Abstract
The postnatal environment, including factors such as weaning and acquisition of the gut microbiota, has been causally linked to the development of later immunological diseases such as allergy and autoimmunity, and has also been associated with a predisposition to metabolic disorders. We show that the very early-life environment influences the development of both the gut microbiota and host metabolic phenotype in a porcine model of human infants. Farm piglets were nursed by their mothers for 1 day, before removal to highly controlled, individual isolators where they received formula milk until weaning at 21 days. The experiment was repeated, to create two batches, which differed only in minor environmental fluctuations during the first day. At day 1 after birth, metabolic profiling of serum by 1H nuclear magnetic resonance spectroscopy demonstrated significant, systemic, inter-batch variation which persisted until weaning. However, the urinary metabolic profiles demonstrated that significant inter-batch effects on 3-hydroxyisovalerate, trimethylamine-N-oxide and mannitol persisted beyond weaning to at least 35 days. Batch effects were linked to significant differences in the composition of colonic microbiota at 35 days, determined by 16 S pyrosequencing. Different weaning diets modulated both the microbiota and metabolic phenotype independently of the persistent batch effects. We demonstrate that the environment during the first day of life influences development of the microbiota and metabolic phenotype and thus should be taken into account when interrogating experimental outcomes. In addition, we suggest that intervention at this early time could provide ‘metabolic rescue' for at-risk infants who have undergone aberrant patterns of initial intestinal colonisation.
Introduction
There is a growing body of evidence which suggests that disease in adulthood can originate in early-life during ‘programming events' (Huh et al., 2012; Vickers, 2014; Langley-Evans, 2015). This term refers to critical points of developmental plasticity where environmental factors, whether stimulus or insult, can have long-term consequences on physiological development (Winick and Noble, 1966). In adult humans, there is a clear association between the intestinal microbiota and metabolic disease (Wen et al., 2008; Koren et al., 2011; Bajaj et al., 2012; Everard et al., 2014). However, the links between the early-life environment and development of a healthy metabolic phenotype are not well understood. There is a great deal of evidence suggesting that microbial acquisition from the early-life environment determines development and function of the immune system (Cebra et al., 1998; Mulder et al., 2009; Lewis et al., 2012). Given the recently identified importance of the interactions between the immune and metabolic systems, here we ask the question–do factors which occur during early-life affect metabolic phenotype in the host as well as the composition of the intestinal microbiota? The long-term effects of differences in early metabolic development, influenced by early-life environment and host-microbial interactions in the gut, may be involved in the pathogenesis of many chronic conditions, including atherosclerosis, diabetes, inflammatory and cardiovascular disease and even obesity (Prentice and Moore, 2005; Wells, 2012; Cox et al., 2014). Indeed evidence suggests that preterm and/or low birth weight and subsequent nutritional intervention predisposes individuals to greater risk of neurodevelopmental conditions and to metabolic syndrome and cardiovascular disease in later life (Barker, 1999; Walhovd et al., 2012; Wang et al., 2012).
The pig is a valuable model for human nutritional studies because it shares many common features of gastrointestinal physiology, microbiology, genetics and diet (Pond and Houpt, 1978; Miller and Ullrey, 1987). Pigs are particularly useful when assessing host microbe interactions because the gut microbiota is substantially more stable over time in both pigs and humans compared with rodent models. Moreover, inter-individual variability is more similar between humans and pigs than between humans and mice; thus, in these respects; the human population is better reflected in this outbred animal species (Thompson et al., 2010). The porcine metabolic phenotype has recently been characterised across a number of biological compartments (Merrifield et al., 2011). Coupled with numerous studies using metabolic profiling to identify the impact of nutrition on health (Bertram et al., 2006; He et al., 2009; Larsen et al., 2010), these studies have validated the use of high-resolution spectroscopic techniques to assess nutritional intervention in this tractable model species. Piglets are precocial; they can be removed from the sow at birth and are therefore easily manipulated from a very early age, unlike human and rodent equivalents.
Early-life and weaning are associated with major shifts in the microbial community of the gut in both humans (Johnson and Versalovic, 2012) and other mammals (Konstantinov et al., 2006). Here, we examine the system-wide metabolic and microbiological consequences of variation in the micro-environment during the first day of life. We also ascertain the subsequent effect of nutritional intervention, in this case weaning on to diets with different protein sources. We propose that this systems approach to exploring early life environment could identify linked patterns of microbial-mammalian dysbiosis and therefore, in the long-term, may provide a platform to identify individual susceptibility to disease. In addition, a systems medicine approach has the potential to assess methods of intervention during an early window of opportunity to limit disease risk by preventing, or correcting, early deviation from intestinal homeostasis.
Materials and methods
Animal husbandry
Animal work was performed according to local ethical guidelines under a UK Home Office License, approved by the local ethical review group. The six large White hybrid, out-bred sows (which had previously had 1–2 litters) used in this experiment were chosen randomly from a commercial herd of approximately 250 animals which were housed all together in a single, open-ended barn. Normal practice on this farm is to artificially inseminate 20 appropriate sows on a weekly basis, and then to move them to one of four farrowing units (temperature controlled at 22 °C plus heat-mats for the piglets) just before the sows are due to give birth. The piglets are normally weaned during their fourth week of life and moved to alternative housing, while their mothers rejoin the herd and are artificially inseminated 4–5 days later. Consequently, farrowing units are run on a 4-week rotation and are steam-cleaned and left standing for 1 day before the next group of sows are brought in to give birth. During this time, the farrowing facility is sprinkled with a dry disinfectant (Naturclean, SCA NuTec, Staffordshire, UK) to reduce damp and microbial load. Experimental piglets were taken from normal sows undergoing this process. There were no symptoms of disease, no antibiotic treatment was given and there were no differences in the diet fed to any of the farm sows during the pregnancy and lactation periods of the specific sows used for these experiments.
During the autumn, three sows (contributing to batch 1) were artificially inseminated using pooled semen from three boars. Eight weeks later, three additional sows (contributing to batch 2) were artificially inseminated using pooled semen from the same three boars. Shortly before sows contributing to batch 1 were due to farrow, they were removed to a farrowing unit and two piglets from each of the resulting litters (n=6) were removed to the University of Bristol's piglet isolator facility at the age of 1 day. This isolator provided a highly controlled, standardised environment: specific pathogen-free, high efficiency particulate air-filtered, positive pressure environment, which is thoroughly cleaned and fumigated with formaldehyde gas (Alphagen Prills pellets, Antec AH International, Sudbury, UK) for 12 h before each experimental run. The piglets were housed together within this isolator for 1 day while they learnt to drink bovine-based formula milk (Piggimilk,Volac Ltd, Sleaford, Lincolnshire, UK). The nutritional composition of the formula is summarised in Supplementary Table S1 and is similar to that given to human infants. To minimise the time taken for all of the piglets to learn to drink from bowls, piglets were housed in groups to learn by emulation. At the age of 2 days, the piglets were moved to individual units, within the same isolator, and fed hourly for 14 days. At 14 days of age, the piglets were litter- and gender-matched into two groups before weaning at 21 days, to separate the effects of weaning from those due to the stress of mixing. One group of piglets were weaned onto a diet in which the protein source was egg, while the other group received a soya-based diet. Both diets were nutritionally balanced and supplemented with appropriate levels of vitamins and minerals (Volac Ltd). The nutritional composition of each of these diets is detailed in Supplementary Table S1. There were no traces of egg in the soya diet, nor soya in the egg diet; a biosecurity barrier was established between the two groups to avoid food crossover. Four weeks after batch 1 sows vacated the farrowing unit, batch 2 sows entered the same unit in preparation for farrowing. An additional group of sows had occupied the unit between batch 1 and batch 2. Again, two piglets were removed from each of the three batch 2 sows at the age of 1 day and underwent exactly the same process as batch 1 piglets. A summary of the experimental design is depicted in Figure 1a and a more detailed schematic is available in Supplementary Figure S3.
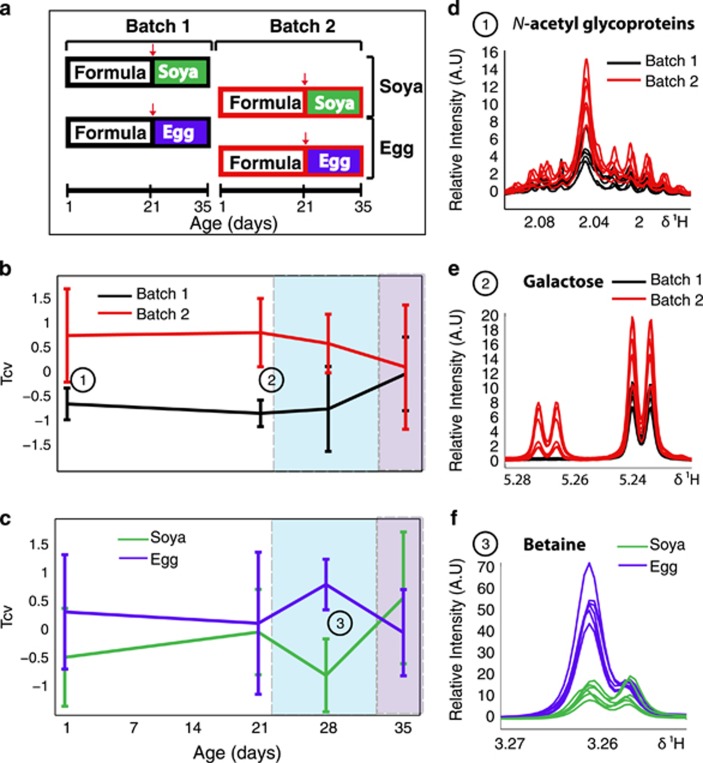
Figure 1.
The temporal effects of early-life environment and weaning diet on the metabolic profile of porcine blood serum. (a) Represents the experimental setup, where batch 1 and 2 correspond to the experimental replicate. Animals were fed formula at 1 day until 21 days, at which point they were weaned onto either an egg- or soya-based diet (weaning is denoted by red arrow). At each time-point, a pairwise comparison was made by O-PLS-DA (mean-centred scaling), between either the two experimental batches; (b) (n=6 per group) or the two weaning diets; (c) (n=6 per group) and the mean of the cross-validated scores (Tcv) from each model (± s.d.) are plotted at 1, 21, 28 and 35 days. The spectral regions derived from 600MHz Carr-Purcell-Meiboom-Gill (CPMG) metabolic profiles of serum of selected discriminatory metabolites at each time-point have been plotted; (d) shows increased serum N-acetyl glycoproteins in batch 2 at 1 day; (e) shows increased galactose in batch 2 at 21 days and (f) depicts increased in betaine in the egg-fed animals at 28 days.
This experimental design resulted in n=6, gender-matched, litter-matched piglets in the treatment groups (weaned onto either an egg diet or soya diet) and n=6, gender-matched piglets in each replicate (batch 1 and batch 2). Replicates could not be litter-matched because they were born 8 weeks apart. Detailed weather conditions, including temperature, wind speed, rainfall, hours of sunshine and global radiation, throughout the whole experiment are shown in Supplementary Figure S4, with the start date of each replicate indicated.
All piglets were bled by venipuncture at the age of 1 day (upon removal from the mother), 21 days (immediately before weaning) and 28 days and samples were stored at −80 °C. In addition, fresh faecal samples were also obtained at 21 days, just prior to weaning, snap-frozen in liquid nitrogen and stored at −80 °C. At 35 days, piglets were killed with an overdose of pentobarbitone (Euthatal, Merial Animal Health, Essex, UK). At post-mortem, heart blood, colonic (descending colon, adjacent to the colo-rectal junction) contents and mucosal scrapings, mesenteric lymph node (MLN) draining the distal jejunum and liver (tip of the same lobe in each piglet) were recovered; tissues, colon content and mucosal scrapings were snap-frozen at post-mortem and stored at −80 °C. Urine was removed directly from the bladder via a sterile syringe, snap-frozen and stored at −40 °C pending nuclear magnetic resonance (NMR) analysis.
Sample preparation for 1H NMR
Serum
Blood serum samples were prepared by the addition of 350 μl 0.9% saline solution containing 10% D2O (to act as a spectrometer field frequency lock) to 200 μl of serum. This mix was then vortexed, centrifuged at 12 000 g for 20 min and transferred into 5 mm outer diameter NMR tubes in preparation for analysis.
Urine
Urine samples were prepared by the addition of 220 μl of a 1 mM 3-trimethylsilyl-1-[2,2,3,3,-2H4] propionate, 3 mM sodium azide (NaN3) and 80/20 (v/v) H2O/D2O phosphate buffer solution (pH 7.4) to 440 μl of urine. This mixture was vortexed, left to stand for 20 min and then centrifuged at 12 000 g for 20 min. Six hundred microlitres of the supernatant was transferred to 5 mm outer diameter NMR tubes (Beckonert et al., 2007).
Tissues
For high-resolution magic angle spinning NMR spectroscopy, intact tissue samples from above (~15 mg) were immersed in a 0.9% saline D2O solution within a zirconium oxide 4mm diameter rotor, and an insert was used to make a spherical sample volume up to 25 μl.
1H NMR spectroscopy
Serum
Serum samples were collected longitudinally throughout the experiment as previously described. The 1H NMR metabolic profiles of these samples were acquired on an Avance 600 MHz NMR spectrometer (Bruker Biospin, Billerica, MA, USA) and spectra were collected using a BACS autosampler with a Bruker 5 mm TXI triple resonance probe at 300 K. A total of 256 scans (16 dummy scans) were collected into 64 k data points over a 20 p.p.m. spectral width using a Carr-Purcell-Meiboom-Gill spin echo sequence, incorporating an echo time of 200 μs to highlight contributions from low-molecular weight moieties.
Urine
The metabolic profile of urine from each of the piglets (collected at 35 days) was obtained by NMR spectroscopy. All spectra were acquired on an Avance 600 MHz NMR spectrometer (Bruker Biospin) and spectra were collected using a BACS autosampler with a Bruker 5 mm TXI triple resonance probe at 300 K. A total of 256 scans (16 dummy scans) were collected into 64 k data points over a 20 p.p.m. spectral width using the first increment of a standard one dimensional experiment with water presaturation during the relaxation delay of 2 s and during the mixing time of 100 ms.
Tissues
Sections from the liver and the MLN were analysed by 1H high-resolution magic angle spinning-NMR as described previously (Merrifield et al., 2011) using a 4 mm g-high-resolution magic angle spinning HPCD probe at 283 K. A total of 256 scans (16 dummy scans) were collected into 64 k data points over a 20 p.p.m. spectral width using a Carr-Purcell-Meiboom-Gill spin echo sequence, incorporating an echo time of 200 μs to highlight contributions from low-molecular weight moieties.
Quantification of the gastrointestinal microbiota using 16S DNA pyrosequencing
DNA was extracted from faecal and colonic content samples using the Qiagen EZ1 kit (Qiagen, Hilden, Germany) after mechanical disruption as follows: for colon content, 100 mg of material was placed in lysozyme buffer (50 mg ml−1) with 0.3 g of glass beads, disrupted in a bead-beater at maximal speed for 1 min and then incubated at 37 °C for 15 min; for colon tissue, 200 mg were placed in Lysing Matrix Tubes D with lysozyme buffer and homogenised with FastPrep-24 (MP Biomedicals, Solon, OH, USA), 2 × 20 s speed 6, with 5-min break. After centrifugation (5 min, 200 g), supernatant was transferred into tubes with 0.3 g of glass beads, disrupted in a bead-beater at 4 °C at maximal speed for 1 min and then incubated at 37 °C for 15 min.
The V1, V2 and V4 regions of the 16 S genes contained in the DNA extracts were amplified, sequenced in multiplex using the GS FLX System (Roche, Rotkreuz, Switzerland), and analysed as previously described (Claus et al., 2011), except for the confidence threshold of the RDP Classifier which was set to 60%, as in previous published studies (Dominguez-Bello et al., 2011; Koren et al., 2011). All data analysis was conducted on the mean relative proportion of bacteria (%). Owing to the high proportion (~35%) of unclassified bacteria of pig origin identified at the genus level, we have used the family level as a trade-off between the coverage of the microbial population and the depth of taxonomical analysis.
Data analysis
An exponential window function with a line broadening of 0.3 Hz was applied prior to Fourier transformation to all one-dimensional NMR spectra. The resultant spectra were phased, baseline-corrected and calibrated to 3-trimethylsilyl-1-[2,2,3,3,-2H4] propionate (δ 0.0, urine) or the α anomeric resonance of glucose (δ 5.23, blood serum and tissues) using an in-house routine (developed by Dr JM Fonville); these were then manually checked using Topspin (2.0a, Bruker BioSpin 2006) as a visualiser. The spectra were subsequently imported into Matlab then down-sampled to 13k data points by linear interpolation (R2009b, The MathsWorks Inc., Cambridge, UK) and the regions containing the water resonance (δ 4.6–5.2) and for urine, urea (δ 4.6–5.9), were removed. The spectra were aligned using an in-house algorithm (Veselkov et al., 2009) to abrogate the effects of peak shifting due to variation in pH. The data were then normalised by the probabilistic quotient method, using the median spectra as the reference (Dieterle et al., 2006) to minimise the effects of inter-sample variation due to phenomena such as dilution or sample volume. Orthogonal partial least squares discriminant analysis (O-PLS-DA) and Wilcoxon rank sum tests were performed in a Matlab environment using in-house algorithms (Cloarec et al., 2005) and the Matlab statistical toolbox. To generate the serum trajectory, a series of pairwise O-PLS-DA models was constructed for each time-point using mean-centred scaling and the cross-validated scores (Tcv) extracted for each model. Results were then normalised by dividing each value by the total Tcv standard deviation for each model. The scores corresponding to the class definition in the pairwise comparison at each time point were represented using their respective mean and standard deviations. For each O-PLS-DA model depicting urinary and microbial differences, it was determined whether the Q2Y value (predictive ability) of the model was significantly different to the Q2Y value calculated from up to 2000 random permutations of Y using the cumulative probability value determined at the 95th quartile (in-house algorithm written by Dr O Cloarec). Models were scaled to unit variance such that each variable had equal weight in the model, and were subject to a sevenfold cross-validation step. The spectral regions or bacteria highlighted by the discriminant analysis as having a correlation coefficient (r) of >+/−0.6 were selected for display via heat maps to provide a global view of metabolic and bacterial variation in response to early-life environment and weaning diet. To assess the effects of diet independently of batch, the models investigating dietary differences include all animals from both batches (n=6 per group), and similarly, to assess the effects of batch independently of diet, animals on each diet (n=6 per group) were included in the model irrespective of batch. For analysis of the tissue, betaine in the liver and MLN and taurine in the liver were found to be significantly different in the soya- and egg-based diets; these metabolites were therefore integrated and analysed independently of the rest of the spectra. Integration of selected metabolite regions was performed in a Matlab environment using an in-house algorithm (Dr R Cavill). The microbial-metabolic interaction map was created using a hierarchical clustering and correlation script in Matlab using an in-house algorithm (Dr JTM Pearce) and fitted with a 50% false-discovery rate (FDR) to correct for multiple testing, the hierarchical clustering algorithm used the Pearson product-moment correlation coefficient. We acknowledge that this seems to be a high FDR, but this is a deliberate compromise considering the small number of observations. Moreover, the FDR calculation assumes that variables are independent and without correlation. Biological process involves intrinsic correlation between metabolites and the nature of NMR spectroscopy brings another level of correlation between variables. All these reasons make the real FDR difficult to estimate and thus justify the acceptance of such a high FDR.
Results
Differences in the first day of life caused a sustained shift in the metabolic phenotype of serum
Serum profiles acquired for two experimental replicates (n=6) of piglets at the age of 1 day (immediately after removal from the mother) and 21 days showed significant metabolic differences, despite the fact that from day 2 onwards, all animals were raised in a standardised, controlled environment (Figure 1b). O-PLS-DA between the two experimental replicates showed that the spectral region of N-acetylglycoproteins varied between the two batches at day 1 (Figure 1d) and that the serum myo-inositol concentration was also increased in batch 2 at this time point. At 21 days, the piglets in the two batches still exhibited significant differences in their serum metabolic profiles; one of the major discriminatory metabolites was galactose (Figure 1e) which was higher in batch 2. Additionally, serum formate levels were also increased in batch 2. However, after weaning (28 days), the metabolic profiles converged and at the end of the experiment (35 days), no significant metabolic differences between the two batches could be detected in serum. A summary of the O-PLS-DA loadings describing the batch-dependant variation of the metabolic serum profiles is provided in the Supplementary Information (Supplementary Figure S1).
Weaning onto different diets caused a transient divergence of metabolic phenotype in serum
The effect of different weaning regimens (egg- vs soya-based diet) was significant 7 days after the introduction of the weaning diet (Figure 1c). This difference was attributed to a transient increase in serum betaine in the egg-fed animals (Figure 1f). Importantly, by the end of the experiment (35 days), there were no significant differences in the serum metabolic profiles of these animals associated either with the early environmental variation (batch) or with weaning diet.
Weaning diet and experimental batch confer significant, but differential, effects on the urinary metabolic phenotype at 35 days
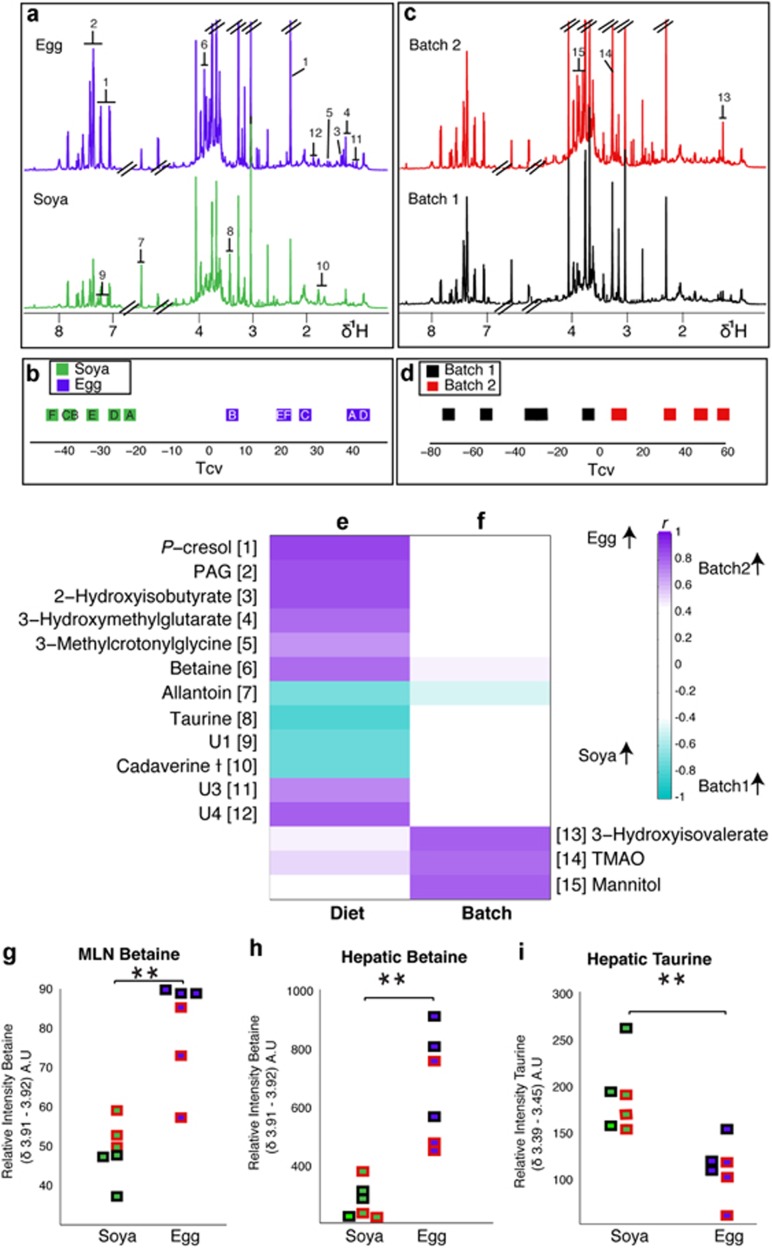
We observed clear and significant differences in urinary metabolite profiles between experimental replicates (Figures 2c and d). This difference was apparent 34 days after environmental standardisation: animals in the second experimental batch excreted relatively more 3-hydroxyisovalerate, trimethylamine-N-oxide (TMAO) and mannitol (Figure 2f), suggesting a sustained alteration of the metabolic phenotype. Urine metabolites of animals weaned onto different diets were also significantly different (Figure 2a). O-PLS-DA analysis shows a difference at 35 days between the animals weaned onto an egg-based diet and those weaned onto a soya-based diet (Tcv, Figure 2b). These differences were generally distinct from differences relating to batch. Diet-induced differences included increased excretion of P-cresol glucuronide, phenylacetylglycine, 2-hydroxyisobutyrate, 3-methylcrotonylglycine, betaine and unknown metabolites U3 (U3 (δ1H 1.44; δ13C 16.5, (s))) and U4 (U4 (δ1H 1.88 (s) and δ1H 2.04 (s)) assigned to the same molecule by statistical total correlation spectroscopy. The 13C shifts could not be unambiguously assigned for these resonances.) in the egg-based diet with a concomitant decrease in urinary allantoin, taurine, U1 (U1 (δ1H 7.09; δ13C 121 (d) & δ1H 7.27; δ13C 133.3 (d)) is likely to be a para-substituted phenolic compound but shows no clear similarity to any reported compounds) and cadaverine (Figure 2e).
Figure 2.
Weaning diet and early environment both affect the urinary metabolic phenotype of the young pig. Median spectra from each group fed either an egg-based (top, n=6) or soya-based (bottom, n=6) diet (a) with numbers corresponding to the listed metabolites (e) and (f). The groups underwent comparison by pairwise O-PLS-DA analysis conducted with one predictive and one orthogonal component (Q2Y: 0.81, R2Y: 0.97, R2X: 0.54) and the cross-validated scores (Tcv) are shown (b), each point represents one animal and the letters correspond to litter. Discriminating metabolites are shown in (e). (c) Represents median spectra from each experimental batch with numbers corresponding to the listed metabolites in (e) and (f). The Tcv from an O-PLS-DA model constructed with one predictive and one orthogonal component (Q2Y: 0.72, R2Y: 0.92, R2X: 0.50) are shown in (d). The discriminatory metabolites from this model are highlighted in (f). Plots of integral regions calculated from 600 MHz HR-MAS-NMR spectra of tissues from each animal; (g) relative betaine concentration in the MLN, where the points on the left represent the animals on the soya diet and those on the right the egg diet. Animals in batch 1 are outlined in black, whereas those in batch 2 are outlined in red. Integral regions of hepatic betaine (h) and hepatic taurine (i). ** denotes P<0.001 (Wilcoxon rank sum).
Metabolic profiles in systemic tissues
Pigs fed an egg-based diet had significantly (P<0.001) higher levels of betaine in the MLN and liver than those fed a soya-based diet (Figures 2g–i). In addition, hepatic taurine levels were significantly (P<0.001) decreased in the egg-fed animals compared with the soya-fed animals.
Significant differences are observable because of experimental batch in the faecal microbiota at 21 days
The faecal and mucosa-associated microbiota was determined by 16 S pyrosequencing. After quality filtering, a total of 2 33 342 sequencing reads were obtained (254 bp average read length covering the V1–V2 and V4 regions of the 16 S gene). Per sample, 1919 sequences (median value) were tentatively classified into 70 families. Analysis of faecal samples collected at 21 days showed significant (P<0.03) microbial differences between the experimental batches (Supplementary Figure S2). The primary difference in microbial composition was in levels of Clostridia Incertae Sedis XN which was significantly increased in batch 2 (r=0.75). At this stage in the experiment, all the animals were on a formula diet and no significant differences between the animals subsequently fed a soya-based or egg-based diet were observed at this time point (as shown by a negative Q2Y). This is consistent with the batch differences pre-existing weaning.
Weaning diet and first day environment significantly affect composition of the colonic microbiota at 35 days
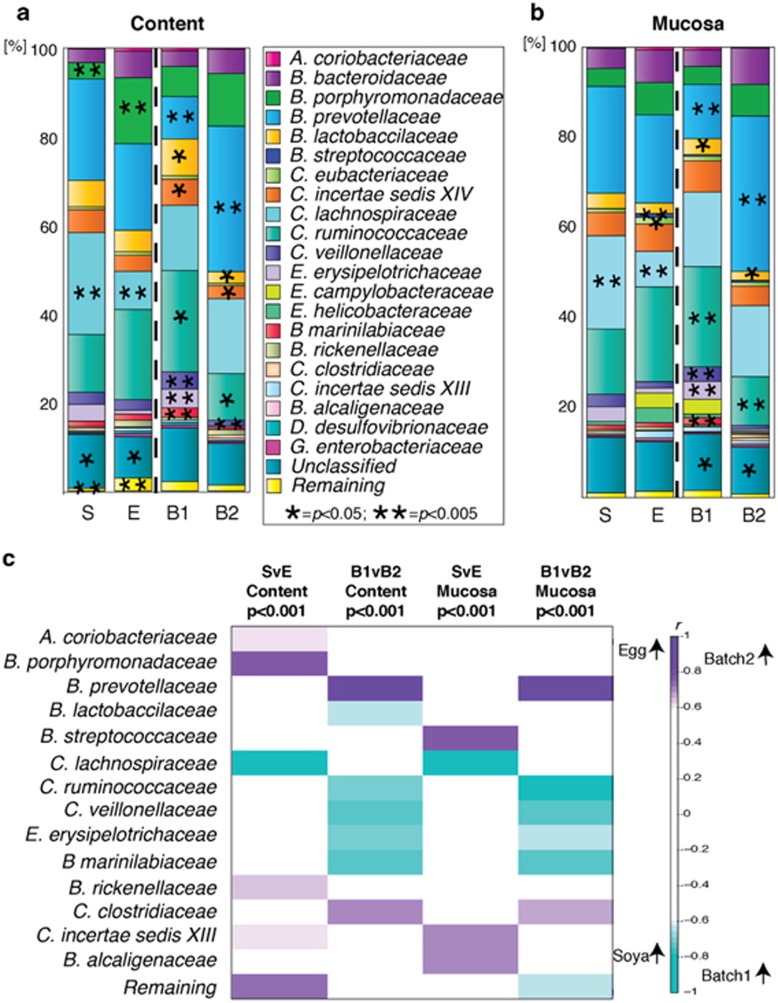
Bacteria belonging to the classes Bacteroidia and Clostridia were dominant in communities of all animals (Figure 3). Significant differences (P<0.05) between the two diets were clearly identifiable for each site; differences in bacterial abundance were observed for three bacterial families in the colonic content and for five families in the mucosa. Inter-batch differences were also observed; nine bacterial families showed significant differences in the colonic content and nine families had different abundances in the mucosa (Figures 3 a and b). Supervised multivariate statistical comparison of the recorded bacterial families was carried out using O-PLS-DA and those groups found to be significantly different (r>±0.6) because of either weaning diet or batch are summarised in Figure 3c by a correlation coefficient heat map. Bacteria of the families Ruminococcaceae, Veillonellaceae, Erysipelotrichaceae and Marinilabiaceae were increased in both the content and mucosa of batch 1 compared with batch 2, whereas Prevotellaceae was increased in the content and mucosa of batch 2. The bacterial response to diet showed less concordance between colonic and mucosal community changes than the batches did and included increased Porphyromonadaceae in the colonic content of piglets fed an egg-based diet. Additionally, there were increased mucosal proportions of Streptococcaceae and Alcaligenaceae.
Figure 3.
Weaning diet and early environment affect the composition of microbiota in the colonic content and mucosa of the young pig. The mean relative proportion (%) of bacterial families measured by 16 S DNA sequencing (V1+V2 and V4 regions) is summarised for each intervention in the colonic content (a) and for the colonic mucosa (b). Each figure has a colour-coded representation of the relative proportion of bacteria as summarised by the key and is separated into animals on a soya-based diet (S, n=6), an egg-based diet (E, n=6), those in batch 1 (B1, n=6) and those in batch 2 (B2, n=6). The diets (soya vs egg) were statistically compared using the Wilcoxon rank sum test for each bacterial family, as were the batches (batch 1 vs batch 2). Statistical significance of P<0.05 is indicated by (*) and P<0.005 by (**). The O-PLS-DA correlation coefficients for a series of four separate O-PLS-DA models, constructed on the mean relative proportions of bacteria, denoted at the top of the column (cmk. Full bacterial family names are: Actinobacteria coriobacteriaceae; Bacteroidia bacteroidaceae; Bacteroidia porphyromonadaceae; Bacteroidia prevotellaceae; Bacilli lactobacillaceae; Bacilli streptococcaceae; Clostridia eubacteriaceae; Clostridia incertae sedis XN; Clostridia lachnospiraceae; Clostridia ruminococcaceae; Clostridia veillonellaceae; Erysipelotrichi erysipelotrichaceae; Erysipelotrichi campylobacteraceae; Epsilonproteobacteria helicobactereaceae; Bacteroidia marinilabiaceae; Bacteroidia rickenellaceae; Clostridia clostridiaceae; Clostridia incertae sedis XIII; Betaproteobacteria alcaligenaceae; Deltaproteobacteria desulfovibrionaceae; Gammaproteobacteria enterobacteriaceae.
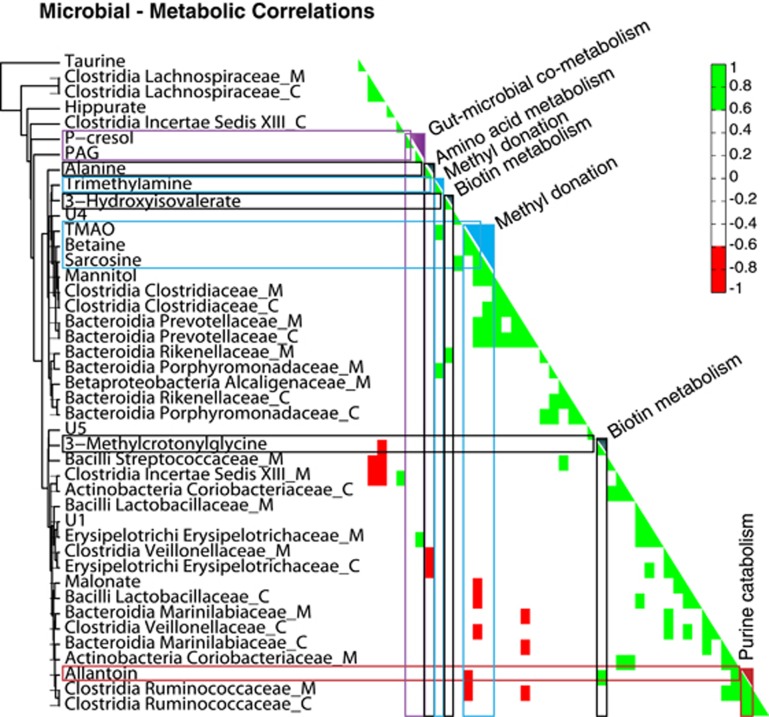
Microbial-metabolic interactions affect a number of metabolic pathways
We identified several statistically significant microbial-metabolic associations (Figure 4). An established gut microbial by-product of dissimilatory amino acid metabolism, phenylacetylglycine, was found to be positively correlated with Erysipelotrichaceae isolated from the colonic mucosa. Metabolites involved in methyl donation, such as betaine and sarcosine, were positively correlated with mucosal and colonic Prevotellaceae, and betaine was negatively correlated with colonic Veillonellaceae and Lactobacillaceae. Importantly, 3-methylcrotonlyglycine, associated with biotin deficiency, was negatively correlated with Lachnospiraceae in the colonic content, whereas, 3-hydroxyisovalerate also associated with biotin metabolism, was positively associated with colonic mucosal Rikenellaceae.
Figure 4.
Microbial-metabolic interactions are visible across multiple metabolic pathways. A hierarchical clustering and correlation map between bacterial families and selected metabolic integrals. Positive correlations are shown in green and negative correlations in red. Gut microbial metabolites are highlighted in purple, metabolites related to methyl donation in blue, metabolites related to biotin in grey and those related to purine metabolism in red. The suffix ‘_C' denotes bacteria isolated from the colonic content and the suffix ‘_M' denotes those isolated from the colonic.
Discussion
We have used a tightly controlled experimental system to explore the effect of the environment during the first 24 h of life and to interrogate the subsequent weaning strategy on the microbiota and metabolism of newborn piglets. We show that minor variations during the first day of life can have long-term effects on patterns of intestinal colonisation and the urinary metabolic phenotype of piglets. Furthermore, piglets subsequently weaned onto either egg- or soya-based diets displayed reliably altered microbiota and metabolic parameters despite the persistent batch-effect observed between experimental replicates.
It is well documented that the microbiota is acquired from both the maternal vaginal canal and the immediate neonatal external environment (Mulder et al., 2011; Schmidt et al., 2011). Previous studies have used early-life interventions to demonstrate that successive microbial colonisation can be affected during infancy (Krishnan and Ramakrishna, 1998; Kirjavainen and Gibson, 1999). The pattern of colonisation appears to be a programming event, which influences both composition and function of the adult microbiota, which subsequently impacts on metabolism (Dominguez-Bello et al., 2011). It is also known that variation in early-life events can be linked to increased disease risk in later life (Dabelea and Crume, 2011; Aiken and Ozanne, 2014). It is very rare, however, that a single bacterial species can be identified as a specific marker or predictor for disease in the absence of a known pathogen.
Significant metabolic effects between the two experimental batches were observed in the serum metabolic profile at the age of 1 day, indicating that metabolic differences between the experimental replicates were established during the first day of life. Serum N-acetyl glycoprotein concentrations have been positively correlated with an acute-phase reaction in humans (Bell et al., 1987), so the observation that at the age of 1 day, piglets in batch 2 had increased levels of N-acetyl glycoproteins despite having different mothers could indicate that animals in this batch were undergoing greater antigenic challenge than those in batch 1. No pathology was observed either on the farm or in the isolator unit during either experimental run, so an acute phase response may have been in response to minor environmental fluctuations. At 21 days after birth, batch differences were still evident in the serum metabolite profiles and this was primarily due to galactose concentrations, which were virtually undetectable in batch 1 but significantly elevated in batch 2. Because there was no variation between batches in the time between feeding and serum sample collection, this could indicate inter-batch variation in intestinal absorption or enzymatic conversion of lactose to galactose (Holmes et al., 1990). By the time the piglets were 35 days old, no difference in the metabolic profile of serum was observed due to either experimental replicate or weaning diet, but strong batch and diet differences were apparent in the microbiome and the urinary metabolic profile. Although the lack of a significant difference in serum metabolic profiles between batches and diet groups by the end of the experiment could indicate the lack of a sustained metabolic effect of these parameters, blood serum is kept under tight homeostatic control. We have shown previously that there is a comparative lack of NMR-detectable metabolites of microbial origin in porcine serum relative to urine (Merrifield et al., 2011). Additionally, we have observed differences in the urinary metabolic profiles in pigs due to weaning diets up to 8 weeks after weaning, despite there being no differences in the serum metabolic profile (Merrifield et al., 2013). Thus, the differences in microbial composition observed at this time point are unlikely to be reflected in this serum, and the observed differences early on in the experiment suggest marked systemic differences between the batches.
It was not feasible for faecal material to be reliably collected from these piglets at a similar time point on day 1 because of inherent differences in production of faecal matter in the neonatal pig; therefore, we can only speculate about differential microbial composition at this time point, however, we suspect that the batch differences observed throughout the experiment are likely to be due to differential early microbial acquisition based on the differing microbial composition observed between batches at later time points.
The significantly higher level of faecal Clostridiae incertae sedis XIV in batch 2 at 21 days supports the hypothesis that the two batches had differential microbial composition, as these samples were collected before the animals were weaned onto either an egg- or soya-based diet. Additionally, both the batch and weaning diet had a sustained effect on the urinary profile, the 16 S DNA profile of both colonic mucosal scrapings and on colonic content observed at 35 days. Gut microbiota can be classified as either ‘autochthonous' (residing in the gut) or ‘allocthonous' (passing through the gut). It is thought that autochthonous bacteria are more permanent members of the microbiota, whereas allocthonous bacteria may be transient and related to diet. The observation made in our experiment that both the composition of mucosal, which may represent autochthonous bacteria, and the luminal content,which may represent allocthonous bacteria, were altered by both the batch and weaning diet, suggests that both early-life environment and weaning diet affect not just the bacterial composition of the gut content, which might be expected, but also the resident colonic bacterial population. These animals were kept under identical environmental conditions following the first day of life and received the same batch of feed between replicates, thus any changes between the batches are indicative of differences in microenvironment during the first 24 h of life, or in the maternal microbiota passed on to the piglets. Given that the batch effect derived from two groups, each containing three sows, it is unlikely that the individual vaginal microbiota was a strong contributory factor to the batch differences. There was minor seasonal variation in outside temperature during the study (Supplementary Figure 2), but we cannot be certain that this is sufficient to affect the maternal/neonatal metabolism of these animals.
Several of the metabolites associated with batch and weaning diet in this study derive from either microbial metabolism or microbial-mammalian co-metabolism, and several of these demonstrate a correlation with bacteria as measured by 16 S DNA, suggesting that both the environment during the first day of life and the weaning diet can induce alteration of microbial composition. We have shown previously that weaning diet has a sustained effect on the urinary metabolic profile in the pig, despite subsequent dietary standardisation (Merrifield et al., 2013), and here, we demonstrate that the early-life environment creates a similar set-point in acquisition of microbiota and development of metabolic phenotype. Importantly, the increase in P-cresol glucuronide and phenylacetylglycine excretion in the egg-fed animals implies a diet-induced alteration of gut microbial activity or ecology, because these metabolites are known end-products of proteolytic fermentation in the distal gut (Yokoyama et al., 1982; Smith and Macfarlane, 1996). Our previous work has demonstrated that piglets initially fed an egg-based diet excreted comparably more phenylacetylglycine and p-cresol glucuronide than animals initially fed a soya-based diet, even after 4 weeks of dietary standardisation by feeding a fish-based diet (Merrifield et al., 2013), implying that these metabolites are a product of altered microbial metabolism rather than of differential dietary protein.
Biotin metabolism correlates to differences in both first day environmental exposure and diet
Differential excretion patterns of the urinary organic acids 3-hydroxyisovalerate and 3-methylcrotonylglycine relating to diet and batch are of interest as these are putative markers of biotin deficiency. Biotin, also known as vitamin H or coenzyme R, is primarily involved in carbohydrate metabolism. Biotin is a cofactor for five mammalian carboxylases, and decreased activity of these leads to increased urinary excretion of 3-hydroxyisovalerate, 3-methylcrotonylglycine and other organic acids (Mock et al., 2004). It is also a cofactor for fatty acid synthesis, gluconeogenesis (Sawamura et al., 2007), and because it is intimately involved with epigenetic regulation of epithelial proliferation (O'Keefe et al., 2009), it has also been linked to the development of colorectal cancers. Biotin is a fermentation product of elements of the microbiome (Hill, 1997) and can also be derived from dietary protein. Despite egg yolk containing high concentrations of biotin, feeding an egg-based diet is reported to reduce the bioavailability of biotin because egg white contains avidin, a protein with a strong affinity for biotin that inhibits uptake across the gastrointestinal tract. However, the cooking process used to prepare the experimental diets destroys avidin activity and, in addition, these specifically formulated diets were supplemented with biotin (200 mg, Volac Ltd).
In this experiment, increased urinary excretion of 3-methycrotonylglycine was observed in the animals fed an egg-based diet, whereas increased urinary excretion of 3-hydroxyisovalerate was seen in batch 2, irrespective of diet. Together, these differences suggest that bioavailability of biotin is more likely to be due to differential microbial production and/or microbial or mammalian metabolism than to dietary intake. Cross-correlation of bacteria and metabolic profiles in this study (Figure 4) also supports this idea. We demonstrate a positive correlation between 3-hydroxyisovalerate and mucosal Rikenellaceae, which belong in the order Bacteroidales. The Bacteroides are also included in this order and members of the Bacteroides genus are known to require biotin for growth (Macy and Probst, 1979), and thus, could theoretically reduce the bioavailability of biotin.
Choline metabolism is affected by both diet and first day environmental exposure
Several studies have suggested that gut bacteria affect the bioavailability of dietary choline to the host, and that microbially derived metabolites of choline have been implicated in important disease processes such as atherosclerosis (Koeth et al., 2013; Spencer et al., 2011). Betaine concentration in the serum was significantly increased in the egg-fed animals 7 days after weaning. Because eggs contain relatively more choline than soya beans (Zeisel et al., 2003), and because plasma concentrations of betaine have been found to correlate with oral doses of betaine (Atkinson et al., 2009), this is to be expected. In this study, although plasma betaine levels normalised 2 weeks after weaning, at the end of the experiment (35 days), hepatic MLN and urinary betaine concentrations were significantly increased in the animals weaned onto an egg-based diet. Whilst these observations are consistent with increased bioavailability of betaine due to dietary intake, urinary betaine was shown to be significantly associated with a number of colonic bacteria. Importantly, urinary TMAO is a host metabolic product of trimethylamine, which is produced by the microbiota from choline (Zeisel et al., 1983, 1989; al-Waiz et al., 1992). Although urinary TMAO was found in higher concentrations in batch 2 animals, hepatic levels of TMA and TMAO were not significantly different between batches, indicating that this difference was not due to increased flavine monooxygenase activity in the liver. Increased levels of TMAO have been related to increased atherogenesis (Koeth et al., 2013). Animals in batch 2 had increased acute phase serum N-acteylglycoproteins at the age of 1 day, so it would be of interest to investigate whether inflammation in early life predisposes to increased TMAO formation and hence atherogenesis.
Choline metabolism in the gastrointestinal tract is intimately associated with gut microbiota, and urinary metabolic end-products of choline degradation, including betaine and sarcosine, were statistically strongly associated with bacteria in the colonic mucosa and content. Urinary betaine and sarcosine were positively correlated with Bacteroida Prevotellaceae in both the mucosa and colonic content. The Prevotellaceae were also significantly increased in batch 2; these bacteria are known to produce H2 and have been found in high concentrations in the gastrointestinal tracts of obese humans (Zhang et al., 2009).
Host-microbial metabolism is influenced by both diet and early-life environmental exposure
Overall, our results are consistent with the hypothesis that there are at least two critical points during early life which affect establishment of gut microbial communities: immediately following birth and at weaning. Our data show that the batch effects initiated in the first day of life were still strongly represented in the microbiota and in urinary metabolites of piglets at the age of 35 days, and that the effects of different diets introduced at 21 days on the urinary metabolite profiles were also sustained. These data also confirm that differences in metabolic phenotype attributed to diet and intestinal microbiota are more readily detected in urine than in serum at 35 days. We have previously shown that weaning diet confers a sustained shift in metabolic phenotype (Merrifield et al., 2013). The results reported here suggest that this may have resulted from an altered intestinal microbiota during the weaning period, as evidenced by the significant differences observed by 16 S DNA sequencing. This is probably a result of the weaning diet and occurred independently of the batch effects.
In adults, the relative stability of the intestinal microbiota means that modulations such as those achieved by prebiotics, probiotics and some antibiotics are dependent on continuous consumption because the microbiota tends to revert to its original profile on cessation of the intervention (Moore and Moore, 1995; Lozupone et al., 2012). Here, we demonstrate that early-life events can cause persistent changes in the microbiome, of at least 5 weeks duration, with equally persistent effects on metabolic function. However, we have shown previously that weaning confers a sustained effect on the urinary metabolic phenotype of at least 8 weeks duration. We propose that variations in the early neonatal environment should be taken into account when designing experimental trials. As batch, or differences in the microenvironment, has a significant effect on both the microbiome and metabolome, it is important to establish whether changes are truly a result of an intervention or a reflection of the early postnatal environment.
In conclusion, we suggest that the early post-natal period may represent a time of physiological plasticity during which future therapeutic interventions could potentially minimise the effect of environmental perturbations, thus impacting positively on human and animal health. This may lower the risk of developing metabolic dysfunction in later life and is especially relevant to infants who have been exposed to known risk factors including premature birth, caesarean delivery, time in the neonatal intensive care unit and those born at term with low birth weights.
Acknowledgments
This work was funded, in part, by Nestec Ltd.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aiken CE, Ozanne SE. (2014). Transgenerational developmental programming. Hum Reprod Update 20: 63–75. [DOI] [PubMed] [Google Scholar]
- al-Waiz M, Mikov M, Mitchell SC, Smith RL. (1992). The exogenous origin of trimethylamine in the mouse. Metabolism 41: 135–136. [DOI] [PubMed] [Google Scholar]
- Atkinson W, Slow S, Elmslie J, Lever M, Chambers ST, George PM. (2009). Dietary and supplementary betaine: effects on betaine and homocysteine concentrations in males. Nutr Metab Cardiovasc Dis 19: 767–773. [DOI] [PubMed] [Google Scholar]
- Bajaj JS, Hylemon PB, Younossi Z. (2012). The intestinal microbiota and liver disease. Am J Gastroenterol Suppl 1: 9–14. [Google Scholar]
- Barker DJP. (1999). Early growth and cardiovascular disease. Arch Dis Child 80: 305–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckonert O, Keun HC, Ebbels TMD, Bundy J, Holmes E, Lindon JC et al. (2007). Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2: 2692–2703. [DOI] [PubMed] [Google Scholar]
- Bell JD, Brown JCC, Nicholson JK, Sadler PJ. (1987). Assignment of resonances for 'acute-phase' glycoproteins in high resolution proton NMR spectra of human blood plasma. FEBS Lett 215: 311–315. [DOI] [PubMed] [Google Scholar]
- Bertram HC, Knudsen KEB, Serena A, Malmendal A, Nielsen NC, Frette XC et al. (2006). NMR-based metabonomic studies reveal changes in the biochemical profile of plasma and urine from pigs fed high-fibre rye bread. Brit J Nutr 95: 955–962. [DOI] [PubMed] [Google Scholar]
- Cebra JJ, Periwal SB, Lee G, Lee F, Shroff KE. (1998). Development and maintenance of the gut-associated lymphoid tissue (GALT): the roles of enteric bacteria and viruses. Dev Immunol 6: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J et al. (2011). Colonization-induced host-gut microbial metabolic interaction. mBio 2: e00271–00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloarec O, Dumas ME, Craig A, Barton RH, Trygg J, Hudson J et al. (2005). Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal Chem 77: 1282–1289. [DOI] [PubMed] [Google Scholar]
- Cox AJ, West NP, Cripps AW. (2014). Obesity, inflammation, and the gut microbiota. Lancet Diabetes Endocrinol 3: 207–215. [DOI] [PubMed] [Google Scholar]
- Dabelea D, Crume T. (2011). Maternal environment and the transgenerational cycle of obesity and diabetes. Diabetes 60: 1849–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle F, Ross A, Schlotterbeck G, Senn H. (2006). Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem 78: 4281–4290. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. (2011). Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology 140: 1713–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Gaia N, Johansson M, Stahlman M, Backhed F et al. (2014). Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J 8: 2116–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He QH, Kong XF, Wu GY, Ren PP, Tang HR, Hao FH et al. (2009). Metabolomic analysis of the response of growing pigs to dietary l-arginine supplementation. Amino Acids 37: 199–208. [DOI] [PubMed] [Google Scholar]
- Hill MJ. (1997). Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev 6: S43–S45. [DOI] [PubMed] [Google Scholar]
- Holmes MA, Arthur PG, Hartmann PE. (1990). Changes in the concentrations of glucose and galactose in the peripheral blood of sucking piglets. J Dairy Res 57: 331–337. [DOI] [PubMed] [Google Scholar]
- Huh SY, Rifas-Shiman SL, Zera CA, Edwards JWR, Oken E, Weiss ST et al. (2012). Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Child 97: 610–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CL, Versalovic J. (2012). The human microbiome and its potential importance to pediatrics. Pediatrics 129: 950–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirjavainen PV, Gibson GR. (1999). Healthy gut microflora and allergy: factors influencing development of the microbiota. Ann Med 31: 288–292. [DOI] [PubMed] [Google Scholar]
- Koeth RA, Wang ZE, Levison BS, Buffa JA, Org E, Sheehy BT et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19: 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinov SR, Awati AA, Williams BA, Miller BG, Jones P, Stokes CR et al. (2006). Post-natal development of the porcine microbiota composition and activities. Environ Microbiol 8: 1191–1199. [DOI] [PubMed] [Google Scholar]
- Koren O, Spor A, Felin J, Fåk F, Stombaugh J, Tremaroli V et al. (2011). Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA 108: 4592–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Ramakrishna BS. (1998). Butyrate and glucose metabolism in isolated colonocytes in the developing rat colon. J Pediatr Gastroenterol Nutr 26: 432–436. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC. (2015). Nutrition in early life and the programming of adult disease: a review. J Hum Nutr Diet 28: 1–14. [DOI] [PubMed] [Google Scholar]
- Larsen FH, Jorgensen H, Engelsen SB, Laerke HN. (2010). Metabolic profiling of lymph from pigs fed with beta-glucan by high-resolution H-1 NMR spectroscopy. Livest Sci 133: 38–41. [Google Scholar]
- Lewis MC, Inman CF, Patel D, Schmidt B, Mulder I, Miller B et al. (2012). Direct experimental evidence that early-life farm environment influences regulation of immune responses. Pediatr Allergy Immunol 23: 265–269. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. (2012). Diversity, stability and resilience of the human gut microbiota. Nature 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macy JM, Probst I. (1979). Biology of gastro-intestinal bacteroides. Annu Rev Microbiol 33: 561–594. [DOI] [PubMed] [Google Scholar]
- Merrifield CA, Lewis M, Claus SP, Beckonert OP, Dumas ME, Duncker S et al. (2011). A metabolic system-wide characterisation of the pig: a model for human physiology. Mol Biosyst 7: 2577–2588. [DOI] [PubMed] [Google Scholar]
- Merrifield CA, Lewis MC, Claus SP, Pearce JTM, Cloarec O, Duncker S et al. (2013). Weaning diet induces sustained metabolic phenotype shift in the pig and influences host response to Bifidobacterium lactis NCC2818. Gut 62: 842–851. [DOI] [PubMed] [Google Scholar]
- Miller ER, Ullrey DE. (1987). The pig as a model for human nutrition. Annu Rev Nutr 7: 361–382. [DOI] [PubMed] [Google Scholar]
- Mock DM, Henrich-Shell CL, Carnell N, Stumbo P, Mock NI. (2004). 3-Hydroxypropionic acid and methylcitric acid are not reliable indicators of marginal biotin deficiency in humans. J Nutr 134: 317–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WEC, Moore LH. (1995). Intestinal floras of populations that have a high-risk of colon-cancer. Appl Environ Microbiol 61: 3202–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder IE, Schmidt B, Stokes CR, Lewis M, Bailey M, Aminov RI et al. (2009). Environmentally-acquired bacteria influence microbial diversity and natural innate immune responses at gut surfaces. BMC Biol 7: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder IE, Schmidt B, Lewis M, Delday M, Stokes CR, Bailey M et al. (2011). Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity. PLoS One 6: e28279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe SJD, Ou JH, Aufreiter S, O'Connor D, Sharma S, Sepulveda J et al. (2009). Products of the colonic microbiota mediate the effects of diet on colon cancer risk. J Nutr 139: 2044–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond W, Houpt K (eds). (1978) The Biology of the Pig. Comstock Publishing Associates: New York, pp 371. [Google Scholar]
- Prentice AM, Moore SE. (2005). Early programming of adult diseases in resource poor countries. Arch Dis Child 90: 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura H, Fukuwatari T, Shibata K. (2007). Effects of excess biotin administration on the growth and urinary excretion of water-soluble vitamins in young rats. Biosci Biotechnol Biochem 71: 2977–2984. [DOI] [PubMed] [Google Scholar]
- Schmidt B, Mulder IE, Musk CC, Aminov RI, Lewis M, Stokes CR et al. (2011). Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS One 6: e28284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Macfarlane GT. (1996). Enumeration of human colonic bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol 81: 288–302. [DOI] [PubMed] [Google Scholar]
- Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. (2011). Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140: 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Hofer MJ, Campbell IL, Holmes AJ. (2010). Community dynamics in the mouse gut microbiota: a possible role for IRF9-regulated genes in community homeostasis. PLoS One 5: e10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselkov KA, Lindon JC, Ebbels TM, Crockford D, Volynkin VV, Holmes E et al. (2009). Recursive segment-wise peak alignment of biological (1)h NMR spectra for improved metabolic biomarker recovery. Anal Chem 81: 56–66. [DOI] [PubMed] [Google Scholar]
- Vickers MH. (2014). Early life nutrition, epigenetics and programming of later life disease. Nutrients 6: 2165–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Brown TT, Kuperman JM, Chung Y, Hagler DJ et al. (2012). Long-term influence of normal variation in neonatal characteristics on human brain development. Proc Natl Acad Sci USA 109: 20089–20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KCW, Botting KJ, Padhee M, Zhang S, McMillen IC, Suter CM et al. (2012). Early origins of heart disease: Low birth weight and the role of the insulin-like growth factor system in cardiac hypertrophy. Clin Exp Pharmacol Physiol 39: 958–964. [DOI] [PubMed] [Google Scholar]
- Wells J (ed). (2012) The Concept of Phenotypic Induction: Programming and Implications for Growth. Springer Science: New York, NY, USA, pp 13–27. [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC et al. (2008). Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature 455: 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winick M, Noble A. (1966). Cellular response in rats during malnutrition at various ages. J Nutr 89: 300–307. [DOI] [PubMed] [Google Scholar]
- Yokoyama MT, Tabori C, Miller ER, Hogberg MG. (1982). The effects of antibiotics in the weanling pig diet on growth and the excretion of volatile phenolic and aromatic bacterial metabolites. Am J Clin Nutr 35: 1417–1424. [DOI] [PubMed] [Google Scholar]
- Zeisel SH, Wishnok JS, Blusztajn JK. (1983). Formation of methylamines from ingested choline and lecithin. J Pharmacol Exp Ther 225: 320–324. [PubMed] [Google Scholar]
- Zeisel SH, DaCosta KA, Youssef M, Hensey S. (1989). Conversion of dietary choline to trimethylamine and dimethylamine in rats: dose-response relationship. J Nutr 119: 800–804. [DOI] [PubMed] [Google Scholar]
- Zeisel SH, Mar MH, Howe JC, Holden JM. (2003). Concentrations of choline-containing compounds and betaine in common foods. J Nutr 133: 1302–1307. [DOI] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y et al. (2009). Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA 106: 2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.