Abstract
A promoter that enabled high-level expression of the target gene during the stationary phase in the absence of an inducer would facilitate the efficient production of heterogeneous proteins at a low cost. In this study, a genome-scale microarray-based approach was employed to identify promoters that induced high-level expression of the target genes in Bacillus subtilis from the late log phase to the stationary phase without an inducer. Eleven candidate promoters were selected based on B. subtilis microarray data and the quantitative PCR analysis. Among the selected promoters, Pylb exhibited the highest activity with the reporter bgaB during the stationary phase. Compared with P43 (a commonly used constitutive promoter), promoter Pylb could express two reporter genes (egfp and mApple), and the expression levels of EGFP and RFP were 7.8- and 11.3-fold higher than that of P43, respectively. This finding was verified by overexpression of the genes encoding pullulanase and organophosphorus hydrolase, the activities of which were 7.4- and 2.3-fold higher, respectively, when driven by Pylb compared with P43. Therefore, our results suggest that the Pylb promoter could be used to overexpress target genes without an inducer; this method could facilitate the identification and evaluation of attractive promoters in the genome.
As a Gram-positive bacterium, Bacillus subtilis is an attractive host for the production of heterologous secretory proteins. B. subtilis is a nonpathogenic bacterium that can efficiently secrete a target protein into the culture medium. Much information concerning large-scale fermentation and production technology using this bacterium is available1,2,3. In addition, the genome and transcriptome in different conditions of B. subtilis have been determined4,5,6.
Promoters are important genomic regulatory elements that directly affect gene expression levels. In bacteria, RNA polymerases and associated sigma factors recognize promoters and are recruited by the binding of regulatory proteins to specific sites within promoters. To date, three types of promoter have been used for the high-level expression of heterologous proteins in B. subtilis: constitutive promoters, inducer-specific promoters, and auto-inducible promoters1,7,8,9,10. Although inducer-specific promoters (e.g., Pspac and Pxyl) are the most widely used type, the requirement for inducer compounds—such as IPTG and xylose—could increase the cost of their industrial application11,12,13,14,15,16. Constitutive promoters, such as P43, are not suitable for production of toxic proteins. Auto-inducible promoters are ideal for large-scale protein production. Such promoters induce expression of the target gene from the late log phase to the stationary phase with no requirement for an inducer, which facilitates efficient production of heterogeneous proteins at a low cost. However, low activity1,17 is a barrier to the widespread use of the auto-inducible promoters available at present. Therefore, there is a need to identify novel auto-inducible promoters with high activity.
Here, we described a genome-wide microarray-based approach to identifying auto-inducible promoters in B. subtilis based on the report by Evert-Jan et al.18. A total of 58 stable phase-preferred and highly expressed genes were identified by microarray analysis. Among these genes (some of which were multicistronic), 21 stably phase-overexpressed genes with expression levels higher than that of cdd under the control of the P43 promoter (a commonly used constitutive promoter)3,19,20 were further evaluated by quantitative reverse transcriptase-PCR. Among the selected promoters, promoter Pylb was the most potential promoter. Circularized RNA reverse-transcription PCR indicated that the transcription initiation site of the Pylb promoter was an adenine, and the key elements (−10 box and −35 box) of the promoter were determined by site-directed mutagenesis. These results indicated Pylb to be an attractive and highly active promoter.
Results
SAM analysis and real-time PCR
To identify the target promoter, B. subtilis genome-wide microarray data were downloaded from NCBI (Accession No. GSE19831) and analyzed using the SAM software (Figure S5). The microarray data covered a total of 4,169 B. subtilis genes and 40 time points. The time points (1–4, 5–17, 19–26, and 27–40) represented the ‘lag’, log, early stationary, and late stationary growth phases, respectively. As shown in Figure S5A, a number of genes were highly expressed during the early and (or) late stationary growth phase. The expression profiles of those genes were shown in Figure S5B. A total of 58 genes were highly expressed during early and (or) late stationary growth phase, and their expression levels were higher than that of cdd gene which controlled by P43 (indicated by an arrowhead). The P43 promoter was selected as the positive control as it was commonly used strong promoter for B. subtilis21,22,23,24.
qRT-PCR was used to verify the expression levels of the 58 genes. RNA was extracted after 6, 12, 18, and 24 h (Figure S1). The first two time points (6 and 12 h) represented the log phase, and the later two (18 and 24 h) represented the stationary growth phase. The 16S rRNA gene was used as the reference gene, and the cdd gene under the control of the P43 promoter was used as a positive control. The results showed that the selected 21 genes could be divided into three categories according to their expression level (Figure S6). The transcript levels of genes argD, yceC, yuaF, msmX, pbpE, rapA, sigW, yvlA, yxbB, ylbP, yobJ, yqeZ and yddF were higher than that of cdd (Figure S6B,C), whereas the transcript levels of the other 8 genes were lower than that of cdd (Figure S6A). Among the selected genes, the transcript levels of rapA and yddF were highest (Figure S6C). However, their transcript levels were unstable during the stationary phase. In addition, ylbP was an attractive gene, as it exhibited very high activity during the stationary phase of growth but low activity in log phase. Therefore, we focused on the Pylb promoter which controlled ylbP in this study.
Evaluation of the selected promoters from B. subtilis
Eleven candidate genes’ promoters (Pydc, PyddA, Pmsm, Ppbp, Prap, Psig, Pyvl, Pylb, Pyob, Pyqe and PyddF) were selected based on the results of SAM and qRT-PCR. Among the eleven promoters, the gene transcriptional levels driven by Ppbp, Prap, Psig, Pylb, Pyqe and PyddF were higher than that of P43, the gene transcriptional levels driven by Pmsm, Pyvl and Pyob were near to that of P43, and the gene transcriptional levels driven by Pydc and PyddA were lower than that of P43. To assess the activity of the eleven candidate promoters, 500–600 base pairs (bp)25 located upstream of the start codons were cloned from the genomic DNA of B. subtilis WB600 and inserted into the region upstream of the reporter gene bgaB. The promoter P43 was used as a positive control. The β-galactosidase activities of these eleven recombinants after culture for 18 h were shown in Fig. 1, and the results of the SDS-PAGE analysis of BgaB in crude extract were shown in Figure S7. Among the selected promoters, the β-galactosidase activity driven by Pylb was highest, 8.2-fold higher than that of P43. However, the β-galactosidase activity driven by PyddF was only 2.4-fold higher than that driven by P43. This may be because PyddF is a log-phase-specific promoter; Indeed, it resulted in the highest β-galactosidase activity in a log phase, 7.2-fold higher than that driven by P43 (Figure S8). Moreover, the β-galactosidase activities driven by Pmsm, Ppbp, Pyvl, Pyob and Pyqe were ~2.0, 1.6, 1.7, 1.8 and 1.3-fold, respectively, of those driven by P43. In contrast, the Pydc and PyddA promoters resulted in negligible β-galactosidase activity. However, the clones of Prap and Psig promoters could be observed blue on the LB agar containing 10 μg/mL kanamycin with x-gal (Figure S9). But it was difficultly to detect the β-galactosidase activity on several different time points in liquid LB medium. All of the results indicated that the promoter Pylb was a good candidate promoter to express the target gene.
Figure 1. β-Galactosidase activities of the eleven candidate promoter clones in B. subtilis.
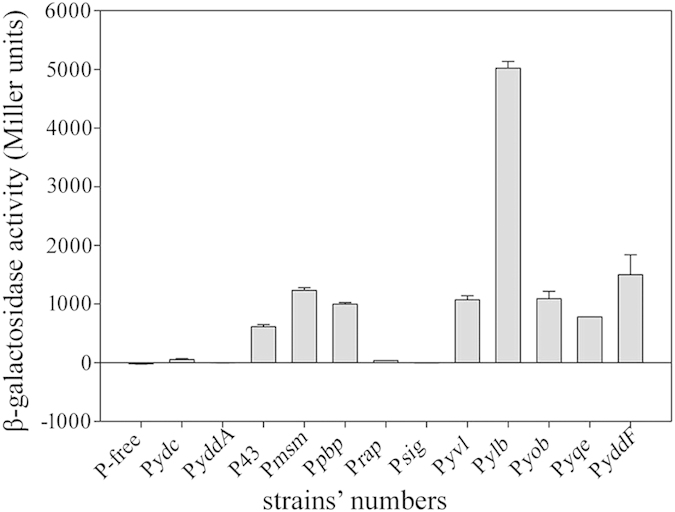
β-Galactosidase production by the eleven candidate promoter clones in B. subtilis WB600 after 18 h of culture. Data are averages of three independent experiments. P-free: B. subtilis WB600 containing the P-free-bgaB-pUBC19 plasmid as a negative control. P43: B. subtilis WB600 containing the P43-bgaB-pUBC19 plasmid as a positive control. Pydc, PyddA, Pmsm, Ppbp, Prap, Psig, Pyvl, Pylb, Pyob, Pyqe and PyddF: B. subtilis WB600 strains containing the Pydc-bgaB-pUBC19, PyddA-bgaB-pUBC19, Pmsm-bgaB-pUBC19, Ppbp-bgaB-pUBC19, Prap-bgaB-pUBC19, Psig-bgaB-pUBC19, Pyvl-bgaB-pUBC19, Pylb-bgaB-pUBC19, Pyob-bgaB-pUBC19, Pyqe-bgaB-pUBC19 and PyddF-bgaB-pUBC19 plasmids, respectively.
P43 and Pylb-driven fluorescent protein expression B. subtilis
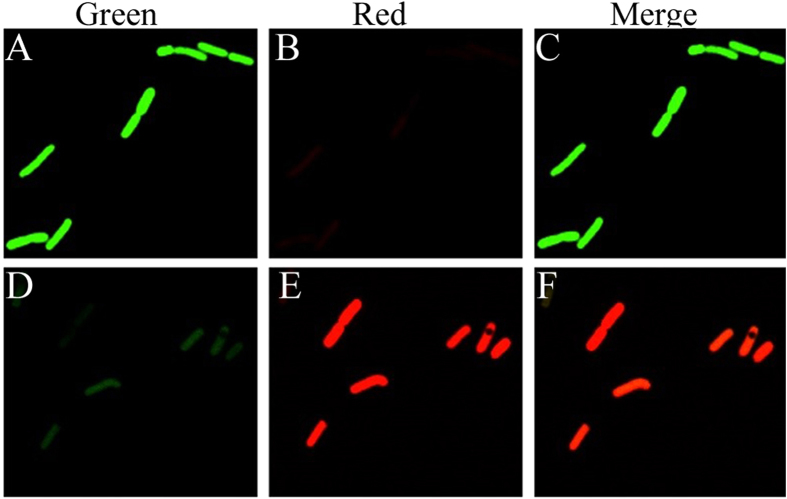
To compare the activity level of the Pylb promoter to that of P43, we used the green (EGFP) and red (mApple) fluorescent proteins as reporters. To prevent any influence of plasmid copy number and cell concentration on promoter activity, three fusion plasmids (G-R-pUBC19, Pylb-R-P43-G-pUBC19, and Pylb-G-P43-R-pUBC19), each of which contained two promoters and two reporter genes, were constructed and transformed into B. subtilis WB600. EGFP and mApple have similar fluorescence intensities; These proteins exhibit relative brightness levels of 100 and 109, respectively26. In the plasmid Pylb-R-P43-G-pUBC19, the Pylb and P43 promoters drove the mApple and egfp reporter genes, respectively. Green and red fluorescence were measured in the recombinant strain harboring the Pylb-R-P43-G-pUBC19 plasmid. If the activity of the Pylb promoter were greater than that of P43, the red fluorescence intensity would be greater than that of green fluorescence. The recombinant strain containing the plasmid E-R-pUBC19 was used as a negative control; no fluorescence was detected in this strain. As shown in Fig. 2, the strains containing the plasmids Pylb-G-P43-R-pUBC19 and Pylb-R-P43-G-pUBC19 exhibited green and red fluorescence after culture for 18 h. These results indicated that the activity of the Pylb promoter was greater than that of P43.
Figure 2. Fluorescence micrographs of B. subtilis WB600 harboring the Pylb-G-P43-R-pUBC19 and Pylb-R-P43-G-pUBC19 plasmids.
(A) Green fluorescence micrograph, (B) Red fluorescence micrograph and (C) Merge of green fluorescence and red fluorescence of B. subtilis WB600 harboring the Pylb-G-P43-R-pUBC19 plasmid after 18 h of culture. (D) Green fluorescence micrograph, (E) Red fluorescence micrograph and (F) Merge of green fluorescence and red fluorescence of B. subtilis WB600 harboring the Pylb-R-P43-G-pUBC19 plasmid after 18 h of culture. Images were acquired using a Nikon microscope with a 100 × objective.
To quantitatively assess the activity of the promoters, the fluorescence intensity of strains containing the Pylb-G-P43-R-pUBC19 or Pylb-R-P43-G-pUBC19 plasmid was monitored spectrophotometrically every hour. The results of the strain containing the recombinant plasmid Pylb-G-P43-R-pUBC19 indicated that the green fluorescence intensity driven by Pylb was higher than the red fluorescence intensity driven by P43 (Fig. 3A), and the strain containing the recombinant plasmid Pylb-R-P43-G-pUBC19 showed that the red fluorescence intensity driven by Pylb was higher than the green fluorescence intensity driven by P43 (Fig. 3B). These results indicated that the Pylb promoter could induce high-level expression of GFP and RFP.
Figure 3. Fluorescence intensity and growth curves of B. subtilis harboring the Pylb-G-P43-R-pUBC19 and Pylb-R-P43-G-pUBC19 plasmids.
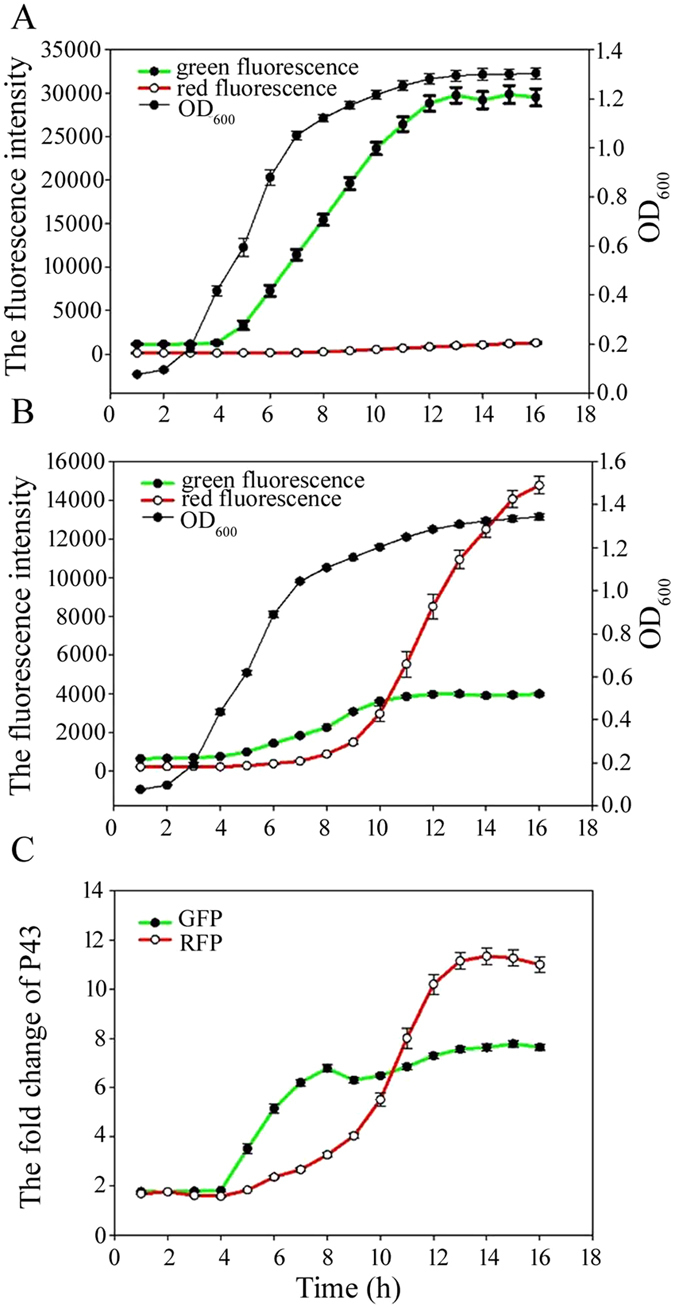
(A) Fluorescence intensity and growth curve of B. subtilis WB600 harboring the Pylb-G-P43-R-pUBC19 plasmid. (B) Fluorescence intensity and growth curve of B. subtilis WB600 harboring the Pylb-R-P43-G-pUBC19 plasmid. (C) Fold-change in Flu/OD600 (fluorescence units per OD600) for Pylb relative to P43. B. subtilis WB600 containing the Pylb-G-P43-R-pUBC19 or Pylb-R-P43-G-pUBC19 fusion plasmid was grown in LB medium containing 10 μg/mL kanamycin. Data are averages of three independent experiments.
We also determined the time point at which the Pylb promoter was expressed and compared it with P43. The Pylb promoter resulted in EGFP and mApple reporter protein expression beginning during log and stationary phases, respectively (Fig. 3A,B). Therefore, the Pylb promoter is capable of inducing expression of its target gene from the log phase to the stationary phase. As the folding rates may vary among proteins27, the time at which a target gene is expressed may be different. The reporter protein expression ratio (EGFP and mApple) between the Pylb and P43 promoters at various time points was shown in Fig. 3C. The reporter protein (EGFP and mApple) expression level was similar during the lag and early log phases. However, in the late log and stationary phases, the reporter protein expression level driven by the Pylb promoter was significantly higher than that driven by P43. Pylb-induced EGFP and mApple levels of expression after 15 h of culture were 7.8 and 11.3-fold, respectively, higher than that of P43. These results indicated that the Pylb promoter was capable of inducing expression of the target protein from the late log phase to the stationary phase with no requirement for an inducer and that the Pylb promoter exhibited higher activity than P43.
Determination of the transcription start site of the Pylb promoter
In the B. subtilis genome, the Pylb promoter controls the expression of ylbP, which is located at 1,576,129–1,576,710 on the complementary strand, upstream of gerR and downstream of panE. Our analyses revealed that the ylbP gene was monocistronic (Fig. 4A). The CR-RT-PCR method was used to determine the transcription start site of the Pylb promoter. RNA was extracted from B. subtilis WB600 containing the F0-bgaB-pUBC19, F3-bgaB-pUBC19, and F4-bgaB-pUBC19 fusion plasmids after 18 h of culture. Sequencing analysis revealed that the transcriptional start site was an adenine (Fig. 4B). To experimentally analyze the core element of the Pylb 5′-flanking region, analysis of the effect of Pylb deletion was performed using the bgaB fusion reporter system. The β-galactosidase activities of the resulting constructs were measured (Fig. 4C), and SDS-PAGE analysis of BgaB was performed (Figure S10). The B. subtilis strain harboring the wild-type Pylb promoter (F0) and those with deletions (F1, F2, F3 and F4) exhibited similar β-galactosidase activities. However, the B. subtilis strain containing the F5 fusion plasmid exhibited negligible β-galactosidase activity. These results revealed that the F4 strain contained the main core element, but F5 was deficient in the −35 element. Therefore, the putative recognition element (−35 TTGGAT −30) and a −10 box (−12 TACAAT −7) were presented in the Pylb promoter.
Figure 4. Map of the ylbP gene, and the promoter sequence and β-galactosidase activity of the deleted promoters clones in B. subtilis.
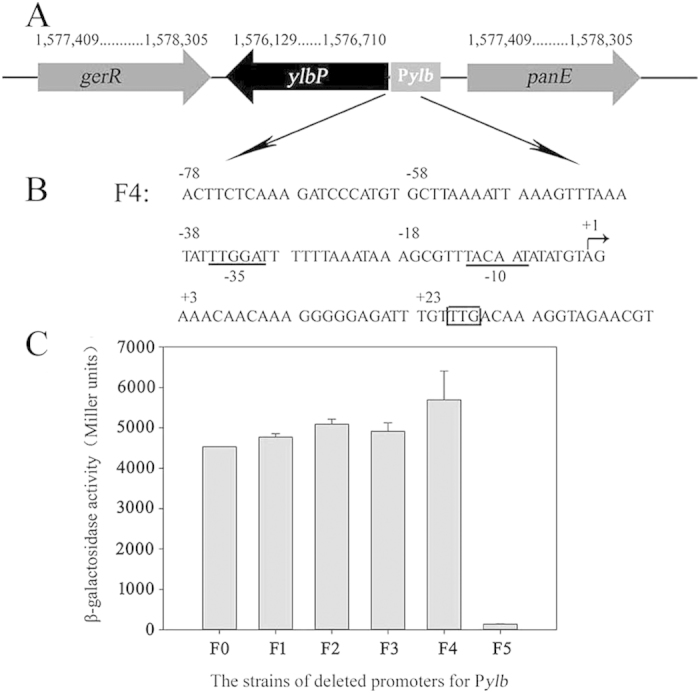
(A) Map showing the ylbP gene in the B. subtilis genome, and ylbP encodes an N-acetyltransferase. (B) Sequence of the Pylb promoter. The start codon of ylbP is shown in the square. The arrow indicates the transcription start site of the Pylb promoter. The −10 box and −35 element are indicated by underlines. (C) The β-galactosidase activity of the B. subtilis strains in which the Pylb promoter was deleted. Data are averages of three independent experiments.
Construction of mutants of promoter Pylb by site-directed mutagenesis
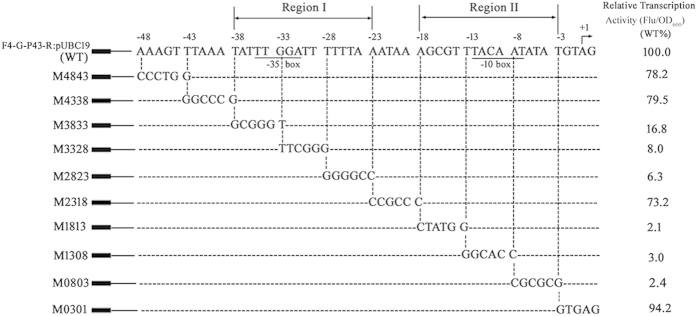
The above results demonstrated that the shortest functional sequence of Pylb was F4 and revealed its putative recognition element (−35 box and −10 box). To experimentally analyze the cis-acting elements of Pylb in detail, site-directed mutagenesis was performed using the EGFP fusion reporter system with the F4-egfp-pUC19 plasmid (see Materials and methods). Thus the promoter region from bp −48 to −1 of F4 was analyzed using site-directed scanning mutagenesis to pinpoint the regions that mediate the transcriptional activity of Pylb. The resulting constructs were named pM4843 to pM0301; the numbers indicated the mutated region. For example, pM4843 indicated mutagenesis from bp −48 to −43 relative to the transcription start site of Pylb. These plasmids were transformed into B. subtilis WB600 to generate the reporter strains M4843 to M0301, which exhibited green fluorescence (Fig. 5). Mutations in the putative −35 box (−35 TTGGAT −30) regions (M3833 and M3328) and −10 element (−12 TACAAT −7) regions (M1308 and M0803) led to complete loss of transcriptional activity. In addition, the region from bp −28 to −23 and that from bp −18 to −13 might be directly involved in the transcriptional activity of Pylb. Notably, the green fluorescence intensity did not significantly change the transcriptional levels of the mutants M4843, M4338, M2318, and M0301, indicating that the corresponding regions in these mutants were not directly involved in the transcriptional activity of Pylb. These results revealed that the promoter regions I (bp −38 to −23) and II (bp −18 to −3) were particularly important for the transcriptional activity of Pylb.
Figure 5. Site-directed mutagenesis from bp −48 to −1 of the Pylb promoter.
The DNA sequence of wild-type Pylb is shown at the top (WT). The transcription start site (TSS) of Pylb was identified by CR-RT-PCR. The mutated nucleotides of the M4843 to M0301 mutants are shown below the WT promoter sequence. Green fluorescence intensity was determined using a microplate reader. The fold-changes in Flu/OD600 (fluorescence units per OD600) relative to the WT (set to 100%) indicate the transcriptional activities of the mutants. The significant fold-changes in strains M3833, M3328, M1308, and M0803 were due to large reductions in fluorescence intensity. Data are averages of three independent experiments, each of which comprised four replicates.
The application prospect of the promoter Pylb
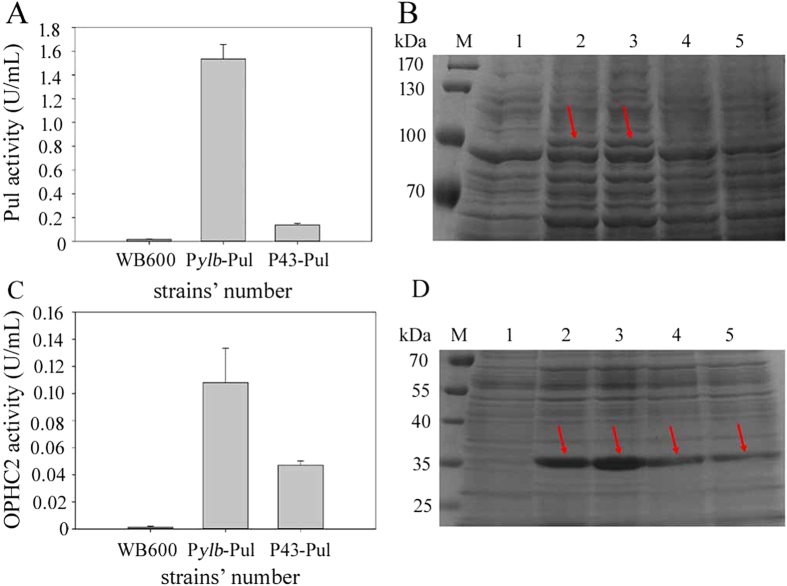
To evaluate the Pylb promoter’s potential for application, Pylb-ZDs/P43-ZDs (containing the promoter and signal peptide of amylase from Bacillus amyloliquefaciens) and two foreign genes, pul/ophc2, were cloned into the shuttle vector pUBC19. The Pul activities of the Pylb-ZDs-Pul-pUBC19 and P43-ZDs-Pul-pUBC19 recombinants after 18 h of culture were shown in Fig. 6A, and the OPHC2 activities were presented in Fig. 6C. Our results confirmed that the Pylb promoter induced significantly higher levels of expression of Pul and OPHC2 than did P43. The Pylb-driven Pul activity was 7.4-fold higher than that driven by P43, and the Pylb-driven OPHC2 activity was 2.3-fold higher than that driven by P43. The results of SDS-PAGE analysis of Pul and OPHC2 in the culture supernatant after concentrated about 15 folds were shown in Fig. 6B,D, respectively. Moreover, high levels of expression of heterologous proteins did not significantly affect the growth of the recombinant B. subtilis WB600 strains (data not shown). In addition, cultivations were also performed in SB medium, as shown the supplementary material. The results of Pul and OPHC2 activity and the SDS-PAGE analysis were shown in Figure S11. It also indicated that the activity of Pylb promoter was stronger than that of P43 when driving Pul and OPHC2. Thus, the Pylb promoter may have potential for the overexpression of proteins in B. subtilis.
Figure 6. Pullulanse and OPHC2 activities of B. subtilis strains harboring various promoters after 18 h of culture.
(A) Production of pullulanse by B. subtilis WB600 harboring the recombinant plasmid Pylb-ZDs-Pul-pUBC19 (indicated by Pylb-Pul), P43-ZDs-Pul-pUBC19 (P43-Pul), or no recombinant plasmid (WB600) after 18 h of culture. (B) SDS-PAGE analysis of pullulanse activity in the culture supernatant that was concentrated about 15 folds after 18 h of culture. M: protein molecular mass marker. 1: B. subtilis WB600 as a negative control. 2 and 3: Pylb-Pul-1, Pylb-Pul-2. 4 and 5: P43-Pul-1 and P43-Pul-2, and the arrows indicate target protein pullulanse. (C) Production of OPHC2 by B. subtilis WB600 harboring the recombinant plasmid Pylb-ZDs-OPHC2-pUBC19 (indicated by Pylb-OPHC2), P43-ZDs-OPHC2-pUBC19 (P43- OPHC2), and no recombinant plasmid (WB600) after 18 h of culture. (D) SDS-PAGE analysis of OPHC2 in the culture supernatant that was concentrated about 15 folds after 18 h of culture. M: protein molecular mass marker. 1: B. subtilis WB600 as a negative control. 2 and 3: Pylb-OPHC2-1, Pylb-OPHC2-2. 4 and 5: P43-OPHC2-1 and P43-OPHC2-2, and the arrows indicate the target protein OPHC2. Data are averages of three independent experiments.
Discussion
One of the key factors for achieving high-level expression of heterologous genes is the use of a strong and controllable promoter. Despite the fact that several strong promoters have been reported7,10,28,29,30,31,32, industrial production using B. subtilis is usually mediated by inducer-specific promoters. In comparison, auto-inducible promoters have a distinct advantage in terms of not requiring an inducer compound, which simplifies the industrial production of the target protein and reduces the cost. Thus, identification of potential auto-inducible promoters would be beneficial.
Nowadays, much information about transcriptome data of many species is available on line, which will help to identify the useful promoters6,18. However, in addition to the strength of the promoter, there are many variables affecting expression levels, including the stability of the mRNA, the culture conditions, growth situation of the cell and so on. Therefore, for the aim to isolate the useful promoter, we need not only analyze the transcriptome date, but also use the reporter genes and systematically study the function of the promoter.
Our results indicated that the Pylb promoter induced sufficiently high levels of expression in B. subtilis of active β-galactosidase, EGFP, RFP, pullulanase, and organophosphorus hydrolase from the late log phase to the stationary phase. In addition, the production of those foreign proteins was higher than that induced by the P43 promoter, which has been used widely in B. subtilis. Moreover, the high expression level of heterologous proteins did not significantly affect the growth of B. subtilis. Thus, the Pylb promoter may have potential for the overexpression of useful proteins in B. subtilis.
According to the B. subtilis genome sequence, the Pylb promoter controls the ylbP gene, which has a length of 483 bp and encodes an N-acetyltransferase. The gene could be expressed in the stationary stage, which has also been confirmed by the transcriptome study in the different conditions, as shown in Subwiki6. This enzyme encoded by ylbP is capable of catalyzing the transfer of acetyl groups between acetyl coenzyme A and amines. Thus, it may be involved in post-translational modification in B. subtilis during the stationary phase. However, the function of and the reason that the ylbP gene was highly expressed from the late log phase to the stationary phase remain unclear.
This study is an example of the discovery of an interesting promoter using a genome-scale microarray-based approach. Genomic and gene expression profile data contain a large quantity of biological information, which could be exploited in various fields. Compared with the traditional promoter trap method32, the technique described herein is more effective in terms of identifying attractive promoters and will used to this end in future studies.
Materials and Methods
Bacterial strains, plasmids and growth conditions
B. subtilis WB600 and Bacillus amyloliquefaciens were stored in our laboratory. Escherichia coli Trans1-T1 was used as the host strain for the cloning experiment (Transgen Biotech, Beijing). The plasmids used in this study are listed in Table S1 in the supplemental material. The bacterial strains were cultured in Luria–Bertani (LB) medium at 37 °C. The following concentrations of antibiotics were used for selection: 100 μg/mL ampicillin (Amp), 10 μg/mL kanamycin (Kana), and 5 μg/mL tetracycline (Tet).
Analysis of B. subtilis DNA microarray data
The B. subtilis microarray data (NCBI Accession No. GSE19831) were reported by Evert-Jan et al.18. B. subtilis exhibits four distinct growth phases: the lag phase, the log growth phase, and the early and late stationary growth phases. The microarray data were grouped into four growth phases, and the Significant Analysis of Microarray software (SAM; Stanford University) was employed to identify genes specifically expressed from the late log growth phase to the stationary phase.
RNA extraction and qRT-PCR
B. subtilis WB600 was cultured at 37 °C overnight in LB medium. Then, a 1:100 volume of a fresh overnight B. subtilis culture was inoculated into 50 mL LB medium in a 100 mL flask and cultured at 37 °C with shaking at 200 rpm. Growth was monitored by measurement of the optical density at 550 nm (OD550) (Figure S1). B. subtilis cells were collected after 6, 12, 18, and 24 h of incubation and subjected to RNA extraction using an RNA Prep Pure Cell/Bacteria Kit (Tiangen Biotech, Beijing) according to the manufacturer’s instructions. To remove DNA contamination, 1 μg of diluted RNA was digested with DNase I (New England Biolabs, USA). Specific primers for the target DNA regions (Table S2) and suitable concentrations of the cDNA templates were used for qRT-PCR. Details of the qRT-PCR method are provided in the Materials and methods section of the supplementary material.
Construction of a promoter trap vector with bgaB as the reporter gene
All primers used in this study are listed in Table S2. First, a promoter trap vector, P-free-bgaB-pUBC19, was constructed as described in the supplementary material. The selected alternative promoters were amplified from the genomic DNA of B. subtilis WB600 using the respective primer pairs, and the commonly used strong constitutive promoter P43 was amplified as the control promoter. The amplified promoters were digested with Pst I and Xho I and inserted into the plasmid P-free-bgaB-pUBC19, which had been treated with the same restriction endonucleases, and then transformed into E. coli strain Trans1-T1 to construct the corresponding fusion expression plasmid Promoter-bgaB-pUBC19. The fusion plasmid Promoter-bgaB-pUBC19 was constructed by means of the experimental procedure shown in Figure S2.
Construction of binary expression vectors
Two termination sequences were added at the ends of the egfp (GenBank Accession No. JQ627826.1) and mApple (GenBank Accession No. HM771700.1) genes, and one termination sequence was added between the two genes. Moreover, the direction of the two genes was reversed. Multiple cloning sites were then added to the ends of egfp and mApple (Figure S3 in the supplemental material). After codon optimization based on the B. subtilis genome, the egfp-mApple gene was synthesized by Genscript Corporation (Nanjing, China). The synthetic egfp-mApple gene was located on the pUC57-Simple plasmid. The fusion egfp-mApple-pUC57 Simple plasmid was digested with Hind III and BamH I and inserted into the plasmid pUBC19, which had been treated with the same restriction endonucleases, and then transformed into E. coli Trans1-T1 to construct the corresponding fusion plasmid (G-R-pUBC19). Next, the promoters P43 and Pylb were inserted upstream of the egfp and mApple, respectively, and then transformed into E. coli Trans1-T1 to construct the corresponding fusion plasmids (Pylb-R-P43-G-pUBC19). Moreover, the promoters P43 and Pylb were inserted prior to mApple and egfp, respectively, and then transformed into E. coli Trans1-T1 to construct the corresponding fusion plasmids (Pylb-G-P43-R-pUBC19, see Figure S3 in the supplemental material).
Construction of fusion plasmids for the truncated 5′-flanking sequence of the Pylb promoter
To analyze its characteristics in detail, five truncated fragments of the Pylb promoter (named F0) promoter −381– + 43 (F1), −233– + 43 (F2), −154– + 43 (F3), −78– + 43 (F4), and −21– + 43 (F5)—were amplified using the cognate primer pairs F1-up, F2-up, F3-up, F4-up, F5-up and Pylb-down (see Table S2 in the supplemental material). The PCR products of the five fragments were digested with Pst I and Xho I and inserted into the P-free-bgaB-pUBC19 plasmid to produce the corresponding fusion plasmids.
Construction of scanning site-directed plasmids harboring the Pylb promoter
To analyze the regulatory region of the Pylb promoter, scanning site-directed mutants were constructed. Briefly, the fragment F4-egfp (containing the −78– + 43 fragment of the Pylb promoter and egfp) was amplified from the plasmid Pylb-G-P43-R-pUBC19 using the primers F4-egfp-up and F4-egfp-down. The amplified product F4-egfp was digested with Xba I and Hind III and inserted into the plasmid pUC19, which had been treated with the same restriction endonucleases, to construct the fusion plasmid F4-egfp-pUC19. Based on the plasmid F4-egfp-pUC19, 10 scanning site-directed mutational plasmids of the F4 fragment were constructed using a previously described two-step PCR method33. After confirmation by sequencing, the 10 scanning site-directed mutational F4-egfp and wild-type F4-egfp fragments were cleaved with Xba I and Hind III and inserted into the plasmid Pylb-G-P43-R-pUBC19, which had been treated with the same restriction endonucleases, to construct mutational plasmids (pM4843 to pM0301).
Construction of plasmids for gene overexpression
The pullulanase and organophosphorus hydrolase (ophc2) genes were selected to evaluate the Pylb promoter’s potential for application in B. subtilis. The signal peptide sequence (ZDs) of α-amylase from B. amyloliquefaciens was inserted into the region upstream of the target genes. Four recombinant plasmids were constructed; the fusion plasmids Pylb-ZDs-Pul-pUBC19 and P43-ZDs-Pul-pUBC19 contained the F4 and P43 promoters, respectively, together with the pullulanase gene from Bacillus naganoensis. The other two fusion plasmids, Pylb-ZDs-OPHC2-pUBC19 and P43-ZDs-OPHC2-pUBC19, contained the organophosphorus hydrolase gene (ophc2) from Pseudomonas pseudoalcaligenes. The details of fusion plasmid construction are provided in the supplemental material (Figure S4).
Transformation of plasmids into B. subtilis WB600
After confirmation by sequencing, the plasmids were extracted from E. coli Trans1-T1 and transformed into B. subtilis WB600 as described previously34. Transformants were harvested by screening the clones on LB agar containing 10 μg/mL kanamycin.
Circularized RNA reverse-transcription-PCR
Circularized RNA reverse transcription PCR (CR-RT-PCR) was used to identify the transcription start site of the Pylb promoter. Extraction and circularization of RNA from B. subtilis WB600 harboring the F0-bgaB-pUBC19, F3-bgaB-pUBC19, and F4-bgaB-pUBC19 fusion plasmids were carried out as described previously35,36. Self-ligated RNA was reverse transcribed using the specific reverse primer PB1 (Table S2 in the supplemental material). The cDNA was first amplified using the specific primers PB2-up and PB2-down. Thereafter, to enhance the specificity, a second PCR was performed using the PB3-up and PB3-down inner primers. The purified products of the second PCR were cloned into the TA vector pGM-T (TianGen Biotech, Beijing) according to standard procedures, and 20 clones were subjected to sequencing.
Determination of enzyme activity
The methods for determination of pullulanase, organophosphorus hydrolase, and β-galactosidase activities are described in the supplementary material.
Fluorescence assay
B. subtilis WB600 cells containing the eGFP or mApple reporter gene were cultured for ~18 h, washed three times, and resuspended in 0.9% NaCl. A 5 μL volume of cells at an appropriate dilution in 0.9% NaCl was placed on a microscope slide and covered with a coverslip. Fluorescence microscopy was performed using a 100 × oil immersion lens (Nikon, Japan) to visualize cell fluorescence. To measure the fluorescence intensity, the constructed B. subtilis WB600 strains harboring the Pylb-R-P43-G-pUBC19 and Pylb-G-P43-R-pUBC19 plasmids were cultured in black, 96-well plates with a clear, flat bottom (Corning, USA) at 37 °C with shaking at 750 rpm in incubator 1000 (Heidolph, Germany). The fluorescence intensity was measured at 1-h intervals using a SpectraMax M2 instrument (Molecular Devices, USA). The excitation and emission wavelengths were 484 and 507 nm and 586 and 592 nm, for enhanced green fluorescent protein (EGFP)26 and red fluorescent protein (RFP)37, respectively. The values shown are the averages of three independent experiments.
Additional Information
How to cite this article: Yu, X. et al. Identification of a highly efficient stationary phase promoter in Bacillus subtilis. Sci. Rep. 5, 18405; doi: 10.1038/srep18405 (2015).
Supplementary Material
Acknowledgments
This work was supported by the National High Technology Research and Development Program of China (863 Program, AA021303). This work was supported by the National Natural Science Foundation of China (NSFC, Grant no. 31500064).
Footnotes
Author Contributions X.Y., J.T., N.W. and Y.F. conceived and coordinated the study and wrote the paper. X.Y., J.X. and P.W. designed, performed and analyzed the experiments. X.L., X.C. and N.W. provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
References
- Schumann W. Production of recombinant proteins in Bacillus subtilis. Adv. Appl. Microbiol. 62, 137–190 (2007). [DOI] [PubMed] [Google Scholar]
- Schallmey M., Singh A. & Ward O. P. Developments in the use of Bacillus species for industrial production. Can. J. Microbiol. 50, 1–17 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang X.-Z., Cui Z.-L., Hong Q. & Li S.-P. High-level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl. Environ. Microbiol. 71, 4101–4103 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. R. & Wipat A. Sequencing and functional analysis of the genome of Bacillus subtilis strain 168. FEBS Lett. 389, 84–87 (1996). [DOI] [PubMed] [Google Scholar]
- Kunst F. et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390, 249–256 (1997). [DOI] [PubMed] [Google Scholar]
- Nicolas P. et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106 (2012). [DOI] [PubMed] [Google Scholar]
- Lee S.-J., Pan J.-G., Park S.-H. & Choi S.-K. Development of a stationary phase-specific autoinducible expression system in Bacillus subtilis. J. Biotechnol. 149, 16–20 (2010). [DOI] [PubMed] [Google Scholar]
- Nijland R., Lindner C., Van Hartskamp M., Hamoen L. W. & Kuipers O. P. Heterologous production and secretion of Clostridium perfringens β-toxoid in closely related Gram-positive hosts. J. Biotechnol. 127, 361–372 (2007). [DOI] [PubMed] [Google Scholar]
- Jan J., Valle F., Bolivar F. & Merino E. Construction of protein overproducer strains in Bacillus subtilis by an integrative approach. Appl. Microbiol. Biotechnol. 55, 69–75 (2001). [DOI] [PubMed] [Google Scholar]
- Wenzel M., Müller A., Siemann-Herzberg M. & Altenbuchner J. Self-inducible Bacillus subtilis expression system for reliable and inexpensive protein production by high-cell-density fermentation. Appl. Environ. Microbiol. 77, 6419–6425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L., Mogk A. & Schumann W. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- Bhavsar A. P., Zhao X. & Brown E. D. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl. Environ. Microbiol. 67, 403–410 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl B., Wehrl W., Wiegert T., Homuth G. & Schumann W. Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J. Bacteriol. 183, 2696–2699 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yansura D. G. & Henner D. J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 81, 439–443 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. T. P., Nguyen H. D. & Schumann W. Novel plasmid-based expression vectors for intra-and extracellular production of recombinant proteins in Bacillus subtilis. Protein Expres. and purif. 46, 189–195 (2006). [DOI] [PubMed] [Google Scholar]
- Phan T. T. P., Nguyen H. D. & Schumann W. Development of a strong intracellular expression system for Bacillus subtilis by optimizing promoter elements. J. Biotechnol. 157, 167–172 (2012). [DOI] [PubMed] [Google Scholar]
- Panahi R., Vasheghani-Farahani E., Shojaosadati S. A. & Bambai B. Induction of Bacillus subtilis expression system using environmental stresses and glucose starvation. Ann. Microbiol. 64, 879–882 (2014). [Google Scholar]
- Blom E. J., Ridder A. N., Lulko A. T., Roerdink J. B. & Kuipers O. P. Time-resolved transcriptomics and bioinformatic analyses reveal intrinsic stress responses during batch culture of Bacillus subtilis. PLoS One 6, e27160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Z. & Doi R. H. Overlapping promoters transcribed by bacillus subtilis sigma 55 and sigma 37 RNA polymerase holoenzymes during growth and stationary phases. J. Biol. Chem. 259, 8619–8625 (1984). [PubMed] [Google Scholar]
- Ye R. Q. et al. High-level secretory production of intact, biologically active staphylokinase from Bacillus subtilis. Biotechnol. Bioeng. 62, 87–96 (1999). [PubMed] [Google Scholar]
- Zhao Q. X., Ding R. R., Kang Y. J. & Chen J. Expression of pectate lyase A from Aspergillus nidulans in Bacillus subtilis. World J. Microb. Biot. 24, 2607–2612 (2008). [Google Scholar]
- Zhou H., Yang Y., Nie X., Yang W. & Wu Y. Comparison of expression systems for the extracellular production of mannanase Man23 originated from Bacillus subtilis B23. Microb. Cell Fact. 12,78 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lu F., Sun T. & Du L. Expression of ksdD gene encoding 3-ketosteroid-Delta1-dehydrogenase from Arthrobacter simplex in Bacillus subtilis. Lett. Appl. Microbiol. 44, 563–568, (2007). [DOI] [PubMed] [Google Scholar]
- Ying Q. et al. Secreted expression of a hyperthermophilic alpha-amylase gene from Thermococcus sp. HJ21 in Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 22, 392–398, (2012). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. High-throughput identification of promoters and screening of highly active promoter-5′-UTR DNA region with different characteristics from Bacillus thuringiensis. PLoS One 8, e62960 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C., Steinbach P. A. & Tsien R. Y. A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909, (2005). [DOI] [PubMed] [Google Scholar]
- Ivankov D. N. & Finkelstein A. V. Prediction of protein folding rates from the amino acid sequence-predicted secondary structure. Proc. Natl. Acad. Sci. USA 101, 8942–8944, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L., Mogk A. & Schumann W. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181, 71–76 (1996). [DOI] [PubMed] [Google Scholar]
- Ming Y. M., Wei Z. W., Lin C. Y. & Sheng G. Y. Development of a Bacillus subtilis expression system using the improved Pglv promoter. Microb. Cell Fact. 9, 55 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechet M. et al. Production of a novel mixture of mycosubtilins by mutants of Bacillus subtilis. Bioresour. Technol. 145, 264–270, (2013). [DOI] [PubMed] [Google Scholar]
- Toymentseva A. A., Schrecke K., Sharipova M. R. & Mascher T. The LIKE system, a novel protein expression toolbox for Bacillus subtilis based on the liaI promoter. Microb. Cell Fact. 11, 143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingming Yang W. Z., Shengyue Ji. et al. Generation of an Artificial Double Promoter for Protein Expression in Bacillus subtilis through a Promoter Trap System. PLoS One 8, e56321 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuzlakoglu Ozturk M., Akbulut N., Issever Ozturk S. & Gumusel F. Ligase-independent cloning of amylase gene from a local Bacillus subtilis isolate and biochemical characterization of the purified enzyme. Appl. Biochem. Biotechnol. 171, 263–278, (2013). [DOI] [PubMed] [Google Scholar]
- Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44, 1072–1078 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J. & Binder S. RT-PCR analysis of 5′ to 3′-end-ligated mRNAs identifies the extremities of cox2 transcripts in pea mitochondria. Nucleic Acids Res. 30, 439–446 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneis M., Hering O., Lange C. & Soppa J. Experimental characterization of Cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet. 3, e229, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C. et al. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 5, 545–551, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.