Abstract
During antibiotic treatment, antibiotic concentration gradients develop. Little is know regarding the effects of antibiotic gradients on populations of nonresistant bacteria. Using a microfluidic device, we show that high-density motile Escherichia coli populations composed of nonresistant bacteria can, unexpectedly, colonize environments where a lethal concentration of the antibiotic kanamycin is present. Colonizing bacteria establish an adaptively resistant population, which remains viable for over 24 h while exposed to the antibiotic. Quantitative analysis of multiple colonization events shows that collectively swimming bacteria need to exceed a critical population density in order to successfully colonize the antibiotic landscape. After colonization, bacteria are not dormant but show both growth and swimming motility under antibiotic stress. Our results highlight the importance of motility and population density in facilitating adaptive resistance, and indicate that adaptive resistance may be a first step to the emergence of genetically encoded resistance in landscapes of antibiotic gradients.
Introduction
Nonresistant bacteria that are refractory to antibiotic treatment are thought to have an important role in the recalcitrance of bacterial infections (Dhar and McKinney, 2007; Mulcahy et al., 2010; Fauvart et al., 2011; Cohen et al., 2013; Lebeaux et al., 2014). The mechanisms underlying the non-inherited antibiotic refractoriness of bacteria are diverse, and only partially understood (Hogan and Kolter, 2002; Lewis, 2010; Balaban et al., 2013; Orman and Brynildsen, 2013). Some forms of tolerance to lethal doses of antibiotics, for example, persistence, are associated with a strongly reduced metabolic activity, whereas others rely on active responses that confer (collective) adaptive resistance (Balaban et al., 2004; Butler et al., 2010; Lewis, 2010; Nguyen et al., 2011; Wakamoto et al., 2013; Meredith et al., 2015). Little is known regarding the role of antibiotic concentration gradients in facilitating the survival of nonresistant bacteria. Antibiotic concentration gradients develop during treatment because of the tissue-dependent drug-penetration and periodic administration of the drug (Mukhopadhyay et al., 1994; Kaiser et al., 2014). Such gradients may act as safe havens for susceptible bacteria by providing regions that (transiently) support growth (Baquero and Negri, 1997; Fernández et al., 2011; Meredith et al., 2015). In addition to clinical settings, antibiotic gradients also arise in, for example, soil communities where narrow-spectrum antibiotics are locally released by antibiotic-producing bacteria establishing short-lived gradients (Romero et al., 2011; Vetsigian et al., 2011; Hol et al., 2014).
To investigate the influence of antibiotic gradients on populations of nonresistant bacteria, we devised a well-controlled two-compartment antibiotic landscape in which nonresistant Escherichia coli bacteria face a concentration gradient of the aminoglycoside kanamycin, which increases from 0- to 50-fold of the minimum inhibitory concentration (MIC) over a range of 2 mm. Strikingly, we observed that motile nonresistant E. coli bacteria can actively colonize environments where the kanamycin concentration is well above the lethal concentration (50-fold the MIC) and establish a viable population there.
Materials and methods
Strains and growth conditions
The strains used in this study (JEK1036 and JEK1037) have been described before in Keymer et al. (2008) and Hol et al. (2013). Both strains are E. coli W3110, JEK1036 is labeled with lacYZ::GFPmut2 (green fluorescent protein) and JEK1037 is labeled with lacYZ::mRFP (red fluorescent protein), respectively. Five out of ten experiments were performed with strain JEK1036 only, three experiments were performed with a 50/50 mix of both strains and two experiments were performed with JEK1037 only. Before all experiments, cells were taken from a −80 °C glycerol stock and grown overnight (37 °C, 200 r.p.m.) in lysogeny broth (LB). Cells were back diluted 1/100 in LB supplemented with 100 μM β-d-1-thiogalactopyranoside and inoculated into the microfluidic device at mid-log phase.
Fabrication and preparation of the microfluidic device
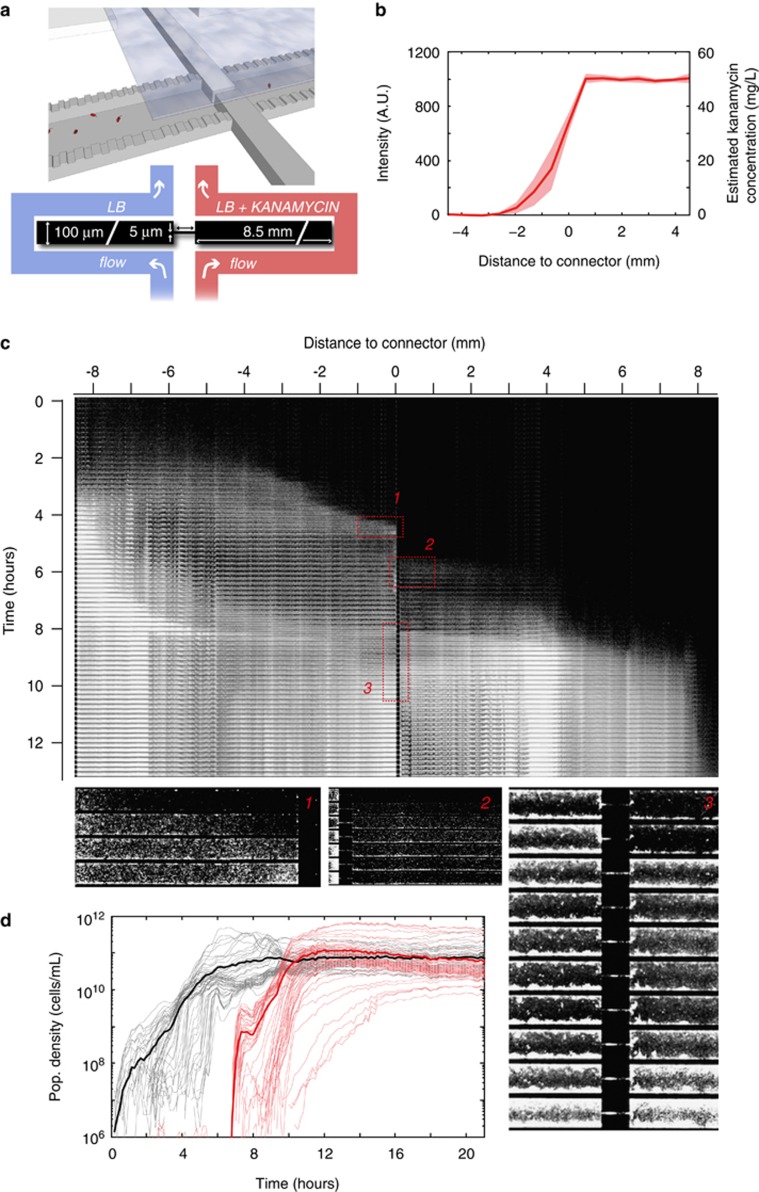
Devices were fabricated in silicon following a previously published protocol (Keymer et al., 2006; Hol et al., 2014). The devices consist of two compartments each having a volume of 8.5 nl (8500 × 100 × 10 μm3) connected by a corridor (100 × 5 × 10 μm3). The compartments are connected to separate flow channels (300 μm wide, 10 μm deep) by 180 nm shallow slits (Figure 1a). Each compartment is connected to the flow channels by 770 slits that are 20 μm wide and spaced 2 μm apart. The shallow slits prevent bacteria from entering the flow channels but allow the diffusion of, for example, fresh medium, antibiotics and waste. A port to inoculate the bacteria was drilled through the silicon at the end of the antibiotic-free compartment opposing the connector. Microhabitats were closed by bonding to polydimethylsiloxane-coated coverslips. Ports to connect tubing to the flow channels were drilled through the silicon, and PEEK tubing was connected using custom-made clamps. A syringe pump was used to flow medium at 3 μl per hour, refreshing the entire volume of the flow channels approximately every 20 min. Fresh LB medium supplemented with 100 μM β-d-1-thiogalactopyranoside flowed through the channel connected to the antibiotic-free compartment, fresh LB medium supplemented with 100 μM β-d-1-thiogalactopyranoside, 50 mg l−1 kanamycin and 2.5 mg l−1 Alexa 350 dye flowed through the channel connected to the kanamycin compartment. To ensure a stable kanamycin gradient, bacteria were inoculated after at least 18 h of steady flow. To assess the concentration profile of Alexa 350, the gradient was imaged before inoculation of the bacteria and again ~2 h after inoculation. During later phases of the experiment, the stability of the gradient was verified by visual inspection every 6–10 h. To estimate the kanamycin concentration, the intensity of the blue fluorescent channel (Alexa 350 emits at ∼450 nm) was measured near the inlet of the kanamycin flow channel (intensity corresponding to the maximum kanamycin concentration of 50 mg l−1) and near the inlet of the antibiotic-free channel (intensity corresponding to no kanamycin). Alexa 350 and kanamycin have comparable molecular weights and therefore are expected to have similar diffusion constants. A linear relation between the intensity of the Alexa dye and the kanamycin concentration was assumed. Figure 1b shows the concentration profile measured 2 h after inoculation of the bacteria and indicates the variation between experiments. After each experiment, we tested the activity of the used kanamycin medium. When an experiment was terminated, the remaining medium in the LB+kanamycin syringe was inoculated with wild-type E. coli bacteria and incubated at 37 °C. This tube was monitored for growth for 2 days. Growth was never observed, demonstrating that kanamycin was indeed present at bactericidal concentrations for the full duration of the experiment.
Figure 1.
Nonresistant E. coli bacteria in an antibiotic gradient. (a) Cartoon and schematic of the two-compartment microfluidic device. The two compartments (8500 × 100 × 10 μm3 each) are connected by a corridor (100 × 5 × 10 μm3). The device is sealed from the top using a polydimethylsiloxane-coated coverslip (displayed partially in the cartoon). Both compartments are connected to separate flow reservoirs by 180 nm shallow, 20 μm wide slits (770 slits per compartment). The slits facilitate the diffusion of continuously flowing fresh medium into the compartments (medium is supplemented with kanamycin in the right compartment). Diffusion of kanamycin through the connecting corridor establishes a concentration gradient. (b) A fluorescent tracer (Alexa 350, left y axis) is used to estimate the kanamycin concentration profile (right y axis). The line shows the average concentration profile of four independent experiments, the shaded area represents the standard deviation. (c) Kymograph showing the population dynamics of a typical experiment. The entire device is imaged every 15 min, and images are stacked vertically to show the population dynamics. The intensity of fluorescently labeled cells is scaled logarithmically. Migrating populations can be seen as laterally moving fronts. Magnified area 1 shows a migrating population that arrests movement when it reaches the connector. Approximately 2 h later, a population invades the kanamycin compartment as shown in magnified area 2. This low-density invasion does not establish a growing population. A subsequent high-density population at t=8.25 h, however, does. Magnified area 3 shows the connector region during this invasion, see Supplementary Movie 1 for a movie of the connector region. (d) Population density curves in the antibiotic-free (black) and kanamycin (red) compartments. Thin lines show local growth measured at a spatial resolution of 130 × 100 × 10 μm3 voxels; fat lines show the average population density for both compartments. Note that the maximum population density in both compartments is similar.
Image acquisition and analysis
Microfluidic devices were imaged at 15 min intervals using an inverted Olympus IX81 microscope equipped with a x20 (numerical aperture=0.75) objective, a Neo sCMOS camera (Andor, Belfast, UK) and a motorized stage (Marzhauser, Wetzlar, Germany) controlled using μManager software (Edelstein et al., 2010). The sample was illuminated using an X-cite 120 Q (Lumen Dynamics, Mississauga, ON, Canada) light source. The microscope was enclosed in an environmental chamber warmed to 37 °C. Images were processed in MatLab (Mathworks, Natick, MA, USA) using a custom script. Particle tracking to obtain swimming trajectories was performed in ImageJ (NIH, Bethesda, MD, USA).
Estimation of population density in the microfluidic device
Background subtraction was performed on all images to set the pixel values of areas not occupied by bacteria to zero. To estimate population density, the average total fluorescence intensity of individual cells was calculated by taking advantage of the fact that single cells can be distinguished in the initial phase of the experiment when population density is low. A custom Matlab script was used to determine the average intensity of several thousands of individual cells during the low-population-density phase. The average total intensity of an individual cell was determined separately for each experiment. Using the obtained average total intensity per cell, the number of cells per voxel (130 × 100 × 10 μm3) was estimated by dividing the total background-corrected fluorescence intensity of a voxel by the determined average total intensity per cell. We verified that there was no significant difference between the brightness of bacteria in the kanamycin compartment versus the antibiotic-free compartment by comparing the intensity distribution of 364 cells in the kanamycin compartment with the intensity distribution of 649 cells in the antibiotic-free compartment (P>0.05 Wilcoxon ranks sum test).
Isolation of bacteria from the microfluidic device
After image acquisition, the device was taken off the microscope stage and the antibiotic-free and antibiotic-containing compartments were separated using a wafer dicer. The antibiotic compartment was diced in small fragments, which were placed in a Falcon tube containing 1 ml fresh liquid LB medium (without kanamycin). Tubes were vortexed and incubated for 30 min. Subsequently, bacteria were harvested by plating 200 μl aliquots on fresh LB agar plates (without kanamycin), which were incubated overnight. Up to 23 single colonies were randomly selected for MIC determination and downstream analysis.
Determination of the MIC
The MIC of cells isolated from the kanamycin compartment was determined using the eTest strip assay (bioMerieux, Marcy l'Étoile, France) following the manufacturer's directions. The ancestral wild-type E. coli strain had an MIC of 1 mg l−1; cells isolated from microfluidic devices 15, 18 and 29 h post invasion also had an MIC of 1 mg l−1. In the experiment where cells were isolated 61 h after invading the kanamycin compartment, a mutant emerged that had an MIC of 25 mg l−1. The MIC of the resistant mutant and wild-type E. coli (ancestor) was verified in liquid culture medium using a dilution assay in a 96-well format. We inoculated four replicates of the wild-type ancestral strain, and two replicates of three clones originating from the single experiment in which resistance emerged, in 200 μl LB medium containing 0, 10, 20, 25 and 50 mg l−1 kanamycin, respectively. After incubation overnight at 37 °C (shaken at 500 r.p.m.), all replicates of all clones originating from the experiment in which a resistant mutant emerged showed growth in 0, 10, 20 and 25 mg l−1 kanamycin (not in 50 mg l−1), whereas all replicates of the ancestral strain only showed growth in 0 mg l−1 kanamycin (not in 10, 20, 25 and 50 mg l−1).
Whole-genome sequencing of the resistant mutant
Whole-genome sequencing was performed on an Illumina MiSeq machine (Illumina, San Diego, CA, USA) using 300 bp paired-end reads. Using the Illumina Nextera XT DNA sample preparation kit (Illumina), DNA libraries were prepared for the pooled sequencing of nine strains. The nine pooled samples included: one clone of the ancestral strain (JEK1036), one clone isolated from each of the three experiments in which resistance did not emerge and five clones originating from the single experiment in which resistance did emerge. The obtained reads were mapped onto the E. coli W3110 reference genome using the in-house developed software Antonie (Antonie is free open source software available at: http://ds9a.nl/antonie/). The average read depth of the nine samples was 105 (range: 80–137). Single-nucleotide polymorphisms were called when nucleotides differed from the ancestral genome in at least 50% of the reads and had a read depth of at least 40. Using these criteria, a C to T mutation in cydA (locus 772891, gi 388476123, NC_007779.1) was found in all five clones that were isolated during the experiment, in which resistance emerged, no other single-nucleotide polymorphisms were detected in any of the experimental isolates. In the five mutant clones, the C to T mutation was present in, respectively, 108/108, 172/173, 114/114, 119/119 and 189/189 reads at position 772 891. The C to T mutation in cydA was never observed (that is, all reads showed a C) in the ancestral wild-type strain, or in any of the three sequenced clones isolated during experiments in which resistance did not emerge. All sequences have been deposited under the study number PRJEB7521 at the European Nucleotide Archive.
Results and Discussion
To study the population dynamics of nonresistant E. coli bacteria in the presence of an antibiotic gradient in a controlled manner, we microfabricated an antibiotic landscape consisting of two compartments (each 8500 × 100 × 10 μm3) filled with liquid medium and connected by a corridor (100 × 5 × 10 μm3) that facilitates cellular migration between the compartments. Using microfluidic flow channels, fresh LB medium is continuously supplied to one compartment, whereas LB medium supplemented with a lethal dose of kanamycin (50 mg l−1) is continuously supplied to the other (see Figure 1a for a schematic of the microfluidic device). Diffusion of kanamycin through the corridor connecting the compartments creates a concentration gradient increasing from 0 to 50 mg l−1 (∼50 × MIC) over a region of ∼2 mm centered on the connector (Figures 1a and b). The device constitutes a well-defined antibiotic landscape in which bacterial population dynamics can be tracked at high spatiotemporal resolution using fluorescence microscopy (Keymer et al., 2006; Hol et al., 2014; Hol and Dekker, 2014). After each experiment, bacteria can be isolated from the device for further analyses.
Nonresistant E. coli bacteria can colonize a lethal antibiotic gradient
We performed 10 independent experiments in which fluorescently labeled E. coli were inoculated into the antibiotic-free compartment at the side opposite to the connector (far left in Figures 1a and c). Figure 1c shows a kymograph (depicting space horizontally and time vertically) of the typical population dynamics of an experiment. After inoculation, E. coli swim into the habitat and establish a population in the antibiotic-free compartment. This population grows, and occasionally gives rise to collectively migrating population waves, visible as laterally migrating fronts in the kymograph (migrating population waves in microfabricated habitats are extensively discussed in, for example, van Vliet et al., 2014). The first population wave visible in Figure 1c halts when it reaches the connector separating the two compartments (t≈4.5 h, magnified area 1 in Figure 1c). Approximately 2 h later, a group of cells swims through the connector and invades the antibiotic zone (t≈6 h, Figure 1c, magnified area 2). This invasion consists of a planktonic subpopulation of ~5 × 103 cells, which disperse into the antibiotic compartment. The invading cells attach to the surface, and subsequently show no growth or motility. As kanamycin gains its bactericidal action from inhibiting protein synthesis, cells that are affected by kanamycin do not necessarily lyse. Although some of the cells that originated from this first invasion remain visible in the antibiotic compartment for an extended period, these cells do not show growth and apparently are not viable.
Several hours later (t≈8.25 h, magnified area 3 in Figure 1c), a subsequent population wave, which is an order of magnitude denser (5 × 104 cells, peak density 3.3 × 1010 cells per ml), invades the antibiotic compartment. Surprisingly, this invading population establishes a sizable population in the antibiotic compartment, which even expands while exposed to the lethal concentration of kanamycin. Figure 1d (red curve) shows that after the successful invasion of the antibiotic compartment, the population sustains growth for ~4 h and reaches a population density comparable to the population density in the antibiotic-free compartment (the black and red lines in Figure 1d saturate at the same level). A successful colonization of the antibiotic compartment was observed in 4 out of 10 independent experiments (Supplementary Movies 1 and 2 show two examples of successful invasions); whereas in 2 out of 10 independent experiments population waves did invade the antibiotic compartment, yet failed to establish a growing population there; in the remaining 4 out of 10 experiments, no invasions of the antibiotic compartment occurred.
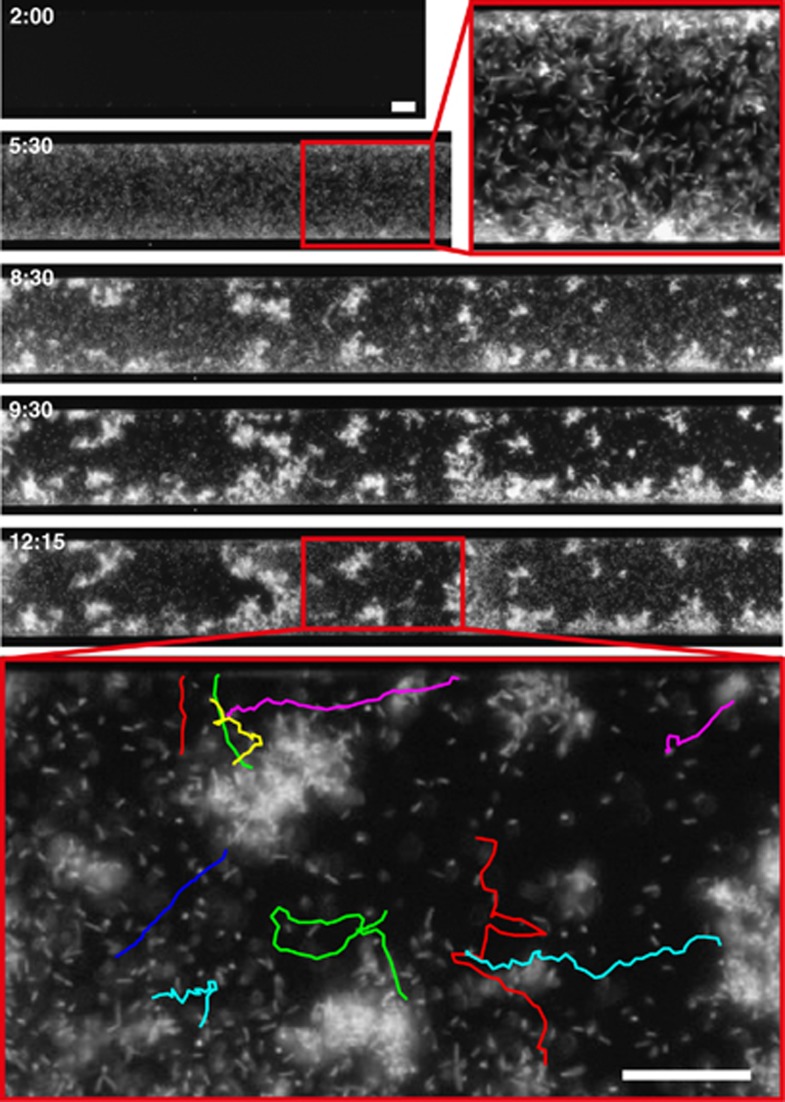
In all four experiments where the antibiotic compartment was successfully invaded, the invading population remained viable for days while exposed to kanamycin and reached local population densities equal to the population density in the antibiotic-free compartment. Furthermore, individual planktonic cells could often be distinguished swimming vigorously through the antibiotic compartment, as can be seen in Figure 2 and Supplementary Movie 3. The observation of individual swimming cells, together with the observed population growth, shows that the invading bacteria do not adopt a dormant persister state but remain active and even attain high densities, similar to the population in the antibiotic-free compartment. Planktonic bacteria generally have a lower tolerance for antibiotics when compared with their surface-associated counterparts. It is therefore remarkable that in addition to surface-associated growth, viable planktonic bacteria can be observed in the kanamycin compartment.
Figure 2.
E. coli aggregate formation and motility in the kanamycin compartment. Snapshots (labeled in hours, top left) of a successful colonization of the kanamycin compartment (Supplementary Movie 2). At t=5:30 h, a high-density population of planktonic cells (maximum density 4.75 × 101 cells per ml) invades the kanamycin compartment. At later time points, cells aggregate and grow in a surface-associated manner. Even after hours of antibiotic exposure, individual planktonic cells can be observed swimming vigorously between cellular aggregates. The bottom panel shows single-cell swimming trajectories (8 s, see Supplementary Movie 3). Scale bars are 25 μm.
Invading population waves need to exceed a critical population density to successfully colonize the antibiotic compartment
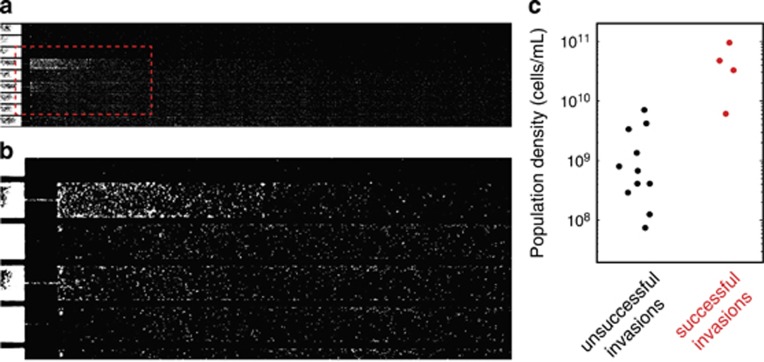
Next, we hypothesized that the density of the invading population could be a decisive factor in determining whether an invading population establishes a viable and growing population in the antibiotic compartment. To test this, we measured the maximum local population density of invading population waves. Figure 3c shows the measured population densities of invading waves that did (‘successful invasions', red dots) and waves that did not (‘unsuccessful invasions', black dots) establish a viable and growing population in the antibiotic compartment. The data show a striking contrast between successful and unsuccessful invasions: all invading populations that established a population in the kanamycin compartment had a density >5 × 109 cells per ml, conversely, the 11 low-density invasions that did not lead to the successful establishment of a population in the antibiotic compartment all had lower densities (that is, ≲5 × 109 cells per ml). The unsuccessful invasions occurred both as low-density invasions preceding a successful high-density invasion (in the four experiments in which the kanamycin compartment was colonized), and in the two experiments where the kanamycin compartment was invaded but not successfully colonized. This observation suggests that to successfully invade the kanamycin compartment, the invading E. coli population has to reach a threshold density. Only migrating populations that exceed this critical density can establish a population in the presence of the antibiotic.
Figure 3.
Invasion success is density dependent. (a) A low-density population invades the kanamycin compartment but fails to establish a growing population there. (b) Magnification of the area indicated with the red dashed line in a. (c) Maximum population density of invasions. Black dots represent invasions that did not establish a population in the kanamycin compartment (cf. a), whereas red dots represent invasions that successfully colonized the kanamycin compartment. These data show that a critical population density is required for a successful colonization of the kanamycin compartment. The densities of unsuccessful and successful invasions are significantly different (P=0.003 using a Wilcoxon rank sum test).
Colonization of the antibiotic compartment is not caused by genetically encoded resistance
To assess whether successful invasions are caused by the acquisition of genetically encoded resistance within the invading populations, we isolated a sample of the population that colonized the kanamycin compartment and measured their MIC (see Materials and methods section for details). Strikingly, in three out of the four independent experiments in which a growing population was established in the antibiotic compartment, the MIC of the bacteria isolated from the kanamycin compartment was identical to the MIC as measured before the experiment. In other words, despite the fact that the invading cells are sensitive to kanamycin, they are able to establish a viable and growing population in the presence of a lethal dose of kanamycin. The unaltered MIC indicates that the kanamycin resistance of the invading populations is caused by an adaptive mechanism conferring phenotypic resistance, instead of being genetically encoded. Most previously described antibiotic-tolerant phenotypes are based on dormancy (Balaban et al., 2004; Lewis, 2010; Fridman et al., 2014) or reported the transient resistance of matured biofilms that were formed in the absence of antibiotics and exposed only after maturation (Spoering and Lewis, 2001; Anderl et al., 2000; Stone et al., 2002; Amini et al., 2011; Nguyen et al., 2011; Kirby et al., 2012). In contrast, the density-dependent adaptive resistance observed here enables planktonic E. coli to found a population in an antibiotic landscape, and permits swimming motility and population expansion in the presence of a lethal concentration of the antibiotic.
This type of adaptive resistance is reminiscent of the non-genetic resistance that some bacterial species exhibit when engaging in collective swarming motility (Kim et al., 2003; Overhage et al., 2008; Butler et al., 2010; Roth et al., 2013). Nonresistant swarming Salmonella enterica populations were previously shown to migrate from an antibiotic-free agar surface onto an agar surface containing lethal doses of an antibiotic (Butler et al., 2010). The internal structure of swarming populations was suggested to facilitate swarming-based adaptive resistance (Butler et al., 2010), as only the bottom layer of a multilayered swarm is exposed to the antibiotic and may thus shield higher layers. Our observations are different in this respect as they concern swimming motility in which bacteria migrate in the liquid antibiotic-containing medium, rather than on top of it. We therefore do not expect shielding to have a major effect in swimming populations. Although the detailed mechanism underlying swarming adaptive resistance remains elusive, it was shown to crucially depend on local population density and motility. We observed similar dependencies in our experiments, which indicates that swarming motility may not be essential and suggests that a more general mechanism may underlie the adaptive resistance of mobile high-density bacterial populations.
Both the adaptively resistant swarming populations described previously (Butler et al., 2010), and the swimming populations described here, attain population densities at least an order of magnitude higher than their broth-grown counterparts in shaken flask (maximum densities in our system are similar to those in the human gut; Guarner and Malagelada, 2003). Spatially structured habitats furthermore allow swarming and swimming populations to achieve these high densities locally, whereas population growth can be sustained through spatial expansion into less dense regions. The combination of very high population density and simultaneous growth is not possible in unstructured environments like shaken flasks, where saturating densities (by definition) do not leave room for growth and spatial heterogeneities in density are abolished by mixing. It is thus possible that spatially structured environments facilitate adaptive resistance by allowing high-density populations to form in permissive environments, which subsequently emit migrant subpopulations that invade a harsh environment after having reached a critical density (Patra and Klumpp, 2014).
Antibiotic exposure of an adaptively resistant population may facilitate the emergence of heritable resistance
In the four independent experiments in which the kanamycin compartment was successfully colonized, bacteria were isolated from the kanamycin compartment, respectively, 15, 18, 29 and 61 h after the invasion. Solely, the experiment in which cells were isolated 61 h after the antibiotic compartment was invaded yielded a resistant mutant; the other three experiments in which bacteria were isolated earlier did not. Whole-genome sequencing of the resistant mutant revealed a C to T point mutation at locus 772891 (gi 388476123, NC_007779.1) in the coding sequence for cydA. The product of the cydA gene is part of the cytochrome bd-I terminal oxidase complex, which functions as the high-affinity terminal oxidase of the oxygen respiratory chain (Govantes et al., 2000). Kanamycin uptake requires a proton-motive force (Taber et al., 1987), and it was previously shown that a cydA knockout mutant has an increased survival upon aminoglycoside treatment because of a reduced proton-motive force (Allison et al., 2011). As the C to T mutation changes a glutamine residue into a stop codon, it is likely that a similar loss of function is responsible for the increased MIC of the mutant that emerged in our experiment.
Measuring the MIC of the isolated mutant showed a 25-fold increase of the MIC compared with the ancestral strain used to inoculate the habitat. The MIC of the bacteria isolated 15, 18 and 29 h post invasion was identical to that of the ancestral wild-type E. coli strain. It is plausible that in the experiment where a resistant mutant was isolated 61 h after the invasion, prolonged exposure of an initially adaptively resistant population led to the emergence of the mutant. This finding highlights the importance of adaptive resistance as a route for bacteria to acquire heritable resistance. The hypothesis that adaptively resistant bacteria serve as an evolutionary reservoir from which genotypic mutants may emerge has recently gained ground (Cohen et al., 2013). An antibiotic gradient or the rudimentary version of that—a two patch source-sink system—may act on such a reservoir and facilitate the evolution of resistance. In contrast to a spatially uniform system, a heterogeneous landscape may constitute an effective filter for genetically resistant mutants to evolve (Hermsen and Hwa, 2010; Hermsen et al., 2012), or for adaptively resistant subpopulations to emerge, from the phenotypic heterogeneity present in a clonal population inhabiting a nonuniform environment (Patra and Klumpp, 2014).
Our results, furthermore, suggest that the observation of population growth in landscape zones where antibiotics are present at concentrations above the MIC may not be an accurate proxy to accurately time the evolution of (genetically encoded) antibiotic resistance as has been reported previously (Zhang et al., 2011), because it does not allow one to distinguish non-inheritable adaptive resistance from genetically encoded resistance. Time-resolved MIC measurements will be necessary to support such observations.
Summing up, we have shown that high-density E. coli populations can migrate along an antibiotic concentration gradient and establish an adaptively resistant population in the presence of a lethal dose of kanamycin. The established population is composed of planktonic and surface-associated cells, expands while exposed to kanamycin at 50-fold the MIC and reaches population densities similar to bacteria in an antibiotic-free region. Quantitative analysis of multiple colonization events of the antibiotic landscape demonstrates that collectively swimming nonresistant bacteria need to exceed a critical population density in order to successfully establish a viable population under lethal antibiotic stress.
Acknowledgments
We thank F Wu, S v Vliet, M Voges, B Beaumont and other members of the Department of Bionanoscience for fruitful discussions, and T Vazquez Faci and E v Rijn for technical assistance. This research was supported by the European Research Council Advanced Grant #247072 to CD and by the Netherlands Organisation for Scientific Research (NWO/OCW), as part of the Frontiers of Nanoscience program.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allison KR, Brynildsen MP, Collins JJ. (2011). Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amini S, Hottes AK, Smith LE, Tavazoie S. (2011). Fitness landscape of antibiotic tolerance in Pseudomonas aeruginosa biofilms. PLoS Pathog 7: e1002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderl JN, Franklin MJ, Stewart PS. (2000). Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother 44: 1818–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Gerdes K, Lewis K, McKinney JD. (2013). A problem of persistence: still more questions than answers? Nat Rev Microbiol 11: 587–591. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. (2004). Bacterial persistence as a phenotypic switch. Science 305: 1622–1625. [DOI] [PubMed] [Google Scholar]
- Baquero F, Negri M-C. (1997). Challenges: Selective compartments for resistant microorganisms in antibiotic gradients. Bioessays 19: 731–736. [DOI] [PubMed] [Google Scholar]
- Butler MT, Wang Q, Harshey RM. (2010). Cell density and mobility protect swarming bacteria against antibiotics. Proc Natl Acad Sci USA 107: 3776–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NR, Lobritz MA, Collins JJ. (2013). Microbial persistence and the road to drug resistance. Cell Host Microbe 13: 632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar N, McKinney JD. (2007). Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol 10: 30–38. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. (2010). Computer control of microscopes using μmanager. Curr Protoc Mol Biol Chapter 14: 14.20.1–14.20.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauvart M, De Groote VN, Michiels J. (2011). Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol 60: 699–709. [DOI] [PubMed] [Google Scholar]
- Fernández L, Breidenstein E, Hancock RE. (2011). Creeping baselines and adaptive resistance to antibiotics. Drug Resistance Updates 14: 1–21. [DOI] [PubMed] [Google Scholar]
- Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. (2014). Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513: 418–421. [DOI] [PubMed] [Google Scholar]
- Govantes F, Albrecht JA, Gunsalus RP. (2000). Oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB operon: roles of multiple promoters and the Fnr-1 and Fnr-2 binding sites. Mol Microbiol 37: 1456–1469. [DOI] [PubMed] [Google Scholar]
- Guarner F, Malagelada J-R. (2003). Gut flora in health and disease. Lancet 361: 512–519. [DOI] [PubMed] [Google Scholar]
- Hermsen R, Deris JB, Hwa T. (2012). On the rapidity of antibiotic resistance evolution facilitated by a concentration gradient. Proc Natl Acad Sci USA 109: 10775–10780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsen R, Hwa T. (2010). Sources and sinks: a stochastic model of evolution in heterogeneous environments. Phys Rev Lett 105: 248104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D, Kolter R. (2002). Why are bacteria refractory to antimicrobials? Curr Opin Microbiol 5: 472–477. [DOI] [PubMed] [Google Scholar]
- Hol FJ, Galajda P, Nagy K, Woolthuis RG, Dekker C, Keymer JE. (2013). Spatial structure facilitates cooperation in a social dilemma: empirical evidence from a bacterial community. PLoS ONE 8: e77042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol FJ, Voges MJ, Dekker C, Keymer JE. (2014). Nutrient-responsive regulation determines biodiversity in a colicin-mediated bacterial community. BMC Biol 12: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hol FJH, Dekker C. (2014). Zooming in to see the bigger picture: microfluidic and nanofabrication tools to study bacteria. Science 346: 1251821. [DOI] [PubMed] [Google Scholar]
- Kaiser P, Regoes RR, Dolowschiak T, Wotzka SY, Lengefeld J, Slack E et al. (2014). Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol 12: e1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer JE, Galajda P, Lambert G, Liao D, Austin RH. (2008). Computation of mutual fitness by competing bacteria. Proc Natl Acad Sci USA 105: 20269–20273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer JE, Galajda P, Muldoon C, Park S, Austin RH. (2006). Bacterial metapopulations in nanofabricated landscapes. Proc Natl Acad Sci USA 103: 17290–17295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Killam T, Sood V, Surette MG. (2003). Swarm-cell differentiation in Salmonella enterica serovar typhimurium results in elevated resistance to multiple antibiotics. J Bacteriol 185: 3111–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby AE, Garner K, Levin BR. (2012). The relative contributions of physical structure and cell density to the antibiotic susceptibility of bacteria in biofilms. Antimicrob Agents Chemother 56: 2967–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeaux D, Ghigo J-M, Beloin C. (2014). Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78: 510–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. (2010). Persister cells. Annu Rev Microbiol 64: 357–372. [DOI] [PubMed] [Google Scholar]
- Meredith HR, Srimani JK, Lee AJ, Lopatkin AJ, You L. (2015). Collective antibiotic tolerance: mechanisms, dynamics and intervention. Nat Chem Biol 11: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Staddon G, Eastman C, Palmer M, Davies ER, Carswell F. (1994). The quantitative distribution of nebulized antibiotic in the lung in cystic fibrosis. Respir Med 88: 203–211. [DOI] [PubMed] [Google Scholar]
- Mulcahy LR, Burns JL, Lory S, Lewis K. (2010). Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. J Bacteriol 192: 6191–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334: 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman MA, Brynildsen MP. (2013). Dormancy is not necessary or sufficient for bacterial persistence. Antimicrob Agents Chemother 57: 3230–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J, Bains M, Brazas MD, Hancock RE. (2008). Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190: 2671–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra P, Klumpp S. (2014). Phenotypically heterogeneous populations in spatially heterogeneous environments. Phys Rev E 89: 030702. [DOI] [PubMed] [Google Scholar]
- Romero D, Traxler MF, López D, Kolter R. (2011). Antibiotics as signal molecules. Chem Rev 111: 5492–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D, Finkelshtein A, Ingham C, Helman Y, Sirota-Madi A, Brodsky L et al. (2013). Identification and characterization of a highly motile and antibiotic refractory subpopulation involved in the expansion of swarming colonies of Paenibacillus vortex. Environ Microbiol 15: 2532–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AL, Lewis K. (2001). Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol 183: 6746–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone G, Wood P, Dixon L, Keyhan M, Matin A. (2002). Tetracycline rapidly reaches all the constituent cells of uropathogenic Escherichia coli biofilms. Antimicrob Agents Chemother 46: 2458–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber HW, Mueller J, Miller P, Arrow A. (1987). Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev 51: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet S, Hol FJ, Weenink T, Galajda P, Keymer JE. (2014). The effects of chemical interactions and culture history on the colonization of structured habitats by competing bacterial populations. BMC Microbiol 14: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetsigian K, Jajoo R, Kishony R. (2011). Structure and evolution of streptomyces interaction networks in soil and in silico. PLoS Biol 9: e1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S et al. (2013). Dynamic persistence of antibiotic-stressed mycobacteria. Science 339: 91–95. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung C-k et al. (2011). Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333: 1764–1767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.