Abstract
Arbuscular mycorrhizal fungi (AMF) occur in the rhizosphere and in plant tissues as obligate symbionts, having key roles in plant evolution and nutrition. AMF possess endobacteria, and genome sequencing of the endobacterium Candidatus Glomeribacter gigasporarum revealed a reduced genome and a dependence on the fungal host. To understand the effect of bacteria on fungal fitness, we used next-generation sequencing to analyse the transcriptional profile of Gigaspora margarita in the presence and in the absence of its endobacterium. Genomic data on AMF are limited; therefore, we first generated a gene catalogue for G. margarita. Transcriptome analysis revealed that the endobacterium has a stronger effect on the pre-symbiotic phase of the fungus. Coupling transcriptomics with cell biology and physiological approaches, we demonstrate that the bacterium increases the fungal sporulation success, raises the fungal bioenergetic capacity, increasing ATP production, and eliciting mechanisms to detoxify reactive oxygen species. By using TAT peptide to translocate the bioluminescent calcium reporter aequorin, we demonstrated that the line with endobacteria had a lower basal intracellular calcium concentration than the cured line. Lastly, the bacteria seem to enhance the fungal responsiveness to strigolactones, the plant molecules that AMF perceive as branching factors. Although the endobacterium exacts a nutritional cost on the AMF, endobacterial symbiosis improves the fungal ecological fitness by priming mitochondrial metabolic pathways and giving the AMF more tools to face environmental stresses. Thus, we hypothesise that, as described for the human microbiota, endobacteria may increase AMF innate immunity.
Introduction
Arbuscular mycorrhizal fungi (AMF) are crucial drivers of plant evolution: as the most widespread component of the plant microbiota, they occupy a double niche, thriving in the soil (pre-symbiotic phase) and inside root tissues (symbiotic phase). In both niches, they have a key role in nutrient cycling and plant health by taking up minerals such as phosphorous, and delivering them to their host plants (Bonfante and Genre, 2010). Notwithstanding their massive presence in the soil, AMF live as obligate biotrophs, which require organic carbon from their host plant. The spores and hyphae of AMF contain thousands of nuclei, making classical genetic approaches unsuitable. Also, many AMF contain endobacteria in their cytoplasm, leading to a further, unexpected increase in their genetic complexity (Bonfante and Anca, 2009). One type of AMF endosymbiont is the rod-shaped, Gram-negative beta-proteobacterium (Bonfante et al., 1994) Candidatus Glomeribacter gigasporarum (CaGg), which forms symbioses only with members of the Gigasporaceae family (Bianciotto et al., 2003; Mondo et al., 2012). The CaGg genome sequence (Ghignone et al., 2012) revealed that Glomeribacter endobacteria are nutritionally dependent on the fungal host (Ghignone et al., 2012); however, their contribution to host fitness remains unclear. Removal of CaGg from the host AMF causes limited changes in spore morphology and no evident impact on mycorrhization (Lumini et al., 2007). The CaGg endobacteria have small genomes, physiological dependence on their hosts and vertical transmission, suggesting that these endobacteria live as mutualistic associates of the AMF. These features convincingly reveal that the endobacterium depends on the fungus, but do not explain why the host fungus maintains the endobacterium, despite its energetic cost (Ghignone et al., 2012).
Here, we used next-generation sequencing to produce a genome-wide transcriptional profile of Gigaspora margarita in the presence and in the absence of its endobacterium. This analysis, based on our de novo assembly of the transcriptome of G. margarita, showed large-scale changes in gene expression related to the presence of the endobacterium and revealed effects mostly targeting the mitochondrion. Our results, confirmed by cell biology approaches and physiological measurements, indicate that the endobacterium increases the environmental fitness of the fungus, raising its bioenergetic capacity and potentially acting as a driver for priming the fungal innate immune response.
Materials and methods
Biological material
Fungal isolate and spore production
Spores of Gigaspora margarita Becker and Hall (BEG 34, deposited at the European Bank of Glomeromycota) containing (B+) or not (B−) the CaGg endobacteria were used in this study. B− spores were obtained from B+ spores as described in Lumini et al. (2007). The absence of endobacteria in the B− line was routinely checked following the protocol described in Salvioli et al. (2008). All the spores were maintained and propagated by using white clover (Trifolium repens) as trap plant. Briefly, clover plants were inoculated with 100–150 spores and after 3 months, new spores were generated and collected by the wet sieving technique (Gerdemann and Nicolson, 1963). To monitor spore production, soil samples from selected pots were sampled three to four times each year, and the spores were collected and counted.
Spore germination and mycorrhization
Spores were divided in batches of 100, surface-sterilised with Chloramine T (3% P/V) and streptomycin sulphate (0.03% P/V); some batches were germinated in 1 ml of sterile distilled water for 10 days in the dark at 30 °C (germinating spores), while others were germinated in 1 ml of sterile distilled water for 3 days in the dark at 30 °C and in a solution 10−7 M of the synthetic strigolactone (SL) analogue GR24 (Chiralix, Nijmegen, The Netherlands) for 7 more days (SL-treated). After 10 days, the germinated spores and their germinating mycelium were collected, immediately frozen in liquid nitrogen, and crushed with a pestle and mortar for further RNA extraction.
Six hundred B+ and 600 B− spores were used to produce mycorrhizal seedlings of Lotus japonicus (Regel) K. Larsen by using the 'Millipore sandwich' method (Novero et al., 2002). After 4 weeks, L. japonicus roots were observed under a stereomicroscope and extraradical mycelium and mycorrhizal roots were sampled and frozen in liquid nitrogen for RNA extraction (symbiotic stage).
Treatment with oxidant agent and SLs
Sterilised spores (B+ and B−) were placed in a multi-well plate (30 spores in each well) and treated with different concentrations of H2O2 (100 mM, 10 mM, 2 mM, 1 mM 0.75 mM, 0.5 mM, 0.3 mM and 0.25 mM), GR24 (10−7 M) or sterile distilled water. Spores were observed under a stereomicroscope after 3 days of treatment at 30 °C to check the germination rate. For each treatment, at least 90 spores belonging to three different wells were observed. To understand whether the oxidant agent can also lead to early transcriptional changes, a new set of sterilised spores were treated with H2O2 0.3 mM, GR24 (10−7M) or sterile distilled water for 3 days at 30 °C, frozen in liquid nitrogen, and used for RNA extraction. The extracted material was processed as described above.
Molecular analyses
RNA extraction and sample preparation for sequencing
Total RNA was extracted using the RNeasy Microarray Tissue Mini Kit (Qiagen, Hilden, Germany). The concentration and quality of the nucleic acids were assessed with a Nanodrop1000 (Thermo Scientific, Wilmington, NC, USA), and the integrity was checked with the Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA, USA). For details, see Supplementary Text.
Real time q-PCR assays
For RT-qPCR validation, RNA was extracted as previously described and treated with the TURBO DNA-free kit (Life Technologies, Carlsbad, CA, USA). The samples were then reverse-transcribed using Superscript II Reverse Transcriptase (Life Technologies). Quantitative real-time PCR experiments and data analysis were carried out as described in Salvioli etal. (2012), using as a reference gene for transcript normalisation the G. margarita elongation factor (Tef). The primer names and corresponding sequences are listed in Supplementary Table S10.
Generation of data, bioinformatics and phylogenetic analyses
Generation of Data Sets 1 and 2 and de novo transcriptome assembly
In the absence of a reference genome, a de novo assembly was generated using reads from four in vitro normalised paired-end libraries (Data Set 1, see below) obtained from the B+ line of G. margarita containing the endobacterium and sampled at four stages of the fungal life cycle (quiescent spores, germinating spores, spores treated with SL and extraradical mycelium), without replicates, and 14 single-end libraries (Data Set 2, see below) obtained from both the B+ strain and the cured line (B− line) sampled at three stages of the fungal life cycle (germinating spores, spores treated with SL and symbiotic mycelium thriving inside the roots). In total, 18 libraries were produced (Supplementary Table S11). Data Set pre-process is described in Supplementary Materials and methods. The de novo assembly of Data Set 1 and 2 libraries was performed on a 60 core and 256 GB RAM machine, running Ubuntu server 12.04 LTS, using Trinity v.Trinityrnaseq_r20131110 (Grabherr et al., 2011). Detailed description of the assembly process is provided in Supplementary Materials and methods.
The G. margarita BEG34 Transcriptome Shotgun Assembly project (Bioproject PRJNA267628; Biosamples SAMN03216569-SAMN03216586) has been deposited at DDBJ/EMBL/GenBank under the accession GBYF00000000. The version described in this paper is the first version, GBYF01000000.
Downstream analyses performed on the assembled transcripts are detailed in Supplementary Materials and methods.
Calling differentially expressed genes
For differentially expressed gene (DEG) identification, DESEq2 1.2.8 Bioconductor package was run with local fit and betaPrior parameter set to TRUE. Independent filtering was enabled (Anders and Huber, 2010; Love et al., 2013). A false discovery rate of 0.05 was set as threshold for DEG calling. The number of DEGs for each contrast is reported in Supplementary Figure S4. According to current standards for RNA-seq (RNA standards v1, May 2011; www.encodeproject.org), at least two biological replicates for each condition were used. Sample clustering (Supplementary Figure S11) was performed on rlog-transformed data (Love et al., 2013).
GO enrichment analyses and KEGG maps
GO enrichment analyses were conducted with the goseq bioconductor package version 1.14.0, while KEGG pathway pictures, KO (KEGG Orthology) mappings were first obtained from KAAS (KEGG Automatic Annotation Server; http://www.genome.jp/kegg/kaas/) using as query trinity-assembled Gigaspora transcripts against the KEGG GENES database. Details are provided in the Supplementary Material.
Phylogenetic analysis
A phylogenetic tree based on the comparison of whole proteomes of G. margarita and various fungal genomes was constructed using CVTree v3 using the default parameters (Xu and Hao, 2009). Fungal proteomes were those available as built-in fungal database proteomes in CVTree. Additional proteomes not available in CVtree databases were added for the analysis, namely Tuber melanosporum version ASM15164v1.21 (retrieved from ftp://ftp.ensemblgenomes.org/), Rhizophagus irregularis and Mucor circinelloides (retrieved from NCBI queries).
Other bioinformatic techniques
Unless otherwise stated, further graphical outputs were generated with scripts as available in DESEQ2 package vignette (Love et al., 2013).
Cellular and physiological analyses
Confocal and ultrastructural analysis
Single G. margarita spores from the B+ and B− lines were placed on microscope slides in 20 μl of Bacteria Counting Kit component A (B-7277; Molecular Probes, Eugene, OR, USA) diluted 1:1000 according to the manufacturer's directions. The Bacteria Counting Kit contains the SYTO BC bacterial stain which is a high-affinity nucleic acid stain that easily penetrates both Gram-positive and Gram-negative bacteria. The spores were then crushed with a coverslip, incubated in the dark for at least 5 min, and observed under a Leica (Wetzlar, Germany) TCSSP2 confocal microscope (excitation 488 nm; emission 520 nm) to detect the endobacteria. To perform ultrastructural observations, parallel sets of single spores were processed by high-pressure freezing followed by freeze substitution, as described in Desirò et al. (2014). After cutting and counterstaining, thin sections were observed under a transmission electron microscope Philips CM10 (Amsterdam, The Netherlands).
Mitochondria staining with MitoTracker Green FM
Mitochondria were stained with the fluorescent probe MitoTracker Green (Life Technologies). Five sterilised spores were placed on a microscope slide along with 50 μl of MitoTracker Green 1 μM and propidium iodide (50 μg ml−1); spores were crushed with a coverslip and observed with a Leica TCSSP2 confocal microscope (excitation 490 nm; emission 515 nm) after 10 min of incubation in the staining solution. At least 20 spores were observed for the B+ and B− lines. Images taken at the same magnification were used to measure the mitochondrial diameter: 89 mitochondria belonging to 8 B+ spores and 76 mitochondria belonging to 7 B− spores were evaluated. Data were subjected to statistical analysis using the Kruskal–Wallis test for nonparametric data.
TAT-aequorin-based Ca2+ measurements
Ca2+ measurements were carried out in germinated spores of G. margarita (samples of 200 spores, germinated for 10 days) of both B+ and B− lines as described (Moscatiello et al., 2014).
ATP detection assay
Total levels of ATP were determined by using a Luminescent ATP Detection Assay kit (Abcam, Cambridge, UK) according to the manufacturer's instructions. The ATP assay is based on the production of light caused by the reaction of ATP with added luciferase and D-luciferin. The emitted light is proportional to the cellular ATP concentration. Luminescence was determined in 3 days germinating spores of G. margarita (B+ and B− lines) and total amounts of cellular ATP were calculated by using an ATP standard dilution series.
Phosphate measurements in roots and shoots from mycorrhizal clover plants
The phosphorous contents were determined as described in Supplementary Material.
Results and Discussion
To perform a comprehensive analysis of the effect of CaGg on the transcriptome of Gigaspora margarita, in the absence of a reference genome for G. margarita, we first created a de novo assembly of the G. margarita transcriptome. To this end, we developed a two-step strategy, where high-throughput sequencing of cDNA (RNA-seq) from the strain of G. margarita containing the endobacterium (B+ line) was performed, creating Data Set 1. We used these data to generate a preliminary de novo assembly of the G. margarita transcriptome. We then conducted a second RNA-seq experiment to compare the B+ line and the cured line (B− line) at three stages of the fungal life cycle (germinating spores, spores treated with SL and symbiotic mycelium thriving inside roots, Supplementary Figure S1), creating Data Set 2. These two sets of data were used to generate the final de novo assembly.
The Gigaspora transcriptome
Analysis of Data Sets 1 and 2 with the Trinity program created a combined de novo assembly comprising 86 183 transcripts (isoforms), with length greater than 350 bp, corresponding to 35 029 total potential genes. This gene count is in line with the about 28 000 genes reported for R. irregularis, the closest relative of G. margarita so far sequenced (Tisserant et al., 2013; Lin et al., 2014). The G. margarita transcripts had an average GC content of 31.96%, a median contig length of 781 bp, and an average contig length of 1185.77 bp; the contig N50 based on all transcripts was 1683 bp.
Only 10 936 contigs (12.7%) were successfully annotated with Blast2GO (E-value filter= 1.0E-6; Annot_cutoff=55; GO _weight=5; HSP-Hit_Cov_cutoff=0), and BLAST searches identified 4904 contigs (5.7%) that matched only sequences in the refseq_protein database (cut-off e-value of 1e-5), but were not further annotated.
The top BLAST hits included genes from species sharing similarities with G. margarita, such as Laccaria bicolor (Basidiomycota) and Tuber melanosporum (Ascomycota), two fungi with symbiotic lifestyles (Martin and Selosse, 2008; Martin et al., 2010), along with saprotrophic or pathogenic fungi such as Schizophyllum commune, Coprinopsis cinerea, Postia placenta, Cryptococcus neoformans and Ustilago maydis (Supplementary Figure S2). The complete collection of G. margarita and R. irregularis transcripts was used to search for their Best Reciprocal Hits. We found that the two organisms share 6276 transcripts (5901 with annotation; Supplementary Table S1) and G. margarita expressed most of the transcripts described as key features of the R. irregularis genome. These transcripts include phosphate transporters, saccharide transporters, chitinases, chitin synthases and HMG-box transcription factors putatively involved in mating-type recognition (see Supplementary Table S2 for a comprehensive list). AM fungi are considered to be asexual, but the huge number of mating type genes in R. irregularis (Tisserant et al., 2013) and in G. margarita opens the question of their biological meaning. Consistent with the analysis of the R. irregularis genome, the G. margarita transcriptome has cobalamin-dependent and cobalamin-independent methionine synthases, both sharing very high identity with the homologous genes in the R. irregularis genome. Both types of methionine synthases are expressed at comparably high levels during the presymbiotic stage of fungal development. As CaGg possesses the operon for synthesis of vitamin B12, the finding that G. margarita produces a transcript for a putative lysosomal cobalamin transporter potentially involved in vitamin B12 import, is of particular interest. Also in agreement with one of the key features of R. irregularis genome, we found no plant-cell wall degrading enzymes among the G. margarita transcripts. Pfam analysis identified 3838 domains (Supplementary Table S3). As observed in Rhizophagus (Tisserant et al., 2013), G. margarita showed an overrepresentation of proteins involved in signalling pathways and ubiquitin-related metabolism. Interestingly, for both fungi, the most abundant domains occurred in genes encoding tyrosine kinases and Sel1 (see Supplementary data). In addition, we also identified 177 transcripts that belong to CaGg, on the basis of their sequences (Ghignone et al., 2012).
Thus, this first glimpse at the G. margarita transcript-repertoire reveals that R. irregularis and G. margarita (notwithstanding their deep differences in phylogeny, life cycle and ecological strategies) have a strict genetic relatedness, as shown by a cladogram based on the comparison of the whole proteome of G. margarita with other fungal genomes (Supplementary Figure S3).
CaGg affects the G. margarita transcriptomic profile most during the fungal presymbiotic phase
Owing to the complexity of the experimental design (two fungal lines investigated at three stages, Supplementary Figure S1), we first used DESeq2 to identify DEGs. As a validation test, we randomly selected some DEGs to check by RT-qPCR (Supplementary Table S4) and found that the RT-qPCR confirmed the differential expression detected by RNA-seq. We then compared the numbers of DEGs in different conditions to understand whether the main driver of gene expression in G. margarita depends on the life cycle stage or on the presence/absence of the endobacterium (see also Supplementary data). We identified the most DEGs (9609) when comparing the germinating spores containing the bacterium (B+) with the germinating spores in the cured B− line (G condition). Comparison of the germinating B+ spores with B− spores treated with the synthetic SL GR24 (SL condition) identified 3427 differentially expressed transcripts. We found very few transcripts that were differentially expressed during the symbiotic phase (Supplementary Figure S4); this could be due to the fungal dilution in the root tissues and/or to a limited effect of the bacteria on the fungus during the symbiosis. This second option is consistent with our quantification of mycorrhizal colonisation in clover, which found no difference between the B+ and B− lines (Supplementary Figure S5), as previously shown for sorghum and carrot (Lumini et al., 2007).
The presence of CaGg inside the fungal cytoplasm seems to be crucial for AMF function during the presymbiotic phase, when the fungus develops in the rhizosphere, outside the protection offered by the plant cell. Confocal and electron microscopy suggest that bacteria are diluted along the intraradical symbiotic mycelium (Bianciotto et al., 1996), while spores (Figure 1) act as a reservoir of these microbial communities (Desirò et al., 2014). This result opens the question of what advantages the endobacterium provides to its fungal host during this phase.
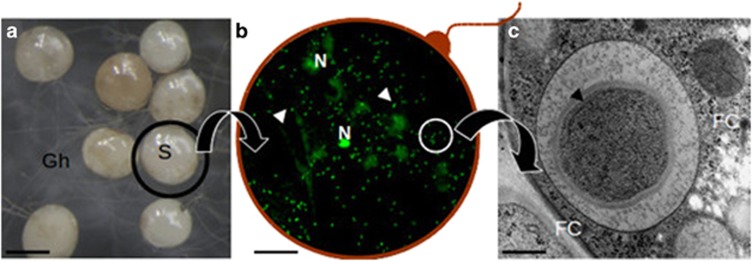
Figure 1.
A composite picture illustrating the location of CaGg. (a) G. margarita spores (S) observed under a stereomicroscope produce a network of germinating hyphae (Gh); (b) a squashed spore reveals the fungal nuclei (N) and a multitude of endobacteria (arrows) after staining with Bacteria Counting Kit component A and observation by confocal microscopy. The red line is drawn to suggest the fungal wall; (c) a bacterium observed by electron microscopy reveals the multilayered Gram-negative wall; it is located inside the fungal cytoplasm (FC), limited by a membrane of fungal origin. Bars correspond to 230 μm in a, 13 μm in b and 0.35 μm in c.
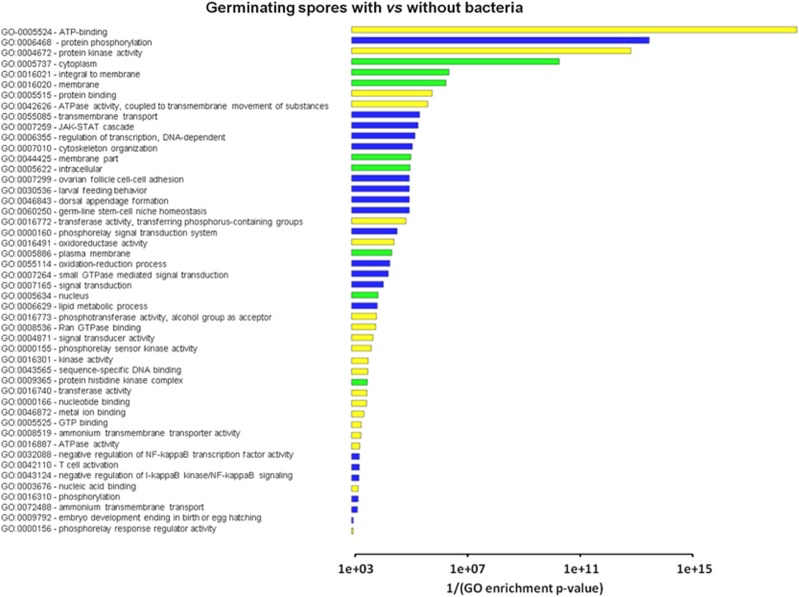
As a second step, we used the DEGs identified by DESeq to estimate Gene Ontology (GO) term enrichment, with GOseq (Young et al., 2010), using a threshold false discovery rate=0.1. Consistent with the presence of more DEGs in the G condition, the GOseq analysis yielded 48 and 5 enriched GO terms in the G and in the SL conditions, respectively. For the G condition, the most relevant enriched GO terms deal with membrane processes and transport, regulation by phosphorylation, signal transduction and oxidoreductase activity (Figure 2). By contrast, in the SL condition, only the ATP binding and protein phosphorylation terms were particularly relevant. These data further suggest that the endobacterial presence exerts a more relevant influence on the fungus at the germinating spore stage. However, specific changes also occur in the SL condition, indicating that the bacterial presence and the SL treatment could interact in influencing the G. margarita transcript profile.
Figure 2.
Gene Ontology (GO) term enrichment as estimated for the comparison B+ versus B− cured germinating spores. The analysis was performed with the Goseq package (Bioconductor), using a threshold false discovery rate=0.1.
CaGg affects the expression of genes involved in growth, development and transport in its fungal host
Among the enriched GO categories in the G condition, the presence of the endobacterium affected membrane processes, and in particular chitin metabolism (Supplementary Table S5). Several differentially expressed chitin synthase transcripts were identified, and five of them were upregulated in the B+ fungus in the G condition, while only one putative chitinase was downregulated. Also, several transcripts related to putative chitin deacetylases, that is, the enzymes that catalyse the deacetylation of chitin in chitosan, were downregulated.
In addition to the remodelling of the fungal wall, the endobacterium affects the expression of genes containing the mating type domains (Supplementary Table S1 and S2), raising the question whether these genes could be involved in the production of the asexual conidia, as reported for Penicillium chrysogenum (Bohm et al., 2013). To test whether the presence of the bacteria affected spore formation, we monitored the spore production of G. margarita over 3 years (Supplementary Table S6) and found that the cured line produced only the 50% of the spores produced by the G. margarita line containing the bacteria.
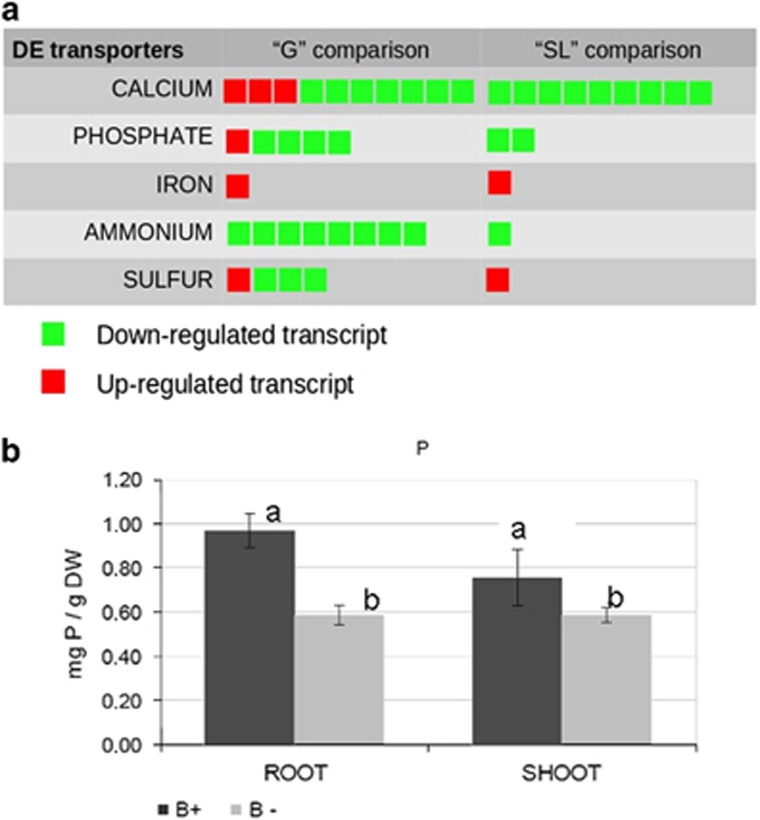
The presence of the endobacterium also affected genes in the transport category (Figure 3a). For example, a plasma membrane iron permease containing an FTR1 domain was among the most upregulated genes in the B+ line, in both the G and SL conditions, as also confirmed by RT-qPCR validation assays (Supplementary Table S4). By contrast, genes related to nitrogen transport were downregulated in the B+ line; in particular, eight putative ammonium transporters were downregulated (Figure 3a). Phosphate uptake is a crucial trait of AM fungi and, accordingly, many transcripts identified in the G. margarita transcriptome encode different phosphate transporters (Supplementary Table S7). Among them, we identified a full-length sequence coding for a phosphate:H symporter belonging to the major facilitator superfamily. This sequence shares the highest similarity with the phosphate transporter described in the extraradical mycelium of Glomus intraradices (Maldonado-Mendoza et al., 2001). Another major facilitator superfamily high affinity P transporter (comp36913_c0), previously described in G. margarita and present in GenBank (GI:591140015), showed the highest expression and upregulation. The closest characterised relative of such sequence is Pho84, which has been demonstrated to function as a transceptor in yeast (Popova et al., 2010).
Figure 3.
(a) Graphical representation of the regulated transcripts referring to the transporter category, as identified in the comparison B+ vs B− under germinating (G) and SL-treated (SL) conditions. Each square corresponds to a regulated transcript. (b) Phosphorous mineral content in clover plants (roots and shoots) colonised by B+ and B− G. margarita lines. Data are mean±s.d. of three biological replicates. Statistically supported differences are indicated with different letters according to a Kruskal–Wallis non-parametric test at P<0.05.
These results open new questions on the potential effect of CaGg on phosphorous transport from the fungus to the host plant. The endobacterium receives phosphate from its fungal host (Ghignone et al., 2012); the resulting stronger Pi-gradient probably causes more sustained Pi uptake by the fungus from the soil, and more phosphate flow towards both the bacterium and the plant. To understand whether this had a positive or negative impact on the plant, we measured the phosphate in roots and shoots of clover plants colonised by G. margarita and found that the plants colonised by the B+ line contained significantly more Pi than the plants colonised by the B− cured line (Figure 3b).
Taken as a whole, the impact of the endobacterium on the fungal transcriptome during the presymbiotic phase leads to functional changes that increase the fungal ecological fitness and may also have deep consequences for the third partner in the interaction, that is, the plant.
CaGg affects transcription of the fungal mitochondrial genes and interferes with the SL response
To better describe the effect of the endobacterial presence on the fungal presymbiotic phase, we assigned KO (KEGG Orthology) terms to the transcript datasets through the KAAS server and then used the Pathview tool to map the pathways (Luo and Brouwer, 2013). This allowed us to illustrate the pathways represented in the fungal transcriptome, and to examine the effects of the endobacterium.
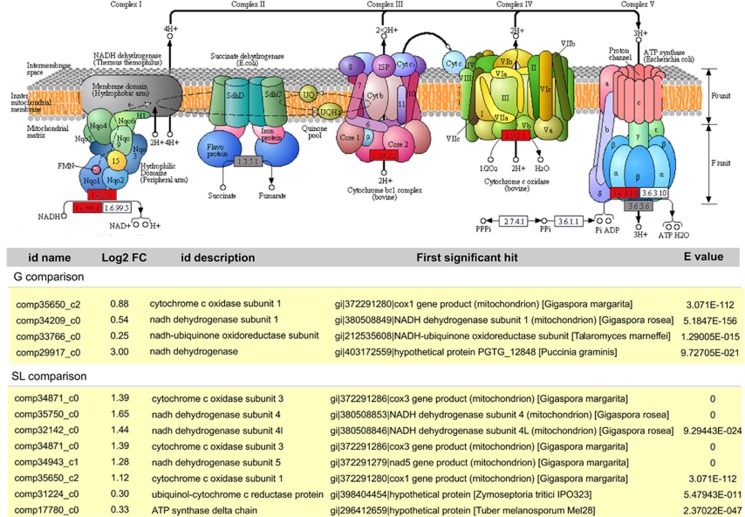
The most impressive changes in pathways in the fungal host involved oxidative phosphorylation, which was strongly induced by the presence of endobacteria (Figure 4). In particular, mitochondrial genes (Pelin et al., 2012), such as those encoding NADH dehydrogenase and Cytochrome oxidase I, were upregulated (Table 1). The transcriptome data therefore point to increased ATP production in the presence of the endobacteria. To validate this observation, we used a luminescence assay to quantify cellular ATP concentrations. Indeed, the cured line of G. margarita produced significantly less ATP (71.18±10.38%) when compared with the line containing the endobacterium (Student's t-test, P<0.05). By contrast, the cured line showed enhanced pentose phosphate metabolism, suggesting that an alternative pathway produces reducing power in the absence of the endobacterium (Supplementary Figure S6a).
Figure 4.
Top: KEGG-based depiction of mitochondrial oxidative phosphorylation in G. margarita. Functions supported by upregulated or non-regulated transcripts are shown in red or grey, respectively. Bottom: list of the transcripts related to mitochondrial oxidative phosphorylation as upregulated by the endobacterial presence under G and SL conditions.
Table 1. Mitochondrial genes upregulated in B+ versus B− germinating (G)- and strigolactone treated (SL) spores.
| ID name | Log2 Fold change | ID description | First significant hit | E value |
|---|---|---|---|---|
| G condition | ||||
| comp35650_c2 | 0.88 | cytochrome c oxidase subunit 1 | gi|372291280|ref|YP_005088168.1|cox1 gene product (mitochondrion) [Gigaspora margarita] | 3.071E-112 |
| comp34209_c0 | 0.54 | nadh dehydrogenase subunit 1 | gi|380508849|ref|YP_005352678.1|NADH dehydrogenase subunit 1 (mitochondrion) [Gigaspora rosea] | 5.185E-156 |
| comp33766_c0 | 0.25 | nadh-ubiquinone oxidoreductase subunit | gi|212535608|ref|XP_002147960.1|NADH-ubiquinone oxidoreductase subunit B17.2, putative [Talaromyces marneffei ATCC 18224] | 1.2901E-15 |
| comp29917_c0 | 3.00 | nadh dehydrogenase | gi|403172559|ref|XP_003331683.2|hypothetical protein PGTG_12848 [Puccinia graminis f. sp. tritici CRL 75-36-700-3] | 9.7271E-21 |
| SL condition | ||||
| comp34871_c0 | 1.39 | cytochrome c oxidase subunit 3 | gi|372291286|ref|YP_005088183.1|cox3 gene product (mitochondrion) [Gigaspora margarita] | 0 |
| comp35750_c0 | 1.65 | nadh dehydrogenase subunit 4 | gi|380508853|ref|YP_005352682.1|NADH dehydrogenase subunit 4 (mitochondrion) [Gigaspora rosea] | 0 |
| comp32142_c0 | 1.44 | nadh dehydrogenase subunit 4l | gi|380508846|ref|YP_005352675.1|NADH dehydrogenase subunit 4L (mitochondrion) [Gigaspora rosea] | 9.2944E-24 |
| comp34871_c0 | 1.39 | cytochrome c oxidase subunit 3 | gi|372291286|ref|YP_005088183.1|cox3 gene product (mitochondrion) [Gigaspora margarita] | 0 |
| comp34943_c1 | 1.28 | nadh dehydrogenase subunit 5 | gi|372291279|ref|YP_005088167.1|nad5 gene product (mitochondrion) [Gigaspora margarita] | 0 |
| comp35650_c2 | 1.12 | cytochrome c oxidase subunit 1 | gi|372291280|ref|YP_005088168.1|cox1 gene product (mitochondrion) [Gigaspora margarita] | 3.071E-112 |
| comp31224_c0 | 0.30 | ubiquinol-cytochrome c reductase protein | gi|398404454|ref|XP_003853693.1|hypothetical protein MYCGRDRAFT_108469 [Zymoseptoria tritici IPO323] | 5.4794E-11 |
| comp17780_c0 | 0.33 | atp synthase delta chain | gi|296412659|ref|XP_002836040.1|hypothetical protein [Tuber melanosporum Mel28] | 2.3702E-47 |
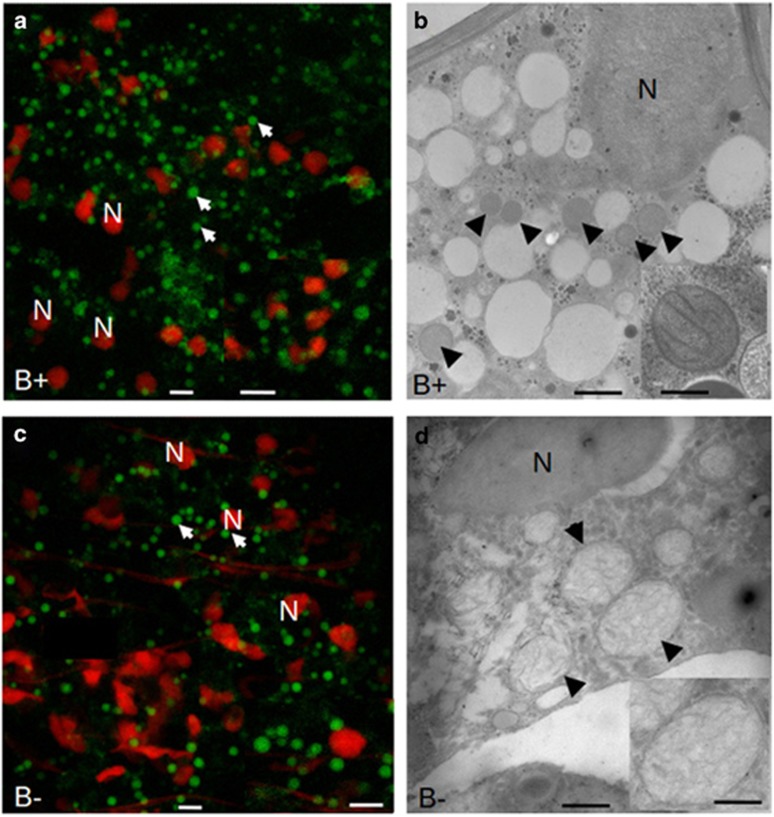
As complex I is responsible for mitochondrial proliferation, mitochondria were monitored by Mitotracker staining in the spores of G. margarita containing, or not containing, the endobacteria (Figure 5). The two conditions showed roundish mitochondria in similar numbers; however, mitochondria from the cured line, at 2.4 μm in diameter, were significantly larger than those from the B+ line, at 1.7 μm in diameter (Supplementary Figure S7). Transmission electron microscopy showed that the mitochondrial cristae were flattened in both lines, but the matrix of mitochondria in the cured line was more electron-transparent. Interestingly, in Alzheimer's disease patients, where mitochondrial dysfunction has an early and preponderant role in the disease, oxidative injury may disturb the structure and fission of mitochondria, resulting in enlarged mitochondria (Moreira et al., 2010). The results suggest that the absence of the endobacterium leads to changes that affect mitochondrial morphology.
Figure 5.
Confocal (left) and electron microscopy (right) images illustrating the shape and morphology of mitochondria from B+ and cured (B−) G. margarita spores. Mitochondria (arrow) were detected by staining with MitoTracker Green, and fungal nuclei (N) with propidium iodide. The inset on the right column reveals the cristae organisation and the differences in the matrix. Bars correspond to 5 μm in a, c and their insets, to 0.7 μm in b and to 0.16 μm in the inset, and to 0.7 μm in d and to 0.4 μm in the inset.
The impact of the endobacterium on fungal mitochondrial genes was further enhanced in the SL condition (Table 1). SLs stimulate mitochondrial activity in the germinating spores of the AMF Gigaspora rosea, which does not contain endobacteria (Besserer et al., 2006). Our analysis surprisingly reveals that the endobacterium and the SL treatment lead to a similar transcriptomic response under the tested conditions (7 days after germination), that is, upregulation of the genes encoding NADH dehydrogenase and Cytochrome oxidase I (COX 1) in the fungus.
According to the concept of the dual effects of mitochondria (Moreira et al., 2010), we wondered whether the higher mitochondrial activity might, on the one hand, lead to increased production of ATP and reactive oxygen species (ROS) or, on the other hand, might give the fungus the capacity to better face oxidative stresses.
G. margarita and its obligate endobacterium both express ROS-scavenger genes
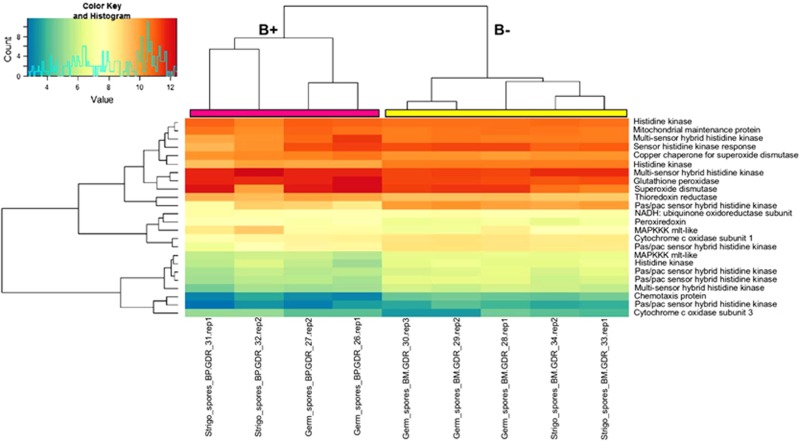
To test the hypothesis of crosstalk between the fungal oxidative response, the presence of the bacterium and the treatment with the SL analogue GR24, the transcriptome data were screened to identify genes involved in the detoxification of ROS. Some of these ROS-related genes were differentially expressed (Figure 6 and Supplementary Table S8). In detail, thioredoxin reductase, peroxiredoxins and glutathione peroxidase, which are ubiquitous molecules involved in ROS detoxification, were upregulated by the presence of the endobacterium. Selenocompound metabolism, which is known to be involved in antioxidant functions, was also upregulated in both the G and SL conditions (Supplementary Figure S6b). Another class of ROS-detoxifying molecules, the superoxide dismutases (SOD), was also affected by the endobacterial presence: two copper-zinc SODs were upregulated in the G condition, and a copper-zinc SOD already characterised in G. margarita, and reported as upregulated during spore germination (Lanfranco et al., 2005) was upregulated by the presence of the endobacterium in the G condition. Interestingly, the copper-zinc SOD was also detected as one of the most expressed proteins in a preliminary analysis comparing the fungal proteome in the presence and absence of the endobacterium (Salvioli et al., 2010). Also, RT-qPCR validated the constitutive expression of some of these respiratory and ROS-related genes (Supplementary Table S4), confirming their higher basal expression in the presence of the endobacterium. Interestingly a screen of the bacterial genome (Ghignone et al., 2012) also revealed ROS-related genes, including a copper-Zn SOD and a thioredoxin peroxidase, in addition to the genes involved in oxidative phosphorylation (Supplementary Table S9).
Figure 6.
Clustering and heatmap analysis of ROS-related genes. DESeq2-normalised and rlog-transformed expression data for differentially expressed ROS-related genes in germinating spores and SL-treated spores (both B+ and B−): samples were clustered with the heatmap.2 function in the gplots 2.14.2 Bioconductor package. Pink and yellow bars at the top of the heatmap were added to mark, respectively, the B+ and B− major tree branches as generated by clustering algorithm.
In conclusion, the availability of the G. margarita transcriptome, its mitochondrial genome and the CaGg genome allowed us to show that the AM fungus with its endobacterium has more constitutive tools to face oxidative stress, expressing a double set of ROS-scavenger genes. Irrespective of the presence of the endobacterium, GR24 treatment enhanced both the respiratory activities and the ROS-related responses, suggesting that the fungus perceives SL as a xenobiotic, and that its effects mostly target the mitochondrion.
The fungal–bacterial association enables a rapid transcriptional response to oxidative stress and SL treatment
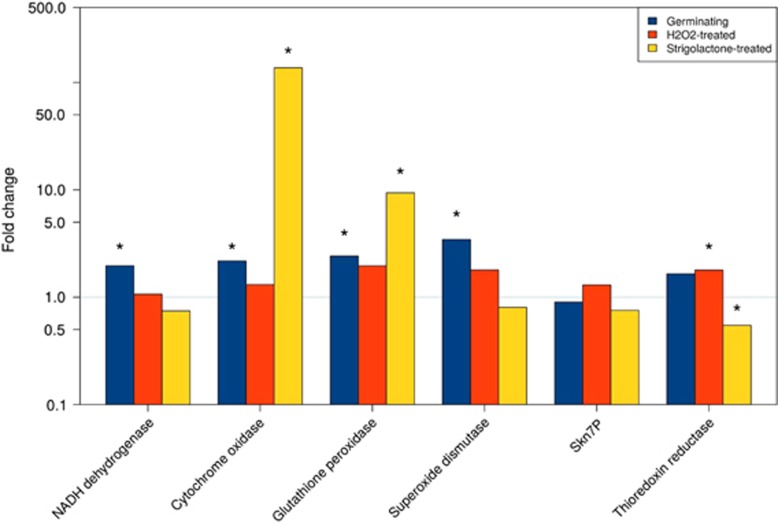
To examine the hypothesis that fungi with the endobacteria react to oxidative stress more actively than the fungi without the endobacteria, we treated the B+ and B− fungal lines with different concentrations of the ROS hydrogen peroxide (H2O2). To understand whether this ROS causes rapid transcriptional changes, and whether SL can mimic these stress-induced changes, we used RT-qPCR to measure the transcript levels of the mitochondrial and ROS-related genes that we identified as DEGs in the RNAseq experiment (Figure 7). Assays were performed on RNA extracted from B+ and B− spores treated with 0.3 mM H2O2 or GR24, after 3 h of incubation. A set of control spores, treated only with water and collected at the same timepoint, was also included. The control experiment confirmed the trend detected in the RNA-seq data, that is, that fungal genes encoding NADH dehydrogenase and COX 1 were expressed to higher levels in the B+ line than in the cured line, as well as glutathione peroxidase, which detoxifies H2O2, and Cu/Zn SOD, which transforms ROS into H2O2 (Figure 7). Hydrogen peroxide treatment stimulated the expression of some of the ROS-related transcripts in both lines (Supplementary Figure S8). However, the fold ratio between the two lines points to a higher expression in the B+ spores, with a significant value for thioredoxin reductase (Figure 7). These transcriptomic results suggest that the B+ line is constitutively more equipped to face oxidative stress conditions.
Figure 7.
Relative quantification of gene expression as obtained for a set of mitochondrial and ROS-related genes. Fold change is calculated for each gene in the B+ versus B− line in germinating, H2O2- and SL-treated spores with the basal expression recorded in the B− condition (threshold line at fold change=1). Statistically significant data (Kruskal–Wallis non-parametric test, P<0.05) are marked with an asterisk.
In agreement with previous observations (Besserer et al., 2006), the 3-h treatment with GR24 increased the transcription of genes encoding NADH Dehydrogenase and COX 1 in both the lines (Supplementary Figure S8), with a significantly higher fold-change ratio in the B+ line for COX1 (Figure 7). By contrast, the results were less clear-cut when ROS-related genes were considered: only the transcript of glutathione peroxidase, which uses H2O2 as a substrate, was strongly upregulated in the B+ spore, when compared with the cured line. On the other hand, the Cu/Zn SOD transcripts were higher after the GR24 treatment in the cured line (Supplementary Figure S8), suggesting that, irrespective of the bacterium, SL affects fungal ROS homeostasis.
To check whether the endobacterium directly perceives oxidative stress and SL, we measured the expression of some genes involved in ROS detoxification (Supplementary Figure S9), under the same conditions of brief H2O2/GR24 treatment. Although no significant changes were detected following GR24 treatment, an expression decrease was observed after the H2O2 ROS treatment, in comparison with the germinating spores, probably due to the antimicrobial effects of the H2O2. This result demonstrates that the bacterium itself does not cooperate with the fungus in ROS detoxification after H2O2/GR24 treatment, but its presence and its constitutively expressed ROS-related genes contribute to the ROS homeostasis of G. margarita. A regulated synthesis of ROS has a role not only in plant communication (Gilroy et al., 2014), but also in fungal morphogenesis, growth and development, as suggested for fungi interacting with plants or animals (Abbà et al., 2009; Ryder et al., 2013; Mu et al., 2014).
The presence of the endobacterium affects the basal intracellular calcium concentration
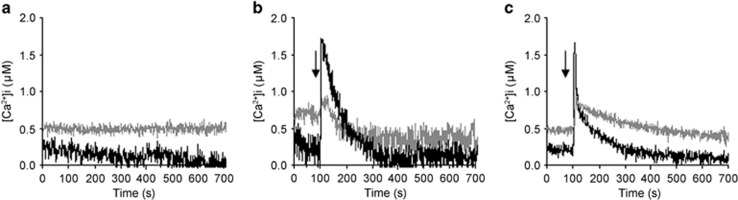
One of the most investigated aspects of the AMF–plant interaction is the calcium-mediated signalling pathway, which is elicited in the plant host by molecules from the AMF, by the symbiotic pathway (Genre et al., 2013; Oldroyd, 2013). To examine intracellular calcium, we used the TAT peptide, which acts as a potent nanocarrier to translocate macromolecules into living cells, to deliver the bioluminescent calcium reporter aequorin inside the AMF. Previous work with this system demonstrated that G. margarita responds to environmental stresses, as well as symbiotic signals, with transient changes in the intracellular calcium concentration ([Ca2+]i) (Moscatiello et al., 2014). We hypothesised that the B+ and B− lines of G. margarita perceive environmental and/or symbiotic signals differently. Using TAT-aequorin, we monitored [Ca2+]i in germinating spores under different conditions (basal status, cold shock and GR24 treatment), using the [Ca2+]i as a fast readout of the fungal response. The cured line in the three conditions showed an unexpected behaviour: the basal [Ca2+]i was significantly higher than that in the B+ fungus (Figure 8), suggesting that calcium constitutively accumulates in the cured line. Cold shock thus elicited a comparatively weaker response in the cured line: the calcium concentration started from a higher level, caused a lower peak, and then remained higher than in the B+ line throughout the experiment. Interestingly, GR24 is considered a symbiotic molecule, and GR24 treatment elicited a calcium signature comparable with that induced by an environmental stress condition, causing a fast and transient [Ca2+]i increase in the B+ line (Moscatiello et al., 2014). However, as observed for the cold shock, the cured line showed a weaker response, because the amplitude of the change in [Ca2+]i, calculated as the difference between the peak and the resting level (Δ[Ca2+]i), appeared reduced. Calcium has a crucial role in the establishment of symbiotic events: plants perceive microbial signals from rhizobia and AMF, activating oscillations in calcium concentration, which persist from minutes to hours (Oldroyd, 2013). Rhizobia and AMF also perceive plant signals, eliciting a transient calcium elevation (Moscatiello et al., 2010; Moscatiello et al., 2014). The relevance of calcium signalling as a mediator of extra- and intracellular stimuli is well described for many filamentous fungi (Bencina et al., 2005), and for Rhizophagus (Liu et al., 2013).
Figure 8.
TAT-aequorin-based Ca2+ measurements in G. margarita germinated spores. Ca2+ assays were performed in germinated fungal spores (10 days) after 1 h incubation with 30 μM TAT-aequorin. Intracellular free Ca2+ concentration was monitored in resting conditions (a) and after treatment (arrow) with a cold shock (b) or SL GR24 (10−6 M) (c). The reported traces represent typical observed responses (n=3). Black trace, B+ line. Grey trace, cured line.
A permanent increase in intracellular calcium has generally been considered as harmful for the cell (Zhivotovsky and Orrenius, 2011). We hypothesise that the absence of the bacterium affects calcium signalling, leading to an increase in the basal [Ca2+]i, which negatively affects fungal cellular metabolism, possibly by interfering with ATP synthesis. Indeed, the low solubility of Ca2+ with phosphates may interfere with ATP-based metabolism, which is an essential feature of all living cells (Case et al., 2007).
Our experiments further indicate that the bacterium may interfere with the regulatory signalling network, which also includes ROS (Gilroy et al., 2014); the endobacteria influence the intracellular calcium level in a still-unknown way, and potentially alter fungal ATP synthesis. It could be speculated that the endobacterium functions as an additional Ca2+ store in both hyphae and spores, thereby cooperating in fine-tuning the [Ca2+]i in the AMF. Irrespective of this, the TAT experiment also shows that GR24 causes a reaction in the fungus similar to the reaction to environmental stress.
Conclusion
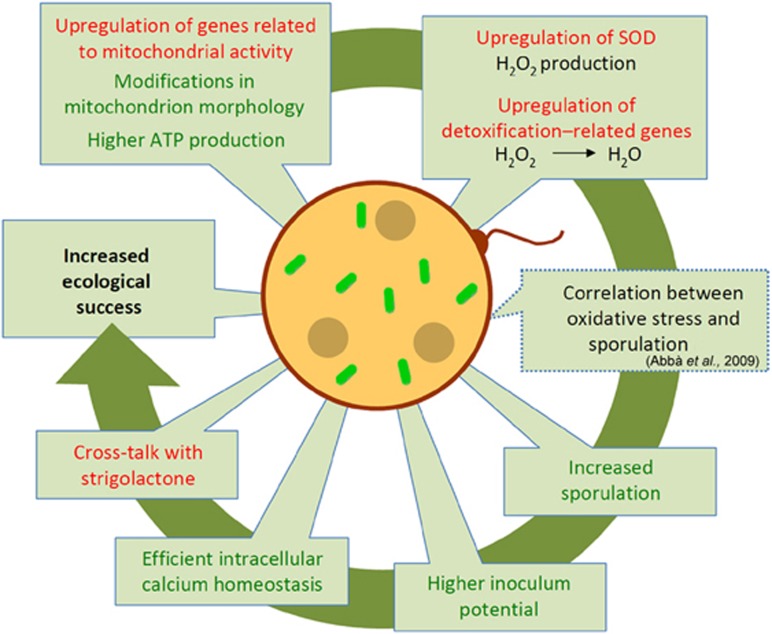
In summary, here, we used a combination of molecular, cellular and physiological approaches to reveal that the obligate endobacterium CaGg affects the biology of the AMF G. margarita, and that its effect is most relevant during the presymbiotic phase, when the fungus, unprotected by its plant host, encounters the biotic and abiotic stimuli in the soil (Figure 9).
Figure 9.
Schematic summary of the mechanisms by which the endobacterium affects G. margarita metabolism. Starting from the left on the top, the drawing illustrates how in the presence of the endobacterium, the fungus G. margarita upregulates genes involved in respiration, leading to higher ATP production. This status is mirrored by morphological changes in the mitochondrial ultrastructure. Thanks to the upregulation of SOD, H2O2 is produced and further detoxified to water by ROS-scavenger enzymes, like glutathione peroxidase. Intracellular calcium homeostasis is efficiently maintained. Treatment with GR24, a synthetic SL induces some transcriptional responses that mimic the endobacterium presence, suggesting that there is interference between the microbe presence and the plant hormone treatment. The oxidative burst is correlated with higher spore production in many fungi: this feature is indeed present in G. margarita, which contains endobacteria. As G. margarita exclusively colonises plants starting from spores, this feature represents an evolutionary benefit. Red font, transcriptome results; green font, data from biochemical, cellular and physiological experiments; interrupted line, hypotheses based on the literature. A full colour version of this figure is available at The ISME Journal online.
The changes we detected in the fungal transcriptional landscape suggest that the presence of the endobacterium tunes a huge number of metabolic pathways, including spore production, fungal wall remodeling and mineral nutrient uptake and transport, also leading to an unexpected positive impact on the phosphate content of plant roots and shoots. However, among the many DEGs, the most interesting indicated that the mitochondrion is a primary target of the effects of the presence of the endobacteria. These effects enhance the positive and negative effects of the mitochondria: on one hand, in the fungal line with its ancient endobacterium (Mondo et al., 2012), activation of respiration leads to a higher ATP production. On the other hand, this line also shows stronger activation of genes involved in ROS generation and detoxification than the cured line. As the endobacterium also constitutively activates respiratory and ROS-scavenger genes, we suggest that the AMF hosting the endobacterium has twice the tools to face environmental stresses. Thus, we hypothesise that, as described for the human microbiota (Chu and Mazmanian, 2013), the intracellular endobacteria may prime the level of innate immunity in the AMF. However, differently from other eukaryotes, experimental data exploring the existence of innate immunity in fungi do not seem to be available. The control of the intracellular concentration of calcium could be one of the key processes, first regulating the bioenergetic status of the fungus. Lastly, the comparison of the fungal transcriptome after treatment with a synthetic SL allowed us to reveal that both the endobacterium and SL produced a comparable response in the fungal mitochondria, possibly because the prokaryote and GR24 (Besserer et al., 2006) have a similar effect on the expression of respiratory genes. We suggest that SLs, which are usually considered as plant symbiotic signals, are first perceived by the AMF as foreign molecules (xenobiotics), activating a fast and transient [Ca2+]i increase, as well as elevated respiration, eventually leading to ROS production and detoxification. In the absence of the bacterium, and in line with the overall slower metabolism shown by the cured line, all these processes are attenuated.
Taken as a whole, our data indicate that the endobacterium improves the fitness of its fungal host during the pre-symbiotic rhizospheric phases. Unlike Rhizophagus (Hempel et al., 2007; Varela-Cervero et al., 2015), Gigaspora mostly produces spores, as pre-symbiotic propagules, which represent the exclusive structures for the success of colonisation, and Gigaspora has taken advantage of the endobacterium. This symbiosis has proven beneficial to the AMF, from an evolutionary point of view, thus potentially offsetting its nutritional cost for the fungus.
Acknowledgments
This paper is dedicated to the memory of Professor Aldo Fasolo, his inexhaustible scientific curiosity, support and inspiration. We wish to thank: Luisa Lanfranco (University of Torino) and Nicholas Corradi (University of Vancouver) for their contributions in the first phases of the project; Andrea Genre (University of Torino) for drawing Figure 9; Mauro degli Esposti (IIT-Genova) for the discussion on bionergetics and for having suggested us to use the Mytotracker staining; Matteo Chialva (University of Torino) for the Pi measurements in plants; Antonella Faccio (IPSP-CNR) for her assistance in electron microscopy; Paola Giannini, Marco Malavasi (University of Piemonte Orientale), Giancarlo Birello (IRCrES-CNR, IT Office) and Raffaele Calogero (University of Torino) for their bioinformatics support; Maria Teresa Della Beffa and Sara Torrielli (University of Torino) for the taking care of the spore production, and Jennifer Mach for the critical reading. Research was funded by the Ateneo Project (ex 60%) to PB. AS fellowship was granted by Piedmont Region and University of Torino.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abbà S, Khouja HR, Martino E, Archer DB, Perotto S. (2009). SOD1-targeted gene disruption in the ericoid mycorrhizal fungus Oidiodendron maius reduces conidiation and the capacity for mycorrhization. Mol Plant Microbe Interact 22: 1412–1421. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencina M, Legisa M, Read ND. (2005). Cross-talk between cAMP and calcium signalling in Aspergillus niger. Mol Microbiol 56: 268–281. [DOI] [PubMed] [Google Scholar]
- Besserer A, Puech-Pages V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S et al. (2006). Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto V, Bandi C, Minerdi D, Sironi M, Tichy HV, Bonfante P. (1996). An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. App Environ Microbiol 62: 3005–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto V, Lumini E, Bonfante P, Vandamme P. (2003). 'Candidatus glomeribacter gigasporarum' gen. nov., sp. nov., an endosymbiont of arbuscular mycorrhizal fungi. Int J Syst Evol Microbiol 53: 121–124. [DOI] [PubMed] [Google Scholar]
- Bohm J, Hoff B, O'Gorman CM, Wolfers S, Klix V, Binger D et al. (2013). Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc Natl Acad Sci USA 110: 1476–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P, Anca IA. (2009). Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu Rev Microbiol 63: 363–383. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Balestrini R, Mendgen K. (1994). Storage and secretion processes in the spore of Gigaspora-margarita Becker and Hall as revealed by high-pressure freezing and freeze-substitution. New Phytol 128: 93–101. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A. (2010). Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat Comm 1: 48. [DOI] [PubMed] [Google Scholar]
- Case RM, Eisner D, Gurney A, Jones O, Muallem S, Verkhratsky A. (2007). Evolution of calcium homeostasis: From birth of the first cell to an omnipresent signalling system. Cell Calcium 42: 345–350. [DOI] [PubMed] [Google Scholar]
- Chu HT, Mazmanian SK. (2013). Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 14: 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desirò A, Salvioli A, Ngonkeu EL, Mondo SJ, Epis S, Faccio A et al. (2014). Detection of a novel intracellular microbiome hosted in arbuscular mycorrhizal fungi. ISME J 8: 257–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech-Pages V, Novero M, Rey T et al. (2013). Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198: 179–189. [DOI] [PubMed] [Google Scholar]
- Gerdemann JW.and Nicolson TH. (1963). Spores of mycorrhizal Endogone extracted from soil by wet sieving and decanting. Trans Brit Mycol Soc 46: 235–244. [Google Scholar]
- Ghignone S, Salvioli A, Anca I, Lumini E, Ortu G, Petiti L et al. (2012). The genome of the obligate endobacterium of an AM fungus reveals an interphylum network of nutritional interactions. ISME J 6: 136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Suzuki N, Miller G, Choi WG, Toyota M, Devireddy AR et al. (2014). A tidal wave of signals: calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci 19: 623–630. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–U130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel S, Renker C, Buscot F. (2007). Differences in the species composition of arbuscular mycorrhizal fungi in spore, root and soil communities in a grassland ecosystem. Environ Microbiol 9: 1930–1938. [DOI] [PubMed] [Google Scholar]
- Lanfranco L, Novero M, Bonfante P. (2005). The mycorrhizal fungus Gigaspora margarita possesses a CuZn superoxide dismutase that is up-regulated during symbiosis with legume hosts. Plant Physiol 137: 1319–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Limpens E, Zhang ZH, Ivanov S, Saunders DGO, Mu DS et al. (2014). Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet 10: e1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gianinazzi-Pearson V, Arnould C, Wipf D, Zhao B, Van Tuinen D. (2013). Fungal genes related to calcium homeostasis and signalling are upregulated in symbiotic arbuscular mycorrhiza interactions. Fungal Biol 117: 22–31. [DOI] [PubMed] [Google Scholar]
- Love M, Anders S, Hubert W. (2013), Differential analysis of count data - the DESeq2 package http://www.bioconductor.org/packages/2.13/bioc/vignettes/deseq2/inst/doc/deseq2.pdf.
- Lumini E, Bianciotto V, Jargeat P, Novero M, Salvioli A, Faccio A et al. (2007). Presymbiotic growth and sporal morphology are affected in the arbuscular mycorrhizal fungus Gigaspora margarita cured of its endobacteria. Cell Microbiol 9: 1716–1729. [DOI] [PubMed] [Google Scholar]
- Luo WJ, Brouwer C. (2013). Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29: 1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Mendoza IE, Dewbre GR, Harrison MJ. (2001). A phosphate transporter gene from the extra-radical mycelium of an arbuscular mycorrhizal fungus Glomus intraradices is regulated in response to phosphate in the environment. Mol Plant Microbe Interact 14: 1140–1148. [DOI] [PubMed] [Google Scholar]
- Martin F, Kohler A, Murat C, Balestrini R, Coutinho PM, Jaillon O et al. (2010). Perigord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464: 1033–1038. [DOI] [PubMed] [Google Scholar]
- Martin F, Selosse MA. (2008). The Laccaria genome: a symbiont blueprint decoded. New Phytol 180: 296–310. [DOI] [PubMed] [Google Scholar]
- Mondo SJ, Toomer KH, Morton JB, Lekberg Y, Pawlowska TE. (2012). Evolutionary stability in a 400-million-year-old heritable facultative mutualism. Evolution 66: 2564–2576. [DOI] [PubMed] [Google Scholar]
- Moreira PI, Carvalho C, Zhu X, Smith MA, Perry G. (2010). Mitochondrial dysfunction is a trigger of Alzheimer's disease pathophysiology. Biochim Biophys Acta 1802: 2–10. [DOI] [PubMed] [Google Scholar]
- Moscatiello R, Sello S, Novero M, Negro A, Bonfante P, Navazio L. (2014). The intracellular delivery of TAT-aequorin reveals calcium-mediated sensing of environmental and symbiotic signals by the arbuscular mycorrhizal fungus Gigaspora margarita. New Phytol 203: 1012–1020. [DOI] [PubMed] [Google Scholar]
- Moscatiello R, Squartini A, Mariani P, Navazio L. (2010). Flavonoid-induced calcium signalling in Rhizobium leguminosarum bv. viciae. New Phytol 188: 814–823. [DOI] [PubMed] [Google Scholar]
- Mu DS, Li CY, Zhang XC, Li XB, Shi L, Ren A et al. (2014). Functions of the nicotinamide adenine dinucleotide phosphate oxidase family in Ganoderma lucidum: an essential role in ganodermic acid biosynthesis regulation, hyphal branching, fruiting body development, and oxidative-stress resistance. Environ Microbiol 16: 1709–1728. [DOI] [PubMed] [Google Scholar]
- Novero M, Faccio A, Genre A, Stougaard J, Webb KJ, Mulder L et al. (2002). Dual requirement of the LjSym4 gene for mycorrhizal development in epidermal and cortical cells of Lotus japonicus roots. New Phytol 154: 741–749. [DOI] [PubMed] [Google Scholar]
- Oldroyd GED. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11: 252–263. [DOI] [PubMed] [Google Scholar]
- Pelin A, Pombert JF, Salvioli A, Bonen L, Bonfante P, Corradi N. (2012). The mitochondrial genome of the arbuscular mycorrhizal fungus Gigaspora margarita reveals two unsuspected trans-splicing events of group I introns. New Phytol 194: 836–845. [DOI] [PubMed] [Google Scholar]
- Popova Y, Thayumanavan P, Lonati E, Agrochao M, Thevelein JM. (2010). Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc Natl Acad Sci USA 107: 2890–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder LS, Dagdas YF, Mentlak TA, Kershaw MJ, Thornton CR, Schuster M et al. (2013). NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proc Natl Acad Sci USA 110: 3179–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvioli A, Chiapello M, Fontaine J, Hadj-Sahraoui AL, Grandmougin-Ferjani A, Lanfranco L et al. (2010). Endobacteria affect the metabolic profile of their host Gigaspora margarita, an arbuscular mycorrhizal fungus. Environ Microbiol 12: 2083–2095. [DOI] [PubMed] [Google Scholar]
- Salvioli A, Lumini E, Anca IA, Bianciotto V, Bonfante P. (2008). Simultaneous detection and quantification of the unculturable microbe Candidatus Glomeribacter gigasporarum inside its fungal host Gigaspora margarita. New Phytol 180: 248–257. [DOI] [PubMed] [Google Scholar]
- Salvioli A, Zouari I, Chalot M, Bonfante P. (2012). The arbuscular mycorrhizal status has an impact on the transcriptome profile and amino acid composition of tomato fruit. BMC Plant Biol 27: 12–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R et al. (2013). Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci USA 110: 20117–20122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela-Cervero S, Vasar M, Davison J, Barea JM, Opik M, Azcon-Aguilar C. (e-pub ahead of print 11 February 2015). The composition of arbuscular mycorrhizal fungal communities differs among the roots, spores and extraradical mycelia associated with five Mediterranean plant species. Environ Microbiol. [DOI] [PubMed]
- Xu Z, Hao BL. (2009). CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res 37: W174–W178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11: R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S. (2011). Calcium and cell death mechanisms: A perspective from the cell death community. Cell Calcium 50: 211–221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.