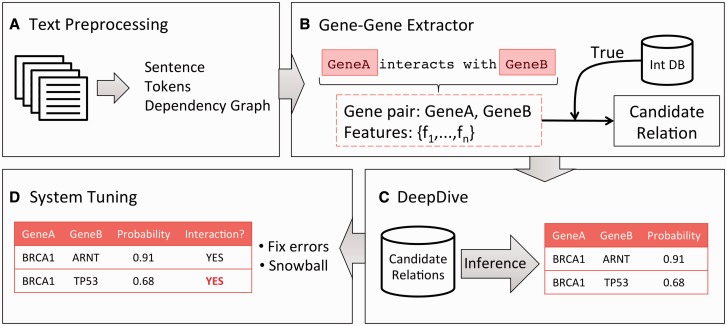
Fig. 1.
Gene–gene extraction pipeline. (A) We performed text pre-processing to parse documents into sentences and tokens and to construct dependency graphs between tokens in the sentences. This parsed data were stored in a sentences database. (B) The gene–gene extractor constructed candidate relations from the sentences and deposited them into a database. These relations composed of a pair of genes and features from the sentence. (C) DeepDive calculated probabilities that the candidate relation was an interaction using inference rules based on the features. (D) We performed system tuning to identify and correct system errors. Furthermore, we performed a snowball technique where we input correct relations as new training examples in the next system iteration