Abstract
Sirtuins or Sir2 family of proteins are a class of NAD+ dependent protein deacetylases which are evolutionarily conserved from bacteria to humans. Some sirtuins also exhibit mono-ADP ribosyl transferase, demalonylation and desuccinylation activities. Originally identified in the yeast, these proteins regulate key cellular processes like cell cycle, apoptosis, metabolic regulation and inflammation. Humans encode seven sirtuin isoforms SIRT1-SIRT7 with varying intracellular distribution. Apart from their classic role as histone deacetylases regulating transcription, a number of cytoplasmic and mitochondrial targets of sirtuins have also been identified. Sirtuins have been implicated in longevity and accumulating evidences indicate their role in a spectrum of diseases like cancer, diabetes, obesity and neurodegenerative diseases. A number of studies have reported profound changes in SIRT1 expression and activity linked to mitochondrial functional alterations following hypoxic-ischemic conditions and following reoxygenation injury. The SIRT1 mediated deacetylation of targets such as PGC-1α, FOXO3, p53 and NF-κb has profound effect on mitochondrial function, apoptosis and inflammation. These biological processes and functions are critical in life-span determination and outcome following injury. Aging is reported to be characterized by declining SIRT1 activity and its increased expression or activation demonstrated prolonged life-span in lower forms of animals. A pseudohypoxic state due to declining NAD+ has also been implicated in aging. In this review we provide an overview of studies on the role of sirtuins in aging and injury.
Keywords: SIRT1, mitochondria, ischemia/reperfusion, hypoxia
Introduction
Protein acetylation is a post translational modification that regulates key cellular functions including DNA recognition, protein–protein interaction, catalytic activity and protein stability [1–3]. The protein acetylation and deacetylation at N-epsilon lysine residues are catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs) respectively [4]. There are four classes of HDACs, Classes I–IV, based on phylogenetic analysis of all HDAC-related proteins [5]. Sirtuins are classified as Class III HDACs that are homologous to yeast transcriptional repressor, Sir2 [4]. The major functional difference between sirtuins and other HDACs is that sirtuins catalyze deacetylation of substrate proteins in a reaction that consumes NAD+. These protein modifying enzymes play significant roles in diverse cellular processes like apoptosis [6, 7], mitochondrial biogenesis [8], lipid metabolism [9], fatty acid oxidation [9, 10], cellular stress response [11–14], insulin secretion [15], aging [16–19] and inflammation [20].
Biochemistry of sirtuins
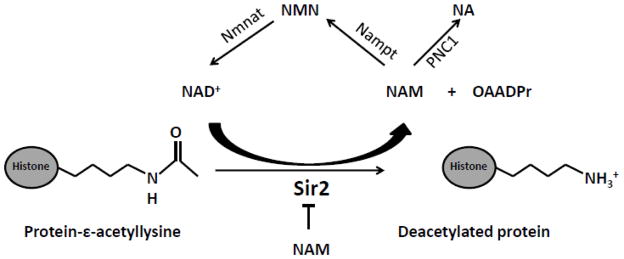
Sir2 (silent information regulator2) was the first sirtuin identified, from studies on mating type regulation in yeast Saccharomyces cerevisiae [21, 22]. The first evidences of the enzymatic activity of sirtuins came from studies on Sir2. Using 32P-labelled NAD it was shown that human ortholog of yeast Sir2 can transfer 32P from NAD+ to bovine serum albumin, suggesting their role in mono ADP ribosylation of proteins [23]. The observed enzymatic activity of Sir2 was found to be critical for the transcriptional repression at the silent mating–type loci, telomeric DNA regions, and the rDNA repeats [24]. The role of sirtuins as histone deacetylases was better characterized following molecular analysis of lysine residues of specific histone subunits [25–27]. Imai et al analyzed the product formed from Sir2 reaction by HPLC and mass spectrometry, and together with supporting data from mutational studies on conserved residues of the core domain of Sir2 concluded that NAD+ dependent deacetylase activity rather than ADP-ribosyltransferase activity account for Sir2 functions in vivo [25]. The histone deacetylation by Sir2 is coupled to NAD breakdown resulting in the formation of deacetylated protein, nicotinamide (NAM) and O-acetyl-ADP-ribose (OAADPr) [28]. The deacetylation reaction catalyzed by Sir2 is represented in Figure 1. The absolute requirement of NAD+ in the reaction, unlike the reactions catalyzed by other known protein deacetylases, makes their chemistry complex and energetically more demanding. However the benefit of this seemingly expensive reaction is its intricate regulation in multiple ways including by its own reaction products. The catalytic activity of Sir2 function is regulated by dynamic changes in cellular NAD+ concentration or the NAD+/NADH ratio [29, 30]. Cellular levels of nicotinamide phosphoribosyltransferase (Nampt), the rate-limiting enzyme for NAD+ synthesis varies during different pathophysiological conditions and hence affect sirtuin activity [31]. NAM is a potent inhibitor of sirtuin-mediated deacetylation [32]. PNC1 (pyrazinamidase/nicotinamidase 1), which encodes an enzyme that deamidates nicotinamide, converts NAM to nicotinic acid by a salvage pathway and regulates NAM accumulation [33]. Therefore both PNC1 and NAM can modulate Sir2 activity in the cells. In addition, OAADPr, another product of Sir2 mediated deacetylation, is also increasingly recognized as an important metabolic by-product [34, 35]. OAADPr was found to regulate gene silencing by facilitating the assembly and loading of the Sir2-4 silencing complex onto nucleosomes [36, 37]. Moreover, it is also a substrate for deacetylation by cellular macrodomain proteins like human MacroD1, human MacroD2, Escherichia coli YmdB, and the sirtuin-linked MacroD-like protein from Staphylococcus aureus [38]. Therefore the deacetylation catalyzed by Sir2 serves critical functions in cellular homeostasis.
Figure 1. Protein Deacetylation by Sir2.
Sir2 catalyzes the transfer of acetyl group from the protein to ADP-ribose moiety of NAD+ to form O-acetyl-ADP-ribose (OAADPr) and nicotinamide (NAM). Nicotinamide phosphoribosyltransferase (Nampt), the rate limiting enzyme in NAD biosynthetic pathway converts NAM to nicotinamide mononucleotide (NMN). NMN is then converted to NAD by nicotinamide mononucleotide adenylyltransferase (Nmnat). Another NAD salvage pathway component, Pnc1, converts NAM to nicotinic acid.
Sir2 gene family is highly conserved from bacteria to humans suggesting a common mechanism of gene silencing across the phylogenetic domains [39]. The bacterial and archaeal sirtuins play important roles in regulating transcription and cellular processes. For instance bacterial sirtuin CobB is an enzyme involved in propionate catabolism and cobalamin biosynthesis [40]. It deacetylates and activates acetyl-CoA synthetase in an NAD-dependent manner [41] and regulates E. coli chemotaxis by deacetylating CheY [42]. These findings suggest that both eukaryotes and prokaryotes carry out lysine acetylation as a common regulatory mechanism [41]. Studies also suggest a role for protein acetylation and deacetylation in bacterial stress response systems [43]. The archael sir2 homolog from Sulfolobus solfataricus P2 has both NAD-dependent deacetylase and mono-ADP-ribosyl transferase activities and regulate the binding of DNA binding protein alba [44].
Crystal structures of bacterial, yeast and mammalian sirtuins have been elucidated and reveal a highly conserved core domain made of a larger region with Rossmann-fold structure and a smaller variable region with zinc ribbon motif [45–47]. The sirtuins also contain N-and C-terminal extensions outside the catalytic core which are not conserved [47]. Prokaryotes generally contain one or two sirtuin genes whereas eukaryotes encode multiple isoforms. Yeasts contain the founding member Sir2 and four homologs of Sir2 (HST1-4). Mammalian sirtuin system is composed of seven genes; SIRT1 to SIRT7 among which SIRT1 has the highest sequence similarity to yeast Sir2 [48]. Sirtuins from a diverse number of organisms were phylogenetically analyzed and organized in to 5 major classes (I, II, III, IV and U) [48]. Class 1 comprises 5 yeast sirtuins (Sir2 and HST proteins) and human sirtuins SIRT1, SIRT2 and SIRT3. Class II has mammalian SIRT4 and sirtuins from other eukaryotes and bacteria. Class III has mammalian SIRT5 as well as bacterial and archael sirtuins. Most bacterial sirtuins belong to Class III. Class IV includes mammalian SIRT6 and 7. Class U contains sirtuins from gram positive bacteria and Thermotoga maritima.
An Overview of Mammalian Sirtuins
Mammalian sirtuins differ in their subcellular localization and function. SIRT1, SIRT6 and SIRT7 are mainly nuclear proteins with distinct subnuclear compartmentalization [49]. SIRT3, SIRT4 and SIRT5 are localized to mitochondria whereas SIRT2 is predominantly cytoplasmic [49]. While SIRT1-3 have strong deacetylase activity, SIRT4-7 are reported to have weak or no detectable deacetylase activity [50, 51] [52]; SIRT4 has predominantly ADP ribosyl transferase activity [53]. Table 1 represents the different mammalian sirtuins, their localization, and intracellular targets.
Table 1.
| Sirtuin | Localization | Substrates | Enzymatic activity |
|---|---|---|---|
| SIRT1 | Nuclear, cytoplasmic | Histone H1 [54], Histone H3[54], Histone H4 [55], p53 [56], NF-κB [57], FOXO4 [58], PGC1α [59], HIF1α [60], HIF2α [61], CTIP2 [62], Tat [63], p300 [64], LXR [65], FXR [66], eNOS [67], MEF2 [68], Notch1 [69], Ku70 [70], XPA[71], WRN [72], NBS1 [73], LKB1 [74], AceCS1 [75], HMGCS1 [76], c-Myc [77], androgen receptor [78], SUV39H1 [79], BMAL1 [80], PER2 [81], DNMT1 [82], hMOF [83], TIP60 [83], cortactin [84], PARP1 [85], SREBP-1C [64], SATB1 [86], RFX-5 [87], TDG [88], FOXA2 [89], IRF-1 [90], HMGB1 [91], PGAM1 [92]; CRABPII [93], TopBP1 [94], PML [95]. | Deacetylation |
| SIRT2 | Nuclear, cytoplasmic | Histone H4 [96], Histone H3 [97], Tubulin [98], FOXO1 [99], FOXO3A [100], p53 [101], p300 [102], p65 [103], PEPCK1 [104], Par-3 [105], CDK9 [106], HIF1α [107], G6PD [108], PGAM [109], ALDH1A1 [110], TUG [111], BubR1 [112], beta-secretase 1 [113]. | Deacetylation, Demyristoylase |
| SIRT3 | Mitochondrial | AceCS2 [75], HMGCS2 [114], LCAD [115], SDH [116], Ku70 [117], SOD2 [118], IDH2 [119], GDH [120], LKB1 [121], MRPL10 [122], LCAD [10]; ATP synthase F1 [123], Cyclophilin D [124], OTC [125], ALDH2 [126], Skp2 [127], FOXO3 [128], PDH [129], OGG1 [130], OPA1 [131], Hsp10 [132], GOT2 [133], MDH [113], Aconitase 2 [113]. | Deacetylation |
| SIRT4 | Mitochondrial | GDH [53], MCD [134], PDH [135], Hsp60 [113], Stress-70 [113], Nnt [113]. | ADP-Ribosylation, Deacetylation, Lipoamidase |
| SIRT5 | Mitochondrial | Cytochrome C [120], CPS1 [136], SOD1 [137], Urate oxidase [138], PML [95], VLCAD [139], Prx-1 [113], HMGCS2 [113], Hsp70 [113], MCAD [113]. | Deacetylation, Demalonylation, Desuccinylation, Deglutarylation |
| SIRT6 | Nuclear | TNFα [140], Histone H3 [51], CtIP [141], PARP1 [142], GCN5 [143], KAP1 [144], GEN1 [113], Kup86 [113], p70 [113]. | Deacetylation, ADP-ribosylation |
| SIRT7 | Nucleolus | Histone H3 [145], PAF53 [146], GABPβ1 [147], p53 [148], MEF-2C [113], DNA-PK [113] | Deacetylation |
Abbreviations: AceCS, acetyl-CoA Synthase; ALDH, aldehyde dehydrogenase; CDK9, cyclin-dependent kinase 9; CPS1, carbamoyl phosphate synthetase 1; CtIP, C-terminal binding protein; CTIP2, chicken ovalbumin upstream promoter transcription factor interacting protein 2; CRABPII, cellular retinoic acid binding protein II; DNA-PK, DNA-dependent protein kinase; DNMT1, DNA methyltransferase 1; eNOS, endothelial nitric oxide synthase; FOX, forkhead transcription factor; FXR, farnesoid X receptor; GABPβ1, GA binding protein 1; GCN5, General Control Non-repressed Protein 5; GDH, glutamate dehydrogenase; G6PD, glucose-6-phosphate dehydrogenase; GOT2, glutamate oxaloacetate transaminase 2; HMGB1, high-mobility group box 1; HMGCS, 3-hydroxy-3-methylglutaryl CoA synthase; hMOF, human ortholog of the Drosophila males-absent-on-the-first; Hsp10, heat shock protein 10; IDH2, isocitrate dehydrogenase 2; IRF-1, interferon regulatory factor 1; KAP1, KRAB-associated protein 1; LCAD, long-chain acyl coenzyme A dehydrogenase; LKB1, liver kinase B1; LXR, liver X receptor; MCAD, medium-chain acyl-CoA dehydrogenase; MCD, malonyl CoA decarboxylase; MDH, malate dehydrogenase; MEF2, myocyte enhancer factor 2; NBS, nijmegen breakage syndrome; Nnt, nicotinamide nucleotide transhydrogenase; OGG1, 8-oxoguanine-DNA glycosylase 1; OPA1, optic atrophy 1; PAF53, polymerase-associated factor 53; PARP1, poly(ADP-ribose) polymerase 1; PDH, pyruvate dehydrogenase; PEPCK1, phosphoenolpyruvate carboxykinase; PER2, period 2; PGAM, phosphoglycerate mutase; PML, Prx-1, peroxiredoxin 1; RFX-5, regulatory factor for X-box; SATB1, special AT-rich sequence-binding protein-1; Skp2, S-phase kinase associated protein 2; SOD2, superoxide dismutase 2; SUV39H1, suppressor of variegation 3–9 homolog 1; TDG, thymine DNA glycosylase; TIP60, HIV-1 TAT-interacting protein of 60 kDa; TopBP1, DNA topoisomerase 2-binding protein 1; XPA, xeroderma pigmentosum group A
SIRT1
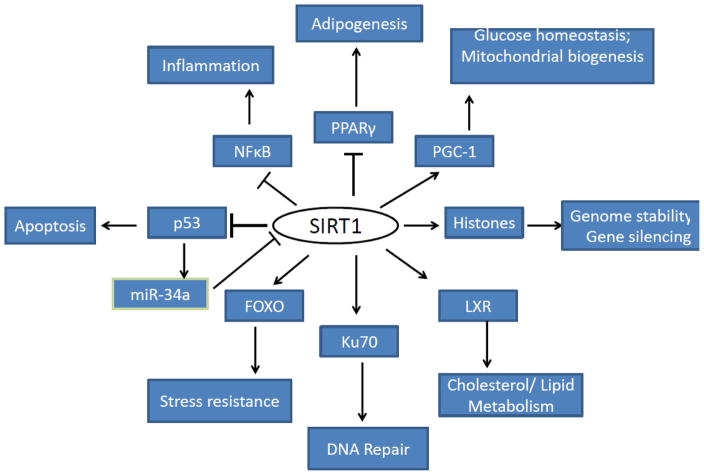
SIRT1, the mammalian ortholog of yeast Sir2, is the most studied mammalian sirtuin. SIRT1 plays important roles in embryonic development [149, 150] and skeletal muscle differentiation [151]. It has diverse functions in the cells ranging from chromatin modification and epigenetics to roles in metabolic pathways, inflammation and stress response (Figure 2) [152]. It interacts with and deacetylates histones and a number of non-histone substrates. SIRT1 preferentially deacetylates specific residues in histone subunits like the lysine 16 of histone 4 (H4K16), lysine 9 of histone 3 (H3K9), lysine 56 of histone 3 (H3K56) [153] and lysine 26 of histone 1 to promote heterochromatin formation and transcriptional silencing [54, 55]. The non-histone protein substrates of SIRT1 include but not limited to tumor suppressor p53, nuclear factor-κB (NF-κB), peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), fork-head box protein O (FOXO) transcription factors, liver X receptor (LXR), PARP, Ku70 and hypoxia-inducible factor (HIF)-1α [154] [11, 58], [60], [61], [65] [70, 85, 155]). SIRT1 plays a predominant role in regulating apoptosis through deacetylating p53 and inhibiting p53 dependent transcription during cellular stress [56, 150, 156]. It also controls inflammation through regulating NF-κB signaling by deacetylating the p65 subunit of the complex thereby inhibiting NF-κB signaling. On the contrary, NF-κB signaling diminishes SIRT1 activity by modulating expression of miR-34a, IFNγ, and reactive oxygen species [57]. miR-34a, a tumor suppressor, has been reported to bind directly to 3′-UTR of SIRT1 thereby repressing its expression and enhancing p53 mediated apoptosis [157]. It has been also known that miR-34a is a transcriptional target of p53 [157, 158]. Furthermore, Kim et al demonstrated that p53 is influenced in miR-34a-mediated repression of SIRT1 in cisplatin-induced cytotoxicity [159]. The antagonistic crosstalk between SIRT1 and NF-κB signaling is apparent in many inflammatory diseases and aging [160–163]. Moreover, inflammation associated increase in nitric oxide (NO) production results in S-Nitrosylation and inhibition of SIRT1 activity which further heightens the inflammatory response through increased acetylation of p65 [164]. In addition, SIRT1 has demonstrated role as a tumor suppressor in rodent studies which likely involves its ability to deacetylate and inhibit beta-catenin transcriptional activity [165, 166].
Figure 2. SIRT1 Regulation in Health and Disease.
Increased SIRT1 activity results in deacetylation of histone and nonhistone substrates affecting multiple cellular processes.
SIRT1 plays a major role in metabolic regulation and PGC-1a deacetylation is one of the important events in this process. Hepatic SIRT1 has been suggested to play an important role in glucose and lipid metabolism in during fasting [167]. SIRT1 has a vital role in maintaining lipid homeostasis through PPARα mediated beta-oxidation of fatty acids in the liver [9] and mobilization of fat from white adipocytes during fasting [168]. In addition, SIRT1 controls mitochondrial biogenesis though regulating PGC-1α pathway [59]. SIRT1 negatively regulates the expression and phosphorylation of signal transducer and activator of transcription 3 (STAT3) and STAT3 mediated cellular respiration [169]. A noteworthy aspect of SIRT1 mediated metabolic control is its regulation by AMPK which is also known as the cellular fuel gauge. AMPK activation has been reported to enhance SIRT1 activity by increasing NAD+ levels resulting in deacetylation of SIRT1 targets [170]. Conversely, SIRT1 activation is reported to induce AMPK activation by deacetylation of AMPK upstream kinase, LKB1 [74]. Adipokines like adiponectin have been shown to stimulate AMPK-SIRT1-PGC-1α pathway and increase mitochondrial content in myocytes [74]. Another key feature of SIRT1 is its positive regulation on insulin secretion in pancreatic β cells by repressing uncoupling protein2 (UCP2) gene [171]. The SIRT1 localization and/or enzymatic activity is subjected to regulation by post-translational modifications like phosphorylation [172], SUMOylation [173], S-nitrosylation [164] and carbonylation [174]. The pleiotropic roles of SIRT1 in the cells make it an important intracellular marker protein in aging as well as different diseases like cardiovascular diseases, diabetes, cancer, neurodegenerative diseases and other conditions in health and disease (Figure 3) [152].
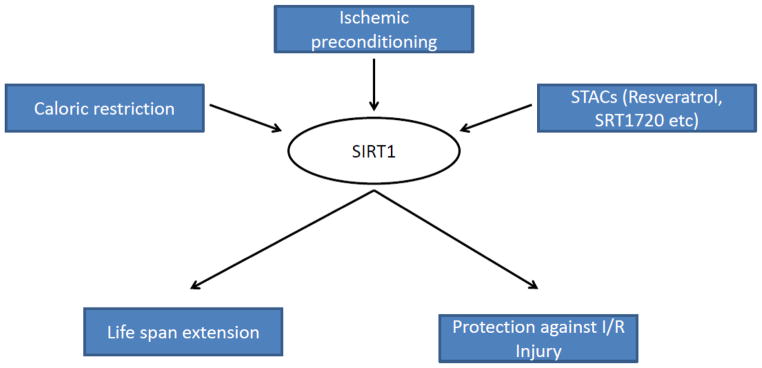
Figure 3. SIRT1 Modulation in Aging and Injury.
SIRT1 expression and/or activity are upregulated with caloric restriction, ischemic preconditioning and STACS resulting in longevity and protection following injury.
SIRT2
SIRT2 is the mammalian ortholog of yeast Hst2 and is predominantly cytoplasmic. It deacetylates α-tubulin [98]. SIRT2 protein levels often fluctuate during cell cycle, with a marked increase in expression and phosphorylation during the mitotic and G2/M transition phase playing a role in mitotic exit during cell cycle [175]. A characteristic feature of SIRT2 is its migration to the nucleus during G2/M transition to modulate chromatin condensation by histone H4 deacetylation [96]. SIRT2 can also deacetylate FOXO1 and promote FOXO1 binding to PPARγ resulting in the suppression of PPARγ activity and hence adipocyte differentiation [99]. It also deacetylates FOXO3A to promote expression of antioxidant and pro-apoptotic molecules under cellular stress [100]. A recent study suggests that cells sense extracellular oxidative stimuli to decrease acetylation of a key enzyme in the pentose phosphate pathway, glucose-6-phosphate dehydrogenase, in a SIRT2-dependent manner [108]. SIRT2 has been implicated in a number of diseases like cancer and neurodegenerative disorders like Parkinson’s and Huntington’s disease [176].
SIRT3
SIRT3 is an NAD+ dependent deacetylase predominantly located in the mitochondrial matrix. Inactive SIRT3 has been shown to be converted to an active form in the matrix following its cleavage by mitochondrial matrix processing peptidase [177]. It is highly expressed in tissues rich in mitochondria [178] and has a role in brown adipose tissue thermogenesis [179]. Another group reported the presence of enzymatically active full length SIRT3 in the nucleus which gets transported to the mitochondria under conditions of cellular stress [180]. However a later study by Cooper et al demonstrated an exclusive mitochondrial localization of human SIRT3 [180]. Consistent with its localization in mitochondria, loss of SIRT3 in a mouse model markedly elevated mitochondrial lysine acetylation [178]. SIRT3 loss resulted in a reduction in basal ATP levels in multiple organs, hyperacetylation of complex I components and reduction in complex I activity [181]. The first SIRT3 substrate identified was mitochondrial enzyme acetyl-CoA synthetase 2, the deacetylation of which leads to its activation [182]. SIRT3 acts as a stress responsive protein in the cardiomyocytes by deacetylating Ku70 and promoting its interaction with proapoptotic protein Bax thereby preventing Bax translocation to the mitochondria [117]. Another SIRT3 interacting partner is mitochondrial complex I protein NDUFA9 [181]. Two critical targets of SIRT3 deacetylation are mitochondrial superoxide dismutase (SOD2) and isocitrate dehydrogenase 2 (IDH2), the enhanced activity of these redox enzymes prevents accumulation of toxic ROS. By regulating ROS production, SIRT3 suppresses the hypoxia inducible factor 1α (HIF-1α) transcriptional activity and hence may act as a tumor suppressor [183]. SIRT3 has been reported to promote mitochondrial fatty acid oxidation by deacetylating long-chain acyl coenzyme A dehydrogenase, key enzyme involved in the oxidation of long-chain substrates [10]. A recent study suggests that a natural polyphenolic, Honokiol, blocks and reverses cardiac hypertrophy in mice by activating mitochondrial SIRT3 [184]. Taken together, it may be concluded that SIRT3 plays a significant role in maintaining mitochondrial homeostasis.
SIRT4
SIRT4 is an NAD+ dependent ADP-ribosyl transferase localized to the mitochondria. Insulin secretion in pancreatic β cells is subjected to modulation by SIRT4 as secretion of insulin granules is triggered by local increase in ATP concentration by enzymes like glutamate dehydrogenase (GDH) that undergoes inhibition by SIRT4 ADP-ribosylation [53]. Another notable function of SIRT4 is its regulatory control over glutamine metabolism to facilitate DNA damage responses and to prevent tumorogenesis [185]. Consistently, mTORC1 pathway which is activated in proliferating tumor cells downregulates SIRT4 expression [186]. Its deacetylase activity is reported to regulate hepatic lipid homeostasis [134]. SIRT4 was recently shown to possess lipoamidase function that inhibits the activity of pyruvate dehyrogenase, an enzyme that links glycolysis to citric acid cycle [135]. Together, these studies highlight a major role for SIRT4 in regulating cellular metabolism and preventing tumorogenesis.
SIRT5
Mitochondrial SIRT5 is an NAD+ dependent protein lysine demalonylase, desuccinylase [52] and a deglutarylase [187]. It plays a pivotal role in ammonia detoxification by deacetylating carbamoyl phosphate synthetase 1(CPS1), the rate limiting step in urea cycle [136]. In fact SIRT5 knockout mice had higher ammonia levels in blood during fasting or with a high protein diet [136]. CPS1 is also a target of desuccinylation [52] and deglutarylation by SIRT5 [187]. Other key targets of SIRT5 include SOD1 [137] and mitochondrial urate oxidase [138]. A recent study has reported its role in desuccinylating a key fatty acid oxidizing enzyme and promoting fatty acid oxidation [139]. Although loss of SIRT5 leads to hypersuccinylation of several metabolic pathway components [188], it is considered a dispensable isoform in terms of regulating metabolism [189].
SIRT6
SIRT6 is localized to nucleus and its enzymatic activities include deacetylation of histones, ADP ribosylase and lysine deacylase [190]. SIRT6 mediated lysine deacylation of tumour necrosis factor-α (TNF-α) promotes its secretion from macrophages [140]. SIRT6 is reported to have a crucial role in maintaining genomic stability and repairing DNA damages by different mechanisms [190]. It has a profound effect on glucose metabolism by suppressing the expression of HIF-1α and other glycolytic genes [191]. Hence a loss of SIRT6 resulted in severe hypoglycemia due to increased glucose uptake in muscle and fat tissues. It also exerts control over hepatic gluconeogenesis by regulating PGC-1α acetylation in an indirect manner through modifying the activity of acetyltransferase GCN5 [143]. Additionally, SIRT6 is involved in lipid metabolism by negatively regulating triglyceride synthesis [192]. Like SIRT1, SIRT6 expression is correlated with longevity; its expression decreased with age in human dermal fibroblasts [193] and overexpression in male mice increased its life span [18]. SIRT6 function has also been implicated in several different types of cancers [190]. Collectively, these studies reveal major roles for SIRT6 in metabolism and aging.
SIRT7
SIRT7 has been shown to be expressed in the nucleolus where it interacts with histones and RNA polymerase I to positively regulate ribosomal gene (rDNA) transcription [194]. It is phosphorylated during mitosis when the rDNA transcription is repressed and undergoes dephosphorylation and conformational changes to resume rDNA transcription at the exit from mitosis [195]. It is also proposed to play a key role in connecting the chromatin remodeling complexes to RNA Pol I machinery during transcription [196]. Initially an anti-proliferative role for SIRT7 was described in a mouse model and murine cell lines [197]. In stark contrast, it was shown to deacetylate and repress transcription of genes linked to tumor suppression, thus maintaining oncogenic transformation [145]. The oncogenic potential of SIRT7 was further demonstrated in human hepatocarcinomas [198] and human colorectal cancer [199]. Other notable functions of SIRT7 include its role in ribosome biogenesis and protein synthesis [200], inhibition of hypoxia-inducible factor HIF-1α and HIF-2α [201], cofactor for transcriptional repression by Myc [202] and hepatic lipid metabolism [203]. A recent study reported its positive influence on mitochondrial homeostasis by regulating acetylation of GABPβ1, a master regulator of nuclear-encoded mitochondrial genes [147]. SIRT7 is considered a potential target for cancer therapy and studies continue to unravel novel roles for SIRT7.
Role of Sirtuins in Aging
The role of sirtuins in extending organism life span has been a topic of great interest among aging researchers. The functional relevance of sirtuins in mitochondrial bioenergetics and oxidative stress coupled with the observation that some of the sirtuins prolonged life prompted extensive studies on sirtuin family of proteins in aging biology. Though most of the studies on sirtuins were focused towards elucidating the functional role of SIRT1, other members of this family are also being studied to understand their role in aging, health and disease [204].
The initial results on the effect of Sir2 on life-span in yeast was further extrapolated into other model organisms like Drosophila, Caenorhabditis elegans and rodents [16, 18, 205, 206]. In yeast, Sir2 mutation shortened lifespan owing to accumulation of toxic extrachromosomal rDNA circles (ERC) whereas Sir2 overexpression extended life span by silencing HM loci and inhibiting ERC formation [205]. Sir2 was found to mediate the life span extension in yeast induced by caloric restriction [207] by increasing mitochondrial oxidation and respiration [208]. Life span may be extended by limiting activity of glucose-sensing cyclic-AMP-dependent kinase (PKA) which requires Sir2 and NAD [208]. Studies have also uncovered the role of nicotinamide clearance by PNC1 in regulating longevity by CR in yeast [33, 209]. Aging is reported to decrease tissue levels of NAD resulting in declined SIRT1 activity and decreased NAD+/NADH leading to increased ROS formation in mitochondria [210]. Another theory is the suppression of target of rapamycin (TOR) signaling pathway by CR [211] which leads to inhibition of ribosome biogenesis and relocalization of the transcription factors Msn2p and Msn4p from the cytoplasm to the nucleus thereby increasing expression of PNC1 [212].
Consistent with a role for Sir2, its activator resveratrol was found to mimic CR induced longevity in the yeast [2] and sirtuin activating compounds (STACs) delayed aging in metazoans [213]. However the involvement of Sir2 or other homologs [214] in CR induced longevity has also been questioned in some studies [215–217]. For instance, CR was reported to enhance longevity in yeast cells lacking Sir2, implying a Sir2 independent mechanism [215]. Conforming to this observation, Sir2 homolog Hst2 was shown to mediate SIR2-independent life-span extension by CR [214]. Contrary to the above, another report precludes the involvement of any Sir2 family members in lifespan extension by CR [216]. Another study reports absence of a role for Sir2 in chronological aging (long term survival of non-dividing cells) in yeast unlike its role in replicative aging [218]. Increased Sir2 gene content also extended life span in Caenorhabditis elegans [16] and Drosophila [206]. In C. elegans, SiR2.1 activated DAF-16 [219], a forkhead transcription factor, that mediates life span regulation by insulin/IGF-1 signaling pathway [220]. Activating autophagy is one of the underlying mechanisms suggested to be behind life span extension benefits of Sir2 [221]. In Drosophila, overexpression of Sir2 in adult fat body but not in muscle promoted longevity indicating tissue specific effects of Sir2 expression [222, 223]. In humans, SIRT1 expression and activity are abrogated in aged arteries suggesting its role in vascular aging [224]. SRT1720, a small molecule activator of SIRT1, improved health and life span of mice, further suggesting the role of SIRT1 [225]. Interestingly a positive correlation exists between mitotic activity and SIRT1 levels in mammalian tissues [226]. The anti-aging effect of SIRT1 likely involves p53, as SIRT1 was found to antagonize promyelocytic leukemia protein (PML) induced acetylation of p53 and cellular senescence in primary mouse embryo fibroblasts [227]. Another plausible mechanism is through repressing PPARγ thereby attenuating adipogenesis and promoting fat mobilization in white adipocytes [168].
SIRT2 is reportedly elevated in the white adipose tissue and kidney of caloric restricted mice where it deacetylates FOXO transcription factors and increases expression of FOXO target genes, p27(Kip1), manganese superoxide dismutase and Bim [100]. Cohen and colleagues recently showed that SIRT6 overexpression in male mice extended life span compared to wild type mice and this was associated with lower serum levels of insulin-like growth factor 1 (IGF1) [18]. They also had observed an increased SIRT6 levels following caloric restriction in rats [228]. Accumulating evidences also suggest a role for SIRT3 in age related pathologies [229, 230]. Interestingly both mammalian SIRT3 and bacterial CobB regulate acetyl-CoA synthetase through its deacetylation suggesting an evolutionary conservation of the mechanism. Although sirtuins possess anti-aging functions from yeast to mammals, the underlying mechanisms have also evolved to meet the complexity of higher order species. In yeast, sirtuins mainly act through suppressing genomic instability (recombination mainly at the ribosomal DNA locus) where as in mammals they affect multiple pathways to regulate aging.
Aging has been described to be characterized by declining NAD+, delinking PGC-1α/β from mitochondrial control and the emergence of a pseudo-hypoxic state [210, 231]. Further, the pseudo-hypoxic state was compared to Warburg reprogramming and was suggested to be reflected in SIRT1 deficiency which may be restored with NAD+ augmentation. This has parallels to injury conditions manifested by hypoxia and nutrient deprivation in downstream tissues and therefore SIRT1 mediated metabolic regulation in tissue injury and repair draws attention for investigations.
Role of Sirtuins in Tissue Injury and Repair
Sirtuins are key physiological modulators controlling a number of critical metabolic pathways and functions including cell death and repair. These physiological regulations occur by virtue of their direct enzymatic action on target proteins as well as due to alterations in the level of metabolites related to the reaction. Though not all sirtuins are robust deacetylases, the network of proteins that include PGC-1α, SIRT1 and AMPK are considered to be a critical part of the energy sensing network in cells [232]. SIRT1 and AMPK act as metabolic sensors by their ability to deacetylate and phosphorylate, respectively, the mitochondrial biogenesis factor PGC-1α. Therefore the actions of SIRT1 are closely linked to enhancement of mitochondrial function and biogenesis and mitigation of redox injury making this protein an attractive target in molecular therapeutics [233]. It is further substantiated by the observation that following cellular stress SIRT1 activity is altered and modulation of the activity and or expression of SIRT1 following cellular injury is important in restoring cellular homeostasis, repair and death.
Sirtuins and cardiac injury
Several studies, including that from our lab, have demonstrated the importance of sirtuins in improving organ function and survival following tissue injury [234–236]. In post myocardial infarction (MI) patients, ischemia/reperfusion (I/R) injury remains the major cause for cardiac remodeling and heart failure [237]. Therefore methods to prevent I/R injury become instrumental in reducing mortality in post MI patients. Numerous studies have been reported on the role of sirtuins, specifically SIRT1, in managing I/R injury [234, 238, 239].
I/R injury was associated with a reduction in SIRT1 mRNA and protein [234]. Using transgenic mice with cardiac specific over expression of SIRT1, Hsu et al clearly demonstrated a significant reduction in myocardial infarction area and a greater recovery after reperfusion of isolated hearts, compared to wild type. Conversely, cardiac specific knockdown of SIRT1 resulted in increased size of myocardial infarction/area. The observed effects of SIRT1 overexpression is attributed to suppression of oxidative stress and apoptosis by FOXO1 mediated upregulation of antioxidant molecules like manganese superoxide dismutase and down regulation of proapoptotic molecules [234]. One of the prominent changes that occurs during cardiac hypertrophy is the shift in myosin isoform from α- to β-myosin heavy chain (MHC) [240]. Interestingly, fructose feeding was shown to have a protective effect on the heart following I/R injury by inducing cardiac α-MHC expression. Fructose feeding also stimulated cardiac NAD+ and SIRT1 levels and these effects were mimicked by resveratrol [241]. The role of SIRT1 in α-MHC expression was further confirmed by cardiac specific overexpression studies. It is interesting to note that both a direct agonist of SIRT1 (resveratrol) and its indirect activation by NAD+ levels could cause similar physiological effects in tested animals. However the mechanism of SIRT1 mediated induction of α-MHC expression is still unclear.
Ischemic preconditioning (IPC) is effective in limiting cardiac damage occurring during prolonged occlusion and reperfusion [237]. Nadtochiy et al studied the role of SIRT1 in cardioprotective effects of acute IPC using SIRT1 deficient and SIRT1 overexpressing mice. Consistent with a role for SIRT1, IPC induced cytosolic lysine deacetylation in wild type hearts whereas SIRT1 deficient hearts had more cytosolic lysine acetylation and were refractive to preconditioning. Conforming to these results, SIRT1 overexpressing mice exhibited decreased cytosolic acetylation and endogenous protection against I/R injury [238]. Both IPC induced lysine deacetylation and cardiac protection was inhibited by SIRT1 inhibitor splitomycin [238]. Nicotinamide phosphoribosyltransferase (Nampt), a key enzyme in the salvage pathway of NAD synthesis, is a critical regulator of energy status and survival in cardiac myocytes [242]. Nampt was found to play a crucial role in mediating the protective effect of IPC against ischemia and reperfusion, which was also mimicked by exogenous nicotinamide mononucleotide (NMN), a product of Nampt in the NAD+ salvage pathway. On the other hand, Nampt can be secreted from cardiomyocytes to act as a proinflammatory cytokine. The exogenous Nampt was found to be a positive regulator of cardiac hypertrophy and adverse ventricular remodeling [243]. Therefore, although intracellular Nampt is essential to the cardiac myocyte survival, exogenous Nampt could be detrimental demanding fine balance between its synthesis and secretion. Interestingly, the cardiac protection conferred by IPC deteriorates with age [244]. The lack of ischemic tolerance in aged hearts is mainly accounted for by a reduced SIRT1 expression and activity although one study has ruled out the role of SIRT1 [244].
Caloric restriction (CR) was previously shown to increase longevity in yeast and other species [207, 245, 246]. Shinumura et al studied the effects of short-term [247] and long-term caloric restriction [248] on ischemic tolerance and ischemic preconditioning (IPC) in aged rats. Short-term CR improved left ventricular function in both young and aged rats which was associated with an increase in AMPK phosphorylation [247]. Long term caloric restriction also improved recovery of left ventricular function and reduced infarct size after ischemia-reperfusion. However these changes were not associated with any changes in expression of myocardial total or phosphorylated AMPK. Strikingly, long-term CR induced cardiac protection was associated with nitric oxide-dependent increase in nuclear SIRT1 content [248]. CR mediated protection against I/R injury was associated with Nampt upregulation whereas the protective effect was abrogated in SIRT1−/− mice, suggesting a Nampt-SIRT1 axis [249]. Analysis of the molecular changes underlying CR induced cardioprotection revealed an overall reduction in acetylated mitochondrial proteins with CR. Consistent with a role for sirtuins, deacetylation of specific proteins of the electron transport chain was observed, which preserves mitochondrial integrity by preventing accumulation of toxic ROS [250]. Deacetylation of mitochondrial proteins also implies an involvement of mitochondrial sirtuins apart from nuclear/cytoplasmic sirtuins like SIRT1.
Studies from our laboratory using a hemorrhagic shock model showed that when 60% of the blood volume was removed from rats and subjected to a prolonged shock phase, there was a significant decline in SIRT1 and PGC-1α protein levels in the heart at two hours following resuscitation [235]. It is unclear whether this is the effect of oxidative stress associated with resuscitation or initiated by the hypoxic/ischemic condition due to hemorrhagic shock. However, administration of resveratrol, a SIRT1 activator and antioxidant, along with resuscitation fluid proved to be beneficial in improving left ventricular function and cardiac contractility, and prolonged lifespan in the absence of resuscitation [235, 251]. Furthermore, hemorrhagic shock induced a shift in the metabolic process towards glycolysis, consistent with a mitochondrial functional decline which was restored by resveratrol administration [252]. Other investigators have also observed the beneficial effect of resveratrol following hemorrhagic shock, though methodologies varied [253–255]. Resveratrol pretreatment was also effective in reducing IR-induced arrhythmias and mortality in rats [80]. Resveratrol was protective against myocardial injury in a rat model of autoimmune myocarditis [81]. The SIRT1 activator resveratrol has been extensively used in aging and injury conditions and at least part of the health effect of resveratrol is likely through activation of SIRT1.
SIRT1 exerts its beneficial effects at both transcriptional level and posttranslational levels. At the transcriptional level, it affects the expression of many antioxidant genes and apoptotic molecules by stimulating the transcriptional activity of FOXO1 [234]. Rui-Hong Wang et al found that hepatic SIRT1 deficiency in mice can impair mTorc2/Akt signaling leading to oxidative damage and insulin resistance [256]. At the post-translational level, SIRT1 influences acetylation and activity of a number of proteins. Likewise the regulation of SIRT1 exists at multiple levels. Aldehyde mediated carbonyl stress is considered as yet another factor contributing to increased susceptibility of aging heart to I/R injury [174]. Carbonyl modification of SIRT1 during aging impairs its activity and causes myocardial ischemic intolerance which could be restored by cardiac aldehyde dehydrogenase activation [174]. Therefore more studies are required to address the posttranslational changes in SIRT1 and other sirtuins that may affect their compartmentalization and function inside the cells. In cardiac myocytes SIRT1 expression is regulated by microRNA, miR199a. miR199a itself is markedly down regulated during cardiac ischemia which favors rapid accumulation of HIF-1α by preventing its degradation [257].
Several endogenous and exogenous molecules have been shown to exercise cardio protective effects through SIRT1 regulation. For example locally acting insulin-like growth factor-1 isoform has been shown to protect cardiomyocytes from oxidative/hypertrophic stress through SIRT1 activation [258]. SIRT1 induction by resveratrol is reported to have a modulatory effect on mitogen-activated protein kinase (MAPK) pathway which is commonly upregulated under stress [259]. Likewise sidenaphil, a phosphodiesterase-5 inhibitor improves I/R injury and a concomitant SIRT1 activation was observed [260]. Cardiomyocyte apoptosis is a characteristic feature of heart failure. Silibinin, a plant flavonoid was found to ameliorate β-adrenergic agonist isoproterenol-induced injury in cultured rat neonatal cardiac myocytes through mechanisms including but not limited to upregulation of SIRT1 [261]. However, in one study it has been shown that constitutive SIRT1 overexpression resulted in impaired cardiac mitochondria and cardiac dysfunction in response to pressure overload [262].
Other sirtuins are also gaining prominence with respect to their roles in managing the detrimental effects of hypoxic/ischemic and reperfusion injury. One of the mitochondrial sirtuins, SIRT4 was shown to play a protective role in hypoxia induced apoptosis in H9c2 cardiomyoblast cells [263]. Similarly, another mitochondrial sirtuin, SIRT5 undergoes marked downregulation in cardiomyocyes upon oxidative stress [264]. Both SIRT4 and SIRT5 knockdown significantly increased apoptosis in cardiomyocytes [263, 264]. The mitochondrial sirtuin, SIRT3 is increasingly recognized as an important molecule in preserving mitochondrial integrity and improving cardiac function. Case studies on post MI patients indicate that exercise training exerts beneficial effects on improving cardiac functions [265, 266]. Jiang et al studied the molecular basis of cardioprotection by aerobic interval training (AIT) exercise in rat models. They found increased mitochondrial biogenesis in AIT rats accompanied by AMPK phosphorylation and increased SIRT3 levels [267]. A recent report shows the protective effect of resveratrol in combating oxidative stress by upregulating SIRT3 expression in the mitochondria of human vascular endothelial cells [268]. Flavanoids like rhamnetin also exhibit cyto protective effects against oxidative stress in H9c2 cardiomyoblasts by SIRT3 and SIRT4 induction [269]. Consistent with these observations, SIRT3 deficient hearts were less tolerant to I/R injury with greater infarct size [270]. This decline in function is attributed to inhibition of enzymatic activities of SIRT3 targets Cx1 and Mn SOD. SIRT3 knockdown in H9C3 cardiac cells made them more vulnerable to oxidative damage [270]. Together these results suggest that SIRT3 may be an important component of damage control in I/R and other forms of injury and demand more attention. SIRT7 is yet another sirtuin with a profound effect on preventing apoptosis and inflammatory cardiomyopathy; the observed effects likely mediated through its effect on p53 deacetylation [148].
Sirtuins and Neuronal injury
The neuroprotective effects of SIRT1 have been demonstrated in different models of traumatic brain injury, ischemic injury and in a number of neurodegenerative disorders. Ischemic brain damage often leads to fatal outcomes unless managed in a narrow window of time [271, 272]. Therefore studies to identify novel strategies to manage ischemic damage would have important clinical implications [273] [274]. Like in hearts, ischemic preconditioning is found to be an effective strategy in protecting neurons from lethal ischemia. An in vitro model of cerebral ischemia using hippocampal slice cultures subjected to oxygen-glucose deprivation (OGD) is a useful alternative to in vivo ischemia [275]. Initial studies by Raval and colleagues using this in vitro model confirmed neuroprotective action of SIRT1 agonist resveratrol in cerebral ischemia [272, 276]. Both IPC and resveratrol preconditioning induced neuroprotection were accompanied by SIRT1 activation and concomitant reduction in uncoupling protein 2 levels [239]. Ischemic brain injury leads to brain cell apoptosis by poly(ADP-ribose)polymerase (PARP) activation which depletes intracellular NAD+ [277]. NAD+ replenishment could reduce ischemic injury by OGD in in vitro cultures of primary neurons [278] and also in rat model of focal ischemia [279]. The positive effects of NAD+ repletion is mimicked by overexpression of Nampt. Nampt overexpression induced neuroprotection was dependent on SIRT1 mediated LKB1 deacetylation and AMPK activation [74]. A later study also showed the involvement of autophagy in the neuroprotection conferred by Nampt in cerebral ischemia. Overexpression of Nampt enhanced autophagy in a SIRT1 dependent manner through TSC2-mTOR-S6K1 signaling pathway [280]. Analyzing the signaling pathways modulated by resveratrol in the ischemic brain concluded an increase in Akt and p38MAPK phosphorylation and a decrease in ERK1/2 phosphorylation. Additionally the expression of SIRT1, PGC-1α and phosphorylation of cyclic AMP-response-element-binding protein were augmented with a reduction in anti-apoptotic Bcl2 transcription [281]. A direct role for SIRT1 in stroke is evident from studies that show greater infarct size in SIRT1 knockout mice that underwent middle cerebral artery occlusion compared to wild type mice [282, 283]. The infarct volume was also affected by pharmacologic modulation of SIRT1; SIRT1 activator A3 decreased infarct volume whereas sirtinol increased the infarct volume. An increase in acetyation of p53 and NFκB explains the exacerbated injury upon SIRT1 deletion [283]. Recent reports highlight the role of SIRT1 in preserving cerebral blood flow following cerebral hypoperfusion injury [284, 285]. These vasculoprotective effects of SIRT1 are likely mediated through its deacetylation of brain endothelial nitric oxide synthase. Traumatic brain injury (TBI) is another domain where SIRT1 action comes to the limelight. SIRT1 induction seen in TBI is crucial in preventing neuronal apoptosis and this protection is lost with SIRT1 inhibition [286]. Taken together these studies indicate an indispensable role for SIRT1 activation in reversing brain damage.
Several natural products and neuroprotective agents have been shown to act through SIRT1. For instance, SIRT1 up regulation is involved in the neuroprotective effects of 2,3,5,4′-tetrahydroxystilbene-2-O-beta-D-glucoside (TSG) [287] and icarin [288] against ischemic brain injury. Epigallocatechin-3-gallate, a component of tea polyphenols conferred protection in an in vitro model of neuronal cell injury by stimulating SIRT1 and PGC-1α levels and suppressing ROS production [289]. SIRT1 is also augmented with vitamin E supplementation that alleviates oxidative damage following mild traumatic brain injury [290]. Adipokine leptin showed neuroprotection in permanent middle cerebral artery occlusion accompanied by increased SIRT1 expression [291]. Erythropoietin is yet another hormone which protects against brain injury by SIRT1 activation [292]. In fact SIRT1 behaves like a sensor that helps the cells adapt to environmental changes. Several dietary modifications which cause cellular oxidative imbalances have been linked with changes in SIRT1 levels. For example a high fat diet diminished SIRT1 expression in hippocampus and cerebral cortex which was reversed by vitamin E supplementation [293]. SIRT1 expression in hippocampus was reduced in mild traumatic brain injury and was restored by omega-3 fatty acids supplementation [294]. Similar to hearts, caloric restriction offers protection against ischemia-induced neurodegeneration. Rats subjected to short term food restriction displayed improved recovery in terms of spatial learning and memory following ischemia [295]. However the role of sirtuins in these protective effects is yet to be determined.
The SIRT1 protective effects are implicated in neurodegenerative diseases like Alzheimer’s [296, 297], Parkinson’s [298], Huntington’s disease [299], Amyotrophic lateral sclerosis [300], Multiple sclerosis [301] and prion diseases [302]. Alzheimer’s and Parkinson’s are often associated with axonal degeneration which is an active process of self-destruction. Rapid Wallerian degeneration is observed in axons and their synapses distal to an injury whereas Wallerian degeneration slow (wlds) mice are protected from axonal degeneration [303]. Nicotinamide mononucleotide adenylyltransferase1 (Nmnat1) is a key enzyme involved in the NAD biosynthetic pathway in the nucleus. An increased Nmnat1 activity and SIRT1 activation is accounted for the axonal protection in wlds mice [274]. These studies clearly underscore a prominent role for SIRT1 in conferring neuroprotection. However SIRT1 also sensitizes neurons to oxidative damage by deacetylating IRS-2 and reducing activation of the Ras/ERK1/2 pathway, hinting at a pro-aging role [304].
Apart from cardio and neuroprotective effects, sirtuins have to be shown to play important role in injuries to other organs. Renal specific overexpression of SIRT1 conferred considerable protection from cisplatin induced acute kidney injury (AKI). These protective effects of SIRT1 were mediated through a reversal of peroxisome number and function, mitochondrial function, attenuation of ROS and apoptosis [305]. Conforming to the above study, SIRT1−/− mice were more susceptible to unilateral ureteral obstruction (UUO) model of kidney injury. The increased susceptibility to injury in SIRT1 deficient mice is attributed to diminished Cox2 expression and increased apoptosis and fibrosis. Opposite effects were observed with SIRT1 pharmacologic activation with resveratrol or SRT2183 [306]. Resveratrol also protected mouse proximal tubular cells from cisplatin induced renal injury through p53 deacetylation and apoptosis, further confirming the role of SIRT1 in p53 mediated apoptosis [307]. Not only does cisplatin increases acetylation of p53 but also induces acetylation of p65 subunit of NF-κB both of which accounts for the cytotoxic effects of cisplatin. Overexpression of SIRT1 in renal proximal tubule cells significantly attenuated the cytotoxic effects of cisplatin by NF-κB deacetylation [308]. Although, beneficial effects of resveratrol are well evident in diabetic nephropathy, it is also suggested to be independent of SIRT1 [309]. Resveratrol feeding in db/db mice significantly improved mitochondrial oxidative stress and associated pathologies but failed to enhance AMPK activation or SIRT1 expression in the kidney precluding a role for SIRT1 [309]. Since this study has not tested SIRT1 activity in the kidney, the role of SIRT1 in mediating resveratrol effects cannot be ruled out. In stark contrast another study clearly shows the involvement of AMPK-SIRT1-PGC1α axis in the salutary effects of resveratrol in diabetic nephropathy [310].
Sirtuin family of proteins are important physiological modulators and play critical roles in cellular homeostasis. Though SIRT1 is among the most studied sirtuins, the role of other sirtuins and small molecule modulators of sirtuins in cell survival, growth, proliferation and death is being investigated by many laboratories and the information gained will allow us to better understand the molecular processes in aging and injury. The profound effect of some of the sirtuins, such as SIRT1 and SIRT3, in regulating mitochondrial function and cellular energetics makes these proteins important players in determining outcome following cell, tissue and organ injury. Declining mitochondrial function is a hallmark of both aging and injury and therefore, further studies on the regulation of sirtuin family of proteins remain important.
Highlights.
Sirtuins are a family of evolutionary conserved proteins.
Sirtuin-mediated deacetylation of critical proteins modulate mitochondrial function.
Sirtuins play important functions in metabolism, immune response and longevity.
Sirtuins are also important in tissue injury and repair.
Acknowledgments
The corresponding author (RR) acknowledges financial support from the National Institute of General Medical Sciences (R01 GM 101927) and laboratory start up assistance from the Georgia Regents University, Augusta, GA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? The EMBO journal. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 3.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends in endocrinology and metabolism: TEM. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 5.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. Journal of molecular biology. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 7.Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circulation research. 2004;95:971–980. doi: 10.1161/01.RES.0000147557.75257.ff. [DOI] [PubMed] [Google Scholar]
- 8.Brenmoehl J, Hoeflich A. Dual control of mitochondrial biogenesis by sirtuin 1 and sirtuin 3. Mitochondrion. 2013;13:755–761. doi: 10.1016/j.mito.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell metabolism. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 12.Dai SH, Chen T, Wang YH, Zhu J, Luo P, Rao W, Yang YF, Fei Z, Jiang XF. Sirt3 attenuates hydrogen peroxide-induced oxidative stress through the preservation of mitochondrial function in HT22 cells. International journal of molecular medicine. 2014;34:1159–1168. doi: 10.3892/ijmm.2014.1876. [DOI] [PubMed] [Google Scholar]
- 13.Kiran S, Anwar T, Kiran M, Ramakrishna G. Sirtuin 7 in cell proliferation, stress and disease: Rise of the Seventh Sirtuin! Cellular signalling. 2015;27:673–682. doi: 10.1016/j.cellsig.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Maksin-Matveev A, Kanfi Y, Hochhauser E, Isak A, Cohen HY, Shainberg A. Sirtuin 6 protects the heart from hypoxic damage. Experimental cell research. 2015;330:81–90. doi: 10.1016/j.yexcr.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 15.Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, Permutt MA, Imai S. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell metabolism. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 17.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 18.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 19.Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, Yamada KA, Imai S. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell metabolism. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu TF, McCall CE. Deacetylation by SIRT1 Reprograms Inflammation and Cancer. Genes & cancer. 2013;4:135–147. doi: 10.1177/1947601913476948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klar AJ, Fogel S, Macleod K. MAR1-a Regulator of the HMa and HMalpha Loci in SACCHAROMYCES CEREVISIAE. Genetics. 1979;93:37–50. doi: 10.1093/genetics/93.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochemical and biophysical research communications. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 24.Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 25.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 26.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes & development. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 30.Pillai JB, Isbatan A, Imai S, Gupta MP. Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2alpha deacetylase activity. The Journal of biological chemistry. 2005;280:43121–43130. doi: 10.1074/jbc.M506162200. [DOI] [PubMed] [Google Scholar]
- 31.Garten A, Petzold S, Korner A, Imai S, Kiess W. Nampt: linking NAD biology, metabolism and cancer. Trends in endocrinology and metabolism: TEM. 2009;20:130–138. doi: 10.1016/j.tem.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. The Journal of biological chemistry. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 33.Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Molecular and cellular biology. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borra MT, O’Neill FJ, Jackson MD, Marshall B, Verdin E, Foltz KR, Denu JM. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. The Journal of biological chemistry. 2002;277:12632–12641. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- 35.Rafty LA, Schmidt MT, Perraud AL, Scharenberg AM, Denu JM. Analysis of O-acetyl-ADP-ribose as a target for Nudix ADP-ribose hydrolases. The Journal of biological chemistry. 2002;277:47114–47122. doi: 10.1074/jbc.M208997200. [DOI] [PubMed] [Google Scholar]
- 36.Tong L, Denu JM. Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochimica et biophysica acta. 2010;1804:1617–1625. doi: 10.1016/j.bbapap.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liou GG, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121:515–527. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 38.Chen D, Vollmar M, Rossi MN, Phillips C, Kraehenbuehl R, Slade D, Mehrotra PV, von Delft F, Crosthwaite SK, Gileadi O, Denu JM, Ahel I. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. The Journal of biological chemistry. 2011;286:13261–13271. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes & development. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 40.Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. The Journal of biological chemistry. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 41.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 42.Li R, Gu J, Chen YY, Xiao CL, Wang LW, Zhang ZP, Bi LJ, Wei HP, Wang XD, Deng JY, Zhang XE. CobB regulates Escherichia coli chemotaxis by deacetylating the response regulator CheY. Mol Microbiol. 2010;76:1162–1174. doi: 10.1111/j.1365-2958.2010.07125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Q, Wood TK. Protein acetylation in prokaryotes increases stress resistance. Biochemical and biophysical research communications. 2011;410:846–851. doi: 10.1016/j.bbrc.2011.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- 45.Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- 46.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nature structural biology. 2001;8:621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 47.Sanders BD, Jackson B, Marmorstein R. Structural basis for sirtuin function: what we know and what we don’t. Biochimica et biophysica acta. 2010;1804:1604–1616. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochemical and biophysical research communications. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 49.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Molecular biology of the cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. The Journal of biological chemistry. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 51.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 54.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Molecular cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 55.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 56.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 57.Kauppinen A, Suuronen T, Ojala J, Kaarniranta K, Salminen A. Antagonistic crosstalk between NF-kappaB and SIRT1 in the regulation of inflammation and metabolic disorders. Cellular signalling. 2013;25:1939–1948. doi: 10.1016/j.cellsig.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 58.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) The Journal of biological chemistry. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 59.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} The Journal of biological chemistry. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 60.Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Molecular cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 61.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324:1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 62.Senawong T, Peterson VJ, Avram D, Shepherd DM, Frye RA, Minucci S, Leid M. Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. The Journal of biological chemistry. 2003;278:43041–43050. doi: 10.1074/jbc.M307477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, Hruby H, Jung M, Verdin E, Ott M. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS biology. 2005;3:e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, Chiang CM, Veenstra TD, Kemper JK. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. The Journal of biological chemistry. 2010;285:33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Molecular cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 66.Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu SY, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell metabolism. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mattagajasingh I, Kim CS, Naqvi A, Yamamori T, Hoffman TA, Jung SB, DeRicco J, Kasuno K, Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Molecular and cellular biology. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guarani V, Deflorian G, Franco CA, Kruger M, Phng LK, Bentley K, Toussaint L, Dequiedt F, Mostoslavsky R, Schmidt MH, Zimmermann B, Brandes RP, Mione M, Westphal CH, Braun T, Zeiher AM, Gerhardt H, Dimmeler S, Potente M. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473:234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jeong J, Juhn K, Lee H, Kim SH, Min BH, Lee KM, Cho MH, Park GH, Lee KH. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Experimental & molecular medicine. 2007;39:8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 71.Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Molecular cell. 2010;39:247–258. doi: 10.1016/j.molcel.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 72.Li K, Casta A, Wang R, Lozada E, Fan W, Kane S, Ge Q, Gu W, Orren D, Luo J. Regulation of WRN protein cellular localization and enzymatic activities by SIRT1-mediated deacetylation. The Journal of biological chemistry. 2008;283:7590–7598. doi: 10.1074/jbc.M709707200. [DOI] [PubMed] [Google Scholar]
- 73.Yuan Z, Seto E. A functional link between SIRT1 deacetylase and NBS1 in DNA damage response. Cell cycle. 2007;6:2869–2871. doi: 10.4161/cc.6.23.5026. [DOI] [PubMed] [Google Scholar]
- 74.Wang P, Xu TY, Guan YF, Tian WW, Viollet B, Rui YC, Zhai QW, Su DF, Miao CY. Nicotinamide phosphoribosyltransferase protects against ischemic stroke through SIRT1-dependent adenosine monophosphate-activated kinase pathway. Annals of neurology. 2011;69:360–374. doi: 10.1002/ana.22236. [DOI] [PubMed] [Google Scholar]
- 75.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hirschey MD, Shimazu T, Capra JA, Pollard KS, Verdin E. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging. 2011;3:635–642. doi: 10.18632/aging.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mao B, Zhao G, Lv X, Chen HZ, Xue Z, Yang B, Liu DP, Liang CC. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. The international journal of biochemistry & cell biology. 2011;43:1573–1581. doi: 10.1016/j.biocel.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 78.Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, Powell MJ, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, Pestell TG, Wang F, Quong AA, Wang C, Pestell RG. Hormonal control of androgen receptor function through SIRT1. Molecular and cellular biology. 2006;26:8122–8135. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 80.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 82.Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, Koomen J, Chen J, Lane WS, Seto E. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Molecular and cellular biology. 2011;31:4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peng L, Ling H, Yuan Z, Fang B, Bloom G, Fukasawa K, Koomen J, Chen J, Lane WS, Seto E. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Molecular and cellular biology. 2012;32:2823–2836. doi: 10.1128/MCB.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Zhang M, Dong H, Yong S, Li X, Olashaw N, Kruk PA, Cheng JQ, Bai W, Chen J, Nicosia SV, Zhang X. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28:445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 85.Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Molecular and cellular biology. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xue Z, Lv X, Song W, Wang X, Zhao GN, Wang WT, Xiong J, Mao BB, Yu W, Yang B, Wu J, Zhou LQ, Hao DL, Dong WJ, Liu DP, Liang CC. SIRT1 deacetylates SATB1 to facilitate MAR HS2-MAR epsilon interaction and promote epsilon-globin expression. Nucleic acids research. 2012;40:4804–4815. doi: 10.1093/nar/gks064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xia J, Wu X, Yang Y, Zhao Y, Fang M, Xie W, Wang H, Xu Y. SIRT1 deacetylates RFX5 and antagonizes repression of collagen type I (COL1A2) transcription in smooth muscle cells. Biochemical and biophysical research communications. 2012;428:264–270. doi: 10.1016/j.bbrc.2012.10.043. [DOI] [PubMed] [Google Scholar]
- 88.Madabushi A, Hwang BJ, Jin J, Lu AL. Histone deacetylase SIRT1 modulates and deacetylates DNA base excision repair enzyme thymine DNA glycosylase. The Biochemical journal. 2013;456:89–98. doi: 10.1042/BJ20130670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang RH, Xu X, Kim HS, Xiao Z, Deng CX. SIRT1 deacetylates FOXA2 and is critical for Pdx1 transcription and beta-cell formation. International journal of biological sciences. 2013;9:934–946. doi: 10.7150/ijbs.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang H, Lee SM, Gao B, Zhang J, Fang D. Histone deacetylase sirtuin 1 deacetylates IRF1 protein and programs dendritic cells to control Th17 protein differentiation during autoimmune inflammation. The Journal of biological chemistry. 2013;288:37256–37266. doi: 10.1074/jbc.M113.527531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rabadi MM, Xavier S, Vasko R, Kaur K, Goligorksy MS, Ratliff BB. High-mobility group box 1 is a novel deacetylation target of Sirtuin1. Kidney international. 2015;87:95–108. doi: 10.1038/ki.2014.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hallows WC, Yu W, Denu JM. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. The Journal of biological chemistry. 2012;287:3850–3858. doi: 10.1074/jbc.M111.317404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tang S, Huang G, Fan W, Chen Y, Ward JM, Xu X, Xu Q, Kang A, McBurney MW, Fargo DC, Hu G, Baumgart-Vogt E, Zhao Y, Li X. SIRT1-mediated deacetylation of CRABPII regulates cellular retinoic acid signaling and modulates embryonic stem cell differentiation. Molecular cell. 2014;55:843–855. doi: 10.1016/j.molcel.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang RH, Lahusen TJ, Chen Q, Xu X, Jenkins LM, Leo E, Fu H, Aladjem M, Pommier Y, Appella E, Deng CX. SIRT1 deacetylates TopBP1 and modulates intra-S-phase checkpoint and DNA replication origin firing. International journal of biological sciences. 2014;10:1193–1202. doi: 10.7150/ijbs.11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guan D, Lim JH, Peng L, Liu Y, Lam M, Seto E, Kao HY. Deacetylation of the tumor suppressor protein PML regulates hydrogen peroxide-induced cell death. Cell death & disease. 2014;5:e1340. doi: 10.1038/cddis.2014.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes & development. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Eskandarian HA, Impens F, Nahori MA, Soubigou G, Coppee JY, Cossart P, Hamon MA. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341:1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- 98.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Molecular cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 99.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Molecular biology of the cell. 2009;20:801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 101.Jin YH, Kim YJ, Kim DW, Baek KH, Kang BY, Yeo CY, Lee KY. Sirt2 interacts with 14-3-3 beta/gamma and down-regulates the activity of p53. Biochemical and biophysical research communications. 2008;368:690–695. doi: 10.1016/j.bbrc.2008.01.114. [DOI] [PubMed] [Google Scholar]
- 102.Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Molecular cell. 2008;32:449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. Journal of cell science. 2010;123:4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- 104.Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, Xiong Y, Guan KL, Zhao S. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Molecular cell. 2011;43:33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beirowski B, Gustin J, Armour SM, Yamamoto H, Viader A, North BJ, Michan S, Baloh RH, Golden JP, Schmidt RE, Sinclair DA, Auwerx J, Milbrandt J. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E952–961. doi: 10.1073/pnas.1104969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang H, Park SH, Pantazides BG, Karpiuk O, Warren MD, Hardy CW, Duong DM, Park SJ, Kim HS, Vassilopoulos A, Seyfried NT, Johnsen SA, Gius D, Yu DS. SIRT2 directs the replication stress response through CDK9 deacetylation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13546–13551. doi: 10.1073/pnas.1301463110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seo KS, Park JH, Heo JY, Jing K, Han J, Min KN, Kim C, Koh GY, Lim K, Kang GY, Uee Lee J, Yim YH, Shong M, Kwak TH, Kweon GR. SIRT2 regulates tumour hypoxia response by promoting HIF-1alpha hydroxylation. Oncogene. 2015;34:1354–1362. doi: 10.1038/onc.2014.76. [DOI] [PubMed] [Google Scholar]
- 108.Wang YP, Zhou LS, Zhao YZ, Wang SW, Chen LL, Liu LX, Ling ZQ, Hu FJ, Sun YP, Zhang JY, Yang C, Yang Y, Xiong Y, Guan KL, Ye D. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. The EMBO journal. 2014;33:1304–1320. doi: 10.1002/embj.201387224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu Y, Li F, Lv L, Li T, Zhou X, Deng CX, Guan KL, Lei QY, Xiong Y. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer research. 2014;74:3630–3642. doi: 10.1158/0008-5472.CAN-13-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun YP, Xiong Y, Guan KL, Lei QY. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. The Journal of clinical investigation. 2014;124:5453–5465. doi: 10.1172/JCI76611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Belman JP, Bian RR, Habtemichael EN, Li DT, Jurczak MJ, Alcazar-Roman A, McNally LJ, Shulman GI, Bogan JS. Acetylation of TUG protein promotes the accumulation of GLUT4 glucose transporters in an insulin-responsive intracellular compartment. The Journal of biological chemistry. 2015;290:4447–4463. doi: 10.1074/jbc.M114.603977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.North BJ, Rosenberg MA, Jeganathan KB, Hafner AV, Michan S, Dai J, Baker DJ, Cen Y, Wu LE, Sauve AA, van Deursen JM, Rosenzweig A, Sinclair DA. SIRT2 induces the checkpoint kinase BubR1 to increase lifespan. The EMBO journal. 2014;33:1438–1453. doi: 10.15252/embj.201386907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, Kambach C, Becker CF, Zerweck J, Schutkowski M, Steegborn C. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nature communications. 2013;4:2327. doi: 10.1038/ncomms3327. [DOI] [PubMed] [Google Scholar]
- 114.Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell metabolism. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bharathi SS, Zhang Y, Mohsen AW, Uppala R, Balasubramani M, Schreiber E, Uechi G, Beck ME, Rardin MJ, Vockley J, Verdin E, Gibson BW, Hirschey MD, Goetzman ES. Sirtuin 3 (SIRT3) protein regulates long-chain acyl-CoA dehydrogenase by deacetylating conserved lysines near the active site. The Journal of biological chemistry. 2013;288:33837–33847. doi: 10.1074/jbc.M113.510354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Finley LW, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP, Haigis MC. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PloS one. 2011;6:e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Molecular and cellular biology. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell metabolism. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 119.Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. The Journal of biological chemistry. 2012;287:14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. Journal of molecular biology. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 121.Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A, Gupta MP. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. The Journal of biological chemistry. 2010;285:3133–3144. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, Palacios OM, Montier L, Bai Y, Tong Q, Koc EC. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. The Journal of biological chemistry. 2010;285:7417–7429. doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vassilopoulos A, Pennington JD, Andresson T, Rees DM, Bosley AD, Fearnley IM, Ham A, Flynn CR, Hill S, Rose KL, Kim HS, Deng CX, Walker JE, Gius D. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxidants & redox signaling. 2014;21:551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. Journal of cell science. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 125.Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Molecular cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xue L, Xu F, Meng L, Wei S, Wang J, Hao P, Bian Y, Zhang Y, Chen Y. Acetylation-dependent regulation of mitochondrial ALDH2 activation by SIRT3 mediates acute ethanol-induced eNOS activation. FEBS letters. 2012;586:137–142. doi: 10.1016/j.febslet.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 127.Wang Z, Inuzuka H, Zhong J, Liu P, Sarkar FH, Sun Y, Wei W. Identification of acetylation-dependent regulatory mechanisms that govern the oncogenic functions of Skp2. Oncotarget. 2012;3:1294–1300. doi: 10.18632/oncotarget.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tseng AH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free radical biology & medicine. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 129.Jing E, O’Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, Bain JR, Lee KY, Verdin EM, Newgard CB, Gibson BW, Kahn CR. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62:3404–3417. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cheng Y, Ren X, Gowda AS, Shan Y, Zhang L, Yuan YS, Patel R, Wu H, Huber-Keener K, Yang JW, Liu D, Spratt TE, Yang JM. Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell death & disease. 2013;4:e731. doi: 10.1038/cddis.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Samant SA, Zhang HJ, Hong Z, Pillai VB, Sundaresan NR, Wolfgeher D, Archer SL, Chan DC, Gupta MP. SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Molecular and cellular biology. 2014;34:807–819. doi: 10.1128/MCB.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lu Z, Chen Y, Aponte AM, Battaglia V, Gucek M, Sack MN. Prolonged fasting identifies heat shock protein 10 as a Sirtuin 3 substrate: elucidating a new mechanism linking mitochondrial protein acetylation to fatty acid oxidation enzyme folding and function. The Journal of biological chemistry. 2015;290:2466–2476. doi: 10.1074/jbc.M114.606228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang H, Zhou L, Shi Q, Zhao Y, Lin H, Zhang M, Zhao S, Yang Y, Ling ZQ, Guan KL, Xiong Y, Ye D. SIRT3-dependent GOT2 acetylation status affects the malate-aspartate NADH shuttle activity and pancreatic tumor growth. The EMBO journal. 2015 doi: 10.15252/embj.201591041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Laurent G, de Boer VC, Finley LW, Sweeney M, Lu H, Schug TT, Cen Y, Jeong SM, Li X, Sauve AA, Haigis MC. SIRT4 represses peroxisome proliferator-activated receptor alpha activity to suppress hepatic fat oxidation. Molecular and cellular biology. 2013;33:4552–4561. doi: 10.1128/MCB.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, Jin W, Huang HH, Chen X. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochemical and biophysical research communications. 2013;441:191–195. doi: 10.1016/j.bbrc.2013.10.033. [DOI] [PubMed] [Google Scholar]
- 138.Nakamura Y, Ogura M, Ogura K, Tanaka D, Inagaki N. SIRT5 deacetylates and activates urate oxidase in liver mitochondria of mice. FEBS letters. 2012;586:4076–4081. doi: 10.1016/j.febslet.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 139.Zhang Y, Bharathi SS, Rardin MJ, Uppala R, Verdin E, Gibson BW, Goetzman ES. SIRT3 and SIRT5 Regulate the Enzyme Activity and Cardiolipin Binding of Very Long-Chain Acyl-CoA Dehydrogenase. PloS one. 2015;10:e0122297. doi: 10.1371/journal.pone.0122297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, Hang HC, Hao Q, Lin H. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dominy JE, Jr, Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, Ruan HB, Feldman J, Pierce K, Mostoslavsky R, Denu JM, Clish CB, Yang X, Shulman GI, Gygi SP, Puigserver P. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Molecular cell. 2012;48:900–913. doi: 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nature communications. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, Struhl K, Garcia BA, Gozani O, Li W, Chua KF. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen S, Seiler J, Santiago-Reichelt M, Felbel K, Grummt I, Voit R. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Molecular cell. 2013;52:303–313. doi: 10.1016/j.molcel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 147.Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO, Schoonjans K, Auwerx J. A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell metabolism. 2014;20:856–869. doi: 10.1016/j.cmet.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 148.Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circulation research. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 149.McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Molecular and cellular biology. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Molecular cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 152.Nakagawa T, Guarente L. Sirtuins at a glance. Journal of cell science. 2011;124:833–838. doi: 10.1242/jcs.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annual review of biochemistry. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 155.Kobayashi J, Antoccia A, Tauchi H, Matsuura S, Komatsu K. NBS1 and its functional role in the DNA damage response. DNA repair. 2004;3:855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 156.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 157.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 159.Kim HJ, Oh GS, Shen A, Lee SB, Choe SK, Kwon KB, Lee S, Seo KS, Kwak TH, Park R, So HS. Augmentation of NAD(+) by NQO1 attenuates cisplatin-mediated hearing impairment. Cell death & disease. 2014;5:e1292. doi: 10.1038/cddis.2014.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhang Z, Lowry SF, Guarente L, Haimovich B. Roles of SIRT1 in the acute and restorative phases following induction of inflammation. The Journal of biological chemistry. 2010;285:41391–41401. doi: 10.1074/jbc.M110.174482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Salminen A, Kauppinen A, Suuronen T, Kaarniranta K. SIRT1 longevity factor suppresses NF-kappaB-driven immune responses: regulation of aging via NF-kappaB acetylation? BioEssays : news and reviews in molecular, cellular and developmental biology. 2008;30:939–942. doi: 10.1002/bies.20799. [DOI] [PubMed] [Google Scholar]
- 162.Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. American journal of respiratory and critical care medicine. 2008;177:861–870. doi: 10.1164/rccm.200708-1269OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schug TT, Xu Q, Gao H, Peres-da-Silva A, Draper DW, Fessler MB, Purushotham A, Li X. Myeloid deletion of SIRT1 induces inflammatory signaling in response to environmental stress. Molecular and cellular biology. 2010;30:4712–4721. doi: 10.1128/MCB.00657-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shinozaki S, Chang K, Sakai M, Shimizu N, Yamada M, Tanaka T, Nakazawa H, Ichinose F, Yamada Y, Ishigami A, Ito H, Ouchi Y, Starr ME, Saito H, Shimokado K, Stamler JS, Kaneki M. Inflammatory stimuli induce inhibitory S-nitrosylation of the deacetylase SIRT1 to increase acetylation and activation of p53 and p65. Science signaling. 2014;7:ra106. doi: 10.1126/scisignal.2005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kabra N, Li Z, Chen L, Li B, Zhang X, Wang C, Yeatman T, Coppola D, Chen J. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. The Journal of biological chemistry. 2009;284:18210–18217. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, Hahn WC, Guarente LP, Sinclair DA. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PloS one. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bernier M, Paul RK, Martin-Montalvo A, Scheibye-Knudsen M, Song S, He HJ, Armour SM, Hubbard BP, Bohr VA, Wang L, Zong Y, Sinclair DA, de Cabo R. Negative regulation of STAT3 protein-mediated cellular respiration by SIRT1 protein. The Journal of biological chemistry. 2011;286:19270–19279. doi: 10.1074/jbc.M110.200311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS biology. 2006;4:e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nasrin N, Kaushik VK, Fortier E, Wall D, Pearson KJ, de Cabo R, Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PloS one. 2009;4:e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tong C, Morrison A, Mattison S, Qian S, Bryniarski M, Rankin B, Wang J, Thomas DP, Li J. Impaired SIRT1 nucleocytoplasmic shuttling in the senescent heart during ischemic stress. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2013;27:4332–4342. doi: 10.1096/fj.12-216473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gu C, Xing Y, Jiang L, Chen M, Xu M, Yin Y, Li C, Yang Z, Yu L, Ma H. Impaired cardiac SIRT1 activity by carbonyl stress contributes to aging-related ischemic intolerance. PloS one. 2013;8:e74050. doi: 10.1371/journal.pone.0074050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Molecular and cellular biology. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.de Oliveira RM, Sarkander J, Kazantsev AG, Outeiro TF. SIRT2 as a Therapeutic Target for Age-Related Disorders. Frontiers in pharmacology. 2012;3:82. doi: 10.3389/fphar.2012.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. The Journal of cell biology. 2002;158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Molecular and cellular biology. 2007;27:8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. The Journal of biological chemistry. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 180.Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes & development. 2007;21:920–928. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pillai VB, Samant S, Sundaresan NR, Raghuraman H, Kim G, Bonner MY, Arbiser JL, Walker DI, Jones DP, Gius D, Gupta MP. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nature communications. 2015;6:6656. doi: 10.1038/ncomms7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, Xu X, Li C, Wang RH, Lee J, Csibi A, Cerione R, Blenis J, Clish CB, Kimmelman A, Deng CX, Haigis MC. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer cell. 2013;23:450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell metabolism. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens RD, Ilkayeva OR, Newgard CB, Jacobson MP, Hellerstein M, Goetzman ES, Gibson BW, Verdin E. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell metabolism. 2013;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS, Lin H, Schoonjans K, Auwerx J. Metabolic characterization of a Sirt5 deficient mouse model. Scientific reports. 2013;3:2806. doi: 10.1038/srep02806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kugel S, Mostoslavsky R. Chromatin and beyond: the multitasking roles for SIRT6. Trends in biochemical sciences. 2014;39:72–81. doi: 10.1016/j.tibs.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, Clish CB, Vaitheesvaran B, Iliopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, Vazquez-Ortiz G, Jeong WI, Park O, Ki SH, Gao B, Deng CX. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell metabolism. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sharma A, Diecke S, Zhang WY, Lan F, He C, Mordwinkin NM, Chua KF, Wu JC. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. The Journal of biological chemistry. 2013;288:18439–18447. doi: 10.1074/jbc.M112.405928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes & development. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. Journal of cell science. 2009;122:489–498. doi: 10.1242/jcs.042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Molecular & cellular proteomics : MCP. 2012;11:M111 015156. doi: 10.1074/mcp.M111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E. Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. Journal of physiology and pharmacology : an official journal of the Polish Physiological Society. 2008;59(Suppl 9):201–212. [PubMed] [Google Scholar]
- 198.Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS, Lee JY, Borlak J, Nam SW. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 199.Yu H, Ye W, Wu J, Meng X, Liu RY, Ying X, Zhou Y, Wang H, Pan C, Huang W. Overexpression of sirt7 exhibits oncogenic property and serves as a prognostic factor in colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:3434–3445. doi: 10.1158/1078-0432.CCR-13-2952. [DOI] [PubMed] [Google Scholar]
- 200.Tsai YC, Greco TM, Cristea IM. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Molecular & cellular proteomics : MCP. 2014;13:73–83. doi: 10.1074/mcp.M113.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hubbi ME, Hu H, Kshitiz, Gilkes DM, Semenza GL. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. The Journal of biological chemistry. 2013;288:20768–20775. doi: 10.1074/jbc.M113.476903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, Li P, Cheng HL, Murphy AJ, Valenzuela DM, Luo H, Kapahi P, Krauss R, Mostoslavsky R, Yancopoulos GD, Alt FW, Chua KF, Chen D. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell reports. 2013;5:654–665. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, Go C, Tasaki M, Uchimura K, Kadomatsu T, Tian Z, Smolka C, Sawa T, Takeya M, Tomizawa K, Ando Y, Araki E, Akaike T, Braun T, Oike Y, Bober E, Yamagata K. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell metabolism. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 204.Sacks FM. The crucial roles of apolipoproteins E and C-III in apoB lipoprotein metabolism in normolipidemia and hypertriglyceridemia. Current opinion in lipidology. 2015;26:56–63. doi: 10.1097/MOL.0000000000000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes & development. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Whitaker R, Faulkner S, Miyokawa R, Burhenn L, Henriksen M, Wood JG, Helfand SL. Increased expression of Drosophila Sir2 extends life span in a dose-dependent manner. Aging. 2013;5:682–691. doi: 10.18632/aging.100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 208.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 209.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kim HJ, Oh GS, Choe SK, Kwak TH, Park R, So HS. NAD(+) Metabolism in Age-Related Hearing Loss. Aging Dis. 2014;5:150–159. doi: 10.14336/AD.2014.0500150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 212.Medvedik O, Lamming DW, Kim KD, Sinclair DA. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS biology. 2007;5:e261. doi: 10.1371/journal.pbio.0050261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 214.Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, Wang C, Lin SJ, Sinclair DA. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 215.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS biology. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, Oakes JA, Kennedy BK, Kaeberlein M. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- 217.Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Comment on “HST2 mediates SIR2-independent life-span extension by calorie restriction”. Science. 2006;312:1312. doi: 10.1126/science.1124608. author reply 1312. [DOI] [PubMed] [Google Scholar]
- 218.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 219.Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 220.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nature genetics. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 221.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, Criollo A, Galluzzi L, Malik SA, Vitale I, Michaud M, Madeo F, Tavernarakis N, Kroemer G. The life span-prolonging effect of sirtuin-1 is mediated by autophagy. Autophagy. 2010;6:186–188. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- 222.Banerjee KK, Ayyub C, Ali SZ, Mandot V, Prasad NG, Kolthur-Seetharam U. dSir2 in the adult fat body, but not in muscles, regulates life span in a diet-dependent manner. Cell reports. 2012;2:1485–1491. doi: 10.1016/j.celrep.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 223.Hoffmann J, Romey R, Fink C, Yong L, Roeder T. Overexpression of Sir2 in the adult fat body is sufficient to extend lifespan of male and female Drosophila. Aging. 2013;5:315–327. doi: 10.18632/aging.100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bai B, Vanhoutte PM, Wang Y. Loss-of-SIRT1 function during vascular ageing: hyperphosphorylation mediated by cyclin-dependent kinase 5. Trends in cardiovascular medicine. 2014;24:81–84. doi: 10.1016/j.tcm.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 225.Mitchell SJ, Martin-Montalvo A, Mercken EM, Palacios HH, Ward TM, Abulwerdi G, Minor RK, Vlasuk GP, Ellis JL, Sinclair DA, Dawson J, Allison DB, Zhang Y, Becker KG, Bernier M, de Cabo R. The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell reports. 2014;6:836–843. doi: 10.1016/j.celrep.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sasaki T, Maier B, Bartke A, Scrable H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging cell. 2006;5:413–422. doi: 10.1111/j.1474-9726.2006.00235.x. [DOI] [PubMed] [Google Scholar]
- 227.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. The EMBO journal. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kanfi Y, Shalman R, Peshti V, Pilosof SN, Gozlan YM, Pearson KJ, Lerrer B, Moazed D, Marine JC, de Cabo R, Cohen HY. Regulation of SIRT6 protein levels by nutrient availability. FEBS letters. 2008;582:543–548. doi: 10.1016/j.febslet.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zeng L, Yang Y, Hu Y, Sun Y, Du Z, Xie Z, Zhou T, Kong W. Age-related decrease in the mitochondrial sirtuin deacetylase Sirt3 expression associated with ROS accumulation in the auditory cortex of the mimetic aging rat model. PloS one. 2014;9:e88019. doi: 10.1371/journal.pone.0088019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tao R, Vassilopoulos A, Parisiadou L, Yan Y, Gius D. Regulation of MnSOD enzymatic activity by Sirt3 connects the mitochondrial acetylome signaling networks to aging and carcinogenesis. Antioxidants & redox signaling. 2014;20:1646–1654. doi: 10.1089/ars.2013.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Current opinion in lipidology. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Canto C, Auwerx J. Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacological reviews. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hsu CP, Zhai P, Yamamoto T, Maejima Y, Matsushima S, Hariharan N, Shao D, Takagi H, Oka S, Sadoshima J. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2010;122:2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jian B, Yang S, Chaudry IH, Raju R. Resveratrol improves cardiac contractility following trauma-hemorrhage by modulating Sirt1. Molecular medicine. 2012;18:209–214. doi: 10.2119/molmed.2011.00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. The Journal of clinical investigation. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Yang X, Cohen MV, Downey JM. Mechanism of cardioprotection by early ischemic preconditioning. Cardiovascular drugs and therapy/sponsored by the International Society of Cardiovascular Pharmacotherapy. 2010;24:225–234. doi: 10.1007/s10557-010-6236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Nadtochiy SM, Yao H, McBurney MW, Gu W, Guarente L, Rahman I, Brookes PS. SIRT1-mediated acute cardioprotection. American journal of physiology Heart and circulatory physiology. 2011;301:H1506–1512. doi: 10.1152/ajpheart.00587.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Della-Morte D, Dave KR, DeFazio RA, Bao YC, Raval AP, Perez-Pinzon MA. Resveratrol pretreatment protects rat brain from cerebral ischemic damage via a sirtuin 1-uncoupling protein 2 pathway. Neuroscience. 2009;159:993–1002. doi: 10.1016/j.neuroscience.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Mercadier JJ, Lompre AM, Wisnewsky C, Samuel JL, Bercovici J, Swynghedauw B, Schwartz K. Myosin isoenzyme changes in several models of rat cardiac hypertrophy. Circulation research. 1981;49:525–532. doi: 10.1161/01.res.49.2.525. [DOI] [PubMed] [Google Scholar]
- 241.Pillai JB, Chen M, Rajamohan SB, Samant S, Pillai VB, Gupta M, Gupta MP. Activation of SIRT1, a class III histone deacetylase, contributes to fructose feeding-mediated induction of the alpha-myosin heavy chain expression. American journal of physiology Heart and circulatory physiology. 2008;294:H1388–1397. doi: 10.1152/ajpheart.01339.2007. [DOI] [PubMed] [Google Scholar]
- 242.Hsu CP, Oka S, Shao D, Hariharan N, Sadoshima J. Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circulation research. 2009;105:481–491. doi: 10.1161/CIRCRESAHA.109.203703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Pillai VB, Sundaresan NR, Kim G, Samant S, Moreno-Vinasco L, Garcia JG, Gupta MP. Nampt secreted from cardiomyocytes promotes development of cardiac hypertrophy and adverse ventricular remodeling. American journal of physiology Heart and circulatory physiology. 2013;304:H415–426. doi: 10.1152/ajpheart.00468.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Adam T, Sharp S, Opie LH, Lecour S. Loss of cardioprotection with ischemic preconditioning in aging hearts: role of sirtuin 1? Journal of cardiovascular pharmacology and therapeutics. 2013;18:46–53. doi: 10.1177/1074248412458723. [DOI] [PubMed] [Google Scholar]
- 245.Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- 246.Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Shinmura K, Tamaki K, Bolli R. Short-term caloric restriction improves ischemic tolerance independent of opening of ATP-sensitive K+ channels in both young and aged hearts. Journal of molecular and cellular cardiology. 2005;39:285–296. doi: 10.1016/j.yjmcc.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 248.Shinmura K, Tamaki K, Bolli R. Impact of 6-mo caloric restriction on myocardial ischemic tolerance: possible involvement of nitric oxide-dependent increase in nuclear Sirt1. American journal of physiology Heart and circulatory physiology. 2008;295:H2348–2355. doi: 10.1152/ajpheart.00602.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Yamamoto T, Byun J, Zhai P, Ikeda Y, Oka S, Sadoshima J. Nicotinamide mononucleotide, an intermediate of NAD+ synthesis, protects the heart from ischemia and reperfusion. PloS one. 2014;9:e98972. doi: 10.1371/journal.pone.0098972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Shinmura K, Tamaki K, Sano M, Nakashima-Kamimura N, Wolf AM, Amo T, Ohta S, Katsumata Y, Fukuda K, Ishiwata K, Suematsu M, Adachi T. Caloric restriction primes mitochondria for ischemic stress by deacetylating specific mitochondrial proteins of the electron transport chain. Circulation research. 2011;109:396–406. doi: 10.1161/CIRCRESAHA.111.243097. [DOI] [PubMed] [Google Scholar]
- 251.Ayub A, Poulose N, Raju R. Resveratrol Improves Survival and Prolongs Life Following Hemorrhagic Shock. Molecular medicine. 2015 doi: 10.2119/molmed.2015.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Jian B, Yang S, Chaudry IH, Raju R. Resveratrol restores sirtuin 1 (SIRT1) activity and pyruvate dehydrogenase kinase 1 (PDK1) expression after hemorrhagic injury in a rat model. Molecular medicine. 2014;20:10–16. doi: 10.2119/molmed.2013.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Powell RD, Swet JH, Kennedy KL, Huynh TT, McKillop IH, Evans SL. Resveratrol attenuates hypoxic injury in a primary hepatocyte model of hemorrhagic shock and resuscitation. The journal of trauma and acute care surgery. 2014;76:409–417. doi: 10.1097/TA.0000000000000096. [DOI] [PubMed] [Google Scholar]
- 254.Wang H, Guan Y, Widlund AL, Becker LB, Baur JA, Reilly PM, Sims CA. Resveratrol ameliorates mitochondrial dysfunction but increases the risk of hypoglycemia following hemorrhagic shock. The journal of trauma and acute care surgery. 2014;77:926–933. doi: 10.1097/TA.0000000000000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yu HP, Hwang TL, Hsieh PW, Lau YT. Role of estrogen receptor-dependent upregulation of P38 MAPK/heme oxygenase 1 in resveratrol-mediated attenuation of intestinal injury after trauma-hemorrhage. Shock. 2011;35:517–523. doi: 10.1097/SHK.0b013e318209e931. [DOI] [PubMed] [Google Scholar]
- 256.Wang RH, Kim HS, Xiao C, Xu X, Gavrilova O, Deng CX. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. The Journal of clinical investigation. 2011;121:4477–4490. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circulation research. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Vinciguerra M, Santini MP, Claycomb WC, Ladurner AG, Rosenthal N. Local IGF-1 isoform protects cardiomyocytes from hypertrophic and oxidative stresses via SirT1 activity. Aging. 2010;2:43–62. doi: 10.18632/aging.100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Becatti M, Taddei N, Cecchi C, Nassi N, Nassi PA, Fiorillo C. SIRT1 modulates MAPK pathways in ischemic-reperfused cardiomyocytes. Cellular and molecular life sciences : CMLS. 2012;69:2245–2260. doi: 10.1007/s00018-012-0925-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Shalwala M, Zhu SG, Das A, Salloum FN, Xi L, Kukreja RC. Sirtuin 1 (SIRT1) activation mediates sildenafil induced delayed cardioprotection against ischemia-reperfusion injury in mice. PloS one. 2014;9:e86977. doi: 10.1371/journal.pone.0086977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zhou B, Wu LJ, Li LH, Tashiro S, Onodera S, Uchiumi F, Ikejima T. Silibinin protects against isoproterenol-induced rat cardiac myocyte injury through mitochondrial pathway after up-regulation of SIRT1. Journal of pharmacological sciences. 2006;102:387–395. doi: 10.1254/jphs.fpj06005x. [DOI] [PubMed] [Google Scholar]
- 262.Kawashima T, Inuzuka Y, Okuda J, Kato T, Niizuma S, Tamaki Y, Iwanaga Y, Kawamoto A, Narazaki M, Matsuda T, Adachi S, Takemura G, Kita T, Kimura T, Shioi T. Constitutive SIRT1 overexpression impairs mitochondria and reduces cardiac function in mice. Journal of molecular and cellular cardiology. 2011;51:1026–1036. doi: 10.1016/j.yjmcc.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 263.Liu B, Che W, Xue J, Zheng C, Tang K, Zhang J, Wen J, Xu Y. SIRT4 prevents hypoxia-induced apoptosis in H9c2 cardiomyoblast cells. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2013;32:655–662. doi: 10.1159/000354469. [DOI] [PubMed] [Google Scholar]
- 264.Liu B, Che W, Zheng C, Liu W, Wen J, Fu H, Tang K, Zhang J, Xu Y. SIRT5: a safeguard against oxidative stress-induced apoptosis in cardiomyocytes. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology. 2013;32:1050–1059. doi: 10.1159/000354505. [DOI] [PubMed] [Google Scholar]
- 265.Wan W, Powers AS, Li J, Ji L, Erikson JM, Zhang JQ. Effect of post-myocardial infarction exercise training on the renin-angiotensin-aldosterone system and cardiac function. The American journal of the medical sciences. 2007;334:265–273. doi: 10.1097/MAJ.0b013e318068b5ed. [DOI] [PubMed] [Google Scholar]
- 266.Godfrey R, Theologou T, Dellegrottaglie S, Binukrishnan S, Wright J, Whyte G, Ellison G. The effect of high-intensity aerobic interval training on postinfarction left ventricular remodelling. BMJ case reports. 2013;2013 doi: 10.1136/bcr-2012-007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Jiang HK, Miao Y, Wang YH, Zhao M, Feng ZH, Yu XJ, Liu JK, Zang WJ. Aerobic interval training protects against myocardial infarction-induced oxidative injury by enhancing antioxidase system and mitochondrial biosynthesis. Clinical and experimental pharmacology & physiology. 2014;41:192–201. doi: 10.1111/1440-1681.12211. [DOI] [PubMed] [Google Scholar]
- 268.Zhou X, Chen M, Zeng X, Yang J, Deng H, Yi L, Mi MT. Resveratrol regulates mitochondrial reactive oxygen species homeostasis through Sirt3 signaling pathway in human vascular endothelial cells. Cell death & disease. 2014;5:e1576. doi: 10.1038/cddis.2014.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Park ES, Kang JC, Jang YC, Park JS, Jang SY, Kim DE, Kim B, Shin HS. Cardioprotective effects of rhamnetin in H9c2 cardiomyoblast cells under H(2)O(2)-induced apoptosis. Journal of ethnopharmacology. 2014;153:552–560. doi: 10.1016/j.jep.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 270.Porter GA, Urciuoli WR, Brookes PS, Nadtochiy SM. SIRT3 deficiency exacerbates ischemia-reperfusion injury: implication for aged hearts. American journal of physiology Heart and circulatory physiology. 2014;306:H1602–1609. doi: 10.1152/ajpheart.00027.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Dasuri K, Zhang L, Keller JN. Oxidative stress, neurodegeneration, and the balance of protein degradation and protein synthesis. Free radical biology & medicine. 2013;62:170–185. doi: 10.1016/j.freeradbiomed.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 272.Raval AP, Lin HW, Dave KR, Defazio RA, Della Morte D, Kim EJ, Perez-Pinzon MA. Resveratrol and ischemic preconditioning in the brain. Current medicinal chemistry. 2008;15:1545–1551. doi: 10.2174/092986708784638861. [DOI] [PubMed] [Google Scholar]
- 273.Jaber S, Polster BM. Idebenone and neuroprotection: antioxidant, pro-oxidant, or electron carrier? Journal of bioenergetics and biomembranes. 2015;47:111–118. doi: 10.1007/s10863-014-9571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Araki T, Sasaki Y, Milbrandt J. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 275.Hassen GW, Tian D, Ding D, Bergold PJ. A new model of ischemic preconditioning using young adult hippocampal slice cultures. Brain research Brain research protocols. 2004;13:135–143. doi: 10.1016/j.brainresprot.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 276.Raval AP, Dave KR, Perez-Pinzon MA. Resveratrol mimics ischemic preconditioning in the brain. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2006;26:1141–1147. doi: 10.1038/sj.jcbfm.9600262. [DOI] [PubMed] [Google Scholar]
- 277.Endres M, Wang ZQ, Namura S, Waeber C, Moskowitz MA. Ischemic brain injury is mediated by the activation of poly(ADP-ribose)polymerase. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 278.Wang S, Xing Z, Vosler PS, Yin H, Li W, Zhang F, Signore AP, Stetler RA, Gao Y, Chen J. Cellular NAD replenishment confers marked neuroprotection against ischemic cell death: role of enhanced DNA repair. Stroke; a journal of cerebral circulation. 2008;39:2587–2595. doi: 10.1161/STROKEAHA.107.509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Ying W, Wei G, Wang D, Wang Q, Tang X, Shi J, Zhang P, Lu H. Intranasal administration with NAD+ profoundly decreases brain injury in a rat model of transient focal ischemia. Frontiers in bioscience : a journal and virtual library. 2007;12:2728–2734. doi: 10.2741/2267. [DOI] [PubMed] [Google Scholar]
- 280.Wang P, Guan YF, Du H, Zhai QW, Su DF, Miao CY. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy. 2012;8:77–87. doi: 10.4161/auto.8.1.18274. [DOI] [PubMed] [Google Scholar]
- 281.Shin JA, Lee KE, Kim HS, Park EM. Acute resveratrol treatment modulates multiple signaling pathways in the ischemic brain. Neurochemical research. 2012;37:2686–2696. doi: 10.1007/s11064-012-0858-2. [DOI] [PubMed] [Google Scholar]
- 282.Liu AJ, Guo JM, Liu W, Su FY, Zhai QW, Mehta JL, Wang WZ, Su DF. Involvement of arterial baroreflex in the protective effect of dietary restriction against stroke. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2013;33:906–913. doi: 10.1038/jcbfm.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Hernandez-Jimenez M, Hurtado O, Cuartero MI, Ballesteros I, Moraga A, Pradillo JM, McBurney MW, Lizasoain I, Moro MA. Silent information regulator 1 protects the brain against cerebral ischemic damage. Stroke; a journal of cerebral circulation. 2013;44:2333–2337. doi: 10.1161/STROKEAHA.113.001715. [DOI] [PubMed] [Google Scholar]
- 284.Hattori Y, Okamoto Y, Maki T, Yamamoto Y, Oishi N, Yamahara K, Nagatsuka K, Takahashi R, Kalaria RN, Fukuyama H, Kinoshita M, Ihara M. Silent information regulator 2 homolog 1 counters cerebral hypoperfusion injury by deacetylating endothelial nitric oxide synthase. Stroke; a journal of cerebral circulation. 2014;45:3403–3411. doi: 10.1161/STROKEAHA.114.006265. [DOI] [PubMed] [Google Scholar]
- 285.Hattori Y, Okamoto Y, Nagatsuka K, Takahashi R, Kalaria RN, Kinoshita M, Ihara M. SIRT1 attenuates severe ischemic damage by preserving cerebral blood flow. Neuroreport. 2015;26:113–117. doi: 10.1097/WNR.0000000000000308. [DOI] [PubMed] [Google Scholar]
- 286.Zhao Y, Luo P, Guo Q, Li S, Zhang L, Zhao M, Xu H, Yang Y, Poon W, Fei Z. Interactions between SIRT1 and MAPK/ERK regulate neuronal apoptosis induced by traumatic brain injury in vitro and in vivo. Experimental neurology. 2012;237:489–498. doi: 10.1016/j.expneurol.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 287.Wang T, Gu J, Wu PF, Wang F, Xiong Z, Yang YJ, Wu WN, Dong LD, Chen JG. Protection by tetrahydroxystilbene glucoside against cerebral ischemia: involvement of JNK, SIRT1, and NF-kappaB pathways and inhibition of intracellular ROS/RNS generation. Free radical biology & medicine. 2009;47:229–240. doi: 10.1016/j.freeradbiomed.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 288.Zhu HR, Wang ZY, Zhu XL, Wu XX, Li EG, Xu Y. Icariin protects against brain injury by enhancing SIRT1-dependent PGC-1alpha expression in experimental stroke. Neuropharmacology. 2010;59:70–76. doi: 10.1016/j.neuropharm.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 289.Ye Q, Ye L, Xu X, Huang B, Zhang X, Zhu Y, Chen X. Epigallocatechin-3-gallate suppresses 1-methyl-4-phenyl-pyridine-induced oxidative stress in PC12 cells via the SIRT1/PGC-1alpha signaling pathway. BMC complementary and alternative medicine. 2012;12:82. doi: 10.1186/1472-6882-12-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Aiguo W, Zhe Y, Gomez-Pinilla F. Vitamin E protects against oxidative damage and learning disability after mild traumatic brain injury in rats. Neurorehabilitation and neural repair. 2010;24:290–298. doi: 10.1177/1545968309348318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Avraham Y, Davidi N, Porat M, Chernoguz D, Magen I, Vorobeiv L, Berry EM, Leker RR. Leptin reduces infarct size in association with enhanced expression of CB2, TRPV1, SIRT-1 and leptin receptor. Current neurovascular research. 2010;7:136–143. doi: 10.2174/156720210791184943. [DOI] [PubMed] [Google Scholar]
- 292.Hou J, Wang S, Shang YC, Chong ZZ, Maiese K. Erythropoietin employs cell longevity pathways of SIRT1 to foster endothelial vascular integrity during oxidant stress. Current neurovascular research. 2011;8:220–235. doi: 10.2174/156720211796558069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wu A, Ying Z, Gomez-Pinilla F. Oxidative stress modulates Sir2alpha in rat hippocampus and cerebral cortex. The European journal of neuroscience. 2006;23:2573–2580. doi: 10.1111/j.1460-9568.2006.04807.x. [DOI] [PubMed] [Google Scholar]
- 294.Wu A, Ying Z, Gomez-Pinilla F. Omega-3 fatty acids supplementation restores mechanisms that maintain brain homeostasis in traumatic brain injury. Journal of neurotrauma. 2007;24:1587–1595. doi: 10.1089/neu.2007.0313. [DOI] [PubMed] [Google Scholar]
- 295.Roberge MC, Hotte-Bernard J, Messier C, Plamondon H. Food restriction attenuates ischemia-induced spatial learning and memory deficits despite extensive CA1 ischemic injury. Behavioural brain research. 2008;187:123–132. doi: 10.1016/j.bbr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 296.Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. The Journal of biological chemistry. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 297.Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C, Meyers D, Cole PA, Ott M, Gan L. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Pallas M, Pizarro JG, Gutierrez-Cuesta J, Crespo-Biel N, Alvira D, Tajes M, Yeste-Velasco M, Folch J, Canudas AM, Sureda FX, Ferrer I, Camins A. Modulation of SIRT1 expression in different neurodegenerative models and human pathologies. Neuroscience. 2008;154:1388–1397. doi: 10.1016/j.neuroscience.2008.04.065. [DOI] [PubMed] [Google Scholar]
- 299.Ho DJ, Calingasan NY, Wille E, Dumont M, Beal MF. Resveratrol protects against peripheral deficits in a mouse model of Huntington’s disease. Experimental neurology. 2010;225:74–84. doi: 10.1016/j.expneurol.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 300.Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, Puigserver P, Sinclair DA, Tsai LH. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. The EMBO journal. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Shindler KS, Ventura E, Dutt M, Elliott P, Fitzgerald DC, Rostami A. Oral resveratrol reduces neuronal damage in a model of multiple sclerosis. Journal of neuro-ophthalmology : the official journal of the North American Neuro-Ophthalmology Society. 2010;30:328–339. doi: 10.1097/WNO.0b013e3181f7f833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Seo JS, Moon MH, Jeong JK, Seol JW, Lee YJ, Park BH, Park SY. SIRT1, a histone deacetylase, xregulates prion protein-induced neuronal cell death. Neurobiology of aging. 2012;33:1110–1120. doi: 10.1016/j.neurobiolaging.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 303.Mack TG, Reiner M, Beirowski B, Mi W, Emanuelli M, Wagner D, Thomson D, Gillingwater T, Court F, Conforti L, Fernando FS, Tarlton A, Andressen C, Addicks K, Magni G, Ribchester RR, Perry VH, Coleman MP. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nature neuroscience. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 304.Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell metabolism. 2008;8:38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Hasegawa K, Wakino S, Yoshioka K, Tatematsu S, Hara Y, Minakuchi H, Sueyasu K, Washida N, Tokuyama H, Tzukerman M, Skorecki K, Hayashi K, Itoh H. Kidney-specific overexpression of Sirt1 protects against acute kidney injury by retaining peroxisome function. The Journal of biological chemistry. 2010;285:13045–13056. doi: 10.1074/jbc.M109.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.He W, Wang Y, Zhang MZ, You L, Davis LS, Fan H, Yang HC, Fogo AB, Zent R, Harris RC, Breyer MD, Hao CM. Sirt1 activation protects the mouse renal medulla from oxidative injury. The Journal of clinical investigation. 2010;120:1056–1068. doi: 10.1172/JCI41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Kim DH, Jung YJ, Lee JE, Lee AS, Kang KP, Lee S, Park SK, Han MK, Lee SY, Ramkumar KM, Sung MJ, Kim W. SIRT1 activation by resveratrol ameliorates cisplatin-induced renal injury through deacetylation of p53. American journal of physiology Renal physiology. 2011;301:F427–435. doi: 10.1152/ajprenal.00258.2010. [DOI] [PubMed] [Google Scholar]
- 308.Jung YJ, Lee JE, Lee AS, Kang KP, Lee S, Park SK, Lee SY, Han MK, Kim DH, Kim W. SIRT1 overexpression decreases cisplatin-induced acetylation of NF-kappaB p65 subunit and cytotoxicity in renal proximal tubule cells. Biochemical and biophysical research communications. 2012;419:206–210. doi: 10.1016/j.bbrc.2012.01.148. [DOI] [PubMed] [Google Scholar]
- 309.Kitada M, Kume S, Imaizumi N, Koya D. Resveratrol improves oxidative stress and protects against diabetic nephropathy through normalization of Mn-SOD dysfunction in AMPK/SIRT1-independent pathway. Diabetes. 2011;60:634–643. doi: 10.2337/db10-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kim MY, Lim JH, Youn HH, Hong YA, Yang KS, Park HS, Chung S, Ko SH, Shin SJ, Choi BS, Kim HW, Kim YS, Lee JH, Chang YS, Park CW. Resveratrol prevents renal lipotoxicity and inhibits mesangial cell glucotoxicity in a manner dependent on the AMPK-SIRT1-PGC1alpha axis in db/db mice. Diabetologia. 2013;56:204–217. doi: 10.1007/s00125-012-2747-2. [DOI] [PubMed] [Google Scholar]