Abstract
Understanding the microbial ecology of a system requires that the observed population dynamics can be linked to their metabolic functions. However, functional characterization is laborious and the choice of organisms should be prioritized to those that are frequently abundant (core) or transiently abundant, which are therefore putatively make the greatest contribution to carbon turnover in the system. We analyzed the microbial communities in 13 Danish wastewater treatment plants with nutrient removal in consecutive years and a single plant periodically over 6 years, using Illumina sequencing of 16S ribosomal RNA amplicons of the V4 region. The plants contained a core community of 63 abundant genus-level operational taxonomic units (OTUs) that made up 68% of the total reads. A core community consisting of abundant OTUs was also observed within the incoming wastewater to three plants. The net growth rate for individual OTUs was quantified using mass balance, and it was found that 10% of the total reads in the activated sludge were from slow or non-growing OTUs, and that their measured abundance was primarily because of immigration with the wastewater. Transiently abundant organisms were also identified. Among them the genus Nitrotoga (class Betaproteobacteria) was the most abundant putative nitrite oxidizer in a number of activated sludge plants, which challenges previous assumptions that Nitrospira (phylum Nitrospirae) are the primary nitrite-oxidizers in activated sludge systems with nutrient removal.
Introduction
Environmental microbial communities are an important resource for improved bioprocesses such as wastewater treatment and soil remediation, and new products such as biomaterials and bioenergy (Curtis et al., 2003; Rittmann et al., 2006). Exploiting these microbial resources optimally requires an understanding of not just which organisms are present, but also their metabolic function and ecology.
The activated sludge process is used to treat wastewater in most towns and cities across the developed world and continues to be rolled out for public sanitation across the rest of the world. The basic activated sludge process employs a mixed microbial consortium under aerobic conditions to remove carbon and nitrify ammonia. More advanced configurations also remove nitrate through denitrification and phosphate through the action of polyphosphate-accumulating organisms. The modern understanding of the ecology of the activated sludge has, for example, led to the optimization of P removal by identifying and favoring selection of P-removing bacteria in favor of other competing bacteria (Oehmen et al., 2010) and solved the most common bulking and foaming problems caused by the overgrowth of particular filamentous organisms (Nielsen et al., 2009).
Many organisms present in activated sludge have been characterized using enrichment cultures or in situ methods, such as microautoradiography and conceptual models of their individual substrate specificities, and interactions have been proposed (Nielsen et al., 2010). Thirty eight of these approximate genus-level probe-defined groups were detected in all of the 25 Danish plants, using direct microscopic counting with fluorescence in situ hybridization (Mielczarek et al., 2013). Many of the same taxa were also observed in plants from China and North America using 16S ribosomal RNA (rRNA) amplicon sequencing (Zhang et al., 2012). This suggests that the knowledge about their in situ ecology, derived from the few well-studied treatment plants, may be generally relevant to activated sludge ecosystems.
Microbial diversity is large (Quince et al., 2008) and hence in situ functional characterization is practical for only a small fraction of the species in a given system. Therefore, the core community concept (Grime, 1998; Gibson et al., 1999) might be useful to identify putatively important organisms. However, a core community may include organisms that are consistently present only in low abundance and that are sampled as a result of the increased sequencing depth of modern amplicon sequencing methods.
Transient organisms are of interest because some of these organisms will be among those abundant organisms that cause process problems and some may be selected by characteristic properties of the plants. Either way identifying and understanding the ecology of such transient organisms will be important for the optimization and design of processes.
The selection of a cutoff for defining high abundance is a somewhat arbitrary affair owing to the continuous nature of the abundance distribution, but we propose a method that is based on putative carbon turnover. Methodological biases aside, the relative carbon removal activity of the organisms should be reflected in their relative abundance as most carbon removal in activated sludge is due to aerobic and denitrifying heterotrophs and these functional guilds have similar growth yields of 0.6 and 0.5 g-biomass g-substrate−1, respectively (Henze et al., 2002). In line with Verstraete et al. (2007), we define abundant organisms as those that constitute the top 80% of reads obtained by amplicon sequencing in a certain sample as these relatively few organisms account for most (~80%) of the carbon turnover.
The influent to wastewater treatment plants contains high concentration of microbes and the cells coming with the wastewater might be observed as abundant organisms despite being inactive in the activated sludge system, simply due to their constant and high rate of immigration. The known flow rates and residence time within activated sludge systems should make it possible to calculate the impact of immigration.
This paper presents detailed examination of the microbial community composition in 13 activated sludge plants from across Denmark and characterizes a core community of only 63 genera that were frequently highly abundant and made up 68% of the organisms observed. We also present an evaluation of the importance of the incoming microbes by the wastewater using mass balances to calculate those being inactive in the plants. This work thus extends simple ‘core' community approaches and identifies putatively important activated sludge organisms that should be further studied.
Materials and methods
Plants and sampling
A cross-section of 13 wastewater treatment plants was selected across Denmark, a region of ~300 km across (Supplementary Figure S1). Two samples from each plant were taken in consecutive years during august (summer; n=26). Aalborg West was additionally sampled periodically over 6 years (n=13, including two samples from the cross-section) as a time series within a single plant. Influent from three plants, Aalborg East & West and Hjørring were sub-sampled from 24-h flow proportional samples kept on ice. Two influent samples from each plant were sampled ~1 month apart along with corresponding activated sludge samples, representing a time span of about one sludge residence time. The sewers in Aalborg East & West are separate systems servicing parts of the same city.
All plants were performing carbon and nitrogen removal, and all additionally had enhanced biological phosphorus removal, except Viborg and Odense NW. Data from the routine process monitoring using standard methods (Rice et al., 2012) were collected from the collaborating utilities (Supplementary Table S1).
Samples for microbiological analysis of activated sludge were taken as 0.5 l grab samples from the aeration tank (that is, during vigorous mixing of the biomass) and influent wastewater was taken as 24 h flow proportional samples at the inflow to the process tanks, that is, after primary clarification if this process was employed at the plant. All samples were homogenized and frozen at −80 °C.
16S rRNA gene sequencing
DNA was extracted from 250 μl of sample with the Fast DNA Soil kit (Q-BioGene, Carlsbad, CA, USA) according to the manufacturer's instructions, except with bead-beating in 80 °C phenol. PCR was made with Platinum High Fidelity Taq Polymerase (Invitrogen, Carlsbad, CA, USA) in two-steps to avoid barcode bias (Berry et al., 2011). An initial 20 cycle PCR with 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with 1–10 ng of DNA, was followed by a second PCR with 7 cycles using 2 μl of the first product amplified the same primers with a 12 nt barcode and Illumina adaptors (Caporaso et al., 2010) producing a 250-bp amplicon. DNA was measured with QuantIT (Molecular Probes, Eugene, OR, USA) and pooled in eqimolar amounts and sequenced on a HiSeq 2000 (Illumina, Carlsbad, CA, USA) producing 2 × 150 bp paired-end reads. Raw sequence data can be accessed from GenBank (PRJEB5095).
Microbial diversity analysis
The paired-end reads were merged using pandaseq v.2.0 (Masella et al., 2012) and sequencing noise removed by discarding unique reads observed less than three times in the total data set and subsequently reformatted for QIIME using the script pandaseq.to.qiime.pl. Preprocessing was made with QIIME v1.5.0 (Caporaso et al., 2010) using uclust for de novo operational taxonomic unit (OTU) clustering (Edgar, 2010), representative sequence picking and RDP Classifier for taxonomic assignment (Wang et al., 2007) against the Greengenes taxonomy, v. Oct 2012 (DeSantis et al., 2006), manually curated to include genus names for most organisms previously observed in activated sludge (McIlroy et al., 2015). Comparisons of relative abundance between samples were made after data sets rarefied to 40 k reads per sample. Analysis was made using R packages, phyloseq (McMurdie and Holmes, 2013) and ggplot2. Correspondence analysis compared Bray–Curtis distances. All data and analysis scripts to generate all figures in the paper can be accessed at www.github.com/aaronsaunders/DKsludge.
The raw sequences from the 94% OTUs that were classified to the genus Tetrasphaera were clustered at 99% identity using uclust, aligned using MUSCLE (Edgar, 2004) and used to calculate a maximum likelihood tree using FastTree (http.//microbesonline.org/fasttree) with Jukes-Cantor evolutionary model, comparing 250 nt. The relative read abundance of the OTUs, which made up more than 0.1% of the total Tetrasphaera reads were visualized using the interactive Tree of Life tool (itol.embl.de). Sequences were attributed to Tetrasphaera clades (Nguyen et al., 2011) by inserting the sequences into the Greengenes reference tree of full-length sequences using the ARB parsimony insertion tool (Ludwig et al., 2004).
Calculation of net growth rates from amplicon data
Net growth rates were calculated in R using mass balance (see Supplementary Methods). In brief, we assumed that growth and decay could be described by first-order processes (equation (1)) and that the system was at steady state, such that there was no net change in the number of cells in the activated sludge biomass (Nx,AS). See Supplementary Methods for a detailed explanation of the calculation.
 |
where:
Nx,AS number of cells of organism x in the activated sludge.
K rate constant (d−1) (positive for net growth, negative for net decay).
nx,ww number of cells of organism x entering per unit time (d−1).
nx,SP number of cells of organism x removed (surplus sludge) per unit time (d−1).
Results
The abundant core community of 13 Danish wastewater treatment plants
The influent wastewater composition, nutrient-loading rates, plant design and operating conditions were largely similar for all the plants studied, and all were performing well with similarly low-effluent concentrations (Supplementary Table S1).
The microbial community composition within the activated sludge was characterized using barcoded amplicons resulting in at least 40 000, non-chimeric, quality-filtered reads per sample and more than two million reads in total. The amplicons were clustered into OTUs at 94%, 97% and 99% identity, which for readability are henceforth referred to as genus-level, species-level and subspecies-level OTUs, respectively (Stackebrandt and Goebel, 1994; Větrovský and Baldrian, 2013). The number of reads in each OTU, or read abundance, was for simplicity treated as reflective of the actual numerical abundance of that organism in the system. However, the reader is reminded that the observed read abundance may have been affected by differences between organisms in DNA extraction efficiency, 16S rRNA gene copy number and primer specificity (see Supplementary Methods).
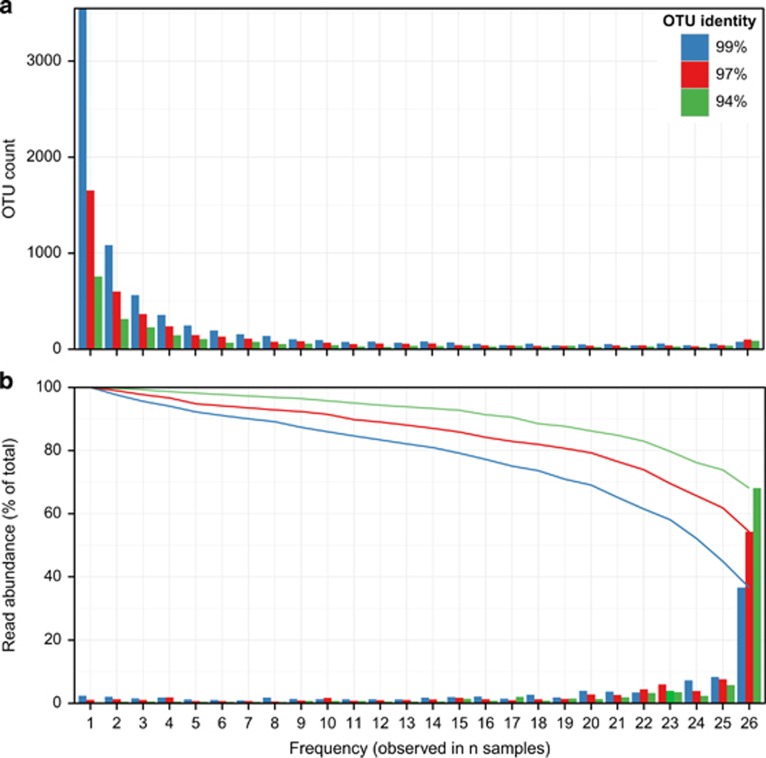
To determine the core community we compared a cross-section of 13 plants across Denmark during consecutive summers, 26 samples in total. The frequency with which each OTU was observed showed a skewed bi-modal distribution with a large number of OTUs observed in only one or few samples, and a second smaller peak of OTUs observed in all 26 samples (Figure 1a). The more frequently observed OTUs tended to be more abundant and the 86 genus-level OTUs observed in every sample accounted for 68% of the total reads (Figure 1b, green bars). The core community increased to 86% of the total reads if OTUs observed in 20 or more samples were included (Figure 1b, green line). Conversely, the 1546 genus-level OTUs observed in five or fewer samples only accounted for 2.3% of the total reads.
Figure 1.
Frequency distribution of OTUs across samples. Core OTUs common to all 26 samples. Core and frequently observed OTUs made up a larger fraction of the total reads than transiently observed OTUs. The number of samples in which each OTU is observed against (a) the number of OTUs observed at each frequency and (b) the bars denote the relative read abundance of OTUs observed at each frequency and the lines present the cumulative total these frequencies from most- to least-frequently observed (lines). Colors denote OTUs clustered at genus-level (green, 94%), species-level (red, 97%), subspecies-level (blue, 99%).
A similar comparison of the temporal stability of the microbial community within a single plant was investigated in the Aalborg West plant in 13 quarters over 6 years, including two of the samples included in the cross-section (Supplementary Figure S2). Ninety-three percent (±3%) of the reads was from the 190 genus-level OTUs observed in every sample in the time series. This demonstrated that the diversity within the plant was stable through time and indicated that the variation between plants in the cross-section was likely to be greater than within a single plant over time.
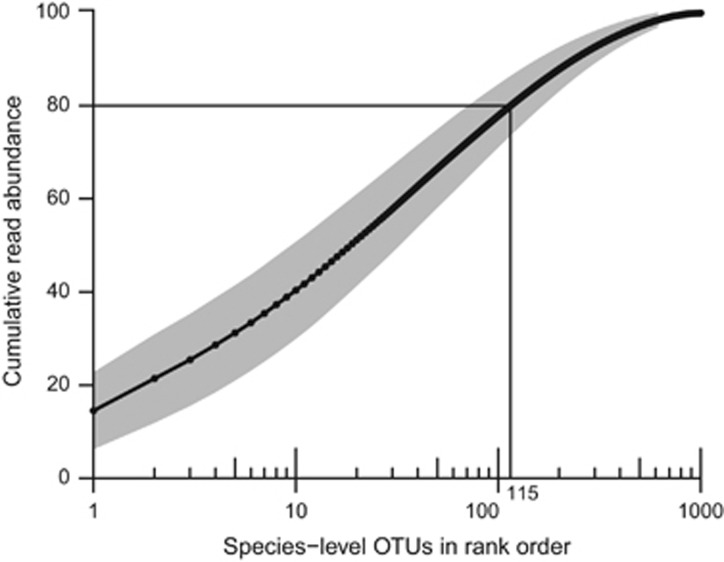
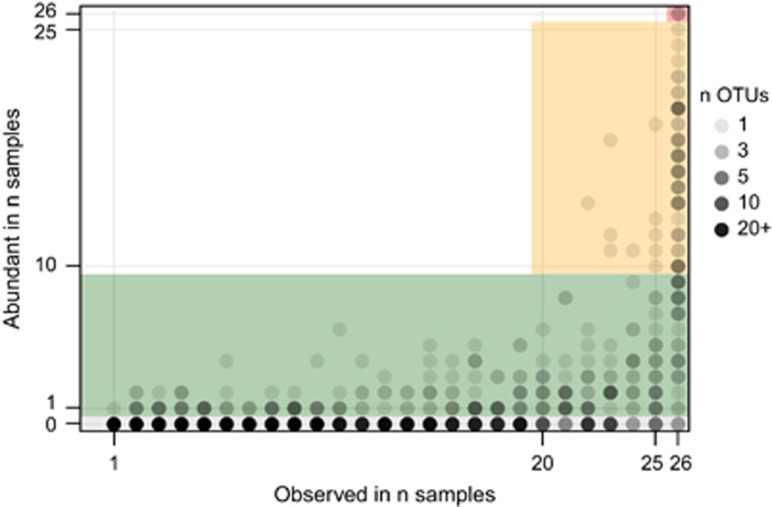
Within each sample, a few abundant OTUs made up a large proportion of the reads. We defined OTUs as abundant when they made up the top 80% of the reads in a sample when ranked by decreasing OTU abundance (Figure 2). OTUs were then binned into four groups based on the frequency with which they were observed and the number of samples in which they were abundant (Figure 3).
Figure 2.
Cumulative read abundance across 26 activated sludge samples. Cumulative read abundance (mean±SD) of species-level OTUs plotted in rank order for the 26 samples. In each sample, the 10 most abundant OTUs made up 40% (±10%) of the total reads on average and the 100 most abundant OTUs made up 78% (±6.8%). OTUs were considered abundant in a sample when they were among the OTUs making up the top 80% of reads.
Figure 3.
Comparison of observation frequency and frequency of high abundance. Genus-level OTUs were plotted with slight transparency (alpha=0.2) such that darker points indicate more OTUs with those characteristics at that position. The OTUs were then classified as (a) Group 1: always abundant (26 samples, red), (b) Group 2: frequently abundant (⩾10 samples, yellow), (c) Group 3: transiently abundant in ⩾1 sample, green) and (d) Group 4: not abundant in any sample (gray). This was compared with the frequency at which the OTU was observed: transient (<20 samples), frequent (⩾20 samples) or core (26 samples).
Group 1 organisms were abundant in every sample and consisted of only three genera that made up 26% of the total reads (Figure 3, red). Group 2 organisms were frequently observed and abundant in at least 10 samples and made up 43% of the total reads (Figure 3, yellow). Group 3 organisms were abundant in <10 samples and made up 23% of the reads (Figure 3, green). At last, the remaining 1982 OTUs (Group 4) were always present in low abundance and thus made up only a minor fraction (8%) of the total reads. Most Group 3 and 4 OTUs were observed in only few samples but some were observed more frequently.
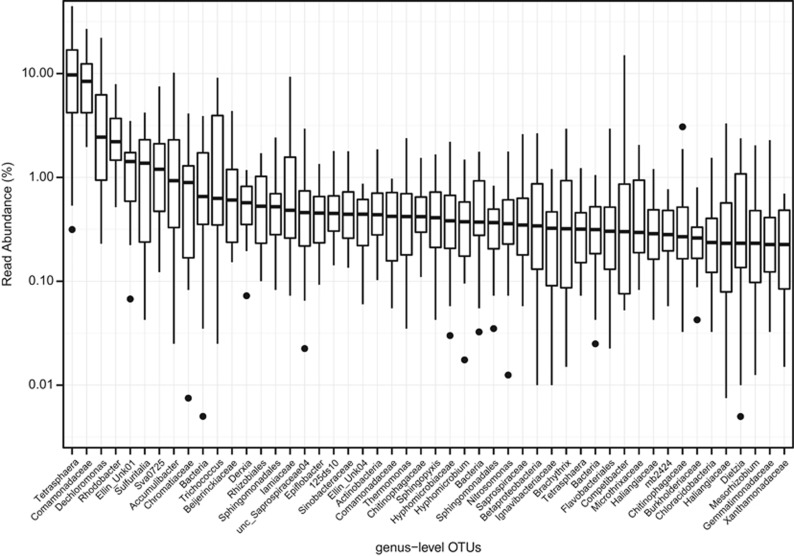
Groups 1 and 2 (Figure 3) were both frequently observed (20+ samples) and frequently abundant (10+ samples), and a made up a total of 64 genera (69% of the total reads).These organisms were the most abundant organisms of those observed in 20+ samples (86% of reads; Figure 1b). The ecological function in activated sludge of 34 of these abundant core genera has been characterized to some extent, but the in situ physiology of the remaining genera is not known and their consistent abundance suggests that they are important to carbon removal in these systems. For example, in the 10 most abundant core genera there were three OTUs classified to Sulfuritalea (class Betaproteobacteria), one to family Chromatiaceae (class Gammaproteobacteria) and another to phylum Acidobacteria (order ‘Sva0725'), respectively, and we have no knowledge of the function of any of these genera. In addition, two other abundant core genera were identified from an undescribed order ‘Ellin' within the Betaproteobacteria for which there is no functional information. Three genera in the top 50 lacked any close relatives in the database and classified only as domain Bacteria.
The genus-level OTUs considered thus far were clustered at 94% sequence identity, and could each potentially contain considerable diversity. However, each OTU typically contained one or few abundant sequence types that were frequently observed in many samples. This is illustrated by the considerable proportion of frequently observed OTUs seen for the OTUs clustered at species-level or subspecies-level (Figure 1; blue and red bars, respectively). For example, 36% (±10%) of the reads in the cross-section samples came from just 77 subspecies-level OTUs, and the subspecies-OTUs observed in at least 14 samples made up 80% of the total reads (Figure 1b; blue line).
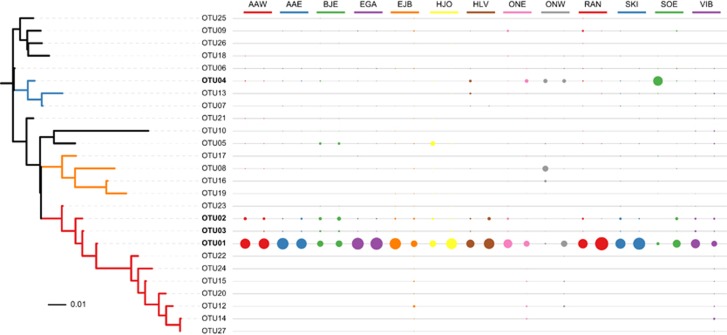
Another illustration of the apparently high degree of subspecies conservation is the sequences assigned to the most abundant genus, Tetrasphaera (Figure 4). The sequences encompassed 156 subspecies-level OTUs, but a single OTU accounted for most of the Tetrasphaera in all but two of the samples, one from Odense West and one from Søholt (Figure 5). Within the time series in Aalborg West over 6 years, the same subspecies-level OTU was dominant (Supplementary Figure S3). These abundant OTUs were from Tetrasphaera clade 3, which is consistent with studies using FISH that found that Tetrasphaera clade 3 was the most abundant in Danish plants (Nguyen et al., 2011; Mielczarek et al., 2013).
Figure 4.
Boxplot of the abundance of the top 50 genus-level OTUs (by median). The upper and lower bounds of boxes denote the 25th and 75th percentiles and the lines denote the max and min values, outliers are shown as dots. OTU labels are the lowest assigned taxonomic rank.
Figure 5.
Relative abundance of 99% OTUs Tetrasphaera across 13 plants. The relationship between the OTUs is presented as a maximum likelihood phylogenetic tree; the branches are colored to denote OTUs from Clade 1 (blue), Clade 2 (yellow) and Clade 3 (red). The size of the circles denotes the relative abundance of each OTU.
The effect of immigration
To investigate to what extent the cells arriving with the influent wastewater directly contributed to the observed diversity in the activated sludge plants, we analyzed the relative abundance of each species-level OTU in the influent wastewater and the corresponding biomass in three activated sludge plants at two time points with a 1-month interval.
The wastewater samples from the three independent sewer systems shared 157 genus-level OTUs accounting for 93% of the reads (Supplementary Figure S4; blue), including 309 subspecies-level OTUs making up 83% of the reads (Supplementary Figure S4; red). The 10 most abundant genera alone accounted for 63% (±11, n=6) of the reads in each sample. These included Arcobacter, Trichococcus, Acinetobacter and one genus-level OTU that could be best classified to the family Comamonadaceae (Supplementary Figure S5). In addition, two genera of Pseudomonadaceae were abundant in the wastewater influent to both Aalborg plants, whereas the genus Lactococcus was abundant in the wastewater at the Hjørring plant.
These few abundant genera arriving with the wastewater appeared to affect the observed diversity in the activated sludge. Thirty-five percent of the OTUs (62% of the reads) in the activated sludge were also observed in the incoming wastewater, and correspondence analysis of the relative abundance of the genus-level OTUs showed that a large component of the variance (correspondence analysis 1, 28.9%) separated the samples of influent wastewater from the samples of the activated sludge (Supplementary Figure S6). However, a considerable component of the variance (correspondence analysis 2, 14.9%) was a component common to the wastewater and activated sludge samples that separated the two Aalborg plants from the Hjørring plant.
To evaluate the degree to which incoming species-level OTUs were growing, and thus contributing to the function of the activated sludge community, their net growth rate was calculated from the numbers entering and leaving the system using a mass balance. A typical plant receives a volume of wastewater approximately equal to its volume per day (that is, hydraulic residence time of 1 day). The wastewater contains ~108 cells ml−1 and the activated sludge biomass within the plant about 1010 cells ml−1, so each day the plants receive a large number of cells, approximately equivalent to 1% of the cells within the plant (Supplementary Figure S7).
To keep a constant amount of biomass in the plant, a small fraction of the biomass is removed daily as surplus sludge. Plants in Denmark with relatively low temperatures and low nutrient loads have a relatively long SRT (solids retention time) of 30 days—thus 1/30 or ~3% the biomass is removed each day. Thus, to counter this removal and persist in the system, organisms must be ‘added' at a rate of 1/SRT per day or 0.033 per day (for 30-day SRT). For most organisms, this addition will mostly be due to net growth, however organisms that are highly abundant in the influent may have sufficient cells entering the plant with the influent that their apparent net growth rate is higher than their actual net growth rate owing to immigration. Indeed, if the immigration is high enough, the population can be sustained in the system even if the actual net growth rate is negative.
In an ecosystem, growth and decay occur at the same time, and the net growth rate (observed change in abundance) is the in situ growth rate minus the decay rate. The average decay rate of 0.1–0.2 per day estimated from oxygen consumption rates (Henze et al., 2002) suggests that organisms with a positive in situ growth rate that is lower than the decay rate can actually have a negative net growth rate, despite in situ growth.
Therefore, we conservatively state that organisms with a net growth rate between −0.1 and 0 are putatively slow growing.
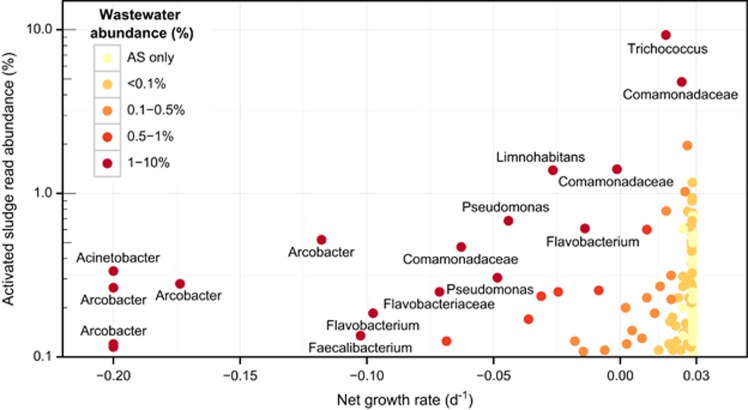
The vast majority of OTUs (90±3% of the reads, n=6) were growing in the system (Supplementary Figure S8). Two OTUs from this group—genus Trichococcus and a particular OTU from the family Comamonadaceae—had a high net growth rate despite being abundant in the influent (Figure 6), indicating that they were indeed active in the plant.
Figure 6.
Comparison of the read abundance in the activated sludge and the net growth rate for the Aalborg East plant. Points are colored by their abundance in the influent wastewater, and taxa >1% in the wastewater are named. Few abundant species-level OTUs had a low net growth rate, indicating that their abundance was due to contribution from the influent bacteria.
The non-growing fraction accounted for only 2.5% (±1%, n=6) of the total reads, whereas 7.5% (±2%, n=6) were putatively slowly growing. These organisms were abundant in the influent and while some were also abundant in the sludge, they were not as numerous as they should have been if they were also growing actively, and were present primarily due to immigration with the wastewater. Arcobacter, Faecalibacterium, Flavobacterium, Pseudomonas and Limnohabitans were the non-growing organisms most abundant in the wastewater. However, Acinetobacter was the only organism in the abundant core community (0.3% of the total reads, and over 1% in two samples) but which was removed based on its slow growth. So, after excluding acinetobacter, the core community of total of 64 such genera (69% of the total reads) (Groups 1 and 2; Figure 3) is reduced to 63 abundant core genus-level OTUs that were actively growing (68% of the total reads).
The transient genera
We identified 252 genera (23% of total reads) that were abundant in at least one but <10 samples (Group 3, Figure 3) and these were classified transient. A number of the transient organisms have been functionally characterized and many of these are important to the activated sludge process as a cause of sporadic problems in the plants. For example, the filamentous organisms Kouleothrix, Anaerolinea, Microthrix and Thiothrix occasionally cause problems with foaming and bulking (Nielsen et al., 2009), and Defluviicoccus may be deleterious for enhanced biological phosphate removal by competing with the polyphosphate-accumulating organisms that facilitate this process (Oehmen et al., 2010).
The transient genus Methylotenera made up 13% and 10% of the reads in the Viborg plant but were not abundant in other plants. This genus contains isolates that are denitrifying methylotrophs, and Viborg was unique among plants that it added methanol as an additional carbon source for denitrification.
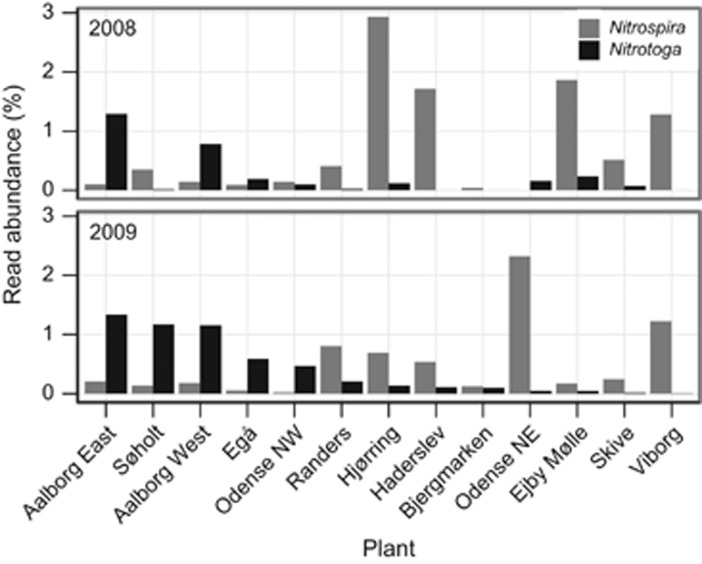
Among the nitrite-oxidizing bacteria (NOB), the well-described genus Nitrospira (phylum Nitrospirae) was common in the activated sludge but not as an abundant core organism, being absent from a number of samples. However, the newly recognized and phylogenetically distinct NOB genus Nitrotoga (Betaproteobacteria; Alawi et al., 2007) was also transient and had a higher read abundance than Nitrospira in nine samples (Figure 7), suggesting that they might make an important contribution to nitrification. The relative read abundance of the two NOBs was not stable and the dominant NOB switched in consecutive years in Odense NE and Søholt (Figure 7). The time series for Aalborg West also showed a switch in the dominance (Supplementary Figure S9), with Nitrotoga being most abundant from 2006 to 2009, followed by a switch to Nitrospira in 2010–2011.
Figure 7.
Read abundance of nitrite-oxidizing bacteria. Nitrospira (gray) and Nitrotoga (black) in activated sludge samples from 13 plants taken during summer of 2008 and 2009. The NOBs in most plants were dominated by Nitrospira, but Nitrotoga dominated in some cases.
Of the three plants for which we could compare the NOB in the influent and activated sludge samples, both genera were present in most of the influent samples though in all cases greatly enriched in the activated sludge. However, Nitrotoga only constituted a considerable fraction of the NOB in Aalborg East (Supplementary Figure S10). The same subspecies-level OTUs were observed in all plants and in both the influent and activated sludge and their relative read abundance was mirrored in the influent.
These examples illustrate the potential importance of transient organisms and underline the need to study the remaining uncharacterized transient organisms further. The 41 transient OTUs were particularly abundant (>1%) in at least one sample (Supplementary Table S2) should be the first selected for further characterization.
Discussion
We propose to classify microorganisms in activated sludge ecosystems as abundant core or abundant transient, and that they are not primarily present due to immigration but to growth within the system. This is more informative than a simple core community of shared OTUs, as the number of shared OTUs will also be a function of the depth of sequencing. The microorganisms identified in this way will not be sensitive to sequencing depth, provided the depth is sufficient to reproducibly sample organisms around the cutoff used to classify organisms as abundant in a sample. These microorganisms are putatively making the greatest contribution to carbon turnover in the system and their characterization should be prioritized.
The investigated Danish wastewater treatment plants consisted of 63 abundant core genus-level OTUs that were actively growing and these organisms alone made up 68% of the total reads. Many of the same genera were also detected in activated sludge systems in China and the USA (Zhang et al., 2012), which suggests that the factors driving the formation of an abundant core community are general for wastewater treatment plants.
A large abundant core community was also present within the wastewater of the three independent sewer ecosystems. Only one other study of wastewater microbial diversity has been made (in Milwaukee, USA; McLellan et al., 2010), and many of the same genera, for example, Acinetobacter, Pseudomonas and Trichococcus, were also abundant. This suggests that the microbial diversity within sewers may be similar across the world.
Immigration had a measurable but modest impact on the observed community. OTUs abundant in the influent with a low net growth rate made up 5–10% of the observed reads in the activated sludge. Owing to high immigration, some Acinetobacter were members of the abundant core community despite having a low net growth rate. These results reaffirm the necessity for validating that organisms are active. The use of mass balance-based calculation of the net growth rate of each OTU individually exploits the known solids residence time of the activated sludge plant and thus may not be widely applicable, but the use of other methods such as reverse-transcribed rRNA (Foesel et al., 2014), may also be applicable for differentiating inactive cells.
The mechanisms driving the assembly of the abundant core community in activated sludge plants were not investigated in our work. The concept of core populations originally arose out of metacommunity approaches to the understanding of biological diversity (Grime, 1998), and our observations are consistent with the hypothesis that core communities are caused by regionally abundant populations. Neutral community assembly was shown to have a role in the shaping of the diversity of ammonia-oxidizing bacteria in wastewater treatment plants (Ofiteru et al., 2010), and these mechanisms may also have a role in shaping the diversity among functional guilds within the community. Neutral processes may also drive the apparently stochastic oscillations in the abundance of Nitrospira and Nitrotoga.
Niche-based selection is likely to also have a role in shaping diversity as we know also that the characterized genera in activated sludge have specific and non-overlapping substrate specificities (Kindaichi et al., 2013), which potentially gives them different selective regimes that diverge from neutrality. The example of the Methylotenera in Viborg WWTP presented in this paper appears to be due to the selection based on the addition of methanol to the sludge. Further investigations are needed to determine the relative importance of these factors in shaping the diversity in a wastewater treatment plants.
The finding that Nitrotoga were numerically abundant putative nitrifiers corroborates reports of their presence in some activated sludge plants in Austria (Lücker et al., 2014) and challenges previous assumptions that Nitrospira are the dominant nitrite-oxidizers in activated sludge. The mean summer water temperature in Danish plants was 17.8 °C (±1.6), which is consistent with enrichment cultures where Nitrotoga were competitive with Nitrospira below 17 °C (Alawi et al., 2009). However, fluctuations in the relative read abundance of the two NOBs even within single plants suggest that other factors are also important. The similar relative abundance of Nitrospira and Nitrotoga in the activated sludge and wastewater influent may indicate that selection already begins in the sewers or that these organisms are acting as a seed for selection in the plants.
We see the classification of OTUs as abundant core or transient organisms as a means to prioritize studies to determine the role of uncharacterized genera, for example, by targeting these organisms for genome sequencing from environmental samples (Albertsen et al., 2013), or using in situ techniques to test their in situ metabolic and ecological niche (Wagner et al., 2006). The identification of abundant core and transient organisms is also being used to select organisms for manual curation of the taxonomy used for classification of amplicon data. This can improve the fraction of OTUs classified to the genus-level with a consistent name, which can be linked to its putative function. This work is on-going in the MiDAS Field Guide project (McIlroy et al., 2015; www.midasfieldguide.org), where current knowledge about the putative physiology of all abundant core and transient organisms in activated sludge is being collected and summarized.
Organisms that are not abundant may still have an important role in ecosystem function, but other criteria are needed to determine in what sense they should be considered important. Low-abundance organisms may act as a seed bank for organisms to grow to transient abundance under conditions not experienced during the sampling period, which may in turn improve functional stability under environmental oscillations. Alternatively, low abundance organisms may have a specialized but important niche, for example, for the degradation of low concentration micropollutants. The 80% threshold for abundance is arbitrary and suited our stated purpose of prioritizing genera for functional characterization. Other purposes might be served with a different cutoff or a range of values to represent the gradual rather than binary natural of the abundance distribution. For example, for our purpose of prioritizing the in situ characterization of organisms, we may consider additional categories of organisms accounting for the top 80–90% or even 80–99% of reads when the ecology of the abundant core and transient organisms has been characterized.
All the plants in this study were meeting carbon, nitrogen and phosphorus effluent limits and have done so for years. If further research were to identify key differences in the communities within poorly performing plants then microbial surveys may be a useful diagnostic of some process problems. Furthermore, times series studies of the effect of strong perturbations in wastewater composition or plant operation may provide a better understanding about factors selecting for the different abundant species.
Acknowledgments
Thanks to the municipal wastewater treatment plants for contributing their process data and experience, to Marianne Stevenson for contributions in the lab, and to Aalborg University, Niels Due Jensen Foundation, Grundfos and the Innovation Fund Denmark (EcoDesign) for funding.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alawi M, Lipski A, Sanders T, Spieck E. (2007). Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. ISME J 1: 256–264. [DOI] [PubMed] [Google Scholar]
- Alawi M, Off S, Kaya M, Spieck E. (2009). Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Rep 1: 184–190. [DOI] [PubMed] [Google Scholar]
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Berry D, Ben Mahfoudh K, Wagner M, Loy A. (2011). Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol 77: 7846–7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis TP, Head IM, Graham DW. (2003). Theoretical ecology for engineering biology. Environ Sci Technol 37: 64–70. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16 S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Foesel BU, Nägele V, Naether A, Wüst PK, Weinert J, Bonkowski M et al. (2014). Determinants of Acidobacteria activity inferred from the relative abundances of 16 S rRNA transcripts in German grassland and forest soils. Environ Microbiol 16: 658–675. [DOI] [PubMed] [Google Scholar]
- Gibson DJ, Ely JS, Collins SL. (1999). The core-satellite species hypothesis provides a theoretical basis for Grime's classification of dominant, subordinate, and transient species. J Ecol 87: 1064–1067. [Google Scholar]
- Grime J. (1998). Benefits of plant diversity to ecosystems: immediate, filter and founder effects. J Ecol 86: 902–910. [Google Scholar]
- Henze M, Harremoës P, la Cour Jansen J, Arvin E. (2002) Wastewater treatment, 3rd edn. Springer: New York, NY, USA. [Google Scholar]
- Kindaichi T, Nierychlo M, Kragelund C, Nielsen JL, Nielsen PH. (2013). High and stable substrate specificities of microorganisms in enhanced biological phosphorus removal plants. Environ Microbiol 15: 1821–1831. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker S, Schwarz J, Gruber-Dorninger C, Spieck E, Wagner M, Daims H. (2014). Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants. ISME J 9: 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 13: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy SJ, Saunders AM, Albertsen M, Nierychlo M, McIlroy B, Hansen AA, Karst SM, Nielsen JL, Nielsen PH. (2015). MiDAS: The field guide to the microbes of activated sludge. Database 2015: bav062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan SL, Huse SM, Mueller-Spitz SR, Andreishcheva EN, Sogin ML. (2010). Diversity and population structure of sewage-derived microorganisms in wastewater treatment plant influent. Environ Microbiol 12: 378–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. Watson, M (ed). PLoS One 8: e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielczarek AT, Nguyen HTT, Nielsen JL, Nielsen PH. (2013). Population dynamics of bacteria involved in enhanced biological phosphorus removal in Danish wastewater treatment plants. Water Res 47: 1529–1544. [DOI] [PubMed] [Google Scholar]
- Nguyen HTT, Le VQ, Hansen AA, Nielsen JL, Nielsen PH. (2011). High diversity and abundance of putative polyphosphate-accumulating Tetrasphaera-related bacteria in activated sludge systems. FEMS Microbiol Ecol 76: 256–267. [DOI] [PubMed] [Google Scholar]
- Nielsen PH, Kragelund C, Seviour RJ, Nielsen JL. (2009). Identity and ecophysiology of filamentous bacteria in activated sludge. FEMS Microbiol Rev 33: 969–998. [DOI] [PubMed] [Google Scholar]
- Nielsen PH, Mielczarek AT, Kragelund C, Nielsen JL, Saunders AM, Kong Y et al. (2010). A conceptual ecosystem model of microbial communities in enhanced biological phosphorus removal plants. Water Res 44: 5070–5088. [DOI] [PubMed] [Google Scholar]
- Nielsen PH, Saunders AM, Hansen AA, Larsen P, Nielsen JL. (2012). Microbial communities involved in enhanced biological phosphorus removal from wastewater - a model system in environmental biotechnology. Curr Opin Biotechnol 23: 452–459. [DOI] [PubMed] [Google Scholar]
- Oehmen A, Carvalho G, Lopez-Vazquez CM, van Loosdrecht MCM, Reis MAM. (2010). Incorporating microbial ecology into the metabolic modelling of polyphosphate accumulating organisms and glycogen accumulating organisms. Water Res 44: 4992–5004. [DOI] [PubMed] [Google Scholar]
- Ofiteru ID, Lunn M, Curtis TP, Wells GF, Criddle CS, Francis CA et al. (2010). Combined niche and neutral effects in a microbial wastewater treatment community. Proc Natl Acad Sci USA 107: 15345–15350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Curtis TP, Sloan WT. (2008). The rational exploration of microbial diversity. ISME J 2: 997–1006. [DOI] [PubMed] [Google Scholar]
- Rice EW, Baird RB, Eaton AD, Clesceri LS. (2012) Standard Methods For The Examination Of Water And Wastewater, 22nd edn, American Water Works Association: MD, USA. [Google Scholar]
- Rittmann BE, Hausner M, Loffler F, Love NG, Muyzer G, Okabe S et al. (2006). A vista for microbial ecology and environmental biotechnology. Environ Sci Technol 40: 1096–1103. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. (1994). Taxonomic note: A place for DNA-DNA reassociation and 16 S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44: 846–849. [Google Scholar]
- Verstraete W, Wittebolle L, Heylen K, Vanparys B, de Vos P, van de Wiele T et al. (2007). Microbial resource management: the road to go for environmental biotechnology. Eng Life Sci 7: 117–126. [Google Scholar]
- Větrovský T, Baldrian P. (2013). The variability of the 16 S rRNA gene in bacterial genomes and its consequences for bacterial community analyses. PLoS One 8: e57923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Nielsen PH, Loy A, Nielsen JL, Daims H. (2006). Linking microbial community structure with function: fluorescence in situ hybridization-microautoradiography and isotope arrays. Curr Opin Biotechnol 17: 83–91. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Shao M-F, Ye L. (2012). 454 pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J 6: 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.