Abstract
Altered nitric oxide (•NO) metabolism underlies cancer pathology, but mechanisms explaining many •NO-associated phenotypes remain unclear. We have found that cellular exposure to •NO changes histone posttranslational modifications (PTMs) by directly inhibiting the catalytic activity of JmjC-domain containing histone demethylases. Herein, we describe how •NO exposure links modulation of histone PTMs to gene expression changes that promote oncogenesis. Through high-resolution mass spectrometry, we generated an extensive map of •NO-mediated histone PTM changes at 15 critical lysine residues on the core histones H3 and H4. Concomitant microarray analysis demonstrated that exposure to physiologic •NO resulted in the differential expression of over 6,500 genes in breast cancer cells. Measurements of the association of H3K9me2 and H3K9ac across genomic loci revealed that differential distribution of these particular PTMs correlated with changes in the level of expression of numerous oncogenes, consistent with epigenetic code. Our results establish that •NO functions as an epigenetic regulator of gene expression mediated by changes in histone PTMs.
Keywords: Nitric oxide, Epigenetics, Cancer, Histone posttranslational modifications, ChIP-sequencing
Introduction
It is increasingly clear that aberrant epigenetic modifications play major roles in cancer progression (1). Recent pre-clinical and clinical research has provided evidence that tumor-associated nitric oxide (•NO, nitrogen monoxide) production and nitric oxide synthase (NOS) expression are associated with the development and progression of various cancers (2–6). Despite these correlations, however, current signaling mechanisms of •NO are insufficient to explain a significant proportion of its bioactivity in cancer. Histone posttranslational modifications (PTMs) play a major role in the organization of chromatin structure and subsequent regulation of gene expression. The number of unique histone PTMs being discovered is continually increasing, however, methylation and acetylation of lysine residues on the core histones remain among the most well-studied. While lysine (K) acetylation (ac) is almost exclusively associated with increased gene expression, methylation (me) can either increase or decrease gene expression depending on the degree of methylation (mono, di, tri) as well as its location and proximity to other modifications (7).
The notion that •NO can directly affect the epigenetic landscape is novel. Most studies that have elucidated epigenetic functions of •NO describe indirect mechanisms of action including regulation of NOS isoform expression and activity (8,9), regulation of transcription factor activities via S-nitrosation (10) or regulation of the expression or functions of histone modifying enzymes (11–14). Previous studies have also demonstrated regulation of microRNAs by •NO as an important disease related epigenetic event (15). Our recent findings, however, provided a direct mechanistic link between cellularly-derived •NO and significant changes in histone PTMs by demonstrating its ability to inhibit the catalytic activity of JmjC-domain containing histone demethylases (16). Although these studies demonstrated a key role for •NO as an endogenously produced regulator of global histone PTM levels, it remains to be understood whether •NO-mediated changes in gene expression could be attributed to specific alterations in the histone code.
Herein, we provide a comprehensive description of the magnitude of histone PTM changes on the core histones H3 & H4 in response to •NO. Critically modified lysine residues were identified using high-resolution mass spectrometry. ChIP-sequencing of key histone PTMs revealed characteristic patterns of their association at specific genomic loci, which correlated to changes in gene expression, transcription factor binding, and phenotypic responses (proliferation and migration). These results introduce a novel signaling mechanism of •NO, which is distinctly different from currently well-accepted models: i.e. direct interactions of •NO with heme proteins such as guanylyl cyclase (sGC), or via the formation of higher nitrogen oxides that form protein adducts containing nitrogen oxide functional groups (i.e. S-nitrosothiols) (17,18). These results, which force us to rethink classical •NO signaling paradigms are discussed in the context of cancer biology.
Materials and Methods
Cell Culture
MDA-MB-231 human triple negative breast cancer cells were obtained from American Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin in a controlled environment (37°C and 5% CO2). Cell line authentication was performed by Short Tandem Repeat (STR) profiling at the University of Illinois Research Resource Center before use. Prior to experiments, cells were grown to 80% confluency and serum starved overnight prior to treatment with DETA/NO.
Western blotting
Histones were acid extracted from cultured cells and incubated overnight with either H3K9me2 (Cell Signaling #4658) or H3K9ac (Cell Signaling #9649) primary antibody prior to imaging (Supplementary Materials and Methods).
Quantitative mass spectrometry analysis of histone PTMs
Isolated histone proteins were derivatized by propionic anhydride and digested with trypsin as described previously (19). Peptides were then derivatized by propionic anhydride and desalted by C18 Stage-tips. Histone peptides were loaded to a 75 μm I.D. × 15 cm fused silica capillary column packed with Reprosil-Pur C18-AQ resin (3 μm; Dr. Maisch GmbH, Germany), and resolved by an EASY-nLC 1000 HPLC system (Thermo Scientific, Odense, Denmark). The HPLC gradient was 2–35% solvent B (A = 0.1% formic acid in water; B = 0.1% formic acid in acetonitrile) in 40 min and from 35% to 98% solvent B in 20 minutes at a flow-rate of 300 nL/min. HPLC was coupled to an LTQ-Orbitrap Elite (Thermo Fisher Scientific, Bremen, Germany). Full MS spectrum (m/z 290–1400) was performed in the Orbitrap with a resolution of 60,000 (at 400 m/z), and the 10 most intense ions were selected for MS/MS performed with collision-induced dissociation (CID) with normalized collision energy of 35 in the ion trap. AGC targets of full MS and MS/MS scans are 1×106 and 1×104, respectively. Precursor ion charge state screening was enabled and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 30 s. Lock mass calibration in full MS scan is implemented using polysiloxane ion 371.10123. Histone peptide abundances were calculated from the acquired raw data by EpiProfile program (in revision).
Microarrays
RNA was extracted using the RNAqueous 4PCR kit (Ambion, for mRNA) or the miRNeasy kit (Qiagen, for microRNA) and prepared for hybridization onto Affymetrix GeneChip® PrimeView™ Human Gene Expression Arrays (for mRNA) or Affymetrix GeneChip® miRNA 4.0 Arrays (for microRNA). mRNA and microRNA samples were labeled using the Affymetrix GeneChip® 3′ IVT Expression Kit and the Affymetrix FlashTag™ Biotin HSR RNA Labeling Kit respectively as per manufacturer’s protocols. The GeneChip® Hybridization, Wash, and Stain Kit was used to prepare samples for hybridization as per manufacturer’s protocols. The GeneChip® Fluidics Station 450 was used for washing and staining. Microarray chips were read on the GeneChip® Scanner 3000 7G with AGCC version 4.0.0.1567. RNA quality was determined using the Agilent 2100 Bioanalyzer. Data has been deposited online in the GEO database and is in accordance with prescribed MIAME guidelines (Accession number: GSE66324).
Analysis of gene expression data from microarrays
Raw microarray data was normalized by RMA using the affy package in R (20). Differential expression between treated and untreated conditions was assessed using a t-test.
Real-time cell proliferation and migration assays
The xCELLigence® DP system (ACEA Biosciences Inc.) was used to measure the proliferative and migratory potential of MDA-MB-231 cells (+/− 250 μM DETA/NO) (Supplementary Materials and Methods).
Chromatin Immunoprecipitation sequencing (ChIP-seq)
Chromatin immunoprecipitation and subsequent library preparation was performed as described in Supplementary Materials and Methods. Briefly, MDA-MB-231 cells in culture were crosslinked, lysed and sheared to 200–500 bp by sonication. Chromatin from 2 million cells was used for each ChIP reaction. Immunoprecipitation was performed by incubating diluted chromatin with Dynabeads G (Invitrogen, pre-washed with BSA/PBS) and the following antibodies overnight at 4°C: H3K9me2 (Abcam, ab1220; 2 μg per ChIP) or H3K9ac (Millipore #07-352; 3.5 μl per ChIP). DNA was purified using the MinElute PCR Purification Kit (Qiagen). Input and ChIP DNA libraries were made using the Diagenode MicroPlex Library Preparation™ kit and sequenced on the Illumina HiSeq 2000. Raw sequencing data has been deposited online in the GEO database (Accession number: GSE66324). Raw reads were trimmed for sequence quality, aligned to human reference genome hg19, and alignments were filtered for quality and uniqueness prior to peak calling. Peak calls for H3K9ac and H3K9me2 data were created with MACS2 and SICER algorithms respectively.
Gene Ontology analysis
Gene Ontology analysis was conducted using the Database for Annotation, Visualization and Integrated Discovery (DAVID) web tool (21). Significant terms with FDR<5% were selected for visualization using bar plots or the Enrichment Map (22) plugin for Cytoscape version 3.2.0 for network analysis (23) (www.cytoscape.org/).
Analysis of transcription factor binding motifs
Upregulated genes associated with a new •NO-induced promoter H3K9ac peak were analyzed for the presence of motifs from the JASPAR core database (24) using CLOVER (25). Sequences from the promoters of upregulated genes (TSS +/− 2 kb) were used as the query sequence, and sequences from all annotated promoters in hg19 (UCSC refseq annotation) were used as background for enrichment testing. Motifs with a p-value < 0.01 were considered enriched.
Results
Nitric oxide alters global levels of posttranslational modifications (PTMs) at numerous sites on core histones
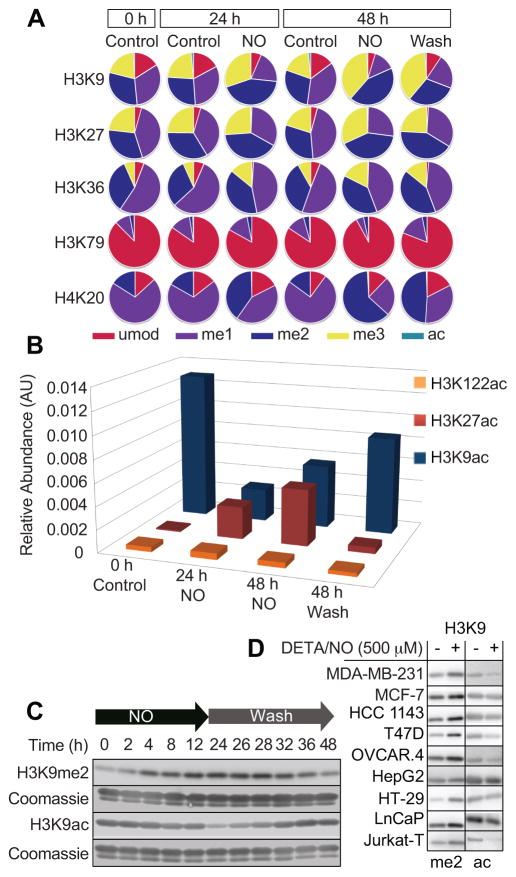
Previously, we elucidated several mechanisms by which physiological and pathological amounts of •NO (~5 – 400 nM) altered histone methylation patterns at key lysine residues (16). In addition to methylation, however, it is well-known that lysines can be modified by acetylation. To comprehensively characterize the full magnitude of PTM alterations in response to •NO, we quantified simultaneous changes in acetylation and methylation at critical lysine residues on the core histones H3 and H4. This was accomplished by exposing MDA-MB-231 triple negative breast cancer cells to a continuous, steady-state, physiologic •NO concentration for either 24 or 48 h (500 μM of the •NO-donor DETA/NO, ~250 nM [•NO]ss) (16). To determine whether these modifications were reversible, we examined the permanence of •NO-mediated changes in histone PTMs following removal of the •NO source. For this group, cells were treated with •NO for 24 hours, the •NO source was removed (wash), and then maintained in culture for an additional 24 hours (48 h total). Following these treatments, histones were isolated and PTMs were identified and quantified by high-resolution mass spectrometry. We focused on lysines 9, 27, 36, 79, and 122 on histone H3 and lysine 20 on histone H4, as acetylation and methylation of these residues are all known to have important gene regulatory functions (Fig. 1A, B) (26,27). All lysine residues were substantially modified in response to •NO and we noted significant changes in the majority of the 38 PTMs that were analyzed (Fig. S1A, B).
Figure 1. Nitric oxide changes histone posttranslational modifications at numerous sites on core histones.
All cells were treated with the •NO-donor DETA/NO (500 μM) and histones were isolated for PTM analysis. (A, B) Relative changes in PTMs at key sites on the core histones H3 and H4 measured by high-resolution mass spectrometry. MDA-MB-231 cells were cultured in the presence or absence of •NO and histones were isolated at the indicated time points (0, 24, 48 h). The “wash” column shows data collected from cells that were treated with •NO for 24 h, followed by 24 h of incubation in •NO-free media (histones were collected 48 h after initial treatment). Significant differences and standard error of mean are provided in Fig. S1 and Table S1. (C) Temporal changes at H3K9me2/ac in response to •NO. MDA-MB-231 cells were exposed to •NO for 24 h, followed by 24 h of incubation in •NO-free media. At the indicated time points, histones were isolated and immunoblots were conducted for H3K9me2/ac. (D) •NO-mediated changes in H3K9me2/ac in 9 different cell types. All cells were exposed to •NO for 24 hours, histones were extracted and immunoblotted for H3K9me2/ac.
Quantitative analysis of these data revealed that methyl modifications of lysine residues were far more abundant (7–99%) than acetyl modifications (0–3%), both before and after •NO exposure (Fig. 1A and Fig. S1A, B). This was true for all lysines examined with the exception of H3K122, where methylation could not be detected under any condition. Nevertheless, changes in global methylation and acetylation in response to •NO were similar in magnitude (~2 fold). In some cases, however, changes in acetylation (~20 fold) far exceeded changes in methylation (~2 fold). Substantial changes in methylation could be seen following •NO exposure at several lysine residues known to have critical gene regulatory functions (H3K9me2 +63%, H3K27me2 +24%, H3K36me3 +100%, H4K20me3 +267%) (27). Additionally, the levels of some methyl modifications persisted long after the •NO source was removed (H3K27me2, 48 h •NO vs. Wash, Fig. 1A, Fig. S1B), whereas at other lysine residues basal levels were restored within 24 hours (H3K9me2, 48 h •NO vs. Wash, Fig. 1A, Fig. S1B).
Similarly, the direction and extent of acetylation changes at H3K9, H3K27 and H3K122 were significantly altered following •NO exposure (Fig. 1B, Fig. S1A, B and Table S1). While •NO decreased acetylation at H3K9 (−76%), the opposite effect was seen at H3K27 (+2,118%). At certain lysine residues such as H3K122, acetylation levels were unaffected by •NO treatment (Fig. 1B). Together, these results highlight the scope of •NO-induced epigenetic changes at the histone level, and demonstrate that differential outcomes depend on the location of lysine residues as well as the exposure conditions of •NO.
H3K9 was one of the most heavily modified residues following •NO treatment (Fig. 1A). Therefore, this residue was selected to further examine the temporal relationship between •NO exposure and the kinetics of methylation and acetylation changes (Fig. 1C). Acetylation and methylation are mutually exclusive for any given lysine residue; therefore, global changes in the amounts of one modification could differentially affect the other. Consistent with our earlier studies (16), continued •NO exposure resulted in increased H3K9me2 over a period of 24 hours. During the same time period, we observed a concurrent decrease in global H3K9ac levels. When the •NO source was removed, both acetyl and methyl modifications returned to their basal levels within 24 hours, suggesting that the effects of •NO on these histone PTMs were persistent yet reversible. We also asked whether the ability of •NO to regulate epigenetic changes could be extended to other cell types. Fig. 1D demonstrates that •NO similarly decreased global H3K9ac levels and increased dimethylation at H3K9 in all 8 cell lines examined. This suggests that the underlying mechanisms governing •NO-mediated changes in histone PTMs are strongly conserved.
Nitric oxide is a principal regulator of transcriptional changes across the genome
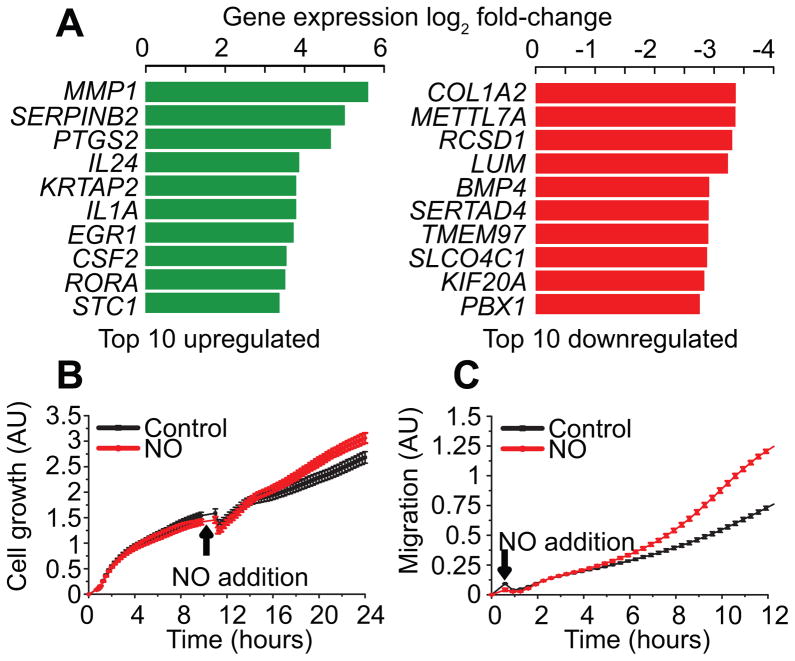
In order to investigate the impact of histone PTM changes on gene transcription, it was necessary to first identify all genes that were differentially expressed in response to •NO. As there has never been a comprehensive gene array study for the expression analysis of •NO-regulated genes in breast cancer, we treated MDA-MB-231 cells with •NO for 24 h and isolated total RNA for hybridization onto microarrays. Out of a total of 44,854 transcripts examined (mRNA and microRNA (miRNA)), an astonishing number of genes were changing in response to •NO. Specifically, we observed upregulation of 2,965 genes (2,920 mRNA, 45 miRNA) and a downregulation of 3,888 genes (3,816 mRNA, 72 miRNA) at a 1.5 fold change (FC) cut-off (Table S2). The most highly upregulated mRNA transcripts in this data set including MMP1 (28), PTGS2 (29), IL-1(30), EGR1 (31) and STC1 (32) are all associated with increased breast cancer proliferation and metastasis (Fig. 2A). Similarly, many of the most strongly downregulated genes such as COL1A2 (33) and BMP4 (34) have been correlated to disease progression and a worse prognosis when silenced. Gene Ontology analysis (FDR<0.05) of microarray data using Database for Annotation, Visualization and Integrated Discovery (DAVID) broadly revealed that downregulated genes were involved in mediating cell adhesion processes, whereas upregulated genes were functionally associated with pathways regulating cell growth, migration, development of vasculature, and inflammatory response (Fig. S2, Dataset S1).
Figure 2. Nitric oxide induces changes in gene expression and cellular phenotype.
(A) List of the 10 most up- and downregulated genes in response to •NO. MDA-MB-231 cells were treated with 500 μM DETA/NO for 24 hours. mRNA was extracted and prepared for hybridization onto GeneChip® PrimeView™ Human Gene Expression Arrays (Affymetrix). Microarray data was analyzed to determine differential gene expression in the presence or absence of •NO. (B) Real-time measurement of cell proliferation (+/− •NO) using the xCELLigence® DP system. MDA-MB-231 cells were seeded in E-plates containing 10% serum and allowed to adhere for 12 h before the addition of •NO (250μM DETA/NO). Cell proliferation was measured for 12 hours following •NO treatment. (C) Real-time measurement of cell migration (+/− •NO) using the xCELLigence® DP system. MDA-MB-231 cells were seeded in CIM plates containing 10% serum as a migratory stimulant. Cell migration was measured for 12 hours following addition of •NO (250μM DETA/NO).
To examine whether •NO-mediated changes in gene expression conferred tumor-associated cell phenotypes, we measured rates of cell proliferation and migration. Fig. 2B demonstrates that the growth rate of MDA-MB-231 cells significantly increased upon exposure to •NO. Concomitantly, the migratory potential of •NO-exposed cells was higher than untreated controls (Fig. 2C). Together, these results are consistent with previously published studies supporting the notion that •NO-associated gene expression changes in breast cancer favors an oncogenic phenotype (4,35).
Nitric oxide alters the distribution of H3K9me2 and H3K9ac across genomic loci
As shown in Fig. 1, methylation and acetylation of H3K9 were heavily modified in response to •NO. To investigate whether these modifications correlated with transcriptional changes, MDA-MB-231 cells were treated with •NO for 24 hours and both H3K9me2- and H3K9ac-associated DNA were isolated using standard chromatin immunoprecipitation (ChIP) protocols. ChIP-seq antibodies were validated for specificity prior to use (Fig. S3A) as per ENCODE guidelines. In addition to immunoblot testing of antibodies, H3K9me2/H3K9ac enrichment was confirmed at known control gene regions by PCR using the standard percent input method (Fig. S3B, primer sequences listed in Table S3).
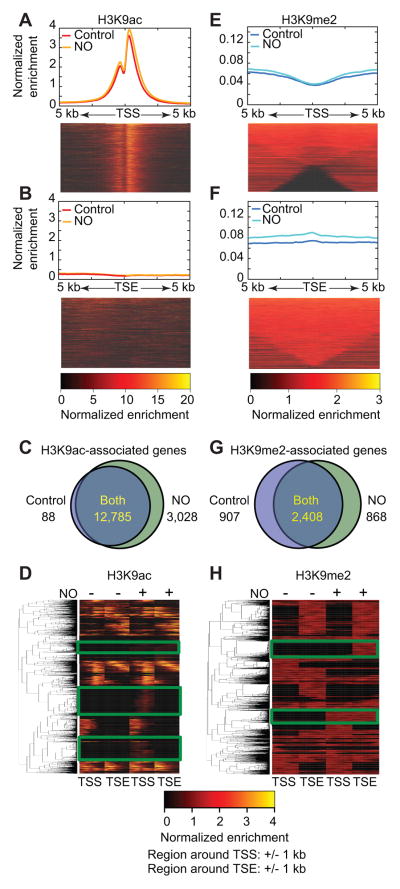
Analysis of the ChIP-seq data revealed striking differences between the distribution patterns of H3K9me2 and H3K9ac across the genome. As expected, the majority of H3K9ac peaks were centered at the transcriptional start site (27) (TSS) (+/− 5 kb) (Fig. 3A), with very little enrichment around the transcriptional end site (TSE) (Fig. 3B). Heatmaps depict the general patterning of H3K9ac in the untreated control samples; histograms illustrate differences in enrichment between control and •NO-treated samples (Fig. 3A, B). When we examined genes enriched for H3K9ac across the genome, we noted that a staggering 3,028 genes gained H3K9ac following •NO exposure, while only 88 genes lost this mark upon treatment (Fig. 3C and Fig. S3C, D). The majority of genes (12,785) retained H3K9ac enrichment following •NO treatment, however, the associated peaks became wider to cover more bases (Fig. 3C and Fig. S3E). A closer look at changes in the enrichment of H3K9ac at specific genes revealed that many of them gained H3K9ac at the TSS in response to •NO (Fig. 3D; TSS with new peaks highlighted in green).
Figure 3. Nitric oxide results in differential patterns of H3K9ac and H3K9me2 distribution across genomic loci.
MDA-MB-231 cells were treated with •NO (500 μM DETA/NO) for 24 hours; H3K9me2/ac ChIP-DNA was isolated using validated antibodies and sequenced to appropriate depths. MACS2 (narrow peak caller) and SICER (broad peak caller) algorithms were used for peak calling within H3K9ac and H3K9me2 enriched regions respectively. (A, B) Heatmaps represent the distribution of H3K9ac in untreated controls. Histograms represent differences in the average H3K9ac enrichment profiles around the TSS and TSE in control and •NO-exposed cells. (C) Number of genes associated with H3K9ac peaks that occur within +/− 5 kb of TSS in control and •NO-treated cells. (D) Representative heatmaps demonstrating the distribution patterns of H3K9ac across genomic loci at TSS and TSE following •NO exposure. Example genes associated with •NO-induced increases in H3K9ac around the TSS are highlighted by green boxes. (E, F) Heatmaps represent the distribution of H3K9me2 in untreated controls. Histograms represent differences in the average H3K9me2 enrichment profiles around the TSS and TSE in control and •NO-exposed cells. (G) Number of genes associated with H3K9me2 peaks that occur within +/− 200 kb of TSS in control and •NO-treated cells. (H) Representative heatmaps demonstrating the distribution patterns of H3K9me2 across genomic loci at TSS and TSE following •NO exposure. Example genes associated with •NO-induced increases in H3K9me2 around the TSE are highlighted by green boxes.
Analysis of the distribution of H3K9me2 revealed enriched regions away from the TSS, and in particular along the gene body, which became maximal around TSEs (Fig. 3E, F). Proportionally, the majority of H3K9me2 peaks (+/− •NO) were located near gene deserts and distal regions (+/− 200 kb from the TSS) (Fig. S3C, D). This is not surprising because H3K9me2 has been shown to influence enhancer driven transcriptional responses via long-range interactions (36). Approximately equal number of genes gained or lost H3K9me2 upon •NO treatment (Fig. 3G). Since methyl peaks tended to be much broader (>20 kb) than acetyl peaks, only marginal increases in H3K9me2 peak widths was observed upon •NO exposure (Fig. S3F). This indicates that •NO-induced diffusive spreading of H3K9me2 across the genome was less pronounced compared to its effect on the distribution of H3K9ac. Lastly, we noted striking increases in H3K9me2 around TSEs following •NO exposure at numerous gene loci, which emerged as a dominant trend across this ChIP-seq data set (Fig. 3F, 3H; highlighted in green).
H3K9ac is enriched at promoter sites of genes that are upregulated by •NO
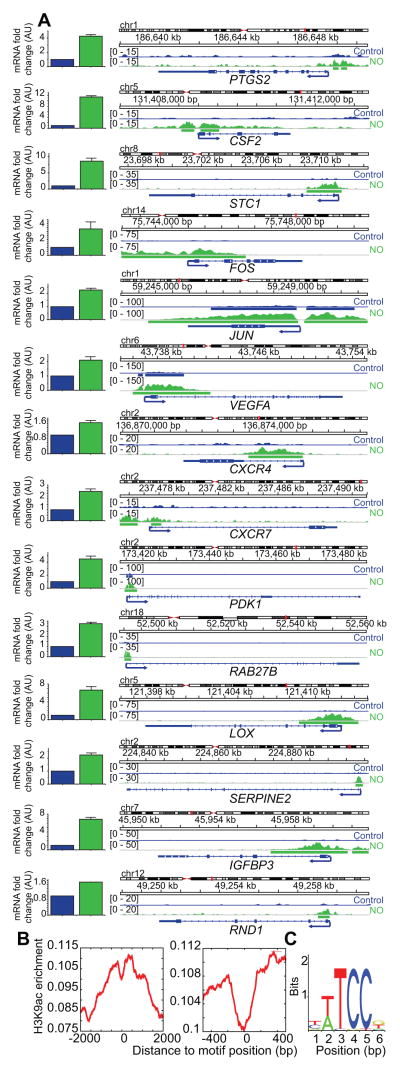
Acetylation of lysine residues is associated with increased gene expression (27). Not surprisingly, 74% of all •NO-upregulated genes (2,187 out of 2,965 genes) showed higher levels of promoter-associated H3K9 acetylation. This included the appearance of new H3K9ac peaks as well as increases in the heights of preexisting H3K9ac peaks around gene promoters after •NO exposure. Many of the most •NO-upregulated genes in our array (Fig. 2A), for example, PTGS2 (encodes for COX2), STC1, and CSF2 showed significant H3K9ac enrichment around their promoters specifically after •NO exposure (Fig. 4A). Additionally, •NO increased H3K9 acetylation at promoters of several genes known to be commonly upregulated in breast cancer including FOS, JUN, VEGFA, CXCR4, CXCR7, PDK1, RAB27B, LOX, SERPINE2, IGFBP3 and RND1 (Fig. 4A) (37–39). GO analysis revealed that upregulated genes with a new •NO-induced promoter H3K9ac peak were involved in a variety of diverse processes, some being critical to tumorigenesis such as migration, DNA replication, and hypoxic responses (Fig. S4, Dataset S2).
Figure 4. Genes upregulated by •NO gain promoter H3K9ac enrichment.
(A) Visualization of increased H3K9ac around gene promoters following •NO exposure using Integrative Genomics Viewer (IGV). Bar plots represent expression changes of corresponding mRNA transcripts as measured by GeneChip® PrimeView™ Human Gene Expression Arrays (Affymetrix). (B) Motif analysis from the JASPAR core database using CLOVER within the same gene set revealed multiple Ets-1 transcription factor binding sites around H3K9ac-associated promoters emergent following •NO treatment. Histogram represents average H3K9ac enrichment around potential Ets-1 binding sites. The “dip” at 0 represents Ets-1 binding and nucleosome exclusion. Right panel is a magnified version of the left panel. (C) Ets-1 binding motif sequence.
Next, we set out to identify transcription networks that interact with H3K9ac enriched promoter regions, as these would have a high likelihood of mediating the upregulation of •NO-associated target genes. To this end, we conducted motif analysis on upregulated genes that gained new promoter H3K9ac peaks after •NO exposure. We used the JASPAR core database and scanned for the presence of binding motifs corresponding to over 400 transcription factors around the TSS (+/− 2 kb) of these genes using CLOVER. We detected 32 transcription factor binding motifs exclusively within this gene set (Table S4). This included 4,477 binding instances for Ets-1, a proto-oncogene in breast cancer that is known to activate transcription of genes involved in matrix remodeling, angiogenesis etc (40). It was previously reported that Ets-1 transcriptional activation by •NO contributed to the increased migratory potential of MDA-MB-231 cells (41). Our microarray data revealed that ETS1 was upregulated by •NO and Fig. 4B demonstrates the distribution of H3K9ac around the Ets-1 binding motif. The Ets-1 binding motif is centered at the “0” position. It is important to note the “dip” in the enrichment of H3K9ac around the “0” position (Fig. 4B, C), which suggests nucleosome exclusion corresponding to Ets-1 binding. We also noted a gradual decrease in H3K9ac enrichment as we moved away from the “0” position of the Ets-1 binding motif. This indicated that putative Ets-1 binding occurred at H3K9ac peaks, suggesting direct cross talk with the histone mark. In addition to Ets-1, CLOVER analysis also revealed other transcription factor binding motifs such as Gata-1 and Sox-2 (Table S4) near new H3K9ac peaks of •NO-upregulated genes. These are well-known to be important in activating genes responsible for driving cancer cell stemness and proliferation (42,43). Together these findings demonstrate that not only does •NO alter the distribution of histone PTMs, but these changes can also affect transcriptional networks and result in measurable down-stream phenotypic effects.
Changes in H3K9me2 correlate to corresponding changes in gene expression
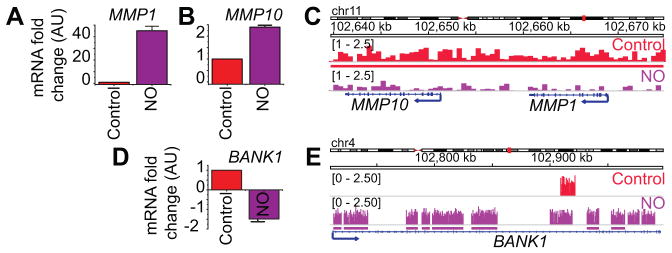
H3K9me2 is generally considered to be a gene-silencing mark (7). Although our data revealed that the majority of H3K9me2 was distributed around the TSE (Fig. 3F), a significant proportion was also found at the promoter regions of specific genes. We examined candidate genes where changes in promoter-associated H3K9me2 correlated with changes in gene expression consistent with the epigenetic code. For example MMP1, the most upregulated gene in our microarray dataset, along with MMP10, had significant loss of H3K9me2 around their promoter regions following •NO exposure (Fig. 2A, 5A–C). Conversely, appearance of new H3K9me2 peaks around the BANK1 promoter correlated with downregulation of its transcript (Fig. 5D, E). Although these are representative examples, MMP1, MMP10 and BANK1 are all known to be involved in various aspects cancer etiology. Specifically, matrix metalloproteinases (MMPs) are involved in breast cancer invasion and metastasis because of their ability to degrade extracellular matrix proteins (28). BANK1, a tumor suppressor, is often silenced in certain cancers and its downregulation is associated with increased breast-brain metastasis (44). Together these results demonstrate that gene expression changes in response to •NO could arise from alterations in different histone PTMs and that •NO-mediated transcriptional responses are consistent with the induction of an oncogenic phenotype (Fig. 2).
Figure 5. •NO-induced gene expression correlates to changes in H3K9me2 around their promoters.
(A, B, D) Changes in the expression levels of three •NO-regulated cancer genes measured using GeneChip® PrimeView™ Human Gene Expression Arrays (Affymetrix). (C, E) Visualization of H3K9me2 enrichment at the candidate gene loci using Integrative Genomics Viewer (IGV).
Discussion
One of the most important aspects to emerge from this study is a deeper appreciation for the true magnitude of •NO signaling. We also introduce novel epigenetic signaling mechanisms that may underlie many •NO-mediated gene expression changes. Our results demonstrate that cellular exposure to physiologically relevant concentrations of •NO alters the expression of thousands of genes. Additionally, these studies are the first to comprehensively characterize alterations in numerous histone PTMs in response to •NO. Over the past 30 years, the majority of research on •NO signaling has focused on the activation or inactivation of discrete proteins and the subsequent down-stream consequences of these events. Our data links changes in gene expression to alterations in specific histone posttranslational modifications and suggests that epigenetic regulation represents an overarching signaling mechanism of •NO. This model could have profound implications on how we think about •NO as a pleiotropic signaling molecule as well as its role as an endogenously produced epigenetic regulator.
We previously elucidated several mechanisms by which •NO alters global histone methylation patterns (16). It was established that •NO could directly inhibit the catalytic activity of JmjC-domain containing histone demethylases by binding to the non-heme iron cofactor in the active site. This mechanism has recently been corroborated by studies that elucidated a crystal structure of the JmjC demethylase, KDM2A, with •NO bound to the catalytic iron (45). Indirectly, we found that •NO could inhibit demethylation by changing the expression levels of methyl modifying enzymes (demethylases/methyltransferases) as well by reducing iron cofactor availability via formation of dinitrosyliron complexes.
On the other hand, mechanisms explaining •NO-mediated changes in histone acetylation remain obscure. The levels of steady-state histone acetylation are a function of the concerted activities of acetyltransferases (HATs) and deacetylases (HDACs). Alterations in histone acetylation levels by •NO imply disruption of the equilibrium between the forward (acetylation) and reverse (deacetylation) reactions. On the basis of current literature describing the structural composition and catalytic activities of HATs and HDACs, these enzymes do not appear to be logical direct targets for •NO. Nevertheless, several important studies have demonstrated disruption of intrinsic HDAC activity via •NO-dependent S-nitrosation of critical cysteine residues (12,14). Another potential explanation is that •NO could affect the activity of certain classes of HDACs by disrupting their zinc finger moieties. Additional indirect mechanisms include the ability of •NO to alter substrate availability (acetyl CoA) as well as induce transcriptional changes in the expression of acetyl-modifying enzymes (unpublished observation). Regardless, methylation and acetylation are mutually exclusive modifications at any specific lysine residue (7); therefore, it is not surprising that changes in methylation at a given location are directly tied to corresponding changes in acetylation. Although our findings do not conclusively elucidate the molecular pathways upstream of •NO-induced histone acetylation changes, these alterations are significant and have demonstrable phenotypic effects.
Our results clearly show that •NO has a significant effect on differentially altering global levels of acetyl- and methyl-lysine modifications at numerous locations on the core histones H3/H4 (Fig. 1). The ChIP-seq data demonstrated that a redistribution of histone marks across the genome is equally important as changes in their overall abundance. Although western blotting revealed quantitative increases in global H3K9me2 and a decrease in H3K9ac in response to •NO, the genomic location of these changes is a more important determinant of their overall effect on transcription. For example, although we observed a significant decrease in acetylation around exons and gene deserts following •NO exposure (Fig. S3C, D), we observed an enhanced association of H3K9ac around gene promoters following •NO treatment. Therefore, •NO-mediated decreases in global H3K9ac (Fig. 1), as evaluated by western blots, may not necessarily result in repressing a significant portion of the genome.
In many cases, the magnitude of •NO-induced PTM changes is large. For example, the extent of dimethylation at H3K9 almost doubles after 24 h of •NO exposure. At H4K20, me2 levels double after 24 h of •NO treatment, and are further elevated almost three fold after 48 h of sustained •NO exposure. Assuming that the average number of histones (H3 and H4) in the genome do not change, this means that twice as many become dimethylated in response to •NO after 24 hours. As the amount of DNA associated with these histone modifications also doubles, this could lead to significant cumulative effects on chromatin structure, gene transcription, and overall cellular phenotype. It is interesting that some of these modifications persist after •NO removal (H3K27me2), whereas others return to baseline within 24 hours (H3K9me2, H3K9ac). This suggests that differential gene expression arising from certain histone PTM changes will potentially mirror the duration of •NO exposure, yet in other instances transcriptional changes could persist long after •NO synthesis has ceased. The tissue and cell type distributions of NOS isoforms are varied, with subsequent fluctuations in the amount and duration of •NO production (5). Therefore, our findings suggest that dissimilar cellular phenotypes could be observed in varying microenvironments since the genetic makeup of cells and the extent of epigenetic regulation are expected to be different. The persistence of many of these modifications (48 h) implies that they represent true heritable epigenetic changes.
It is clear that the histone epigenetic code is an important determinant of cellular phenotype (46). H3K9ac is associated with open chromatin and gene activation, while H3K9me2 is a silencing mark associated with condensed chromatin (7). Our data demonstrated that for numerous genes, their patterns of regulation by •NO abided by this code. For example, •NO enriched H3K9ac around promoter regions of upregulated genes (Fig. 4A). We also noted loss of promoter H3K9me2 for candidate upregulated genes (Fig. 5C) and gain of H3K9me2 at certain silenced loci (Fig. 5E). Not all gene expression changes, however, were consistent with alterations in the epigenetic code. Although histone acetylation is explicitly associated with open chromatin and gene activation, we observed enrichment of H3K9ac around the TSS of downregulated genes as well. The simplest explanation is that regulation of these genes is occurring via non-•NO-mediated mechanisms or by •NO through non-epigenetic means. This study only examined the functional consequences of two histone PTMs (H3K9ac/me2), yet it is clear that changes in numerous other histone PTMs are occurring simultaneously in response to •NO (Fig. 1). Combinatorial PTM patterns are well-known to have significant functional consequences on gene expression (27,46,47), therefore, it is not surprising that the differential distribution of one histone modification does not correlate with the transcriptional responses of all its associated genes. Moreover, bivalent histone marks as well as crosstalk between PTMs within the same histone tail or between different histones (cis or trans) can differentially influence gene expression (48,49). Furthermore, long-range interactions via enhancers or the effects of increased H3K9me2 at TSEs in response to •NO may have important yet unrealized consequences (Fig. 3F, H). What our data also revealed was that the expression levels of numerous miRNAs are significantly changing in response to •NO (Table S2). This may be another means of epigenetic regulation by •NO as the expression levels of miRNAs within a tumor will affect amounts of target mRNAs and their down-stream protein products. As miRNAs can serve as tumor biomarkers, elucidating their expression profiles could have diagnostic or therapeutic value in •NO-associated cancers (50). Regardless, our data provides compelling evidence to establish epigenetic regulation via histone PTM changes as a vital upstream mechanism driving •NO-induced gene expression changes.
Ets-1 is a transcription factor known to activate numerous oncogenes (40) and has been implicated in the mechanism of •NO-induced proliferation of MDA-MB-231 cells (41). Interestingly, we noted upregulation of ETS1 by •NO in our microarray as well as the enhanced proliferation and migration of MDA-MB-231 cells (Fig. 2B, C). Additionally, we found the presence of Ets-1 binding motifs around the TSS of numerous upregulated genes with a new •NO-induced H3K9ac peak. More specifically, our analysis indicated that Ets-1 binding likely centered within a given H3K9ac peak. Therefore, communicative signals between Ets-1 and the acetylated histone could be important in activating transcription of the gene at the TSS (Fig. 4B). In this manner, not only do our results corroborate previously established findings, but they also provide additional important insights into the potential upstream mechanisms driving these phenotypic observations.
In general, tumor-associated nitric oxide (•NO) production and elevated nitric oxide synthase (NOS) expression are associated with the development and progression of more aggressive cancers. Similarly, it is being increasingly realized that epigenetic regulation is an important component in cancer development. Our findings establish a relationship between epigenetics and •NO in the context of tumor etiology by demonstrating that •NO exposure results in significant alterations in numerous histone PTMs that go on to alter cellular transcription and phenotype. This represents a novel alternate molecular mechanism to explain signaling actions of •NO in cancer (and other settings). To summarize this model: •NO is synthesized by or around tumor cells, it then inhibits surrounding JmjC demethylases, which directly changes the distribution of histone methylation and also indirectly influences other histone PTMs. These histone PTMs function in combination with microenvironmental factors, which ultimately favor an oncogenic gene expression profile and cellular phenotype. Although our findings do not explain all upstream mechanisms driving •NO-mediated gene expression changes, the notion that a substantial amount may be occurring through epigenetic means is novel. In additional to the numerous biological roles of •NO, it should now be considered an endogenous epigenetic regulatory molecule by virtue of its ability to impact gene expression through significant changes in the histone architecture.
Supplementary Material
Acknowledgments
Financial Support: This project was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award number R01GM085232. The project was also supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1TR000050. B.A.G acknowledges funding from the NIH through an Innovator award (DP2OD007447) and grant R01GM110174. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors also acknowledge ongoing support from the UIC Cancer Center.
Footnotes
Conflict of Interest: The authors declare no conflicts of interest.
References
- 1.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Developmental cell. 2010;19(5):698–711. doi: 10.1016/j.devcel.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Burke AJ, Sullivan FJ, Giles FJ, Glynn SA. The yin and yang of nitric oxide in cancer progression. Carcinogenesis. 2013;34(3):503–12. doi: 10.1093/carcin/bgt034. [DOI] [PubMed] [Google Scholar]
- 3.Chang CF, Diers AR, Hogg N. Cancer cell metabolism and the modulating effects of nitric oxide. Free radical biology & medicine. 2015;79C:324–36. doi: 10.1016/j.freeradbiomed.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinecke JL, Ridnour LA, Cheng RY, Switzer CH, Lizardo MM, Khanna C, et al. Tumor microenvironment-based feed-forward regulation of NOS2 in breast cancer progression. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(17):6323–8. doi: 10.1073/pnas.1401799111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hickok JR, Thomas DD. Nitric oxide and cancer therapy: the emperor has NO clothes. Curr Pharm Des. 2010;16(4):381–91. doi: 10.2174/138161210790232149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vasudevan D, Thomas DD. Insights into the diverse effects of nitric oxide on tumor biology. Vitamins and hormones. 2014;96:265–98. doi: 10.1016/B978-0-12-800254-4.00011-8. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes & development. 2001;15(18):2343–60. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 8.Re A, Aiello A, Nanni S, Grasselli A, Benvenuti V, Pantisano V, et al. Silencing of GSTP1, a prostate cancer prognostic gene, by the estrogen receptor-beta and endothelial nitric oxide synthase complex. Molecular endocrinology. 2011;25(12):2003–16. doi: 10.1210/me.2011-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross TJ, Kremens K, Powers LS, Brink B, Knutson T, Domann FE, et al. Epigenetic silencing of the human NOS2 gene: rethinking the role of nitric oxide in human macrophage inflammatory responses. Journal of immunology. 2014;192(5):2326–38. doi: 10.4049/jimmunol.1301758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall HE, Stamler JS. Nitrosative stress-induced apoptosis through inhibition of NF-kappa B. The Journal of biological chemistry. 2002;277(37):34223–8. doi: 10.1074/jbc.M201638200. [DOI] [PubMed] [Google Scholar]
- 11.Sen N, Snyder SH. Neurotrophin-mediated degradation of histone methyltransferase by S-nitrosylation cascade regulates neuronal differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(50):20178–83. doi: 10.1073/pnas.1117820108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455(7211):411–5. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 13.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, et al. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nature cell biology. 2008;10(7):866–73. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colussi C, Mozzetta C, Gurtner A, Illi B, Rosati J, Straino S, et al. HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(49):19183–7. doi: 10.1073/pnas.0805514105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okayama H, Saito M, Oue N, Weiss JM, Stauffer J, Takenoshita S, et al. NOS2 enhances KRAS-induced lung carcinogenesis, inflammation and microRNA-21 expression. International journal of cancer Journal international du cancer. 2013;132(1):9–18. doi: 10.1002/ijc.27644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickok JR, Vasudevan D, Antholine WE, Thomas DD. Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases. The Journal of biological chemistry. 2013;288(22):16004–15. doi: 10.1074/jbc.M112.432294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ignarro LJ. Nitric oxide as a unique signaling molecule in the vascular system: a historical overview. Journal of physiology and pharmacology : an official journal of the Polish Physiological Society. 2002;53(4 Pt 1):503–14. [PubMed] [Google Scholar]
- 18.Smith BC, Marletta MA. Mechanisms of S-nitrosothiol formation and selectivity in nitric oxide signaling. Current opinion in chemical biology. 2012;16(5–6):498–506. doi: 10.1016/j.cbpa.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods in enzymology. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 21.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research. 2007;35(Web Server issue):W169–75. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PloS one. 2010;5(11):e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, et al. A travel guide to Cytoscape plugins. Nature methods. 2012;9(11):1069–76. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic acids research. 2014;42(Database issue):D142–7. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frith MC, Fu Y, Yu L, Chen JF, Hansen U, Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic acids research. 2004;32(4):1372–81. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nature reviews Molecular cell biology. 2014;15(11):703–8. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Kato Y, Erzinger SA, Kiriakova GM, Qian Y, Palmieri D, et al. The role of MMP-1 in breast cancer growth and metastasis to the brain in a xenograft model. BMC Cancer. 2012;12:583. doi: 10.1186/1471-2407-12-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Kong X, Zhang J, Luo Q, Li X, Fang L. MiRNA-26b inhibits proliferation by targeting PTGS2 in breast cancer. Cancer Cell Int. 2013;13(1):7. doi: 10.1186/1475-2867-13-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nozaki S, Sledge GW, Jr, Nakshatri H. Cancer cell-derived interleukin 1alpha contributes to autocrine and paracrine induction of pro-metastatic genes in breast cancer. Biochem Biophys Res Commun. 2000;275(1):60–2. doi: 10.1006/bbrc.2000.3241. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell A, Dass CR, Sun LQ, Khachigian LM. Inhibition of human breast carcinoma proliferation, migration, chemoinvasion and solid tumour growth by DNAzymes targeting the zinc finger transcription factor EGR-1. Nucleic acids research. 2004;32(10):3065–9. doi: 10.1093/nar/gkh626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang AC, Doherty J, Huschtscha LI, Redvers R, Restall C, Reddel RR, et al. STC1 expression is associated with tumor growth and metastasis in breast cancer. Clin Exp Metastasis. 2015;32(1):15–27. doi: 10.1007/s10585-014-9687-9. [DOI] [PubMed] [Google Scholar]
- 33.Loss LA, Sadanandam A, Durinck S, Nautiyal S, Flaucher D, Carlton VE, et al. Prediction of epigenetically regulated genes in breast cancer cell lines. BMC Bioinformatics. 2010;11:305. doi: 10.1186/1471-2105-11-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ampuja M, Jokimaki R, Juuti-Uusitalo K, Rodriguez-Martinez A, Alarmo EL, Kallioniemi A. BMP4 inhibits the proliferation of breast cancer cells and induces an MMP-dependent migratory phenotype in MDA-MB-231 cells in 3D environment. BMC Cancer. 2013;13:429. doi: 10.1186/1471-2407-13-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glynn SA, Boersma BJ, Dorsey TH, Yi M, Yfantis HG, Ridnour LA, et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. The Journal of clinical investigation. 2010;120(11):3843–54. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature reviews Genetics. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 37.Gasparini G. Prognostic value of vascular endothelial growth factor in breast cancer. The oncologist. 2000;5(Suppl 1):37–44. doi: 10.1634/theoncologist.5-suppl_1-37. [DOI] [PubMed] [Google Scholar]
- 38.Jiao X, Katiyar S, Willmarth NE, Liu M, Ma X, Flomenberg N, et al. c-Jun induces mammary epithelial cellular invasion and breast cancer stem cell expansion. The Journal of biological chemistry. 2010;285(11):8218–26. doi: 10.1074/jbc.M110.100792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mukherjee D, Zhao J. The Role of chemokine receptor CXCR4 in breast cancer metastasis. American journal of cancer research. 2013;3(1):46–57. [PMC free article] [PubMed] [Google Scholar]
- 40.Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi: 10.1186/1476-4598-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Switzer CH, Cheng RY, Ridnour LA, Glynn SA, Ambs S, Wink DA. Ets-1 is a transcriptional mediator of oncogenic nitric oxide signaling in estrogen receptor-negative breast cancer. Breast Cancer Res. 2012;14(5):R125. doi: 10.1186/bcr3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boidot R, Vegran F, Jacob D, Chevrier S, Cadouot M, Feron O, et al. The transcription factor GATA-1 is overexpressed in breast carcinomas and contributes to survivin upregulation via a promoter polymorphism. Oncogene. 2010;29(17):2577–84. doi: 10.1038/onc.2009.525. [DOI] [PubMed] [Google Scholar]
- 43.Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31(11):1354–65. doi: 10.1038/onc.2011.338. [DOI] [PubMed] [Google Scholar]
- 44.Salhia B, Kiefer J, Ross JT, Metapally R, Martinez RA, Johnson KN, et al. Integrated genomic and epigenomic analysis of breast cancer brain metastasis. PloS one. 2014;9(1):e85448. doi: 10.1371/journal.pone.0085448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Z, Cheung P, Kuo AJ, Yukl ET, Wilmot CM, Gozani O, et al. A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes & development. 2014;28(16):1758–71. doi: 10.1101/gad.246561.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 47.Garske AL, Oliver SS, Wagner EK, Musselman CA, LeRoy G, Garcia BA, et al. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nature chemical biology. 2010;6(4):283–90. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Current opinion in cell biology. 2003;15(2):172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 49.Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes & development. 2013;27(12):1318–38. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.