Summary
Human palatine tonsils are oropharyngeal lymphoid tissues containing multiple invaginations (crypts) in which the continuity of the outer surface epithelium is disrupted and the isolated epithelial cells intermingle with other cell types. We now show that primitive epithelial cells detectable in vitro in 2D colony assays and in a 3D culture system are CD44+NGFR+ and present in both surface and crypt regions. Transcriptome analysis indicated a high similarity between CD44+NGFR+ cells in both regions, although those isolated from the crypt contained a higher proportion of the most primitive (holo)clonogenic cells. Lentiviral transduction of CD44+NGFR+ cells from both regions with human papillomavirus 16-encoded E6/E7 prolonged their growth in 2D cultures and caused aberrant differentiation in 3D cultures. Our findings therefore reveal a shared, site-independent, hierarchical organization, differentiation potential, and transcriptional profile of normal human tonsillar epithelial progenitor cells. They also introduce a new model for investigating the mechanisms of their transformation.
Keywords: tonsil, epithelial progenitors, HPV, oropharyngeal cancer
Graphical Abstract
Highlights
-
•
Tonsillar surface and crypt epithelial progenitor cells are detected similarly
-
•
Both surface and crypt epithelial progenitors in the tonsil are CD44+NGFR+
-
•
Stratified epithelium can be regenerated from primitive tonsillar crypt cells
-
•
HPV16 E6/E7 deregulates crypt epithelial progenitor growth and differentiation
In this article, Eaves and colleagues show that the dispersed epithelial population in the crypts of normal human tonsils mimics the structurally distinct, continuous epithelium on the outer surface in the shared possession of primitive progenitors with similar phenotypic, transcriptional, and functional properties that are altered by lentivirally introduced human papillomavirus E6/E7 oncogenes.
Introduction
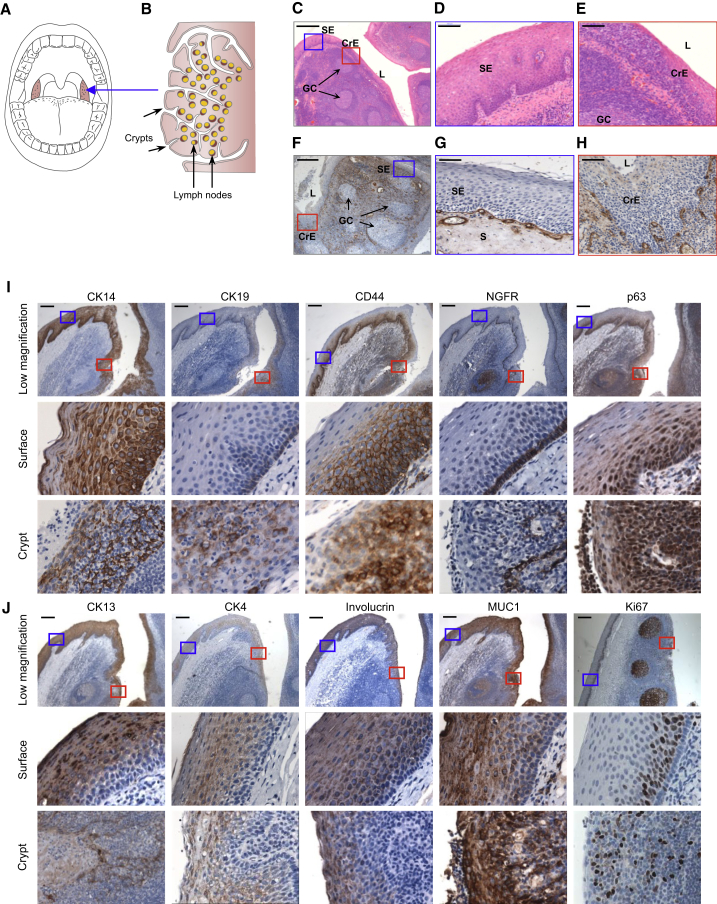
The human palatine tonsil is part of a circular collection of specialized B-lymphoid structures located in the oropharynx at the opening of the aerodigestive tract (Figure 1A). The outer surface of the human palatine tonsil is lined with a stratified squamous epithelium that invaginates into the underlying lymphoid tissue to form tonsillar crypts, enlarging the tonsillar surface area by up to 300 cm2 (Howie, 1980; Figure 1B). The stratified squamous epithelium overlying the surface of the human tonsil is similar to many epithelial linings of human organs and tissues in its continuous nature and separation from the underlying connective tissue by a basement membrane (Figures 1C and 1D). Any discontinuity in this surface structure is regarded as pathological and is referred to as an ulcer or erosion. In contrast, the discontinuous arrangement of the epithelial cells present in the crypt is normal and appears rather sieve-like, allowing ready access of tonsillar lymphocytes to antigens from the external environment (Figures 1C and 1E; Perry and Whyte, 1998).
Figure 1.
Cellular Composition of the Human Palatine Tonsil
(A) A drawing showing paired human palatine tonsils on either side of the oropharynx.
(B) A schematic depicting a cross-section of a human palatine tonsil with multiple crypts and lymph nodes.
(C–E) H&E-stained FFPE tonsil tissue sections showing stratified surface epithelium with an organized multi-layer structure similar to oral mucosa (blue box, C and D), and the thinning epithelium overlying tonsillar crypts heavily infiltrated by lymphocytes (red box, C and E). SE, surface epithelium; CrE, crypt epithelium; S, stroma; GC, germinal center; L, lumen). Scale bars, 500 μm (C) and 100 μm (D and E).
(F–H) IHC staining of collagen IV in FFPE tonsil tissue sections showing the presence of a continuous basement membrane in the surface epithelium (blue box, F and G) and a discontinuous and indistinct basement membrane in the crypt epithelium (red box, F and H). Scale bars, 500 μm (F) and 100 μm (G and H).
(I and J) In situ IHC analysis using a panel of antibodies against membrane or intracellular antigens. Top rows: low-magnification images showing both surface (blue box) and crypt (red box) epithelia, portions of which are enlarged in the center and bottom rows. Scale bar, 200 μm.
Human tonsillar epithelial cells are thought to derive during embryogenesis from the endoderm of the second pharyngeal pouch (Perry and Whyte, 1998), but little is known about their lifelong maintenance, turnover, and differentiation. The lack of a counterpart in mice or other rodents (Casteleyn et al., 2011) may be one of the reasons why these cells have not been well characterized. Cells with long-term tissue-specific regenerative potential have now been identified and characterized in several human epithelial tissues (Van Keymeulen and Blanpain, 2012). In many cases, this has led to evidence of a differentiation hierarchy composed of prospectively separable subsets of cells with different tissue-sustaining and differentiation abilities. The histology of the surface epithelium of the tonsil suggests that a similar hierarchy exists in this tissue, but direct evidence for its organization has not yet been reported. It is also not clear whether or how this paradigm applies to the dispersed population of epithelial cells found in the tonsillar crypts. This latter issue is of particular interest because the majority of human papillomavirus (HPV)-positive head and neck cancers appear to originate in this site (Begum et al., 2005, El-Mofty and Patil, 2006, Gillison et al., 2000). However, so far, very little is known about the oncogenic process that leads to the formation of these tumors (Begum et al., 2005).
We now describe conditions for the sensitive and quantitative detection, isolation, and characterization of different subsets of tonsillar surface and crypt epithelial cells using 2D colony assay and 3D culture systems, flow cytometry, immunohistochemical (IHC) staining, and transcriptome analyses. We also report the utility of these methods to reveal the presence of primitive epithelial cells within the crypt region and an ability of experimentally introduced HPV16-E6/E7 oncogenes to alter their growth and differentiation activities.
Results
Human Tonsillar Surface and Crypt Epithelial Populations Contain Different Representations of Similar Phenotypes
IHC staining of different regions of tonsillar tissue in formalin-fixed, paraffin-embedded samples revealed a continuous collagen IV-positive basement membrane separating the surface epithelium from the underlying stroma (Figures 1F and 1G). In contrast, in the crypt, collagen IV staining was weakly detectable and discontinuously distributed (Figures 1F and 1H). Cytokeratin-14 (CK14) was stained strongly in cells throughout the surface epithelial layer and on putative epithelial cells in the crypt (Figure 1I; Table S1), suggesting its likely utility as a pan-tonsillar epithelial marker, consistent with previous reports of its ubiquitous expression in keratinocytes (Cooper et al., 1985, Moll et al., 1982). Cell-surface antigens including CD44, MUC1, and nerve growth factor receptor (NGFR) and intracellular antigens CK19, p63, CK13, and CK4 were all expressed, but variably, in the surface epithelium, where their location could be associated with an increasingly differentiated state from the basal layer to the outer surface. For example, CK19 and NGFR were exclusive to the basal layer of the surface epithelium (Figure 1I, center row), whereas p63, a widely annotated marker of basal epithelial cells (Pellegrini et al., 2001, Rock et al., 2009, Senoo et al., 2007), and CD44, a receptor for hyaluronic acid, were seen not only in the basal layers but also in the parabasal layers, extending up to the mid-layers of the surface epithelium (Figure 1I, center row). CK13, CK4, involucrin, and cell-surface MUC1 were detected only in the parabasal and upper layers of the surface epithelium and not in any basal cells (Figure 1J, center row). Cells expressing the proliferation marker Ki67 were also absent from the basal layer of the surface epithelium and found only in cells in the overlying three to five parabasal layers. In the crypt regions, all of the same markers were expressed in some cells, but these appeared to be more randomly organized because of their dispersed distribution (Figure 1J, bottom row).
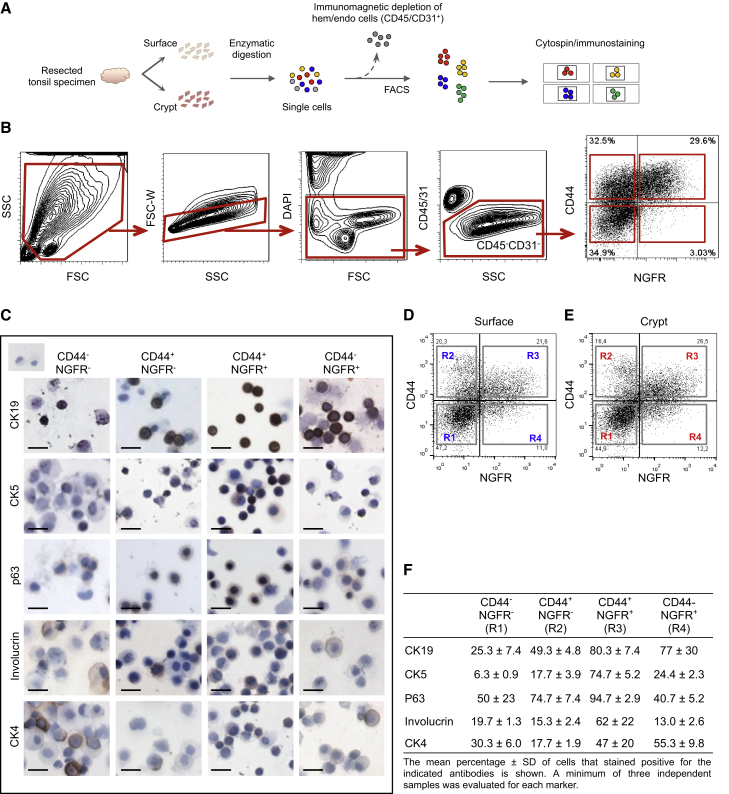
To address this difference between the surface and crypt epithelia, we first dissected out surface and crypt tissue samples with a scalpel to analyze viable cell suspensions prepared from each site (Figure 2A). The majority of hematopoietic (CD45+) and endothelial (CD31+) cells were then removed immunomagnetically by EasySep, and the negatively selected cells were co-stained with antibodies to CD44 and NGFR for multiparameter flow cytometry analysis (Figure 2B). Most of the CD45−CD31− cells from both sites were distributed among three subsets: CD44+NGFR+, CD44−NGFR−, and CD44+NGFR− cells, with very few CD44−NGFR+ cells detected (Figures 2D and 2E). Cells from each of these four subsets were then isolated at >90% purity by fluorescence-activated cell sorting (FACS) and analyzed for expression of intracellular markers in fixed cytospin preparations (Figure 2C and 2F). This analysis showed that the CD44+NGFR+ subpopulation isolated from both sites was the most highly enriched for cells strongly expressing CK19 and CK5/p63 (markers of the basal and mid-layers of the surface epithelium). In contrast, fewer CD44+NGFR+ cells from either site were positive for involucrin or CK4, both of which appear as differentiation markers in the surface epithelium. The proportion of involucrin-positive or CK4+ cells varied significantly in a site-dependent manner. 20% and 28% of CD44+NGFR+ cells in the surface epithelium expressed involucrin and CK4, respectively (Table S2), with much higher proportions of these (69% and 86%, respectively) in the CD44+NGFR+ crypt cells. CD44+NGFR− and CD44−NGFR+ subpopulations expressed lower levels of CK5, CK19, and p63. In CD44−NGFR+ cells, p63 expression was detected in the cytoplasm and appeared to be absent in the nucleus. CD44−NGFR− cells showed low expression of the basal markers and strong expression of the markers of differentiated cells. These findings suggest that phenotypically similar subsets of epithelial cells are present on the surface and in the crypts of the human tonsil, with some variability in their distribution.
Figure 2.
Phenotypic Characterization of Tonsillar Epithelial Cells According to Their Surface Expression of CD44 and NGFR
(A) Experimental protocol for analyzing single CD45− (non-hematopoietic) and CD31− (non-endothelial) cells according to their expression of CD44 and NGFR.
(B) FACS gating strategies. SSC, side scatter; FSC, forward scatter; FSC-W, FSC width.
(C) IHC analysis of CD45−CD31−cells isolated by FACS at purities of >90% according to their expression of CD44 and NGFR and cytospun prior to fixation and staining with antibodies to CK19, CK5, p63, and CK4. Inset, negative control. Scale bar, 20 μm.
(D) FACS plots for CD45−CD31− surface cells.
(E) FACS plots for CD45−CD31− crypt cells.
(F) Percentages of cells positive for the indicated intracellular proteins. Values shown are the mean ± SD. At least three independent samples were evaluated for each marker.
Different Types of Clonogenic Tonsillar Epithelial Progenitor Cells Can Be Detected In Vitro
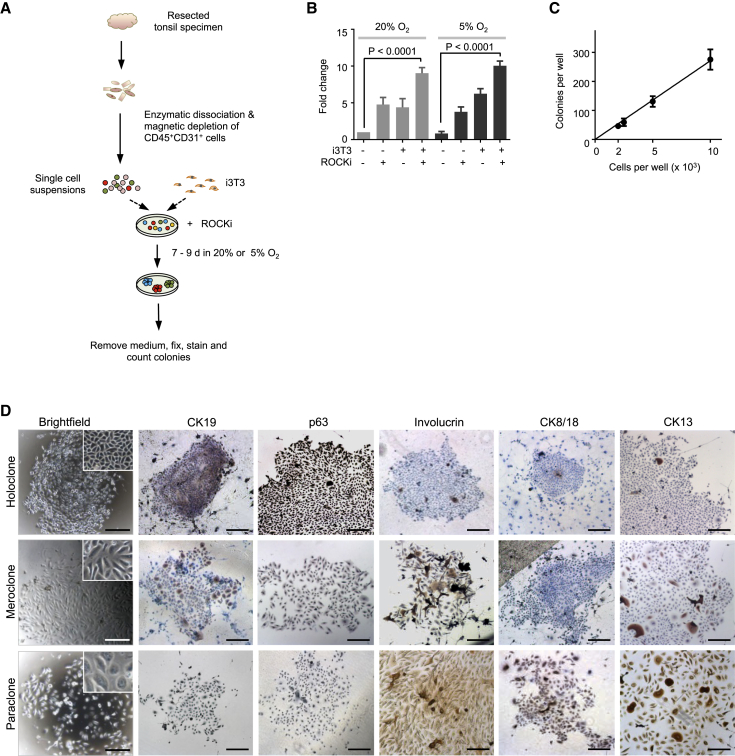
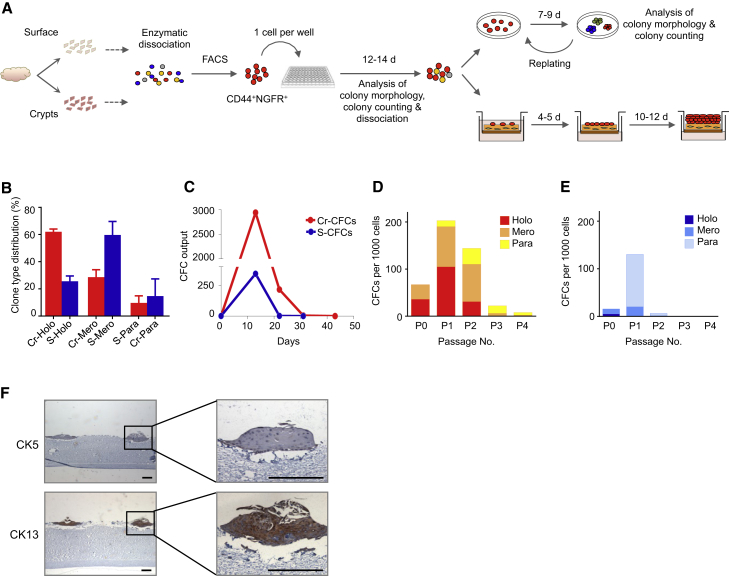
In preliminary experiments, we found that enzymatically dissociated suspensions of tonsillar cells (depleted of CD45+ cells and CD31+ cells by magnetic cell separation) form discrete adherent colonies of >50 cells within 7–9 days in vitro (Figure 3A) under conditions similar to those developed for other tissue sources of epithelial cells (Van Keymeulen and Blanpain, 2012). Examination of several aspects of the culture protocol revealed that the highest plating efficiencies were obtained when the cells were co-cultured with irradiated fibroblasts in ScienCell medium supplemented with 10 μM Rho-associated protein kinase (ROCK) inhibitor and incubated in an atmosphere of reduced (5%) O2 (Figure 3B). The latter two conditions were tested based on their ability to enhance the cloning efficiency in vitro of embryonic stem cells and cells from various epithelial tissues (Chapman et al., 2010, Makarem et al., 2013, Matusik et al., 2008, Watanabe et al., 2007, Zhang et al., 2011). Under these conditions, a positive linear relationship was obtained reproducibly between the number of cells seeded per well and the number of colonies generated, validating the use of such cultures to quantify progenitor frequencies (overall ∼2.7%; Figure 3C).
Figure 3.
Optimization of the CFC Assay
(A) Schematic showing the experimental design used to optimize conditions to detect CFCs. Single cells obtained by enzymatic dissociation of normal tonsil specimens were cultured with or without irradiated fibroblasts (i3T3) or ROCK inhibitor (ROCKi), in either 20% or 5% O2, and colonies were scored in dishes fixed 7–9 days (d) later.
(B) Enhancing effects of the use of feeders, a ROCK inhibitor, and a reduced O2 level on colony growth, calculated by comparing the increased numbers of colonies obtained from a given number of cells under each test condition relative to the control condition (no added i3T3 cells or ROCKi). Values shown are the mean ± SEM, derived from ten experiments.
(C) Demonstration of a linear relationship between the number of input cells plated per well and colony yields under optimized CFC assay conditions. Values shown are the mean ± SEM, derived from three experiments.
(D) IHC characterization of the different types of colonies produced. Holoclones expressed high levels of CK19 and p63 (markers of basal cells) but infrequently or rarely expressed involucrin, CK8/18, and CK13 (markers of differentiated tonsillar epithelial cells). Many of the cells in mero/paraclones expressed differentiation markers but rarely expressed any basal markers. Scale bars, 1 mm.
Under the same conditions, we observed a wide range of colony morphologies that could be categorized into the three types described previously for epidermal colonies (Barrandon and Green, 1987). These are referred to as holoclones (tightly clustered large colonies with a smooth perimeter in which less than 5% of cells express terminally differentiated features), paraclones (highly irregularly shaped colonies in which all cells appear to be terminally differentiated), and meroclones (colonies of intermediate size and composition) (Figure 3D). IHC analysis showed most of the cells within holoclones to be strongly positive for CK19 and p63, markers of basal/parabasal cells, with little evidence of the differentiation markers involucrin, CK8/18, and CK13. In contrast, paraclones were negative for CK19 and p63 and generally positive for the differentiation markers. Meroclones showed a heterogeneous staining pattern with varying percentages of positive cells for all markers examined.
Human Tonsillar Epithelial Progenitor Cells Are CD44+NGFR+
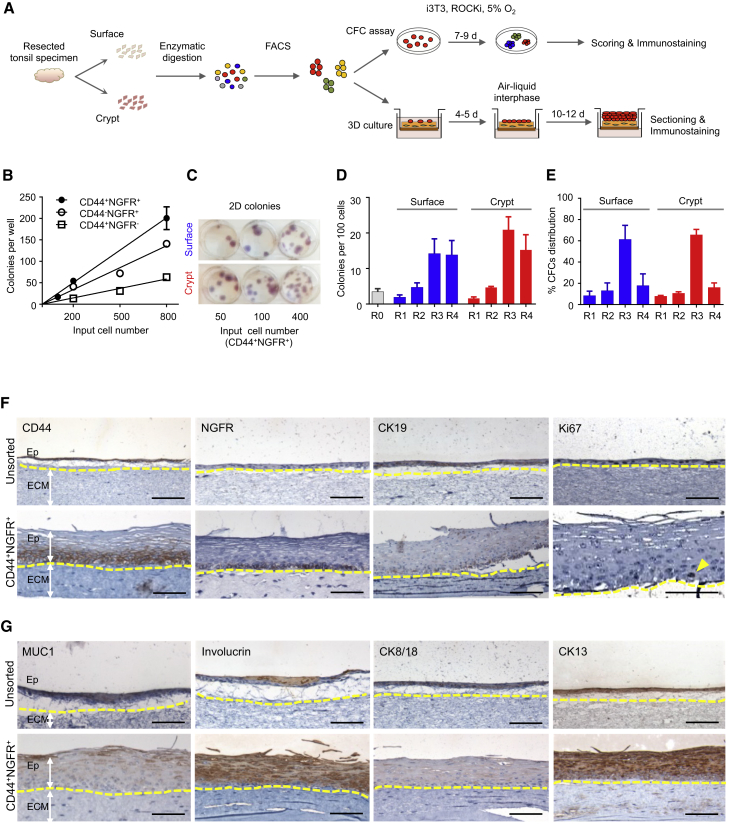
We then isolated the same four CD45/CD31-depleted epithelial cell subsets (according to their different cell surface levels of expression of CD44 and NGFR) and measured the frequency of adherent colony-forming cells (CFCs) in each. The results showed the highest enrichment of CFCs in the CD44highNGFRhigh subset (Figure S1). We then repeated this study on separately isolated surface and crypt regions (Figure 4A). The results showed that colony yields from the purified cells from both sites remained linearly related to the number of cells plated per well (Figures 4B and 4C), with the highest frequencies of CFCs measured in the CD44+NGFR+ fraction from both sites (14% ± 4% and 21% ± 4% CFCs for the surface and crypt, respectively (n = 4) (Figure 4D). This subset from both sites also contained the majority of the CFCs in that region (64% ± 10% and 72% ± 3% for surface and crypt samples, respectively( n = 4) (Figure 4E).
Figure 4.
Functional Comparison of Surface and Crypt CD44+NGFR+ Cells
(A) Experimental protocol for studying the activities of FACS-purified CD44+NGFR+ cells in 2D CFC and organotypic 3D culture systems.
(B) Linear relationship between the number of cells seeded and the number of colonies formed per well in CFC assays.
(C) Hematoxylin staining of 14-day colonies generated in CFC assays.
(D) Number of CFCs detected per 100 input cells from each of the four subpopulations analyzed (R1, R2, R3, and R4, as shown in Figures 2D and 2E) in four experiments.
(E) Distribution of CFCs in each of the four surface and crypt subpopulations analyzed in four experiments.
(F and G) IHC staining of epithelial (Ep) tissue layers generated in organotypic 3D cultures of unsorted (top) and purified CD44+NGFR+ cells (bottom). Shown is the expression of markers specific for basal and/or parabasal cells (F) or differentiated cells (G). Arrowhead, Ki67+ cell. Scale bars, 100 μm.
See also Figure S1.
We also assessed the growth and differentiation activities of FACS-purified CD44+NGFR+ cells in organotypic 3D cultures (Okawa et al., 2007). In this 3D system, dissociated suspensions of cells are seeded on top of an extracellular matrix (ECM) layer containing collagen I and fibroblasts and incubated at an air-liquid interface (Figure 4A). Unseparated CD45−CD31− cells from the crypt produced a patchy monolayer in such cultures (initiated with 1.25 × 105 cells/culture; Figures 4F and 4G, top), whereas the same number of CD44+NGFR+ crypt cells formed a complete sheet of multilayered epithelium (Figures 4G and 4H, bottom), with a similar result obtained in cultures initiated with purified CD44+NGFR+ surface epithelial cells (data not shown). IHC analysis of the cells in the 3D structures produced by epithelial cells from either tonsillar region indicated that the expression pattern of CD44, NGFR, CK19, Ki67, MUC1, involucrin, CK8/18, and CK13 recapitulated the immunophenotype typical of the surface epithelium of the tonsil (Figures 4F and 4G, bottom rows). These findings extend the results of the 2D assays, indicating that the majority of primitive tonsillar epithelial cells have a CD44+NGFR+ phenotype and that they are highly enriched in that subset.
Crypt and Surface Regions Are Differentially Enriched in the Most Primitive Subsets of Epithelial Progenitors
To interrogate more deeply the functional equivalence of CD44+NGFR+ cells in the crypt and surface sites, we used a more discriminating experimental design to compare the proliferative potential of CD44+NGFR+ cells (Figure 5A). Single CD44+NGFR+ cells were plated individually into each well of a 96-well plate and cultured in 5% O2 with irradiated fibroblasts and 10 μM ROCK inhibitor. At the end of 12–14 days, a higher proportion of holoclones was seen in the wells seeded with a CD44+NGFR+ cell from crypt compared with surface isolates (62% ± 2% versus 26% ± 4%, p < 0.0011; Figure 5B). Individual serial replating of their clonal progeny showed that, on average, those derived from crypt cells could be passaged longer than those derived from surface cells (Figure 5C). This outcome was accompanied by a greater capacity of CD44+NGFR+ crypt CFCs to generate daughter CFCs that produced holoclones in later passages (Figures 5D and 5E). The crypt-derived clones were also able to produce multilayered epithelial structures in 3D cultures at lower input cell doses (down to 1,400 cells obtained from five primary colonies; i.e., 100-fold fewer than the number of freshly isolated CD44+NGFR+ cells used). Differentiation of the cells generated in these 3D assays of cells produced in vitro was again evident from IHC staining, which showed the presence of cells expressing CK5, a marker of basal/parabasal cells, and CK13, a marker of differentiated cells (Figure 5F).
Figure 5.
Crypt-Derived CD44+NGFR+ Cells Contain More Primitive CFCs Than Their Surface Counterparts
(A) Experimental protocol to compare the long-term proliferative and differentiation activity of surface- and crypt-derived CD44+NGFR+ CFCs seeded initially in single cell cultures.
(B) Proportion of single (S) surface- and crypt (Cr)-derived CD44+NGFR+ cells that produced holoclones (Holo), meroclones (Mero), and paraclones (Para) after 10–14 days of 2D cultures initiated with a single cell each. Data were pooled from three independent experiments, and one of three donor samples were used each time.
(C) CFC outputs in serially passaged CFC cultures initiated with single CD44+NGFR+ cells (colonies were dissociated and replated every 7–9 days over a period of 6 weeks).
(D) Changes in the proportion of holoclones, meroclones, and paraclones produced by the CFCs in harvests of serially cultured cells originating from initial CD44+NGFR+ crypt CFCs.
(E) Changes in colony types produced as in (D) but from initial CD44+NGFR+ surface epithelial CFCs.
(F) IHC staining of CK5+ and CK13+ cells present in an organotypic 3D culture initiated from 1,400 cells produced from five crypt-derived CFCs. Scale bars, 200 μm.
CD44+NGFR+ Tonsillar Epithelial Cells Have a Unique, Site-Independent Transcriptional Profile
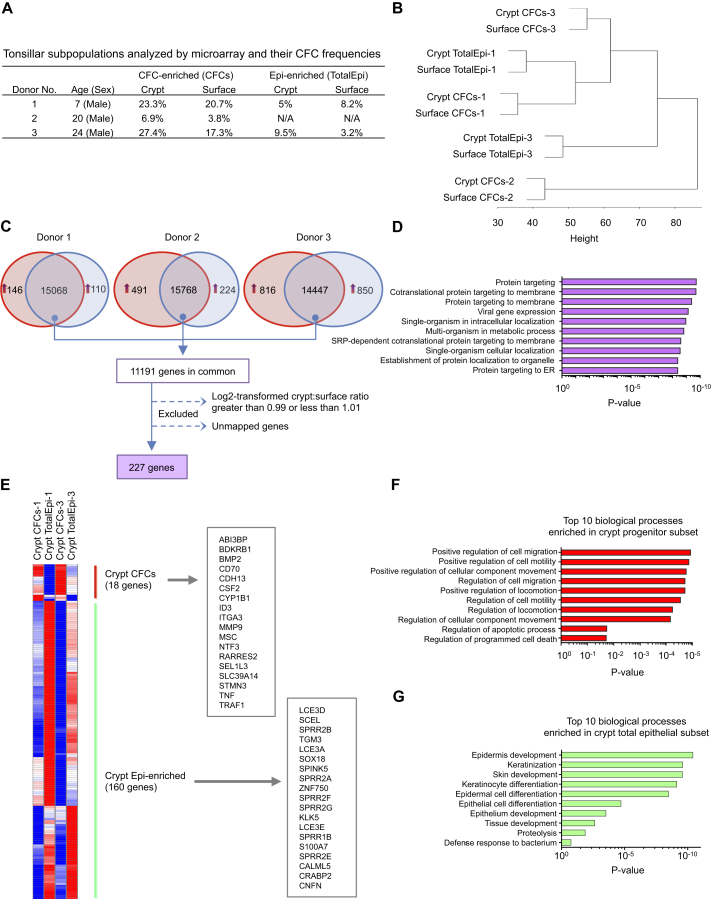
To further characterize the progenitor-enriched CD44+NGFR+ cells present in the crypt and surface regions of the tonsil, we isolated this subset from each site as well as the total epithelial (CD45−CD31−) populations, extracted RNA directly from each of these freshly isolated samples, and then used the Illumina HumanHT-12 V4 BeadChip to characterize their transcriptomes (Figure 6A). After quality control filtering and quantile normalization, 47,320 probes were included for further analyses. The results showed that transcript profiles were more tightly clustered by donor than by the region of the tonsil (crypt or surface) from which the epithelial cells had been isolated (Figure 6B). Comparison of the transcript profiles of the CD44+NGFR+ cells from all three donors from which these cells were isolated revealed a subset of shared transcripts (Figure 6C). To obtain a “site-independent” transcriptome signature of this tonsillar CFC-enriched subset, we identified genes that were the most similarly expressed by applying an additional filter: crypt-to-surface ratio of log2-transformed expression values close to 1 (i.e., ≥0.99 and ≤1.01, n = 227). Gene ontology (GO) analysis of the 227 genes so identified showed a significant association of these with protein targeting and localization and viral gene expression (p < 0.05; Figure 6D). However, when we compared them by site, only eight and seven genes were commonly overexpressed in all three paired crypt and surface samples, respectively (Figures S2A and S2B), suggesting a wide variability between individuals, possibly because of confounding effects of different (and difficult-to-ascertain) levels of inflammation dictating tonsil removal.
Figure 6.
Transcriptome Analysis of Different Subsets of Tonsillar Epithelial Cells
(A) The age and sex of the donors and colony-forming efficiencies of the freshly isolated CD44+NGFR+ cells (CFCs) and CD45−CD31− total epithelial cells (TotalEpi) used for the transcriptome analyses.
(B) Illumina transcriptome analyses identifying the relatedness of all four subsets of tonsillar cells derived from an unsupervised hierarchical clustering analysis.
(C) Number of genes expressed in crypt CD44+NGFR+ cells and donor-matched surface TotalEpi subsets obtained from three donors. The number of genes more highly expressed in crypt or in surface by at least a 1.5-fold differential is shown in the red and blue areas, respectively. The number in the overlapping region represents the number of transcripts that displayed a <1.5-fold differential expression between the two sites. Those with crypt-to-surface ratios of log2-transformed values ≥0.99 and ≤1.01 were considered to be equally expressed and used for analysis.
(D) The top ten biological processes associated with genes equally expressed in surface and crypt CD44+NGFR+ subsets based on gene ontology assignment. SRP, signal recognition particle.
(E) Hierarchical cluster analysis of CD44+NGFR+ cells and their respective TotalEpi subsets generated by identifying the genes that displayed a 2-fold or higher differential expression in the Illumina data comparisons. Shown in red are genes that were highly overexpressed in CD44+NGFR+ cells in comparison with the TotalEpi populations. Shown in green are those more highly expressed in the two TotalEpi populations included in this comparison (n = 2, p ≤ 0.05).
(F) The top ten biological GO categories found to be upregulated in crypt CD44+NGFR+ cells compared with the TotalEpi populations.
(G) The top ten biological GO categories upregulated in the TotalEpi populations compared with CD44+NGFR+ crypt cells.
See also Figure S2.
Comparison of the transcripts in the CD44+NGFR+ cells with those in the matched total epithelial cell isolates using a threshold difference in expression of ≥2-fold identified a group of genes that were more highly expressed in the crypt CFC-enriched cells from two donors (compared with their total crypt epithelial counterparts) and another gene set that was more highly expressed in the total crypt epithelial cell population. Analysis of both of these sets of differentially expressed genes by hierarchical clustering and GO (Figures 6E–6G) showed that the first set of genes that were more highly expressed in the crypt CD44+NGFR+ cells were associated with cell migration and motility (Figure 6F) and that those showing higher expression in both the surface and crypt epithelial cells were linked to epidermal development and keratinization (Figure 6G; Figure S2E).
HPV16 E6/E7 Oncoproteins Alter the In Vitro Growth of CD44+NGFR+ Human Tonsillar Epithelial Cells
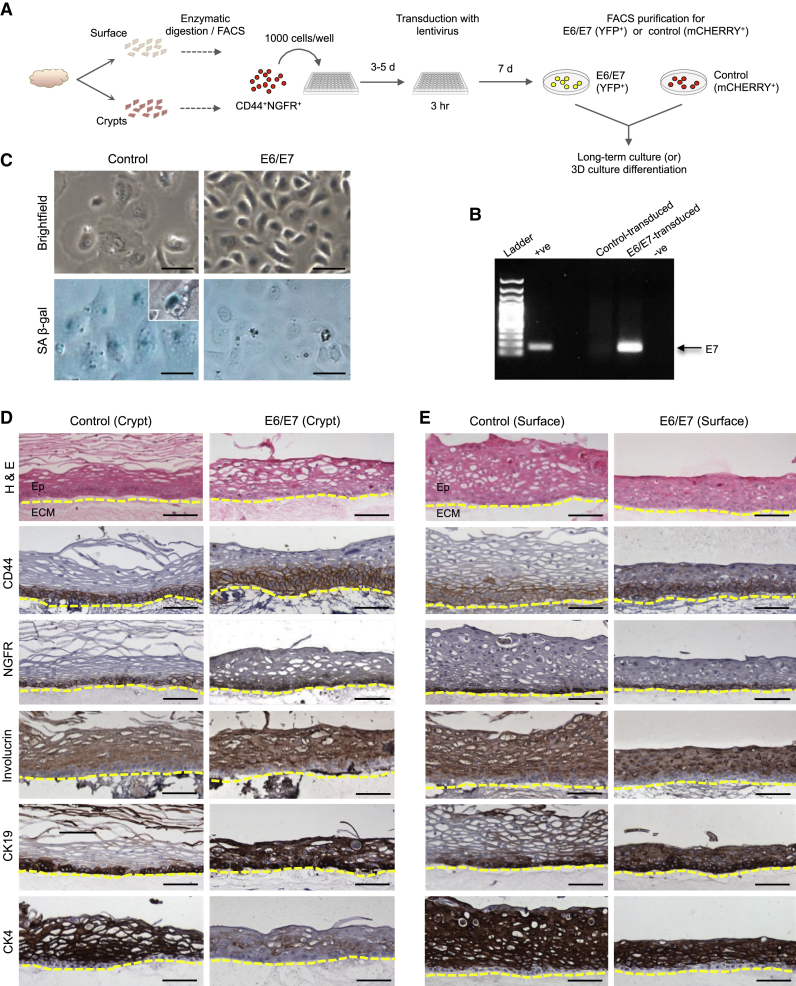
To determine whether the E6/E7 oncoproteins encoded by HPV16 can alter the growth of tonsillar epithelial cells, we produced an E6/E7 yellow fluorescent protein (YFP) lentivirus and a control lentivirus encoding a fluorescent mCHERRY reporter and then used these to transduce purified CD44+NGFR+ surface and crypt cells (at ∼80% efficiency; Figure S3). Expression of the E6/E7 cassette in FACS-purified YFP+ (E6/E7-transduced) cells and mCHERRY+ (control-transduced) cells (Figure 7A) was demonstrated by PCR amplification of the E7 sequence (Figure 7B). Both surface- and crypt-derived control cells maintained in 2D cultures displayed characteristics of senescence within four passages in vitro (i.e., acquisition of a large “fried egg” morphology and positive staining for senescence-associated β-galactosidase (SA β-gal) within 22–25 days; Figure 7C). In contrast, both surface and crypt E6/E7-transduced cells were able to proliferate for many more passages (>10 and >14 weeks, respectively) before any sign of senescence became apparent (Figure 7B).
Figure 7.
Effect of HPV16-E6/E7 Expression in CD44+NGFR+ Crypt Cells
(A) Experimental design.
(B) PCR showing stable expression of Lenti-HPV16-E6/E7 in transduced crypt cells after 13 days in culture (third passage). The ethidium bromide-stained bands represent the E7 gene. +ve, positive; -ve, negative.
(C) SA β-gal staining of the cells produced after 22 days in vitro under 2D CFC assay conditions by control and E6/E7-expressing cells. Scale bars, 50 μm.
(D) IHC of cells produced in cultures of control- and E6/E7-transduced crypt CD44+NGFR+ cells seeded at 4 × 105 cells/well in 3D differentiation cultures. Scale bars, 100 μm.
(E) IHC of cells produced in cultures of control- and E6/E7-transduced surface CD44+NGFR+ cells seeded at 4 × 105 cells/well in 3D differentiation cultures. Scale bar, 100 μm.
See also Figure S3.
To investigate whether E6/E7 oncoproteins would also affect the differentiation activity of tonsillar epithelial cells, we plated E6/E7-expressing (YFP+) surface and crypt cells and their (mCHERRY+) controls in 3D cultures at 4 × 105 cells/well. Multilayered epithelial populations were generated from all cell sources, but the structures produced by the E6/E7-expressing crypt cells showed an obviously less differentiated morphology than those produced in the control cultures, as evidenced by an absence of keratinization, fewer cell layers, an expansion of CD44-positive cells (normally basal/parabasal cell-restricted), a pan-epithelial expression of CK19 (normally a basal cell marker), and a negative or equivocal expression of CK4 (a differentiation marker of the cells normally expressed very strongly in the suprabasal cell layers) (Figure 7D). In the cultures of E6/E7-transduced surface epithelial cells, however, an induced pan-epithelial expression of CK19 was also seen, but not the changes in CD44 and CK4 expression seen in the cultures of E6/E7-transduced crypt cells (Figure 7E).
Discussion
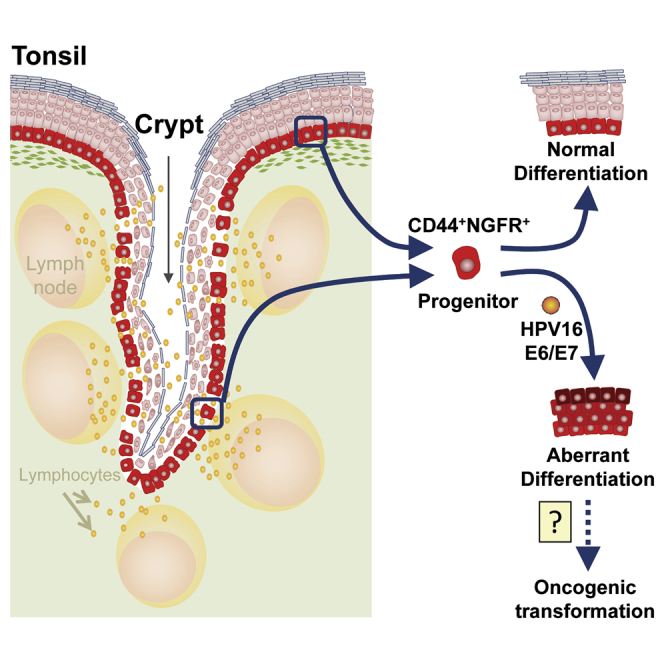
The epithelial cells in the crypts of human tonsils form a “reticulated” epithelium, a term that denotes their unique mesh-like organization in the crypt regions where they are interspersed with lymphocytes (Stohr, 1884), although the degree of reticulation varies markedly between individuals (Reibel and Sørensen, 1991). The non-epithelial cells in the crypt are thought to induce multiple changes in the neighboring epithelial cells, including a rearranged cytoskeleton of the tonofilaments as seen in squamous epithelial cell populations, increased numbers of ribosomes and mitochondria, redistributed desmosomal contacts, and changes in the expression of keratin (Perry, 1994, Sato et al., 1988). These effects have been described historically as “lymphoepithelial symbiosis” (Fioretti, 1957) and have been thought to induce a loss of many of the typical properties of epithelial cells (Perry, 1994). However, very little attention has been devoted to investigating the progenitor cells that maintain the different features of the epithelial cells located on the surface and in the crypts of the tonsil or how the different sources of epithelial cells might differ in their growth and differentiation potential, regulation, and susceptibility to HPV E6/E7-mediated oncogenic changes.
Here we show that epithelial progenitor cells can be viably isolated from both crypt and surface regions of normal human tonsils using a highly efficient quantitative 2D clonogenic assay protocol and a 3D culture system that also promotes their differentiation into a multi-layered organotypic structure. We further show that the progenitors identified using these two assays share a common CD44+NGFR+ phenotype that allows them to be selectively purified; CD44+NGFR+ subpopulations obtained from surface and crypt are transcriptionally similar. CD44 is a well established marker of basally located cells in many epithelia (Desai et al., 2000, Guo et al., 2012, Louderbough et al., 2011, Paik et al., 2012). Likewise, NGFR (also referred to as p75[NTR] or CD271) has been reported to characterize esophageal keratinocyte stem cells (Okumura et al., 2003) and show restricted expression in the basal layers of oral mucosal epithelium (Gemenetzidis et al., 2010). Co-expression of these two receptors on tonsillar epithelial progenitors indicates that these cells express a site-independent receptor for hyaluronan (CD44) that is known to be important for cell-cell interactions, cell adhesion, homing, and migration and one for NGF that can regulate cell survival and proliferation, at least in neural cells (Smith et al., 1994).
The finding that CD44+NGFR+ cells isolated from both surface and crypt sites contain most of the clonogenic progenitor activity in 2D assays and are also highly enriched in their content of these cells indicates that they are also biologically similar. This finding was further supported by the demonstration of a molecular similarity of both subsets, as indicated by their similar transcriptomes. Further support for this conclusion was provided by the finding that both sources of CD44+NGFR+ cells can recapitulate, in vitro, the complex differentiation programs exclusively displayed in the surface epithelium of the tonsil in vivo. Therefore, the possession of this developmental potential is not site-dependent whereas its expression appears to be. On the other hand, it is interesting to note that this subset of crypt epithelial cells appears to be more highly enriched in progenitors with greater proliferative potential, as shown by the reduced propensity and speed with which their progeny became incapable of further growth and acquired other features of differentiated epithelial cells.
Finally, we show that tonsillar epithelial progenitors can be readily transduced with standard lentiviral vectors and show the ability of this approach to analyze the effect of forced expression of the HPV16 E6/E7 oncogenes on their growth and differentiation in vitro. Tonsillar epithelium has been given increasing attention over the past decade because of the recent rise in HPV-associated oropharyngeal carcinoma (Chaturvedi et al., 2008). A recently published large-scale cohort study showed that HPV16-encoded E6 antigens were present in “prediagnostic” sera of approximately one-third of individuals who were later diagnosed with oropharyngeal cancer (Kreimer et al., 2013). Unlike HPV-associated cervical carcinoma, where exposure of potential target cells requires damage to the stratified epithelial tissue (Roberts et al., 2007), the discontinuous organization of epithelial progenitors in the crypt of the tonsil is thought to make these cells more accessible to HPV infection than their counterparts on the surface. HPV16 DNA is frequently detected by in situ hybridization in tonsillar squamous cell carcinoma, and expression of p16, the most commonly used marker of biologically active E6/E7, is generally confined to the crypt epithelium (Begum et al., 2005, Kim et al., 2007). However, the actual cellular targets of HPV infection in the human tonsil have not been defined (Begum et al., 2005, El-Mofty and Patil, 2006). Here we provide initial proof-of-principle evidence that the E6/E7 oncogenes encoded by HPV16 are sufficient to induce changes in the growth and differentiation control of the primitive progenitor-enriched population of CD44+NGFR+ epithelial cells present in normal human tonsillar crypts. These findings demonstrate the power of the systems described here for isolating, characterizing, and genetically manipulating these progenitors. They therefore set the stage for further interrogation of the mechanisms by which the biology of these primitive human tonsillar cells is normally controlled and how it may be oncogenically altered by HPV infection.
Experimental Procedures
Cell Preparation
We obtained human tonsil specimens from patients undergoing routine tonsillectomy for chronic/recurrent tonsillitis (n = 38, ages 17–45), and children undergoing tonsillectomy (n = 10, ages 4–13), with informed consent procedures approved by the University of British Columbia Research Ethics Board. Healthy tonsils are sites of permanent activation of lymphoid cells. Therefore, any non-malignant tonsils determined by the absence of active pathological lesions were considered “normal” for the purposes of this study. Resected tonsils were first examined grossly, crypt and surface regions were dissected separately using a scalpel as required, and the tissues were then immediately placed in complete dissociation medium (CDM) containing DMEM (Life Technologies) supplemented with 5% fetal bovine serum (FBS), 10 μg/ml ciprofloxacin (Life Technologies), and a 1:50 dilution of antibiotic-antimycotic (100×, Life Technologies) on ice until processed. Tissue was then finely minced with scalpels in a glass petri dish and then enzymatically dissociated in CDM supplemented with 1 mg/ml collagenase (Life Technologies) and 500 μg/ml dispase (STEMCELL Technologies) for 7–12 hr at 37°C. The resultant cell suspension was washed with CDM twice by centrifugation at 1,200 rpm for 4 min. We then treated the pellet with 1× red blood cell (RBC) lysis buffer (BioLegend) for 5 min on ice to remove contaminating RBCs. The pellet obtained by centrifugation at 1,500 rpm for 4 min was enzymatically and mechanically dissociated into single cells in 0.05% Trypsin/EDTA and DNase I (Roche) for 4–6 min, passed through a 100-μm filter (BD Biosciences) to remove debris, washed with PBS (Life Technologies) containing 2% FBS (2% PBS supplemented with 2% FBS [PF]), and centrifuged at 1,500 rpm for 4 min. This final pellet was resuspended in 2% PF and passed through a 40-μm filter to remove remaining cell aggregates and debris. In some experiments, red blood cells and granulocytes were removed by Ficoll-density centrifugation.
To remove lymphocytes and endothelial cells, we labeled 108 cells/ml with biotinylated antibodies against CD45 (clone HI30, BD Pharmingen) at 1:25 and CD31 (clone WM59, eBioscience) at 1:50 and incubated the suspension for 15 min at room temperature. We then added EasySep biotin selection cocktail at 100 μl/ml cells, incubated for another 15 min at room temperature, followed by addition of EasySep magnetic nanoparticles (STEMCELL) at 50 μg/ml and 10 min of incubation at room temperature. We then brought the total volume to 2.5 ml by adding 2% PF, gently mixed the cells with a pipette, and let the suspension stand in the magnet at room temperature for 5 min to allow the magnetized CD45+ cells and CD31+ cells to bind to the wall of the tube. The unbound cell suspension was then poured off into a new tube. This incubation/pour-off step was repeated twice.
To further fractionate the cells, we restained the immunomagnetically CD45 and CD31-depleted cells with streptavidin-allophycocyanin (APC, BD Pharmingen) or streptavidin-fluorescein isothiocyanate (FITC, BD Pharmingen) and then anti-CD44-phycoerythrin (PE, clone 515, BD Pharmingen) or anti-PE-Cy7, anti-CD271(NGFR)-APC, or anti-FITC (clone ME20.4-1.H4, MACS) to subfractionate the CD45−CD31− cells as described. If biotin pre-selection was omitted, then CD45 and CD31 were labeled with anti-CD45-pacific blue and anti-CD31-pacific blue. Cells were then sorted using an Influx II, FACSAria II, or FACSAria III (BD Biosciences), and data were analyzed using FlowJo.
Immunohistochemical Analysis
A tissue array was constructed from formalin-fixed, paraffin-embedded (FFPE) blocks containing 10 normal tonsillectomy samples. We microscopically examined the H&E slides of tissue cut from blocks to identify the region that contains both tonsillar surface epithelium and crypts. We then excised the selected area (typically 0.7–0.9 cm in diameter) from each block using scalpels and re-embedded the isolated tissue to create a block of five cases (hereafter referred to as macroarray). The blocks were cut into 4- to 5-μm sections, deparaffinized in xylene, and serially hydrated in alcohol solutions, followed by 20-min heat-induced epitope retrieval in Dako target retrieval solution. Endogenous peroxidase was blocked using Dako blocking buffer for 10 min, followed by 10% PF for 15 min. Primary antibodies were diluted in Dako antibody diluent, and tissue was incubated at room temperature for 2–3 hr or at 4°C overnight. Primary antibodies were detected using their horseradish peroxidase (HRP)-linked secondary antibody diluted previously and included in the Dako mouse-specific or rabbit-specific EnVision+ kits. Slides were washed three times for 5 min each in PBS between incubations. HRP activity was detected using the Dako liquid 3,3′-diaminobenzidine (DAB+) substrate chromagen system. Tissue was then stained with hematoxylin (Surgipath), coverslipped, and stored at room temperature. Antibodies used were as follows: PE-conjugated anti-CD44 (clone 515, BD Pharmingen), anti-NGFR-FITC (clone ME20.4-1.H4, Miltenyi Biotec), anti-CD227 (MUC1, clone human metapneumovirus [HMPV], BD Pharmingen), anti-CK4 (clone 6B10, Santa Cruz Biotechnology), anti-CK5 (clone 3E2F1, Santa Cruz), anti-CK13 (clone EPR3671, Novus Biologicals), anti-CK14 (clone SP53, Cell Marque), anti-CK8/18 (clone 4546, Cell Signaling Technology), anti-CK19 (clone A53-B/A2-26, Cell Marque), anti-involucrin (clone SPM259, Santa Cruz), anti-Ki67 (clone MIB-1, Dako), anti-p63 (clone 4A4, Santa Cruz), and anti-Collagen IV (clone CIV22, Dako).
Immunocytochemical Analyses
FACS-purified subpopulations were spun independently onto glass slides at 700 rpm for 3 min, dried at room temperature, and fixed in a 1:1 v/v mixture of methanol and acetone for 15 min at −20°C. The fixed cells were either stained immediately for intracellular proteins as described above or kept at −80°C until used later.
2D CFC Assay Cultures
We seeded each well of a 6-well plate with test cells combined with 105 irradiated (50 Gy of 300 peak kilovoltage [kVp] X-rays) mouse embryo fibroblast NIH 3T3 cells in tonsil epithelial cell medium (TEpiCM, ScienCell) supplemented with 5% FBS. Cultures were incubated at 37°C and 5% CO2 for 2 days and then changed to serum-free TEpiCM. In the later stage of the study, we supplemented the medium with 10 μM ROCK inhibitor (Y-27632, Cellagen Technology). Cultures were incubated in either 20% or 5% O2 depending on the experiments. After 7–10 days, we fixed wells in a 1:1 v/v mixture of methanol and acetone at −20°C for 15 min. The fixed colonies were stained immediately with hematoxylin for scoring or with various antibodies for IHC analysis as described above. Remaining colonies were kept in PBS at 4°C for a maximum of 4 weeks until used for IHC.
Organotypic 3D Cultures
A 3D collagen matrix consisting of a bottom acellular layer and a top cellular layer was prepared by mixing a rat tail collagen I solution (BD Biosciences) with DMEM (Life Technologies) and reconstitution buffer, FBS, and NaOH to give a 75% collagen solution in DMEM containing 5% FBS. For creationg of the acellular layer, 1 ml of the collagen solution was added into each insert in a 6-well transwell plate (Corning) and placed in a 37°C incubator until the collagen solidified. For creation of the cellular layer, mouse fibroblasts were added to the remaining collagen solution to give a final concentration of 5,000 cells/ml, 3 ml of which was added to each well already containing a solidified acellular layer. These collagen matrices were allowed to contract for 5–7 days in DMEM supplemented with 10% FBS at 37°C, and then freshly dissociated human tonsillar epithelial cells were seeded on top of the matrices. For the first 4 days after addition of test cells, cultures were fed with epithelialization (Ep 1) medium containing DMEM:Ham’s F12 (3:1) supplemented with 180 μM adenine, 5 μg/ml insulin, 5 μg/ml transferrin, 20 pM T3, 0.4 μg/ml hydrocortisone, and 0.2% FBS. For the following 2–3 days, the medium was replaced with Ep 2 medium, which was identical to Ep 1 medium except that it contained 2% FBS instead of 0.2% FBS. The medium was then removed from the inserts to create an air-liquid interface and cultured in fresh Ep 2 medium for 10–12 days, after which cultures were harvested by fixing in 10% neutralized buffered formalin (Fisher Scientific) and, later, paraffin-embedded, sectioned, stained, and analyzed.
Transcriptome Analysis
Total RNA was extracted using the AllPrep DNA/RNA micro kit (QIAGEN). Based on the RNA quality assessment with the Agilent Bioanalyzer 2100 (Agilent Technologies) using the Agilent RNA 6000 nanokit or picokit (Agilent Technologies), samples with an RNA integrity number of 7.0 or higher were selected for gene expression profiling. Quality control and gene expression profiling were performed at The Centre for Applied Genomics. Briefly, 70 ng of RNA was labeled with the TotalTM-96 RNA amplification kit (Ambion), and cRNA was hybridized to the Illumina HumanHT-12 V4 BeadChip according to the manufacturer’s specifications for the whole genome gene expression direct hybridization assay workflow. RNA was amplified using the Ambion Illumina TotalTM-96 RNA amplification kit and biotinylated using Biotin-NTP mix. Samples were incubated for 18 hr at 58°C for hybridization. Slides were subsequently washed and scanned using the Illumina BeadArray reader obtain raw image and data files. Raw signals were processed with GenomeStudio (Illumina, version V20011.1) and analyzed further with the Partek Genome Suite (Partek). Unsupervised hierarchical clustering was performed on log2-transformed data using Gene-E (http://www.broadinstitute.org/cancer/software/GENE-E/index.html), whereas principal component analysis (PCA) was performed in log2-transformed data using R statistical software version 3.1.2 using the “prcomp” function (R Development Core Team, 2014). Transcript level differences of ≥1.5 with p ≤ 0.05 were considered significant.
Lentiviral Preparations
Human papillomavirus E6 and E7 oncogenes were cloned from pLXSN-HPV16-E6/E7 (Addgene, plasmid ID 52394; Halbert et al., 1991) into the pLKO.1 YFP lentiviral vector (Lenti-HPV16-E6/E7-YFP) and sequence-verified. Virus-containing supernatants were produced and used to transduce test cells as described previously (Imren et al., 2004), with minor adaptations for tonsil cells. Briefly, the freshly sorted CFC-enriched populations were infected in 100 μl of TEpiCM containing 5% FBS, 10 μM ROCK inhibitor, and lentiviral supernatant (either Lenti-HPV16-E6/E7-YFP or Lenti-mCHERRY) for 3 hr at 37°C. Subsequently, cells were washed with serum-free TEpiCM supplemented with 10 μM ROCK inhibitor and incubated for 4–5 days in the same medium before isolation of the transduced (YFP+ or mCHERRY+) cells. Stable expression of oncogenes in the transduced cells was verified by PCR using the following primers: forward, GAACCGGACAGAGCCCATTA; reverse, TCTGAGAACAGATGGGGCAC.
Statistical Analysis
GraphPad Prism 6 software was used for generation of all graphs and statistical analyses. Numerical data are shown as mean ± SEM or SD and are based on a minimum of three repeats for each independent biological sample. p Values were generated using Student’s t test.
Author Contributions
S.Y.C.K., N.K., C.J.E., and M.P.R conceptualized the project. S.Y.C.K. and N.K. designed the experiments. S.Y.C.K. performed the experiments. All authors contributed to the analysis and interpretation of the data. S.Y.C.K. and C.J.E. wrote the manuscript. All authors edited the manuscript and approved the final version.
Acknowledgments
The authors thank the surgeons, pathologists, and technologists associated with the THINC study (HPV in Tonsils: Natural History of Infection and Role in Oral Cancers): Drs. K. Berean, R. O’Connor, M.J. Dickson, S. Durham, P. Lee, P. Mick, and D. Mintz; the technologists S. Ramji and M. Lok for assistance with obtaining tonsillectomy samples; D. Ko and W. Xu for assistance with flow cytometry (Flow Cytometry Facility, Terry Fox Laboratory); I. Sun for excellent technical support; and M. Hale and G. Edin for assistance with the production of lentivirus containing HPV16-E6/E7. N.K. was supported by CBCF-BC/Yukon and MITACS Elevate postdoctoral fellowships. This work was supported by grants from the Canadian Cancer Society Research Institute (to M.P.R) and the Canadian Cancer Research Society (to C.J.E.).
Published: October 29, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes three figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.09.020.
Accession Numbers
The accession number for the microarray data reported in this paper is NCBI GEO: GSE69767.
Supplemental Information
References
- Barrandon Y., Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum S., Cao D., Gillison M., Zahurak M., Westra W.H. Tissue distribution of human papillomavirus 16 DNA integration in patients with tonsillar carcinoma. Clin. Cancer Res. 2005;11:5694–5699. doi: 10.1158/1078-0432.CCR-05-0587. [DOI] [PubMed] [Google Scholar]
- Casteleyn C., Breugelmans S., Simoens P., Van den Broeck W. The tonsils revisited: review of the anatomical localization and histological characteristics of the tonsils of domestic and laboratory animals. Clin. Dev. Immunol. 2011;2011:472460. doi: 10.1155/2011/472460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman S., Liu X., Meyers C., Schlegel R., McBride A.A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Invest. 2010;120:2619–2626. doi: 10.1172/JCI42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A.K., Engels E.A., Anderson W.F., Gillison M.L. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J. Clin. Oncol. 2008;26:612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- Cooper D., Schermer A., Sun T.-T. Biology of disease. Lab. Invest. 1985;52:243–255. [PubMed] [Google Scholar]
- Desai S., Lim S.D., Jimenez R.E., Chun T., Keane T.E., McKenney J.K., Zavala-Pompa A., Cohen C., Young R.H., Amin M.B. Relationship of cytokeratin 20 and CD44 protein expression with WHO/ISUP grade in pTa and pT1 papillary urothelial neoplasia. Mod. Pathol. 2000;13:1315–1323. doi: 10.1038/modpathol.3880241. [DOI] [PubMed] [Google Scholar]
- El-Mofty S.K., Patil S. Human papillomavirus (HPV)-related oropharyngeal nonkeratinizing squamous cell carcinoma: characterization of a distinct phenotype. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2006;101:339–345. doi: 10.1016/j.tripleo.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Fioretti A. Deca; 1957. La Tonsilla Palatina. [Google Scholar]
- Gemenetzidis E., Elena-Costea D., Parkinson E.K., Waseem A., Wan H., Teh M.T. Induction of human epithelial stem/progenitor expansion by FOXM1. Cancer Res. 2010;70:9515–9526. doi: 10.1158/0008-5472.CAN-10-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison M.L., Koch W.M., Capone R.B., Spafford M., Westra W.H., Wu L., Zahurak M.L., Daniel R.W., Viglione M., Symer D.E. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J. Natl. Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- Guo C., Liu H., Zhang B.H., Cadaneanu R.M., Mayle A.M., Garraway I.P. Epcam, CD44, and CD49f distinguish sphere-forming human prostate basal cells from a subpopulation with predominant tubule initiation capability. PLoS ONE. 2012;7:e34219. doi: 10.1371/journal.pone.0034219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert C.L., Demers G.W., Galloway D.A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie A.J. Scanning and transmission electron microscopy on the epithelium of human palatine tonsils. J. Pathol. 1980;130:91–98. doi: 10.1002/path.1711300205. [DOI] [PubMed] [Google Scholar]
- Imren S., Fabry M.E., Westerman K.A., Pawliuk R., Tang P., Rosten P.M., Nagel R.L., Leboulch P., Eaves C.J., Humphries R.K. High-level beta-globin expression and preferred intragenic integration after lentiviral transduction of human cord blood stem cells. J. Clin. Invest. 2004;114:953–962. doi: 10.1172/JCI21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Koo B.S., Kang S., Park K., Kim H., Lee K.R., Lee M.J., Kim J.M., Choi E.C., Cho N.H. HPV integration begins in the tonsillar crypt and leads to the alteration of p16, EGFR and c-myc during tumor formation. Int. J. Cancer. 2007;120:1418–1425. doi: 10.1002/ijc.22464. [DOI] [PubMed] [Google Scholar]
- Kreimer A.R., Johansson M., Waterboer T., Kaaks R., Chang-Claude J., Drogen D., Tjønneland A., Overvad K., Quirós J.R., González C.A. Evaluation of human papillomavirus antibodies and risk of subsequent head and neck cancer. J. Clin. Oncol. 2013;31:2708–2715. doi: 10.1200/JCO.2012.47.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louderbough J.M.V., Brown J.A., Nagle R.B., Schroeder J.A. CD44 Promotes Epithelial Mammary Gland Development and Exhibits Altered Localization during Cancer Progression. Genes Cancer. 2011;2:771–781. doi: 10.1177/1947601911428223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarem M., Kannan N., Nguyen L.V., Knapp D.J.H.F., Balani S., Prater M.D., Stingl J., Raouf A., Nemirovsky O., Eirew P., Eaves C.J. Developmental changes in the in vitro activated regenerative activity of primitive mammary epithelial cells. PLoS Biol. 2013;11:e1001630. doi: 10.1371/journal.pbio.1001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matusik R.J., Jin R.J., Sun Q., Wang Y., Yu X., Gupta A., Nandana S., Case T.C., Paul M., Mirosevich J. Prostate epithelial cell fate. Differentiation. 2008;76:682–698. doi: 10.1111/j.1432-0436.2008.00276.x. [DOI] [PubMed] [Google Scholar]
- Moll R., Franke W.W., Schiller D.L., Geiger B., Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Okawa T., Michaylira C.Z., Kalabis J., Stairs D.B., Nakagawa H., Andl C.D., Johnstone C.N., Klein-Szanto A.J., El-Deiry W.S., Cukierman E. The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation. Genes Dev. 2007;21:2788–2803. doi: 10.1101/gad.1544507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura T., Shimada Y., Imamura M., Yasumoto S. Neurotrophin receptor p75(NTR) characterizes human esophageal keratinocyte stem cells in vitro. Oncogene. 2003;22:4017–4026. doi: 10.1038/sj.onc.1206525. [DOI] [PubMed] [Google Scholar]
- Paik D.Y., Janzen D.M., Schafenacker A.M., Velasco V.S., Shung M.S., Cheng D., Huang J., Witte O.N., Memarzadeh S. Stem-like epithelial cells are concentrated in the distal end of the fallopian tube: a site for injury and serous cancer initiation. Stem Cells. 2012;30:2487–2497. doi: 10.1002/stem.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini G., Dellambra E., Golisano O., Martinelli E., Fantozzi I., Bondanza S., Ponzin D., McKeon F., De Luca M. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA. 2001;98:3156–3161. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M.E. The specialised structure of crypt epithelium in the human palatine tonsil and its functional significance. J. Anat. 1994;185:111–127. [PMC free article] [PubMed] [Google Scholar]
- Perry M., Whyte A. Immunology of the tonsils. Immunol. Today. 1998;19:414–421. doi: 10.1016/s0167-5699(98)01307-3. [DOI] [PubMed] [Google Scholar]
- R Development Core Team . R Foundation for Statistical Computing; 2014. R: A language and environment for statistical computing. [Google Scholar]
- Reibel J., Sørensen C.H. Association between keratin staining patterns and the structural and functional aspects of palatine tonsil epithelium. APMIS. 1991;99:905–915. doi: 10.1111/j.1699-0463.1991.tb01278.x. [DOI] [PubMed] [Google Scholar]
- Roberts J.N., Buck C.B., Thompson C.D., Kines R., Bernardo M., Choyke P.L., Lowy D.R., Schiller J.T. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat. Med. 2007;13:857–861. doi: 10.1038/nm1598. [DOI] [PubMed] [Google Scholar]
- Rock J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., Hogan B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Wake K., Watanabe I. Changes in cell shapes and cytokeratins of epithelial cells during the infiltration of lymphocytes in the human palatine tonsils. Acta Otolaryngol. Suppl. 1988;454:48–52. doi: 10.3109/00016488809125004. [DOI] [PubMed] [Google Scholar]
- Senoo M., Pinto F., Crum C.P., McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Smith C.A., Farrah T., Goodwin R.G. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- Stohr P. Über Mandeln und Balgdrüsen. Virchows Arch. 1884;97:211–236. [Google Scholar]
- Van Keymeulen A., Blanpain C. Tracing epithelial stem cells during development, homeostasis, and repair. J. Cell Biol. 2012;197:575–584. doi: 10.1083/jcb.201201041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K., Sasai Y. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Zhang L., Valdez J.M., Zhang B., Wei L., Chang J., Xin L. ROCK inhibitor Y-27632 suppresses dissociation-induced apoptosis of murine prostate stem/progenitor cells and increases their cloning efficiency. PLoS ONE. 2011;6:e18271. doi: 10.1371/journal.pone.0018271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.