Summary
The derivation of genetically modified induced pluripotent stem (iPS) cells typically involves multiple steps, requiring lengthy cell culture periods, drug selection, and several clonal events. We report the generation of gene-targeted iPS cell lines following a single electroporation of patient-specific fibroblasts using episomal-based reprogramming vectors and the Cas9/CRISPR system. Simultaneous reprogramming and gene targeting was tested and achieved in two independent fibroblast lines with targeting efficiencies of up to 8% of the total iPS cell population. We have successfully targeted the DNMT3B and OCT4 genes with a fluorescent reporter and corrected the disease-causing mutation in both patient fibroblast lines: one derived from an adult with retinitis pigmentosa, the other from an infant with severe combined immunodeficiency. This procedure allows the generation of gene-targeted iPS cell lines with only a single clonal event in as little as 2 weeks and without the need for drug selection, thereby facilitating “seamless” single base-pair changes.
Highlights
-
•
Episomal reprogramming system is enhanced by expression of miR302/367
-
•
Gene targeting and reprogramming can be combined in a simple one-step procedure
-
•
Clonal gene-corrected iPS cell lines can be obtained in as little as 2 weeks
In this article, Howden and colleagues show that reprogramming and gene targeting can be combined in a simple one-step procedure. Reporter lines or genetically corrected iPS cell lines are derived from patient fibroblasts with relatively high efficiencies in the absence of drug selection in as little as 2 weeks.
Introduction
Induced pluripotent stem (iPS) cells, generated by introducing defined factors to reprogram terminally differentiated somatic cells, offer enormous potential for the development of autologous or customized cellular therapies to treat or correct many inherited and acquired diseases (Takahashi et al., 2007, Yu et al., 2007). Complications associated with immunorejection can be avoided through the generation and subsequent disease correction of patient-specific iPS cells, which can be differentiated into relevant cell types for the repopulation and regeneration of a defective tissue or organ. Gene targeting by homologous recombination is the ideal approach for the correction of genetic defects as it enables replacement of the defective allele with a normal functional one without disturbing the remaining genome. The generation of a genetically modified iPS cell line typically involved multiple procedures that required the cells to be in culture for an extensive period, drug selection, and several clonal events (Hockemeyer et al., 2009, Howden et al., 2011, Liu et al., 2011, Zou et al., 2011). In the first step, somatic cells are reprogrammed, and several clones are expanded and characterized. Gene targeting constructs are then introduced, and cells are usually subjected to drug selection to isolate and identify correctly modified iPS cell colonies. Once successfully targeted clones are identified, it is preferable to excise the drug selectable marker, commonly flanked by loxP or FRT sites. Taken together, the multiple steps required for the generation of genetically modified iPS cell lines typically require cells to be in culture for several months, which is not compatible for patients for whom urgent medical intervention is imperative. Furthermore, there is evidence to suggest that increased culture times are associated with undesirable changes in genomic integrity, such as duplications of oncogenic genes (Laurent et al., 2011) and other karyotypic abnormalities (Chen et al., 2008). Here we report that reprogramming and gene targeting can be performed together in a one-step procedure that requires only a single electroporation. Multiple gene-targeted iPS cell clones can be generated from patient cells in as little as 2 weeks, requiring only a single clonal event. The procedure also does not require the use of drug selection and permits the generation of clones that contain “seamless” single base-pair changes, without leaving residual loxP or FRT sites in the host genome.
Results
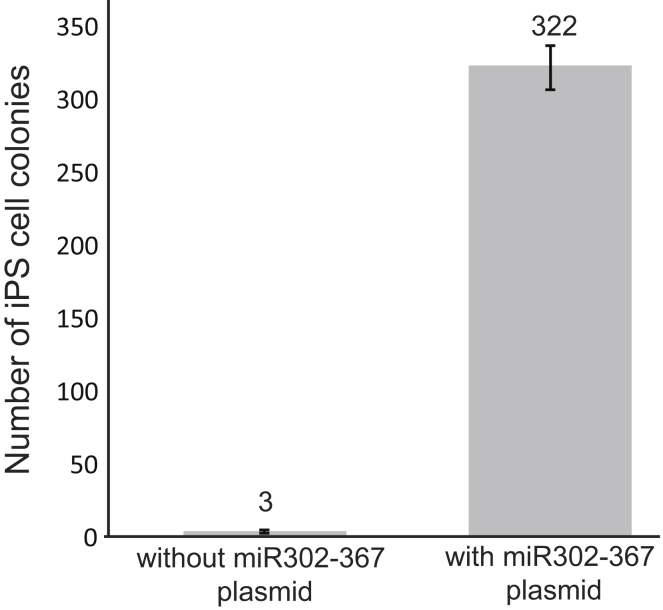
We used an enhanced episomal-based reprogramming system to generate iPS cell lines that would eventually be free of vector sequences. In addition to the seven factors (OCT4, SOX2, NANOG, c-MYC, KLF4, LIN28, and the SV40 Large T-Antigen) encoded by the three oriP-based vectors previously reported to induce pluripotency (Yu et al., 2009), we also forced expression of the micro RNA (miR) 302/367 cluster, which is known to facilitate reprogramming and maintenance of pluripotency (Lin et al., 2008, Miyoshi et al., 2011). The inclusion of an additional episomal vector encoding miR 302/367 resulted in a substantial increase (more than 100-fold) in the total number of iPS cell colonies in human fibroblasts (Figure 1). This plasmid was included in all subsequent reprogramming experiments and was necessary to obtain sufficient iPS cell colony numbers when combining gene targeting and reprogramming in a single step.
Figure 1.
Episomal Reprogramming System Is Enhanced with Inclusion of Plasmid Encoding the miR302/367 Cluster
Reprogramming experiments were performed with and without inclusion of the miR302/367 expression plasmid using a normal male fibroblast line. Data represent an average of three independent experiments ± SD.
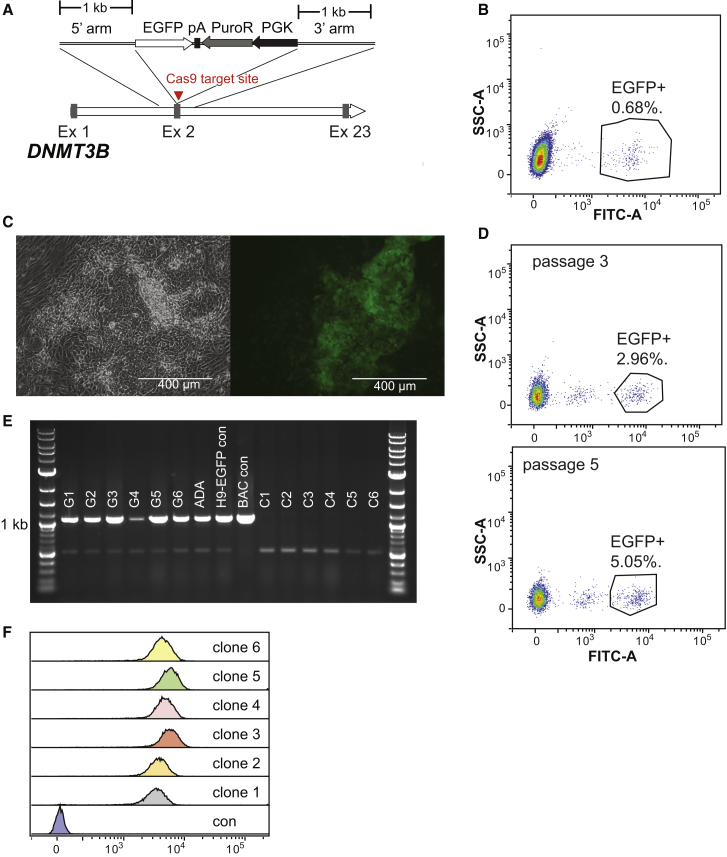
In our initial attempts at simultaneous reprogramming and gene targeting of somatic cells, we chose to target the DNMT3B gene with an EGFP reporter, since DNMT3B is highly expressed in pluripotent cells and quickly downregulated following differentiation, allowing targeted iPS cell colonies to be easily identified by fluorescent microscopy. To facilitate homologous recombination at the DNMT3B locus, we used in vitro transcribed mRNA encoding the Cas9 protein derived from N. meningitides (Hou et al., 2013), a plasmid encoding a DNMT3B-specific short-guide (sg)RNA and a donor template encoding an EGFP reporter and puromycin resistance gene flanked by 1-kb homology arms specific to sequences upstream and downstream of the DNMT3B start codon (Figure 2A). We first evaluated targeting of DNMT3B using this system in the embryonic stem cell line H9 and routinely obtained a gene-targeting efficiency of 0.5%–0.9% (Figure 2B). We next co-transfected the reprogramming plasmids along with the DNMT3B-specific gene-targeting factors into a fibroblast line derived from a patient with autosomal dominant retinitis pigmentosa. Although iPS cell colonies first emerged as early as 10 days following transfection, we observed the vast majority emerge between 2 and 3 weeks postelectroporation, at which point the culture was routinely analyzed by fluorescent microscopy. Of three independent experiments we identified 8, 13, and 44 iPS cell colonies that stably expressed the EGFP reporter, indicative of a successful gene-targeting event at the DNMT3B locus (Figure 2C). We obtained a large number of iPS cell colonies (>1,000) from each of these experiments, making it difficult to accurately assess gene-targeting efficiency. Thus, to estimate targeting efficiency in the pool of iPS cells, we passaged cells from a single representative experiment approximately 3 weeks post-transfection using EDTA to selectively remove iPS cells from the residual fibroblasts before re-plating. As measured by the number of EGFP-expressing cells, targeting efficiency was approximately 3% and 5% following flow cytometric analysis of the total cell population after three and five passages, respectively (Figure 2D). An increase in the number of EGFP-expressing cells is most likely due to a further loss of the residual parental fibroblast population, and we did not observe any further increase in the number of EGFP-expressing cells after five passages. Using the reprogramming experiments that were not passaged, we randomly selected and expanded six EGFP-expressing and six EGFP-non-expressing colonies for further analysis. Gene targeting of the DNMT3B locus was confirmed in all six EGFP-expressing clones by PCR using primers that flank the recombination junction site, but not in any of the EGFP-non-expressing clones (Figure 2E). Flow cytometry analysis also revealed a uniform level of EGFP expression in > 95% of the cell population with similar fluorescence intensities observed in all six clones (Figure 2F). Although targeting of DNMT3B in H9 cells with the same donor template routinely yielded numerous puromycin-resistant colonies, confirming functionality of the phosphoglycerate kinase (PGK) promoter in pluripotent stem cells, EGFP-expressing iPS cell lines generated by simultaneous reprogramming and targeting of DNMT3B exhibited puromycin sensitivity. This suggests the PGK promoter was transcriptionally silenced during the reprogramming process, making drug selection of simultaneously reprogrammed and gene-targeted clones infeasible. Conversely, when we used a one-step procedure to generate iPS cell lines with an EGFP reporter fused to the OCT4 coding region using the Cas9/CRISPR system described previously (Hou et al., 2013) (Figure S1), these clones did exhibit resistance to puromycin in the culture media. In this case the puromycin resistance gene is fused to the EGFP reporter via a 2A sequence and is thereby driven from the endogenous OCT4 regulatory region rather than the minimal and non-specific PGK promoter.
Figure 2.
Gene Targeting of the DNMT3B Locus
(A) Schematic diagram of the DNMT3B locus and the donor template used for gene targeting. pA, polyA signal; Ex, exon.
(B) Flow cytometric analysis of H9 cells 3 days following the transfection of DNMT3B-specific donor template, mRNA encoding Cas9 protein, and plasmid carrying DNMT3B-specific sgRNA.
(C) Representative phase contrast and fluorescent images of an iPS cell colony generated after simultaneous reprogramming and gene targeting of DNMT3B in fibroblasts derived from a patient with retinitis pigmentosa, acquired 2.5 weeks post-electroporation.
(D) Flow cytometric analysis of the total cell population after EDTA passaging, initiated approximately 3 weeks post-electroporation. Analysis was performed after three and five passages.
(E) PCR analysis across recombination junction to confirm gene targeting of DNMT3B, using primers specific to sequence upstream of 3′ homology arm and EGFP. Analysis was performed on 6 EGFP-expressing iPS cell clones (lanes G1–G6) and 6 EGFP-non-expressing iPS cell clones (lanes C1–C6). An H9-derived DNMT3B-EGFP knockin and BAC carrying EGFP inserted at DNMT3B start codon (used to generate donor template) were included as positive controls.
(F) Flow cytometric analysis of EGFP-expressing iPS cell lines generated after simultaneous reprogramming and gene targeting of the DNMT3B locus.
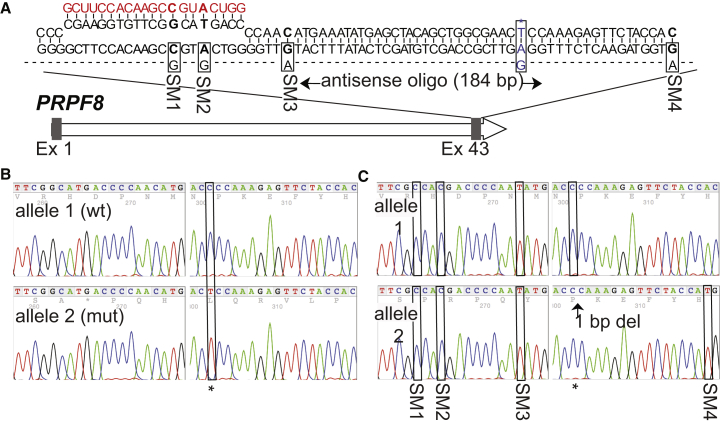
We have also successfully used our one-step protocol to simultaneously reprogram and genetically correct the disease-causing mutation in the patient fibroblasts, an autosomal dominant C > T transition in exon 42 of the PRPF8 gene. This was achieved using a plasmid encoding the Cas9 protein from S. pyogenes (Mali et al., 2013b), a plasmid encoding a PRPF8-specific sgRNA that binds 33 bp upstream of the disease-causing mutation, and a 184-bp single-stranded oligodeoxynucleotide (ssODN) (Figure 3A). The ssODN was engineered to contain four synonymous mutations to minimize the possibility of Cas9 protein re-cutting following homologous recombination and to aid in the identification of clones that had undergone a gene-targeting event (Figure 3A). Approximately 3 weeks post-transfection, we randomly isolated and expanded a total of 72 iPS cell colonies for further analysis. A PCR product encoding the region of interest was amplified from the genomic DNA of all 72 clones using primers flanking the target site, which was subsequently analyzed by Sanger sequencing. Cas9-induced modification of one or both PRPF8 alleles was observed in 22 (31%) of the clones analyzed, most commonly detected as a nonhomologous end joining (NHEJ) event within the intended cut site. Homologous recombination at the target site could be detected in 6 (8%) clones, as evidenced by the loss of the disease-causing mutation or the presence of one or more synonymous mutations carried by the corrective ssODN (Table 1). Genetic correction of the autosomal dominant patient-specific mutation was observed in 2 clones, while targeting of the wild-type allele was observed in 4 clones. We were unable to determine which allele had undergone gene targeting in 1 clone (P.57) due to a 151-bp deletion spanning the site of the mutation. Surprisingly, 1 clone (P.50) appeared to have undergone bi-allelic homologous recombination, as evidenced by correction of the patient-specific mutation and the presence of ssODN-specific synonymous mutations on both alleles (Figures 3B and 3C). However, this clone also contained a 1-bp deletion approximately 50 bp upstream of the intended site of the Cas9-induced double-stranded break. We hypothesize that this is most likely due to homologous recombination with an incorrectly synthesized ssODN rather than an additional mutation caused by NHEJ, which normally occurs at the site of the double-stranded break.
Figure 3.
Simultaneous Reprogramming and Genetic Correction of the PRPF8 Gene in Fibroblasts from a Patient with Retinitis Pigmentosa
(A) Schematic diagram of the PRPF8 gene, with mutation in exon 42. The Cas9 target site (red), the patient-specific mutation (blue), and antisense single-stranded DNA template used for gene repair are shown.
(B) Sequencing analysis of exon 42 of the PRPF8 gene in the genomic DNA from uncorrected patient-specific iPS cells. Both wild-type and mutant alleles are shown.
(C) Sequencing analysis of genomic DNA from a single iPS cell clone following successful simultaneous reprogramming and genetic correction of patient-specific fibroblasts. Both alleles appear to have undergone homologous recombination with the corrective ssODN as evidenced by the presence of the ssODN-specific synonymous mutations (SM 1-4) on both alleles. One allele also has a single base-pair deletion, which is most likely caused by an ssODN that was incorrectly synthesized. The location of the patient-specific mutation and synonymous mutations introduced by the repair ssODN are marked by black boxes.
Table 1.
Analysis of Gene-Targeted iPS Cell Clones Derived from Patient with Retinitis Pigmentosa
| Clone | Modification Observed at PRPF8 Target Site (Exon 42) |
|---|---|
| P.16 | no correction of mutation but presence of SM1 and SM2 on mutant allele; wild-type allele unmodified |
| P.50 | one allele contains SMs 1–3; other allele contains SMs 1–4 and 1-bp deletion ≈30 bp upstream of Cas9 target site |
| P.57 | one allele contains SMs 1–3; other allele contains 151-bp deletion (spanning 124 bp downstream and 7 bp upstream of Cas9 target site) and 105-bp insertion |
| P.71 | correction of mutant allele, but no SMs present; wild-type allele has a 2-bp insertion within Cas9 target site |
| P.72 | wild-type allele contains SMs 1–4; mutant allele has 1-bp deletion |
| P.73 | wild-type allele contains SMs 1–3; mutant allele has 2-bp deletion within Cas9 target site |
SM, synonymous mutation.
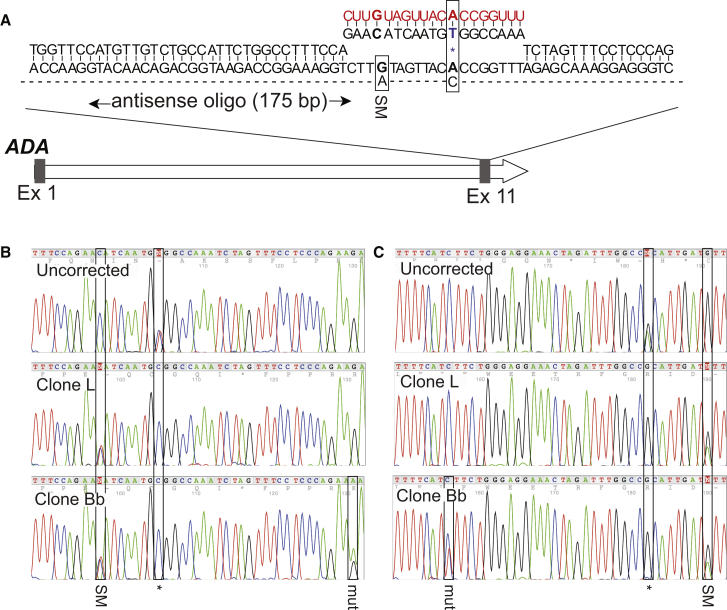
Next we attempted to correct the disease-causing mutation in a fibroblast line isolated from an infant with severe combined immunodeficiency (SCID), caused by mutations in the gene encoding adenosine deaminase (ADA). SCID patients could particularly benefit from a one-step protocol that facilitates the expedited generation of gene-corrected iPS cells because without early intervention, such as a bone marrow transplant, patients typically die within the first 1 to 2 years of life. We first attempted to simultaneously reprogram and target DNMT3B in ADA-SCID fibroblasts and identified one EGFP-expressing colony (0.9%) out of a total of 108 iPS cell colonies (Figure S2). PCR analysis confirmed targeting of the DNMT3B locus (see Figure 2E). We next attempted to simultaneously reprogram and correct one of the disease-causing mutations in the ADA-SCID fibroblasts using our one-step protocol. The fibroblasts were derived from a patient who is a compound heterozygote: one allele has a C > T transition in exon 11 of the ADA gene (1,081C > T), and the second allele has an A > G transition in the 3-prime splice site of intron 3, resulting in a deletion of exon 4 from mature mRNA. We chose to correct the C > T transition in exon 11 using an sgRNA specific to the mutant, but not wild-type, exon 11 sequence of the ADA gene (Figure 4A). We hypothesized that this would minimize Cas9 cutting in both alleles, as seen in the majority of the PRPF8 gene-targeted iPS cell lines, where only 1 out of the 6 clones did not have a second allele modified, either by NHEJ or a second homologous recombination event. To facilitate gene correction we used a 175-bp single-stranded corrective ssODN, which was engineered to contain a single synonymous mutation within the Cas9 target site (Figure 4A). A total of 55 colonies were expanded and screened, with Cas9-induced modification of ADA exon 11 observed in 20 (36%) clones, as determined by Sanger sequencing of a 1.4-kb PCR product amplified from genomic DNA using primers flanking the target site. Gene targeting was detected in 3 (5%) clones, as evidenced by the loss of the disease-causing mutation and the presence of the synonymous mutation carried by the corrective ssODN. Genetic correction of the patient-specific mutation in exon 11 was observed in all three clones, without modification of the second allele, indicating that Cas9 preferentially favored the mutant exon 11 sequence (over wild-type). This was also evident upon analysis of the uncorrected clones that had undergone NHEJ at the target site. Interestingly, in one gene-corrected iPS cell line, we also detected a G > A transition approximately 35 bp downstream of the intended site of the Cas9-induced double-stranded break (Figure 4B). Again, due to its location relative to the Cas9 target site, we hypothesize that this is most likely the result of homologous recombination with an incorrectly synthesized ssODN. Expression of corrected ADA mRNA was also confirmed in all three gene-corrected iPS cell lines following total RNA extraction and RT-PCR to amplify the complete ADA transcript, which was then sequenced (Figure 4C). The two gene-corrected lines that did not carry the additional G > A transition were characterized further by teratoma formation, G-banding, and PCR analysis to detect residual reprogramming plasmids. Both clones formed teratomas comprising all three primary germ layers following injection into immunocompromised mice (Figure S3A), exhibited normal karyotypes (Figure S3B), and were found to be free of residual plasmid sequences (Figure S3C).
Figure 4.
Simultaneous Reprogramming and Genetic Correction of ADA-SCID Fibroblasts
(A) Schematic diagram of the ADA gene, with mutation in exon 11. The Cas9 target site (red), the patient-specific mutation (blue), and antisense single-stranded DNA template used for gene repair are shown.
(B) Sequencing analysis of exon 11 of the ADA gene in the genomic DNA of an uncorrected and two gene-corrected iPS cell lines derived from ADA-SCID fibroblasts. One of the gene-corrected lines (clone Bb) was also found to carry a G > A transition approximately 35 bp downstream of the intended DNA double-stranded break, and most likely introduced by an incorrectly synthesized ssODN.
(C) Sequencing analysis of the ADA transcript amplified from the cDNA of an uncorrected and two gene-corrected iPS cell lines. The location of the patient-specific mutation, synonymous mutation, and G > A transition introduced by the repair ssODN are marked by black boxes.
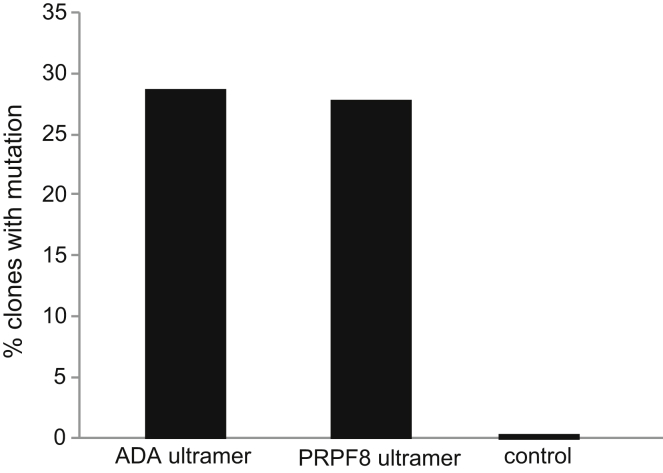
To further investigate the possibility that incorrectly synthesized oligonucleotides may introduce additional mutations following homologous recombination with the host genome, we performed sequencing analysis of the ADA- and PRPF8-specific ssODNs used for gene repair in the experiments described previously. ssODNs were PCR amplified using primers that bind the 5′ and 3′ ends and inserted into a plasmid vector. We then analyzed the oligonucleotide-specific sequences in individual clones by Sanger sequencing. In a control experiment, to estimate polymerase error rate, PRPF8-specific ssODN sequence from one representative clone was PCR amplified, re-cloned and subsequently analyzed by Sanger sequencing. No mutations were detected in any of the 30 clones analyzed from the control experiment. In contrast, mutations were detected in approximately 28% (9/32) and 29% (10/34) of clones harboring the PRPF8 and ADA ssODN sequences, respectively (Figure 5). Although single nucleotide deletions were the most common type of mutation observed, single base insertions, single base substitutions, and deletions up to three bases in size were also detected (Table S1). Furthermore, mutations were observed throughout the ssODN and did not appear to exhibit any significant positional bias. Interestingly, a mutation within one ADA-specific ssODN sequence was also detected at the same location as that observed in the ADA gene-corrected iPS line with the additional G > A transition. Since the ssODNs used in our study were > 150 bases in size and purified by standard desalting, perhaps the incidence of additional mutations following gene repair could be reduced by using ssODNs that are smaller in length or purified by PAGE or high-pressure liquid chromatography (HPLC) following synthesis.
Figure 5.
Sequencing Analysis of Oligonucleotides Used in Gene Repair Experiments
The number of plasmid clones (expressed as percentage of total clones analyzed) carrying oligonucleotide sequence with at least one mutational event is shown. As a control, the PRPF8-specific oligonucleotide sequence from a single plasmid clone was PCR-amplified and re-cloned to assess polymerase error rate.
Discussion
We have demonstrated the feasibility of performing reprogramming and gene correction together in a simple one-step procedure that enables the generation of multiple gene-corrected and uncorrected iPS cell lines in as little as 2 weeks, requiring considerably less time and resources compared to conventional multi-step protocols that can take several months to complete. In a therapeutic context this should facilitate transplantation medicine by making gene-corrected cells available to patients in a more timely manner, while potentially minimizing the risks associated with extended cell culture, drug selection, and multiple clonal events. In addition, we anticipate that comparisons between corrected and matched uncorrected control iPS cell lines generated from a single experiment will also be extremely useful for disease modeling and understanding the underlying molecular mechanisms governing disease, because any observed differences between corrected and uncorrected cells can be attributed to the patient-specific mutation rather than differences in genetic background.
However, it is important to note that a number of studies have demonstrated that iPS cell lines derived from skin biopsies typically harbor a unique subset of de novo genetic abnormalities, either in the form of copy-number variation or single base-pair changes (Abyzov et al., 2012, Gore et al., 2011) and that iPS cell lines generated from the same parental line can vary significantly with respect to whole-genome gene expression in the differentiated state (Reinhardt et al., 2013). Nonetheless, it is reasonable to expect that the confounding effects arising from the variations that exist across different iPS cell clones may be minimized by comparing multiple gene-corrected or gene-targeted clones with multiple uncorrected clones. In this regard a consistent difference that is observed exclusively in the corrected versus uncorrected lines can most likely be attributed to the patient-specific mutation rather than variations that may exist from one clone to the next. In the current study we routinely observed targeting efficiencies of > 5%, enabling the generation of multiple gene-targeted and “matched” uncorrected clones from a single experiment.
The relatively high gene-targeting frequencies obtained using our one-step protocol may in part be attributed to the possibility that the iPS cell colonies themselves act as a form of selection. This is based on the assumption than an iPS cell colony that has taken up the plasmids required for reprogramming will have also taken up the DNA constructs required for gene targeting. Following simultaneous reprogramming and targeting of the DNMT3B locus in the fibroblast line derived from a patient with retinitis pigmentosa, the estimated targeting efficiency in the total iPS cell population was approximately 5%, more than 5-fold higher than that observed in the embryonic stem cell line H9. With respect to the ADA and PRPF8 gene correction experiments, we obtained targeting efficiencies of 5% and 8%, respectively. This is notably higher than the frequencies of gene targeting that have previously been reported in pluripotent stem cells using ssODNs, without the aid of drug selection, where correctly targeted cells typically comprise less than 1% of the total population (Soldner et al., 2011, Yang et al., 2013). Another notable advantage of our one-step protocol over conventional approaches is that it does not require additional steps for the clonal isolation of iPS cells. In the absence of selection, this is most often performed using fluorescently activated cell sorting or limiting cell dilution to expand a clonal population from a single iPS cell, which is inefficient and cumbersome because human pluripotent stem cells exhibit poor survivability in the absence of appropriate cell-to-cell contacts.
Although bi-allelic Cas9-induced modification was a common outcome in the iPS cell lines generated in this study, we show that this can be minimized by designing sgRNAs that specifically target patient-specific mutations. This approach is feasible for patients with autosomal dominant and compound heterozygous mutations, but not for mutations that are autosomal recessive. However, since autosomal recessive diseases normally require only a single functional allele, bi-allelic modification resulting in correction of one allele and mutagenic NHEJ in the other, is less of a concern. It will also be important to evaluate the use of Cas9 variants that have improved DNA cleavage specificity in the protocol described here. Use of the paired nickase (Cas9-D10A) (Mali et al., 2013a, Ran et al., 2013) and dCas9-Fok1 fusions (fCas9) (Guilinger et al., 2014), for example, should minimize the potential of off-target effects that are largely associated with the wild-type form of the Cas9 protein. Keeping background damage to a minimum will be essential not only for retaining the downstream therapeutic potential of the cells but also for an accurate recapitulation of the disease and corrected (wild-type) phenotype, which will be important for disease modeling and drug discovery purposes. Finally, there is also significant value in adapting our protocol to permit the generation of gene-corrected iPS cell lines from alternative cell sources such as blood, which involve less invasive procedures and may harbor fewer somatic mutations compared with fibroblasts derived from skin biopsies. Since expansion of fibroblasts from an initial skin biopsy can take several weeks, the ability to simultaneously reprogram and genetically repair cells isolated from blood draws will also serve to expedite the process of generating gene-corrected iPS cell lines even further.
Experimental Procedures
Plasmid DNA and mRNA
The episomal vectors (pEP4EO2SEN2L, pEP4EO2SET2K, and pEP4EO2SEM2K) carrying seven factors for reprogramming have been described previously (Yu et al., 2009). An additional plasmid carrying the miR 302/367 cluster was generated by PCR—amplifying an approximately 1.2-kb fragment from genomic DNA (extracted from human embryonic stem cell line H1) and cloned into the pSimpleII plasmid (an OriP-containing plasmid) under the control of the elongation factor-1α promoter. The plasmids used for targeting OCT4 have been previously described (Hou et al., 2013). The plasmid hCas9 was a gift from George Church (Addgene plasmid #41815). All sgRNA targeting the DNMT3B, PRPF8, or ADA genes were driven from a U6 promoter cloned into pstBlue-1 (Novagen). The DNMT3B targeting vector was generated by inserting a DNA cassette encoding an EGFP reporter, PGK-promoter-driven puromycin gene, and kanamycin gene flanked by FRT sites into a bacterial artificial chromosome (BAC) clone containing the complete DNMT3B coding region (CTD-2608L15) using the Red-ET recombination system (Genebridges). The kanamycin resistance gene was subsequently removed from the gene-targeting vector with Flpe-recombinase (Genebridges). All plasmids were prepared by cesium chloride gradient extraction. In vitro transcribed mRNA encoding Cas9 and EBNA1 was generated using the SP6 mMessage mMachine kit (Life Technologies). ssODNs (Ultramers) were purchased from Integrated DNA Technologies.
Fibroblast Culture
Fibroblasts derived from a patient with retinitis pigmentosa were obtained from the Peirce Lab, Penn State University, and ADA-SCID fibroblasts were obtained from Coriell Laboratories (ID no. GM02824). Fibroblasts were cultured in DMEM (Invitrogen) supplemented with 15% fetal bovine serum (FBS) (HyClone) at 37°C, 5% CO2, and 5% O2.
Flow Cytometry
Cells were analyzed for EGFP expression by flow cytometry using a FACSCanto II flow cytometer (BD Biosciences). Data acquisition and analysis were performed using FACsDiva software version 6.1.3 (BD Biosciences) and FlowJo (Tree Star) software version 9.5.1. Gating was performed on live cells based on forward and side scatter analysis.
Reprogramming/Gene Correction
Fibroblasts were harvested 2 days after passaging and resuspended in OptiMEM (Life Technologies) at a final concentration of 4–6 × 106 cells/ml. 500 μl of the cell suspension was added to a 0.4-cm cuvette containing the reprogramming and gene-targeting DNA constructs and electroporated (220 V, 1,000 μF) using the Gene Pulser II (BioRad). See Table S2 for DNA concentrations used for each experiment. In vitro transcribed mRNA encoding a truncated version of the EBNA1 protein was also included to enhance nuclear uptake of the oriP-containing reprogramming vectors (Chen et al., 2011, Howden et al., 2006). Following electroporation, cells were plated on a single 10-cm Matrigel-coated (Corning) plate and maintained in fibroblast media until 4 days post-transfection, and then they were switched to E7 medium (E8 medium without transforming growth factor β) with 100 μM sodium butyrate and changed every other day as described previously (Chen et al., 2011). Sodium butyrate was removed from the media after the appearance of the first iPS cell colonies at around day 10. After isolation, iPS cells were maintained and expanded in E8 medium with daily media changes and passaged every 3–4 days with EDTA in 1× PBS as previously described (Chen et al., 2011).
PCR, RT-PCR, and Sanger Sequencing
Total genomic DNA was extracted by using the DNeasy Blood and Tissue Kit (QIAGEN). Total RNA was extracted by using the RNeasy Plus Mini Kit (QIAGEN). cDNA was synthesized using SuperScript III (Life Technologies) according to the manufacturer’s protocol. PCR was performed by using GoTaq Green PCR Mastermix (Promega). Prior to sequencing, PCR products were treated with rAPid Alkaline Phosphatase (Roche Life Science) and Endonuclease I (NEB) for 30 min at 37°C followed by heat inactivation at 80°C for 15 min. Sequencing reactions were performed using BigDye Terminator v3.1 (Life Technologies). Reactions were then purified and sequenced by the Genome Center, University of Wisconsin. For sequencing analysis of single alleles, PCR products amplified from genomic DNA extracted from each of the PRPF8 gene-corrected clones were cloned into the pUC19 vector (Life Technologies). Plasmid DNA was extracted from 12 randomly selected colonies for each of the 6 cloned PCR products and sequenced as described above.
Oligonucleotide Sequences
For lists of primers used in this study for PCR amplification and generation of sgRNA plasmids, see Table S3.
Author Contributions
S.E.H. and J.A.T. conceived the experiments and wrote the paper; J.P.M. and B.M.D. performed teratoma injection and harvesting. A.G.E. and E.G.S. assisted with ssODN analysis experiments.
Acknowledgments
We thank Eric Peirce for providing the fibroblast line derived from a patient with retinitis pigmentosa. This work was supported by a Wynd-Gund Translation Research Award from the Foundation Fighting Blindness. S.E.H. is supported by a National Health and Medical Research Council (NHMRC) Overseas Biomedical Fellowship. A.G.E. an E.G.S. are senior research fellows of the NHMRC.
Published: November 12, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes three figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.10.009.
Supplemental Information
References
- Abyzov A., Mariani J., Palejev D., Zhang Y., Haney M.S., Tomasini L., Ferrandino A.F., Rosenberg Belmaker L.A., Szekely A., Wilson M. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Ye Z., Yu X., Zou J., Mali P., Brodsky R.A., Cheng L. Trophoblast differentiation defect in human embryonic stem cells lacking PIG-A and GPI-anchored cell-surface proteins. Cell Stem Cell. 2008;2:345–355. doi: 10.1016/j.stem.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A., Li Z., Fung H.L., Young J.E., Agarwal S., Antosiewicz-Bourget J., Canto I., Giorgetti A., Israel M.A., Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z., Zhang Y., Propson N.E., Howden S.E., Chu L.F., Sontheimer E.J., Thomson J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden S.E., Wardan H., Voullaire L., McLenachan S., Williamson R., Ioannou P., Vadolas J. Chromatin-binding regions of EBNA1 protein facilitate the enhanced transfection of Epstein-Barr virus-based vectors. Hum. Gene Ther. 2006;17:833–844. doi: 10.1089/hum.2006.17.833. [DOI] [PubMed] [Google Scholar]
- Howden S.E., Gore A., Li Z., Fung H.L., Nisler B.S., Nie J., Chen G., McIntosh B.E., Gulbranson D.R., Diol N.R. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc. Natl. Acad. Sci. USA. 2011;108:6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L.C., Ulitsky I., Slavin I., Tran H., Schork A., Morey R., Lynch C., Harness J.V., Lee S., Barrero M.J. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.L., Chang D.C., Chang-Lin S., Lin C.H., Wu D.T., Chen D.T., Ying S.Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–2124. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.H., Suzuki K., Qu J., Sancho-Martinez I., Yi F., Li M., Kumar S., Nivet E., Kim J., Soligalla R.D. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell. 2011;8:688–694. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N., Ishii H., Nagano H., Haraguchi N., Dewi D.L., Kano Y., Nishikawa S., Tanemura M., Mimori K., Tanaka F. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P., Schmid B., Burbulla L.F., Schöndorf D.C., Wagner L., Glatza M., Höing S., Hargus G., Heck S.A., Dhingra A. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Soldner F., Laganière J., Cheng A.W., Hockemeyer D., Gao Q., Alagappan R., Khurana V., Golbe L.I., Myers R.H., Lindquist S. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Yang L., Guell M., Byrne S., Yang J.L., De Los Angeles A., Mali P., Aach J., Kim-Kiselak C., Briggs A.W., Rios X. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Mali P., Huang X., Dowey S.N., Cheng L. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.