The mitochondrial Ca2+ uptake protein At-MICU shapes mitochondrial Ca2+ dynamics, providing molecular in vivo evidence for the existence and function of a mitochondrial uniporter complex in plants.
Abstract
Plant organelle function must constantly adjust to environmental conditions, which requires dynamic coordination. Ca2+ signaling may play a central role in this process. Free Ca2+ dynamics are tightly regulated and differ markedly between the cytosol, plastid stroma, and mitochondrial matrix. The mechanistic basis of compartment-specific Ca2+ dynamics is poorly understood. Here, we studied the function of At-MICU, an EF-hand protein of Arabidopsis thaliana with homology to constituents of the mitochondrial Ca2+ uniporter machinery in mammals. MICU binds Ca2+ and localizes to the mitochondria in Arabidopsis. In vivo imaging of roots expressing a genetically encoded Ca2+ sensor in the mitochondrial matrix revealed that lack of MICU increased resting concentrations of free Ca2+ in the matrix. Furthermore, Ca2+ elevations triggered by auxin and extracellular ATP occurred more rapidly and reached higher maximal concentrations in the mitochondria of micu mutants, whereas cytosolic Ca2+ signatures remained unchanged. These findings support the idea that a conserved uniporter system, with composition and regulation distinct from the mammalian machinery, mediates mitochondrial Ca2+ uptake in plants under in vivo conditions. They further suggest that MICU acts as a throttle that controls Ca2+ uptake by moderating influx, thereby shaping Ca2+ signatures in the matrix and preserving mitochondrial homeostasis. Our results open the door to genetic dissection of mitochondrial Ca2+ signaling in plants.
INTRODUCTION
For plants to grow, develop, and survive under changeable conditions, constant sensing of their environment is critical. At the cellular level, external stimuli have to be perceived and integrated to trigger both rapid responses and long-term acclimation. Rapid adjustment is particularly important for the endosymbiotic organelles, i.e., the mitochondria and the chloroplasts, since perturbation of the finely tuned metabolic, redox, and chemiosmotic processes can severely affect cellular homeostasis or even trigger programmed cell death (Kim et al., 2006; Logan, 2007). Dynamic changes in free Ca2+ concentrations have been recognized to act in the transduction within the cell of both endogenous and external signals in order to regulate biotic and abiotic stress responses (Knight et al., 1991; Kiegle et al., 2000; Whalley et al., 2011), developmental programs (Foreman et al., 2003; Monshausen et al., 2008), stomatal dynamics (Gilroy et al., 1991; Allen et al., 2000; Pei et al., 2000; Kim et al., 2010), protein import into cell compartments (Chigri et al., 2006), and symbiotic plant-microbe interactions (Wais et al., 2000; Miller et al., 2013).
Mechanistically, free Ca2+ transients in the cellular compartments are produced by the interplay between influx, buffering capacity and export from a compartment. While in mammalian cells, the extracellular space and the endoplasmic reticulum act as main stores for Ca2+ release into the cytosol and the organelles in turn, and this situation may be more complex in plants. In the endoplasmic reticulum lumen of Arabidopsis thaliana, Ca2+ increases follow cytosolic transients, albeit with distinct dynamics, suggesting that the Ca2+ contributing to the transients is released from another source (Bonza et al., 2013). The vacuole and the thylakoid lumen have both been implicated as likely intracellular Ca2+ stores in this respect (Rocha and Vothknecht, 2012; Nomura and Shiina, 2014).
The study of cytosolic Ca2+ transients has delivered valuable insights into signaling and regulation, but the complex makeup of the plant cell, with numerous subcellular compartments, is currently only loosely integrated into this picture. However, precise spatial organization is central if Ca2+ transients are to act as specific signals. Like mammals (Kirichok et al., 2004; Santo-Domingo and Demaurex, 2010), plants contain a mitochondrial uptake system for Ca2+, which has been investigated extensively by biochemical methods for half a century (Hanson et al., 1965; Dieter and Marmé, 1980; Akerman and Moore, 1983; Zottini and Zannoni, 1993). However, conflicting observations in vitro on the Ca2+ uptake of isolated mitochondria from different plant species and distinct characteristics from those of mammalian mitochondria have led to a long-standing debate about the dominating influx mechanism in plants.
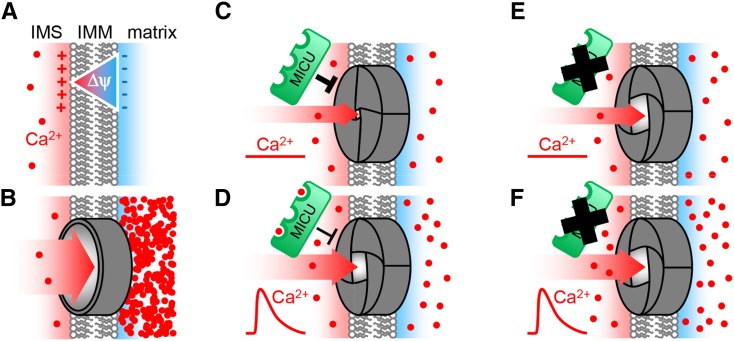
Differential Ca2+ dynamics between the cytosol and chloroplasts and mitochondria have been observed in vivo using the aequorin probe and, more recently, sensors of the Cameleon family (Sai and Johnson, 2002; Logan and Knight, 2003; Loro et al., 2012, 2013; Mehlmer et al., 2012). Mitochondrial Ca2+ transients coincide with those in the cytosol overall, yet their signature is typically distinct, suggesting specific modulation. The ability of Ca2+ to enter the mitochondria requires tight regulation due to the electrical potential ΔΨ of about −180 mV across the inner mitochondrial membrane, which, according to the Nernst equation, would lead to an accumulation of divalent Ca2+ ions in the matrix by ∼106-fold. Thus, even at steady state conditions when cytosolic Ca2+ concentrations are low (∼100 to 200 nM; Logan and Knight, 2003; Nomura and Shiina, 2014), equilibration across the inner mitochondrial membrane would lead to a matrix concentration of ∼100 to 200 mM, triggering dysfunction and/or permeability transition in turn (Arpagaus et al., 2002; Scott and Logan, 2008; Bernardi and Di Lisa, 2015; Halestrap and Richardson, 2015).
Mitochondria and chloroplasts share a requirement for semiautonomous regulation, and a regulatory role of Ca2+ is likely mediated by a variety of Ca2+ binding proteins and Ca2+-modulated processes in both organelles. In mammals, mitochondrial Ca2+ can modulate the activity of different tricarboxylic acid cycle dehydrogenases, coupling overall mitochondrial energy output to demand or trigger apoptosis in extreme cases (Griffiths and Rutter, 2009). In plants, Ca2+ binding EF-hand proteins, including a glutamate dehydrogenase, a family of MCF ATP/Pi transporters (APC1-3), and two alternative NAD(P)H dehydrogenases (NDB1 and NDB2), localize to mitochondria and show Ca2+-dependent activity (although the latter are regulated by Ca2+ from the intermembrane space side of the inner mitochondrial membrane; Møller et al., 1981; Rasmusson and Møller, 1991; Turano et al., 1997; Geisler et al., 2007; Stael et al., 2011; Monné et al., 2015). Furthermore, the EF-hand protein LETM1 and the calmodulin-like protein CML30 have recently been found to be localized in mitochondria, and additional proteins with Ca2+ binding motifs have been detected in proteomic data sets of plant mitochondria (Chigri et al., 2012; Zhang et al., 2012; Salvato et al., 2014). Moreover, there is evidence for Ca2+ dependence of mitochondrial permeability transition, protein import, and respiration in plants (Miernyk et al., 1987; Arpagaus et al., 2002; Kuhn et al., 2009; Manzoor et al., 2012). Nevertheless, it remains unclear if metabolic flux through pyruvate dehydrogenase and the tricarboxylic acid cycle can be regulated by Ca2+ in vivo (Møller and Rasmusson, 1998).
The recent identification of several protein families making up the mitochondrial Ca2+ uniporter complex (MCUC) in mammals, including the pore-forming MCU (MCU and MCUb; Baughman et al., 2011; De Stefani et al., 2011; Raffaello et al., 2013), and the regulatory, associated MICUs (MICU1, 2, and 3; Perocchi et al., 2010; Plovanich et al., 2013), EMRE (Sancak et al., 2013), and MCUR1/CCDC90A (although potentially not directly; Mallilankaraman et al., 2012b; Paupe et al., 2015), has spurred intense research across the biomedical disciplines (Drago et al., 2012; Mallilankaraman et al., 2012a; Csordás et al., 2013; Hoffman et al., 2013; Marchi et al., 2013; Pan et al., 2013; Raffaello et al., 2013; Kamer and Mootha, 2014; Kovács-Bogdán et al., 2014; Logan et al., 2014; Patron et al., 2014; Wang et al., 2014). While some genes of the complex components appear not to be present in plants (e.g., MCUb and EMRE), others have multiple homologs (six for the functional pore-forming subunit MCU in Arabidopsis [Stael et al., 2012] and maize [Zea mays; Meng et al., 2015] and two for MCUR1/CCDC90A in Arabidopsis), suggesting potential functional modification and/or differentiation. The homologous genes present in plants are good candidates for an involvement in mitochondrial Ca2+ homeostasis, regulation, and signaling, but experimental evidence is lacking.
In this work, we initiate the genetic and physiological analysis of mitochondrial Ca2+ regulation in plants. We focus on At-MICU, the only Arabidopsis homolog of the mammalian MICU gene family, and employ functional imaging to dissect the role of MICU in living roots. Our observations provide molecular in vivo evidence for the existence and function of a MCUC in plants. They further suggest that MICU specifically shapes mitochondrial Ca2+ dynamics in Arabidopsis by negative regulation of matrix influx.
RESULTS
MICU Is Conserved in Plants with a Single Homolog in Arabidopsis
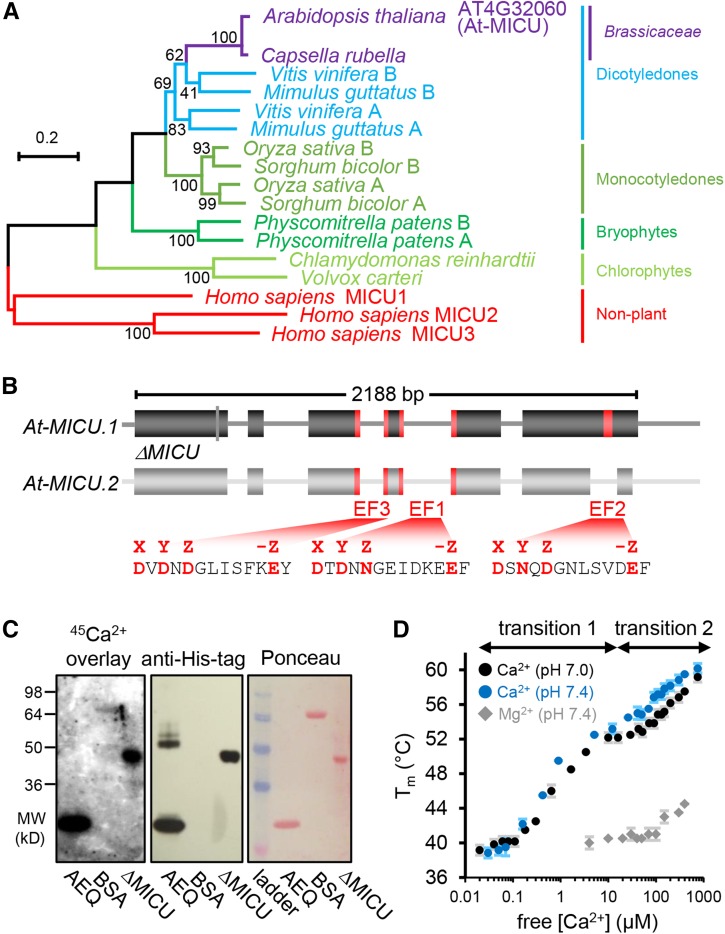
The discovery of MICU1 in mammals was achieved by a phylogenetic approach, making use of the conservation of mitochondrial Ca2+ uniport in most eukaryotes (Perocchi et al., 2010). We performed a refined phylogenetic analysis for plants, confirming that MICU is conserved throughout the plant kingdom (Figure 1A; Supplemental Data Set 1 and Supplemental File 1), which suggests an important functional role in plants (Stael et al., 2012). Most plant species included in the analysis contained at least two MICU homologs. Only one homolog was found in a small number of specific clades across the phylogeny including the Chlorophytes and the Brassicacea. This indicates that, although diversification can occur, one MICU locus is sufficient and Arabidopsis represents this minimal configuration. Furthermore, homologs of the MICU1 isoforms MICU2 and 3 that are present in vertebrates (Plovanich et al., 2013; Kamer and Mootha, 2014) could not be found in Arabidopsis. The amino acid sequence of Arabidopsis MICU (At-MICU; AT4G32060; predicted splicing variant 1: At-MICU.1) showed 45% similarity and 25% identity to its homolog in human. The two Ca2+ binding EF hand motifs present in mammalian MICU1 are conserved, while there is an additional third EF-hand predicted in Arabidopsis (Figure 1B; Supplemental Figure 1). All three EF-hand motifs feature all characteristics required for functionality, including the critical charged residues for Ca2+ binding. A second predicted splicing variant, At-MICU.2, lacks the C-terminal EF-hand. We sought to exploit this genetic situation as a tangible model to dissect the physiological role of MICU in plants.
Figure 1.
Phylogenetic Properties of MICU in Plants, Gene Model in Arabidopsis, and Ca2+ Binding.
(A) Phylogenetic tree of homologs from representative plant species to human MICU proteins. The scale bar represents 20 substitutions per 100 amino acids. Bootstrap values (on nodes) were calculated using 1000 replicates. The alignments are shown in Supplemental File 1.
(B) Gene model of Arabidopsis MICU variants. MICU.1 and MICU.2 represent predicted splice forms; ΔMICU, a variant of MICU.1 cDNA where nucleotides coding for 117 N-terminal amino acids were removed to use in recombinant protein expression. Exons and introns are illustrated as boxes and lines, respectively. Nucleotides encoding predicted Ca2+ binding EF-hand motifs are highlighted in red. All three EF-hands are conserved among plants. EF1 and EF2 are also conserved in mammalian MICUs, while EF3 is not (Supplemental Figure 1). The amino acid sequences that include all critical charged residues (red) predict genuine Ca2+ binding capability for all three EF-hands.
(C) 45Ca2+ overlay assays of recombinantly purified ΔMICU protein (Supplemental Figure 2) compared with recombinant aequorin (AEQ) and BSA (BSA) as positive and negative control, respectively. The presence of protein was validated by immunodetection of the His-tags of ΔMICU and AEQ, and Ponceau S staining of separate control membranes. 45Ca2+ binding was visualized by autoradiography.
(D) Thermostabilization of ΔMICU through Ca2+. Protein melting points (Tm) were recorded in the presence of Ca2+/EGTA-adjusted free Ca2+ at pH 7.0 (black circles) and 7.4 (blue circles), and Mg2+/EDTA-adjusted free Mg2+ concentrations at pH 7.4 (gray diamonds). Concentration ranges of free Ca2+ associated with clear Tm transition are indicated. n = 3 (2 for Mg2+); error bars = sd.
MICU Binds Ca2+ in Vitro
To assess if At-MICU is indeed able to bind Ca2+, as predicted by the presence of the EF-hand motifs, we performed Ca2+ overlay assays and measured the effect of free Ca2+ concentrations on protein folding stability by ligand-induced thermal stabilization assays. For that we used a recombinantly expressed and purified truncated version of At-MICU.1 (∆MICU; Figure 1B; Supplemental Figure 2A). ∆MICU lacks the first 117 amino acids, thereby eliminating the signal peptide and putative transmembrane/disordered sequences, and carries an N-terminal His-tag. All predicted EF-hand motifs remain intact. Using 45Ca2+ overlay after gel-based protein separation (Kato et al., 1998), strong 45Ca2+ signals were present at the predicted molecular weights of ΔMICU and aequorin (positive control), but not BSA (negative control; Figure 1C). Based on the hypothesis that Ca2+ binding alters the structural stability of ΔMICU, we further investigated the affinity range of Ca2+ binding by differential scanning fluorimetry in the presence of increasing free Ca2+ concentrations and physiological buffer pH (Figure 1D) and observed thermal stabilization with increasing free Ca2+ concentrations. Two sigmoid transitions, indicating stabilization by Ca2+ binding, could be clearly distinguished. The first occurred between ∼200 nM and 5 μM and the second between ∼25 and 500 μM free Ca2+. Since it is plausible that more than one binding site contributes to one transition, this points to the presence of at least two Ca2+ binding sites with apparent binding affinities around 1 and 100 μM, respectively. The Mg2+ control showed poor stabilization only starting above around 100 μM, suggesting that Ca2+ binding is selective. The first Ca2+-induced transition coincides with the physiological concentration range of intracellular free Ca2+ and is lower than the binding constants estimated for mammalian MICU1 (Wang et al., 2014). While the binding affinities measured must be considered qualitative estimates, they clearly indicate that ΔMICU can bind Ca2+ in vitro at physiological concentrations with impact on the structural properties of the protein.
MICU Is Expressed in Green and Non-Green Tissues and Localizes to Mitochondria
Public transcriptomic data indicate that MICU is ubiquitously expressed across Arabidopsis tissues (Zimmermann et al., 2004; Stael et al., 2012). Expression in root, shoot, floral bud, and pollen is also supported by two shotgun proteomic screens of whole-tissue extracts (AtPeptide MS/MS and Pep2Pro MS/MS; Joshi et al., 2011). Experimental evidence on the subcellular localization of MICU has been lacking, however. Bioinformatics algorithms provide ambiguous targeting predictions with an overall priority for mitochondria or plastids (Joshi et al., 2011; Tanz et al., 2013; Hooper et al., 2014).
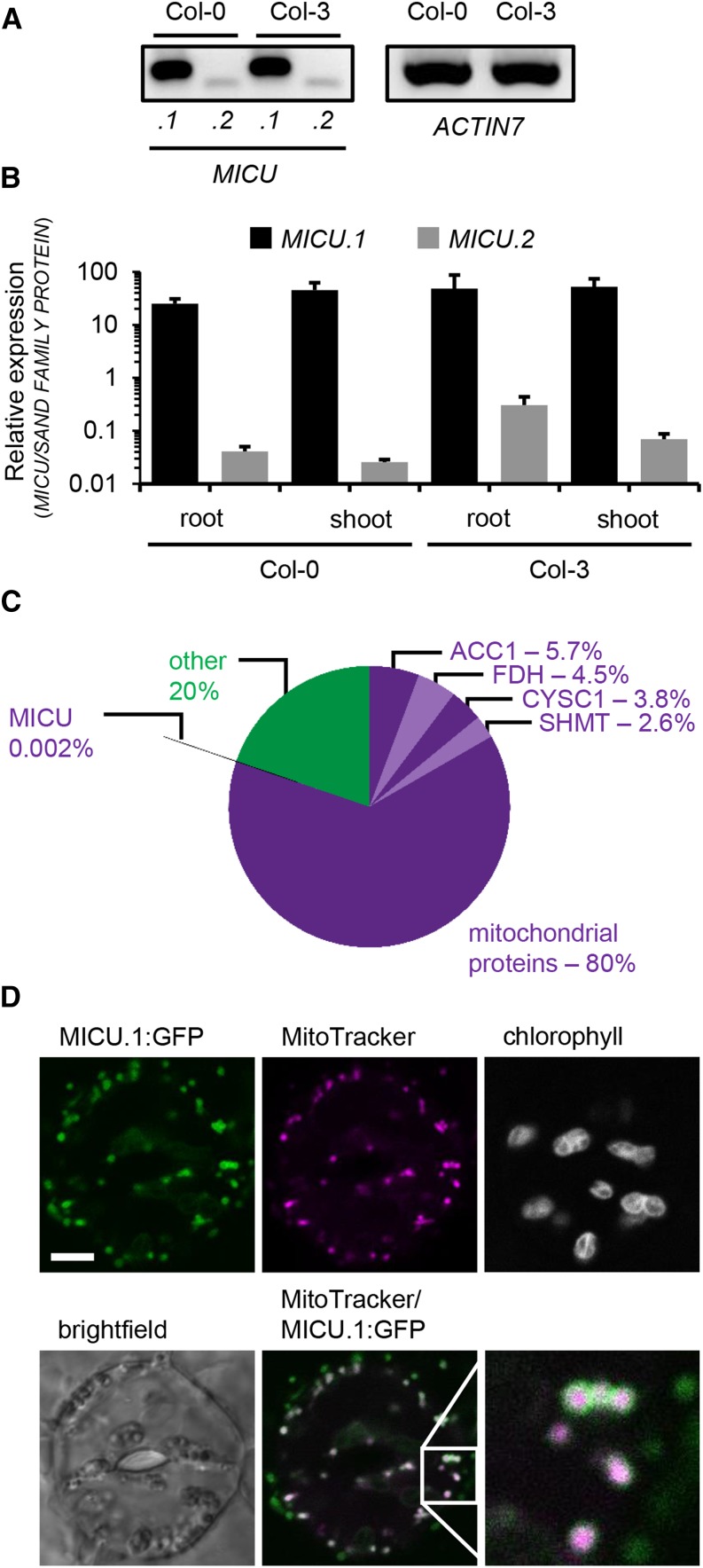
RT-PCR analysis of the full-length MICU.1 transcript in whole-plant extracts of the accessions Col-0 and Col-3 suggested transcript levels similar to the ACTIN7 control (Figure 2A). A predicted splicing variant, MICU.2, could be detected at much lower levels. This was refined by quantitative RT-PCR for roots and rosettes separately, showing MICU.1 expression of comparable magnitude in both green and non-green tissue. The second splicing variant MICU.2 was present at >100 times lower abundance compared with the main transcript MICU.1 (Figure 2B).
Figure 2.
Expression and Subcellular Localization of MICU in Arabidopsis.
(A) Splicing variant-specific RT-PCR for MICU.1 and MICU.2 in extracts of whole Arabidopsis seedlings (Col-0 and Col-3). ACTIN7 was used as reference transcript.
(B) qRT-PCR of MICU.1 and MICU.2 in root and shoot tissue and SAND FAMILIY PROTEIN as a reference. Values are mean from three biological and three technical replicates; error bars = sd of biological replicates.
(C) Quantitative proteomic assessment of mitochondria isolated from Arabidopsis seedlings. iBAQ-based protein abundances annotated as mitochondrial (purple colors) and non-mitochondrial (green; Supplemental Data Set 2) are shown. In addition, the four most abundant proteins detected (all annotated mitochondrial; different shades of purple) and MICU are indicated with their respective abundances. ACC1, ATP/ADP carrier 1; FDH, formate dehydrogenase; CYSC1, cysteine synthase C1; SHMT, serine hydroxymethyl transferase 1.
(D) Representative image of an Arabidopsis seedling stably expressing MICU.1:GFP. Leaf was stained with MitoTracker Orange, and the epidermis was imaged by confocal laser scanning microscopy. The lower right image represents a magnified view of the region in the square. GFP, green; MitoTracker, magenta. Bar = 5 μm.
We detected MICU in the proteome of our mitochondrial preparations from Arabidopsis (Figure 2C; Supplemental Data Set 2) as one out of 653 identified Arabidopsis proteins at a protein and peptide false discovery rate of 1%. Of the protein preparation, 80% was annotated as mitochondrial (based on SUBAcon of the SUBA3 database; Tanz et al., 2013; Hooper et al., 2014), suggesting reasonable mitochondrial purity. Identified proteins covered five orders of magnitude in abundance, as estimated by the iBAQ value, which is based on peptide ion intensity. MICU was detected with only one unique peptide with an Andromeda score of 40 (0 to ∼500 as typical score range; 11 to 216 for this data set; good confidence from a score >35), a posterior probability of 0.01, and abundance ranked at 590st, with a relative protein abundance of 0.002% among all identified proteins. The low abundance of MICU identified in the proteomic experiment may explain the absence of MICU from previous plant mitochondrial proteome studies and strengthened the hypothesis of MICU as a mitochondrial protein. Additional support comes from the presence of a homolog of MCU (AT1G57610) in our data set (Supplemental Data Set 2), the pore-forming component of the mitochondrial Ca2+ uniporter in mammals. This is consistent with the recent detection of an MCU homolog in an extension of the study of the potato tuber mitochondrial proteome (Salvato et al., 2014; I.M. Møller, personal communication).
For independent validation, we raised a polyclonal antibody against MICU.1 protein, which we applied to probe for MICU directly. However, the purified antibody did not label a clear band at the expected size for MICU.1 (∼51 kD, based on cleavage site prediction at position 41 by TargetP; Emanuelsson et al., 2007) in whole-cell extracts or isolated mitochondria from Col-0 wild-type plants, although a 35S overexpressed MICU.1:GFP fusion protein appeared as a band of the predicted size at ∼81 kD in protein extracts from isolated mitochondria (Supplemental Figures 2B and 2C). This suggests specific MICU recognition by the antibody, but with insufficient affinity for detection of endogenous MICU, even in enriched preparations under the conditions used.
As an orthogonal approach, we analyzed N- and C-terminal GFP fusions of the full-length coding sequence of MICU.1 under the control of a 35S promoter by confocal microscopy. Transient expression of the C-terminal fusion construct MICU.1:GFP in wild tobacco (Nicotiana benthamiana) leaf epidermis showed GFP fluorescence that colocalized with the mitochondrial marker MitoTracker Orange, indicating mitochondrial localization. By contrast, the N-terminal fusion construct GFP:MICU.1 displayed cytosolic and nuclear localization, suggesting that the N terminus, which is blocked by the N-terminal GFP-fusion, is critical for mitochondrial targeting (Supplemental Figure 3).
To rule out potential localization artifacts, we generated stable Arabidopsis transformants using the C-terminal GFP fusion construct. Three independent lines were assessed in leaf epidermis, hypocotyl, and rhizodermis. GFP signal was consistently observed in small, round-to-elongated, highly dynamic subcellular structures and colocalized with MitoTracker Orange and/or tetramethyl rhodamine methyl ester (TMRM) (Figure 2D; Supplemental Figure 4; Schwarzländer et al., 2012a), confirming the transient expression data in wild tobacco. Minor signal in the GFP channel that was occasionally observed in chloroplast regions of leaf samples was equally observed in controls, suggesting minor spectroscopic bleed-through of chlorophyll fluorescence.
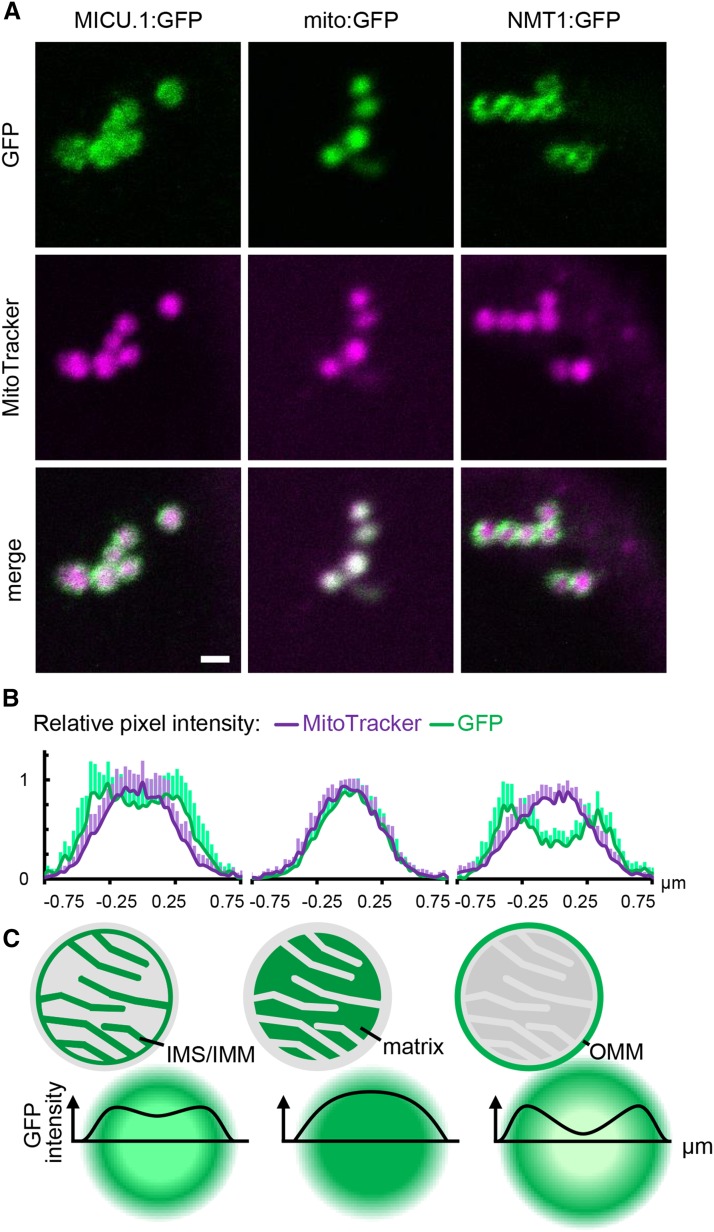
When imaging larger mitochondria of the stable 35S:MICU.1:GFP lines, it was possible to resolve GFP fluorescence in ring-like structures. The GFP signal did not strictly colocalize with the MitoTracker signal but instead appeared consistently broader (Figure 3A). Since resolution of subcompartments in individual mitochondria is close to the resolution limit of classical light microscopy, we determined and merged the pixel intensity distributions of multiple mitochondria to improve signal-to-noise statistics (Figure 3B) (Elgass et al., 2009). The highest GFP intensity pixels surrounded the MitoTracker signal maximum, suggesting that MICU was localized in a different mitochondrial subcompartment to MitoTracker. Using GFP targeted to the matrix (mito-GFP; Logan and Leaver, 2000) as a control showed an intensity distribution that resembled that of MitoTracker, ruling out chromatic aberration effects, while the outer mitochondrial membrane (labeled with NETWORK1/ELM1-GFP; Logan et al., 2003; Arimura et al., 2008) showed two pronounced intensity maxima, with the MitoTracker maximum in between. Both intensity distributions differed from that of MICU.1:GFP, indicating that the protein localizes to a mitochondrial subcompartment other than the matrix and the outer mitochondrial membrane. This points to presence of At-MICU at/in the inner mitochondrial membrane (IMM) and/or in the intermembrane space (IMS; Figure 3C), which is in agreement with a majority of MICU localization studies in mammalian systems.
Figure 3.
Fine Localization of MICU in Arabidopsis Mitochondria.
(A) Confocal laser scanning microscopy images of Arabidopsis seedlings expressing MICU.1:GFP and stained with MitoTracker Orange. GFP fusions of the Nicotiana plumbaginifolia β-ATPase mitochondrial presequence (mito:GFP) and NETWORK1/ELM1 (NMT1) were used as markers for the mitochondrial matrix and outer mitochondrial membrane (OMM), respectively. GFP, green; MitoTracker, magenta. Bar = 1 μm.
(B) Pixel intensities in the GFP and MitoTracker channels plotted across individual mitochondria. n = 9; error bars = sd.
(C) Schematic representation of intensity distributions as predicted for different mitochondrial subcompartments due to the limit of resolution for conventional light microscopy (Abbe limit).
Two T-DNA Insertion Lines Lack MICU Expression
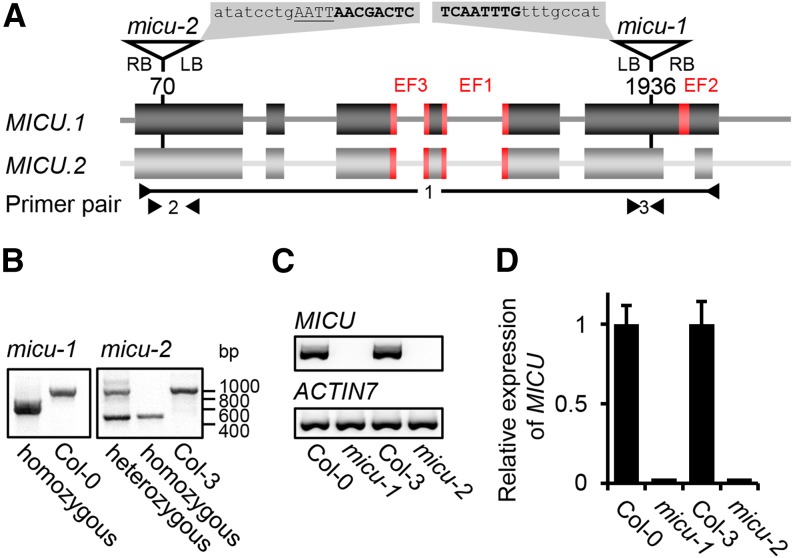
Based on the EF-hand motifs, the ability to bind Ca2+, the homology to mammalian MICU proteins, and the mitochondrial localization, we hypothesized that At-MICU is involved in mitochondrial Ca2+ dynamics in planta and chose a reverse genetic approach to test this hypothesis. Two independent Arabidopsis lines with T-DNA insertions in either the sixth (micu-1) or the first (micu-2) exon of the MICU locus were chosen (Figure 4A), and the precise positions of the respective insertion sites were determined by sequencing. The T-DNA cassette was integrated 1936 or 70 bp downstream of the MICU start codon in micu-1 or micu-2, respectively. Homozygous lines were selected and validated using PCR-based genotyping (Figure 4B). Transcript analysis by quantitative as well as standard RT-PCR showed a complete loss of MICU transcript (both splicing variants) in both insertion lines compared with their corresponding wild-type controls (Col-0 for micu-1 and Col-3 for micu-2; Figures 4C and 4D). This provided the basis for a genetic dissection of MICU function in vivo.
Figure 4.
Molecular Characterization of T-DNA Insertions in the MICU Locus.
(A) Triangles and corresponding nucleotide positions indicate T-DNA insertions in the MICU gene model for the micu-1 and micu-2 lines. Sequences on gray background show the precise insertion sites determined by sequencing. T-DNA in lowercase, endogenous sequence in bold capitals, and additional base pairs in underlined capitals.
(B) Genotyping of the T-DNA insertions in both micu lines.
(C) and (D) MICU transcript analysis in both micu lines by standard PCR using primer pair 1 (P2886/P2893) and ACTIN7 (P364/P436) as reference gene (C) and by quantitative RT-PCR with primer pairs 2 (P3529/P3530) and 3 (P3389/P3390) and SAND FAMILIY PROTEIN (P2455/P2456) as reference (D). Values are mean from three biological and three technical replicates; error bars = sd of biological replicates.
Generation of Sensor Lines for in Vivo Measurements of Matrix Ca2+ Dynamics
To obtain a specific measure of free Ca2+ concentration in the mitochondrial matrix in planta, we used the genetically encoded sensor Yellow Cameleon 3.6 (YC3.6; Nagai et al., 2004; Palmer and Tsien, 2006; Krebs et al., 2012), which was recently adapted for dynamic measurement of Ca2+ dynamics in the mitochondrial matrix of Arabidopsis (Loro et al., 2012; Loro and Costa, 2013; Wagner et al., 2015). Reversible Ca2+ binding by the sensor induces a conformational change that can be detected by a change in the Förster resonance energy transfer (FRET) efficiency between the two fluorophores that are part of the probe (ECFP and cpVenus) such that the FRET ratio (cpVenus:ECFP) increases with increasing free Ca2+ concentration, thereby providing a quantitative readout. The sensor construct for mitochondrial targeting was transformed into homozygous micu-1 and micu-2 and at least five positive independent T1 individuals were selected by sensor fluorescence for each. Suitable fluorescence intensity (i.e., expression levels) as well as correct mitochondrial localization of the sensor, by colocalization with MitoTracker Orange, were confirmed by confocal laser scanning microscopy (Figure 5A). For all measurements, homozygous T3 or T4 seedlings of at least two independent lines were used for both micu-1 and micu-2. This provided a robust system to assess the impact that lack of MICU has on mitochondrial Ca2+ dynamics in vivo.
Figure 5.
Root Ca2+ Levels in the Mitochondrial Matrix of micu and the Wild Type in Vivo.
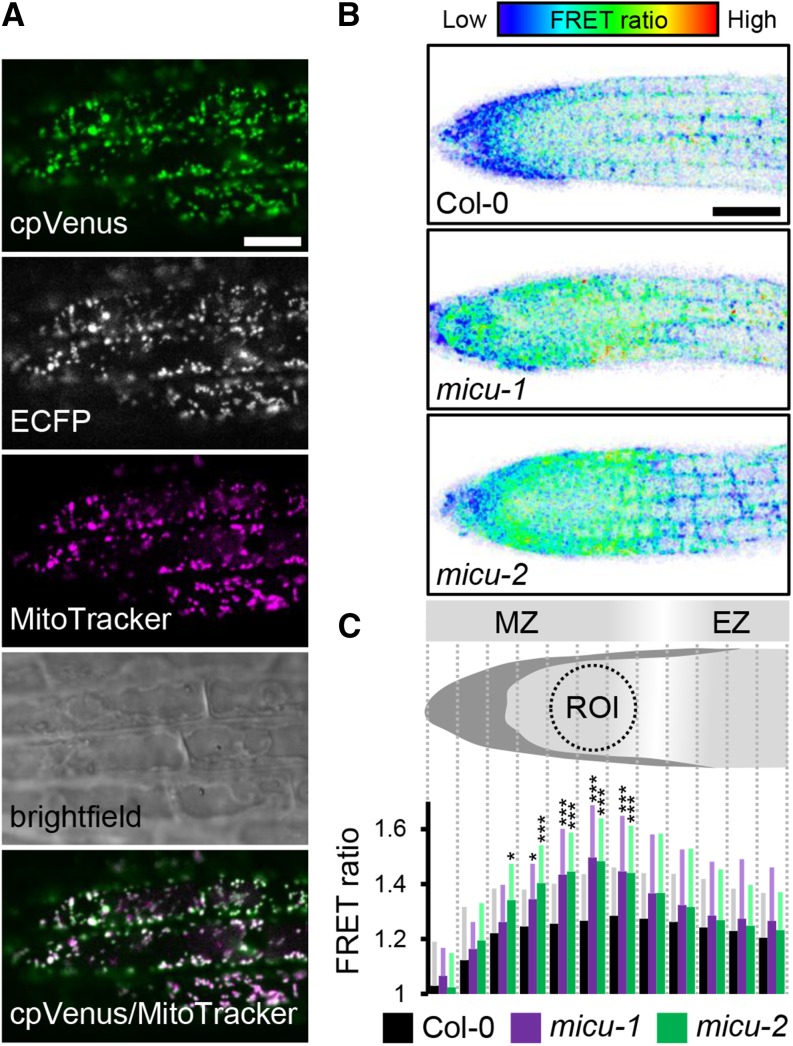
(A) Arabidopsis seedlings stably expressing mitochondrial Yellow Cameleon (4mt-YC3.6) stained with MitoTracker Orange and imaged by confocal laser scanning microscopy. cpVenus signal, green; MitoTracker signal, magenta. Bar = 10 μm.
(B) Representative false-color images illustrate FRET ratios along the root tip of roots expressing 4mt-YC3.6 in Col-0, micu-1, and micu-2 backgrounds. Low FRET ratios indicating low Ca2+ are shown in blue and high FRET ratios indicating high Ca2+ are shown in red. Bar = 50 μm.
(C) Schematic representation of the Arabidopsis root tip depicting 12 sections along the longitudinal root axis into which the ratio images were subdivided for quantitative analysis of FRET ratios. MZ, meristematic zone; EZ, elongation zone. Resulting FRET ratios for Col-0 (black), micu-1 (purple), and micu-2 (green) backgrounds for each section. n = 20; error bars = sd (for each data point indicated in gray, light purple, and light green). *P ≤ 0.05 and ***P ≤ 0.001 (t test). A dotted circle indicates the ROI for the dynamic analyses.
Resting Levels of Free Ca2+ in the Mitochondrial Matrix Are Strongly Increased in micu Roots
For in vivo imaging of free Ca2+, we focused on roots, where we have experimentally demonstrated MICU expression (Figure 2B). Root tissue offers favorable properties for in vivo imaging, including strong sensor signal, the opportunity to record data in a submerged state that is not dissimilar from the physiological state on culture plates, no risk of interference from chlorophyll autofluorescence, and established treatment procedures to induce mitochondrial Ca2+ transients (Loro et al., 2012; Loro and Costa, 2013; Wagner et al., 2015). We analyzed the confocal data sets in a ratiometric manner, generating pseudo-color images to represent the FRET ratios across the root tip. This indicated that ratio values for the micu lines were increased compared with the control, suggesting an elevated free Ca2+ concentration in the matrix (Figure 5B). The increase was similar in both micu lines and very consistent between experiments (and research labs). Nevertheless, the ratio images suggested substantial steady state heterogeneity in matrix calcium between the different tissue types in the root tip. Values in the root cap were consistently lower than those around the meristematic zone, with a decrease toward the elongation zone (Figure 5B). To account for this baseline heterogeneity and to assess quantitatively where the apparent FRET ratio changes were most pronounced in the micu lines, we systematically subdivided the ratio images into 12 equal sections starting from the tip (Figures 5B and 5C). While the sections in the cap area and the start of the elongation zone only showed minor differences between the lines, they were most pronounced and highly significant in the upper meristematic zone. To minimize the impact of tissue heterogeneity, we focused on a region of interest (ROI) in this area for the subsequent assessment of Ca2+ dynamics (Figure 5C). Semiquantitative assessment of intact root tips with the cationic lipophilic red fluorescent probe TMRM did not show any significant differences between the lines in the ROI (Supplemental Figure 5). TMRM reversibly accumulates in the mitochondrial matrix in response to the electrical potential ΔΨ across the inner membrane. Due to the Nernstian response characteristics of TMRM, even small changes in ΔΨ lead to pronounced changes in probe fluorescence (Schwarzländer et al., 2012a), suggesting that changes in ΔΨ were unlikely to account for the observed elevation in free matrix Ca2+. Increased FRET ratios of the Cameleon sensor suggest that matrix Ca2+ activity is constitutively elevated in root tissues of the micu lines and that MICU plays a role in maintaining matrix Ca2+ levels at low concentrations in vivo.
Mitochondrial Ca2+ Transients Occur Faster and Are Shifted toward Higher Maxima in micu Roots
To investigate if MICU is also involved in active Ca2+ dynamics, we next analyzed matrix Ca2+ transients in roots. Since the stimulation of well defined and strictly reproducible Ca2+ transients in complex plant tissue is technically challenging, we employed a custom perfusion setup for in vivo fluorescence microscopy of Arabidopsis roots. We applied auxin in the form of 10 μM 1-naphthaleneacetic acid (NAA), for which we have observed reliable induction of mitochondrial Ca2+ transients. Remarkable reproducibility of the observed response allowed quantitative analysis of bona fide in vivo Ca2+ dynamics (Figures 6A and 6B; Supplemental Figure 6A and Supplemental Movie 1). The analysis is based on FRET ratios for maximal rigor and avoidance of additional assumptions related to the transformation to absolute free Ca2+ concentrations. Nevertheless, meaningful estimation of free Ca2+ concentrations is possible (Supplemental Figure 7).
Figure 6.
Matrix Ca2+ Transients in Root Tips of micu and Wild-Type Seedlings.
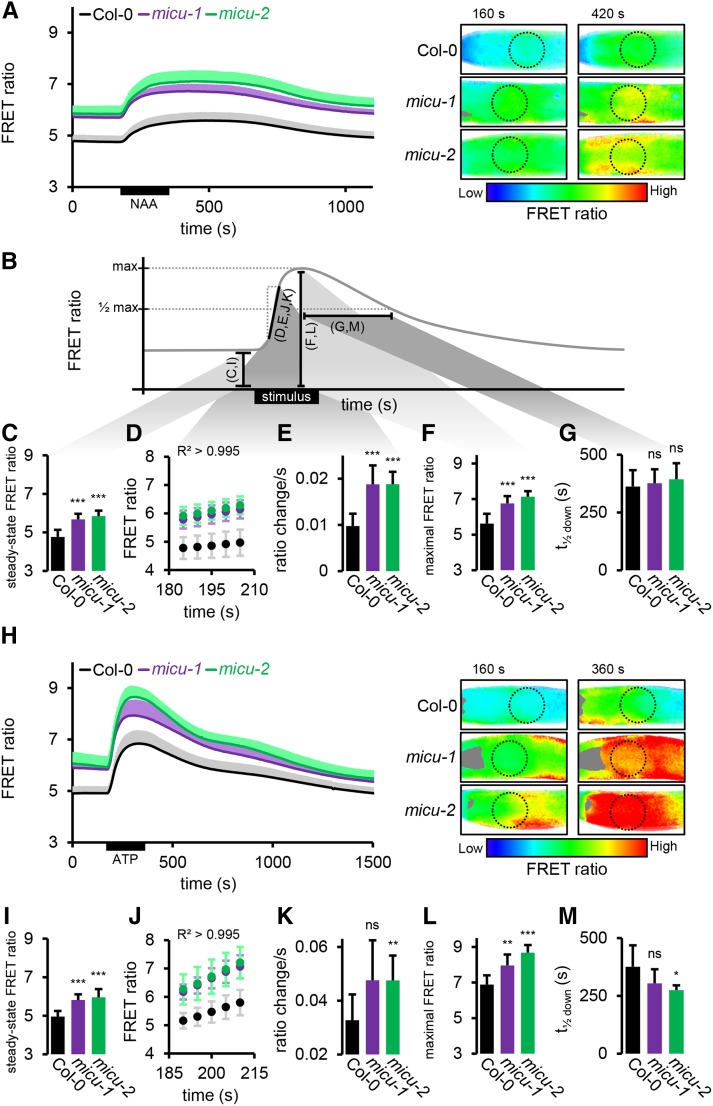
(A) Root tips of seedlings expressing 4mt-YC3.6 in control (Col-0) imaged under continuous perfusion and treated with 10 μM NAA for 3 min. FRET ratios of the ROI optimized in Figure 5C (dotted circles) are plotted over time. Representative ratiometric images are shown for two selected time points with low FRET ratios, indicating low Ca2+, in blue and high FRET ratios, indicating high Ca2+, in red.
(B) Schematic representation of the extracted kinetic parameters of the sensor response shown in the individual panels. Black bars and letters in parentheses indicate the individual parameters analyzed in (C) to (G) and (I) to (M). This relationship is further illustrated by gray shading between black bars and by (C) to (G).
(C) Steady state FRET ratios preceding NAA application (averaged over 35-s time window).
(D) FRET ratio increase following NAA application; linear region was selected with R2 > 0.995.
(E) Linear rate representing slope of regression of (D).
(F) Peak FRET ratio after NAA application.
(G) Time to pass half-maximal ratio during recovery.
(H) to (M) Analogous measurements as for (A) to (F) of treatments with 0.1 mM ATP instead of NAA. n ≥ 7; error bars = sd (for each data point of the time lapse curves indicated in gray, light purple, and light green). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 (t test).
The baseline ratio was stable in all lines, but significantly elevated in the micu lines (4.76 ± 0.37 in Col-0, 5.68 ± 0.29 in micu-1, and 5.85 ± 0.28 in micu-2; Figures 6A to 6C), confirming the steady state measurements by confocal imaging (see Figure 5; note that only relative, not absolute, FRET ratios can be compared between the two imaging techniques). NAA application caused an instantaneous increase in FRET ratios in all lines, reaching a relatively stable plateau for ∼300 s followed by recovery to baseline. The pronounced steady state shift between Col-0 and both micu lines remained significant throughout the entire transient. This suggests that (1) the maximal free Ca2+ concentrations reached in the matrix during the transient are much higher in the micu background and (2) that the baseline concentrations in the micu lines are comparable to those reached in the wild type upon excitation by the auxin stimulus. Although the transients of the micu lines appeared slightly different from one another, with micu-2 reaching a higher plateau than micu-1, this was not significant at any time point. The shape of the transients was also consistently altered in both micu lines relative to the wild type. The initial, linear rate of FRET ratio increase (R2 > 0.995) was nearly doubled (0.011 ± 0.003 s−1 in Col-0, 0.019 ± 0.004 s−1 in micu-1, and 0.019 ± 0.003 s−1 in micu-2), suggesting more rapid Ca2+ accumulation in the matrix (Figures 6B, 6D, an 6E). The plateau was reached earlier, demonstrated by shortened times to pass the half-maximal ratio (t1/2 up: 61.8 ± 7.2 s in Col-0, 32.1 ± 11 s in micu-1, and 45 ± 7.6 s in micu-2) and a higher peak FRET ratio was reached (5.62 ± 0.55 in Col-0, 6.76 ± 0.41 in micu-1, and 7.13 ± 0.31 in micu-2), representing higher maximal Ca2+ concentrations (Figures 6B and 6F). In contrast, the time to pass the half-maximal ratio on the recovery was not significantly altered (t1/2 down: 688.2 ± 70.9 s in Col-0, 636.4 ± 60.8 s in micu-1, and 682.5 ± 70 s in micu-2; Figures 6B and 6G), indicating unchanged kinetics of matrix Ca2+ clearance after the NAA stimulus.
To test if the observed changes in organellar Ca2+ kinetics were specific to NAA or can be generalized, we selected extracellular ATP (eATP; 0.1 mM) as an independent and well-established stimulus (Tanaka et al., 2010; Krebs et al., 2012; Loro et al., 2012; Behera et al., 2013; Choi et al., 2014). Importantly, our measurements in the control demonstrated that eATP triggers mitochondrial Ca2+ transients with a different signature to that of NAA (Figure 6H; Supplemental Figure 6B and Supplemental Movie 2).
The resting ratios were very similar to those measured prior to the NAA time courses, confirming elevated ratios in the micu lines (4.92 ± 0.27 in Col-0, 5.81 ± 0.3 in micu-1, and 5.96 ± 0.43 in micu-2; Figures 6B, 6H, and 6I). eATP gave rise to a transient increase in free Ca2+ in all lines, with a clear main maximum, higher than that of NAA, before declining again (Figure 6H). Similar to NAA, the shift between Col-0 and both micu lines remained significant throughout the entire transient, and the shape of the transients was distinct. The initial, linear rate, which was 3 times faster for eATP than for NAA in Col-0, was increased even further in the micu lines (0.032 ± 0.008 s−1 in Col-0, 0.044 ± 0.012 s−1 in micu-1, and 0.047 ± 0.009 s−1 in micu-2; Figures 6B, 6J, and 6K). Consistently, the maximum was also reached sooner (t1/2 up: 40.9 ± 7 s in Col-0, 29.2 ± 5.4 s in micu-1, and 37.1 ± 7 s in micu-2) and higher peak FRET ratios were reached (6.89 ± 0.52 in Col-0, 7.96 ± 0.62 in micu-1, and 8.68 ± 0.43 in micu-2; Figures 6B and 6L). In addition, the recovery was quicker in the micu lines (t1/2 down: 527.7 ± 92.4 s in Col-0, 426.7 ± 61 s in micu-1, and 407.1 ± 20.8 s in micu-2; Figure 6M), suggesting increased clearance rates after the eATP-induced transient, in contrast with observations for NAA.
The consistent behavior of the micu lines upon treatment with either auxin or eATP suggests that the mechanisms responsible for the different mitochondrial Ca2+ transients measured between micu lines and Col-0 are independent of stimulus identity. The only exception is the recovery rate that was unchanged in response to NAA, but increased in the mutants in response to eATP. It is possible that the high absolute matrix Ca2+ concentration in the micu lines in response to eATP (Supplemental Figure 7) may have additional and potentially independent regulatory impact.
Cytosolic Ca2+ Transients Appear Unchanged in micu Roots
Given the pronounced changes in matrix Ca2+ dynamics, we aimed to explore how those alterations are connected to subcellular Ca2+ dynamics in other compartments. To investigate cytosolic Ca2+ transients in the mutant micu lines, we expressed the Cameleon sensor YC3.6 in the cytosol (Krebs et al., 2012; Loro et al., 2012) in an analogous manner to the generation of the mitochondrial sensor lines, thereby enabling assessment of cytosolic Ca2+ dynamics under identical conditions to those used for mitochondrial Ca2+ measurements (Figures 7A and 7F; Supplemental Figures 6C and 6D and Supplemental Movies 3 and 4).
Figure 7.
Cytosolic Ca2+ Transients in Root Tips of micu and Wild-Type Seedlings.
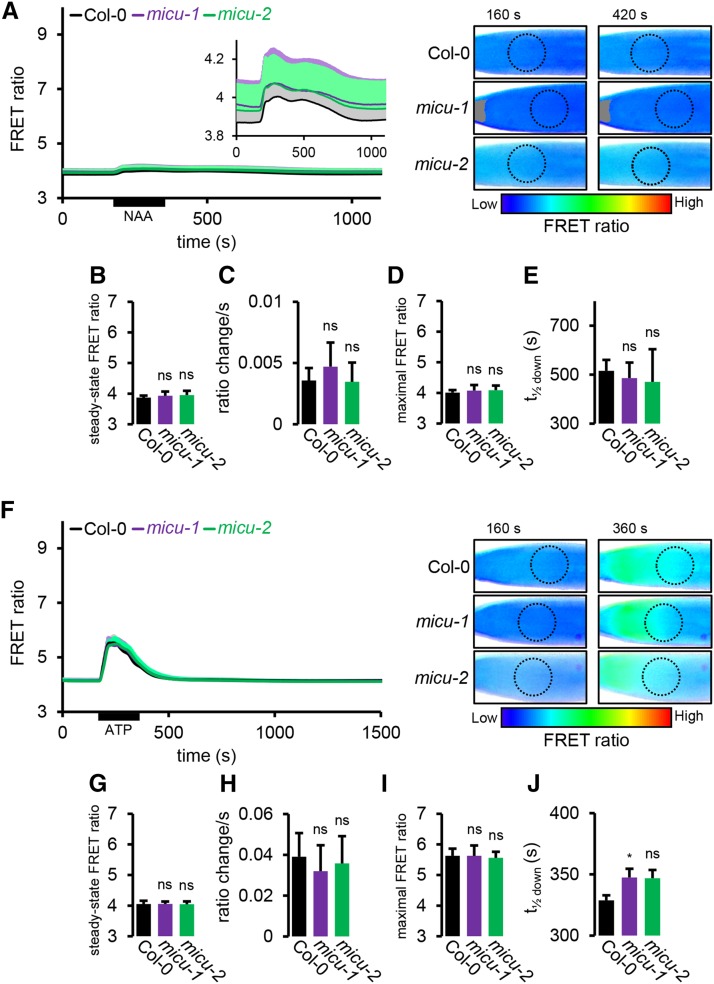
(A) Root tips of seedlings expressing NES-YC3.6 in control (Col-0) imaged under the same continuous perfusion conditions as in Figure 6A. FRET ratios of the ROI optimized in Figure 5C (dotted circles) are plotted over time. An inset serves to magnify the mild response using a different FRET ratio scale. Representative ratiometric images are shown for two selected time points with low FRET ratios, indicating low Ca2+, in blue and high FRET ratios, indicating high Ca2+, in red.
(B) Steady state FRET ratios preceding NAA application (averaged over 35-s time window).
(C) Linear rate of FRET ratio increase following NAA application (R2 > 0.995).
(D) Peak FRET ratio after NAA application.
(E) Time to pass half-maximal ratio during recovery.
(F) to (J) Analogous measurements as for (A) to (E) of treatments with 0.1 mM ATP instead of NAA. n ≥ 7; error bars = sd (for each data point of the time lapse curves indicated in gray, light purple, and light green). ns: P > 0.05 and *P ≤ 0.05 (t test).
Under steady state conditions, FRET ratios were consistently lower in the cytosol compared with the mitochondrial matrix, indicating lower free Ca2+ concentrations. Interestingly, the micu mutants did not differ significantly from the control (Figures 7A, 7B, 7F, and 7G). Similar behavior for control and micu lines was also found for cytosolic Ca2+ dynamics in response to both NAA and eATP, and there was no significant difference in FRET ratios found at any time point. NAA induced a much milder transient in the cytosol than in the mitochondrial matrix that was also distinct in shape, containing at least two components (Figure 7A). eATP triggered a cytosolic spike that was shorter lived and lower in amplitude compared with the mitochondrial matrix. The micu lines showed no alteration in any of the kinetic parameters that were changed in the matrix (see Figure 6B; Figures 7C, 7D, and 7H to 7J; Supplemental Table 1). To make sure that the Cameleon sensor was technically functional in the cytosol and responses were comparable with those in the mitochondrial matrix, we triggered a more pronounced Ca2+ transient as a positive control. We used a 20-fold higher concentration of eATP (2 mM) as a stimulus, which, in agreement with previous observations of dose dependence (Loro et al., 2012, 2013; Bonza et al., 2013), resulted in a pronounced increase in Ca2+ amplitude (Supplemental Figure 6D, inset), adding additional weight to the validity of the quantitative sensing setup.
Those observations suggest that while MICU shapes Ca2+ dynamics in the mitochondrial matrix, the impact appears to be mitochondria specific and does not have any major influence on the shape of cytosolic Ca2+ transients in planta.
Impacts on Mitochondrial Ultrastructure and Respiratory Characteristics in the micu Lines
To investigate if the regulatory impact of MICU on mitochondrial Ca2+ dynamics can be connected to structural properties of cellular organization, we performed transmission electron microscopy of root tips. A proportion of the mitochondrial populations observed showed decreased electron density and disorganization toward the center of their matrix. Yet, cristae were still clearly visible toward the periphery and appeared more tubular compared with the control, where cristae generally showed typical saccular configuration (Douce, 1985). Mitochondria with such abnormal internal ultrastructure were significantly enriched in the elongation zone of the micu lines (33% and 38%) compared with their corresponding controls (13 and 24%; Figure 8A; Supplemental Figure 8A and Supplemental Table 2). In the root apex, the number of abnormal mitochondria was generally low, although micu-2 also contained significantly more translucent mitochondria in this area compared with its corresponding wild type (Supplemental Figure 8B and Supplemental Table 2). No change in mitochondrial shape or size was evident.
Figure 8.
Root Mitochondrial Ultrastructure, Respiration, and Whole-Plant Development of micu Lines.
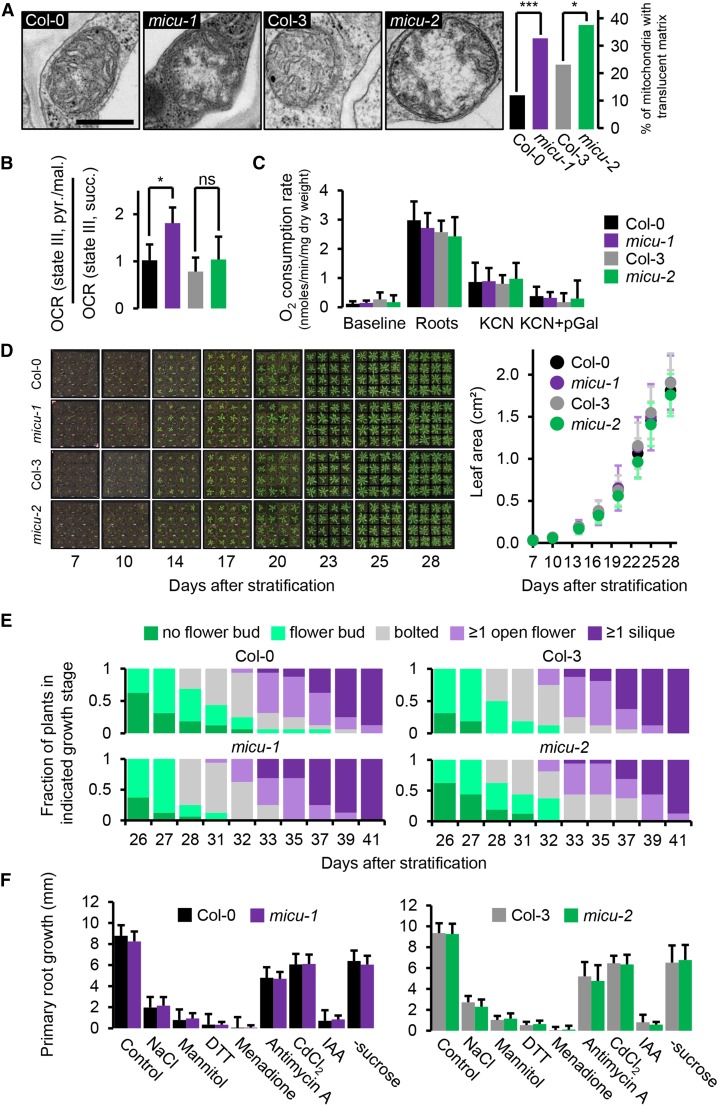
(A) Mitochondrial ultrastructure in cells of the root elongation zone of Arabidopsis seedlings. Exemplary electron micrographs of micu-1, micu-2, and their respective wild types (Col-0 and Col-3; see Supplemental Figure 8A for more images) to illustrate the overrepresentation of mitochondria with translucent matrices in the micu lines. Percentages of mitochondria scored by matrix appearance are shown as a bar graph; ≥185 mitochondria from three different seedlings were scored per line. Col-0 (black), micu-1 (purple), Col-3 (gray), and micu-2 (green). The analysis was repeated twice with consistent observations. *P ≤ 0.01 and ***P ≤ 0.001 (χ2 test; Supplemental Table 2). Bar = 0.5 μm.
(B) OCRs under pyruvate/malate-driven state III respirations over OCR under succinate-driven state III respiration of purified mitochondria of micu-1, micu-2, and their respective wild-type seedlings (Supplemental Figure 9). *P ≤ 0.05 and (ns) P > 0.05 (t test).
(C) Root respiration rates of micu-1, micu-2, and their respective wild-type lines and after addition of KCN (4 mM) and KCN (4 mM) + pGal (propylgallate; 0.2 mM). n = 5; error bars = sd.
(D) Vegetative rosette growth of micu-1, micu-2, and their respective wild types (Col-0 and Col-3) between 7 and 28 d after stratification as measured by total leaf area. n = 16; error bars = sd.
(E) Distribution of reproductive growth stages over time. n = 16.
(F) Root growth under abiotic stress treatments. Seedlings were grown on vertical 0.5× MS + 1% (w/v) sucrose medium plates for 5 d before being carefully transferred onto media supplemented with stress-inducing additives or lacking sucrose: NaCl (150 mM), mannitol (300 mM), DTT (1 mM), menadione (60 μM), antimycin A (20 μM), CdCl2 (100 μM), and IAA (indole-3-acetic acid; 1 μM). Primary root growth 2 d after seedling transfer is shown. n ≥ 24 per genotype and condition; error bars = sd.
None of the pairwise comparisons for (C), (D), and (F) (micu versus the wild type; t test) indicated any significant (P < 0.05) change. All phenotyping experiments were repeated at least twice with similar results.
To check if the ultrastructural changes indicated mitochondrial function being more generally altered in the micu lines, we assessed respiration of purified, intact mitochondria from whole seedlings. This allows a detailed appraisal of the mitochondrial respiratory machinery, at the cost of losing tissue specific effects, by averaging over large numbers of mitochondria from all tissues. For all preparations, outer membrane integrity was >90%, as measured by cytochrome c latency. We could not detect significant changes for the oxygen consumption rates (OCRs) of micu mitochondria in state I-V (with one exception) and in response to inhibition of KCN-sensitive transport and alternative oxidase nor for respiratory control and P:O ratio (Supplemental Figure 9). This indicated that the respiratory machinery remained functional and efficient in the micu lines.
Yet, we observed a tendency of decreased maximal rates (state III and V, as induced by carbonyl cyanide m-chlorophenyl hydrazone) in the micu mitochondria under succinate respiration (Supplemental Figure 9B). By contrast, this was not the case under pyruvate/malate respiration, where this trend appeared even inverted, i.e., increased maximal rates in micu mitochondria. Since the different substrates provide electrons to the ubiquinone pool of the electron transport chain via distinct import and dehydrogenase systems [succinate, predominantly via succinate dehydrogenase/complex II; pyruvate/malate, predominantly via NAD(P)H generated by malate dehydrogenase and the pyruvate dehydrogenase complex, and, in turn, complex I and the internal alternative NAD(P)H-dehydrogenases], respiratory capacities may be independently adjusted, with limitations becoming apparent in oxygen consumption at maximal rates. To test this hypothesis, we assessed the ratio of OCRs under pyruvate/malate-driven compared with succinate-driven state III respiration (Figure 8B). This ratio was consistently increased in all six micu mitochondrial preparations compared with their corresponding wild-type mitochondria (1.8× for micu-1 versus Col-0; 1.2× for micu-2 versus Col-3 on average) but only reached significance for micu-1 versus Col-0.
Alterations in electron flux stoichiometry suggest changes in the respiratory machinery that are permanent enough to be preserved in isolated organelles under the in vitro conditions employed. Since those conditions do not preserve physiological Ca2+ status and cannot be straightforwardly compared with the in vivo situation, we also assayed respiration in intact roots. Oxygen consumption rates were unchanged between the lines under control conditions and after successive treatment with the cytochrome c oxidase inhibitor KCN and the alternative oxidase inhibitor propylgallate (Figure 8C). Those data suggest no overall changes in root respiratory rate and in the capacities of both terminal oxidases at whole-tissue level. They do not provide details on the mechanistic organization of respiration nor on specific tissue areas or cell types, however, which deserve detailed investigation in the future.
Absence of MICU Does Not Cause Pronounced Developmental Alterations of the Whole Plant
To find out if the observed functional and structural phenotypes at mitochondrial and cellular level carry through to the whole plant, we compared the micu-1 and micu-2 lines against their specific wild-type backgrounds for developmental phenotypes. Employing a stringent and highly reproducible quantitative assessment regime, we could not observe any consistent differences throughout the life cycle of soil-grown plants (Figures 8D and 8E; Supplemental Figures 10A to 10C). Precedence for the importance of Ca2+ regulation, mitochondrial function, and environmental stimuli for root development (Kiegle et al., 2000; Finkemeier et al., 2005; Monshausen et al., 2008; Morgan et al., 2008) prompted us to also quantify primary root growth performance of seedlings on media plates under normal growth conditions and a set of abiotic stress conditions. All stress conditions led to decreased root growth, yet none triggered any significant change in growth between micu and the corresponding wild-type plants within 2 d of stress exposure (Figure 8F; Supplemental Figure 10D). Although our phenotype analysis cannot be comprehensive, it suggests that absence of MICU has a pronounced impact on mitochondrial Ca2+ and is associated with alterations of the mitochondrial ultrastructure and the respiratory machinery but does not lead to detectable changes in overall root respiration or visible alterations of whole plant development.
DISCUSSION
The Physical Basis of Mitochondrial Ca2+ Dynamics in Plants
Our study reopens the investigation of mitochondrial Ca2+ regulation in plants. It ties in with a large body of work performed over the last decades on various plant species and tissues investigating the functional properties of Ca2+ uptake into the matrix, mostly in vitro, i.e., in isolated mitochondria (Hanson et al., 1965; Dieter and Marmé, 1980; Akerman and Moore, 1983; Yamaya et al., 1984; Zottini and Zannoni, 1993). Convincing evidence has been provided for two different Ca2+ uptake models, either by uniport or by exchange. Yet, the observation of different uptake characteristics compared with isolated mammalian mitochondria has led to a conception that a uniporter system related to that in mammals may not exist in plants. Indeed, different systems may contribute to mitochondrial Ca2+ dynamics, but the physical players involved are still largely unknown. Recently, the glutamate receptor 3.5 (GLR3.5) was identified as a putative plant mitochondrial cation transporter (Teardo et al., 2015). However, despite Ca2+ being a potential substrate, no pronounced changes in mitochondrial Ca2+ dynamics could be detected. This may be explained by the existence of a sophisticated, multiplayer network for mitochondrial Ca2+ regulation, in which the loss of individual components can be, at least partially, compensated for. Our finding that loss of MICU indeed deregulates mitochondrial Ca2+ dynamics in a physiological in vivo situation (Figures 5 and 6) hence provides strong evidence in favor of a Ca2+ uniporter system related to that in mammals being present and functional in plants. The pronounced changes observed suggest that the uniport plays a major role in regulating matrix Ca2+ dynamics that cannot be fully compensated for by other channels or transporters. Our data do not rule out, and indeed support, the hypothesis that additional systems are operational in parallel.
Arabidopsis as a Model for Functional Analysis of MICU and the Mitochondrial Ca2+ Uniporter Complex in Eukaryotes
The Arabidopsis nuclear genome contains a single homolog of the mammalian MICU gene family (Figures 1A and 1B), allowing us to combine the advantage of the minimal genetic complexity of MICU with a reverse genetics approach in the intact organism using stable and independent T-DNA insertion lines (Figure 4). In contrast, the majority of genetic work to achieve loss of function in mammalian cells has been based on posttranscriptional knockdown strategies (Perocchi et al., 2010; Csordás et al., 2013; Patron et al., 2014) with only few studies employing knockout systems (Kamer and Mootha, 2014; Logan et al., 2014). Arabidopsis offers a favorable genetic situation to dissect MICU function in conjunction with substantial phylogenetic distance to the mammalian models, while the sessile and photoautotrophic lifestyle of plants places the system in a fundamentally different physiological context compared with mammalian cells. Use of closely related models, including mouse and a variety of different human and other mammalian cell lines, can hamper a clear distinction of fundamental, conserved players and mechanisms from variation within the mammalian class, species, or even cell lines. For instance, three MICU isoforms of distinct function operate in mammalian cells, necessitating a complex working model of MCU regulation (Plovanich et al., 2013). Yet, the number of MICU homologs varies in eukaryotes, pointing to a specific situation in mammals that cannot be straightforwardly generalized to other species. The Arabidopsis model provides an important step toward such an integrated understanding of MCUC across eukaryotes.
Localization of MICU Provides the Basis for Its Function
Subcellular proteomics and GFP fusion assays independently suggest that At-MICU localizes to the mitochondria in plants, in agreement with the situation in mammals (Figure 2). Moreover, the fluorescence distribution of MICU:GFP in Arabidopsis mitochondria (Figure 3) sheds light on the controversy of IMS versus matrix localization of MICU (Perocchi et al., 2010; Csordás et al., 2013; Hoffman et al., 2013; Patron et al., 2014; Pendin et al., 2014). Our data add weight to the hypothesis that the C terminus of MICU is localized in the IMS and that MICU is not a matrix protein. A conclusive verdict would necessitate independent evidence, such as immunodetection of MICU in subfractionated mitochondria or immunogold labeling of MICU in conjunction with transmission electron microscopy, which has been hampered by the lack of sensitivity of our antibody. Yet, together with the majority of studies that have presented evidence for IMS localization, it appears justified to consider MICU as exposed to the IMS in plants. The topology is important since it provides the basis for the physiological context in which the Ca2+ binding and potentially regulatory EF-hand motifs of MICU operate. Ca2+-dependent regulation of MICU from the IMS side is consistent with mitochondrial Ca2+ uniport activity being gated by cytosolic Ca2+, which was described before the physical components of the MCUC were identified (Moreau et al., 2006). Our in vitro observation that ΔMICU can bind Ca2+ within the intracellular physiological concentration range (Figures 1C and 1D), with higher affinity than the mammalian MICU homologs (Wang et al., 2014), and without the need to predict microdomains of high local Ca2+ concentrations, makes Ca2+-dependent regulation of MICU a realistic possibility.
Toward an Understanding of Subcellular Ca2+ Dynamics
Our assessment of subcellular Ca2+ dynamics using the genetically encoded Cameleon sensor YC3.6 provides depth and specificity to the analysis of intracellular physiology in planta (Figures 5 to 7). Stimulation of cytosolic Ca2+ transients by auxin has been reported recently (Monshausen et al., 2011), and critical roles in mediating auxin responses have been suggested for intracellular Ca2+ (Vanneste and Friml, 2013). Therefore, it is interesting to note that the mitochondrial matrix response to NAA was far more pronounced than that of the cytosol when measured in the root tip, a hot spot of auxin regulation (Figures 6 and 7). In addition, both steady state concentrations and responses of matrix Ca2+ appeared strongly tissue type specific. Generally, side-by-side comparison of cytosol and mitochondria in root tip cells indicates higher steady state concentrations of free Ca2+ in the matrix compared with the cytosol (Supplemental Figure 7). This is in agreement with early work based on aequorin and rhod2 measurements in Arabidopsis seedlings and maize suspension-cultured cells (Subbaiah et al., 1998; Logan and Knight, 2003). By contrast, higher intracellular Ca2+ concentrations, with less clearly defined subcellular differences, have been reported from tobacco suspension cells (Manzoor et al., 2012). Our observations further refine the understanding of the relationship of Ca2+ dynamics in the cytosol and the mitochondrial matrix. Plant mitochondrial Ca2+ dynamics reflect cytosolic transients, yet with different amplitude and longer recovery time (Figures 6 and 7), which is in agreement with previous observations (Logan and Knight, 2003; Loro et al., 2012). This justifies the interpretation that plant mitochondria rapidly accumulate Ca2+ from the cytosol driven by the membrane potential ΔΨ across the inner membrane, yet in a controlled manner, since concentrations do not rise to Nernstian equilibrium. Clearance of matrix Ca2+ is slow compared with the cytosol, which may be explained by the need to export Ca2+ against the electrochemical gradient using proton-coupled transport. Hence, mitochondria maintain a “memory” of cytosolic Ca2+ transients even after a steady state concentration has been reestablished in the cytosol. The differential responses of the cytosol and the matrix to auxin and eATP indicate differential translation of the cytosolic signature at the inner mitochondrial membrane. Amplitude, duration, and number of components of the cytosolic transients appear to modulate the characteristics of matrix transients. Despite this connection, a nonlinear relationship between Ca2+ dynamics in the cytosol and matrix appears to give rise to distinct, compartment-specific Ca2+ signatures (Figures 6 and 7). Significant integration must occur at the inner mitochondrial membrane by mechanisms that are currently not understood. Our data show that MICU plays an important part in the choreography of matrix signatures without affecting that of cytosolic transients in any major manner in the analyzed tissues. They further provide evidence for cell-type-specific differences in the root tip (Figures 5B and 5C), which deserve future investigation.
Absence of MICU Specifically Deregulates Matrix Ca2+ Dynamics in Planta
The role of MCUC components in regulating intracellular Ca2+ levels has been investigated in several studies employing forward and reverse genetics in mammalian cells (Mallilankaraman et al., 2012a; Csordás et al., 2013; Pan et al., 2013; Logan et al., 2014; Patron et al., 2014; Petrungaro et al., 2015). Nevertheless, the critical step toward the bona fide in vivo situation, i.e., assessing Ca2+ responses in the intact tissue of a multicellular organism such as a living, anesthetized mouse (Breckwoldt et al., 2014), has not been made. Here, we address that shortcoming. We have found steady state concentrations in the matrix to be strongly increased and transients to be shifted toward higher Ca2+ concentrations in root tips of Arabidopsis seedlings lacking MICU (Figures 5 and 6). In addition, the stimulus-induced rate of Ca2+ accumulation in the matrix is accelerated. Both effects may be interpreted as a result of decreased Ca2+ influx control: If lack of MICU increased Ca2+ leakage via MCUC, driven by the electrical gradient, this would be expected to cause a shift in the dynamic equilibrium between Ca2+ influx and export toward an increased steady state concentration of matrix Ca2+. The same reasoning may apply to the uptake rate upon onset of a cytosolic Ca2+ elevation, leading to accelerated Ca2+ accumulation in the matrix. This points to a role of MICU in controlling matrix Ca2+ by constraining the flux through MCUC driven by the steep negative membrane potential ΔΨ. As such, our data provide strong evidence that the mammalian mitochondrial Ca2+ uniporter system is not only functionally conserved in plants, but also consists, at least partially, of homologous subunits. The changes in matrix Ca2+ dynamics indicate that, despite likely redundancy of the Ca2+ regulatory network, the uniporter system plays a prominent role in shaping Ca2+ transients and setting the Ca2+ concentration in the matrix. However, the question if, and which of the plant homologs of MCU, the pore-forming protein of the mammalian MCUC also form a channel that can be regulated by MICU will have to be addressed in future studies.
Interestingly, no clear changes in Ca2+ dynamics were detectable in the cytosol of the micu lines in our hands (Figure 7). This is in contrast with observations in mammalian cells where mitochondria have been suggested to buffer cytosolic Ca2+ to shape transients, and interference with uniporter components, including MICU1, strongly modify cytosolic patterns (Drago et al., 2012; Logan et al., 2014). The specific modification of Ca2+ dynamics in the matrix of Arabidopsis mitochondria further supports the concept of the mitochondrion as a site of active Ca2+ signaling, rather than a passive buffering compartment for Ca2+ during cytosolic transients.
A Working Model of MICU Function
Based on our observations, we propose a speculative working model for MICU function in Arabidopsis (Figure 9): MICU localizes to the mitochondria and is exposed to the IMS (Figures 2C, 2D, and 3). A relatively low abundance is common for channel proteins and their regulators. At the IMM, it interacts (stably or transiently) with the IMM-spanning channel, presumably composed of MCU proteins. This channel is prone to spontaneous Ca2+ influx due to a steep negative membrane potential ΔΨ. MICU acts as a throttle to prevent this by an unknown mechanism, which may include the induction of a conformational change at the channel to inhibit Ca2+ flow. Upon a rise in free Ca2+ levels of the cytosol/IMS, specific EF-hands of MICU bind Ca2+ (Figures 1C and 1D). This may partially overcome the inhibitory effect of MICU on the pore by a conformational change and/or by dissociation (Petrungaro et al., 2015). In the absence of MICU, the pore lacks stringent sealing, resulting in steady leakage of Ca2+ from the IMS into the matrix. This leads to an increased steady state concentration in the matrix (Figures 5 and 6) that cannot be fully compensated for by increased Ca2+ buffering or export. Upon a rise in the cytosolic/IMS Ca2+ levels in micu plants (Figure 7), the MCUC is primed for Ca2+ uptake, without the inhibitory effect of MICU, leading to an increased uptake rate and an additive Ca2+ accumulation on top of the already elevated steady state concentration (Figure 6).
Figure 9.
Model of the Role of MICU in Arabidopsis.
(A) and (B) The impact of a hypothetical mitochondrial inner membrane that is nonpermeable (A) or fully permeable (B) to Ca2+ ions. Lack of permeability would prevent Ca2+ presence, while full permeability would cause Nernstian accumulation in the matrix driven by the steep electrical gradient of the mitochondrial membrane potential ΔΨ. Control of matrix Ca2+ requires an intermediate state between those extremes.
(C) A selective Ca2+ channel, likely to be composed of MCU proteins, provides restricted passage. MICU is localized in the IMS and acts as a throttle for Ca2+ flux by inhibiting the channel. Under steady state conditions, this inhibition allows relatively low Ca2+ levels to be maintained in the matrix, despite the coexistence of the channel and the large membrane potential ΔΨ.
(D) and (E) The presence of three EF-hands and modification of protein conformation by Ca2+ binding provides a mechanistic basis for the inhibitory effect of MICU to be tuned during Ca2+ transients (D). In the absence of MICU, its inhibitory effect on the channel is lifted, resulting in elevated steady state levels of free Ca2+ in the matrix based on an increased influx rate (E).
(F) When Ca2+ levels rise in the cytosol and IMS, e.g., by a physiological stimulus, uptake into the matrix can occur at an increased rate and free Ca2+ accumulates at higher levels. The remaining control is likely to be provided by buffering and export systems and the biophysical properties as well as abundance of the channel itself. Additional channel regulators may also contribute, but no homologs of mammalian candidates exist in Arabidopsis. Our data support this model both in vitro and in the bona fide in vivo situation of intact Arabidopsis root tissue.
MICU Provides Functional Regulation to Mitochondrial Ca2+ Uptake That Differs from the Mammalian System
The model outlined in the previous section and in Figure 9 is in overall agreement with several characteristics that have been proposed for MCUC regulation in mammalian cells, although it should be emphasized that no clear consensus on the function of the mammalian MICU proteins has been reached yet (Kamer et al., 2014; Kevin Foskett and Madesh, 2014). Taking this shortcoming into account, the situation in Arabidopsis still differs from that in mammals in several respects. Those differences may at least be partially attributed to the presence of one MICU isoform in Arabidopsis, rather than three in mouse and human (Figure 1A; Supplemental Figure 1). First, increased steady state levels of Ca2+ that have been observed upon both removal and overexpression of MICU1 have been interpreted as a “gatekeeping” function of mammalian MICU (Mallilankaraman et al., 2012a; Csordás et al., 2013). This inhibitory function that sets a threshold for flux through the MCU pore has since been refined and is now specifically attributed to the MICU1 paralog MICU2 (Kamer and Mootha, 2014; Patron et al., 2014). The impact of MICU1 on baseline Ca2+ levels may be due to the fact that MICU1 forms a heterodimer, stabilizing MICU2 and mediating its interaction with the MCU pore multimer (Harrington and Murphy, 2015; Petrungaro et al., 2015). Thus, increased matrix Ca2+ levels upon loss of MICU in Arabidopsis (Figures 5 and 6) resemble the functional impact of either MICU1 or MICU2 removal in mammalian cells. Second, in the absence of MICU1, but not MICU2, decreased amplitudes of Ca2+ transients have been found in mammalian cells (Kamer and Mootha, 2014; Patron et al., 2014; Harrington and Murphy, 2015). This points to the MICU system as a stimulatory regulator of the MCU pore during Ca2+ transients, mediated by MICU1. Yet, no inhibition of the transient amplitude was observed in the Arabidopsis micu lines compared with the control (Figure 6). Hence, our data do not justify the interpretation of an activating, cooperative effect of MICU on flux during a transient in Arabidopsis, as proposed for MICU1 in mammals. Instead, they suggest that At-MICU acts as a functional homolog of mammalian MICU2, but not MICU1, despite its higher sequence homology to MICU1. It needs to be pointed out that alternative explanations may account for our observation that the peak Ca2+ levels reached during a transient are increased in the micu lines (Figure 6), which appears in contrast with observations made in mammalian systems. For instance, acclimation of matrix Ca2+ buffering and/or Ca2+ export properties in the micu lines by a yet unknown mechanism remains conceivable.
Given the complex and not yet reliably understood interaction between MICU1 and 2 in mammals, with opposing functional effects, it is currently not clear if and how only a single MICU protein can deliver regulatory sophistication for mitochondrial Ca2+ uptake in plants. A key question to address will be that of potential interaction partners of At-MICU, including the nature of the putative regulatory interaction with the MCU pore. The presence of the two MICU splicing variants that we found to be highly differentially expressed, and to contain different numbers of EF-hand motifs (Figures 1B, 2A, and 2B), may offer intriguing possibilities for fine tuning.
Phenotypes and the Regulatory Impact of MICU in the Context of the Physiological Network
At the cellular level, a clear phenotype in both mitochondrial Ca2+ physiology (Figures 5 and 6) and mitochondrial ultrastructure occurred in the root elongation zone of micu mutants (Figure 8A). The nature of the electron-translucent areas in the matrix of those abnormal mitochondria is not clear. Ca2+ phosphate deposits as a result of Ca2+ overload would be expected to give crystalline, electron-dense areas (Peverly et al., 1974). Instead, the fine filaments within the translucent matrix are reminiscent of mitochondrial DNA (Douce, 1985). Increased mitochondrial DNA copy numbers have been associated with dysfunction-induced mitochondrial biogenesis in animals. Tubular, as opposed to saccular cristae, which are typical for root mitochondria, suggest limited bioenergetic activity, consistent with ongoing synthesis of the respiratory machinery and/or acclimation to respiratory induction by constitutive elevation of matrix Ca2+ (Manzoor et al., 2012).
In the light of altered Ca2+ dynamics and mitochondrial ultrastructure and respiratory characteristics in the mutants (Figures 8A and 8B), the absence of any obvious developmental phenotype at the whole plant level appears surprising at first glance (Figures 8D and 8E). Yet, it provides a remarkable parallel to the situation in mammals: Against all expectations, fully viable and healthy mice lacking the pore-forming MCU protein could be generated that showed loss of rapid mitochondrial Ca2+ uptake (Pan et al., 2013; Kwong et al., 2015; Luongo et al., 2015), although this appears to depend on the configuration of the genetic background (Murphy et al., 2014). Similarly, human individuals lacking MICU1 protein are fully viable, despite strong alterations in cytosolic and mitochondrial Ca2+ dynamics (Logan et al., 2014). Nevertheless, proximal myopathy, learning difficulties, and a progressive extrapyramidal movement disorder appear to be associated with the mutation. Yet, our finding that the Arabidopsis micu lines do not show any obvious change in development appears to match the general tendency of the absence of a severe developmental phenotype (Kamer and Mootha, 2015). In all systems, pronounced changes in intracellular Ca2+ physiology and mitochondrial function appear to not, or only mildly, carry through to the whole organism.
Hot spots of ATP demand in mammals, e.g., the active muscular and neuronal tissues, strictly depend on energy supply by mitochondrial respiration. It is not clear if a comparable situation exists in plants. Plant cells with high energetic demands and particularly active Ca2+ dynamics, such as fast-growing root hairs or pollen tubes (Colaço et al., 2012), have been shown to be able to maintain their function without mitochondrial ATP supply (Rounds et al., 2010). Instead of being streamlined for maximal efficiency in energy conversion, plant mitochondria show features, such as alternative electron transport pathways (Rasmusson et al., 2004), that indicate key roles in maintaining metabolic and energetic balance. This is likely an adaptation of plants to their sessile lifestyle in changing environments and may also underpin the evolution of plant mitochondrial Ca2+ regulation, whereby the organelle is constantly fine-tuning its function to be best tailored to a fluctuating environment. Our observation that most key parameters of respiration were unaltered in isolated micu mitochondria (Supplemental Figure 9), with preliminary evidence that the relative capacities of pyruvate/malate- and succinate-driven respiration are specifically shifted into opposite directions (Supplemental Figure 9; Figure 8B), fits such a concept of plant mitochondrial regulation. Yet, the potential shift in respiratory capacities can only be regarded as a first indication and will require future validation before interpretation with respect to the relationship between mitochondrial Ca2+ and bioenergetics metabolism. This may require orthogonal approaches, such as respiratory protein complex biochemistry, and benefit from tissue- or even cell-specific bioenergetic assessments to avoid dilution of localized changes. In particular, respiratory analysis specifically in the upper meristematic zone of the root tip will be desirable, where changes in matrix calcium dynamics were prominent in the micu lines.
Intracellular Ca2+ transients can be triggered by several stress treatments in plants (Knight et al., 1991; Kiegle et al., 2000; Logan and Knight, 2003; Loro et al., 2012), and recently it was reported that, similar to mammalian systems, an increased Ca2+ concentration in the matrix can coincide with stimulated mitochondrial respiration in plant cells (Manzoor et al., 2012). In addition, stress-inducible membrane potential pulses in single Arabidopsis mitochondria have been associated with mitochondrial Ca2+ dynamics. Pulsing is conserved in other eukaryotes and leads to respiratory uncoupling at the level of the individual mitochondrion (Duchen et al., 1998; Schwarzländer et al., 2012a, 2012b; Hou et al., 2013). Yet, we could not detect any clear stress-induced root growth phenotype of the micu lines under standardized culture conditions (Figure 8F), which may hint to more specific roles of mitochondrial Ca2+ and/or regulatory backup for plant growth and survival. The absence of a pronounced developmental phenotype in the micu lines may be advantageous for further mechanistic dissection of mitochondrial Ca2+ signaling since pleiotropic effects, although impossible to avoid completely, may be less dominant, allowing a meaningful dissection of specific and physiological Ca2+-related functions in planta.
In conclusion, we dissected subcellular Ca2+ dynamics in plants genetically. The Arabidopsis micu lines provide a conclusive case for the genetic modification of organellar Ca2+ physiology in vivo with much potential to facilitate a mechanistic understanding of how physiology is dynamically orchestrated at a subcellular level in plants. Our observations demonstrate that MICU regulates mitochondrial Ca2+ dynamics in planta by restricting influx. Bona fide in vivo imaging of the free Ca2+ concentration in Arabidopsis roots lacking MICU expression shows Ca2+ accumulation in the mitochondrial matrix under steady state conditions and modified mitochondrial Ca2+ transients, while the cytosolic Ca2+ concentration remains unaffected. As such, At-MICU displays functional parallels to mammalian MICU2, negatively regulating Ca2+ uptake.
METHODS
Plant Materials, Transformation, and Culture
The Arabidopsis thaliana micu-1 (SALK_064052; Col-0 background; Alonso et al., 2003) and micu-2 (SAIL_359_H07; Col-3 background; Sessions et al., 2002) lines were obtained from the Nottingham Arabidopsis Stock Centre and validated by genotyping and sequencing. Arabidopsis ecotypes Col-0 and Col-3 (segregated out from the T-DNA line) were used as wild-type lines. To generate the stable sensor and GFP-fusion lines, Agrobacterium tumefaciens-mediated transformation of Arabidopsis was performed by floral dip (Clough and Bent, 1998). Primary transformants were selected by antibiotic resistance and/or fluorescence. Homozygous lines were selected by segregation analysis using fluorescence as a selection marker. Where not indicated otherwise, surface-sterilized seeds were grown on vertical culture plates containing half-strength Murashige and Skoog (MS) agar (Murashige and Skoog, 1962) with 0.1% (w/v) sucrose, under long-day conditions after stratification at 4°C in the dark. For mitochondrial isolations, seedlings were cultured in hydroponic pots for 14 d as described before (Sweetlove et al., 2007). For localization studies in wild tobacco, Agrobacterium suspensions were infiltrated into leaves of 4- to 5-week old Nicotiana benthamiana plants. Transformed plants were returned to growth chambers for 2 to 4 d before further analysis.
Molecular Biology
A plasmid containing the full-length cDNA for At4g32060 (clone U20031) was obtained from the ABRC and used as template for PCR using Gateway attB-flanked primers to enable recombination into vectors pMDC45 and pMDC83 (Curtis and Grossniklaus, 2003) to generate GFP:MICU.1 and MICU.1:GFP, respectively. Both constructs were transformed into Agrobacterium strain AGL1 by electroporation. The agrobacteria strains for Arabidopsis transformation with the 4mt-YC3.6 and NES-YC3.6 sensor constructs were described previously (Loro et al., 2012). Selection of homozygous individuals of the micu lines was performed by PCR-based genotyping. Sequencing of the T-DNA insertion sites was performed using primers LBb1.3 and LB1mod. For expression analysis by RT-PCR, total RNA was isolated from seedling tissue using the NucleoSpin RNA II kit (Macherey-Nagel). RNA was reverse transcribed to cDNA using an M-MLV reverse transcriptase (Life Technologies) with oligo(dT) or random hexamer primers and used for standard and quantitative PCR using the primer combinations listed in Supplemental Table 3. qRT-PCR was performed on a CFX96 system (Bio-Rad) with PerfeCTa SYBR Green FastMix (Quanta Biosciences).
Phylogenetic Analysis
Protein sequences of representative plant species were retrieved from http://phytozome.jgi.doe.gov using human MICU1 (UniProt identifier Q9BPX6) as query sequence. Following manual curation, sequences were aligned with Muscle (Edgar, 2004) in MEGA 6 (Tamura et al., 2013) with default parameters (Supplemental File 1) and clustered in an unrooted maximum likelihood tree using the JTT model (Jones et al., 1992) and uniform rates among sites (MEGA6). The tree with the highest log likelihood (-9511.9518) is shown in Figure 1A and Supplemental Figure 1. Bootstrap values reported on nodes in Figure 1A were derived from 1000 repetitions.
Purification of ΔMICU
The coding sequence of At-MICU lacking its first 117 amino acids (ΔMICU) was PCR amplified from Arabidopsis cDNA with primers ΔMICU_fw and ΔMICU_rev. The PCR product was digested with NheI/XhoI and cloned into pET28a (Novagen). Following transformation into Escherichia coli ArcticExpress DE3 (Agilent Technologies) or E. coli Rosetta(DE3), expression of His:ΔMICU was induced with 0.5 mM isopropyl-β-thiogalactopyranoside for 16 h at 20°C. Cells were lysed in lysis buffer (25 mM Tris, 150 mM NaCl, and 5% [v/v] glycerol, pH 7.5) supplemented with 0.1 mM DTT. After centrifugation, total soluble proteins were loaded onto a Ni2+ chromatography column (GE Healthcare) and washed with lysis buffer containing 1 M NaCl and lysis buffer containing 15 mM imidazole before proteins were eluted with a 100 to 500 mM imidazole gradient (Supplemental Figure 2A).
Ca2+ Binding Assays
Ca2+ overlay assays were performed according to Maruyama et al. (1984) with 3 μg purified ΔMICU (∼46 kD), 2.5 μg recombinant aequorin (∼22 kD), and 5 μg BSA (∼66 kD). Ligand-induced thermal stabilization was measured using the ThermoFluor assay to analyze the Ca2+ and Mg2+ binding properties of recombinant ∆MICU (Matulis et al., 2005). Purified ∆MICU at 0.7 to 1 μM in 25 mM Tris, 150 mM NaCl, 100 mM imidazole, and 5% (v/v) glycerol, pH 7.4, or 25 mM Bis-Tris, 150 mM NaCl, 100 mM imidazole, and 5% (v/v) glycerol, pH 7.0, was mixed with 5 μL of 50× SYPRO-Orange dye (Life Technologies). Free Ca2+ and Mg2+ concentrations were calculated using Maxchelator software (maxchelator.stanford.edu) and adjusted in the presence of 0.25 mM EGTA or 0.1 mM EDTA, respectively. Melting curves were recorded on an Applied Biosystems 7500 RT-PCR cycler with band-pass TAMRA filter applying a temperature gradient between 15 and 95°C (1°C min−1). Melting point calculations were done in GraphPad Prism 6.
Antibody Production and Immunoblotting
Purified ΔMICU was separated by SDS-PAGE and isolated from the gel before being used to immunize rabbits with poly(A)-poly(U) as adjuvant. ΔMICU was then immobilized on CNBr-activated Sepharose 4B (GE Healthcare) to affinity purify the ΔMICU-specific IgG antibody from total rabbit serum. For immunoblots, proteins were transferred onto polyvinylidene fluoride membranes. Blocked membranes were probed over night at 4°C with ΔMICU-specific primary antibody at a 1:100 dilution. After applying a secondary anti-rabbit antibody conjugated to horseradish peroxidase, chemiluminescent signal was detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Mitochondrial Proteomics
Arabidopsis mitochondria were isolated from 2-week-old Col-0 seedlings (expressing the mitochondrial Ca2+ sensor YC3.6) as previously described (Sweetlove et al., 2007; Schwarzländer et al., 2011). Mitochondrial protein (800 μg) was isolated and trypsinated, and 1 μg of peptides was analyzed using an EASY-nLC 1000 (Thermo Scientific) coupled to a Q Exactive Plus mass spectrometer (Thermo Scientific) with column and gradient as described (König et al., 2014). Mass spectra were acquired in a data-dependent manner, with an automatic switch between MS and MS/MS scans using a top 12 method. MS spectra were acquired in the Orbitrap analyzer, with a mass range of 300 to 1750 m/z, at a resolution of 70,000 and a target value of 106 ions. Peptide fragmentation was performed with the HCD method, and MS/MS spectra were acquired in the Orbitrap analyzer and a target value of 105 ions at a resolution of 17,500. Raw MS files were processed using MaxQuant software (version 1.4.1.2; http://www.maxquant.org/) with iBAQ enabled (Cox and Mann, 2008; Cox et al., 2014). MS/MS spectra were searched by the Andromeda search engine against the decoy Arabidopsis TAIR10_pep_20101214 database including 248 common contaminant proteins. The list of typical contaminants in proteomic sample preparations is an integral part of the MaxQuant software package (Cox et al., 2009). Peptides and proteins were identified at a false discovery rate of 1%, with trypsin specificity and a maximum of two missed cleavages. Carbamidomethylation of cysteine residues was set as fixed, and oxidation of methionine and acetylation of the N terminus were set as variable modifications. The iBAQ algorithm was used for ranking of the absolute abundance of different proteins within a single sample (Schwanhäusser et al., 2009)
Confocal Laser Scanning Microscopy
Confocal imaging was performed using a Zeiss LSM780 confocal microscope and a ×40 (C-Apochromat, 1.20 numerical aperture, water immersion) or ×63 lens (Plan-Apochromat, 1.40 numerical aperture, oil immersion). For colocalization studies with MitoTracker, plant material was vacuum-infiltrated for 30 min with 200 nM MitoTracker Orange (Invitrogen). GFP was excited at 488 nm, and MitoTracker Orange was excited at 543 nm. Fluorescence of GFP and MitoTracker were measured at 495 to 545 nm and 570 to 620 nm, respectively, with the pinhole set to 1 airy unit. Chlorophyll was recorded at the 633-nm laser excitation line and emission at 650 to 600 nm. Pixel intensities of the GFP and MitoTracker channel were extracted using Zeiss ZEN software. Ca2+ imaging of root tips of intact 7- to 8-d-old seedlings was performed as recently described (Loro and Costa, 2013; Wagner et al., 2015). YC3.6 was excited at 458 nm, and emission of FRET pair proteins ECFP and cpVenus was collected at 465 to 500 and 525 to 560 nm, respectively. Membrane potential imaging was performed in living root tissue of 5-d-old seedlings expressing 4mt-YC3.6 in the presence of 50 nM TMRM as described previously (Schwarzländer et al., 2012a). TMRM was excited at 543 nm and cpVenus was excited at 514 nm; emission was collected at 570 to 620 and 525 to 560 nm, respectively.
Time-Lapse Imaging by Fluorescence Microscopy
Seven- to eight-day-old seedlings were carefully placed in a custom perfusion chamber and stabilized with wet cotton wool to continuously perfuse the roots with the imaging solution (5 mM KCl, 10 mM MES, and 10 mM CaCl2, pH 5.8, adjusted with Tris; Behera et al., 2013). The shoot was not submerged. Since Ca2+ transients can be readily triggered by mechanical stimuli, seedlings were rested under continuous perfusion for 10 min after mounting and before ratiometric image acquisition, to allow potential handling-related Ca2+ transients to settle. Consecutive acquisition after another 10 min did not show any change in FRET ratio, indicating that stable Ca2+ levels were reached at the time of measurement. For stimulation, the perfusion medium was supplemented with NAA (10 μM) or Na2ATP (0.1 mM; for positive control 2 mM) added to the chamber by perfusion with the same solution. Treatment solution was applied for 3 min and then washed out. Seedling roots expressing the Cameleon sensor in the mitochondria or the cytosol (Krebs et al., 2012; Loro et al., 2012) were imaged using a Nikon Ti-E inverted fluorescence microscope with a CFI PLAN APO 20× VC dry objective. Excitation light was produced by a Prior Lumen 200 PRO fluorescent lamp (Prior Scientific) at 440 nm (436/20 nm) set to 20%. Images were collected with a Hamamatsu Dual CCD Camera ORCA-D2. For Cameleon analysis, the FRET CFP/YFP optical block A11400-03 (emission 1 483/32 nm for CFP and emission 2 542/27 nm for the FRET) with a Hamamatsu dichroic 510 nm mirror was used for the simultaneous CFP and cpVenus acquisitions. Exposure times were 100 ms with a 4×4 pixel binning in the case of roots of seedlings expressing the mitochondrial Cameleon and 100 ms with a 2×2 CCD binning in the case of roots of seedlings expressing the cytosolic Cameleon. Images were acquired every 5 s. Filters and dichroic mirrors were purchased from Chroma Technology. A Nikon NIS-Element was used as a platform to control microscope, illuminator, camera, and postacquisition analyses.
Ratiometric Image Data Analysis
Individual ratiometric images and time-lapse imaging data of YC3.6 were analyzed using a custom MatLab program package (Fricker, 2015). Errors provided represent sd.
Transmission Electron Microscopy
Images were obtained from 2-week-old seedlings as described previously (Carraretto et al., 2013).
Respiration Assays
Oxygen consumption of intact Arabidopsis roots and isolated mitochondria were measured in Oxytherm Clark-type electrodes (Hansatech). Whole roots from 16-d-old seedlings were cut below the hypocotyl-root junction and assayed in 5 mM KCl, 10 mM MES, and 10 mM CaCl2, pH 5.8, adjusted with Tris. Arabidopsis mitochondria from 2-week-old seedlings were isolated as described by Sweetlove et al. (2007) and Schwarzländer et al. (2011). Oxygen consumption and integrity of the outer mitochondrial membrane were assayed as described before (Sweetlove et al., 2002).
Whole-Plant Phenotyping
Plants were grown under long-day conditions (17°C, 16 h at 65 μmol photons m−2 s−1, 8 h dark) in individual pots randomly distributed among standard greenhouse flats. Development of the leaf rosette was precisely documented photographically, and rosette parameters (area, solidity, and length of the major rosette axis) were analyzed with the Leaf Lab tool (version 1.41). Critical reproductive growth stages (Supplemental Figure 10C) were documented after inflorescence emergence. Surface-sterilized seeds were vertically grown on half-strength MS medium (Murashige and Skoog, 1962) + 1% (w/v) sucrose + 0.8% (w/v) Phytagel for 5 d under long-day conditions (16 h at 120 μmol photons m−2 s−1 at 22°C, 8 h dark at 18°C). Seedlings were then transferred onto half-strength MS medium + 1% (w/v) sucrose + 0.8% (w/v) Phytagel supplemented with stress-inducing compounds. Root growth and development under long-day conditions was documented for 5 d after transfer.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MICU, AT4G32060; NMT1/ELM1, AT5G22350; ACTIN7, AT5G09810; and SAND FAMILY PROTEIN, AT2G28390.
Supplemental Data
Supplemental Figure 1. Predicted features of At-MICU.
Supplemental Figure 2. Purification of ΔMICU and immunoblot with At-MICU antibody.
Supplemental Figure 3. MICU.1:GFP and GFP:MICU.1 localization in wild tobacco leaf epidermis.
Supplemental Figure 4. Localization of MICU.1:GFP in different Arabidopsis tissues.
Supplemental Figure 5. Assessment of mitochondrial membrane potential in root tips.
Supplemental Figure 6. Reproducibility of Ca2+ transients.
Supplemental Figure 7. Considerations on absolute free Ca2+ concentrations.
Supplemental Figure 8. Mitochondrial ultrastructure in cells of the root elongation zone and apex.
Supplemental Figure 9. Analysis of respiratory parameters in purified mitochondria.
Supplemental Figure 10. Experimental design of micu phenotyping.
Supplemental Table 1. Kinetic parameters of cytosolic calcium transients in Col-0 and micu lines after stimulus with either NAA or eATP.
Supplemental Table 2. Analysis of mitochondrial ultrastructure.
Supplemental Table 3. List of primers.
Supplemental Data Set 1. Sequence information used for phylogenetic analysis.
Supplemental Data Set 2. Proteins identified in Arabidopsis mitochondrial extracts.
Supplemental Movie 1. Mitochondrial Ca2+ transient induced by NAA.
Supplemental Movie 2. Mitochondrial Ca2+ transient induced by eATP.
Supplemental Movie 3. Cytosolic Ca2+ transient induced by NAA.
Supplemental Movie 4. Cytosolic Ca2+ transient induced by eATP.
Supplemental File 1. Sequence alignments for phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Jan Riemer (University of Cologne, Germany), Stefanie Müller (University of Freiburg, Germany), and Christopher Grefen (University of Tübingen, Germany) for important discussions. We also thank Jutta Baldauf and Heiko Schoof (both University of Bonn, Germany) for advice on the statistics. S.W. and T.N. thank the Deutscher Akademischer Austauschdienst for travel support. I.F. and M.F. acknowledge funding by the Max Planck Gesellschaft. L.C., L.N., and I.S. thank the University of Padova for support through Progetto di Ateneo. I.S. is grateful for funding by the Ministero dell’Istruzione, dell’Università e della Ricerca through the PRIN project (2010CSJX4F) and A.C. through the FIRB 2010 programme (RBFR10S1LJ_001). M.S. thanks the Deutsche Forschungsgemeinschaft for funding through the Emmy-Noether programme (SCHW1719/ 1-1).
AUTHOR CONTRIBUTIONS
S.W. designed and performed the research, analyzed the data, and cowrote the article. S.B. and F.G.D. performed time-lapse imaging. S.D.B., L. Cendron, and L.N. carried out Ca2+ binding analyses. D.C.L. generated the GFP fusion constructs and lines for subcellular localization. P.F. assisted with gene expression analysis, respiratory analysis, phenotyping, and image analysis. L. Carraretto performed the transmission electron microscopy analysis. E.T. generated the MICU antibody. T.N. cogenerated sensor lines and assisted in the optimization of in vivo sensing. M.F. and I.F. performed the proteomic analysis. M.D.F. contributed the custom imaging analysis software and consulted on the data analysis. O.V.A. assisted in stress phenotyping. A.J.M. provided imaging expertise and facilities. I.S. cosupervised the research, specifically the transmission electron microscopy analysis, and the Ca2+ binding analyses and assisted in writing the article. A.C. cosupervised the research, designed and performed time-lapse imaging, and assisted in writing the article. M.S. conceived, performed, and supervised the research and cowrote the article.
Glossary
- MCUC
mitochondrial Ca2+ uniporter complex
- IMM
inner mitochondrial membrane
- IMS
intermembrane space
- YC3.6
Yellow Cameleon 3.6
- FRET
Förster resonance energy transfer
- ROI
region of interest
- TMRM
tetramethyl rhodamine methyl ester
- NAA
1-naphthaleneacetic acid
- eATP
extracellular ATP
- OCR
oxygen consumption rate
- MS
Murashige and Skoog
Footnotes
Articles can be viewed online without a subscription.
References
- Akerman K.E., Moore A.L. (1983). Phosphate dependent, ruthenium red insensitive CA2+ uptake in mung bean mitochondria. Biochem. Biophys. Res. Commun. 114: 1176–1181. [DOI] [PubMed] [Google Scholar]
- Allen G.J., Chu S.P., Schumacher K., Shimazaki C.T., Vafeados D., Kemper A., Hawke S.D., Tallman G., Tsien R.Y., Harper J.F., Chory J., Schroeder J.I. (2000). Alteration of stimulus-specific guard cell calcium oscillations and stomatal closing in Arabidopsis det3 mutant. Science 289: 2338–2342. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Arimura S., Fujimoto M., Doniwa Y., Kadoya N., Nakazono M., Sakamoto W., Tsutsumi N. (2008). Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites. Plant Cell 20: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpagaus S., Rawyler A., Braendle R. (2002). Occurrence and characteristics of the mitochondrial permeability transition in plants. J. Biol. Chem. 277: 1780–1787. [DOI] [PubMed] [Google Scholar]
- Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., Bao X.R., Strittmatter L., Goldberger O., Bogorad R.L., Koteliansky V., Mootha V.K. (2011). Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476: 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behera S., Krebs M., Loro G., Schumacher K., Costa A., Kudla J. (2013). Ca2+ imaging in plants using genetically encoded Yellow Cameleon Ca2+ indicators. Cold Spring Harb. Protoc. 2013: 700–703. [DOI] [PubMed] [Google Scholar]
- Bernardi P., Di Lisa F. (2015). The mitochondrial permeability transition pore: molecular nature and role as a target in cardioprotection. J. Mol. Cell. Cardiol. 78: 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonza M.C., Loro G., Behera S., Wong A., Kudla J., Costa A. (2013). Analyses of Ca2+ accumulation and dynamics in the endoplasmic reticulum of Arabidopsis root cells using a genetically encoded Cameleon sensor. Plant Physiol. 163: 1230–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckwoldt M.O., et al. (2014). Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat. Med. 20: 555–560. [DOI] [PubMed] [Google Scholar]
- Carraretto L., Formentin E., Teardo E., Checchetto V., Tomizioli M., Morosinotto T., Giacometti G.M., Finazzi G., Szabó I. (2013). A thylakoid-located two-pore K+ channel controls photosynthetic light utilization in plants. Science 342: 114–118. [DOI] [PubMed] [Google Scholar]
- Chigri F., Flosdorff S., Pilz S., Kölle E., Dolze E., Gietl C., Vothknecht U.C. (2012). The Arabidopsis calmodulin-like proteins AtCML30 and AtCML3 are targeted to mitochondria and peroxisomes, respectively. Plant Mol. Biol. 78: 211–222. [DOI] [PubMed] [Google Scholar]
- Chigri F., Hörmann F., Stamp A., Stammers D.K., Bölter B., Soll J., Vothknecht U.C. (2006). Calcium regulation of chloroplast protein translocation is mediated by calmodulin binding to Tic32. Proc. Natl. Acad. Sci. USA 103: 16051–16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Tanaka K., Cao Y., Qi Y., Qiu J., Liang Y., Lee S.Y., Stacey G. (2014). Identification of a plant receptor for extracellular ATP. Science 343: 290–294. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colaço R., Moreno N., Feijó J.A. (2012). On the fast lane: mitochondria structure, dynamics and function in growing pollen tubes. J. Microsc. 247: 106–118. [DOI] [PubMed] [Google Scholar]
- Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J.V., Mann M. (2009). A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4: 698–705. [DOI] [PubMed] [Google Scholar]
- Csordás G., et al. (2013). MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca²⁺ uniporter. Cell Metab. 17: 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Stefani D., Raffaello A., Teardo E., Szabò I., Rizzuto R. (2011). A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476: 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter P., Marmé D. (1980). Ca2+ transport in mitochondrial and microsomal fractions from higher plants. Planta 150: 1–8. [DOI] [PubMed] [Google Scholar]
- Douce R. (1985). Mitochondria in Higher Plants: Structure, Function and Biogenesis. (Orlando, FL: Academic Press; ). [Google Scholar]
- Drago I., De Stefani D., Rizzuto R., Pozzan T. (2012). Mitochondrial Ca2+ uptake contributes to buffering cytoplasmic Ca2+ peaks in cardiomyocytes. Proc. Natl. Acad. Sci. USA 109: 12986–12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen M.R., Leyssens A., Crompton M. (1998). Transient mitochondrial depolarizations reflect focal sarcoplasmic reticular calcium release in single rat cardiomyocytes. J. Cell Biol. 142: 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgass K., Caesar K., Schleifenbaum F., Stierhof Y.D., Meixner A.J., Harter K. (2009). Novel application of fluorescence lifetime and fluorescence microscopy enables quantitative access to subcellular dynamics in plant cells. PLoS One 4: e5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971. [DOI] [PubMed] [Google Scholar]
- Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L.J., Dietz K.J. (2005). The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 280: 12168–12180. [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D., Davies J.M., Dolan L. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446. [DOI] [PubMed] [Google Scholar]
- Fricker M.D. (2015). Quantitative redox imaging software. Antioxid. Redox Signal. http://dx.doi.org/10.1089/ars.2015.6390. [DOI] [PubMed] [Google Scholar]
- Geisler D.A., Broselid C., Hederstedt L., Rasmusson A.G. (2007). Ca2+-binding and Ca2+-independent respiratory NADH and NADPH dehydrogenases of Arabidopsis thaliana. J. Biol. Chem. 282: 28455–28464. [DOI] [PubMed] [Google Scholar]
- Gilroy S., Fricker M.D., Read N.D., Trewavas A.J. (1991). Role of calcium in signal transduction of commelina guard cells. Plant Cell 3: 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E.J., Rutter G.A. (2009). Mitochondrial calcium as a key regulator of mitochondrial ATP production in mammalian cells. Biochim. Biophys. Acta 1787: 1324–1333. [DOI] [PubMed] [Google Scholar]
- Halestrap A.P., Richardson A.P. (2015). The mitochondrial permeability transition: a current perspective on its identity and role in ischaemia/reperfusion injury. J. Mol. Cell. Cardiol. 78: 129–141. [DOI] [PubMed] [Google Scholar]
- Hanson J.B., Malhotra S.S., Stoner C.D. (1965). Action of calcium on corn mitochondria. Plant Physiol. 40: 1033–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington J.L., Murphy E. (2015). The mitochondrial calcium uniporter: mice can live and die without it. J. Mol. Cell. Cardiol. 78: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N.E., et al. (2013). MICU1 motifs define mitochondrial calcium uniporter binding and activity. Cell Reports 5: 1576–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper C.M., Tanz S.K., Castleden I.R., Vacher M.A., Small I.D., Millar A.H. (2014). SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30: 3356–3364. [DOI] [PubMed] [Google Scholar]
- Hou T., Zhang X., Xu J., Jian C., Huang Z., Ye T., Hu K., Zheng M., Gao F., Wang X., Cheng H. (2013). Synergistic triggering of superoxide flashes by mitochondrial Ca2+ uniport and basal reactive oxygen species elevation. J. Biol. Chem. 288: 4602–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Joshi H.J., et al. (2011). MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol. 155: 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer K.J., Mootha V.K. (2014). MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep. 15: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer K.J., Mootha V.K. (2015). The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 16: 545–553. [DOI] [PubMed] [Google Scholar]
- Kamer K.J., Sancak Y., Mootha V.K. (2014). The uniporter: from newly identified parts to function. Biochem. Biophys. Res. Commun. 449: 370–372. [DOI] [PubMed] [Google Scholar]
- Kato M., Watanabe Y., Iino S., Takaoka Y., Kobayashi S., Haga T., Hidaka H. (1998). Cloning and expression of a cDNA encoding a new neurocalcin isoform (neurocalcin alpha) from bovine brain. Biochem. J. 331: 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevin Foskett J., Madesh M. (2014). Regulation of the mitochondrial Ca2+ uniporter by MICU1 and MICU2. Biochem. Biophys. Res. Commun. 449: 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegle E., Moore C.A., Haseloff J., Tester M.A., Knight M.R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23: 267–278. [DOI] [PubMed] [Google Scholar]
- Kim M., Lim J.H., Ahn C.S., Park K., Kim G.T., Kim W.T., Pai H.S. (2006). Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 18: 2341–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61: 561–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirichok Y., Krapivinsky G., Clapham D.E. (2004). The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427: 360–364. [DOI] [PubMed] [Google Scholar]
- Knight M.R., Campbell A.K., Smith S.M., Trewavas A.J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 352: 524–526. [DOI] [PubMed] [Google Scholar]
- König A.C., et al. (2014). The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol. 164: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács-Bogdán E., Sancak Y., Kamer K.J., Plovanich M., Jambhekar A., Huber R.J., Myre M.A., Blower M.D., Mootha V.K. (2014). Reconstitution of the mitochondrial calcium uniporter in yeast. Proc. Natl. Acad. Sci. USA 111: 8985–8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M., Held K., Binder A., Hashimoto K., Den Herder G., Parniske M., Kudla J., Schumacher K. (2012). FRET-based genetically encoded sensors allow high-resolution live cell imaging of Ca2+ dynamics. Plant J. 69: 181–192. [DOI] [PubMed] [Google Scholar]
- Kuhn S., Bussemer J., Chigri F., Vothknecht U.C. (2009). Calcium depletion and calmodulin inhibition affect the import of nuclear-encoded proteins into plant mitochondria. Plant J. 58: 694–705. [DOI] [PubMed] [Google Scholar]
- Kwong J.Q., Lu X., Correll R.N., Schwanekamp J.A., Vagnozzi R.J., Sargent M.A., York A.J., Zhang J., Bers D.M., Molkentin J.D. (2015). The mitochondrial calcium uniporter selectively matches metabolic output to acute contractile stress in the heart. Cell Reports 12: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C.V., et al.; UK10K Consortium (2014). Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat. Genet. 46: 188–193. [DOI] [PubMed] [Google Scholar]
- Logan D.C. (2007). The mitochondrial compartment. J. Exp. Bot. 58: 1225–1243. [PubMed] [Google Scholar]
- Logan D.C., Knight M.R. (2003). Mitochondrial and cytosolic calcium dynamics are differentially regulated in plants. Plant Physiol. 133: 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan D.C., Leaver C.J. (2000). Mitochondria-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J. Exp. Bot. 51: 865–871. [PubMed] [Google Scholar]
- Logan D.C., Scott I., Tobin A.K. (2003). The genetic control of plant mitochondrial morphology and dynamics. Plant J. 36: 500–509. [DOI] [PubMed] [Google Scholar]
- Loro G., Costa A. (2013). Imaging of mitochondrial and nuclear Ca2+ dynamics in Arabidopsis roots. Cold Spring Harb. Protoc. 2013: 781–785. [DOI] [PubMed] [Google Scholar]
- Loro G., Drago I., Pozzan T., Schiavo F.L., Zottini M., Costa A. (2012). Targeting of Cameleons to various subcellular compartments reveals a strict cytoplasmic/mitochondrial Ca2+ handling relationship in plant cells. Plant J. 71: 1–13. [DOI] [PubMed] [Google Scholar]
- Loro G., Ruberti C., Zottini M., Costa A. (2013). The D3cpv Cameleon reports Ca2+ dynamics in plant mitochondria with similar kinetics of the YC3.6 Cameleon, but with a lower sensitivity. J. Microsc. 249: 8–12. [DOI] [PubMed] [Google Scholar]
- Luongo T.S., et al. (2015). The mitochondrial calcium uniporter matches energetic supply with cardiac workload during stress and modulates permeability transition. Cell Reports 12: 23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K., et al. (2012a). MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell 151: 630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallilankaraman K., et al. (2012b). MCUR1 is an essential component of mitochondrial Ca2+ uptake that regulates cellular metabolism. Nat. Cell Biol. 14: 1336–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoor H., Chiltz A., Madani S., Vatsa P., Schoefs B., Pugin A., Garcia-Brugger A. (2012). Calcium signatures and signaling in cytosol and organelles of tobacco cells induced by plant defense elicitors. Cell Calcium 51: 434–444. [DOI] [PubMed] [Google Scholar]
- Marchi S., et al. (2013). Downregulation of the mitochondrial calcium uniporter by cancer-related miR-25. Curr. Biol. 23: 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K., Mikawa T., Ebashi S. (1984). Detection of calcium binding proteins by 45Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J. Biochem. 95: 511–519. [DOI] [PubMed] [Google Scholar]
- Matulis D., Kranz J.K., Salemme F.R., Todd M.J. (2005). Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 44: 5258–5266. [DOI] [PubMed] [Google Scholar]
- Mehlmer N., Parvin N., Hurst C.H., Knight M.R., Teige M., Vothknecht U.C. (2012). A toolset of aequorin expression vectors for in planta studies of subcellular calcium concentrations in Arabidopsis thaliana. J. Exp. Bot. 63: 1751–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q., Chen Y., Zhang M., Chen Y., Yuan J., Murray S.C. (2015). Molecular characterization and phylogenetic analysis of ZmMCUs in maize. Biologia 70: 599–605. [Google Scholar]
- Miernyk J.A., Fang T.K., Randall D.D. (1987). Calmodulin antagonists inhibit the mitochondrial pyruvate dehydrogenase complex. J. Biol. Chem. 262: 15338–15340. [PubMed] [Google Scholar]
- Miller J.B., Pratap A., Miyahara A., Zhou L., Bornemann S., Morris R.J., Oldroyd G.E. (2013). Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 25: 5053–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller I.M., Johnston S.P., Palmer J.M. (1981). A specific role for Ca2+ in the oxidation of exogenous NADH by Jerusalem-artichoke (Helianthus tuberosus) mitochondria. Biochem. J. 194: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller I.M., Rasmusson A.G. (1998). The role of NADP in the mitochondrial matrix. Trends Plant Sci. 3: 21–27. [Google Scholar]
- Monné M., Miniero D.V., Obata T., Daddabbo L., Palmieri L., Vozza A., Nicolardi M.C., Fernie A.R., Palmieri F. (2015). Functional characterization and organ distribution of three mitochondrial ATP-Mg/Pi carriers in Arabidopsis thaliana. Biochim. Biophys. Acta 1847: 1220–1230. [DOI] [PubMed] [Google Scholar]
- Monshausen G.B., Messerli M.A., Gilroy S. (2008). Imaging of the Yellow Cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 147: 1690–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen G.B., Miller N.D., Murphy A.S., Gilroy S. (2011). Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J. 65: 309–318. [DOI] [PubMed] [Google Scholar]
- Moreau B., Nelson C., Parekh A.B. (2006). Biphasic regulation of mitochondrial Ca2+ uptake by cytosolic Ca2+ concentration. Curr. Biol. 16: 1672–1677. [DOI] [PubMed] [Google Scholar]
- Morgan M.J., Lehmann M., Schwarzländer M., Baxter C.J., Sienkiewicz-Porzucek A., Williams T.C., Schauer N., Fernie A.R., Fricker M.D., Ratcliffe R.G., Sweetlove L.J., Finkemeier I. (2008). Decrease in manganese superoxide dismutase leads to reduced root growth and affects tricarboxylic acid cycle flux and mitochondrial redox homeostasis. Plant Physiol. 147: 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Murphy E., Pan X., Nguyen T., Liu J., Holmström K.M., Finkel T. (2014). Unresolved questions from the analysis of mice lacking MCU expression. Biochem. Biophys. Res. Commun. 449: 384–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Yamada S., Tominaga T., Ichikawa M., Miyawaki A. (2004). Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. USA 101: 10554–10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura H., Shiina T. (2014). Calcium signaling in plant endosymbiotic organelles: mechanism and role in physiology. Mol. Plant 7: 1094–1104. [DOI] [PubMed] [Google Scholar]
- Palmer A.E., Tsien R.Y. (2006). Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 1: 1057–1065. [DOI] [PubMed] [Google Scholar]
- Pan X., et al. (2013). The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat. Cell Biol. 15: 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patron M., Checchetto V., Raffaello A., Teardo E., Vecellio Reane D., Mantoan M., Granatiero V., Szabò I., De Stefani D., Rizzuto R. (2014). MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol. Cell 53: 726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paupe V., Prudent J., Dassa E.P., Rendon O.Z., Shoubridge E.A. (2015). CCDC90A (MCUR1) is a cytochrome c oxidase assembly factor and not a regulator of the mitochondrial calcium uniporter. Cell Metab. 21: 109–116. [DOI] [PubMed] [Google Scholar]
- Pei Z.M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734. [DOI] [PubMed] [Google Scholar]
- Pendin D., Greotti E., Pozzan T. (2014). The elusive importance of being a mitochondrial Ca2+ uniporter. Cell Calcium 55: 139–145. [DOI] [PubMed] [Google Scholar]
- Perocchi F., Gohil V.M., Girgis H.S., Bao X.R., McCombs J.E., Palmer A.E., Mootha V.K. (2010). MICU1 encodes a mitochondrial EF hand protein required for Ca2+ uptake. Nature 467: 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrungaro C., Zimmermann K.M., Küttner V., Fischer M., Dengjel J., Bogeski I., Riemer J. (2015). The Ca2+-dependent release of the Mia40-induced MICU1-MICU2 dimer from MCU regulates mitochondrial Ca2+ uptake. Cell Metab. 22: 721–733. [DOI] [PubMed] [Google Scholar]
- Peverly J.H., Miller R.J., Malone C., Koeppe D.E. (1974). Ultrastructural evidence for calcium phosphate deposition by isolated corn shoot mitochondria. Plant Physiol. 54: 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plovanich M., et al. (2013). MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One 8: e55785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaello A., De Stefani D., Sabbadin D., Teardo E., Merli G., Picard A., Checchetto V., Moro S., Szabò I., Rizzuto R. (2013). The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 32: 2362–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson A.G., Møller I.M. (1991). Effect of calcium ions and inhibitors on internal NAD(P)H dehydrogenases in plant mitochondria. Eur. J. Biochem. 202: 617–623. [DOI] [PubMed] [Google Scholar]
- Rasmusson A.G., Soole K.L., Elthon T.E. (2004). Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu. Rev. Plant Biol. 55: 23–39. [DOI] [PubMed] [Google Scholar]
- Rocha A.G., Vothknecht U.C. (2012). The role of calcium in chloroplasts--an intriguing and unresolved puzzle. Protoplasma 249: 957–966. [DOI] [PubMed] [Google Scholar]
- Rounds C.M., Hepler P.K., Fuller S.J., Winship L.J. (2010). Oscillatory growth in lily pollen tubes does not require aerobic energy metabolism. Plant Physiol. 152: 736–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai J., Johnson C.H. (2002). Dark-stimulated calcium ion fluxes in the chloroplast stroma and cytosol. Plant Cell 14: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvato F., Havelund J.F., Chen M., Rao R.S., Rogowska-Wrzesinska A., Jensen O.N., Gang D.R., Thelen J.J., Møller I.M. (2014). The potato tuber mitochondrial proteome. Plant Physiol. 164: 637–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., et al. (2013). EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342: 1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo-Domingo J., Demaurex N. (2010). Calcium uptake mechanisms of mitochondria. Biochim. Biophys. Acta 1797: 907–912. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B., Gossen M., Dittmar G., Selbach M. (2009). Global analysis of cellular protein translation by pulsed SILAC. Proteomics 9: 205–209. [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., Logan D.C., Fricker M.D., Sweetlove L.J. (2011). The circularly permuted yellow fluorescent protein cpYFP that has been used as a superoxide probe is highly responsive to pH but not superoxide in mitochondria: implications for the existence of superoxide ‘flashes’. Biochem. J. 437: 381–387. [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., Logan D.C., Johnston I.G., Jones N.S., Meyer A.J., Fricker M.D., Sweetlove L.J. (2012a). Pulsing of membrane potential in individual mitochondria: a stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis. Plant Cell 24: 1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M., Murphy M.P., Duchen M.R., Logan D.C., Fricker M.D., Halestrap A.P., Müller F.L., Rizzuto R., Dick T.P., Meyer A.J., Sweetlove L.J. (2012b). Mitochondrial ‘flashes’: a radical concept repHined. Trends Cell Biol. 22: 503–508. [DOI] [PubMed] [Google Scholar]
- Scott I., Logan D.C. (2008). Mitochondrial morphology transition is an early indicator of subsequent cell death in Arabidopsis. New Phytol. 177: 90–101. [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stael S., Rocha A.G., Robinson A.J., Kmiecik P., Vothknecht U.C., Teige M. (2011). Arabidopsis calcium-binding mitochondrial carrier proteins as potential facilitators of mitochondrial ATP-import and plastid SAM-import. FEBS Lett. 585: 3935–3940. [DOI] [PubMed] [Google Scholar]
- Stael S., Wurzinger B., Mair A., Mehlmer N., Vothknecht U.C., Teige M. (2012). Plant organellar calcium signalling: an emerging field. J. Exp. Bot. 63: 1525–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah C.C., Bush D.S., Sachs M.M. (1998). Mitochondrial contribution to the anoxic Ca2+ signal in maize suspension-cultured cells. Plant Physiol. 118: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L.J., Heazlewood J.L., Herald V., Holtzapffel R., Day D.A., Leaver C.J., Millar A.H. (2002). The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32: 891–904. [DOI] [PubMed] [Google Scholar]
- Sweetlove L.J., Taylor N.L., Leaver C.J. (2007). Isolation of intact, functional mitochondria from the model plant Arabidopsis thaliana. Methods Mol. Biol. 372: 125–136. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Swanson S.J., Gilroy S., Stacey G. (2010). Extracellular nucleotides elicit cytosolic free calcium oscillations in Arabidopsis. Plant Physiol. 154: 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz S.K., Castleden I., Hooper C.M., Vacher M., Small I., Millar H.A. (2013). SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 41: D1185–D1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teardo E., Carraretto L., De Bortoli S., Costa A., Behera S., Wagner R., Lo Schiavo F., Formentin E., Szabò I. (2015). Alternative splicing-mediated targeting of the Arabidopsis GLUTAMATE RECEPTOR3.5 to mitochondria affects organelle morphology. Plant Physiol. 167: 216–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano F.J., Thakkar S.S., Fang T., Weisemann J.M. (1997). Characterization and expression of NAD(H)-dependent glutamate dehydrogenase genes in Arabidopsis. Plant Physiol. 113: 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., Friml J. (2013). Calcium: the missing link in auxin action. Plants 2: 650–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Nietzel T., Aller I., Costa A., Fricker M.D., Meyer A.J., Schwarzländer M. (2015). Analysis of plant mitochondrial function using fluorescent protein sensors. Methods Mol. Biol. 1305: 241–252. [DOI] [PubMed] [Google Scholar]
- Wais R.J., Galera C., Oldroyd G., Catoira R., Penmetsa R.V., Cook D., Gough C., Denarié J., Long S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Yang X., Li S., Wang Z., Liu Y., Feng J., Zhu Y., Shen Y. (2014). Structural and mechanistic insights into MICU1 regulation of mitochondrial calcium uptake. EMBO J. 33: 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalley H.J., Sargeant A.W., Steele J.F., Lacoere T., Lamb R., Saunders N.J., Knight H., Knight M.R. (2011). Transcriptomic analysis reveals calcium regulation of specific promoter motifs in Arabidopsis. Plant Cell 23: 4079–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaya T., Oaks A., Matsumoto H. (1984). Stimulation of mitochondrial calcium uptake by light during growth of corn shoots. Plant Physiol. 75: 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., et al. (2012). LETM proteins play a role in the accumulation of mitochondrially encoded proteins in Arabidopsis thaliana and AtLETM2 displays parent of origin effects. J. Biol. Chem. 287: 41757–41773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zottini M., Zannoni D. (1993). The use of fura-2 fluorescence to monitor the movement of free calcium ions into the matrix of plant mitochondria (Pisum sativum and Helianthus tuberosus). Plant Physiol. 102: 573–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.