Abstract
The occurrence of alternative Nod factor (NF)-independent symbiosis between legumes and rhizobia was first demonstrated in some Aeschynomene species that are nodulated by photosynthetic bradyrhizobia lacking the canonical nodABC genes. In this study, we revealed that a large diversity of non-photosynthetic bradyrhizobia, including B. elkanii, was also able to induce nodules on the NF-independent Aeschynomene species, A. indica. Using cytological analysis of the nodules and the nitrogenase enzyme activity as markers, a gradient in the symbiotic interaction between bradyrhizobial strains and A. indica could be distinguished. This ranged from strains that induced nodules that were only infected intercellularly to rhizobial strains that formed nodules in which the host cells were invaded intracellularly and that displayed a weak nitrogenase activity. In all non-photosynthetic bradyrhizobia, the type III secretion system (T3SS) appears required to trigger nodule organogenesis. In contrast, genome sequence analysis revealed that apart from a few exceptions, like the Bradyrhizobium ORS285 strain, photosynthetic bradyrhizobia strains lack a T3SS. Furthermore, analysis of the symbiotic properties of an ORS285 T3SS mutant revealed that the T3SS could have a positive or negative role for the interaction with NF-dependent Aeschynomene species, but that it is dispensable for the interaction with all NF-independent Aeschynomene species tested. Taken together, these data indicate that two NF-independent symbiotic processes are possible between legumes and rhizobia: one dependent on a T3SS and one using a so far unknown mechanism.
Introduction
The legume-rhizobium symbiosis is of immense biological and ecological importance. It is a major contributor to the global nitrogen cycle, and serves as a model to study plant–microbe interactions. A major advance in our understanding of the legume–rhizobium symbiosis was the discovery, in the early 1990s, that this interaction relies on the exchange of diffusible signal molecules between the two partners (Lerouge et al., 1990). Flavonoids secreted by the host plant roots activate in rhizobia the expression of nodulation genes necessary for the synthesis and secretion of lipochitooligosaccharides, named Nod factors (NFs). In return, the perception of NFs by LysM-receptor-like kinase (LysM-RLKs) induces a signal transduction cascade that is required for infection and nodule organogenesis (Oldroyd, 2013). For a long time, this molecular dialogue was assumed to be a universal paradigm for all legume–rhizobium interactions. However, in 2007, it has been shown that some photosynthetic Bradyrhizobium strains that are able to induce nitrogen-fixing nodules on some Aeschynomene species lack the canonical nodABC genes required for NF synthesis (Giraud et al., 2007). This indicated, for the first time, that an alternative symbiotic interaction independent of NFs was possible between a legume and a rhizobium strain.
Since this discovery, two other examples of NF-independent symbiosis have been reported. In the model plant Lotus japonicus, which uses normally a classical NF-dependent symbiotic process, it has been observed that double mutant lines affected in some key determinants of the NF signaling pathway were occasionally able to form infected nodules when the plants were inoculated with a compatible rhizobium strain unable to produce NF (Madsen et al., 2010). More recently, it has been shown that a NF-deficient mutant of Bradyrhizobium elkanii could induce nodules on Glycine max cv. Enrei mutated or not in the LysM-RLK nfr1 Nod factor receptor (Okazaki et al., 2013). In the later case, this NF-independent symbiosis was shown to be only possible if the bacteria possessed a functional type III secretion system (T3SS). This secretory machinery initially identified in animal and plant pathogens permits the delivery of effector molecules inside the host cells that suppress plant defense response system and favor the infection (Hueck, 1998). It is proposed that in B. elkanii, T3SS effectors could also activate host nodulation signaling by bypassing NF recognition (Okazaki et al., 2013).
These two examples of NF-independent symbiosis which were put forward in an artificial system, using plant and bacterial mutant backgrounds, appear weakly efficient. Indeed, the frequency of nodulated plants, the number of nodules formed per plant and the level of infection of these nodules were drastically reduced in comparison with the normal context in which plant and bacteria are not altered in the NF signaling pathway. In addition, these interactions do not involve the formation of infection threads in either root hairs or nodule tissues but use an intercellular infection mode that is considered more ancestral. This suggests that the NF-independent process may constitute the ground state of rhizobium–legume symbiosis which has been maintained in legumes during evolution but which was overridden by the acquisition of a more sophisticated process involving NF signaling and infection thread formation.
In the Aeschynomene genus, only a monophyletic clade encompassing 12 species, including A. indica and A. evenia, were found to be nodulated by nod gene-lacking photosynthetic bradyrhizobia (Chaintreuil et al., 2013). In contrast to other legumes, the NF-independent pathway appears to be dominant in this clade and most probably exclusive because most of the natural symbionts either lack nod genes or they can be inactivated without effect on the efficiency of nodule formation (Bonaldi et al., 2011). Furthermore, besides lacking nod genes, most photosynthetic bradyrhizobia lack a T3SS. Again the ORS285 strain is an exception and probably acquired nod and T3SS genes simultaneously by lateral gene transfer (Mornico et al., 2011). This indicates that photosynthetic bradyrhizobia could interact with Aeschynomene using another pathway than the one operating during the interaction between a NF-deficient B. elkanii mutant and soybean.
To clarify the different alternative NF-independent pathways, we examined in this study the ability of a large collection of rhizobia to interact symbiotically with A. indica. We found that a large number of non-photosynthetic bradyrhizobia are able to induce nodules on A. indica but that the bacterial infection and nitrogen fixation largely differed between species. By mutagenesis, we demonstrated for two strains (B. elkanii and Bradyrhizobium sp. STM6978) that the T3SS has an essential role in nodule organogenesis. In parallel, we examined the symbiotic role of the T3SS in the interaction of Bradyrhizobium sp. ORS285 with various NF-dependent and NF-independent Aeschynomene species. The obtained data indicate that photosynthetic and non-photosynthetic Bradyrhizobium strains can nodulate Aeschynomene species via two NF-independent processes that differ in the requirement of a T3SS.
Materials and methods
Bacterial strains and growth conditions
All tested bacterial strains are listed in Table 1 and Supplementary Table S1. Strains were grown in YM medium at 28 °C (Vincent, 1970). The mutants of B. elkanii USDA61 (BEnodC, BErhcJ and BEttsnod) were previously obtained (Okazaki et al., 2013). The GusA- and GFP-tagged strains of USDA61 were obtained by introducing plasmids pCAM120 (Wilson et al., 1995) and pDG70 (Gage 2002), respectively. For their culture, appropriate antibiotics were added as indicated in (Okazaki et al., 2013). The ORS285ΔT3SS and STM6978ΔT3SS mutants were constructed for this study (see Supplementary Method). For their culture, the medium was supplemented with kanamycin (50 μg ml–1) and cefotaxime (20 μg ml–1), respectively.
Table 1. Symbiotic properties of various Bradyrhizobium strains on Aeschynomene indica.
| Strain | Accession | Nodulationa | Infectionb | ARA activityc | T3SSd |
|---|---|---|---|---|---|
| B. betae | LMG 21987 T | 0 | — | 0 | − |
| B. canariense | LMG 22265 T | 0 | — | 0 | − |
| B. elkanii | USDA61 | 20.6 | Inter | 0 | GS |
| B. elkanii | USDA76 | 14.6 | Inter | 0 | GS |
| B. elkanii | USDA94 | 25 | Inter | 0 | GS |
| B. japonicum | USDA4 | 0 | — | 0 | GS |
| B. japonicum | USDA110 | 0 | — | 0 | GS |
| B. japonicum | USDA38 | 0 | — | 0 | GS |
| B. japonicum | USDA122 | 0 | — | 0 | GS |
| B. japonicum | USDA124 | 0 | — | 0 | GS |
| B. liaoningense | LMG 18230 T | 0 | — | 0 | + |
| B. yuanmingense | LMG 21825 | 0 | — | 0 | + |
| Bradyrhizobium sp. | DOA9 | 0 | — | 0 | + |
| Bradyrhizobium sp. | ORS86 | 19 | ND | 0 | + |
| Bradyrhizobium sp. | ORS101 | 0 | — | 0 | + |
| Bradyrhizobium sp. | ORS110 | 14.4 | ND | 0 | + |
| Bradyrhizobium sp. | ORS111 | 17.6 | ND | 0 | + |
| Bradyrhizobium sp. | ORS112 | 15.6 | ND | 0 | + |
| Bradyrhizobium sp. | ORS121 | 6.6 | inter | 0 | + |
| Bradyrhizobium sp. | ORS149 | 0 | — | 0 | + |
| Bradyrhizobium sp. | ORS159 | 0 | — | 0 | + |
| Bradyrhizobium sp. | ORS162 | 0 | — | 0 | + |
| Bradyrhizobium sp. | ORS163 | 0 | — | 0 | + |
| Bradyrhizobium sp. | ORS186 | 0 | — | 0 | + |
| Bradyrhizobium sp. | ORS187 | 0 | — | 0 | + |
| Bradyrhizobium sp. | ORS278 | 21.6 | intra | 100%-5/5 | − |
| Bradyrhizobium sp. | ORS1824 | 19.6 | ND | 0 | + |
| Bradyrhizobium sp. | ORS1836 | 0 | — | 0 | + |
| Bradyrhizobium sp. | STM3557 | 4.6 | intra | 0 | + |
| Bradyrhizobium sp. | STM3560 | 9.2 | inter | 0 | + |
| Bradyrhizobium sp. | STM3561 | 7.5 | intra | 8.5%-3/5 | + |
| Bradyrhizobium sp. | STM3562 | 10.4 | inter | 0 | + |
| Bradyrhizobium sp. | STM3564 | 0 | — | 0 | − |
| Bradyrhizobium sp. | STM3565 | 0 | — | 0 | − |
| Bradyrhizobium sp. | STM3566 | 0 | — | 0 | + |
| Bradyrhizobium sp. | STM3567 | 10.6 | inter | 0 | + |
| Bradyrhizobium sp. | STM3569 | 25.8 | inter | 0 | + |
| Bradyrhizobium sp. | STM3571 | 9 | intra | 7.2%–3/5 | + |
| Bradyrhizobium sp. | STM6978/ORS3257 | 12 | intra | 14%–4/5 | + |
| Bradyrhizobium sp. | STM6980/ORS3409 | 12.5 | intra | 0 | + |
Number of nodules per plant (mean estimated on five plants).
inter, intercellular; intra, intracellular; ND, no bacteria detected in the nodule tissue; —, No nodule formed.
First, relative acetylene reduction activity (percentage of ORS278) estimated on 5 whole plants; Second, number of plants displaying an acetylene reduction assay (ARA) activity on the five plants tested.
Presence of T3SS (GS, deduced from the genome sequence; +, PCR using universal primers positive; −, PCR using universal primers negative).
Research of the presence of a T3SS by PCR
The presence of a T3SS gene cluster in the various Bradyrhizobium strains was analyzed by PCR. Two pairs of degenerated primers were designed from conserved regions of rhcN genes of B. elkanii USDA61, B. japonicum USDA110 and Bradyrhizobium sp. ORS285: rhcN.univ.f1 (5′-GGRGAACTDTGCYTVYTGCAGGATC-3')/rhcN.univ.r1 (5′-CCATARATTCCRATCCGCTGRCCTTC-3′) and rhcN.univ.f2 (5′-TGGTCGCTYGAGGCBGARGTNATCGGT-3′)/rhcN.univ.r2 (5′- GCTCGCCCATRCCGGCKCGCTCCAAC-3′). In the case of a positive result, the PCR product obtained was sequenced to analyze the amplification specificity.
Molecular phylogeny of internal transcribed spacer
For bacterial strains for which no internal transcribed spacer sequence was available, the 16–23S rDNA internal transcribed spacer region was amplified and sequenced as described by Guye et al., 2009. The resulting sequences were aligned with ClustalX (Thompson et al., 2002), and subsequently used to construct a phylogenetic tree using the Neighbor Joining method (Saitou and Nei, 1987). A bootstrap confidence analysis was performed on 1000 replicates to determine the reliability of the obtained phylogenetic tree (Felsenstein, 1985).
Plant cultivation and symbiotic analysis
The accessions of Aeschynomene species used in this study, their geographic origin and their sources are listed in Supplementary Table S2. Seed sterilization and plant culture was performed as before (Bonaldi et al., 2011). Four days after the transfer of the plantlets to test tubes covered by aluminium foil to protect root system from light, each seedling was inoculated with 1 ml of cell suspension resulting from a 4-day-old bacterial culture washed in buffered nodulation medium and adjusted to reach an optical density of one at 600 nm. Fourteen days after inoculation, five plants were analyzed for the presence of root nodules and nitrogenase enzyme activity as previously described (Bonaldi et al., 2010).
Cytological analysis of the nodules
Semi-thin nodule sections (30–40-μm-thick) were prepared using a vibratome (VT1000S; Leica, Nanterre, France). Nodules elicited by the ORS285 GFP-tagged strain were analyzed as previously described (Bonaldi et al., 2011). For the nodules elicited with the non-tagged strains, the nodule sections were incubated for 15 min in live/dead staining solution (5 μM SYTO 9 and 30 μM propidium iodide in 50 mM Tris pH 7.0 buffer; Live/Dead BacLight, Invitrogen, Carlsbad, CA, USA). Sections were then removed and incubated an additional 15 min in 10 mM phosphate saline buffer containing calcofluor white M2R (Sigma, Munich, Germany) to a final concentration of 0.01% (wt/vol) to stain the plant cell wall (Nagata and Takebe, 1970). Analyses were carried out using a confocal laser-scanning microscope (Carl Zeiss LSM 700, Jena, Germany). Calcofluor was exited at 405 nm with emission signal collection at 405–470 nm. For SYTO 9 and propidium iodide, an excitation wavelength of 488 and 555 nm was used with emission signal collection at 490–522 nm and 555–700 nm, respectively. Images were obtained using the ZEN 2008 software (Zeiss, Oberkochen, Germany).
Results
B. elkanii is able to induce nodules on some NF-independent Aeschynomene species in a T3SS-dependent manner
We first analyzed the ability of B. elkanii USDA61 and its derivatives BEnodC (ΔnodC), BErhcJ (ΔrhcJ, a T3SS-deficient mutant) and the double mutant BEttsnod to nodulate the NF-independent A. indica species. Nodulation was analyzed 14 days post inoculation, a lapse of time during which photosynthetic bradyrhizobia are able to establish a functional symbiosis with this species (Bonaldi et al., 2011). As shown in Figure 1a, the B. elkanii wild-type (WT) strain was able to elicit a comparable number of ‘nodules' on A. indica roots as compared with the photosynthetic Bradyrhizobium ORS278 strain that was used as control for a natural host bacterium. As observed for ORS278, these nodules were formed at the emergence of lateral root but they differed by their size, which was slightly smaller, and their shape, which was not perfectly spherical (Figures 1c and f).
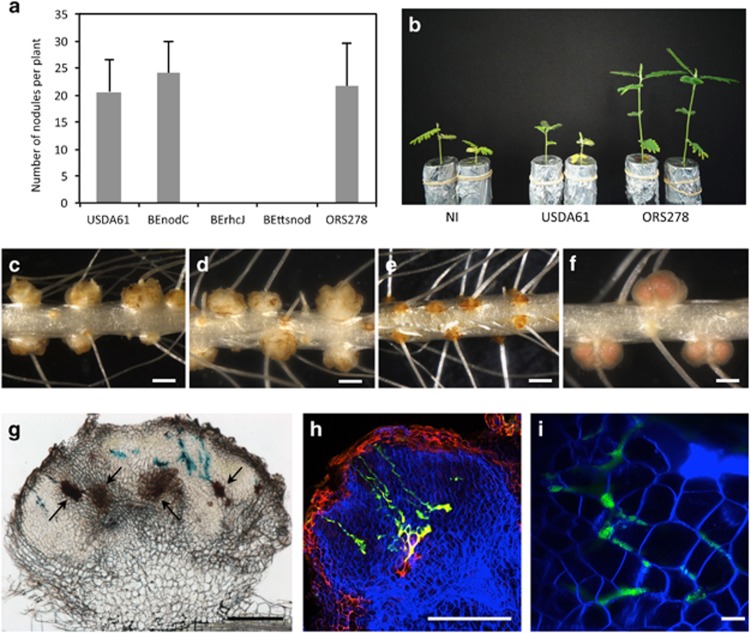
Figure 1.
Bradyrhizobium elkanii is able to induce nodules on Aeschynomene indica in a T3SS-dependent manner. (a) Number of nodules per plant elicited by B. elkanii USDA61, BEnodC, BErhcJ, BEttsnod and Bradyrhizobium ORS278 strain on A. indica plants at 14 days post inoculation (d.p.i.). Error bars represent s.d. (n=5). (b) Comparison of the growth of the plants (aerial part), non-inoculated (NI) or inoculated with B. elkanii USDA61 and Bradyrhizobium ORS278 (at 14 d.p.i.). (c–f) Nodules elicited by B. elkanii USDA61 (c), BEnodC (d), BErhcJ (e), ORS278 (f). Bars, 1 mm. (g) Transversal section of a nodule elicited by B. elkanii tagged with gusA and stained with X-Gluc. Bar, 200 μm. Black arrows indicate the presence of necrotic area. (h and i) Confocal microscopy observation of nodules elicited by B. elkanii tagged with GFP. Bars, 200 μm (h) and 10 μm (i). A full colour version of this figure is available at the ISME journal online.
When we analyzed the BEnodC, no effect of the mutation was detected, that is, the number of nodules and their shape was comparable with that obtained with the WT strain, confirming the absence of a recruitment for NFs during the interaction of B. elkanii with A. indica (Figures 1a and d). In contrast, no nodules were formed when the plants were inoculated with BErhcJ or BEttsnod (Figures 1a and e). This indicates that the T3SS is absolutely required to initiate the symbiotic interaction with A. indica.
Despite the presence of nodules, A. indica plants inoculated by B. elkanii showed typical nitrogen starvation symptoms, that is, foliar chlorosis and reduced plant growth, indicating that the formed nodules were non-effective (Figure 1b). In agreement, no nitrogenase activity could be detected on these plants using the acetylene reduction assay (Table 1). To examine the level of infection of these nodules, we performed cytological analysis of nodules elicited by a GFP or GusA-tagged WT strain using optical and confocal microscopy. As shown in Figures 1g–i, we observed that the nodules were only weakly infected and contained bacteria that proliferated in enlarged intercellular spaces between host cells. Neither intracellular infection nor the formation of infection threads was detected. Noteworthy is the occurrence of brown necrotic areas in almost all the nodules, suggesting the induction of a plant defense response when B. elkanii cells enter the nodule tissue (Figure 1g). Taken together, these data indicate that B. elkanii is able to initiate nodule organogenesis in A. indica plants through a T3SS-dependent-NF-independent process but the bacterial infection is rapidly arrested likely owing to the elicitation of plant defense reactions.
We next addressed the question whether other NF-independent Aeschynomene species would be more permissive and allow a more intimate contact with B. elkanii. For this purpose, 12 additional Aeschynomene accessions corresponding to 9 different species and 4 different ecotypes of A. evenia spp. evenia were inoculated with the GFP-tagged B. elkanii strain. We observed that only 7 out of the 12 tested accessions were nodulated and more unexpectedly among the four A. evenia spp. evenia ecotypes tested, only two were nodulated by B. elkanii (Supplementary Figures 1A and M). Thus, a high specificity of recognition which controls the triggering of nodule organogenesis exists between B. elkanii and the Aeschynomene species. On the other hand, cytological analysis of the nodules elicited on the compatible Aeschynomene species, showed that in all cases, the infection was restricted to the intercellular space (Supplementary Figures 1N and Q). This indicates that B. elkanii does not possess all the keys to establish a functional endosymbiosis with Aeschynomene plants.
The ability to induce nodules on NF-independent Aeschynomene species is widespread among Bradyrhizobium species
The increasing availability of genome sequences has revealed that the presence of a T3SS is frequent among rhizobia (Tampakaki, 2014). We therefore hypothesized that some strains could be able to exploit this T3SS machinery to develop a more compatible symbiotic relationship with Aeschynomene. To examine this possibility, we screened a large collection of rhizobium strains belonging to the Allorhizobium, Azorhizobium, Bradyrhizobium, Rhizobium, Sinorhizobium, Mesorhizobium genus as well as some Beta-rhizobia for their ability to induce nodules on A. indica. Among the 72 strains tested, only strains belonging to the Bradyrhizobium genus were able to elicit nodules (Supplementary Table S1 and Table 1). A phylogenetic tree based on the internal transcribed spacer region showed that the nodulation-positive strains, representing ~50% of the Bradyrhizobium strains tested, do not form a specific clade but are scattered within the genus (Figure 2). This indicates that the ability to nodulate NF-independent Aeschynomene species is a widespread character among the Bradyrhizobium species and that it is not strictly correlated with a specific phylogenetic position. By PCR and sequencing, using universal primers targeting the canonical rhcN gene required for the functioning of the T3SS injectisome, we confirmed that all the positive strains contained a T3SS (Table 1). However, the presence of a T3SS is not the unique criteria required to induce nodules on A. indica because among the strains unable to induce nodules on this species, a large number, including the B. japonicum strains, do contain a T3SS. This suggests that the positive strains, beside a T3SS apparatus, produce specific type III effectors that are necessary to initiate nodule organogenesis.
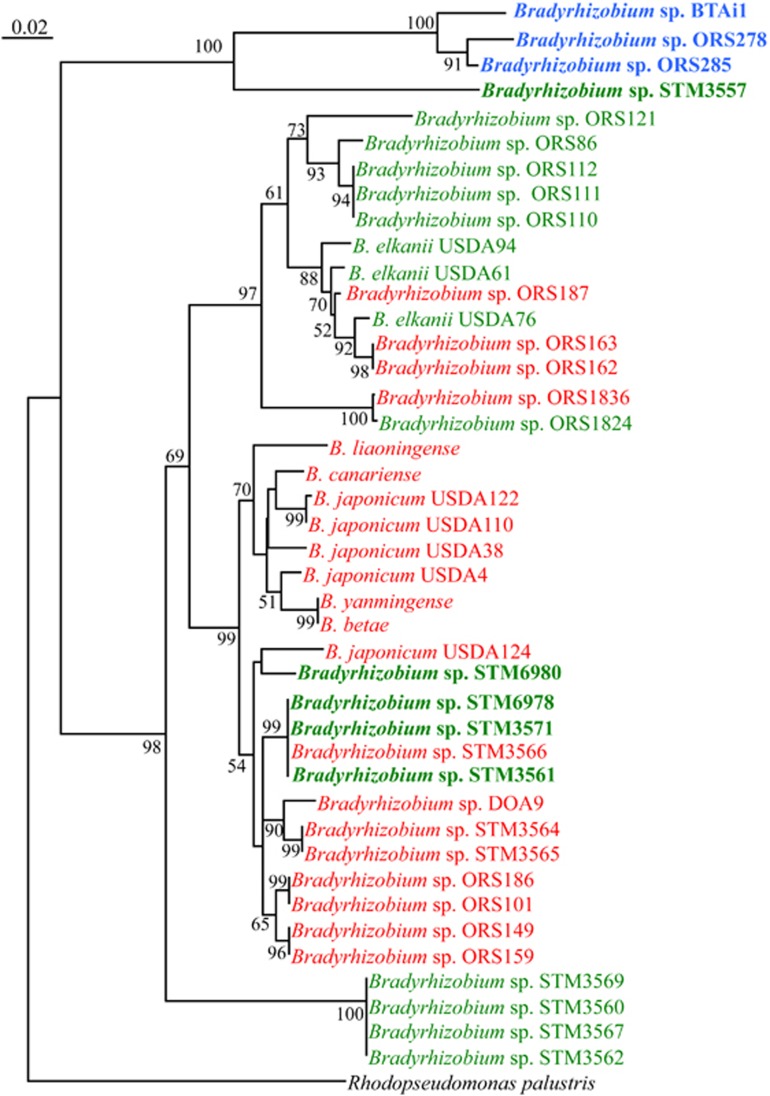
Figure 2.
The ability to induce nodules on NF-independent Aeschynomene species is widespread among Bradyrhizobium species. Neighbor joining phylogenetic relationship among the Bradyrhizobium strains tested for their ability to form nodules on A. indica based upon aligned sequences of 16S-23S rDNA IGS regions. Percent of bootstrap replicates (1000 replicates) are indicated at each tree node (only if >50%). In red, negative strain; in green, positive strains; in blue, photosynthetic Bradyrhizobium strains that are able to induce nitrogen-fixing nodules on A. indica; in bold, strains for which an intracellular infection is observed. A full colour version of this figure is available at the ISME journal online.
Some non-photosynthetic Bradyrhizobium strains induce nitrogen-fixing nodules on A. indica in a T3SS-dependent manner
Among the positive strains, several, like STM6978, induced on A. indica larger nodules than those observed with B. elkanii USDA61 (Figure 3e). These nodules displayed an irregular shape with several deformations suggesting perturbations in nodule development (Figures 3e and f). However, hand-sectioning of those nodules showed, similar as the control ORS278 strain, a central tissue with a light pink color suggesting the presence of leghemoglobin, an indicator of nitrogenase activity (Figures 3i and j). In agreement, acetylene reduction assays showed that some plants inoculated with some of these strains displayed a weak nitrogenase activity that represented at maximum (for strain STM6978) 10–15% of the one obtained with the ORS278 strain (Table 1). Although, this activity was not sufficient to support plant growth (Figure 3d), these data indicate that several non-photosynthetic Bradyrhizobium strains are able to establish a more intimate symbiotic relation with A. indica as compared with B. elkanii USDA61.
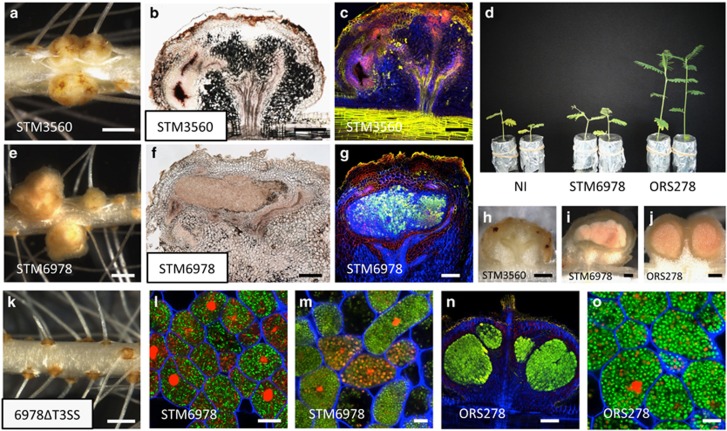
Figure 3.
Symbiotic phenotypes of Bradyrhizobium STM3560 and STM6978 on A. indica. (a and e) Nodules elicited by Bradyrhizobium STM3560 (a) and STM6978 (e). Bars, 1 mm. (b, f and h–j) Transversal section of nodules elicited by Bradyrhizobium STM3560 (b and h), STM6978 (f and i) and ORS278 (j). Bars, 200 μm. (d) Comparison of the growth of the plants (aerial part), non-inoculated (NI) or inoculated with STM6978, and Bradyrhizobium ORS278 (at 14 d.p.i.). (c, g and n) Confocal microscopy observation of nodules elicited by Bradyrhizobium STM3560 (c), STM6978 (g) and ORS278 (n). (l, m and o) Confocal microscopy images showing the morphology of the intracellular bacteria. Note that STM6978 bacteroids showed mainly a rod shape (l) and rarely a spheric shape (m) whereas the ORS278 bacteroids were all spherical (o). Live bacteria are stained green, whereas dead bacteria stain red owing to uptake of propidium iodide. Bars, 200 μm (c, g and n) and 10 μm (l, m and o). (k) A. indica roots inoculated with Bradyrhizobium STM6978ΔT3SS. Bar, 1 mm. A full colour version of this figure is available at the ISME journal online.
Using the live-dead staining protocol (Haag et al., 2011), a systematic cytological analysis was performed by confocal microscopy on the nodules elicited by all the positive strains. As observed previously for B. elkanii USDA61, some strains, such as STM3560, only infected the nodule tissue intercellularly and induced plant defense reactions (Figures 3b, c and h and Table 1). In contrast, other strains, such as STM6978, formed nodules with a central tissue completely infected by bacteria as observed for nodules formed by the ORS278 strain (Figures 3f, g and n and Table 1). The host cells were filled with viable bacteria as indicated by the green SYTO9 staining (Figures 3g and l). However, in contrast to nodules induced by the ORS278 strain, the morphological bacterial transition from a rod to a spherical shape during bacteroid differentiation (Figure 3o) was very rarely observed with the other Bradyrhizobium strains and when it occurred, the spherical bacteroids were stained red by propidium iodide, indicating that the bacteria were dead (Figure 3m).
We also tested whether the STM6978 strain could induce more efficient nodules on other Aeschynomene species. Of the 12 additional species tested, 7 were nodulated and for 4 of them, an intracellular invasion of plant cells could be observed (Supplementary Figure 2). However, only with one species, A. scabra, a nitrogenase activity could be detected. Similary as observed with A. indica, this activity remained weak and the same disorder in bacteroid differentiation was observed (Supplementary Figure 2). This indicates that although the STM6978 strain is recognized as friendly and internalized by the plant host cells in some Aeschynomene species, it is blocked at the bacteroid differentiation step which has been shown to go along with the establishment of nitrogenase activity (Bonaldi et al., 2011).
To confirm that the T3SS is required for the establishment of this more intimate symbiotic relationship, we constructed a T3SS mutant of the STM6978 strain. This mutant strain had completely lost its ability to induce nodules whatever the Aeschynomene species tested (Figure 3k).
The T3SS of the photosynthetic Bradyrhizobium ORS285 strain is not required for NF-independent symbiosis but contributes to the NF-dependent nodulation
Unlike other photosynthetic Bradyrhizobium strains with sequenced genomes, the ORS285 strain contains nod genes and a T3SS gene cluster (Mornico et al., 2011). The composition and organization of this T3SS cluster is exactly the same as found in B. elkanii and B. japonicum USDA110 suggesting that it is functional (Supplementary Figure 3). This raises the question whether this T3SS has a symbiotic role. To examine this, we constructed a 285ΔrhcN mutant and analyzed its symbiotic properties on various NF-dependent and NF-independent Aeschynomene species. On the three NF-independent species tested (A. indica, A. evenia and A. sensitiva), no effect of the T3SS mutation was observed. The number of nodules per plant, the morphology of the nodules and the nitrogenase activity of the plants were comparable between the WT strain and the T3SS mutant (Figure 4a). In contrast, on NF-dependent hosts plants, various effects of the ΔrhcN mutation were observed (Figures 4a and b): (i) no effect on A. nilotica, (Figures 4a–c and h), (ii) a positive effect on A. uniflora, that is, the mutant induced a higher number of nodules per plant which were more developed (Figures 4a, b, d and i)—however, like for the WT strain, nodules induced by the ΔrhcN mutant are not efficient in nitrogen fixation—and (iii) a negative effect on A. afraspera, that is, the mutant induced less nodules as compared with the WT strain (Figure 4a).
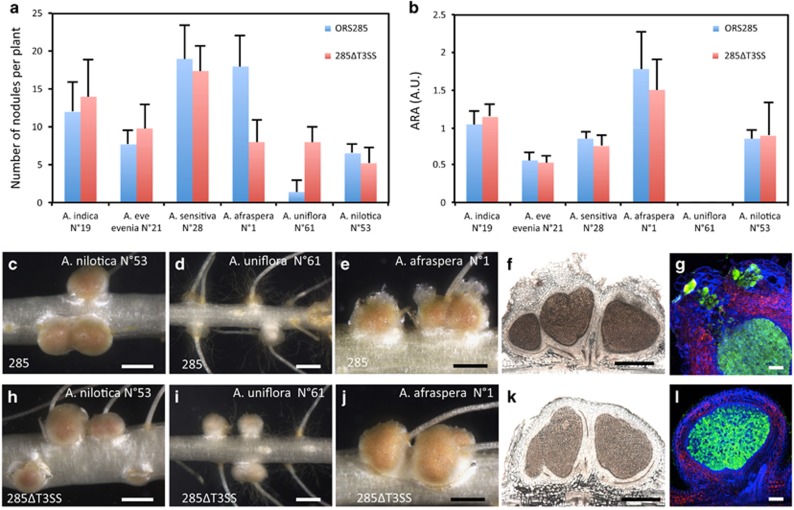
Figure 4.
Comparison of the symbiotic properties of Bradyrhizobium ORS285 and the ΔT3SS mutant on various NF-dependent and NF-independent Aeschynomene species. (a) Number of nodules per plant elicited by Bradyrhizobium ORS285 (blue bars) and ORS285ΔT3SS (red bars) on various Aeschynomene species. (b) Acetylene-reducing activity (ARA) in various Aeschynomene plants inoculated with Bradyrhizobium ORS285 (blue bars) and ORS285ΔT3SS (red bars). A.U., Arbitrary Unit. Error bars in a and b represent s.d. (n=5). (c–e, h–j). Aspect of the nodules elicited by ORS285 and 285ΔT3SS on A. nilotica (c and h), A. uniflora (d and i) and A. afraspera (e and j). Bars, 1 mm. (f–k, g–l) Cytological observations of the A. afraspera nodules elicited by ORS285 (f and g) and 285ΔT3SS (k and l). Bars, 500 μm (f and l) and 100 μm (g and l). A full colour version of this figure is available at the ISME journal online.
On A. afraspera plants, we previously noticed that the ORS285 strain induces atypical nodules characterized by the presence of an outgrowth on the top of the nodules which is constituted by giant plant cells infected by the bacteria (Bonaldi et al., 2011), (Figures 4e–g). In contrast to bacteria in the infected central tissue, the bacteria in the outgrowth remain enclosed in large tubular structures of plant origin that look like permanent infection threads (Bonaldi et al., 2011). Interestingly, besides forming fewer nodules, we observed that the nodules elicited by the 285ΔT3SS mutant lack this unusual infected tissue suggesting that its formation is dependent of the T3SS (Figures 4j–l). We also examined the ability of B. elkanii USDA61 and strain STM6978 to nodulate A. afraspera (Supplementary Figure 4). Both strains were able to induce efficient nodules on this species, but unlike ORS285 and similar to the 285ΔT3SS strain, these nodules lack this unusual outgrowth (Supplementary Figures 4C–E). In addition, inactivation of the T3SS in these two strains did not affect the interaction with A. afraspera (Supplementary Figures 4A and B).
Altogether, these data suggest that the ORS285 strain possesses a specific cocktail of T3SS effectors that can have an impact on the nodulation of NF-dependent hosts but that do not have a crucial role on the nodulation of NF-independent ones.
Discussion
In this study, we demonstrate that NF-independent Aeschynomene species can be nodulated and infected by highly diverse Bradyrhizobium strains via two distinct processes that differ by the requirement of a T3SS.
This observation is rather puzzling considering that in nature, mainly photosynthetic Bradyrhizobium strains, lacking a T3SS, were isolated from root and stem nodules of NF-independent Aeschynomene species (Lorquin et al., 1993; Molouba et al., 1999; Montecchia et al., 2002). Nevertheless, one study reported the isolation of a few non-photosynthetic Bradyrhizobium strains isolated from A. indica species growing in French Guiana (Miché et al., 2010). These strains efficiently induce nodules on both the root and the stem of Aeschynomene and occupy a distinct phylogenetic position between photosynthetic and classical bradyrhizobia. They may, thus, represent a new Bradyrhizobium species. However, genome sequence analysis of one representative strain (STM3843) revealed the absence of the canonical nodABC genes as well as a T3SS gene cluster (Mornico et al., 2011). This suggests that these bacteria use a NF-independent T3SS-independent process, probably similar to the one used by photosynthetic bradyrhizobia, to interact symbiotically with Aeschynomene plants. Therefore, it remains unclear whether the ability of non-photosynthetic bradyrhizobia to nodulate A. indica using a T3SS does exist in natural settings, considering the weak nitrogen fixation activity of the positive strains identified in this study in comparison with the photosynthetic ones.
Several observations emphasize a very high specificity of the NF-independent T3SS-dependent interaction: (i) despite the frequent presence of a T3SS among the rhizobia tested, only Bradyrhizobium strains are found to be able to induce nodules on Aeschynomene, (ii) similarly, not all of the tested Bradyrhizobium strains possessing a T3SS, such as Bradyrhizobium japonicum strains, are able to nodulate A. indica, (iii) the host spectra of B. elkanii and strain STM6978 towards NF-independent Aeschynomene species do not overlap perfectly and (iv) most surprisingly, different ecotypes of the same A evenia spp. evenia species respond differently to the Bradyrhizobium strains. To explain the high specificity of the T3SS-dependent interaction, some parallels with the conventional NF-dependent symbiosis can be advanced. The involvement of plant flavonoids to trigger the expression of the T3SS genes might constitute a first level of specificity. Indeed, it has been demonstrated in rhizobia that the expression of most T3SS genes encoding the transport machinery and secreted effectors is dependent on plant flavonoids which trigger a regulatory cascade controlled by the transcriptional activators NodD1, NodD2, SyrM2 and TtsI (Wassem et al., 2008). We examined this possibility during the interaction of B. japonicum USDA110 with A. indica or A. evenia by adding genistein to the plant culture medium, a flavonoid known to induce T3SS genes in this bacterium (Zehner et al., 2008). However, no effect of genistein was observed, suggesting that B. japonicum is hampered at another level to interact positively with these Aeschynomene species. A second possible level of specificity is the plant immune system that confers to the host the capacity to recognize and respond to bacterial infection (For review, Wang et al., 2012; Gourion et al., 2014). The perception of microbe-associated molecular patterns (MAMPs) by host pattern recognition receptors triggers MAMP-triggered immunity. It is suggested that rhizobial surface polysaccharides (exopolysaccharide, capsular polysaccharide or lipopolysaccharide) could suppress or help to evade this first basal defense (Fraysse and Couderc, 2003; D'Haeze and Holsters, 2004; Aslam et al., 2008). In addition, this MAMP-triggered immunity could be dampened by T3SS effectors delivered into the host cell. In turn, plants also have the ability to recognize these effector proteins through the direct or indirect action of resistance (R) genes to initiate a second barrier of defense, the so-called effector-triggered immunity. The effector-triggered immunity is much stronger and faster than MAMP-triggered immunity and often involves a form of localized cell death called the hypersensitive response (Dodds and Rathjen, 2010). The necrotic tissues observed in the A. indica nodules elicited by B. elkanii and others strains are reminiscent to hypersensitive response and could also be the result of T3SS effectors recognized by R gene products of the host. If that would be the case, the T3SS permits nodulation but, at the same time, inhibits the intracellular colonization by eliciting defense reactions in developed nodules. Taken together, all the different plant and bacterial actors susceptible to modulate the plant immune response could constitute key determinants governing this host specificity.
Besides interfering with the plant immunity system, we hypothesize that the non-photosynthetic Bradyrhizobium strains nodulating Aeschynomene also possess effectors able to activate the symbiotic signaling pathways. In conventional rhizobium–legume symbiosis, Nod factors activate two distinct spatial and temporal programs: bacterial infection and nodule organogenesis (Oldroyd et al., 2011). Among the non-photosynthetic bradyrhizobia able to induce nodules on Aeschynomene through their T3SS, two distinct classes could be distinguished: a first group, including B. elkanii strains, for which the infection remains limited to intercellular spaces, and a second group, including STM6978, for which the host cells are infected intracellularly. It is tempting to speculate that the bacteria of these two groups share in common some effectors that trigger the nodule organogenesis signaling pathway. On the other hand, bacteria of the second group could posses other specific effectors able to trigger an infection program or additional signals such as exopolysaccharide suggested to have a direct signaling role in bacterial release (Kelly et al., 2013).
Although we identified some non-photosynthetic Bradyrhizobium strains able to go very far in the symbiotic relationship with Aeschynomene, we observed that these bacteria are unable to fix nitrogen efficiently. This is surprising because all the conditions appear to be gathered in the nodule for an efficient nitrogen fixation. The host cells are filled with bacteria indicating that the plant furnished appropriate nutrients for bacterial multiplication and leghemoglobin synthesis is observed indicating that the partial oxygen tension should be suitable for a nitrogenase activity. However, our cytological observations indicate that these bacteria are compromised in the differentiation step as in contrast to photosynthetic Bradyrhizobium strains for which the rod-shaped bacteria transform into spherical bacteroids, this morphological change is rarely observed in the nodules infected with the non-photosynthetic Bradyrhizobium strains. Recent data suggested that Aeschynomene species impose a differentiation process on their endosymbionts through the use of a class of nodule-specific cysteine rich peptides (NCR) (Czernic P, personal communication), a modus operandi which is very similar to the one described in Medicago species (Van de Velde et al., 2010). NCRs are related to antimicrobial peptides of innate immunity that target the bacteria and convert them in a differentiated, enlarged and polyploid state (Van de Velde et al., 2010). It is possible that the impairment of non-photosynthetic Bradyrhizobium strains to differentiate in Aeschynomene results from a higher sensitivity of the bacteria to the NCR peptides. In accordance, the few bacterial cells displaying a spherical shape are dead as indicated by their red staining owing to propidium iodide uptake. Alternatively, it is also possible that the NCRs are not imported in the bacterial cell owing to the lack of a specific transporter or that the NCRs do not find the appropriate bacterial targets to trigger the differentiation process. Further studies are necessary to clarify the mode of action of the NCRs and to determine whether they constitute specificity determinants for the nitrogen fixation stage.
How the photosynthetic bradyrhizobia interact symbiotically with Aeschynomene plants remains unknown. Up to date, the different mutagenesis strategies (random or targeted) that have been used have failed in the isolation of a strict nodulation minus mutant (Giraud et al., 2007; Bonaldi et al., 2010). Furthermore, we do not have any experimental evidence indicating that a diffusible bacterial signal molecule triggers early symbiotic responses as observed for NFs. It is therefore tempting to speculate that the triggering of the symbiotic program involves a direct contact between the plant and the bacterium, probably through the perception of a bacterial surface component by a plant receptor. Increasing evidence now suggests that NFs perception might have evolved from plant innate immunity signaling and the possibility has been evocated that the first function of Nod factors was to suppress defense reaction (Liang et al., 2014). The presented results that non-photosynthetic bradyrhizobia can interact symbiotically in the absence of NFs but through a T3SS which is also known to interfere with the plant immunity reinforces the hypothesis that the suppression of the MAMP-triggered immunity constituted the first evolutionary step towards symbiosis. It therefore makes sense to hypothesize that the NF-independent symbiosis between photosynthetic bradyrhizobia and Aeschynomene involves the activation of a common symbiotic program but relies first on a specific mechanism of MAMP-triggered immunity suppression. Two elements led us suppose a co-evolution between photosynthetic Bradyrhizobium strains and NF-independent Aeschynomene species; (i) photosynthetic bradyrhizobia are the main natural symbionts of Aeschynomene and these bacteria were not found to be able to nodulate other legume species and (ii) both photosynthetic Bradyrhizobium strains and NF-independent Aeschynomene species form a monophyletic group (Chaintreuil et al., 2013). We therefore hypothesize that the co-evolution between these two partners has involved the modification of a specific MAMP signal and its cognate plant receptor to avoid the development of plant defense reactions during the interaction.
The deciphering of the molecular basis of these two alternative NF-independent symbiotic processes should shed new light on the evolution of the rhizobium-legume interaction and might deepen our knowledge on how the plant evolved symbiosis from pathogen resistance mechanisms.
Acknowledgments
This work was supported by grant from the French national research agency (ANR-BugsInaCell-13-BSV7-0013) and the Franco-Thai PHC Siam program (project SIAM N°29589XA). We thank P Mergaert (ISV, CNRS, Gif-sur-Yvette, France) for criticisms and corrections of the manuscript. We thank Tatiana Krassova-Wade (LCM, IRD, Dakar, Senegal) for providing some Bradyrhizobium strains.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aslam SN, Newman MA, Erbs G, Morrissey KL, Chinchilla D, Boller T et al. (2008). Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol 18: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Bonaldi K, Gargani D, Prin Y, Fardoux J, Gully D, Nouwen N et al. (2011). Nodulation of Aeschynomeneafraspera and A. indica by photosynthetic Bradyrhizobium sp. strain ORS285: the Nod-dependent versus the Nod-independent symbiotic interaction. Mol Plant Microbe Interact 24: 1359–1371. [DOI] [PubMed] [Google Scholar]
- Bonaldi K, Gourion B, Fardoux J, Hannibal L, Cartieaux F et al. (2010). Large-scale transposon mutagenesis of photosynthetic Bradyrhizobium sp. strain ORS278 reveals new genetic loci putatively important for Nod-independent symbiosis with Aeschynomene indica. Mol Plant Microbe Interact 23: 760–770. [DOI] [PubMed] [Google Scholar]
- Chaintreuil C, Arrighi JF, Giraud E, Miché L, Moulin L, Dreyfus B et al. (2013). Evolution of symbiosis in the legume genus Aeschynomene. New Phytol 200: 1247–1259. [DOI] [PubMed] [Google Scholar]
- D'Haeze W, Holsters M. (2004). Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol 12: 555–561. [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 424–429. [DOI] [PubMed] [Google Scholar]
- Fraysse N, Couderc F, Poinsot V. (2003). Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur J Biochem 270: 1365–1380. [DOI] [PubMed] [Google Scholar]
- Gage DJ. (2002). Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol 184: 7042–7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre JC et al. (2007). Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316: 1307–1312. [DOI] [PubMed] [Google Scholar]
- Gourion B, Berrabah F, Ratet P, Stacey G. (2014). Rhizobium-legume symbioses: the crucial role of plant immunity. Trends Plant Sci pii: S1360–S1385 (14)00297-0. [DOI] [PubMed] [Google Scholar]
- Gueye F, Moulin L, Sylla S, Ndoye I, Béna G. (2009). Genetic diversity and distribution of Bradyrhizobium and Azorhizobium strains associated with the herb legume Zornia glochidiata sampled from across Senegal. Syst Appl Microbiol 32: 387–399. [DOI] [PubMed] [Google Scholar]
- Haag AF, Baloban M, Sani M, Kerscher B, Pierre O, Farkas A et al. (2011). Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol 9: e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62: 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ, Muszyński A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N et al. (2013). Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosis. Mol Plant Microbe Interact 26: 319–329. [DOI] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC et al. (1990). Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature 344: 781–784. [DOI] [PubMed] [Google Scholar]
- Liang Y, Tóth K, Cao Y, Tanaka K, Espinoza C, Stacey G. (2014). Lipochitooligosaccharide recognition: an ancient story. New Phytol 204: 289–296. [DOI] [PubMed] [Google Scholar]
- Lorquin J, Molouba F, Dupuy N, Ndiaye S, Alazard D, Gillis M et al Diversity of photosynthetic Bradyrhizobium strains from stem nodules of Aeschynomene species. In Palacios R, Mora J, Newton WE (Eds). (1993) New Horizons in Nitrogen Fixation. Boston: Kluwer, 17: 683–689. [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS et al. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miché L, Moulin L, Chaintreuil C, Contreras-Jimenez JL, Munive-Hernández JA, Del Carmen Villegas-Hernandez M et al. (2010). Diversity analyses of Aeschynomene symbionts in Tropical Africa and Central America reveal that Nod-independent stem nodulation is not restricted to photosynthetic bradyrhizobia. Environ Microbiol 12: 2152–2164. [DOI] [PubMed] [Google Scholar]
- Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B et al. (1999). Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16 S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol 65: 3084–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecchia MS, Kerber NL, Pucheu NL, Perticari A, García AF. (2002). Analysis of genomic diversity among photosynthetic stem-nodulating rhizobial strains from northeast Argentina. Syst Appl Microbiol 25: 423–433. [DOI] [PubMed] [Google Scholar]
- Mornico D, Miché L, Béna G, Nouwen N, Verméglio A, Vallenet D et al. (2011). Comparative genomics of Aeschynomene symbionts: insights into the ecological lifestyle of Nod-independent photosynthetic bradyrhizobia. Genes 3: 35–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Takebe L. (1970). Cell wall regeneration and cell division in isolated tobacco mesophyll protoplasts. Planta 92: 301–308. [DOI] [PubMed] [Google Scholar]
- Okazaki S, Kaneko T, Sato S, Saeki K. (2013). Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc Natl Acad Sci USA 110: 17131–17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45: 119–144. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Saitou RR, Nei M. (1987). A neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 44: 406–425. [DOI] [PubMed] [Google Scholar]
- Tampakaki AP. (2014). Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front Plant Sci 5: 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Higgins DG. (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2: Unit 2.3. [DOI] [PubMed] [Google Scholar]
- Van de Velde W, Zehirov G, Szatmari A, Debreczeny M, Ishihara H, Kevei Z et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126. [DOI] [PubMed] [Google Scholar]
- Vincent JM. (1970) A Manual for the Practical Study of Root-Nodule Bacteria. Blackwell Scientific Publications: Oxford, United Kingdom. [Google Scholar]
- Wang D, Yang S, Tang F, Zhu H. (2012). Symbiosis specificity in the legume: rhizobial mutualism. Cell Microbiol 14: 334–342. [DOI] [PubMed] [Google Scholar]
- Wassem R, Kobayashi H, Kambara K, Le Quéré A, Walker GC, Broughton WJ et al. (2008). TtsI regulates symbiotic genes in Rhizobium species NGR234 by binding to tts boxes. Mol Microbiol 68: 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KJ, Sessitsch A, Corbo JC, Giller KE, Akkermans AD, Jefferson RA. (1995). β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology 141: 1691–1705. [DOI] [PubMed] [Google Scholar]
- Zehner S, Schober G, Wenzel M, Lang K, Göttfert M. (2008). Expression of the Bradyrhizobium japonicum type III secretion system in legume nodules and analysis of the associated tts box promoter. Mol Plant Microbe Interact 21: 1087–1093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.