Abstract
Purpose
The purpose of this study is to evaluate the influence of follicle-stimulating hormone receptor (FSHR) Asn680Ser polymorphism on the ovarian response to exogenous follicle-stimulating hormone (FSH) and clinical outcomes in women undergoing controlled ovarian hyperstimulation (COH).
Methods
A database search was conducted to identify the eligible studies that investigated the effect of FSHR Asn680Ser polymorphism on ovarian response and clinical outcomes. A pooled analysis was performed with the odds ratio (OR) or weighted mean difference (WMD) and their respective 95 % confidence interval (CI) by the STATA software with random effects model.
Results
Sixteen cohort studies comprising a total of 4287 subjects were included. The number of retrieved oocytes was significantly fewer in subjects with the SS genotype at position 680, compared to subjects with the NN or NS genotype (WMD = −1.36, 95 % CI = −1.85 to −0.87). Lack of association was detected between the genotypes (SS genotype vs. NN or NS genotype) and clinical outcomes such as exogenous FSH dose (WMD = 98.96 IU, 95 % CI = −22.33 to 220.24), poor response (OR = 1.08, 95 % CI = 0.71–1.64), ovarian hyperstimulation syndrome (OHSS) (OR = 1.58, 95 % CI = 0.41–6.07), and clinical pregnancy rate (OR = 1.10, 95 % CI = 0.86–1.40). However, poor ovarian response and number of retrieved oocytes were significantly influenced by the Asn680Ser polymorphism in the Asian subjects. In addition, no publication bias was detected.
Conclusion
FSHR Asn680Ser polymorphism might be a significant biomarker for predicting the number of retrieved oocytes and poor response, especially in Asian subjects. Other outcomes such as exogenous FSH dose, OHSS, and pregnancy rate were not influenced by FSHR Asn680Ser polymorphism.
Keywords: FSHR, Genetic polymorphism, Ovarian response, Meta-analysis
The prevalence of infertility has significantly increased over the recent decades, affecting about 15 % of all couples at reproductive age [1]. However, this problem was not successfully overcome until the development of in vitro fertilization (IVF) [1]. Today, 2–3 % of all births in developed countries are estimated to be the result of IVF procedures [2]. However, assisted reproduction technique (ART) is a complicated program which requires controlled ovarian hyperstimulation (COH) with exogenous follicle-stimulating hormone (FSH) to achieve maturation of multiple follicles and oocytes. The effectiveness and safety of IVF treatment depend substantially on the ovarian response to exogenous FSH [1]. However, the ovarian response to stimulation with gonadotropin was varied, ranging from poor to high responses [3]. In addition, a standard fixed dose of gonadotropin may not be suitable for all women, and thus selecting a suitable initial dose of gonadotropin plays an important role in determining the outcomes of COH and subsequent IVF. The women with poor response may easily suffer from few or no mature follicle which results in cancellation of IVF procedures. Conversely, women with high response would be at risk of developing potentially life-threatening ovarian hyperstimulation syndrome (OHSS) [4]. In Italy, nearly 4500 cycles were cancelled every year due to abnormal responses to gonadotropin stimulation [5]. Therefore, individualization and optimization of the stimulation protocols were needed to minimize the risk of OHSS while maximizing the probability of live birth. With the rapid development of pharmacogenetics, genetic biomarkers are now considered as a promising approach to improve response rate and minimize adverse events.
FSH and its receptor (FSHR) play a significant role in follicle development and regulation of steroidogenesis within the ovary [6]. The loss-of-function mutation in FSHR gene was found to be associated with ovarian dysfunction [7]. Recently, hundreds of common variants or single-nucleotide polymorphisms (SNPs) of the FSHR gene have been identified. In these variants, two substitutions in exon 10, an asparagine-serine change at position 680 (Asn680Ser) and an alanine-threonine change at position 307 in the amino acid sequence (Ala307Thr), particularly have been proposed to be associated with ovarian dysfunction and alter the effect of COH in women with normal ovarian function [8, 9]. As these two polymorphisms are in near-complete linkage disequilibrium, most previous studies have only focused on the Asn680Ser variant [8]. Most studies evaluating the role of FSHR genetic polymorphisms showed that homozygosity for the serine variant (SS) at position 680 in ovulatory patients was associated with higher baseline FSH levels [10]. In addition to these SNPs, splice variants in FSHR that have been identified in women undergoing ovarian stimulation may also contribute to the variability in ovarian response. However, results from multiple studies are conflicting, and previous meta-analyses have failed to confirm the association between FSHR Asn680Ser polymorphism and the outcomes related to COH [10–29]. A pooled analysis of only four studies, as well as a recent meta-analysis, showed that the 680 SS genotype of FSHR played a role in the ovarian response during stimulation with exogenous gonadotropin [11, 30]. However, our previous meta-analysis concluded that, except for basal FSH levels, none of the COH outcomes in terms of peak estradiol levels, gonadotropin dose, oocytes retrieved, or pregnancy rate was significantly associated with different genotype groups [10]. Therefore, considering the conflicting results from previous meta-analysis and the substantial number of original studies, an updated meta-analysis was necessary to evaluate the role of FSHR Asn680Ser polymorphism in ovarian response and other IVF outcomes in women undergoing COH.
Methods
The meta-analysis was performed according to Meta-analysis Of Observational Studies in Epidemiology (MOOSE) guideline. Formal institutional review board approval was not required because only published data were pooled.
Search strategy
A database search was conducted to identify the relevant studies in PubMed, Cochrane Library, and Web of Science regardless of their language and publication status until April 2015. To investigate the relationship between FSHR Asn680Ser SNP and COH outcomes, the following search strategies were used without any other restrictions: (FSHR OR FSH OR follicle-stimulating hormone OR FSH receptor) AND (polymorphism OR genotype OR genetic OR pharmacogenetics) AND (in-vitro fertilization OR ovulation induction OR IVF OR COH OR controlled ovarian hyperstimulation OR controlled ovarian stimulation OR ICSI). In addition, references of review articles and included trials were manually searched to identify the additional eligible studies.
Study selection
One reviewer initially evaluated articles for eligibility (HT). The selected articles were reevaluated by another reviewer (YY), and the final inclusion decision was based on consensus between the two reviewers. Studies were included if they met the following criteria: (1) patients underwent IVF/ICSI, (2) FSHR genotyping was performed in some or all of the patients, and (3) ovarian response and COH outcomes were presented based on genotypes.
Data extraction and quality assessment
Data from all eligible studies were extracted and summarized independently by two reviewers (HT and YY). The following background information was extracted from the studies: (1) design, (2) region, (3) procedure, (4) cause of infertility, and (5) treatment protocol. In addition, the following outcomes were extracted: (1) total gonadotropin used, (2) number of retrieved oocytes, (3) clinical pregnancy rate, and (4) ovarian response including incidence of poor response and OHSS. If the minimum, median, and maximum values were provided instead of the mean and standard deviation (SD), the mean and SD were estimated by the use of a method described elsewhere [31, 32]. In addition, if needed, two subgroups (such as NN, NS) were combined into a single group (NN + NS) according to a method described previously [31]. Any discrepancy was resolved by consensus. If possible, the original authors were contacted for the more detailed information by e-mail.
The quality of included studies was assessed by two reviewers (HT and WS) independently through a checklist derived from the Strengthening the Reporting of Genetic Association (STREGA) recommendations for reports on genetic association studies [33] and modified according to the quality checklist described elsewhere [34, 35].
Statistical analysis
STATA version 12.0 (Austin, TX, USA) was used to calculate the weighted mean difference (WMD) for continuous data and the odds ratio (OR) for dichotomous data with their 95 % confidence intervals (CIs), respectively. Heterogeneity for each outcome analysis was assessed by the I2 statistic, with I2 ≤ 25 %, 25 % < I2 < 50 %, and I2 ≥ 50 % considered as low, moderate, and high degree of heterogeneity, respectively. Considering the clinical heterogeneity across the included studies, a random effects model, rather than a fixed effect model, was used to pool the data for each outcome. In addition, a meta-regression was carried out to explore the reasons for heterogeneity across all eligible trials and a subgroup analysis was also performed to assess whether the pooled outcomes could vary by patient characteristics (such as region). Finally, publication bias was assessed by using Egger’s or Begg’s test.
Results
Identification of studies and quality assessment
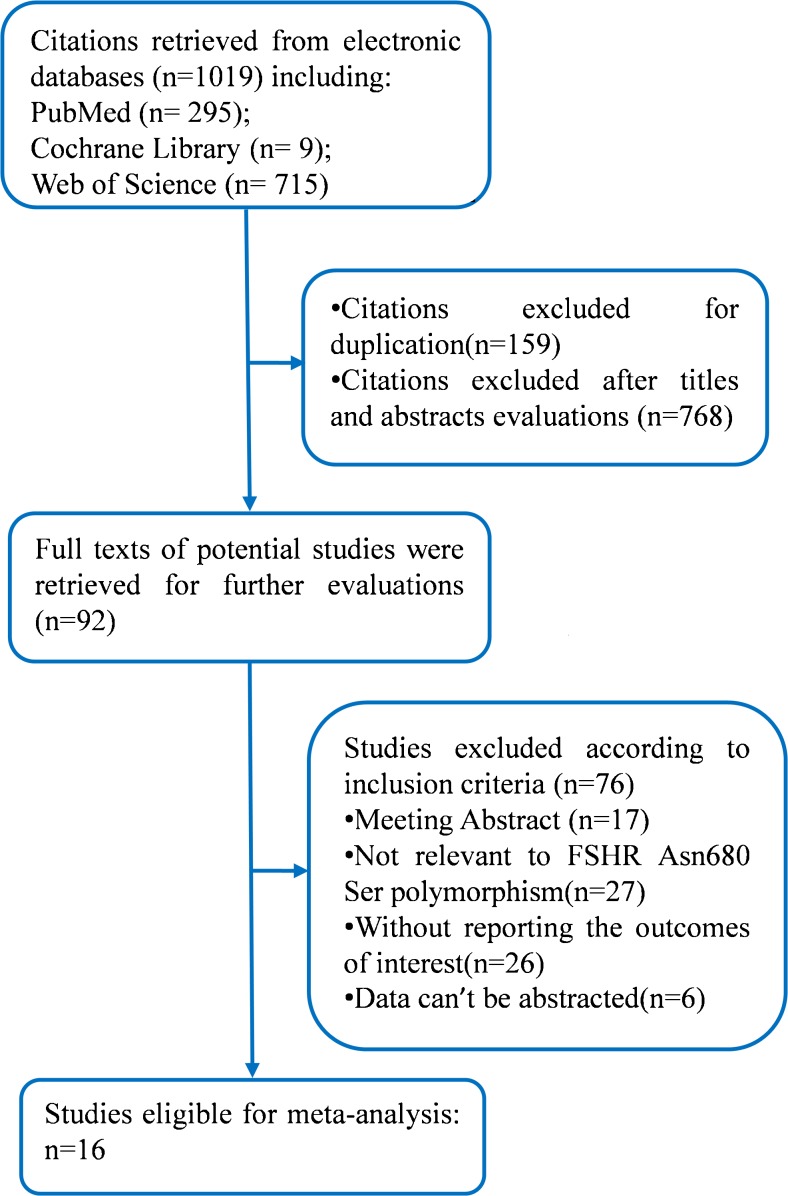
A total of 1019 citations were initially retrieved with our search strategy, in which 1003 citations were carefully excluded. Only 16 studies involving 4287 patients were included in our meta-analysis [12–27]. The process of identification of the eligible studies and the reasons for exclusion were presented in Fig. 1. In addition, Table 1 presented the characteristics of the 16 included studies. All these studies were published between 2000 and 2015, and most of the studies were performed in Europe and Asia. The number of patients involved in each study varied from 20 to 1052. The quality of the included studies was presented in Table 2.
Fig. 1.
Flow chart of the identification of eligible studies
Table 1.
Characteristics of studies included in the meta-analysis
| Study | Design | Region | No. of patients | Age (mean/median) | Procedure | Treatment protocol | Frequency of Asn680Ser (%) | Outcome reported | ||
|---|---|---|---|---|---|---|---|---|---|---|
| NN | NS | SS | ||||||||
| Perez Mayorga 2000 [12] | RC | Germany | 161 | 32.4 | ICSI | GnRH agonist/FSH | 46 | 72 | 43 | ①② |
| Sodu 2002 [13] | RC | Japan | 58 | 31 | IVF | GnRH agonist/FSH | 19 | 28 | 11 | ① |
| De Castro 2003 [14] | RC | Spain | 102 | 33 | IVF or ICSI | FSH | 83 | 19 | ①②③ | |
| Behre 2005 [15]a | PC | Germany | 68 | 33 | IVF or ICSI | GnRH agonist or antagonist/FSH | 44 | 24 | ①②⑤ | |
| Jun 2006 [16] | PC | Korea | 263 | 32.6 | IVF | GnRH agonist or antagonist/FSH | 110 | 120 | 33 | ①②⑤ |
| Klinkert 2006 [17] | RC | Netherlands | 105 | 36.9 | IVF | GnRH agonist/FSH | 40 | 47 | 18 | ①②③⑤ |
| Loutradis 2006 [18] | RC | Greece | 125 | 30.2 | IVF or ICSI | GnRH agonist/FSH | 34 | 49 | 42 | ①②③⑤ |
| Achrekar 2009 [19] | RC | India | 50 | 31 | IVF or ICSI | GnRH agonist/FSH | 21 | 23 | 6 | ①②④ |
| Huang 2010 [20] | RC | China | 136 | 30.33 | IVF or ICSI | GnRH agonist/FSH | 38 | 64 | 34 | ①②⑤ |
| Sheikhha 2011 [21] | RC | Iran | 108 | 30 | IVF | GnRH agonist/FSH | 22 | 71 | 15 | ①②③⑤ |
| Boudjenah 2012 [22] | PC | France | 427 | 31.2 | ICSI | GnRH agonist /FSH | 142 | 191 | 94 | ①③ |
| Genro 2012 [23] | PC | Caucasian | 81 | 35 | IVF | GnRH agonist/FSH | 29 | 40 | 12 | ①②⑤ |
| Mohiyiddeen 2013 [24] | PC | United Kingdom | 421 | 33.5 | IVF | GnRH agonist or antagonist/FSH | 128 | 214 | 79 | ①②③④⑤ |
| Mohiyiddeen 2013 [25] | PC | United Kingdom | 212 | 33.5 | ICSI | GnRH agonist or antagonist/FSH | 72 | 106 | 34 | ①②⑤ |
| Yan 2013 [26] | PC | China | 450 | 32 | IVF | GnRH agonist/FSH | 211 | 197 | 42 | ①② |
| Huang 2015 [27] | PC | China | 1250 | 31 | IVF | GnRH agonist/FSH | 506 | 572 | 172 | ①②③⑤ |
① Total exogenous FSH dose, ② number of retrieved oocytes, ③ incidence of poor response, ④ incidence of OHSS, ⑤ clinical pregnancy rate
RC retrospective cohort, PC prospective cohort, IVF in vitro fertilization, ICSI intracytoplasmic sperm injection, GnRH gonadotropin releasing hormone, FSH follicle-stimulating hormone
aOnly included the group of 120 IU/day
Table 2.
Quality assessment of studies included in the meta-analysis
| Study | Clear statement of objectives and hypothesis | Clear eligibility criteria for study participants | Clear definition of all variables | Clear definition of the outcome | Credible genetic testing method | Replicability of statistical methods | Assessment of Hardy-Weinberg equilibrium | Sufficient descriptive demographic data | Clear report of dropout and reasons | Statement of genotype frequencies and outcome data |
|---|---|---|---|---|---|---|---|---|---|---|
| Perez Mayorga 2000 [12] | + | + | + | − | + | + | + | + | + | + |
| Sodu 2002 [13] | + | − | + | − | + | + | + | + | ? | + |
| De Castro 2003 [14] | + | + | + | − | + | + | + | + | + | + |
| Behre 2005 [15]a | + | + | + | − | + | + | − | + | + | + |
| Jun 2006 [16] | + | + | + | − | + | + | + | + | + | + |
| Klinkert 2006 [17] | + | + | + | − | + | + | + | + | + | + |
| Loutradis 2006 [18] | + | + | + | − | + | + | − | + | + | + |
| Achrekar 2009 [19] | + | + | + | − | + | + | + | + | + | + |
| Huang 2010 [20] | + | + | + | − | + | + | + | + | + | + |
| Sheikhha 2011 [21] | + | + | + | + | + | + | + | + | + | + |
| Boudjenah 2012 [22] | + | + | + | + | + | + | + | + | + | + |
| Genro 2012 [23] | + | + | + | − | + | + | + | + | + | + |
| Mohiyiddeen 2013 [24] | + | + | + | + | + | + | + | + | + | + |
| Mohiyiddeen 2013 [25] | + | + | + | + | + | + | + | + | + | + |
| Yan 2013 [26] | + | + | + | − | + | + | + | + | + | + |
| Huang 2015 [27] | + | + | + | − | + | + | + | + | + | + |
Total dose of exogenous FSH
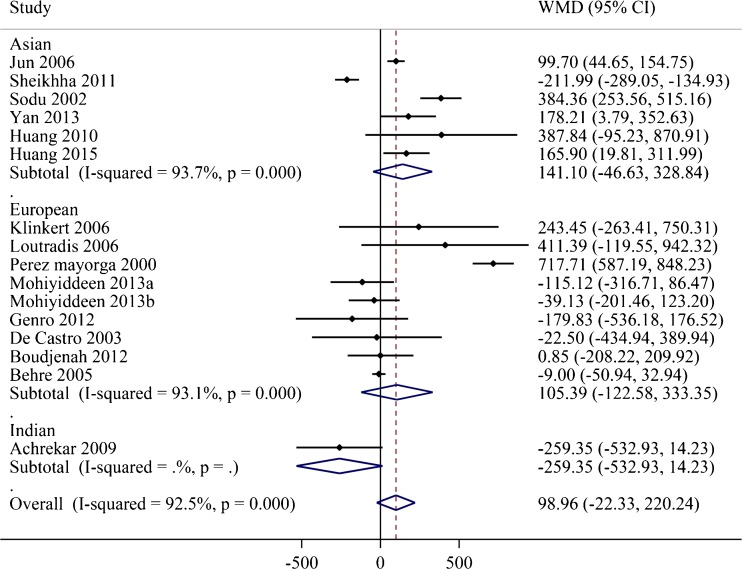
The total exogenous FSH requirement during the COH was evaluated based on the 16 studies [12–27]. As shown in Fig. 2, there was no significant difference between the subjects with SS genotype and those with NN or NS genotype (WMD = 98.96 IU, 95 % CI = −22.33 to 220.24, P = 0.153) with a significant heterogeneity (I2 = 92.5 %). In addition, a subgroup analysis by region was conducted to explore the source of heterogeneity. No significant difference was detected between SS and (NN + NS) groups in Asian (WMD = 141.10 IU, 95 % CI = −46.63 to 328.84, P = 0.218), European (WMD = 105.39 IU, 95 % CI = −122.58 to 333.35, P = 0.365), and Indian subgroups (WMD = −259.35 IU, 95 % CI = −532.93 to 14.23, P = 0.063), but the heterogeneity across the studies could not be completely eliminated by subgroup analysis (I2 > 50 %).
Fig. 2.
Forest plots for the relationships between the FSHR Asn680Ser polymorphism and total exogenous FSH dose sub-grouped by regions. The black dots and horizontal lines represent the study-specific MD and 95 % CI. The diamonds represent the pooled MD and 95 % CI. WMD weighted mean difference, CI confidence interval
Number of retrieved oocytes
Fourteen studies evaluating the association between FSHR Asn680Ser genotype and number of retrieved oocytes were included [12, 14–21, 23–27]. As shown in Fig. 3, a significant difference was found in oocyte number when comparing SS with (NN + NS) group (WMD = −1.36, 95 % CI = −1.85 to −0.87, P < 0.001) with high heterogeneity (I2 = 61.9 %). Subgroup analysis by region showed that there was a significant difference in Asian group (WMD = −1.85, 95 % CI = −2.08 to −1.63, P < 0.001), but not in European (WMD = −0.83, 95 % CI = −1.67 to 0.01, P = 0.052) or Indian groups (WMD = −2.77, 95 % CI = −9.51 to 3.97, P = 0.420).
Fig. 3.
Forest plots for the relationships between the FSHR Asn680Ser polymorphism and number of retrieved oocytes sub-grouped by regions. The black dots and horizontal lines represent the study-specific MD and 95 % CI, respectively. The diamonds represent the pooled MD and 95 % CI. WMD weighted mean difference, CI confidence interval
Ovarian response
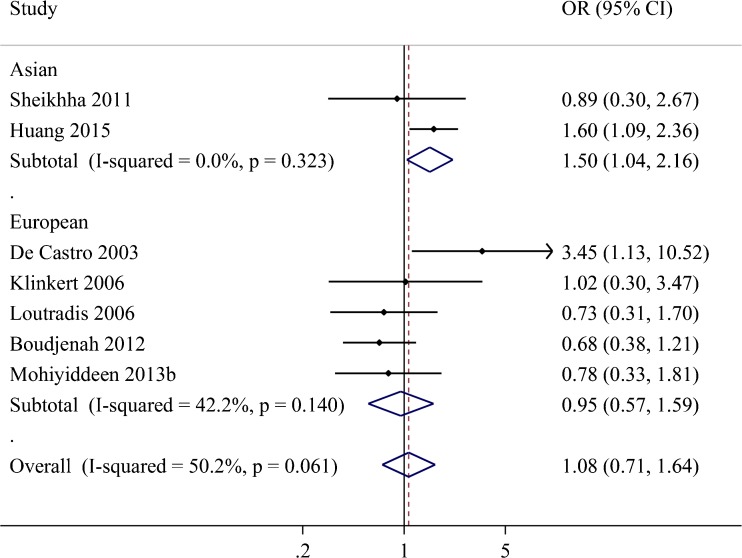
Seven studies provided the data about the ovarian response in terms of poor response outcome [3, 17, 18, 21, 22, 24, 27]. As shown in Fig. 4, there was no significant difference in poor response rate between the subjects with SS genotype and those with NN or NS genotype (OR = 1.08, 95 % CI = 0.71–1.64, P = 0.139) with high heterogeneity (I2 = 50.2 %), which was confirmed in the European subgroup (OR = 0.95, 95 % CI = 0.57–1.59, P = 0.847). However, there was a significant difference in the Asian subgroup (OR = 1.50, 95 % CI = 1.04–2.16, P = 0.028): subjects with SS genotype had a higher risk of poor ovarian response than those with (NN + NS) genotype.
Fig. 4.
Forest plots for the relationships between the FSHR Asn680Ser polymorphism and ovarian response sub-grouped by regions. The black dots and horizontal lines are the study-specific OR and 95 % CI, respectively, and the diamonds represent the pooled OR and 95 % CI. OR odds ratio, CI confidence interval
Only two studies evaluated the incidence of OHSS [19, 24]. The results of meta-analysis showed that there was no significant difference between SS group and (NN + NS) group (OR = 1.58, 95 % CI = 0.41–6.07, P = 0.504) with no significant heterogeneity (I2 = 0 %), which indicated that FSHR Asn680Ser could not be a genetic biomarker to predict the OHSS.
Rate of clinical pregnancy
Ten studies provided the data regarding the clinical pregnancy rate [15–18, 20, 21, 23–25, 27]. As shown in Fig. 5, there was no significant difference between SS and (NN + NS) groups (OR = 1.10, 95 % CI = 0.86–1.40, P = 0.454) with low heterogeneity (I2 = 6.6 %). Subgroup analysis did not show any difference in the subgroup of European (OR = 1.37, 95 % CI = 0.90–2.09, P = 0.145) or Asian group (OR = 0.95, 95 % CI = 0.71–1.28, P = 0.753).
Fig. 5.
Forest plots for the relationships between the FSHR Asn680Ser polymorphism and clinical pregnancy rates sub-grouped by regions. The black dots and horizontal lines correspond to the study-specific OR and 95 % CI. The diamonds represent the pooled OR and 95 % CI. OR odds ratio, CI confidence interval
Meta-regression and publication bias
Meta-regression analysis confirmed that none of the considered factors (such as design, region, procedure, and protocol) was the main source of heterogeneity (all P values >0.05). There was no publication bias detected by Egger’s or Begg’s test in any of the associations reported above (all P values >0.05).
Discussion
The identification of the variants of FSHR has facilitated the research regarding their value as predictors of ovarian response to an exogenous stimulation in women undergoing COH. In recent years, most of the FSHR genotype-based studies focused on the Asn680Ser polymorphism. However, results from previous meta-analysis and individual studies about the association between this polymorphism and COH outcomes were still inconsistent. Based on evidence from 16 recent studies, FSHR Asn680Ser polymorphism might influence the number of retrieved oocytes, but significance of this polymorphism to COH outcomes was poor, in terms of clinical parameters such as exogenous FSH dose, incidence of ovarian response, and clinical pregnancy rate. An interesting finding in our studies is that FSHR Asn680Ser might be a promising genetic marker for predicting the ovarian response in Asian patients.
In IVF programs, exogenous FSH is administered for ovarian stimulation. Determining the dose of FSH to attain optimum response is still one of the ongoing challenges in the field of infertility management in IVF clinics. To identify genetic markers for guiding personalized dosing of exogenous gonadotropin, numerous studies have been carried out to evaluate individual variability in the ovarian response to gonadotropin [12–28]. Consistent with our previous meta-analysis, as well as many original studies, no significant difference was observed in total amount of exogenous FSH when comparing subjects harboring the SS genotype with those harboring the NN or NS genotype, but the subjects with the SS genotype displayed a trend to need more exogenous FSH (WMD = 98.96 IU, 95 % CI = −22.33 to 220.24). However, the subgroup analysis by region revealed a different trend: unlike the subjects from Asia and Europe, the subjects carrying the SS genotype from India tend to need less exogenous FSH. These results showed that the ovarian response to exogenous FSH might differ between ethnicities. However, only one study involving 50 Indian subjects was included and studies larger in scale are needed. Further analysis regarding the number of oocytes retrieved showed that fewer oocytes were retrieved in the SS group than those in the NN or NS group (WMD = −1.36, 95 % CI = −1.85 to −0.87), suggesting that patients with the SS genotype had lower sensitivity to FSH. Therefore, same exogenous FSH dosage given to the patients without considering the genetic background might result in better outcomes in the subjects with the NS or NN genotype than those with the SS genotype. Similar results were found in the Asian subgroup, but not in European or Indian subgroups. Therefore, to achieve adequate ovarian stimulation and retrieve enough oocytes, patients carrying the SS genotype might need a higher dosage of exogenous FSH than those carrying the NN or NS genotype, especially in Asian subjects. Furthermore, FSHR polymorphism may represent a predictive marker for the response to exogenous FSH during COH treatment. The results from two meta-analyses showed that the SS genotype was associated with a poor response during COH [11]. However, our study found no significant association between Asn680Ser polymorphism and ovarian response without considering racial background. In contrast, Asian subjects with the SS genotype had a higher risk of poor ovarian response than those with the NN or NS genotype (OR = 1.50, 95 % CI = 1.04–2.16), and this result was consistent with the genotype-specific difference in the number of oocytes retrieved. However, in the term of OHSS, a lack of significant difference was observed between SS and (NN + NS) groups (OR = 1.58, 95 % CI = 0.41–6.07).
The rate of pregnancy in an IVF cycle is considered as a critical measure to determine the IVF outcome. However, there is a lack of consistent evidence regarding this outcome in recent association studies. In the included studies, only the study by Klinkert et al. reported that the subjects with the SS genotype were more likely to have a higher pregnancy rate when compared with those with the NN genotype [17]. However, our meta-analysis from 10 studies comprising a total of 2762 subjects showed that there was a lack of association between the FSHR Asn680Ser polymorphism and the rate of pregnancy (OR = 1.10, 95 % CI = 0.86–1.40). When taking ethnic background into account, subgroup analysis revealed a difference between Asian (OR = 0.95, 95 % CI = 0.71–1.28) and European (OR = 1.37, 95 % CI = 0.90–2.09), which might be interpreted by the fewer oocytes retrieved and poor response in Asian subjects with the SS genotype. Further studies are necessary to confirm whether higher exogenous FSH could influence the number of oocytes and eventually improve the clinical pregnancy rate.
Admittedly, there were some limitations in the present meta-analysis. Firstly, our work only focused on the data about FSHR Asn680Ser polymorphism. Other FSHR polymorphisms, such as Ala307Thr, -29G >A, and -211G >T, might also play a role in modulating ovarian response to gonadotropin administration [19, 36]. However, the role of these polymorphisms was not assessed in our study, because the available studies about these polymorphisms are few and further studies are necessary to confirm their clinical associations. Secondly, although a meta-regression and a subgroup analysis were conducted to explore the source of heterogeneity across the included studies, we cannot exclude the possibilities of other confounding factors, such as the ethnicity. Multiple ethnicities were included and analyzed in the original trials, which limits the sub-analysis by region rather than ethnicity. Thirdly, insufficient subjects (2767 subjects) and few frequency of SS genotype (506 subjects, about 19 % of all subjects included) detected in the population limited the power to provide a reliable and conclusive suggestion. Further, more well-designed trials are warranted to confirm the findings.
In summary, our meta-analysis of current available studies suggested that FSHR Asn680Ser polymorphism might be a significant biomarker for predicting the number of retrieved oocytes and poor response, especially in Asian subjects. Other outcomes such as exogenous FSH dose, OHSS, and pregnancy rate were not influenced by FSHR Asn680Ser polymorphism. However, it does not translate into statistically significant differences in these clinical outcomes, possibly due to insufficient sample size in the meta-analysis. Further investigations will be required to confirm these findings.
Acknowledgments
The authors are thankful to Prof. Hongguang Xie and Dr Shenyu Zhai, who provided valuable feedback about the manuscript
Compliance with ethical standards
Formal institutional review board approval was not required because only published data were pooled.
Conflict of interests
The authors declare that they have no competing interests.
Footnotes
Capsule The FSHR Asn680Ser polymorphism might be a significant biomarker for predicting the number of retrieved oocytes and poor response, especially in Asian subjects.
Huilin Tang and Yingying Yan contributed equally to this work.
References
- 1.Altmae S, Hovatta O, Stavreus-Evers A, Salumets A. Genetic predictors of controlled ovarian hyperstimulation: where do we stand today? Hum Reprod Update. 2011;17(6):813–28. doi: 10.1093/humupd/dmr034. [DOI] [PubMed] [Google Scholar]
- 2.Gearhart J, Coutifaris C. In vitro fertilization, the Nobel Prize, and human embryonic stem cells. Cell Stem Cell. 2011;8(1):12–5. doi: 10.1016/j.stem.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 3.de Castro F, Moron FJ, Montoro L, Real LM, Ruiz A. Pharmacogenetics of controlled ovarian hyperstimulation. Pharmacogenomics. 2005;6(6):629–37. doi: 10.2217/14622416.6.6.629. [DOI] [PubMed] [Google Scholar]
- 4.Enskog A, Henriksson M, Unander M, Nilsson L, Brannstrom M. Prospective study of the clinical and laboratory parameters of patients in whom ovarian hyperstimulation syndrome developed during controlled ovarian hyperstimulation for in vitro fertilization. Fertil Steril. 1999;71(5):808–14. doi: 10.1016/S0015-0282(99)00090-4. [DOI] [PubMed] [Google Scholar]
- 5.La Marca A, Sunkara SK. Individualization of controlled ovarian stimulation in IVF using ovarian reserve markers: from theory to practice. Hum Reprod Update. 2014;20(1):124–40. doi: 10.1093/humupd/dmt037. [DOI] [PubMed] [Google Scholar]
- 6.Simoni M, Gromoll J, Nieschlag E. The follicle-stimulating hormone receptor: biochemistry, molecular biology, physiology, and pathophysiology. Endocr Rev. 1997;18(6):739–73. doi: 10.1210/edrv.18.6.0320. [DOI] [PubMed] [Google Scholar]
- 7.Aittomaki K, Herva R, Stenman UH, Juntunen K, Ylostalo P, Hovatta O, et al. Clinical features of primary ovarian failure caused by a point mutation in the follicle-stimulating hormone receptor gene. J Clin Endocrinol Metab. 1996;81(10):3722–6. doi: 10.1210/jcem.81.10.8855829. [DOI] [PubMed] [Google Scholar]
- 8.Simoni M, Nieschlag E, Gromoll J. Isoforms and single nucleotide polymorphisms of the FSH receptor gene: implications for human reproduction. Hum Reprod Update. 2002;8(5):413–21. doi: 10.1093/humupd/8.5.413. [DOI] [PubMed] [Google Scholar]
- 9.Simoni M, Tempfer CB, Destenaves B, Fauser BC. Functional genetic polymorphisms and female reproductive disorders: part I: polycystic ovary syndrome and ovarian response. Hum Reprod Update. 2008;14(5):459–84. doi: 10.1093/humupd/dmn024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao Y, Ma CH, Tang HL, Hu YF. Influence of follicle-stimulating hormone receptor (FSHR) Ser680Asn polymorphism on ovarian function and in-vitro fertilization outcome: a meta-analysis. Mol Genet Metab. 2011;103(4):388–93. doi: 10.1016/j.ymgme.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Moron FJ, Ruiz A. Pharmacogenetics of controlled ovarian hyperstimulation: time to corroborate the clinical utility of FSH receptor genetic markers. Pharmacogenomics. 2010;11(11):1613–8. doi: 10.2217/pgs.10.156. [DOI] [PubMed] [Google Scholar]
- 12.Perez Mayorga M, Gromoll J, Behre HM, Gassner C, Nieschlag E, Simoni M. Ovarian response to follicle-stimulating hormone (FSH) stimulation depends on the FSH receptor genotype. J Clin Endocrinol Metab. 2000;85(9):3365–9. doi: 10.1210/jcem.85.9.6789. [DOI] [PubMed] [Google Scholar]
- 13.Sudo S, Kudo M, Wada S, Sato O, Hsueh AJ, Fujimoto S. Genetic and functional analyses of polymorphisms in the human FSH receptor gene. Mol Hum Reprod. 2002;8(10):893–9. doi: 10.1093/molehr/8.10.893. [DOI] [PubMed] [Google Scholar]
- 14.de Castro F, Ruiz R, Montoro L, Perez-Hernandez D, Sanchez-Casas Padilla E, Real LM, et al. Role of follicle-stimulating hormone receptor Ser680Asn polymorphism in the efficacy of follicle-stimulating hormone. Fertil Steril. 2003;80(3):571–6. doi: 10.1016/S0015-0282(03)00795-7. [DOI] [PubMed] [Google Scholar]
- 15.Behre HM, Greb RR, Mempel A, Sonntag B, Kiesel L, Kaltwasser P, et al. Significance of a common single nucleotide polymorphism in exon 10 of the follicle-stimulating hormone (FSH) receptor gene for the ovarian response to FSH: a pharmacogenetic approach to controlled ovarian hyperstimulation. Pharmacogenet Genomics. 2005;15(7):451–6. doi: 10.1097/01.fpc.0000167330.92786.5e. [DOI] [PubMed] [Google Scholar]
- 16.Jun JK, Yoon JS, Ku SY, Choi YM, Hwang KR, Park SY, et al. Follicle-stimulating hormone receptor gene polymorphism and ovarian responses to controlled ovarian hyperstimulation for IVF-ET. J Hum Genet. 2006;51(8):665–70. doi: 10.1007/s10038-006-0005-5. [DOI] [PubMed] [Google Scholar]
- 17.Klinkert ER, te Velde ER, Weima S, van Zandvoort PM, Hanssen RG, Nilsson PR, et al. FSH receptor genotype is associated with pregnancy but not with ovarian response in IVF. Reprod Biomed Online. 2006;13(5):687–95. doi: 10.1016/S1472-6483(10)60660-8. [DOI] [PubMed] [Google Scholar]
- 18.Loutradis D, Patsoula E, Minas V, Koussidis GA, Antsaklis A, Michalas S, et al. FSH receptor gene polymorphisms have a role for different ovarian response to stimulation in patients entering IVF/ICSI-ET programs. J Assist Reprod Genet. 2006;23(4):177–84. doi: 10.1007/s10815-005-9015-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Achrekar SK, Modi DN, Desai SK, Mangoli VS, Mangoli RV, Mahale SD. Follicle-stimulating hormone receptor polymorphism (Thr307Ala) is associated with variable ovarian response and ovarian hyperstimulation syndrome in Indian women. Fertil Steril. 2009;91(2):432–9. doi: 10.1016/j.fertnstert.2007.11.093. [DOI] [PubMed] [Google Scholar]
- 20.Huang SY, Yang J, Yin TL, Li X, Li J, Xu WM. Association of gene polymorphism of follicle stimulating hormone receptor with ovarian response in IVF cycles. Med J Wuhan Univ. 2010;31(3):334–8. [Google Scholar]
- 21.Sheikhha MH, Eftekhar M, Kalantar SM. Investigating the association between polymorphism of follicle-stimulating hormone receptor gene and ovarian response in controlled ovarian hyperstimulation. J Hum Reprod Sci. 2011;4(2):86–90. doi: 10.4103/0974-1208.86089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudjenah R, Molina-Gomes D, Torre A, Bergere M, Bailly M, Boitrelle F, et al. Genetic polymorphisms influence the ovarian response to rFSH stimulation in patients undergoing in vitro fertilization programs with ICSI. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0038700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genro VK, Matte U, De Conto E, Cunha-Filho JS, Fanchin R. Frequent polymorphisms of FSH receptor do not influence antral follicle responsiveness to follicle-stimulating hormone administration as assessed by the Follicular Output RaTe (FORT) J Assist Reprod Genet. 2012;29(7):657–63. doi: 10.1007/s10815-012-9761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohiyiddeen L, Newman WG, Cerra C, Horne G, Mulugeta B, Byers H, et al. FSH receptor genotype does not predict metaphase-II oocyte output or fertilization rates in ICSI patients. Reprod Biomed Online. 2013;27(3):305–9. doi: 10.1016/j.rbmo.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Mohiyiddeen L, Newman WG, Cerra C, McBurney H, Mulugeta B, Roberts SA, et al. A common Asn680Ser polymorphism in the follicle-stimulating hormone receptor gene is not associated with ovarian response to gonadotropin stimulation in patients undergoing in vitro fertilization. Fertil Steril. 2013;99(1):149–55. doi: 10.1016/j.fertnstert.2012.08.037. [DOI] [PubMed] [Google Scholar]
- 26.Yan Y, Gong Z, Zhang L, Li Y, Li X, Zhu L, et al. Association of follicle-stimulating hormone receptor polymorphisms with ovarian response in Chinese women: a prospective clinical study. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0078138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X, Li L, Hong L, Zhou W, Shi H, Zhang H, et al. The Ser680Asn polymorphism in the follicle-stimulating hormone receptor gene is associated with the ovarian response in controlled ovarian hyperstimulation. Clin Endocrinol (Oxf) 2015;82(4):577–83. doi: 10.1111/cen.12573. [DOI] [PubMed] [Google Scholar]
- 28.Mohiyiddeen L, Newman WG, McBurney H, Mulugeta B, Roberts SA, Nardo LG. Follicle-stimulating hormone receptor gene polymorphisms are not associated with ovarian reserve markers. Fertil Steril. 2012;97(3):677–81. doi: 10.1016/j.fertnstert.2011.12.040. [DOI] [PubMed] [Google Scholar]
- 29.Lledo B, Guerrero J, Turienzo A, Ortiz JA, Morales R, Ten J, et al. Effect of follicle-stimulating hormone receptor N680S polymorphism on the efficacy of follicle-stimulating hormone stimulation on donor ovarian response. Pharmacogenet Genomics. 2013;23(5):262–8. doi: 10.1097/FPC.0b013e32835fe813. [DOI] [PubMed] [Google Scholar]
- 30.Pabalan N, Trevisan CM, Peluso C, Jarjanazi H, Christofolini DM, Barbosa CP, et al. Evaluating influence of the genotypes in the follicle-stimulating hormone receptor (FSHR) Ser680Asn (rs6166) polymorphism on poor and hyper-responders to ovarian stimulation: a meta-analysis. J Ovarian Res. 2014;7:285. doi: 10.1186/s13048-014-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins JPT, Green S. Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 [updated March 2011]. The Cochrane Collaboration. 2011. www.cochrane-handbook.org. Accessed 3 Jun 2015.
- 32.Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. 2005;5:13. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the STROBE statement. PLoS Med. 2009;6(2) doi: 10.1371/journal.pmed.1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Qian W, Li J, Zhang P, Yang Z, Chen W, et al. Who are at risk for thromboembolism after arthroplasty? A systematic review and meta-analysis. Thromb Res. 2013;132(5):531–6. doi: 10.1016/j.thromres.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Terrazzino S, Cargnin S, Del Re M, Danesi R, Canonico PL, Genazzani AA. DPYD IVS14 + 1G > A and 2846A > T genotyping for the prediction of severe fluoropyrimidine-related toxicity: a meta-analysis. Pharmacogenomics. 2013;14(11):1255–72. doi: 10.2217/pgs.13.116. [DOI] [PubMed] [Google Scholar]
- 36.Dolfin E, Guani B, Lussiana C, Mari C, Restagno G, Revelli A. FSH-receptor Ala307Thr polymorphism is associated to polycystic ovary syndrome and to a higher responsiveness to exogenous FSH in Italian women. J Assist Reprod Genet. 2011;28(10):925–30. doi: 10.1007/s10815-011-9619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]