Abstract
Modern definitions of epigenetics incorporate models for transient but biologically important changes in gene expression that are unrelated to DNA code but responsive to environmental changes such as injury-induced stress. In this scheme, changes in oxygen levels (hypoxia) and/or metabolic co-factors (iron deficiency or diminished 2-oxoglutarate levels) are transduced into broad genetic programs that return the cell and the organism to a homeostatic set point. Over the past two decades, exciting studies have identified a superfamily of iron-, oxygen-, and 2-oxoglutarate-dependent dioxygenases that sit in the nucleus as modulators of transcription factor stability, co-activator function, histone demethylases, and DNA demethylases. These studies have provided a concrete molecular scheme for how changes in metabolism observed in a host of neurological conditions, including stroke, traumatic brain injury, and Alzheimer’s disease, could be transduced into adaptive gene expression to protect the nervous system. We will discuss these enzymes in this short review, focusing primarily on the ten eleven translocation (TET) DNA demethylases, the jumonji (JmJc) histone demethylases, and the oxygen-sensing prolyl hydroxylase domain enzymes (HIF PHDs).
Keywords: Stroke, brain, epigenetics, metabolism, 2-oxoglutarate-dependent dioxygenases, neuroprotection
1. Introduction
Epigenetics has been classically defined as heritable changes in gene expression that are not encoded within the genome. More recent definitions account for growing recognition that transcription is highly dynamic and can mediate short- and long-term changes in gene expression via epigenetic proteins in response to environmental signals and stresses. Simply, epigenetic proteins can modify DNA or histone proteins to govern the interaction between these regulators of chromatin structure. Accordingly, epigenetic proteins can favor a transcriptionally competent state (euchromatin) or a transcriptionally incompetent state (heterochromatin). As such, they are uniquely poised to modulate cell phenotypes in response to stress to create cells and thus organisms that have greater fitness or reserve to withstand pathologies.
Evidence that epigenetic proteins are modulated to foster adaptation to genetic and environmental stresses in the nervous system dates back less than 15 years ago. Seminal studies from Leslie Thompson’s and Larry Marsh’s labs demonstrated that the mutant huntingtin protein could interact with and suppress the histone acetyltransferase activity of established co-activators, including CREB-binding protein (CBP) and P300/CBP-associated factor (PCAF) (Steffan et al., 2001). Accordingly, they found that global histone acetylation was diminished by mutant huntingtin in fly models and that non-selective histone deacetylase inhibitors (HDACi) could reverse not only changes in histone acetylation but also neurodegeneration. The findings precipitated an avalanche of studies, including some from our own laboratory, that have examined the effects of non-selective inhibitors of zinc-dependent HDACs on neuronal death in a host of models (Langley et al., 2008; Ryu et al., 2003; Ryu et al., 2005; Sleiman et al., 2014).
Zinc-dependent HDACs modify the N-terminal tails of histones by deacetylating them, thus enhancing electrostatic interactions between DNA and histone proteins to favor a transcriptionally incompetent state. Accordingly, inhibitors of HDACs lead to hyperacetylation of histones, thus neutralizing electrostatic interactions between DNA and protein and favoring a transcriptionally competent state. Non-selective inhibitors of HDACs have been found to protect neurons, enhance behavior, and extend lifespan in almost every neurological disease model in which they have been tested (Table 1). In some cases, the evidence that these agents are targeting HDACs to mediate their salutary effects is compelling; in other cases, off-target effects should be considered (Olson et al., 2015). Nevertheless, these studies have provided fuel and momentum to the notion that epigenetic proteins are ripe therapeutic targets for neuroprotection.
Table 1.
HDAC inhibitors in rodent models of neurological diseases.
| Disease | Animal Model | HDAC Inhibitor | Outcome | References |
|---|---|---|---|---|
| Alzheimer’s Disease | APP23 mouse | VPA | Ameliorated memory deficits, reduced plaque formation | (Qing et al., 2008) |
| APPSWE mouse | GHB | Ameliorated cognitive deficits | (Klein et al., 2015) | |
| APPSWE mouse | PBA | Reduced plaque severity, improves memory performance | (Wiley et al., 2011) | |
| SAMP8 mouse | EGCG | Reduced spatial learning and memory deficits | (Chang et al., 2015) | |
| Tg6799 mouse | VPA | Ameliorated memory deficits, increased number of cholinergic neurons in the medial septum | (Noh and Seo, 2014) | |
| Amyotrophic lateral sclerosis | SOD1-G93A mouse | PBA | Prolonged survival, improved motor function, reduced brain atrophy and neuron loss | (Ryu et al., 2005, Petri et al., 2006, Del Signore et al., 2009) |
| SOD1-G93A mouse | TSA | Attenuated motorneuron death, axonal degeneration, denervation of muscular junctions, skeletal muscle atrophy, ameliorated motor function and survival | (Yoo and Ko, 2011) | |
| SOD1-G93A mouse | VPA | Prolonged survival | (Sugai et al., 2004) | |
| SOD1-G93A mouse | VPA | Prolonged survival, delayed disease onset, improved motor function | (Feng et al., 2008) | |
| Dentatorubral-pallidoluysian atrophy | ATN1-118Q mouse | SB | Prolonged survival, improved motor function | (Ying et al., 2006) |
| Friedreich’s ataxia | YG8R FRDA mouse | pimelic o-aminobenzamide compounds 106 and 109 | Improved motor function | (Sandi et al., 2011) |
| Huntington’s disease | N171-82Q mouse | HDACi 4b | Improved body weight, motor function, exploratory and cognitive behavior | (Jia et al., 2012) |
| N171-82Q, mouse | PBA | Prolonged survival, reduced brain atrophy | (Gardian et al., 2005) | |
| N171-82Q, mouse | VPA | Prolonged survival, improved exploratory behavior | (Zadori et al., 2009) | |
| N171-82Q mouse, YAC128 mouse | VPA | Prolonged survival, improved motor function and anxiety-/depressive-like behavior | (Chiu et al., 2011) | |
| R6/2 mouse | HDACi 4b | Improved motor function and reduced weight loss and striatal neuron atrophy | (Thomas et al., 2008) | |
| R6/2 mouse | SAHA | Improved motor function | (Hockly et al., 2003) | |
| R6/2 mouse | SB | Prolonged survival, improved motor function, reduced striatal neuron atrophy | (Ferrante et al., 2003) | |
| Multiple Sclerosis | EAE adoptive transfer, mouse | PBA, SPA | Reduced symptoms of adoptive EAE/neurological impairment, brain inflammation | (Dasgupta et al., 2003) |
| EAE, mouse | TSA | Reduced EAE scores, peak of remission phase, and disease index, brain inflammation and demyelination | (Camelo et al., 2005) | |
| EAE, mouse | SAHA | Reduced disease incidence and onset, cumulative and maxium EAE scores, brain inflammation and demyelination | (Ge et al., 2013) | |
| EAE, rat | VPA | Reduced EAE duration, cumulative and EAE scores, brain inflammation and demyelination | (Castelo- Branco et al., 2014) | |
| Neuropathic pain | Chronic constriction injury, rat | SB | Attenuated mechanical and thermal hypersensitivity | (Kukkar et al., 2014) |
| L5 spinal nerve ligation, rat | Baicalin | Attenuated mechanical and thermal hypersensitivity | (Cherng et al., 2014) | |
| Sciatic nerve ligation, mouse | TSA, VPA, SAHA | Reversed c-fiber hyposensitivity | (Matsushita et al., 2013) | |
| Sciatic nerve ligation, rat | MS-275, MGCD0103 | Attenuated mechanical and thermal hypersensitivity | (Denk et al., 2013) | |
| Parkinson’s Disease | 6-OHDA, rat | VPA | Reduced degeneration of dopaminergic neurons, tyrosine hydroxylase expression | (Monti et al., 2012) |
| A30P + A53T human α-synuclein mouse | PBA | Improved motor function, reduced loss in tyrosine hydroxylase-positive neurons, attenuated reduction in dopamine levels | (Ono et al., 2009) | |
| MPTP, mouse | PBA | Reduced loss in tyrosine hydroxylase-positive neurons, attenuated reduction in dopamine and its metabolites | (Gardian et al., 2004) | |
| MPTP acute and chronic, mouse | PBA | Reduced loss in tyrosine hydroxylase-positive neurons, attenuated reduction in levels of dopamine and its metabolites, improved motor function | (Roy et al., 2012) | |
| MPTP, mouse | VPA | Reduced loss in tyrosine hydroxylase-positive neurons, increased striatal dopamine levels | (Kidd and Schneider, 2011) | |
| MPTP and Y39C human α-synuclein mouse | PBA | Reduced loss in striatal tyrosine hydroxylase and dopamine levels, tyrosine hydroxylase-positive neurons, improved learning and motor function | (Zhou et al., 2011) | |
| MPTP, rat | VPA | Ameliorated short- term memory and olfactory discrimination impairments, dopamine depletion in striatum and olfactory bulb | (Castro et al., 2012) | |
| Rotenone, mouse | PBA | Reduced loss in striatal tyrosine hydroxylase and dopamine levels, tyrosine hydroxylase-positive neurons, improved learning and motor function | (Inden et al., 2007) | |
| Rotenone, rat | VPA | Reduced loss of tyrosine hydroxylase expression, death of nigral neurons, dopamine levels | (Monti et al., 2010) | |
| Spinal bulbar muscular atrophy | AR-97Q mouse | SB | Ameliorated muscle atrophy, gait disturbance, motor function, body weight, prolonged survival | (Minamiyama et al., 2004) |
| Spinal cord injury | T9 clip compression, rat | VPA | Improved motor function, reduced lesion sizes | (Yu et al., 2012) |
| T9 contusion injury, mouse | VPA | Improved motor function | (Abematsu et al., 2010) | |
| T9 contusion injury, rat | VPA | Improved motor function, reduced lesion volume | (Lv et al., 2012) | |
| T10 contusion injury, rat | VPA | Improved motor function, reduced lesion volume | (Lee et al., 2012, Lu et al., 2013) | |
| T10 contusion injury, rat | VPA | Improved motor function, reduced motor neuron loss and myelin sheet damage | (Hao et al., 2013) | |
| Spinal muscular atrophy | SMA mouse | SB | Prolonged survival, reduced muscular bundle atrophy | (Chang et al., 2001) |
| SMA mouse | TSA | Prolonged survival, weight gain, improved motor function, retained structurally intact motor units, increased size of muscle fibers | (Avila et al., 2007, Narver et al., 2008) | |
| SMA mouse | VPA | Improved motor function, attenuates motor neuron death | (Tsai et al., 2006) | |
| US-SMA mouse, Taiwanese-SMA mouse | SAHA | Prolonged survival, weight gain, improved motor function, reduced degeneration of motor neurons, increased size of muscle fibers | (Riessland et al., 2010) | |
| Spinocerebellar ataxia | ATXN3-79Q mouse | SB | Prolonged survival, prevented weight loss, delayed onset of neurological deficits and improved motor function | (Chou et al., 2011) |
| Stroke | Global ischemia, rat | VPA | Reduced neuronal death, spatial cognitive deficiencies | (Xuan et al., 2012) |
| Intracerebral hemorrhage, collagenase infusion, rat | VPA | Reduced hemorrhage volume and brain atrophy, improved sensorimotor function | (Sinn et al., 2007) | |
| Neonatal lipopolysaccharide-sensitized hypoxia-ischemia | TSA | Reduced grey and white matter damage, improved learning | (Fleiss et al., 2012) | |
| Permanent MCAO and transient hypoxia, mouse | PBA | Reduced infarct volumes, hemispheric swelling, and neurological deficit scores | (Qi et al., 2004) | |
| Permanent MCAO, mouse | SAHA | Reduced infarct volumes | (Faraco et al., 2006) | |
| Permanent MCAO, mouse | TSA | Reduced infarct volumes and neurological deficit scores | (Wang et al., 2012) | |
| Permanent MCAO, rat | SB | Reduced infarct volumes | (Langley et al., 2008) | |
| Permanent MCAO, rat | SB, TSA, VPA | Reduced infarct volumes, Improved motor, sensory, and reflex performance | (Kim et al., 2007, Kim et al., 2009) | |
| Permanent MCAO, rat | VPA | Improved sensorimotor function | (Liu et al., 2012) | |
| Transient MCAO, mouse | TSA | Reduced infarct volumes and neurological deficit scores | (Yildirim et al., 2008) | |
| Transient MCAO, rat | VPA | Reduced infarct volumes, improved neurological deficit scores | (Ren et al., 2004) | |
| Traumatic brain injury | CCI, mouse | SB | Improves learning and memory | (Dash et al., 2009) |
| CCI, mouse | VPA | Reduced lesion volumes, improved motor function | (Yu et al., 2013) | |
| CCI, rat | VPA | Reduced lesion volumes, improved motor function and spatial learning and memory | (Dash et al., 2010) | |
| CHI, mouse | ITF2357 | Accelerated neurobehavioral recovery, reduced lesion volumes and number of degenerating neurons | (Shein et al., 2009) |
6-OHDA – 6-hydroxydopamine, CCI – controlled cortical impact, CHI – closed head injury, EAE –experimental autoimmune encephalomyelitis, EGCG – (−)-Epigallocatechin-3-gallate, GHB – γ-hydroxybutyrate, MCAO – middle cerebral artery occlusion, MPTP – 1-methyl 1-4-phenyl-1,2,3,6-tetrahydropyridine, PBA – phenylbutyrate, SAHA – suberoylanilide hydroxamic acid, SB – sodium butyrate, SPA – sodium phenylacetate, TSA – Trichostatin A, VPA – Valproic Acid
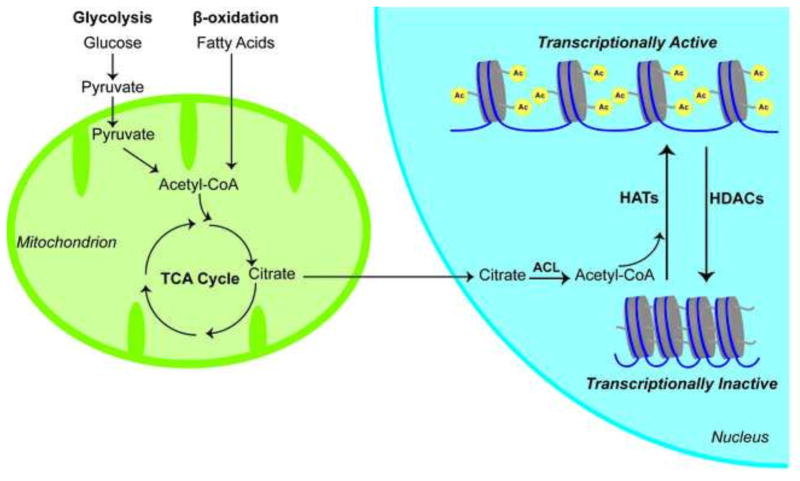
Recent studies from yeast have shown that activity of histone acetyltransferases (HATs), which put acetyl groups on histones to counteract the effects of HDACs, is dependent on adequate levels of acetyl-CoA, a metabolic substrate (Fig. 1) (Takahashi et al., 2006; Wellen et al., 2009). Indeed, in yeast, elevated levels of acetyl-CoA are sufficient to drive the transcription of pro-growth genes via effects on histone acetylation (Cai et al., 2011). These and other studies have supported the notion that acetyl-CoA levels can fluctuate almost ten-fold depending on the state of the cell, and that these fluctuations would be sufficient to influence HAT activity given the known Km of the enzymes for acetyl-CoA. This emphasizes the function of histone acetylation and gene transcription as an integrator of cellular acetyl-CoA levels. Cytosolic and nuclear acetyl-CoA levels are produced largely from the metabolism of glucose via glycolysis and the TCA cycle to produce citrate. ATP citrate lyase then converts citrate into acetyl-CoA. In this scheme, gene transcription is directly regulated by glucose availability via acetyl-CoA and the activity of HATs.
Fig. 1. Acetyl-CoA can be produced through either glycolysis or beta-oxidation and is subsequently used by mitochondria in the TCA cycle.
Citrate, one of the intermediates of the TCA cycle, can be imported in to the nucleus and metabolized to Acetyl-CoA by ATP citrate lyase (ACL). The HATs then utilize Acetyl-CoA to add acetyl groups on to the histone tails to increase transcriptional activity. This process can be undone by the HDACs, which repress transcription.
2. Epigenetic and transcriptional regulators as sensors that transduce metabolic changes into adaptive changes in gene expression: the superfamily of 2-oxoglutarate-dependent dioxygenases
There is growing recognition that a number of epigenetic proteins besides HATs have the potential to directly transduce changes in metabolism into gene responses via their properties as metabolic sensors. A sensor detects changes in the environment and transduces those changes into a cellular response. The superfamily of oxygen-, iron-, and 2-oxoglutarate-dependent dioxygenases includes several families of proteins, which are known to be epigenetic modulators or transcriptional regulators. Under some circumstances, these proteins can sense not only changes in oxygen tension but also changes in iron availability and 2-oxoglutarate levels. In this review, we will discuss three members of this family: TET DNA demethylases, jumonji C domain histone demethylases, and HIF PHDs, with specific attention to their ability to sense metabolic changes in neurons and other cell types in the CNS.
2.1. Ten eleven translocation (TET) proteins
TET proteins were initially identified in rare cases of acute myeloid and lymphocytic leukemia (Hill et al., 2014; Ko et al., 2015; Pastor et al., 2013). This translocation fuses the mixed lineage leukemia gene (MLL1) on chromosome 10 with the TET1 protein on chromosome 11. Clues to TET function came from domain mapping and evolution studies of the three mammalian isoforms TET1, TET2, and TET3. These isoforms evolved in jaw-containing vertebrates from a single gene in phylogenetically older organisms. Specifically, the TET proteins evolved from J-binding proteins in Trypanosome brucei, the causative parasite of African sleeping sickness in humans.
J-binding proteins (JBP) belong to the large AlkB family of oxygen-, iron-, and 2-oxoglutarate-dependent dioxygenases that remove aberrant methylation from damaged DNA by an oxidative mechanism. They produce “base J” in Trypanosome brucei by oxidizing the methyl group on thymidine. Computational analysis of the JBP proteins identified similar putative nucleic acid modifying enzymes, including JBP-TET proteins. This protein then fused with a CXXC domain (a motif capable of binding methylcytosines) to found the TET subfamily of DNA modifying dioxygenases. Of note, TET proteins are present in all species that possess methylcytosines. For example, the worm C. elegans lacks methylcytosines and also lacks TET proteins. The ability of TET proteins to bind methylcytosines and the similarity between thymine and methylcytosine oxidation inspired the notion that TET proteins might catalyze not only oxidation of 5-methylcytosine but also, ultimately, demethylation. This is supported by data from multiple converging lines of inquiry (Tan and Shi, 2012).
For many years, it has been thought that DNA methylation is a mechanism for silencing parasitic elements and unwanted genes in the genome during the developmental establishment of cell identity. Characterization of DNA methyltransferases without clear presence of demethylases gave rise to the notion that methylation was an important, irreversible mechanism in silencing transcription. The ability of some methyl-binding proteins to bind to methylated DNA (CpG islands) and recruit co-repressors substantiated this view.
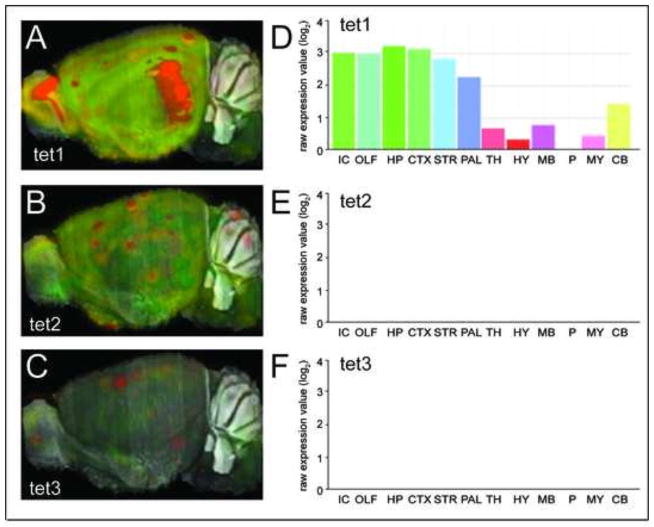
This view was challenged by the discovery that the TET proteins can demethylate DNA, showing that DNA methylation is highly dynamic during an organism’s lifetime (Guo et al., 2011). The discovery revealed a series of intermediate DNA modifications, each with the potential to bind and recruit distinct proteins and thus function as novel epigenetic marks. In situ hybridization of individual TET isoforms (taken from the Allen mouse brain atlas) illustrates the heterogeneous expression of the isoforms in the brain (Fig. 2). TET1 is most highly expressed in the isocortex (IC), olfactory bulb (OLF), hippocampus (HP), striatum (STR), and pallidum (PAL), moderately expressed in the cerebellum (CB), thalamus (TH), midbrain (MB), and cortex as well as to a lower extent in the hypothalamus. By contrast, the basal expression of TET2 and TET3 expression is low in all the brain regions. It is important to note that these studies reflect expression under steady state conditions.
Fig. 2. Ten Eleven Translocation (TET) family message expression in mouse brain.
Gene expression levels of TET1 (A), TET2 (B), TET3 (C), and quantitation of region specific expression of TET1 (D), TET2 (E), TET3 (F) in the mouse brain. Reproduced from Allen Mouse Brain Atlas, Seattle (WA): Allen Institute for Brain Science. Available from: http://mouse.brain-map.org.
2.2. TET proteins catalyze DNA modification via their iron-, oxygen-, and 2-oxoglutarate activity
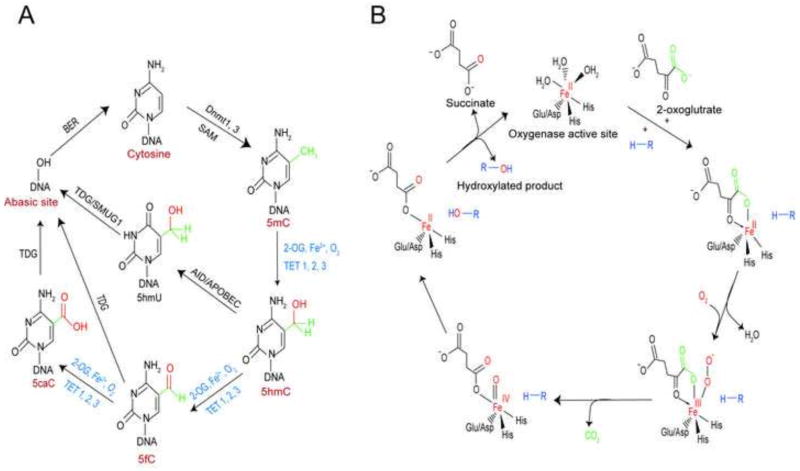
TET proteins catalyze the formation of 5-hydroxymethylcytosine (5hmC) from 5-methyl cytosine (5mC). 5hmC can serve as a substrate for TET enzymes and generate 5-formylcytosine (5fC). 5fC can serve as a substrate for TET proteins and generate 5-carboxylcytosine (5caC) (Fig. 3A). Full demethylation of cytosines occurs when 5fC and 5caC act as substrates for thymine-DNA glycosylase enzymes via base excision repair (Hahn et al., 2013; Mellen et al., 2012). An important and possibly selective role for TET proteins in the brain is supported by observations that 5hmC is higher in the brain than in any other organ. Depletion of TET2 or TET3 reduces the proliferation of neural progenitor cells (Li et al., 2015), and recently TET3 has been shown to underlie activity-dependent homeostatic plasticity (Yu et al., 2015).
Fig. 3. TETs could act as stress sensors and catalyze DNA modification via their iron-, oxygen- and 2-oxoglutarate-dependent dioxygenase activity.
(A) Catalysis of DNA modification by TET proteins. DNA methyltransferase (Dnmt) in presence of methyl donor S-adenosyl methionine (SAM) modifies cytosine (C) into 5-methylcytosine (5mC), which is oxidized sequentially into 5-hydroxymethylcytosine (5hmC), 5-formyl cytosine (5fC), and 5-carboxyl cytosine (5caC), respectively by TETs in presence of 2-oxoglutarate, Fe2+ and O2. Demthylation can also involve deamination of 5hmC to 5-hydroxymethyl uridine (5hmU) by an AID/APOBEC enzyme. 5hmU, in turn, generates an abasic site through base excision repair (BER) mediated by DNA glycosylase SMUG1 or Thymine DNA glycosylase (TDG). 5fc or 5caC is excised by thymine DNA glycosylase (TDG) producing an abasic site as part of BER that, in turn, regenerates unmodified cytosine. (B) Catalytic mechanism of 2-oxoglutarate, Fe2+ and O2-dependent dioxygenases.
2.3. TET proteins as stress sensors?
An exciting but underdeveloped area of investigation is whether the TET proteins can serve as stress sensors, modulating their activity in response to changes in oxygen tension, cellular iron levels or 2-oxoglutarate levels. While the family of 2-oxoglutarate-dependent dioxygenases is large and diverse, the catalytic cycle of this family is highly conserved. This conservation is attributed to a core structural motif composed of 8 β-strands arranged like a “jelly roll” surrounded by α-helices. The jelly roll contains the active site with a non-heme iron coordinated by a triad of two histidines and one glutamate/aspartate. As iron has six coordination sites, labile water molecules loosely coordinate the other three sites. Accordingly, the iron in the active site is relatively exposed and, therefore, may be susceptible to auto-oxidation as well as coordination by drugs. Whether this exposed iron is also vulnerable to bulk changes in iron concentrations has not been explored systematically but will be discussed later in this review (Loenarz and Schofield, 2011; Ozer and Bruick, 2007).
The catalytic cycle of all the 2-oxoglutarate-dependent dioxygenases is similar and begins with the binding of 2-oxoglutarate and the entry of the prime substrate (to be hydroxylated) with the concomitant displacement of a water molecule from iron. The binding of oxygen to iron leads to the oxidative decarboxylation of 2-oxoglutarate to form succinate. The result is the formation of a highly reactive ferryl-oxo species that hydroxylates the prime substrate. In this process, Fe3+ is generated in the active site, and it must be reduced back to Fe2+ for additional hydroxylations to occur. It is believed that ascorbate plays the important role of regenerating reduced iron in the active site (Kuiper and Vissers, 2014). In this catalytic cycle, one molecule of oxygen goes to succinate, and the other goes to hydroxylate the prime substrate (Fig. 3B).
2.4. Does oxygen availability modulate the activity of TET enzymes in the nervous system?
As oxygen is an essential cofactor in the hydroxylation of methylcytosines by TET enzymes, it is reasonable to predict that the activity of the enzyme might be diminished under conditions of hypoxia. While a number of groups have established that hypoxia can transcriptionally regulate TET function, no group to date has examined whether hypoxia can influence the level of DNA methylation via suppression of TET enzyme activity.
A challenge in this analysis is that hypoxia, through its ability to inhibit or suppress mitochondrial oxidative phosphorylation, can influence the amount of 2-oxoglutarate. 2-oxoglutarate is generated in the mitochondrial tricarboxylic acid (TCA) cycle by isocitrate dehydrogenase and thus can theoretically negatively regulate TET enzymes and other 2-oxogtutarate dehydrogenases. In the absence of hypoxia, acquired or inherited dysfunction of mitochondrial TCA cycle enzymes, observed in diseases such as Alzheimer’s disease or brain gliomas, might also affect the level of 2-oxoglutarate and consequently the function of enzymes that use this cosubstrate for hydroxylation reactions.
The emergence of metabolomics along with DNA methylation analysis with greater coverage should improve our understanding of how changes in cellular cofactors for TET enzymes influence both their function in a single cell type and across various organs and tissues. The model predicts that under conditions of hypoxia, ischemia, or mitochondrial dysfunction, TET function is inhibited, leading to transient hypermethylation of DNA in coding regions of genes involved in ATP consumption, reactive oxygen species (ROS) generation, and cell death. All aspects of this model have yet to undergo experimental testing. The hypermethylated DNA would recruit methyl-binding proteins with their associated co-repressors to create a chromatin state unfavorable for transcription.
3. Jumonji domain demethylases: 2-oxoglutarate-dependent dioxygenases that demethylate lysines on histones
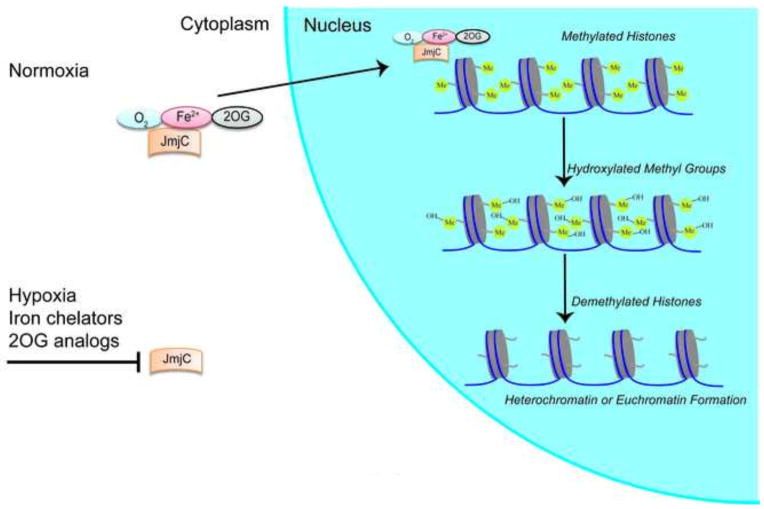
Another group of enzymes that require oxygen, 2-oxoglutarate, and iron is the jumonji (JmJc) family of histone demethylases (Hancock et al., 2015). The jumonji domain was so named because of the abnormal shape of the neural grooves in the developing neural plate of the jumonji mutant mice. (Jumonji is the word for cruciform or cross in Japanese, which was used to describe the cross-shaped morphology.) The JmJc domain is found in all proteins in the family. By homology to the AlkB protein, the JmJc was predicted and then demonstrated to demethylate lysine residues via hydroxylation (Fig. 4). Distinct isoforms of the family have been found to demethylate specific residues of histones H3 or H4 (Shmakova et al., 2014).
Fig. 4. Epigenetic regulation of histone methylation by Jumonji.
Under normoxia, Jumonjis (JmjCs) in the presence of oxygen (O2), Iron (Fe2+), and 2-oxogluterate (2OG) selectively hydroxylates methylated lysines in N-terminal histone tails. Hydroxylated methyl groups are demethylated leading to either heterochromatin or euchromatin formation. During hypoxia, iron insufficiency or 2OG depletion, JmjCs may be inhibited, but experimental support for this type of regulation does not exist.
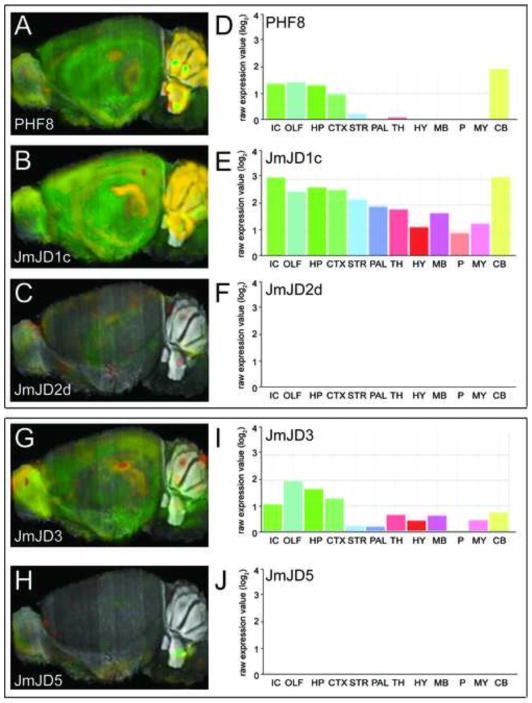
Jumonji-containing histone demethylases are differentially expressed in adult microglia, neurons, and astrocytes. PHF8, JmJD1c, and JmJD2d are expressed in all three cell types, although the expression is highest in neurons. JMJD3 and JMJD5 are highly expressed in microglia (Smith et al., 2014). In situ hybridization of individual JmJD isoforms illustrates the heterogeneous expression of the isoforms in the brain. At basal levels, expression of JmJD1c in the brain is high, that of PHF8 and JmJD3 is moderate, and expression of JmJD2d and JmJD5 is low. As JmJDs are hypoxia-regulated, the distribution of expression is likely to change after stress. However, this has not been formally evaluated (Fig. 5).
Fig. 5. Jumonji domain demethylases expression in mouse brain.
Gene expression levels of PHF8 (A), JmJD1c (B), JmJD2d (C), and quantitation of region specific expression of PHF8 (D), JmJD1c (E), JmJD2d (F) in the mouse brain. Gene expression levels of JmJD3 (G), JmJD5 (H), and quantitation of region specific expression of JmJD3 (I), JmJD5 (J) in the mouse brain. Reproduced from Allen Mouse Brain Atlas, Seattle (WA): Allen Institute for Brain Science. Available from: http://mouse.brain-map.org.
Covalent modifications of histones on lysines and arginines are well established to involve monomethylation, dimethylation, and trimethylation. These modifications occur on lysines 4, 9, 27, 36, and 79 on histone 3, and at lysines 8, 12, 16, 20, 31, 44, 59, and 79 on histone 4. While certain marks are commonly associated with activation or repression, there is growing recognition that specific marks can be repressive or activating, depending on the context, such as cell type or expression of reader proteins (Fig. 5) (Isles, 2015). This has important implications for how we think metabolism might influence histone methylation.
With the number of jumonji-containing histone demethylases expressed in neurons, astrocytes, and microglia (Smith et al., 2014), it is difficult to imagine a coherent coordinated response if they were all equally sensitive to changes in oxygen, 2-oxoglutarate, and metal concentrations. More likely, there are certain jumonji family members whose activity is more sensitive to changes in these cofactors. Those family members, whether expressed under steady state or induced by stress, could mediate important activation or silencing as part of the adaptive response to stress. Indeed, recently published studies confirm data from our own laboratory that hypoxia can induce expression of histone demethylase JMJD3 in neurons in a HIF-dependent manner (Lee et al., 2014).
To address the relevance of this induction to the hypoxia response and to see whether stress alone influences jumonji function, individual methylation marks will need to be examined at specific gene loci using ChIP-Seq and RNA-Seq under conditions in which oxygen tension, iron levels, and 2-oxoglutarate levels are manipulated carefully.
4. HIF prolyl hydroxylases: canonical sensors of hypoxia, iron deficiency and 2-oxoglutarate dysmetabolism and novel epigenetic modulators?
The most well studied subfamily of the 2-oxoglutarate-dependent dioxygenases are the HIF-prolyl hydroxylases (HIF PHDs) (Bruick and McKnight, 2001). These enzymes are best known for their ability to regulate the stability of the stress-responsive transcription factor, hypoxia-inducible factor-1 (HIF-1). HIF-1, a heterodimer consisting of a regulated HIF-1α subunit and a constitutively expressed HIF-1β subunit, was purified by Greg Semenza and colleagues at Johns Hopkins following the identification of a hypoxia response element in the 3′ untranslated region of the erythropoietin gene (Semenza and Wang, 1992). HIF response elements are found in non-coding regions of all the genes, including Erythropoietin (Epo), vascular endothelial growth factor (VEGF), and glycolytic enzymes induced by hypoxia. This coordinated gene cassette has improved our understanding of how all cells, including neurons, adapt to changes in oxygen tension.
Epo is one of more than 100 genes that are known to be transcriptionally induced by decreased cellular oxygen tension in order to increase red blood cell mass and to enhance oxygen delivery to tissues. Hypoxia-induced changes in Epo are known to underlie the erythrocytosis associated with low-to-high altitude transit in humans. Other well-known transcriptional responses include VEGF, an angiogenic growth factor that increases blood supply to hypoxic tissues, and glycolytic enzymes (including lactate dehydrogenase), which facilitate the enhancement of ATP generation in the absence of oxygen. Of note, although Epo is induced by hypoxia in the kidney, liver and astrocytes, HIF-1 is ubiquitously expressed in all tissues of the body (Semenza, 2014).
In 2001, Bill Kaelin’s group and Peter Ratcliffe’s group independently identified the HIF PHDs as oxygen-, iron-, and 2-oxoglutarate-dependent enzymes that can regulate HIF stability in normoxia (Kaelin and Ratcliffe, 2008). Since then, based on the work of many laboratories, HIF regulation by hypoxia has been elucidated. The HIF PHDs began as single genes in flies and worms and diverged to three isoforms in mammalian cells, HIF PHD1, HIF PHD2, and HIF PHD3 (Eltzschig et al., 2014). Each of these isoforms has distinct subcellular localization and distinct tissue distribution.
Under conditions of normoxia where iron and 2-oxoglutarate are at steady state, physiological levels, HIF PHDs hydroxylate prolines 402 and 564 in HIF1α. This post-translational modification leads to the recruitment of the E3 ubiquitin ligase (the Von Hippel-Lindau protein), the polyubiquitination of HIF-1α, and its degradation by the proteasome. Accordingly, in each cell of the human body, HIF is being constitutively synthesized and degraded.
If oxygen, iron or 2-oxoglutarate levels fall below a critical threshold, the HIF PHDs cease to function, and HIF-1α is not hydroxylated and thus not degraded. Stabilized HIF-1α can dimerize with HIF-1β to transduce an adaptive response to hypoxia. Prior studies from our laboratory have shown that this adaptive response can include pro-survival genes and pro-death genes, such as the BH3-only family member, BNIP3, a gene also noted to induce mitophagy. Pro-survival and pro-death responses mediated by HIF-1α can happen in the same cell type depending on the type of stimulus, thus highlighting the importance of context in determining whether a cell dies or survives once HIF-1α is activated (Aminova et al., 2005). It is now clear that astrocytic versus neuronal HIF-1 can also mediate divergent responses (Ruscher et al., 2002; Vangeison et al., 2008).
Despite this complexity in HIF biology, inhibition of the HIF PHDs molecularly or pharmacologically in diverse cell types has been almost universally associated with neuroprotection and observed in more than a half dozen laboratories. Indeed, initial studies from our laboratory over 15 years ago suggested that the broad, salutary effects of the iron chelator, desferoxamine, could be attributed to inhibition of the HIF PHDs and suppression of proline hydroxylation of specific proteins rather than inhibiting iron catalyzed generation of free radicals. Since then, we have used selective chemical tools, peptide inhibitors, and short interfering RNAs to demonstrate that HIF PHD1 is a critical target for neuroprotection in neurons (Aleyasin et al., 2015; Aminova et al., 2008; Karuppagounder and Ratan, 2012; Karuppagounder et al., 2013; Siddiq et al., 2005; Siddiq et al., 2009; Speer et al., 2013; Zaman et al., 1999).
4.1. Regulation of HIF PHD activity by changes in neuronal metabolism
Unlike TET or jumonji containing proteins, HIF PHDs are well established to be modulated by changes in oxygen tension. An elegant study by the group of Joachim Fandrey systematically manipulated oxygen tension in non-neural cells, and they found that nuclear PHD activity is higher than cytoplasmic activity with oxygen concentrations above 100 μM (Berchner-Pfannschmidt et al., 2008). Below 100 μM, the steepest decline in PHD activity happens between 30 μM and 80 μM, in the dynamic range of oxygen concentrations for most tissues. Monolayer cultures can vary between 0 and 170 μM, thus creating a significant amount of variability of PHD activity between laboratories even in the same cell type. Brain oxygen concentrations are in the range of 40–100 μM in cortex from a host of species, but are substantially lower in white matter, suggesting that HIF PHD activity may vary widely depending on the region of the brain and the subcellular locale examined.
The ability of HIF PHD inhibition to lead to the HIF-dependent transcriptional induction of the HIF PHDs themselves suggests a mechanism for system reset once oxygen concentrations have been restored. In many publications regarding HIF stabilization in the brain, ischemia and hypoxia have been used interchangeably as stresses that could lead to loss of HIF PHD function and stabilization of HIF in the central nervous system. Our data suggest that they are not congruent when it comes to HIF stabilization in human neuronal cells in vitro. Concentration-dependent reductions in glucose lead to translational repression of HIF-1α, which offsets any enhanced stability of the protein (Karuppagounder et al., 2013).
4.2. Regulation of HIF PHDs by 2-oxoglutarate dynamics in living cells
2-oxoglutarate is produced by oxidative decarboxylation of isocitrate via isocitrate dehydrogenase (IDH) (Reitman and Yan, 2010). IDH exists as five isoforms that can be divided into two separate classes. The first class uses NADP+ as the electron acceptor, the second class uses NAD+ as an electron acceptor. All of the NAD+-dependent IDH isoforms are present in the mitochondrial matrix, while the NADP+ isoforms are divided between the cytoplasm (peroxisome) and the mitochondria (Ronnebaum et al., 2006). The cytosolic form, IDH1 appears to be an important source of NADPH and may be involved in glucose sensing and antioxidant defense. IDH2 is a mitochondrial protein and may be an important regulator of the TCA cycle via its ability to substitute for IDH3 functionally or drive the reaction in the reverse direction.
Recent studies have looked at endogenous levels of 2-oxoglutarate in distinct areas of the brain and in neuronal cell lines. Of note, the levels are significantly higher in the substantia nigra (2.7 mM) than in the striatum (1.85 mM), which were both higher than the cortex (0.64 mM) (Thirstrup et al., 2011), but it is unclear to what extent that measurement reflects distinct numbers or distribution of neurons versus glia. Indeed, transformed “neuron like cell lines”, including PC12 (rat pheochromocytoma) and SH-SY5Y (human neuroblastoma), have levels of 2-oxoglutarate above 2 mM (Thirstrup et al., 2011). These findings suggest that mean levels of 2-oxoglutarate are significantly higher than the Km for most of the HIF PHDs, which is approximately 1 mM. This makes it unlikely that large changes in 2-oxoglutarate levels are able to directly influence HIF PHD activity unless there are similarly large changes in fumarate or succinate, which have been shown to act as competitive inhibitors of the HIF PHDs (Isaacs et al., 2005). Prior studies from our lab have not shown that fumarate or succinate can modulate HIF PHD activity or HIF stability in neurons (Niatsetskaya et al., 2010).
Of course, it is formally possible that 2-oxoglutarate levels vary depending on the neural compartment, but no group has as yet examined 2-oxoglutarate levels in the nucleus versus the cytoplasm versus the axon or dendrite. In this scheme, it is possible that nuclear 2-oxoglutarate levels fluctuate to a greater degree than in other compartments particularly given the number of 2-oxoglutarate dependent dioxygenases in the nucleus.
4.3. HIF PHDs as iron sensors and mediators of neuroprotective responses
Our laboratory’s interest in the HIF PHDs evolved nearly a decade before they were molecularly identified via an unexpected result in a somewhat mundane experiment (Zaman et al., 1999). Iron chelators had been known to protect neurons from a host of insults, but the precise mechanism by which these agents acted to mediate their salutary effects was unknown. In fact, the protective effects of iron chelators had been tacitly linked to the direct reduction in free radical production that could be catalyzed by metals (Winterbourn and Kettle, 2013).
Through a serendipitous series of observations, we correlated the protective effects of iron chelators in vitro and in vivo with the stabilization of HIF1 and activation of HIF1 dependent genes (Zaman et al., 1999). In this study, we found that the addition of cobalt chloride, a transition metal without intrinsic antioxidant activity could like iron chelation, stabilize HIF-1 and protect neurons. Our findings suggested but did not prove that the mechanism of protection of iron chelators could be linked to oxygen sensing.
Frank Sharp and colleagues extended these in vitro findings to in vivo studies in a model of neonatal ischemia. They showed that cobalt chloride (60 mg/kg) or desferoxamine (200 mg/kg) could induce HIF-1 expression in the brain and precondition the brain against neurotoxic insults (Bergeron et al., 2000). Subsequent observations demonstrated that forced expression of HIF-1 in vitro potentiated some mediators of cell death (oxidative stress), while protecting other forms of cell death (ER stress) (Aminova et al., 2005; Aminova et al., 2008).
This led many groups, including our own, to focus on the mechanism of oxygen sensing as the target for protection by iron chelators. The discovery of the HIF PHDs by Bill Kaelin’s group and Peter Ratcliffe’s group as iron-dependent dioxygenase provided a bona fide target for iron chelation in the protection of neurons from a host of insults (Kaelin and Ratcliffe, 2008).
4.4. Iron metabolism and HIF PHD activity in brain
The requirement of HIF PHDs for iron and the ability of iron chelators to inhibit the HIF PHDs to stabilize HIF and possibly other proteins in neurons led to a straightforward model. Agents that deplete iron should lead to inhibition of HIF PHDs, stabilization of HIF-1, and shift cellular metabolism away from oxidative phosphorylation.
As many proteins in the mitochondrial TCA cycle and electron transport chain are iron sulfur cluster proteins, these proteins would be inhibited by low levels of cellular iron (Maio and Rouault, 2015). Depletion of iron should lead to loss of function of the mitochondria and provide the driving force for enhancement of aerobic glycolysis by HIF. In fact, direct inhibition of the iron sulfur cluster machinery in mitochondria leads to an adaptive response that includes the reduction of genes involved in mitochondrial respiration and an increase in genes involved in glycolysis.
Accordingly, HIF PHDs may have evolved as iron-dependent dioxygenases because of the importance of iron to mitochondrial function (Hausmann et al., 2008). The mechanism by which global iron depletion leads to HIF PHD inhibition is still not fully resolved, as the affinity of iron in the active site is very high and would not be expected to be lost by changes in cellular iron. Rather, the effect may be a slow one, which requires turnover of existing proteins and depletion of iron in chaperones that normally deliver iron to the HIF PHDs (Nandal et al., 2011). Currently our understanding of how iron overload conditions affect HIF PHD function is limited. Future studies should systemically modulate cellular iron homeostasis to provide a clearer picture of how iron-dependent dioxygenases are influenced by changes in this important cofactor.
5. Conclusion
Pharmacological and molecular data from a host of laboratories have shown that inhibition of PHDs can confer protection to the neurons in a dish and in the brain in response to a host of insults. Discussion of the mechanisms of protection by these agents is beyond the scope of this review but suffice it to say that the protective effects are well established in multiple laboratories and multiple models. Future studies will explore the role of other 2-oxoglutarate-dependent dioxygenases such as Tet DNA demethylases and jumanji histone demethylases in cellular fitness.
We believe the breadth of insults that can be abrogated by inhibition of HDACs or HIF PHDs reflects the broad adaptive transcriptional response triggered by manipulating these targets. The ability to activate and/or repress broad programs of transcription means that inhibition of these enzymes provides the coordinated potential benefit of many drugs acting at cellular, local, and systemic levels to restore homeostasis. Current studies are elucidating alternate pathways by which these important stress sensors are working to effect both transcriptional and epigenetic pathways to induce fitness in the brain.
Highlights.
Homeostatic epigenetics
HATs sense acetyl-CoA
Chromatin modifiers sense Fe2+, 2-OG and O2
Acknowledgments
This work was supported by the National Institute of Health (P01 NIA AG014930, Project 1 to R.R.R.), the Dr. Miriam and Sheldon G. Adelson Program in Neurorehabilitation and Neural Repair, the Sperling Center for Hemorrhagic Stroke Recovery at the Burke Medical Research Institute and New York State Department of Health Center for Research Excellence in Spinal Cord Injury (Grant DOHC019772). This work was also supported by grant TL1TR000459 of the Clinical and Translational Science Center at Weill Cornell Medical College.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aleyasin H, et al. Antihelminthic benzimidazoles are novel HIF activators that prevent oxidative neuronal death via binding to tubulin. Antioxid Redox Signal. 2015;22:121–34. doi: 10.1089/ars.2013.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminova LR, et al. Prosurvival and prodeath effects of hypoxia-inducible factor-1alpha stabilization in a murine hippocampal cell line. J Biol Chem. 2005;280:3996–4003. doi: 10.1074/jbc.M409223200. [DOI] [PubMed] [Google Scholar]
- Aminova LR, Siddiq A, Ratan RR. Antioxidants, HIF prolyl hydroxylase inhibitors or short interfering RNAs to BNIP3 or PUMA, can prevent prodeath effects of the transcriptional activator, HIF-1alpha, in a mouse hippocampal neuronal line. Antioxid Redox Signal. 2008;10:1989–98. doi: 10.1089/ars.2008.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchner-Pfannschmidt U, et al. Nuclear oxygen sensing: induction of endogenous prolyl-hydroxylase 2 activity by hypoxia and nitric oxide. J Biol Chem. 2008;283:31745–53. doi: 10.1074/jbc.M804390200. [DOI] [PubMed] [Google Scholar]
- Bergeron M, et al. Role of hypoxia-inducible factor-1 in hypoxia-induced ischemic tolerance in neonatal rat brain. Ann Neurol. 2000;48:285–96. [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–40. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Cai L, et al. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Molecular cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltzschig HK, Bratton DL, Colgan SP. Targeting hypoxia signalling for the treatment of ischaemic and inflammatory diseases. Nat Rev Drug Discov. 2014;13:852–69. doi: 10.1038/nrd4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, et al. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–8. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock RL, et al. Epigenetic regulation by histone demethylases in hypoxia. Epigenomics. 2015:1–21. doi: 10.2217/epi.15.24. [DOI] [PubMed] [Google Scholar]
- Hausmann A, et al. Cellular and mitochondrial remodeling upon defects in iron-sulfur protein biogenesis. J Biol Chem. 2008;283:8318–30. doi: 10.1074/jbc.M705570200. [DOI] [PubMed] [Google Scholar]
- Hill PW, Amouroux R, Hajkova P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics. 2014;104:324–33. doi: 10.1016/j.ygeno.2014.08.012. [DOI] [PubMed] [Google Scholar]
- Isaacs JS, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–53. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Isles AR. Neural and behavioral epigenetics; what it is, and what is hype. Genes Brain Behav. 2015;14:64–72. doi: 10.1111/gbb.12184. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Karuppagounder SS, Ratan RR. Hypoxia-inducible factor prolyl hydroxylase inhibition: robust new target or another big bust for stroke therapeutics? J Cereb Blood Flow Metab. 2012;32:1347–61. doi: 10.1038/jcbfm.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppagounder SS, et al. In vitro ischemia suppresses hypoxic induction of hypoxia-inducible factor-1alpha by inhibition of synthesis and not enhanced degradation. J Neurosci Res. 2013;91:1066–75. doi: 10.1002/jnr.23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, et al. TET proteins and 5-methylcytosine oxidation in hematological cancers. Immunol Rev. 2015;263:6–21. doi: 10.1111/imr.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper C, Vissers MC. Ascorbate as a co-factor for fe- and 2-oxoglutarate dependent dioxygenases: physiological activity in tumor growth and progression. Front Oncol. 2014;4:359. doi: 10.3389/fonc.2014.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley B, et al. Pulse inhibition of histone deacetylases induces complete resistance to oxidative death in cortical neurons without toxicity and reveals a role for cytoplasmic p21waf1/cip1 in cell cycle-independent neuroprotection. The Journal of Neuroscience. 2008;28:163–176. doi: 10.1523/JNEUROSCI.3200-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, et al. HIF-1-dependent induction of Jumonji domain-containing protein (JMJD) 3 under hypoxic conditions. Mol Cells. 2014;37:43–50. doi: 10.14348/molcells.2014.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, et al. Critical Role of Tet3 in Neural Progenitor Cell Maintenance and Terminal Differentiation. Molecular Neurobiology. 2015;51:142–154. doi: 10.1007/s12035-014-8734-5. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Schofield CJ. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem Sci. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Maio N, Rouault TA. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim Biophys Acta. 2015;1853:1493–1512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, et al. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–30. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandal A, et al. Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metab. 2011;14:647–57. doi: 10.1016/j.cmet.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niatsetskaya Z, et al. HIF prolyl hydroxylase inhibitors prevent neuronal death induced by mitochondrial toxins: therapeutic implications for Huntington’s disease and Alzheimer’s disease. Antioxid Redox Signal. 2010;12:435–43. doi: 10.1089/ars.2009.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson DE, et al. Hydroxamate-Based Histone Deacetylase Inhibitors Can Protect Neurons from Oxidative Stress via a Histone Deacetylase-Independent Catalase-Like Mechanism. Chemistry & biology. 2015;22:439–445. doi: 10.1016/j.chembiol.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozer A, Bruick RK. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol. 2007;3:144–53. doi: 10.1038/nchembio863. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–56. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–41. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnebaum SM, et al. A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J Biol Chem. 2006;281:30593–602. doi: 10.1074/jbc.M511908200. [DOI] [PubMed] [Google Scholar]
- Ruscher K, et al. Erythropoietin is a paracrine mediator of ischemic tolerance in the brain: evidence from an in vitro model. J Neurosci. 2002;22:10291–301. doi: 10.1523/JNEUROSCI.22-23-10291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, et al. Histone deacetylase inhibitors prevent oxidative neuronal death independent of expanded polyglutamine repeats via an Sp1-dependent pathway. Proceedings of the National Academy of Sciences. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, et al. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. 2005;93:1087–98. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–54. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- Shmakova A, et al. Chromatin and oxygen sensing in the context of JmjC histone demethylases. Biochem J. 2014;462:385–95. doi: 10.1042/BJ20140754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq A, et al. Hypoxia-inducible factor prolyl 4-hydroxylase inhibition. A target for neuroprotection in the central nervous system. J Biol Chem. 2005;280:41732–43. doi: 10.1074/jbc.M504963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiq A, et al. Selective inhibition of hypoxia-inducible factor (HIF) prolyl-hydroxylase 1 mediates neuroprotection against normoxic oxidative death via HIF- and CREB-independent pathways. J Neurosci. 2009;29:8828–38. doi: 10.1523/JNEUROSCI.1779-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleiman S, et al. Hydroamic based histone deacetylase (HDAC) inhibitors can mediate neuroprotection independent of HDAC inhibition. Journal of Neuroscience. 2014 doi: 10.1523/JNEUROSCI.1010-14.2014. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Kimyon RS, Watters JJ. Cell-type-specific Jumonji histone demethylase gene expression in the healthy rat CNS: detection by a novel flow cytometry method. ASN Neuro. 2014;6:193–207. doi: 10.1042/AN20130050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer RE, et al. Hypoxia-inducible factor prolyl hydroxylases as targets for neuroprotection by “antioxidant” metal chelators: From ferroptosis to stroke. Free Radic Biol Med. 2013;62:26–36. doi: 10.1016/j.freeradbiomed.2013.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Takahashi H, et al. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Molecular cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirstrup K, et al. Endogenous 2-oxoglutarate levels impact potencies of competitive HIF prolyl hydroxylase inhibitors. Pharmacol Res. 2011;64:268–73. doi: 10.1016/j.phrs.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Vangeison G, et al. The good, the bad, and the cell type-specific roles of hypoxia inducible factor-1 alpha in neurons and astrocytes. J Neurosci. 2008;28:1988–93. doi: 10.1523/JNEUROSCI.5323-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn CC, Kettle AJ. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal. 2013;18:642–60. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- Yu H, et al. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat Neurosci. 2015;18:836–43. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman K, et al. Protection from oxidative stress-induced apoptosis in cortical neuronal cultures by iron chelators is associated with enhanced DNA binding of hypoxia-inducible factor-1 and ATF-1/CREB and increased expression of glycolytic enzymes, p21(waf1/cip1), and erythropoietin. J Neurosci. 1999;19:9821–30. doi: 10.1523/JNEUROSCI.19-22-09821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]