Abstract
Young/adolescent humans demonstrate many microorganisms associated with periodontal disease in adults and substantial gingival inflammatory responses. However, younger individuals do not demonstrate the soft and hard tissue destruction that hallmark periodontitis. This study evaluated responses to the oral microbial ecology in gingival tissues from clinically healthy young Macaca mulatta (<3 years old) compared to older animals (5-23 years old). Global transcriptional profiling of four age groups revealed a subset of 159 genes that were differentially expressed at least across one of the age comparisons. Correlation metrics generated a relevance network abstraction of these genes. Partitioning of the relevance network revealed seven distinct communities comprising functionally related genes associated with host inflammatory and immune responses. A group of genes were identified that were selectively increased/decreased or positively/negatively correlated with gingival profiles in the animals. A Principal Components Analysis created metagenes of expression profiles for classifying the 23 animals. The results provide novel system-level insights into gene expression differences in healthy young tissues weighted towards host responses that were associated with anti-inflammatory biomolecules or those linked with T cell regulation of responses. The combination of the regulated microenvironment may help to explain the apparent “resistance” of younger individuals to developing periodontal disease.
Keywords: Keywords: nonhuman primates, periodontitis, inflammation, aging
Introduction
The nonhuman primate has been documented as a model of periodontitis that demonstrates extensive similarities in clinical, microbiological and immunological features of human periodontitis (1-7). The human subgingival ecology has been shown to exhibit over 700 species of bacteria (8) and differs both qualitatively and quantitatively in health, gingivitis, and periodontitis (9). Recent studies have demonstrated a very similar microbiota inhabiting the oral cavity of rhesus monkeys (Macaca mulatta) (B. Paster, AADR 2014, abst. #1588, personal communication).
It is clear that the oral microbiome is acquired early in life and varies among individuals including the types of commensal bacteria, as well as varies in the quality and quantity of proposed opportunistic pathogens that trigger periodontitis later in life (10-13). There is minimal evidence that these pathogens are acquired exogenously in adults that develop periodontitis, thus, research has attempted to focus identification of risk by examining local inciting or environmental factors that would help trigger the bacterial changes in the disease and identifying genetic polymorphisms that could contribute to dysfunctional responses in the periodontium to the microbial challenge.
However, opportunistic pathogens can emerge in the ecology leading to chronic immunoinflammatory lesions and tissue destructive events. It has been recognized that an individual's oral microbiome is acquired early in life, evolves and matures over some time interval, but clearly becomes an intra-individual autochthonous ecology. In a subset of the human population, and our data support in nonhuman primates as well, this ecological changes either trigger a local disease process in the periodontium, or reflect changes in the oral environment that select for more pathogenic biofilms. Routinely when this process occurs it is as an adult or aged individual. However, we have negligible information regarding the ontogeny of the various innate immune, inflammatory, and adaptive immune response pathways in gingival tissues of young individuals, nor is there any data available that describes how variations in the evolving oral microbiome “drive”, not only the maturation of these pathways at the mucosal sites, but also how these microbial variations can result in dissimilarities in the maturation of host response capabilities in the tissues.
We have been using the nonhuman primate model of periodontitis to explore functional genomics that would be involved in creating a local environment in the gingival milieu related to health, or disease, or increased risk for disease. As such we have been targeting specific molecular pathways to determine the transcriptome in gingival tissues, as a representative mucosal tissue, obtained from animals representing young individuals (approximately 10 year old humans) to aged individuals (approximately 70-80 year old humans) (1, 14-16). These studies have shown significant differences in apoptosis pathway gene expression profiles associated with aging, even in healthy gingival tissues (15, 16). Differences were also noted in inflammasome gene pathways, including both receptors critical for signaling, and downstream effector functions (17), and in antigen processing and presentation pathways (18), all focused on changes with aging that could presage disease risk and account for the increased incidence and severity of disease in aged individuals.
Based upon the existing literature that supports that young individuals and adolescents harbor many of the oral microorganisms considered to contribute to periodontitis in adults (12, 19, 20), and generally demonstrate a high prevalence of gingivitis, it is very infrequent that they develop destructive periodontitis (20). This report posits that the lack of progression of chronic inflammation in young individuals to a tissue destructive process that is the hallmark of periodontitis, will be reflected by differential expression of genes in response to the bacterial biofilm challenge that are more tissue protective and help maintain the integrity of the tissues even in the presence of persistent inflammation.
Methods
Nonhuman primate model and Oral Clinical Evaluation
Rhesus monkeys (Macaca mulatta) (n=34; 14 females and 20 males) housed at the Caribbean Primate Research Center (CPRC) at Sabana Seca, Puerto Rico, were used in these studies. Healthy animals (5-7/group) were distributed by age into four groups: ≤3 years (young), 3-7 years (adolescent), 12-16 years (adult) and 18-23 years (aged). A protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Puerto Rico, enabled anesthetized animals to be examined for clinical measures of periodontal health including probing pocket depth (PPD), and bleeding on probing (BOP) as we have described previously (21). Health was defined as mean mouth values of PPD <3mm and BOP <1.
The nonhuman primates were typically fed a 20% protein, 5% fat, and 10% fiber commercial monkey diet (diet 8773, Teklad NIB primate diet modified: Harlan Teklad). The diet was supplemented with fruits and vegetables, and water was provided ad libitum in an enclosed corral setting.
Tissue sampling and gene expression microarray analysis
A buccal gingival sample from healthy sites from the premolar/molar maxillary region of each animal was taken using a standard gingivectomy technique, and maintained frozen in RNAlater solution. Total RNA was isolated from each gingival tissue using a standard procedure as we have described and tissue RNA samples submitted to the microarray core to assess RNA quality analyze the transcriptome using the GeneChip® Rhesus Macaque Genome Array (Affymetrix) (16, 22). Individual samples were used for gene expression analyses.
Based upon the microarray outcomes we selected 5 genes and performed a qPCR analysis using a standard technique in our laboratory employing a Roche 480 LightCycler (23). qPCR primers were designed using software PrimerQuest at Integrated DNA Technologies website (www.idtdna.com) and were synthesized by Integrated DNA Technologies, Inc (Coralville, IA). Primers were prepared for PSMB8 (forward - GGCGCTGTCATCGATTTCTT; reverse – ATGGCTTTGTAGACGCCTTTC; amplicon 103 bp), IL1A (forward – CTGAAGAAGAGACGGTTGAGTT; reverse – CGACCTGGGCTTGATGATT; amplicon 99 bp), IL22 (forward – GAGCGCTGCTAT CTGATGAA; reverse – GCACCACCTCCTGCATATAA; amplicon 100 bp), IL17F (forward - ATCTCCATGAATTCCGTTCCC; reverse – AACAGTCACCAGCACCTTC; amplicon 105 bp), TNFSRSF17 (forward – GGCAGGACTGGTGATGAAA; reverse – GTGGAAAGCAATGGTCAGAATC; amplicon 118 bp) and GAPDH (forward – GGTGTGAACCATGAGAAGTATGA; reverse – GAGTCCTTCCACGATACCAAAG; amplicon 123 bp) genes. The level of message was determined according to our previously published methods (23) and those levels compared across the RNA samples prepared from each of the healthy groups.
Data analysis
Normalization and background subtraction was accomplished using the RMA approach (24). Parametric t-test (α = 0.01) was subsequently to determine genes that changed significantly across a given pair of groups. A fold change cut-off (≥ 2-fold) was subsequently imposed to eliminate noisy expression profiles. Differentially expressed genes that satisfied the fold-change cut-off at least across one of the six pair-wise comparisons was chosen for subsequent analysis. JMP (version 10.0, SAS Inc., Cary, NC) was used to create metagenes independently of group classification using principal components based on the correlation matrix. The plots are of the first two PCA scores across the healthy tissues. The variability is explained by each of the scores indicated on the plots. The data has been uploaded into the ArrayExpress data base (www.ebi.ac.uk) under accession number: E-MTAB-1977.
Results
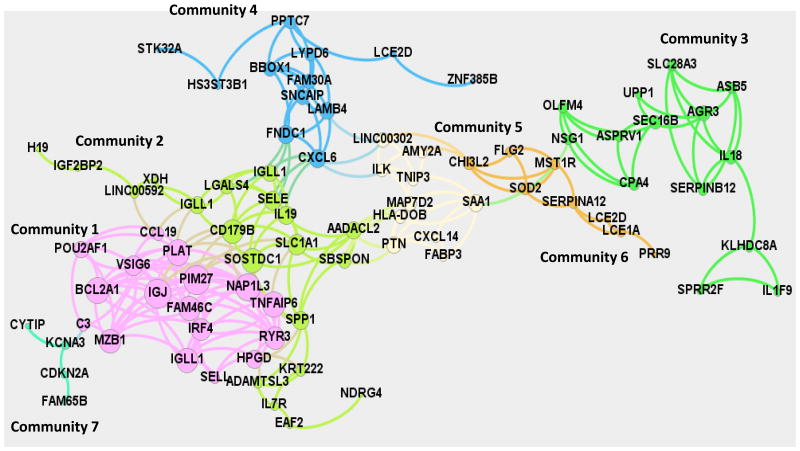
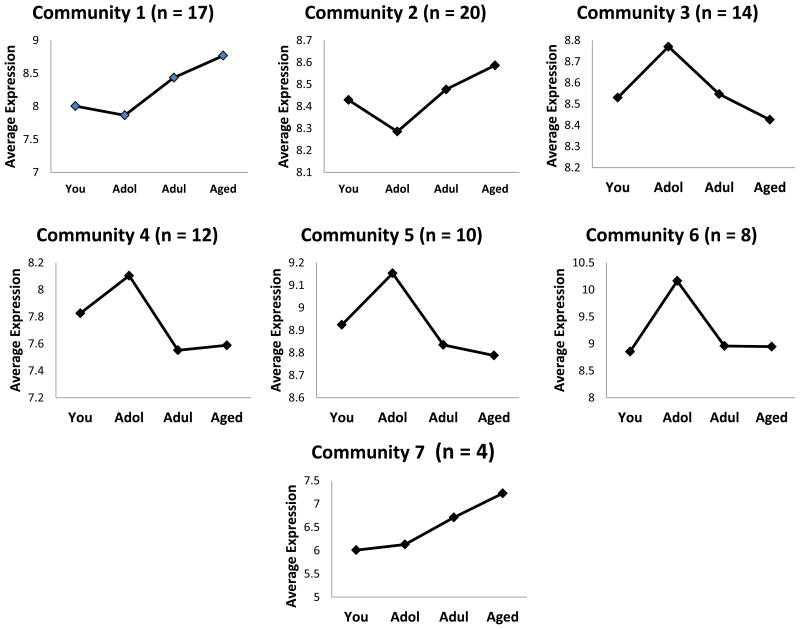
Differential gene expression analysis using parametric t-test (p-value < 0.01, fold change ≥ 2) across the 4 groups resulted in 159 genes. Relevance network (25) abstraction of the 159 genes was subsequently by connecting the highly correlated genes (Pearson-Correlation, p < 0.01) by an undirected edge. Duplicate genes and those transcripts that were not annotated were dropped from the relevance network abstraction. Yifan-Hu visualization of the relevance network is shown in Figure 1. The giant component of the network comprised of 85 nodes and 235 edges where each node is connected to the other directly or indirectly was subsequently partitioned into distinct communities using the Louvain method for community structure detection (26) implemented in Gephi 8.1 (27). Seven communities with varying connectivity and number of genes were observed (Table 1, Figure 1) in the giant component. Community 1 consisted of 17 genes including PIM1, IGJ and NAP1L3 with degree centralities of 14, 14 and 13, respectively. Interestingly, the IGJ gene codes for the immunoglobulin J polypeptide, which is the linker protein for IgA and IgM polypeptides. It also contributes to binding these immunoglobulins to secretory component at mucosal surfaces. PIM1 oncogene belongs to the serine/threonine protein kinase family and is expressed primarily in B-lymphoid cells and contributes to both cell proliferation and survival (28). NAP1L3 (nucleosome assembly protein 1-like 3) has been suggested to contribute to the RIG-I-like receptor signaling pathway and may have some role in the mucosal immune network (http://immunet-dev.princeton.edu/genes/detail/homo-sapien/NAP1L3/). The average degree centrality of Community 1 was the largest among all the communities (∼10) indicating a densely connected community and dominant players in the network. The average expression profile of the genes in Community 1 is shown in Figure 2 and exhibits an increasing trend as a function of age. Community 2 consists of 20 genes with average degree centrality (∼6). Genes with a high degree of centrality in Community 2 include (SOSTDC1, CD179B, SLC1A1) with degree centralities of 11, 11 and 9, respectively. SOSTDC1 (sclerostin domain containing 1) is a member of the sclerostin family and functions as a bone morphogenetic protein (BMP) antagonist, as well as enhancing Wnt signaling and inhibiting TGFβ signaling (29). CD179b (IGLL1; immunoglobulin lambda-like polypeptide 1) is a receptor found on the surface of pro-B and pre-B cells. It transduces signals for cellular proliferation, differentiation, and allelic exclusion at the Ig heavy chain gene locus, was well as promoting Ig light chain gene rearrangements (30). The SLC1A1 [solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1] gene encodes a member of the high-affinity glutamate and aspartate transporters. The SLC1A1 protein provides cysteine uptake for GI epithelial, neuronal, and immune cells, and its activity is decreased during oxidative stress and, thus, it has been implicated in the intestinal immune network for IgA production (31). The average expression profile of the members in Community 2 is similar to that of Community 1 with a general decrease with adolescence and increasing levels of expression in adult and aged gingival tissues (Figure 2). Of the 17 genes in Community 1, 10 are associated with host immune responses, and Community 2 contains 12 genes with 5 related to host responses and inflammation within this network. The average expression profile across Community 7 also exhibited an increasing trend somewhat similar to that of Communities 1 and 2. In contrast, Communities 3, 4, 5, and 6 exhibit an increased expression in adolescence, and then show a decreasing trend in the average expression profile through adult and aged tissues to levels comparable to young animals (Figure 2). Within these 4 communities, only Community 5 showed 8/12 genes that were related to host immune responses. Finally, a group of 3 genes (IL1A, MUC4, DEFB4A) was also identified as an isolated community that was connected to the giant component. The average expression profile was similar to that of Community 2. Interestingly all three of these genes are intimately associated with host responses in the oral cavity, and were also identified as altered in exploring the immunology array of genes.
Figure 1.
Yifan-Hu visualization of the relevance network stratified into eight distinct communities. Each point denotes a gene and the number of lines signify the strength of the association of expression across all 4 age groups. Genes within each of the communities are represented by the same color.
Table 1.
Listing of genes that were networked in the various communities based upon differences among the age groups in healthy gingival tissues. The asterisk (*) denotes those genes related to host responses and immune functions.
| Gene ID | Function |
|---|---|
| COMMUNITY 1 | |
| PIM27 | *Pim-2 oncogene; cell survival |
| IGJ | *Ig J (joining) chain |
| NAP1L3 | *Nucleosome assembly protein 1-like 3 |
| BCL2A1 | *BCL2-related protein; apoptosis IL-3 |
| TNFAIP6 | *TNFa induced protein 6; inflammation |
| PLAT | *Tissue plasminogen activator |
| MZB1 | *Marginal zone B/B1 cell protein |
| FAM46C | *Family with sequence similarity 46, member C; interferon/viral regulation |
| IGLL1 | *Ig lambda-like polypeptide 1 |
| RYR3 | Ryanodine receptor 3; Ca+2 homeostasis |
| IRF4 | *Interferon regulatory factor 4 (LOC10042412) |
| VSIG6 | V-set and Ig domain containing 6 |
| HPGD | Hydroxyprostaglandin dehydrogenase 15-(NAD) |
| POU2AF1 | *POU class 2 associating factor 1; B cells |
| CCL19 | *Chemokine (C-C motif), ligand 19 |
| C3 | *Complement component 3 |
| SELL | *L-selectin |
| COMMUNITY 2 | |
| SOSTDC1 | *Sclerostin domain containing 1; enhances Wnt/inhibits TGFβ |
| CD179B | *IGLL1, preB cell receptor |
| SLC1A1 | Solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
| SELE | *E-selectin |
| IL19 | *Interleukin 19 |
| SPP1 | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein 1) |
| AADACL2 | *Arylacetamide deacetylase-like 2 |
| IGLL1 | *Immunoglobulin lambda-like polypeptide 1 precursor, isoform 7 |
| IGLL1 | *Immunoglobulin lambda-like polypeptide 1-like, isoform 4 |
| SBSPON | *Somatomedin B and thrombospondin, type 1 domain containing |
| LGALS4 | Lectin, galactoside-binding, soluble, 4 |
| ADAMTSL3 | A Disintegrin-Like And Metalloprotease Domain With Thrombospondin Type I –like 3 (LOC714346) |
| LINC00592 | Long intergenic non-protein coding RNA 592 |
| KRT222 | Ketatin 222 |
| IL7R | *Interleukin 7 receptor |
| XDH | Xanthine dehydrogenase |
| EAF2 | ELL (elongation factor RNA polymerase II) associated factor 2 |
| IGF2BP2 | Insulin-like growth factor 2 mRNA binding protein 2 |
| H19 | H19, imprinted maternally expressed transcript (non-protein coding) |
| NDRG4 | N-Myc Downstream-Regulated Gene 4 (LOC712742) |
| COMMUNITY 3 | |
| AGR3 | Similar to breast cancer membrane protein 11 |
| IL18 | *Interleukin 18 |
| SEC16B | SEC16 homolog B (similar to regucalcin gene promotor region related protein) |
| SLC28A3 | Solute carrier family 28, member 3 |
| ASB5 | Ankyrin repeat and SOCS box-containing 5 |
| SERPINB12 | Serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| CPA4 | Carboxypeptidase A4 |
| OLFM4 | *Olfactomedin 4 |
| KLHDC8A | Kelch domain containing 8A |
| ASPRV1 | Aspartic peptidase, retroviral-like 1 |
| NSG1 | Neuron specific gene family member 1 (D4S234E) |
| IL1F9 (IL-36g) | *Interleukin 1 family, member 9 |
| SPRR2F | Small proline rich protein 2F (LOC717894) |
| UPP1 | Uridine phosphorylase 1 |
| COMMUNITY 4 | |
| CXCL6 | *Chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) |
| FNDC1 | Fibronectin type III domain containing 1 |
| LAMB4 | Laminin, beta 4 |
| SNCAIP | Synuclein, alpha interacting protein |
| FAM30A (KIAA0125) | Family with sequence similarity 30, member A |
| BBOX1 | Gamma-butyrobetaine hydroxylase 1 |
| PPTC7 | *T-cell activation protein phosphatase |
| LYPD6 | LY6/PLAUR domain containing 6 |
| LCE2D | Late cornified envelope 2D (LOC100423831) |
| HS3ST3B1 | Heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
| ZNF385B | Zinc finger protein 385B-like |
| STK32A | Serine/threonine kinase 32A |
| COMMUNITY 5 | |
| SAA1 | *Serum amyloid A1 |
| PTN | *Pleiotrophin |
| LINC00302 | Long-intergenic non-protein coding RNA 302 |
| MAP7D2 | MAP7 domain containing 2 |
| HLA-DOB | *Major histocompatibility complex, class II, DO beta |
| TNIP3 | *Tumor necrosis factor alpha induced protein 3 (TNFAIP3) interacting protein 3 |
| ILK | *Epithelial Integrin-linked kinase |
| AMY2A | Amylase, alpha 2A (pancreatic) |
| FABP3 | Fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| CXCL14 | *Chemokine (C-X-C motif) ligand 14 |
| COMMUNITY 6 | |
| CHI3L2 | *Chitinase3-like 2 |
| MST1R | *Macrophage stimulating 1 receptor (LOC100423330) |
| SERPINA12 | Serpin peptidase inhibitor, clade A (α1 antiproteinase) |
| SOD2 | *Manganese, superoxide dismutase |
| LCE1A | Late cornified envelope 1A (LOC713600) |
| FLG2 | Filaggrin family member 2 |
| LCE2D | Late cornified envelope 2D (LOC100423831) |
| PRR9 | Proline rich 9 |
| COMMUNITY 7 | |
| KCNA3 | Potassium voltage-gated channel, shaker-related subfamily, member 3 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (LOC709988) |
| FAM65B | Family with sequence similarity 65, member B (LOC715354) |
| CYTIP | Cytohesin 1 interacting protein |
Figure 2.
Average expression profiles of the genes across the age groups of healthy gingival tissues. Points denote mean expression for each group for the genes in each Community.
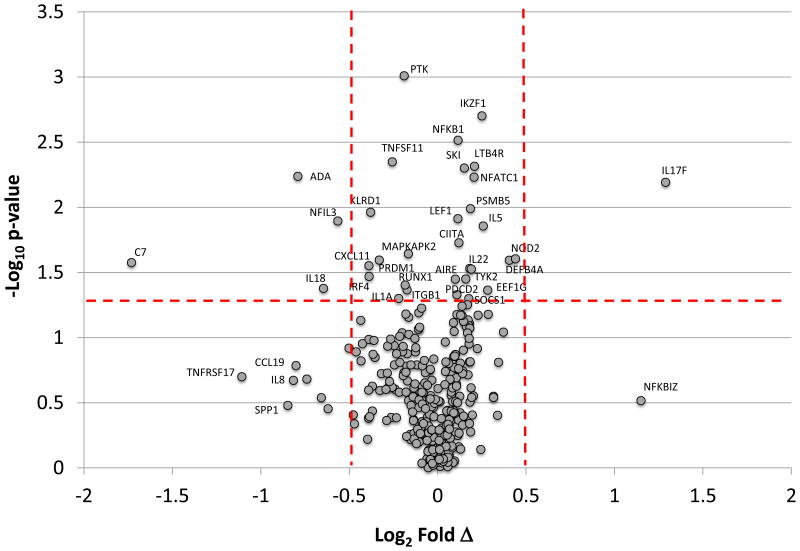
Based upon the features of the immune system network of differentially expressed genes in Communities 1, 2, and 5, we explored an array of 511 genes reflecting host innate immune, inflammatory, and adaptive immune responses (target set derived from Human Immunology Kit, NanoString Technologies; http://www.nanostring.com/media/pdf/PDS_nCounter_Human_Immunology.pdf). The results of this targeted gene identification in Figure 3 are displayed in a Volcano Plot that identifies the immune system genes that were differentially expressed in gingival tissues from young animals compared to the other age groups.
Figure 3.
Volcano plot identifying gene expression profiles between young tissues and all other combined age groups based upon p-value and fold expression. The red horizontal dashed line denotes p-value <0.05 and the vertical dashed lines denote differences in expression between young and other age groups at ±1.4 fold (log2 = +0.5).
Table 2 is a summary of the differentially expressed and aging correlated immune system genes in the healthy gingival tissues. From this analysis we identified 97 genes that were lower in the young [under-expressed and/or significantly positively (p<0.05) correlated] and 26 genes that were higher in the young gingival tissues [over-expressed and/or negatively correlated]. Some striking observations can be discerned from this catalogues of changes. First, evaluation of the cytokine/chemokine differences support a more anti-inflammatory milieu in the gingival tissues of the young animals. This is evidenced by elevated expression of anti-inflammatory cytokines/receptors such as IL22, IL17F, IL5, and TGFB1, with decreased expression of a range of pro-inflammatory cytokines/receptors (ie. IL18, IL1A, IL6, TNFSF13B, CCR5,). Chemokines related to inflammatory and more tissue destructive potential (eg. CXCL11, CCL5, CXCL13, CCL19) were all decreased in expression in the young tissues. Decreased transcription factor gene expression was identified for AIRE, CIITA, NFATC1, NFKB1, SKI, SOCS1, and TP53, in the young tissues.
Table 2.
Genes that were significantly over- or under-expressed in gingival tissues between young and other age groups of animals, and those that were significantly positively correlated (decreased in young) or negatively correlated (increased in young) with aging.
| Gene | Over | Under | Positive | Negative |
|---|---|---|---|---|
| Cytokines/Chemokines | ||||
| CCL19 | 0.6140 | |||
| CCL20 | 0.4322 | |||
| CCL3 | 0.4597 | |||
| CCL5 | 0.5583 | |||
| CCL8 | 0.5277 | |||
| CXCL11 | 0.020 | 0.5550 | ||
| CXCL13 | 0.5584 | |||
| IL16 | 0.5861 | |||
| IL17F | 0.007 | |||
| IL18 | 0.032 | |||
| IL1A | 0.040 | 0.6732 | ||
| IL1B | 0.033 | |||
| IL1RN | 0.4761 | |||
| IL22 | 0.033 | |||
| IL5 | 0.013 | |||
| IL6 | 0.6274 | |||
| IL7 | 0.5367 | |||
| PPBP | 0.5351 | |||
| TGFB1 | -0.4540 | |||
| TNFSF13B | 0.5694 | |||
| Transcription | ||||
| AIRE | 0.035 | -0.6258 | ||
| CIITA | 0.018 | |||
| ETS1 | 0.4680 | |||
| GFI1 | 0.6021 | |||
| IKBKB | 0.6028 | |||
| IKZF1 | 0.5819 | |||
| IRF1 | 0.4252 | |||
| IRF4 | 0.033 | 0.6231 | ||
| IRF5 | 0.5550 | |||
| LEF1 | 0.6124 | |||
| NFATC1 | 0.005 | -0.4607 | ||
| NFIL3 | 0.012 | |||
| NFKB1 | 0.003 | -0.4359 | ||
| RELA | 0.4361 | |||
| RUNX1 | 0.039 | |||
| SKI | 0.004 | |||
| SOCS1 | -0.4269 | |||
| STAT2 | 0.049 | |||
| STAT4 | 0.4234 | |||
| TP53 | -0.4908 | |||
| Receptors | ||||
| CCR5 | 0.4600 | |||
| CD14 | 0.4853 | |||
| CD164 | 0.5668 | |||
| CD2 | 0.6286 | |||
| CD27 | 0.5856 | |||
| CD44 | -0.5563 | |||
| CD46 | 0.4300 | |||
| CD48 | 0.6025 | |||
| CD53 | 0.6063 | |||
| CD81 | -0.5143 | |||
| CD82 | 0.4877 | |||
| CD86 | 0.5423 | |||
| CD9 | 0.4258 | |||
| CLEC4E | 0.5048 | |||
| CSF2RB | 0.5327 | |||
| CSF3R | 0.4318 | |||
| CTLA4 | 0.4918 | |||
| CXCR4 | 0.4365 | |||
| ICOS | 0.5962 | |||
| IFNAR2 | 0.4696 | |||
| IL1R2 | 0.4802 | |||
| IL2RG | 0.5369 | |||
| IL4R | 0.043 | |||
| KLRD1 | 0.010 | 0.6280 | ||
| LTB4R | 0.004 | |||
| LTBR | 0.5662 | |||
| LY96 | 0.6142 | |||
| MASP2 | 0.4263 | |||
| NOD2 | 0.0248 | -0.4654 | ||
| PRDM1 | 0.5863 | |||
| PSMB5 | 0.010 | |||
| PSMD7 | 0.4541 | |||
| PTAFR | 0.7595 | |||
| TLR2 | 0.4581 | |||
| TLR4 | 0.4865 | |||
| TMEM173 | 0.6165 | |||
| TNFRSF17 | 0.5393 | |||
| Signaling | ||||
| MAPKAPK2 | 0.022 | 0.6809 | ||
| PTPN22 | 0.4734 | |||
| SH2D1A | 0.4304 | |||
| SMAD5 | -0.7383 | |||
| SYK | 0.4723 | |||
| TAGAP | 0.6237 | |||
| TRAF3 | 0.4566 | |||
| TYK2 | 0.029 | |||
| ZAP70 | 0.6092 | |||
| Cell Communication | ||||
| ICAM1 | 0.4560 | |||
| ICAM2 | 0.5432 | |||
| ICAM3 | 0.5057 | |||
| ITGA6 | 0.4463 | |||
| ITGB1 | 0.042 | |||
| ITGB2 | 0.5712 | |||
| NCAM1 | -0.6141 | |||
| PDGFB | -0.5024 | |||
| PTK2 | 0.0009 | 0.4439 | ||
| SELE | 0.5393 | |||
| SELL | 0.5939 | |||
| SPP1 | 0.5130 | |||
| TNFAIP6 | 0.4670 | |||
| Cell Development | ||||
| ADA | 0.003 | 0.6407 | ||
| BATF | 0.7221 | |||
| CSF1 | 0.4530 | |||
| EEF1G | 0.036 | -0.5507 | ||
| G6PD | 0.4359 | |||
| HFE | 0.4957 | |||
| LCP2 | 0.5376 | |||
| MME | 0.4448 | |||
| MS4A1 | 0.4275 | |||
| OAZ1 | 0.6009 | |||
| TNFSF11 | 0.004 | 0.6345 | ||
| Complement | ||||
| C1QA | 0.5234 | |||
| C1S | 0.4498 | |||
| C7 | 0.026 | 0.7253 | ||
| Apoptosis | ||||
| CASP10 | 0.4783 | |||
| CLU | 0.4359 | |||
| PDCD1LG2 | 0.4224 | |||
| PDCD2 | 0.050 | -0.4793 | ||
| TNFSF10 | -0.4293 | |||
| TNFSF15 | -0.4691 | |||
| Antimicrobial | ||||
| CAMP | 0.4756 | |||
| CTSC | 0.5175 | |||
| CYBB | 0.4818 | |||
| DEFB4A | 0.025 | |||
| IFIT2 | -0.5051 | |||
| IFNB1 | 0.5326 |
NOD2, a pattern recognition receptor for intracellular infections and linked to proteasome function, levels were increased in young gingival tissues. Proteasome molecules, PSMB5 (proteasome subunit, beta type, 5), a catalytic subunit that is not present in the immunoproteasome and is replaced by catalytic subunit PSMB8 was also increased in the young healthy tissues. Generally genes related to intracellular signaling molecules were decreased in the young tissues, except SMAD5 (SMAD family member 5), involved in TGFβ signaling pathway and TYK 2 (component of both the type I and type III interferon signaling pathways) were elevated in young tissues. Similarly cell communication molecules were generally decreased in young tissues with only NCAM1 and PDGFB decreasing from young to aged tissues. Gene expression of molecules associated with cell development were decreased in young tissues except for EEF1G (eukaryotic translation elongation factor 1 gamma), which is responsible for the enzymatic delivery of aminoacyl tRNAs to the ribosome. All of the complement components that were differentially expressed in the healthy tissues were increased with aging. Changes in apoptosis related genes demonstrated that both PDCD2 (programmed cell death 2), encoding a nuclear protein that responds to BCL6 in regulating apoptosis, TNFSF10 (tumor necrosis factor ligand superfamily, member 10, TRAIL) that preferentially induces apoptosis in transformed and tumor cells, and TNFSF15 that acts as an autocrine factor to promote activation of caspases inducing apoptosis (particularly in endothelial cells) were elevated in tissues from the young animals. Finally, a range of antimicrobial peptides were altered in the gingival tissues, with both DEFB4A (human beta defensin 2) and IFIT2 (interferon-induced protein with tetratricopeptide repeats 2) were increased in young gingival tissues compared to the other age groups.
Table 3 provides an evaluation comparing the differences in gene expression using the microarray to those obtained from a set of genes analyzed using qPCR. The results demonstrate that the expression profiles exhibited identical changes indirection of expression with some variation in the absolute magnitude of difference in the young gingival tissues using these two independent assessments.
Table 3.
Comparison of gene expression profiles using qPCR and microarray analyses. Values represent fold-difference comparing Young Healthy to Adult Healthy tissue message levels assigned a value of 1.0.
| Gene ID | Fold Difference |
|---|---|
| PSMB5 | |
| qPCR | 1.50 ± 0.15 |
| GeneChip | 1.38 ± 0.11 |
| IL1A | |
| qPCR | -1.69 ± 0.11 |
| GeneChip | -2.36 ± 0.08 |
| IL22 | |
| qPCR | 11.94± 5.53 |
| GeneChip | 3.59 ± 0.33 |
| IL17F | |
| qPCR | 20.30 ± 12.72 |
| GeneChip | 2.43 ± 1.17 |
| TNFSRSF17 | |
| qPCR | -1.88 ± 2.04 |
| GeneChip | -2.15 ± 0.14 |
| CXCR4 | |
| qPCR | 2.01 ± 0.38 |
| GeneChip | 1.38 ± 0.12 |
| SAA1 | |
| qPCR | 3.56 ± 0.21 |
| GeneChip | 1.81 ± 0.16 |
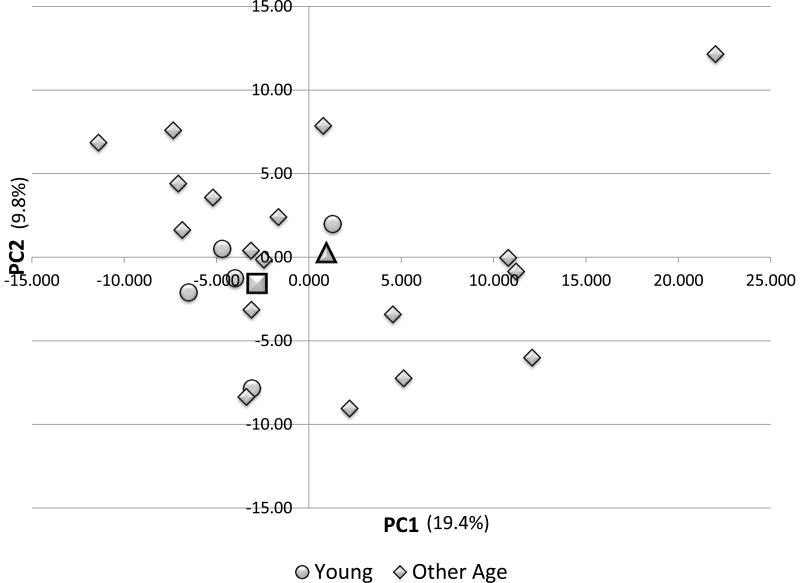
Figure 4 provides the results of a Principal Components Analysis of the immune system genes in comparing the patterns of gene expression in healthy gingival tissues from the young animals versus the other age groups. The graph suggests a grouping of the young animals based upon this composite metagene; however, the animals from the other age groups tended to be spread across the various quadrants of the plot, suggesting many similarities in gene expression. This is evidenced in that only 29% of the variation in expression is related to the age distribution for expression in healthy gingival tissues. The crucial gene profile determinants of the PC1 and PC2 were evaluated and are displayed in Table 4. The loading values of 69/319 genes (PC1) showed an elevated correlation in distributing the young animals compared to other age groups. Similarly, 41/319 genes primarily contributed to the PC2 variation. Of the genes in PC1 59/69 were highly positively correlated with substantial representation of cyotkines/chemokines, transcription factors, receptors, and cell communication molecules. In contrast, PC2 genes were primarily negatively correlated (23/41) displaying a different set of cytokines/chemokines and receptors.
Figure 4.
Principal Components Analysis of immunology gene set for young and other age groups of animals. Each point denotes the PC1 and PC2 metagene position for an animal. The square denotes the mean PC values for the young group and the triangle signifies the mean for the other age groups.s
Table 4.
Gene expression contribution to Principal Component separation of gingival tissue profiles in young versus other age groups of animals. The values denote loading values for the PC analysis and are listed from highest positive to lowest negative value in each category. Data are presented on genes derived from all 319 evaluated with positive values ≥0.6 and negative values ≤-0.4
| Gene ID | PC1 | Gene ID | PC2 |
|---|---|---|---|
| Cytokines/Chemokines | Cytokines/Chemokines | ||
| CCL19 | 0.8780 | CXCL12 | 0.7486 |
| IL16 | 0.8559 | IL1B | 0.7296 |
| TNFSF13B | 0.8558 | IL2 | 0.6187 |
| IL6 | 0.8216 | IL28B | 0.6058 |
| CXCL13 | 0.7823 | IL1A | -0.4395 |
| CCL5 | 0.6851 | CXCL11 | -0.5044 |
| TNFSF8 | 0.6403 | IL1RN | -0.5748 |
| IL7 | 0.6270 | IL7 | -0.6532 |
| IL12B | 0.6265 | Transcription Factors | |
| CXCL10 | 0.6053 | IRF4 | 0.6059 |
| Transcription Factors | TBP | 0.6573 | |
| IRF8 | 0.9001 | NFKBIZ | -0.4875 |
| IKZF1 | 0.8905 | Receptors | |
| GFI1 | 0.7589 | CD58 | 0.8584 |
| IRF1 | 0.7531 | IL11RA | 0.7479 |
| STAT4 | 0.6977 | IL2RA | 0.7279 |
| LEF1 | 0.6913 | FCGRT | 0.6945 |
| STAT2 | 0.6376 | FCER1A | 0.6427 |
| JAK2 | -0.5575 | RORC | 0.6294 |
| Receptors | TGFBR2 | 0.6291 | |
| CD53 | 0.9094 | TGFBR1 | 0.6094 |
| CTLA4 | 0.9032 | IL1R1 | -0.4092 |
| IL2RG | 0.8815 | B2M | -0.4193 |
| LY96 | 0.8763 | CD82 | -0.4461 |
| CD74 | 0.8747 | PSMB10 | -0.4668 |
| ICOS | 0.8617 | TNFRSF1B | -0.4859 |
| TNFRSF17 | 0.8486 | PSMB5 | -0.6201 |
| CCR7 | 0.8464 | PSMD7 | -0.6984 |
| CD2 | 0.8218 | TLR7 | -0.7420 |
| IFNAR2 | 0.8183 | Signaling | |
| CD27 | 0.8135 | IRAK1 | 0.6093 |
| CXCR4 | 0.7999 | IL1RAP3 | 0.6052 |
| TMEM173 | 0.7605 | SMAD3 | -0.5263 |
| KLRD1 | 0.7374 | MAP4K4 | -0.7219 |
| CLEC4E | 0.7113 | UBE2L3 | -0.5158 |
| CSF1R | 0.6909 | Cell Communication | |
| TLR4 | 0.6841 | ICAM3 | -0.6066 |
| CXCR3 | 0.6469 | Cell Development | |
| CD209L2 | 0.6260 | FYN | 0.6619 |
| CD83 | 0.6059 | KIT | 0.6189 |
| PSMB5 | -0.4194 | OAZ1 | -0.4615 |
| CD44 | -0.5552 | Apoptosis | |
| CD36 | -0.6934 | CDKN1A | -0.4657 |
| Signaling | BCL3 | -0.4719 | |
| MAPKAPK2 | 0.7262 | PDCD2 | -0.5063 |
| PTPN22 | 0.7410 | Antimicrobial | |
| ARHGDIB | 0.6972 | CTSG | -0.4105 |
| TAGAP | 0.8666 | DEFB4A | -0.4847 |
| SMAD5 | -0.4312 | ||
| Cell Communication | |||
| ICAM2 | 0.8101 | ||
| ITBG2 | 0.8869 | ||
| SPP1 | 0.7957 | ||
| ENTPD1 | 0.7123 | ||
| SELE | 0.6757 | ||
| SELL | 0.6010 | ||
| ITGA6 | -0.4206 | ||
| NCAM1 | -0.6459 | ||
| Cell Development | |||
| LCP2 | 0.8750 | ||
| MS4A1 | 0.7718 | ||
| BTK | 0.8517 | ||
| BATF | 0.6428 | ||
| EEF1G | -0.4622 | ||
| Complement | |||
| C1S | 0.7640 | ||
| C1QA | 0.8634 | ||
| SERPING1 | 0.7395 | ||
| C5 | -0.6314 | ||
| Apoptosis | |||
| BCL3 | 0.6031 | ||
| PDCD1LG2 | 0.6668 | ||
| PCDC2 | -0.4150 | ||
| Antimicrobial | |||
| CYBB | 0.9140 | ||
| IFITM1 | 0.8173 | ||
| IFI35 | 0.7135 |
Discussion
Periodontal disease manifests as a persistent inflammatory response of the local tissues that has been suggested to reflect changes in the characteristics of the subgingival microbial ecology at diseased sites (32-34). Additional findings in studies of periodontitis report the increased frequency and severity of disease with aging (35-37), leading to the consideration that periodontitis is a disease of aging related to altered immune functions that occur with increasing prevalence coincident with decades of life in the general population (38), or potentially a reflection of changing oral environments that select for a microbial ecology with greater pathogenic potential.
Of additional interest is the other side of the aging pendulum in which it has been described that gingivitis is nearly universal in children and adolescents, and generally responds well to improved oral hygiene and periodic professional care (20). Gingivitis is the most common and prevalent disease form of the periodontium among children and adolescents with the incidence and severity increasing from childhood to adolescence, reaching a peak prevalence of 80% at 11– 13 years of age (39). However, beyond the small percentage (eg. about 0.5-1%) of children/adolescents that express a rather unique form of periodontitis that has been termed localized/generalized juvenile periodontitis, early onset periodontitis, or localized/generalized aggressive periodontitis (40, 41), the destructive form of this chronic inflammation of the gingiva does not generally occur in young individuals. This observation contrasts with the age dependent inflammatory reaction of the gingival tissues that has been related to changes in the qualitative and quantitative microbiome of the dental biofilms, the characteristics of immune responses, hormonal changes, and morphological differences in the periodontium that have been shown to increase the frequency of transition from the reversible inflammation of gingivitis to the irreversible tissue destruction of periodontitis in adults. Of particular interest is that beyond the clinical features of inflammation in the gingiva of children, available data demonstrate the existence of oral bacterial species identified as critical to pathogenic biofilms for periodontitis in the supra- and subgingival plaque of many children (42, 43). Thus, the microbial stimuli for triggering periodontitis are in the ecology, and the individuals respond to accumulation of these bacteria with gingival inflammation, but uniformly do not progress to periodontitis. However, 3 decades later, a large percentage of these children/adolescents will develop periodontitis based upon current epidemiologic evidence (44, 45), and apparently in the absence of extrinsic acquisition of new oral pathogens (46, 47). One interpretation of these observations and the hypothesis to be tested in this study is that the localized response of the gingival tissues in children to the microbial challenge is molecularly different than those responses in adults and results in a non-destructive management of the bacterial population.
The results of this study demonstrated a range of genes related to innate immunity, inflammation, and adaptive immunity were expressed in gingival tissues of the young nonhuman primates. Using a network analysis strategy on a set of 159 genes from the total microarray analysis, we identified distinctive patterns of communities of networked genes that were differentially expressed in young gingival tissues. These communities demonstrated a high representation of components of immunologic pathways that were expressed in healthy young gingival tissues compared to healthy tissues from other age groups. We then targeted, more specifically a framework of a set of about 511 gene probes that are linked to innate immune, inflammatory, and adaptive immune responses. From this we identified an array of approximately 123 that demonstrated differential expression in young healthy tissues and/or showed a significant correlation related to healthy gingival tissues across the lifespan.
Generally, these gene profiles were identified for cytokines/chemokines, transcription factors, receptors, signaling molecules, cell communication factors, cell development molecules, complement components, apoptosis pathway molecules and antimicrobial peptides. Of the 123 genes, 79% were decreased in young tissues compared with healthy gingiva from other age groups. A high frequency of expression of immune related genes was related to transcription factors where ∼35% of the genes that were differentially expressed were increased in the young tissues. These included AIRE (role in immunity by regulating the expression of autoantigens and negative selection of autoreactive T-cells), CIITA (essential for transcriptional activity of the HLA class II promoter), NFATC1 (plays a role in the inducible expression of cytokine genes in T-cells, especially in the induction of the IL-2 or IL-4 gene transcription), NFKB1 (pleiotropic transcription factor present in almost all cell types responds to a vast array of stimuli for many biological processes, including inflammation, immunity, and apoptosis), SKI (a repressor of TGFβ signaling), SOCS1 (part of a classical negative feedback system that regulates cytokine signal transduction), and TP53 (regulates expression of target genes related to cell cycle arrest, apoptosis, and senescence). While the exact relationship among this array of transcription factors is not obvious, it does appear that they relate to T cell regulation of responses, control of anti- and pro-inflammatory responses, and can contribute to increased apoptosis, which we have noted previously in young tissues (16, 48). This was also observed with multiple pro-apoptotic genes that were elevated in the young tissues (PDCD2, TNFSF10, TNFSF15).
A limited array of cytokines/chemokines were at elevated levels in the young gingival tissues. IL17F is expressed by activated T cells, and stimulates the production of cytokines, including IL-6, IL-8, and GM-CSF. IL-5 is anti-inflammatory cytokine synthesized by Th2 immune cells and acts as a growth and differentiation factor for B cells and eosinophils. IL-22 is a member of the IL-10 family of anti-inflammatory cytokines. It plays a role in coordinating adaptive and innate immune responses and primary targets are non-hematopoietic cells including epithelial cells. TGFB1 (transforming growth factor beta 1) is a member of the TGF-beta family of cytokines that regulate proliferation, differentiation, adhesion, and migration of many cell types. It is a potent stimulator of osteoblast functions and considered an anti-inflammatory cytokine. Thus, it appears that some of these unique features of the responses in young gingival tissues are also related to controlling T cell functions and creating a more prominent anti-inflammatory regulatory microenvironment.
These findings suggest that novel gene patterns could provide some guidance regarding the apparent “resistance” of the periodontium in the young in response to a microbial challenge eliciting an inflammatory response but lacking progression to destructive periodontitis, even in the presence of this clinical/molecular gingival inflammation. However, we still have little understanding of how the acquisition of the oral microbiome contributes to the development and maturation of the immune response repertoire in gingival tissues (49). Knowledge of this process will potentially help to clarify the early tissue alterations that could translate into longer-term risk for disease, as well as focusing efforts on approaches to effectively modulate the microbial acquisition by children to improve long term oral health.
Acknowledgments
This work was supported by National Institute of Health grants P20GM103538 and UL1TR000117. We express our gratitude to the Caribbean Primate Research Center (CPRC) supported by grant P40RR03640, and the Microarray Core of University Kentucky for their invaluable technical assistance. We thank M. Kirakodu for data management support. The authors have no financial conflicts related to these studies.
Footnotes
The authors have no financial conflict with these studies.
References
- 1.Gonzalez O, Tobia C, Ebersole J, Novak MJ. Caloric restriction and chronic inflammatory diseases. Oral Dis. 2012;18:16–31. doi: 10.1111/j.1601-0825.2011.01830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oz HS, Puleo DA. Animal models for periodontal disease. Journal of biomedicine & biotechnology. 2011;2011:754857. doi: 10.1155/2011/754857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Struillou X, Boutigny H, Soueidan A, Layrolle P. Experimental animal models in periodontology: a review. Open Dent J. 2010;4:37–47. doi: 10.2174/1874210601004010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Persson GR, Engel LD, Whitney CW, et al. Macaca fascicularis as a model in which to assess the safety and efficacy of a vaccine for periodontitis. Oral Microbiol Immunol. 1994;9:104–111. doi: 10.1111/j.1399-302x.1994.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 5.Schou S, Holmstrup P, Kornman KS. Non-human primates used in studies of periodontal disease pathogenesis: a review of the literature. J Periodontol. 1993;64:497–508. doi: 10.1902/jop.1993.64.6.497. [DOI] [PubMed] [Google Scholar]
- 6.Madden TE, Caton JG. Animal models for periodontal disease. Methods in enzymology. 1994;235:106–119. doi: 10.1016/0076-6879(94)35135-x. [DOI] [PubMed] [Google Scholar]
- 7.Graves DT, Kang J, Andriankaja O, Wada K, Rossa C., Jr Animal models to study host-bacteria interactions involved in periodontitis. Frontiers of oral biology. 2012;15:117–132. doi: 10.1159/000329675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahn J, Yang L, Paster BJ, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One. 2011;6:e22788. doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paster BJ, Dewhirst FE. Molecular microbial diagnosis. Periodontol 2000. 2009;51:38–44. doi: 10.1111/j.1600-0757.2009.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18:109–120. doi: 10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]
- 12.Xin B, Luo A, Qin J, et al. Microbial diversity in the oral cavity of healthy Chinese Han children. Oral Dis. 2012 doi: 10.1111/odi.12018. [DOI] [PubMed] [Google Scholar]
- 13.Diaz PI. Microbial diversity and interactions in subgingival biofilm communities. Frontiers of oral biology. 2012;15:17–40. doi: 10.1159/000329669. [DOI] [PubMed] [Google Scholar]
- 14.Ebersole JL, Cappelli D, Mathys EC, et al. Periodontitis in humans and non-human primates: oral-systemic linkage inducing acute phase proteins. Annals of periodontology / the American Academy of Periodontology. 2002;7:102–111. doi: 10.1902/annals.2002.7.1.102. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez O, Novak MJ, Orraca L, Martinez-Gonzalez J, Stromberg AJ, Ebersole JL. Apopotosis gene expression in healthy and oral mucosal tissues with aging. Apoptosis. 2013 doi: 10.1007/s10495-013-0806-x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez OA, Stromberg AJ, Huggins PM, Gonzalez-Martinez J, Novak MJ, Ebersole JL. Apoptotic genes are differentially expressed in aged gingival tissue. J Dent Res. 2011;90:880–886. doi: 10.1177/0022034511403744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez OA, Kirakodu S, Novak MJ, et al. Effects of aging in the expression o fNOD-like receptors adn inflammasome-related genes in the oral mucosa. Microbes and Infection. 2014 doi: 10.1111/omi.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez OA, Novak MJ, Kirakodu S, et al. Comparative analysis of gingival tissue antigen presentation pathways in ageing and periodontitis. J Clin Periodontol. 2014;41:327–339. doi: 10.1111/jcpe.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bimstein E, Ram D, Irshied J, Naor R, Sela MN. Periodontal diseases, caries, and microbial composition of the subgingival plaque in children: a longitudinal study. ASDC J Dent Child. 2002;69:133–137. 123. [PubMed] [Google Scholar]
- 20.Bimstein E, Huja PE, Ebersole JL. The potential lifespan impact of gingivitis and periodontitis in children. The Journal of clinical pediatric dentistry. 2013;38:95–99. doi: 10.17796/jcpd.38.2.j525742137780336. [DOI] [PubMed] [Google Scholar]
- 21.Ebersole JL, Steffen MJ, Gonzalez-Martinez J, Novak MJ. Effects of age and oral disease on systemic inflammatory and immune parameters in nonhuman primates. Clin Vaccine Immunol. 2008;15:1067–1075. doi: 10.1128/CVI.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meka A, Bakthavatchalu V, Sathishkumar S, et al. Porphyromonas gingivalis infection-induced tissue and bone transcriptional profiles. Mol Oral Microbiol. 25:61–74. doi: 10.1111/j.2041-1014.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirakodu SS, Govindaswami M, Novak MJ, Ebersole JL, Novak KF. Optimizing qPCR for the Quantification of Periodontal Pathogens in a Complex Plaque Biofilm. Open Dent J. 2008;2:49–55. doi: 10.2174/1874210600802010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Butte AJ, Kohane IS. Mutual information relevance networks: functional genomic clustering using pairwise entropy measurements. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2000:418–429. doi: 10.1142/9789814447331_0040. [DOI] [PubMed] [Google Scholar]
- 26.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008;10:P10008. [Google Scholar]
- 27.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks International AAAI Conferecne on Weblogs and Social Media. 2009 [Google Scholar]
- 28.Zhu N, Ramirez LM, Lee RL, Magnuson NS, Bishop GA, Gold MR. CD40 signaling in B cells regulates the expression of the Pim-1 kinase via the NF-kappa B pathway. J Immunol. 2002;168:744–754. doi: 10.4049/jimmunol.168.2.744. [DOI] [PubMed] [Google Scholar]
- 29.Henley KD, Gooding KA, Economides AN, Gannon M. Inactivation of the dual Bmp/Wnt inhibitor Sostdc1 enhances pancreatic islet function. American journal of physiology Endocrinology and metabolism. 2012;303:E752–761. doi: 10.1152/ajpendo.00531.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiyokawa N, Sekino T, Matsui T, et al. Diagnostic importance of CD179a/b as markers of precursor B-cell lymphoblastic lymphoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2004;17:423–429. doi: 10.1038/modpathol.3800079. [DOI] [PubMed] [Google Scholar]
- 31.Waly MI, Hornig M, Trivedi M, et al. Prenatal and Postnatal Epigenetic Programming: Implications for GI, Immune, and Neuronal Function in Autism. Autism research and treatment. 2012;2012:190930. doi: 10.1155/2012/190930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji S, Choi Y. Innate immune response to oral bacteria and the immune evasive characteristics of periodontal pathogens. J Periodontal Implant Sci. 2013;43:3–11. doi: 10.5051/jpis.2013.43.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Redlich K, Smolen JS. Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat Rev Drug Discov. 2012;11:234–250. doi: 10.1038/nrd3669. [DOI] [PubMed] [Google Scholar]
- 34.Kinane DF, Bartold PM. Clinical relevance of the host responses of periodontitis. Periodontol 2000. 2007;43:278–293. doi: 10.1111/j.1600-0757.2006.00169.x. [DOI] [PubMed] [Google Scholar]
- 35.Hajishengallis G. Too old to fight? Aging and its toll on innate immunity Mol Oral Microbiol. 2010;25:25–37. doi: 10.1111/j.2041-1014.2009.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huttner EA, Machado DC, de Oliveira RB, Antunes AG, Hebling E. Effects of human aging on periodontal tissues. Special care in dentistry : official publication of the American Association of Hospital Dentists, the Academy of Dentistry for the Handicapped, and the American Society for Geriatric Dentistry. 2009;29:149–155. doi: 10.1111/j.1754-4505.2009.00082.x. [DOI] [PubMed] [Google Scholar]
- 37.Gonsalves WC, Wrightson AS, Henry RG. Common oral conditions in older persons. American family physician. 2008;78:845–852. [PubMed] [Google Scholar]
- 38.Chung HY, Lee EK, Choi YJ, et al. Molecular inflammation as an underlying mechanism of the aging process and age-related diseases. J Dent Res. 2011;90:830–840. doi: 10.1177/0022034510387794. [DOI] [PubMed] [Google Scholar]
- 39.Dibart S. Children, adolescents and periodontal diseases. J Dent. 1997;25:79–89. doi: 10.1016/s0300-5712(96)00019-x. [DOI] [PubMed] [Google Scholar]
- 40.Song HJ. Periodontal considerations for children. Dent Clin North Am. 2013;57:17–37. doi: 10.1016/j.cden.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 41.Armitage GC, Cullinan MP. Comparison of the clinical features of chronic and aggressive periodontitis. Periodontol 2000. 2010;53:12–27. doi: 10.1111/j.1600-0757.2010.00353.x. [DOI] [PubMed] [Google Scholar]
- 42.Kinane DF, Podmore M, Murray MC, Hodge PJ, Ebersole J. Etiopathogenesis of periodontitis in children and adolescents. Periodontol 2000. 2001;26:54–91. doi: 10.1034/j.1600-0757.2001.2260104.x. [DOI] [PubMed] [Google Scholar]
- 43.Bimstein E, Ebersole JL. Serum antibody levels to oral microorganisms in children and young adults with relation to the severity of gingival disease. Pediatric dentistry. 1991;13:267–272. [PubMed] [Google Scholar]
- 44.Eke PI, Thornton-Evans G, Dye B, Genco R. Advances in surveillance of periodontitis: the Centers for Disease Control and prevention periodontal disease surveillance project. J Periodontol. 2012;83:1337–1342. doi: 10.1902/jop.2012.110676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. Cdc Periodontal Disease Surveillance workgroup: James Beck GDRP. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- 46.Genco RJ, Borgnakke WS. Risk factors for periodontal disease. Periodontol 2000. 2013;62:59–94. doi: 10.1111/j.1600-0757.2012.00457.x. [DOI] [PubMed] [Google Scholar]
- 47.Nibali L, Henderson B, Sadiq ST, Donos N. Genetic dysbiosis: the role of microbial insults in chronic inflammatory diseases. Journal of oral microbiology. 2014:6. doi: 10.3402/jom.v6.22962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez OA, John Novak M, Kirakodu S, et al. Effects of aging on apoptosis gene expression in oral mucosal tissues. Apoptosis. 2013;18:249–259. doi: 10.1007/s10495-013-0806-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebersole JL, Holt SC, Delaney JE. Acquisition of Oral Microbes and Associated Systemic Responses of Newborn Nonhuman Primates. Clinical and vaccine immunology. 2014;21:21–28. doi: 10.1128/CVI.00291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]