Abstract
Modern microbial mats are potential analogues of some of Earth's earliest ecosystems. Excellent examples can be found in Shark Bay, Australia, with mats of various morphologies. To further our understanding of the functional genetic potential of these complex microbial ecosystems, we conducted for the first time shotgun metagenomic analyses. We assembled metagenomic next-generation sequencing data to classify the taxonomic and metabolic potential across diverse morphologies of marine mats in Shark Bay. The microbial community across taxonomic classifications using protein-coding and small subunit rRNA genes directly extracted from the metagenomes suggests that three phyla Proteobacteria, Cyanobacteria and Bacteriodetes dominate all marine mats. However, the microbial community structure between Shark Bay and Highbourne Cay (Bahamas) marine systems appears to be distinct from each other. The metabolic potential (based on SEED subsystem classifications) of the Shark Bay and Highbourne Cay microbial communities were also distinct. Shark Bay metagenomes have a metabolic pathway profile consisting of both heterotrophic and photosynthetic pathways, whereas Highbourne Cay appears to be dominated almost exclusively by photosynthetic pathways. Alternative non-rubisco-based carbon metabolism including reductive TCA cycle and 3-hydroxypropionate/4-hydroxybutyrate pathways is highly represented in Shark Bay metagenomes while not represented in Highbourne Cay microbial mats or any other mat forming ecosystems investigated to date. Potentially novel aspects of nitrogen cycling were also observed, as well as putative heavy metal cycling (arsenic, mercury, copper and cadmium). Finally, archaea are highly represented in Shark Bay and may have critical roles in overall ecosystem function in these modern microbial mats.
Introduction
Microbial mats have persisted for ~85% of the geological history of the Earth and dominate parts of the fossil record (Walter and Buick, 1980; Dupraz and Visscher, 2005). Microbial mats are laminated organo-sedimentary biofilms, with distinctive microorganisms characterising each of the layers, their structures often stabilised through the presence of exopolymeric substances (EPS) (Dupraz and Visscher, 2005). Some microbial mats can lead to the formation of microbially induced mineral precipitations known as microbialites (Schneider et al., 2013), although the process of microbialite formation is still not completely understood (Dupraz et al., 2009). Microbialites can be delineated by their carbonate macrostructure, those with a laminated formation are often referred to as stromatolites, while their non-laminated clotted counterparts are referred to as thrombolites (Mobberley et al., 2013).
The oldest fossilised microbialites are found in Western Australia, dating back ~3.5 Gyr ago (Kranendonk et al., 2008). The dominance of marine stromatolites collapsed ~542 Myr ago, possibly due to eukaryotic grazing (Bernhard et al., 2013). These communities thrive at the intersection of abiotic and biotic factors that promote organomineralisation (Dupraz and Visscher, 2005; Dupraz et al., 2009). A host of biological factors are also necessary, such as the presence of EPS in cyanobacterial mats (Marvasi et al., 2010), which serve as a location of mineral nucleation, while providing a heterotrophic microenvironment favourable for organomineralisation via a range of community metabolisms, including dissimilatory sulphate reduction (Visscher et al., 2000; Dupraz and Visscher, 2005; Dupraz et al., 2009). Microbial mats have influenced Earth's evolution by adopting biologically coupled redox conditions leading to the process of oxygenic photosynthesis (Dupraz et al., 2009). Delineating modern microbial mat microbial assemblages provides further understanding of early complex microbial ecosystems and primordial biogeochemical cycling.
Despite several in-depth studies of freshwater mats and microbialites (for example, Breitbart et al., 2009; Nitti et al., 2012), marine microbial mat and microbialite metagenomic studies have been constrained to Highbourne Cay in the Bahamas, leaving a large knowledge gap of the metabolic potential and community structure in other marine mat systems. Shark Bay offers a more extreme modern analogue, as it is hypersaline (>60 psu salinity) compared with Highbourne Cay that has standard marine salinity (33 to 35 psu) (Reid et al., 2000; Burns et al., 2004). Shark Bay in Western Australia harbours modern analogues consisting of several morphotypes, including lithifying and non-lithifying microbial mats (smooth and pustular, respectively), and mature columnar stromatolites (Goh et al., 2009; Jahnert et al., 2013). Previous polyphasic approaches combining bacterial culture-dependent and independent approaches by our group and others reported a high diversity of bacterial and haloarchaeal OTUs in Shark Bay stromatolites and microbial mats (Burns et al., 2004; Papineau et al., 2005; Allen et al., 2009; Goh et al., 2009). In addition, recent work has documented for the first time extensive eukaryotic diversity in mat systems including Shark Bay (Edgecomb et al., 2014), as well as detailed analyses of solute distribution in the Shark Bay mats (Pages et al., 2014).
The use of next-generation sequencing platforms in early-Earth ecosystems is still limited; however, one study indicated that there are characteristic functional profiles in different metagenomes in the Bahaman stromatolites (Dinsdale et al., 2008). A range of virulence genes were also identified that are often necessary to facilitate symbiotic relationships, and thus may provide clues to interactions between microorganisms in these systems. Metagenomic data on Bahaman stromatolites have been enlightening (Desnues et al., 2008; Khodadad and Foster, 2012; Mobberley et al., 2013), revealing a basis of a core microbial community structure (dominated by Cyanobacteria) and photosynthetic-induced carbonate precipitation. Recent in-depth studies have also combined metagenomic, stable isotopic and lipid analyses to study modern microbialites in Mexico (Breitbart et al., 2009; Nitti et al., 2012), indicating these metagenomes are capable of extensive environmental adaptation. These studies illustrate the significant potential of a metagenomic approach to unravel the underlying genetic complexity of these ecosystems; however, to date comparative data on the Shark Bay systems is lacking.
The aim of this study was thus to comprehensively decipher the core functional gene potential and complexity of Shark Bay marine microbial mats and stromatolites. We applied shotgun metagenomic sequencing of distinct Shark Bay mat and stromatolite systems for the first time, and also compared and contrasted the taxonomic and metabolic potential of previously obtained metagenomic data sets from marine stromatolites and thrombolites from Highbourne Cay, Bahamas (Khodadad and Foster, 2012; Mobberley et al., 2013). As early work suggested there were many similarities between the fabric of Shark Bay and Highbourne Cay stromatolites (Reid et al., 2003), delineating (any) differences at the functional metagenomic level may provide distinctions between the two environments. This study has significantly enhanced our understanding of marine mat and microbialite community structure, as well as the potential of fundamental processes of elemental cycling, adaptation and microbial interactions likely critical to ecosystem function.
Materials and methods
Sampling and site description
Microbial mats (smooth and pustular) and stromatolites (columnar) were collected from Nilemah tidal flats (26°25 S, 114°130 E) in April 2011 in Hamelin Pool, Shark Bay, Western Australia (Figure 1), using methods described previously (Burns et al., 2004). Samples were collected at low tide using a sterile spatula to remove mat sections at a depth of ~4 cm from the mat surface. At the time of sampling, the temperature was recorded as 29.7 oC, salinity 68 PSU and pH 8.1. Samples were placed in sterile specimen containers and stored at 4oC during transportation. DNA was extracted immediately upon sample return. Hamelin Pool in Western Australia spans an area of ~1220 km2 with an average tidal range of ~60 cm and is well protected by carbonate banks, which confine sea water inflow. In addition, Nilemah contains some of the most well-developed microbial sedimentary systems in Shark bay (Jahnert et al., 2013).
Figure 1.
Sampling site and sample descriptions. (a) The location of sample site in Hamelin Pool. (b) Shark Bay smooth mat sample. (c) Shark Bay pustular mat sample. (d) Shark Bay columnar stromatolite sample.
DNA extraction
Microbial mats (~3x3x0.5 cm3) were homogenised individually using a sterile mortar and pestle. Total mat DNA was extracted from ~100 mg of homogenised material with five biological replicates with a combination of bead beating and chemical lysis using PowerBiofilm DNA isolation kits (Mobio, Carlsbad, CA, USA) as per manufacturers instructions. One exception to the manufacturer's instruction was during the bead-beating step the FastPrep FP120 B10101 Savant (Qbiogene, Cedex, France) was employed for 30 s at full speed (6.5 scale) instead of the 10 min vortexing step. This allowed for greater DNA quality without added EPS, commonly co-extracted from the mats. DNA from each replicate was pooled and precipitated using standard ethanol precipitation. DNA concentrations were quantified using fluorometric dsDNA binding stain PicoGreen (Invitrogen, Carlsbad, CA, USA). DNA quality was determined by UV absorbance measurements (260/280 and 260/230 ratios) using a Nanodrop-1000 (Thermofisher, Wilmington, DE, USA).
Metagenomic sequencing and data analysis
Whole-shotgun sequencing was carried out with 454 pyrosequencing using Titanium chemistry. Approximately 2 μg of DNA from each sample was used for library construction then barcoded with multiplex identifier tagging. The libraries were pooled then sequenced at the Ramaciotti Centre for Genomic Analysis. For the 454 data, the raw SFF files were converted to FASTQ format and binned by molecular barcode (multiplex identifier). 454 pyrosequencing data were examined for quality using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Shark Bay metagenome barcodes were removed by Tagcleaner (Schmieder et al., 2010), sequences were trimmed for low quality (>Q25), poly-A/T/N tails, de-duplicated (100% extract match), ambiguous bases/sequences (<100 bp) and sequences with complexity (<70) on entropy scales by PRINSEQ (Schmieder and Edwards, 2011). The Shark Bay mats (smooth and pustular) sequenced here, and the Highbourne Cay mats (Type 1 and Type 3; Mobberley et al., 2013) and Highbourne Cay thrombolites (Thr A, ThrB, ThrC; Khodadad and Foster, 2012) sequences obtained from MG-RAST, were all assembled (kmer size: 39) using the Ray DeNovo Assembler (Boisvert et al., 2010, Boisvert et al., 2012; Khodadad and Foster, 2012). The Shark Bay columnar stromatolite and the Highbourne Cay stromatolite data could not be assembled and were compared as reads.
Contigs were annotated by the Metagenomic Rapid Annotations using Subsystems Technology (MG-RAST) server (Meyer et al., 2008). MG-RAST analysis of the contigs and unassembled sequences were compared against the RefSeq and SEED databases using BLAT parameters of only matches of ⩾60% similarity and ⩾15 bp that had an E-value of ⩾10-5. Metapathways pipelines were also used for MetaCyc database annotations using LAST (Local Alignment Search Tool) for matches of ⩾180 bp sequences and ⩾50 alignment score (Kiełbasa et al., 2011; Konwar et al., 2013).
Statistical analysis was completed using Statistical Analysis of Metagenomic Profiles (STAMP) (Parks and Beiko, 2010) and R (R development Core Team 2013). STAMP and R were used to analyse the MG-RAST data for 16 S SSU Greengenes (Phyla Level), RefSeq (Phyla level) and SEED subsystems (level II) results. The OTU cutoff used in this analysis is 97%. PCA analysis of the normalised RefSeq and Greengenes (SSU rRNA) classifications used R libraries Ecodist (dissimilarity-based functions for ecological analysis), and pvclust (hierarchical clustering with P-values via Multiscale Bootstrap Resampling) using ward clustering and Bray-Curtis distance metrics at a thousand replicates. MetaCyc pathways analyses were based on normalised pathways corrected for pathway size (number of genes), and number of ORFs in a given pathway, and the top 50 shared pathways extracted for relative abundance threshold above the size and ORFs. The STAMP Welch's t-test was completed using (two groups), one-sided, CI method Welch's inverted, size and with no multiple test correction using Benjamini-Hochberg FDR. Dotplots of the normalised RefSeq and 16 S SSU classifications were completed using R libraries Reshape2 with the melt function then plotted using ggplot2 (Wickham, 2009). These data were visualised using the R library ggplot2 (Wickham, 2009). Recruitment read analysis was undertaken using FR-hit using default parameters with a minimum identity >70% and an E-value >1e−5 (Niu et al., 2011), then were visualised with the R library ggplot2 (Wickham, 2009).
Metagenomic data depositing
The unassembled sequences and contigs from Shark Bay smooth and pustular mats, and columnar stromatolites are available in MG-RAST. The Ray assembled raw coverage files (.xml) are also available. The MG-RAST identification numbers are Shark Bay smooth reads (4488569.3), pustular reads (4488568.3), columnar reads (4488574.3), smooth contigs (4494617.3), pustular contigs (4494618.3), Highbourne Cay (thrombolites ThrA to C assemblies (4532758.3 to 4532760.3), mats type 1 and type 3 assemblies (4532761.3 to 4532762.3) and stromatolite (4440061.3).
Results and Discussion
Assembly of metagenomic data
To facilitate comparison of 454 FLX titanium pyrosequencing (~250 bp read length) to HiSeq Illumina sequencing (100 bp paired end) we used the metagenomic assembler RayMeta (Boisvert et al., 2012). We also compared other assemblers including Velvet (Zerbino and Birney, 2008), MetaVelvet (Namiki et al., 2012) and AbySS (Simpson et al., 2009) for our Shark Bay pyrosequencing and for the reanalysis of the Highbourne Cay data (data not shown). RayMeta out-performed all other assemblers for total bases assembled, low chimeric sequences and longer over contig length. Comparing short >100 bp read length did not provide the best annotations, and as longer read length provides greater accuracy, assembly is preferred whenever possible (Wommack et al., 2008; Liu et al., 2012).
The metagenomic assembly was comparable, even using different sequencing technologies for the marine mats and microbialites surveyed in this study. With the exception of the stromatolite metagenomes (which could not be assembled), all assembled samples had an average contig length (>400 bp), N50 (>300 bp) and median contig read lengths (>210 bp) (Supplementary Table S1). The Highbourne thrombolites sequenced on Illumina HiSeq had the longest contigs including some greater than 15 kb (Supplementary Table 1). Our assembly greatly increased read length for searchable annotations greater than threefold (~100 to >300 bp) for the Highbourne thrombolite metagenomes (Supplementary Table 1,Mobberley et al., 2013). Owing to this increase in read length sequencing technologies could be directly compared. While assembly numbers are low (>1%), this is quite common and comparable among highly complex heterogeneous metagenomic samples (Howe et al., 2014). The Shark Bay and Highbourne Cay stromatolite metagenomes did not assemble well (<500 contigs) using any de novo assembly platform (for example, AbySS, RayMeta, MetaVelvet), likely due to too few sequences and high genetic heterogeneity in the samples. The Highbourne Cay stromatolite metagenomes were sequenced on Roche 454 GS20 pyrosequencing (circa 2005), before the advent of longer-read 454 chemistries resulting in short-read lengths (<100 bp). Although DNA from stromatolite and thrombolite samples from Highbourne Cay was extracted by others utilising different approaches (Mobberley et al., 2013; Khodadad and Foster 2012), the methods of assembly employed in the present study were the least biased and most independent ways to compare all labs and datasets, supported by rarefaction analyses using protein-coding genes (RefSeq, Supplementary Figure 1). Assembly is an excellent way to obtain high confidence annotation; potentially when contigs from overlapping regions are assembled they effectively become one read, making it difficult to count absolute abundances as those reads are lost. The coverage information of each overlapping contig can be successfully used to recover this information, however, and these are freely available in MG-RAST. De Novo assembly of complex microbial communities remains an ongoing challenge and further advances in read lengths should improve this in the near future.
Sample characteristics and diversity metrics
Shark Bay samples included microbial mats (pustular and smooth) and mature columnar stromatolites (Figure 1). The smooth mat surface was characterised by a smooth, soft layer, whereas pustular mats had an amorphous rough top layer (Figure 1). The mature columnar stromatolites consists of hard layers with no obvious biomass beyond the first 3–5 mm of the top layer, whereas the smooth and pustular mats have significant biomass over a range of layers (Figure 1). Highbourne Cay and Shark Bay surrounding waters have very similar pH (~ 8) and sub-tropical water temperatures (~20–25 °C) (Baumgartner et al., 2009; Allen et al., 2009). Shark Bay differs from Highbourne mainly in salinity, containing nearly double the level (66–72 psu, compared with (33–33.5 psu), respectively (Edgecomb et al., 2014). Furthermore, the Highbourne systems are exposed to higher wave energy compared with their Shark Bay counterparts (Eckman et al., 2008), which may also impact diversity.
The Shark Bay microbial mat metagenomes exhibited higher diversity of protein-coding genes than Highbourne Cay metagenomic assembled samples. The smooth and pustular metagenomes had the highest observed alpha diversity (>700 index units Supplementary Table 2). The Highbourne Cay thrombolites had the lowest observed alpha diversity (>150 index units, Supplementary Table 2). Both the Highbourne and Shark Bay columnar stromatolites had similar alpha diversity. Previous SSU rRNA diversity studies have predicted that the Highbourne Cay stromatolites (all mat types combined) would have higher diversity than Shark Bay columnar stromatolites, likely due to salinity stress in the latter (Supplementary Table 2; Allen et al., 2009; Baumgartner et al., 2009; Mobberely et al., 2013). However, our metagenomic data suggest that both stromatolites, whether from Shark Bay or Highbourne Cay, have similar diversity (272.36 vs 294.25 index units). While diversity metrics using metagenomic data are not as standardised as single-marker genes (for example, SSU rRNA), they are useful metrics to compare whole communities.
Microbial community structure of Shark Bay microbial mats
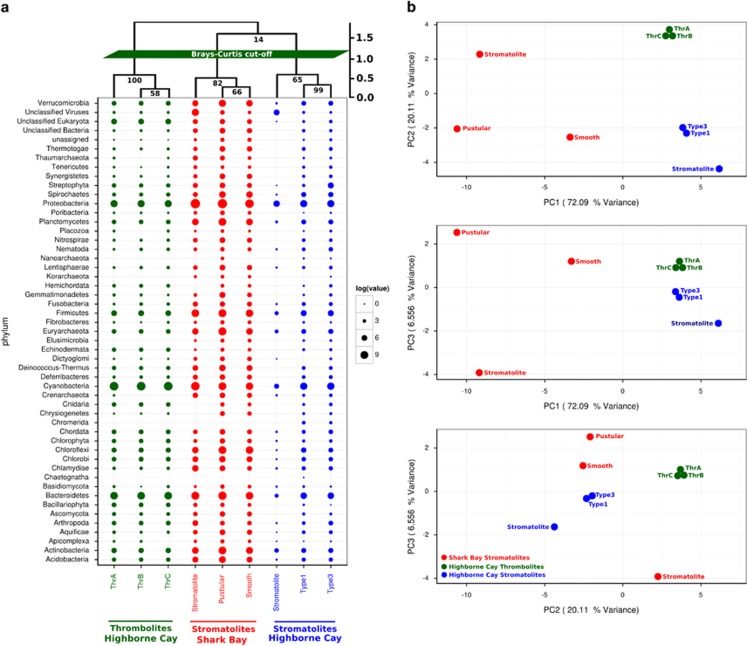
The microbial community structure was relatively similar based on the bacterial composition of Shark Bay communities across all samples, whether microbial mat (smooth and pustular) or mature columnar stromatolites. The Shark Bay bacterial communities were dominated by Proteobacteria, Firmicutes, Cyanobacteria, Bacteroidetes, Actinobacteria, Chloroflexi and Acidobacteria based on protein-coding (RefSeq, Figure 2a) and SSU ribosomal genes (GreenGenes, Figure 3a). RefSeq annotations suggest higher levels of heterotrophic taxa including Proteobacteria, Firmicutes, Chloroflexi, Actinobacteria and Acidobacteria in Shark Bay than in Highbourne Cay (Figure 3a). PCA of both protein-coding and SSU ribosomal genes further revealed that Shark Bay is distinct from both Highbourne Cay stromatolites and thrombolites in terms of microbial community structure (Figures 2b and 3b). SSU rRNA genes extracted directly from the metagenomic contigs (Shark Bay pustular and smooth mats) and metagenomic reads (Shark Bay stromatolite) suggested a greater proteobacterial presence in Shark Bay than in Highbourne Cay (>50%) (Figure 3a). Highbourne Cay thrombolites contrast Shark Bay metagenomes by a higher abundance of Cyanobacteria than Proteobacteria (Figures 2a and 3a). Proteobacterial dominance in Shark Bay has been well documented by previous 16 S rDNA clone libraries of the smooth and pustular mats, as well as for mature columnar stromatolites (Burns et al., 2004; Papineau et al., 2005; Allen et al., 2009; Goh et al., 2009), which was further supported by protein-coding and SSU ribosomal genes directly extracted from the Shark Bay metagenomes in the present study.
Figure 2.
Community composition across microbial mats, thrombolites and stromatolites based on protein-coding genes (RefSeq Database). (a) Composition and abundance of bacteria, archaea and eukaryote sequences in Shark Bay microbial mats and stromatolites, as well as Highbourne Cay stromatolites and thrombolites. (b) PCA plots were constructed from similarity matrices utilising protein coding sequence recruitment. Proportion variance was explained by each component printed next to the axes labels.
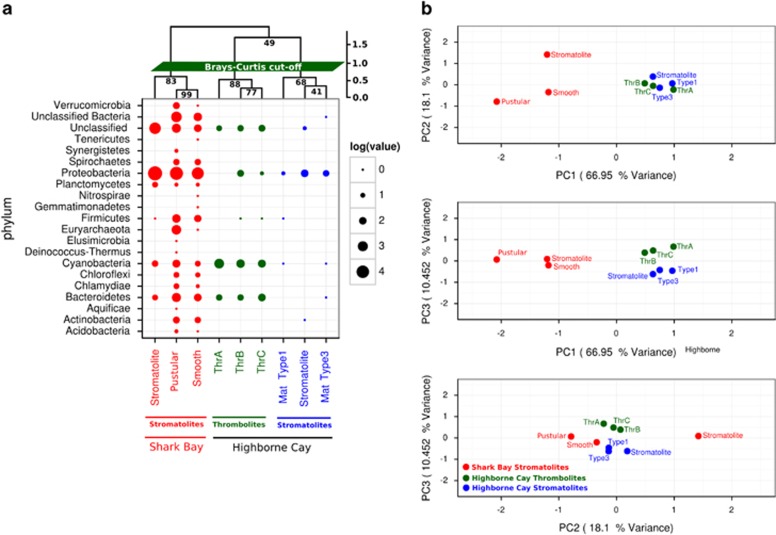
Figure 3.
Community composition across microbial mats, thrombolites and stromatolites based on 16 s rDNA. (a) Bacterial composition through 16 S rDNA recruitment from Shark Bay stromatolites metagenomes as well as Highbourne Cay thrombolites and stromatolites. (b) PCA plots were constructed from similarity matrices utilising 16 S rDNA recruitment. Proportion variance was explained by each component printed next to the axes labels. Refseq databases were used for these analyses.
Proteobacteria (for example, Alphaproteobacteria and Deltaproteobacteria), Cyanobacteria and Bacteroidetes are the core members of the marine mat microbiome. Proteobacteria, Cyanobacteria and Bacteroidetes also appear to be the dominant taxa in marine mats based on protein-coding genes (Figures 2a and 3a). The dominance of Proteobacteria (for example, Alphaproteobacteria and Deltaproteobacteria), Cyanobacteria and Bacteroidetes in marine microbialites of Highbourne Cay is confirmed by our reanalysis of the previously documented data (Baumgartner et al., 2009; Khodadad and Foster, 2012). We confirmed the observations that Highbourne Cay thrombolites are dominated by Cyanobacteria based on the reassembly of short Illumina reads into longer contigs using Ray Meta (Mobberley et al., 2013). Alphaproteobacteria and Deltaproteobacteria have been linked to carbonate precipitation by photoheterotrophic and anoxic dissimilatory sulphate reduction, which could influence the formation of modern mats (Dupraz and Visscher, 2005). Cyanobacteria are fundamental community members as they provide nutrients, structure and directly act in the microbial mat formation processes. Many filamentous cyanobacteria can influence carbonate precipitation through photosynthesis-induced alkalinisation, produce extracellular polymeric substances (EPS) that bind Ca2+ ions and provide nucleation points for precipitation (Dupraz and Visscher, 2005; Dupraz et al., 2009). Moreover, cyanobacteria classified to the order Gloeobacterales were identified in our Shark Bay mat databases, a group that has been identified as a major contributor in calcification (Couradeau et al., 2012). However, the role of this order in Shark Bay microbial mats and microbialites is still to be determined, and will be better clarified with future studies employing FISH with single-cell genomics.
Bacteroidetes have also been found among both marine (for example, Highbourne Cay) and freshwater mats (for example, Lake Van) in clone library surveys (López-García et al., 2005). Although Bacteroidetes have not been functionally linked to carbonate precipitation in microbial mats, certain strains encode urease, which under the geochemistry present in marine mats (for example, high alkalinity, high DIC, alkaline pH), could potentially lithify calcium carbonate (Castanier et al., 1999). Bacteroidetes are potentially dominant members of the heterotrophic microbial consortium that degrade EPS-rich cyanobacterial mats and releasing nutrients (for example, DOC, PO43– and NH3) that could lead to further mineralisation in stromatolites. However, further investigation is needed into the microbialite-specific Bacteroidetes that dominate marine microbialites, to ascertain their exact roles in these systems.
Of particular interest Shark Bay appears to be novel compared with other marine microbialite forming ecosystems in that it contains a relatively high abundance of archaea (Burns et al., 2004; Papineau et al., 2005; Allen et al., 2009; Goh et al., 2009). Analyses in the present study of protein-coding genes (RefSeq classification) confirm this and reveal that Shark Bay metagenomes have similar abundances of Crenarchaeota, although pustular mats have two log units greater abundance of Euryarchaeota (Figure 2a). Interestingly, no archaeal ribosomal sequences were obtained from columnar stromatolite metagenomes, whereas pustular mats had abundant ribosomal sequences from Euryarchaeota (Figure 3a). The archaea in Shark Bay microbial mats are principally halophiles, and many unclassified haloarchaea have been detected from Shark Bay microbial mats and stromatolites and not the surrounding seawater (Goh et al., 2009), indicating that these systems are a potential reservoir for these archaea. Halococcus hamelinensis,Haloferax elongans and Haloferax mucosum are haloarchaea that have been isolated from Shark Bay microbialites (Goh et al., 2006; Allen et al., 2008), and several studies have also characterized the genomes of these archaea (Burns et al., 2012; Lynch et al., 2012; Guddhka et al., 2015). Our data indicate evidence for higher abundance of haloarchaea among the contigs in Shark Bay than in Highbourne Cay contigs. Metagenomic read recruitment analyses undertaken in the present study (Supplementary Figure 3) also indicate H. hamelinensis sequences are present in abundance in our data sets from Shark Bay. Further investigation of the haloarchaea in Shark Bay including potential roles or niche selection due to the prevailing hypersaline conditions is ongoing.
Of potential significance to nitrogen cycling in these systems, archaea from the phylum Thaumarchaeota were relatively abundant in the Shark Bay microbial mats (Figure 2), a group of organisms possessing high nitrification rates in marine systems (Veuger et al., 2012). In contrast, more common nitrifying bacteria, such as Nitrosomomonas and Nitrobacter, were in very low abundance in the Shark Bay metagenomes examined in the present study. Hypersalinity appears to inhibit the growth of nitrifying bacteria (Jeffries et al., 2012), and this may contribute to their scarcity in the Shark Bay systems. Finally, the potential paleobiological significance of a high abundance and diversity of halophiles in the Shark Bay systems is seen with the observation that some Precambrian stromatolites have also been suggested to have formed in shallow, hypersaline settings (Flannery and Walter, 2011).
Metabolic potential of Shark Bay microbial mats
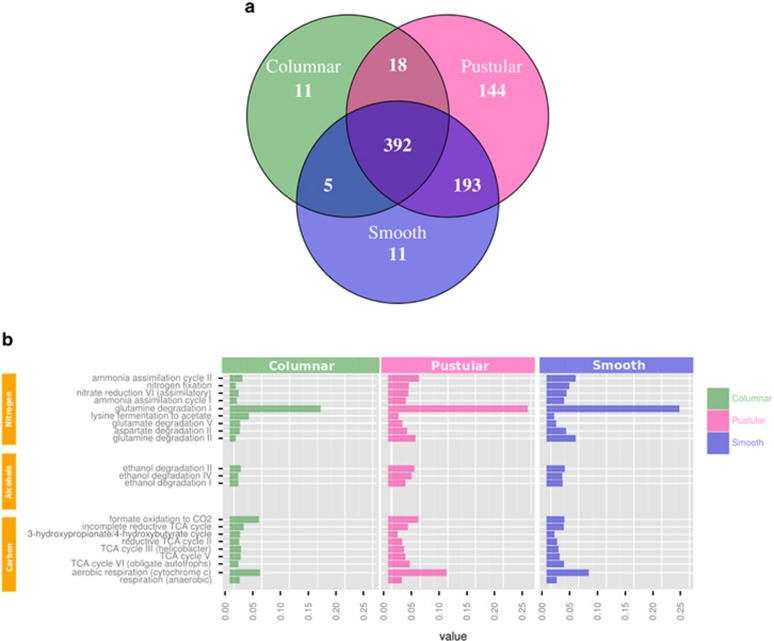
The metabolic potential of hypersaline marine mats and microbialites is relatively unknown. We compared the metabolic potential of stromatolites and microbial mats from Shark Bay against two functional databases, SEED subsystems and MetaCyc pathways, using metapathways (ePGDBs). Among the MetaCyc pathway annotations there were 392 shared pathways relating mainly to cellular homoeostasis (for example, nucleic acid, protein and carbohydrate synthesis and degradation, Figure 4a). Mature stromatolite microbial metagenomes had very few unique pathways (11) and few shared pathways compared with only pustular and/or smooth mats (18 and 5, respectively, Figure 4a). This indicated that the smooth and pustular mats are similar not just by microbial community structure, but also metabolically by sharing more pathways (193, Figure 4a), than directly with columnar stromatolites (18 and 5, Figure 4a). This suggested that while many pathways are shared, very few unique pathways exist among a single microbialite morphotype.
Figure 4.
MetaCyc pathway analysis using Metapathways (ePGDBs). (a) Venn diagram of MetaCyc pathways across Shark Bay mats and stromatolites (pustular, smooth, columnar) (b) Shared abundance of nitrogen, primary ethanol and carbon-related pathway among Shark Bay mats and stromatolites.
Elemental cycling
The cycling of key elements is not only fundamental to maintain the delicate balance between distinct microbial groups in Shark Bay microbial systems, but may also be critical in mediating mineral precipitation/dissolution events in these ecosystems. Pathway annotations of columnar stromatolites, pustular and smooth mats revealed potentially novel pathways relating to carbon fixation and fermentation (Figure 4b). From our analyses, 3-hydroxypropionate/4-hydroxybutyrate (3HP/4HB), reductive TCA II, incomplete reductive TCA II, TCA III, TCA VI (photoautotrophic common carbon fixation) and TCA V are all predicted as the central carbon fixation pathways found within Shark Bay microbial mat metagenomes. 3-hydroxypropionate/4-hydroxybutyrate and reductive TCA cycles are also found among deep-branching phyla including anoxygenic photoautotrophs such as Chloroflexus aurantiacus (Holo, 1989), as well as archaea such as the Thaumarchaeota. Chloroflexus aurantiacus also secretes 3-hydroxypropionate to assimilate acetate and CO2 (Holo, 1989). Whether this mode of carbon fixation also indirectly affects mineral precipitation in Shark Bay systems remains to be established, yet it does suggest that ancient forms of carbon fixation pathways could be active in modern day Shark Bay stromatolitic systems. The 3HP/4HB cycle identified in archaea has also been implicated as important in global carbon cycling (Berg et al., 2007), further indicating the potential significance of this carbon fixation system in Shark Bay microbial mats.
Primary alcohol degradation pathways were also present among the shared MetaCyc pathways. All three ethanol degradation pathways were present among Shark Bay mats and stromatolite (Figure 4b). Fermentation including primary alcohols can lead to dissolution via the increase of acidic by-products (Dupraz and Visscher, 2005). Ethanol degradation I, II and IV yields acetaldehyde and acetate to acetyl CoA as its final metabolic product. Ethanol can also be a sole carbon source for sulphate reducers, including Desulfovibrio (Dilling and Cypionka, 1990). Ethanol, acetate and acetaldehyde can be used as terminal electron acceptors by dissimilatory sulphate reducers as substrates that can precipitate carbonate, and thus could offset carbonate lost by direct fermentation of primary alcohols (Visscher et al., 2000; Visscher and Stolz, 2005; Dupraz and Visscher, 2005; Gallagher et al., 2012).
Among the shared pathways across the Shark Bay microbial mats, nitrogen cycling was well represented. MetaCyc pathways for nitrogen fixation, nitrate reduction VI (assimilatory) and ammonia assimilation cycle 1 and 2 were present in all samples (Figure 4b). Pustular and smooth mats had slightly higher levels of nitrogen fixation and ammonia assimilation cycle II abundance (Figure 4b). Amino-acid biosynthesis related to glutamate, aspartate and glutamine degradation had the highest relative abundance of shared MetaCyc pathways (Figure 4b). Glutamate degradation I, with its end point metabolites of ammonia and L-glutamate, was the most heavily represented pathway in all samples tested. If free ammonia is released within the mat layers it can theoretically increase pH and alkalinity pushing the saturation index towards lithification of carbonates within the local environment (Walter, 1976). Of further interest to ecosystem function, very few bacterial nitrification genes in the Shark Bay metagenomes were observed, supporting the lack of common bacterial nitrifiers (Figures 2 and 3). An incomplete nitrogen cycle may result in a build-up of potentially toxic ammonia, and, as alluded to earlier, may indicate the presence of an alternative nitrifying pathway in Shark Bay microbial mats. We propose that ammonia-oxidising Thaumarchaeota may contribute to nitrification in Shark Bay microbial mats. This group of archaea oxidise ammonia aerobically to nitrite through an unknown pathway (Vueger et al., 2012). Thaumarchaeota has also been shown to increase in abundance as salinity increases in other environments (Xie et al., 2014), and its presence in Shark Bay mats and stromatolites demonstrates that salinity may be an important factor regulating the nitrogen cycle.
Further complementing the community analyses delineating a high abundance of Proteobacteria (Figures 2 and 3), functional annotation of Shark Bay metagenomes revealed a range of diverse metabolic pathways involved in sulphur metabolism. Sulphur oxidation pathways (encoded by the Sox gene cluster) were present in all metagenomes, though data suggest a potentially incomplete Sox pathway (Supplementary Table 3), and are conserved across the Proteobacteria (Friedrich et al., 2001). Sulphate reduction pathways consisting of sulphate adenyltransferase (Sat), and adenylylsulphate reductase (apsAB) were identified across all metagenomes; however, dissimilatory sulphite reductase (dsrAB) was not observed (Supplementary Table 3). Finally, annotated genes involved in organic sulphur assimilation in Shark Bay could be grouped into alkanesulphonate assimilation and utilisation, glutathione utilisation, L-cysteine metabolism, taurine utilisation and dimethylsulphoniopropionate catabolism. The significance of sulphur metabolism in the Shark Bay microbial mats was seen in a recent study examining the distribution of sulphur species in these systems (Pages et al., 2014), identifying specific niches of sulphide production coinciding with cyanobacterial photosynthesis, suggesting metabolic cooperation in these modern systems.
In terms of phosphorous cycling, the Shark Bay metagenomes analysed here contained functional annotations clustering with phosphate, polyphosphate, phosphonate and alkylphosphonate metabolism, as well as cyanobacterial phosphorus accumulation and phosphonate biosynthesis. Interestingly, an early study of phosphorus concentration in Shark Bay sediments indicated phosphate scarcity in the hypersaline area of Shark Bay (Atkinson, 1987). Shark Bay metagenomes have a near complete Pho regulon pathway (an inducible phosphate starvation sensor regulon) in all samples suggesting these communities may be well adapted to phosphate limitation. Shark Bay microbial mats also have a high abundance of alkaline phosphatases, consistent with other microbialite forming ecosystems, such those found in a soda lake in Mexico (Valdespino-Castillo et al., 2014). These alkaline phosphatases potentially allow for a greater dissolved organic phosphate utilisation (Valdespino-Castillo et al., 2014). On the basis of this we suspect that these alkaline phosphatases would allow for higher dissolved organic phosphate utilisation in the hypersaline Shark Bay mat systems. Microbialites from other low-P environments have also been shown to metabolise other sources of P such as phosphonates or polyphosphate (Breitbart et al., 2009). Exopolyphosphatase and polyphosphate kinase genes were identified in all Shark Bay metagenomes, which may facilitate polyphosphate accumulation in low P environments. It has been suggested that polyphosphate was present on early Earth during the prebiotic era, and the regulatory functions of polyphosphate in adaptive responses to high salinity, heavy metal toxicity and oxidative stress have been described (Achbergerova and Nahalka, 2011, Seufferheld et al., 2008). Thus, the presence of polyphosphate metabolism genes in Shark Bay systems also indicates a putative role of polyphosphate as an energy source and, potentially, a precursor of stress adaptation on early Earth.
Stress response
One of the defining features of Shark Bay mats and microbialites is they are constantly subjected to a range of conditions including variable temperature, salinity, UV, desiccation and oxidative stress (Burns et al., 2009). Salinity in particular is regarded as a determining factor affecting the global distribution of microbial communities (Lozupone and Knight, 2007). Hypersaline conditions in Shark Bay are the main stressors that likely select for microbial community structure and may account for the high abundance of halophilic archaea present among the microbial mats. Furthermore, osmoadaptive mechanisms in several cyanobacteria and archaea isolated from Shark Bay stromatolites have been described (Goh et al., 2010, 2011), and the current metagenomic analyses delineated osmoadaptive mechanisms as a whole in these communities. Functional annotation of Shark Bay metagenomes here identified clusters of osmoadaptive traits, with the majority of the sequences categorised as choline and betaine uptake and betaine biosynthesis (Supplementary Table 4). Genes encoding pathways involved in trehalose biosynthesis were also observed, and, although some annotation for osmoadaptive mechanisms involved in potassium uptake were observed, they were in lower abundance compared with compatible solute-related adaptive traits. The data suggest that compatible solutes likely have dominant roles in osmoadaptive mechanisms in these communities, supporting previous studies (Goh et al., 2010; 2011). In addition, both glycine betaine and trehalose constitute a significant carbon source for microorganisms, and glycine betaine can represent up to 20% of total nitrogen in the surface layers of hypersaline sediments (Welsh, 2000). It is possible the cycling of compatible solutes is critical in the Shark Bay systems as both a protective mechanism as well as potential source of carbon and energy in times of stress.
Of potential interest, all Shark Bay metagenomes had functional annotations for a range of pathways involved in heavy metal cycling, including those for zinc, chromium, mercury, copper, cadmium and arsenic (Supplementary Table 5). Arsenic metabolism genes included those encoding arsenate reductase, arsenic resistance proteins and arsenical resistance operon repressors. Arsenic cycling is of particular interest in modern microbial mats, with a recent study providing evidence for arsenic cycling in fossil stromatolites over 2.7 billion years old (Sforna et al., 2014). Although an early study reported cadmium accumulation in several species of molluscs in Shark Bay (McConchie and Lawrance, 1991), further work is needed to elucidate the exact importance of heavy metals in these systems.
Comparative metabolic potential of marine mats and microbialites
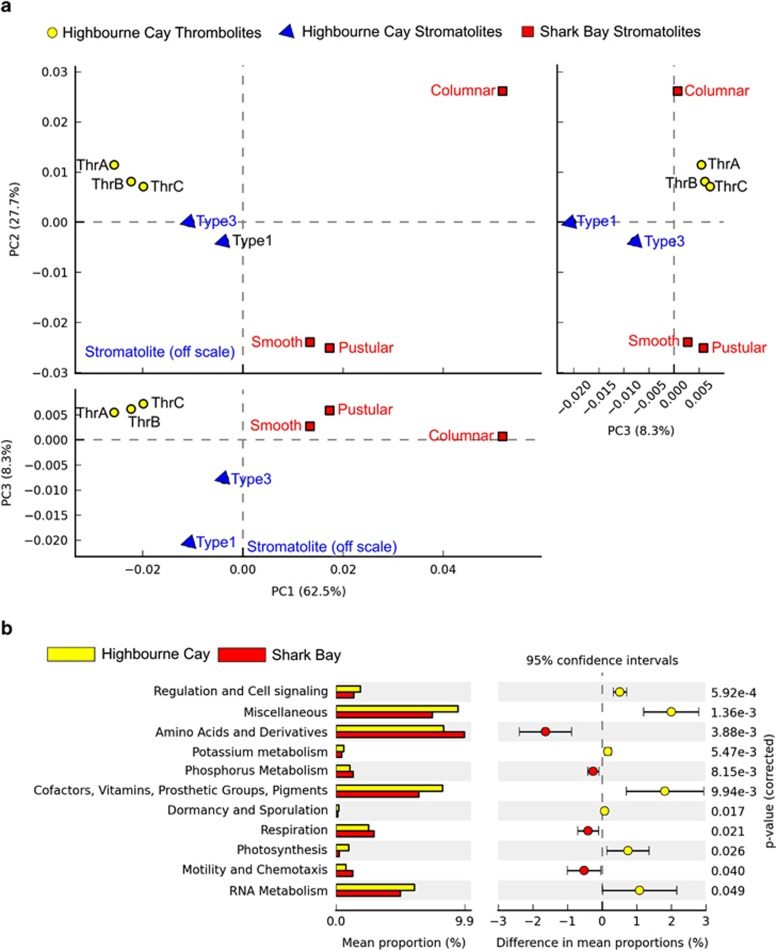
Principal coordinate analyses revealed that the metabolic potential of Shark Bay metagenomes are distinct from those at Highbourne Cay (SEED, Level I annotations; Figure 5a). Phosphorus, amino acid, respiration, motility and chemotaxis metabolic pathways are statistically significantly more abundant in Shark Bay versus Highbourne Cay metagenomes (SEED, Level I annotations; Figure 5b). Photosynthesis is statistically greater in relative abundance in Highbourne Cay compared to Shark Bay metagenomes (Figure 5b). This suggests that Shark Bay carbonate precipitation metabolic potential is more heterotrophic in nature (dissimilatory sulphate/nitrate reduction and/or ammonification from amino acids) than the photosynthetic-based carbonate precipitation at Highbourne Cay. Furthermore, functional level classification at the next hierarchical level (SEED Subsystem Level II) revealed further differences between the two environments (Supplementary Figure 2). In particular, statistically significant categories related to stress adaptation (osmotic stress, periplasmic stress, antiporters) were significantly enriched in Shark Bay over Highbourne Cay (Supplementary Figure 2 P-value<0.01). This is not surprising given the hypersaline setting of Shark Bay; however, it may be that environmental factors such as salinity determine major differences in functional potential between the two settings. Furthermore, both sulphur metabolism and phosphorus and phosphate metabolism pathway abundance was significantly enriched in Shark Bay over Highbourne Cay (Supplementary Figure 2, P-value<0.05).
Figure 5.
Principal coordinate analyses comparison of the metabolic potential of Shark Bay and Highbourne Cay metagenomes. (a) PCA plots show the results from the STAMP ANOVA analyses. ANOVA used multiple groups and a post hoc test (Tukey-Kramer at 0.95), an effect size (Eta-squared), and a q-value (<0.05) of 11 active only features for SEED level I. A multiple-test correction using Benjamini-Hochberg FDR was employed for microbial community structure using class-level classification. (b) The chart depicts post-havoc confidence interval plots (>95%) based on ANOVA parameters including SEED level I for Highbourne Cay vs Shark Bay carried out in STAMP. Similarities and differences between Highbourne Cay and Shark Bay microbial mats are shown.
The greater level of photosynthetic metabolism genes present in Highbourne Cay (especially thrombolites) are likely due to the dominance of the microbial communities by cyanobacteria. As a further indicator of the potential importance of salinity in defining microbial mat and microbialite communities, salt stress has been shown to inhibit the photosynthetic capability of cyanobacteria (Allakhverdiev and Murata, 2008; Hagemann, 2011), which may further explain these metagenomic differences observed. Furthermore, it has been suggested that Pavilion Lake microbialite heterotrophs contribute to the cementation process of thrombolites by subsequent in-filling of microstructures (Omelon et al., 2013), a process that could potentially be occurring in Shark Bay systems.
Conclusions
Modern marine microbial mats represent one of the best windows to the past, and the present study is the first detailed metagenomic analysis of the modern ecosystems in Shark Bay. Shark Bay differs greatly in salinity content from the open marine Highbourne Cay, and salinity may be a major factor influencing ecosystem function in Shark Bay. However, the two environments do contain the same functional groups likely involved in carbonate precipitation, including filamentous cyanobacteria and sulphate-reducers. Shark Bay also appears to have a greater abundance of halophilic archaea not found in other microbialite-forming ecosystems to date and likely a result of higher salinities. Alternative non-rubisco-based carbon fixation including reductive 3-hydroxypropionate/4-hydroxybutyrate pathways is highly represented in Shark Bay metagenomes, and further work may help clarify whether these pathways influence, for example, precipitation processes. The present study has revealed key insights into metagenomic potential in modern marine mats and microbialites, and hinted at the possibility of novel microbial interactions that drive ecosystem function. While sequence data alone will never completely characterise microbial mat and microbialite communities, our work has defined the Shark Bay ‘stromatonome' and provides a platform for further detailed analyses on the impact of eukaryotic and viral diversity, as well as targeted and global gene expression studies under different environmental conditions (for example, season, salinity), to comprehensively understand these complex microbial ecosystems.
Acknowledgments
This work was funded by the Australian Research Council. Metagenomic sequencing was undertaken at the Ramaciotti Centre for Genomics. Special thanks to Niels W Hanson and Kishori M Konwar for python and R scripts for parsing metapathways data.
The authors declare no conflict of interests.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Achbergerova L, Nahalka J. (2011). Polyphosphate-an ancient energy source and active metabolic regulator. Microb Cell Fact 10: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allakhverdiev S, Murata N. (2008). Salt stress inhibits photosystems II and I in cyanobacteria. Photosynthesis Res 98: 529–539. [DOI] [PubMed] [Google Scholar]
- Allen MA, Goh F, Leuko S, Echigo A, Mizuki T, Usami R et al. (2008). Haloferax elongans sp. nov. and Haloferax mucosum sp. nov., isolated from microbial mats from Hamelin Pool, Shark Bay, Australia. Int J Syst Evol Microbiol 58: 798–802. [DOI] [PubMed] [Google Scholar]
- Allen MA, Goh F, Burns BP, Neilan BA. (2009). Bacterial, archaeal and eukaryotic diversity of smooth and pustular microbial mat communities in the hypersaline lagoon of Shark Bay. Geobiology 7: 82–96. [DOI] [PubMed] [Google Scholar]
- Atkinson MJ. (1987). Low phosphorus sediments in a Hypersaline Marine Bay. Estuar Coast Shelf S 24: 335–347. [Google Scholar]
- Baumgartner LK, Spear JR, Buckley DH, Pace NR, Reid RP, Dupraz C et al. (2009). Microbial diversity in modern marine stromatolites, Highbourne Cay, Bahamas. Environ Microbiol 11: 2710–2719. [DOI] [PubMed] [Google Scholar]
- Berg IA, Kockelkorn D, Buckel W, Fuchs G. (2007). A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Science 318: 1782–1786. [DOI] [PubMed] [Google Scholar]
- Bernhard JM, Edgecomb VP, Visscher PT, McIntyre-Wressig A, Summons RE, Bouxsein ML, Louis L et al. (2013). Insights into foraminiferal influences on microfabrics of microbialites at Highbourne Cay, Bahamas. Proc Natl Acad Sci USA 110: 9830–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Laviolette F, Corbeil J. (2010). Ray: simultaneous assembly of reads from a mix of high-throughput sequencing technologies. J Comput Biol 17: 1519–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil JRay Meta: scalable de novo metagenome assembly and profiling. (2012) Genome Biol 13: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hoare A, Nitti A, Siefert J, Haynes M, Dinsdale E et al. (2009). Metagenomic and stable isotopic analyses of modern freshwater microbialites in Cuatro Cienegas, Mexico. Environ Microbiol 11: 16–34. [DOI] [PubMed] [Google Scholar]
- Burns BP, Goh F, Allen M, Neilan BA. (2004). Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ Microbiol 6: 1096–1101. [DOI] [PubMed] [Google Scholar]
- Burns BP, Anitori R, Butterworth P, Henneberger R, Goh F, Allen MA et al. (2009). Modern analogues and the early history of microbial life. Precambrian Res 173: 10–18. [Google Scholar]
- Burns BP, Gudhka R, Neilan BA. (2012). Genome sequence of the halophilic archaeon Halococcus hamelinensis. J Bacteriol 194: 2100–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanier S, Metayer-Levrel GL, Perthuisot JP. (1999). Ca-carbonates precipitation and limestone genesis — the microbiogeologist point of view. Sediment Geol 126: 923. [Google Scholar]
- Couradeau E, Benzerara K, Gerard E, Moreira D, Bernard S, Brown Jr GE et al. (2012). An early-branching microbialite cyanobacterium forms intracellular carbonates. Science 336: 459–462. [DOI] [PubMed] [Google Scholar]
- Desnues C, Rodriguez-Brito B, Rayhawk S, Kelley S, Tran T, Haynes M et al. (2008). Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452: 340–343. [DOI] [PubMed] [Google Scholar]
- Dilling W, Cypionka H. (1990). Aerobic respiration in sulfate-reducing bacteria. FEMS Microbial Lett 71: 123–128. [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452: 629–632. [DOI] [PubMed] [Google Scholar]
- Dupraz C, Visscher PT. (2005). Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol 13: 429–438. [DOI] [PubMed] [Google Scholar]
- Dupraz C, Reid RP, Braissant O, Decho AW, Norman RS, Visscher PT. (2009). Processes of carbonate precipitation in modern microbial mats. Earth Sci Rev 96: 141–162. [Google Scholar]
- Eckman JE, Andres MS, Marinelli RL, Bowlin E, Reid RP, Aspden RJ et al. (2008). Wave and sediment dynamics along a shallow subtidal sandy beach inhabited by modern stromatolites. Geobiology 6: 21–32. [DOI] [PubMed] [Google Scholar]
- Edgecomb VP, Bernhard JM, Summons RE, Orsi W, Beaudoin D, Visscher PT. (2014). Active eukaryotes in microbialites from Highbourne Cay, Bahamas, and Hamelin Pool (Shark Bay), Australia. ISME J 8: 418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery DT, Walter MR. (2011). Archean tufted microbial mats and the great oxidation event: new insights into an ancient problem. Aust J Earth Sci 59: 1–11. [Google Scholar]
- Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, Fischer J. (2001). Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl Environ Microbiol 67: 2873–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Kading TJ, Braissant O, Dupraz C, Visscher PT. (2012). Inside the alkalinity engine: the role of electron donors in the organomineralization potential of sulfate-reducing bacteria. Geobiology 10: 518–530. [DOI] [PubMed] [Google Scholar]
- Goh F, Leuko S, Allen MA, Bowman JP, Kamekura M, Neilan BA et al. (2006). Halococcus hamelinensis sp. nov., a novel halophilic archaeon isolated from stromatolites in Shark Bay, Australia. Int J Syst Evol Microbiol 56: 1323–1329. [DOI] [PubMed] [Google Scholar]
- Goh F, Allen MA, Leuko S, Kawaguchi T, Decho AW, Burns BP et al. (2009). Determining the specific microbial populations and their spatial distribution within the stromatolite ecosystem of Shark Bay. ISME J 3: 383–396. [DOI] [PubMed] [Google Scholar]
- Goh F, Barrow KD, Burns BP, Neilan BA. (2010). Identification and regulation of novel compatible solutes from hypersaline stromatolite-associated cyanobacteria. Arch Microbiol 192: 1031–1038. [DOI] [PubMed] [Google Scholar]
- Goh F, Jeon YJ, Barrow K, Neilan BA, Burns BP. (2011). Osmoadaptive strategies of the archaeon Halococcus hamelinensis isolated from a hypersaline stromatolite environment. Astrobiology 11: 529–536. [DOI] [PubMed] [Google Scholar]
- Gudhka R, Neilan BA, Burns BP. (2015). Adaptation, ecology, and evolution of the halophilic stromatolite archaeon Halococcus hamelinensis inferred through genome analyses. Archaea 2015: 241608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M. (2011). Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35: 87–123. [DOI] [PubMed] [Google Scholar]
- Holo H. (1989). Chloroflexus aurantiacus secretes 3-hydroxypropionate, a possible intermediate in the assimilation of CO2 and acetate. Arch Microbiol 151: 252–256. [Google Scholar]
- Howe AC, Jansson JK, Malfatti SA, Tringe SG, Tiedje JM, Brown CT. (2014). Tackling soil diversity with the assembly of large, complex metagenomes. Proc Natl Acad Sci USA 111: 4904–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnert R, Collins L, Ariztegui D. (2013). Controls on microbial activity and tidal flat evolution in Shark Bay, Western Australia. Sedimentology 60: 1071–1099. [Google Scholar]
- Jeffries TC, Seymour JR, Newton K, Smith RJ, Seuront L, Mitchell JG. (2012). Increases in the abundance of microbial genes encoding halotolerance and photosynthesis along a sediment salinity gradient. Biogeosciences 9: 815–825. [Google Scholar]
- Khodadad C, Foster J. (2012). Metagenomic and metabolic profiling of nonlithifying and lithifying stromatolitic mats of Highborne Cay, The Bahamas. PloS One 7: e38229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa S, Wan R, Sato K, Horton P, Frith M. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res 21: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konwar K, Hanson N, Pagé A, Hallam S. (2013). MetaPathways: a modular pipeline for constructing pathway/genome databases from environmental sequence information. BMC Bioinform 14: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranendonk MJ, Philippot P, Lepot K, Bodorkos S, Pirajno F. (2008). Geological setting of Earth's oldest fossils in the ca. 3.5 Ga Dresser Formation, Pilbara Craton, Western Australia. Precambrian Res 167: 93–124. [Google Scholar]
- Liu KL, Porras-Alfaro A, Kuske CR, Eichorst SA, Xie G. (2012). Accurate, rapid taxonomic classification of fungal large-subunit rRNA genes. Appl Environ Microbiol 78: 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Kazmierczak J, Benzerara K, Kempe S, Guyot F, Moreira D. (2005). Bacterial diversity and carbonate precipitation in the giant microbialites from the highly alkaline Lake Van, Turkey. Extremophiles 9: 263–274. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch EA, Langille MG, Darling A, Wilbanks EG, Haltiner C, Shao KS et al. (2012). Sequencing of seven haloarchael genomes reveals patterns of genomic flux. Plos One 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvasi M, Visscher PT, Casillas Martinez L. (2010). Extracellular polymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett 313: 1–9. [DOI] [PubMed] [Google Scholar]
- McConchie DM, Lawrance LM. (1991). The origin of high cadmium loads in some bivalve mollusks from Shark Bay, Western Australia: a new mechanism for cadmium uptake by filter feeding organisms. Arch Environ Con Tox 21: 303–310. [DOI] [PubMed] [Google Scholar]
- Mobberley JM, Khodadad CLM, Foster JS. (2013). Metabolic potential of lithifying cyanobacteria-dominated thrombolitic mats. Photosynthesis Res 118: 125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki T, Hachiya T, Tanaka H, Sakakibara Y. (2012). MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res 40: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitti A, Daniels CA, Siefert J, Souza V, Hollander D, Breitbart ME. (2012). Spatially resolved genomic, stable isotopic, and lipid analyses of a modern freshwater microbialite from Cuatro Ciénegas, Mexico. Astrobiology 12: 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Zhu Z, Fu L, Wu S, Li W. (2011). FR-HIT, a very fast program to recruit metagenomic reads to homologous reference genomes. Bioinformatics 27: 1704–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelon CR, Brady AL, Slater GF, Laval B, Lim DSS, Southam G. (2013). Microstructure variability in freshwater microbialites, Pavilion Lake, Canada. Palaeogeo Palaeoclim Palaeoecol 392: 62–70. [Google Scholar]
- Pages A, Welsh DT, Teasdale PR, Grice K, Vacher M, Bennet WW et al. (2014). Diel fluctuations in solute distribution and biogeochemical cycling in a hypersaline microbial mat from Shark, WA. Marine Chem 167: 102–112. [Google Scholar]
- Papineau D, Walker JJ, Mojzsis SJ, Pace NR. (2005). Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl Environ Microbiol 71: 4822–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Beiko RG. (2010). Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26: 715–721. [DOI] [PubMed] [Google Scholar]
- Reid RP, Visscher PT, Decho AW, Stolz JF, Bebout BM, Dupraz C et al. (2000). The role of microbes in accretion, lamination and early lithification of modern marine stromatolites. Nature 406: 989–992. [DOI] [PubMed] [Google Scholar]
- Reid RP, James NP, Macintyre IG, Dupraz CP, Burne RV. (2003). Shark Bay stromatolites: microfabrics and reinterpretations of origins. Facies 49: 299–324. [Google Scholar]
- Schmieder R, Lim WY, Rohwer R, Edwards R. (2010). TagCleaner: Identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinformatics 11: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Arp G, Reimer A, Reitner J, Daniel R. (2013). Phylogenetic Analysis of a Microbialite-Forming Microbial Mat from a Hypersaline Lake of the Kiritimati Atoll, Central Pacific. Plos One 8: e66662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufferheld MJ, Alvarez HM, Farias ME. (2008). Role of polyphosphates in microbial adaptation to extreme environments. Appl Environ Microbiol 74: 5867–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sforna MC, Philippot P, Somogyi A, van Zuilen MA, Medjoubi K, Schoepp-Cothenet B et al. (2014). Evidence for arsenic metabolism and cycling by microorganisms 2.7 billion years ago. Nature Geosci 7: 811–815. [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. (2009). ABySS: a parallel assembler for short read sequence data. Genome Res 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdespino-Castillo PM, Alcántara-Hernández RJ, Alcocer J, Merino-Ibarra M, Macek M, Falcón LI. (2014). Alkaline phosphatases in microbialites and bacterioplankton from Alchichica soda lake, Mexico. FEMS Microbiol Ecol 90: 504–519. [DOI] [PubMed] [Google Scholar]
- Veuger B, Pitcher A, Schouten S, Sinninghe Damsté JS, Middelburg JJ. (2012). Nitrification and growth of autotrophic nitrifying bacteria and Thaumarchaeota in the Coastal North Sea. Biogeosciences Dis 9: 16877–16906. [Google Scholar]
- Visscher PT, Reid RP, Bebout BM. (2000). Microscale observation of sulfate reduction: Evidence of microbial activity forming lithified micritic laminae in modern marine stromatolites. Geology 28: 919–922. [Google Scholar]
- Visscher PT, Stolz J. (2005). Microbial communities as biogeochemical reactors. Palaeogeogr Palaeoclimatol Palaeoecol 219: 87–100. [Google Scholar]
- Walter MR. (1976) Developments in Sedimentology – Stromatolites. New York: Elsevier, 20: 123–125. [Google Scholar]
- Walter MR, Buick R, Dunlop JSR. (1980). Stromatolites 3400-3500 Myr old from the North Pole area, Western Australia. Nature 284: 443–445. [Google Scholar]
- Welsh D. (2000). Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol Rev 24: 263–290. [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer: Dordrecht, Heibelberg, London, New York. [Google Scholar]
- Wommack KE, Bhavsar J, Ravel J. (2008). Metagenomics: read length matters. Appl Environ Microbiol 74: 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Zhang C, Zhou X, Wang P. (2014). Salinity-dominated change in community structure and ecological function of Archaea from the lower Pearl River to coastal South China Sea. Appl Microbiol Biotech 98: 7971–7982. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.