Abstract
Background
Silence of the tumor suppressor miR-34c is implicated in the development of colorectal cancer (CRC). For the past few years, Resveratrol (Res) has been introduced to oncotherapies alone or with traditional chemotherapeutic drugs. However, the study of molecular mechanism involved in the anti-CRC effect of Res is still ongoing.
Methods
The anti-CRC effect of Res alone or with Oxaliplatin (Oxa) was determined by cell viability assay, soft agar colony formation assay, flow cytometry and real-time cellular analyzer in HT-29 (p53+) and HCT-116 (p53−) CRC cell lines. Expressions of miR-34c and its targets were detected by qPCR and/or western blot. To evaluate the role of miR-34c in anti-CRC effect by Res alone or with Oxa, miR-34c was up or down-regulated by lentiviral mediation or specific inhibitor, respectively. To investigate how miR-34c was increased by Res, the methylation status of miR-34c promoter was detected by MSP. The tumor bearing mouse model was established by subcutaneous injection of HCT-116 cells to assess anti-CRC effect of Res alone or with Oxa in vivo. IL-6 and TNF-α in xenografts were detected by ELISA.
Results
Res inhibited cell viability, proliferation, migration and invasion as well as promoted apoptosis both in HT-29 and HCT-116 CRC cells. The anti-CRC effect of Res was partially but specifically through up-regulating miR-34c which further knocked down its target KITLG; and the effect was enhanced in the presence of p53 probably through inactivating PI3K/Akt pathway. Besides, Res sensitized CRC cells to Oxa in a miR-34c dependent manner. The xenograft experiments showed that exposure to Res or Oxa suppressed tumor growth; and the efficacy was evidently augmented by the co-treatment of Res and Oxa. Likewise, miR-34c level was elevated in xenografts of Res-treated mice while the KITLG was decreased. Finally, Res clearly reduced IL-6 in xenografts.
Conclusion
Res suppressed CRC by specifically activating miR-34c-KITLG in vitro and in vivo; and the effect was strengthened in the presence of p53. Besides, Res exerted a synergistic effect with Oxa in a miR-34c dependent manner. We also suggested that Res-increased miR-34c could interfere IL-6-triggered CRC progression.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-015-1958-6) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, KITLG, MiR-34c, Oxaliplatin, Resveratrol
Background
The incidence and mortality of colorectal cancer (CRC) rank the top 5 among all malignant neoplasms both in China and western countries. Except for exairesis, chemotherapy is one of the most common treatments for CRC patients, especially those who have distant metastasis. However, long-term use of chemotherapeutic drugs e.g. oxaliplatin (Oxa) can cause several side effects such as hepatotoxicity and neurotoxicity and induce drug resistance [1–3]; thus, it is hard to achieve the expectations. Nowadays, several natural compounds have been introduced in anti-tumor researches and clinical oncotherapies, either alone or combined with traditional chemotherapeutic drugs. Resveratrol (Res) is a natural polyphenolic compound rich in peanuts, red wine and grapes. Apart from the well-documented anti-inflammation and anti-oxidation effects [4, 5], Res has an anti-tumor potential in CRC and other cancers in vitro and in vivo [6–9], excitingly, without apparent side effects.
Efforts have been made to figure out how Res plays the anti-tumor role, which is of importance for the better application of Res in clinic. It has been reported that Res could induce cell apoptosis and cell cycle arrest via p53 pathway and/or caspase/cyclin-CDK pathway to achieve its anti-tumor activities [10, 11]. Identification of the role that microRNAs play in human cancer pathogenesis triggered researches in the regulation of Res on microRNA expressions. Depending on their targets, microRNAs could serve as either oncogenes or tumor suppressing genes. Accumulating evidence showed that Res decreased miR-520 h and subsequently suppressed tumor cell invasion and migration in lung cancer cells [12]. Res also inhibited cancer growth and metastasis of SW480 human CRC cells by inducing miR-663 expression [13]. These observations clearly indicated that microRNAs were involved in the Res-mediated anti-tumor activities.
MiR-34c is suggested to be a candidate of tumor suppressing gene and epigenetically silenced in CRC [14, 15]. We recently found that over-expression of miR-34c induced apoptosis and inhibited proliferation and invasion in CRC cells by silencing its target, stem cell factor (SCF, also known as KITLG) [16], suggesting miR-34c as a promising target for the treatment of CRC patients. Besides, it has been recently raised that Res inhibited human CRC cell growth and induced apoptosis through up-regulating miR-34a, a homologue of miR-34c, implying a possible similar modulation of Res on miR-34c expression [17]. However, whether miR-34c is implicated in the Res-mediated anti-CRC effect has not yet been fully elucidated. Furthermore, how Res synergizes with Oxa in the treatment of CRC needs clarified besides its protection from the Oxa-induced hepatotoxicity and neurotoxicity [18].
In the present study, we provided evidence that Res itself could not only exert significant anti-CRC effect, but also showed a synergistic effect with Oxa in a miR-34c dependent manner.
Methods
Cell culture and reagents
Human CRC cell lines HT-29 (p53+) and HCT-116 (p53−) were purchased from the Cell Bank of Chinese Academy of Sciences. All cell lines were cultured in DMEM medium (Life Technologies, USA) supplemented with 10 % fetal bovine serum (Life Technologies) and 1 % penicillin/streptomycin (Life Technologies). Cells were grown at 37 °C in the presence of 5 % CO2 and treated with Res (Sigma, USA) and/or Oxa (Sigma). DMSO (Sigma) was used as control.
Cell viability assay
Cell viability was detected by cell counting kit-8 (CCK-8, Dojindo Laboratories, Japan) according to the manufacturer’s protocol. Cells were seeded in 96-well plates at 3 × 103 cells per well. 10 μL of the tetrazolium substrate was added to each well. The plates were incubated at 37 °C for 1 h and the absorbance was measured at 450 nm using Multiskan FC (Thermo Scientific, USA). All experiments were done in triplicate and repeated three independent times.
Combination Index (CI) calculation
To assess the drug interactions of Res and Oxa, the CI value defined by median-effect analysis was calculated as follows:
where (D)Res and (D)Oxa are the doses for each drug alone that inhibits 50 % cell viability, and (DX)Res and (DX)Oxa are the doses for Res and Oxa in a combination that inhibits 50 % cell viability. CI < 1 indicates a synergistic effect; CI = 1, additive effect; and CI > 1, antagonistic effect [19].
Soft agar colony formation assay
The bottom of 6-well plate was coated with 0.6 % low melting agarose (Promega, USA) and covered with 0.35 % agarose containing 1000 cells. The plates were incubated at standard incubator condition for 2 weeks. Colonies were counted under inverted phase contrast microscope (Leica DMI3000 B, Germany) in 9 randomly sampled visual fields each well by stereological technique. All experiments were done in triplicate.
Western blot
The harvested cells were suspended in RIPA (Radio-Immunoprecipitation Assay) lysis buffer (Applygen, Beijing, China). After 10 % SDS-PAGE, the proteins were transferred onto PVDF membrane (Merk-Millipore, USA) and blocked with 5 % non-fat dry milk or 5 % bovine serum albumin (Sigma) for 1 h. The membrane was incubated with rabbit anti-KITLG (1:500, Abcam, UK), mouse anti-p53 (1:1000, Cell Signaling Technology, USA), rabbit anti-Akt (1:1000, Cell Signaling Technology) or rabbit anti-p-Akt (1:2000, Cell Signaling Technology) primary antibody at 4 °C overnight. Then, the membrane was incubated with HRP-conjugated secondary goat anti-mouse IgG (1:2000, Santa Cruz, USA) or goat anti-rabbit IgG (1:2000, Santa Cruz) for 1 h at 25 °C. The proteins were detected using ECL chemiluminescence (Thermo Scientific) and viewed in Fusion FX Vilber Lourmat (France). A mouse anti-actin (1:4000, Santa Cruz) antibody was used as an internal control.
Real-time monitoring of cellular proliferation, migration and invasion
We employed real-time cellular analyzer (RTCA, ACEA Biosciences, USA) to monitor cellular proliferation, migration and invasion as we previously described [16]. Briefly, for the proliferation assay, the cells were seeded in E-plates at a density of 8000 cells/well and the cell index (CI) was automatically recorded every 15 min. For the migration assay, the cells were seeded in the upper chambers of CIM-plates in serum-free medium at a density of 20,000 cells/well. The bottom chambers of CIM-plates were filled with serum-containing medium to promote migration across the membranes towards the serum gradient. The CI was registered only from the cells that were capable of migrating through the membranes. The protocol for the invasion assay was identical to that for the migration assay, except that the upper chambers were loaded with 30 μl of 5 % Matrigel (BD Biosciences, USA) solution to create a 3D biomatrix film in each well prior to cell seeding. Four independent experiments were performed respectively. The slope of CI curve was analyzed.
RNA extraction and qPCR
The total microRNA was extracted using miRNApure Mini Kit (CWBiotech, Beijing), according to the manufacturer’s instruction. Reverse transcription was performed using Taqman microRNA RT Kit (Life Technologies) and Taqman microRNA Assay with specific stem-loop primers. Real-time PCR was performed using Taqman Universal Master Mix II (Life Technologies). The reactions were incubated at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min in ABI 7500 real-time PCR system. Results were normalized to the internal control, RNU6B.
The total RNA was extracted using TRIzol reagent (Life Technologies) according to the manufacturer’s instruction. Reverse transcription reactions were performed using High Capacity RNA-to-cDNA Kit (Life Technologies). Real-time PCR was performed in ABI 7500 real-time PCR system using SYBR Green PCR Master Mix (Life Technologies). The primers are listed in Table 1. The reactions were incubated at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min.
Table 1.
Primers
| KITLG | Forward | CAGAGTCAGTGTCACAAAACCATT |
|---|---|---|
| Reverse | TTGGCCTTCCTATTACTGCTACTG | |
| E-cadherin | Forward | TGCCCAGAAAATGAAAAAGG |
| Reverse | GTGTATGTGGCAATGCGTTC | |
| PTEN | Forward | GCTGTGGTTGCCACAAAGTGCC |
| Reverse | GCAGGTAGAAGGCAACTCTGCCA | |
| HOXB3 | Forward | AAAGGCACAAAACACGTTCC |
| Reverse | GGATCTCTCACCATCCCTGA | |
| FGFR1 | Forward | CCCGTAGCTCCATATTGGACA |
| Reverse | TTTGCCATTTTTCAACCAGCG | |
| Pim | Forward | GAGAAGGACCGGATTTCCGAC |
| Reverse | CAGTCCAGGAGCCTAATGACG | |
| GAPDH | Forward | AGAAGGCTGGGGCTCATTTG |
| Reverse | AGGGGCCATCCACAGTCTTC | |
| Unmethylation | Forward | TTTTTATTTGTTTTGTTTTGTGTTTGTTTTG |
| Reverse | CCTAAAACTAACTCTCTCAACCCCA | |
| Methylation | Forward | ATTCGTTTCGTTTCGCGTTCGTTTC |
| Reverse | CTAAAACTAACTCTCTCGACCCCG |
All reverse transcription reactions included no-template controls, and all PCR reactions were run in triplicates. Relative microRNA or mRNA expression was determined using the comparative CT (2-ΔΔCt) method.
Flow cytometry
For cell apoptosis analysis, AlexaFluor® 488 Annexin V/Dead Cell Apoptosis Kit (Life Technologies) was used according to the manufacturer’s instruction. In Brief, cells were seeded in 6-well plates and 1 × 105 cells were resuspended in annexin binding buffer, after which annexin V and propidium iodide (PI, 100 μg/mL) were added and incubated at 25 °C for 15 min in the dark. Cell apoptosis was analyzed in Coulter EPLCS XL (Beckman Coulter, USA). All experiments were done in triplicate.
For cell cycle analysis, 1 × 106 cells were fixed with 70 % ethanol for 2 h at 4 °C, followed by the treatment with phosphate buffered saline containing RNase (50 μg/mL) for 30 min at 37 °C. Cells were stained with PI (500 μg/mL, Sigma) for 30 s in the dark. Cell cycle kinetics were analyzed by measuring the DNA contents in Coulter EPLCS XL. All experiments were done in triplicate.
Over-expression of miR-34c by lentiviral mediation
The full length of pre-miR-34c was chemically synthesized and introduced into GV217 lentiviral vector (GeneChem, Shanghai, China) in the unique EcoRI site to construct a lentivirus encoding miR-34c (Lv-miR-34c). The Lv-miR-34c or its control, Lv-NC, was transfected into CRC cells seeded in 6-well plates when reaching 30 % confluence. After 3 days, the infectious efficiency was evaluated by observing the EGFP-expression with an inverted phase contrast microscope (Leica DMI3000 B, Germany).
MiR-34c knockdown
For knockdown of miR-34c, the specific miR-34c inhibitor was purchased from Ribobio (Guangzhou, China). The inhibitor or its control, inhibitor-NC, was transfected into CRC cells using riboFECT™ CP Transfection Kit (Ribobio) according to the manufacturer’s instruction.
Methylation Specific PCR (MSP)
The genomic DNA of CRC cells was extracted using QIAamp® DNA Mini Kit (Qiagen, USA). 200 ~ 500 ng DNA was subject to bisulfite conversion using EZ DNA Methylation-Gold™ Kit (Zymo Research, USA). The methylation-sensitive PCR was performed using Platinum Taq DNA Polymerase (Life Technologies). The PCR reaction conditions consisted of an initial incubation at 94 °C for 2 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 68 °C for 1 min using verity 96-well thermo cycler (Applied Biosystems). The primers are listed in Table 1. The PCR products were electrophoresed in 0.75 % agarose gel, and visualized by uitraviolet illumination.
Xenograft in BALB/c nude mouse
In order to determine the in vivo anti-CRC effect of Res, the CRC cell xenograft in BALB/c athymic nude mice (3–4 weeks old) were performed. Twenty-eight nude mice were purchased from the Experimental Animal Center in the Capital Medical University and housed under Specific Pathogen Free condition. 5 × 106 HCT-116 cells suspended in 50 μL phosphate buffered saline were subcutaneously injected into the right armpit of the nude mice. Ten days after cell xenograft, the nude mice were randomly grouped (7 mice/group) and received Res (100 mg/kg), Oxa (10 mg/kg), Res (100 mg/kg) + Oxa (10 mg/kg) or DMSO via tail-vein injection every day for 2 weeks based on modification of previous report [18]. The body weight and tumor size were measured every other day. The tumor volume was calculated as: W2 × (L/2), where W represents the tumor width and L the tumor length. The experimental procedures were approved by the Institutional Review Board of the Capital Medical University.
ELISA
The xenografts were lyzed; and the supernatants from xenografts as well as the serum of mice were collected. The concentrations of IL-6 and TNF-α was determined by ELISA according to the manufacturer’s instructions (R&D Systems, USA). The absorbance of each well at 490 nm was read using the microplate reader (Multiskan FC, Thermo Scientific). All experiments were done in triplicates.
Statistics analysis
All statistical analysis were performed using SPSS 13.0 (Chicago, USA). Values are expressed as means ± SEM. Results were analyzed by Student’s t test or one-way ANOVA. The P value < 0.05 was considered to be statistically significant.
Results
In vitro anti-CRC effect of Res
Cell viability was significantly reduced after 24 h-treatment with Res in a dose dependent manner (P < 0.01 ~ 0.001, Fig. 1a). Based on the results, we used 100 μM Res for HT-29 cells and 50 μM Res for HCT-116 cells in the following experiments. Importantly, it has been delineated that Res derivatives had no effect on non-tumor cells (IEC18 intestinal epithelium cells) [20]. By real-time monitoring in RTCA we found that Res decreased cell proliferation by 45.9 % in HT-29 cells (P < 0.05) and 57.4 % in HCT-116 cells (P < 0.05) (Fig. 1b). To find out the mechanism of the Res-induced proliferative suppression, we detected the cell cycle by flow cytometry. HCT-116 cells arrested in G0/G1 phase were elevated by 54.4 % (P < 0.001) after treated with Res, and the percentage of G2/M phase HT-29 cells were elevated by 37.1 % (P < 0.001) (Fig. 1c, Additional file 1: Figure S1), indicating that Res inhibited the proliferation of CRC cells by arresting cell cycle. Res as well increased apoptosis in HT-29 cells by 38.6 % (P < 0.01) and in HCT-116 cells by 45.5 % (P < 0.01) (Fig. 1d, Additional file 2: Figure S2). Effect of Res on the malignant transformation of CRC cells was examined by soft agar colony formation assay. The number and size of colonies in the Res-treated HCT-116 and HT-29 cells were obviously reduced compared with those in controls (P < 0.001, Fig. 1e). Besides, Res significantly suppressed cell migration by 42.2 %, (P < 0.01) and invasion by 51.1 % (P < 0.01) of HCT-116 cells observed by RTCA (Fig. 1f).
Fig. 1.
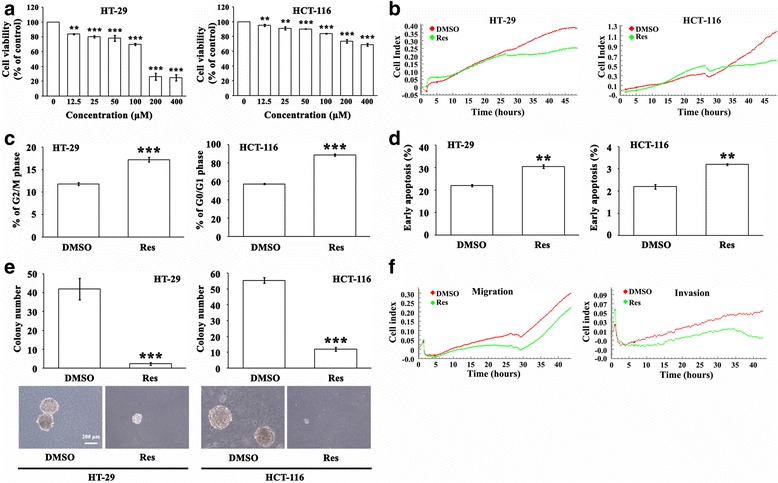
a Twenty-four hr-Res treatment significantly reduced CRC cell viability detected with CCK8 in a dose-dependent manner (12.5 ~ 400 μМ). ** P < 0.01, *** P < 0.001 (b) CRC cell growth was monitored by RTCA for 48 h in the presence of Res or DMSO. Cells treated with Res displayed lower proliferation. c Res induced cell cycle arrest in HT-29 cells (G2/M phase) and HCT-116 cells (G0/G1 phase) analyzed by flow cytometric analysis. *** P < 0.001 (d) Res increased cell apoptosis in CRC cells analyzed by flow cytometric analysis. ** P < 0.01 (e) Soft agar colony formation assay showed that the size and number of colonies in Res-treated CRC cells were reduced. *** P < 0.001 (f) Res inhibited the migratory and invasive capacities of HCT-116 cells monitored by RTCA
Anti-CRC effect of Res is partially through activating miR-34c-KITLG axis
MicroRNAs emerge as potent regulators of numerous oncogenes and anti-oncogenes. We then asked whether the anti-CRC role of Res could be through microRNAs. We selected tumor suppressing microRNA, miR-34c which we have previously reported [16], to investigate the potential anti-CRC mechanism of Res. After exposure to Res for 24 h, miR-34c was strikingly increased 5.4 folds (P < 0.01) in HCT-116 cells and 19.2 folds (P < 0.01) in HT-29 cells (Fig. 2a), while KITLG, a target of miR-34c [16], was evidently decreased in these CRC cells (P < 0.05, Fig. 2b). To consolidate the role of miR-34c in the Res-mediated anti-CRC activity, we knocked down miR-34c in HCT-116 cells by its specific inhibitor (Fig. 2c). The inhibitory effect of Res on KITLG expression was abolished when silencing the endogenous miR-34c (P < 0.05, Fig. 2d). Interestingly, accompanied with the reduced miR-34c the anti-CRC effect of Res were attenuated. The Res-suppressed cell proliferation, migration and invasion were recovered by 14.6% (P < 0.05), 11.9% (P < 0.05) and 49.3% (P < 0.01) respectively in HCT-116 cells in the presence of miR-34c inhibitor (Fig. 2e). Moreover, we detected some other documented oncogenic microRNAs including miR-9 and miR-19a, and tumor suppressing microRNAs including miR-28, miR-33a, miR-34a and miR-214, as well as their targets E-cadherin, PTEN, HoxB3, Pim, KIT and FGFR1, respectively [21–26], to figure out the Res-induced microRNA expression profile. Results showed that despite Res induced miR-28 and miR-34a (P < 0.01, Fig. 2g), their respective targets HoxB3 and KIT were not decreased during Res treatment (Fig. 2g), suggesting that Res probably had a specific effect on miR-34c-KITLG axis in CRC cells. Collectively, these results demonstrated that the anti-CRC effect of Res could be partially through activating the miR-34c-KITLG axis.
Fig. 2.
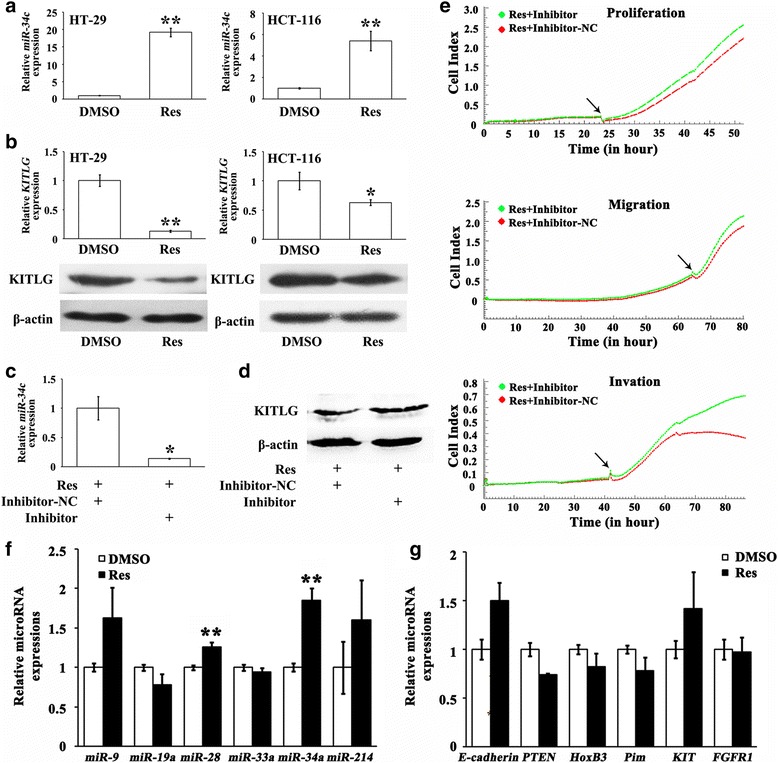
a Res increased miR-34c in CRC cells, which was more prominent in p53 + HT-29 cells than in p53 − HCT-116 cells. ** P < 0.01 (b) KITLG, the target of miR-34c, was significantly decreased at mRNA and protein levels upon the treatment of Res. * P < 0.05, ** P < 0.01 (c) MiR-34c inhibitor efficiently knocked down miR-34c in HCT-116 cells following 24 h-exposure to Res compared with inhibitor-NC. * P < 0.05 (d) Res-decreased KITLG protein was recovered in the presence of miR-34c inhibitor in HCT-116 cells; while inhibitor-NC had no effect. e Res-refrained proliferation, migration and invasion were reversed in HCT-116 cells in the presence of miR-34c inhibitor. Arrows denotes the time when miR-34c inhibitor or inhibitor-NC was added into the medium. f Res elevated miR-28 and miR-34a levels in HCT-116 cells; while other microRNAs remained stable. ** P < 0.01 (g) Res did not alter the target genes of the selected microRNAs in HCT-116 cells
Upregulation of miR-34c by Res is not through demethylation but p53 related
It is well accepted that the promoter of miR-34c is hypermethylated in CRC tissues and cell lines, which lead to silencing of miR-34c [27]. So we carried out MSP to investigate whether the Res-induced miR-34c in CRC cells could be resulted from demethylation of the miR-34c promoter. Unfortunately, we did not find apparent demethylation upon the treatment of Res for 24 h, 48 h or 72 h, indicating the upregulation of miR-34c by Res was not through inducing demethylation of the miR-34c promoter (Fig. 3a).
Fig. 3.
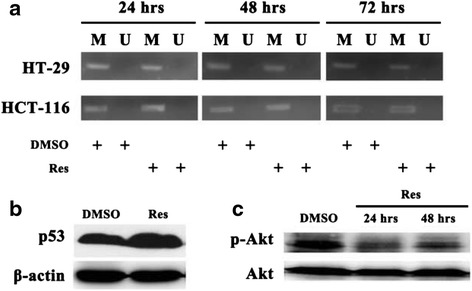
a The methylation state in the miR-34c promoter of CRC cell was not altered in the presence or absence of Res. U: unmethylation, M: methylation (b) Res promoted p53 expression. c Res inhibited phosphorylation of Akt in p53 + HT-29 cells
MiR-34c transcription is under the control of p53 which is hypoexpressed in CRC tissues and cell lines [27]. Could p53 be involved in the Res-induced miR-34c in CRC cells? We used p53+ and p53− CRC cells and found elevated miR-34c in both CRC cell lines, indicating that Res regulated miR-34c in a p53 independent way. However, Res increased p53 protein in p53+ HT-29 cells (Fig. 3b), and the inducement of miR-34c by Res was more prominent in HT-29 cells than that in p53− HCT-116 cells (Fig. 2a), suggesting an involvement of p53 in the Res-induced miR-34c in CRC cells. Liu et al. [28] recently stated that Res inhibited CRC cell proliferation through restraining PI3K/Akt signaling that is involved in p53 degradation. In our study, the phosphorylation of Akt was also attenuated by 24 or 48 h-Res treatment, which was, however, only observed in p53+ HT-29 cells (Fig. 3c).
Res sensitizes CRC cells to Oxa by up-regulating miR-34c
Oxa has been widely used in treating CRC patients. Consistent with previous studies, our result showed that Oxa significantly inhibited CRC cell viability in a dose dependent manner (P < 0.05 ~ 0.001, Fig. 4a). To explore whether there was a synergism of Oxa and Res, the CRC cells were treated with Res and Oxa concurrently for 24 h. Res induced potentiation of cell viability inhibition mediated by Oxa as indicated by CI analysis (CI = 0.66 in HT-29 cells and 0.27 in HCT-116 cells) (P < 0.01 or 0.001, Fig. 4b), suggesting that Res have synergistic anti-CRC effect with Oxa.
Fig. 4.
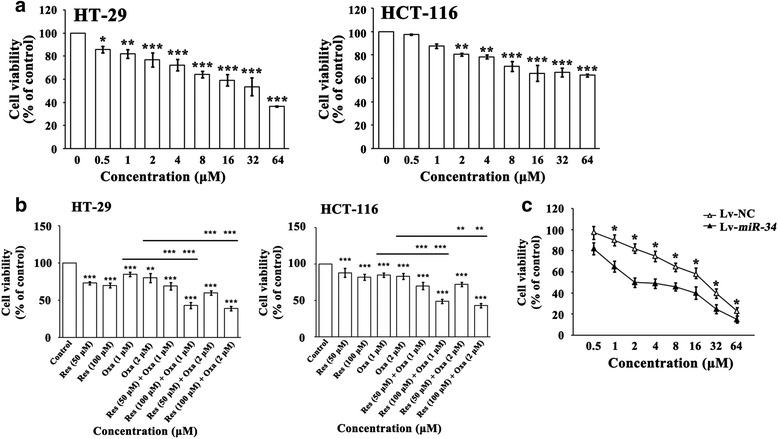
a Twenty-four hr-Oxa treatment evidently inhibited CRC cell viability detected with CCK8 in a dose-dependent manner (0.5 ~ 64 μМ). * P < 0.05, ** P < 0.01, *** P < 0.001 (b) Co-treatment with Res and Oxa elicited significant synergic effects on CRC viability inhibition, according to the CI values which was < 1. ** P < 0.01, *** P < 0.001 (c) Cell viability was deeply weaker in HCT-116 cells overexpressing miR-34c compared with controls in the presence of 2 μM Oxa. * P < 0.05
Next, we wished to find out the underlying mechanism of the synergistic anti-CRC effect of Res and Oxa. Since Res markedly increased miR-34c in CRC cells, we presumed that the up-regulated miR-34c could be a potential contributor. Therefore, we over-expressed miR-34c in HCT-116 cells by lentivirus. Upon the treatment with Oxa for 48 h, the viability of HCT-116 cells overexpressing miR-34c was significantly decreased compared with controls (P < 0.05, Fig. 4c). The result provided evidence that Res sensitized the CRC cells to Oxa chemotherapy probably through up-regulating miR-34c.
In vivo anti-CRC effect of Res
To qualify the anti-CRC efficacy of Res in vivo, we performed HCT-116 xenograft experiment in BALB/c nude mice. As shown in Fig. 5a and Additional file 3: Figure S3, Res or Oxa alone significantly inhibited tumor growth (P < 0.05); and the co-treatment of Res and Oxa elicited a clearly additive effect, indicated by the much slower tumor growth compared with Res or Oxa alone, respectively (P < 0.05). It was noted that 3 out of 7 xenografts vanished during the treatment of Res and Oxa. Since DMSO has chronic toxicity for animal, we measured the body weight during the animal experiment. The body weight of mice in the 4 groups did not significantly changed during experiment (Additional file 4: Figure S4). We further detected microRNA profile in xenograft tumors and/or serum; and the results showed only miR-34c was clearly elevated in tumors but not in serum after exposure to Res for 2 weeks, indicating that Res had a relatively specific effect on miR-34c expression in vivo (P < 0.01, Fig. 5b). Simultaneously, KITLG was down-regulated in xenografts of Res-treated mice (Fig. 5c). Interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) are important proinflammatory cytokines produced by inflammatory cells as well as tumor cells. Clinical data revealed that IL-6 and TNF-α acted as growth factors and were associated with increased risk and advanced stages of CRC [29–32]. Our in vivo study displayed that Res decreased IL-6 secretion in tumors (P < 0.05) while TNF-α remained unchanged (Fig. 5d).
Fig. 5.
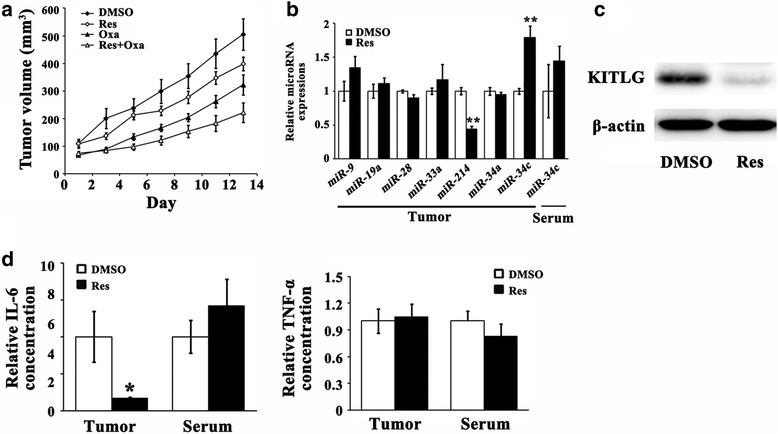
a Res or Oxa could significantly restrict the xenograft growth and the effect was overtly enhanced when co-treated with Res and Oxa. b Res increased miR-34c in tumors but not in serum. MiR-214 was also increased in tumors after Res treatment. ** P < 0.01 (c) KITLG was apparently reduced in tumors of Res-treated mice. d IL-6 in tumors was diminished while TNF-α stayed unvarying
Discussion
Res has been of interest in oncotherapies because of its potent anti-tumor activities and minimal side effects. To better understand the underlying mechanism of Res in treating CRC, we conducted in vitro and in vivo experiments. Consistent with previous reports, Res repressed CRC cell viability, proliferation, capacity of forming colonies, migration and invasion, as well as induced cell cycle arrest and apoptosis. Note that, Res arrested HCT-116 cells in G0/G1 phase while HT-29 cells in G2/M phase, indicating that the effect of Res on the cell cycle arrest was cell type dependent. The most interesting result was that the anti-CRC effect of Res was partially through up-regulating the tumor suppressing microRNA, miR-34c, which silenced its target KITLG. Besides miR-34c, previous studies showed that Res could increase some tumor suppressing microRNAs in CRC. Namely, Res increased miR-663 that targeted TGF-β and potentiated the anti-CRC effect of Res [13]. MiR-129 was suggested to participate in the anti-CRC action of piceatannol, a naturally occurring analog of Res, by targeting Bcl-2 [33]. Nevertheless, though we did detect significant increases of tumor suppressing microRNAs, miR-28 and miR-34a, their respective targets, HoxB3 and KIT, were not decreased, implying that miR-28-HoxB3 and miR-34a-KIT axes may not involve in the anti-CRC activities of Res. But we could not exclude the involvement of miR-28 and miR-34a since they may potentiate the anti-CRC effect of Res by silencing other undetermined targets which we did not detect in this study. Fortunately, miRNA microarrays showed that Res treatment in SW480 CRC cells significantly decreased miR-17, miR-21, miR-25, miR-92a-2, miR-103-1 and miR-103-2 which have been shown to behave as onco-miRNAs [13]. Here, we also showed that Res did not up-regulate oncogenic miR-9 and miR-19a in HCT-116 cells. It was thus inferred that the modulation of Res on the microRNA expressions was distinct: positive on tumor suppressing microRNAs and negative on oncogenic microRNAs. However, the reasons for the distinct effects of Res have been veiled yet. Taken together, we proposed that Res had a robust positive effect on miR-34c-KITLG axis in CRC cells and the effect was, to some extent, specific.
It’s acknowledged that the promoter of miR-34c is hypermethylated in CRC tissues and cell lines which lead to silencing of miR-34c [27]. Therefore, we considered Res-induced demethylation might be responsible for the increased miR-34c. However, we did not observe any demethylated bands by MSP method upon the treatment of Res, indicating demethylation did not account for the Res-induced miR-34c. P53 is a tumor suppressor and drives miR-34c transcript [27]. Could p53 conduced to the Res-stimulated miR-34c expression in CRC cells? Res increased miR-34c expression both in p53+ and p53− CRC cells, suggesting the effect of Res on miR-34c expression was p53 independent. However, it was worth noting that p53 was elevated after treated with Res in HT-29 cells and, simultaneously, the inducement of miR-34c by Res was more prominent in HT-29 cells, indicating p53 facilitated the effect of Res on miR-34c expression. We also found inactivation of PI3K/Akt signaling upon the treatment of Res only in HT-29 cells, which possibly account for the up-regulation of p53 by inhibiting MDM2, a ubiquitin protein ligase controlling the degradation of p53. A recent work provided a new evidence that Res can bind directly and distinctively to miR-33a and miR-122 and divergently modulate their levels in hepatic cells [34]. Whether the increase of miR-34c in CRC was due to the stabilization of Res deserves deep investigation. If so, we hope to find out the underlying mechanism of the distinct effects of Res on the oncogenetic microRNAs and tumor suppressing microRNAs.
Oxa is one of the first choices treating CRC patients currently; but the side effects and drug resistance caused by the long-term use interfere with the therapeutic efficiency. In this study, Res was introduced to facilitate the chemosensitivity to Oxa in CRC cells, which was probably relied on the elevated miR-34c. Moreover, no publications showed that Res was harmful to patients, suggesting Res could be a safe medication in preventing CRC. Lines of evidence suggested Res reversed multidrug resistance by down-regulating MDR-1 gene in breast cancer cells [35, 36]. Wen et al. [37] addressed that PI3K/Akt signaling positively regulated MDR-1 in CRC. According to the previous and present results, we presumed that Res-induced miR-34c in CRC cells might be associated with the reduced MDR-1 or other multidrug resistance-related genes via silencing PI3K/Akt pathway, which, however, needs further investigation.
The in vivo xenograft experiment further consolidated the in vitro results. Res raised the miR-34c expression in tumors but not in serum, giving a clue that the miR-34c-inducing effect of Res was tissue specific instead of systemic. Moreover, the proinflammatory cytokine IL-6 was reduced exposure to the 2-week Res treatment. IL-6 could act as growth factor in multiple tumors by activating the oncogenic STAT3 transcription factor. Rokavec et al. [38] identified that miR-34a/c could target IL-6R by binding to its 3′ untranslated region (3′UTR); thereby disrupted the IL-6/IL-6R-STAT3-miR-34a loop. Here, we hypothesized that the IL-6-triggered CRC progression could be interfered by the Res-increased miR-34c.
Conclusion
In summary, the present study revealed that Res inhibited CRC by activating miR-34c-KITLG in vitro and in vivo; and the effect was strengthened in the presence of p53. In addition, the up-regulated miR-34c by Res sensitized chemosensitivity to Oxa treatment in CRC cells.
Acknowledgements
This study was supported by the National Science Foundation of China (81300285, 81572322 and 31371220), the Specialized Research Fund for the Doctoral Program of Higher Education of China (20121107120020), the Scientific Research Key Program of Beijing Municipal Commission of Education (KZ201310025020), the Open Project of Beijing Key Laboratory of Cancer Invasion and Metastasis Research (2015ZLQX06), the Scientific Research Foundation for Young Teachers in the Capital Medical University (2014PY49) and the Beijing Municipal Science & Technology Commission (Z151100001615039).
Additional files
Representative histograms of the cell cycle detected by flow cytometry. (TIF 5617 kb)
Representative diagrams of the apoptosis detected by flow cytometry. (TIF 2449 kb)
HCT-116 xenografts in nude mice were collected after 2-week treatment of Res, Oxa or Res + Oxa. (TIF 3.44 mb)
There was no apparent body weight lost in any group during the animal experiment, suggesting no toxicity for mice. (TIF 2934 kb)
Footnotes
Shu Yang and Wenshuai Li contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SY and DZ designed experiments. SY and WL performed experiments and wrote the manuscript. HS, BW, FJ, TS, HC, PS and YW performed experiments. DZ gave suggestion on discussion and interpretation on the data. All authors read and approved the final manuscript.
Contributor Information
Shu Yang, Email: sheilayslamb@aliyun.com.
Wenshuai Li, Email: liwenshuailong@126.com.
Haimei Sun, Email: haimei@ccmu.edu.cn.
Bo Wu, Email: wubo2007@ccmu.edu.cn.
Fengqing Ji, Email: jfq@ccmu.edu.cn.
Tingyi Sun, Email: styj211@ccmu.edu.cn.
Huanhuan Chang, Email: changhuan.0704@163.com.
Ping Shen, Email: 1051424476@qq.com.
Yaxi Wang, Email: wangyaxi1@sina.com.
Deshan Zhou, Phone: +86-10-8395-0079, Email: zhoudeshan2008@163.com.
References
- 1.Vincenzi B, Daniele S, Frezza AM, Berti P, Vespasiani U, Picardi A, Tonini G. The role of S-adenosylmethionine in preventing oxaliplatin-induced liver toxicity: a retrospective analysis in metastatic colorectal cancer patients treated with bevacizumab plus oxaliplatin-based regimen. Support Care Cancer. 2012; 20:135-9. [DOI] [PubMed]
- 2.Kagiava A, Theophilidis G, Sargiannidou I, Kyriacou K, Kleopa KA. Oxaliplatin-induced neurotoxicity is mediated through gap junction channels and hemichannels and can be prevented by octanol. Neuropharmacology. 2015;97:289–305. doi: 10.1016/j.neuropharm.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, Wan G, Spizzo R, Ivan C, Mathur R, Hu X, et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol Oncol. 2014;8:83–92. doi: 10.1016/j.molonc.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leiro J, Alvarez E, Arranz JA, Laguna R, Uriarte E, Orallo F. Effects of cis-resveratrol on inflammatory murine macrophages: antioxidant activity and down-regulation of inflammatory genes. J Leukoc Biol. 2004;75:1156–65. doi: 10.1189/jlb.1103561. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y, He XQ, Huang X, Ding L, Xu L, Shen YT, et al. Resveratrol protects mouse oocytes from methylglyoxal-induced oxidative damage. PLoS One. 2013;8:e77960. doi: 10.1371/journal.pone.0077960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji Q, Liu X, Han Z, Zhou L, Sui H, Yan L, et al. Resveratrol suppresses epithelial-to-mesenchymal transition in colorectal cancer through TGF-β1/Smads signaling pathway mediated Snail/E-cadherin expression. BMC Cancer. 2015;15:97. doi: 10.1186/s12885-015-1119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aggarwal BB, Bhardwaj A, Aggarwal RS, Seeram NP, Shishodia S, Takada Y. Role of resveratrol in prevention and therapy of cancer: preclinical and clinical studies. Anticancer Res. 2004;24:2783–840. [PubMed] [Google Scholar]
- 8.Lee HS, Ha AW, Kim WK. Effect of resveratrol on the metastasis of 4T1 mouse breast cancer cells in vitro and in vivo. Nutr Res Pract. 2012;6:294–300. doi: 10.4162/nrp.2012.6.4.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dias SJ, Li K, Rimando AM, Dhar S, Mizuno CS, Penman AD, et al. Trimethoxy-resveratrol and piceatannol administered orally suppress and inhibit tumor formation and growth in prostate cancer xenografts. Prostate. 2013;73:1135–46. doi: 10.1002/pros.22657. [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, Liu B, E C, Liu J, Zhang Q, Liu J, et al. Resveratrol inhibits the proliferation of human melanoma cells by inducing G1/S cell cycle arrest and apoptosis. Mol Med Rep. 2015;11:400–4. doi: 10.3892/mmr.2014.2716. [DOI] [PubMed] [Google Scholar]
- 11.Liu B, Zhou Z, Zhou W, Liu J, Zhang Q, Xia J, et al. Resveratrol inhibits proliferation in human colorectal carcinoma cells by inducing G1/S-phase cell cycle arrest and apoptosis through caspase/cyclin-CDK pathways. Mol Med Rep. 2014;10:1697–702. doi: 10.3892/mmr.2014.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu YH, Chen HA, Chen PS, Cheng YJ, Hsu WH, Chang YW, et al. MiR-520h-mediated FOXC2 regulation is critical for inhibition of lung cancer progression by resveratrol. Oncogene. 2013;32:431–43. doi: 10.1038/onc.2012.74. [DOI] [PubMed] [Google Scholar]
- 13.Tili E, Michaille JJ, Alder H, Volinia S, Delmas D, Latruffe N, et al. Resveratrol modulates the levels of microRNAs targeting genes encoding tumor-suppressors and effectors of TGF-β signaling pathway in SW480 cells. Biochem Pharmacol. 2010;80:2057–65. doi: 10.1016/j.bcp.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 15.Roy S, Levi E, Majumdar AP, Sarkar FH. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012. doi:10.1186/1756-8722-5-58. [DOI] [PMC free article] [PubMed]
- 16.Yang S, Li WS, Dong F, Sun HM, Wu B, Tan J, et al. KITLG is a novel target of miR-34c that is associated with the inhibition of growth and invasion in colorectal cancer cells. J Cell Mol Med. 2014;18:2092–102. doi: 10.1111/jcmm.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumazaki M, Noguchi S, Yasui Y, Iwasaki J, Shinohara H, Yamada N, et al. Anti-cancer effects of naturally occurring compounds through modulation of signal transduction and miRNA expression in human colon cancer cells. J Nutr Biochem. 2013;24:1849–58. doi: 10.1016/j.jnutbio.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Schwingel TE, Klein CP, Nicoletti NF, Dora CL, Hadrich G, Bica CG, et al. Effects of the compounds resveratrol, rutin, quercetin, and quercetin nanoemulsion on oxaliplatin-induced hepatotoxicity and neurotoxicity in mice. Naunyn Schmiedebergs Arch Pharmacol. 2014;387:837–48. doi: 10.1007/s00210-014-0994-0. [DOI] [PubMed] [Google Scholar]
- 19.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzym Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 20.Chalal M, Delmas D, Meunier P, Latruffe N, Vervandier-Fasseur D. Inhibition of cancer derived cell lines proliferation by synthesized hydroxylated stilbenes and new ferrocenyl-stilbene analogs. Comparison with resveratrol. Molecules. 2014;19:7850–68. doi: 10.3390/molecules19067850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L, et al. MicroRNA-9 promotes tumor metastasis via repressing E-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–80. doi: 10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Y, Liu J, Kang Y, He Y, Liang B, Yang P, et al. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J Exp Clin Cancer Res. 2014;33:67. doi: 10.1186/s13046-014-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeida MI, Nicoloso MS, Zeng L, Ivan C, Spizzo R, Gafà R, et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142:886–96. doi: 10.1053/j.gastro.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas M, Lange-Grünweller K, Weirauch U, Gutsch D, Aigner A, Grünweller A, et al. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 2012;31:918–28. doi: 10.1038/onc.2011.278. [DOI] [PubMed] [Google Scholar]
- 25.Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY, et al. Identification of microRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology. 2014;60:598–609. doi: 10.1002/hep.27118. [DOI] [PubMed] [Google Scholar]
- 26.Siemens H, Jackstadt R, Kaller M, Hermeking H. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget. 2013;4:1399–415. doi: 10.18632/oncotarget.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–32. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 28.Liu YZ, Wu K, Huang J, Liu Y, Wang X, Meng ZJ, et al. The PTEN/PI3K/Akt and Wnt/β-catenin signaling pathways are involved in the inhibitory effect of resveratrol on human colon cancer cell proliferation. Int J Oncol. 2014;45:104–12. doi: 10.3892/ijo.2014.2392. [DOI] [PubMed] [Google Scholar]
- 29.Landi S, Moreno V, Gioia-Patricola L, Guino E, Navarro M, de Oca J, et al. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res. 2003;63:3560–6. [PubMed] [Google Scholar]
- 30.Al Obeed OA, Alkhayal KA, Al Sheikh A, Zubaidi AM, Vaali-Mohammed MA, Boushey R, et al. Increased expression of tumor necrosis factor-α is associated with advanced colorectal cancer stages. World J Gastroenterol. 2014;20:18390–6. doi: 10.3748/wjg.v20.i48.18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan YS, Hsu HP, Lai MD, Yen MC, Fang JH, Weng TY, et al. Suppression of mucin 2 promotes interleukin-6 secretion and tumor growth in an orthotopic immune-competent colon cancer animal model. Oncol Rep. 2014;32:2335–42. doi: 10.3892/or.2014.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bigatto V, De Bacco F, Casanova E, Reato G, Lanzetti L, Isella C, et al. TNF-α promotes invasive growth through the MET signaling pathway. Mol Oncol. 2015;9:377–88. doi: 10.1016/j.molonc.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, Jia R, Wang C, Hu T, Wang F. Piceatannol promotes apoptosis via up-regulation of microRNA-129 expression in colorectal cancer cell lines. Biochem Biophys Res Commun. 2014;452:775–81. doi: 10.1016/j.bbrc.2014.08.150. [DOI] [PubMed] [Google Scholar]
- 34.Baselga-Escudero L, Blade C, Ribas-Latre A, Casanova E, Suárez M, Torres JL, et al. Resveratrol and EGCG bind directly and distinctively to miR-33a and miR-122 and modulate divergently their levels in hepatic cells. Nucleic Acids Res. 2014;42:882–92. doi: 10.1093/nar/gkt1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang F, Wu XN, Chen J, Wang WX, Lu ZF. Resveratrol reverses multidrug resistance in human breast cancer doxorubicin-resistant cells. Exp Ther Med. 2014;7:1611–6. doi: 10.3892/etm.2014.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sprouse AA, Herbert BS. Resveratrol augments paclitaxel treatment in MDA-MB-231 and paclitaxel-resistant MDA-MB-231 breast cancer cells. Anticancer Res. 2014;34:5363–74. [PubMed] [Google Scholar]
- 37.Wen F, He S, Sun C, Li T, Wu S. PIK3CA and PIK3CB expression and relationship with multidrug resistance in colorectal carcinoma. Int J Clin Exp Pathol. 2014;7:8295–303. [PMC free article] [PubMed] [Google Scholar]
- 38.Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–67. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]


