Abstract
The mitochondrial alternative oxidase, AOX, carries out the non proton-motive re-oxidation of ubiquinol by oxygen in lower eukaryotes, plants and some animals. Here we created a modified version of AOX from Ciona instestinalis, carrying mutations at conserved residues predicted to be required for chelation of the diiron prosthetic group. The modified protein was stably expressed in mammalian cells or flies, but lacked enzymatic activity and was unable to rescue the phenotypes of flies knocked down for a subunit of cytochrome oxidase. The mutated AOX transgene is thus a potentially useful tool in studies of the physiological effects of AOX expression.
The mitochondrial alternative oxidase, AOX, carries out the non proton-motive re-oxidation of ubiquinol by molecular oxygen. Terminal electron transfer by AOX constitutes a parallel system to that provided by OXPHOS complexes III and IV in plants, fungi, protists and many animal phyla1. AOX is believed to become activated under stress conditions, when the OXPHOS cytochrome chain is overloaded or unavailable.
In many organisms this is achieved, at least in part, via the regulated expression of the AOX gene, which is induced by a variety of stresses relevant to OXPHOS dysfunction2,3. The enzyme is also inherently responsive to the metabolic signature of such stresses in different organisms. Firstly, it is activated by high levels of its reduced substrate, ubiquinol4,5, which is assumed to reflect a lower affinity for the substrate than that exhibited by OXPHOS complex III, with which it competes. Thus, under normal physiological conditions, most of the electron flow from ubiquinol to oxygen is channelled through complexes III and IV, even if AOX is physically present. Only if ubiquinol levels increase, for example, if the enzymatic capacity of complexes III and IV becomes limiting, will AOX become functionally significant. In addition, AOX is allosterically activated in many organisms by metabolites whose levels increase under conditions of OXPHOS insufficiency, for example pyruvate3, as well as by other metabolites indicative of cellular redox state.
Although the AOX gene has been lost, during the course of evolution, in the lineages leading to the most complex and advanced metazoan groups, including mammals1, we reasoned that its reintroduction by transgenesis should enable such animals to buffer many of the pathological stresses resulting from OXPHOS dysfunction6. Thus AOX could become a therapeutic tool for treating mitochondrial diseases and other conditions mediated by OXPHOS dysfunction7. Preliminary tests in model organisms, including cultured human cells8,9, Drosophila10,11 and the mouse12, support this concept. In particular, the expression of AOX from the tunicate Ciona intestinalis, was shown to compensate many of the phenotypes resulting from cytochrome oxidase (COX, complex IV) deficiency in Drosophila, including the knockdown of structurally essential subunits of the complex11. However, if AOX is to be of value in eventual therapy, the mechanism of this compensation needs to be established. The hypothesized enzymatic by-pass is only one of several possible such mechanisms. Expression of an inert transgene, such as GFP, in place of AOX, was unable to rescue the phenotypes produced by engineered deficiency of cytochrome oxidase10,11. However, this control cannot be unambiguously interpreted, since the expressed GFP was not targeted to mitochondria, and even if it were, does not possess other structural features of AOX that enable it to insert into the inner mitochondrial membrane in a specific fashion and interact with other components thereof.
In order to provide a more applicable test of whether the ability of AOX to rescue COX deficiency depends on its primary enzymatic activity, we sought to engineer the AOX in such a way as to destroy this activity, whilst producing only a minimal effect on the overall structure, stability and expression of the protein. To do this, we took advantage of the fact that AOX is well conserved phylogenetically, that the residues contributing to its active site have been characterized in a number of species, and that the structure of a representative AOX, from the protistan parasite Trypanosoma brucei, has recently been published13. Using currently available bioinformatics tools, we modelled the structure of the Ciona intestinalis enzyme against this template, predicted amino-acids required for binding the catalytically essential diiron moiety at the active site, and proceeded via alanine-substitution mutagenesis to create an expressible version of the enzyme expected to lack enzymatic activity, despite being predicted to fold to a similar overall structure. In several different contexts (cultured human and Drosophila cells, as well as whole animals), we found that the mutated AOX was stably expressed but devoid of detectable enzymatic activity. Furthermore, expression of the transgene encoding the mutated AOX was unable to rescue engineered COX deficiency in the fly, confirming that this rescue indeed depends on the enzymatic activity of AOX.
Materials and Methods
Sequence alignments and molecular modelling
The sequences of AOX homologues found by BlastP searching were aligned using the MUSCLE algorithm built into the software MEGA614, with default parameters. A homologous model of the structure of one subunit of the C. intestinalis AOX was generated using the software I-TASSER15, based on the crystal structure of the Trypanosoma brucei AOX (PDB 3VV9:A)13 as template and the multiple sequence alignment described above as input restraint. Other parameters were set as default. Selection of the model was based upon the best accuracy estimations provided by the C-scores, estimated TM-scores and RMSD values. Because the N-terminal region (M1-K103) of the C. intestinalis AOX structure could not be modelled with high accuracy, this region was eliminated from the analysis. The dimeric model of C. intestinalis AOX and the positioning of the two diiron centres (one per subunit) were built by overlapping two copies of the model generated by I-TASSER into the crystal structure of the dimeric T. brucei AOX using Pymol (www.pymol.org). Pymol was also used to analyze all structure models and to produce the figures.
Cloning procedures and mutagenesis
For Drosophila expression, the C. intestinalis AOX coding sequence, including its natural stop codon, was recloned from the pMT/V5-His B vector (Invitrogen), in which it had been previously propagated, into the EcoRI site of pUASTattB16. Based on the multiple sequence alignment shown in Fig. S1, and the results of molecular modelling (see Results), PCR-based alanine substitution mutagenesis and recloning were carried out according to the scheme of Fig. S2. Mutations E239A, H242A, E344A and H347A were introduced, using the plasmid-borne AOX cDNA as template, Pfu DNA polymerase (Stratagene) and oligonucleotides (all shown 5′ to 3′) as follows: GAAGCTGAAAATGcGAGAATGgcCTTAATGACTGCG and CGCAGTCATTAAGgcCATTCTCgCATTTTCAGCTTC to create E239A/H242A, followed by ATCTGAGCTGATGcAGCACATgcCAGATCAGTCAAC and GTTGACTGATCTGgcATGTGCTgCATCAGCTCGGAT to create E344A/H347A (lowercase letters indicate the sites of introduced mutations). For expression in S2 cells, constructs containing the original and mutated AOX cDNA inserts, again using the natural stop codon, were recloned into the EcoRI site of pAc5.1/V5-His B (Invitrogen, USA) to create pAC/AOX17 and pAC/mutAOX. For transient mammalian expression, the wild-type and mutated AOX coding sequences were recloned, respectively, into a pBR322-derived kanR plasmid containing the CAG promoter18 and bovine growth hormone poly(A) signal, together with other elements not relevant to the present study (copies of the tet operator, loxP sites, insulator elements and portions of the porcine Ggta1 gene), to create the expression constructs pCAG-AOX and pCAG-mutAOX. The nucleotide sequences of all clones were confirmed by Sanger sequencing using the Big Dye Terminator v3.1 kit (Life Technologies) and an ABI3130xl Genetic Analyzer, according to the manufacturer’s specifications.
Drosophila stocks and maintenance
Except where stated, flies were maintained and grown on standard medium at 25°C, using a 12 h light/dark cycle, as previously10,19. Balancers, recipient line w1118, the RNAi line for CG9603 (Vienna Drosophila RNAi Center line 106661), the ubiquitous da-GAL4 driver (Bloomington line 8641) and the driver line bearing elavC155-GAL4 on chromosome X and UAS-Dcr2 on chromosome 2 (Bloomington line 25750), were obtained from stock centres. ΦC31 recombinase-mediated-site-directed transgenesis was used to generate transgenic fly lines (service provided by BestGene Inc, Chino Hills, CA), using recipient lines with the following integration sites: attP18 (chromosome X), attP40 (chromosome 2) and attP2 (chromosome 3), according to Pfeiffer et al.20, employing the wild-type and mutated AOX constructs cloned in pUASTattB and pUASTattB itself as empty-vector control. Following characterization, transgenic lines were maintained over balancers appropriate for chromosome X, 2 or 3, bearing standard markers (FM7, CyO, TM3Sb, respectively). Transgenic lines UAS-AOXF24 and UAS-AOXF6 were described previously10.
Cell culture and transfection
HEK293T cells were cultured as previously21. Plates of 3 × 106 cells were transfected with 24 μg of the pCAG-AOX or pCAG-mutAOX plasmids or, as control, empty vector (pWPI, Addgene), using 60 μl Lipofectamine® 2000 (Invitrogen) under manufacturer’s recommended conditions. Drosophila S2 cells were grown and transfected with pAc5.1/V5-His B or derivatives as previously17.
Expression assays
RNA extraction and QRTPCR to measure AOX transcript levels using RpL32 RNA as an internal normalization standard were as previously described10, using RNA from 2 day-old adult male and female flies. Protein extraction from 2 day-old Drosophila adults and Western blots were conducted essentially as by Fernandez-Ayala et al.10, with the following modifications: for females, 1% SDS was used for lysis instead of 1.5% Triton X-100, flies were processed in batches of 30 (females) or 40 (males), SDS-PAGE used Any kD™ Criterion™ TGX™ 18-well gels (Bio-Rad), Prestained Protein Ladder (Thermo-Scientific) and ProSieveTM EX Running and Transfer Buffers (Lonza), and membranes were treated in PBS-Tween® instead of TBS. Primary antibodies used were customized rabbit anti-AOX10 (21st Centrury Biochemicals, 1:10,000), rabbit anti-α-actininin C-20-R (Santa Cruz Biotechnology, 1:5,000) and mouse anti-ATP5A (Abcam, 1:50,000). Secondary antibodies were Peroxidase Goat Anti-rabbit IgG and Horse Anti-mouse IgG (both from Vector Laboratories, 1:10,000). Post-nuclear extracts (PN) from HEK293T cells were prepared according to Cannino et al.21. Protein concentrations were measured using the Bradford assay.
Respirometry
Oxygen consumption of 5 × 106 human cells was measured 48 h after transfection, following permeabilization with 80 μg/ml digitonin, in a Clark-type electrode (Hansatech Oxytherm system) using respiratory buffer A22 at 37 °C. Complex II-driven respiration was measured in the presence of 10 mM ADP and 10 mM succinate. AOX-driven (antimycin-resistant) respiration was measured after the further addition of (60 ng/ml) antimycin A, with subtraction of any residual oxygen consumption after adding 100 μM n-propyl gallate. Respirometry on S2 cells was as described previously17 and was also conducted on homogenates from 1–4 day-old Drosophila males. Briefly, 25 males were gently homogenized in 0.8 ml ice-cold isolation buffer (250 mM sucrose, 5 mM Tris-HCl, 2 mM EGTA, pH 7.4) and muslin-filtered. Respirometry was performed on 150 μl aliquots of this homogenate, mixed with 500 μl assay buffer (120 mM KCl, 5 mM KH2PO4, 3 mM HEPES-KOH, 1 mM EGTA, 1 mM MgCl2, 0.2% BSA, pH 7.2), substrates (15 mM glycerol-3-phosphate and 5 mM ADP) and inhibitors as for permeabilized mammalian cells.
Behavioural assays
Time to eclosion following Drosophila crosses was measured as previously23. Eggs from parents crossed two days earlier were collected over three consecutive nights, and cultured at 25°C. Adults less than 24 h old were collected and sorted on ice, after which batches of 5 male flies were placed in each empty vial. After a 10 min waiting period, flies were tipped down and their subsequent behaviour recorded using a DFK 21AF04 camera (The Imaging Source, Bremen, Germany) and Media Recorder 2 software (Noldus, Wageningen, Netherlands). The climbing index11 for each vial was manually calculated from recordings as the mean number of flies which climbed 6 cm in 10 s in three trials. Climbing indices from different genotypes were compared by one-way ANOVA with Bonferoni adjustment, using SPSS 12. The box plot was drawn with BoxPlotR (boxplot.tyerslab.com), with Tukey style whiskers extending to the data point that is no more than 1.5 × IQR (interquartile range) from the edge of the box24.
Human subjects
The work reported here did not use human subjects or any materials derived from human subjects, other than the freely available cell-line HEK293T.
Results and Discussion
Modelling and creation of mutated AOX transgene
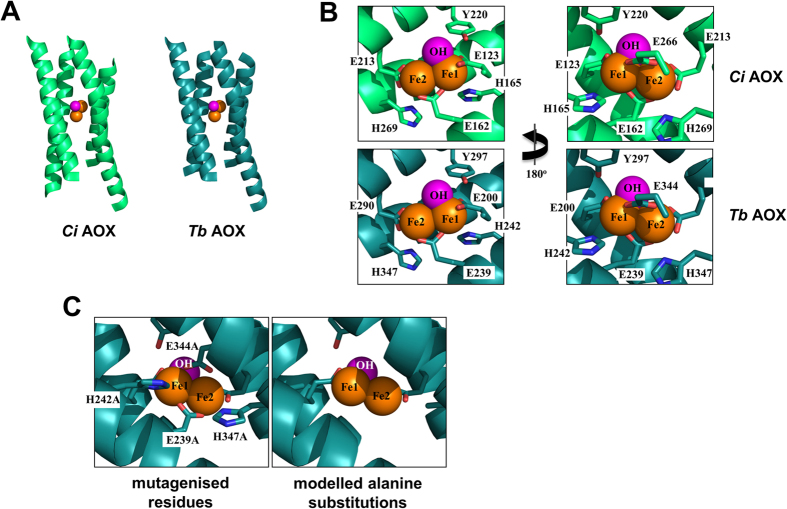
Alignment of the predicted Ciona intestinalis AOX amino-acid sequence with the corresponding protein from other taxa, including Trypanosoma brucei, revealed conservation of residues implicated in the organization of the diiron centre of the enzyme, as previously reported by Shiba et al.13. The four invariant glutamate residues and two histidines correspond in Ciona AOX with E200, E239, E290, E344, H242 and H347 (Fig. S1), numbered from the first methionine of the putative preprotein. In the Trypanosoma AOX structure, the conserved histidines participate in a hydrogen bond network that also includes a conserved tyrosine, Y297 in Ciona AOX (Fig. S1). Structural modelling (Fig. 1) showed that Ciona AOX can fold to an almost identical structure as its Trypanosoma counterpart, ignoring the poorly conserved N-terminal region (residues 1–103 of the Ciona protein, Fig. S1). Four alpha-helices enclose the diiron centre of each protomer of the homodimeric protein, with the conserved glutamate and histidine residues similarly juxtaposed as in the Trypanosoma protein (Fig. 1). Based on this structure, we tested the functional significance of the conserved residues at the predicted diiron centre, by mutating four of them to alanine (E239A, H242A, E344A, H347A), in appropriate transgenic constructs for expression in mammalian cells and Drosophila (Fig. S2). The mutations were predicted to destroy the binding of iron to the active site, whilst only minimally disturbing the overall structure of each subunit.
Figure 1. Structural modelling and mutagenesis of active site of Ciona intestinalis AOX.
(A) Model of the active site of the Ciona (Ci) enzyme, green, compared with the structure of the Trypanosoma brucei (Tb) AOX, blue. In both cases, the diiron site (iron moieties in orange, hydroxyl in pink) is buried in a four alpha-helix bundle. For clarity, only one protomer is shown. (B) Conserved residues binding the diiron centre show an identical arrangement in the Ci model (green) as in the Tb structure (blue). (C) The residues selected for alanine-substitution mutagenesis in the Ci enzyme (here shown in blue), alongside the resulting modelled structure.
Mutated AOX can be stably expressed in mammalian cells and flies
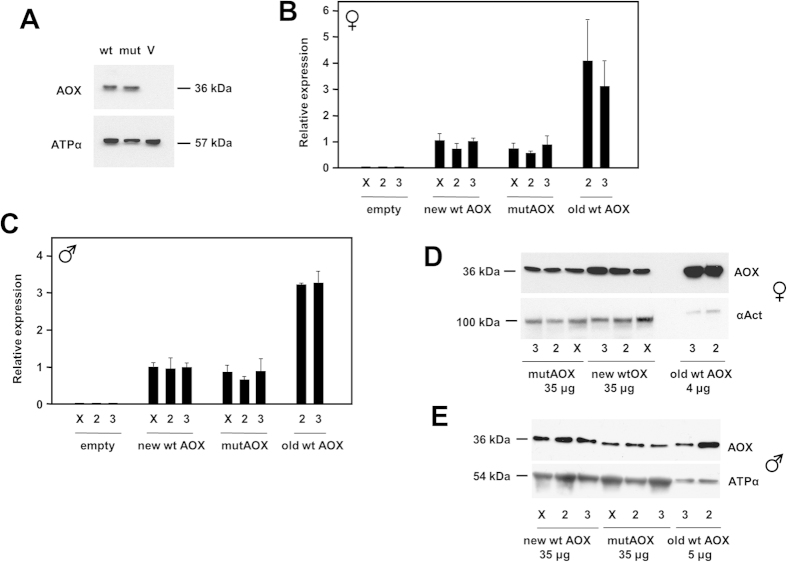
In order to test its functionality, the expression of the mutated AOX construct (mutAOX) was first verified, following transient transfection into cultured human cells. Based on Western blotting (Fig. 2A), the mutAOX protein was the same size and comparably expressed as wild-type AOX. Next, the mutAOX transgene, under the control of the GAL4-dependent UAS promoter, was introduced into the Drosophila genome by targeted insertion at single sites on each chromosome. Parallel control lines were created, containing wild-type AOX and empty vector, inserted at the same sites. Following validation of the insertions by PCR and sequencing, we measured transgene expression directed by the ubiquitous da-GAL4 driver, at both RNA and protein levels, using QRTPCR (Fig. 2B, C) and Western blotting (Fig. 2D, E).
Figure 2. Expression of AOX transgenes in mammalian cells and Drosophila.
(A) Western blot of protein extracts from HEK293T cells transfected with wild-type and mutated AOX constructs (wt, mut) or empty vector (V), probed for AOX and for ATP synthase subunit α as loading control. (B,C) Relative AOX expression at RNA level, based on QRTPCR, in (B) females and (C) males of different Drosophila lines transgenic for wild-type or mutated AOX, or empty vector, inserted on chromosomes X, 2 and 3, as shown, in combination with the ubiquitous da-GAL4 driver. New wt (wild-type) and mutAOX lines were those created by site-specific integration at defined chromosomal sites using the ΦC31 system; old wt AOX lines were UAS-AOXF6 (chromosome 2) and UAS-AOXF24 (chromosome 3). For males, all values were significantly different from empty-vector lines; old wt AOX lines were significantly different from new wt AOX lines (p < 0.001, ANOVA followed by post-hoc Bonferoni-corrected t test), but mutAOX and new wt AOX lines were not significantly different from each other. Statistical analysis for females gave similar results, although greater sample-to-sample variation for old wt AOX lines yielded only p < 0.05 comparing them with new wt or mutAOX lines. (D,E) Western blot of protein extracts from the same flies (amounts as shown), probed for AOX or, as loading control, either ATP synthase subunit α or α-actinin, as indicated.
In both females (Fig. 2B) and males (Fig. 2C), the expression of wild-type and mutAOX were similar at the RNA level, but 3–4 fold less than AOX in the previously created transgenic lines, engineered by random P-element insertion. At the protein level, mutAOX showed slightly lower expression than wild-type AOX in both sexes, and expression was again less than in the previously created lines (Fig. 2D, E).
When expressed ubiquitously using the da-GAL4 driver, the AOX and mutAOX transgenes produced only very small changes in developmental timing, most of them non-significant compared with the corresponding vector-only line (Fig. 3).
Figure 3. Developmental time to eclosion of AOX transgenic flies.
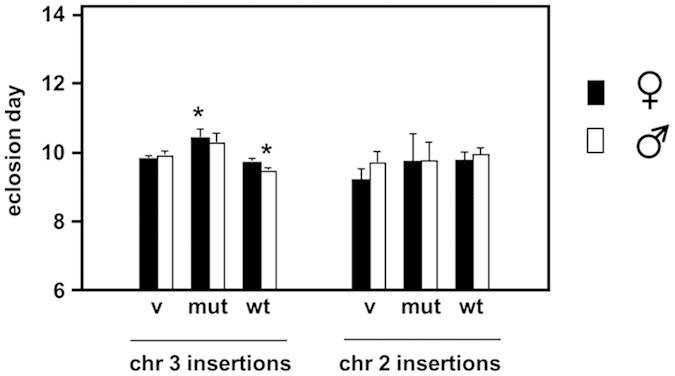
Eclosion day (mean +SD) for females and males of different Drosophila lines transgenic for wild-type (wt) or mutated (mut) AOX, or empty vector (v), inserted on chromosomes X, 2 and 3, as shown, in combination with the ubiquitous da-GAL4 driver. *denotes significant difference from flies of the same sex from the empty vector line on the same chromosome, p < 0.05 (Student’s t test).
Mutated AOX lacks detectable enzymatic activity
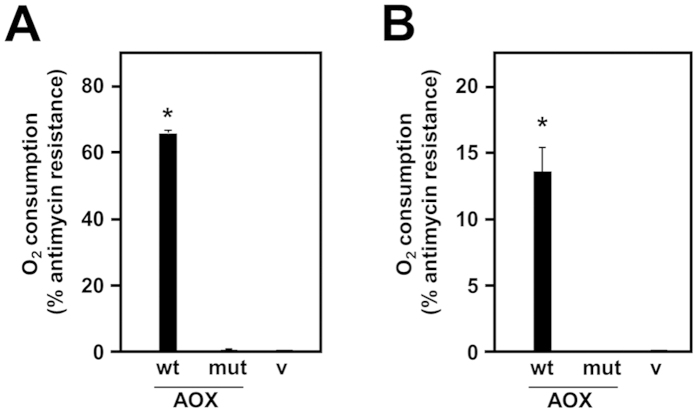
The functionality of the expressed AOX variants was tested by polarography. Permeabilized HEK293T cells, following transient transfection with wild-type AOX, supported approximately 80% of the uninhibited oxygen consumption, in the presence of antimycin. Antimycin-resistant oxygen consumption was undetectable in permeabilized cells transiently transfected with the mutAOX construct or empty vector (Fig. 4A). A similar result was obtained after transfection of Drosophila S2 cells. After transfection with either of two different AOX-expressing constructs, whole-cell respiration in the presence of antimycin was 70–73% of the uninhibited rate, but was undetectable in control cells or cells transfected with the mutAOX construct (Table S1). Finally, in homogenates from male transgenic flies carrying targeted insertions at the same locus (on chromosome 2), induced to express the transgene ubiquitously using the da-GAL4 driver, wild-type AOX supported 14% of the uninhibited substrate oxidation rate in the presence of antimycin (Fig. 4B), whereas mitochondria from mutAOX- or empty vector-transgenic flies showed no antimycin-resistant substrate oxidation. In every polarography experiment, expression of the AOX transgene was verified by Western blotting as per Fig. 2.
Figure 4. Respirometry of AOX-transfected cells and flies.
Oxygen consumption (% resistant to antimycin, as defined in Materials and Methods) of (A) permeabilized, transiently transfected cells, and (B) homogenates from male transgenic flies induced for expression using da-GAL4 driver, expressing wild-type (wt) or mutated (mut) AOX or empty vector (v). The flies had transgenic insertions on chromosome 2. *denotes significant difference from vector-only flies.
Mutated AOX is unable to rescue COX knockdown in flies
The fact that the mutated AOX is devoid of detectable enzymatic activity allowed us to use the newly created transgenic lines to test whether the previously observed phenotypic rescue of flies knocked down for a subunit of cytochrome oxidase (Cox7a) was due to the enzymatic activity of AOX or some other property conferred by the AOX protein, when expressed in Drosophila. Moreover, the fact that the newly created transgenic lines express AOX at only about 30% of the level of the lines previously studied, allowed us to test whether phenotypic rescue was quantitatively dependent on AOX expression level. Ubiquitous knockdown of CG9603, the broadly expressed isogene for Cox7a, was previously shown to produce pupal lethality11, which was rescued by high-level expression of AOX.
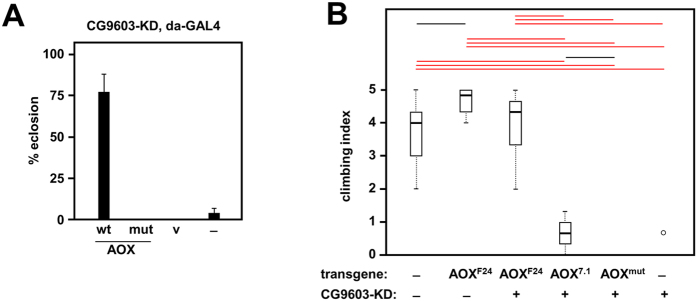
To test the new transgenic lines, we first confirmed that the RNAi line used in the experiment was devoid of the additional insertion previously reported to confer pupal lethality unrelated to specific target knockdown25 (Fig. S3). We then combined the CG9603 RNAi line with AOX and control transgenes, plus the da-GAL4 driver to induce simultaneous transgene expression and Cox7a knockdown. Wild-type AOX rescued the lethality, as previously (Fig. 5A), whereas mutAOX or the empty vector were unable to do so, confirming that AOX enzymatic activity is required for the rescue.
Figure 5. AOX rescue of Cox7a deficiency.
(A) Survival (%) from egg to eclosion of flies of the indicated genotypes, all bearing the da-GAL4 driver and the CG-9603 knockdown (RNAi) construct. Lines tested contained either no additional transgene (–), vector only (v), wild-type (wt) or mutated AOX (mut), in each case on chromosome 3. (B) Boxplot of climbing index of flies of the indicated genotypes. All flies carried the elavC155-GAL4 driver on chromosome X plus UAS-Dcr2 with or without the CG9603 knockdown (RNAi) construct on chromosome 2, and the indicated AOX transgene on chromosome 3 (AOX7.1 is the ΦC31-targeted insertion). Bars indicate medians, boxes show the first and third quartiles percentiles, whiskers are plotted according to the Tukey scheme (Krzywinski and Altman, 2014). Significant differences based on ANOVA are indicated by horizontal lines (black, red) denoting p < 0.05 and 0.001, respectively. A single outlier point is indicated by an open circle.
Next, we investigated the effects of CG9603 knockdown and its potential rescue by AOX, using the neuron-specific driver elavC155-GAL4. Previously, it was shown that this produces a locomotor defect in newly eclosed flies11. To potentiate the phenotype, we included UAS-Dcr2 in the background, so as to increase the penetrance of RNAi. Without concomitant AOX rescue, the resulting flies showed a severe locomotor defect as measured by their inability to climb the walls of the vial, in a standard negative geotaxis assay (Fig. 5B). High-level expression of AOX produced, as before, a clear rescue, whilst lower-level expression using the newly created transgenic lines produced only a modest phenotypic improvement (wild-type AOX), or no improvement at all (mutAOX, Fig. 5B).
Structural conclusions
Alternative oxidases are members of a superfamily of metalloenzymes, characterized by a common catalytic function of activation of molecular oxygen, and by common structural elements defining the catalytic diiron centre, including the four-helix bundle fold and a motif comprising two histidine residues, four carboxylate groups, and a bridging carboxylate group across the diiron centre26,27,28. The crystal structure of the trypanosomal enzyme indicates that it is a homodimer with each monomer comprising six long and four short α-helices13. The subunits interact with each other via α-helices 2, 3 and 4, whereas the hydrophobic region formed by α-helices 1, 2, 4 and 5 is proposed to anchor the protein to the inner surface of the mitochondrial inner membrane. A series of conserved arginine residues, capable of interacting with phospholipid head-groups, may assist inner membrane anchorage13. Our structure modelling of the C. intestinalis AOX suggests that the same structural elements are conserved in animal AOXs, and that the enzyme is also a homodimer inserted into the mitochondrial inner membrane.
In addition, the model predicts that the active site, and therefore the mechanism of oxygen activation, are also conserved in animal AOXs. The four-helix bundle, which acts as a structural platform for the binding of the two iron atoms, buries the active site deep in a hydrophobic environment. In T. brucei AOX, glutamate residues 123, 162, 213 and 266, in addition to a hydroxo-bridge, are responsible for directly coordinating the diiron centre. The centre is further stabilized by a redox-active tyrosine residue29,30, Y220, and two histidine residues (H165 and H269), which are within hydrogen-bond distances of E123, E169 and E213. The C. intestinalis AOX model indicates that the homologous residues E200, E239, E290, E344, Y297, H242 and H347 organize the active site in the same way.
Functional conclusions
In theory, the mutagenesis of a single glutamate residue should be enough to destabilize the diiron centre31. However, taking advantage of the proximity in the DNA sequence of the codons for E239 and H242 and of those for E344 and H347, we were able to create alanine substitutions for four important active site residues simultaneously. According to our model, these mutations should disrupt iron binding, thus generating a mutant devoid of catalytic activity, without any major disturbance to the overall protein structure. These predictions are supported by the fact that the mutant and wild-type proteins were expressed at comparable levels in mammalian cells and in flies, but that no enzymatic activity could be detected.
Importantly, the mutated enzyme was unable to rescue the organismal phenotypes arising from engineered cytochrome oxidase deficiency. In theory, the action of a foreign protein in attenuating such phenotypes could be due to any of several different mechanisms, of which the provision of an enzymatic by-pass for ubiquinol oxidation is only one. In previous work we found that Ciona AOX, when expressed in Drosophila mitochondria, decreased the net production of mitochondrial ROS even under non-inhibited conditions10,32. The mechanism of this remains unknown, but one possibility is that AOX is able to act directly or indirectly as an antioxidant, e.g. by binding and quenching quinone radicals via some other mechanism. Studies in various organisms have supported the idea that a hydrophobic pocket, located between α-helices 2 and 3, binds and channels ubiquinone to the active site33, which might be involved in such an activity.
A second possibility would be a hormetic response to disruption of the inner mitochondrial membrane or its protein complexes by the foreign protein. The induction of a variety of defence pathways to protect cells from increased ROS, disturbed protein, lipid or redox homeostasis, or altered mitochondrial turnover or dynamics, might equip the organism to cope with the additional but related stresses of respiratory insufficiency. Many studies in model organisms support this concept of ‘mitohormesis’34. Whilst we cannot rule out that such effects are material in other contexts, our findings do exclude them in regard to the developmental lethality produced by global cytochrome oxidase knockdown, or the locomotor dysfunction resulting from its knockdown specifically in neurons11. Based on our findings, that mutAOX cannot compensate these phenotypes, we infer that the rescue of these effects of cytochrome oxidase deficiency by AOX is almost certainly due to its enzymatic activity as a quinol oxidase, though formally we cannot exclude other, unknown effects of iron binding. A requirement for enzymatic activity might not be true of every phenotypic feature conferred by AOX in model organisms. Our findings indicate a robust way to test this in regard to all potential such phenotypes, allowing the mechanisms by which AOX acts to be probed, controlled or verified.
Several quantitative issues are also addressed by our findings. The first is that the extent of phenotypic rescue depends in some instances on the AOX expression level, but in other cases, such as the rescue of the developmental lethality caused by ubiquitous COX knockdown, is an all-or-none phenomenon. We suggest that this reflects a threshold effect wherein even the three-fold lower expression level of AOX, when integrated at specific sites by ΦC31-mediated recombination (in comparison with P element-mediated integrants created previously), exceeds a threshold value required to maintain metabolic homeostasis and complete development. In contrast, the lower expression level of the targeted integrants gave a clearly weaker rescue of locomotor dysfunction, when COX was knocked down only in neurons, roughly in proportion to the decreased expression level.
It may also be noted that the amount of antimycin-resistance conferred upon respiration in homogenates from the targeted integrants was still approximately 14%, compared with approximately 20% for the P element-mediated integrants, even though they are expressed at a much higher level. The level of respiratory antimycin-resistance in the fly may vary between tissues, and this 20% maximum may reflect only the properties of the predominant class of mitochondria. Most of the respiratory capacity in adult flies is vested in the flight muscles, where mitochondria make up almost one-third of the total tissue mass35. The apparent upper limit of how much electron flow can be diverted through AOX probably reflects specific features of this tissue and its energetic needs. The limit could be dictated by the constraints of membrane architecture, for example, if much of the ubiquinone pool is channelled directly from complex I to complex III via respiratory supercomplexes, such that it equilibrates only slowly with free ubiquinones available to AOX36. Most of the respiratory activity in adult Drosophila indeed resides in supercomplexes37. Such a phenomenon may account for the inferred threshold effect on the rescue of developmental lethality. Conversely, the organization of the respiratory chain may differ in other tissues, such as in neurons, where a more graded response to the AOX expression level is evident.
In conclusion, mutAOX offers a useful tool for future studies of the mechanism(s) whereby expression of Ciona AOX modifies the phenotypes of model organisms, potentially contributing the eventual development of AOX-based therapies.
Additional Information
How to cite this article: Andjelković, A. et al. Diiron centre mutations in Ciona intestinalis alternative oxidase abolish enzymatic activity and prevent rescue of cytochrome oxidase deficiency in flies. Sci. Rep. 5, 18295; doi: 10.1038/srep18295 (2015).
Supplementary Material
Acknowledgments
We thank Tony Moore for useful discussions, Filippo Scialo for the construction of the original AOX plasmid for expression in S2 cells, Dmitro Gospodaryov for critical reading of the manuscript and Samuli Hartikainen, Eveliina Kaulio, Tea Tuomela, Essi Kiviranta, Outi Kurronen, Merja Jokela and Maarit Myöhänen for technical assistance. Funding was provided by Academy of Finland (CoE grant 272376), the European Research Council (advanced grant 232738 to HTJ), the EU (Marie Curie International Incoming Fellowship 328988 to MTO), Tampere University Hospital Medical Research Fund, and the Sigrid Juselius Foundation.
Footnotes
Author Contributions A.A., M.T.O., H.T.J. and P.R. conceived and planned the project. A.A., M.T.O., G.C., C.Y. and P.K.D. conducted the laboratory work and analysis. H.T.J., M.S. and E.D. supervised the laboratory work and contributed analysis and insights. H.T.J. and M.T.O. compiled the figures and drafted the manuscript.
References
- McDonald A. E., Vanlerberghe G. C. & Staples J. F. Alternative oxidase in animals: unique characteristics and taxonomic distribution. J. Exp. Biol. 212, 2627–2634 (2009). [DOI] [PubMed] [Google Scholar]
- Feng H. et al. Expression and signal regulation of the alternative oxidase genes under abiotic stresses. Acta. Biochim. Biophys. Sin. 45, 985–994 (2013). [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G. C. Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 14, 6805–6847 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefnagel M. H. & Wiskich J. T. Activation of the plant alternative oxidase by high reduction levels of the Q-pool and pyruvate. Arch. Biochem. Biophys. 355, 262–270 (1998). [DOI] [PubMed] [Google Scholar]
- Castro-Guerrero N. A., Krab K. & Moreno-Sánchez R. The alternative respiratory pathway of euglena mitochondria. J Bioenerg. Biomembr. 36, 459–469 (2004). [DOI] [PubMed] [Google Scholar]
- Rustin P. & Jacobs H. T. Respiratory chain alternative enzymes as tools to better understand and counteract respiratory chain deficiencies in human cells and animals. Physiol. Plant 137, 362–370 (2009). [DOI] [PubMed] [Google Scholar]
- El-Khoury R. et al. Engineering the alternative oxidase gene to better understand and counteract mitochondrial defects: state of the art and perspectives. Br. J. Pharmacol. 171, 2243–2249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkaart A., Dassa E. P., Jacobs H. T. & Rustin P. Allotopic expression of a mitochondrial alternative oxidase confers cyanide resistance to human cell respiration. EMBO Rep. 7, 341–345 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassa E. P. et al. Expression of the alternative oxidase complements cytochrome c oxidase deficiency in human cells. EMBO Mol. Med. 1, 30–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ayala D. J. et al. Expression of the Ciona intestinalis alternative oxidase (AOX) in Drosophila complements defects in mitochondrial oxidative phosphorylation. Cell Metab. 9, 449–460 (2009). [DOI] [PubMed] [Google Scholar]
- Kemppainen K. K. et al. Expression of alternative oxidase in Drosophila ameliorates diverse phenotypes due to cytochrome oxidase deficiency. Hum. Mol. Genet. 23, 2078–2093 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoury R. et al. Alternative oxidase expression in the mouse enables bypassing cytochrome c oxidase blockade and limits mitochondrial ROS overproduction. PLoS Genet. 9, e1003182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T. et al. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc. Natl. Acad. Sci. USA 110, 4580–4585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoli A., Tettamanzi A. G. & Zhang Y. Computational protein design and large-scale assessment by I-TASSER structure assembly simulations. J Mol. Biol. 407, 764–776 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F. & Besler K. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312–3317 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoh A. et al. Screen for mitochondrial DNA copy number maintenance genes reveals essential role for ATP synthase. Mol. Syst. Biol. 10, 734 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K. & Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991). [DOI] [PubMed] [Google Scholar]
- Sanz A. et al. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc. Natl. Acad. Sci. USA 107, 9105–9110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer B. D. et al. Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannino G. et al. Glucose modulates respiratory complex I activity in response to acute mitochondrial dysfunction. J Biol. Chem. 287, 38729–38740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chretien D. et al. Reference charts for respiratory chain activities in human tissues. Clin. Chim. Acta. 228, 53–70 (1994). [DOI] [PubMed] [Google Scholar]
- Toivonen J. M. et al. technical knockout, a Drosophila model of mitochondrial deafness. Genetics 159, 241–254 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. & Altman N. Points of Significance: Visualizing samples with box plots. Nature Methods 11, 119–120 (2014). [DOI] [PubMed] [Google Scholar]
- Green E. W., Fedele G., Giorgini F. & Kyriacou C. P. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat. Methods 11, 222–223 (2014). [DOI] [PubMed] [Google Scholar]
- Berthold D. A., Voevodskaya N., Stenmark P., Gräslund A. & Nordlund P. EPR studies of the mitochondrial alternative oxidase. Evidence for a diiron carboxylate center. J Biol. Chem. 277, 43608–43614 (2002). [DOI] [PubMed] [Google Scholar]
- Berthold D. A. & Stenmark P. Membrane-bound di-iron carboxylate proteins. Annu. Rev. Plant Biol. 54, 497–517 (2003). [DOI] [PubMed] [Google Scholar]
- Simone F., Reisner E. & Lippard S. J. Current challenges of modeling diiron enzyme active sites for dioxygen activation by biomimetic synthetic complexes. Chem. Soc. Rev. 39, 2768–2779 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albury M. S., Affourtit C., Crichton P. G. & Moore A. L. Structure of the plant alternative oxidase. Site-directed mutagenesis provides new information on the active site and membrane topology. J Biol. Chem. 277, 1190–1194 (2002). [DOI] [PubMed] [Google Scholar]
- Affourtit C., Albury M. S., Crichton P. G. & Moore A. L. Exploring the molecular nature of alternative oxidase regulation and catalysis. FEBS Lett. 510, 121–126 (2002). [DOI] [PubMed] [Google Scholar]
- Ajayi W. U., Chaudhuri M. & Hill G. C. Site-directed mutagenesis reveals the essentiality of the conserved residues in the putative diiron active site of the trypanosome alternative oxidase. J Biol. Chem. 277, 8187–8193 (2002). [DOI] [PubMed] [Google Scholar]
- Sanz A., Fernández-Ayala D. J., Stefanatos R. K. & Jacobs H. T. Mitochondrial ROS production correlates with, but does not directly regulate lifespan in Drosophila. Aging 2, 200–223 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albury M. S., Elliott C. & Moore A. L. Towards a structural elucidation of the alternative oxidase in plants. Physiol. Plant 137, 316–327 (2009). [DOI] [PubMed] [Google Scholar]
- Yun J. & Finkel T. Mitohormesis. Cell Metab. 19, 757–766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenbook L. & Williams C. M. Mitochondria in the flight muscles of insects III. Mitochondrial cytochrome c in relation to the aging and wing beat frequency of flies. J. Gen. Physiol. 39, 497–512 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova M. L. & Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim. Biophys. Acta. 1837, 427–443 (2014). [DOI] [PubMed] [Google Scholar]
- Celotto A. M., Chiu W. K., Van Voorhies W. & Palladino M. J. Modes of metabolic compensation during mitochondrial disease using the Drosophila model of ATP6 dysfunction. PLoS One 6, e25823 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.