Abstract.
We generated a highly deuterated cholesterol analog (D38-cholesterol) and demonstrated its use for selective vibrational imaging of cholesterol storage in mammalian cells. D38-cholesterol produces detectable signals in stimulated Raman scattering (SRS) imaging, is rapidly taken up by cells, and is efficiently metabolized by acyl-CoA cholesterol acyltransferase to form cholesteryl esters. Using hyperspectral SRS imaging of D38-cholesterol, we visualized cholesterol storage in lipid droplets. We found that some lipid droplets accumulated preferentially unesterified D38-cholesterol, whereas others stored D38-cholesteryl esters. In steroidogenic cells, D38-cholesteryl esters and triacylglycerols were partitioned into distinct sets of lipid droplets. Thus, hyperspectral SRS imaging of D38-cholesterol demonstrates a heterogeneous incorporation of neutral lipid species, i.e., free cholesterol, cholesteryl esters, and triacylglycerols, between individual lipid droplets in a cell.
Keywords: biophotonics, microscopy, nonlinear optics, Raman spectroscopy
1. Introduction
Cholesterol is a ubiquitous molecule involved in multiple physiological pathways. Altered cholesterol metabolism has a major health impact, manifested in a wide range of illnesses from cardiovascular diseases to neurological disorders.1,2 A deep understanding of cholesterol metabolism is key toward developing efficient and targeted drugs for overcoming cholesterol-related disorders.
Cellular cholesterol storage and transport have been visualized mostly using fluorescence techniques. Measurements based on dehydroergosterol, a fluorescent cholesterol analog, have been helpful in elucidating cholesterol-trafficking mechanisms.3 Other fluorescent labels, including filipin,4 which associates with cholesterol in membranes, and fluorescently tagged cholesterols, such as BODIPY-cholesterol,5 have unveiled information on the recruitment of cholesterol to membranes.6 Nonetheless, the use of fluorescent lipids can produce unwanted artifacts,7–9 including altered lipid metabolism and detachment of the fluorescent label from the sterol.
Raman active labels offer an alternative to fluorescent probes. Unlike fluorescent labels, Raman markers are unaffected by photobleaching effects, enabling sustained imaging of cells. More importantly, the spatial extent of Raman active modes is limited to a single or a few chemical bonds, which is much smaller than the chemical structure of fluorescent moieties. Raman labels can be chosen from the spectral signatures of endogenous compounds or from chemically modified probes that enhance the Raman response and selectivity. The Raman spectrum of endogenous cholesterol exhibits several spectral features that can be utilized as marker bands. This notion has been used to selectively visualize cholesterol crystals in tissues using nonlinear Raman imaging techniques, such as coherent anti-Stokes Raman scattering (CARS)10–12 and stimulated Raman scattering (SRS)13,14 microscopy.
However, noncrystalline pools of cholesterol in cells are often complex mixtures of lipophilic constituents, rendering the intrinsic spectral signatures of the compounds insufficient for uniquely identifying cholesterol in intracellular environments. To improve selectivity, chemical analogs of cholesterol, selected for their unique Raman response, may be used.15 Several Raman labels, such as alkyne tags16,17 that exhibit large Raman cross sections and signatures in regions of the spectra free of endogenous signals, have been shown to be tolerated by live cells, enabling the rapid visualization of selected metabolites, nucleotides, and protein residues in cells with SRS imaging.18,19 Recently, this principle has been extended to imaging cholesterol by substituting part of the molecule’s aliphatic tail with a phenyl-capped diyne moiety.20 The phenyl-diyne cholesterol exhibits a very high Raman cross section, and relative to a terminal alkyne group, phenyl-capping gives rise to reduced cytotoxicity.
Nonetheless, although alkyne moieties are structurally small, they represent a chemical group that is foreign to the intracellular environment. While a Raman probe like phenyl-diyne cholesterol may display limited cytotoxicity, it is unknown how the foreign chemical structure may affect various downstream processes relevant to cholesterol homeostasis. In order to minimize artifacts beyond cytotoxicity, Raman probes that display minimal chemical dissimilarities with natural cholesterol are of interest. In this regard, probes based on the substitution of hydrogen by deuterium are particularly attractive, as the chemical properties of deuterated isotopologs closely resemble those of their endogenous counterparts. Deuterium-based Raman labels have been successfully used in nonlinear optical imaging applications to help increase specificity to water dynamics,21 selected phospholipids,22,23 neutral lipids,24–26 and protein metabolism.27,28
Labels based on stable isotopes, including deuterium, have found widespread use in clinical applications. Because the level of toxicity of pharmacological substances and their deuterated analogs is typically identical, deuterated substances are widely used in clinical studies aimed at studying the metabolism and movement of drugs in humans.29 Similarly, deuterated cholesterol has been clinically administered to humans without harmful side effects, underlining the biocompatibility and usefulness of deuterated cholesterol probes.
In this work, we explore the utility of deuterated cholesterol as a Raman probe. Despite the fact that deuterated cholesterol is commercially available, only a maximum of seven hydrogen atoms are replaced by deuterium (D7-cholesterol). Hence, the retrieved Raman response in the spectral range characteristic of the stretching mode is found to be relatively weak, reducing the effectiveness of the probe. To boost the sensitivity, we have optimized a procedure for producing D38-cholesterol, a probe that features an average of 38 bonds and an improved Raman cross section in the spectral range. With this increase in the degree of deuteration, we show that D38-cholesterol can be detected at physiologically meaningful levels in cells using rapid SRS imaging. Using hyperspectral SRS and multivariate analysis, we demonstrate that D38-cholesterol is readily internalized by cells and is naturally esterified and stored in lipid droplets. In addition, we use D38-cholesterol to study heterogeneity in cholesterol storage in steroidogenic cells.
2. Methods
2.1. D38-Cholesterol
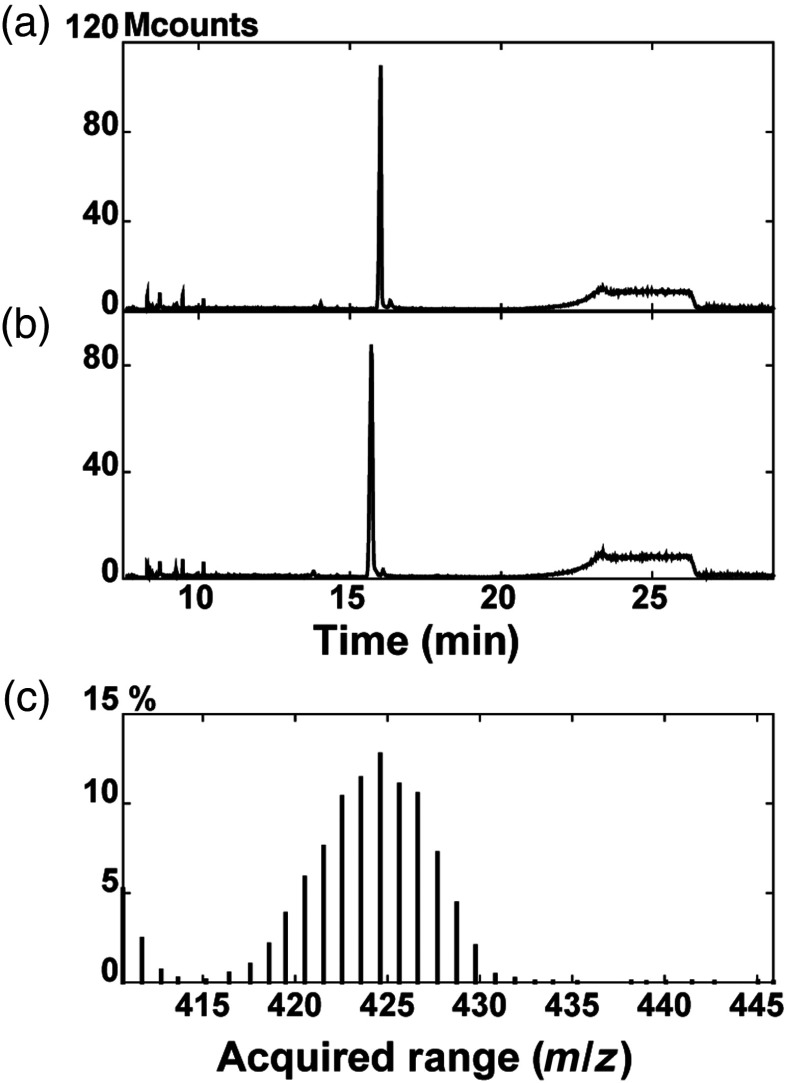
Deuterated cholesterol was produced by a yeast strain (RH6829) engineered to produce cholesterol instead of ergosterol.30 An overview of the cholesterol biosynthetic pathway and principles of biosynthetic isotopic labeling has been published.31 Yeast growth media for isotopic labeling were 0.7% yeast nitrogen base (US Biological), 0.5% yeast extract (BD), 1.25% glucose, and uracil and leucine in deuterium oxide (99.8%, ARMAR, Switzerland). Precultures (1 mL) in this medium were used to inoculate 2 L of media, which were grown to a stationary phase with shaking (3 to 4 days) at 30°C. The deuterated cholesterol was purified from harvested cells and analyzed by gas chromatography-mass spectrometry (GC-MS) as described.30 The yield was approximately 10 mg purified deuterated cholesterol per liter of cell culture. Analysis of the GC-MS profile of the deuterated cholesterol showed a GC profile [Fig. 1(b)] identical to commercial cholesterol [Fig. 1(a)], except that the retention time of deuterated cholesterol was slightly earlier than cholesterol. Examination of the high range of intact ions under the peak [Fig. 1(c)] showed an average of 424 denoting an average substitution of 38 hydrogen atoms by deuterium. Analysis of the deuterated cholesterol by NMR confirmed its purity and showed that all positions were substituted between 70% and 90%, consistent with the average of 82% substitution calculated from the MS profile. D7-cholesterol was obtained from Sigma-Aldrich and used without further purification.
Fig. 1.
GC-MS analysis of D38-cholesterol produced from yeast. Commercial cholesterol (a) and deuterated cholesterol (b) purified from yeast were analyzed by GC-MS and the GC profiles are shown. The higher masses, representing unfragmented deuterated cholesterol (peak in b), are shown in (c), with an average around 424.6, denoting an average replacement of 38 H by D.
2.2. Reagents
Cell culture reagents were obtained from Life Technologies or Lonza. Methyl--cyclodextrin, oleic acid, fatty-acid-free bovine serum albumin (BSA), and cholesterol were purchased from Sigma. D38-cholesterol or cholesterol/methyl--cyclodextrin stock complexes were prepared as described previously,32 using a methyl--cyclodextrin to cholesterol (D38-cholesterol) ratio of 6.2:1 and a final concentration of 45 to 50 mM for D38-cholesterol. Lipoprotein-deprived serum (LPDS) was prepared from fetal bovine serum by potassium bromide density ultracentrifugation.33 Oleic acid was complexed to fatty-acid-free BSA in an 8:1 ratio.34
2.3. Cell Culture and Lipid Administrations
Y1 cells were cultured in Dulbecco’s modified eagle’s medium (DMEM)/F12 (1:1) with 15% horse serum and 2.5% fetal bovine serum. Culture medium was supplemented with penicillin/streptomycin ( each) and l-glutamine (2 mM). For lipid loadings, Y1 cells were seeded in culture medium for 24 h. Cells were washed three times with phosphate buffered saline (PBS) and incubated with DMEM/F12 medium containing 5% LPDS and cholesterol/cyclodextrin, 45- to D38-cholesterol/cyclodextrin, oleic acid/BSA, or oleic acid/BSA plus D38-cholesterol/cyclodextrin for 24 h. Control cells were treated with DMEM/F12 medium containing 5% LPDS for 24 h.
2.4. Lipid Extraction and Quantifications
Lipid extraction and quantifications were performed as described previously.35 Lipids were extracted using chloroform:methanol in a 1:1 ratio. Solvents were evaporated under nitrogen, and dried lipids were dissolved in chloroform:methanol (2:1 ratio). Lipid solutions were spotted on thin layer chromatography (TLC) plates, and cholesteryl ester, triacylglycerols, and free cholesterol were resolved by phase separation using hexane:diethylether:acetic acid (80:20:1 ratio). Cholesteryl ester, triacylglycerols, and free cholesterol were quantified using ImageJ and normalized to protein content, measured by Bio-Rad protein determination.
2.5. Sample Preparation for Stimulated Raman Scattering Microscopy
Cells grown on No. 1.5 borosilicate coverslips were washed with PBS and fixed with 4% paraformaldehyde for 15 min. Coverslips were rinsed several times with PBS, mounted in PBS, and sealed with epoxy glue to prevent cells from drying.
2.6. Hyperspectral Imaging with Stimulated Raman Scattering Microscopy
SRS signals were obtained by combining two laser beams: a Stokes beam fixed at 1064 nm () and a pump beam tuned to the wavelength of interest (Table 1). The system consists of an optical parametric oscillator (OPO; Levante, Emerald OPO, Berlin, Germany) pumped by a 76-MHz mode-locked Nd:Vanadate laser (Picotrain, High-Q, Hohenems, Austria) that delivers a fundamental beam at 1064 nm (Stokes beam) with 7-ps pulses and a second harmonic generated beam at 532 nm. The latter pumps the OPO that generates the pump beam for the SRS process. The tuning of the pump beam was possible by adjusting the crystal temperature, the Lyot filter, and the cavity length of the OPO. The two beams were overlapped both temporally and spatially and sent into a laser scanner (Fluoview 300, Olympus, Center Valley, Pennsylvania), attached to an inverted microscope (IX71, Olympus). The combined beams were then focused through a , 0.75 NA objective lens (UplanS Apo, Olympus) onto the sample. SRS images were obtained by detecting the stimulated Raman loss of the pump beam. The Stokes beam was modulated at 10 MHz with an acousto-optic modulator (AOM; Crystal Technology, Palo Alto, California), and the pump intensity modulation was detected by a photodiode (FDS1010; Thorlabs, Newton, New Jersey). The signal at 10 MHz was demodulated with a home-built lock-in amplifier. The average combined power of Stokes and pump beams at the specimen was kept under 50 mW throughout this study to minimize sample photodamage. Additional spontaneous Raman spectra of pure cholesterol were acquired with a commercial Raman microscope (InVia Confocal; Renishaw, Wotton-under-Edge, Gloucestershire, United Kingdom).
Table 1.
Pump wavelengths corresponding to the Raman shifts of interest for SRS hyperspectral interrogation at the and stretching ranges.
| pump (nm) | Raman shift () | |
|---|---|---|
| CH range | 820 to 803 | 2796.6 to 3054.8 |
| CD range | 880 to 855 | 1965.1 to 2297.4 |
2.7. Spectral Analysis with Vertex Component Analysis
Hyperspectral coherent Raman scattering imaging has been previously used to acquire simultaneous chemical and spatial information of biological samples.13,24,36 In combination with multivariate analysis, we can extract the spectral information from the hyperspectral stacks. In this case, we chose vertex component analysis (VCA) to retrieve the most prominent spectral features in the hyperspectral-SRS stack.37,38 Briefly, the VCA algorithm identifies the main spectral components in the image that are identified as end members. We use three end members that define the vertices of a polygon (a triangle in this case), and each one is assigned a base color (red, green, and blue—RGB, in this case). Each pixel of the hyperspectral stack (each spectrum in the image) is then defined as a linear combination of the vertex spectra. The result can be visualized in an RGB color map in which the colors denote the spectral class of each pixel.
3. Results
3.1. Characterization of D38-Cholesterol
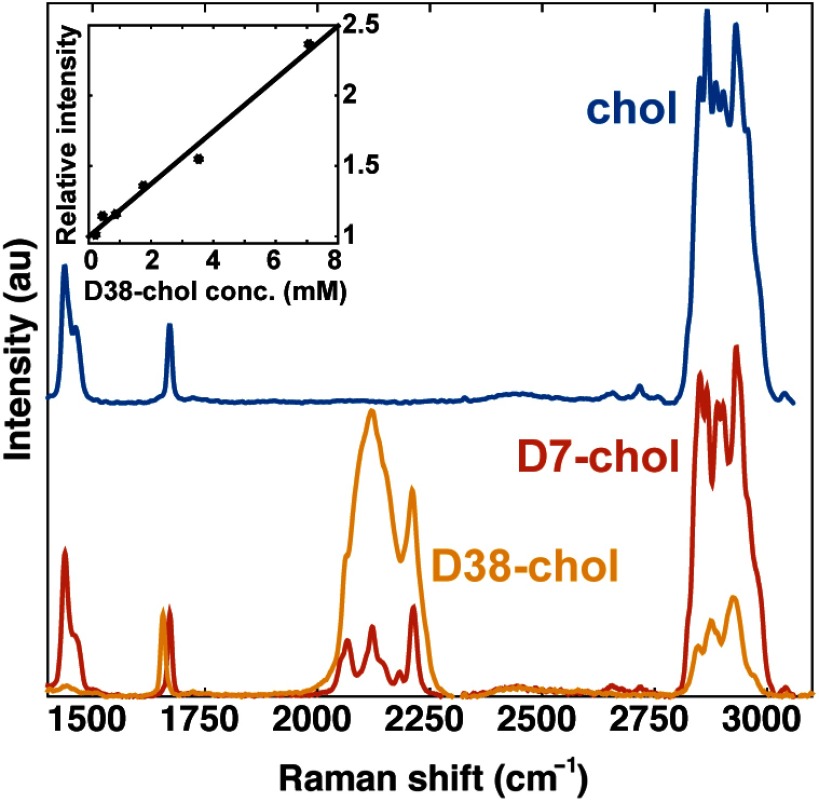
Deuterated lipids increase specificity for Raman, SRS, and CARS imaging due to their specific vibrational frequencies from 2000 to , situated in a silent region of the Raman spectrum and free of vibrational modes from endogenous compounds. The blue line in Fig. 2 shows the Raman spectrum of cholesterol. The strongest peaks in the spectrum are found at 2700 to ( stretching modes), with additional peaks at ( stretching modes) and ( scissoring mode). The Raman spectrum of commercially available D7-cholesterol, which has seven hydrogen atoms replaced by deuterium, is shown in red in Fig. 2. The frequency of the CD modes is shifted relative to the CH stretching band, giving rise to a Raman band in an otherwise silent region of the Raman spectrum from 2000 to . However, because only 7 out of 46 possible positions are substituted with deuterium, the CD spectral features of D7-cholesterol are relatively weak, while the CH region remains the dominant contribution to the spectrum. The yellow line in Fig. 2 corresponds to the Raman spectrum of D38-cholesterol. By substituting 38 sites with deuterium (82.6% deuteration), the response in the 2000 to range is significantly improved, whereas the magnitude of the CH stretching bands is considerably reduced. D38-cholesterol shows two main peaks in the CD band, centered at 2120 and at . This heavily deuterated cholesterol also exhibits a shift of the stretching band to , relative to natural cholesterol. A similar shift has previously been reported for the deuterated fatty acid D6-arachidonic acid.26 Note that besides a significant reduction of the CH band, the spectral profile of the CH stretches of D38-cholesterol is markedly different from the spectral bandshape of natural cholesterol in this range. The contribution of the symmetric stretching mode at in D38-cholesterol is relatively weak, whereas the strongest contribution in this range is found at . This characteristic bandshape provides an additional handle to identify D38-cholesterol and, as shown in Sec. 3.3, can be utilized to discriminate lipid droplets with predominantly esterified cholesterol from lipid droplets with a high concentration of unesterified cholesterol.
Fig. 2.
Spectral properties of cholesterol, D38-cholesterol, and D7-cholesterol. Raman spectra of cholesterol (blue), D38-cholesterol (yellow), and D7-cholesterol (red) are shown in the range from 1400 to . Inset: the SRS intensity of D38-cholesterol in cyclohexane relative to the noise floor increases linearly with the concentration of solute. The detection limit was found to be .
The sensitivity of the SRS microscope to D38-cholesterol is depicted in the inset of Fig. 2. Here, the SRS signal is measured relative to the noise floor of the experiment, as determined by the photothermal signal of glass. At , we find a detection limit of D38-cholesterol in cyclohexane at a pixel dwell time of , when the SRS signal of D38-cholesterol approaches the noise floor. As expected, there is a linear dependence of the SRS signal on D38-cholesterol concentration.39
3.2. D38-Cholesterol: Uptake, Metabolic Processing, and Storage in Lipid Droplets
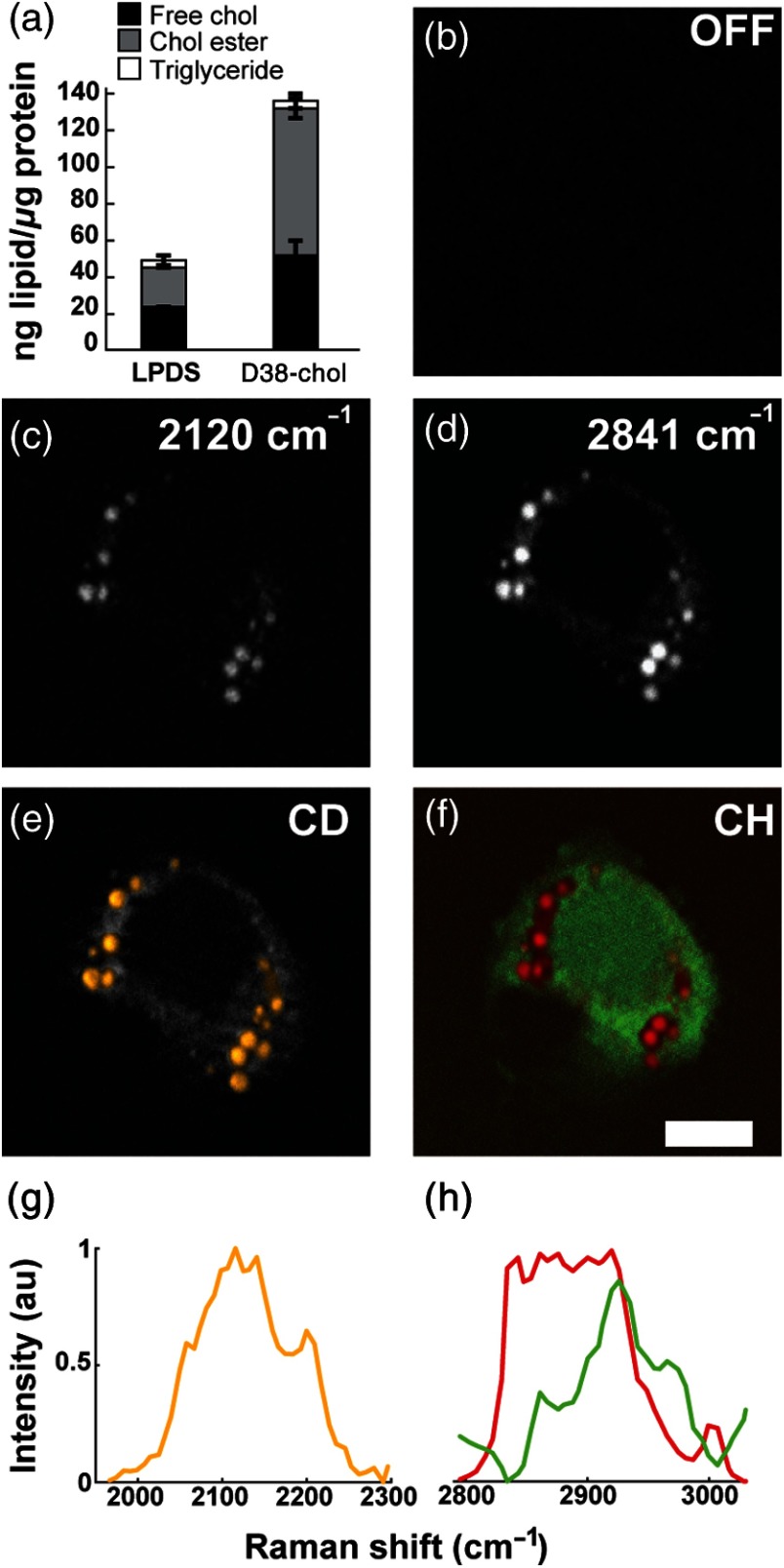
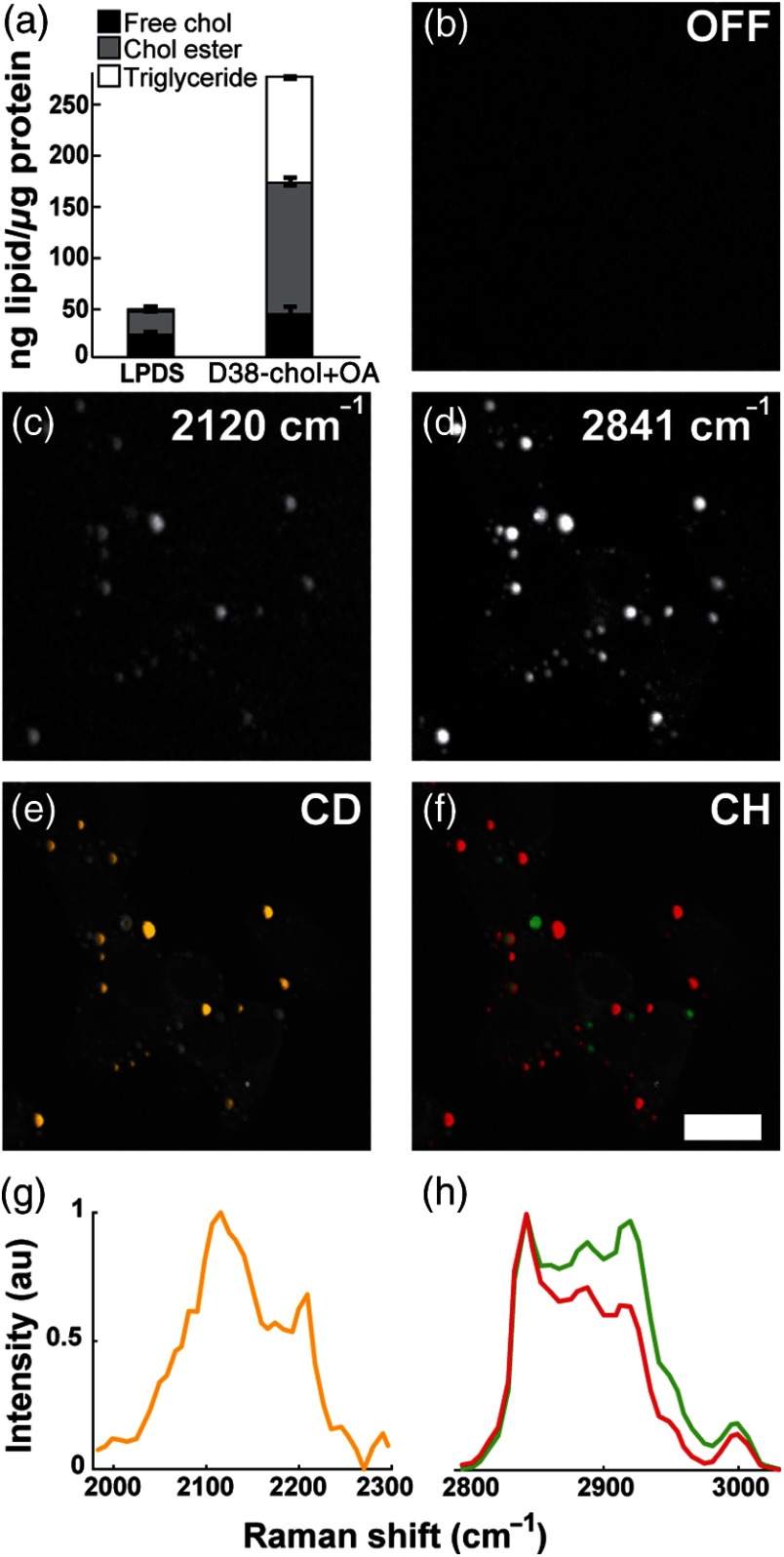
To examine the biocompatibility of D38-cholesterol in mammalian cells, we treated Y1 adrenal cells with D38-cholesterol/cyclodextrin for 24 h. Cyclodextrin forms a complex with cholesterol and transfers it to cells via the plasma membrane.40 This treatment results in efficient cellular uptake of D38-cholesterol with a 2.9-fold increase in total cholesterol and 3.5-fold for cholesteryl esters [Fig. 3(a)] compared to the LPDS control. Using TLC, we determined that approximately 58% of the sequestered D38-cholesterol was esterified [Fig. 3(a)], confirming enzymatic processing of D38-cholesterol and highlighting its superiority to fluorescent sterol analogs in regard to metabolic processing.5,20 Importantly, we find that the levels of esterified D38-cholesterol are similar to the levels of esterified natural cholesterol under similar treatment conditions (Fig. 6), indicating that the deuterated probe does not affect acylCoA:cholesterol acyltransferase (ACAT)-mediated esterification. Next, we performed SRS imaging of D38-cholesterol-loaded cells. Figures 3(b)–3(d) show a cell treated with D38-cholesterol imaged at three different frequencies: (off resonance), ( stretches), and ( symmetric stretches), respectively. The strongest signal at derives from droplet-like structures, as does the more intense signal at , indicative of intracellular lipid droplets. This observation provides evidence that D38-cholesterol has been processed by the cell and is stored in lipid droplets.
Fig. 3.
Cellular imaging of D38-cholesterol uptake, esterification, and storage inside lipid droplets. Y1 adrenal cells were treated with D38-cholesterol/cyclodextrin for 24 h, and cells were used for lipid quantifications (a) or fixed and subjected to SRS imaging (b) , off resonance; (c) , stretches; and (d) , symmetric stretches). The result of hyperspectral SRS imaging and multivariate analysis is depicted in yellow for D38-cholesterol in the CD region (e and g), overlaid on the maximum intensity projection of the CH spectral scan in gray scale, and in red for lipid and green for protein in the CH region (f and h). Normalized spectra. .
To further prove the presence of D38-cholesterol in intracellular lipid droplets, we show the result of a hyperspectral SRS multivariate analysis (VCA) in both the CD and CH spectral regions. The CD range spanned from 1965 to , with a step size of , whereas the CH region ranges from 2797 to , with a step size of . The yellow areas in Fig. 3(e), superimposed on the maximum intensity projection of the spectral scan in the CH stretching region (in gray), exhibit the spectrum depicted in Fig. 3(g). These locations indicate the presence of D38-cholesterol and spatially correlate with the lipid droplet areas as seen in the CH images, confirming that D38-cholesterol is stored in lipid droplets. A corresponding analysis in the CH range shows that the same droplets also contain nondeuterated lipids, whose spectral profile is depicted in red [Figs. 3(f) and 3(h)]. These lipids largely represent fatty acids esterified to D38-cholesterol, some pre-existing nondeuterated cholesterol, and a small fraction of triglycerides (5.1% of total neutral lipids), as confirmed by TLC. The green areas in Figs. 3(f) and 3(h) correspond to the protein-rich content of the cell, which is shown here for context. Hence, these combined results show that D38-cholesterol is processed by the cells, is esterified to a similar degree as natural cholesterol, and is stored in lipid droplets.
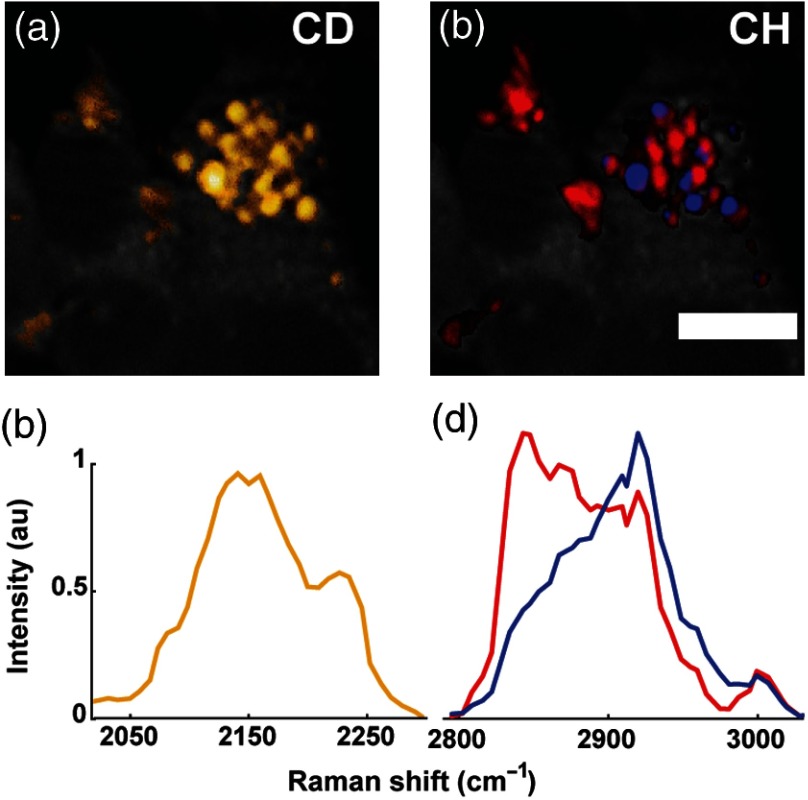
3.3. Hyperspectral Stimulated Raman Scattering Imaging of D38-Cholesterol Identifies Subpopulations of Esterified and Unesterified Cholesterols
Besides identifying cholesterol in lipid droplets, the unique Raman spectrum of D38-cholesterol can be utilized to reveal additional information about the degree of cholesterol esterification. To access this information, we used hyperspectral-SRS imaging combined with VCA. Figure 4 shows a cell that contains two subsets of lipid droplets with different spectral profiles. The CD range reveals droplets that contain D38-cholesterol [Figs. 4(a) and 4(c)]. A subset of these lipid droplets, depicted in blue, exhibit a CH spectrum that resembles the Raman spectrum of unesterified D38-cholesterol. In red, we show the subset of droplets that are represented by a spectrum indicative of both esterified D38-cholesterol and nondeuterated lipids [Figs. 4(b) and 4(d)]. These droplets most likely consist of fatty acids esterified to cholesterol and a small amount of triglycerides and/or pre-existing nondeuterated cholesterol.
Fig. 4.
D38-cholesterol allows visualization of a subset of lipid droplets enriched with free cholesterol. Y1 adrenal cells were treated with D38-cholesterol/cyclodextrin as in Fig. 3. The result of hyperspectral SRS imaging and multivariate analysis is depicted here in yellow for D38-cholesterol in the CD region (a and c), and in red for esterified cholesterol and blue for free D38-cholesterol in the CH region (b and d). Normalized spectra. .
The CD range of the spectrum contains limited information about the state of cholesterol esterification. However, the spectral information acquired in the CH range helps to discriminate between esterified and free cholesterol accumulations. By combining CD imaging with hyperspectral analysis of the CH region, we thus demonstrate that lipid droplet heterogeneity may not only occur at the level of cholesterol ester and triglyceride partitioning, but also at the level of cholesterol esterification. Some lipid droplets show higher enrichment in free cholesterol than others.
3.4. Distinct Cholesteryl Ester and Triacylglycerol Containing Lipid Droplets Visualized by D38-Cholesterol and Oleic Acid Administration
Studies based on fluorescent tracers have suggested that steroidogenic cells can store triglyceride and cholesterol esters in distinct lipid droplets resulting in lipid droplet heterogeneity.32 However, this might result from the altered properties of BODIPY-labeled lipid analogs as compared to natural lipids.24 Here, we demonstrate lipid droplet heterogeneity without fluorescent lipid analogs by means of SRS microscopy and D38-cholesterol.
Y1 cells were treated with D38-cholesterol and oleic acid for 24 h. TLC analysis shows that this treatment results in a 5.8-fold increase in the accumulation of cholesteryl esters ( protein) and a 38-fold increase in the level of triglycerides ( protein) [Fig. 5(a)] with respect to the LPDS-treated cells. Cells subjected to simultaneous D38-cholesterol and oleic acid treatment are depicted in the SRS images shown in Figs. 5(b)–5(f). Lipid droplets containing D38-cholesterol are visualized at the Raman shift in Fig. 5(c). The overall population of lipid droplets is visualized by tuning to the Raman shift of , shown in Fig. 5(d), which marks lipid droplets irrespective of the presence of D38-cholesterol. Comparing the 2120 and images makes it clear that the concentration of D38-cholesterol in the available droplets varies significantly. This is made clearer in the hyperspectral image in Fig. 5(e), which highlights the D38-cholesterol containing lipid droplets in yellow [see the corresponding spectra in Fig. 5(g)]. Some lipid droplets, irrespective of size, contain D38-cholesterol at appreciable levels [yellow droplets in Fig. 5(e)], whereas others appear devoid of D38-cholesterol [gray droplets in Fig. 5(e)]. The hyperspectral image obtained in the CH stretching range, shown in Fig. 5(f), reveals two subsets of droplets with different spectral profiles [Fig. 5(h)]. Most of the droplets depicted in red also contain D38-cholesterol, while the droplets that appear in green in the CH range exhibit little to no CD signal. A third subset contains a mixture of both green and red spectra. The hyperspectral information in both Figs. 5(e) (in yellow) and 5(f) (in red and green) is overlaid on the maximum intensity projection of the hyperspectral scan in the CH stretching region (in gray) to demarcate the cells. These results show that intracellular lipid droplets are heterogeneous in terms of chemical composition and that cholesterol partitions inhomogenously among the available lipid reservoirs.
Fig. 5.
Visualization of lipid droplet heterogeneity in steroidogenic cells using D38-cholesterol. Y1 adrenal cells were treated with D38-cholesterol/cyclodextrin together with oleic acid for 24 h. Cells were either used for lipid quantification (a) or subjected to SRS imaging [(b) , off resonance; (c) , CD2 stretches; and (d) , symmetric stretches]. (Note that LPDS control treatment is the same as in Fig. 3.) The result of hyperspectral SRS imaging and multivariate analysis is depicted in yellow for D38-cholesterol in the CD region (e and g), and in red and green for the lipids in the CH region (f and h). The results are overlaid on the maximum intensity projection of the CH spectral range, shown in gray scale, which outlines the cellular morphology. Normalized spectra. .
4. Discussion
The ability to identify cholesterol and follow its movement in cells is important for understanding changes in cellular cholesterol levels and distribution and its implications in cholesterol-related diseases. The availability of biocompatible probes that minimally interfere with the cell’s natural physiology is a critical component in studying cholesterol homeostasis. Given that fluorescent analogs to cholesterol have been shown to alter key aspects of cholesterol metabolism, most notably the level of ACAT-mediated esterification,5,20 the search for alternative probes has become a relevant research focus. In this regard, although signal levels from Raman labels are generally lower compared to those from fluorescent labels, they offer tangible benefits over their fluorescent counterparts, as Raman labels are insensitive to photobleaching and can be engineered so as to improve biocompatibility while reducing cytotoxicity. Recent successes include the development of phenyl-diyne cholesterol,20 which exhibits an exceptional Raman cross section and was shown to be well tolerated by cells.
Compared to phenyl-diyne cholesterol, the Raman cross section of deuterated cholesterol is weaker. However, deuterated labels remain attractive because they generally exhibit excellent biocompatibility and very low cytotoxicity levels, as underlined by the widespread use of deuterated probes in clinical studies. It is known that deuterated cholesterol is nontoxic to humans,41,42 rendering deuterated cholesterol also a reliable Raman probe for in-depth studies at the cellular and small-organism level. To improve the Raman cross section of deuterated cholesterol, we produced D38-cholesterol using engineered yeast cells, which boost the Raman signal intensity at about five times relative to commercially available deuterated cholesterol. With this improvement, the SRS detection sensitivity of D38-cholesterol is at a pixel dwell time, which allows its detection at physiologically relevant concentrations in the cell. Our production procedure allows for cheap, batch-produced D38-cholesterol in large quantities, which further accentuates the practical utility of this probe.
We have confirmed the biocompatibility of D38-cholesterol. We find that internalized D38-cholesterol is esterified by Y1 cells and stored in lipid droplets. Unlike BODIPY-cholesterol, D38-cholesterol does not appear to alter the ACAT-mediated esterification process. Concentration levels of cholesteryl esters stored in lipid droplets are well within SRS detection levels of D38-cholesterol under all conditions studied here. Therefore, using D38-cholesterol as an SRS probe offers an opportunity to study details of cellular cholesterol-storage processes that have so far remained under-illuminated.
Besides using D38-cholesterol’s biocompatibility and its detectability in SRS imaging, we have extracted additional information about cholesterol storage through examining the spectral SRS content in both the vibrational range of the CD- and CH-stretching modes. In particular, spectral variations in the CH stretching range can be used to distinguish distributions of esterified D38-cholesterol from free D38-cholesterol. In this capacity, we have used hyperspectral SRS imaging of D38-cholesterol to highlight unanticipated heterogeneity in the chemical composition of stored neutral lipids between individual lipid droplets. First, hyperspectral SRS imaging of D38-cholesterol in Y1 adrenal cells revealed the presence of lipid droplets that contained a high concentration of unesterified cholesterol adjacent to lipid droplets containing predominantly esterified cholesterol. Moreover, in these cells, cholesterol and triacylglycerols were deposited in distinct lipid droplets. Note that such heterogeneous distribution of neutral lipids was not detectable by hyperspectral SRS imaging in the absence of deuterated cholesterol.25
Note also that it is not straightforward to discriminate subpopulations of lipid droplets based on the degree of cholesterol esterification by using Raman spectroscopy without the use of the D38-cholesterol probe. The characteristic stretching mode of ester groups at , which serves as a marker for cholesteryl esters,43 is not only weak, but also overlaps spectrally with ester group vibrations of triacylglycerols. Given that triacylglycerols are ubiquitous in lipid droplets, the marker band cannot be readily used as a unique probe for cholesteryl esters. Similarly, in the CH stretching range, the spectral signatures of free cholesterol are insufficiently different from esterified cholesterol to unambiguously discriminate subpopulations of lipid droplets with markedly different levels of esterification. The D38-cholesterol probe, on the other hand, exhibits a unique and characteristic bandshape in the CH stretching range, which enabled the identification of lipid droplet heterogeneity in terms of cholesterol esterification by using hyperspectral SRS imaging.
The nonuniform distribution of lipid-droplet-associated proteins between individual droplets is well appreciated.44 However, the plausible heterogeneity in the distribution of the stored lipids has so far received little attention. This has largely been due to the lack of reliable imaging methods, with the SRS imaging of D38-cholesterol now providing a novel approach. The accumulation of cholesteryl esters in lipid droplets is thought to act as a buffering mechanism to reduce the toxicity of excess free cholesterol in cellular membranes.45 Here, we observed, in addition to cholesteryl esters, free cholesterol deposition in a subset of lipid droplets. It is conceivable that this acts as an additional cholesterol detoxification mechanism, for instance, during atherogenesis when macrophages are challenged with large amounts of cholesterol.
We also report that cells specialized in steroid hormone production from cholesterol store triacylglycerols and cholesteryl esters in different lipid droplets. Whether such lipid sorting occurs in other cell types should be addressed in future studies. Overall, lipid droplet heterogeneity is a highly interesting phenomenon and has important implications for the mechanisms of lipid droplet formation and utilization as well as the biophysical properties of lipid droplets. For instance, we have recently observed that cholesteryl ester enrichment of lipid droplets increases their ordering.35 Furthermore, lipid droplet heterogeneity necessitates the segregation of regulatory proteins, as shown for lipid droplet coat proteins.32 This concept should also apply for the enzymes involved in neutral lipid metabolism.
5. Conclusion
In this work, we have generated D38-cholesterol and shown its utility for SRS imaging studies of cholesterol storage in cells. The main advancements of this work are as follows: (1) A procedure for the efficient and affordable production of D38-cholesterol; (2) Demonstration of the cellular uptake and processing of D38-cholesterol with efficiencies that are similar to those of natural cholesterol; (3) The use of hyperspectral SRS to visualize D38-cholesterol while also providing insight into lipid metabolism through bandshape analysis in the CH-stretching range; and (4) The application of D38-cholesterol as a probe for heterogeneity in cholesterol storage among the pool of lipid droplets. We expect that the availability of D38-cholesterol will lead to new discoveries of cholesterol movement and distribution in cells, as exemplified here for the case of lipid droplet heterogeneity.
Acknowledgments
EOP thanks NIH grant P41-RR01192 (Laser Microbeam and Medical Program, LAMMP) for support. EI thanks the Academy of Finland (Grants 272130, 282192, and 284667) and Sigrid Juselius Foundation for support. SP thanks the Academy of Finland (Grant 275964) and Paulo Foundation for support. HR thanks the Swiss National Science Foundation, the NCCR Chemical Biology, and Damien Jeannerat (Uni Geneva) for NMR analysis.
Biographies
Alba Alfonso-García obtained a BS in physics from University of Barcelona, Spain, and a MSc in optics and photonics from the Karlsruhe Institute of Technology, Germany. She is currently a PhD student in the biomedical engineering program at the University of California, Irvine. She is interested in applying nonlinear optical techniques to image biological processes, with a particular focus on cholesterol metabolism.
Simon G. Pfisterer is an Academy of Finland postdoctoral researcher in the group of Prof. Elina Ikonen at University of Helsinki, Department of Anatomy, Helsinki, Finland. His studies are focused on how lipids are transported inside mammalian cells and stored in lipid droplets.
Howard Riezman is full professor at the University of Geneva and Director of the Swiss National Center of Competence in Research in Chemical Biology. He is well known for the discovery of roles of actin, receptor ubiquitination, and sphingolipids in membrane trafficking. His current research focuses on the metabolism and function of lipids in yeast, worm and animal models using a variety of techniques including biochemistry, genetics, metabolic engineering, mass spectrometry, modeling, and chemical biology.
Elina Ikonen is an Academy of Finland Professor at the Department of Anatomy, University of Helsinki, Finland. She is the director of the Finnish Centre of Excellence in Biomembrane Research (ProLipids) and of the Helsinki Functional Imaging Center. Her research is focused on intracellular cholesterol transport, metabolism and how changes in the cellular cholesterol balance lead to human diseases.
Eric O. Potma is an associate professor in the Department of Chemistry at the University of California, Irvine (UCI). He holds an adjunct position in the Beckman Laser Institute and Medical Clinic at UCI. His research group is active in developing nonlinear optical imaging techniques for the purpose of interrogating biological tissues and nanostructured materials.
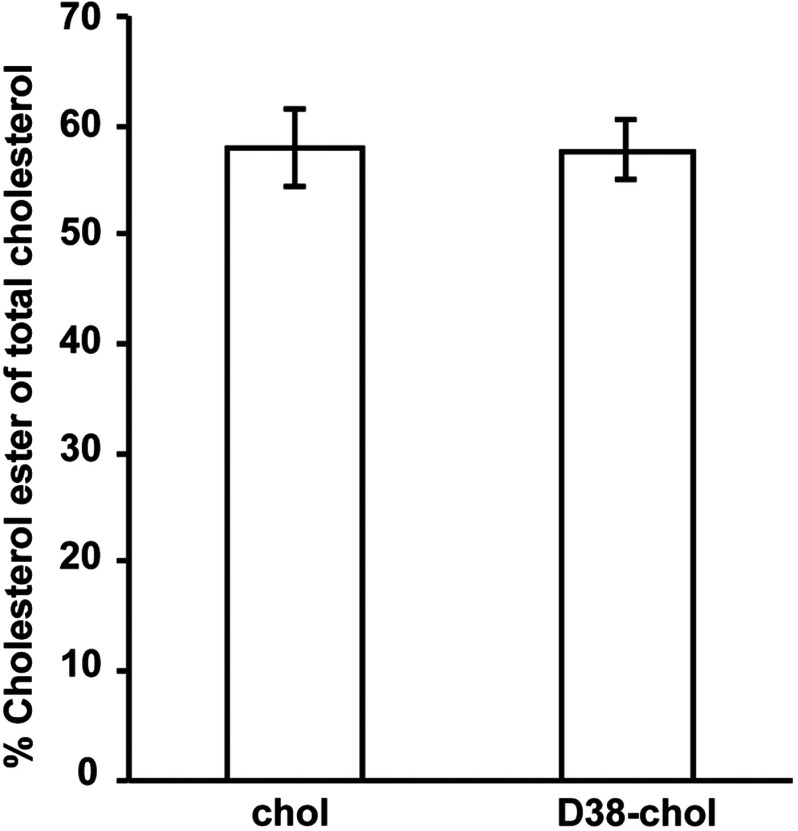
Appendix: Comparison of the Esterification Levels Between Natural Cholesterol and D38-Cholesterol
To compare the levels of esterification between natural cholesterol and D38-cholesterol, Y1 cells were loaded for 24 h with sterol/cyclodextrin complex, and the content of free and esterified cholesterol was analyzed using TLC. The TLC results are shown in Fig. 6. We find that the levels of esterified D38-cholesterol are identical to the levels of esterified natural cholesterol under a similar treatment.
Fig. 6.
Esterification levels of D38-cholesterol match those of natural cholesterol.
References
- 1.Ikonen E., “Mechanisms for cellular cholesterol transport: defects and human disease,” Physiol. Rev. 86(4), 1237–1261 (2006). 10.1152/physrev.00022.2005 [DOI] [PubMed] [Google Scholar]
- 2.Simons K., Ehehalt R., “Cholesterol, lipid rafts, and disease,” J. Clin. Invest. 110(5), 597–603 (2002). 10.1172/JCI0216390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee S., et al. , “Cholesterol distribution in living cells: fluorescence imaging using dehydroergosterol as a fluorescent cholesterol analog,” Biophys. J. 75(4), 1915–1925 (1998). 10.1016/S0006-3495(98)77632-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smejkal G. B., Hoppe G., Hoff H. F., “Filipin as a fluorescent probe of lipoprotein-derived sterols on thin-layer chromatograms,” Anal. Biochem. 239(1), 115–117 (1996). 10.1006/abio.1996.0301 [DOI] [PubMed] [Google Scholar]
- 5.Hölttä-Vuori M., et al. , “BODIPY-cholesterol: a new tool to visualize sterol trafficking in living cells and organisms,” Traffic 9(11), 1839–1849 (2008). 10.1111/j.1600-0854.2008.00801.x [DOI] [PubMed] [Google Scholar]
- 6.Maxfield F. R., Wüstner D., “Analysis of cholesterol trafficking with fluorescent probes,” Chapter 17 in Methods in Cell Biology, Di Paolo G., Wenk M. R., Eds., Vol. 108, pp. 367–393, Academic Press, Amsterdam: (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimpl G., Gehrig-Bruger K., “Cholesterol reporter molecules,” Biosci. Rep. 27, 335–358 (2007). 10.1007/s10540-007-9060-1 [DOI] [PubMed] [Google Scholar]
- 8.Fukumoto S., Fujimoto T., “Deformation of lipid droplets in fixed samples,” Histochem. Cell Biol. 118(5), 423–428 (2002). 10.1007/s00418-002-0462-7 [DOI] [PubMed] [Google Scholar]
- 9.Kinkel A., et al. , “Oil red-O stains non-adipogenic cells: a precautionary note,” Cytotechnology 46(1), 49–56 (2004). 10.1007/s10616-004-3903-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S. H., et al. , “Multiplex coherent anti-stokes Raman spectroscopy images intact atheromatous lesions and concomitantly identifies distinct chemical profiles of atherosclerotic lipids,” Circ. Res. 106(8), 1332–1341 (2010). 10.1161/CIRCRESAHA.109.208678 [DOI] [PubMed] [Google Scholar]
- 11.Lim R. S., et al. , “Identification of cholesterol crystals in plaques of atherosclerotic mice using hyperspectral CARS imaging,” J. Lipid Res. 52(12), 2177–2186 (2011). 10.1194/jlr.M018077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cicchi R., et al. , “Characterization of collagen and cholesterol deposition in atherosclerotic arterial tissue using non-linear microscopy,” J. Biophotonics 7(1–2), 135–143 (2014). 10.1002/jbio.201300055 [DOI] [PubMed] [Google Scholar]
- 13.Suhalim J. L., et al. , “Characterization of cholesterol crystals in atherosclerotic plaques using stimulated Raman scattering and second-harmonic generation microscopy,” Biophys. J. 102(8), 1988–1995 (2012). 10.1016/j.bpj.2012.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P., et al. , “Label-free quantitative imaging of cholesterol in intact tissues by hyperspectral stimulated Raman scattering microscopy,” Angew. Chem. Int. Ed. 52(49), 13042–13046 (2013). 10.1002/anie.201306234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sijtsema N. M., et al. , “Imaging with extrinsic Raman labels,” Appl. Spectrosc. 50(5), 545–551 (1996). 10.1366/0003702963905989 [DOI] [Google Scholar]
- 16.Yamakoshi H., et al. , “Imaging of EdU, an alkyne-tagged cell proliferation probe, by Raman microscopy,” J. Am. Chem. Soc. 133(16), 6102–6105 (2011). 10.1021/ja108404p [DOI] [PubMed] [Google Scholar]
- 17.Yamakoshi H., et al. , “Alkyne-tag Raman imaging for visualization of mobile small molecules in live cells,” J. Am. Chem. Soc. 134(51), 20681–20689 (2012). 10.1021/ja308529n [DOI] [PubMed] [Google Scholar]
- 18.Chen Z., et al. , “Multicolor live-cell chemical imaging by isotopically edited alkyne vibrational palette,” J. Am. Chem. Soc. 136(22), 8027–8033 (2014). 10.1021/ja502706q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei L., et al. , “Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering,” Nat. Methods 11(4), 410–412 (2014). 10.1038/nmeth.2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee H. J., et al. , “Assessing cholesterol storage in live cells and C. elegans by stimulated Raman scattering imaging of phenyl-diyne cholesterol,” Sci. Rep. 5, 7930 (2015). 10.1038/srep07930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potma E. O., et al. , “Real-time visualization of intracellular hydrodynamics in single living cells,” Proc. Natl. Acad. Sci. U. S. A. 98(4), 1577–1582 (2001). 10.1073/pnas.98.4.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potma E. O., Xie X. S., “Direct visualization of lipid phase segregation in single lipid bilayers with coherent anti-stokes Raman scattering microscopy,” ChemPhysChem 6(1), 77–79 (2005). 10.1002/cphc.200400390 [DOI] [PubMed] [Google Scholar]
- 23.Li L., Wang H., Cheng J. X., “Quantitative coherent anti-stokes Raman scattering imaging of lipid distribution in coexisting domains,” Biophys. J. 89(5), 3480–3490 (2005). 10.1529/biophysj.105.065607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu D., et al. , “In vivo metabolic fingerprinting of neutral lipids with hyperspectral stimulated Raman scattering microscopy,” J. Am. Chem. Soc. 136(24), 8820–8828 (2014). 10.1021/ja504199s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu D., et al. , “Quantitative chemical imaging with multiplex stimulated Raman scattering microscopy,” J. Am. Chem. Soc. 134(8), 3623–3626 (2012). 10.1021/ja210081h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stiebing C., et al. , “Complexity of fatty acid distribution inside human macrophages on single cell level using Raman micro-spectroscopy,” Anal. Bioanal. Chem. 406(27), 7037–7046 (2014). 10.1007/s00216-014-7927-0 [DOI] [PubMed] [Google Scholar]
- 27.Wei L., et al. , “Vibrational imaging of newly synthesized proteins in live cells by stimulated Raman scattering microscopy,” Proc. Natl. Acad. Sci. U. S. A. 110(28), 11226–11231 (2013). 10.1073/pnas.1303768110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei L., et al. , “Imaging complex protein metabolism in live organisms by stimulated Raman scattering microscopy with isotope labeling,” ACS Chem. Biol. 10(3), 901–908 (2015). 10.1021/cb500787b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kushner D. J., Baker A., Dunstall T. G., “Pharmacological uses and perspectives of heavy water and deuterated compounds,” Can. J. Physiol. Pharmacol. 77(2), 79–88 (1999). 10.1139/y99-005 [DOI] [PubMed] [Google Scholar]
- 30.Souza C. M., et al. , “A stable yeast strain efficiently producing cholesterol instead of ergosterol is functional for tryptophan uptake, but not weak organic acid resistance,” Metab. Eng. 13(5), 555–569 (2011). 10.1016/j.ymben.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 31.Shivapurkar R., et al. , “An efficient method for the production of isotopically enriched cholesterol for NMR,” J. Lipid Res. 52(5), 1062–1065 (2011). 10.1194/jlr.D014209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh K., et al. , “Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets,” J. Cell Sci. 125(17), 4067–4076 (2012). 10.1242/jcs.104943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldstein J. L., Basu S. K., Brown M. S., “Receptor-mediated endocytosis of low-density lipoprotein in cultured cells,” Methods Enzymol. 98, 241–260 (1982). 10.1016/0076-6879(83)98152-1 [DOI] [PubMed] [Google Scholar]
- 34.Hölttä-Vuori M., et al. , “Alleviation of seipinopathy-related ER stress by triglyceride storage,” Hum. Mol. Genet. 22(6), 1157–1166 (2013). 10.1093/hmg/dds523 [DOI] [PubMed] [Google Scholar]
- 35.Bautista G., et al. , “Polarized THG microscopy identifies compositionally different lipid droplets in mammalian cells,” Biophys. J. 107(10), 2230–2236 (2014). 10.1016/j.bpj.2014.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang D., et al. , “Quantitative vibrational imaging by hyperspectral stimulated Raman scattering microscopy and multivariate curve resolution analysis,” Anal. Chem. 85(1), 98–106 (2013). 10.1021/ac3019119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nascimento J. M. P., Bioucas Dias J. M., “Vertex component analysis: a fast algorithm to unmix hyperspectral data,” IEEE Trans. Geosci. Remote Sens. 43(4), 898–910 (2005). 10.1109/TGRS.2005.844293 [DOI] [Google Scholar]
- 38.Tabarangao J. T., Slepkov A. D., “Mimicking multimodal contrast with vertex component analysis of hyperspectral CARS images,” J. Spectrosc. 2015, 575807 (2015). 10.1155/2015/575807 [DOI] [Google Scholar]
- 39.Alfonso-García A., et al. , “Biological imaging with coherent Raman scattering microscopy: a tutorial,” J. Biomed. Opt. 19(7), 071407 (2014). 10.1117/1.JBO.19.7.071407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christian A. E., et al. , “Use of cyclodextrins for manipulating cellular cholesterol content,” J. Lipid Res. 38(11), 2264–2272 (1997). [PubMed] [Google Scholar]
- 41.Lütjohann D., et al. , “Evaluation of deuterated cholesterol and deuterated sitostanol for measurement of cholesterol absorption in humans,” J. Lipid Res. 34(6), 1039–1046 (1993). [PubMed] [Google Scholar]
- 42.Amiot M. J., et al. , “Phytosterol ester processing in the small intestine: impact on cholesterol availability for absorption and chylomicron cholesterol incorporation in healthy humans,” J. Lipid Res. 52(6), 1256–1264 (2011). 10.1194/jlr.M013730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Czamara K., et al. , “Raman spectroscopy of lipids: a review,” J. Raman Spectrosc. 46(1), 4–20 (2015). 10.1002/jrs.4607 [DOI] [Google Scholar]
- 44.Kory N., et al. , “Protein crowding is a determinant of lipid droplet protein composition,” Dev. Cell 34(3), 351–363 (2015). 10.1016/j.devcel.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng B., et al. , “The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages,” Nat. Cell Biol. 5(9), 781–792 (2003). 10.1038/ncb1035 [DOI] [PubMed] [Google Scholar]