Summary
Members of the miR-290 family are the most abundantly expressed microRNAs (miRNAs) in mouse embryonic stem cells (ESCs). They regulate aspects of differentiation, pluripotency, and proliferation of ESCs, but the molecular program that they control has not been fully delineated. In the absence of Dicer, ESCs fail to express mature miR-290 miRNAs and have selective aberrant overexpression of Hoxa, Hoxb, Hoxc, and Hoxd genes essential for body plan patterning during embryogenesis, but they do not undergo a full differentiation program. Introduction of mature miR-291 into DCR−/− ESCs restores Hox gene silencing. This was attributed to the unexpected regulation of Polycomb-mediated gene targeting by miR-291. We identified the methyltransferase Ash1l as a pivotal target of miR-291 mediating this effect. Collectively, our data shed light on the role of Dicer in ESC homeostasis by revealing a facet of molecular regulation by the miR-290 family.
Highlights
-
•
Silencing of Hox genes in ESCs is defective in the absence of Dicer
-
•
A member of the miR-290 family is sufficient to rescue the Hox gene-silencing defect
-
•
There is widespread Polycomb deregulation in Dicer-deficient ESCs
-
•
miR-290 can restore Polycomb localization by regulating Ash1l
Muljo, Lenardo, and colleagues find that in Dicer-deficient mouse embryonic stem cells (mESCs), there is reduced Polycomb Repressive Complex 2 binding and increased gene expression at many loci, including the Hoxa–Hoxd gene cluster. These defects in mESC-fate programming can be rescued by the miR-290 family and to a lesser extent by knockdown of Ash1l, a putative target of miR-290.
Introduction
Mouse embryonic stem cells (ESCs) that lack microRNAs (miRNAs) due to Dicer1 or Dgcr8 deficiency do not proliferate well and display severe differentiation defects (Kanellopoulou et al., 2005, Murchison et al., 2005, Wang et al., 2008). The most highly expressed miRNAs in mouse ESCs belong to the miR-290 family, a cluster of nine miRNAs (also referred to as miR-290∼295), six of which share the same “seed” sequence (Houbaviy et al., 2003). The orthologous human families are miR-302 and miR-371 (Suh et al., 2004). In mice, the miR-290 cluster is transcribed from a single locus on chromosome 7 by the core ESC transcriptional network (Marson et al., 2008) and can rescue defective proliferation in ESCs that lack miRNAs (Wang et al., 2008). While the importance of the miR-290 family is clear, how it contributes to the gene expression program in ESCs is not fully known.
The Hox family of transcription factors governs the anterior to posterior axial body plan of vertebrates (Pearson et al., 2005). In mouse and human, the Hox genes are found in four chromosomal clusters (A, B, C, and D). Hox genes are transcriptionally inactive in ESCs due to the action of Polycomb repressive complexes (PRC) (Bracken et al., 2006, Lee et al., 2006), but the role, if any, of miRNAs in this process has not been established.
Polycomb group (PcG) proteins are transcriptional repressors that regulate embryonic development and function in ESC pluripotency and induced pluripotent stem cell (iPSC) generation (Bernstein et al., 2006, Boyer et al., 2006, Onder et al., 2012). There are two Polycomb complexes, PRC1 and PRC2, that differ biochemically (Di Croce and Helin, 2013). PRC2 catalyzes the trimethylation of histone H3K27 (H3K27me3), which is recognized by PRC1, although PRC1 can be recruited to chromatin independently of PRC2 and H3K27me3 (Schwartz and Pirrotta, 2014). Overall, despite extensive study, it remains unclear how Polycomb repressive chromatin domains are established in ESCs and reversed during development to allow the expression of differentiation genes.
Both PcG proteins and Dicer are required for ESC proliferation, pluripotency, and differentiation and play key roles in development, but the interplay between the two has not been studied. We observed that in DCR−/− ESCs, Hox genes, which are Polycomb targets, were upregulated, which in turn led to the finding that miR-290 is required for efficient gene repression involving Polycomb targeting.
Results
Collectively, miR-290 miRNAs with the seed sequence 5′-AAGUGC-3′ account for ∼70% of mature miRNAs expressed in ESCs, and these are undetectable in DCR−/− ESCs by Nanostring analysis (Figure S1A) (Calabrese et al., 2007, Houbaviy et al., 2003). To determine the role of the miR-290 family in ESC gene regulation, we transfected a synthetic miR-291a-3p mimic (abbreviated as miR-291 hereafter) into DCR−/− ESCs and performed transcriptome sequencing analysis (RNA-seq). Our RNA-seq analysis revealed that genes belonging to the Hoxa, Hoxb, and Hoxd gene clusters were among the most differentially expressed in DCR−/− ESCs (Figure 1A; Table S1). Transfection of miR-291 dramatically restored Hox gene silencing (Figure 1A). In contrast, previously characterized miR-290 targets, such as Cdkn1a and Rbl2 (Wang et al., 2008) were modestly regulated (Figures 1A and S1B).
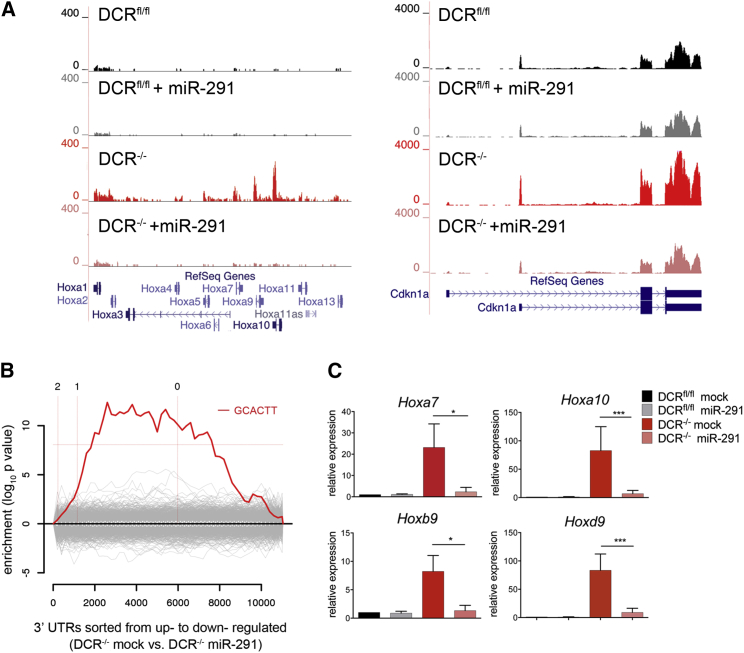
Figure 1.
miR-291 Restores Hox Gene Repression in DCR−/− ESCs
(A) Genome browser screenshot of the RNA-seq reads aligning to the Hoxa and Cdkn1a loci in DCRfl/fl and DCR−/− ESCs ± miR-291.
(B) Sylamer analysis of motifs enriched in the 3′ UTRs of differentially expressed mRNAs after transfection with miR-291. Vertical red dashed lines mark the cut-off for log2-fold change of 2, 1, and 0 as indicated.
(C) qRT-PCR for representative Hox genes, in DCRfl/fl and DCR−/− cells before and after transfection with miR-291. Each bar represents the mean ± SD of three independent experiments.
See also Figure S1.
To demonstrate that miR-291 transfection mimics physiological levels, we measured its relative expression by qRT-PCR. miR-291 levels in transfected DCR−/− ESCs were 6-fold higher than in untransfected DCRfl/fl cells (Figure S1C). Considering that miR-291a is one of six miRNAs with the same seed sequence, some of which are more highly expressed in ESCs (Figure S1A), the miR-291 concentration after transfection in DCR−/− ESCs was actually at or below endogenous aggregate levels for the whole miR-290 family. Thus the observed repression of Hox genes was not due to overexpression of miR-291. In addition, transfection of miR-291 had little effect on the transcriptome of DCRfl/fl cells—only four genes exhibited a greater than 2-fold change (data not shown). Also, the only motif enriched in the 3′ UTRs of transcripts regulated by miR-291 by Sylamer analysis (van Dongen et al., 2008) was complementary to the miR-290 seed sequence, thereby indicating specificity.
We confirmed the suppression of representative Hox genes, including Hoxa7, Hoxa10, Hoxb9, and Hoxd9 upon miR-291 transfection by qRT-PCR (Figure 1C). Similarly, transfection of miR-294, another member of the miR-290 family, potently reduced Hox gene transcripts (Figure S1D). Thus, the miR-290 family is a regulator of Hox gene expression in ESCs.
Hox genes are activated during ESC differentiation. However, Hox gene overexpression did not appear to be the consequence of a broad program of differentiation of DCR−/− ESCs. For example, we did not observe downregulation of the core pluripotency factors Oct4, Sox2, Klf4, and Nanog nor upregulation of differentiation markers such as Brachyury, Fgf5, Gata4, and Gata6 (Figures 2A and S2).
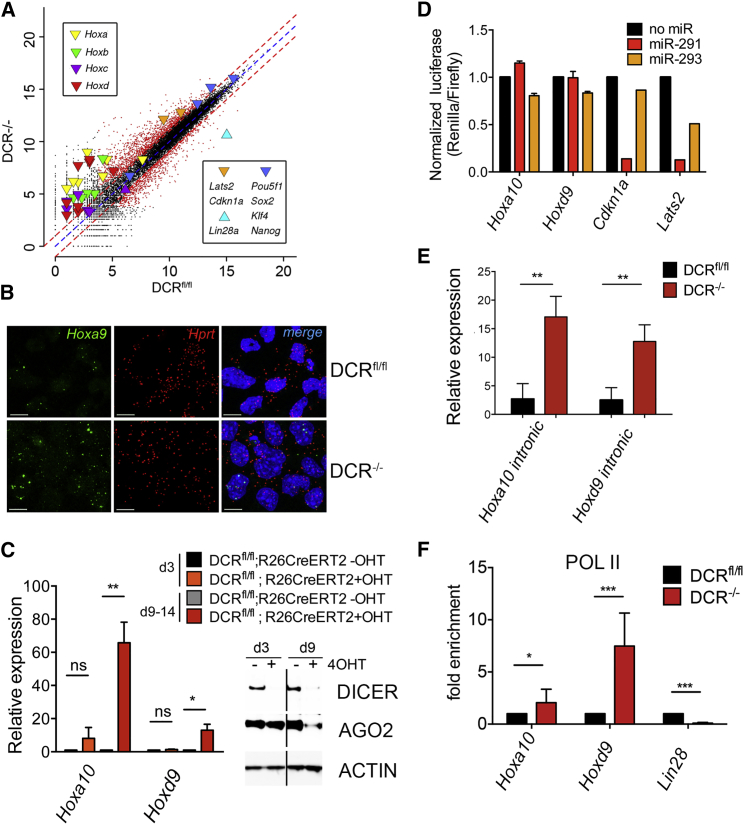
Figure 2.
Homeobox Genes Are Transcriptionally Upregulated in DCR−/− ESCs
(A) Scatterplot of normalized read values of DCRfl/fl versus DCR−/− ESCs from an RNA-seq experiment. Highlighted are genes belonging to the Hox clusters, the core pluripotency factors Oct4, Sox2, Klf4, Nanog, and Lin28a and two miR-290 targets, Cdkn1a and Lats2. Red dashed lines indicate the 2-fold change cut-off for differentially expressed genes (dots in red).
(B) RNA-FISH for Hoxa9 (green) and Hprt (red) in DCRfl/fl and DCR−/− ESCs (scale bar, 10 μm). Nuclei are counterstained by DAPI.
(C) qRT-PCR for representative Hox genes in DCRfl/fl;R26CreERT2 cells with or without the addition of tamoxifen (4OHT). (Bars represent mean ± SD of three independent experiments.) Inset shows western blot analysis of DICER and AGO2 protein levels. ACTIN is shown as a loading control.
(D) Normalized luciferase levels after transfection of DCR−/− ESCs with indicated vectors ± miR-291 or miR-293. Values were normalized to the no miRNA control. Bars represent mean of two (Hoxa10-UTR and Hoxd9-UTR) or one (Cdkn1a and Lats2) independent experiments.
(E) qRT-PCR for representative Hox genes using intronic primers.
(F) ChIP-qPCR of RNA POL II at Hoxa10, Hoxd9, and Lin28a promoters (bars represent mean ± SD of eight independent experiments). Data are represented as fold change of DCR−/− over DCRfl/fl signal.
See also Figure S2.
Since RNA-seq and qRT-PCR analyses cannot provide information at the single-cell level, we performed RNA fluorescence in situ hybridization (RNA-FISH) for Hoxa9 to assess whether the observed upregulation was due to a few cells expressing high levels of Hox transcripts or a general characteristic of DCR−/− ESCs. The RNA-FISH analysis showed more Hoxa9 transcripts in the majority of DCR−/− ESCs, while the housekeeping gene Hprt was not differentially expressed (Figure 2B). The DAPI nuclear signal demarcates individual cells.
To exclude that Hox derepression was due to prolonged culture of DCR−/− ESC clones, we deleted Dicer acutely using a tamoxifen-inducible Cre DCRfl/fl ES cell line (DCRfl/fl; R26CreERT2) (Nesterova et al., 2008). Three days after tamoxifen (4OHT) addition, DICER protein was dramatically reduced, and 9 days later ARGONAUTE2 (AGO2), which is destabilized in the absence of mature miRNAs (Martinez and Gregory, 2013), was also decreased (Figure 2C inset), while Hoxa10 and Hoxd9 transcripts were upregulated.
To test whether miR-291 regulated Hox mRNA stability, we inserted the 3′ UTR of Hoxa9 and Hoxd10 downstream of a luciferase reporter. We observed no effect on luciferase expression when Hox reporter vectors were cotransfected with miR-291 in DCR−/− ESCs in contrast to the observed repression when bona fide direct targets such as Cdkn1a and Lats2 3′ UTRs were tested (Figure 2D). Cotransfection with miR-293 (a non-“seed”-containing miRNA from the miR-290 cluster) was used as a negative control. These observations suggest that the effect of miR-291 on Hox genes is indirect and likely transcriptional; we therefore performed qRT-PCR for Hoxa9 and Hoxd10 using intronic primers. Unspliced primary transcripts, a proxy of ongoing transcription, were upregulated in DCR−/− ESCs (Figure 2E). In addition, RNA polymerase (POL) II occupancy at Hox gene promoters was higher in DCR−/− compared to DCRfl/fl ESCs (Figure 2F). Thus, the increased Hox gene expression appeared to be due to transcriptional regulation.
We next examined how miRNAs could regulate Hox gene transcription. In ESCs, Hox gene promoters are bivalent (Bernstein et al., 2006, Boyer et al., 2006), with both activating and repressive histone H3 modifications at lysines 4 and 27 (H3K4me3 and H3K27me3, respectively), but are maintained transcriptionally silent by PcG proteins. Thus, we performed chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) to determine the localization of the core PRC2 components SUZ12 and JARID2 as well as the H3K27me3 mark catalyzed by this holoenzyme. Using ChIP-seq, and ChIP-qPCR for validation, we found that SUZ12, JARID2, and H3K27me3 were all reduced at Hox gene promoters in DCR−/− ESCs (Figures 3A and 3B). The Lin28a promoter, which is not a Polycomb target, was used as a negative control. We also observed a reduction in the association of the PRC1 subunit RING1b at Hox loci (Figure S3A).
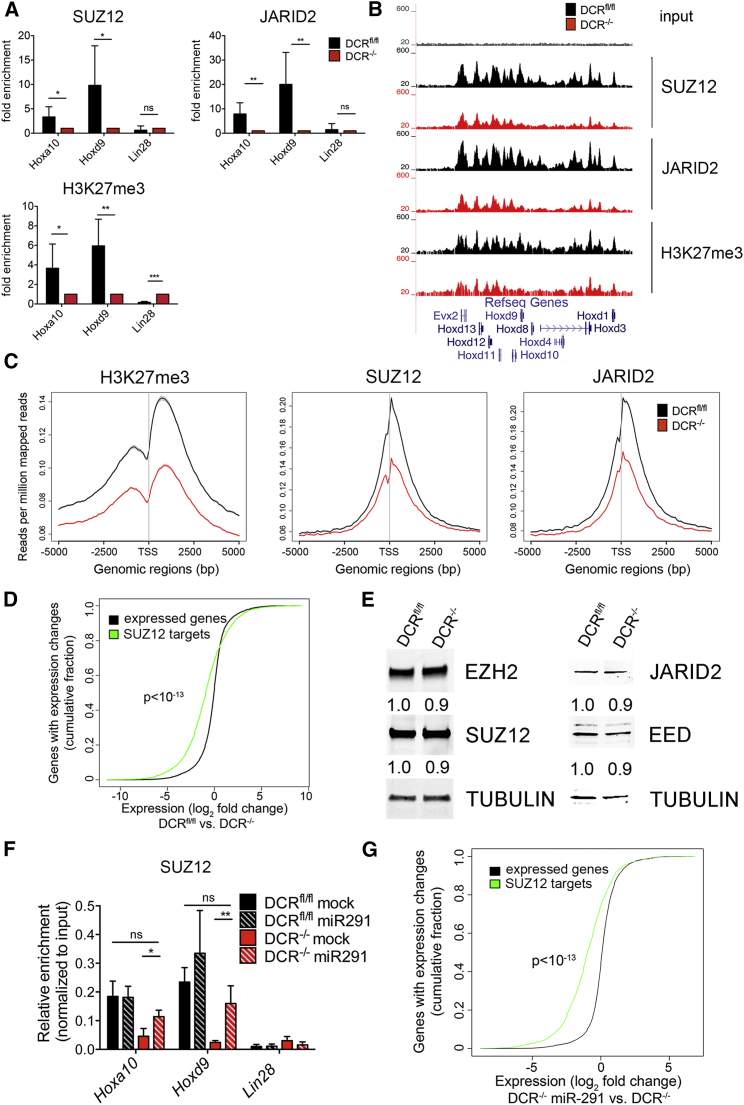
Figure 3.
Defect in PcG Protein Recruitment in DCR−/− ESCs.
(A) ChIP-qPCR analysis of SUZ12, JARID2, H3K27me3 at Hoxa10 and Hoxd9 promoters and the Lin28a promoter as a control (bars represent mean ± SD of at least five independent experiments). Data are represented as fold change of DCRfl/fl over DCR−/− signal.
(B) Genome browser screenshot of the reads aligning to the Hoxd locus after ChIP with anti-SUZ12, anti-JARID2, or anti-H3K27me3 antibodies.
(C) Line plot depicts distribution of H3K27me3 (left), SUZ12 (middle), and JARID2 (right) localization at TSS ± 5 kb of all RefSeq genes.
(D) Cumulative fraction plot of the total number of expressed genes (black line) and genes that have a SUZ12 peak in DCRfl/fl ESCs (green line) sorted by log2-fold change of the reads in DCRfl/fl versus DCR−/− ESCs. P value was calculated using a Kolmogorov-Smirnov test.
(E) Western blot showing the levels of EZH2, SUZ12, JARID2, and EED. Numbers below each lane represent the signal of each band normalized to the signal from tubulin. The normalized signal from the DCRfl/fl lysates is set to 1.
(F) ChIP-qPCR for SUZ12 in DCRfl/fl and DCR−/− cells before and after transfection with miR-291. Bars represent mean ± SD of three independent experiments.
(G) Same as (D), but the log2-fold change of the reads in DCR−/− cells ± miR-291 is plotted. P value was calculated using a Kolmogorov-Smirnov test.
See also Figure S3.
Furthermore, we observed a global reduction in Polycomb at sites throughout the genome. In the absence of Dicer, there was a marked reduction of SUZ12, JARID2, and H3K27me3 at transcriptional start sites (TSSs) (Figure 3C). To assess the impact of reduced Polycomb accumulation on the transcriptome of DCR−/− ESCs, we analyzed differentially expressed genes and found there was a significant enrichment for SUZ12 targets in the genes upregulated in DCR−/− ESCs (Figure 3D). However, not all PRC2 targets with reduced JARID2, SUZ12, and H3K27me3 were transcriptionally upregulated (Figures S3D and S4D). SUZ12, JARID2, EED, and EZH2 protein levels were not decreased nor were mRNA levels for other PcG proteins (Figures 3E and S3C). Thus, it seems that deletion of Dicer1 affects the targeting of PcG proteins to cognate genomic locations rather than the expression levels of PcG proteins.
Since Hox genes are prototypical PcG targets in many organisms (Boyer et al., 2006, Lewis, 1978), we focused on their regulation as a reflection of overall PcG function. We found that miR-291 significantly increased Suz12 binding at the Hoxa10 and Hoxd9 TSSs in DCR−/−, but not in DCRfl/fl ESCs (Figure 3F). Moreover, miR-291 transfection in DCR−/− ESCs significantly reduced Suz12 target gene transcripts (Figure 3G).
To further investigate the mechanism of miR-291 regulation of PcG recruitment at TSSs and Hox gene repression, we examined the differential expression of candidate regulatory genes from RNA-seq analyses. Interestingly, many Trithorax group genes, which are known antagonists of Polycomb, such as mixed lineage leukemia (Mll) were upregulated in DCR−/− ESCs and reduced upon miR-291 transfection (Figure S4A). Since we observed no increase in H3K4me3, catalyzed by MLL proteins, at Hox loci (Figure S4B) or globally (Figure S4C), MLL proteins, despite their potentially important upregulation, are not likely responsible for antagonizing PRC recruitment in DCR−/− ESCs. We also investigated other prominently deregulated genes, such as the histone acetyltransferase Kat2b and Mllt6, which probably helps catalyze H3K79 methylation (Mohan et al., 2010). However, inhibition by garcinol of Kat2b or knockdown of Mllt6 did not restore Hox gene repression (data not shown).
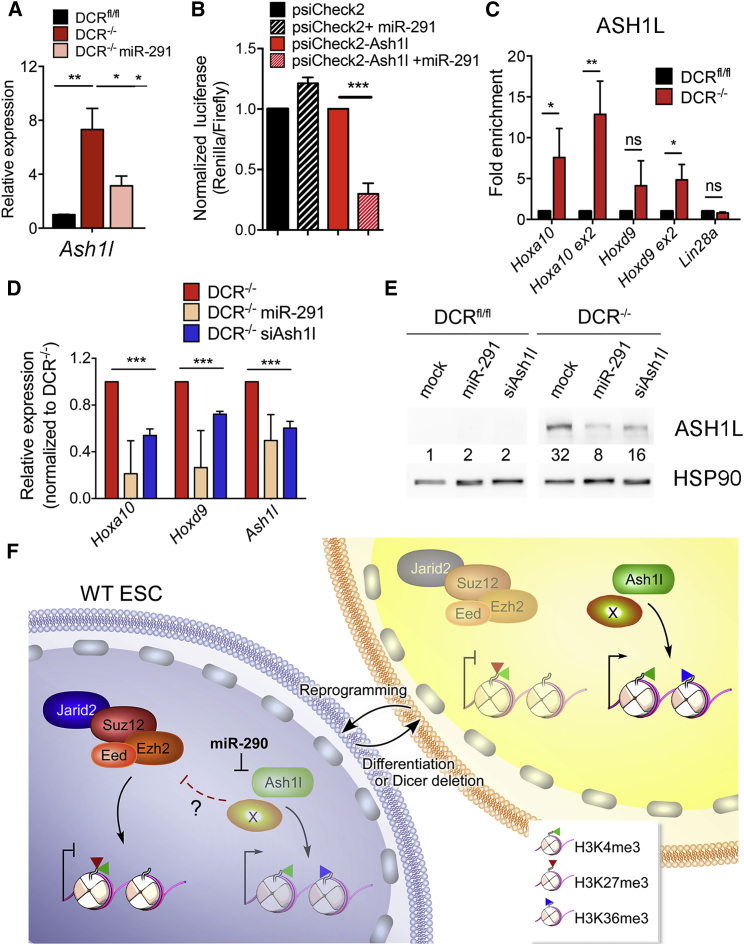
However, we discovered that miR-291 regulated another Trithorax group protein, the H3K36 methyltransferase Ash1l. It was recently reported that H3K36me3 deposition at Hox loci occurs independently of transcription via the action of ASH1L and is sufficient to evict Polycomb (Miyazaki et al., 2013). We observed that Ash1l transcripts and protein were suppressed by miR-291 (Figures 4A and 4D). Since there is a predicted miR-290 site in the 3′ UTR of Ash1l (Lewis et al., 2005), we cloned it into a luciferase reporter vector and assessed its activity upon miR-291 cotransfection. We found that the Ash1l 3′ UTR decreased luciferase levels when cotransfected with miR-291 (Figure 4B). ASH1L was also significantly enriched at Hox coding regions in DCR−/− ESCs by ChIP (Figure 4C). We then tested whether Ash1l knockdowns could silence Hox genes in the absence of Dicer. Although the Ash1l knockdown was partial, it significantly reduced Hoxa10 and Hoxd9 expression in DCR−/− ESCs (Figures 4D and 4E). Notably, the effect of Ash1l knockdown was less dramatic than usually observed with miR-291, which could reflect either the partial knockdown or the possibility of additional miR-291 targets. Since miRNAs usually target a large number of genes to synergistically induce desired cellular phenotypes, it is likely that a combination of factors, including Ash1l, leads to transcriptional derepression and Polycomb eviction at Hox genes and other Polycomb sites.
Figure 4.
Histone Methyltransferase Ash1l Is a Target of miR-291 and Regulates Hox Gene Expression
(A) Relative expression of Ash1l in DCRfl/fl and DCR−/− ESCs ± miR-291.
(B) Normalized luciferase levels after transfection of DCR−/− ESCs with indicated vectors ± miR-291. Values were normalized to the no miRNA control. Bars represent mean ± SD of three independent experiments.
(C) ChIP-qPCR analysis of ASH1L enrichment at indicated loci. Data are represented as fold change of DCR−/− over DCRfl/fl signal.
(D) Relative expression of Hoxa10, Hoxd9, and Ash1l in DCR−/− ESCs after transfection with the indicated miRNA or siRNA. Bars represent mean ± SD of three independent experiments. Data are represented as fold change compared to DCR−/− ESCs.
(E) Western blot showing the levels of ASH1L in DCRfl/fl and DCR−/− ESCs mock transfected or transfected with miR-291 or siRNAs targeting Ash1l (siAsh1l). Numbers below each lane indicate the signal of each band normalized to the signal from HSP90. The normalized signal for DCRfl/fl is set to 1.
(F) Schematic model of miR-290 regulation of PcG targeting and Ash1l.
See also Figure S4.
Discussion
Dicer is essential for the ESC phenotype, although expression of the core pluripotency factors is maintained in DCR−/− ESCs, indicating that miRNAs may control additional determinants of pluripotency. We found that genes associated with ESC differentiation, specifically the Hox family, were overexpressed in DCR−/− ESCs. First, we observed that Hox genes were regulated by miR-290, the most abundantly expressed miRNA family in undifferentiated ESCs. Second, we could attribute this effect to reduced localization of PRC2. This is important because PcG proteins maintain ESCs in a pluripotent state by silencing Hox and other “bivalent” differentiation genes primed for transcription (Bernstein et al., 2006, Boyer et al., 2006). Loss of Dicer altered PRC2 recruitment throughout the genome, illustrating the crucial role of miRNAs in governing the targeting of this silencing complex. Consistently, a significant number of PcG target genes were transcriptionally activated in DCR−/− ESCs (Figures 3D and S4D). However, some genes lost Polycomb binding but were not transcriptionally activated (Figures S3D and S4D). Hence, PRC removal from chromatin is not always a secondary effect of transcriptional activation of differentiation genes in DCR−/− ESCs (Riising et al., 2014). In fact, there is only 20% overlap of genes activated upon Dicer deletion and retinoic-acid-induced differentiation, and most of these genes are Polycomb targets (Figure S4D). Moreover, we found that reduction of PcG proteins at genomic loci could not be attributed to changes in the expression levels of PRC1 and PRC2 subunits (Figures 3E and S3C). Rather, miR-290 members regulate the targeting of PRC1 and PRC2 to appropriate loci in ESCs to maintain their “stemness.”
The miR-290 cluster has been previously implicated in regulation of de novo methyltransferases (Dnmts) (Sinkkonen et al., 2008), and DNA methylation may affect PcG localization (Reddington et al., 2013). It will be interesting to see if the observed reduction in Polycomb binding is partially due to loss of proper DNA methylation in DCR−/− ESCs and redistribution of PRC components across the genome.
We found that miR-291 repressed Ash1l, which can activate Hox genes by evicting Polycomb during differentiation (Miyazaki et al., 2013, Tanaka et al., 2011). Ash1l is a predicted target of miR-291, which we validated using reporter assays. We observed that knockdown of Ash1l reduced Hox gene expression in DCR−/− ESCs, suggesting that reduction of H3K36 methylation is sufficient to partially suppress Hox gene transcription. Thus, our data reveal a circuit of miRNA control of ESC gene expression through Ash1l and targeting of PcG proteins.
It has been reported that the miR-290 family enhances generation and quality of mouse and human iPS cells, but the mechanism is not fully understood (Anokye-Danso et al., 2011, Judson et al., 2009, Liao et al., 2011, Miyoshi et al., 2011). PRC2 is also required for reprogramming (Onder et al., 2012, Pereira et al., 2010), and similar to DCR−/− ESCs, PRC2 mutant ESCs fail to differentiate (Pasini et al., 2007). This could imply that these two phenotypes may be related, since we show that PcG targeting is influenced by miR-290 miRNAs in ESCs. Thus, this regulatory mechanism may affect not only Hox genes but also other factors important for pluripotency. It will be interesting to explore whether there is a broader role of this gene regulatory circuit in pluripotency and differentiation in various types of stem cells and cancer.
Experimental Procedures
Cell Culture and Transfections
All animal work was done in accordance with the guidelines of the Institutional Animal Care and Use Committee. DCRfl/fl ESCs were derived from days post-coitum (DPC) 3.5 embryos. DCR−/− clones were isolated after Adeno-Cre (Harvard Gene Therapy). For acute Dicer deletion, cells were treated with 2.5 μM of 4OHT (Sigma). Prior to transfections, harvesting of cells for RNA, protein, or ChIP, mouse embryonic fibroblasts (MEFs) were removed using MEF removal microbeads (Miltenyi Biotech). For detailed description, please see the Supplemental Experimental Procedures.
Protein Analyses, ChIP, and ChIP-Seq
ChIP was performed according to published protocols. A detailed description is provided in the Supplemental Experimental Procedures. All P values were calculated with an unpaired Student’s t test unless otherwise stated. ∗ p < 0.05; ∗∗ < 0.01; ∗∗∗ p < 0.001.
Western blotting was performed as previously described. Western blots (WBs) for PRC2 components were analyzed using the Li-Cor imaging system and software.
RNA Analyses
RNA was prepared with Trizol (Invitrogen) or RNAzol (MRC) reagent according to the manufacturer’s instructions. All primers were designed using the Primer 3 software (Steve Rozen, Helen J. Skaletsky, http://biotools.umassmed.edu/bioapps/primer3_www.cgi), and sequences are provided in Table S2. RNA-seq libraries were prepared using the TruSeq Stranded mRNA Sample Prep Kit or TruSeq RNA Sample Preparation Kit v2 and analyzed on a GAIIx genome analyzer (Illumina). For RNA FISH, labeled probes (Quantigene RNA VIEW ISH probes) were purchased from Panomics and used according to the manufacturer’s instructions (see also Supplemental Experimental Procedures). miRNA abundance was quantified with the nCounter miRNA Expression Assay (Nanostring Technologies) or with individual Taqman miRNA assays (ABI).
Acknowledgments
We would like to thank Drs. David Livingston, Ron Germain, Kathrin Plath, Markus Hafner, and Carrie Lucas for critically reading this manuscript and Bryan Chim for help with bioinformatics analyses. We also thank Dr. Livingston for his support during the generation of the DCR−/− ESCs, anti-Dicer, and anti-Ago2 antibodies and Drs. Merkenschlager and Graham for sharing the DCRfl/fl;R26CreERT2 cells. We also thank Dr. Chaigne-Delalande and members of the Lenardo and Muljo laboratories for helpful discussions, M. Smelkinson and S. Ganesan at the NIAID Imaging core for assistance with microscopy, and A. Athman, NIAID Visual and Medical Arts Department, for help with Figure 4F. This study utilized NIAID high-performance computing and NIH Biowulf cluster and was supported by the NIH Intramural Research Program of the NIAID.
Published: November 5, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.10.001.
Contributor Information
Michael J. Lenardo, Email: mlenardo@niaid.nih.gov.
Stefan A. Muljo, Email: stefan.muljo@nih.gov.
Accession Numbers
The accession number for the RNA-seq and ChIP-seq data reported in this paper is NCBI GEO: GSE60397.
Supplemental Information
References
- Anokye-Danso F., Trivedi C.M., Juhr D., Gupta M., Cui Z., Tian Y., Zhang Y., Yang W., Gruber P.J., Epstein J.A., Morrisey E.E. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Dietrich N., Pasini D., Hansen K.H., Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese J.M., Seila A.C., Yeo G.W., Sharp P.A. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L., Helin K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray M.F., Sharp P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Judson R.L., Babiarz J.E., Venere M., Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E.B. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liao B., Bao X., Liu L., Feng S., Zovoilis A., Liu W., Xue Y., Cai J., Guo X., Qin B. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J. Biol. Chem. 2011;286:17359–17364. doi: 10.1074/jbc.C111.235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A., Levine S.S., Cole M.F., Frampton G.M., Brambrink T., Johnstone S., Guenther M.G., Johnston W.K., Wernig M., Newman J. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N.J., Gregory R.I. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA. 2013;19:605–612. doi: 10.1261/rna.036434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H., Higashimoto K., Yada Y., Endo T.A., Sharif J., Komori T., Matsuda M., Koseki Y., Nakayama M., Soejima H. Ash1l methylates Lys36 of histone H3 independently of transcriptional elongation to counteract polycomb silencing. PLoS Genet. 2013;9:e1003897. doi: 10.1371/journal.pgen.1003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D.L., Kano Y., Nishikawa S., Tanemura M., Mimori K., Tanaka F. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Mohan M., Herz H.M., Takahashi Y.H., Lin C., Lai K.C., Zhang Y., Washburn M.P., Florens L., Shilatifard A. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–589. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova T.B., Popova B.C., Cobb B.S., Norton S., Senner C.E., Tang Y.A., Spruce T., Rodriguez T.A., Sado T., Merkenschlager M., Brockdorff N. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin. 2008;1:2. doi: 10.1186/1756-8935-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Bracken A.P., Hansen J.B., Capillo M., Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J.C., Lemons D., McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Pereira C.F., Piccolo F.M., Tsubouchi T., Sauer S., Ryan N.K., Bruno L., Landeira D., Santos J., Banito A., Gil J. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Reddington J.P., Perricone S.M., Nestor C.E., Reichmann J., Youngson N.A., Suzuki M., Reinhardt D., Dunican D.S., Prendergast J.G., Mjoseng H. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 2013;14:R25. doi: 10.1186/gb-2013-14-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riising E.M., Comet I., Leblanc B., Wu X., Johansen J.V., Helin K. Gene silencing triggers Polycomb repressive complex 2 recruitment to CpG islands genome wide. Mol. Cell. 2014;55:347–360. doi: 10.1016/j.molcel.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Schwartz Y.B., Pirrotta V. Ruled by ubiquitylation: a new order for polycomb recruitment. Cell Rep. 2014;8:321–325. doi: 10.1016/j.celrep.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Sinkkonen L., Hugenschmidt T., Berninger P., Gaidatzis D., Mohn F., Artus-Revel C.G., Zavolan M., Svoboda P., Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Kawahashi K., Katagiri Z., Nakayama Y., Mahajan M., Kioussis D. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression. PLoS ONE. 2011;6:e28171. doi: 10.1371/journal.pone.0028171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S., Abreu-Goodger C., Enright A.J. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Baskerville S., Shenoy A., Babiarz J.E., Baehner L., Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.