus contrast injections) and/or the MR imaging was performed preoperatively for stereotactic resection and/or biopsy within 1 day after imaging. Subjects were pooled from 2 separate institutions: Barrow Neurological Institute at St. Joseph's Hospital and Medical Center and Mayo Clinic, Arizona. All patient data were anonymized for Health Insurance Portability and Accountability Act compliance. The institutional review board approved our study. All patients undergoing pMRI had estimated glomerular filtration rates of >60 mg/min/1.72 m2.
Perfusion MR Imaging Data Acquisition
Each 3T examination was performed on 1 of 2 MR imaging magnets (Signa HDx; GE Healthcare, Milwaukee, Wisconsin; or Magnetom Skyra; Siemens, Erlangen, Germany). All patients underwent initial preload dose administration that allowed the acquisition of PLD-corrected DSC-pMRI data, which were all acquired via a second GBCA injection (0.05-mmol/kg, gadodiamide or gadobenate dimeglumine) by using previously described methods.8,19 In all patients, the PLD amount totaled 0.1 mmol/kg, administered via either a single bolus injection or 2 separate (0.05-mmol/kg) bolus injections, depending on the departmental protocol at the time of imaging. In a subset of patients, we acquired non-PLD-corrected DSC-pMRI data during the initial PLD bolus injection, by using either 0.05- or 0.1-mmol/kg GBCA injections, depending on the clinical perfusion MR imaging protocol used at the time of acquisition. We performed a separate subanalysis to determine the impact of different injection doses as shown in On-line Table 1.
All DSC data (gradient-echo echo-planar imaging with TR/TE/flip angle = 1500–2000/20 ms/60°, FOV = 24 × 24 cm, matrix = 128 × 128, 5-mm section, no gap) were acquired during 3 minutes with the bolus injection occurring at the 1-minute mark after the start of the DSC sequence. All GBCA injections were via power injector at 3–5 mL/s, followed by a 20-mL normal saline flush. The final GBCA dose for all patients (irrespective of the method of PLD administration) was 0.15 mmol/kg of body weight.
Perfusion MR Imaging Data Analysis
After transferring all MR imaging data to an off-line workstation and removing baseline points collected during the first 5 seconds, we generated whole-brain rCBV maps by using 2 commonly published commercial software packages: nordicICE (Version 2.3.13) and IB Neuro (Version 1.1), both approved by the US Food and Drug Administration. For NICE, we used all available default options and included leakage correction in all cases. Default options consisted of automatic prebolus baseline selection to define the prebolus baseline and integration intervals and subsequent noise threshold adjustment to maximize brain tissue used for CBV calculation. We did not use spatial or temporal smoothing for either software package, to help maintain data integrity and limit potential confounding factors. We performed rCBV calculations with γ variate fitting before leakage correction or without γ variate fitting. For IBN, we used all default options including leakage correction: 1) automated detection of brain tissue mask for voxels used in CBV calculation, 2) automated detection of contrast arrival within brain mask voxels to define the prebolus baseline and integration intervals, and 3) leakage correction based on Boxerman et al.17 For rCBV generated with either NICE or IBN, we coregistered the rCBV maps with stereotactic anatomic images by using registration methods implemented in the Insight Segmentation and Registration Toolkit (www.itk.org) within the IB Suite (Version 1.0.454; Imaging Biometrics), as previously described.17,18,29,30
We normalized all rCBV maps to mean CBV from two 3 × 3 voxel-sized square ROIs within the contralateral frontal and parietal normal-appearing white matter.8,19 To reduce variability, we used identical normal-appearing white matter ROIs for both software package analyses to generate all rCBV metrics. We calculated multiple previously published rCBV metrics including the following: 1) volume fraction of tumor voxels above the rCBV threshold of 1.75 (percentage >1.75); 2) volume fraction of tumor voxels above the rCBV threshold of 1.0, also known as perfusion MR imaging fractional tumor burden (FTB); 3) histogram mean rCBV; and 4) histogram mode rCBV for all tumor voxels. We chose the thresholds of 1.0 and 1.75 because of previous studies reporting the biologic significance of these values.6,8,30 On the basis of the rCBV maps generated from NICE and IBN packages, we calculated volume fraction metrics by using the IB Suite and histogram metrics by using custom code written in Matlab (Version R2012a; MathWorks, Natick, Massachusetts). To reduce variability, we also used identical segmented enhancing tumor volumes for both software analyses and all rCBV metrics (as described below).
Conventional MR Imaging Acquisition and Analysis
For each examination, we acquired routine conventional contrast-enhanced MR imaging that included pre- and postcontrast T1-weighted spoiled gradient-echo (inversion recovery prepped) stereotactic (ie, volumetric) MR imaging datasets (TI/TR/TE = 300/6.8/2.8 ms, matrix = 320 × 224, FOV = 26 cm, section thickness = 2 mm). Tumor volumes were defined as abnormal enhancing tissue by an experienced neuroradiologist (L.S.H.). In nonenhancing glioma, we defined tumor volumes by using T2-weighted stereotactic MR imaging (TR/TE = 4500/82 ms, matrix = 256 × 256, FOV = 26 cm, section thickness = 2 mm).
Stereotactic Biopsy, Image Coregistration, and Histologic Microvessel Analysis
Our cohort included a subset of patients in whom neurosurgeons collected an average of 2–3 tissue specimens from each tumor by using stereotactic surgical localization, following the smallest possible diameter craniotomies to minimize brain shift. Biopsies were performed without knowledge of rCBV analyses. Similar to those in previous studies, biopsy locations and neuronavigational coordinates were recorded and coregistered with MR imaging to enable localized rCBV measurement (3 × 3 voxel-sized ROIs) at corresponding biopsy sites.11,31 Multiple biopsy targets in the same patient were separated by a minimum of 2 cm. The neurosurgeon visually validated stereotactic imaging locations with corresponding intracranial anatomic landmarks, such as vascular structures. Stereotactic biopsy samples were sectioned (10-μm thickness), CD-34 stained, and submitted for quantification of total microvessel area (MVA) by using previously published methods.31–34 Corresponding sections were also stained with hematoxylin-eosin per standard protocol. For each CD-34-stained slide, we measured total microvessel area as previously described.31,32,35 Raw data from 7 of these patients were studied previously.31 The current study differs in the following ways: 1) We used commercial software packages and modeling correction to measure rCBV, 2) we determined test performance differences between packages, and 3) we compared PLD against non-PLD conditions.
Quantification of Histologic Tumor Fraction in Recurrent Glioblastoma Multiforme
Our cohort included a subset of 25 patients with recurrent glioblastoma multiforme, previously treated with the protocol of Stupp et al.36 We enrolled each of these patients at the time of recurrence, at which time they underwent preoperative MR imaging (including pMRI) for surgical debulking of newly developed or enlarging lesions suspicious for recurrence identified on surveillance contrast-enhanced MR imaging.
Following debulking, we fixed all surgical tissue specimens in 10% formalin, embedded them in paraffin, sectioned them (10 μm), and stained them with hematoxylin-eosin per standard diagnostic protocol at our institution. Two neuropathologists quantified glioblastoma multiforme and/or posttreatment effect elements for all specimens without knowledge of DSC-MR imaging, by simultaneously estimating histologic fractional volume of tumor relative to nonneoplastic treatment-related features, as previously described.8,30,37,38 Features of tumor recurrence38 and posttreatment effect37,39 were quantified and used to determine the histologic tumor fraction from surgical resection material to diagnose either tumor progression (histologic tumor fraction of ≥50%) or posttreatment effect (histologic tumor fraction of <50%) on the basis of group median values. Raw data from these 25 patients have been studied previously.8 Like the prior study, the current study measures FTB but with several important differences in experimental design: 1) We used and compared 2 separate modeling algorithm implementations to calculate FTB, 2) we assessed performance differences between methods by comparing test accuracies (with receiver operating characteristic analysis), and 3) we use a simplified classification system to establish the clinical presence/absence of tumor progression.
Statistical Analysis
A biostatistician performed all analyses. We first determined Pearson and Spearman correlations between non-PLD- and PLD-corrected conditions for all rCBV metrics as calculated by IBN and NICE. Second, we used receiver operating characteristic analysis to determine the accuracy of FTB (as measured by IBN and NICE) to diagnose tumor versus posttreatment effect. Finally, we determined Pearson and Spearman correlations between localized rCBV and MVA from corresponding stereotactic biopsies. P < .05 was statistically significant.
Results
Subjects and Tumor Types
We enrolled 52 patients (17 women, 35 men; mean age, 53 years), of whom 87% (45/52) had high-grade gliomas with 78% (35/45) presenting at recurrence after standard multimodal therapy. On-line Table 2 summarizes the tumor types for primary and recurrent cases.
Comparing rCBV Measurements in the Presence and Absence of Preload Dose
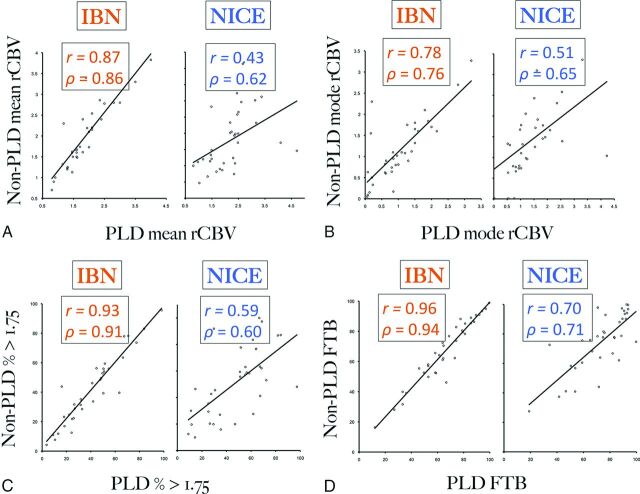
Comparing rCBV between PLD and non-PLD conditions gives an indication of how well modeling implementation corrects T1 leakage errors. We acquired both PLD- and non-PLD-corrected rCBV values in a subset of patients (n = 36) for whom we calculated 4 separate rCBV metrics (mean, mode, percentage >1.75, and FTB) by using both IB Neuro and nordicICE software packages. When we used IBN (Fig 1), rCBV thresholding metrics correlated very highly between non-PLD- and PLD-corrected conditions (FTB: r = 0.96, ρ = 0.94; percentage > 1.75: r = 0.93, ρ = 0.91); correlations were also high for mean rCBV (r = 0.87, ρ = 0.86) and mode rCBV (r = 0.78, ρ = 0.76). With NICE modeling, these correlations dropped substantially (Fig 1) for thresholding metrics (FTB: r = 0.70, ρ = 0.71; percentage > 1.75: r = 0.59, ρ = 0.60), mean rCBV (r = 0.43, ρ = 0.62), and mode rCBV (r = 0.51, ρ = 0.65). When we added γ variate fitting, correlations for mean rCBV by using NICE decreased though the other metrics remained largely unchanged (Table 1). On visual inspection of thresholding maps, non-PLD and PLD-corrected voxels showed greater spatial correspondence when using IBN compared with NICE (Fig 2). Table 1 summarizes correlations for all conditions. Of these 36 patients, 10 received PLD via 2 separate half-dose injections. To assess the potential effects of heterogeneity in PLD administration, we performed a subanalysis (n = 26) excluding these 10 subjects, which showed correlations consistent with the original analysis (On-line Table 1).
Fig 1.
A–D, Scatterplots correlating rCBV metrics with and without preload dosing (PLD), as measured by 2 separate modeling algorithms (IBN, NICE without γ variate fitting). PLD- and non-PLD corrected values are shown in the x- and y-axes, respectively. Overall, IBN measurements demonstrate consistently higher Pearson (r) and Spearman (ρ) correlations for mean rCBV, mode rCBV, fractional tumor burden (FTB), and percentage of voxels > 1.75. The thresholding metrics (FTB, percentage > 1.75) correlate most strongly between PLD- and non-PLD-corrected conditions.
Table 1:
Pearson (r) and Spearman (ρ) correlations between rCBV metrics under PLD-corrected and non-PLD-corrected conditions, as measured by IBN and NICE perfusion software algorithmsa
| rCBV Metric | Non-PLD vs PLD (IBN) | P Value | Non-PLD vs PLD (NICE) with gvf | P Value | Non-PLD vs PLD (NICE) without gvf | P Value |
|---|---|---|---|---|---|---|
| Mean | r = 0.87 | <.001 | r = 0.11 | .54 | r = 0.43 | .01 |
| ρ = 0.86 | <.001 | ρ = 0.42 | .02 | ρ = 0.62 | <.001 | |
| Mode | r = 0.78 | <.001 | r = 0.44 | .01 | r = 0.51 | .01 |
| ρ = 0.76 | <.001 | ρ = 0.65 | <.001 | ρ = 0.65 | <.001 | |
| % < 1.75 | r = 0.93 | <.001 | r = 0.55 | <.001 | r = 0.59 | <.001 |
| ρ = 0.91 | <.001 | ρ = 0.61 | <.001 | ρ = 0.60 | <.001 | |
| FTB | r = 0.96 | <.001 | r = 0.79 | <.001 | r = 0.70 | <.001 |
| ρ = 0.94 | <.001 | ρ = 0.72 | <.001 | ρ = 0.71 | <.001 |
Note:—gvf indicates γ variate fitting.
NICE calculations were performed with and without gvf. IBN modeling shows substantially higher correlation between PLD and non-PLD metrics (compared with NICE), suggesting higher rCBV accuracy in the absence of PLD correction. Statistical significance is P value < .05.
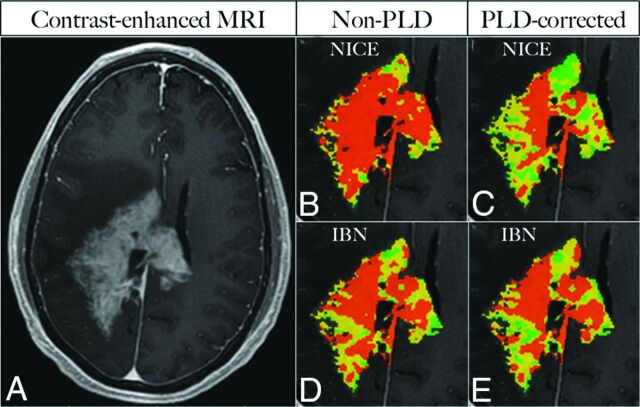
Fig 2.
Image of a representative case in a 39-year-old patient with recurrent high-grade ganglioglioma shows an enhancing mass (A). Color overlay percentage > 1.75 thresholding maps (B–E) depict orange voxels with high rCBV > 1.75, compared with intermediate yellow voxels (rCBV, 1.0–1.75) and low green voxels (rCBV < 1.0). With NICE, both spatial distribution and percentage of orange voxels show high discrepancy between non-PLD- (70%, B) and PLD-corrected (35%, C) maps. With IBN, the percentage of orange voxels on the non-PLD map (54%, D) approximates that on the PLD-corrected map (51%, E) with high spatial congruence.
The Type of Modeling Implementation Impacts the Accuracy of rCBV to Diagnose Tumor versus Pseudoprogression/Radiation Necrosis
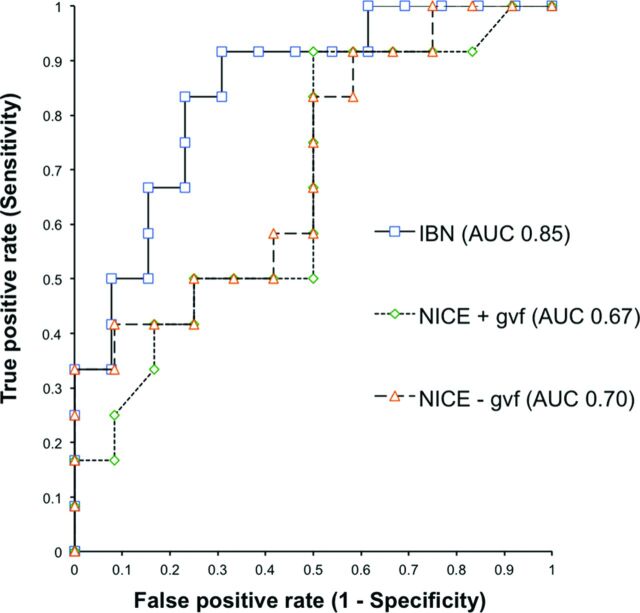
In a subset of patients with recurrent glioblastoma multiforme (n = 25) undergoing surgical debulking for suspected tumor recurrence, we used receiver operating characteristic analysis to determine the accuracy of FTB, as measured by IBN or NICE, to diagnosed tumor versus posttreatment effect (ie, pseudoprogression, radiation necrosis). We used histologic tumor fraction from surgical resection to categorize each subject's diagnosis as either tumor recurrence (histologic tumor fraction of ≥50%) or posttreatment effect (histologic tumor fraction of <50%). We used PLD correction for all cases. The area under the curve for FTB, as measured by IBN (0.85), was significantly higher than that by NICE (0.67; P < .01) (Fig 3).
Fig 3.
Receiver operating characteristic curves for fractional tumor burden to predict histopathology (tumor versus posttreatment effect) in patients with recurrent glioblastoma multiforme (n = 25). FTB by IB Neuro (blue) demonstrates a significantly larger area under the curve (AUC) compared with nordicICE (without γ variate fitting, orange) FTB measurements (0.85 versus 0.70, P < .01), suggesting that different modeling algorithms can impact the accuracy in predicting histopathologic diagnosis. Adding γ variate fitting further reduces NICE estimates of FTB (0.67, green).
Influence of PLD and Modeling Correction on the Correlation of rCBV with Stereotactic Microvessel Area Quantification
We measured localized rCBV values corresponding to coregistered stereotactic biopsy samples (n = 29) in a subset of patients (n = 12). We determined Spearman and Pearson correlations between matched rCBV and histologic microvessel area measurements under multiple conditions, which varied by method of modeling correction or the presence/absence of PLD correction (Table 2). Both PLD correction and IBN modeling were needed to maximize rCBV correlations with MVA (r = 0.64, ρ = 0.58, P = .001).
Table 2:
Correlations between rCBV and fractional MVA under different PLD and modeling conditionsa
| Conditions for rCBV Measurement | Pearson Correlation (r) | P Value | Spearman Correlation (ρ) | P Value |
|---|---|---|---|---|
| Fractional MVA | 1.00 | – | 1.00 | – |
| No PLD (IBN) | 0.46 | .02 | 0.33 | .12 |
| No PLD (NICE + gvf) | 0.51 | .01 | 0.26 | .19 |
| No PLD (NICE − gvf) | 0.35 | .10 | 0.18 | .39 |
| PLD (IBN) | 0.64 | <.001 | 0.58 | .001 |
| PLD (NICE + gvf) | 0.53 | <.01 | 0.28 | .15 |
| PLD (NICE − gvf) | 0.59 | .001 | 0.40 | .04 |
Note:—+ indicates with; −, without; –, not applicable; gvf, γ variate fitting.
Both PLD correction and IBN software modeling were needed to achieve maximal correlation.
Discussion
Relative CBV represents the most robust and widely used perfusion MR imaging metric in neuro-oncology.1–31,40–46 Of the techniques that measure rCBV, DSC is the most commonly used method because of wide availability, straightforward postprocessing, and easy-to-use software programs.40,41 DSC uses the indicator dilution theory based on susceptibility (T2-/T2*-weighted signal drop) from first-pass transit of a single GBCA bolus injection. DSC assumes an intact BBB with no extravascular GBCA leakage or recirculation and thus requires correction methods when these factors occur (discussed below). Dynamic contrast-enhancement MRI and arterial spin-labeling offer alternative approaches to DSC for calculation of rCBV. The theory and limitations of these techniques have been described previously.23,24,40–42
Correctly performing DSC requires several technical considerations based on comparison data from prior studies validating optimal conditions for best practice. First, DSC-pMRI generally necessitates both PLD and mathematic modeling to achieve the highest degree of T1 leakage correction and rCBV accuracy.17–19 Results from our study support this requirement (Table 2). Regarding PLD amount, most groups use a single dose (0.1 mmol/kg) of GBCA,8,9,14–20,22,25,26,42 particularly at 1.5T, though adequate PLD correction could be achieved with a GBCA dose as low as 0.05 mmol/kg at 3T.19 Second, gradient-echo T2*-weighted DSC represents the most preferred and widely published method for DSC. While spin-echo T2-weighted DSC offers a higher signal-to-noise ratio and fewer susceptibility artifacts,45 double or triple GBCA injection doses (0.2–0.3 mmol/kg) are typically needed during the acquisition of spin-echo DSC,2,7,30 to overcome the lower contrast-to-noise ratio (ie, signal drop in response to the GBCA first-pass bolus). Compared with spin-echo, gradient-echo DSC offers advantages such as the following: 1) superior contrast-to-noise ratio (ie, greater signal drop during GBCA first-pass), which allows lower contrast dosage during DSC acquisition (0.05–0.1 mmol/kg) and improves the quality of rCBV data, minimizing the need for signal denoising; 2) greater sensitivity to microvessels of all sizes (including larger tortuous glomeruloid-type vessels commonly observed in high-grade gliomas); and 3) the ability to use flip angles of <90° to minimize T1 leakage effects.11,19,31,42,44,45 Finally, in regard to mathematic modeling, the algorithm published by Boxerman et al17 remains the most highly cited and validated method to date and has been implemented commercially for widespread use.
The study results here underscore the importance of how software programs implement a particular modeling algorithm for rCBV calculation. In this study, we tested 2 widely published FDA-approved commercial packages that offer separate implementations of the Boxerman method,8,9,14–21,24,26–28 and we evaluated how the modeling implementation by each software program would impact T1 leakage correction and rCBV correlation with histologic measures. We minimized potential confounding factors by using identical segmented tumor volumes and regions in normalized white matter to evaluate each implementation method, and we used default settings and leakage correction for both software packages. For NICE, these included automated selection of the prebolus baseline and subsequent noise thresholding to maximize brain tissue for calculation of CBV. The rCBV metrics on IBN (compared with NICE) demonstrated greater consistency between PLD and non-PLD conditions, most notably with mean rCBV (IBN: r = 0.87; NICE: r = 0.43) and percentage > 1.75 (IBN: r = 0.93; NICE: r = 0.59). This suggests that the modeling correction by IBN provides more effective correction of T1 errors, which are most prominent at non-PLD conditions.
While we observed strong correlations between non-PLD and PLD measures (when using IBN), further studies are likely needed to determine the following: 1) whether PLD can or should be omitted, 2) what the optimal conditions would be to allow PLD omission (ie, modeling implementation, 3T field strength), and 3) whether this omission would significantly impact prognostic and diagnostic accuracy. Under PLD conditions, separate experiments confirmed significantly higher FTB accuracy with IBN (area under the curve = 0.84), compared with NICE (area under the curve = 0.67, p < 0.01), in diagnosing histopathologically confirmed tumor versus posttreatment effect (ie, pseudoprogression, radiation necrosis). IBN also provided the highest degree of correlation between localized rCBV and tissue microvessel area (Table 2).
In this study, we chose to validate rCBV measurements against histopathology rather than outcomes. Imaging measurements such as rCBV are most directly related to histologic correlates such as microvessel volume and histologic identity (eg, tumor grade, tumor versus posttreatment effect). How these histologic features (and their imaging correlates) predict survival may be confounded by a number of different factors such as age, molecular markers (isocitrate dehydrogenase), methylation status (eg, O6-methylguanine-DNA methyltransferase), extent of resection, salvage therapy at the time of recurrence, and so forth.6–16,47,48 While clinical outcomes are desirable as end points, they must be correlated with imaging and histologic features together in a controlled trial with a larger patient cohort, which is beyond the scope of this article. Our purpose in this study was simply to determine which method of rCBV measurement (ie, which software package) came closest to informing of underlying tissue features. We think that this context justifies the rationale for validating rCBV against histopathologic benchmarks.
We recognize potential study limitations. First, we limited the scope of the evaluation to 2 specific software packages, though many commercial options exist. We simplified the project to maximize the potential clinical impact because we evaluated the most published and validated modeling algorithm to date (Boxerman et al17) and the 2 most commonly published commercial platforms that implement that algorithm. Future studies may incorporate other vendor packages in a more comprehensive fashion. In fact, the study results here may motivate the development of a framework by which to standardize or evaluate modeling implementation for all vendors. Second, we did not evaluate the factors underlying the differences in modeling implementation because the software code was not made available to us for analysis or modification. Regardless, this comparison study simulates what would be available to end users within the clinical environment and demonstrates that differences between programs can significantly impact rCBV accuracy. Future efforts to develop or use open-source software may help elucidate some of the differences among commercial packages. Third, nonuniformity in PLD administration (single-dose bolus versus 2 separate half-dose injections) may theoretically degrade correlations between non-PLD- and PLD-corrected rCBV. However, strong correlations within the full cohort and subanalysis (On-line Table 1) suggest negligible impact. Fourth, while we used 2 different MR imaging scanners (Signa HDx; Magnetom Skyra), all MR imaging–histologic correlations came from 1 scanner (Signa HDx). Moreover, both scanners used identical field strengths (3T) and DSC parameters (ie, pulse sequence, injection rates, and so forth). These factors, along with strong correlations in a subanalysis (On-line Table 3), suggest a negligible impact on study results. Fifth, the observed strength of rCBV-MVA correlations in this study (at best r = 0.64) may be underestimated because Pathak et al46 have shown that correlations between rCBV and histologic vascular fraction can be further improved when accounting for histologic section thickness as a potential confound. Sixth, there was variability in the TR (1.5 versus 2.0 seconds) of the DSC acquisitions for some patients, depending on the clinical protocol used at the time of imaging. While we do not anticipate this having a significant effect on correlations, we recognize it as a potential limitation. Finally, nonuniformity of the GBCA type (gadodiamide or gadobenate dimeglumine) resulted from protocol changes during the study. Subanalysis (On-line Table 4) based on GBCA type suggested negligible effects.
Conclusions
Different implementations of perfusion MR imaging software modeling can impact the accuracy of leakage correction, rCBV calculation, and correlations with histologic benchmarks. Future decisions about pMRI standardization should incorporate comparison data that have validated rCBV measurements against clinical benchmarks such as histopathology.
Supplementary Material
ABBREVIATIONS:
- FTB
fractional tumor burden
- GBCA
gadolinium-based contrast agents
- IBN
IB Neuro
- MVA
microvessel area
- NICE
nordicICE
- PLD
preload dose
- pMRI
perfusion MR imaging
- rCBV
relative cerebral blood volume
Footnotes
Disclosures: Leland S. Hu, Zachary Kelm, Leslie C. Baxter—RELATED: Grant: National Institutes of Health.* Timothy J. Kaufmann—UNRELATED: Consultancy: SpineThera, Comments: startup company hoping to create a sustained-release epidural corticosteroid preparation. Peter Nakaji—UNRELATED: Board Membership: Foundation for International Education for Neurological Surgery, Barrow Neurological Institute comanagement*; Consultancy: Zeiss, Aesculap, AlloSource; Grants/Grants Pending: Barrow Neurological Foundation*; Payment for Lectures (including service on Speakers Bureaus): Zeiss; Patents (planned, pending or issued): AlloSource, GammaTile, Incubeon; Stock/Stock Options: Microfabrica. Bradley J. Erickson, P. Korfiatis—RELATED: Grant: National Cancer Institute (1U01CA160045).* *Money paid to the institution.
This work was supported by the National Institutes of Health, NS 082609-01, CA 01-16045, and the Mayo Clinic Foundation.
REFERENCES
- 1. Fink JR, Carr RB, Matsusue E, et al. Comparison of 3 Tesla proton MR spectroscopy, MR perfusion and MR diffusion for distinguishing glioma recurrence from posttreatment effects. J Magn Reson Imaging 2012;35:56–63 10.1002/jmri.v35.1 [DOI] [PubMed] [Google Scholar]
- 2. Lev MH, Ozsunar Y, Henson JW, et al. Glial tumor grading and outcome prediction using dynamic spin-echo MR susceptibility mapping compared with conventional contrast-enhanced MR: confounding effect of elevated rCBV of oligodendrogliomas [corrected]. AJNR Am J Neuroradiol 2004;25:214–21; Erratum in: AJNR Am J Neuroradiol 2004;25:B1 [PMC free article] [PubMed] [Google Scholar]
- 3. Knopp EA, Cha S, Johnson G, et al. Glial neoplasms: dynamic contrast-enhanced T2*-weighted MR imaging. Radiology 1999;211:791–98 10.1148/radiology.211.3.r99jn46791 [DOI] [PubMed] [Google Scholar]
- 4. Cha S, Lupo JM, Chen MH, et al. Differentiation of glioblastoma multiforme and single brain metastasis by peak height and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. AJNR Am J Neuroradiol 2007;28:1078–84 10.3174/ajnr.A0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Law M, Yang S, Wang H, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol 2003;24:1989–98 [PMC free article] [PubMed] [Google Scholar]
- 6. Law M, Young RJ, Babb JS, et al. Gliomas: predicting time to progression or survival with cerebral blood volume measurements at dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2008;247:490–98 10.1148/radiol.2472070898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maia AC Jr, Malheiros SM, da Rocha AJ, et al. Stereotactic biopsy guidance in adults with supratentorial nonenhancing gliomas: role of perfusion-weighted magnetic resonance imaging. J Neurosurg 2004;101:970–76 10.3171/jns.2004.101.6.0970 [DOI] [PubMed] [Google Scholar]
- 8. Hu LS, Eschbacher JM, Heiserman JE, et al. Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro Oncol 2012;14:919–30 10.1093/neuonc/nos112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gahramanov S, Muldoon LL, Varallyay CG, et al. Pseudoprogression of glioblastoma after chemo- and radiation therapy: diagnosis by using dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging with ferumoxytol versus gadoteridol and correlation with survival. Radiology 2013;266:842–52 10.1148/radiol.12111472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barajas RF Jr, Chang JS, Segal MR, et al. Differentiation of recurrent glioblastoma multiforme from radiation necrosis after external beam radiation therapy with dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2009;253:486–96 10.1148/radiol.2532090007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu LS, Baxter LC, Smith KA, et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. AJNR Am J Neuroradiol 2009;30:552–58 10.3174/ajnr.A1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sawlani RN, Raizer J, Horowitz SW, et al. Glioblastoma: a method for predicting response to antiangiogenic chemotherapy by using MR perfusion imaging—pilot study. Radiology 2010;255:622–28 10.1148/radiol.10091341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galbán CJ, Chenevert TL, Meyer CR, et al. Prospective analysis of parametric response map-derived MRI biomarkers: identification of early and distinct glioma response patterns not predicted by standard radiographic assessment. Clin Cancer Res 2011;17:4751–60 10.1158/1078-0432.CCR-10-2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. LaViolette PS, Cohen AD, Prah MA, et al. Vascular change measured with independent component analysis of dynamic susceptibility contrast MRI predicts bevacizumab response in high-grade glioma. Neuro Oncol 2013;15:442–50 10.1093/neuonc/nos323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmainda KM, Prah M, Connelly J, et al. Dynamic-susceptibility contrast agent MRI measures of relative cerebral blood volume predict response to bevacizumab in recurrent high-grade glioma. Neuro Oncol 2014;16:880–88 10.1093/neuonc/not216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmainda KM, Zhang Z, Prah M, et al. Dynamic susceptibility contrast MRI measures of relative cerebral blood volume as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 multicenter trial. Neuro Oncol 2015;17:1148–56. 10.1093/neuonc/nou364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boxerman JL, Schmainda KM, Weisskoff RM. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol 2006;27:859–67 [PMC free article] [PubMed] [Google Scholar]
- 18. Paulson ES, Schmainda KM. Comparison of dynamic susceptibility-weighted contrast-enhanced MR methods: recommendations for measuring relative cerebral blood volume in brain tumors. Radiology 2008;249:601–13 10.1148/radiol.2492071659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hu LS, Baxter LC, Pinnaduwage DS, et al. Optimized preload leakage-correction methods to improve the diagnostic accuracy of dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging in posttreatment gliomas. AJNR Am J Neuroradiol 2010;31:40–48 10.3174/ajnr.A1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emblem KE, Bjornerud A, Mouridsen K, et al. T(1)- and T(2)(*)-dominant extravasation correction in DSC-MRI, Part II: predicting patient outcome after a single dose of cediranib in recurrent glioblastoma patients. J Cereb Blood Flow Metab 2011;31:2054–64 10.1038/jcbfm.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu HL, Wu YY, Yang WS, et al. Is Weisskoff model valid for the correction of contrast agent extravasation with combined T1 and T2* effects in dynamic susceptibility contrast MRI? Med Phys 2011;38:802–09 10.1118/1.3534197 [DOI] [PubMed] [Google Scholar]
- 22. Quarles CC, Gochberg DF, Gore JC, et al. A theoretical framework to model DSC-MRI data acquired in the presence of contrast agent extravasation. Phys Med Biol 2009;54:5749–66 10.1088/0031-9155/54/19/006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Law M, Young R, Babb J, et al. Comparing perfusion metrics obtained from a single compartment versus pharmacokinetic modeling methods using dynamic susceptibility contrast-enhanced perfusion MR imaging with glioma grade. AJNR Am J Neuroradiol 2006;27:1975–82 [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson G, Wetzel SG, Cha S, et al. Measuring blood volume and vascular transfer constant from dynamic, T(2)*-weighted contrast-enhanced MRI. Magn Reson Med 2004;51:961–68 10.1002/(ISSN)1522-2594 [DOI] [PubMed] [Google Scholar]
- 25. Batchelor TT, Sorensen AG, di Tomaso E, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell 2007;11:83–95 10.1016/j.ccr.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sorensen AG, Emblem KE, Polaskova P, et al. Increased survival of glioblastoma patients who respond to antiangiogenic therapy with elevated blood perfusion. Cancer Res 2012;72:402–07 10.1158/1538-7445.AM2012-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roder C, Bender B, Ritz R, et al. Intraoperative visualization of residual tumor: the role of perfusion-weighted imaging in a high-field intraoperative magnetic resonance scanner. Neurosurgery 2013;72(2 suppl operative):ons151–58; discussion ons158 [DOI] [PubMed] [Google Scholar]
- 28. Jain R, Poisson L, Narang J, et al. Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology 2013;267:212–20 10.1148/radiol.12120846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emblem KE, Scheie D, Due-Tonnessen P, et al. Histogram analysis of MR imaging-derived cerebral blood volume maps: combined glioma grading and identification of low-grade oligodendroglial subtypes. AJNR Am J Neuroradiol 2008;29:1664–70 10.3174/ajnr.A1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gasparetto EL, Pawlak MA, Patel SH, et al. Posttreatment recurrence of malignant brain neoplasm: accuracy of relative cerebral blood volume fraction in discriminating low from high malignant histologic volume fraction. Radiology 2009;250:887–96 10.1148/radiol.2502071444 [DOI] [PubMed] [Google Scholar]
- 31. Hu LS, Eschbacher JM, Dueck AC, et al. Correlations between perfusion MR imaging cerebral blood volume, microvessel quantification, and clinical outcome using stereotactic analysis in recurrent high-grade glioma. AJNR Am J Neuroradiol 2012;33:69–76 10.3174/ajnr.A2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma S, Sharma MC, Sarkar C. Morphology of angiogenesis in human cancer: a conceptual overview, histoprognostic perspective and significance of neoangiogenesis. Histopathology 2005;46:481–89 10.1111/his.2005.46.issue-5 [DOI] [PubMed] [Google Scholar]
- 33. Leon SP, Folkerth RD, Black PM. Microvessel density is a prognostic indicator for patients with astroglial brain tumors. Cancer 1996;77:362–72 [DOI] [PubMed] [Google Scholar]
- 34. Folkerth RD. Histologic measures of angiogenesis in human primary brain tumors. Cancer Treat Res 2004;117:79–95 10.1007/978-1-4419-8871-3 [DOI] [PubMed] [Google Scholar]
- 35. Wesseling P, van der Laak JA, de Leeuw H, et al. Quantitative immunohistological analysis of the microvasculature in untreated human glioblastoma multiforme: computer-assisted image analysis of whole-tumor sections. J Neurosurg 1994;81:902–09 10.3171/jns.1994.81.6.0902 [DOI] [PubMed] [Google Scholar]
- 36. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 37. Burger PC, Mahley MS Jr, Dudka L, Vogel FS. The morphologic effects of radiation administered therapeutically for intracranial gliomas: a postmortem study of 25 cases. Cancer 1979;44:1256–72 [DOI] [PubMed] [Google Scholar]
- 38. Forsyth PA, Kelly PJ, Cascino TL, et al. Radiation necrosis or glioma recurrence: is computer-assisted stereotactic biopsy useful? J Neurosurg 1995;82:436–44 10.3171/jns.1995.82.3.0436 [DOI] [PubMed] [Google Scholar]
- 39. Louis DN. WHO Classification of Tumors of the Central Nervous System. 4th ed. Lyon, France: International Agency for Research on Cancer, World Health Organization; 2007 [Google Scholar]
- 40. Essig M, Shiroishi MS, Nguyen TB, et al. Perfusion MRI: the five most frequently asked technical questions. AJR Am J Roentgenol 2013;200:24–34 10.2214/AJR.12.9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Falk A, Fahlström M, Rostrup E, et al. Discrimination between glioma grades II and III in suspected low-grade gliomas using dynamic contrast-enhanced and dynamic susceptibility contrast perfusion MR imaging: a histogram analysis approach. Neuroradiology 2014;56:1031–38 10.1007/s00234-014-1426-z [DOI] [PubMed] [Google Scholar]
- 42. Kassner A, Annesley DJ, Zhu XP, et al. Abnormalities of the contrast re-circulation phase in cerebral tumors demonstrated using dynamic susceptibility contrast-enhanced imaging: a possible marker of vascular tortuosity. J Magn Reson Imaging 2000;11:103–13 10.1002/(SICI)1522-2586(200002)11:23.0.CO;2-Z [DOI] [PubMed] [Google Scholar]
- 43. Young GS, Setayesh K. Spin-echo echo-planar perfusion MR imaging in the differential diagnosis of solitary enhancing brain lesions: distinguishing solitary metastases from primary glioma. AJNR Am J Neuroradiol 2009;30:575–77 10.3174/ajnr.A1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sugahara T, Korogi Y, Kochi M, et al. Perfusion-sensitive MR imaging of gliomas: comparison between gradient-echo and spin-echo echo-planar imaging techniques. AJNR Am J Neuroradiol 2001;22:1306–15 [PMC free article] [PubMed] [Google Scholar]
- 45. Schmainda KM, Rand SD, Joseph AM, et al. Characterization of a first-pass gradient-echo spin-echo method to predict brain tumor grade and angiogenesis. AJNR Am J Neuroradiol 2004;25:1524–32 [PMC free article] [PubMed] [Google Scholar]
- 46. Pathak AP, Schmainda KM, Ward BD, et al. MR-derived cerebral blood volume maps: issues regarding histological validation and assessment of tumor angiogenesis. Magn Reson Med 2001;46:735–47 10.1002/mrm.1252 [DOI] [PubMed] [Google Scholar]
- 47. Thuy MN, Kam JK, Lee GC, et al. A novel literature-based approach to identify genetic and molecular predictors of survival in glioblastoma multiforme: analysis of 14,678 patients using systematic review and meta-analytical tools. J Clin Neurosci 2015;22:785–99 10.1016/j.jocn.2014.10.029 [DOI] [PubMed] [Google Scholar]
- 48. Sanai N, Polley MY, McDermott MW, et al. An extent of resection threshold for newly diagnosed glioblastomas. J Neurosurg 2011;115:3–8 10.3171/2011.2.JNS10998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.