Abstract
Non–small cell lung cancers (NSCLCs) frequently express estrogen receptor (ER) β, and estrogen signaling is active in many lung tumors. We investigated the ability of genes contained in the prediction analysis of microarray 50 (PAM50) breast cancer risk predictor gene signature to provide prognostic information in NSCLC. Supervised principal component analysis of mRNA expression data was used to evaluate the ability of the PAM50 panel to provide prognostic information in a stage I NSCLC cohort, in an all-stage NSCLC cohort, and in The Cancer Genome Atlas data. Immunohistochemistry was used to determine status of ERβ and other proteins in lung tumor tissue. Associations with prognosis were observed in the stage I cohort. Cross-validation identified seven genes that, when analyzed together, consistently showed survival associations. In pathway analysis, the seven-gene panel described one network containing the ER and progesterone receptor, as well as human epidermal growth factor receptor (HER)2/HER3 and neuregulin-1. NSCLC cases also showed a significant association between ERβ and HER2 protein expression. Cases positive for HER2 expression were more likely to express HER3, and ERβ-positive cases were less likely to be both HER2 and HER3 negative. Prognostic ability of genes in the PAM50 panel was verified in an ERβ-positive cohort representing all NSCLC stages. In The Cancer Genome Atlas data sets, the PAM50 gene set was prognostic in both adenocarcinoma and squamous cell carcinoma, whereas the seven-gene panel was prognostic only in squamous cell carcinoma. Genes in the PAM50 panel, including those linking ER and HER2, identify lung cancer patients at risk for poor outcome, especially among ERβ-positive cases and squamous cell carcinoma.
Abbreviations: NSCLC, non–small cell lung cancer; ER, estrogen receptor; PGR, progesterone receptor; HR, hazard ratio; HER2, human epidermal growth factor receptor 2; HER3, human epidermal growth factor receptor 3; FFPE, formalin-fixed, paraffin embedded; PCA, principal component analysis; DFS, disease-free survival; PFS, progression-free survival; OS, overall survival; TCGA, The Cancer Genome Atlas; PAM50, prediction analysis of microarray 50 gene set; IHC, immunohistochemistry; IPA, Ingenuity Pathway Analysis
Introduction
Lung cancer is the leading cause of cancer-related mortality in the United States. Complete surgical resection of early stage non–small cell lung cancer (NSCLC) provides the best chance for cure, but there is significant risk of relapse, with a 5-year survival rate for patients with stage IA disease of only 73% and 58% for stage IB disease [1]. Adjuvant therapy for stage I disease is not currently recommended [2]. Identifying patients at high risk of recurrence based on the biology of their lung tumors could identify those in need of adjuvant treatment, and a hormone-associated prognostic signature could suggest the use of adjuvant endocrine therapy.
Lung tumors express estrogen receptors (ERs) [3], [4], [5], [6], [7], [8], [9], [10], [11], are inhibited by antiestrogens [6], [7], and are responsive to the steroid hormones β-estradiol [5], [9], [11] and progesterone [12]. Hormone pathways affect lung cancer survival [13], [14], [15], [16]. NSCLCs are often ERβ positive [13], and β-estradiol both induces proliferation in NSCLCs [9] and promotes lung cancer in animal models [17]. Thus, there are similarities between breast cancer and lung cancer in how estrogen controls tumor progression. We hypothesized that the prediction analysis of microarray 50 (PAM50) gene panel developed for classification of breast cancer into subtypes with different clinical behavior [18], and especially useful in predicting survival of ERα-positive breast cancer [19], may also inform prognosis in lung cancer. We examined the prognostic ability of the PAM50 gene panel in NSCLC using data from a stage I NSCLC cohort, an independent all-stage NSCLC cohort consisting only of ERβ-positive cases, and The Cancer Genome Atlas (TCGA) data. We also investigated the relationship between tumor mRNA and protein expression level for several PAM50 genes, and the relationship of protein expression to detection of the full-length active isoform of ERβ protein (ERβ-1) by immunohistochemistry (IHC). Pathway analysis was used to explore signaling interactions predicted to be involved in the function of informative PAM50 genes.
Material and Methods
Analysis of PAM50 mRNA Gene Expression in NSCLC Patient Cohorts
We examined the relationship between disease-free survival (DFS) and expression of the PAM50 genes [18], [19] in a cohort of 104 stage I NSCLCs (cohort 1, Table 1). Cases were selected from the University of Pittsburgh Lung Cancer SPORE Tissue Bank based on the following criteria: tumor tissue originated from a completely resected primary stage I adenocarcinoma or squamous cell lung cancer diagnosed between 1998 and 2009; smoking history, age at diagnosis, sex, presence or absence of pleural invasion, and outcome were recorded; and no neoadjuvant therapy was given. To minimize survival bias, only tumors resected within 3 months of an incident biopsy-proven lung cancer diagnosis were included. Of 140 stage I cases meeting these criteria, 104 cases (Table 1) had fresh-frozen banked lung cancer tissue available for RNA isolation. Intraoperative brachytherapy status and adjuvant treatment status were known. A minority of patients received these therapies, and they were not significantly associated with prognosis. Age at diagnosis, sex, smoking history, pleural invasion, and histology also did not significantly impact prognosis (data not shown). Outcome data were obtained from the University of Pittsburgh Cancer Institute Cancer Registrar (http://cancerregistrynetwork.upmc.com/). DFS was calculated from date of complete surgical resection to the first documented detectable malignancy related to the resected lung cancer.
Table 1A.
Cohort 1: Stage I NSCLC (N = 104)
| All Cases (N = 104) | ERβ-Positive Cases (N = 35) | |
|---|---|---|
| Age (mean ± SD) | 68.2 ± 9.2 | 68.5 ± 9.5; n.s. |
| Sex | ||
| Female | 57 (54.8%) | 20 (57.1%); n.s. |
| Male | 47 (45.2%) | 15 (42.9%) |
| Stage | ||
| IA | 37 (35.6%) | 8 (22.9%); n.s. |
| IB | 67 (64.4%) | 27 (77.1%) |
| Histology | ||
| Adenocarcinoma | 78 (75.0%) | 17 (48.6%); P = .005 |
| Squamous cell | 26 (25.0%) | 18 (51.4%) |
| Smoking history | ||
| Active | 36 (34.6%) | 18 (51.4%); n.s. |
| Former | 60 (57.7%) | 17 (48.6%) |
| Never | 8 (7.7%) | 0 (0%) |
| Median DFS (years) | ||
| All cases | 6.32 (4.31, Inf)⁎ | 4.31 (3.27, Inf)⁎; n.s. |
| Adenocarcinoma | 6.95 (4.03, Inf)⁎ | 6.30 (3.35, Inf)⁎ |
| Squamous cell | 6.00 (3.54, Inf)⁎ | 3.64 (1.10, Inf)⁎ |
| ERβ Allred score | ||
| High (Allred score > 4) | Not applicable | 20 (57.1%) |
| Low (Allred score ≤ 4) | Not applicable | 15 (42.9%) |
n.s., not significant.
95% Confidence interval.
Sections of each tumor were examined by a board-certified pathologist (S.D.); diagnosis, staging, and adequacy of tumor tissue were confirmed. The American Joint Committee on Cancer seventh edition staging guidelines were followed. RNA was isolated from frozen tumor tissue and, after passing quality checks, was used to assess mRNA expression with the Illumina Human HT-12 v4 BeadChip. Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-3448. mRNA data were subjected to background subtraction and quantile normalization. Supervised principal component analysis (PCA) was conducted, first to evaluate the ability of all the PAM50 genes to associate with DFS and second by using the top genes selected using the marginal P value of .1 as cutoff. All results were subjected to 10-fold cross-validation, including the feature selection step.
A second cohort of 63 lung cancer cases representing all histologies and stages (cohort 2, Table 1B) was used to confirm PCA results from cohort 1. Previously published IHC data of ERβ-1 expression [13] were available for all 63 cases in cohort 2; all were positive for ERβ (Allred score > 0). Outcome data for these patients have previously been used for analysis of survival related to ERβ IHC scores [13]. The 63 cohort 2 cases are a subset of the 183 cases in the published study [13]; they are ERβ-positive cases for which sufficient tumor material remained for RNA isolation. Because cohort 2 contains some patients who were never disease free, progression-free survival (PFS) instead of DFS was used. PFS was calculated from the date of resection to the first documentation of malignancy for disease-free patients or from diagnostic biopsy to first documented date of progression for patients who were never disease free. Microarray data are available in the ArrayExpress database under accession number E-MTAB-3665.
Table 1B.
Cohort 2: All-Stage ERβ Positive NSCLC (N = 63)
| Age (mean ± SD) | 69.38 ± 8.58 |
| Sex | |
| Female | 30 (47.6%) |
| Male | 33 (52.4%) |
| Stage | |
| IA | 12 (19.0%) |
| IB | 18 (28.6%) |
| IIA/IIB | 12 (19.0%) |
| IIIA/IIIB | 15 (23.8%) |
| IV | 6 (9.6%) |
| Histology | |
| Adenocarcinoma | 26 (41.3%) |
| Squamous cell | 29 (46.0%) |
| Large cell/undifferentiated | 6 (9.5%) |
| Other | 2 (3.2%) |
| Smoking history | |
| Active | 24 (38.1%) |
| Former | 32 (50.8%) |
| Never | 2 (3.2%) |
| Unknown | 5 (7.9%) |
| Median PFS (years) | |
| All cases | 1.86 (1.21, Inf)⁎ |
| Adenocarcinoma | 1.71 (1.21, Inf)⁎ |
| Squamous cell | 1.96 (0.997, Inf)⁎ |
| ERβ Allred score | |
| High (Allred score > 4) | 32 (50.8%) |
| Low (Allred score ≤ 4) | 31 (49.2%) |
95% Confidence interval.
Analysis Using TCGA Data Sets
The cBioPortal for Cancer Genomics Web site, http://www.cbioportal.org/public-portal/, was used to access TCGA mRNA expression data. The provisional RNASeq V2 RSEM z-score for each gene was used. The z-score (positive or negative) represents a normalized relative expression level and was used as a surrogate for the mRNA level. Data for all 50 PAM50 genes were available from the TCGA cases. Overall survival (OS) data and tumor stage, if available (adenocarcinoma study), were downloaded. For adenocarcinoma, 203 cases had mRNA expression and OS data; for squamous cell carcinoma, 220 cases had these data. DFS or PFS data were not available for most TCGA cases. The cBioPortal was used to assess correlation of expression among the PAM50 genes and value of individual genes in assessing OS based on a z-score cutoff of + 1.5, + 2.0, or + 3.0. Association of OS with individual PAM50 gene z-scores in TCGA data (by Kaplan-Meier survival analysis and log rank test) was analyzed using the R package on the cBioPortal website. Data were accessed from the cBioPortal between October 1, 2014, and December 15, 2014.
Pathway/Network Analysis
The Ingenuity Pathway Analysis (IPA) software program (version 21901358) was used to explore pathway relationships. Relative expression observed in the poor-survival group compared with the better-surviving group for each gene, as determined in cohort 1, was entered for analysis. Predicted interacting networks among the genes were generated, as well as canonical signaling systems, upstream regulators, functions or diseases affected by these genes, and P values for the likelihood of associations. The IPA program was accessed between December 12, 2014, and December 31, 2014.
Immunohistochemistry
Protein expression was measured by staining formalin-fixed, paraffin embedded (FFPE) tissues with the following antibodies and dilutions: ERβ-1 (AbD Serotec; MCA 1974ST; 1:40; detects the full-length isoform of ERβ), HER2 (Thermo Scientific; MS-730; 1:400), HER3 (Santa Cruz Biotechnology; sc-285; 1:100), c-Myc (Abcam; ab32072; 1:50), and cyclin E (Abcam; ab9517; 1:100). Immunoreactive cells were visualized with diaminobenzidine chromogenic substrate and counterstained with hematoxylin. Positive and negative controls were included. Exclusion of the primary antibody was used for negative controls. Breast carcinoma tissue was used as a positive control. Each section was scored semiquantitatively by a board-certified pathologist (S.D.) under blinded conditions. IHC results of ERβ-1 for cases in cohort 2 were previously reported using the Allred scoring system [13]. The H-score system was used for c-Myc, HER3, and cyclin E staining [20]. HER2 expression was scored 0 to 3 as for breast cancer based on percentage of positively stained tumor cells [21]. Cases used for IHC (N = 91; 42 from cohort 1, 26 from cohort 2, and an additional 23 stage I cases with FFPE tumor tissue that met the eligibility criteria for inclusion in cohort 1 but did not yield mRNA of sufficient quality for gene expression) had sufficient FFPE tumor tissue for analysis and gave interpretable results in scoring for ERβ-1 and at least one other protein (CCNE1, MYC, HER2. and HER3).
Statistical Analysis
To evaluate the prognostic ability of PAM50 genes, we applied PCA to the gene expression matrix. The prognostic values of the top three principal components (PCs) were evaluated. Using the median of each PC score as the cutoff, the cohort was divided into two groups. Log rank tests were used to compare survival of these two groups. To evaluate the prognostic value of top (selected) PAM50 genes, we used supervised PCA as detailed by Bair and Tibshirani [22]: first, the marginal association of each gene is evaluated by Cox proportional hazard; next, only markers that reached a marginal P value of .1 or less are subjected to PCA analysis to determine the prognostic value. To avoid overfitting, all models were evaluated by 10-fold cross-validation. When using only the top genes (evaluated by marginal association of the gene and outcomes), the feature selection step is included in the cross-validation cycles.
To evaluate the association between the ERβ status (positive versus negative) and protein expression of other genes assessed by IHC (positive or negative), two-sided Fisher’s exact tests were used. t tests were used to compare the log mRNA expression level of genes between samples that exhibited high and low protein expression level (assessed by IHC). For this analysis, IHC score of each protein was dichotomized into high versus low groups. Kaplan-Meier survival curves were used to evaluate DFS and PFS. Hazard ratios (HRs) and the corresponding 95% confidence intervals (CIs) were calculated using Cox proportional hazard models. All tests were two sided, and RNA expression analyses were conducted using the R software program (version 3.0.2).
Results
Survival Analysis in Stage I Lung Cancer
In the stage I cohort (cohort 1, N = 104, Table 1A), expression of three PAM50 genes (CXXC5, FGFR4, FOXC1) individually showed significant (P < .05) association with DFS (data not shown). Higher expression of each of these genes was associated with worse survival (Table 2). The other PAM50 genes were not individually associated with survival. The top three PCs that described variation in expression of the PAM50 gene panel were evaluated for association with DFS. Using PC2, the PAM50 gene panel separated cohort 1 based on DFS, but the result was not significant (Table 3). The relationship with DFS was stronger in cohort 1 using only seven PAM50 genes, each marginally associated with DFS at P < .1 in at least five cycles (Table 2) during the 10 × cross-validation (Table 3). These genes (GRB7, MYC, MIA, CXXC5, PGR, FGFR4, and FOXC1) show the strongest association with DFS in the stage I cohort. The HR of the worse prognostic group using the seven genes was 2.08 (95% CI: 1.18-3.66) using PC2, P = .01 (Table 3).
Table 2.
Differential Expression of PAM50 Genes Selected During 10 × Cross-Validation of Survival Analysis⁎
| Gene | Times Selected | Relative Expression in High-Risk Survival Group | Direction |
|---|---|---|---|
| KRT14 | 1 | 1.51 | Upregulated |
| KRT17 | 1 | 1.36 | Upregulated |
| CDH3 | 1 | 1.55 | Upregulated |
| PHGDH | 1 | 0.72 | Downregulated |
| CCNE1 | 1 | 1.10 | Upregulated |
| TMEM45B | 1 | 1.46 | Upregulated |
| SFRP1 | 1 | 1.25 | Upregulated |
| ACTR3B | 1 | 0.51 | Downregulated |
| EXO1 | 1 | 2.11 | Upregulated |
| NAT1 | 2 | 1.98 | Upregulated |
| ERBB2 | 2 | 1.11 | Upregulated |
| MLPH | 4 | 1.67 | Upregulated |
| CDCA1 | 4 | 1.27 | Upregulated |
| GRB7 | 5 | 2.36 | Upregulated |
| MIA | 6 | 1.56 | Upregulated |
| MYC | 7 | 1.29 | Upregulated |
| PGR | 8 | 0.84 | Downregulated |
| FGFR4 | 9 | 1.56 | Upregulated |
| CXXC5 | 9 | 2.06 | Upregulated |
| FOXC1 | 10 | 1.29 | Upregulated |
Using a marginal P < .1 as cutoff.
Table 3.
PCA of Survival of the PAM50 Gene Panel in NSCLC Cohorts
| Group | Model | HR | CI | P Value |
|---|---|---|---|---|
| Cohort 1 Stage I, DFS (N = 104) (discovery Set) |
All genes | 1.51 | (0.86, 2.67) | .15 |
| Seven-gene model | 2.08⁎ | (1.18, 3.66) | .01 | |
| Cohort 1 subgroup Stage I, ERβ positive, DFS (N = 35) |
All genes | 2.47 | (0.89, 6.81) | .080 |
| Seven-gene model | 2.44⁎ | (0.99, 5.99) | .050 | |
| Cohort 2 All stages, PFS ERβ positive (N = 63) |
All genes | 1.84 | (0.91, 3.75) | .091 |
| Seven-gene model | 2.26⁎ | (1.10, 4.65) | .026 | |
| TGCA Adenoca all stages, OS (N = 203) |
All genes | 2.14⁎ | (1.27, 3.61) | .004 |
| Seven-gene model | 1.42 | (0.86, 2.35) | .16 | |
| TGCA Adenoca stage I, OS (N = 101) |
All genes | 3.17⁎ | (1.26, 7.97) | .01 |
| Seven-gene model | 1.39 | (0.56, 3.43) | .48 | |
| TGCA Sq cell ca OS (N = 220) |
All genes | 1.66⁎ | (1.09, 2.54) | .018 |
| Seven-gene model | 1.62⁎ | (1.06, 2.47) | .026 |
Adenoca, adenocarcinoma; Sq cell ca, squamous cell carcinoma.
P < .05.
Directionality of change for the genes selected at least once during cross validation is shown in Table 2. The mRNA expression of GRB7, MIA, MYC, FGFR4, CXXC5, and FOXC1 were upregulated in high-risk cases, whereas PGR mRNA expression was downregulated. Eleven PAM50 genes marginally selected were upregulated and two were downregulated in high-risk compared with low-risk cases. Kaplan-Meier survival curves of cohort 1 using the seven informative genes (not shown) showed that median DFS of the high-risk group was 4.50 years (N = 55, 30 events), whereas median DFS in the low-risk group was 6.95 years (N = 49, 20 events, P = .01). Cohort 1 was not selected for ERβ-positive lung tumors, but based on our previous work [13] and the literature [14], [15], we expect 65% to 75% of NSCLCs to be ERβ positive. We did not find that the PAM50 genes clustered NSCLCs (either adenocarcinoma or squamous cell carcinoma) into subtypes as has been found for breast cancer. Therefore, we could not use the risk score algorithm developed for breast cancer in survival analysis.
Relationship of PAM50 Genes to ERβ-Positive Status
PAM50 genes were subsequently analyzed in a subset of 35 cases from cohort 1 (Table 1A) for which ERβ IHC was found to be positive (Allred score > 0). We have previously found little or no ERα protein in NSCLC when using IHC antibodies validated for ERα determination in breast cancer patients [13]. Therefore, only ERβ was analyzed. The ERβ-positive subset did not significantly differ from the entire cohort by age, sex, stage, smoking history, or survival (Table 1A) but did differ by histology: 48.6% were squamous cell carcinoma compared with 25% in cohort 1 (P < .005). The PAM50 panel was prognostic in the ERβ-positive subset with marginal significance (HR 2.47 [95% CI: 0.89-6.89], P = .080, Table 2), whereas the seven-gene panel significantly associated with DFS in ERβ-positive stage I cases (HR 2.44 [95% CI: 0.99-5.99], P = .050, Table 2). We also stratified the cases by ERβ score into high and low subgroups. Although the ERβ high cases showed better separation of survival groups using either the PAM50 panel or the seven-gene model compared with the ERβ low cases (HR > 6, P < .005), these results were unstable because of small sample size, and this analysis may be overfit (data not shown).
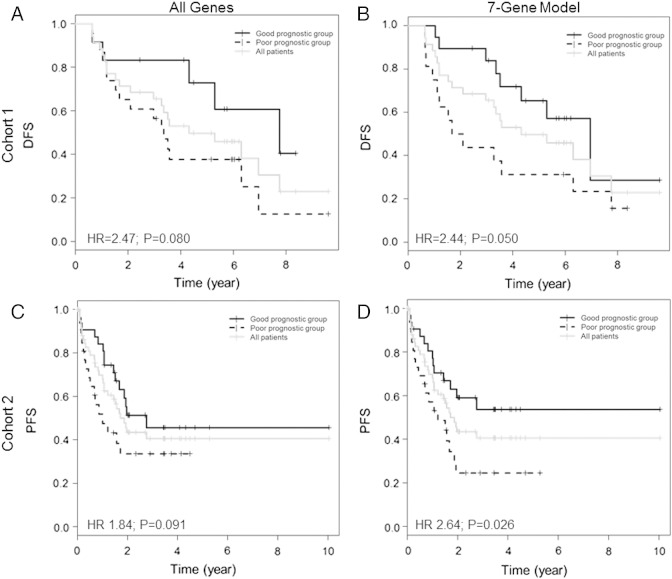
For the ERβ-positive stage I cases, Kaplan-Meier survival curves using all PAM50 genes showed that median DFS in the high-risk group was 3.27 years (N = 18, 16 events), whereas median DFS in the low-risk group was 6.6 years (N = 17, 5 events, Figure 1A). Using the seven-gene model with ERβ-positive stage I cases, median DFS was 2.37 years (N = 14, 13 events) in the high-risk group compared with 6.6 years in the low-risk group (N = 21, 8 events, Figure 1B).
Figure 1.
Kaplan-Meier survival curves of NSCLC cases dichotomized using PCA of PAM50 genes. (A) DFS of ERβ-positive subset from cohort 1 (stage I NSCLC) using all 50 PAM50 genes (N = 35). (B) DFS of ERβ-positive subset from cohort 1 (stage I NSCLC) using the seven-gene model (N = 35). (C) PFS of ERβ-positive subset from cohort 2 (all-stage NSCLC) using all 50 PAM50 genes (N = 63). (D) PFS of ERβ-positive subset from cohort 2 (all-stage NSCLC) using the seven-gene model (N = 63).
Pathway Analysis of Informative PAM50 Genes
Using IPA software, we identified networks likely to be involved in the prognostic signatures. The relative expression of the 20 genes retained at least once in PCA cross-validation (Table 2) was evaluated, as well as the seven-gene signature. Using 20 genes, 3 interacting networks were identified that included cancer, reproductive systems disease, and breast cancer proliferation as top functions (Supplemental Table 1). Neuregulin signaling, E2-mediated S phase entry, and HER2/HER3 signaling were among the top 10 canonical pathways. Using the seven-gene signature, only one interacting network was identified (illustrated in Supplemental Figure 1). Cell migration, proliferation of tumor cells, and cancer were the top three functions for this network, and STAT3, neuregulin signaling, and E2-mediated S phase entry were among the top 10 canonical pathways (Supplemental Table 1).
Supplemental Figure 1.
IPA network described by the seven-gene model. Red: Gene is overexpressed in cases with poor survival. Green: Gene is underexpressed in cases with poor survival.
Likely top upstream regulators for the 20-gene signature were related to hormones and the HER2 or HER3 pathway, and for some regulators, an activation score could be computed. These included R5020 (a progestin; activation score of 1.96), trastuzumab (a neutralizing antibody for HER2), HOXB4 (a homeobox transcription factor that regulates MYC and PGR), NRG-1 (a ligand for HER3/HER4; activation score of 1.98), the ER letrozole (an aromatase inhibitor), and β-estradiol (activation score of 2.14). The cell cycle gene cyclin D (CCND1) was also an upstream regulator. In the seven-gene signature, hormonal agents were found as significant upstream regulators: premarin (an estrogen) as well as the selective ER modulator bazedoxifene. Letrozole, the ER, and NRG1 also remained in the model as predicted top upstream regulators of the seven-gene network (Supplemental Table 1). Activation scores could not be predicted in the seven-gene analysis because of the small number of genes analyzed.
The single network described by the seven PAM50 genes (Supplemental Figure 1) contains the ER, PGR, and the ERBB (HER) family of proteins, as well as β-estradiol, AKT, MAPK, ERK1/2, and JNK, suggesting interactions of hormone receptors that involve cytoplasmic signaling with the ERBB/HER receptors. The upstream regulator analysis suggests the HER3 ligand NRG1 is a driver of this network. Nongenomic signaling involving cross talk between β-estradiol and EGFR/HER1 has been described in NSCLC by us and others [23], [24], [25]; the PAM50 model suggests that other HER family members are also important in cross talk with steroid hormones in NSCLC.
Relationship of PAM50 Gene mRNA Expression to Protein Expression
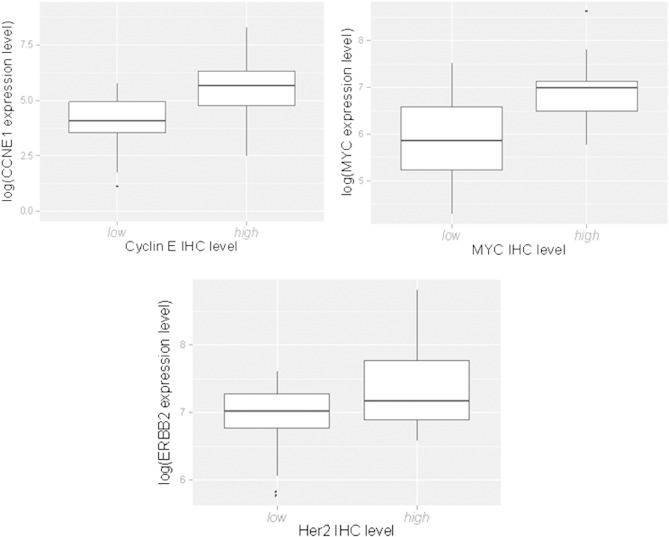
To confirm mRNA expression differences at the protein level, we examined several genes (HER2, CCNE1, and MYC) by carrying out IHC in the same FFPE blocks used for ERβ determination. These proteins were chosen because they contributed to the top differentially expressed genes from Table 2 and because reliable antibodies were available for IHC. Comparing IHC scores to the mRNA level measured by the Illumina Bead Chip array, we observed significant associations between mRNA levels and IHC scores for CCNE1 (P = .001) and MYC (P = .0003, Supplemental Figure 2). For HER2, the mRNA expression level was often very low, and there was a higher mRNA level with IHC scores of 2 (the threshold for an HER2-positive assessment in breast cancer) compared with < 2, but it did not reach statistical significance (Supplemental Figure 2).
Supplemental Figure 2.
log mRNA expression level versus IHC protein expression score in the subset of cohort 1 shown in Table 4 (43 cases) for CCNE1 (P = .001), MYC (P = .0003), and HER2 (P = .15).
Association of ERβ IHC Expression with IHC Expression of PAM50 Genes
We additionally evaluated the relationship between IHC scores for CCNE1, MYC, and HER2 and the status of ERβ protein (ERβ positive, Allred score > 0, compared with ERβ negative, Allred score 0). We observed a significant positive relationship between a detectable level of CCNE1, MYC, and HER2 (IHC scores > 0) and ERβ-positive status. A positive result for staining of CCNE1 was more frequently observed in the ERβ-positive group than the ERβ-negative group (59.0% vs 33.3%, P = .027, Table 4). A similar result was observed for MYC in the ERβ-positive compared with ERβ-negative group (56.1% vs 20.0%, P = .001). For HER2, an IHC score greater than 0 was observed in 70.2% of ERβ-positive cases compared with 34.5% of ERβ-negative cases (P = .002, Table 4). Using the clinical definition of HER positivity (a score of 2 or more), we observed that 35.1% of ERβ-positive cases fell in this group compared with 6.9% of ERβ-negative cases (P = .013, Table 4). Because HER3 was predicted to interact in a network with informative PAM50 genes, we also examined HER3 by IHC, but a relationship with ERβ positive cases was not found. However, when HER2 and HER3 (receptors that often heterodimerize with each other) were considered together, a greater proportion of ERβ-positive cases were both clinically positive for HER2 (score of 2) and positive for HER3 compared with ERβ-negative cases (21.1% vs 3.4%, P = .052). ERβ-positive cases were less likely than ERβ-negative cases to be negative for both HER2 and HER3 (24.6% vs 55.2%, P = .008, Table 4).
Table 4.
Association of IHC Scores for CCNE1, MYC, HER2, and HER3 with ERβ
| ERβ Group |
CCNE1 Positive |
P |
MYC Positive |
P |
HER2 Positive |
P |
HER2 Clinically Positive (+ 2) |
P |
HER3 Positive |
P |
HER2+2 and HER3 Positive (> 0) |
P |
HER2 & HER3 Negative |
P |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | > 0 | 0 | > 0 | 0 | > 0 | < 2 | 2 | 0 | > 0 | No | Yes | No | Yes | ||||||||
| Positive (Allred score > 0) | 25 | 36 59.0% | .027 | 25 | 32 56.1% | .001 | 17 | 40 70.2% | .002 | 37 | 20 35.1% | .004 | 38 | 22 36.7% | .64 | 45 | 12 21.1% | .052 | 43 | 14 24.6% | .008 |
| N = 61 | N = 57 | N = 57 | N = 57 | N = 60 | N = 57 | N = 57 | |||||||||||||||
| Negative (Allred score = 0) | 20 | 10 33.3% | 24 | 6 20.0% | 19 | 10 34.5% | 27 | 2 6.9% | 21 | 9 30.0% | 28 | 1 3.4% | 13 | 16 55.2% | |||||||
| N = 30 | N = 30 | N = 29 | N = 29 | N = 30 | N = 29 | N = 29 | |||||||||||||||
There was also a significant relationship between HER3 staining and HER2 score (P = .026). For a HER2 score of 0, 6 of 37 cases (16.2%) had HER3 scores of > 0. For a HER2 score of 1, 12 of 27 (44.4%) had HER3 scores > 0. For a HER2 score of 2, 13 of 22 cases (59.1%) had HER3 scores > 0.
Performance of PAM50 Panel in All-Stage NSCLC Cohort
To verify the results in cohort 1, we examined PAM50 genes in a separate cohort of 63 NSCLC cases, which included all stages and histologic types (cohort 2, Table 1B). These cases were known to be ERβ positive based on Allred scores > 0 using the ERβ-1-specific antibody, and an example of the staining has been published previously [13]. Compared with cohort 1, cohort 2 had significantly fewer stage I cases (P < .0001) and contained more squamous cell carcinoma (P < .008). Using all 50 PAM50 genes, cohort 2 marginally separated into two survival groups, whereas the seven-gene model was prognostic (HR 2.64 [95% CI: 1.17-5.11], P = .026, Table 3). Kaplan-Meier survival curves using all PAM50 genes showed that in the high-risk group, median PFS was 0.99 years (N = 28, 16 events), whereas in the low-risk group, median PFS was 2.75 years (N = 35, 15 events, Figure 1C). Using the seven-gene model, median PFS was 1.06 years (N = 33, 20 events) in the high-risk group, whereas median PFS was not reached in the low-risk group (N = 30, 11 events, Figure 1D).
Survival Analysis of PAM50 Genes in the TCGA NSCLC Cohorts
In TCGA NSCLC publically available RNASeq V2 RSEM data, the most common alteration reported for PAM50 genes was overexpression (z-score threshold of + 1.5 or higher). Fewer than 4% of TCGA cases had negative z-scores for any of the PAM50 genes. In lung adenocarcinoma, many genes such as MKI67, ANLN, NDC80, and EXO1 showed highly significant correlations (P < .0001 or better) with up to 15 other PAM50 genes, suggesting that these genes are concordantly expressed (data not shown). In lung adenocarcinoma, overexpression of 23 PAM50 genes was independently prognostic for OS by log rank test at P < .05 or better (Supplemental Table 2).
Analysis of TCGA squamous cell carcinoma also showed positive correlations among many PAM50 genes at P < .05. Several genes (MKI67, ANLN, EXO1, and NDC80) showed significant positive correlations with up to 15 other PAM50 genes, suggesting that PAM50 genes are also expressed together in lung squamous cell carcinoma (data not shown). In the squamous cell carcinoma data set, 11 PAM50 genes were independently prognostic for survival at P < .05 or better when overexpressed (Supplemental Table 2).
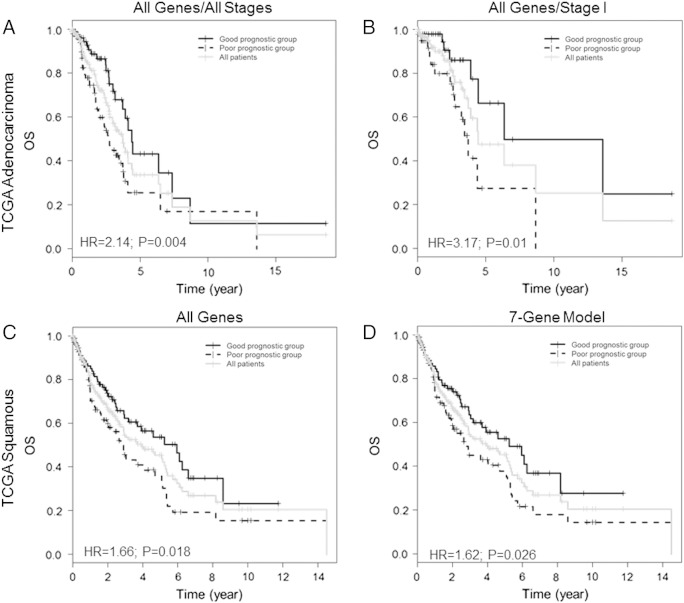
We next examined the PAM50 panel and the seven-gene signature by PCA in the TCGA cohorts using the z-scores as a surrogate for mRNA expression level. There are no data on ERβ IHC scores for these cases, so assessment in an ERβ-positive subgroup could not be done. In adenocarcinoma, the complete PAM50 panel was prognostic for OS (HR 2.14 [95% CI: 1.27-3.61], P = .004, Table 3). Survival curves showed that median OS was 32.7 months for the poor-survival group and 52.5 months for the better-surviving group (Figure 2A). The seven-gene panel was not informative in the TCGA adenocarcinoma cohort (Table 3). Stage I TCGA cases were also analyzed separately. The 50-gene signature was informative (HR 3.17 [95%CI: 1.26-7.97], P = .01), but the 7-gene signature was not. Kaplan-Meier survival curves using all 50 genes (Figure 2B) showed that the poor-surviving stage I group reached median OS at 44.4 months, whereas the better-surviving group reached median OS at 76.2 months. In squamous cell carcinoma, both the all-gene model and the seven-gene model gave significant results in OS associations: HR of 1.66 (95% CI: 1.09-2.54, P = .018) and 1.62 (95% CI: 1.06-2.47, P = .026, Table 3), respectively. Median OS was 32.8 and 71.3 months in the high- and low-risk groups, respectively, by the all-gene model (Figure 2C) and 23.5 and 38.1 months, respectively, in the seven-gene model (Figure 2D).
Figure 2.
Kaplan-Meier survival curves cases in TCGA cohorts dichotomized using PCA of PAM50 genes. (A) OS in the model using all 50 PAM50 genes in all stages of adenocarcinoma (N = 203). (B) OS in the model using all 50 PAM50 genes in stage I adenocarcinoma only (N = 101). (C) OS in the model using all 50 PAM50 genes in all squamous cell carcinoma cases (N = 220). (D) OS using the seven-gene model in all squamous cell carcinoma cases (N = 220).
Discussion
PAM50 genes separate breast cancer into five subtypes with varying biological behavior and outcome [18]. PAM50 genes identify both ERα-positive and -negative breast cancer cases belonging to these subtypes [18], but the panel contains many estrogen-related genes and has proven extremely useful in survival analysis of ERα-positive cases in different therapeutic settings [19], [26], [27]. PAM50 risk scores based on assignment to an intrinsic subtype can predict risk of recurrence and poor survival after endocrine therapy in ERα-positive breast cancer [19], [26]. PAM50 risk scores can also supplement IHC determinations of ER, PGR, and HER2 in breast cancer for better prediction of recurrence [27]. We did not find that PAM50 genes separated NSCLC into different subtypes, and the three genes whose overexpression had the highest relationship to NSCLC survival are markers for three different breast cancer subtypes with different expected outcomes: HER2-enriched (FGFR4), basal-like (FOXC1), and luminal B (CXXC5), suggesting that the breast cancer subtypes are not recapitulated in NSCLC. The prognostic value of PAM50 genes in NSCLC does appear to be related to hormone pathways, however. Performance of PAM50 genes in separating NSCLC by median survival was somewhat better in cases known to be ERβ positive than in unselected cases. ER and PGR pathways were predicted to be involved in interactions among PAM50 genes informative for NSCLC survival. ERβ status was related to protein expression of MYC, CCNE1, and HER2.
Cross-validation of PCA results in stage I NSCLC identified seven genes that most often retained survival associations during the discovery phase. These genes were prognostic in stage I cases unselected for ERβ status and also performed well in the ERβ-positive subset where a two-fold difference in median DFS was observed. In ERβ-positive NSCLC of all stages, the difference in median PFS conferred by these seven genes could not be calculated because the low-risk group did not reach 50% progression.
Pathway analysis suggested that the seven informative PAM50 genes participate in estrogen and HER2/HER3 signaling. HER2 protein was detected more frequently in cases that were ERβ positive. HER2 level was also a factor in the extent of HER3 protein expression. The interactive network involving the seven genes included PGR, β-estradiol, and signaling molecules downstream of HER family proteins. HER2 activation has been reported to phosphorylate cytoplasmic PGR, causing its proteosomal degradation [28]. Low PGR was associated with more aggressive breast cancers that were driven by HER signaling (28), a mechanism that could also be operative in lung cancer. Our data suggest that interactions between hormonal and HER signaling define more aggressive lung tumors.
Many PAM50 genes were individually prognostic in the TCGA data and were expressed concordantly, suggesting that PAM50 genes contain biologically meaningful information in NSCLC. Although no ERβ IHC data are available from these cases, the literature suggests [13], [14], [15] that the majority of TCGA cases would be ERβ positive if analyzed. In these large data sets, the PAM50 model using all genes was prognostic in both data sets, whereas the prognostic ability of the seven-gene signature was not reproduced in TCGA adenocarcinoma data. In squamous cell carcinoma, the 7-gene and 50-gene panels both separated lung tumors into survival groups. The seven-gene signature identified squamous cell carcinoma TCGA patients with a median survival of less than 2 years.
In TCGA data, the proportion of lung adenocarcinomas and squamous cell carcinomas overexpressing genes in the seven-gene signature was similar, as was the proportion of tumors displaying amplified, mutated, or overexpressed ERBB2/HER2, HER3, and NRG1. Thus, the interacting network predicted for the seven-gene signature does not seem to be preferentially dysfunctional in squamous cell carcinoma compared with adenocarcinoma. Although we did not previously find any difference in extent of ERβ between adenocarcinoma and squamous cell carcinoma [13], it is possible the ERβ-HER2/HER3 interaction may be a stronger driver of progression in squamous cell carcinoma than in adenocarcinoma. HER2 has recently been found as a prognostic marker in squamous cell carcinoma of the head and neck [29].
HER2 has no ligand binding domain but heterodimerizes with HER3 after HER3 is activated by ligands such as NRG1. Cross talk between ER and EGFR/HER1 has been reported in both breast and lung cancer [23], [24], occurring through the release of EGFR ligands such as HB-EGF and amphiregulin after being stimulated by estrogen. Because activated HER1 heterodimerizes with HER2 and HER2 heterodimers produce more durable signaling [29], HER2 may be an important contributor in EGFR cross talk with ERβ. Recent reports suggest correlations between ER-positive breast cancer and expression of both HER2 and HER3, whereas cross talk between ER and HER2 is reported to contribute to resistance to endocrine therapies [30]. Our results point to release of NRG1 or other ligands for the HER family of receptors, stimulated through ERβ, in causing HERs to drive NSCLC aggressive behavior. Although ERβ is often thought of as an antiproliferative molecule in breast cancer, it is usually studied in the context of ERα expression [31]. In studies that examined ERα-negative breast cancer expressing ERβ, ERβ was associated with proliferation [31], and ERβ-positive patients lacking ERα responded to tamoxifen [31]. Because most lung tumors have little or no functional ERα [13], ERβ may assume many of the proliferative functions of ERα.
This study provides evidence that genes within the PAM50 signature have prognostic value in lung cancer, especially among squamous cell carcinoma. Strengths of the study include ability to reproduce the results in different NSCLC cohorts and validation of pathway ions at the protein level. Pathways that were delineated by the most prognostic genes also involved biologically plausible interactions that included hormone receptor action. Limitations of the study include use of case cohorts with different stage and histological distributions, need to define survival by different parameters in the different cohorts, and lack of ERβ IHC information in TCGA data. Like most studies that look at DFS and PFS, we assigned time of progression to the visit at which it was detected, which may have biased our results. mRNA profiling data were used in the Pittsburgh cohorts, whereas RNA Seq data were used for TCGA data. These limitations could have contributed to inability to reproduce all findings in the TCGA adenocarcinoma data.
HER2 overexpression warrants further investigation as a target in lung cancer even in the absence of gene amplification. Further study of interactions between HER2 and HER3 proteins and ER signaling in NSCLC, as well as co-targeting the ER and HER pathways in NSCLC, is also warranted. Expression of ERβ and PAM50 genes might also be useful in identifying NSCLC patients most likely to benefit from endocrine therapy in the adjuvant setting.
The following are the supplementary data related to this article
Pathway Analysis of PAM50 Genes
Survival Association of Individual PAM50 Genes in TGCA Adenocarcinoma Data (P < .1)
Survival Association of Individual PAM50 Genes in TGCA Squamous Cell Carcinoma Data (P < .1)
Acknowledgements
Supported by National Institutes of Health grants P50 CA090440 and P30 CA047904, The V Foundation for Cancer Research, and the University of Pittsburgh Medical Center (UPMC) Genomics Initiative. This study used data generated by the TCGA Research Network: http://cancergenome.nih.gov.
Footnotes
Supported by National Institutes of Health grants P50 CA090440 and P30 CA047904, The V Foundation for Cancer Research, and the University of Pittsburgh Medical Center (UPMC) Genomics Initiative.
Conflicts of interest: none.
References
- 1.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM staging groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumors. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 2.Strauss GM, Herndon JE, II, Maddaus MA, Johnstone DW, Johnson EA, Harpole DH, Gillenwater HH, Watson DM, Sugarbaker DJ, Schilsky RL. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non–small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26:5043–5051. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegfried JM. Women and lung cancer: does oestrogen play a role? Lancet Oncol. 2001;2:506–513. doi: 10.1016/S1470-2045(01)00457-0. [DOI] [PubMed] [Google Scholar]
- 4.Stabile LP, Siegfried JM. Estrogen receptor pathways in lung cancer. Curr Oncol Rep. 2004;6:259–267. doi: 10.1007/s11912-004-0033-2. [DOI] [PubMed] [Google Scholar]
- 5.Hershberger PA, Vasquez AC, Kanterewicz B, Land S, Siegfried JM, Nichols M. Regulation of endogenous gene expression in human non–small cell lung cancer cells by estrogen receptor ligands. Cancer Res. 2005;65:1598–1605. doi: 10.1158/0008-5472.CAN-04-2694. [DOI] [PubMed] [Google Scholar]
- 6.Traynor AM, Schiller JH, Stabile LP, Kolesar JM, Eickhoff JC, Dacic S, Hoang T, Dubey S, Marcotte SM, Siegfried JM. Pilot study of gefitinib and fulvestrant in the treatment of post-menopausal women with advanced non–small cell lung cancer. Lung Cancer. 2009;64:51–59. doi: 10.1016/j.lungcan.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siegfried JM, Gubish CT, Rothstein ME, Henry C, Stabile LP. Combining the multitargeted tyrosine kinase inhibitor vandetanib with the antiestrogen fulvestrant enhances its antitumor effect in non–small cell lung cancer. J Thorac Oncol. 2012;7:485–495. doi: 10.1097/JTO.0b013e31824177ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mollerup S, Jorgensen K, Berge G, Haugen A. Expression of estrogen receptors alpha and beta in human lung tissue and cell lines. Lung Cancer. 2002;37:153–159. doi: 10.1016/s0169-5002(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 9.Stabile LP, Davis AL, Gubish CT, Hopkins TM, Luketich JD, Christie N, Finkelstein S, Siegfried JM. Human non–small cell lung tumors and cells derived from normal lung express both estrogen receptor alpha and beta and show biological responses to estrogen. Cancer Res. 2002;62:2141–2150. [PubMed] [Google Scholar]
- 10.Wu CT, Chang YL, Shih JY, Lee YC. The significance of estrogen receptor beta in 301 surgically treated non–small cell lung cancers. J Thorac Cardiovasc Surg. 2005;130:979–986. doi: 10.1016/j.jtcvs.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Marquez-Garban DC, Chen HW, Fishbein MC, Goodglick L, Pietras RJ. Estrogen receptor signaling pathways in human non–small cell lung cancer. Steroids. 2007;72:135–143. doi: 10.1016/j.steroids.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishibashi H, Suzuki T, Suzuki S, Niikawa H, Lu L, Miki Y, Moriya T, Hayashi S, Handa M, Kondo T. Progesterone receptor in non–small cell lung cancer—a potent prognostic factor and possible target for endocrine therapy. Cancer Res. 2005;65:6450–6458. doi: 10.1158/0008-5472.CAN-04-3087. [DOI] [PubMed] [Google Scholar]
- 13.Stabile LP, Dacic S, Land SR, Lenzner DE, Dhir R, Acquafondata M, Landreneau RJ, Grandis JR, Siegfried JM. Combined analysis of estrogen receptor beta-1 and progesterone receptor expression identifies lung cancer patients with poor outcome. Clin Cancer Res. 2011;17:154–164. doi: 10.1158/1078-0432.CCR-10-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz AG, Wenzlaff AS, Prysak GM, Murphy V, Cote ML, Brooks SC, Skafar DF, Lonardo F. Reproductive factors, hormone use, estrogen receptor expression and risk of non small-cell lung cancer in women. J Clin Oncol. 2007;25:5785–5792. doi: 10.1200/JCO.2007.13.3975. [DOI] [PubMed] [Google Scholar]
- 15.Kawai H. Estrogen receptors as the novel therapeutic biomarker in non–small cell lung cancer. World J Clin Oncol. 2014;5:1020–1027. doi: 10.5306/wjco.v5.i5.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mah V, Seligson DB, Li A, Marquez DC, Wistuba II, Elshimali Y, Fishbein MC, Chia D, Pietras RJ, Goodglick L. Aromatase expression predicts survival in women with early-stage non small cell lung cancer. Cancer Res. 2007;67:10484–10490. doi: 10.1158/0008-5472.CAN-07-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang H, Liao Y, Xu L, Zhang C, Liu Z, Deng Y, Jiang Z, Fu S, Chen Z, Zhou S. Estrogen and insulin-like growth factor 1 synergistically promote the development of lung adenocarcinoma in mice. Int J Cancer. 2013;133:2473–2482. doi: 10.1002/ijc.28262. [DOI] [PubMed] [Google Scholar]
- 18.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, Ferree S, Storhoff J, Schaper C, Cuzick J. Comparison of PAM50 risk of recurrence score with oncotype DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol. 2013;31:2783–2790. doi: 10.1200/JCO.2012.46.1558. [DOI] [PubMed] [Google Scholar]
- 20.Detre S, Saclani Jotti G, Dowsett M. A "quickscore" method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Press OA, Guzman R, Cervantes M, Santiago A, Press MF. Characterization of HER2 status by fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) Methods Mol Biol. 2014;1180:181–207. doi: 10.1007/978-1-4939-1050-2_10. [DOI] [PubMed] [Google Scholar]
- 22.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stabile LP, Lyker JS, Gubish CT, Zhang W, Grandis JR, Siegfried JM. Combined targeting of the estrogen receptor and the epidermal growth factor receptor in non–small cell lung cancer shows enhanced antiproliferative effects. Cancer Res. 2005;65:1459–1470. doi: 10.1158/0008-5472.CAN-04-1872. [DOI] [PubMed] [Google Scholar]
- 24.Pietras RJ, Marquez-Garban DC. Membrane-associated estrogen receptor signaling pathways in human cancers. Clin Cancer Res. 2007;13:4672–4676. doi: 10.1158/1078-0432.CCR-07-1373. [DOI] [PubMed] [Google Scholar]
- 25.Hershberger PA, Stabile LP, Kanterewicz B, Rothstein ME, Gubish CT, Land S, Shuai Y, Siegfried JM, Nichols M. Estrogen receptor beta (ERbeta) subtype-specific ligands increase transcription, p44/p42 mitogen activated protein kinase (MAPK) activation and growth in human non–small cell lung cancer cells. J Steroid Biochem Mol Biol. 2009;116:102–109. doi: 10.1016/j.jsbmb.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sestak I, Cuzick J, Dowsett M, Lopez-Knowles E, Filipits M, Dubsky P, Cowens JW, Ferree S, Schaper C, Fesl C. Prediction of late distant recurrence after 5 years of endocrine treatment: a combined analysis of patients from the Austrian breast and colorectal cancer study group 8 and arimidex, tamoxifen alone or in combination randomized trials using the PAM50 risk of recurrence score. J Clin Oncol. 2015;33:916–922. doi: 10.1200/JCO.2014.55.6894. [DOI] [PubMed] [Google Scholar]
- 27.Chia SK, Bramwell VH, Tu D, Shepherd LE, Jiang S, Vickery T, Mardis E, Leung S, Ung K, Pritchard KI. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res. 2012;18:4465–4472. doi: 10.1158/1078-0432.CCR-12-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23:7721–7735. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Pollock NI, Grandis JR. HER2 as a therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res. 2015;21:526–533. doi: 10.1158/1078-0432.CCR-14-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giuliano M, Trivedi MV, Schiff R. Bidirectional crosstalk between the estrogen receptor and human epidermal growth factor receptor 2 signaling pathways in breast cancer: molecular basis and clinical implications. Breast Care. 2013;8:256–262. doi: 10.1159/000354253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skliris GP, Leygue E, Watson PH, Murphy LC. Estrogen receptor alpha negative breast cancer patients: estrogen receptor beta as a therapeutic target. J Steroid Biochem Mol Biol. 2008;109:1–10. doi: 10.1016/j.jsbmb.2007.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pathway Analysis of PAM50 Genes
Survival Association of Individual PAM50 Genes in TGCA Adenocarcinoma Data (P < .1)
Survival Association of Individual PAM50 Genes in TGCA Squamous Cell Carcinoma Data (P < .1)