Abstract
Background
Cardiomyocytes are rich in mitochondria which are situated in spatially-distinct subcellular regions including those under the plasma membrane, subsarcolemmal mitochondria; and those between the myofibrils, interfibrillar mitochondria. We previously observed subpopulation-specific differences in mitochondrial proteomes following diabetic insult. The objective of this study was to determine whether mitochondrial genome-encoded proteins are regulated by microRNAs inside the mitochondrion and whether subcellular spatial location or diabetes mellitus influences the dynamics.
Methods and Results
Using microarray technology coupled with cross-linking immunoprecipitation and next generation sequencing, we identified a pool of mitochondrial microRNAs, termed mitomiRs that are redistributed in spatially-distinct mitochondrial subpopulations in an inverse manner following diabetic insult. Redistributed mitomiRs displayed distinct interactions with the mitochondrial genome requiring specific stoichiometric associations with RISC constituents argonaute-2 (Ago2) and fragile X mental retardation–related protein 1 (FXR1) for translational regulation. In the presence of Ago2 and FXR1, redistribution of mitomiR-378 to the IFM following diabetic insult led to down regulation of mitochondrially-encoded F0 component ATP6. Next generation sequencing analyses identified specific transcriptome and mitomiR sequences associated with ATP6 regulation. Overexpression of mitomiR-378 in HL-1 cells resulted in its accumulation in the mitochondrion and down-regulation of functional ATP6 protein, while antagomir blockade restored functional ATP6 protein and cardiac pump function.
Conclusions
We propose mitomiRs can translationally regulate mitochondrially-encoded proteins in spatially-distinct mitochondrial subpopulations during diabetes mellitus. The results reveal the requirement of RISC constituents in the mitochondrion for functional mitomiR translational regulation and provide a connecting link between diabetic insult and ATP synthase function.
Keywords: microRNA, mitochondria, diabetes mellitus
Introduction
Emerging technologies are likely to aid in the discovery of the underlying gene regulation linking the mitochondrion to disease initiation and progression. Cardiac tissue is rich in mitochondria with spatially-distinct subpopulations which include subsarcolemmal mitochondria (SSM), located beneath the cell membrane, and interfibrillar mitochondria (IFM), situated between the myofibrils1, 2. A wealth of studies have indicated that spatially-distinct cardiac mitochondrial subpopulations are influenced differentially by pathological and physiological stresses3. We have observed distinct effects on cardiac IFM following type 1 diabetic insult with a more limited effect on SSM2, 4. Type 1 diabetes mellitus influences cardiac contractility of which mitochondrial dysfunction is central2, 4–6. The complete mitochondrial genomic sequence has been elucidated7 with each mitochondrion possessing between two and ten copies8. The mitochondrial genome is comprised of 37 genes, 13 of which code for proteins that form parts of the oxidative phosphorylation machinery, while the remaining genes code for 22 transfer RNAs and two ribosomal RNAs9. Absence of introns renders the mitochondrial DNA compact while lack of histones influences mutation rates and DNA damage repair mechanisms8. As such, the development and progression of many human diseases involves regulation of the mitochondrial genome10, 11.
MicroRNAs (miRNAs) have been shown to play an integral role in regulating gene expression through translational repression or degradation of target messenger RNAs (mRNA). MiRNAs are non-coding RNAs, which influence post-transcriptional gene regulation by binding to the 3'-untranslated region (3'-UTR) of target mRNAs. Regulation of gene expression at the post-transcriptional level occurs through binding of the miRNA to complementary sites on the target mRNA. Association with the RNA-induced silencing complex (RISC) has been shown to occur in the cytoplasm, thereby regulating translation and/or degradation of the target mRNA12. Target mRNAs can contain AU rich elements (ARE) which, in conjunction with a given miRNA, will enhance recruitment of the RISC components, including nuclear-encoded argonaute-2 (Ago2) and fragile X mental retardation–related protein 1 (FXR1), thereby influencing translation13. Mature miRNAs have been observed in the nucleus and specialized processing bodies14, as well as in the mitochondrion15–19 though the mechanisms contributing to their presence is limited. A recent review supporting the existence of miRNAs in the mitochondrion have utilized the terminology mitomiR in reference to mitochondrial miRNAs20. Pharmacological inhibition of cardiac-specific miRNA enhances metabolic energy homeostasis suggestive of a link between miRNA regulation and the mitochondrion21. Further, mice lacking specific miRNAs, including miR-378, exhibit enhanced mitochondrial fatty acid metabolism and elevated oxidative capacity22. Though increasing evidence indicates that mitochondrial subpopulations are differentially influenced by physiological stressors4, 23–26, no study has reported specific enrichment patterns of mitomiRs in spatially-distinct subpopulations or whether distribution patterns are influenced by pathological states, including diabetes mellitus. The results of our studies suggest a dynamic system requiring the presence of RISC constituents in the mitochondrion, which function to regulate the mitochondrial genome during diabetic insult in a spatially-distinct manner.
Methods
Experimental Animals and Diabetes Induction
The experiments in this study conformed to the NIH Guide for the Care and Use of Laboratory Animals (8th Edition) and were approved by the West Virginia University Animal Care and Use Committee. Male FVB mice (The Jackson Laboratory, Bar Harbor, ME) were housed in the West Virginia University Health Sciences Center animal facility and given food and water ad libitum. Type 1 diabetes mellitus was induced in 5 week old mice using multiple low-dose streptozotocin (STZ; Sigma, St. Louis, MO) injections as previously described2, 4. Mice that served as vehicle controls were given the same volume per body weight of sodium citrate buffer. One week post-injections, hyperglycemia was confirmed by measuring fasting blood glucose levels using a commercially available kit (Bayer, Mishiwaka, IN). Blood glucose levels greater than 250 mg/dL were considered diabetic.
Preparation of Individual Mitochondrial Subpopulations
At 5 weeks post-hyperglycemia onset, mice were sacrificed and their hearts excised. SSM and IFM subpopulations were isolated as previously described1 with minor modifications2, 4, 27–29. Following isolation, SSM and IFM were further purified by percoll gradient (23%, 15%, 10% and 3% percoll solution) and centrifuged in a Beckman Optima™ MAX-XP Ultracentrifuge (Beckman Coulter, Fullerton, CA) at 32,000 × g for 8 minutes. Mitochondrial subpopulation pellets were resuspended in a sucrose based SEM buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS, pH 7.2) or mitochondrial extraction buffer (Biovision, Mountain View, CA) depending upon the assay to be performed. Mitochondrial protein concentrations were determined using the Bradford method and bovine serum albumin as a standard30.
RNA Isolation and Microarray Experimentation
Total RNA was extracted from isolated mitochondrial subpopulations using a Vantage Total RNA Purification Kit (Origene, Rockville, MD). Sample labeling and hybridization was performed using an Agilent miRNA Complete Labeling and Hybridization Kit (Agilent Technologies, Santa Clara, CA) following the manufacturer’s protocol. Briefly, 80 ng of total RNA was labeled and subsequently hybridized overnight (20 hours at 55°C) to an Agilent Mouse microRNA Microarray 8x60K v.16 (1080 targets) using Agilent’s recommended hybridization chamber and oven. The microarrays were washed once with Agilent Gene Expression Wash Buffer 1 (5 minutes at room temperature) followed by a second wash with preheated Agilent Gene Expression Wash Buffer 2 (5 minutes at 37°C). Fluorescence signals were detected using Agilent’s Microarray Scanner System (Agilent Technologies, Santa Clara, CA). Signal intensities from the raw data lists were normalized using the 90% percentile. To discriminate questionable results from relevant results normalized data lists were generated and filtered only for those mitomiRs which had at least in one sample a signal intensity ≥ 1 light unit, a signal intensity of ≥ 10 light unit and a signal intensity of ≥ 100 light unit. MiRNA expression FES derived output data files were analyzed using Gene Spring GX software (Agilent Technologies, Santa Clara, CA). Candidate miRNAs were identified and analyzed statistically.
QRT-PCR of Mature miRNAs
Briefly, 50 ng of total RNA was reverse transcribed into cDNA using a First-strand cDNA Synthesis Kit for miRNA (Origene, Rockville, MD) as per the manufacturer’s protocol. Equal amounts of cDNA from control and diabetic mouse mitochondrial subpopulations were subjected to qRT-PCR. For HL-1 and miR-378 overexpressing HL-1 cell lines, isolated mitochondria were subjected to qRT-PCR. Custom primers were designed for miR-378 (Origene, Rockville, MD) and pre-miR-378 (Integrated DNA Technologies, Coralville, IA) and utilized for the quantification of cDNA replicates using QSTAR SYBR master mix (Origene, Rockville, MD). The sequences of the miR-378 primers were the following: forward sequence 5’-CTG ACT CCA GGT CCT GTG-3’; reverse sequence 5’-GAA CAT GTC TGC GTA TCT C-3’. The sequences for the pre-miR-378 primers were the following: forward sequence 5’-CTC CAG GTC CTG TGT GTT ACC-3’; reverse sequence 5’-GCC TTC TGA CTC CAA GTC CA-3’. The sequences for the RNA polymerase II (AMA) primers were the following: forward sequence 5’-GCG CAG AGA GTA TCC TGG AC-3’’ reverse sequence: 5’-GGA GCT CGA GTG GAT CTT CG-3’. QRT-PCR assays were performed in duplicate using an Applied Biosystems 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). All miRNA expression levels were normalized to U6 RNA content or AMA content for pre-miR-37831.
Western Blot Analyses
SDS-PAGE was run on 4–12% gradient gels, as previously described2, 32, with equal amounts of protein loaded for each study treatment. Relative amounts of subpopulation-specific mitochondrial localized RISC complex proteins, Ago2 and FXR1, as well as biotinylated proteins were quantified using the following primary antibodies; anti-Ago2 rabbit antibody (product #2897, Cell Signaling, Danvers, MA), anti-FXR1 rabbit antibody (product #4264S, Cell Signaling, Danvers, MA) and anti-biotin rabbit antibody (product #5571, Cell Signaling, Danvers, MA). Mitochondrial loading control was assessed using an anti-COX IV rabbit antibody (product #ab16056, Abcam, Cambridge, MA). Relative amounts of specific proteins were quantified using the following antibodies: anti-ATP6 rabbit antibody (product #ab102573, Abcam, Cambridge, MA), anti-GAPDH rabbit antibody (product #ab8245, Abcam, Cambridge, MA), anti-calnexin rabbit antibody (product #ADI-SPA-860, Enzo, Farmingdale, NY), and anti-ATPB mouse antibody (product# ab14730, Cambridge, MA). The secondary antibody used in the analyses was a goat anti-rabbit IgG HRP conjugate (product #10004301, Cayman Chemical, Ann Arbor, MI) or anti-mouse IgG HRP Conjugate (product #31430, Thermo Scientific, Pittsburgh, PA). Detection and quantitation of chemiluminescent signals were performed using a G:BOX (Syngene, Frederick, MD), and the data expressed as arbitrary optical density units (ODU). Secondary confirmation of protein loading and equal protein transfer was performed using Ponceau staining.
ATP Synthase Activity
ATP synthase activity was determined by following the decrease in NADH absorption at 340 nm as previously described29 and final values expressed as micromoles consumed per minute per milligram of protein30.
UV Cross-Linking Immunoprecipitation (CLIP)
Mouse heart tissue was minced in a 2 ml volume of ice-cold PBS, transferred to a petri dish with a suspension depth of approximately 1 mm. Samples were irradiated 5 times with 400 mJ/cm2 (~15 cm distance from the UV source) on ice using a CL-1000 Ultraviolet Crosslinker (UVP, Upland, CA), and mixed between each irradiation33. After irradiation, samples were pelleted by centrifugation (2500 rpm for 5 minutes at 4°C) and the supernatant removed. SSM and IFM were isolated as described above and resuspended in IP buffer (20mM Tris pH 8.0; 137mM NaCl; 10% Glycerol; 1% Nonidet P-40 (NP-40); 2mM EDTA; 1/100 protease inhibitor cocktail). Protein concentrations were determined as above. Two hundred µg of resuspended protein was added to IP buffer up to 1 ml and the RNA digested by addition of 10 µl of RNase I (Life Technologies, Grand Island, NY) (1:500 dilution). The complex was incubated (3 minutes at 37°C) with constant shaking (1,100 rpm) then transferred to ice. Fifty µl of Dynabeads Protein A (Life Technologies, Grand Island, NY) were washed 3 times with IP buffer and then resuspended in a 100 µl of solution of IP buffer and 5 µl of a specific antibody (anti-Ago2 or anti-FXR1). The antibody was allowed to bind to the beads by rotating the tubes (1 hour at room temperature). The beads were then washed three times with IP buffer, and cross-linked mitochondrial subpopulation lysates added followed by tube rotation (2 hours at 4°C). Beads were then washed three times with IP buffer. A portion of the sample was utilized to check antibody specificity using high RNase I (1:50 dilution), while the other portion was used for RNA 3'-end biotinylation labeling (Thermo Scientific, Pittsburgh, PA) using T4 RNA ligase to attach a single biotinylated nucleotide to the 3' terminus of an RNA strand. Samples were incubated overnight at 16°C and then washed with IP buffer. Thirty µl of loading buffer without denaturing agent was added to both the biotinylated and high RNAse I treated beads and heated for 10 minutes at 70°C with shaking (1000 rpm). Samples were submitted to SDS-PAGE as described above. The biotinylated (low RNAse I) sample bands were cut with a clean scalpel and the nitrocellulose membrane placed into a microcentrifuge tube. One hundred µl of proteinase K solution was added to each tube, incubated for 20 minutes at 37°C with shaking (1000 rpm), and then the RNA extracted using a Vantage Total RNA Purification Kit (Origene, Rockville, MD) as above. To determine mitomiR and mitochondrial mRNA abundances in the mitoRISCome, we utilized next generation sequencing (see below) as well as complementary qRT-PCR analyses on targets that were the focus of the current study. These complementary approaches were undertaken to provide confirmational evidence of reproducibility of the next generation sequencing. QRT-PCR was performed using custom primers designed for targets of mouse miR-378 as described above as well as mitochondrially-encoded genome primer duplex targets (Origene, Rockville, MD). Data were normalized to CLIP total RNA input and RISC proteins (Ago2 and FXR1).
Next Generation Sequencing
Four groups of pooled CLIP-RNA samples (control and diabetic; SSM and IFM; n = 5 per group) were used for high-throughput sequencing. MRNA and miRNA library preparation was performed using Illumina TrueSeq mRNA Library and Illumina TrueSeq small mRNA Library Preparation kits (Illumina, San Diego, CA), respectively. Briefly, as per the manufacturer’s instructions, initial RT–PCR, and barcoded products were run on a 2% agarose gel and 200- to 400-bp products purified using QiaEx II beads (Qiagen, Valencia, CA). Gel-purified material was amplified for 12 cycles with fusion Illumina adaptor primers and the DNA was subjected to next generation sequencing. The sequencing of CLIP tags was performed with barcoded libraries using the Illumina Sequencing (Mi-Seq) Adaptor primers. Base calling, library sorting by barcode, and mapping to the transcriptome were performed, subsequently.
Bioinformatic Analysis of CLIP Tags
CLIP tags were aligned to the mm10 genome with mitochondrial genome and analyzed with Avadis NGS which is an integrated platform that provides tools for extensive workflows for alignment, analysis, management and visualization of next-generation sequencing data (http://www.avadis-ngs.com/). To crosscheck the Avadis NGS results, we employed several other platforms including Galaxy (http://galaxy.psu.edu), the UCSC genome browser (http://genome.ucsc.edu/), and miRBASE (http://microrna.sanger.ac.uk). RNA abundance from the paired-end RNA-Seq data was expressed as Fragments Per Kilobase of transcript per Million mapped reads (FPKM). To assess RISCome enrichment and depletion scores between the diabetic and control SSM and IFM, diabetic RISCome FPKM values were normalized to control RISCome FPKM values with a cut off ≥1.5. To estimate the false negative rate, we utilized DESeq for identifying differentially expressed gene miRNAs. Multiple test corrections were performed using Benjamin Hochberg FDR (cut off 0.05). Results were compared by examining the number of Ago2-mRNA clusters with no predicted seeds in the top 20 Ago2-miRNAs identified by next generation sequencing. To generate Ago2-mitomiR ternary maps, we examined all mitomiR seeds in the Ago2 footprint region analysis. From these data we looked for the greatest enrichment of miRNA seeds in the Ago2-mRNA clusters that were present within the top 20 identified miRNAs. We calculated the frequency of conserved mitomiR seed matches observed in the Ago2-mRNA footprint regions.
Cell Culture and Stable Cell Line Development
Cell culture and stable cell line development were carried out using the mouse cardiomyocyte cell line (HL-1) which maintains a cardiac-specific phenotype following repeated passaging34 and HEK293 cells as previously described35. Cells were grown at 37°C in a humidified atmosphere of 5% CO2 / 95% air and maintained in either Claycomb media (HL-1) (Sigma Aldrich, St. Louis, MO) with 10% fetal bovine serum (Sigma, St. Louis, MO) and other supplements as previously described34 or Dulbecco’s Modified Eagle Medium (Cellgro, Manassas, VA)(HEK293). MiRNA expression plasmid (pCMV-MIR) which contains coral GFP for cellular tracking and assessment of transfection efficiency, housing the mouse precursor sequence of miR-378 (pre-miR-378) was utilized for stable cell line development (Product # sc401025; Origene, Rockville, MD). The pre-miR-378 precursor sequence was the following: 5′-AGGGCTCCTG ACTCCAGGTC CTGTGTGTTA CCTAGAAATA GCACTGGACT TGGAGTCAGA AGGCCT-3′, where the mature sequence is underlined. The pre-miR-378 sequence was situated at the 293 bp – 358 bp of a 656 bp insert. The total 656 bp sequence was situated in the multicloning site of the pCMV-MIR plasmid between Sgf I and Mlu I restriction sites.
Cells were seeded at a density of ~1–3 × 105 cells/35-mm dishes and transfected at 60–70% confluence using an X-tremeGENE HP DNA Transfection Kit per the manufacturer’s protocol (Roche, Indianapolis, IN). Forty-eight hours post-transfection, stable cell lines were selected in media containing G418 (Gibco, Grand Island, NY). After, 4–5 passages, cells were washed with PBS and harvested in either 1X RIPA Buffer (Sigma, St. Louis, MO) for total lysate or purified mitochondria isolated using a mitochondrial isolation kit (Biovision, Milpitas, CA) for protein and enzymatic analyses. Citrate synthase activity was measured using a commercially available kit (Sciencell, San Diego, CA).
miR Targeting Luciferase Assay
The human ATP6 gene was purchased in the pMIR-Target vector and utilized for miRNA binding analyses (Origene, Rockville, MD). 100 ng of ATP6 pMIR-Target vector was co-transfected as described previously35 with 100 ng of human miR-378, human miR-200c, or empty pCMV-MIR in triplicate in 96-well culture plates. 24 hours post-transfection, luciferase activities were assayed with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) per manufacturer’s protocol using a Flexstation 3 Luminometer (Molecular Devices, Sunnyvale, CA). Luciferase activity was quantified and reported as the ratio of Firefly luciferase to Renilla luciferase.
In Vivo Antagomir Delivery in the Diabetic Heart
To determine whether manipulation of miR-378 in vivo influences ATP6 levels, ATP synthase functionality and ultimately, cardiac function, we utilized an antagomir delivery approach. Antagomirs are anti-sense oligonucleotide sequences conjugated with cholesterol which can be delivered by injection, systemically36, 37. Antagomirs provide sustained miRNA blockade and are currently in Phase II trials to treat a number of pathologies38, 39. Linked nucleic acid (LNA) antagomirs are unique in that their sugar ring is locked with a methylene bridge placing the miRNA into an optimal end conformation and creating a more stable duplex leading to greater specificity for a given nucleic acid target. Four groups were utilized for the studies; 1) LNA-miR-378 treatment (Seq: 5’TGACTCCAAGTCCAG3’) (Exiqon, Woburn, MA) which was injected following confirmation of hyperglycemia; 2) LNA-scrambled control (Seq: 5’ACGTCTATACGCCCA3’) (Exiqon, Woburn, MA) which was injected following confirmation of hyperglycemia; 3) diabetic control which was injected with STZ; and 4) vehicle control which was injected with 0.9% NaCl. Five week old male FVB mice were imaged by echocardiography (described below) as a baseline after which type 1 diabetes mellitus was induced using multiple low-dose STZ injections as described above. Antagomir and vehicle treatment groups were intraperitoneally-injected once weekly for 4 weeks at a volume of 250 µl per injection and a concentration of 25 mg antagomir/kg body weight. This concentration and dosing schedule was chosen based on previous literature using similar LNA-miR compounds40. One week following the last injection (5 weeks total treatment time; 11 weeks of age) mice were reimaged by echocardiography. Mice were then euthanized and cardiac mitochondrial subpopulations isolated as described above.
Cardiac Function
Transthoracic echocardiography was performed as previously described by our laboratory41, 42 and others43. Briefly, mice were lightly anesthetized with inhalant isofluorane and transferred to dorsal recumbency. Using the Vevo 2100 Imaging System (Visual Sonics, Toronto, Canada) and a 32- to 55-MHz linear array transducer, micro-ultrasound images were acquired. M-mode images were captured via the parasternal short axis at midpapillary level with all images acquired at the highest possible frame rate (233–401 frames/s). Ejection fraction and fractional shortening were obtained from the scanned images.
Statistical Analyses
Means and standard errors (SE) are reported for all data sets. A Mann-Whitney non-parametric test was utilized when evaluating differences between two groups, and a Kruskal-Wallis analysis of variance (ANOVA) non-parametric test followed by a Dunn’s Multiple Comparison test was utilized when evaluating differences between three or more groups (GraphPad Software Inc., La Jolla, CA). For cardiac functional parameters, data were analyzed with a pair-wise Student’s t-test. P<0.05 was considered significant across all testing.
Results
Mitochondrial Purity and Quality Control
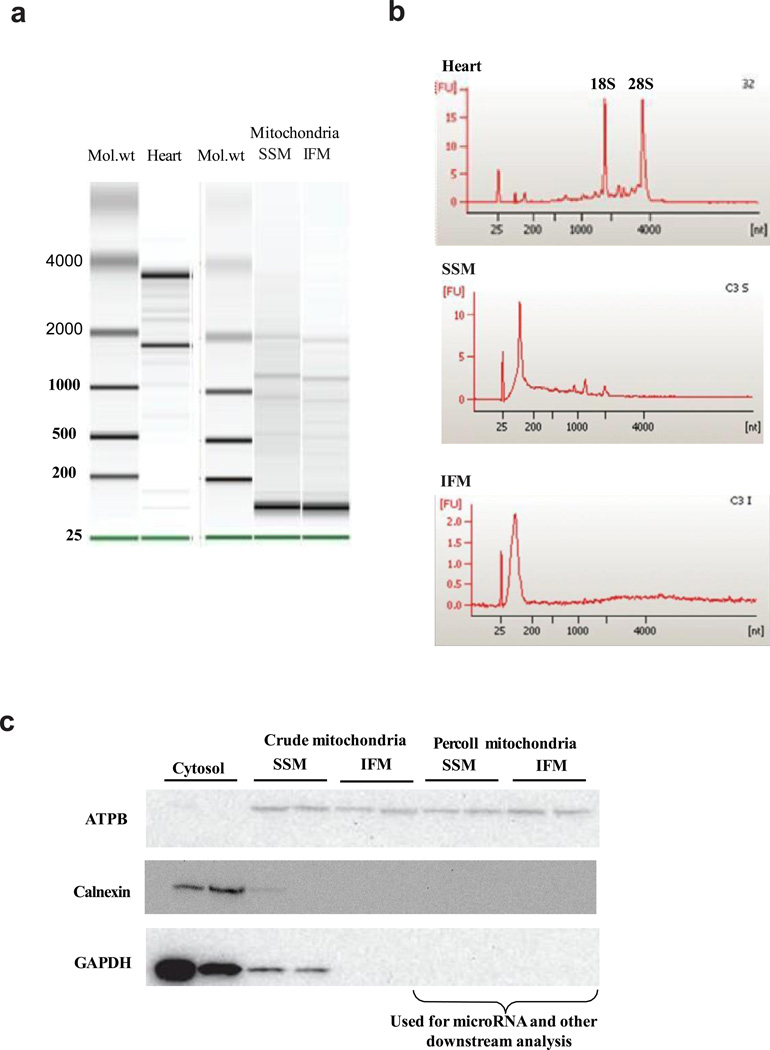
Isolation of purified SSM and IFM subpopulations from control and diabetic mouse heart was performed using a multi-step approach. Mitochondrial subpopulation isolation was followed by percoll density gradient centrifugation to eliminate extramitochondrial contamination. This approach has been proven to yield enriched mitochondria with a high degree of purity and elimination of extra-organelle contamination in a number of different tissue types, including muscle44–46. Confirmation of mitochondrial purity was assessed by gel electrophoresis and electropherograms identifying ribosomal RNAs (18S and 28S) in whole heart which were not identified in mitochondrial subpopulations (Figures 1a and 1b). The presence of a large peak between 25 and 200 nt in the mitochondrial samples was reflective of small RNAs including miRNAs (Figure 1b). Further confirmation of mitochondrial purity was carried out by assessing the presence of the cytosolic protein GAPDH, the endoplasmic reticulum protein calnexin, and the mitochondrial protein ATPB. Figure 1c indicates the presence of ATPB solely in mitochondrial preparations, crude as well as percoll gradient, with the presence of GAPDH and calnexin solely in the cytosol and crude SSM preparations. Absence of GAPDH and calnexin in percoll gradient mitochondria confirmed mitochondrial purity. Percoll gradient mitochondria was used for the experiments.
Figure 1.
Quality control assessment. (A) Gel electrophoresis of RNA isolated from whole heart and RNA from mitochondrial subpopulations. The lowest migrating green band is an internal standard. (B) Electropherograms of total RNA from whole heart and from isolated mitochondria. Scaling of the y-axis is done automatically, relative to the strongest signal within a single run. (C) Western blot analyses for GAPDH, calnexin, and ATPB protein content in cytosol, crude mitochondria, and percoll gradient purified mitochondria.
Redistribution of MitomiRs in the Diabetic Heart
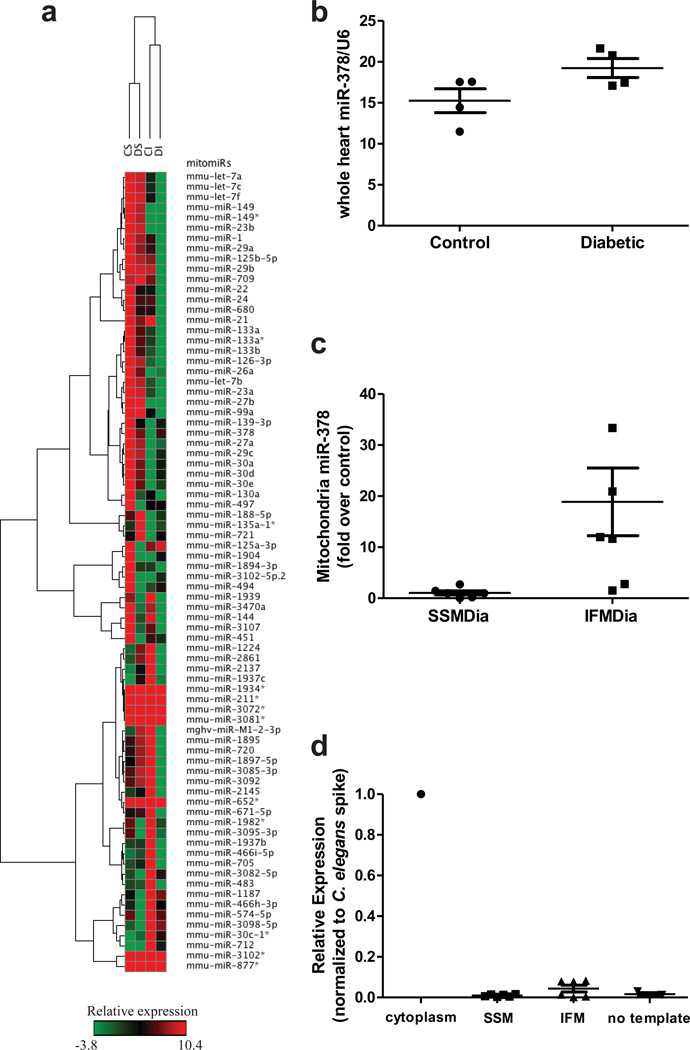
We profiled the expression levels of miRNAs in diabetic and control mouse cardiac SSM and IFM using a high-throughput miRNA microarray. Profiling of the mitomiRs, revealed a significantly altered inverse expression profile following diabetes mellitus insult for many of the mitomiRs when comparing the two subpopulations. The miRNA array analyses indicated that 78 mitomiRs were differentially regulated in the two mitochondrial subpopulations following type 1 diabetic insult relative to control (Figure 2a). Confirmation of the microarray analyses was performed by selecting a mitomiR displaying significantly increased expression levels in diabetic IFM relative to control (mitomiR-378). Evaluation of miR-378 presence in whole heart tissue by qRT-PCR revealed no significant differences relative to control (Figure 2b). In contrast, the observed change in expression levels of mitomiR-378 was validated in IFM by qRT-PCR (Figure 2c), suggesting that the increased presence of mitomiR-378 in the IFM was a function of redistribution as opposed to an increase in mass flux to the heart. It should be pointed out the absolute differences in fold change of mitomiR-378 differed between microarray and qRT-PCR platforms which may be a function of the previously noted underestimation in microarray approaches47. Pre-miR-378 was not detected at a significant level in isolated mitochondria as reflected by similar levels to no template controls suggesting that its origination and processing into a mature form occurs primarily in the cytosol (Figure 2d).
Figure 2.
Relative normalized mitomiR expression patterns in cardiac mitochondrial subpopulations. (A) Hierarchical clustering heat map for the microarray analysis of mitomiR expression profiles in cardiac mitochondrial subpopulations. CS = control SSM, DS = diabetic SSM, CI = control IFM, DI = diabetic IFM; n = 4. All mitomiRs reported in the heat map are expressed as normalized intensities. (B) QRT-PCR analyses of miR-378 in control and diabetic whole heart tissue. Values are represented as mean ± SE, n = 4. U6 mRNA served as control. (C) QRT-PCR analyses of mitomiR-378 in SSM and IFM diabetic mitochondrial subpopulations as compared with control. Values are represented as mean ± SE, n = 6. U6 mRNA served as control. (D) QRT-PCR analyses for pre-miR-378 in SSM and IFM diabetic mitochondrial subpopulations as compared with control. Values are represented as mean ± SE, n = 6 for SSM and IFM. AMA mRNA served as a reference control; cytoplasm served as a positive control; no template served as a negative control.
MitomiR Predicted Mitochondrial Targets and Sequence Specific Motifs
To determine whether resident mitomiRs were associated with the regulation of mitochondrial genome-encoded proteins, we utilized bioinformatic tools to predict potential mitomiR interactions with the mitochondrial genome. Using the target-prediction algorithms from various miRNA databases (RNAhybrid, MicroCosm and miRWalk), we analyzed the seed sequences from each identified mitomiR (7 seed nucleotides) and evaluated their probability of interacting with sequences from the mitochondrial genome by assessing the free energy value that characterizes the stability of the miRNA/mRNA interaction (greater than −20 kcal/mol)48. Among the 78 mitomiRs identified, 46 were predicted to bind directly to mRNA sequences from the mitochondrial genome suggesting potential regulation of translation and/or influence on mRNA stability. Using Ingenuity Pathway Analysis software to analyze mitochondrial genome targets, we observed potential interactions between all 13 mitochondrial genome-encoded electron transport chain proteins (Supplementary Figure 1a). Of the 46 mitomiRs predicted to bind to mRNA from the mitochondrial genome, 20 indicated potential binding to a single mitochondrial genome target (Supplementary Figure 1a; in red with red lines), while 26 displayed the potential to bind to multiple mitochondrial genome targets (Supplementary Figure 1a; in blue with black lines). Of interest was a predicted interaction between mitomiR-378 and ATP6 (Supplementary Figure 1a; orange line). We determined whether any specific conserved nucleotide sequence motifs existed among the 46 mitomiRs predicted to bind to mRNA from the mitochondrial genome and identified 40 mitomiRs with specific tri-nucleotide motifs suggesting sequence commonality among the majority of identified mitomiRs predicted to bind to the mitochondrial transcriptome. Specific enriched motifs included AGG, UGG and CCU tri-nucleotide sequences (Supplementary Figure 1b). Of particular interest was the observed high enrichment of UGG and AGG motifs in the 7 nucleotide seeding region (Supplementary Figure 1b). It would be of interest to determine whether the existence of these motifs in a given miRNA may be predictive of its potential for distribution to the mitochondrion. A similar type of phenomenon based upon the possession of specific sequence elements has been reported for the subcellular localization of miRNAs into other organelles49.
Functional Consequences of MitomiR-378 Targeting to the Mitochondrial Genome
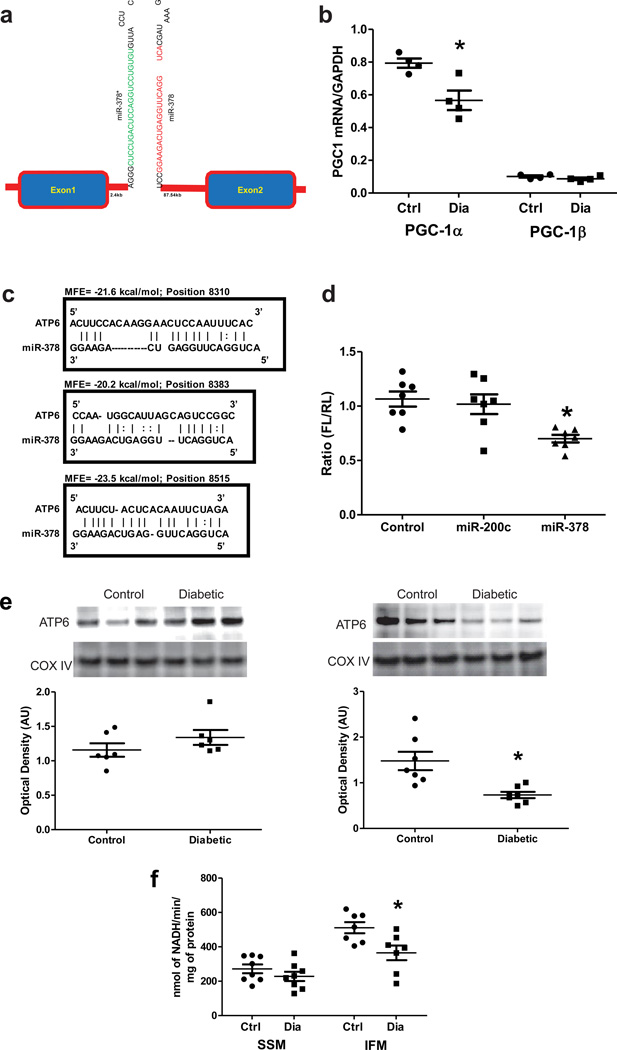
Because it displayed a robust increase in diabetic IFM relative to control IFM and was identified as a potential regulator of the ATP synthase, we performed additional analyses on mitomiR-378. MiR-378 is of nuclear origin and originates from the first intron between exons 1 and 2 of the PPARGC1β (PGC-1β) gene which generates a hairpin structure (Figure 3a)50. In the type 1 diabetic heart PGC-1α mRNA expression was significantly down-regulated while PGC-1β remained unchanged (Figure 3b). These results are in agreement with a previous report22. Bioinformatic analyses indicated that mitomiR-378 was predicted to bind to the mRNA sequence of ATP synthase F0 subunit 6 (ATP6), a constituent of the F0 component of the ATP synthase, which plays a role in the translocation of protons across the inner mitochondrial membrane. Interestingly, mitomiR-378 was indicated as potentially binding to three different regions of the mitochondrial genome at starting positions 8,310, 8,383 and 8,515 on the ATP6 mRNA with high affinity based upon the minimum free energy (MFE) value characterizing the interaction between miRNA and target (Figure 3c). To confirm these prediction analyses we performed a cell-based luciferase reporter assay in which the ATP6 full-length sequence was downstream of the firefly luciferase gene in the pMIR-Target plasmid. To determine specificity of miR-378, the luciferase reporter plasmid was cotransfected into HEK293 cells with either miR-378, miR-200c which is not predicted to bind with the ATP6 mRNA, or pCMV-MIR control. MiR-378 was capable of significantly reducing luciferase activity by 25%, whereas the control miRNA plasmid (pCMV-MIR) and miR-200c could not (Figure 3d). These results indicate that ATP6 is a potential target of miR-378 and recognizes the putative recognition sites. Western blot analyses indicated significant decreases in ATP6 of diabetic IFM relative to control IFM (Figure 3e). Evaluation of ATP synthase activities indicated a significant decrease in diabetic IFM with no significant effect to diabetic SSM, relative to respective controls (Figure 3f). These data support the hypothesis that enhanced mitomiR-378 expression in diabetic IFM may account for decreases in F0 ATP synthase subunits which lead ultimately to a decreased ability to drive the ATP synthase and generate ATP for cardiac pump function.
Figure 3.
MitomiR-378 and its targeting to the mitochondrial genome. (A) Schematic representation of miR-378 location in the PPARGC1b (PGC-1b) gene. (B) QRT-PCR quantification of PGC1α and PGC1β transcripts in control and diabetic hearts. GAPDH served as control. Values are represented as mean ± SE; n = 4 for each group. *P < 0.05 for Control vs. Diabetic. (C) Evaluation of the minimum free energy (MFE) value that characterizes the stability of mitomiR-378/mRNA interaction, identified by MicroCosm and RNAhybrid. (D) Relative luciferase activities of ATP6 reporter co-expressed with miR-378, miR-200c, or plasmid (pCMV-MIR) control in HEK293 cells 24 hours post transfection. Firefly luciferase activity was normalized to Renilla luciferase activity. Values are represented as mean ± SEM; n = 7 per group. *p<0.05 for miR-378 vs. all other groups. (E) Western blot analysis of ATP6 in control and diabetic SSM and IFM. Cox IV serves as a loading control. Values are represented as mean ± SE; n = 6. *p<0.05 for Control vs. Diabetic. (F) ATP synthase activity expressed in activity/min/mg protein. Values are means ± SE; n = 8 for each group. *P < 0.05 for Control IFM vs. Diabetic IFM.
Redistribution of RISC Components in the Mitochondrion
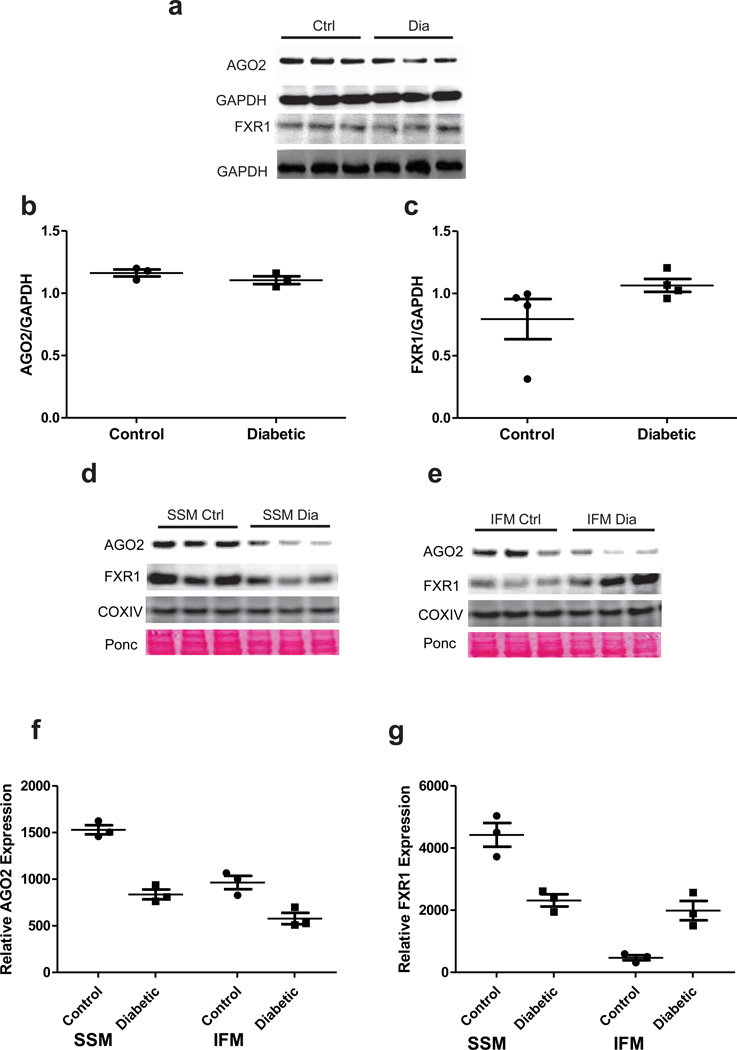
Small RNAs can interact with the RISC which contains proteins that include Argonautes, and FXR1, as well as other proteins and cellular factors51, 52. Assembly of the RISC and its subsequent association with a miRNA has been implicated in its biological function including regulation of mRNA expression53, 54. MiRNA-mediated inhibition of translation depends largely upon the degree of complementarity between a specific miRNA and its mRNA target55. RISC components such as FXR1 and Argonautes such as Ago2 interact with miRNAs during pathological conditions suggesting that stress-induced activation of the RISC contributes to its assembly56. Examination of RISC components Ago2 and FXR1 in whole heart homogenate revealed no significant differences during diabetic insult indicating that the absolute levels of these proteins are not changed as a result of the pathology (Figures 4a–c). Interestingly, examination of Ago2 and FXR1 protein contents in the mitochondrion revealed that both proteins were present within the two subpopulations (Figures 4d–g), whereas the presence of Dicer, a RISC-loading protein, and GW182, a cytoplasmic miRNA docking protein, were not present (Supplementary Figure 2). Following diabetic insult, both proteins displayed redistributed patterns with Ago2 levels decreasing in both SSM (46% decrease) and IFM (41% decrease) while FXR1 levels were decreased only in the SSM (48% decrease) (Figures 4d–g). In contrast, FXR1 protein contents were dramatically increased in diabetic IFM (423% increase) (Figures 4e and 4g). It is unclear why FXR1 was increased solely in diabetic IFM. Among FXR1’s proposed subcellular locales is the Z-disc where it colocalizes with proteins that play an integral role in cytoskeleton maintenance in part through regulating local mRNA targets19. Because IFM and FXR1 are both situated in close proximity to the contractile apparatus (myofibril) there may be an enhanced potential for FXR1 localization to the IFM. Future studies addressing this finding are warranted. Taken together, these results indicate that RISC components Ago2 and FXR1 are present in both mitochondrial subpopulations. Further, following diabetic insult, both proteins display subpopulation-specific differential distribution patterns.
Figure 4.
Redistribution of RISC components in the mitochondrion. (A) Western blot analyses of Ago2 and FXR1 in control and diabetic whole heart. GAPDH serves as loading control. (B) Quantification of Ago2 protein content in control and diabetic whole heart. Values are means ± SE; n = 3. (C) Quantification of FXR1 protein content in control and diabetic whole heart. Values are means ± SE; n = 4. GAPDH served as loading control. Western blot analyses of Ago2 and FXR1 in Control and Diabetic (D) SSM and (E) Control and Diabetic IFM. Quantification of (F) Ago2 and (G) FXR1 protein content in control and diabetic mitochondrial subpopulations. Values are means ± SE; n = 3. COX IV serves as a loading control.
MitomiR and RISC Constituent Interactions Influence Mitochondrial Genome-Encoded Protein Abundance
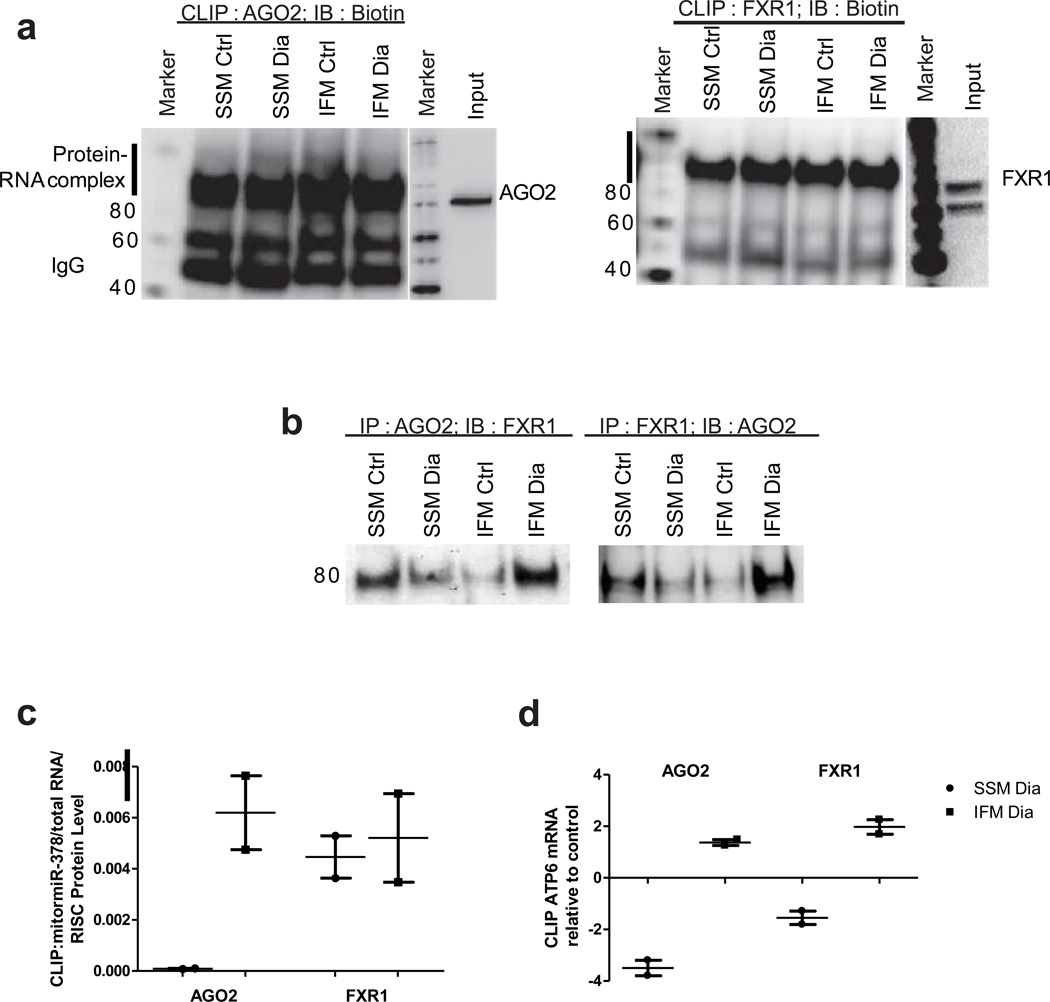
Based on our broad-scale mitomiR analyses, we determined how a particular mitomiR and mRNA target interact. Chi et al. identified Ago2-miRNA-mRNA complexing using high-throughput sequencing of RNAs isolated by cross-linked immunoprecipitation (HITS-CLIP) technology in mouse brain33. Using a similar approach we determined the endogenous interactions between mitomiRs, mRNAs and mitochondrial RISC components Ago2 and FXR1 in control and diabetic mitochondrial subpopulations. Cross-linked immunoprecipitation (CLIP) was utilized to determine the presence of mitomiRs and mRNAs enriched with RISC components Ago2 and FXR1 in an effort to identify and detail the presence of a functional regulatory complex in the mitochondrion (Supplementary Figure 3). Ago2-RNA and FXR1-RNA complexes were identified in both control and diabetic states and displayed molecular weight range shifts from 85 kDa to between 95 and 110 kDa indicating binding to the mitomiR-mRNA complex (Figure 5a). To confirm the interaction between RISC components Ago2 and FXR1, the complex was treated with high RNAse I and both RISC components immunoprecipitated with one another revealing interaction between the two proteins which was altered by the diabetic phenotype. Specifically, diabetic IFM displayed an increased interaction between Ago2 and FXR1 relative to control in both immunoprecipitation experiments which was not observed in diabetic SSM (Figure 5b). To determine the specific mitomiR and mRNAs present in the CLIP, portions of the membrane in which the protein-RNA complex resided were isolated and RNA extracted after which the reverse transcribed product was utilized for cDNA synthesis. CDNA libraries were analyzed to identify enriched quantities of mitomiR-378 by qRT-PCR analysis and its association with both Ago2 and FXR1. These analyses indicated that in diabetic IFM, both RISC constituents Ago2 and FXR1 were highly associated with mitomiR-378 (Figure 5c). In contrast, diabetic SSM displayed little association between mitomiR-378 and Ago2 yet a strong association between mitomiR-378 and FXR1 (Figure 5c). These observations indicate that in the diabetic IFM, mitomiR-378 is highly associated with components Ago2 and FXR1 from the mitochondrial RISC. In contrast, diabetic SSM display low enrichment of mitomiR-378 such that the association of Ago2 with this mitomiR is limited despite the presence of FXR1. These data suggest that the regulation of mitochondrial genome-encoded proteins by mitomiRs requires the presence of a given mitomiR and the appropriate amount of mitochondrial RISC proteins Ago2 and FXR1. In the absence of either protein and/or the mitomiR, regulation of mitochondrial gene expression by the mitomiR may be limited. Complementary analyses were carried out by examining the interaction between RISC constituents Ago2 and FXR1 with mitochondrially-encoded ATP6 mRNA using qRT-PCR (Figure 5d). The analyses indicated that mitochondrially-encoded ATP6 mRNA was enriched in the diabetic IFM relative to control IFM mitoRISCome regardless of whether immunoprecipitating with Ago2 or FXR1. In contrast, this phenomenon was not apparent in the diabetic SSM which displayed decreases in ATP6 mRNA in both Ago2 and FXR1 immunoprecipitated mitoRISComes (Figure 5d). These data confirm that mitochondrial RISC constituent Ago2 interacts in a ternary complex with both the specific mitomiR (mitomiR-378) and targeted mitochondrial genome mRNA (ATP6) providing a regulatory platform for the control of mitochondrial gene translation.
Figure 5.
Crosslinked immunoprecipitation (CLIP) in cardiac mitochondrial subpopulations and MitomiR-378 RISC constituent interactions with the mitochondrial genome. (A) Western blots of biotinylated RNA from CLIP-Ago2 and CLIP-FXR1 reactions illustrating crosslinked protein/RNA and the associated gel shift from 80 kDa to 95–110 kDa. (B) Western blot analyses of CLIP-Ago2 and CLIP-FXR1 subjected to RNAase I treatment at 1:50 dilution (high RNAase) illustrating interaction between the two proteins in the absence of RNA. (C) CLIP-Ago2 and CLIP-FXR1 associated enrichment analyses of mitomiR-378 analyzed by qRT-PCR in control and diabetic cardiac mitochondrial subpopulations. Values are presented as means ± SE; n = 2 where each individual sample represents a pool of 5 individual animals. (D) CLIP-Ago2 and CLIP-FXR1 associated enrichment analysis of transcripts for mitochondrial encoded ATP6 mRNA levels as assessed by qRT-PCR analysis in control and diabetic cardiac mitochondrial subpopulations. Values are presented as means ± SE; n = 2 where each individual sample represents a pool of 5 individual animals.
Next Generation Sequencing of Mitochondrial RISCome Association with Mitochondrial mRNA and MitomiRs
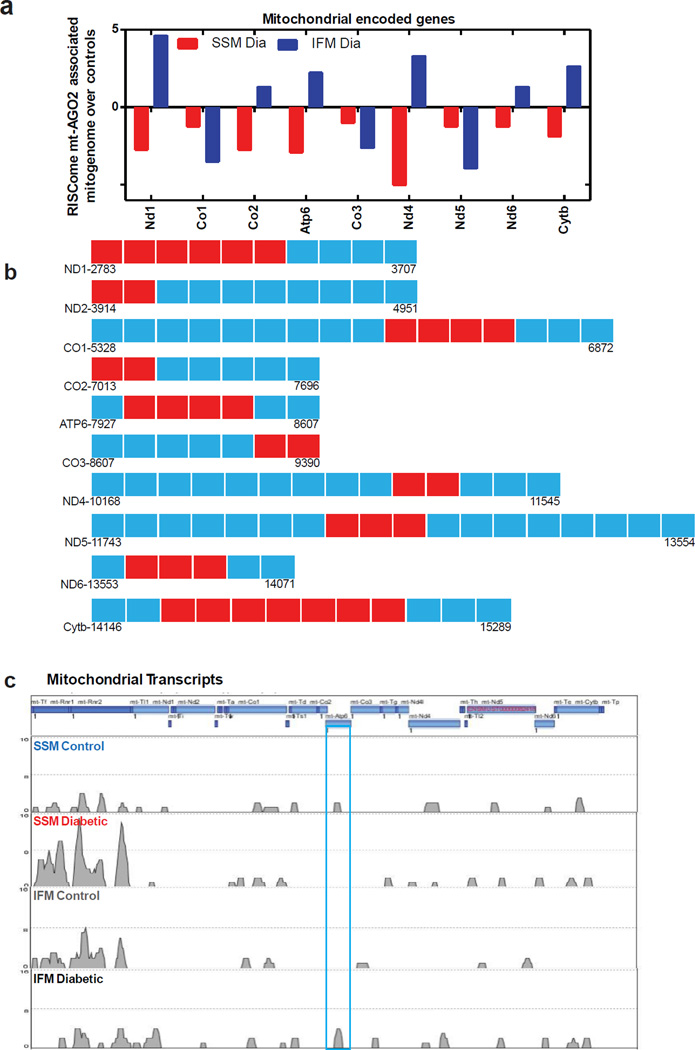
Our assessment of RISC component associations with mitochondrial miRNA and mRNA revealed differential binding patterns with Ago2 such that functional association was lost in the complex with diabetic SSM, while a strong association existed with the diabetic IFM. Thus, our analyses suggested that Ago2 presence was requisite for translational control of mitochondrial genome protein expression via mitomiR regulatory mechanisms. To capture the distinct features of the mitochondrial transcriptome functional complex, we assessed CLIP-Ago2 (mitoRISCome)/mRNA complex products utilizing true RNA-seq library preparations to establish mitochondrial mRNA transcripts through sequencing with a high-throughput Mi-seq platform. Total reads and uniquely matched reads are reported in Supplementary Table 1. Specific associations between RISC component (Ago2) and each of the 13 mitochondrially-encoded proteins under control and diabetic conditions are indicated in Figure 6c. The presence of mitochondrial mRNAs in the different groups varied depending upon treatment and subpopulation type. Of particular interest was the presence of mRNA encoding ATP6 in the mitoRISCome of diabetic IFM which was not present in the control IFM. This finding was confirmatory of those in the mitoRISCome analyses (Figure 5d). Our analyses identified 9 mitochondrially-encoded mRNAs in the diabetic subpopulations which were differentially expressed in the mitoRISCome relative to controls (Figure 6a). Further, 6 of the differentially expressed mRNAs displayed opposite expression patterns from their subpopulation counterpart (Figure 6a). Mapping of the mitochondrial mRNAs identified in the mitoRISCome next generation sequencing enabled identification of specific cluster regions in each of the 13 mRNA transcripts which were present in the mitoRISCome (Figure 6b; blocks indicated in red).
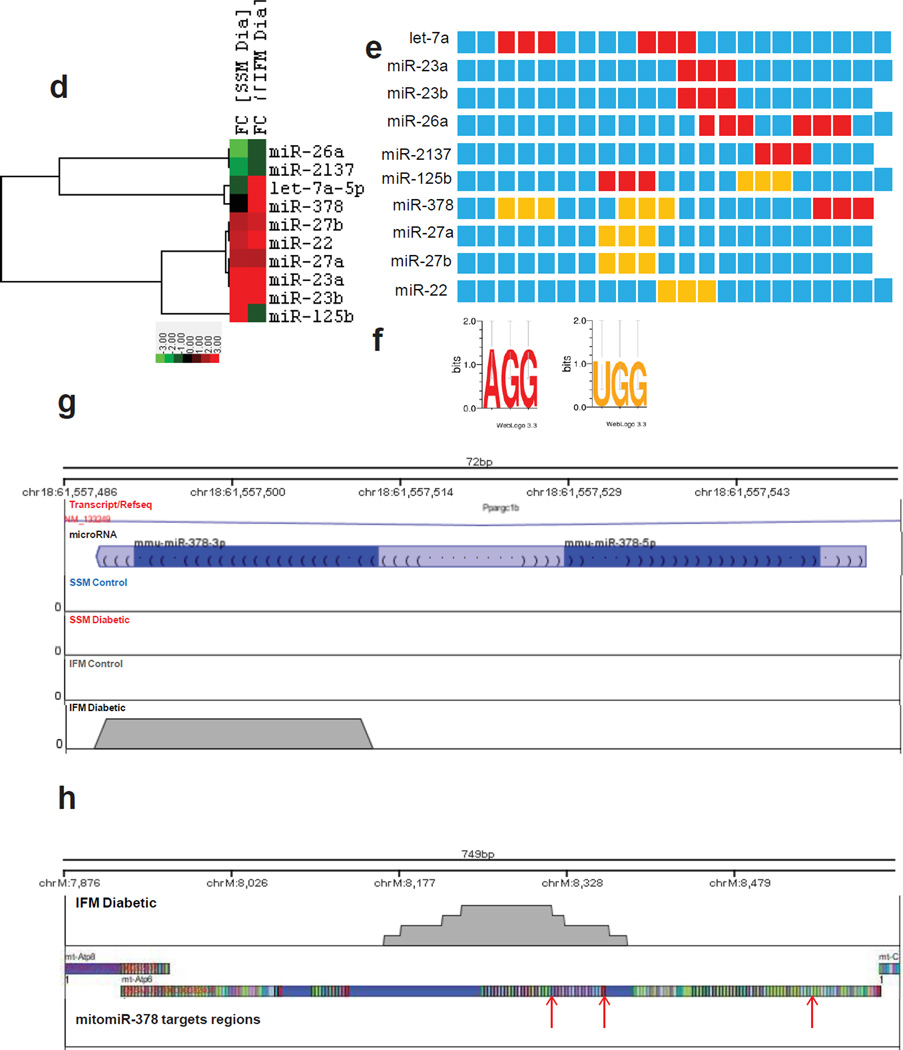
Figure 6.
Genomic sequencing analyses of mitochondrial RISCome association with mitochondrial mRNA and mitomiRs. (A) Mitochondrially-encoded mRNAs in the diabetic subpopulations which were differentially expressed in the mitoRISCome relative to controls. (B) Mapping of the footprint regions of mitochondrial mRNA sequence identified by next generation sequencing which were present in the mitoRISCome. (C) Genome browser illustration of mitochondrial RNA-seq reads mapping to rRNA, tRNA, mRNAs; histogram indicates RNA-seq read distribution which includes the transcript region. (D) MitomiR heat map derived from next generation sequencing of small RNAs identifying enrichment and depletion patterns within the mitoRISCome of diabetic SSM and diabetic IFM, relative to respective controls. (E, F) Mapping of the location of the footprint regions of mitomiR sequence identified by next generation sequencing which were present in the mitoRISCome and were enriched with tri-nucleotide motifs, AGG (red blocks) or UGG (yellow blocks). (G) Mapping of mitoRISCome associated mitomiR-378 enrichment clusters sites in control and diabetic SSM and IFM. (H) MitoRISCome:mitomiR378 ternary maps for the ATP6 target in the diabetic IFM.
To capture the distinct features of the mitomiR signatures in the mitoRISCome functional complex we assessed mitoRISCome/mitomiR complex products utilizing small RNA-seq library preparation to establish mitomiR transcripts through sequencing with a Mi-seq platform. High throughput sequencing of small RNAs in the mitoRISCome identified 37 mitomiRs which were differentially regulated as a result of diabetic insult in mitochondrial subpopulations (Supplementary Figure 4). Of the 37 mitomiRs identified, 10 (Figure 6d) were also observed in our initial microarray experiments (Figure 2a) and underwent prediction analyses (Supplementary Figure 1a). Interestingly, our high throughput sequencing of small RNAs in the mitoRISCome revealed the presence of a number of mitomiRs that were not identified in our initial microarray analyses. Several variables may have accounted for the discrepancy including the lack of miRNA probes on the microarray (miRNA- 3963, -5102, -5105, -5108, -5109, -5112, -5115, -5119, and -5131). Thus, our microarray data potentially underestimate the absolute number of mitomiRs and could be considered a limitation of the studies. As microarray technologies continue to advance, it is probable that a greater number of miRNAs will be identified in the mitochondrion. Further, studies suggest comparing results from microarray and next generation sequencing shows good correlation between expression levels for a given gene between the two platforms, and the use of both can provide complementary information. However, issues such as sensitivity, background noise interference and hybridization saturation can lead to differences in results57. Among the identified mitomiRs in the mitoRISCome was mitomiR-378 (Figure 6d). Of particular interest was the observation that the microarray and snapshot mitoRISCome analyses, though identifying similar mitomiRs, revealed differential signature patterns, such that the relative ratio of mitomiR in the mitochondrion (Figure 2a) did not necessarily match the data from the mitoRISCome analyses (Figure 6d). These data suggest that the presence of the mitomiR in the mitochondrion does not necessarily reflect it as possessing a functionally active regulatory role on the mitochondrial transcriptome at a given time. Such a phenomenon is in agreement with other studies58, 59. Moreover, our current study revealed that functional miRNA:mRNA interactions are not limited to the regulation of nuclear-encoded genes, but may also play a role in the regulation of mitochondrially-encoded genes. Diabetic IFM displayed the greatest response in terms of increased functional mitomiRs relative to control (Figure 6d). Mapping of the mitomiRs identified in the mitoRISCome next generation sequencing which were also identified in the microarray analyses enabled identification of specific regions in each of the 10 mitomiRs present in the mitoRISCome (Figure 6e). As with the mRNA analyses, we performed an unbiased search of associated sequences in the 3–22 nucleotide regions of these 10 mitomiRs to determine whether specific sequence motifs were common. Our results revealed enriched nucleotide motifs, AGG (in red) and UGG (in yellow) in all 10 mitomiRs, and these motifs were highly enriched in mitomiR-378 (Figures 6e and 6f). To confirm the presence of mitomiR-378 in the mitoRISCome we determined whether the mature mitomiR-378 was present. Mapping of the next generation sequencing indicated the presence of mitomiR-378 solely in the diabetic IFM of which only the mature transcript was apparent (Figure 6g). Sequence scanning analysis of the ternary complex revealed mitomiR-378 binding sites were present within the footprint region of the ATP6 transcriptome at starting positions 8,310 with a secondary binding site at position 8,383 (Figure 6h). These results were consistent with the previous bioinformatically-predicted targets for mitomiR-378 (Figure 3c).
miR-378 Overexpression in an In Vitro Cellular System
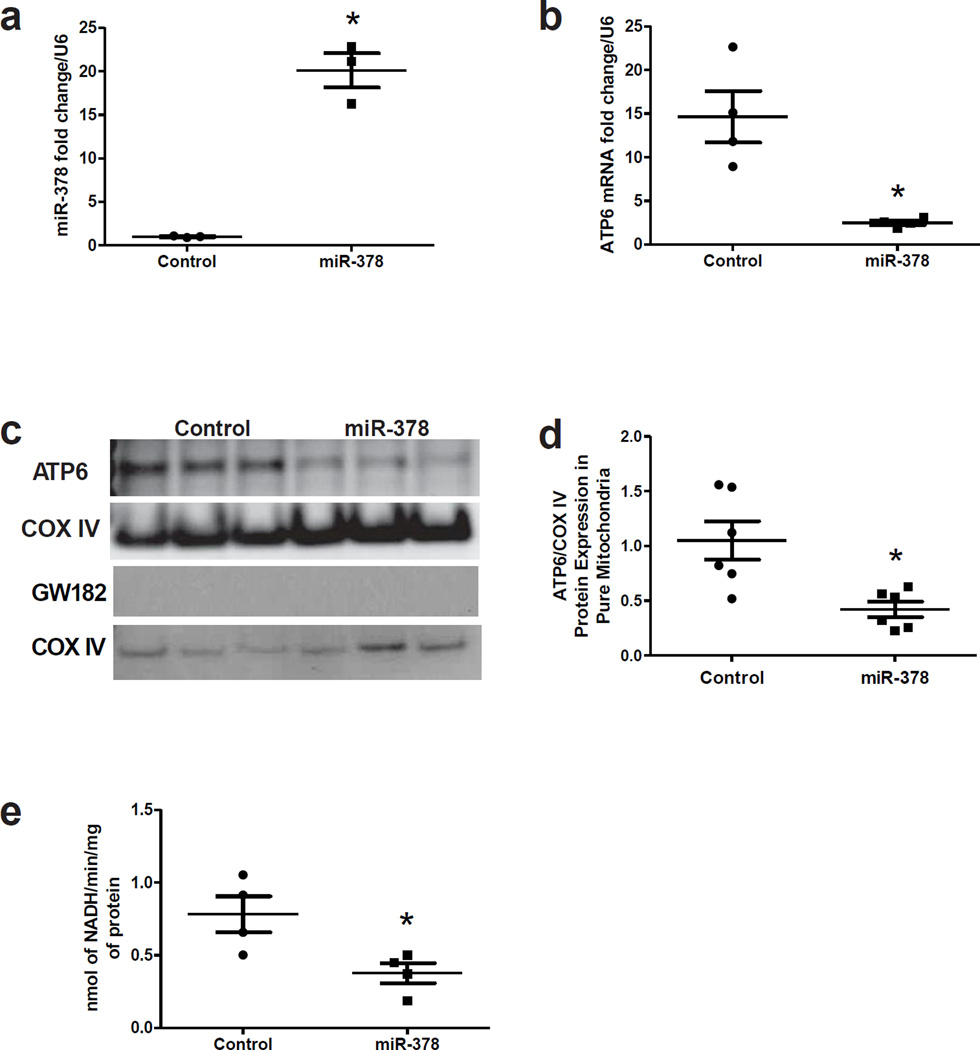
To validate the regulatory activity of mitomiR-378 on ATP6, we generated a stable HL-1 cell line overexpressing the mouse miR-378 which contained a GFP coding sequence to enable cellular tracking. To determine whether down-regulation of ATP6 and ATP synthase functional decrements were associated with miR-378 presence in the mitochondrion, mitochondria was isolated from miR-378 overexpressing HL-1 cells and the presence of mitomiR-378 examined by qRT-PCR analyses. Our data revealed an approximately 22 fold increase of mitomiR-378 in isolated mitochondria from the stable HL-1 cell line overexpressing miR-378 as compared to control HL-1 cells (Figure 7a). QRT-PCR analyses revealed a significant decrease in ATP6 mRNA abundance (Figure 7b) with concomitant decreases in ATP6 protein content (Figures 7c and 7d) and ATP synthase activity (Figure 7e). Further, GW182 presence was not detected in the mitochondrion (Figure 7c), suggesting that its presence was not required for miR-378 regulation of ATP6 in our cellular model. It should be noted that the potential impact of GW182 on cytosolic miR-378 targets was not assessed in our cellular system and as a result is a limitation of the studies. With that being said, no differences were observed in citrate synthase activities (HL-1, 0.67 ± 0.21 units × mg protein−1 vs. miR-378, 0.69 ± 0.27 units × mg protein−1) suggesting that the decrease in ATP6 protein observed in the face of miR-378 overexpression was not a function of changes in mitochondrial content. Taken together, these data provide complementary evidence that in a model of enhanced miR-378 expression, translocation to the mitochondrion can occur which is associated with a decrease in mitochondrial genome-encoded ATP6 and a concomitant reduction in the functionality of the ATP synthase.
Figure 7.
Validation of miR-378 mitochondrial targeting in vitro. (A) RT-PCR analyses for miR-378 levels in isolated mitochondria from HL-1 (Control) and miR-378 cells; n = 3. (B) QRT-PCR analyses for ATP6 mRNA in isolated mitochondria from Control and miR-378 cells; n = 4. (C) Representative Western blot analyses of ATP6 and GW182 protein levels in isolated mitochondria from Control and miR-378 cells. COX IV protein expression is utilized as a loading control. (D) Quantitative analysis of ATP6 protein levels in isolated mitochondria from Control and miR-378 cells. Values are expressed per COX IV protein levels; n = 6. (e) ATP synthase activity in control HL-1 cells and in miR-378 cells; n = 4. Values are means ± SE. *P < 0.05 for Control vs. miR-378.
Manipulation of miR-378 In Vivo with Targeted Antagomir Delivery
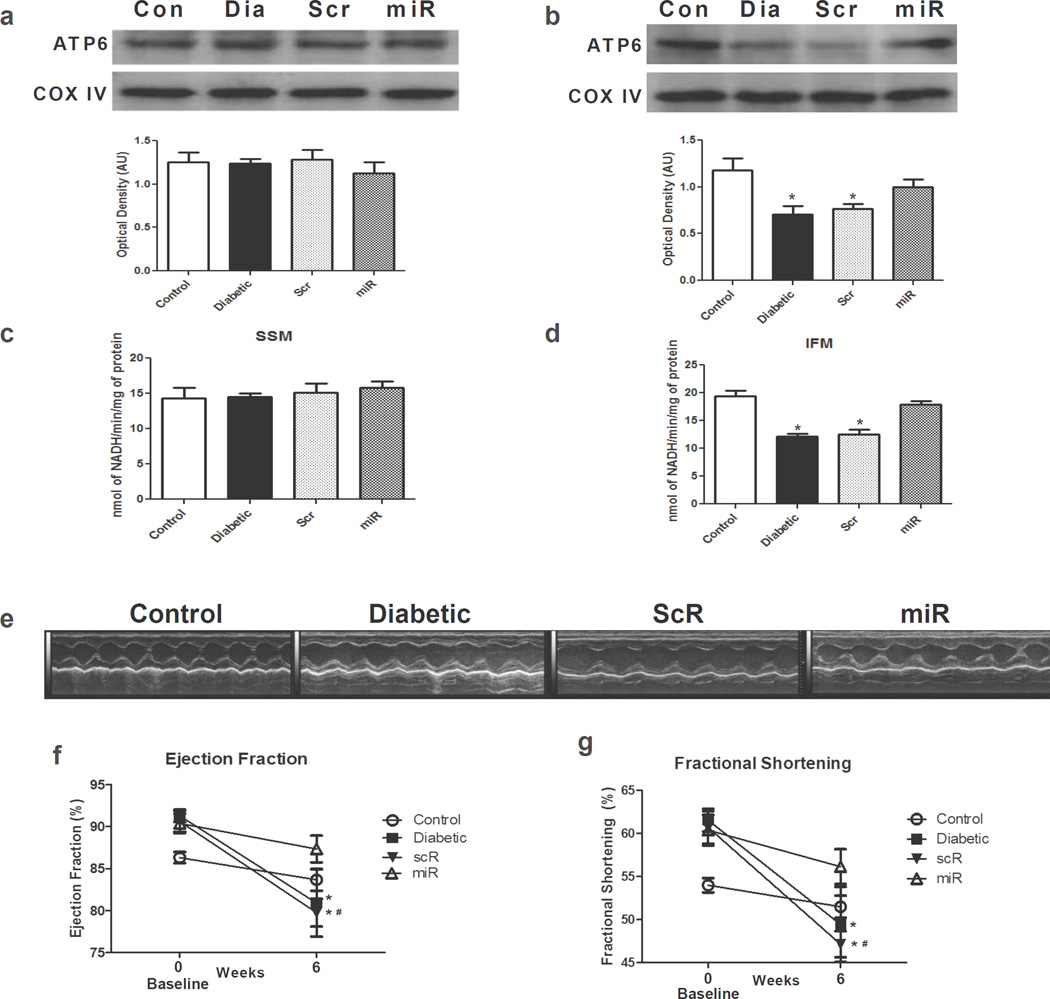
To determine whether manipulation of miR-378 in vivo could influence ATP generating capacity through ATP6 preservation, we utilized an LNA-linked antagomiR delivery approach targeting miR-378 (LNA-miR-378). Repeated injection of LNA-miR-378 over a five week period led to preservation of ATP6 protein levels in diabetic IFM which were similar to non-diabetic controls, whereas diabetic and scrambled miR diabetic groups displayed significant decreases in ATP6 protein levels relative to control (Figure 8b). In contrast, no significant differences in ATP6 protein levels were observed among any group in the SSM (Figure 8a). To determine whether preservation of ATP6 protein levels impacted ATP generating capacity, we assessed ATP synthase activities following LNA-miR-378 intervention and diabetes mellitus. As with ATP6 protein levels, ATP synthase activities displayed similar changes in diabetic IFM, with LNA-miR-378 treatment providing restoration of ATP synthase activity which was significantly decreased in diabetic and scrambled miR diabetic groups (Figure 8d). Further, no significant differences in ATP synthase activities were observed among any group in the SSM (Figure 8c). Because preservation of ATP synthase activity ultimately influences cardiac pump function by providing appropriate ATP levels, we assessed ejection fraction and fractional shortening changes prior to and following LNA-miR-378 intervention and diabetes mellitus. Our results revealed preservation of %ejection fraction (Figures 8e and 8f) and %fractional shortening (Figure8e and 8g) in LNA-miR-378 diabetic hearts which was significantly decreased in diabetic and scrambled miR diabetic hearts. Taken together, these findings provide in vivo confirmation of an LNA-miR-378 impact on diabetic IFM which manifests as preservation of ATP6 protein content, ATP synthase levels and cardiac pump function.
Figure 8.
Validation of miR-378 mitochondrial targeting in vivo. Quantitative analyses by Western blot of ATP6 protein levels following LNA-miR-378 treatment and diabetic induction in (A) SSM and (B) IFM. COX IV protein expression is utilized as a loading control and values are expressed per COX IV. ATP synthase activities levels following LNA-miR-378 treatment and diabetic induction in (C) SSM and (D) IFM. For ATP6 and ATP synthase analyses. Values are means ± SE, n = 5. *P < 0.05 for Control vs. Diabetic and Control vs. Scrambled. (E) Representative M-mode images of Control, Diabetic, Scrambled and LNA-miR-378 treated hearts 5 weeks following diabetes mellitus induction. (F) Quantitative summary of % ejection fraction and (G) % fractional shortening prior to (baseline) and 5 weeks following diabetes mellitus induction. Values are means ± SE, n = 5. *P < 0.05 for baseline vs. 5 weeks; #P < 0.05 for LNA-miR-378 vs. Scrambled. ScR = Scrambled; miR = LNA-miR-378.
Discussion
Our findings indicate for the first time that mitomiRs are differentially distributed in spatially-distinct cardiac mitochondrial subpopulations. Upon diabetic insult, cardiac mitomiR distribution patterns are dramatically altered and in many cases display opposite/reciprocal arrangements. Type 1 diabetes mellitus is associated with cardiac functional deficits, which may result from a decreased ability for cardiac mitochondria to generate ATP. Among the genes encoded for by the mitochondrial genome are 13 proteins of the electron transport chain that are constituents of complexes I, III, IV, and V. Thus, regulation of the mitochondrial genome by mitomiRs may have profound effects on the expression of key components driving ATP synthesis and ultimately influencing cardiac function.
Using a number of bioinformatics tools, we identified 46 mitomiRs that were predicted to bind to mitochondrial genome-encoded mRNAs. Over half of the identified mitomiRs displayed the potential for binding multiple mitochondrial-encoded genome mRNAs. Such a finding is intriguing and suggests the potential for an individual mitomiR to regulate the translation of multiple mitochondrial genome products. Such a dynamic is consistent with proposed mechanisms of action for other miRNAs60, 61 and as a result, mitomiR manipulation for therapeutic intervention represents an attractive strategy for addressing pathological conditions, such as diabetes mellitus, as it may offer the ability to rectify proteomic loss of multiple mitochondrial genome-encoded proteins simultaneously. Additional bioinformatic analyses of the nucleotide sequences in the 46 identified mitomiRs indicated that ~ 85% possessed specific conserved tri-nucleotide motifs which included AGG, UGG, and CCU. Of particular interest was an observed concentration of AGG and UGG motifs contained in the 2–7 nucleotide seeding region of a number of mitomiRs. We speculate that the existence of these motifs in the mitomiR may influence its translocation into mitochondria and that their presence may predict the potential for a given miRNA to exist in the mitochondrion. Further investigation is needed to determine the relative importance of these particular motifs.
Because it displayed a dynamic change in its content following diabetic insult and a reciprocal response in spatially-distinct mitochondrial subpopulations with an enhanced presence solely in IFM, the mitochondrial subpopulation most negatively impacted by type 1 diabetic insult2, 4, 28, 35, 41, we chose to examine mitomiR-378 for our subsequent studies. MiR-378 plays an important regulatory role in cardiac remodeling, cell survival62, 63 and its complimentary pair, miR-378*, mediates a metabolic shift to oxidative phosphorylation22, 50. The goal of the current study was tease out specific effects on metabolism that were impacted by mitochondrial miR-378 presence. Though it displayed no significant change in the heart following diabetic insult, miR-378 was dramatically increased in the IFM suggesting that its enhanced presence may be of particular relevance specifically to the mitochondrion. Identification of a mitochondrial genome target for mitomiR-378, ATP6 and the associated decrease in ATP synthase activity specifically in the IFM, suggest a coordinated regulatory role for mitomiR-378 in type 1 diabetic IFM. Manipulation of this axis may contribute to the development of diabetic cardiomyopathy by compromising the generation of ATP. Indeed, overexpression of miR-378 in a cell type that maintains a cardiac phenotype revealed increased mitochondrial presence and a concomitant decrease in ATP6 content and function in an in vitro model. Further, utilization of an antagomir approach directed at miR-378 demonstrated preservation of ATP6 protein content, ATP synthase activity, and cardiac pump function confirming this axis in vivo. Taken together our findings suggest enhanced mitomiR-378 presence in the type 1 diabetic IFM which is the mitochondrial subpopulation most impacted by the pathology2, hence contributing to bioenergetic deficits occurring in the face of the diabetic phenotype as a result of inadequate provision of ATP for cardiac function.
Perhaps the most interesting finding in the current study was the identification of RISC presence in the mitochondrion which contained key components, Ago2 and FXR1. It appears that the mitochondrial RISC functions similarly to its cytoplasmic counterpart, influencing translation. Reports of changes in miRNA expression, has provided indirect evidence for translational repression, yet evidence for a direct effect of the miRNA on the mRNA is limited. Use of the CLIP experimental approach provides a clear means of identifying direct miRNA/mRNA/protein interactions confirming direct association between nucleotide and protein components33. Following type 1 diabetes mellitus, mitochondrial RISC components were differentially expressed in the mitochondrion relative to control and the response was subpopulation-specific in nature. Our results indicate that translational regulation of ATP6 by mitomiR-378 requires both Ago2 and FXR1 presence in the RISC, as well as the necessary mitomiR (mitomiR-378) as observed in diabetic IFM. When Ago2 content is limiting and the mitomiR (mitomiR-378) is not present, regulation via the mitochondrial RISC does not occur, as observed in diabetic SSM (Supplementary Figure 5). Our results are in agreement with literature suggesting the relative importance of FXR1 presence, in that its knockout is lethal in mice and in zebrafish as a result of cardiomyopathy and muscular dystrophy64. Reports indicate a requirement of both FXR1 and Ago2 in the RISC for translational activation13, 65–67, via AU-rich elements. Our current data extend these findings by identifying a similar phenomenon within the mitochondrion that occurs in a subpopulation-specific manner during diabetic insult and requires specific sequence motifs (AGG and UGG) in the miRNA sequence. Thus, redistribution of mitomiRs and the presence of specific mitochondrial RISC component stoichiometry, contributes to the formation of a functional complex which is associated with loss of mitochondrially-encoded proteins in the diabetic heart.
Recent crystallographic and NMR structural analyses have indicated that all three molecules (mRNA, miRNA and protein) require close proximity and direct interaction to facilitate a functional cytoplasmic ternary regulatory complex33, 58, 68–70. Nevertheless, identification of a functional RISC in the mitochondrion is highly challenging. Recent reports suggest that RNA can be imported into the mitochondrion71, 72 which supports our current findings. The use of CLIP technology coupled with next generation sequencing offers an experimental platform for determining binding maps and sequence motifs necessary for miRNAs to exert regulatory influence on mRNA expression/translation. Our CLIP analyses confirmed that mitomiR-378 targets two different sites on the ATP6 mRNA, which was bioinformatically predicted prior to sequencing analyses. These results were interesting in that miRNAs have been identified as binding to the 3'-untranslated region of mRNA to exert regulatory activity. Nevertheless, the mitochondrial genome does not possess 3’-untranslated regions, rather the sequence identity from the bioinformatic and next generation sequencing analyses predicted binding directly to the coding region. Such a finding is consistent with previous studies demonstrating that the mouse Nanog, Oct4 and Sox2 genes were targeted by miRNA in their amino acid coding sequence resulting in translational repression73. Our results indicate that mitomiR-378 targets the ATP6 coding region in a similar manner to control translational activity.
In summary, we propose that mitomiR translational regulation of mitochondrially-encoded proteins occurs in spatially-distinct mitochondrial subpopulations in response to type 1 diabetic insult. Redistribution of mitomiRs and the presence of a specific mitochondrial RISC component stoichiometry, contributes to the formation of a functional ternary complex which is associated with loss of mitochondrially-encoded proteins in the diabetic heart. These results suggest a dynamic system that can regulate the mitochondrial proteome during pathological states in a spatially-distinct manner.
Supplementary Material
Acknowledgments
We would like to acknowledge Dr. William Claycomb for the HL-1 cells utilized in the cell culture experiments. We would like to acknowledge Dr. Dustin Long in the WVU Biostatistics Department for his help with the statistical analyses. Next generation sequencing experiments were carried out in conjunction with the West Virginia University Genomics Core Facility. Echocardiography experiments were carried out in conjunction with the West Virginia University Animal Models of Imaging Core Facility.
Funding Sources: This work was supported by the National Institutes of Health from the National Institutes of Diabetes and Digestive and Kidney Diseases [DP2DK083095] awarded to J.M.H. This work was supported by the National Institute of General Medical Sciences [U54GM104942] awarded to J.M.H through the WVCTSI. This work was supported by a Grant-In-Aid from the American Heart Association [0855484D] awarded to J.M.H. This work was supported by the American Cancer Society [IRG0906104] awarded to I.M. W.A.B. is a recipient of an American Heart Association Predoctoral Fellowship [10PRE3420006]. C.E.N. is a recipient of an American Heart Association Predoctoral Fellowship [13PRE16850066]. D.L.S. is a recipient of an American Heart Association Predoctoral Fellowship [14PRE19890020]. W.A.B, T.L.C., and D.L.S. are recipients of NIH Predoctoral Fellowships [T32HL090610]. C.E.N. is a recipient of a National Science Foundation Integrative Graduate Education and Research Traineeship [DGE1144676]. Ingenuity Pathway Analyses were supported by a WV-INBRE Grant [8P20GM10343412]. Next Generation Sequencing Analyses were supported by WVU Genomics Core Facility Grant awarded to J.M.H that was funded by NIH CoBRE grant [GM103503] to the WVU Center for Neuroscience.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Palmer JW, Tandler B, Hoppel CL. Biochemical properties of subsarcolemmal and interfibrillar mitochondria isolated from rat cardiac muscle. J Biol Chem. 1977;252:8731–8739. [PubMed] [Google Scholar]
- 2.Dabkowski ER, Williamson CL, Bukowski VC, Chapman RS, Leonard SS, Peer CJ, et al. Diabetic cardiomyopathy-associated dysfunction in spatially distinct mitochondrial subpopulations. Am J Physiol Heart Circ Physiol. 2009;296:H359–H369. doi: 10.1152/ajpheart.00467.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollander JM, Thapa D, Shepherd DL. Physiological and structural differences in spatially distinct subpopulations of cardiac mitochondria: Influence of cardiac pathologies. Am J Physiol Heart Circ Physiol. 2014;307:H1–H14. doi: 10.1152/ajpheart.00747.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baseler WA, Dabkowski ER, Williamson CL, Croston TL, Thapa D, Powell MJ, et al. Proteomic alterations of distinct mitochondrial subpopulations in the type 1 diabetic heart: Contribution of protein import dysfunction. Am J Physiol Regul Integr Comp Physiol. 2011;300:R186–R200. doi: 10.1152/ajpregu.00423.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bugger H, Boudina S, Hu XX, Tuinei J, Zaha VG, Theobald HA, et al. Type 1 diabetic akita mouse hearts are insulin sensitive but manifest structurally abnormal mitochondria that remain coupled despite increased uncoupling protein 3. Diabetes. 2008;57:2924–2932. doi: 10.2337/db08-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bugger H, Chen D, Riehle C, Soto J, Theobald HA, Hu XX, et al. Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes. 2009;58:1986–1997. doi: 10.2337/db09-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 8.Luft R. The development of mitochondrial medicine. Proc Natl Acad Sci U S A. 1994;91:8731–8738. doi: 10.1073/pnas.91.19.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maechler P, Wollheim CB. Mitochondrial function in normal and diabetic beta-cells. Nature. 2001;414:807–812. doi: 10.1038/414807a. [DOI] [PubMed] [Google Scholar]
- 10.Kwong JQ, Beal MF, Manfredi G. The role of mitochondria in inherited neurodegenerative diseases. J Neurochem. 2006;97:1659–1675. doi: 10.1111/j.1471-4159.2006.03990.x. [DOI] [PubMed] [Google Scholar]
- 11.McDermott-Roe C, Ye J, Ahmed R, Sun XM, Serafin A, Ware J, et al. Endonuclease g is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. Micrornas: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: Micrornas can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 14.Huang L, Mollet S, Souquere S, Le Roy F, Ernoult-Lange M, Pierron G, et al. Mitochondria associate with p-bodies and modulate microrna-mediated rna interference. J Biol Chem. 2011;286:24219–24230. doi: 10.1074/jbc.M111.240259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrey E, Saint-Auret G, Bonnamy B, Damas D, Boyer O, Gidrol X. Pre-microrna and mature microrna in human mitochondria. PloS one. 2011;6:e20220. doi: 10.1371/journal.pone.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das S, Ferlito M, Kent OA, Fox-Talbot K, Wang R, Liu D, et al. Nuclear mirna regulates the mitochondrial genome in the heart. Circ Res. 2012;110:1596–1603. doi: 10.1161/CIRCRESAHA.112.267732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. Micrornas identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sripada L, Tomar D, Prajapati P, Singh R, Singh AK, Singh R. Systematic analysis of small rnas associated with human mitochondria by deep sequencing: Detailed analysis of mitochondrial associated mirna. PloS one. 2012;7:e44873. doi: 10.1371/journal.pone.0044873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, et al. Microrna directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bandiera S, Mategot R, Girard M, Demongeot J, Henrion-Caude A. Mitomirs delineating the intracellular localization of micrornas at mitochondria. Free Radic Biol Med. 2013;64:12–19. doi: 10.1016/j.freeradbiomed.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, et al. A cardiac microrna governs systemic energy homeostasis by regulation of med13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, et al. Control of mitochondrial metabolism and systemic energy homeostasis by micrornas 378 and 378*. Proc Natl Acad Sci U S A. 2012;109:15330–15335. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurd BJ, Holloway GP, Yoshida Y, Bonen A. In mammalian muscle, sirt3 is present in mitochondria and not in the nucleus; and sirt3 is upregulated by chronic muscle contraction in an adenosine monophosphate-activated protein kinase-independent manner. Metabolism. 2012;61:733–741. doi: 10.1016/j.metabol.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Heather LC, Cole MA, Tan JJ, Ambrose LJ, Pope S, Abd-Jamil AH, et al. Metabolic adaptation to chronic hypoxia in cardiac mitochondria. Basic Res Cardiol. 2012;107:268. doi: 10.1007/s00395-012-0268-2. [DOI] [PubMed] [Google Scholar]
- 25.Ritov VB, Menshikova EV, He J, Ferrell RE, Goodpaster BH, Kelley DE. Deficiency of subsarcolemmal mitochondria in obesity and type 2 diabetes. Diabetes. 2005;54:8–14. doi: 10.2337/diabetes.54.1.8. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka-Esposito C, Chen Q, Lesnefsky EJ. Blockade of electron transport before ischemia protects mitochondria and decreases myocardial injury during reperfusion in aged rat hearts. Transl Res. 2012;160:207–216. doi: 10.1016/j.trsl.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dabkowski ER, Williamson CL, Hollander JM. Mitochondria-specific transgenic overexpression of phospholipid hydroperoxide glutathione peroxidase (gpx4) attenuates ischemia/reperfusion-associated cardiac dysfunction. Free Radic Biol Med. 2008;45:855–865. doi: 10.1016/j.freeradbiomed.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 28.Williamson CL, Dabkowski ER, Baseler WA, Croston TL, Alway SE, Hollander JM. Enhanced apoptotic propensity in diabetic cardiac mitochondria: Influence of subcellular spatial location. Am J Physiol Heart Circ Physiol. 2010;298:H633–H642. doi: 10.1152/ajpheart.00668.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dabkowski ER, Baseler WA, Williamson CL, Powell M, Razunguzwa TT, Frisbee JC, et al. Mitochondrial dysfunction in the type 2 diabetic heart is associated with alterations in spatially distinct mitochondrial proteomes. Am J Physiol Heart Circ Physiol. 2010;299:H529–H540. doi: 10.1152/ajpheart.00267.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular rnas are a large class of animal rnas with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 32.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 33.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute hits-clip decodes microrna-mrna interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. Hl-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baseler WA, Thapa D, Jagannathan R, Dabkowski ER, Croston TL, Hollander JM. Mir-141 as a regulator of the mitochondrial phosphate carrier (slc25a3) in the type 1 diabetic heart. Am J Physiol Cell Physiol. 2012;303:C1244–C1251. doi: 10.1152/ajpcell.00137.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of micrornas in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 37.Montgomery RL, Hullinger TG, Semus HM, Dickinson BA, Seto AG, Lynch JM, et al. Therapeutic inhibition of mir-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGregor RA, Choi MS. Micrornas in the regulation of adipogenesis and obesity. Curr Mol Med. 2011;11:304–316. doi: 10.2174/156652411795677990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wahid F, Shehzad A, Khan T, Kim YY. Micrornas: Synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta. 2010;1803:1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 40.Boon RA, Seeger T, Heydt S, Fischer A, Hergenreider E, Horrevoets AJ, et al. Microrna-29 in aortic dilation: Implications for aneurysm formation. Circ Res. 2011;109:1115–1119. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 41.Baseler WA, Dabkowski ER, Jagannathan R, Thapa D, Nichols CE, Shepherd DL, et al. Reversal of mitochondrial proteomic loss in type 1 diabetic heart with overexpression of phospholipid hydroperoxide glutathione peroxidase. Am J Physiol Regul Integr Comp Physiol. 2013;304:R553–R565. doi: 10.1152/ajpregu.00249.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thapa D, Nichols CE, Lewis SE, Shepherd DL, Jagannathan R, Croston TL, et al. Transgenic overexpression of mitofilin attenuates diabetes mellitus-associated cardiac and mitochondria dysfunction. J Mol Cell Cardiol. 2015;79:212–223. doi: 10.1016/j.yjmcc.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bauer M, Cheng S, Jain M, Ngoy S, Theodoropoulos C, Trujillo A, et al. Echocardiographic speckle-tracking based strain imaging for rapid cardiovascular phenotyping in mice. Circ Res. 2011;108:908–916. doi: 10.1161/CIRCRESAHA.110.239574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson MF, Sims NR. Improved recovery of highly enriched mitochondrial fractions from small brain tissue samples. Brain research. Brain Res Brain Res Protoc. 2000;5:95–101. doi: 10.1016/s1385-299x(99)00060-4. [DOI] [PubMed] [Google Scholar]
- 45.Glancy B, Balaban RS. Protein composition and function of red and white skeletal muscle mitochondria. Am J Physiol Cell Physiol. 2011;300:C1280–C1290. doi: 10.1152/ajpcell.00496.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo D, Nguyen T, Ogbi M, Tawfik H, Ma G, Yu Q, et al. Protein kinase c-epsilon coimmunoprecipitates with cytochrome oxidase subunit iv and is associated with improved cytochrome-c oxidase activity and cardioprotection. Am J Physiol Heart Circ Physiol. 2007;293:H2219–H2230. doi: 10.1152/ajpheart.01306.2006. [DOI] [PubMed] [Google Scholar]
- 47.Etienne W, Meyer MH, Peppers J, Meyer RA., Jr Comparison of mrna gene expression by rt-pcr and DNA microarray. BioTechniques. 2004;36:618–620. doi: 10.2144/04364ST02. 622, 624-616. [DOI] [PubMed] [Google Scholar]
- 48.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microrna/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microrna nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 50.Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, et al. Mir-378(*) mediates metabolic shift in breast cancer cells via the pgc-1beta/errgamma transcriptional pathway. Cell Metab. 2010;12:352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 51.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, et al. Trbp recruits the dicer complex to ago2 for microrna processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Filipowicz W. Rnai: The nuts and bolts of the risc machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 53.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human risc couples microrna biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 54.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of rnai. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 55.He L, Hannon GJ. Micrornas: Small rnas with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 56.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of argonaute protein reveals microrna-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sirbu A, Kerr G, Crane M, Ruskin HJ. Rna-seq vs dual- and single-channel microarray data: Sensitivity analysis for differential expression and clustering. PloS one. 2012;7:e50986. doi: 10.1371/journal.pone.0050986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu Y, Matkovich SJ, Hecker PA, Zhang Y, Edwards JR, Dorn GW., 2nd Epitranscriptional orchestration of genetic reprogramming is an emergent property of stress-regulated cardiac micrornas. Proc Natl Acad Sci U S A. 2012;109:19864–19869. doi: 10.1073/pnas.1214996109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW., 2nd Risc rna sequencing for context-specific identification of in vivo microrna targets. Circ Res. 2011;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Law PT, Ching AK, Chan AW, Wong QW, Wong CK, To KF, et al. Mir-145 modulates multiple components of the insulin-like growth factor pathway in hepatocellular carcinoma. Carcinogenesis. 2012;33:1134–1141. doi: 10.1093/carcin/bgs130. [DOI] [PubMed] [Google Scholar]
- 61.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knezevic I, Patel A, Sundaresan NR, Gupta MP, Solaro RJ, Nagalingam RS, et al. A novel cardiomyocyte-enriched microrna, mir-378, targets insulin-like growth factor 1 receptor: Implications in postnatal cardiac remodeling and cell survival. J Biol Chem. 2012;287:12913–12926. doi: 10.1074/jbc.M111.331751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee DY, Deng Z, Wang CH, Yang BB. Microrna-378 promotes cell survival, tumor growth, and angiogenesis by targeting sufu and fus-1 expression. Proc Natl Acad Sci U S A. 2007;104:20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whitman SA, Cover C, Yu L, Nelson DL, Zarnescu DC, Gregorio CC. Desmoplakin and talin2 are novel mrna targets of fragile x-related protein-1 in cardiac muscle. Circ Res. 2011;109:262–271. doi: 10.1161/CIRCRESAHA.111.244244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mortensen RD, Serra M, Steitz JA, Vasudevan S. Posttranscriptional activation of gene expression in xenopus laevis oocytes by microrna-protein complexes (micrornps) Proc Natl Acad Sci U S A. 2011;108:8281–8286. doi: 10.1073/pnas.1105401108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. Microrna-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vasudevan S, Steitz JA. Au-rich-element-mediated upregulation of translation by fxr1 and argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chi SW, Hannon GJ, Darnell RB. An alternative mode of microrna target recognition. Nat Struct Mol Biol. 2012;19:321–327. doi: 10.1038/nsmb.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frank F, Sonenberg N, Nagar B. Structural basis for 5'-nucleotide base-specific recognition of guide rna by human ago2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target rna duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.von Ameln S, Wang G, Boulouiz R, Rutherford MA, Smith GM, Li Y, et al. A mutation in pnpt1, encoding mitochondrial-rna-import protein pnpase, causes hereditary hearing loss. Am J Hum Genet. 2012;91:919–927. doi: 10.1016/j.ajhg.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang G, Shimada E, Zhang J, Hong JS, Smith GM, Teitell MA, et al. Correcting human mitochondrial mutations with targeted rna import. Proc Natl Acad Sci U S A. 2012;109:4840–4845. doi: 10.1073/pnas.1116792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. Micrornas to nanog, oct4 and sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.