Abstract
Fetal development could be considered a sensitive period wherein exogenous insults and changes to the maternal milieu can have long-term impacts on developmental programming. The placenta provides the fetus with protection and necessary nutrients for growth, and responds to maternal cues and changes in nutrient signaling through multiple epigenetic mechanisms. The X-linked enzyme O-linked-N-acetylglucosamine transferase (OGT) acts as a nutrient sensor that modifies numerous proteins to alter various cellular signals, including major epigenetic processes. This review describes epigenetic alterations in the placenta in response to insults during pregnancy, the potential links of OGT as a nutrient sensor to placental epigenetics, and the implications of placental epigenetics in long-term neurodevelopmental programming. We describe the role of placental OGT in the sex-specific programming of hypothalamic-pituitary-adrenal (HPA) axis programming deficits by early prenatal stress as an example of how placental signaling can have long-term effects on neurodevelopment.
Keywords: Placenta, epigenetics, OGT, neurodevelopment, fetal development, nutrition, stress
1. Introduction
Prenatal development is a particularly vulnerable period in life when tissues are rapidly developing and are susceptible to shifts in programming. Across gestation, fetal needs change to accommodate the trajectory of tissue development, making specific windows of pregnancy particularly important for tissue growth, and allowing environmental perturbations to have long-term effects on these developing systems. Throughout pregnancy, the placenta acts as the command post for incoming and outgoing messages to and from maternal and fetal compartments. Epigenetic responses to maternal and fetal signals are an obvious candidate for transforming early life inputs into long-term programmatic outcomes. We have gained a broader understanding of how specific insults during development impact the fetal brain, and are beginning to understand the importance of placental signaling in neurodevelopmental programming. This review summarizes evidence for the role of placental epigenetics in neurodevelopmental programming with a focus on the integration of nutrient signals into chromatin changes and the sex specificity of these findings.
2. The placenta is a diplomat for maternal-fetal relations
The placenta is a dynamic endocrine tissue displaying robust responses to alterations in the maternal milieu. As the fetal sustenance delivery system, placental health is critical for fetal growth and development, and acts at the interface to communicate maternal nutritional status and environmental disruptions. Studies in both humans and rodents have demonstrated that a wide array of maternal inputs, such as over and under-nutrition, smoking, drug and alcohol intake, infection and stress can induce marked transformations in placental physiology ranging from alterations in aspects of gross placental morphology such as obvious changes in placental weight, to the more subtle changes in placental gene expression that may predict altered transport of important signals to the fetus (not exhaustive: (Amankwah and Kaufmann, 1984; Eguchi et al., 1989; Ganapathy, 2011; Gheorghe et al., 2010; Godfrey et al., 1996; Godfrey and Barker, 1995; Jauniaux and Burton, 2007; Kennedy, 1984; La Torre et al., 2006; Mairesse et al., 2007; Pastrakuljic et al., 1999; Zdravkovic et al., 2005)). Many of these insults result in decreases in both placental and fetal growth, which can have long-term impacts on offspring health.
Across the timeline of in utero development, the primary placental function is to transfer nutrients and gases required for fetal development. The placenta serves as an arbitrator between the mother and fetus, guaranteeing fetal needs while concurrently negotiating with the maternal immune system. The dominant cell type within the placenta is the fetally-derived trophoblast cell. Although there are developmental and structural differences between the mouse and human placenta (as reviewed in Malassine et al., 2003), they share remarkably conserved profiles of mRNA and protein expression, and mouse models are widely accepted as proxies for many aspects of human placental function (Cox et al., 2009). Trophoblast cells are the first to differentiate post fertilization, comprising the outer layer of the blastocyst and eventually giving rise to all placental layers excluding the maternal decidua (see Rossant and Cross, 2001 for review of placental development). During early pregnancy, these cells initiate vascular remodeling during trophoblast invasion of the uterus, allowing the placenta to establish vascular inputs from the mother. This occurs during the first trimester in humans, but not until mid-gestation in rodents. Establishing robust connections with the maternal circulation is critical for gaining access to sufficient nutrients and has a direct impact on fetal and placental growth. In the mouse, this is achieved by a placental structure with two major zones: the labyrinth and junctional zones. Maternal-fetal exchange occurs within the branching labyrinth of cytotrophoblast cells encompassed by the multinucleated syncytiotrophoblast layer that compartmentalizes maternal and fetal blood. The labyrinth is separated from the maternal decidua by a junctional zone containing trophoblastic giant cells and spongiotrophoblastic cells. In humans, placental villi extend directly into maternal blood. These villi consist of a synctiotrophoblast layer and villous cytotrophblast cells that are anchored to the maternal decidua by extravillous cytotrophblast cells (comparisons reviewed comprehensively in Carter, 2007; Georgiades et al., 2002; Malassine et al., 2003).
The fetal origin of the trophoblastic cell lineage prompts the maternal immune system to recognize these cells as foreign entities. Fortunately, evolution has designed mechanisms to suppress this maternal host response and defend the semi-allogenic fetus (reviewed in Munoz-Suano et al., 2011). The critical placental role in suppression of the maternal immune system has led some to consider it an immune organ, while others categorize it as an endocrine tissue due to its production and release of numerous hormones and factors into maternal and fetal circulation (Fowden et al., 2005a; Murphy et al., 2006; Petraglia et al., 1996; Simpson and MacDonald, 1981). Placental hormones come in all varieties including steroids, peptides, eicosanoids and gylcoproteins (Fowden et al., 2005b). Interaction of the immune and endocrine systems occurs through placental production of the steroid hormone, progesterone, which is necessary for immunosuppression and anti-inflammatory processes involved in maintenance of pregnancy (Siiteri et al., 1977). In addition to mediating immunosuppression, placental-derived hormones are responsible for a range of necessary physiological changes during pregnancy, including uterine expansion, mammary tissue development, and eventually, the initiation of parturition (Linzer and Fisher, 1999). Importantly, during early pregnancy placental lactogen and progesterone signal the need for maternal resources to be dispatched for use by the fetus, altering maternal metabolism to increase fetal access to glucose (Fowden et al., 2006).
2.1 Sex differences in the placenta
As trophoblast cells originate from the embryo, they reflect fetal sex as either XX or XY, allowing for sex differences in placental biochemistry, function, and signaling. Although studies in rodent models identified preferential inactivation of the paternal X chromosome in the female placenta via imprinting (Takagi and Sasaki, 1975; Wang et al., 2001), analysis of the human female placenta revealed random patterns of X inactivation (de Mello et al., 2010; Looijenga et al., 1999). Further, silenced X chromosomes in the placenta are under less stringent epigenetic repression relative to those in somatic tissues, allowing for reactivation of the inactive X chromosome and non-random X inactivation within the placenta in response to intrauterine conditions (Migeon et al., 2005). This plasticity in X-inactivation in the placenta may be an important contributor to sex-differences in response to environmental perturbations during gestation, whereby females may be buffered from detrimental conditions to a greater degree than males due to increased expression of important X-linked genes.
In addition to sex differences determined by sex chromosomes, multiple studies have identified sex differences in autosomal gene expression in the placenta at both the mRNA and protein level (reviewed in Clifton, 2010), with striking female-biased expression of several key immune regulators in the human placenta (Sood et al., 2006), suggesting that sex dictates how the placenta negotiates with the maternal immune system. In addition to sex differences in immune system communication, fetal sex can also govern nutrient allocation from the mother. On average, male fetuses are larger than females (Forsen et al., 1999; Thomas et al., 2000), suggesting greater nutrient requirements in males during fetal growth. David Barker postulated that male fetuses are more dependent on maternal sources of nutrition allowing for this enhanced fetal growth. His group argued that male placentas are more efficient than female placentas at extracting nutrients, whereas female placentas may have a greater capacity to store energy (Eriksson et al., 2010). Studies in humans have shown that fetal sex may modulate nutritional input to the placenta/fetus, particularly during the second trimester of pregnancy wherein women carrying male fetuses have higher energy intake than those pregnant with female fetuses (Tamimi et al., 2003).
3. Nutritional requirements during fetal development
Macronutrients, gases and metabolites are transferred by the placenta into fetal circulation via passive (urea & carbon dioxide out, fatty acids & oxygen in) and facilitative diffusion (glucose, lactate, fatty acids), active transport (amino acids), and endo- and exocytosis (Watson and Cross, 2005). Glucose is the primary fuel for the fetus and placenta. During early pregnancy the fetus produces very small amounts of glucose, necessitating glucose transfer from maternal blood (Hay et al., 1984; Marconi et al., 1996). Not surprisingly, in human pregnancies low maternal blood glucose levels lead to small for gestation age (SGA) neonates, whereas hyperglycemia results in fetal macrosomia, which makes blood glucose during in utero development a potential predictive factor of later health and disease (Cianfarani et al., 2003; Combs et al., 1992). Fetal nutrient requirements change over the course of pregnancy, and studies in humans suggest that nutritional intake during early pregnancy is particularly critical for directing normal fetal growth and development (Moore et al., 2004).
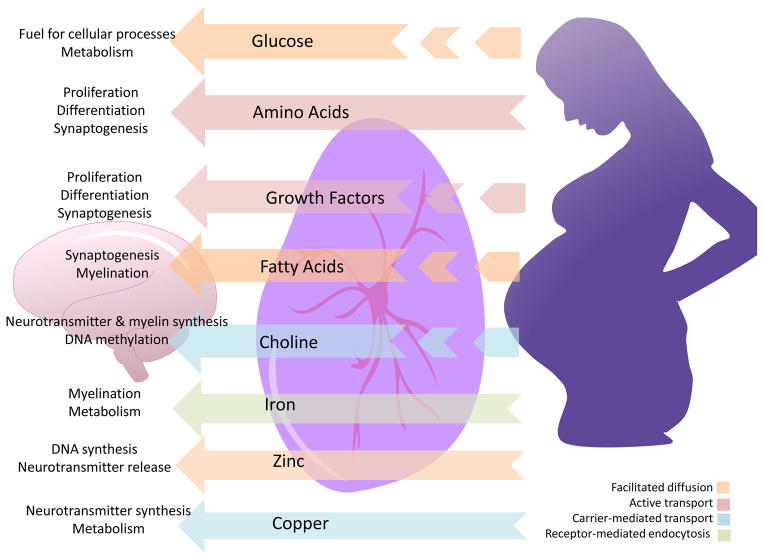
The developing brain is a nutritionally-demanding tissue, and is particularly sensitive to insufficiencies or overabundance of specific nutrients and growth factors. Although the initial stages of nervous system development occur as early as 2–3 weeks post fertilization in humans, neuronal proliferation occurs later in the first trimester, extending into the second trimester. Neural migration and synaptogenesis occur predominantly in the late second and third trimesters (reviewed in Tau and Peterson, 2010). During early pregnancy the brain is extremely plastic but vulnerable to broad environmental fluctuations that can impact long-term programming. After 24 weeks of gestation, wherein myelination and synapse formation occur, nutrient requirements in the developing human brain become critical (Figure 1; reviewed in Georgieff, 2007). Of course, the developmental timeline for specific brain regions differs, such that certain rapidly developing regions may be more sensitive to nutrient availability at particular gestational stages than others that are slower to develop or that have already been established. The hypothalamus, the brain’s command center for neuroendocrine function, begins to develop around mid-gestation in humans and continues to mature until adolescence (Gunnar et al., 2009; Markakis, 2002), forming its most critical connections during late pregnancy. It is important to note that the rate and timing of development, and therefore sensitivity, differs between the hypothalamic sub-regions (Bouret, 2010).
Figure 1.
Nutritional requirments for brain development. During late gestation, specific nutritional inputs (center) to the brain are necessary for specific neurodevelopmental processes (left). These nutrients come from either maternal circulation and the placenta combined (broken arrows) or maternal circulation alone (solid arrows). Their mechanism of transport across the placenta also differs (indicated by arrow color).
3.1 Placental energy balance as a mediator of developmental programming
Barker’s Developmental Origins of Health and Disease hypothesis has been widely accepted and supported by research across various systems within the field of developmental biology (Barker, 2007, 2004; Godfrey et al., 2007; Wadhwa et al., 2009). This hypothesis states that developmental programming prepares offspring for conditions outside of the womb by taking external factors into account during development. The most famous examples of this hypothesis are seen in cases of maternal under- or over-nutrition, which program metabolic dysregulation in offspring preparing them for predicted conditions of feast or famine through alterations in placental signaling (Wadhwa et al., 2009). Studies in both humans and animal models demonstrate that while caloric-restriction is associated with decreases in nutritional signals, such as IGF-1, insulin, and leptin, obesity increases levels of these factors in maternal circulation. Receptors for insulin, IGF-1, and leptin are present at the maternal-fetal interface, and their signaling stimulates amino acid transporter activity in trophoblast cells (Lager and Powell, 2012), directly coupling maternal nutritional status to placenta function and undoubtedly altering the availability of nutrients crossing into the fetal circulation. Changes in the availability of these factors for the fetus and/or placenta promote cellular signaling events important for long-term metabolic programming. Numerous studies in animal models and humans have reported strong associations between maternal nutrition and alterations in postnatal adiposity, appetite, metabolism, and brain function (Bayol et al., 2007; Breton, 2013; Jones et al., 2009; Muhlhausler et al., 2006; Sullivan et al., 2010; Taylor and Poston, 2007). Leptin drives the formation of neural projections during hypothalamic development that are critical for feeding behaviors and metabolic regulation throughout life (Bouret et al., 2004), providing a potential molecular mechanism by which maternal over or under nutrition and adiposity can program offspring long-term food intake and weight gain. Another recent study found an association between gestational diabetes and an increased risk for offspring autism spectrum disorder (ASD; Xiang et al., 2015), once again highlighting the importance of maternal energy balance in brain development
4. Epigenetics: energy availability and long-term programming
Epigenetics are a means by which environmental stimuli drive short- and long-term gene expression patterns. As a mediator of maternal and environmental signals to the developing fetus, epigenetic processes within the placenta are particularly powerful such that alterations of placental gene expression, downstream function, and signaling during fetal development have the potential for dramatic changes in developmental programming. As the brain is an energetically-expensive organ, consuming massive amounts of maternal resources during its development, it is also a particularly vulnerable site dependent on and susceptible to changes in placental function. For example, increased methylation of the leptin receptor gene in the human placenta is associated with increased lethargy and hypotonicity in male, but not female newborns (Lesseur et al., 2014), supporting not only a connection between epigenetic regulation of the leptin receptor and the relay of important information to the fetal brain, but also a sex-specificity in these outcomes.
At present, the three major epigenetic players are post-translational histone modifications, DNA methylation, and small noncoding RNAs, such as microRNAs (miRNAs). All of these processes are closely intertwined. For example, changes in histone marks, such as acetylation or methylation, are necessary for DNA methylation and demethylation to occur (Cedar and Bergman, 2009). DNA methylation can regulate miRNA expression and vice versa (Han et al., 2007; Wu et al., 2010), and miRNA frequently target and regulate levels of the histone-modifying enzymes deacetylases and methyltransferases (Guil and Esteller, 2009; Tuddenham et al., 2006; Varambally et al., 2008; Wong and Tellam, 2008). Modifications made to the N-terminal tails of histone proteins that change protein charge and attract or repel DNA, trigger the tightening or loosening of coiled DNA, ultimately impacting the accessibility of particular genomic regions for transcription (Berger, 2002). Histone tails are subject to several types of modification that typically occur on lysine, arginine or serine residues. At present, the most well-established histone modifications include phosphorylation, acetylation, methylation, ubquitation, sumoylation, biotinylation, and glycosylation, although additional modifications will likely be discovered as epigenetic research continues to expand (Peterson and Laniel, 2004). The exact location, titer and combination of these modifications form the so-called histone code, which ultimately determines small- and large-scale chromatin conformational changes (Jenuwein and Allis, 2001). DNA methylation is the direct addition of methyl groups to DNA nucleotides. The most well-known function for DNA methylation is transcriptional silencing by recruitment of histone modifying complexes to collapse chromatin structure or direct inhibition of transcription factor binding to promoter start sites (Bird, 1985). However, DNA methylation can also direct alternative promoter usage, or be a vestigial mark of previous transcriptional activity (Hon et al., 2013; Jones, 2012). microRNAs (miRNAs) are short, single-stranded non-coding RNAs that typically mediate post-transcription gene silencing (Filipowicz et al., 2008). Most miRNAs are transcribed into primary miRNAs (pri-miRNAs) by RNA polymerase II, although the largest human miRNA cluster, C19MC, which has received particular attention in the placenta, is transcribed by RNA polymerase III (Borchert et al., 2006). pri-miRNAs, precursors that can give rise to many mature miRNAs, are further processed into pre-miRNAs by the endonuclease activity of the enzyme Drosha, which cleaves hairpin pri-miRNAs leaving a two-nucleotide overhang on the 3′ end that is recognized by the nucleocytoplasmic shuttle, exportin-5 (Lee et al., 2003; Yi et al., 2003). Once in the cytoplasm, pre-miRNAs are further cleaved by the enzyme Dicer, resulting in mature miRNAs that are enabled for loading into the RNA-induced silencing complex (RISC) (Tijsterman and Plasterk, 2004). miRNAs target one or multiple mRNAs for degradation via complementary binding. miRNAs have more recently been shown to direct DNA methylation and histone modifications after being shuttled back into the nucleus via specific transporters (Benetti et al., 2008; Guan et al., 2011; Noh et al., 2013).
4.1 Histone modifications in the placenta
Histone modifications have a unique function in the placenta as the primary mediators of imprinting in some genomic regions. In fact, DNA methylation is not necessary to maintain imprinting in one location on chromosome 7, as it is chiefly maintained by H3K9me2 and H3K27me3 through the actions of the histone methyltransferase EZH2, a member of the Polycomb repressive complex (Lewis et al., 2004; Umlauf et al., 2004). Genomic compartments such as enhancers, promoters, and silenced regions are typically associated with the histone marks H3K4me1/H3K27ac, H3K4me3, and H3K27me3, respectively. Using ChIP-Seq against these marks, Julie Baker and colleagues discovered that enhancer regions in trophoblast cells are enriched for endogenous retroviruses, which are suggested to be important contributors to species-specific placental evolution (Chuong et al., 2013). Despite their intriguing role in early placental development, changes in the histone code in response to changes in the maternal milieu have not been currently examined.
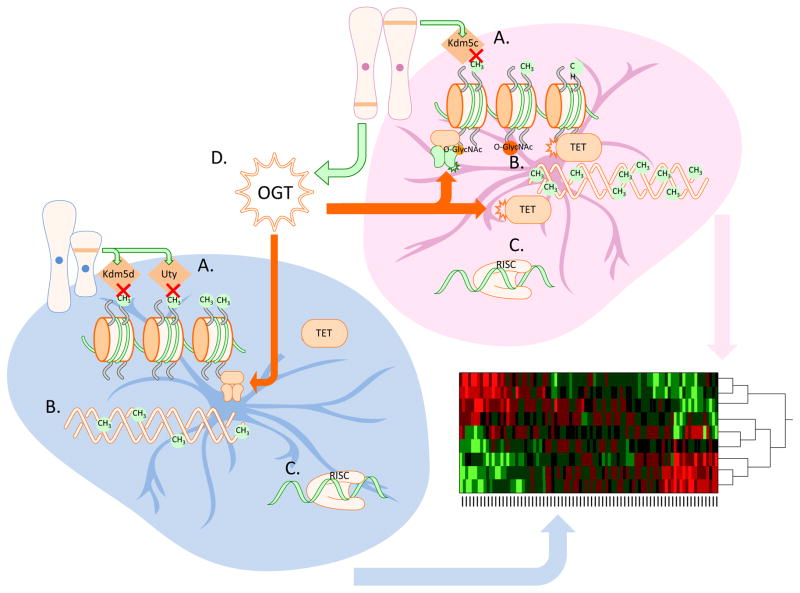
Sex differences in several epigenetic regulators have been identified across placental development. Expression of the X-linked H3K4me3 demethylase, Kdm5c, is higher in the female placenta, whereas expression of the Y-linked genes for the H3K27me3 and H3K4me3 demethylases, Uty and Kdm5d respectively, are higher in males (Figure 2A.; Howerton et al., 2013). These differences suggest that epigenetic control of gene expression within the placenta may lead to sex differences in placental function, and perhaps lead to known gender biases in sensitivity to insults associated with neurodevelopmental disorders.
Figure 2.
Sex differences in epigenetic regulation of placental gene expression. A. Sex chromosome complement leads to male biased expression of the Y-linked histone demethylases Kdm5d and Uty, and female biased expression of the histone demethylase Kdm5c. B. DNA in the female placenta is more heavily methylated than in the male placenta. C. To date, there have not been reports of sex difference in miRNA expression in the placenta. D. Expression of the X-linked nutrient sensing enzyme O-linked N-acetlyglucosamine (O-GlcNAc) transferase (OGT) is higher in the female placenta, leading to numerous sex differences in chromatin regulatory processes.
4.2 DNA methylation in the placenta
Changes in DNA methylation in the placenta have only recently been explored with regard to sex and developmental programming. In mice, female placental tissue has higher levels of global DNA methylation compared to male placentas (Figure 2B; Gallou-Kabani et al., 2010), which may provide females additional protection from dynamic changes in gene expression resulting from environmental insults. In addition, tighter regulation of gene expression in females provides a plausible mechanism by which males are preferentially affected by exogenous insults, such as maternal stress during gestation. This sex difference in DNA methylation likely accounts for aspects of the reported sex differences in gene expression in the placenta described in numerous studies (Gallou-Kabani et al., 2010; Howerton et al., 2013; Mao et al., 2010; Osei-Kumah et al., 2011; Sood et al., 2006). Although a recent paper reported finding no sex differences in mRNA levels of the enzymatically active DNMT isoforms (Gabory et al., 2012), DNMT enzyme activity might differ in the placenta by sex as it does in the neonatal brain (Nugent et al., 2015).
DNA methylation likely plays an important role in temporal control of gene expression across pregnancy in order to meet the changing needs of the developing fetus. Overall levels of DNA methylation increase toward the end of pregnancy (Chavan-Gautam et al., 2011; Novakovic et al., 2011). Genome-wide analysis of human placental tissue across gestation revealed widespread differences in methylation across trimesters, likely reflecting changes in cellular composition and differentiation across gestation (Novakovic et al., 2011). Most of the genes displaying differential methylation between the first and third trimesters were associated with immune related pathways and corresponded with gene expression patterns previously described across gestation (Mikheev et al., 2008; Winn et al., 2007).
Genomic imprinting is the epigenetic process whereby specific genes are epigenetically silenced based on parent of origin via DNA methylation and histone modifications. Although the best known examples of the developmental functions of imprinted genes stem from the well-know conflict theory of genomic imprinting, wherein paternally-derived genes are thought to promote pre- and postnatal growth, while maternally-expressed genes suppress growth (Hurst and McVean, 1997), regulation of cellular differentiation within the placenta is likely a more critical function of genomic imprinting (Miri and Varmuza, 2009). Elevated expression of the majority of known imprinted genes within the placenta, compared to embryonic and adult tissues, signifies the importance of imprinted genes in placental development and function (as thoroughly reviewed in Coan et al., 2005). Imprinted genes are not found in non-mammalian species leading some to theorize the co-evolution of genomic imprinting and placentation.
4.3 miRNAs in the placenta
miRNAs are undoubtedly the most well-studied epigenetic regulators of placental function in relation to maternal and fetal health and disease (Mouillet et al., 2011), most likely due to the relative ease of quantifying their expression in placental tissue as well as in maternal circulation. miRNA expression differs significantly between the fetal and maternal sides of the placenta, and mRNA targets of these differentially expressed miRNAs suggest their importance in epigenetic regulation of maternal blood vessel development and fetal neuronal differentiation (Wessels et al., 2013). Analysis of miRNA expression from first and third-trimester human placentas has identified distinct miRNA-cluster expression profiles across pregnancy (Gu et al., 2013). This study found that miRNAs epigenetically regulate the expression of gene sets associated with both adaptive and innate immune responses throughout pregnancy, while miRNAs controlling oncogenic, angiogenic and anti-apoptotic genes appear dominant during the first trimester, and miRNAs promoting cell differentiation are highly expressed in late pregnancy (Gu et al., 2013).
In addition to their role in controlling gene expression necessary for normal placental development and function, several studies have examined the effects of maternal health and environmental insults on placental miRNA expression. Comparisons of miRNA expression in human placenta samples from patients with preeclampsia and fetal growth restriction have revealed several miRNAs associated with these conditions (Enquobahrie et al., 2011; Higashijima et al., 2013; Zhu et al., 2009). In preeclampsic tissue, miRNAs regulating genes involved in cardiovascular and reproductive system development, immunological function, and other cellular processes were significantly altered, suggesting, not surprisingly, that epigenetic processes are disrupted following abnormal placenta implantation (Enquobahrie et al., 2011; Zhu et al., 2009). Although the impact of prenatal nutrition on placental miRNA expression has yet to be thoroughly examined, specific miRNAs have been associated with birth weight, and suggest that decreased expression of miR-16 and miR-21 strongly increase the risk for being considered small for gestation age (Maccani et al., 2011). Interestingly, there are currently no reports of basal sex differences in miRNA expression in the placenta to our knowledge.
A study in humans has recently linked miRNA signaling in the placenta to neurodevelopmental programming (Maccani et al., 2013). This study quantified the expression of six placental miRNAs previously shown to be responsive to environmental perturbations (Maccani et al., 2010), correlating their expression with infant neurobehavioral outcomes as measured by the NICU Network Neurobehavioral Scales (NNNS) (Lester et al., 2004). They found that expression of miR-16 in the placenta was negatively correlated with neonatal attention scores, while miR-146a and miR-182 were positively correlated with movement scores. These results suggest a potential link between placental miRNA signaling and neurodevelopment, although long-term follow-up and reassessment of neurodevelopmental outcomes is needed.
An aspect of placental miRNA signaling that makes these epigenetic regulators particularly intriguing in early life programming research is their ability to be detected in maternal blood (Chim et al., 2008), making them potential biomarkers for complications such as preeclampsia and ectopic pregnancy (Gunel et al., 2011; Miura et al., 2015; Zhao et al., 2013, 2012), as well as one day potentially for other maternal or fetal insults predictive of risk for neurodevelopmental disorders, such as autism spectrum disorders which have recently been associated with preeclampsic pregnancies (Walker et al., 2015). Although blood is rich in RNAses, miRNAs can travel through the maternal bloodstream complexed with Argonaute2 (Ago2), a key component of the RISC complex, or within extracellular vesicles termed exosomes providing protection from degradation (Arroyo et al., 2011; Fevrier and Raposo, 2004). Exosomes are small vesicles secreted by most cell types in the body that contain various signaling factors thought to be important in short- and long-term communication between cells and tissues. Although the function of circulating miRNAs is not well understood, they are presumed to direct gene expression in their recipient cells/tissues. Placentally-derived exosomes containing miRNAs have been identified in circulation during human pregnancy (Luo et al., 2009), although their maternal targets and their potential to reach the fetus is not currently understood. In a study using primary trophoblast cultures, exosome-mediated transfer of specific miRNAs from trophoblasts to non-placental cells conferred viral resistance to recipient cells (Delorme-Axford et al., 2013), suggesting an important role for placental exosome communication for immune system regulation during pregnancy.
5. OGT: a transceiver of nutrient signals
Various intra- and extra-cellular signals communicate the need to regulate chromatin repression/activation, determining which genes can be readily transcribed, and which mRNAs to target for destruction prior to protein translation. Signals related to nutrient availability are of particular importance for epigenetic programming in the placenta. The enzyme O-linked N-acetlyglucosamine (O-GlcNAc) transferase (OGT) is at the crossroads of nutritional signals and chromatin regulation. OGT catalyzes the addition of O-linked N-acetlyglucosamine (O-GlcNAc) to serine and threonine residues on thousands of intracellular proteins to extensively alter cellular signaling events in a nutrient-responsive manner. Discovered over 30 years ago in lymphocytes (Torres and Hart, 1984), the pervasiveness of this monosaccharide post-translation protein modification has only recently been widely appreciated, as we now know that O-GlcNAclyation impacts myriad signaling functions. O-GlcNAc competes with serine/threonine phosphorylation, affording OGT enormous potential to dynamically alter cellular signaling. In addition, O-GlcNAclyation can enhance proteosomal activity and protein stability and structure, as well as directly alter gene expression by guiding transcription factor localization and influencing RNA polymerase II activity (reviewed in Bond & Hanover, 2013). The OGT is located on the X-chromosome and escapes X-inactivation in the placenta resulting in higher OGT expression and O-GlcNAclyation in females, adding an additional level of importance to its role in sex-specific signaling.
Nutrient excess stimulates O-GlcNAc transfer to recipient proteins as the final step of the hexosamine biosynthetic pathway, a key mediator of glucose metabolism (Love and Hanover, 2005; Marshall et al., 1991). As nutrient transfer in the placenta from maternal sources to the developing fetus is of paramount importance, placental OGT affectively serves as a molecular “canary in a coal mine”, sensing changes in maternal energy and altering placental signaling to ultimately impact fetal programing. Recently, OGT’s domain has been expanded to epigenetics with the discovery that O-GlcNAclyation regulates several critical epigenetic links, including DNA demethylases and histone marks (reviewed by Dehennaut, Leprince, & Lefebvre, 2014).
5.1. OGT’s influence on epigenetics
The core histone proteins (H2A, H2B, H3, & H4) are directly modified by OGT, establishing O-GlcNAclyation as an important regulator of the histone code (Fujiki et al., 2011; Sakabe et al., 2010; Zhang et al., 2011). In addition, OGT’s widespread impact on cellular signaling can promote changes in other histone modifications, such as histone methylation.
The Polycomb Group proteins are typically associated with histone modifications important from early embryogenesis throughout life, most notably histone H3 di- and tri-methylation at lysine 27 (H3K27me3). Mutations in polycomb repressive complex 1 (PRC1) proteins are associated with abnormalities in neurological, skeletal, and stem cell development, whereas polycomb repressive complex 2 (PCR2) mutations result in embryonic lethality (reviewed in Kerppola, 2009). Studies in Drosophila initially identified OGT’s importance for maintenance of Polycomb transcriptional repression (Gambetta et al., 2009), a finding that has since been expanded to mammals. OGT mediated O-GlcNAcylation of EZH2, a H3K27 histone methyltransferase integral to PRC2, is necessary for stabilization of EZH2’s structure. OGT knockout reduces H3K27me3 without altering other histone methylation sites on H3, presumably due to decreased EZH2 function (Chu et al., 2014).
In addition to mediating epigenetic silencing through its interactions with PRC complexes as discussed above, OGT can facilitate permissive epigenetic marks through its association with the ten eleven translocation (TET) family of proteins (Figure 2D). The TET proteins are best known for their role in DNA demethylation (reviewed in Pastor et al., 2013). OGT uses TET2 and TET3 as scaffolding proteins, allowing OGT to modify histone H2B at ser112 (Chen et al., 2013), a site-specific modification that is associated with transcriptional activation (Fujiki et al., 2011). Further, the transcriptional activation mark H3K4me3 is dependent on O-GlcNAcylation of host cell factor 1 (HCF1), a key component of the SET1/COMPASS histone methylatransferase complex, in a TET2/3 dependent manner (Deplus et al., 2013). Although the above studies described the control of OGT activity by TET proteins, a reciprocal interaction wherein OGT regulates TET protein activity was only recently identified (Bauer et al., 2015). As O-GlcNAcylation competes with phosphorylation at serine and threonine residues, OGT activity can prevent phosphorylation at specific loci, altering protein regulation and signaling. TET proteins are typically highly phosphorylated in their N-terminal regulatory regions and OGT’s interaction with TET proteins is associated with decreased phosphorylation and enhanced O-GlcNAcylation, which might be important for dynamic regulation of these enzymes and thus potentially important for DNA methylation.
6. Prenatal stress alters placental function and signaling: impact on neurodevelopment
Studies in both human populations and animal models suggest a strong relationship between maternal stress and altered offspring physiological and psychiatric outcomes (reviewed in Bale et al., 2010; Talge et al., 2007; Weinstock, 2005).
6.1 Fetal programming by prenatal stress
A hallmark of stress exposure is increased production and circulation of cortisol (CORT; corticosterone in rodents). During pregnancy, high levels of maternal CORT are converted to inactive cortisone in the placenta by the enzyme 11 β-hydroxysteroid dehydrogenase type 2 (11β-HSD2)(Benediktsson et al., 1997). Early prenatal stress reduces 11β-HSD2 in the female mouse placenta (Pankevich et al., 2009), and stress in mid-gestation decreases 11β-HSD2 mRNA in both sexes in rat placenta (Peña et al., 2012). Decreased placental 11β-HSD2 expression in response to stress is associated with increased 11β-HSD2 promoter methylation and DNMT3a expression in the placenta (Peña et al., 2012), providing an epigenetic link between prenatal stress exposure and long-term gene programming in the placenta. The opposite effect of stress on 11β-HSD2 methylation was reported in the human placenta in women exposed to the stress of socioeconomic hardship during pregnancy. Methylation of 11β-HSD2 was lowest in placental tissue from women classified as having experienced high levels of socioeconomic adversity during pregnancy, although this study did not assess circulating CORT or 11β-HSD2 mRNA expression, and methylation was quantified at only 4 CpG sites. Male placentas were particularly vulnerable to adversity-associated decreases in this methylation. The authors concluded that low levels of 11β-HSD2 methylation may be an adaptive mechanism to increase placental 11β-HSD2 expression and buffer the fetus from maternal CORT (Appleton et al., 2013).
Other studies have examined methylation of key genes controlling CORT signaling in the placenta in association with infant neurobehavioral measures, an indication of future mental health and neurological function (Liu et al., 2010; Tronick and Lester, 2013). Impairments in infant neurobehavior (assessed by measuring reflex symmetry, excitability, and habituation to stimuli) were associated with increased methylation of Nr3c1, which codes for the glucocorticoid receptor (GR), in placentas from mothers who reported depression, and increased methylation of 11β-HSD2 in placentas from mothers who reported anxiety during pregnancy (Conradt et al., 2013). In a separate cohort of healthy patients, methylation at specific CpG sites on the Nr3c1 promoter in the placenta was positively correlated with infant quality of movement and attention scores (Bromer et al., 2013). High levels of methylation in the promoter region of placental 11β-HSD2 were associated with low infant birth weight and reduced quality movement scores (Marsit et al., 2012). Further, infants with high placental Nr3c1 methylation combined with low 11β-HSD2 methylation had greater reflex asymmetry, and infants with low placental Nr3c1 methylation and high 11β-HSD2 methylation showed lower excitability. Infants with high methylation in both genes had impairments in stimulus habituation, suggesting that specific patterns of methylation of these critical regulators of CORT in the placenta give rise to divergent endophenotypes (Appleton et al., 2015). High methylation of FKBP5, a CORT binding protein that negatively regulates CORT function, is associated with decreased expression of FKBP5 mRNA in the placenta and increased risk for infant arousal deficits (Paquette et al., 2014). Combined, these studies suggest that the placenta actively responds to exogenous hormonal cues, including those of maternal stress, via altered DNA methylation patterns that are associated with infant neurobehavioral outcome.
6.2 OGT regulates sex-specific programming of prenatal stress
Recently, our lab examined gene expression profiles in the male and female placenta in response to early prenatal stress across gestation (Howerton et al., 2013). Our model of early prenatal stress (EPS), in which pregnant mice are subject to chronic variable stress for the first 7 days of gestation, produces male offspring with endophenotypes characterized by hypothalamic mitochondrial dysfunction, HPA axis hypersensitivity, and enhanced stress responsiveness (Howerton and Bale, 2014; Howerton et al., 2013; Mueller and Bale, 2008). While the expression levels of thousands of placental genes dramatically changed across gestation, only 8 genes displayed significant sex differences across development. The X-linked OGT was significantly lower in the male placenta compared to the female placenta, and was further decreased following EPS, distinguishing it a potential mediator of the sex-specific effects of EPS on offspring programming. As detailed above, OGT is a perfect candidate to be the mediator of environmental signals from the maternal milieu to control placental homeostasis, ultimately directing the relay of information to the developing fetal brain. Importantly, the sex difference in OGT was accompanied by widespread sex differences in O-GlcNAcylation in the placental proteome, and EPS decreased O-GlcNAcylation levels extensively. These findings are exciting because they suggest widespread differences in OGT-mediated cellular signaling and potential differences in epigenetic regulation between the sexes and in response to stress, and indicate that OGT action in the placenta may be a key molecular mechanism for male-biased vulnerabilities to neurodevelopmental insults.
This hypothesis was supported by placental-specific knockout of OGT (pl-OGT), which completely recapitulated the EPS phenotype in male offspring (Howerton and Bale, 2014). As adults, pl-OGT males had decreased body weights and a hypersensitive HPA axis, as previously described in EPS males (Howerton et al., 2013; Mueller and Bale, 2008; Pankevich et al., 2009). In addition, both EPS and pl-OGT males showed similar placental gene expression patterns, with alterations in gene sets associated with endocrine and immune signals, suggesting that OGT regulates these genes in response to EPS and that these outcomes may be involved in signaling in the developing brain. Finally, when looking at hypothalamic function, both pl-OGT and EPS males had deficits in mitochondrial function, demonstrating that changes in placental signaling associated with stress can program long-term changes in the brain impacting neuroendocrine functioning. As OGT has numerous effectors in the proteome, the exact mechanism by which OGT programs gene expression in response to EPS is unknown. However, OGT’s well-described interactions with critical epigenetic mediators suggests that sex and stress-related differences in OGT levels likely lead to lasting differences in placental gene expression via epigenetic mechanisms (Bauer et al., 2015; Chu et al., 2014; Dehennaut et al., 2014; Deplus et al., 2013; Gambetta et al., 2009; Olivier-Van Stichelen et al., 2014; Sakabe et al., 2010).
7. Conclusions
Regulation of nutrient and metabolic sensing within the placenta is a critical aspect in homeostatic maintenance and guidance of appropriate signaling to the developing fetus. As with any biological system, the precise coordination required to produce a healthy brain is remarkable. Placental transmission of nutrients is merely a small part of this highly complex process, although the importance of its contribution is illuminated in instances of poor nutrition, stress or other environmental insults, which have long-lasting effects on offspring development. Metabolic programming is particularly vulnerable to these perturbations. Placental OGT is one known contributor to fetal programming, since it transduces environmental signals into epigenetic changes that can ultimately alter placental gene expression and function. While the literature reviewed above gives us a fundamental picture of the placenta’s role in neurodevelopmental programming, it is by no means exhaustive and the intricacies of how placental function influences brain development will undoubtedly become increasingly clear with additional mechanistic studies aided by modern technologies, particularly powerful transgenic mouse models and Next-Generation Sequencing.
Highlights.
The placenta is a critical mediator of maternal and fetal signals
Nutrient cues and maternal insults alter placental function
Alterations in placental epigenetics can have lasting impacts on fetal programming
Fetal sex also influence placental function
O-linked-N-acetylglucosamine transferase (OGT) is a key mediator of sex differences and stress responsivity in the placenta
Acknowledgments
Work in the Bale lab is supported by supported by MH091258, MH087597, MH099910, MH104184 to TLB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Bridget M. Nugent, Email: bnugent@vet.upenn.edu.
Tracy L. Bale, Email: tbale@vet.upenn.edu.
References
- Amankwah KS, Kaufmann RC. Ultrastructure of human placenta: effects of maternal drinking. Gynecol Obstet Invest. 1984;18:311–316. doi: 10.1159/000299099. [DOI] [PubMed] [Google Scholar]
- Appleton AA, Armstrong DA, Lesseur C, Lee J, Padbury JF, Lester BM, Marsit CJ. Patterning in placental 11-B hydroxysteroid dehydrogenase methylation according to prenatal socioeconomic adversity. PLoS One. 2013;8:e74691. doi: 10.1371/journal.pone.0074691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton AA, Lester BM, Armstrong DA, Lesseur C, Marsit CJ. Examining the joint contribution of placental NR3C1 and HSD11B2 methylation for infant neurobehavior. Psychoneuroendocrinology. 2015;52:32–42. doi: 10.1016/j.psyneuen.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL, Baram TZ, Brown AS, Goldstein JM, Insel TR, McCarthy MM, Nemeroff CB, Reyes TM, Simerly RB, Susser ES. Early life programming and neurodevelopmental disorders. Biol Psychiatry. 2010;68:314–319. doi: 10.1016/j.biopsych.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJP. The origins of the developmental origins theory. J Intern Med. 2007;261:412–417. doi: 10.1111/j.1365-2796.2007.01809.x. [DOI] [PubMed] [Google Scholar]
- Barker DJP. The developmental origins of adult disease. J Am Coll Nutr. 2004;23:588S–595S. doi: 10.1080/07315724.2004.10719428. [DOI] [PubMed] [Google Scholar]
- Bauer C, Gobel K, Nagaraj N, Colantuoni C, Wang M, Muller U, Kremmer E, Rottach A, Leonhardt H. Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the glycosyltransferase OGT. J Biol Chem. 2015 doi: 10.1074/jbc.M114.605881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayol SA, Farrington SJ, Stickland NC. A maternal “junk food” diet in pregnancy and lactation promotes an exacerbated taste for “junk food” and a greater propensity for obesity in rat offspring. Br J Nutr. 2007;98:843–851. doi: 10.1017/S0007114507812037. [DOI] [PubMed] [Google Scholar]
- Benediktsson R, Calder AA, Edwards CRW, Seckl JR. Placental 11β-hydroxysteroid dehydrogenase: a key regulator of fetal glucocorticoid exposure. Clin Endocrinol (Oxf) 1997;46:161–166. doi: 10.1046/j.1365-2265.1997.1230939.x. [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, Schoeftner S, Murchison E, Andl T, Chen T, Klatt P. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15:268–279. doi: 10.1038/nsmb.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1985;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bond MR, Hanover JA. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Bouret SG. Role of early hormonal and nutritional experiences in shaping feeding behavior and hypothalamic development. J Nutr. 2010;140:653–657. doi: 10.3945/jn.109.112433. [DOI] [PubMed] [Google Scholar]
- Bouret SG, Draper SJ, Simerly RB. Trophic Action of Leptin on Hypothalamic Neurons That Regulate Feeding. Sci. 2004;304:108–110. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- Breton C. The hypothalamus–adipose axis is a key target of developmental programming by maternal nutritional manipulation. J Endocrinol. 2013;216:R19–R31. doi: 10.1530/JOE-12-0157. [DOI] [PubMed] [Google Scholar]
- Bromer C, Marsit CJ, Armstrong DA, Padbury JF, Lester B. Genetic and epigenetic variation of the glucocorticoid receptor (NR3C1) in placenta and infant neurobehavior. Dev Psychobiol. 2013;55:673–683. doi: 10.1002/dev.21061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AM. Animal models of human placentation–a review. Placenta. 2007;28:S41–S47. doi: 10.1016/j.placenta.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Chavan-Gautam P, Sundrani D, Pisal H, Nimbargi V, Mehendale S, Joshi S. Gestation-dependent changes in human placental global DNA methylation levels. Mol Reprod Dev. 2011 doi: 10.1002/mrd.21296. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chim SSC, Shing TKF, Hung ECW, Leung T, Lau T, Chiu RWK, Lo YMD. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, Kang ML, Wong CH, Juan LJ. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci. 2014;111:1355–1360. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MAK, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 2013;45:325–329. doi: 10.1038/ng.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfarani S, Maiorana A, Geremia C, Scirè G, Spadoni GL, Germani D. Blood glucose concentrations are reduced in children born small for gestational age (SGA), and thyroid-stimulating hormone levels are increased in SGA with blunted postnatal catch-up growth. J Clin Endocrinol Metab. 2003;88:2699–2705. doi: 10.1210/jc.2002-021882. [DOI] [PubMed] [Google Scholar]
- Clifton VL. Review: Sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta. 2010;31(Suppl):S33–9. doi: 10.1016/j.placenta.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Coan PM, Burton GJ, Ferguson-Smith AC. Imprinted genes in the placenta--a review. Placenta. 2005;26(Suppl A):S10–20. doi: 10.1016/j.placenta.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Combs CA, Gunderson E, Kitzmiller JL, Gavin LA, Main EK. Relationship of fetal macrosomia to maternal postprandial glucose control during pregnancy. Diabetes Care. 1992;15:1251–1257. doi: 10.2337/diacare.15.10.1251. [DOI] [PubMed] [Google Scholar]
- Conradt E, Lester BM, Appleton AA, Armstrong DA, Marsit CJ. The roles of DNA methylation of NR3C1 and 11β-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics. 2013;8:1321–1329. doi: 10.4161/epi.26634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B, Kotlyar M, Evangelou AI, Ignatchenko V, Ignatchenko A, Whiteley K, Jurisica I, Adamson SL, Rossant J, Kislinger T. Comparative systems biology of human and mouse as a tool to guide the modeling of human placental pathology. Mol Syst Biol. 2009:5. doi: 10.1038/msb.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mello JCM, de Araujo ESS, Stabellini R, Fraga AM, de Souza JES, Sumita DR, Camargo AA, Pereira LV. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS One. 2010;5:e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehennaut V, Leprince D, Lefebvre T. O-GlcNAcylation, an Epigenetic Mark. Focus on the Histone Code, TET Family Proteins, and Polycomb Group Proteins. Front Endocrinol (Lausanne) 2014;5:155. doi: 10.3389/fendo.2014.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme-Axford E, Donker RB, Mouillet JF, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE. Human placental trophoblasts confer viral resistance to recipient cells. Proc Natl Acad Sci. 2013;110:12048–12053. doi: 10.1073/pnas.1304718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, Shih AH, Levine RL, Bernard O, Mercher T, Solary E, Urh M, Daniels DL, Fuks F. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi Y, Yamamoto M, Arishima K, Shirai M, Wakabayashi K, Leichter J, Lee M. Histological changes in the placenta induced by maternal alcohol consumption in the rat. Neonatology. 1989;56:158–164. doi: 10.1159/000243117. [DOI] [PubMed] [Google Scholar]
- Enquobahrie DA, Abetew DF, Sorensen TK, Willoughby D, Chidambaram K, Williams MA. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204:178.e12–21. doi: 10.1016/j.ajog.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson JG, Kajantie E, Osmond C, Thornburg K, Barker DJP. Boys live dangerously in the womb. Am J Hum Biol. 2010;22:330–335. doi: 10.1002/ajhb.20995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Forsen T, Eriksson JG, Tuomilehto J, Osmond C, Barker DJP. Growth in utero and during childhood among women who develop coronary heart disease: longitudinal study. Bmj. 1999;319:1403–1407. doi: 10.1136/bmj.319.7222.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowden AL, Giussani DA, Forhead AJ. Endocrine and metabolic programming during intrauterine development. Early Hum Dev. 2005a;81:723–734. doi: 10.1016/j.earlhumdev.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Giussani DA, Forhead AJ. Endocrine and metabolic programming during intrauterine development. Early Hum Dev. 2005b;81:723–734. doi: 10.1016/j.earlhumdev.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Ward JW, Wooding FPB, Forhead AJ, Constancia M. Programming placental nutrient transport capacity. J Physiol. 2006;572:5–15. doi: 10.1113/jphysiol.2005.104141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480:557–560. doi: 10.1038/nature10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A, Ferry L, Fajardy I, Jouneau L, Gothié JD, Vigé A, Fleur C, Mayeur S, Gallou-Kabani C, Gross MS, Attig L, Vambergue A, Lesage J, Reusens B, Vieau D, Remacle C, Jais JP, Junien C. Maternal Diets Trigger Sex-Specific Divergent Trajectories of Gene Expression and Epigenetic Systems in Mouse Placenta. PLoS One. 2012;7:e47986. doi: 10.1371/journal.pone.0047986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallou-Kabani C, Gabory A, Tost J, Karimi M, Mayeur S, Lesage J, Boudadi E, Gross MS, Taurelle J, Vigé A. Sex-and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One. 2010;5:e14398. doi: 10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Müller J. Essential Role of the Glycosyltransferase Sxc/Ogt in Polycomb Repression. Sci. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- Ganapathy V. Drugs of abuse and human placenta. Life Sci. 2011;88:926–930. doi: 10.1016/j.lfs.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- Georgieff MK. Nutrition and the developing brain: nutrient priorities and measurement. Am J Clin Nutr. 2007;85:614S–620S. doi: 10.1093/ajcn/85.2.614S. [DOI] [PubMed] [Google Scholar]
- Gheorghe CP, Goyal R, Mittal A, Longo LD. Gene expression in the placenta: maternal stress and epigenetic responses. Int J Dev Biol. 2010;54:507. doi: 10.1387/ijdb.082770cg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey K, Robinson S, Barker DJP, Osmond C, Cox V. Maternal nutrition in early and late pregnancy in relation to placental and fetal growth. Bmj. 1996;312:410. doi: 10.1136/bmj.312.7028.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey KM, Barker DJP. Maternal nutrition in relation to fetal and placental growth. Eur J Obstet Gynecol Reprod Biol. 1995;61:15–22. doi: 10.1016/0028-2243(95)02148-l. [DOI] [PubMed] [Google Scholar]
- Godfrey KM, Lillycrop KA, Burdge GC, Gluckman PD, Hanson MA. Epigenetic mechanisms and the mismatch concept of the developmental origins of health and disease. Pediatr Res. 2007;61:5R–10R. doi: 10.1203/pdr.0b013e318045bedb. [DOI] [PubMed] [Google Scholar]
- Gu Y, Sun J, Groome LJ, Wang Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am J Physiol - Endocrinol Metab. 2013 doi: 10.1152/ajpendo.00660.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan YJ, Yang X, Wei L, Chen Q. MiR-365: a mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J. 2011;25:4457–4466. doi: 10.1096/fj.11-185132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Gunel T, Zeybek YG, Akcakaya P, Kalelioglu I, Benian A, Ermis H, Aydinli K. Serum microRNA expression in pregnancies with preeclampsia. Genet Mol Res. 2011;10:4034–4040. doi: 10.4238/2011.November.8.5. [DOI] [PubMed] [Google Scholar]
- Gunnar MR, Wewerka S, Frenn K, Long JD, Griggs C. Developmental changes in hypothalamus–pituitary–adrenal activity over the transition to adolescence: normative changes and associations with puberty. Dev Psychopathol. 2009;21:69–85. doi: 10.1017/S0954579409000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Witmer PDW, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1290–1294. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- Hay WW, Sparks JW, Wilkening RB, Battaglia FC, Meschia G. Fetal glucose uptake and utilization as functions of maternal glucose concentration. Am J Physiol Metab. 1984;246:E237–E242. doi: 10.1152/ajpendo.1984.246.3.E237. [DOI] [PubMed] [Google Scholar]
- Higashijima A, Miura K, Mishima H, Kinoshita A, Jo O, Abe S, Hasegawa Y, Miura S, Yamasaki K, Yoshida A, Yoshiura K, Masuzaki H. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat Diagn. 2013;33:214–222. doi: 10.1002/pd.4045. [DOI] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howerton CL, Bale TL. Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc Natl Acad Sci. 2014;111:9639–9644. doi: 10.1073/pnas.1401203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howerton CL, Morgan CP, Fischer DB, Bale TL. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc Natl Acad Sci. 2013;110:5169–5174. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, McVean GT. Growth effects of uniparental disomies and the conflict theory of genomic imprinting. Trends Genet. 1997;13:436–443. doi: 10.1016/S0168-9525(97)01273-0. [DOI] [PubMed] [Google Scholar]
- Jauniaux E, Burton GJ. Morphological and biological effects of maternal exposure to tobacco smoke on the feto-placental unit. Early Hum Dev. 2007;83:699–706. doi: 10.1016/j.earlhumdev.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science (80-) 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones HN, Woollett LA, Barbour N, Prasad PD, Powell TL, Jansson T. High-fat diet before and during pregnancy causes marked up-regulation of placental nutrient transport and fetal overgrowth in C57/BL6 mice. FASEB J. 2009;23:271–278. doi: 10.1096/fj.08-116889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kennedy LA. Changes in the term mouse placenta associated with maternal alcohol consumption and fetal growth deficits. Am J Obstet Gynecol. 1984;149:518–522. doi: 10.1016/0002-9378(84)90028-0. [DOI] [PubMed] [Google Scholar]
- Kerppola TK. Polycomb group complexes – many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Torre R, Nigro G, Mazzocco M, Best AM, Adler SP. Placental enlargement in women with primary maternal cytomegalovirus infection is associated with fetal and neonatal disease. Clin Infect Dis. 2006;43:994–1000. doi: 10.1086/507634. [DOI] [PubMed] [Google Scholar]
- Lager S, Powell TL. Regulation of nutrient transport across the placenta. J Pregnancy. 2012;2012:179827. doi: 10.1155/2012/179827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lesseur C, Armstrong DA, Murphy MA, Appleton AA, Koestler DC, Paquette AG, Lester BM, Marsit CJ. Sex-specific associations between placental leptin promoter DNA methylation and infant neurobehavior. Psychoneuroendocrinology. 2014;40:1–9. doi: 10.1016/j.psyneuen.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester BM, Tronick EZ, Brazelton TB. The Neonatal Intensive Care Unit Network Neurobehavioral Scale procedures. Pediatrics. 2004;113:641–667. [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Linzer DIH, Fisher SJ. The placenta and the prolactin family of hormones: regulation of the physiology of pregnancy. Mol Endocrinol. 1999;13:837–840. doi: 10.1210/mend.13.6.0286. [DOI] [PubMed] [Google Scholar]
- Liu J, Bann C, Lester B, Tronick E, Das A, Lagasse L, Bauer C, Shankaran S, Bada H. Neonatal Neurobehavior Predicts Medical and Behavioral Outcome. Pediatr. 2010;125:e90–e98. doi: 10.1542/peds.2009-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Verkerk AJ, van Putten WL, Oosterhuis JW. Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am J Hum Genet. 1999;64:1445–52. doi: 10.1086/302382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Hanover JA. The Hexosamine Signaling Pathway: Deciphering the “O-GlcNAc Code”. Sci Signal. 2005;2005:re13–re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, Takizawa T, Shigihara T, Goto T, Izumi A. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- Maccani MA, Avissar-Whiting M, Banister CE, McGonnigal B, Padbury JF, Marsit CJ. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics. 2010;5:583–589. doi: 10.4161/epi.5.7.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani MA, Padbury JF, Lester BM, Knopik VS, Marsit CJ. Placental miRNA expression profiles are associated with measures of infant neurobehavioral outcomes. Pediatr Res. 2013;74:272–278. doi: 10.1038/pr.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccani MA, Padbury JF, Marsit CJ. miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS One. 2011;6:e21210. doi: 10.1371/journal.pone.0021210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairesse J, Lesage J, Breton C, Bréant B, Hahn T, Darnaudéry M, Dickson SL, Seckl J, Blondeau B, Vieau D. Maternal stress alters endocrine function of the feto-placental unit in rats. Am J Physiol Metab. 2007;292:E1526–E1533. doi: 10.1152/ajpendo.00574.2006. [DOI] [PubMed] [Google Scholar]
- Malassine A, Frendo J, Evain-Brion D. A comparison of placental development and endocrine functions between the human and mouse model. Hum Reprod Update. 2003;9:531–539. doi: 10.1093/humupd/dmg043. [DOI] [PubMed] [Google Scholar]
- Mao J, Zhang X, Sieli PT, Falduto MT, Torres KE, Rosenfeld CS. Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc Natl Acad Sci. 2010;107:5557–5562. doi: 10.1073/pnas.1000440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marconi AM, Paolini C, Buscaglia M, Zerbe G, Battaglia FC, Pardi G. The impact of gestational age and fetal growth on the maternal-fetal glucose concentration difference. Obstet Gynecol. 1996;87:937–942. doi: 10.1016/0029-7844(96)00048-8. [DOI] [PubMed] [Google Scholar]
- Markakis EA. Development of the neuroendocrine hypothalamus. Front Neuroendocrinol. 2002;23:257–291. doi: 10.1016/s0091-3022(02)00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- Marsit CJ, Maccani MA, Padbury JF, Lester BM. Placental 11-beta hydroxysteroid dehydrogenase methylation is associated with newborn growth and a measure of neurobehavioral outcome. PLoS One. 2012;7:e33794. doi: 10.1371/journal.pone.0033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Axelman J, Jeppesen P. Differential X reactivation in human placental cells: implications for reversal of X inactivation. Am J Hum Genet. 2005;77:355–64. doi: 10.1086/432815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheev AM, Nabekura T, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF, Unadkat JD. Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reprod Sci. 2008;15:866–877. doi: 10.1177/1933719108322425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri K, Varmuza S. Imprinting and extraembryonic tissues-mom takes control. Int Rev Cell Mol Biol. 2009;276:215–62. doi: 10.1016/S1937-6448(09)76005-8. [DOI] [PubMed] [Google Scholar]
- Miura K, Higashijima A, Mishima H, Miura S, Kitajima M, Kaneuchi M, Yoshiura K, Masuzaki H. Pregnancy-associated microRNAs in plasma as potential molecular markers of ectopic pregnancy. Fertil Steril. 2015;103:1202–1208. doi: 10.1016/j.fertnstert.2015.01.041. [DOI] [PubMed] [Google Scholar]
- Moore VM, Davies MJ, Willson KJ, Worsley A, Robinson JS. Dietary composition of pregnant women is related to size of the baby at birth. J Nutr. 2004;134:1820–1826. doi: 10.1093/jn/134.7.1820. [DOI] [PubMed] [Google Scholar]
- Mouillet J, Chu T, Sadovsky Y. Expression patterns of placental microRNAs. Birth Defects Res Part A Clin Mol Teratol. 2011;91:737–743. doi: 10.1002/bdra.20782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlhausler BS, Adam CL, Findlay PA, Duffield JA, McMillen IC. Increased maternal nutrition alters development of the appetite-regulating network in the brain. FASEB J. 2006;20:1257–1259. doi: 10.1096/fj.05-5241fje. [DOI] [PubMed] [Google Scholar]
- Munoz-Suano A, Hamilton AB, Betz AG. Gimme shelter: the immune system during pregnancy. Immunol Rev. 2011;241:20–38. doi: 10.1111/j.1600-065X.2011.01002.x. [DOI] [PubMed] [Google Scholar]
- Murphy VE, Smith R, Giles WB, Clifton VL. Endocrine regulation of human fetal growth: the role of the mother, placenta, and fetus. Endocr Rev. 2006;27:141–169. doi: 10.1210/er.2005-0011. [DOI] [PubMed] [Google Scholar]
- Noh JH, Chang YG, Kim MG, Jung KH, Kim JK, Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH. MiR-145 functions as a tumor suppressor by directly targeting histone deacetylase 2 in liver cancer. Cancer Lett. 2013;335:455–462. doi: 10.1016/j.canlet.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Novakovic B, Yuen RK, Gordon L, Penaherrera MS, Sharkey A, Moffett A, Craig JM, Robinson WP, Saffery R. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics. 2011;12:529. doi: 10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent BM, Wright CL, Shetty AC, Hodes GE, Lenz KM, Mahurkar A, Russo SJ, Devine SE, McCarthy MM. Brain feminization requires active repression of masculinization via DNA methylation. Nat Neurosci. 2015;18 doi: 10.1038/nn.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier-Van Stichelen S, Abramowitz LK, Hanover JA. X marks the spot: does it matter that O-GlcNAc Transferase is an X-linked gene? Biochem Biophys Res Commun. 2014;453:201–7. doi: 10.1016/j.bbrc.2014.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osei-Kumah A, Smith R, Jurisica I, Caniggia I, Clifton VL. Sex-specific differences in placental global gene expression in pregnancies complicated by asthma. Placenta. 2011;32:570–578. doi: 10.1016/j.placenta.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Pankevich DE, Mueller BR, Brockel B, Bale TL. Prenatal stress programming of offspring feeding behavior and energy balance begins early in pregnancy. Physiol Behav. 2009;98:94–102. doi: 10.1016/j.physbeh.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette AG, Lester BM, Koestler DC, Lesseur C, Armstrong DA, Marsit CJ. Placental FKBP5 Genetic and Epigenetic Variation Is Associated with Infant Neurobehavioral Outcomes in the RICHS Cohort. PLoS One. 2014;9:e104913. doi: 10.1371/journal.pone.0104913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrakuljic A, Derewlany LO, Koren G. Maternal cocaine use and cigarette smoking in pregnancy in relation to amino acid transport and fetal growth. Placenta. 1999;20:499–512. doi: 10.1053/plac.1999.0418. [DOI] [PubMed] [Google Scholar]
- Peña CJ, Monk C, Champagne FA. Epigenetic effects of prenatal stress on 11β-hydroxysteroid dehydrogenase-2 in the placenta and fetal brain. PLoS One. 2012;7:e39791. doi: 10.1371/journal.pone.0039791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Petraglia F, Florio P, Nappi C, Genazzani AR. Peptide Signaling in Human Placenta and Membranes: Autocrine, Paracrine, and Endocrine Mechanisms*. Endocr Rev. 1996;17:156–186. doi: 10.1210/edrv-17-2-156. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: Lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Sakabe K, Wang Z, Hart GW. β-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siiteri PK, Febres F, Clemens LE, Chang RJ, Gondos B, Stites D. PROGESTERONE AND MAINTENANCE OF PREGNANCY: IS PROGESTERONE NATURE’S IMMUNOSUPPRESSANT?*. Ann N Y Acad Sci. 1977;286:384–397. doi: 10.1111/j.1749-6632.1977.tb29431.x. [DOI] [PubMed] [Google Scholar]
- Simpson ER, MacDonald PC. Endocrine physiology of the placenta. Annu Rev Physiol. 1981;43:163–188. doi: 10.1146/annurev.ph.43.030181.001115. [DOI] [PubMed] [Google Scholar]
- Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Natl Acad Sci. 2006;103:5478–5483. doi: 10.1073/pnas.0508035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan EL, Grayson B, Takahashi D, Robertson N, Maier A, Bethea CL, Smith MS, Coleman K, Grove KL. Chronic consumption of a high-fat diet during pregnancy causes perturbations in the serotonergic system and increased anxiety-like behavior in nonhuman primate offspring. J Neurosci. 2010;30:3826–3830. doi: 10.1523/JNEUROSCI.5560-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. 1975. [DOI] [PubMed] [Google Scholar]
- Talge NM, Neal C, Glover V. Antenatal maternal stress and long-term effects on child neurodevelopment: how and why? J Child Psychol Psychiatry. 2007;48:245–261. doi: 10.1111/j.1469-7610.2006.01714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamimi RM, Lagiou P, Mucci LA, Hsieh CC, Adami HO, Trichopoulos D. Average energy intake among pregnant women carrying a boy compared with a girl. Bmj. 2003;326:1245–1246. doi: 10.1136/bmj.326.7401.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tau GZ, Peterson BS. Normal development of brain circuits. Neuropsychopharmacology. 2010;35:147–168. doi: 10.1038/npp.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PD, Poston L. Developmental programming of obesity in mammals. Exp Physiol. 2007;92:287–298. doi: 10.1113/expphysiol.2005.032854. [DOI] [PubMed] [Google Scholar]
- Thomas P, Peabody J, Turnier V, Clark RH. A new look at intrauterine growth and the impact of race, altitude, and gender. Pediatrics. 2000;106:e21–e21. doi: 10.1542/peds.106.2.e21. [DOI] [PubMed] [Google Scholar]
- Tijsterman M, Plasterk RHA. Dicers at RISC: the mechanism of RNAi. Cell. 2004;117:1–3. doi: 10.1016/s0092-8674(04)00293-4. [DOI] [PubMed] [Google Scholar]
- Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- Tronick E, Lester BM. Grandchild of the NBAS: the NICU network neurobehavioral scale (NNNS): a review of the research using the NNNS. J Child Adolesc Psychiatr Nurs. 2013;26:193–203. doi: 10.1111/jcap.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuddenham L, Wheeler G, Ntounia-Fousara S, Waters J, Hajihosseini MK, Clark I, Dalmay T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science (80-) 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa PD, Buss C, Entringer S, Swanson JM. Developmental origins of health and disease: brief history of the approach and current focus on epigenetic mechanisms, in: Seminars in Reproductive Medicine. NIH Public Access. 2009:358. doi: 10.1055/s-0029-1237424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CK, Krakowiak P, Baker A, Hansen RL, Ozonoff S, Hertz-Picciotto I. Preeclampsia, placental insufficiency, and autism spectrum disorder or developmental delay. JAMA Pediatr. 2015;169:154–162. doi: 10.1001/jamapediatrics.2014.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- Watson ED, Cross JC. Development of structures and transport functions in the mouse placenta. Physiology. 2005;20:180–193. doi: 10.1152/physiol.00001.2005. [DOI] [PubMed] [Google Scholar]
- Weinstock M. The potential influence of maternal stress hormones on development and mental health of the offspring. Brain Behav Immun. 2005;19:296–308. doi: 10.1016/j.bbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Wessels JM, Edwards AK, Khalaj K, Kridli RT, Bidarimath M, Tayade C. The MicroRNAome of Pregnancy: Deciphering miRNA Networks at the Maternal-Fetal Interface. PLoS One. 2013;8:e72264. doi: 10.1371/journal.pone.0072264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M, Feng K-TV, Bernlohr DA, McDonagh S, Pereira L, Sali A, Fisher SJ. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology. 2007;148:1059–1079. doi: 10.1210/en.2006-0683. [DOI] [PubMed] [Google Scholar]
- Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Xiang AH, Wang X, Martinez MP, Walthall JC, Curry ES, Page K, Buchanan TA, Coleman KJ, Getahun D. Association of maternal diabetes with autism in offspring. JAMA. 2015;313:1425–1434. doi: 10.1001/jama.2015.2707. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdravkovic T, Genbacev O, McMaster MT, Fisher SJ. The adverse effects of maternal smoking on the human placenta: a review. Placenta. 2005;26:S81–S86. doi: 10.1016/j.placenta.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Zhang S, Roche K, Nasheuer HP, Lowndes NF. Modification of histones by sugar beta-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J Biol Chem. 2011;286:37483–37495. doi: 10.1074/jbc.M111.284885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Moley KH, Gronowski AM. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin Biochem. 2013;46:953–960. doi: 10.1016/j.clinbiochem.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhao Q, Warrick J, Lockwood CM, Woodworth A, Moley KH, Gronowski AM. Circulating microRNA miR-323-3p as a biomarker of ectopic pregnancy. Clin Chem. 2012;58:896–905. doi: 10.1373/clinchem.2011.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Han T, Sargent IL, Yin G, Yao Y. Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am J Obstet Gynecol. 2009;200:661.e1–7. doi: 10.1016/j.ajog.2008.12.045. [DOI] [PubMed] [Google Scholar]