Summary
Insertion of reporter cassettes into the Lgr5 locus has enabled the characterization of mouse intestinal stem cells (ISCs). However, low cell surface abundance of LGR5 protein and lack of high-affinity anti-LGR5 antibodies represent a roadblock to efficiently isolate human colonic stem cells (hCoSCs). We set out to identify stem cell markers that would allow for purification of hCoSCs. In an unbiased approach, membrane-enriched protein fractions derived from in vitro human colonic organoids were analyzed by quantitative mass spectrometry. Protein tyrosine pseudokinase PTK7 specified a cell population within human colonic organoids characterized by highest self-renewal and re-seeding capacity. Antibodies recognizing the extracellular domain of PTK7 allowed us to isolate and expand hCoSCs directly from patient-derived mucosa samples. Human PTK7+ cells display features of canonical Lgr5+ ISCs and include a fraction of cells that undergo differentiation toward enteroendocrine lineage that resemble crypt label retaining cells (LRCs).
Highlights
-
•
Ex vivo cultured human colon stem cells (hCoSCs) express the surface marker PTK7
-
•
hCoSCs with highest PTK7 surface abundance display highest self-renewal capacity
-
•
Antibodies against PTK7 enable purification of hCoSCs from patient samples
-
•
A fraction of PTK7+ hCoSCs display genetic features of crypt label retaining cells
In this article, Batlle and colleagues show that the protein pseudo-kinase 7 (PTK7) represents a highly specific marker for human colonic stem cells (hCoSCs). PTK7 surface abundance can be used to enrich for hCoSCs from in vitro colonic organoid cultures and patient-derived mucosa samples. PTK7+ hCoSC fractions display genetic features of canonical Lgr5+ intestinal stem cells and slow cycling crypt label retaining cells (LRCs).
Introduction
The epithelium that lines the small intestine and the colon is in constant self-renewal with a life cycle of 3–4 days (Clevers, 2013a). Regeneration occurs from units embedded within the submucosa known as crypts, each containing about 6–10 intestinal stem cells (ISCs) (Clevers, 2013a). Differentiated epithelial cells reside at the surface. The biology of mouse ISCs has been intensively studied in recent years. They express Lgr5, a receptor for the WNT pathway agonist R-Spondin 1 (R-SPO) (Barker et al., 2007, de Lau et al., 2011, Leushacke and Barker, 2014). Mouse Lgr5+ cells are rapidly dividing multipotent stem cells that reside at the bottommost positions of the small intestine and colonic crypts. Under culturing conditions that simulate the stem cell niche, mouse Lgr5+ cells form 3D structures in vitro, known as organoids (Sato et al., 2009).
The progeny of ISCs, transit amplifying cells or progenitors, will eventually give rise to the majority of differentiated cells as they migrate upward along the crypt axis. Recently it was shown that a fraction of Lgr5+ cells remains relatively slow proliferating and differentiates directly into less abundant differentiated lineages such as Paneth or enteroendocrine cells. This Lgr5+ cell subpopulation can be experimentally recognized by the retention of molecules that label DNA and therefore has been known as label retaining cells (LRCs) (Clevers, 2013b, Potten et al., 1974). They are characterized by co-expression of ISC-specific genes and markers of Paneth cells and enteroendocrine lineages (Buczacki et al., 2013, Grün et al., 2015). LRCs are fated to differentiate; nevertheless, under certain conditions, such as damage of the cycling ISC pool, they act as facultative stem cells and contribute to the regeneration of the intestinal epithelium (Buczacki et al., 2013). Mouse LRCs also expand as in vitro organoids (Buczacki et al., 2013).
There is a growing interest in understanding the biology of human colonic stem cells (hCoSCs), as it is widely believed that they play key roles in various human intestinal disorders (Barker et al., 2009). Furthermore, a better understanding of this cell population is key to develop therapeutic approaches to regenerate disease-affected tissue (Yui et al., 2012). We recently showed that in media containing WNT3a, R-SPO, EGF, and Noggin (WREN media) human colonic crypts form organoids that expand as undifferentiated 3D cell cultures (Jung et al., 2011). However, only around 5%–6% of the cells within these cultures remain as multipotent stem cells. We searched for surface markers that identify bona fide hCoSCs from human organoid cultures. We characterize protein tyrosine pseudokinase 7 (PTK7) as a hCoSC marker that allows purification and optimized ex vivo expansion of self-renewing multipotent hCoSCs.
Results
Quantitative Mass Spectrometry Analyses of Human Colon Organoid-Derived Membrane Proteins
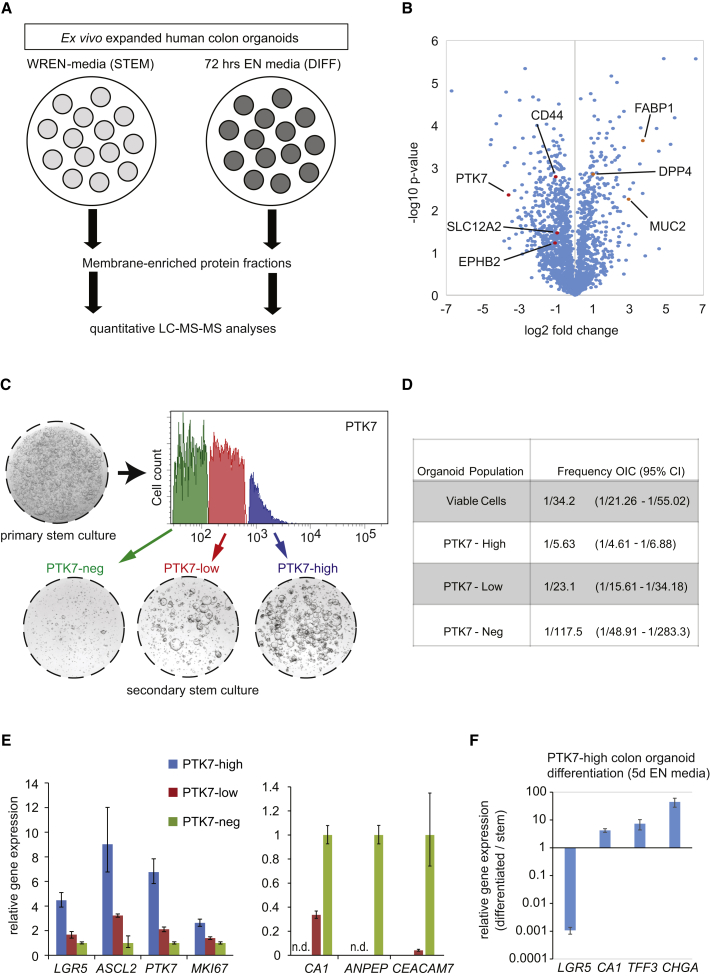
We searched for surface markers that would enable discrimination of distinct cell populations in human colon organoids (Jung et al., 2011). By quantitative mass spectrometry (q-LC-MS-MS) analyses, we compared plasma membrane-enriched protein fractions of human colon crypts cultured in stem (WREN) versus differentiated (media containing EGF and Noggin [EN]) conditions (Jung et al., 2011) (Figure 1A). Whereas this experimental setting may not exactly reflect lineage specification and differentiation present in vivo, we reasoned that a subset of ISC marker genes would be differentially expressed between the two conditions. We identified 261 proteins that were enriched more than 2-fold in stem cells and 119 proteins in differentiated cells (t test, p value < 0.01; Table S1). Proteins significantly enriched in WREN or EN human colonic organoid cells (t test significance labeled as “+” in Table S1) were also enriched in membrane proteins when compared with the human proteome (Table S2). Multiple markers of enterocytes, mucosecreting, and enteroendocrine cells were upregulated in EN organoid cells, including MUC2, FABP1, or DPP4 (Figure 1B; Table S1). Concomitantly, several previously identified ISC markers such as CD44 (Gracz et al., 2013), SLC12A2 (Whissell et al., 2014), and EPHB2 (Merlos-Suárez et al., 2011) were upregulated under WREN conditions (Figure 1B; Table S1). We focused our attention on PTK7, a transmembrane pseudokinase that regulates canonical and non-canonical WNT signaling during embryo development (Puppo et al., 2011) (Figure 1B). PTK7 was also found in a second independent proteomic study (Figure S1A; Table S1).
Figure 1.
Quantitative Mass Spectrometry Reveals PTK7 Is Enriched in Ex Vivo Cultured hCoSCs and Specifies Organoid Cells with Highest Self-Renewal Capacity
(A) Experimental scheme: Human colonic organoids were cultured under stem cell (WREN, STEM) or differentiation (EN, DIFF) conditions. Differentiation was induced for 72 hr. After isolation of membrane-enriched protein fractions, samples were subjected to quantitative mass spectrometry (LC-MS-MS).
(B) Volcano plot of statistical significance (−log10 p value) against fold-change (log2 fold change) between human colonic organoid cultures maintained in either WREN or EN media. Negative fold changes represent organoid stem cell-enriched proteins (see Table S1 for all data).
See Figure S1 and Table S1 for an additional experiment.
(C) Fractionation of single cells from in vitro cultured organoids by FACS according to PTK7 surface abundance (blue: PTK7-High, red: PTK7-Medium, green: PTK7-Neg) (upper). Same cell numbers of each fraction were seeded, and the corresponding organoid-forming capacity was assessed (bottom).
(D) Estimated frequencies of organoid-initiating cells (OIC) in different subpopulations of in vitro grown human colonic organoid cells isolated according to PTK7 surface abundance. 40, 150, and 400 cells of each fraction were plated, and frequencies were calculated by applying the maximum-likelihood estimation method of limiting dilutions assay (Hu and Smyth, 2009). CI, confidence interval.
(E) Quantitative real-time PCR (qRT-PCR) analysis of stem cell, proliferation (left), and differentiation marker (right) gene expression on different fractions of organoid cells purified according to PTK7 levels in (C).
Error bars indicate SD (n = 3 technical replicates, see Figures 2 and S2 for supporting results). n.d., not detectable.
(F) qRT-PCR analysis of LGR5 and differentiation marker gene expression in organoid cells purified according to PTK7 levels in (C), expanded in WREN media, and maintained under EN media for 5 days. Error bars indicate SD (n = 3 biological replicates).
High PTK7 Expression Levels Segregate with High Self-Renewal Capacity in Ex Vivo Cultured Human Colonic Crypts
In a previous study, we observed that only around 5%–6% of viable cells within human colonic organoids cultured in WREN media hold the capacity to reform organoids upon serial passaging (Jung et al., 2011). Our data suggested that only a subpopulation of organoid cells remains as bona fide stem cells (Jung et al., 2011). Flow-cytometry analysis of these cultures revealed heterogeneous surface expression of PTK7 (Figure 1C, upper). We found that cells with high PTK7 surface abundance exhibited a higher re-seeding capacity when compared with PTK7-low, or PTK7-negative cells (Figures 1C, 2B, S1B, and S2B). To determine the frequency of organoid re-initiating cells (OIC) of each PTK7 population, we plated different cell numbers (40, 150, and 400 viable cells) and counted the number of organoids after 1 week. We then calculated OIC frequencies by applying the maximum-likelihood estimation method for limiting dilutions assay (Hu and Smyth, 2009). Simply re-plating the total of viable cells yielded a frequency of one OIC every 34 cells (3%), in line with our previous observations (Jung et al., 2011). We calculated an OIC of 1 in 6 for PTK7-high (16.7%), 1 in 23 for PTK7-low (4.3%), and 1 in 118 (0.85%) for PTK7-negative cells (Figure 1D). Furthermore, PTK7-high cells expressed elevated levels of bona fide ISC marker genes such as LGR5 and ASCL2 (Figure 1E, left), whereas PTK7-low and PTK7-negative WREN-cultured organoid cells expressed relatively higher levels of intestinal differentiation genes, including ANPEP, CEACAM7, and CA1 (Figure 1E, right). Upon removal of growth factors, secondary organoid cultures derived from PTK7-high cells underwent multilineage differentiation (Figure 1F).
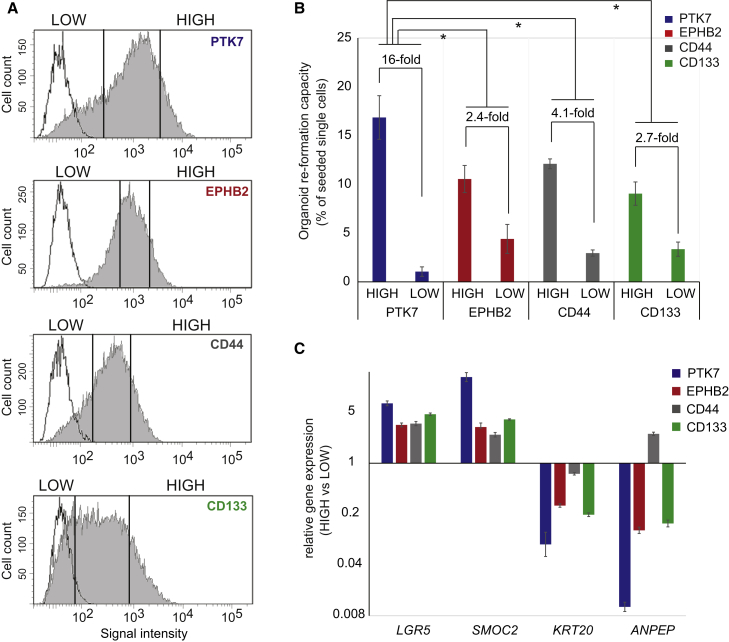
Figure 2.
Comparison of PTK7 to Other Stem Cell Markers Proofs Enhanced Capacity to Enrich for Self-Renewing Organoid Cells
(A) Human colonic organoids maintained in WREN media were disaggregated and stained against the indicated surface markers. Via FACS, 9%–10% of cells showing the brightest staining intensity (HIGH) and 20%–21% of cells showing the weakest intensity (LOW) were isolated. Control (transparent profile) and specific staining (gray profile) are shown in each histogram.
See Figures S2A–S2C for an equivalent experiment derived from a different individual.
(B) Organoid formation capacity of cellular fractions isolated via FACS as shown in (A). Each cellular fraction was seeded four times (biological replicate), and the frequency of organoid formation was assessed 8 days after seeding. Error bars indicate SEM. Significance of PTK7 high versus low enrichment over EPHB2, CD44, or CD133 performance was assessed using the F-test (Supplemental Experimental Procedures). ∗p < 0.02.
(C) qRT-PCR analysis of stem cell and differentiation marker gene expression on organoid cells HIGH and LOW for the indicated marker surface abundance according to (A) (see also Figure S2). Gene expression was assessed three times (technical replicate), and error bars indicate SD.
We and others have previously identified surface markers for purification of mouse and human adult stem cells from intestinal samples. These include EPHB2 (Jung et al., 2011, Merlos-Suárez et al., 2011), CD44 (Gracz et al., 2013, Wang et al., 2013), and Prominin-1/CD133 (Hou et al., 2011, Zhu et al., 2009). These markers showed heterogeneous surface expression in human colonic organoid cultures maintained in WREN media (Figures 2A and S2A). We thus assessed organoid-forming capacity of cells isolated using these surface markers by fluorescence-activated cell sorting (FACS). We compared the 10% more positive versus 20% more negative population for each of these markers within the same organoid culture. This experiment demonstrated PTK7-high cells contained significantly more organoid forming cells than equivalent populations purified with the other ISC markers (Figure 2B). In addition, PTK7-high cells displayed higher expression levels of ISC markers genes (LGR5, SMOC2) and decreased expression of differentiation markers (KRT20, FABP2) compared with EPHB2-high, CD44-high, and CD133-high cells (Figure 2C). We obtained equivalent results using organoids derived from a different individual (Figure S2). Our data indicate that human colonic organoids in WREN media contain phenotypically distinct cell populations and that high PTK7 surface levels specify bona fide stem cells in these cultures.
PTK7 Enriches in Self-Renewing, Multipotent Stem Cells of the Human Colon
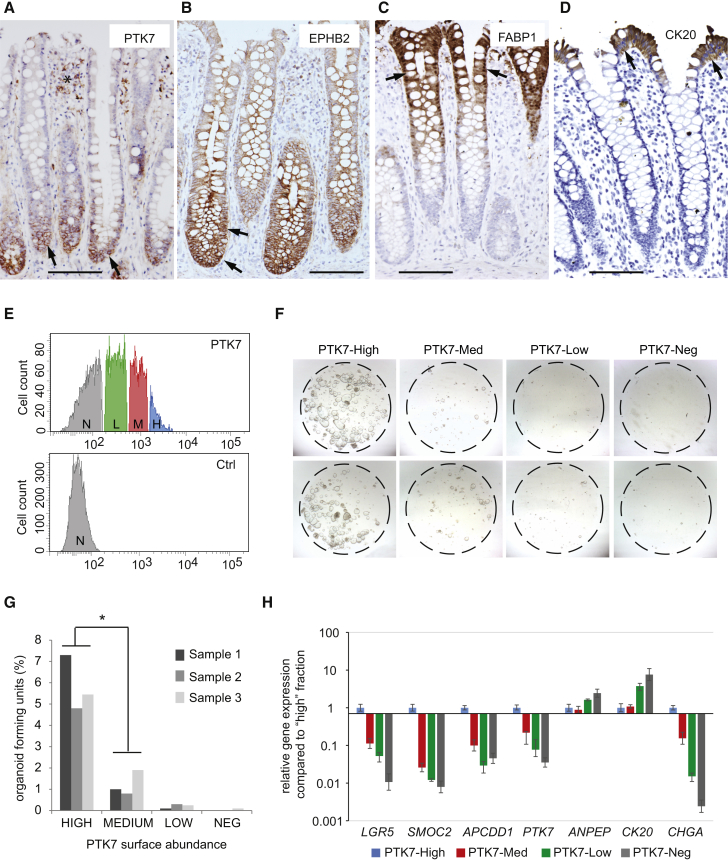
Immunohistochemical analysis confirmed that PTK7 antibodies stained the bottom third of human colonic crypts with the highest intensity observed at the bottommost positions where stem cells reside (Figures 3A and S3A). PTK7 expression was more restricted to the crypt base than that of EPHB2 (Figure 3B), which is the standard surface marker used in the field for the purification of hCoSCs (Jung et al., 2011). In addition, PTK7+ cells were negative for KRT20, MUC2, or FABP1 and CA2 (pan, mucosecreting, or absorptive differentiation markers, respectively) (Figures 3C, 3D, S4A, and S4B). These results encouraged us to isolate different cell fractions based on PTK7 surface abundance directly from human samples of patients undergoing colectomy. As in organoid cultures, human epithelial colonic cells (EpCAM-positive and CD11/CD31/CD45-negative) displayed heterogeneous PTK7 surface expression (Figure 3E). We counted the number of organoids generated by PTK7-high, PTK7-med, PTK7-low, and PTK7-neg intestinal epithelial cells in WREN media (Figure 3F). This experiment demonstrated that highest de novo organoid-forming capacity co-segregated with elevated PTK7 surface levels (n = 3 independent mucosa cultures; Figure 3G). When cultured and serially passaged for several weeks under WREN conditions, PTK7-high single cell-derived human colonic organoids maintained the ability to differentiate into absorptive and secretory lineages (Figure S3B). PTK7-high cells expressed elevated levels of the ISC markers LGR5 and ASCL2 (Figures 3H and S3C). Therefore, PTK7 expression is highest in human CoSCs and can be utilized to purify this cell population from human mucosal samples.
Figure 3.
PTK7 Specifies Self-Renewing, Multipotent Stem Cells of the Human Colon
(A–D) Immunohistochemistry analysis of (A) PTK7, (B) EPHB2, (C) FABP1, and (D) CK20 expression on serial sections of normal human colonic mucosa. Black arrows indicate specific staining. Scale bar represents 100 μm.
(E) FACS profile of single-cell suspensions from primary human colon stained with antibody to PTK7 (APC-conjugate) (upper). Only epithelial cells were included (EpCAM+) and non-epithelial cells were excluded (CD11−/CD31−/CD45−) from the analysis. Control staining was performed to define PTK7-negative fraction (lower). H, PTK7 high; M, PTK7 medium; L, PTK7 low; N, PTK7 negative.
(F) Representative images of organoid cultures 10 days after seeding equal cell numbers.
(G) Quantification of organoid-forming capacity of colon cells sorted in (E). Shown are data collected from three independent 3D cultures (biological replicates). t test, ∗p value of the average PTK7 high versus average medium lower than 0.05.
(H) qRT-PCR analysis of gene expression on primary human colon epithelial cells purified by FACS according to different PTK7 surface levels. Error bars indicate SD (n = 3 technical replicates, see Figure S3C for an additional supporting experiment).
PTK7-High hCoSCs Display Genetic Features of Canonical ISCs and of LRCs
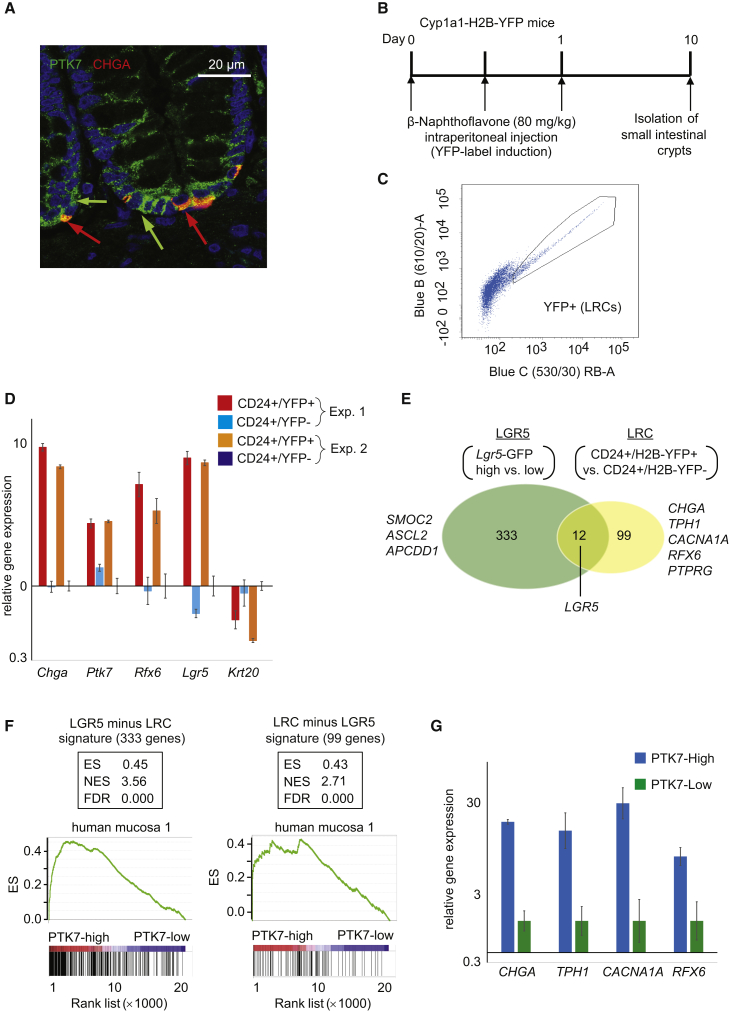
Transcriptomic profiling of PTK7-high cells directly purified from fresh human mucosa samples confirmed enrichment in most genes characteristic of mouse ISCs, including LGR5 (Barker et al., 2007), ASCL2 (van der Flier et al., 2009), and SMOC2 (Muñoz et al., 2012) (Table S3). Yet, we also noticed that several markers of the enteroendocrine lineage such as CHGA and CHGB were upregulated in human PTK7-high cells (Figures 3H and S4E; Table S3). Immunofluorescence on human mucosa tissue sections confirmed that a small proportion (9% on average) of PTK7+ cells positioned at the crypt base co-expressed CHGA (n = 76 crypts analyzed from two different individuals; data not shown), with some crypts showing up to four PTK7/CHGA double-positive cells at their base (Figures 4A and S4C). Recent characterization of the mouse ISC niche has identified a subpopulation of Lgr5+ cells in the small intestine undergoing direct differentiation toward enteroendocrine and Paneth cell lineages (Buczacki et al., 2013, Grün et al., 2015). They are termed LRCs because, as a result of slower proliferation rates, they do not dilute DNA labeling molecules. We thus sought to clarify whether PTK7 also marks LRCs in the mouse intestine. None of the commercially available anti-PTK7 antibodies recognized the mouse protein, which precluded isolation of PTK7+ cells by flow cytometry from mouse tissues (data not shown). Therefore, we used Cyp1a1-H2B-YFP mice, which upon induction with beta-Naphthoflavone, express histone H2B-YFP fusion protein in intestinal cells (Buczacki et al., 2013). In pulse-chase experiments, around 8%–10% of CD24+/Lectin− crypt cells retained the H2B-YFP label 10 days after induction (Figures 4B and 4C). Mouse SI LRCs (CD24+/H2B−YFP+) expressed Lgr5, Chga, and Rfx6 compared with non-LRCs (CD24+/H2B−YFP−). This experiment revealed that LRCs also expressed high levels of Ptk7 (Figure 4D).
Figure 4.
PTK7+ Human CoSCs Show Mixed Features of Canonical CoSCs and LRCs
(A) Immunofluorescence staining on formalin-fixed paraffin sections of DAPI (blue), PTK7 (green), and CHGA (red). Scale bar size is indicated.
(B) Experimental scheme. Mice were given three consecutive intraperitoneal injections of 80 mg/kg β-Naphthoflavone (β-NF) within 24 hr. At day 10, animals were killed and small intestinal (SI) crypt cells isolated.
(C) FACS profile of single cell suspension derived from Cyp1a1-H2B-YFP mouse small intestinal crypts. CD24+/lectin-negative cells were divided into YFP-positive label-retaining cells (LRCs) and YFP-negative cells (Non-LRCs).
(D) qRT-PCR expression analysis showing mouse SI label-retaining cells (CD24+/YFP+) are enriched in Chga, Rfx6, Lgr5, and Ptk7 gene expression compared to CD24+/YFP− cells. Shown are two independent experiments (biological replicates) 1 and 2. Each sample was analyzed three times (technical replicate), and error bars indicate SD.
(E) Venn diagram showing genes that overlap between the humanized LGR5 stem cell signature (Hans Clevers Lab) and the humanized label-retaining cell (LRC) signature (Doug Winton Lab).
(F) GSEA analysis was performed using the specific canonical (LGR5) and label-retaining (LRC) cell signatures (see Table S4 and Supplemental Experimental Procedures) for our PTK7 hCoSC microarray datasets (Table S3).
See Figure S4D for an independent experiment performed on a different individual.
(G) qRT-PCR expression analysis showing that PTK7-high human colonic cells are enriched in LRC gene expression when compared with PTK7-low cells. Samples derived from one patient were analyzed three times (technical replicate). Error bars indicate SD (n = 3).
See Figures S4E and S4F for additional supporting data.
To extend our analysis, we studied the expression of genes characteristic of mouse Lgr5+ ISCs (Muñoz et al., 2012) or of LRCs (Buczacki et al., 2013) in PTK7+ hCoSCs. We derived an ISC (LGR5) signature comparing the transcriptomic profiles of Lgr5-GFP-high versus Lgr5-GFP-low crypt cells purified from Lgr5-GFP knockin mice (Muñoz et al., 2012) (Table S4). This gene set contained many WNT targets, including the ISC marker genes LGR5, SMOC2, ASCL2, and APCDD1. To generate a specific LRC signature, we selected genes upregulated in H2B-YFP-retaining crypt cells (H2B−YFP+; CD24+) compared with crypt non-retaining cells (H2B−YFP−; CD24+) (Table S4; Supplemental Experimental Procedures for details) (Buczacki et al., 2013). Only 12 genes overlapped between the LGR5 and LRC gene sets (Figure 4E). Gene Set Enrichment Analysis (GSEA) showed that both signatures were highly enriched in PTK7-high hCoSCs cells (Figures 4F and S4D). We confirmed by qRT-PCR that genes specific of LRCs, such as CHGA, TPH1, RFX6, and CACNA1A, were upregulated in PTK7-high cells compared with PTK7-low cells purified from human colon samples (Figure 4G). In contrast, EPHB2-high or CD44-high cells purified from these samples were not enriched in LRC-specific genes (Figures S4E and S4F). These results indicate that the PTK7+ human colonic epithelial cell population expresses gene programs of both canonical ISCs and of LRCs.
Discussion
Insertion of reporter cassettes into the Lgr5 locus has enabled characterization of mouse ISCs (Barker et al., 2007, Barker et al., 2010) and development of ex vivo organoid culture systems (Sato et al., 2009). Subsequent transplantation assays using fetal or adult ISCs have paved the way for regenerative therapies of intestinal injuries (Fordham et al., 2013, Yui et al., 2012). However, low cell surface abundance of the LGR5 protein and lack of high-affinity anti-LGR5 antibodies represent a roadblock to isolate human colon stem cells using this marker gene. Our data show that PTK7 is a surrogate of LGR5 in human colon. The methodology developed herein enables the isolation of a cell population largely enriched in CoSCs from human mucosa samples. Remarkably, the possibility of purifying the cell population with highest self-renewal capacity from organoid cultures through PTK7 could facilitate the development of therapeutic approaches based on CoSC transplantation.
Identification of a pool of slow cycling ISCs committed to differentiate toward Paneth and enteroendocrine lineages has clarified the long-lasting debate about the existence of quiescent stem cells in the intestine (Clevers, 2013b). One of the defining features of intestinal LRCs is the ability to gain multipotent growth capacity upon tissue injury (Buczacki et al., 2013). However, it remains unclear whether a similar cell type is also present in the human intestine. Expression profiling of PTK7-high cell population suggests the existence of LRC-like cells in the human colonic epithelium and implies phenotypic and functional heterogeneity within the hCoSC compartment similar to that present in mouse crypts. Importantly, Buczacki et al. (2013) demonstrated that mouse LRCs maintain the ability to form intestinal organoids when plated in the culture dish. However, under these conditions, LRCs lose their quiescent/slow-cycling character and revert into actively proliferating ISCs (Buczacki et al., 2013). Similarly, we were unable to identify LRCs in human colon organoids (data not shown). Future efforts should aim to optimize organoid culture conditions in order to promote the concomitant propagation of canonical stem cells and LRCs in vitro. The biology of these reserve stem cells in humans holds great interest as they might be responsible for regeneration of human colon mucosa upon certain insults such as chemotherapy.
Experimental Procedures
Organoid Cultures from Colonic Crypts
Colonic tissue pieces were incubated in 10 mM DTT for 5 min at room temperature and transferred to 8 mM EDTA for 60 min at 4°C, and vigorous shaking yielded supernatants enriched in colonic crypts. Crypts were either directly cultured, or single-cell suspensions were obtained by incubation with 0.4 mg/ml Dispase (GIBCO). Crypts or single cells were mixed with 50 μl matrigel (BD Biosciences) and plated on 24-well culture dishes. Embedded cells were overlaid with WREN medium (Wnt3a-conditioned medium and ADF 50:50, Glutamax, 10 mM HEPES, N-2 [1×], B-27 without retinoic acid [1×], 10 mM Nicotinamide, 1 mM N-acetyl-L-cysteine, 1 μg/ml RSPO1, 50 ng/ml human epidermal growth factor [EGF], 100 ng/ml human Noggin, 1 μg/ml Gastrin, and 0.05 μM PGE2, 0.1 μM A83-01, 10 μM p38 inhibitor SB202190, 10 μM Y27632). Medium was replaced with fresh WREN medium every other day. Biological samples were obtained from individuals treated at the Hospital del Mar (Barcelona, Spain) or from Hospital Clinic (Barcelona, Spain) under informed consent. Experiments were approved by the ethics committee of IRB/Hospital Clinic (projectERC-208488/CRCprogramme).
Author Contributions
P.J. designed, planned, and performed experiments and analyzed the results. C.S. and M.S. performed LC-MS-MS analysis and analyzed results. F.M.B. provided crucial assistance with experiments and analyzed data. S.J.B. and D.J.W. contributed transgenic animals and contributed crucial experimental protocols. X.H.-M. provided assistance with transgenic animals. M.S. provided assistance with immunohistochemistry. M.D.-F. and P.A. analyzed data. E.B., with the assistance of P.J., conceptualized and supervised the project, analyzed results, and wrote the manuscript.
Acknowledgments
We thank all members of the Batlle laboratory for support and discussions. We are grateful for the assistance by the IRB Barcelona core facilities for Flow Cytometry, Histopathology, Functional Genomics, Protein Expression, Biostatistics, and Advanced Digital Microscopy. We also thank Isabella Dotti and Azucena Salas from Hospital Clínic and Mar Iglesias Coma from Hospital del Mar in Barcelona for support. This work has been supported by grants from ERC-2007-StG208488, La Marató de TV3 (proj 122930) and by the Fundación Botín and Banco Santander through its Santander Universities Global Division.
Published: November 5, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.10.003.
Accession Numbers
The accession number for the microarray dataset reported in this paper is GEO GSE68340.
Supplemental Information
p < 0.05.
See Buczacki et al. (2013).
References
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H., Sato T., Stange D.E., Begthel H., van den Born M. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- Buczacki S.J., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., Winton D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Clevers H. Stem Cells: A unifying theory for the crypt. Nature. 2013;495:53–54. doi: 10.1038/nature11958. [DOI] [PubMed] [Google Scholar]
- de Lau W., Barker N., Low T.Y., Koo B.K., Li V.S., Teunissen H., Kujala P., Haegebarth A., Peters P.J., van de Wetering M. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- Fordham R.P., Yui S., Hannan N.R., Soendergaard C., Madgwick A., Schweiger P.J., Nielsen O.H., Vallier L., Pedersen R.A., Nakamura T. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracz A.D., Fuller M.K., Wang F., Li L., Stelzner M., Dunn J.C., Martin M.G., Magness S.T. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells. 2013;31:2024–2030. doi: 10.1002/stem.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H., van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- Hou N.Y., Yang K., Chen T., Chen X.Z., Zhang B., Mo X.M., Hu J.K. CD133+ CD44+ subgroups may be human small intestinal stem cells. Mol. Biol. Rep. 2011;38:997–1004. doi: 10.1007/s11033-010-0195-y. [DOI] [PubMed] [Google Scholar]
- Hu Y., Smyth G.K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Jung P., Sato T., Merlos-Suárez A., Barriga F.M., Iglesias M., Rossell D., Auer H., Gallardo M., Blasco M.A., Sancho E. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- Leushacke M., Barker N. Ex vivo culture of the intestinal epithelium: strategies and applications. Gut. 2014;63:1345–1354. doi: 10.1136/gutjnl-2014-307204. [DOI] [PubMed] [Google Scholar]
- Merlos-Suárez A., Barriga F.M., Jung P., Iglesias M., Céspedes M.V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Muñoz P. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Muñoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten C.S., Kovacs L., Hamilton E. Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet. 1974;7:271–283. doi: 10.1111/j.1365-2184.1974.tb00907.x. [DOI] [PubMed] [Google Scholar]
- Puppo F., Thomé V., Lhoumeau A.C., Cibois M., Gangar A., Lembo F., Belotti E., Marchetto S., Lécine P., Prébet T. Protein tyrosine kinase 7 has a conserved role in Wnt/β-catenin canonical signalling. EMBO Rep. 2011;12:43–49. doi: 10.1038/embor.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., van Gijn M.E., Hatzis P., Kujala P., Haegebarth A., Stange D.E., Begthel H., van den Born M., Guryev V., Oving I. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- Wang F., Scoville D., He X.C., Mahe M.M., Box A., Perry J.M., Smith N.R., Lei N.Y., Davies P.S., Fuller M.K. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145:383–395.e1. doi: 10.1053/j.gastro.2013.04.050. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whissell G., Montagni E., Martinelli P., Hernando-Momblona X., Sevillano M., Jung P., Cortina C., Calon A., Abuli A., Castells A. The transcription factor GATA6 enables self-renewal of colon adenoma stem cells by repressing BMP gene expression. Nat. Cell Biol. 2014;16:695–707. doi: 10.1038/ncb2992. [DOI] [PubMed] [Google Scholar]
- Yui S., Nakamura T., Sato T., Nemoto Y., Mizutani T., Zheng X., Ichinose S., Nagaishi T., Okamoto R., Tsuchiya K. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- Zhu L., Gibson P., Currle D.S., Tong Y., Richardson R.J., Bayazitov I.T., Poppleton H., Zakharenko S., Ellison D.W., Gilbertson R.J. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
p < 0.05.
See Buczacki et al. (2013).