Abstract
Autophagy is an important lysosomal degradation pathway that aids in the maintenance of cellular homeostasis by breaking down and recycling intracellular contents. Dysregulation of autophagy is linked to a growing number of human diseases. The Beclin 1-Vps34 protein-protein interaction network is critical for autophagy regulation and is therefore essential to cellular integrity. Manipulation of autophagy, in particular via modulation of the action of the Beclin 1-Vps34 complexes, is considered a promising route to combat autophagy-related diseases. Here we summarize recent findings on the core components and structural architecture of the Beclin 1-Vps34 complexes, and how these findings provide valuable insights into the molecular mechanisms that underlie the multiple functions of these complexes and for devising therapeutic strategies.
Keywords: Beclin 1, Vps34, Nrbf2, complex, structure, CX-MS, EM, inhibitor, drug design
1. Introduction
1.1. Autophagy and its molecular machinery
Autophagy is an intracellular degradation process which, often initiated by stressors, functions to break down and recycle damaged organelles, long-lived or aggregated proteins, and pathogens. This key function of autophagy is evolutionarily conserved from yeast to higher eukaryotes, and many of the autophagy-related (Atg) yeast genes have mammalian orthologs [1,2]. In mammals, there are three pathways for autophagic recycling: microautophagy [3,4], chaperone-mediated autophagy [5], and macroautophagy [6,7]. Microautophagy and chaperone-mediated autophagy eliminate cytoplasmic materials by directly depositing cargoes into late endosomes or lysosomes, whereas macroautophagy involves the formation of unique double-membraned autophagosomes that later fuse with late endosomes or lysosomes to recycle cellular molecules and organelles. Of the three pathways, the majority of the research to date has been focused on macroautophagy (hereafter called autophagy), which is the emphasis of this review.
The autophagic process is composed of a series of sequential steps: induction/cargo recognition, phagophore formation, vesicle expansion and completion, autophagosome-lysosome fusion, cargo degradation, and recycling of essential components (Figure 1). This process is orchestrated by a myriad of autophagy proteins, which form functional multiprotein complexes: the Ulk1 complex, the Beclin 1-Vps34 complexes, the Atg2-WIPI2 complex, and both Atg12-Atg5·Atg16L1 and LC3-phosphotidylethanolamine (PE) ubiquitin-like conjugation systems [8,9]. In particular, assembly of the Beclin 1-Vps34 multiprotein complexes that control both autophagosome biogenesis and maturation is crucial to cellular maintenance. Functional anomalies (whether instigated by genetic deficiencies or signaling defects) in the molecular components of the Beclin 1-Vps34 complexes cause disruption in cellular homeostasis, and are associated with many human diseases, including but not limited to infection, aging, neurodegeneration, and cancer [10] (see more discussion below). Therefore, understanding 1) the entirety of the Beclin 1-Vps34 complex components including the structure of each component, 2) the molecular architecture of the protein complexes as a whole including how the protein complex components interact and assemble, and 3) how compromise of these protein complexes causes specific pathologies, is at the forefront of much autophagy research.
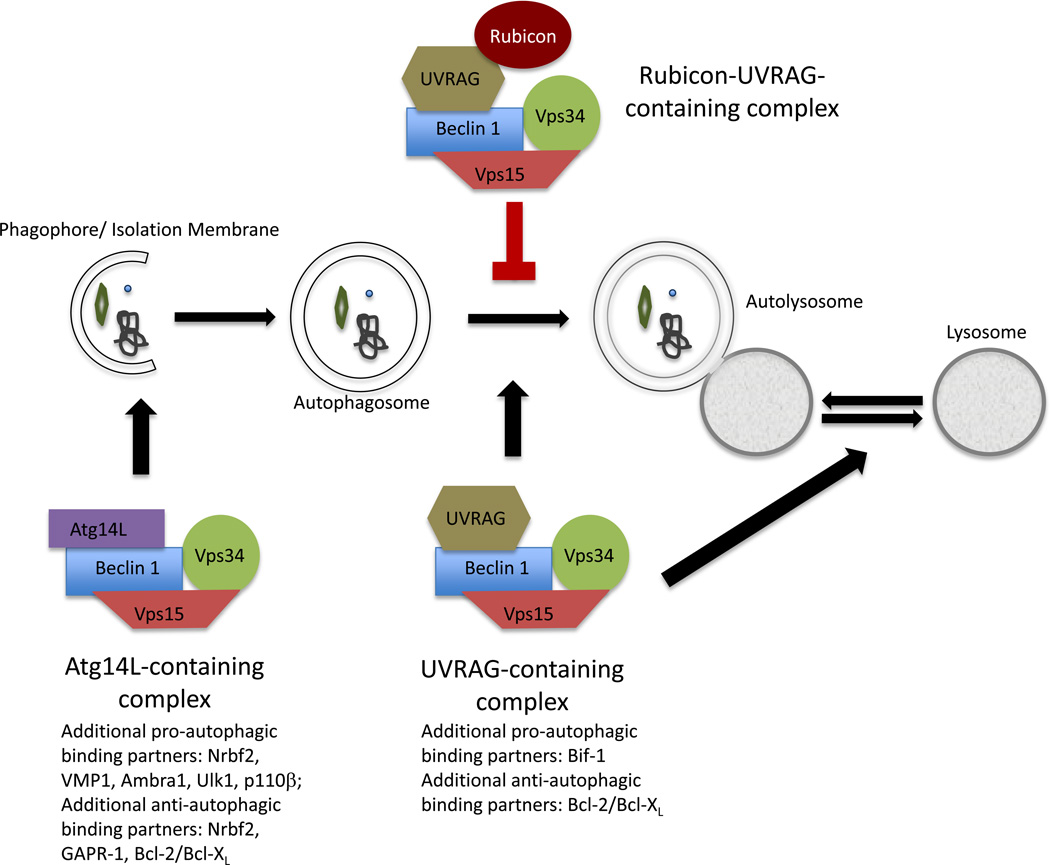
Figure 1.
A schematic of the autophagy pathway with the involvement of the Beclin 1-Vps34 class III PI(3)K complexes identified. The core class III PI(3)K complexes consist of Beclin 1, Vps34, and Vps15. The Atg14L-containing Beclin 1-Vps34 complex marks the sites at which the phagophores/isolation membranes form to enclose the cytoplasmic contents to be degraded. The phagophores subsequently elongate and seal off to form mature autophagosomes. Autophagosome maturation, particularly autophagosome fusion with the lysosome, is facilitated by the UVRAG-containing Beclin 1-Vps34 complex. The inner membranes of the autophagosomes and their contents are degraded and recycled to maintain cellular homeostasis. The interaction of Rubicon with the UVRAGcontaining Beclin 1-Vps34 complex blocks the fusion between autophagosomes and lysosomes. In addition, the UVRAG-containing Beclin 1-Vps34 complex also regulates the tubule scission step during autophagic lysosome reformation.
1.2. Beclin 1-Vps34 complexes in infection
There are numerous infectious diseases linked to the autophagy process. Studies show that as an important self-defense mechanism, host cells can clear intracellular pathogens (e.g., bacteria and viruses) through xenophagy, a specific type of cargo-selective autophagy; conversely, pathogens can also target the autophagic machinery, including the Beclin 1-Vps34 complex components, to invade cells and enhance their virulence [11–13].
Beclin 1 has been shown to be critical for immunity. Multiple viral proteins, including α-herpes simplex virus 1 (HSV-1)-encoded neurovirulence factor ICP34.5 [14] and the γ-herpesvirus Bcl-2 homologs (i.e., KSHV v-Bcl-2 and murine γ-HV68 M11) [15–17], bind Beclin 1 and prevent autophagosome formation. The human immunodeficiency virus (HIV) type-1 Nef protein [18] and influenza A M2 protein [19] bind Beclin 1 and block autophagosome maturation. HIV-1 infection also downregulates Beclin 1 at the transcriptional level and inhibits autophagy [20]. Despite its susceptibility to be hijacked by viruses, Beclin 1 can promote innate antimicrobial immune responses by directly interacting with cyclic GMP-AMP synthetase (cGAS). This interaction suppresses cyclic GMP-AMP production and releases the inhibitory action of RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein (Rubicon [21,22]) on the UV-irradiation resistance-associated gene (UVRAG [23])-containing Beclin 1-Vps34 complex, thus allowing for induction of autophagic removal of cytosolic pathogen DNA [24]. Moreover, Beclin 1 is important for immune cell development, as Beclin 1 deficiency results in dramatic depletion of T and B cell precursors [25,26].
Other components of the Beclin 1-Vps34 complexes may also be important for fine-tuning the immune response. For example, Rubicon is utilized by the Kaposi’s sarcoma-associated herpesvirus (KSHV) to inhibit autophagosome maturation; specifically, KSHV promotes the interaction of Rubicon with the UVRAG-containing Beclin 1-Vps34 complex, thus blocking Vps34 catalytic activity [27]. Rubicon is also shown to function as a regulator of the NADPH-oxidase-containing phagocytosis complex [28] and the CARD9-containing signaling complex [29], connecting this Beclin 1-interacting protein to the innate immune response against various microbial infections.
1.3. Beclin 1-Vps34 complexes in aging and neurodegeneration
Autophagy can be induced by starvation. Coincidentally, dietary restriction is the most effective means to decelerate aging [30,31]. Studies show that rapamycin, which induces autophagy through inhibition of mammalian target of rapamycin complex 1 (mTORC1), prolongs lifespan in mice [32]; therefore, dietary restriction, which inhibits mTORC1, may be critical for decelerating aging also through inducing autophagy.
Specifically, Beclin 1 and its interacting proteins are important in aging and age-related neurodegenerative diseases such as Huntington’s disease (HD), Alzheimer’s disease (AD), Parkinson’s disease (PD), and Amyotrophic lateral sclerosis (ALS). Inactivation of bec-1, the Caenorhabditis elegance ortholog of Becn1 (the gene encoding Beclin 1), reversed the lifespan extension of the insulin-like tyrosine kinase receptor daf-2 loss-of-function mutation [33]. Beclin 1 levels decline with age in the human brain [34], consistent with the ideas that (1) reduced Beclin 1 levels lead to a decline in autophagic activity and (2) declined autophagic activity is probably an important factor contributing to aging [35–37]. Moreover, Beclin 1 is recruited to the cytoplasmic Huntingtin (Htt) inclusions in the brain of the R6/2 HD mouse model and accumulation of mutant Htt is highly sensitive to decreased Beclin 1 levels, suggesting that accumulation of mutant Htt in the aged brain is likely a consequence of age-dependent reduction of Beclin 1 levels and autophagic activity [34]. It is also reported that a decline in Beclin 1 expression in the brains of AD patients can result in reduced Vps34 protein levels, leading to neurotoxic accumulation of autophagosomes as well as impaired amyloid precursor protein (APP) processing and turnover [38,39]. Microglia isolated from AD brains also show significantly reduced Beclin 1 levels, which may lead to impaired retromer trafficking and receptor-mediated phagocytosis, contributing to AD pathology [40]. Furthermore, PINK1, a key neuroprotective protein in PD, interacts with Beclin 1 to promote both basal and starvation-induced autophagy [41]. Recent studies also reveal two roles for Beclin 1, through its interaction with another PD protein, PARK2, in the translocation of PARK2 to mitochondria and the initiation of mitophagy prior to formation of autophagosomes [42]. This study reported additional interactions of PARK2 with the Beclin 1-interacting proteins Vps34 and autophagy/Beclin 1 regulator 1 (AMBRA1), which are enhanced upon treating cells with carbonyl cyanide mchlorophenylhydrazone (CCCP) [42]. Interestingly, an earlier study showed that lentivirus-mediated overexpression of Beclin 1 induced autophagy, and reduced the accumulation of abnormal α-synuclein aggregates and related neurodegenerative pathology in α-synuclein models of PD [43]. In addition, Beclin 1 is associated with ALS; however, in this context, Beclin 1 reduction protects against ALS development [44].
1.4. Beclin 1-Vps34 complexes in cancer
Mono-allelic loss of BECN1 in 40–75% of sporadic human breast, ovarian, and prostate cancers was first reported in 1999, linking autophagy deficiency to cancer [45,46]. Subsequently, mouse genetic studies show that heterozygous disruption of Becn1 increased the frequency of spontaneous tumors (e.g., mammary hyperplasia, liver and lung carcinomas, and lymphomas), implicating a haploinsufficient tumor suppressor function of Beclin 1 [47,48]. However, with the improvement of human tumor sequencing and gene expression databases, a recent study reveals close proximity of BECN1 to the BRAC1 gene on chromosome 17q21, and the presence of deletions encompassing both BRCA1 and BECN1, or deletions of only BRCA1, but not deletions of only BECN1, in breast and ovarian cancers, challenging Beclin 1’s tumor suppressor role [49]. In addition, a study using tissue-specific Atg5 and Atg7 knockout mice shows that autophagy deficiency can lead to benign tumors in liver, but not in other tissues [50]. These new findings leave the mechanism underlying Beclin 1’s role as a tumor suppressor under contention. Further data focusing on the direct impact of Beclin 1 loss in a tissue specific manner, notably in breast, ovarian, and prostate, is necessary to corroborate its context-dependent role as a tumor suppressor. However, the connections between UVRAG mutation and colon/gastric cancers remain uncontested [23,51], and a recent human genetic study draws a new link between Nrbf2 and breast cancer [52], relating the Beclin 1 interactome to cancer.
1.5. Synopsis of this review
Pharmacological modulators of autophagy that are currently in clinical trials are limited to sirolimus (mTOR inhibitor) and hydroxychloroquine (ClinicalTrials.gov), with the target of hydroxychloroquine yet to be identified. As summarized in Section 1.2–1.4, the functionality and regulation of the Beclin 1-Vps34 complexes are important in autophagy-related pathologies. Therefore, the Beclin 1-Vps34 complexes provide promising targets for therapeutics to treat these autophagy-related diseases. Structure-based drug design is essential for successful creation of highly potent and target-specific drugs, examples of which include the design of small-molecule inhibitors targeting three major classes of antiapoptotic proteins — anti-apoptotic B cell lymphoma 2 (Bcl-2), inhibitor of apoptosis proteins (IAPs), and murine double-minute 2 (MDM2) [53,54]. This review covers a growing number of studies that determine the Beclin 1 interactome and elucidate the structural architecture of the Beclin 1-Vps34 complexes, employing techniques such as affinity purification followed by mass spectrometry, nuclear magnetic resonance (NMR), X-ray crystallography, crosslinking mass spectrometry (CX-MS), and electron microscopy (EM). This review also takes an in-depth look at the most recently identified Beclin 1-Vps34 complex component, Nrbf2, the high resolution structures of the Beclin 1-Vps34 complex core components, and the architecture of the Beclin 1-Vps34 complexes. Lastly, this review uses an example of the most recent breakthrough in structure-based design for inhibitors of the Vps34 catalytic site to illustrate how understanding the identity, function, structure of each component of the Beclin 1-Vps34 complexes, as well as the architecture of the Beclin 1-Vps34 complexes as a whole, can facilitate the design of pharmacological interventions that specifically target autophagy.
2. Beclin 1 interactome
2.1. Core components
In mammalian autophagy, phosphatidylinositol 3-phosphate (PtdIns(3)P) production is essential to recruit effectors for autophagosome formation [55–59] and autophagosome/endosome maturation [60–62]. PtdIns(3)P is produced primarily by the class III phosphatidylinositide-3-kinase (PI(3)K) Vps34 and its levels are also affected by class II PI(3)K and phosphoinositide 3-phosphatases (e.g., Jumpy [63] and MTMR3 [64]). The class III PI(3)K complexes consist of three core subunits: Beclin 1 (the yeast Atg6/Vps30 ortholog), Vps34, and Vps15 [65,66]. All Beclin 1 forms complexes with Vps34, whereas a considerable portion of Vps34 does not interact with Beclin 1, Atg14L or UVRAG [66,67]. The Beclin 1-Vps34-Vps15 core complex regulates the autophagy pathway as well as the endosomal pathway, depending on additional components [22,68,69]. Loss of Beclin 1 destabilizes the Beclin 1-Vps34-Vps15 core complex and leads to an impairment in Vps34 activity, autophagic flux, and endocytic trafficking [22,68–70]. Beclin 1 significantly enhances the interaction between UVRAG and Rubicon, the two additional class III PI(3)K complex components that regulate the endosomal pathway [22,68]. It is also reported that depletion of Vps15 function results in a reduction in Vps34 and Beclin 1 protein levels, indicating a dependency of the stability of the core complex on Vps15 [68].
2.2. Two complexes, distinct functions
The yeast Atg6/Vps30-Vps34-Vps15 core complex forms two distinct complexes and functions in autophagy and vacuolar enzyme sorting, by interacting with the adapter proteins Atg14 and Vps38, respectively [71]. Likewise, the mammalian Beclin 1-Vps34-Vps15 core complex forms at least two complexes with distinct functions. One Beclin 1-Vps34 complex contains Atg14L (Atg14-like, the yeast Atg14 ortholog, also named Atg14 or Barkor for Beclin-1-associated autophagy-related key regulator) which positively regulates autophagy by promoting autophagosome formation [21,22,72,73]. In particular, Atg14L localizes to the endoplasmic reticulum (ER) [74], more specifically to the ER-mitochondria contact sites [75], in a process dependent on the Ulk1 complex [74,76] and the ER SNARE, syntaxin 17 [75]. ER-localized Atg14L then recruits the core Beclin 1-Vps34-Vps15 complex to the autophagosome initiation sites to generate PtdIns(3)P [74,76]. Atg14L is also required for phosphorylation of Beclin 1 by AMP-activated protein kinase (AMPK) and this phosphorylation event is necessary for optimal autophagic activity [67,77]. Only very recently, Atg14L was reported to function beyond autophagosome biogenesis: Atg14L can self-associate, bind to the syntaxin 17-synaptosomal-associated protein, 29 kDa (SNAP29) complex, and promote autophagosome-endolysosome fusion [78].
The second Beclin 1-Vps34 protein complex contains UVRAG, which shows 10% identity and 32% similarity to yeast Vps38 [73]. The binding of UVRAG and ATG14L to the core complex is mutually exclusive [21,69,72,73]. In association with the core complex, UVRAG promotes autophagosome maturation [73,79,80]. In particular, UVRAG interacts with class C Vps and Rab7 to promote fusion of autophagosomes with late endosomes/lysosomes [79]. The interaction of UVRAG with the C-Vps tethering complex, which accelerates autophagosome maturation and endocytic vesicle trafficking, occurs independent of Beclin 1 [70,79]. In contrast, the negative autophagy regulator Rubicon binds UVRAG when S498 of UVRAG is phosphorylated by mTORC1, inhibiting autophagosome maturation [21,22,80]. Despite consensus on the role of UVRAG in endosomal trafficking and autophagosome maturation, as well as the recent exciting findings of autolysosome tubule scission [81], insulin sensing and organismal glucose homeostasis regulation [82] by the UVRAG-containing Beclin 1-Vps34 complex, it remains under debate whether the UVRAG-containing Beclin 1-Vps34 complex plays a role in autophagosome formation. For example, some evidence shows that UVRAG associates with Bif-1 and Beclin 1 to promote autophagosome formation [23,83]; conversely, other groups were unable to detect autophagy impairment upon UVRAG deficiency [73,84].
Collectively, the Atg14L-containing and UVRAG-containing class III PI(3)K complexes appear to diverge the Beclin 1-Vps34-Vps15 core complex for autophagy and endocytic trafficking, respectively. It will be interesting to see if the association of Beclin 1 with UVRAG is attenuated and Beclin 1 is switched to Atg14L-containing class III PI(3)K complex when cells undergo environmental changes that induce autophagy.
2.3. Nrbf2: A novel component of the Atg14L-containing Beclin 1-Vps34 complex
Recent affinity purification- and mass spectrometry-based proteomic analyses of the mammalian autophagy protein-protein interaction network confirmed previously reported binding partners of the Beclin 1-Vps34-Vps15 core complex, and revealed a novel component of this complex, nuclear receptor binding factor 2 (Nrbf2, also named COPR2 for comodulator of proliferator-activated receptor (PPAR) and retinoid X receptor (RXR) isoform 2) [69,85–87]. Nrbf2 was originally characterized as a regulator of nuclear receptors including PPARα, retinoic acid receptor (RAR), and RXRα [88,89] without any known function in autophagy.
The interaction of Nrbf2 with the Beclin 1-Vps34-Vps15 core complex is mediated by Atg14L, revealing a functional role for cytoplasmic Nrbf2 [69,87]. Nrbf2 was found to have either no [69,86] or weak [85,87] interaction with UVRAG. These results place the Nrbf2 protein primarily as a component of the Atg14L-containing class III PI(3)K complex that regulates autophagosome biogenesis; this inference was confirmed by the colocalization of Nrbf2 with FIP200, Ulk1, and Atg5 [69], as well as the requirement of full length Nrbf2 for WIPI2-positive phagophore formation [87]. Detailed protein-protein interaction analyses using immunoprecipitation and Western blot analyses further revealed that Nrbf2 is important for linking the Beclin 1-Atg14L to the Vps34-Vps15 sub-complexes [69,87]. In addition to the mammalian Nrbf2 studies, an independent study in yeast revealed that the protein encoded by Atg38, the presumptive Nrbf2 ortholog, is a component of the yeast Atg6/Vps30-Vps34-Vps15 complex, interacts with Atg14, but not Vps38, links the Atg6/Vps30-Atg14 to the Vps34-Vps15 subcomplexes, and colocalizes with Atg17 [90]. Taken together, these results suggest that the protein-protein interaction and localization of Atg38/Nrbf2 during autophagy is evolutionarily conserved.
Despite the consensus on the protein-protein interactions that involve Atg38/Nrbf2 and on the colocalization of Atg38/Nrbf2 with pre-autophagosomal structures (PAS)/isolation membranes, the functional significance of Atg38/Nrbf2 as a component of the Atg14/Atg14L-containing class III PI(3)K complex in autophagy control is under debate. In the human RPE-1 cell line, Nrbf2 siRNA treatment resulted in increased autophagic flux (as monitored by p62 levels, LC3II in the absence versus presence of a lysosomal inhibitor, and long-lived protein degradation), and increased total cellular PtdIns(3)P levels [69]. In contrast, Nrbf2−/− mouse embryonic fibroblast (MEF) cells [87], HEK293A cells stably expressing GFP-LC3 [86], Nrbf2 siRNA treated U2OS GFP-LC3 cells [85], and Atg38Δ and Atg38Δ GFP-Atg14 yeast cells [90] showed defective autophagy. Nrbf2−/− MEF cells also showed defective Atg14L-associated Vps34 activity [87]. These discrepancies are presumably rooted in the context-dependency of the Beclin 1-Vps34 protein-protein interaction network [69].
2.4. Other Beclin 1-interacting proteins
Beclin 1 is known to interact with additional protein partners to elicit specific autophagic responses, including but not limited to negative regulators (e.g., Bcl-2, Bcl-XL and Golgi-associated plant pathogenesis-related protein 1 (GAPR-1, also called GLIPR2 for glioma pathogenesis-related protein 2)) and positive regulators (e.g., Vacuole membrane protein 1 (VMP1), Ambra1, Dapper1, class IA PI(3)K p110β, and serine-threonine kinase 38 (STK38)). The anti-apoptotic Bcl-2 family members interact with Beclin 1 and block Beclin 1 interaction with the other components of the Beclin 1-Vps34 complexes, leading to inhibition of autophagy and reduction in UVRAG-mediated endocytic trafficking [15,91,92]. Phosphorylation of Beclin 1 at residue T119 in its Bcl-2 homology (BH3) domain by death-associated protein kinase (DAP-kinase) [93], and phosphorylation of Bcl-2 at residues T69, S70, and S87 of the non-structured loop by c-Jun N-terminal protein kinase 1 (JNK-1) [94] independently disrupt the interaction between Beclin 1 and Bcl-2 and enhance autophagic activity in the cells. Moreover, the epidermal growth factor receptor (EGFR) tyrosine kinase binds to Beclin 1, leading to its phosphorylation at residues Y229, Y233 and Y352, which in turn enhances Beclin 1 interaction with Bcl-2 to inhibit autophagic activity [95]. Similarly, GAPR-1, which binds a region (residues 267–284) of the evolutionarily conserved domain (ECD) of Beclin 1, also negatively regulates autophagy by tethering Beclin 1 to the Golgi apparatus [96].
VMP1 was originally discovered in pancreatic tissues undergoing pancreatitis-induced autophagy, where VMP1 expression results in autophagosome formation [97]. Studies show that VMP1, via its C-terminal Atg domain, directly binds the BH3 domain of Beclin 1, promoting dissociation of Bcl-2 from Beclin 1; these activities result in the Beclin1-Vps34 complex formation and autophagy [97,98].
Ambra1, an essential protein for neural tube development, is not only required for starvation-induced autophagy [99], but also involved in mitochondrial dynamics and degradation [100–102]. Ambra1 binds Beclin 1 and serves its pro-autophagic role by assisting in the regulation of the Beclin 1-Vps34 complexes [103]. Through its association with Ulk1 kinase and the E3 ligase TRAF6, Ambra1 localizes the Beclin 1-Vps34-Vps15 core complex to omegasomes [104]. In this transient association, Ulk1 phosphorylates Ambra1 and Beclin 1; phosphorylated Ambra1 promotes the stability and ubiquitinylation of Ulk1 by TRAF6, leading to autophagy induction [104]. In addition, under normal cellular homeostatic conditions, Ambra1 is a substrate of mTORC1, resulting in inhibition of Ambra1’s pro-autophagic effects [104].
Dapper1, a Dishevelled-interacting protein known as a modulator of Wnt signaling through promoting lysosomal degradation of Dishevelled, was recently found to bind Atg14L and Beclin 1, enhancing Atg14L-containing Beclin 1-Vps34 complex formation, Vps34 activity, autophagosome biogenesis, and autophagic flux [105]. Loss of Dpr1, the gene encoding Dapper1, specifically in the mouse CNS resulted in motor coordination impairment and CNS pathologies resembling autophagy-deficient mice [105].
Besides its canonical role in inhibiting autophagy via the Akt-mTORC1 axis, p110β binds the autophagy-promoting Atg14L-containing Beclin 1-Vps34 complex and facilitates the generation of cellular PtdIns(3)P [106]. This complex also contains Rab5 [107] and acts as a molecular sensor for growth factor availability [108].
Last but not least, most recently, STK38 has been reported to bind to Beclin 1 and promote autophagosome biogenesis through enhancing the autophagy-inducing, exocyst component Exo84-RalB interaction and recruitment of the Atg14L-containing Beclin 1-Vps34 complex to Exo84 [109,110].
In short, Beclin 1 has been found to interact with a growing number of binding partners, which either promote autophagy/endocytosis or inhibit autophagy. Future research into the molecular mechanisms surrounding these interacting partners will provide more insights into the roles that the Beclin 1-Vps34 complexes play in regulating different cellular trafficking pathways under various nutrient and stress conditions.
3. Structures of the Beclin 1-Vps34 complex components
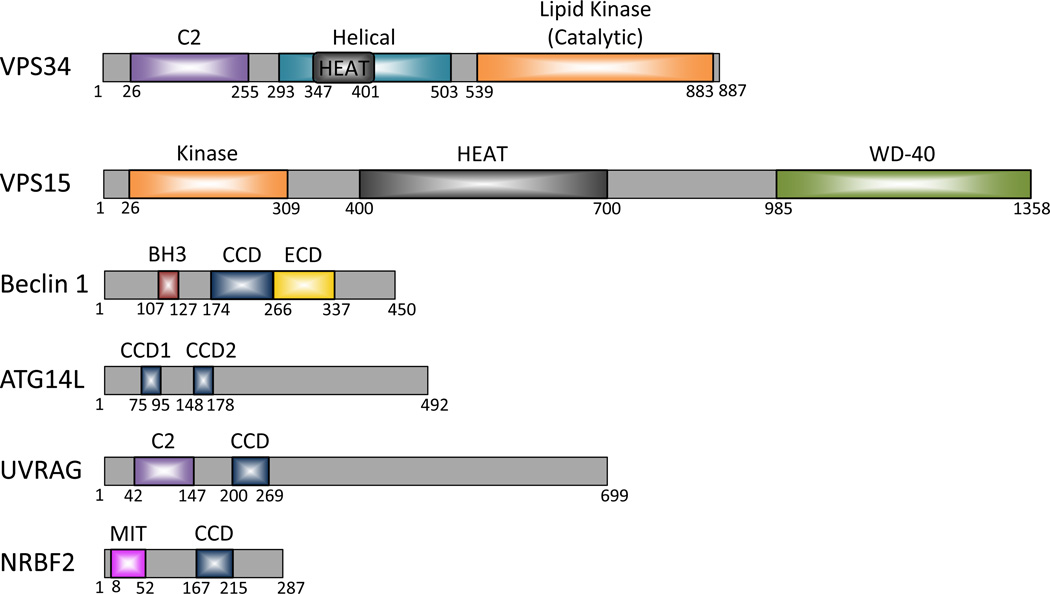
The domains of the Beclin 1-Vps34 complex components are largely identified (Figure 2). However, despite significant efforts towards resolving structures of the Beclin 1-Vps34 complex components, to date high resolution structures are available only for the Beclin 1 BH3, coiled-coil (CCD), and ECD (or BARA for β-α repeated, autophagy-specific) domains, Nrbf2 microtubule interacting and trafficking (MIT) domain (PDB database entry only), Vps34 helical and catalytic (HELCAT) domains, and Vps15 WD-40 domain (see below and summarized in Table 1 and Figure 3).
Figure 2.
Domain structures of the proteins associated with the human Beclin 1-VPS34 complexes. Abbreviations: C2, protein kinase C conserved region 2; HEAT, huntingtin, EF3, a subunit of PP2A, TOR1; WD-40 repeats, Trp-Asp (W-D) or beta-transducin repeats; BH3, Bcl-2 homology 3; CCD, coiled-coil domain; ECD, evolutionarily conserved domain; MIT, microtubule-interacting and trafficking.
Table 1.
Summary of reported structures for Beclin 1-Vps34 complex components.
| Component | Domain | Species | Residues in the constructs |
Detection Assay | Reference | PDB code |
|---|---|---|---|---|---|---|
| Beclin 1 | BH3 domain | H. sapiens | 107–135 | X-ray crystallography | (Oberstein et al. 2007) | 2P1L |
| H. sapiens | 104–131 | NMR | (Feng et al. 2007) | 2PON | ||
| M. musculus | 106–124 | X-ray crystallography | (Ku et al. 2008) | 3BL2* | ||
| H. sapiens | 105–130 | X-ray crystallography | (Sinha et al. 2008) | 3DVU* | ||
| H. sapiens | 107–130 | X-ray crystallography | (Su et al. 2014) | 4MI8* | ||
| CCD | R. norvegicus | 174–266 | X-ray crystallography | (Li et al. 2012) | 3Q8T | |
| ECD/BARA | S. cerevisiae | BARA 320–539 | X-ray crystallography | (Noda et al. 2012) | 3VP7 | |
| H. sapiens | ECD 248–450 | X-ray crystallography | (Huang et al. 2012) | 4DDP | ||
| Vps34 | C2 domain | |||||
| Helical domain & Lipid kinase domain | D. melanogaster | 258–949 | X-ray crystallography | (Miller et al. 2010) | 2X6H, 2X6F**, 2X6I**, 2X6J**, 2X6K** |
|
| H. sapiens | 293–887 | X-ray crystallography | (Dowdle et al. 2014) | 4PH4** | ||
| H. sapiens | 282–879 | X-ray crystallography | (Ronan et al. 2014) | 4OYS** | ||
| H. sapiens | 282–879 | X-ray crystallography | (Pasquier et al. 2015) | 4UWF**, 4UWG**, 4UWH**, 4UWK**, 4UWL** |
||
| Vps15 | Kinase domain | |||||
| HEAT domain | ||||||
| WD-40 domain | S. cerevisiae | 1027–1454 | X-ray crystallography | (Heenan et al. 2009) | 3GRE | |
| Nrbf2 | MIT | M. musculus | 1–97 | NMR | (Suetake et al. 2015) | 2CRB |
| CCD |
Notes:
Co-crystal structure in complex with γ-herpesvirus 68 M11, the viral Bcl-2 homolog.
Co-crystal structure in complex with VPS34 inhibitor.
References:
Dowdle, W. E., B. Nyfeler, J. Nagel, R. A. Elling, S. Liu, E. Triantafellow, S. Menon, Z. Wang, A. Honda, G. Pardee, J. Cantwell, C. Luu, I. Cornella-Taracido, E. Harrington, P. Fekkes, H. Lei, Q. Fang, M. E. Digan, D. Burdick, A. F. Powers, S. B. Helliwell, S. D'Aquin, J. Bastien, H. Wang, D. Wiederschain, J. Kuerth, P. Bergman, D. Schwalb, J. Thomas, S. Ugwonali, F. Harbinski, J. Tallarico, C. J. Wilson, V. E. Myer, J. A. Porter, D. E. Bussiere, P. M. Finan, M. A. Labow, X. Mao, L. G. Hamann, B. D. Manning, R. A. Valdez, T. Nicholson, M. Schirle, M. S. Knapp, E. P. Keaney and L. O. Murphy (2014). "Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo." Nat Cell Biol 16(11): 1069–1079.
Feng, W., S. Huang, H. Wu and M. Zhang (2007). "Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1." J Mol Biol 372(1): 223–235.
Heenan, E. J., J. L. Vanhooke, B. R. Temple, L. Betts, J. E. Sondek and H. G. Dohlman (2009). "Structure and function of Vps15 in the endosomal G protein signaling pathway." Biochemistry 48(27): 6390–6401.
Huang, W., W. Choi, W. Hu, N. Mi, Q. Guo, M. Ma, M. Liu, Y. Tian, P. Lu, F. L. Wang, H. Deng, L. Liu, N. Gao, L. Yu and Y. Shi (2012). "Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein." Cell Res 22(3): 473–489.
Ku, B., J. S. Woo, C. Liang, K. H. Lee, H. S. Hong, X. E, K. S. Kim, J. U. Jung and B. H. Oh (2008). "Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68." PLoS Pathog 4(2): e25.
Li, X., L. He, K. H. Che, S. F. Funderburk, L. Pan, N. Pan, M. Zhang, Z. Yue and Y. Zhao (2012). "Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG." Nat Commun 3 662.
Miller, S., B. Tavshanjian, A. Oleksy, O. Perisic, B. T. Houseman, K. M. Shokat and R. L. Williams (2010). "Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34." Science 327(5973): 1638–1642.
Noda, N. N., T. Kobayashi, W. Adachi, Y. Fujioka, Y. Ohsumi and F. Inagaki (2012). "Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy." J Biol Chem 287(20): 16256–16266.
Oberstein, A., P. D. Jeffrey and Y. Shi (2007). "Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein." J Biol Chem 282(17): 13123–13132.
Pasquier, B., Y. El-Ahmad, B. Filoche-Romme, C. Dureuil, F. Fassy, P. Y. Abecassis, M. Mathieu, T. Bertrand, T. Benard, C. Barriere, S. El Batti, J. P. Letallec, V. Sonnefraud, M. Brollo, L. Delbarre, V. Loyau, F. Pilorge, L. Bertin, P. Richepin, J. Arigon, J. R. Labrosse, J. Clement, F. Durand, R. Combet, P. Perraut, V. Leroy, F. Gay, D. Lefrancois, F. Bretin, J. P. Marquette, N. Michot, A. Caron, C. Castell, L. Schio, G. McCort, H. Goulaouic, C. Garcia-Echeverria and B. Ronan (2015). "Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)- 3,4-dihydro-2Hpyrimido[ 1,2-a]pyrimidin-6-one: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors." J Med Chem 58(1): 376–400.
Ronan, B., O. Flamand, L. Vescovi, C. Dureuil, L. Durand, F. Fassy, M. F. Bachelot, A. Lamberton, M. Mathieu, T. Bertrand, J. P. Marquette, Y. El-Ahmad, B. Filoche-Romme, L. Schio, C. Garcia-Echeverria, H. Goulaouic and B. Pasquier (2014). "A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy." Nat Chem Biol 10(12): 1013–1019.
Sinha, S., C. L. Colbert, N. Becker, Y. Wei and B. Levine (2008). "Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11." Autophagy 4(8): 989–997.
Su, M., Y. Mei, R. Sanishvili, B. Levine, C. L. Colbert and S. Sinha (2014). "Targeting gamma-herpesvirus 68 Bcl-2-mediated down-regulation of autophagy." J Biol Chem 289(12): 8029–8040.
Suetake, T., F. Hayashi and S. Yokoyama (2015). "Solution structure of MIT domain from mouse Nrbf2." (To be published).
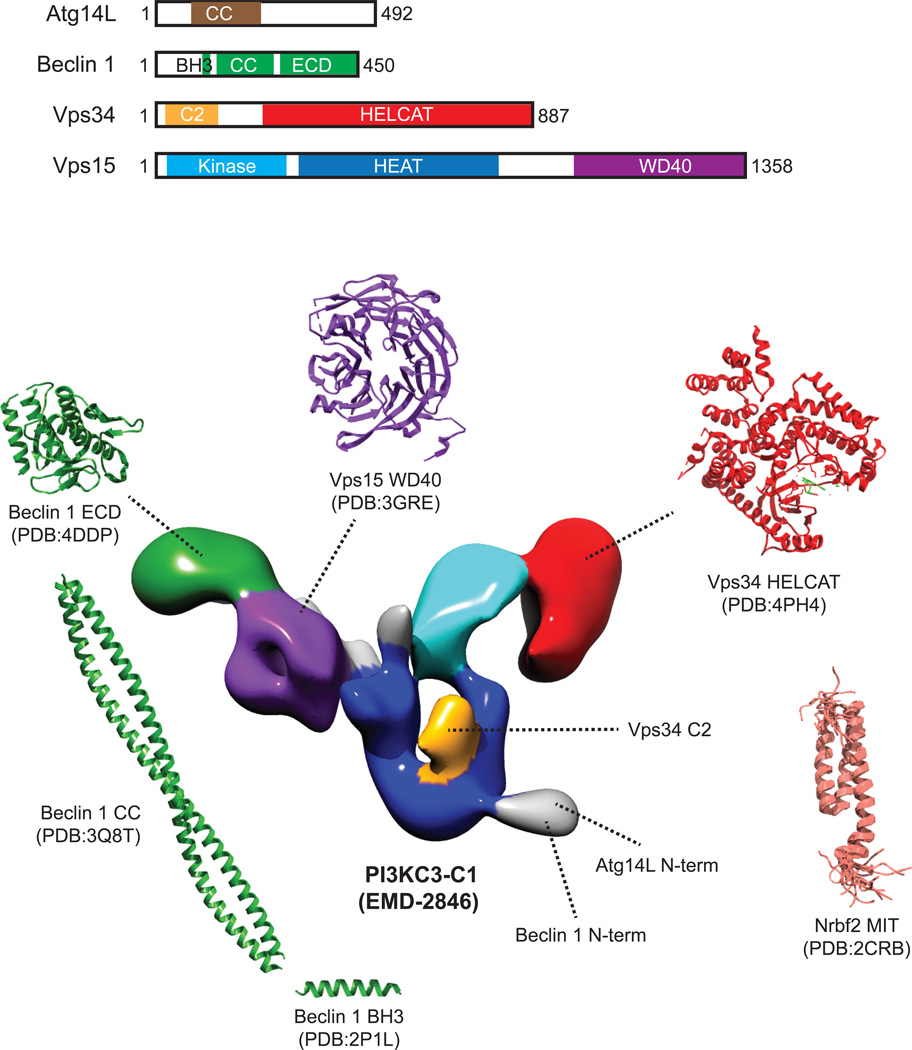
Figure 3.
Currently available structural information of the Atg14L-containing, autophagic Beclin 1-Vps34 class III PI(3)K complex I (PI3KC3-C1). PI3KC3-C1 consists of four components: Atg14L, Beclin 1, Vps34, and Vps15. The V-shaped 3D reconstruction of human PI3KC3-C1 was determined by single-particle EM (EMDB: EMD-2846). Vps15 is thought to be the scaffolding subunit and the locations of its protein kinase (cyan), HEAT (blue), and WD-40 (purple) domains are colored in the EM-based 3D model. Thus far, the only available high-resolution structural information for this protein came from crystallographic analysis of yeast Vps15 WD-40 domain (PDB: 3GRE, residues 1031–1454). The catalytic subunit of PI3KC3-C1 is Vps34, which produces PtdIns(3)P at the autophagosome, endosome and lysosome membranes. The crystal structure of the helical and catalytic domains (“HELCAT”) of human VPS34 in complex with the selective inhibitor PIK-III has been recently determined (PDB:4PH4, residues 293–871 in red, inhibitor in green). Beclin1, which plays a central role in regulating the activity of PI3KC-C1, contains three functional domains: the N-terminal BH3 domain (PDB: 2P1L, residues 107–135 for human), the central CCD (PDB: 3Q8T, residues 144–269 for rat), and the ECD (PDB: 4DDP, residues 248–450 for human). The approximate location of the Beclin 1 ECD in the context of the full complex, as determined by EM-based labeling, is colored in green in the 3D model. The positions of Vps34 C2 domain and the N-termini of Atg14L and Beclin 1 are colored in orange and grey, respectively. Nrbf2, although recently shown to be a subunit of the PI3KC-C1, was not included in the single-particle EM study. However, the structure of the N-terminal MIT domain of mouse Nrbf2 has previously been determined by NMR spectroscopy (PDB: 2CRB, residues 4–86).
3.1. Beclin 1 structure
Beclin 1, first identified as a Bcl-2-interacting protein [111], is composed of 450 amino acids for human and 448 amino acids for mouse. Beclin 1 serves as a platform molecule for the class III PI(3)K complexes by protein-protein interactions through three domains: the BH3 domain (corresponding to residues 107–127 for human) [112,113], the CCD (residues 174–266 for rat; residues 187–319 for Saccharomyces cerevisiae) [114,115], and the C-terminal evolutionarily conserved domain (ECD, residues 244–337 for human) or the β-α repeated, autophagy-specific (BARA) domain (residues 320–557 for Saccharomyces cerevisiae Atg6/Vps30) [112,115–117].
The short N-terminal BH3 motif of Beclin 1 is known to bind Bcl-2 family proteins, including cellular Bcl-2 [15,92] and Bcl-XL [92,112,113,118], KSHV v-Bcl-2 [15], and the γ-herpesvirus 68 Bcl-2 homolog M11 [16,17,118]. The Beclin 1 BH3 domain forms an amphipathic α-helix and fits into a hydrophobic pocket in the Bcl-XL, as revealed by both X-ray crystallography [112] and NMR spectroscopy [113]. Important residues in Beclin 1 BH3 domain for binding to the Bcl-2 family proteins include L110 and F121 for mouse Beclin 1 [17] and correspondingly L112 and F123 for human Beclin 1 [16] as well as G120 and D121 for human Beclin 1 [118]. Su and colleagues also identified a selective Beclin 1 BH3 domain-derived, inhibitory peptide that attenuates the M11-mediated down-regulation of autophagy without binding to cellular Bcl-2 homologs [118]. As mentioned before in Section 2.4, phosphorylation of Beclin 1 in its BH3 domain at T119 by DAP-kinase [93] and phosphorylation of the unstructured loop of Bcl-2 at residues T69, S70, and S87 by JNK-1 [94] independently disrupt the interaction between Beclin 1 and Bcl-2 and enhance autophagic activity in the cells; and the EGFR tyrosine kinase binds to Beclin 1 and phosphorylates Beclin 1 at residues Y229, Y233 and Y352, leading to enhanced Beclin 1 interaction with Bcl-2 and inhibition of autophagic activity [95]. These studies provide the molecular mechanisms by which autophagy can be regulated by the Bcl-2 family proteins, which are important for understanding the crosstalks between apoptosis and autophagy as well as how viruses can subvert host autophagy for infection.
Interactions with factors other than Bcl-2 family proteins at the Beclin 1 BH3 domain also impact autophagy regulation. For example, binding of the VMP1 C-terminal Atg domain at the Beclin 1 BH3 domain releases Beclin 1 from Bcl-2 to form the Beclin1-Vps34 complexes and induce autophagy [97,98]. The interaction between Ambra1 and Beclin 1 also occurs via the BH3 domain region (residues 141–150) [101], although Ambra 1 is capable of interacting with Bcl-2 independent of Beclin 1. It is thought that when cells are faced with acute stresses, Ambra1 disassociates from Bcl-2 and subsequently binds to Beclin 1 to promote autophagy. The exact structural context behind the Ambra1-Beclin 1 interaction awaits further investigation.
The CCD is a common structural motif for hydrophobic protein-protein interactions. Beclin 1, Atg14L and UVRAG all possess CCD [21–23,72,114]. Beclin 1 is known to self-associate [21]. Recent X-ray crystallographic analysis revealed that the Beclin 1 CCD (rat, residues 174–266) consists of a dimeric anti-parallel coiled-coil, interacting in a head-to-tail fashion with an unstable interface [114]. The Beclin 1 CCD is also necessary for assembly of the Atg14L- [21,22,72,114] and UVRAG-containing [21,23,114] Beclin 1-Vps34 complexes that regulate autophagic and endocytic pathways, respectively. The homodimeric Beclin 1 is metastable, and the Beclin 1 CCD interface allows division of the two Beclin 1 molecules, and assembly of the more stable Beclin 1-Atg14L or Beclin 1-UVRAG heterodimers. The Beclin 1-UVRAG interaction is stronger than the Beclin 1-Atg14L interaction, as shown by in vitro assays and cell culture analysis [114]. It is thought that the functionally inactive Beclin 1 homodimers serve as a reservoir for new Beclin 1-Vps34 complex formation when required by changes in cellular conditions. The structure of the Beclin 1 CCD revealed a population of polar or charged residues in the hydrophobic interface of Beclin 1 dimers that render the Beclin 1 homodimers unstable and promote stable interactions between Beclin 1 and Atg14L/UVRAG [114]. However, the mechanism by which Atg14L or UVRAG manipulates these residues to form stable coiled-coil assemblies is still unclear. Much will be deduced about the molecular mechanism of these specific interactions when the structure of each Beclin 1-Vps34 complex is revealed.
The ECD is located at the C-terminal half of Beclin 1 without sequence homology to any known structure; nonetheless, this region of the protein is indispensable for Beclin 1 functionality [116]. Recent studies examining the crystal structure of the C-terminal region of Beclin 1 that encompasses the ECD led to the identification of a novel class of membrane-binding domain [116]. This stable domain consists of three superimposable internal repeats, each comprised of a pair of short β-strands followed by an α-helix. The three α-helices form a helical bundle, surrounded by three β-sheets, six loops and an N-terminal α-helix [116]. Specifically, hydrophobic protrusion in the surface loop L4, consisting of three aromatic residues (F359, F360, and W361), facilitates Beclin 1 binding to lipid membranes enriched in cardiolipin. Although the mechanism and significance of cardiolipin preference are not understood, this aromatic hydrophobic finger is necessary for the insertion of the protein into lipid membranes and the resultant deformation of the liposomes that resembles membrane curvature. Mutations that disrupt this aromatic finger compromise omegasome formation and autophagy [116]. It will be interesting to see if the Beclin 1 ECD aromatic finger residues facilitate interaction with endosomal membranes. It is speculated that the Beclin 1 ECD hydrophobic finger is masked as a result of Beclin 1 homodimerization, and that inhibition is relieved upon Beclin 1-Atg14L/UVRAG heterodimerization [116]. In contrast, mutations in the hydrophobic finger did not cause drastic alteration in the Beclin 1-Atg14L/UVRAG/Vps34 interactions. Adjacent to the aromatic finger in the Beclin 1 ECD is a deep cleft, but the functionality of this feature remains unclear.
Interestingly, studies examining the C-terminal region of the yeast Atg6/Vps30 protein revealed a globular fold comprised of three β-sheet-α-helix repeats [115] that are similar to the mammalian Beclin 1 ECD structure; this finding, along with sequence homology, suggests an evolutionarily conserved feature of this domain [115,116]. However, the aromatic finger residues found in mammalian Beclin 1 are not conserved in yeast Atg6/Vps30 (F430, R431, and K432 in our own alignment and in [115]). The lack of hydrophobic finger in yeast Atg6/Vps30 also poses the question as to whether and how Atg6/Vps30 binds membranes. Truncation analysis of Atg6/Vps30 C-terminal domain determined that, like Beclin 1 ECD, Atg6/Vps30 ECD is not required for the assembly of either Atg14- or Vps38-containing Atg6/Vps30-Vps34 complex; rather it is required for targeting the Atg14-containing Atg6/Vps30-Vps34 complex to the PAS, but is not necessary for Vps38-mediated vacuolar protein sorting [115]. Thus, the yeast Atg6/Vps30 C-terminal domain is termed BARA domain, short for β-α repeated, autophagy-specific domain. It will be interesting to see if the Beclin 1 ECD is dispensable for endocytic trafficking.
3.2. Vps34 structure
Vps34 was originally identified in yeast as part of the PI(3)K complex, responsible for control of intracellular protein trafficking [119,120], with substrate specificity for Ptdlns but not PtdIns4P or Ptdlns(4,5)P2 [119]. Human Vps34 was subsequently cloned and found to specifically phosphorylate Ptdlns to PtdIns(3)P [65]. Common to the catalytic subunits of other PI(3)Ks, Vps34 is composed of an N-terminal C2 domain (residues 1–255), a middle helical domain (residues 293–530), and a Cterminal catalytic kinase domain (residues 533–887) [121]. Vps34 interacts with the membrane via the C2 domain, as well as the extreme C-terminal helix that is also required for lipid kinase activity in both yeast [122] and human [123,124]. It is thought that the C2 domain binds Beclin 1 [23], although it has no role in regulating enzyme catalytic activity [124]. The Vps34 C-terminal kα11 and kα12 helices bind Vps15 [124]. The last 11 amino acids at the C terminus of Vps34 are required for PI(3)K activity, although they are not required for binding to Vps15 [122].
The crystal structure of Drosophila melanogaster Vps34 (DmVps34) HELCAT domains (i.e., Δ1–257), either alone or in complex with inhibitors, shows a compact unit with a regulatory C-terminal helix as well as a solenoid helical domain packed against a classical bilobal catalytic domain characteristic of protein kinases [124]. This crystal structure features (1) a hook-shaped, completely ordered, phosphoinositide-binding loop (activation loop) for recognizing PtdIns, (2) a phosphate-binding loop (P-loop) for binding ATP, and (3) a catalytic loop that catalyzes the transfer of the ATP γ-phosphate to the 3-hydroxyl of PtdIns [124]. The crystal structures of the human Vps34 HELCAT domains were later determined in the apo form and in the presence of newly developed Vps34 inhibitors [125–127].
Mutational analysis of the DmVps34 activation loop key residues proposed to bind phosphoinositide headgroup (e.g., K833, P832 and Y826; corresponding to human K771, P770 and Y764, respectively) shows either significantly impaired enzyme activity or complete inactivity. The Vps34 catalytic loop contains a conserved DRH motif (Dm 805–807; Hs 743–745) and two aspartic acid residues (Dm 805 and 823; Hs 743 and 761), all of which are important for the enzymatic reaction. Interestingly, deletion or mutations (e.g., H879A and W885A) of the C-terminal helix greatly diminish ATPase activity in the presence of the PtdIns:PS vesicles, whereas deletion or mutations (e.g., W885A and Y884A) of the C-terminal helix or mutations in the activation loop (e.g., K771A) increase basal ATPase activity in the absence of vesicles, suggesting dual roles of the C-terminal helix in activation (on the membrane) and auto-inhibition (off the membrane) [124]. These results are confirmed by the structure which reveals that the C-terminal helix blocks the catalytic loop when Vps34 is not associated with lipid membrane, but upon association with lipid membrane, the obstruction is displaced, allowing for Vps34 catalytic activity.
The ATP-binding site of Vps34 is unique in regions critical to inhibitor binding, such as the P-loop, hinge region and gatekeeper amino acids [126]. The Vps34 ATP-binding pocket is significantly smaller in volume than that of other PI(3)Ks (e.g., the class I p110γ), as a result of distinct structural characteristics of Vps34 including the inward-curling P loop. Also unique to Vps34, a bulky residue in the P loop (Hs F612; Dm F673) packs against the aromatic hinge residue (Hs F684; Dm Y746), likely leading to a more rigid and constrained ATP-binding pocket in Vps34 as compared to in other Class I PI(3)Ks. As a result, the smaller and more rigid and constrained ATP-binding pocket of Vps34 restricts the binding of typical PI(3)K inhibitors, making it very difficult to develop potent and high-affinity inhibitors specific for Vps34.
The structure of Vps34 (either apo-protein or in complex with 3-methyladenine (3-MA) or any of the new Vps34 inhibitors) provides a better understanding of the Vps34 ATP-binding pocket, leading to successful structure-based design of improved (high affinity and highly specific) compounds targeting the Vps34 activity (see Section 5.1) [125–127]. In addition, the unique C-terminal helix may also represent a target for specific, non-ATP-competitive Vps34 inhibitors.
3.3. Vps15 structure
Vps15 was originally identified in yeast as a 160 kDa protein serine/threonine kinase associated with Vps34 [128,129]. The membrane localization of Vps34 and activation of Vps34 lipid kinase activity are dependent on the Vps15 serine/threonine kinase activity [130]. Similarly, human Vps15 (formerly called p150, 166 kDa) associates with Vps34 in an approximately equimolar ratio [65] and markedly increases the in vitro Vps34 lipid kinase activity [131,132].
Experimental data from yeast [71], fly [133], and human [132] suggest that Vps15 plays a role in Vps34-dependent autophagy. Notably, regulation of Vps34 activity by autophagy-related proteins and by nutrients requires the presence of Vps15 [132]. In particular, Vps15 enhances the binding of Beclin 1/UVRAG to Vps34, as well as Beclin 1/UVRAG-stimulated or amino acid/glucose-sensitive Vps34 activity; and Beclin 1/UVRAG enhances binding of Vps15 to Vps34 without increasing the total Vps34 and Vps15 levels [132].
Human Vps15 consists of an N-terminal serine/threonine kinase domain (residues 11–262), a C-terminal WD-40 domain (residues 1000–1300), and an intermediate domain (residues 400–700) containing several HEAT repeats that connect the kinase and WD-40 domains [131,134]. Moreover, both yeast Vps15 [128,129] and human Vps15 [131] are myristoylated at their N-termini ((M)G1A2Q3L4S5L6 (yeast) and (M)G1N2Q3L4A5G6 (human)). Myristoylation anchors Vps15 to the target membranes such as endosomal membranes and isolation membranes (or pre-autophagosomal site (PAS) in yeast), facilitates translocation of Vps34 to these membranes, and activates the essential PI(3)K activity of Vps34 [132]. The WD-40 domain of Vps15 binds activated Rab5 to target the Vps34-Vps15 sub-complex to early endosomes [135,136]. The WD-40 domain of Vps15 also binds Rab7 and the PtdIns3P phosphatases myotubularin 1 and 2 (MTM1 and MTM2) mutual exclusively on late endosomes [137,138].
The crystal structure of the S. cerevisiae Vps15 WD-40 domain features a sevenbladed propeller resembling that of typical Gβ proteins [134]. The WD-40 domain serves as a scaffold to assemble proteins such as Atg14. Co-immunoprecipitation studies using full length and truncation mutants of Vps15 show that the WD-40 domain alone and the kinase domain alone pull down Atg14 [134]. In addition, mutagenesis studies show that the kinase domain together with the intermediate domain of Vps15, but neither the WD-40 domain alone nor the WD-40 domain together with the intermediate domain, partially retain vacuolar sorting of carboxypeptidase Y [134]; however, putative substrates of the kinase domain of Vps15 have yet to be identified.
4. Beclin 1-Vps34 Complex Architecture
Understanding how protein complexes come together to regulate cellular processes is beneficial for development of potent and improved therapies to control diseases which these complexes influence. However, isolation of endogenous multi-protein complexes is technically challenging due to the dynamic nature, low abundance, and heterogeneity associated with such assemblies. Here we summarize the current understanding of Beclin 1-Vps34 complex architecture using affinity purification (AP) in combination with Western blot analysis, direction interaction detection by in vitro binding assays, crosslinking mass spectrometry, and single-particle electron microscopy.
4.1. Affinity purification in combination withWestern blot analysis
Beclin 1-Vps34 complex architecture has been assessed by affinity purification in combination with Western blot analysis, which examines pairwise protein-protein interactions (summarized in Table 2). Most data indicate that the binding of Beclin 1/Atg6 to Atg14L/Atg14 and UVRAG/Vps38 is mutually exclusive [21,69,71–73], suggesting that there are two major Beclin 1-Vps34 complexes.
Table 2.
Summary of reported interactions among the Beclin 1-Vps34 complex key components including Beclin 1, Vps34, Vps15, Atg14L, UVRAG and Nrbf2.
| Component #1 |
Domain of Component #1 |
Component #2 | Direct interaction | Detection Assay | Reference | Confirmed by CX-MS1 |
Consistent with EM2 |
|---|---|---|---|---|---|---|---|
| Beclin 1 (Atg6/Vps 30 in yeast) | Full length (h, 1–450) | Beclin 1 Vps34 Vps15 Atg14L UVRAG/Vps38 Nrbf2 |
Direct interaction -- -- Direct interaction Direct interaction -- |
Y2H AP AP AP, Y2H AP, Y2H AP |
3 3–14 3,5–9,13,15 3,5–9,13,15,16 3,5–9,12,13,15 5,6,8–11 |
||
| BH3 domain | |||||||
| CCD | Beclin 1 | Direct interaction | AP, analytical ultracentrifugation, in vitro binding, bimolecular fluorescence complementation assay | 17 | |||
| Vps34 | -- | AP | 12 | Y | Y | ||
| Atg14L | Direct interaction | AP, analytical ultracentrifugation, in vitro binding | 3,5,7,17 | Y | |||
| UVRAG | Direct interaction | AP, analytical ultracentrifugation, in vitro binding | 3,7,12,17 | Y | |||
| ECD/BARA | Atg14L Vps34 |
-- -- |
AP AP |
5 4,12 18 |
Y | ||
| Vps34 | Full length (1–887) | Beclin1 Vps15 Atg14L UVRAG Nrbf2 |
-- Direct interaction -- -- -- |
AP, in vitro binding AP, crosslinking AP AP AP |
3–14 3,6,8,10,11,14,19–22 3,6–8,10,11,13,15 3,6,8,10,11,14,15 6,8–11 |
||
| C2 domain | Beclin 1 | -- | AP | 12,15 | Y | Y | |
| Atg14L | -- | AP | 15 | Y | Y | ||
| UVRAG | -- | AP | 15 | Y | Y | ||
| Helical domain | |||||||
| Lipid kinase domain | Vps15 | -- | Y2H, functional assays (e.g., CPY sorting or maturation, PtdIns(3)P levels) | (yeast)23 | Y | ||
| Vps15 | Full length (1–1358) | Beclin 1 Vps34 Atg14L UVRAG Nrbf2 |
-- Direct interaction -- -- Direct interaction |
AP AP, Y2H, crosslinking AP AP AP, in vitro binding |
3,5–8,13,15 3,6,8,10,14,21,22, (yeast)3,11,19,20,23 3,6,8,15 6 6,8–11 |
||
| Kinase domain | Vps34 | Direct interaction | Y2H Crosslinking |
(yeast)23 (yeast)20 |
Y | ||
| Atg14 | -- | AP | (yeast)24 | ||||
| HEAT domain | Vps34 | -- | Y2H | (yeast)23 | |||
| WD-40 domain | Atg14 Nrbf2 |
-- -- |
AP In vitro binding |
(yeast)24 10 |
|||
| Atg14L (Atg14 in yeast) | Full length (1–492) | Beclin 1 Vps34 Vps15 Nrbf2 |
Direct interaction -- -- Direct interaction |
AP, Y2H AP AP AP, in vitro binding |
3,5–8,13,15,16 3,6–8,10,11,13,15 3,6,8,15 6,8–11 |
||
| Zinc finger | |||||||
| CCD1 (75–95) | Beclin 1 Vps34 Nrbf2 |
-- -- -- |
AP AP AP |
5,9 5 8,9 |
Y | ||
| CCD2 (148–178) | Beclin 1 | -- | AP | 5,9 | Y | ||
| CCD (71–184 or 88–178) | Beclin 1 Vps34 |
Direct interaction -- |
AP, analytical ultracentrifugation, in vitro binding AP |
3,7,15,17 15 |
Y Y |
||
| UVRAG (Vps38 in yeast) | Full length (1–698) | Beclin 1 Vps34 Vps15 Nrbf2 |
Direct interaction -- -- -- |
AP, Y2H AP AP -- |
3,5–8,12,13,15 3,6,8,10,11,14,15 3,6 only shown in9 |
||
| C2 domain | |||||||
| CCD | Beclin 1 Vps34 |
Direct interaction -- |
AP, analytical ultracentrifugation, in vitro binding | 7,12,17 | Y Y |
||
| Nrbf2 (Atg38 in yeast) | Full length (1–287) | Beclin 1 Vps34 Vps15 Atg14L UVRAG Nrbf2/Atg38 |
Weak direct Weak direct Direct interaction Direct interaction -- Direct interaction |
AP, Y2H (weak) AP, Y2H (weak) AP, in vitro binding AP, in vitro binding, Y2H AP AP, Y2H |
5,6,8–11 6,8–11 8–11 6,8–11 only shown in9 8,11 |
||
| MIT domain (8–52) Yeast 1–80 |
Beclin 1 Atg14L Atg14 |
-- -- Direct interaction |
AP AP Y2H |
8 9 11 |
|||
| Yeast 1–120 | Atg14 Vps34 |
Direct interaction Direct interaction |
Y2H, in vitro binding Y2H, in vitro binding |
11 11 |
|||
| CCD (167–215) Or yeast 121–226 |
Nrbf2 Vps34 |
Direct interaction Direct interaction |
AP, Y2H Y2H |
8,11 11 |
Notes:
Abbreviations: AP – affinity purification; Y2H – yeast-two-hybrid; CX-MS – crosslinking mass spectrometry; EM – single-particle electron microscopy; Y – Yes.
Mutual exclusiveness of Atg14L and UVRAG was reported in binding to Beclin 17,17, Nrbf28,10, or binding to each other3,7–9,15,17. Ectopically expressed Atg14L and UVRAG were co-IP’ed5.
Residue numbers follow selected references, as they are not exactly the same in different references.
References:
. Shi Y, Pellarin R, Fridy PC, et al. A strategy for dissecting the architectures of native macromolecular assemblies. Nat Methods. 2015(Accepted 20 August, published online 5 October, 2015).
. Baskaran S, Carlson LA, Stjepanovic G, et al. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife. 2014;3:e05115.
. Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396.
. Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1(1):46–52.
. Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476.
. Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76.
. Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105(49):19211–19216.
. Zhong Y, Morris DH, Jin L, et al. Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels. J Biol Chem. 2014;289(38):26021–26037.
. Lu J, He L, Behrends C, et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity.Nat Commun. 2014;5:3920–3934.
. Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM. NRBF2 regulates macroautophagy as a component of Vps34 complex I. Biochem J. 2014;461:315–322.
. Araki Y, Ku WC, Akioka M, et al. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203(2):299–313.
. Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8(7):688–699.
. Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152(3):519–530.
. Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J. 2009;417(3):747–755.
. Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372.
. Kametaka S, Okano T, Ohsumi M, Ohsumi Y. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem. 1998;273(35):22284–22291.
. Li X, He L, Che KH, et al. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012;3:662.
. Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagyrelated protein 6 and its specific role in autophagy. J Biol Chem. 2012;287(20):16256–16266.
. Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12(5):2195–2204.
. Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129(2):321–334.
. Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. J Biol Chem. 1997;272(4):2477–2485.
. Volinia S, Dhand R, Vanhaesebroeck B, et al. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14(14):3339–3348.
. Budovskaya YV, Hama H, DeWald DB, Herman PK. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J Biol Chem. 2002;277(1):287–294.
. Heenan EJ, Vanhooke JL, Temple BR, Betts L, Sondek JE, Dohlman HG. Structure and function of Vps15 in the endosomal G protein signaling pathway. Biochemistry. 2009;48(27):6390–6401.
Genetic knockout or transient/stable knockdown of one component of the Beclin 1-Vps34 complex has been a widely used experimental approach to determine the protein component(s) that are critical for complex assembly and stability. For example, this approach was used to demonstrate that Beclin 1, Vps34 or Vps15 stabilizes both Atg14L and UVRAG; Beclin 1 stability depends on Vps34, Vps15, Atg14L, UVRAG and Nrbf2, while the stabilities of Vps34 and Vps15 depend on each other [68,69,71,73].
Moreover, this approach has also been utilized to show that in RPE-1 cells, Beclin 1 is required for Vps34→Vps15 and Vps34→Nrbf2 interactions, where the arrows point from bait to prey; Atg14L is required for Nrbf2→Vps34, Nrbf2→Vps15, and Beclin 1↔Nrbf2 interactions; and Nrbf2 is required for Atg14↔Vps34, Atg14L→Vps15, and Vps34→Beclin 1 interactions [69]. Several of these requirements for inter-molecular interactions within the Beclin 1-Vps34 complexes were confirmed in Nrbf2−/− MEF cells and mouse brain models: e.g., Atg14L is required for the Beclin 1→Nrbf2 interaction, and Nrbf2 is required for the Atg14↔Vps34, Atg14L→Vps15, Vps34→Vps15, and Vps34→Beclin 1 interactions, and the total and Atg14L-linked Vps34 activities [87]. Moreover, the regulation of Vps34 kinase activity by Nrbf2 requires Vps15 [87]. Similarly, in yeast, Atg14 is required for Atg38 to bind Vps34, Vps15 and Atg6/Vps30, and Atg38 is required for Atg14 to bind Vps34 and Vps15, but not Atg6/Vps30 [90]. In addition, in yeast, Vps38 is required for Atg6/Vps30 to interact with Vps34 and Vps15 [71]. Taken together, these studies suggest that Atg38/Nrbf2 is an integral structural component that brings Vps34-Vps15 and Beclin 1-Atg14L (or Atg6/Vps30-Atg14 in yeast) sub-complexes together.
Another experimental approach to assess Beclin 1-Vps34 complex architecture by affinity purification in combination with Western blot analysis involves utilizing ectopically expressed, tagged full length and truncation mutants of Beclin 1 complex components to identify protein domains critical for complex assembly (summarized in Table 2). Using this approach, it has been shown that the Beclin 1 CCD (residues 144–269) is sufficient to bind to UVRAG and Vps34 [21,23], and the Beclin 1 CCD is both sufficient and necessary to bind to Atg14L [22,21,72]. In a similar fashion, the Beclin 1 ECD domain (residues 244–337) has been shown to be required for the Beclin 1-Vps34 interaction and Beclin 1-associated Vps34 lipid kinase activity [23,117].
For Vps34, the Vps34 N-terminal C2 domain (residues 1–282) is sufficient to bind to Beclin 1 [23], and this domain also interacts with Atg14L and UVRAG [73]. The WD-40 domain (residues 1027–1454) of Vps15 alone and the kinase domain (residues 1–294) alone pull down Atg14 in yeast [134]. Endogenous immunoprecipitation with anti-Vps15 antibody further shows that point mutations in the Vps15 kinase domain (e.g., D165R and E200R) greatly weaken the Vps34-Vps15 association [139]. In addition, both the WD-40 domain and the other regions of Vps15 bind to Nrbf2 [86].
Both UVRAG and Atg14L utilize their CCD to bind to the Beclin 1-Vps34-Vps15 core complex. The UVRAG CCD (residues 200–269) is sufficient and necessary to bind to Beclin 1 [23]. Deletion of the first or both CCD domains of Atg14L abolished the Atg14L-Nrbf2 interaction [69,87]. In agreement with this finding, Atg38 can only be pulled down by full-length yeast Atg14, but not truncated mutants, suggesting that a rather large portion of Atg14 is necessary for its binding to Atg38 [90]. In addition, deletion of Atg14L CCD (residues 71–184) abolished both the Atg14L-Beclin 1 and Atg14L-Vps34 interactions without compromising Atg14L localization to the isolation membranes. However, Atg14L ΔCCD mutant could not restore autophagy in Atg14L siRNA-treated cells. These result suggest that Atg14L CCD is required for autophagy but not for Atg14L isolation membrane translocation [21,22,72,73].
Nrbf2 contains an N-terminal MIT domain (residues 8–52). MIT domains are known to form an asymmetric three-helix bundle involved in protein-protein interactions, and are conserved among Vps4 [140–142], spastin [143], katanin, and a sorting nexin, which may play a role in intracellular trafficking [144]. The Nrbf2 MIT domain is required for Nrbf2-Atg14L, Atg14L-Vps34, and Atg14L-Vps15 interactions, the total and Atg14L-linked Vps34 activities, and the formation of WIPI2-associated phagophores upon autophagy induction [87]. Moreover, both Nrbf2 MIT domain and CCD (residues 168–209) facilitate Vps34-Vps15 interactions and Vps15-enhanced Vps34 activity [87]. In addition, Atg38/Nrbf2 appears to form a homodimer via the CCD, facilitating the stability of the Atg14L/Atg14-containing complex [69,90].
4.2. Direct interaction detection
Co-immunoprecipitation of two proteins from cell cultures or tissues does not necessarily guarantee direct interactions between these components. Thus, recombinantly purified or in vitro translated proteins are often used to determine if two proteins directly bind to one another. Protein pairs within the Beclin 1-Vps34 complexes for which direct interactions have been reported include: Vps34 (via helices kα11 and kα12)-Vps15 [124], Vps34 (via C2 domain)-Beclin 1 [124], Vps15 (via either WD-40 domain alone or other regions)-Nrbf2 [86], Beclin 1 (via CCD)-Beclin 1 (via CCD) [114], Beclin 1 (via CCD)-Atg14L (via CCD) [114], Beclin 1 (via CCD)-UVRAG (via CCD) [23,114], Atg14LNrbf2 [87], and Atg38 (via residues 1–120)-Vps34/Vps15 [90] (summarized in Table 2). Interestingly, Beclin 1 forms, via its CCD, a metastable homodimer that readily dissociates to form more stable heterodimers through the CCDs of Atg14L and UVRAG [114]. Beclin 1 mutants defective in homodimer formation (e.g., L178A/L259A, L178A/L192A, and L178A/L196A) retain WT capability for the Beclin 1-Atg14L or Beclin 1-UVRAG heterodimer formation; whereas Beclin 1 mutants that form “stabilized” homodimers (e.g., E189L/A255L or E189L/A217L/E224L/A255L) show impaired interaction [114].
Yeast-two-hybrid (Y2H) is another widely-used experimental approach to identify direct interactions. For example, both the N-terminal protein kinase domain (residues 1–117 or longer) and the HEAT domain (which contains three tandem repeats of about 39 amino acids each) of Vps15 are required for interaction with Vps34 (via a 28-amino acid region encompassing residues 837–864) [122]. Y2H also demonstrates Beclin 1 homodimer formation as well as direct Beclin 1-Atg14L and Beclin 1-UVRAG interactions [21]. Y2H further shows that in yeast, Atg38 self-associates via residues 120–226 and this self-association is required for the association between the Vps34-Vps15 and Atg14-Atg6/Vps30 sub-complexes. Additionally, the Atg38 MIT (residues 1–80) domain directly binds Atg14, Atg38 weakly binds Atg6/Vps30 and Vps34, and the Atg38-Vps34 binding is independent of the MIT domain [90] (Table 2).
4.3. Crosslinking mass spectrometry (CX-MS)
Crosslinking with dithiobis(succinimidylpropionate) (DSP) followed by immunoprecipitation with anti-Vps15 antibody has previously been used to demonstrate Vps34-Vps15 interaction [130] and impairment of this interaction due to point mutations in the Vps15 kinase domain, e.g., D165R and E200R [139].
We recently developed a novel and robust proteomic strategy that provides the requisite sensitivity for hybrid structural dissection of large, multi-subunit native complexes [145]. This strategy utilizes engineered, ultra-high affinity GFP VHH nanobodies for efficient and pristine affinity capture of native complexes, and features direct on-bead crosslinking (by amine-specific disuccinimidyl suberate crosslinker) of the affinity captures followed by bottom-up proteomics and high-resolution MS to identify the cross-linked peptides. These cross-linked peptides are used as “chemical rulers” for measuring inter-atomic distances to determine the architectures of the complexes [145]. We applied this technology to study the molecular architecture of the Beclin 1-Vps34 complexes from a single Becn1-EGFP/+ transgenic mouse liver. Our study revealed multiple crosslinks between the Beclin 1 CCD (residues 142–267) and Vps34 N-terminal C2 domain (residues 1–255), including Beclin 1 (201)-Vps34 (36), Beclin 1 (204)-Vps34 (33), Vps34 (36)-Beclin 1 (204), and Beclin 1 (204)-Vps34 (29) [145]. This crosslinking information enriches the limited structural information that is currently available on these native complexes.
Besides confirming previously identified interacting domains (Table 2), the CX-MS strategy is also useful for identifying the proximities of specific cross-linked amino acids within and between the various subunits comprising the protein complexes. The novel inter-molecular crosslinks that we identified include Beclin 1 (52)-UVRAG (217), Vps15 (280)-Vps34 (287), Vps34 (287)-Nrbf2 (85), Vps15 (1028)-Atg14L (343), Vps15 (1028)-UVRAG (405), UVRAG (401)-Vps15 (1028), Vps15 (982)-UVRAG (395), UVRAG (395)-Vps15 (982), Vps15 (993)-Atg14L (184), Vps15 (801)-Vps34 (79), Vps15 (816)-Vps34 (92), Vps15 (888)-Vps34 (33), and Vps15 (951)-Vps34 (180) [145]. These findings suggest that the Vps34 C2 domain and a region between the HEAT and the C-terminal WD-40 domains of Vps15 are brought into close proximity in the fully assembled complex. The possibility of direct interaction between the Vps34 N-terminal C2 domain and the region between the HEAT and WD-40 domains of Vps15 provides additional possible contact points between these two subunits, which differs from the observation in yeast, in which the C-terminal lipid kinase domain of Vps34 interacts with kinase-HEAT domains of Vps15 [122]. The architecture of native mouse Beclin 1-Vps34 complexes revealed by CX-MS are in agreement with the human recombinant complexes solved by single particle electron microscopy (EM, see Section 4.4).
Interestingly, the mapped crosslinks show that Atg14L and UVRAG may competitively interact for the same lysine residues on the Beclin 1 (at residue 235, in the CCD), Vps34 (at residues 29 and 209, in the C2 domain), and Vps15 (at residue 1028) triad. These crosslinks include UVRAG (284, in the CCD)-Beclin 1 (235), Beclin 1 (235)-Atg14L (161 and 164, in the CCD), Atg14L (136)-Vps34 (29), Vps34 (29)-UVRAG (255, in the CCD), UVRAG (266, in the CCD)-Vps34 (29), Vps34 (209)-Atg14L (136), UVRAG (266)-Vps34 (209), Vps15 (1028)-Atg14L (343), Vps15 (1028)-UVRAG (405), and UVRAG (401)-Vps15 (1028). Although the Beclin 1 CCD is known to bind Atg14L and UVRAG in a mutually exclusive manner, neither the Vps34 N-terminal C2 domain or the C-terminus of Vps15 have been previously reported to bind Beclin 1 CCD mutual exclusively, except that the Vps34 C2 domain was shown to interact with both Atg14L and UVRAG [73].
4.4. Single-particle electron microscopy (EM) and hydrogen-deuterium exchange
Recent single-particle EM and hydrogen-deuterium exchange studies provide the first view on the overall architecture and dynamics of the Atg14L- and UVRAG-containing Beclin 1-Vps34 complexes in their fully functional states [146]. A three dimensional reconstruction of the Atg14L-containing Beclin 1-Vps34 complex was obtained at 28 Å resolution, which shows a V-shaped, loosely connected, elongated structure. Complementary EM-based N- and C-terminal maltose binding protein (MBP) localization studies enabled the mapping of the positions of the subunit termini in the complex. Additional docking analysis of known crystal structures or homology models onto the density permitted generation of a three-dimensional model of the complex [146] (Figure 3). In this model, Vps15 is positioned throughout the V-shaped structure and appears to organize the complex. The Vps15 N-terminus is located near the C-terminus of Vps34 in the right arm of the V, the Vps15 C-terminal WD-40 domain forms a donut shaped region in the left arm of the V, and the HEAT repeat domain forms an arch-shaped region at the junction of the V, where the Vps34 N-terminal C2 domain, Beclin 1 N-terminus, and Atg14L N-terminus appear near the junction. The close proximity of the Beclin 1 and Atg14L N-termini suggests that the Atg14L and Beclin 1 coiled coils are parallel to each other, in contrast to the previously reported antiparallel coiled-coil for the Beclin 1 homodimer [114]. Beclin 1 is positioned along the left arm of the V, with its C-terminal BARA domain (or ECD) at the tip of the V and above the Vps15 WD-40 domain. Two long, narrow tubes of electron density (the first one located between the Beclin 1 BARA and Vps15 WD-40 domains; the second one located between the Vps15 WD-40 and HEAT domains) are presumed to account for the coiled-coil dimer of Atg14L and Beclin 1. This assumption is consistent with the newly identified contacts in our CX-MS study (i.e., Vps15 (801)-Vps34 (79), Vps15 (816)-Vps34 (92), Vps15 (888)-Vps34 (33), and Vps15 (951)-Vps34 (180)) (see Section 4.3) [145]. These new data suggest that the Vps34 C2 domain interacts with the Beclin 1 and Atg14L CCDs and with the region between the HEAT and WD-40 domains of Vps15 (Table 2).
A single-particle EM structure of the UVRAG-containing Beclin 1-Vps34 complex was also obtained, which shows essentially identical overall conformation and architecture to those of the Atg14L-containing Beclin 1-Vps34 complex, with an additional electron density for the UVRAG N-terminal C2 domain near the junction of the V and the Atg14L N-terminus [146]. The newly identified crosslinks in our CX-MS study (i.e., Vps15 (1028)-Atg14L (343), Vps15 (1028)-UVRAG (405), UVRAG (401)-Vps15 (1028)) (see Section 4.3) [145] further suggest that both Atg14L and UVRAG, like Beclin 1, also align the left arm of the V, likely “glued” to Beclin 1 by parallel CCDs. In addition, the newly identified Vps34 (287)-Nrbf2 (85) crosslink in our CX-MS study suggests that Nrbf2 is located at the region between the C2 and helical domains of Vps34 (likely where the UVRAG C2 domain is), which is consistent with the reports that Nrbf2 and UVRAG are also mutually exclusive [69,90,86].
The dynamics of the full complex was analyzed by hydrogen deuterium exchange with MS readout (HDX-MS), which revealed a conserved 20-residue ordered region between the C2 and helical domains of Vps34 (i.e., residues 218–237, termed the C2-Helix Internal Linker (CHIL) motif) [146]. It is postulated that this linker provides an anchor for a limited displacement of the Vps34 HELCAT domains. Furthermore, HDX-MS data also show that both the linker regions between the Vps34 C2 and helical domains (e.g., around residue 287) and between the Vps15 HEAT and WD-40 domains are highly disordered, consistent with lack of electron density in these regions. Structurally homogeneous subsets of EM images show Vps15 to pivot at the base of the V structure, between the WD-40 domain and the C-terminus of the HEAT repeat. This, assisted by additional flexibility between the HEAT and kinase domain, allows movement of the Vps15 kinase domain relative to the WD-40 domain. The exact role that this movement plays in regulating the Beclin 1-Vps34 complex functions is unclear, although the Beclin 1 N-terminus is in close proximity to the pivot point. In addition, our CX-MS results show crosslinks between this pivot point to Atg14L and UVRAG, i.e., Vps15 (982)-UVRAG (395), UVRAG (395)-Vps15 (982), and Vps15 (993)-Atg14L (184). As the N-termini of Beclin 1 and Atg14L appear to be a convergent region for signaling inputs (e.g., phosphorylation by Ulk1 and AMPK, binding to Bcl-2, and ER-targeting), it is possible that the pivot point of the V receives signals while the right arm of the V transmits signals through the aforementioned intra-complex long range allosteric movement.
Results from single-particle EM and HDX-MS analyses of the reconstituted Atg14- and UVRAG-containing Beclin 1-Vps34 complexes [146], together with those from CX-MS analysis of native Beclin 1-Vps34 complexes [145], collectively lead to a model of subunit architecture and their dynamics (see Figure 3 for the architecture of the Atg14L-containing Beclin 1-Vps34 complex). These approaches provide exciting and valuable ways for better analyzing multi-protein complexes like the Beclin 1-Vps34 complexes. These approaches, which allow analysis of endogenous protein complexes, permit modeling of structures that contain disordered or flexible regions, which is problematic for most other structural analysis platforms. Resolving protein complexes by these approaches will be useful in the design of drugs to target these complexes more effectively.
5. Drug design targeting the Beclin 1-Vps34 complexes
Strong evidence linking autophagy to many human diseases suggests that the autophagy pathway is an important target for drug intervention. Therapeutic possibilities that aim at the Beclin 1-Vps34 complexes may prove beneficial in the treatment of these diseases. Once a promising drug target is identified, structure-based design is often utilized for novel drug discovery during the hit-to-lead stage [53]. Moreover, structure-based design has also been shown to be critical for developing new generation of drugs that overcome drug resistance [53]. One structure-based design strategy proven very successful is the targeting of catalytic domains of enzymes, ion channels, or receptors. For example, STI571 (imatinib; Gleevec, Novartis, Basel, Switzerland), a competitive inhibitor at the ATP-binding site of the oncogenic BCR-ABL, was developed through structure-based design in combination with in vitro screening for tyrosine kinase inhibitors [147]. Additional structure-based design led to development of second generation BCR-ABL inhibitors (e.g., dasatinib and nilotinib) that inhibit imatinib-resistant mutant BCR-ABL [148,149]. A second structure-based design strategy targets allosteric sites that are independent of the enzyme-active site, but cause conformational changes to alter enzyme function. A third design strategy targets protein-protein interactions, which has recently been shown to successfully influence previously non-druggable proteins or pathways due to hot spots or adaptability of the targeted interfaces. This type of drugs induce biochemical perturbations, including post-translational modification, allosteric transition, protein degradation or de novo protein synthesis, that can change protein-protein interactions [150]. Examples of structure-based drug design using this strategy include the small molecule BH3 mimetics (e.g., ABT-737) which competitively displace the pro-apoptotic Bcl-2 family proteins from binding to the anti-apoptotic Bcl-2 family proteins and lead to apoptosis, the SMAC mimetics which bind to anti-apoptotic proteins (e.g., XIAP, cIAP1 and cIAP2) and cause the release of apoptotic SMAC and caspases, and the MDM2 antagonists (e.g., Nutlin-3) which mimick the p53 residues for MDM2 binding and competitively displace p53 from the MDM2 binding site, promoting p53 stabilization and activation, and apoptosis in cancer cells with wildtype p53 [54].
Phosphorylation of PtdIns to generate PtdIns(3)P is the primary function of Vps34 thus the Beclin 1-Vps34 complexes. Structure-based drug design can target the Vps34 PI(3)K activity either directly at the Vps34 catalytic site or indirectly via modulating the Beclin 1-Vps34 complex stability and architecture.
5.1. Drug design directly targeting Vps34
Vps34 is the sole class III lipid kinase, and it is known to regulate many cellular processes, including vesicular trafficking in the endosomal/lysosomal system [119,139], autophagy [71], nutrient sensing in the mTOR pathway [151–157], and trimeric G-protein signaling to mitogen-activated protein kinase (MAPK) [158]. Wortmannin [159], 3-MA [160,161] and LY294002 [162] are commonly used to inhibit Vps34 PI(3)K activity, even though they also inhibit class I PI(3)K [163].
With the crystal structure of Drosophila Vps34 in place [124], development of Vps34 inhibitors to modulate Vps34 PI(3)K activity is at the forefront of much biomedical research. Recently, multiple research groups generated potent and highly selective small molecular inhibitors of Vps34 (summarized in Table 3), including Vps34-IN1 (IC50=25 nM) [164], PIK-III (IC50=18 nM) [125], SAR405 (IC50=1.2 nM) [126], and compound 31 (IC50=2 nM) [127]. These compounds are specific to Vps34 and few offtarget effects have been observed against other PI(3)K isoforms or other lipid kinases [164,125–127].
Table 3.
Summary of reported Vps34 inhibitors.
| Vps34 inhibitor | Molecular Structure |
IC50 (in vitro) |
IC50 (in vivo) | IC50 for other lipid kinases | IC50 for protein kinases |
Reference | Patent by |
|---|---|---|---|---|---|---|---|
| Wortmannin |  |
1.8 ~ 4.0 nM |
6 µM by counting GFP- 2xFYVE puncta; 30 nM by overall proteolysis in rat hepatocytes |
Wortmannin and its analogues are not inhibitors of Ptdlns-4-kinase (at conc. up to 32 µM). |
Wortmannin and its analogues are not inhibitors of protein kinase C (at conc. up to 22 µM), c-Src tyrosine kinase (at conc. up to 5.5 µM), and phosphoinositide- specific phospholipase C (at conc. up to 0.27 mM). |
[1–4] | |
|
3-methyladenine (3-MA) |
 |
~ 1 mM (at 50 µM ATP) |
616 µM by counting GFP- 2xFYVE puncta; 5 mM 3-MA inhibited 60% proteolysis in hepatocytes. |
[1,3,5] | |||
|
LY294002 (2-(4-morpholinyl)-8- phenylchromone) |
 |
1.4 ~ 4.2 µM |
10 µM by overall proteolysis in rat hepatocytes |
[2,3,6] | |||
| PIK-III |  |
18 nM | 55 µM by counting GFP- 2xFYVE puncta; |
>1 µM for PI(3)Kα, PI(3)Kβ, PI(3)Kγ, PI(3)Kδ, PI4Kβ. |
>9.1 µM for mTOR | [1] | Novartis |
|
VPS34-IN1 (1-[{2-[(2- chloropyridin- 4yl)amino]-4’- (cyclopropylmethyl)- [4,5’-bipyrimidin]-2’- yl}amino]-2- methylpropan- 2-ol) (CAS registry number 1383716-33-3) |
 |
25 nM | 100 nM by counting GFP- 2xFYVE puncta; 100 nM ~ 1 µM by SGK3 activity and T- loop/hydrophobic motif phosphorylation |
>1 µM for other lipid kinases in the Dundee panel (19 lipid kinases, includes class I PI(3)Ks), the AstraZeneca panel (8 lipid kinases, includes class I) and ProQuinase panel (13 lipid kinases, includes class I and class II PI(3)Ks); did not significantly inhibit any of the lipid kinases tested including class I (p110α, p110β p110γ and p110δ) and all three members of the class II (PI(3)KC2α, PI(3)KC2β, PI(3)KC2γ) PI(3)Ks. |
1 µM did not significantly inhibit the activity of any of the protein kinases in the Dundee panel (140 kinases) and the ProQinase panel (300 kinases) |
[7] | Novartis (WO 201208 5815 A1) |
|
SAR405 ((8s)-9-[(5- Chloranylpyridin-3- yl)methyl]-2-[(3r)-3- Methylmorpholin-4- yl]-8- (Trifluoromethyl)- 6,7,8,9a-Tetrahydro- 3h-Pyrimido[1,2- A]pyrimidin-4-One) (CAS registry number 1523406-39-4) |
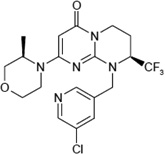 |
1.2 nM (KD=1.5 µM) |
27 nM by counting GFP- 2xFYVE puncta; 419 nM by counting % of cells with GFP- LC3 dots in response to starvation |
In vitro > 10 µM for class I PI(3)Ks (p110α, p110β, p110γ and p110δ) and class II PI(3)Ks (PI(3)KC2α, PI(3)KC2β and PI(3)KC2γ); In vivo: 1 µM leads to 39%, 68%, and 63% inhibition of p110α, p110β and p110δ, respectively. |
In vitro > 10 µM for mTOR; In vivo: 1 µM leads to 52% inhibition of SMG1, but not any other protein kinases. 10 µM did not affect Akt, S6, p53, and phosphorylation. |
[2] | Sanofi |
|
Compound 31 ((2S)-8-[(3R)-3- Methylmorpholin-4- yl]-1-(3-methyl-2- oxobutyl)-2- (trifluoromethyl)-3,4- dihydro-2Hpyrimido[ 1,2-a]pyrimidin-6- one) |
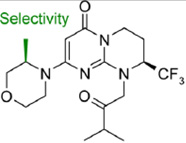 |
2 nM (KD= 2.7 ± 0.9 nM) |
82 nM by counting GFP- 2xFYVE puncta; |
In vitro > 2 µM for class I PI(3)Ks (p110α, p110β, p110γ and p110δ); and > 10 µM for class II PI(3)Ks (PI(3)KC2α and PI(3)KC2γ); |
In vitro > 10 µM for mTOR; In vivo: 8.5 µM for Akt (@Ser473); |
[8] | Sanofi |
References:
. Dowdle, W.E., B. Nyfeler, J. Nagel, R.A. Elling, S. Liu, E. Triantafellow, … L.O. Murphy, Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol, 2014. 16(11): p. 1069–1079.
. Ronan, B., O. Flamand, L. Vescovi, C. Dureuil, L. Durand, F. Fassy, … B. Pasquier, A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol, 2014. 10(12): p. 1013–1019.
Blommaart, E.F., U. Krause, J.P. Schellens, H. Vreeling-Sindelarova, and A.J. Meijer, The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem, 1997. 243(1–2): p. 240–246.
. Powis, G., R. Bonjouklian, M.M. Berggren, A. Gallegos, R. Abraham, C. Ashendel, … et al., Wortmannin, a potent and selective inhibitor of phosphatidylinositol-3-kinase. Cancer Res, 1994. 54(9): p. 2419–2423.
. Seglen, P.O. and P.B. Gordon, 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A, 1982. 79(6): p. 1889–1892.
. Vlahos, C.J., W.F. Matter, K.Y. Hui, and R.F. Brown, A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem, 1994. 269(7): p. 5241–5248.
. Bago, R., N. Malik, M.J. Munson, A.R. Prescott, P. Davies, E. Sommer, … D.R. Alessi, Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J, 2014. 463(3): p. 413–427.
. Pasquier, B., Y. El-Ahmad, B. Filoche-Romme, C. Dureuil, F. Fassy, P.Y. Abecassis, … B. Ronan, Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors. J Med Chem, 2015. 58(1): p. 376–400.
The development of these new Vps34 inhibitors subsequently led to the identification of novel Vps34 targets and clarification of the kinase-dependent biological functions of Vps34 [164,125–127]. The catalytic function of Vps34 remained elusive until a recent report by Bago et al. [164]. Utilizing the specific Vps34 inhibitor Vps34-IN1, Bago and colleagues showed that Vps34 controls the activity of SGK3 (serum- and glucocorticoid-regulated protein kinase 3), a PtdIns(3)P effector, by targeting SGK3 to the endosome where it is subsequently phosphorylated [164]. In a second study, using the specific Vps34 inhibitor PIK-III, Dowdle and colleagues revealed a novel autophagy substrate, NCOA4, which is required for ferritin degradation in lysosomes and for iron homeostasis [125]. A third ATP-competitive inhibitor of Vps34, SAR405, was reported to block starvation- and the ATP-competitive mTOR inhibitor AZD8055-induced autophagy by preventing autophagosome formation, as monitored by GFP-LC3 puncta and LC3II levels in the presence of hydroxychloroquine [126]. SAR405 was also reported to block vesicle trafficking, as shown by swollen LysoTracker- and Lamp1-positive late endosomes/lysosomes as well as impaired cathepsin D maturation without altering cellular uptake of dextran [126]. Further investigations are necessary to determine if inhibition of Vps34 PI(3)K activity utilizing these inhibitors will provide a useful therapeutic strategy for clinical use, either alone or in combination with other therapeutic agents. For example, evidence reveals a role for Vps34 in mTOR activation [151–157], providing the potential to combine a Vps34 inhibitor with a mTOR inhibitor to better combat disease [126].
5.2. Drug design targeting the stability of the Beclin 1-Vps34 complexes
Besides directly targeting the Vps34 lipid kinase activity as described above, drugs can be designed to allosterically impair the Vps34 PI(3)K activity. One such approach is to modulate the stability of the Beclin 1-Vps34 complex. For example, spautin-1 administration is shown to block the activities of the ubiquitin-specific peptidase 10 and 13 (i.e., USP10 and USP13, which promote the de-ubiquitinylation and stability of Beclin 1), thereby leading to degradation of Beclin 1 and the Beclin 1-Vps34 complexes thus inhibition of autophagy [165].
5.3. Drug design targeting protein-protein interactions within the Beclin 1-Vps34 complexes
A common approach to alter Vps34 activity is to modulate the Beclin 1-Vps34 protein complex architecture, such as the availability of Beclin 1 for complex formation. A general mechanism of autophagy regulation repeatedly emerging from protein-protein interaction studies is the sequestration of Beclin 1 by negative regulators until autophagy induction signals are received. These negative regulators include Bcl-2 [15,92], Bcl-XL [92,112,113], M11 [16,17], and GAPR-1 [96].
Viral interference with the autophagic process prevents the antimicrobial host defense mechanism, and allows the virus to flourish. In some cases, virulence gene products block autophagy initiation through their interaction with Beclin 1 [13,166]. Mounting evidence has already suggested that development of pharmacological agents aimed at increasing autophagy for treatment of bacterial, parasitic and viral infections may prove to be an effective therapeutic strategy. In these instances, BH3 mimetics disrupt the inhibitory interaction between the BH3 domain of Beclin 1 and Bcl-2/Bcl-XL, leading to Beclin 1-dependent allosteric activation of the pro-autophagic lipid kinase Vps34, ultimately leading to autophagy induction [167]. Similarly, Tat-Beclin 1 peptide disrupts the inhibitory interaction between GAPR-1 and Beclin 1, leading to Beclin 1-dependent Vps34 activation and autophagy induction. These processes have been shown to decrease the in vitro replication of several pathogens, including HIV-1, Sindbis virus, chikungunya virus, West Nile virus, and the intracellular bacterium, Listeria monocytogenes, and have reduced mortality in mice infected with chikungunya or West Nile virus [96].
In other cases, bacteria exploit the host autophagy initiation proteins, e.g., Ulk1, Beclin 1, Atg14L, and Vps34, to survive [168]. In these instances, inhibitors that target either the pathogen’s interaction with the Beclin 1-Vps34 complexes, or individual proteins that comprise the complexes, might attenuate the pathogen’s attack on the autophagy process. In addition, post-translational modifications of Beclin 1-Vps34 complex components (e.g., Beclin 1, Vps34, Atg14L) also modulate Beclin 1 complex architecture so as to regulate autophagy.
5.4. Challenges
Despite intense interest in targeting the Beclin 1-Vps34 complexes and autophagy for therapeutics, challenges remain for such drug design. For example, the Beclin 1-Vps34 complex components, structure, and architecture, as well as the dynamics and regulatory mechanisms of the complexes have not been fully elucidated. In addition, because Beclin 1-Vps34 complexes have multiple functions in autophagosome biogenesis, autophagosome maturation, and trafficking, the design of drugs that specifically target just one of the functions is likely to be challenging. One strategy to cope with this difficulty is to design drugs that specifically target interactions between Atg14L (or UVRAG) and the Beclin 1-Vps34-Vps15 core complex. Moreover, although counting GFP-2xFYVE puncta is a commonly used readout for PtdIns(3)P production, a more autophagy-specific readout (e.g., counting GFP-DFCP1 or GFPWIPI2 puncta) may be needed for monitoring Atg14L-containing Beclin 1-Vps34 complex activity and for structure-based design in combination with high throughput screen.
There are also challenges that are common for targeting autophagy. For example, therapeutic success is contingent on the timing of the treatment, as cancer development goes through two phases with distinct autophagy contributions. During tumorigenesis, autophagy eliminates oncogenic abnormalities [50], thus needs to be up-regulated to eliminate cancer cells; however at later stages, tumor cells activate autophagy as a survival mechanism, therefore blockage of autophagy is important to suppress tumor growth and metastasis [169,170]. As most cancer treatment is aimed at pre-existing tumors, use of an autophagy inhibitor in combination with chemotherapeutic agents and ionizing radiation might constitute a more promising therapeutic strategy. To date, drugs aimed at specifically inhibiting the Beclin 1-Vps34 complexes do not exist in the clinic. Studies that test the newly developed Vps34 inhibitors should be valuable. Caution is also necessary when designing drugs to target autophagy, as they might possess autophagy-independent effects. Due to autophagy’s important role in controlling infection and immunity, drugs that inhibit autophagy might cause adverse secondary symptoms.
As a deeper understanding of the role of autophagy in disease unfolds, it is expected that the protein interactions that regulate the relevant process will be more fully elucidated. Manipulation of these autophagy proteins (e.g., the Beclin 1-Vps34 complex components) or their gene expression has the potential to combat human diseases. With specific protein-protein interactions known and the development of more powerful computational capabilities, including protein-protein docking studies, the structural basis required to attain optimal drug design for complex protein-protein networks like the Beclin 1-interactome is coming into view [171]. With evolving technologies, development of synthetic peptides or small molecule drugs that selectively target specific binding/modification sites on the proteins mentioned in this review could prove beneficial in therapies that either up- or down-regulate autophagy.
Acknowledgment
This work was supported, in whole or in part, by the Ellison Medical Foundation (to Q.J.W.), by the University of Kentucky Research Postdoctoral Fellowship (to D.H.M.), by National Institutes of Health Grants P41 GM103314 and GM109824 (to B.T.C.), and by an operating grant from the Canadian Institutes of Health Research (MOP-126126) and startup funds from the University of British Columbia (to C.K.Y.). We apologize to those authors whose work could not be cited due to space limitations.
Footnotes
Note added in proof
While this review was in proof, Roger Williams' group reported the 4.4 Å crystal structure of the yeast Vps38-containing Atg6/Vps30-Vps34 complex [172]. This new work provides exciting atomic-resolution insights into the architectures of the Beclin 1-Vps34 complexes and future structure-based drug design.
Compliance with ethics guidelines
Deanna Morris, Calvin Yip, Yi Shi, Brian Chait and Qingjun Wang declare that they have no conclict of interest. This manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee.
References
- 1.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 2.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 3.Kunz JB, Schwarz H, Mayer A. Determination of four sequential stages during microautophagy in vitro. J Biol Chem. 2004;279(11):9987–9996. doi: 10.1074/jbc.M307905200. [DOI] [PubMed] [Google Scholar]
- 4.Uttenweiler A, Schwarz H, Neumann H, Mayer A. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol Biol Cell. 2007;18(1):166–175. doi: 10.1091/mbc.E06-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massey AC, Kaushik S, Cuervo AM. Lysosomal chat maintains the balance. Autophagy. 2006;2(4):325–327. doi: 10.4161/auto.3090. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581(11):2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 7.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Hurley JH, Schulman BA. Atomistic autophagy: the structures of cellular self-digestion. Cell. 2014;157(2):300–311. doi: 10.1016/j.cell.2014.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chew LH, Yip CK. Structural biology of the macroautophagy machinery. Frontiers in Biology. 2014;9(1):18–34. doi: 10.1007/s11515-014-1293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 11.Arroyo DS, Gaviglio EA, Peralta Ramos JM, Bussi C, Rodriguez-Galan MC, Iribarren P. Autophagy in inflammation, infection, neurodegeneration and cancer. Int Immunopharmacol. 2014;18(1):55–65. doi: 10.1016/j.intimp.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munz C. Beclin-1 targeting for viral immune escape. Viruses. 2011;3(7):1166–1178. doi: 10.3390/v3071166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudchodkar SB, Levine B. Viruses and autophagy. Rev Med Virol. 2009;19(6):359–378. doi: 10.1002/rmv.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orvedahl A, Alexander D, Talloczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1(1):23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Sinha S, Colbert CL, Becker N, Wei Y, Levine B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy. 2008;4(8):989–997. doi: 10.4161/auto.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ku B, Woo JS, Liang C, Lee KH, Hong HS, E X, Kim KS, Jung JU, Oh BH. Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathog. 2008;4(2):e25. doi: 10.1371/journal.ppat.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong CS, Wu L, Kominami E, Ueno T, Yamamoto A, Federico M, Panganiban A, Vergne I, Deretic V. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. Journal of Cell Biology. 2009;186(2):255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, Garcia-Sastre A, Munz C. Matrix Protein 2 of Influenza A Virus Blocks Autophagosome Fusion with Lysosomes. Cell Host & Microbe. 2009;6(4):367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou DJ, Spector SA. Human immunodeficiency virus type-1 infection inhibits autophagy. Aids. 2008;22(6):695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, Akira S, Noda T, Yoshimori T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 22.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8(7):688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 24.Liang Q, Seo GJ, Choi YJ, Kwak MJ, Ge J, Rodgers MA, Shi M, Leslie BJ, Hopfner KP, Ha T, Oh BH, Jung JU. Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014;15(2):228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arsov I, Li X, Matthews G, Coradin J, Hartmann B, Simon AK, Sealfon SC, Yue Z. BAC-mediated transgenic expression of fluorescent autophagic protein Beclin 1 reveals a role for Beclin 1 in lymphocyte development. Cell Death Differ. 2008;15(9):1385–1395. doi: 10.1038/cdd.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arsov I, Adebayo A, Kucerova-Levisohn M, Haye J, MacNeil M, Papavasiliou FN, Yue Z, Ortiz BD. A role for autophagic protein beclin 1 early in lymphocyte development. J Immunol. 2011;186(4):2201–2209. doi: 10.4049/jimmunol.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang Q, Chang B, Brulois KF, Castro K, Min CK, Rodgers MA, Shi M, Ge J, Feng P, Oh BH, Jung JU. Kaposi's sarcoma-associated herpesvirus K7 modulates Rubicon-mediated inhibition of autophagosome maturation. J Virol. 2013;87(22):12499–12503. doi: 10.1128/JVI.01898-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang CS, Lee JS, Rodgers M, Min CK, Lee JY, Kim HJ, Lee KH, Kim CJ, Oh B, Zandi E, Yue Z, Kramnik I, Liang C, Jung JU. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe. 2012;11(3):264–276. doi: 10.1016/j.chom.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang CS, Rodgers M, Min CK, Lee JS, Kingeter L, Lee JY, Jong A, Kramnik I, Lin X, Jung JU. The autophagy regulator Rubicon is a feedback inhibitor of CARD9-mediated host innate immunity. Cell Host Microbe. 2012;11(3):277–289. doi: 10.1016/j.chom.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergamini E, Cavallini G, Donati A, Gori Z. The role of autophagy in aging: its essential part in the anti-aging mechanism of caloric restriction. Ann N Y Acad Sci. 2007;1114:69–78. doi: 10.1196/annals.1396.020. [DOI] [PubMed] [Google Scholar]
- 31.Cuervo AM. Calorie restriction and aging: the ultimate "cleansing diet". J Gerontol A Biol Sci Med Sci. 2008;63(6):547–549. doi: 10.1093/gerona/63.6.547. [DOI] [PubMed] [Google Scholar]
- 32.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301(5638):1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 34.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, MacDonald M, Yankner B, Yuan J. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281(20):14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 35.Terman A. The effect of age on formation and elimination of autophagic vacuoles in mouse hepatocytes. Gerontology. 1995;41(Suppl 2):319–326. doi: 10.1159/000213753. [DOI] [PubMed] [Google Scholar]
- 36.Terman A, Gustafsson B, Brunk UT. The lysosomal-mitochondrial axis theory of postmitotic aging and cell death. Chem Biol Interact. 2006;163(1–2):29–37. doi: 10.1016/j.cbi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur J Biochem. 2002;269(8):1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 38.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118(6):2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaeger PA, Pickford F, Sun CH, Lucin KM, Masliah E, Wyss-Coray T. Regulation of Amyloid Precursor Protein Processing by the Beclin 1 Complex. PLoS One. 2010;5(6) doi: 10.1371/journal.pone.0011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucin KM, O'Brien CE, Bieri G, Czirr E, Mosher KI, Abbey RJ, Mastroeni DF, Rogers J, Spencer B, Masliah E, Wyss-Coray T. Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer's disease. Neuron. 2013;79(5):873–886. doi: 10.1016/j.neuron.2013.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arena G, Gelmetti V, Torosantucci L, Vignone D, Lamorte G, De Rosa P, Cilia E, Jonas EA, Valente EM. PINK1 protects against cell death induced by mitochondrial depolarization, by phosphorylating Bcl-xL and impairing its pro-apoptotic cleavage. Cell Death Differ. 2013;20(7):920–930. doi: 10.1038/cdd.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choubey V, Cagalinec M, Liiv J, Safiulina D, Hickey MA, Kuum M, Liiv M, Anwar T, Eskelinen EL, Kaasik A. BECN1 is involved in the initiation of mitophagy: it facilitates PARK2 translocation to mitochondria. Autophagy. 2014;10(6):1105–1119. doi: 10.4161/auto.28615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, Gindi R, Adame A, Wyss-Coray T, Masliah E. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson's and Lewy body diseases. J Neurosci. 2009;29(43):13578–13588. doi: 10.1523/JNEUROSCI.4390-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nassif M, Valenzuela V, Rojas-Rivera D, Vidal R, Matus S, Castillo K, Fuentealba Y, Kroemer G, Levine B, Hetz C. Pathogenic role of BECN1/Beclin 1 in the development of amyotrophic lateral sclerosis. Autophagy. 2014;10(7):1256–1271. doi: 10.4161/auto.28784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59(1):59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 46.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 47.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Mol Cancer Res. 2014;12(4):485–490. doi: 10.1158/1541-7786.MCR-13-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25(8):795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim MS, Jeong EG, Ahn CH, Kim SS, Lee SH, Yoo NJ. Frameshift mutation of UVRAG, an autophagy-related gene, in gastric carcinomas with microsatellite instability. Hum Pathol. 2008;39(7):1059–1063. doi: 10.1016/j.humpath.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 52.Darabi H, McCue K, Beesley J, Michailidou K, Nord S, Kar S, Humphreys K, Thompson D, Ghoussaini M, Bolla MK, Dennis J, Wang Q, Canisius S, Scott CG, Apicella C, Hopper JL, Southey MC, Stone J, Broeks A, Schmidt MK, Scott RJ, Lophatananon A, Muir K, Beckmann MW, Ekici AB, Fasching PA, Heusinger K, Dos-Santos-Silva I, Peto J, Tomlinson I, Sawyer EJ, Burwinkel B, Marme F, Guenel P, Truong T, Bojesen SE, Flyger H, Benitez J, Gonzalez-Neira A, Anton-Culver H, Neuhausen SL, Arndt V, Brenner H, Engel C, Meindl A, Schmutzler RK, B. German Consortium of Hereditary. Ovarian C, Arnold N, Brauch H, Hamann U, Chang-Claude J, Khan S, Nevanlinna H, Ito H, Matsuo K, Bogdanova NV, Dork T, Lindblom A, Margolin S, kConFab AI, Kosma VM, Mannermaa A, Tseng CC, Wu AH, Floris G, Lambrechts D, Rudolph A, Peterlongo P, Radice P, Couch FJ, Vachon C, Giles GG, McLean C, Milne RL, Dugue PA, Haiman CA, Maskarinec G, Woolcott C, Henderson BE, Goldberg MS, Simard J, Teo SH, Mariapun S, Helland A, Haakensen V, Zheng W, Beeghly-Fadiel A, Tamimi R, Jukkola-Vuorinen A, Winqvist R, Andrulis IL, Knight JA, Devilee P, Tollenaar RA, Figueroa J, Garcia-Closas M, Czene K, Hooning MJ, Tilanus-Linthorst M, Li J, Gao YT, Shu XO, Cox A, Cross SS, Luben R, Khaw KT, Choi JY, Kang D, Hartman M, Lim WY, Kabisch M, Torres D, Jakubowska A, Lubinski J, McKay J, Sangrajrang S, Toland AE, Yannoukakos D, Shen CY, Yu JC, Ziogas A, Schoemaker MJ, Swerdlow A, Borresen-Dale AL, Kristensen V, French JD, Edwards SL, Dunning AM, Easton DF, Hall P, Chenevix-Trench G. Polymorphisms in a Putative Enhancer at the 10q21.2 Breast Cancer Risk Locus Regulate NRBF2 Expression. Am J Hum Genet. 2015;97(1):22–34. doi: 10.1016/j.ajhg.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martell RE, Brooks DG, Wang Y, Wilcoxen K. Discovery of novel drugs for promising targets. Clin Ther. 2013;35(9):1271–1281. doi: 10.1016/j.clinthera.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Bai L, Wang S. Targeting apoptosis pathways for new cancer therapeutics. Annu Rev Med. 2014;65:139–155. doi: 10.1146/annurev-med-010713-141310. [DOI] [PubMed] [Google Scholar]
- 55.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55(2):238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6(4):506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 57.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci U S A. 2011;108(19):7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simonsen A, Birkeland HC, Gillooly DJ, Mizushima N, Kuma A, Yoshimori T, Slagsvold T, Brech A, Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004;117(Pt 18):4239–4251. doi: 10.1242/jcs.01287. [DOI] [PubMed] [Google Scholar]
- 60.Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, Zhong Q. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011;286(1):185–191. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A. 2010;107(45):19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188(2):253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009;28(15):2244–2258. doi: 10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010;11(4):468–478. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- 65.Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, Zvelebil MJ, Domin J, Panaretou C, Waterfield MD. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14(14):3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2(4):330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152(1–2):290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thoresen SB, Pedersen NM, Liestol K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res. 2010;316(20):3368–3378. doi: 10.1016/j.yexcr.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 69.Zhong Y, Morris DH, Jin L, Patel MS, Karunakaran SK, Fu YJ, Matuszak EA, Weiss HL, Chait BT, Wang QJ. Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels. J Biol Chem. 2014;289(38):26021–26037. doi: 10.1074/jbc.M114.561134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McKnight NC, Zhong Y, Wold MS, Gong S, Phillips GR, Dou Z, Zhao Y, Heintz N, Zong WX, Yue Z. Beclin 1 is required for neuron viability and regulates endosome pathways via the UVRAG-VPS34 complex. PLoS Genet. 2014;10(10):e1004626. doi: 10.1371/journal.pgen.1004626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152(3):519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190(4):511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 76.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6(6):764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fogel AI, Dlouhy BJ, Wang C, Ryu SW, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol. 2013;33(18):3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, Pfuetzner RA, Brunger AT, Zhong Q. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520(7548):563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol. 2008;10(7):776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim YM, Jung CH, Seo M, Kim EK, Park JM, Bae SS, Kim DH. mTORC1 Phosphorylates UVRAG to Negatively Regulate Autophagosome and Endosome Maturation. Mol Cell. 2015;57(2):207–218. doi: 10.1016/j.molcel.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Munson MJ, Allen GF, Toth R, Campbell DG, Lucocq JM, Ganley IG. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J. 2015;34(17):2272–2290. doi: 10.15252/embj.201590992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nemazanyy I, Montagnac G, Russell RC, Morzyglod L, Burnol AF, Guan KL, Pende M, Panasyuk G. Class III PI3K regulates organismal glucose homeostasis by providing negative feedback on hepatic insulin signalling. Nat Commun. 2015;6:8283. doi: 10.1038/ncomms9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mule JJ, Pledger WJ, Wang HG. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9(10):1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takats S, Pircs K, Nagy P, Varga A, Karpati M, Hegedus K, Kramer H, Kovacs AL, Sass M, Juhasz G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25(8):1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM. NRBF2 regulates macroautophagy as a component of Vps34 complex I. Biochem J. 2014;461:315–322. doi: 10.1042/BJ20140515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu J, He L, Behrends C, Araki M, Araki K, Wang QJ, Catanzaro JM, Friedman SL, Zong WX, Fiel MI, Li M, Yue Z. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun. 2014;5:3920–3934. doi: 10.1038/ncomms4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Flores AM, Li L, Aneskievich BJ. Isolation and functional analysis of a keratinocyte-derived, ligand-regulated nuclear receptor comodulator. J Invest Dermatol. 2004;123(6):1092–1101. doi: 10.1111/j.0022-202X.2004.23424.x. [DOI] [PubMed] [Google Scholar]
- 89.Yasumo H, Masuda N, Furusawa T, Tsukamoto T, Sadano H, Osumi T. Nuclear receptor binding factor-2 (NRBF-2), a possible gene activator protein interacting with nuclear hormone receptors. Biochim Biophys Acta. 2000;1490(1–2):189–197. doi: 10.1016/s0167-4781(99)00244-4. [DOI] [PubMed] [Google Scholar]
- 90.Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203(2):299–313. doi: 10.1083/jcb.201304123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Noble CG, Dong JM, Manser E, Song H. Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin1. J Biol Chem. 2008;283(38):26274–26282. doi: 10.1074/jbc.M804723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26(10):2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10(3):285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30(6):678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, Grishin NV, Peyton M, Minna J, Bhagat G, Levine B. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013;154(6):1269–1284. doi: 10.1016/j.cell.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, Kinch L, Wilkins AD, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NV, Spector SA, Kaloyanova DV, Levine B. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494(7436):201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, Seux M, Nowak J, Gonzalez CD, Iovanna JL, Vaccaro MI. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem. 2007;282(51):37124–37133. doi: 10.1074/jbc.M706956200. [DOI] [PubMed] [Google Scholar]
- 98.Molejon MI, Ropolo A, Re AL, Boggio V, Vaccaro MI. The VMP1-Beclin 1 interaction regulates autophagy induction. Sci Rep. 2013;3:1055. doi: 10.1038/srep01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T, Tooze SA. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 2012 doi: 10.1038/emboj.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Nardacci R, Piacentini M, Campanella M, Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2014;22(3):419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, Piacentini M, Levine B, Cecconi F. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30(7):1195–1208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Van Humbeeck C, Cornelissen T, Hofkens H, Mandemakers W, Gevaert K, De Strooper B, Vandenberghe W. Parkin interacts with Ambra1 to induce mitophagy. J Neurosci. 2011;31(28):10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447(7148):1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 104.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, Cecconi F. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15(4):406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 105.Ma B, Cao W, Li W, Gao C, Qi Z, Zhao Y, Du J, Xue H, Peng J, Wen J, Chen H, Ning Y, Huang L, Zhang H, Gao X, Yu L, Chen YG. Dapper1 promotes autophagy by enhancing the Beclin1-Vps34-Atg14L complex formation. Cell Res. 2014;24(8):912–924. doi: 10.1038/cr.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dou Z, Chattopadhyay M, Pan JA, Guerriero JL, Jiang YP, Ballou LM, Yue Z, Lin RZ, Zong WX. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol. 2010;191(4):827–843. doi: 10.1083/jcb.201006056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ravikumar B, Imarisio S, Sarkar S, O'Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121(Pt 10):1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dou Z, Pan JA, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, Chen JS, Liang Z, Li G, Backer JM, Lin RZ, Zong WX. Class IA PI3K p110beta subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell. 2013;50(1):29–42. doi: 10.1016/j.molcel.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, Camonis JH, Yeaman C, Levine B, White MA. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144(2):253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Joffre C, Dupont N, Hoa L, Gomez V, Pardo R, Goncalves-Pimentel C, Achard P, Bettoun A, Meunier B, Bauvy C, Cascone I, Codogno P, Fanto M, Hergovich A, Camonis J. The Pro-apoptotic STK38 Kinase Is a New Beclin1 Partner Positively Regulating Autophagy. Curr Biol. 2015;25(19):2479–2492. doi: 10.1016/j.cub.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72(11):8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282(17):13123–13132. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 113.Feng W, Huang S, Wu H, Zhang M. Molecular basis of Bcl-xL's target recognition versatility revealed by the structure of Bcl-xL in complex with the BH3 domain of Beclin-1. J Mol Biol. 2007;372(1):223–235. doi: 10.1016/j.jmb.2007.06.069. [DOI] [PubMed] [Google Scholar]
- 114.Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, Zhang M, Yue Z, Zhao Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J Biol Chem. 2012;287(20):16256–16266. doi: 10.1074/jbc.M112.348250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, Deng H, Liu L, Gao N, Yu L, Shi Y. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22(3):473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1(1):46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 118.Su M, Mei Y, Sanishvili R, Levine B, Colbert CL, Sinha S. Targeting gamma-herpesvirus 68 Bcl-2-mediated down-regulation of autophagy. J Biol Chem. 2014;289(12):8029–8040. doi: 10.1074/jbc.M113.515361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260(5104):88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- 120.Herman PK, Emr SD. Characterization of VPS34, a gene required for vacuolar protein sorting and vacuole segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10(12):6742–6754. doi: 10.1128/mcb.10.12.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410(1):1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- 122.Budovskaya YV, Hama H, DeWald DB, Herman PK. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J Biol Chem. 2002;277(1):287–294. doi: 10.1074/jbc.M109263200. [DOI] [PubMed] [Google Scholar]
- 123.Siddhanta U, McIlroy J, Shah A, Zhang Y, Backer JM. Distinct roles for the p110alpha and hVPS34 phosphatidylinositol 3'-kinases in vesicular trafficking, regulation of the actin cytoskeleton, and mitogenesis. J Cell Biol. 1998;143(6):1647–1659. doi: 10.1083/jcb.143.6.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, Williams RL. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327(5973):1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, Cantwell J, Luu C, Cornella-Taracido I, Harrington E, Fekkes P, Lei H, Fang Q, Digan ME, Burdick D, Powers AF, Helliwell SB, D'Aquin S, Bastien J, Wang H, Wiederschain D, Kuerth J, Bergman P, Schwalb D, Thomas J, Ugwonali S, Harbinski F, Tallarico J, Wilson CJ, Myer VE, Porter JA, Bussiere DE, Finan PM, Labow MA, Mao X, Hamann LG, Manning BD, Valdez RA, Nicholson T, Schirle M, Knapp MS, Keaney EP, Murphy LO. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16(11):1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 126.Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, Marquette JP, El-Ahmad Y, Filoche-Romme B, Schio L, Garcia-Echeverria C, Goulaouic H, Pasquier B. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014;10(12):1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 127.Pasquier B, El-Ahmad Y, Filoche-Romme B, Dureuil C, Fassy F, Abecassis PY, Mathieu M, Bertrand T, Benard T, Barriere C, El Batti S, Letallec JP, Sonnefraud V, Brollo M, Delbarre L, Loyau V, Pilorge F, Bertin L, Richepin P, Arigon J, Labrosse JR, Clement J, Durand F, Combet R, Perraut P, Leroy V, Gay F, Lefrancois D, Bretin F, Marquette JP, Michot N, Caron A, Castell C, Schio L, McCort G, Goulaouic H, Garcia-Echeverria C, Ronan B. Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors. J Med Chem. 2015;58(1):376–400. doi: 10.1021/jm5013352. [DOI] [PubMed] [Google Scholar]
- 128.Herman PK, Stack JH, DeModena JA, Emr SD. A novel protein kinase homolog essential for protein sorting to the yeast lysosome-like vacuole. Cell. 1991;64(2):425–437. doi: 10.1016/0092-8674(91)90650-n. [DOI] [PubMed] [Google Scholar]
- 129.Herman PK, Stack JH, Emr SD. A genetic and structural analysis of the yeast Vps15 protein kinase: evidence for a direct role of Vps15p in vacuolar protein delivery. EMBO J. 1991;10(13):4049–4060. doi: 10.1002/j.1460-2075.1991.tb04981.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12(5):2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. J Biol Chem. 1997;272(4):2477–2485. doi: 10.1074/jbc.272.4.2477. [DOI] [PubMed] [Google Scholar]
- 132.Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J. 2009;417(3):747–755. doi: 10.1042/BJ20081865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lindmo K, Brech A, Finley KD, Gaumer S, Contamine D, Rusten TE, Stenmark H. The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy. 2008;4(4):500–506. doi: 10.4161/auto.5829. [DOI] [PubMed] [Google Scholar]
- 134.Heenan EJ, Vanhooke JL, Temple BR, Betts L, Sondek JE, Dohlman HG. Structure and function of Vps15 in the endosomal G protein signaling pathway. Biochemistry. 2009;48(27):6390–6401. doi: 10.1021/bi900621w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1(4):249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 136.Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic. 2002;3(6):416–427. doi: 10.1034/j.1600-0854.2002.30605.x. [DOI] [PubMed] [Google Scholar]
- 137.Cao C, Backer JM, Laporte J, Bedrick EJ, Wandinger-Ness A. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol Biol Cell. 2008;19(8):3334–3346. doi: 10.1091/mbc.E08-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Cao C, Laporte J, Backer JM, Wandinger-Ness A, Stein MP. Myotubularin lipid phosphatase binds the hVPS15/hVPS34 lipid kinase complex on endosomes. Traffic. 2007;8(8):1052–1067. doi: 10.1111/j.1600-0854.2007.00586.x. [DOI] [PubMed] [Google Scholar]
- 139.Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129(2):321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449(7163):735–U711. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 141.Stuchell-Brereton MD, Skalicky JJ, Kieffer C, Karren MA, Ghaffarian S, Sundquist WI. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449(7163):740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 142.Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Developmental Cell. 2008;15(1):62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 143.Ciccarelli FD, Proukakis C, Patel H, Cross H, Azam S, Patton MA, Bork P, Crosby AH. The identification of a conserved domain in both spartin and spastin, mutated in hereditary spastic paraplegia. Genomics. 2003;81(4):437–441. doi: 10.1016/s0888-7543(03)00011-9. [DOI] [PubMed] [Google Scholar]
- 144.Phillips SA, Barr VA, Haft DH, Taylor SI, Haft CR. Identification and characterization of SNX15, a novel sorting nexin involved in protein trafficking. Journal of Biological Chemistry. 2001;276(7):5074–5084. doi: 10.1074/jbc.M004671200. [DOI] [PubMed] [Google Scholar]
- 145.Shi Y, Pellarin R, Fridy PC, Fernandez-Martinez J, Thompson MK, Li Y, Wang QJ, Sali A, Rout MP, Chait BT. A strategy for dissecting the architectures of native macromolecular assemblies. Nat Methods. 2015 doi: 10.1038/nmeth.3617. (Accepted 20 August, published online 5 October, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baskaran S, Carlson LA, Stjepanovic G, Young LN, Kim do J, Grob P, Stanley RE, Nogales E, Hurley JH. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. Elife. 2014;3:e05115. doi: 10.7554/eLife.05115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2(5):561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 148.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305(5682):399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 149.Burgess MR, Skaggs BJ, Shah NP, Lee FY, Sawyers CL. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc Natl Acad Sci U S A. 2005;102(9):3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6(7):569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 151.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280(38):33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 152.Xu L, Salloum D, Medlin PS, Saqcena M, Yellen P, Perrella B, Foster DA. Phospholipase D mediates nutrient input to mammalian target of rapamycin complex 1 (mTORC1) J Biol Chem. 2011;286(29):25477–25486. doi: 10.1074/jbc.M111.249631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yoon MS, Du G, Backer JM, Frohman MA, Chen J. Class III PI-3-kinase activates phospholipase D in an amino acid-sensing mTORC1 pathway. J Cell Biol. 2011;195(3):435–447. doi: 10.1083/jcb.201107033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102(40):14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7(5):456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, Zong WX. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109(6):2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhou X, Takatoh J, Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PLoS One. 2011;6(1):e16358. doi: 10.1371/journal.pone.0016358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Slessareva JE, Routt SM, Temple B, Bankaitis VA, Dohlman HG. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell. 2006;126(1):191–203. doi: 10.1016/j.cell.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 159.Powis G, Bonjouklian R, Berggren MM, Gallegos A, Abraham R, Ashendel C, Zalkow L, Matter WF, Dodge J, Grindey G, et al. Wortmannin, a potent and selective inhibitor of phosphatidylinositol-3-kinase. Cancer Res. 1994;54(9):2419–2423. [PubMed] [Google Scholar]
- 160.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243(1–2):240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 161.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79(6):1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269(7):5241–5248. [PubMed] [Google Scholar]
- 163.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010;285(14):10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, Alessi DR. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J. 2014;463(3):413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, Jin M, Zhu Z, Wang H, Yu J, Hao Y, Choi A, Ke H, Ma D, Yuan J. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147(1):223–234. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, Geneste O, Kroemer G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3(4):374–376. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 168.Starr T, Child R, Wehrly TD, Hansen B, Hwang S, Lopez-Otin C, Virgin HW, Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11(1):33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mathew R, White E. Autophagy in tumorigenesis and energy metabolism: friend by day, foe by night. Curr Opin Genet Dev. 2011;21(1):113–119. doi: 10.1016/j.gde.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, Kalaany NY, Jacks T, Chan CS, Rabinowitz JD, White E. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4(8):914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vakser IA. Protein-protein docking: from interaction to interactome. Biophys J. 2014;107(8):1785–1793. doi: 10.1016/j.bpj.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350(6257):aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]