Components of heterotrimeric G-proteins directly interact with the soybean Nod factor receptors, which phosphorylate the regulatory RGS protein, thereby affecting G-protein cycle-dependent nodulation.
Abstract
Signaling pathways mediated by heterotrimeric G-protein complexes comprising Gα, Gβ, and Gγ subunits and their regulatory RGS (Regulator of G-protein Signaling) protein are conserved in all eukaryotes. We have shown that the specific Gβ and Gγ proteins of a soybean (Glycine max) heterotrimeric G-protein complex are involved in regulation of nodulation. We now demonstrate the role of Nod factor receptor 1 (NFR1)-mediated phosphorylation in regulation of the G-protein cycle during nodulation in soybean. We also show that during nodulation, the G-protein cycle is regulated by the activity of RGS proteins. Lower or higher expression of RGS proteins results in fewer or more nodules, respectively. NFR1 interacts with RGS proteins and phosphorylates them. Analysis of phosphorylated RGS protein identifies specific amino acids that, when phosphorylated, result in significantly higher GTPase accelerating activity. These data point to phosphorylation-based regulation of G-protein signaling during nodule development. We propose that active NFR1 receptors phosphorylate and activate RGS proteins, which help maintain the Gα proteins in their inactive, trimeric conformation, resulting in successful nodule development. Alternatively, RGS proteins might also have a direct role in regulating nodulation because overexpression of their phospho-mimic version leads to partial restoration of nodule formation in nod49 mutants.
INTRODUCTION
Biological nitrogen fixation has a major effect on the global nitrogen cycle. Leguminous plants such as soybean (Glycine max) develop specialized organs, the nodules, on their roots to host symbiotic rhizobia for the fixation of atmospheric nitrogen to ammonia. Nodule formation is an exquisitely complex cellular and developmental event that involves intricate crosstalk between symbiotic bacteria and the host plant. The process begins when the flavonoids secreted by the host plants are sensed by rhizobia, which synthesize lipochitooligosaccharides, known as Nod factors. A series of signaling and developmental events follow, resulting in successful nodulation (Oldroyd et al., 2011). Nod factors are sensed at the epidermal root hair cells of the host plants by specific Nod factor receptors (NFRs). Upon Nod factor perception, individual root hair cells curl to form a niche for the rhizobial microcolony that results in formation of an infection thread, which grows toward the root cortex. At the onset of infection, the root cortex cells undergo dedifferentiation to form nodule primordia. When the infection thread reaches the nodule primordia, rhizobia are released into organelles called symbiosomes, where they differentiate into nitrogen fixing bacteroids. The dividing cortex cells enclosing bacteroids develop into functional nodules (Oldroyd and Downie, 2008; Desbrosses and Stougaard, 2011; Oldroyd et al., 2011; Popp and Ott, 2011).
Nodule formation is an energetically demanding process and is therefore precisely controlled by the host plants. Several of these regulatory events, from Nod factor perception to downstream changes in gene expression, have been characterized in multiple leguminous plants (Cullimore et al., 2001; Kouchi et al., 2004; Mitra et al., 2004; Smit et al., 2005; Libault et al., 2009). It has been confirmed that the lysM (lysine) motif family of receptor-like kinases (NFR1 and NFR5) present at the plasma membrane of the epidermal cells directly bind Nod factors to initiate nodulation (Madsen et al., 2003; Radutoiu et al., 2003, 2007; Broghammer et al., 2012). One of the important downstream events involves changes in calcium spiking in and around the cell nucleus; these changes are sensed by a calcium/calmodulin-dependent protein kinase (CCaMK). Activation of CCaMK is central to the regulation of nodule development as its constitutive activation leads to spontaneous nodule formation (Tirichine et al., 2006; Hayashi et al., 2010; Liao et al., 2012; Takeda et al., 2012; Routray et al., 2013). Active CCaMK phosphorylates transcriptional activator CYCLOPS, which transactivates NODULE INCEPTION to initiate nodule development (Marsh et al., 2007; Singh et al., 2014). CCaMK also induces transcription factors of the GRAS domain family, such as NSP1 and NSP2, which bind to the promoters of early nodulation (Enod) genes to regulate root hair deformation and nodule formation (Udvardi and Scheible, 2005; Gleason et al., 2006; Hirsch et al., 2009). Additional proteins involved in actin rearrangement and protein degradation, as well as hormone perception and signaling, are also involved in nodule development. Proteins of the nuclear pore complex (NENA), an ankyrin protein Vapyrin, an ARID domain-containing protein (SIP1), and HMGR1, have been shown to act in conjunction with CCaMK and have a role in nodule formation in Lotus japonicus and Medicago truncatula (Kevei et al., 2007; Zhu et al., 2008; Groth et al., 2010; Hayashi et al., 2010; Murray et al., 2011).
While the events following the activation of CCaMK have been explored relatively extensively, how the signal perception at the plasma membrane is transduced to changes in the nucleus remains poorly defined. Specifically, the identity of proteins acting directly downstream of the receptors remains unknown. Biochemical, pharmacological, and genetic approaches have identified several possible candidates that can act as secondary messengers connecting events at the plasma membrane to nuclear responses. These include phospholipase C and D proteins, which can generate lipid secondary messengers (den Hartog et al., 2001; Munnik, 2001). It has been proposed that the lipid secondary messengers directly affect the calcium channels present at the nuclear membrane, resulting in the activation of CCaMK (Delmas et al., 2005; Oldroyd and Downie, 2006; Downie, 2014). Additional signaling proteins that have been shown to affect nodule formation include members of the mitogen-activated protein kinase cascade, 14-3-3 proteins, monomeric GTPases of Rab and Rac family, and the heterotrimeric GTP binding proteins (Fernandez-Pascual et al., 2006; Blanco et al., 2009; Chen et al., 2012; Ke et al., 2012; Radwan et al., 2012; Choudhury and Pandey, 2013). Of these, the components of heterotrimeric G-protein complex are especially interesting as these are traditionally known to interact with the receptors at the plasma membrane and relay the information to intracellular targets in a wide range of signaling pathways in all eukaryotes. Furthermore, heterotrimeric G-protein signaling has been linked to changes in calcium signature, mitogen-activated protein kinase activity, regulation of monomeric GTPases, and phospholipase C- and D-mediated signaling, all of which are involved during nodulation (Park et al., 1993; Zhu and Birnbaumer, 1996; Lopez-Ilasaca, 1998; Perfus-Barbeoch et al., 2004; Qi and Elion, 2005; Currie, 2010).
The heterotrimeric G-protein complex is composed of α, β, and γ subunits and the regulator of G-protein signaling (RGS) protein. In the classical signaling paradigm, GDP-bound Gα interacts with Gβγ and is associated with a cell surface G-protein-coupled receptor (GPCR), representing its inactive stage. Signal perception by GPCR leads to an exchange of GTP for GDP on Gα, which results in the generation of active Gα⋅GTP and freed Gβγ, both of which can interact with different effectors to propagate the signal. The intrinsic GTPase activity of Gα returns it to its inactive form (Cabrera-Vera et al., 2003; Offermanns, 2003). RGS proteins are one of the key regulators of the G-protein cycle. These act as GTPase activity accelerating proteins (GAPs) by enhancing the rate of GTP hydrolysis by Gα. The G-protein cycle can be modified, genetically or biochemically, to favor the presence of active or inactive states (McCudden et al., 2005; Siderovski and Willard, 2005; Lambert et al., 2010). While the basic G-protein components and their overall biochemical activities are conserved between plant and mammalian systems, the plant G-protein cycle seems to be regulated differently. The plant Gα proteins are relatively slower GTPases in comparison to the mammalian Gα proteins and are thought to be constitutively active (Urano et al., 2012a). Therefore, the RGS protein-mediated acceleration of GTP hydrolysis has been proposed to be the key regulatory step of plant G-protein signaling in contrast to mammalian systems where the GDP/GTP exchange mediated by the GPCRs is the rate-limiting step of the G-protein cycle (Johnston et al., 2007; Urano et al., 2012a).
Plants also possess relatively fewer G-protein subunits when compared with the mammalian systems (Chen et al., 2003; Perfus-Barbeoch et al., 2004; Chakravorty et al., 2011). The most elaborate plant G-protein network identified to date is present in soybean where recent genome duplication has led to existence of 4 Gα, 4 Gβ, 12 Gγ, and 2 RGS proteins (Bisht et al., 2011; Choudhury et al., 2011, 2012). Detailed characterization of G-proteins from soybean has offered the opportunity to test their direct role in signaling during nodulation. We previously reported that decreased expression of Gβ and group I Gγ genes leads to a significant decrease in nodule number, whereas the converse is true for the overexpression of specific Gβ and Gγ genes. We have also shown that the Gα proteins interact with the Nod factor receptors NFR1α and NFR1β, even though we did not see an effect of decrease in Gα level on nodule development (Choudhury and Pandey, 2013). Our data suggested two likely possibilities: Either the Gβγ proteins are directly involved in regulation of nodule formation with no input from the Gα proteins, or the RNAi-mediated suppression of Gα proteins was not sufficient to result in change in nodulation phenotype.
In this work, we used a combination of genetics and biochemistry to uncover the mechanism of G-protein cycle-dependent regulation of nodule formation. Our data show that the active Gα proteins are negative regulators of nodule formation in soybean. Changing the availability of free, active Gα proteins by modulating the level of the regulatory RGS proteins results in significantly altered nodule numbers. We further demonstrate that the RGS proteins directly interact with, and are phosphorylated by, the NFR1 proteins. Phosphorylation of RGS proteins has important physiological consequences as overexpression of phospho-dead or phospho-mimic versions of RGS proteins results in a significant effect on nodule formation. Our data support a model where the components of heterotrimeric G-proteins and its regulator act downstream of NFR1 to control nodulation.
RESULTS
Active Gα Proteins Are Negative Regulators of Nodule Formation
The inherent nature of the G-protein cycle entails specific modes of regulation where active Gα and freed Gβγ proteins can either transduce the signal individually with no input or effect on the other subunit or in combination where the availability or proper localization of the subunits is dependent on each other. Less common possibilities, such as signaling by an intact heterotrimer, also exist. These regulatory modes allow for an extremely high degree of plasticity in signal response coupling by G-proteins (Pandey et al., 2010).
In our previous experiments, we observed no significant effect of lower expression of Gα genes on nodulation phenotypes of plants (Choudhury and Pandey, 2013). This could be due to two possible scenarios: One, the signaling is mediated exclusively by freed Gβγ subunits with no input from Gα proteins, as has been observed during regulation of primary root growth in Arabidopsis thaliana (Chen et al., 2006); or two, Gα proteins are in fact involved during this process, but their effect is not obvious because of incomplete suppression of their expression, which is significantly high in nodules and in hairy roots (Choudhury and Pandey, 2013). To address these two possibilities, we generated additional Gα-RNAi constructs driven by a Figwort mosaic virus (FMV) promoter targeting the highly conserved switch region that is required for its activity, or the C-terminal region (Supplemental Figure 1), and evaluated their effect on nodule development in a soybean hairy root transformation system. Transcript levels of each of the four Gα genes were significantly decreased in both these RNAi lines (Supplemental Figures 2A and 2B). We recorded the number of nodules formed on the hairy roots 32 d postinoculation (dpi) with Bradyrhizobium japonicum. Interestingly, a significant increase in nodule number was observed for the Gα-RNAi lines compared with the empty vector (EV) containing control lines using both these RNAi constructs (Figures 1A and 1B). On an average ∼17 nodules were formed per hairy root in EV control lines compared with ∼23 nodules per hairy root in Gα-RNAi plants (Figures 1A and 1B). These results implied that the effective suppression of Gα gene expression promotes nodulation. This phenotype is opposite of what we had observed previously in the Gβ- and Gγ-RNAi lines (Choudhury and Pandey, 2013), where the suppression of Gβ or Gγ genes resulted in the formation of fewer nodules on transgenic roots. Such opposite regulatory mechanisms by different G-protein subunits have also been seen in Arabidopsis Gα or Gβ mutants during the lateral root formation (Chen et al., 2006) or stomatal development (Zhang et al., 2008), although in both these cases, the Gβ protein acts as a negative regulator of signaling.
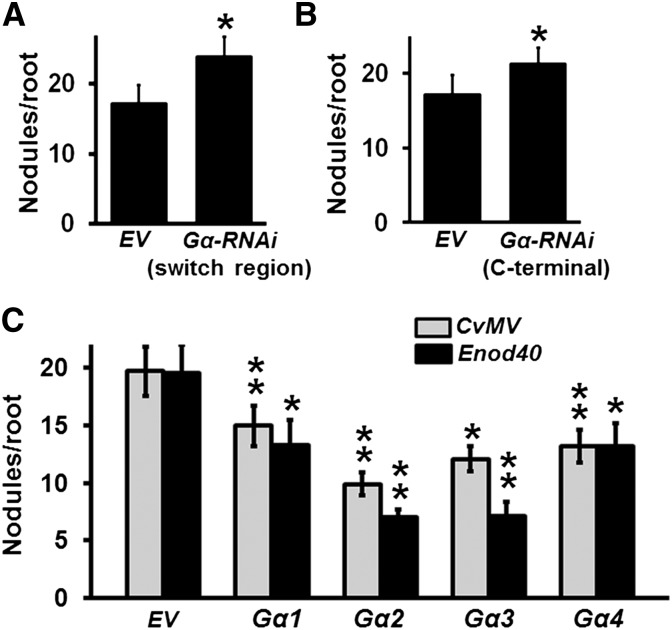
Figure 1.
Negative Regulation of Nodule Formation by Gα Proteins.
(A) Nodulation phenotype of soybean Gα-RNAi transgenic hairy roots driven by FMV promoter using a construct targeting the switch region of Gα proteins.
(B) Nodulation phenotype of Gα-RNAi hairy roots driven by FMV promoter using a construct targeting the C-terminal region of Gα proteins.
(C) Nodulation phenotype of soybean hairy roots overexpressing individual Gα genes driven by CvMV or Enod40 promoter. Nodule numbers on transgenic hairy roots were counted at 32 dpi with B. japonicum and were compared with their respective EV-transformed control hairy roots. The data are average values from three biological replicates (40 to 50 individual plants/biological replicate containing transgenic nodulated roots). Asterisks indicate statistically significant differences compared with EV control (*P < 0.05; **P < 0.01; Mann-Whitney U test).
To corroborate the role of Gα proteins in regulation of nodule development, we took a gain of function approach. We generated Gα overexpression lines where expression of each of the four Gα genes was driven by a constitutively active Cassava vein mosaic virus (CvMV) promoter or by a nodule-specific Enod40 promoter (Supplemental Figure 1). The transcript level of each Gα gene in overexpression lines was higher compared with their respective EV control lines (Supplemental Figure 3A). The nodulation phenotype of transformed hairy roots was observed after 32 dpi with B. japonicum. Overexpression of individual Gα genes led to a decrease in nodule number per plant (Figure 1C). The phenotype was more pronounced upon overexpression of group II Gα genes (Gα 2 and 3) compared with what was observed with group I Gα genes (Gα 1 and 4). On average, ∼13 and ∼8 nodules were observed per hairy root upon overexpression of group I and group II Gα, respectively, compared with ∼19 nodules per hairy root in EV controls (Figure 1C). In additional experiments, we made use of known point mutant versions of Gα proteins that have distinct effects on their activity. A mutation in a conserved glutamine residue, demonstrated to be important for the GTPase activity of Gα proteins (Q223L in Gα1), results in a GTPase activity-lacking, constitutively active protein (Roy Choudhury et al., 2014). Similarly, we have previously shown that a glycine-to-serine mutation in Gα protein (G196S in soybean Gα1; corresponds to G302S of yeast Gpa1) makes it nearly incapable of being deactivated by RGS protein (Roy Choudhury et al., 2014). Overexpression of Gα1Q223L and Gα1G196S, as confirmed by evaluating the transcript level of the transformed genes (Supplemental Figure 3B), also resulted in a significant decrease in nodule number per transformed root (Supplemental Figure 3C), similar to what was observed with the overexpression of Gα1. These results confirmed a negative role of active Gα proteins during soybean nodulation.
RGS Proteins Affect Nodulation
The role of RGS proteins as key regulators of the G-protein cycle is well established. Since the inherent GTPase activity of plant Gα proteins is thought to be slow, RGS proteins have been proposed to be absolutely required for the generation of GDP-bound Gα proteins and consequently the formation of heterotrimer (Urano et al., 2012a). We have previously shown that both RGS genes of soybean are expressed in roots, hairy roots, and in nodules (Choudhury et al., 2012). We evaluated the transcript levels of RGS1 and RGS2 in response to B. japonicum infection at different time points as well as their expression levels in non-nodulating nod49 and in supernodulating nts382 soybean mutants. In wild-type roots, the transcript levels of both RGS genes increased significantly after B. japonicum infection compared with the noninfected roots at each of the time points tested (Figure 2A). The infected roots also had noticeably more RGS protein compared with its level in noninfected roots (Figure 2A, inset). Furthermore, compared with the wild-type Bragg variety, RGS1 and RGS2 exhibited lower transcript levels in nod49 mutant roots (Supplemental Figures 4A and 4B). Conversely, in nts382 mutant roots, higher transcript levels of both RGS genes were observed at each of the time points tested (Supplemental Figures 4A and 4B).
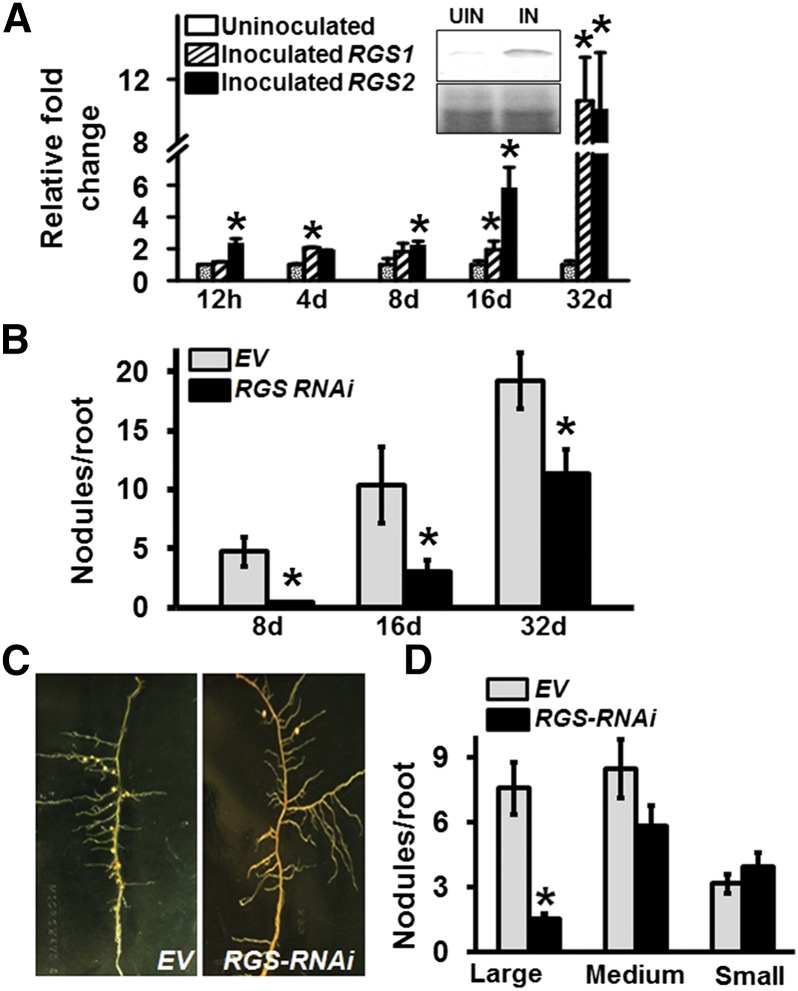
Figure 2.
The Role of RGS Proteins in Regulating Soybean Nodulation.
(A) The transcript level of RGS genes was measured at different time points (12 h, 4 d, 8 d, 16 d, and 32 d) after inoculation with B. japonicum. Data are normalized to Actin gene expression. Fold change represents transcript levels of RGS genes in comparison to the control uninoculated roots, which was set at 1. Data are averaged from three biological replicates of three different experiments. Error bars represent the se of means. Asterisks indicate statistically significant differences compared with EV control (*P < 0.05; Student’s t test). Inset shows RGS protein level (upper panel) as determined by immunoblotting with Arabidopsis RGS1 antibody of microsomal protein fractions isolated from B. japonicum inoculated (IN) or noninoculated (NIN) hairy roots. Lower panel shows the protein gel showing equal protein loading.
(B) Nodule number in EV-transformed and RGS-RNAi hairy roots at 8, 16, and 32 d after inoculation with B. japonicum.
(C) Representative picture of hairy roots and nodules of EV and RGS-RNAi lines.
(D) Nodule phenotypes (large, >2 mm in diameter; medium, 0.5 to 2 mm in diameter; small/immature, <0.5 mm in diameter) for EV and RGS-RNAi lines on soybean hairy roots at 32 dpi with B. japonicum. The data in (B) and (D) represent average of three biological experiments and 40 to 50 individual plants/biological replicate containing transgenic nodulated roots. Error bars represent ±se. Asterisks indicate statistically significant differences compared with EV control (*P < 0.05; Mann-Whitney U test).
To test the effect of altered expression of RGS proteins during nodule development, RNAi and overexpression approaches were followed. Due to the high sequence identity between two RGS genes, a single construct driven by the FMV promoter was used for RNAi-mediated silencing. The transcript levels of both RGS genes were significantly reduced in RGS-RNAi lines compared with the EV control lines (Supplemental Figure 5A). RGS-RNAi roots displayed significantly less root hair deformation (Supplemental Figure 5B) and lower number of nodules from early on. Approximately 40% reduction in nodule number compared with EV containing roots was observed at 32 dpi (Figures 2B and 2C). Moreover, significantly lower numbers of large, mature nodules were observed on RGS-RNAi hairy roots and ∼35% of the nodules were small, pale, and immature (Figure 2D).
Both Early Perception and Late Developmental Events Are Affected by the Lower Expression of Gα and RGS Genes
A complex series of signaling and developmental events leads to nodule formation. To evaluate whether the altered expression of Gα and RGS genes affects the early signaling and perception events, or only the later developmental stages resulting in altered nodule numbers, we analyzed a set of early events during nodule formation. We determined the expression levels of two early nodulation marker genes, Enod40 (Figure 3A) and cytokinin oxidase (Figure 3B), in the hairy roots of Gα-RNAi and RGS-RNAi lines at 6, 12, and 24 h postinoculation with B. japonicum and compared it to their expression levels in control hairy roots containing empty vectors. Both these genes displayed the expected increase in transcript levels in response to B. japonicum infection at each of the time points tested in the EV control plants. Enod40 exhibited overall higher expression in Gα-RNAi and lower expression in RGS-RNAi lines compared with its expression in the EV control plants. The expression of cytokinin oxidase increased to a similar level in both EV control lines and in the Gα-RNAi lines, whereas its expression was significantly lower in the RGS-RNAi lines than that of the EV control at each of the time points tested. These data suggest that the early gene expression changes in response to B. japonicum infection are affected by the altered expression of Gα and RGS genes.
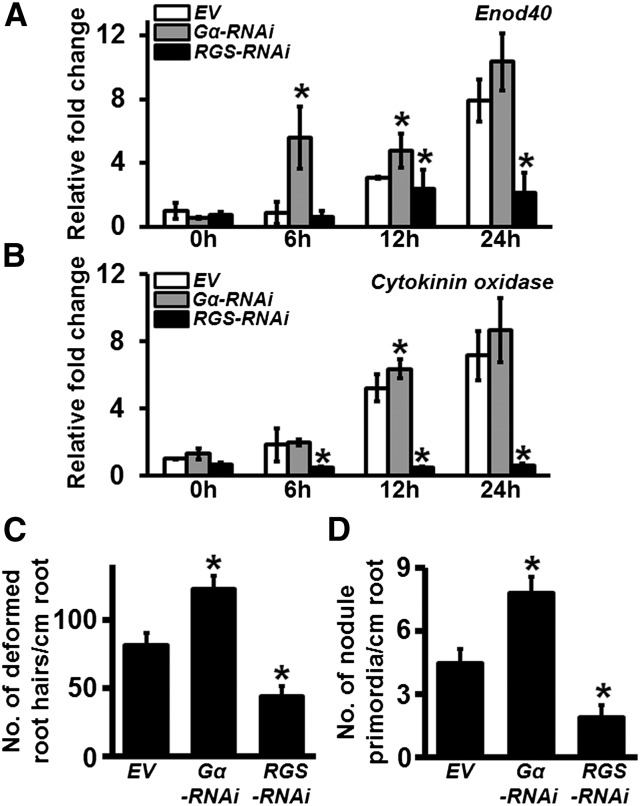
Figure 3.
Quantification of Early Changes in Response to B. japonicum Infection in Gα-RNAi and RGS-RNAi Lines.
(A) and (B) Relative expression of early nodulation marker genes in soybean Gα-RNAi and RGS-RNAi hairy roots. Gene-specific primers were used to amplify and quantify the transcript levels of Enod40 (Glyma01g03470.1) (A) and Cytokinin oxidase (Glyma.17g054500.1) (B) at 6, 12, and 24 h postinoculation with B. japonicum. Two biological replicates with three technical replicates each were used for expression analysis and data were averaged. The expression values across different samples are normalized to Actin expression. Error bars represent the se of the mean. Asterisks indicate statistically significant differences compared with EV control (*P < 0.05; Student’s t test).
(C) Quantification of deformed root hairs/centimeter transgenic roots in Gα-RNAi and RGS-RNAi lines.
(D) Quantification of nodule primordia/centimeter transgenic roots in Gα-RNAi and RGS-RNAi lines. The data in (C) and (D) are average values from three independent experiments (n = 10 to 12 plant each replicate). Error bars represent sd. Asterisks indicate statistically significant differences compared with EV control (*P < 0.05; Mann-Whitney U test).
We also evaluated the number of deformed root hairs in both Gα-RNAi and RGS-RNAi hairy roots at 4 dpi with B. japonicum. Gα-RNAi exhibited significantly higher whereas RGS-RNAi exhibited significantly lower root hair deformation, respectively, compared with the EV control hairy roots (Figure 3C). A similar trend was observed with the developing nodule primordia, where higher and lower numbers of nodule primordia were observed due to lower expression of Gα and RGS genes, respectively, at 6 dpi with B. japonicum (Figure 3D) compared with roots containing EV constructs. Cross-sectional views of the similar sized nodules from EV and RGS-RNAi roots exhibited a severe reduction of bacterial infection and bacteroids in the nodules formed on RGS-RNAi roots (Figures 4A to 4C). No additional effect on overall root length or lateral root formation was seen in Gα-RNAi or RGS-RNAi roots (Supplemental Figures 6C and 6D).
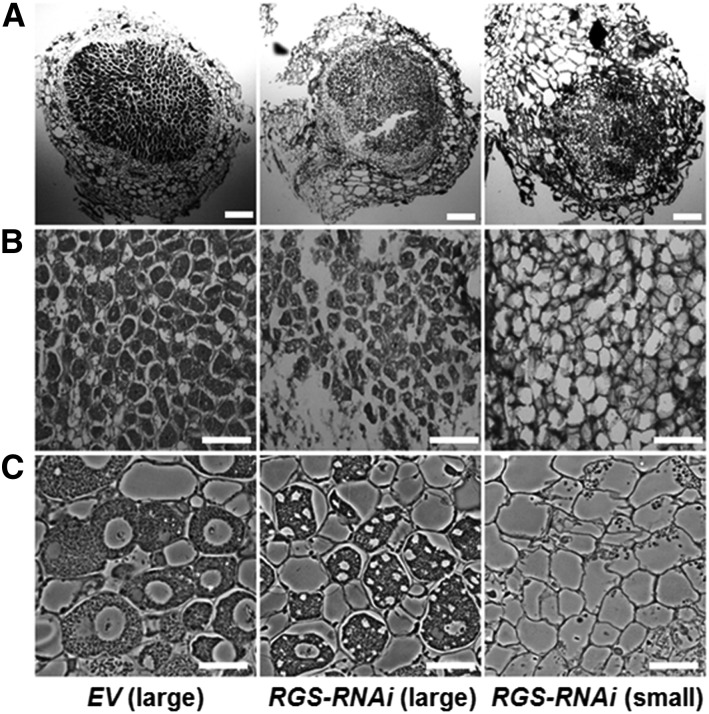
Figure 4.
Altered Morphology of Nodules Formed on RGS-RNAi Hairy Roots.
(A) Light micrographs (4×) of soybean root nodule sections of EV and RGS-RNAi lines. Semithin (5 mm thickness) wax sections, obtained using a microtome, were observed under a light microscope. Bars = 200 μm.
(B) Light micrographs of soybean root nodule at 20× magnification (partial sectional view). Bars = 100 μm.
(C) Nodule sections (0.5 µm thickness) from resin-embedded material were observed using a phase contrast light microscope at 60× magnification. Bars = 15 μm.
Mature large nodules of EV and mature large and small nodules of RGS-RNAi lines (32 dpi with B. japonicum) were used for microscopic studies and representative images obtained using at least 8 to 10 nodules of three independent experiments are shown. The infected cells in EV-transformed nodules are heavily packed with bacteroids, while the cells in RGS-RNAi lines are only partially filled.
RGS Proteins Are Positive Regulators of Nodule Formation
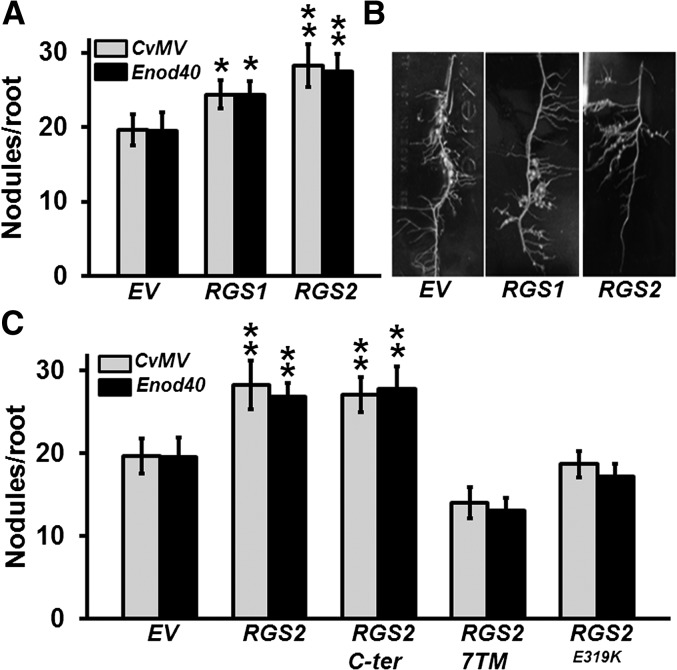
To confirm the positive regulation of nodule formation by RGS proteins, we generated RGS1 and RGS2 overexpression lines using constructs driven by the CvMV and Enod40 promoters. Transcript levels of both genes were analyzed in the transformed hairy roots to ascertain their higher expression (Supplemental Figure 7A). Clear differences in nodule numbers were observed, as overexpression of RGS1 and RGS2 led to ∼20 and 40% more nodules, respectively (Figures 5A and 5B).
Figure 5.
The Effect of Overexpression of Specific RGS Genes on Nodule Formation.
(A) Nodule number on transgenic hairy roots at 32 dpi due to the CvMV and Enod40 promoter-driven expression of RGS1 and RGS2 genes compared with EV control lines.
(B) Representative picture of transgenic hairy roots overexpressing RGS1 and RGS2 genes compared with EV control lines at 32 dpi with B. japonicum.
(C) Nodulation phenotype of transgenic soybean hairy roots overexpressing native full-length RGS2, the RGS domain containing C-terminal region of RGS2 (RGS2 C-ter), N-terminal transmembrane-spanning region of RGS2 (RGS2-7TM), and GAP activity-dead version of RGS2 (RGS2E319K) driven by CvMV or Enod40 promoter. Nodule number was counted at 32 dpi with B. japonicum.
The data in (A) and (C) are average of three biological replicates and each replicate consisted of 40 to 50 transgenic root samples per construct. Error bars represent the se of means. Asterisks denote significant difference, *P < 0.05 and **P < 0.01, respectively, using Mann-Whitney U test compared with EV control.
Plant RGS proteins are chimeric as they contain an N-terminal seven-transmembrane domain that is fused with the C-terminal RGS box-containing domain. This C-terminal domain is responsible for the GAP activity of RGS proteins and for their interaction with Gα protein (Chen et al., 2003). To test whether the nodulation-related effects of altered expression of RGS is related to its biochemical activity and consequently its influence on the regulation of G-protein cycle, we overexpressed the N-terminal (1 to 250 amino acids) and C-terminal domains (251 to 464 amino acids) of RGS2 protein in soybean hairy roots. Overexpression of the C-terminal region resulted in a significant increase in nodule number per hairy root, similar to the full-length RGS proteins, whereas no statistically significant effect was seen due to the overexpression of the N-terminal region only (Figure 5C). This implies that the biochemically active domain of the RGS protein is responsible and sufficient for the regulation of nodule development. Finally, to determine whether this effect is linked to the modulation of G-protein cycle and not due to some yet undefined G-protein independent role of RGS proteins in plants, we made use of a GAP activity-dead version of RGS protein. A single point mutation in RGS protein that changes a conserved glutamate to glutamine, lysine, or alanine (E319K/Q/A) abrogates its GAP activity on Gα protein (Choudhury et al., 2012) (see also Figure 10). Overexpression of RGS2E319K had no effect on nodule development, confirming that the effect of RGS protein during nodule development is via its regulation of G-protein cycle (Figure 5C). The transcript levels of native and mutated versions of individual genes were tested in all overexpression lines to ascertain their higher expression levels (Supplemental Figure 7B).
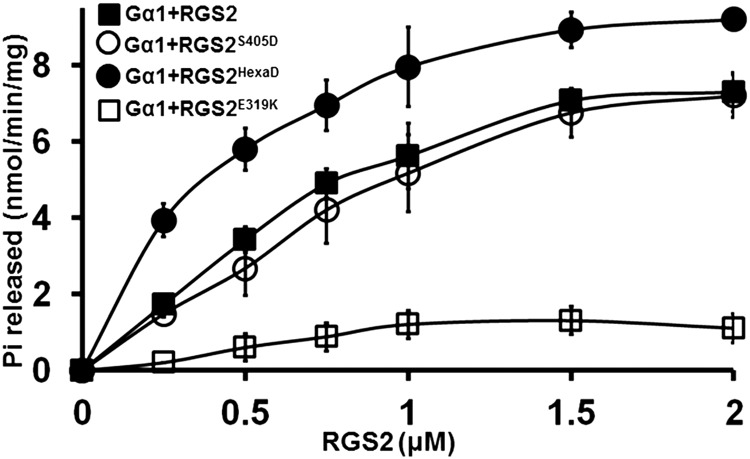
Figure 10.
Effect of Phospho-Mimic Mutant Versions of RGS2 Protein on Its GAP Activity toward Gα1.
Rate of Pi release due to the GTPase activity of Gα1 in the presence of varying concentrations of native and mutant RGS2 proteins. RGS2E319K mutant, which displays no GAP activity, was used as a control. Experiments were repeated three times, and data were averaged. Error bars represent the mean ± se.
Expression of a Subset of Nodulation Marker Genes Corresponds to the RGS-Dependent Regulation of G-Protein Cycle
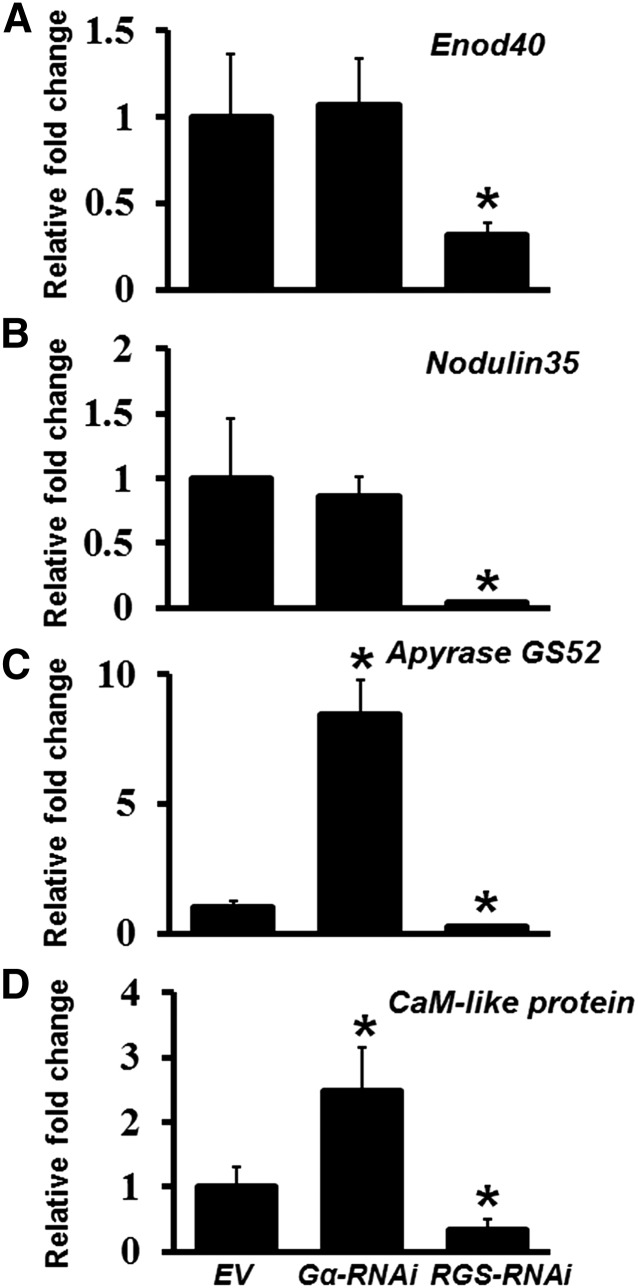
A number of nodulation marker genes have been characterized in soybean (Bergmann et al., 1983; Yang et al., 1993; Govindarajulu et al., 2009; Libault et al., 2009, 2010; Um et al., 2013). We have previously shown that the expression of many of these genes is altered in the Gβ- and Gγ-RNAi lines (Choudhury and Pandey, 2013). Since the phenotype of Gα-RNAi roots is opposite to that of the RGS-RNAi roots, we assessed the transcript level of a set of nodulation marker genes in both these backgrounds to determine the extent to which their expression is affected by G-protein cycle. The transcript levels were analyzed at 32 dpi and were compared with the EV control roots. Clear differences in the gene expression patterns were observed. Specifically, the expression of each of the genes tested was significantly downregulated in the RGS-RNAi lines (Figure 6), similar to what was observed before for the Gβ- and group I Gγ-RNAi lines (Choudhury and Pandey, 2013). Some of the nodulation marker genes such as Enod40 and Nodulin35 did not show a difference in their expression, whereas genes such as Apyrase GS52 and a CaM-like protein exhibited opposite regulation of transcript levels, when compared between Gα- and RGS-RNAi roots. It should be noted that we have previously analyzed the levels of additional nodulation-related genes in Gα-RNAi plants (which did not show an effect on nodule formation due to incomplete silencing) and some of those genes were shown to be downregulated in Gα-RNAi lines, similar to Gβ- and Gγ -RNAi lines (Choudhury and Pandey, 2013). This suggests that complex gene regulatory patterns exist during nodule formation and the expression of at least a subset of nodulation marker genes is regulated in a G-protein-dependent manner.
Figure 6.
Relative Expression of Nodulation-Related Genes in Soybean Gα- and RGS-RNAi Hairy Roots.
Gene-specific primers were used to amplify and quantify the transcript levels of Enod40 (Glyma01g03470.1) (A), Nodulin35 (Glyma10g23790.1) (B), Apyrase GS52 (Glyma16g04750) (C), and Calmodulin-like protein (Glyma02g06680) (D) in Gα-RNAi and RGS-RNAi hairy roots at 32 dpi with B. japonicum. Two biological replicates with three technical replicates each were used for expression analysis and data were averaged. The expression values across different samples are normalized against soybean Actin gene expression. Expression in EV lines was set at 1. Error bars represent the se of the mean. Asterisks indicate statistically significant differences compared with EV control (*P < 0.05) using Student’s t test.
RGS Proteins Interact with NFR1 Receptors
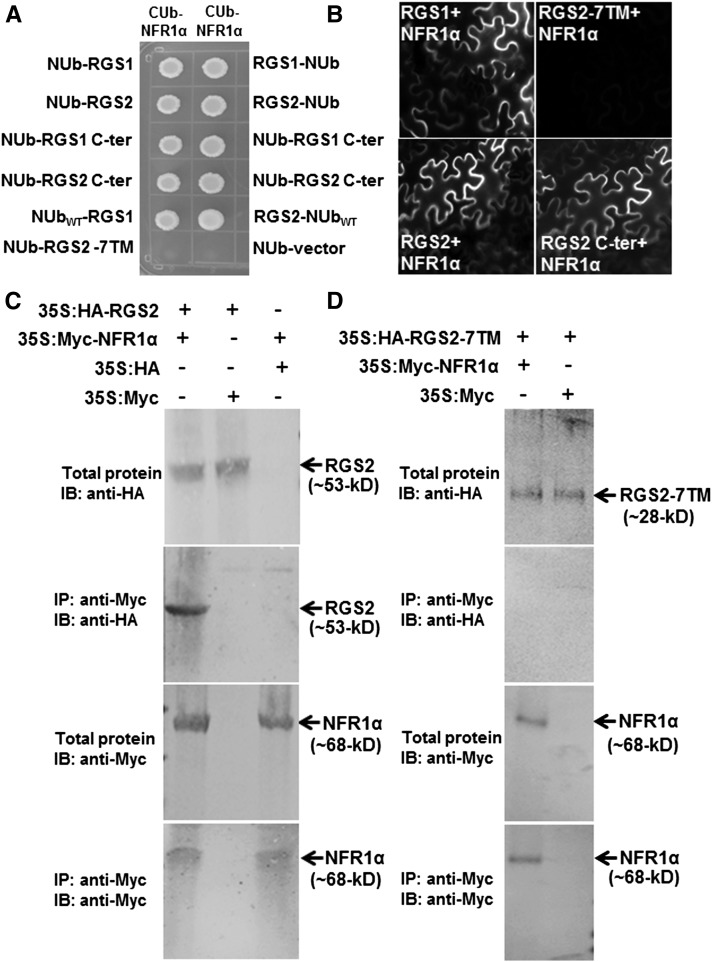
In metazoan systems, Gα proteins are known to interact with GPCRs, whereas data from various plant species suggest that the plant Gα proteins may couple with additional cell surface-localized receptors such as LRR family receptor kinases (Bommert et al., 2013; Liu et al., 2013; Ishida et al., 2014). Toward this, we have previously shown that the soybean Gα proteins interact with the NFR1 receptors (Choudhury and Pandey, 2013). However, the mechanism by which the receptor might affect Gα activity or its interaction with other proteins remains unknown at this time. Since G-proteins and their regulatory proteins usually exist in large macromolecular complexes and the plant RGS proteins are plasma membrane localized, we assessed the direct interaction of RGS proteins with the NFR receptors. The soybean genome encodes two copies of NFR1 and NFR5 proteins, NFR1α and NFR1β, and NFR5α and NFR5β. These are lysM (lysine) motif receptor kinases, with one or two transmembrane domains (Supplemental Figure 8). We cloned full-length NFR1α, NFR1β, NFR5α, and NFR5β genes from soybean nodule cDNA and tested their interaction with the RGS proteins in a split ubiquitin-based interaction system. For these interactions, the full-length or the kinase domain containing the C-terminal region of NFR proteins was expressed as CUb fusions and the RGS proteins were expressed as NUb fusions in both orientations (NUb-RGS and RGS-NUb). NUb fusions were made with full-length, N-terminal, and C-terminal regions of RGS proteins. RGS proteins (full-length as well as C-terminal domain) interacted with NFR1α (Figure 7A) and NFR1β (Supplemental Figure 9A), but not with NFR5α and NFR5β (data not shown), as evaluated by yeast growth on media lacking Leu, Trp, His, and Ade, in the presence of 200 µM Met. The N-terminal region of RGS protein (RGS 7TM) did not interact with NFR1α protein (Figure 7A; Supplemental Figure 9). Interaction between the two proteins was also assessed by bimolecular fluorescence complementation (BiFC) analysis. Each RGS protein was expressed as an N-terminal fusion to the C terminus of YFP (RGS-cYFP), and NFR1 was expressed as an N-terminal fusion to the N terminus of YFP (NFR1-nYFP). The interaction was confirmed by reconstitution of YFP fluorescence in tobacco (Nicotiana tabacum) leaves coinfiltrated with both the expression constructs. Strong YFP fluorescence was observed in all four possible combinations indicating that both soybean RGS proteins can interact with NFR1α and NFR1β proteins (Figure 7B; Supplemental Figure 10A). Similar to yeast based assays, the N-terminal region of RGS2 did not interact with NFR1α, whereas the C-terminal region of RGS2 interacted with NFR1α with similar efficiency as wild-type RGS2 (Figure 7B). The interaction between RGS and NFR1 was further confirmed by in vivo coimmunoprecipitation (co-IP) assay. Tobacco leaves expressing HA-tagged RGS2 and Myc-tagged NFR1α were immunoprecipitated with anti-Myc antibodies. Immunoblotting with anti-HA antibodies showed the presence of RGS2 from plants expressing Myc-tagged NFR1α but not from plants expressing empty vectors (Figure 7C). No interaction was observed when the N-terminal region of RGS2 was used in co-IP assay under identical conditions (Figure 7D). In additional assays, the C-terminal region of NFR1 interacted with the C-terminal region of RGS with similar efficiency as the full-length proteins, in both yeast-based assays (Supplemental Figures 9B to 9E) and in BiFC assays (Supplemental Figure 10B). The N-terminal region of NFR1 did not interact with RGS proteins (Supplemental Figure 10B).
Figure 7.
Soybean RGS Proteins Interact with NFR1 Receptors.
(A) Interaction between RGS and NFR using a split ubiquitin-based interaction assay. The picture shows yeast growth on selective media with 200 µM methionine. In all cases, full-length RGS proteins, the N-terminal seven-transmembrane region (7TM), and the C-terminal RGS domain containing RGS proteins were used as NUb fusions in both orientations (NUb-RGS denoting NUb fused to the N terminus of RGS and RGS-NUb denoting NUb fused to the C terminus of RGS). NFR1α was used for the CUb fusion. NUbwt and NUb-vector fusion constructs were used as positive and negative controls, respectively. Two biological replicates of the experiment were performed with identical results.
(B) Interaction between RGS (in 77-nEYFP-N1) and NFR1 (in 78-cEYFP-N1) proteins using BiFC assay. Agrobacteria containing different combinations of RGS1 and RGS2 and NFR1α were infiltrated in tobacco leaves, and reconstitution of YFP fluorescence due to protein-protein interaction was visualized under a Nikon Eclipse E800 microscope with epifluorescence modules. At least four independent infiltrations were performed for each protein combination with similar results.
(C) Interaction between RGS and NFR1α protein using an in vivo co-IP assay. Anti-Myc antibody can pull down HA-tagged RGS2 from total protein extracts of plants expressing 35S:HA-RGS2 and 35S:Myc-NFR1α (lane 1) but not from total protein extracts from plants expressing 35S:HA-RGS2 and Myc-tagged EV (lane 2) or 35S:Myc-NFR1α and HA-tagged EV (lane 3).
(D) The N-terminal seven-transmembrane domain of RGS2 did not interact with NFR1α in a similar in vivo co-IP assay.
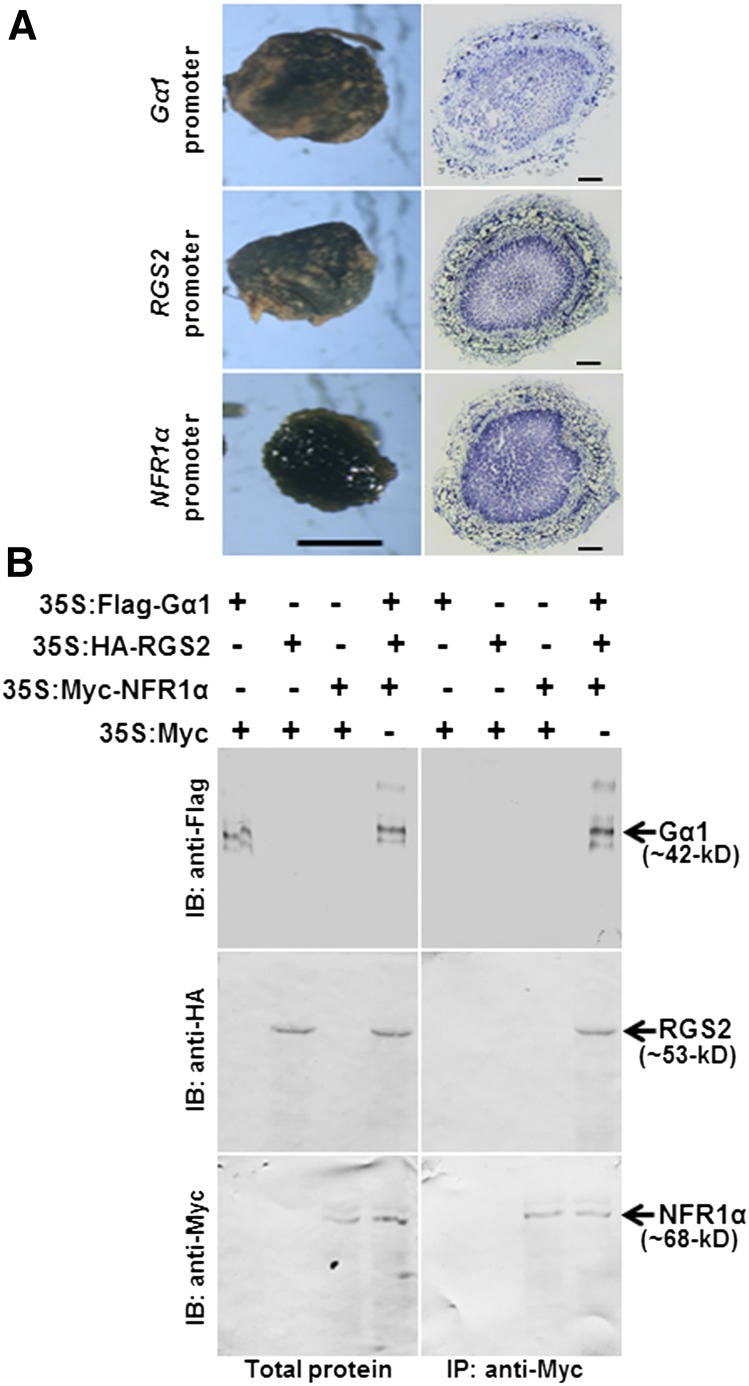
We have previously shown that NFR1 interacts with Gα. Moreover, Gα protein also interacts with RGS. All three proteins are expressed in nodules and in hairy roots (Figure 8A; Supplemental Figure 11). To evaluate the possibility that a tripartite complex might exist between NFR1, Gα, and RGS proteins, we performed additional co-IP assays. Flag-tagged Gα1, HA-tagged RGS2, and Myc-tagged NFR1α proteins were infiltrated in tobacco leaves. The proteins were immunoprecipitated with anti-Myc antibodies. Immunoblotting with Myc, HA, and Flag antibodies showed the protein bands corresponding to the tagged versions of Gα1, RGS2, and NFR1α (Figure 8B), suggesting that the three proteins likely exist as a complex in vivo.
Figure 8.
NFR1α, Gα, and RGS Exhibit Overlapping Expression Patterns in Nodules and Form a Tripartite Complex in Vivo.
(A) GUS staining of proGmGα1:GUS, proGmRGS2:GUS, and proGmNFR1α:GUS in mature nodules (bar = 1 mm) and nodule cross sections (bar = 200 µm).
(B) In vivo co-IP suggests the presence of three proteins as a complex. Total proteins were extracted from tobacco leaves infiltrated with 35S:Flag-Gα1, 35S:HA-RGS2, 35S:Myc-NFR1α, and 35S:Myc-tagged EV in different combinations. Anti-Myc antibody was used for immunoprecipitation. The total protein extracts and Myc antibody immunoprecipitated proteins were further immunoblotted with Flag, HA, and Myc antibodies to detect the Gα1, RGS2, and NFR1α proteins, respectively.
NFR1 Receptors Phosphorylate RGS Proteins
NFR1 receptors are active kinases (Madsen et al., 2011; Wang et al., 2014a) and phosphorylate NFR5 coreceptors, but whether there are additional phosphorylation substrates in vivo is not known. The interaction between RGS proteins and the Gα proteins with NFR1 receptors prompted us to evaluate if either of these proteins could be the in vivo substrates of NFR1 receptors. In the Arabidopsis sugar signaling pathway, WNK8 kinase phosphorylates RGS1 and a phosphatase inhibitor calyculin A increases RGS1 phosphorylation, suggesting that RGS1 can undergo steady state phosphorylation and dephosphorylation (Urano et al., 2012b). Based on sequence identity with Arabidopsis RGS1, soybean RGS proteins are predicted to have 50 potential phosphorylation sites at their C-terminal region. Multiple potential phospho-sites are predicted in Gα proteins as well, based on its homology with the Arabidopsis Gα protein.
We performed in vitro phosphorylation assays using recombinant Gα protein or C-terminal RGS proteins as substrates and the C-terminal domain of NFR1α protein as a kinase. NFR1α was able to effectively phosphorylate RGS1 and RGS2 (Figure 9A). No phosphorylation was observed when Gα was used as substrate either in its native or constitutively active form (Figure 9A). NFR1 was also autophosphorylated under these assay conditions (Figure 9A).
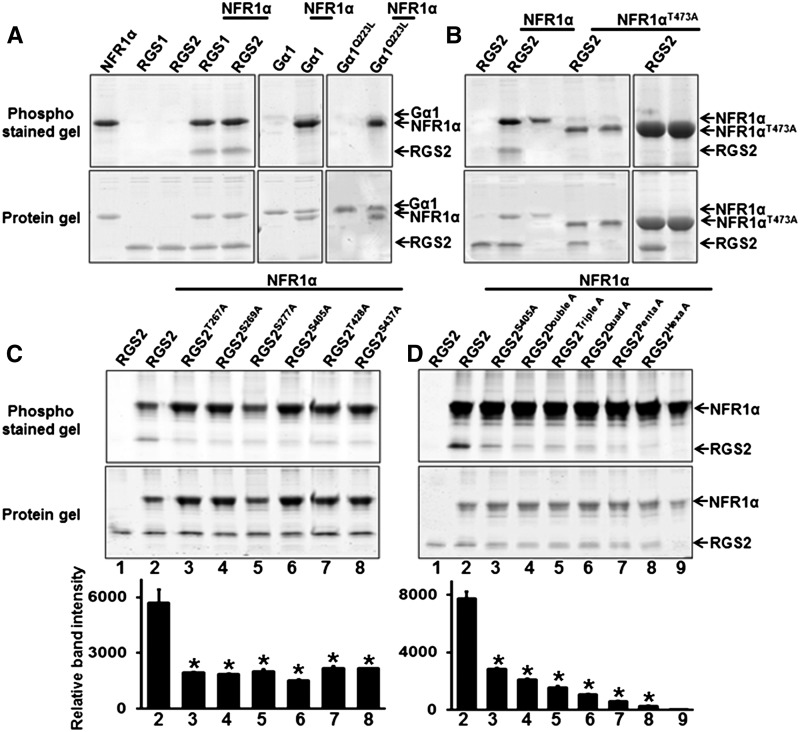
Figure 9.
NFR1α Autophosphorylates and Transphosphorylates RGS Proteins.
(A) The indicated proteins (recombinant, purified NFR1α C-terminal, RGS1 C-terminal, RGS2 C-terminal, Gα, and GαQ223L) were subjected to in vitro phosphorylation assay either alone or in combination.
(B) Effect of a point mutation in the active site of NFR1α (NFR1αT473A) on RGS protein phosphorylation.
(C) Phosphorylation assay using different single point mutant versions of RGS2 proteins (based on the information from LC-MS/MS data).
(D) Phosphorylation assay using higher order mutations in RGS2 protein (RGS2S405A, double, RGS2S405A, S269A; triple, RGS2S405A, S269A, S277A; quad, RGS2S405A, S269A, S277A, S405A; penta, RGS2S405A, S269A, S277A, S405A, T428A; hexa, RGS2S405A, S269A, S277A, S405A, T428A, S437A).
Recombinant purified proteins were incubated with NFR1α protein for in vitro phosphorylation assay and gel image was visualized after Pro-Q Diamond Phosphoprotein gel staining. In all cases, the upper panel represents Pro-Q Diamond-stained phosphoprotein gel and the lower panel shows the same gel stained with Sypro Ruby to visualize protein profiles. The bottom panel of (C) and (D) shows the quantification of band intensity of phosphorylated RGS2 protein by Image J. Asterisks indicate statistically significant differences compared with native RGS2 control (*P < 0.05; Student’s t test).
A single tyrosine residue present in the activation loop of the L. japonicus homolog of NFR1 has been shown to be important for its kinase activity (Madsen et al., 2011). This amino acid corresponds to Tyr-473 in soybean NFR1α. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis identified 14 phosphorylation sites at minimum localization threshold of 99% within the C-terminal region of phosphorylated NFR1α. Tyr-473 was identified as one of these sites (Supplemental Figure 12). To confirm the specificity of phosphorylation of RGS proteins by NFR1 receptors, we generated its kinase-dead version, NFR1αT473A, and assessed its ability to phosphorylate RGS. No phosphorylation of RGS was observed with the NFR1αT473A even when significantly higher (5×) protein quantities were used (Figure 9B). However, in contrast to the mutation in L. japonicus protein, which results in abrogation of its autophosphorylation activity as well as its substrate phosphorylation activity, the soybean NFR1αT473A mutation had no effect on its autophosphorylation activity. Incidentally, the autophosphorylated NFR1αT473A migrates faster on gels compared with the native NFR1α (Figure 9B).
LC-MS/MS analysis identified five phosphorylation sites in NFR1α phosphorylated RGS2 at a minimum localization threshold of 99%: Thr-267, Ser-269, Ser-277, Ser-405, and Thr-428. An additional site, Ser-437, was identified when the minimum localization threshold was decreased to 80% (Supplemental Figures 13 and 14). The Ser-437 phosphosite has been shown to be critical during the RGS1-dependent regulation of the Arabidopsis sugar signaling pathway (Urano et al., 2012b). To validate LC-MS/MS data and to identify the exact residue(s) phosphorylated by NFR1α, we made point mutations at each of the six residues to change the serine or threonine to alanine in RGS2 protein. The recombinant mutant proteins (RGS2T267A, RGS2S269A, RGS2S277A, RGS2S405A, RGS2T428A, and RGS2S437A) were purified and tested for their ability to be phosphorylated by NFR1α. Each of the mutant versions of RGS2 exhibited a strong reduction in phosphorylation, with the strongest effect observed for RGS2S405A (Figure 9C). To further assess the effect of multiple potential phosphorylation sites in RGS, we generated phospho-dead versions of the protein at additional sites in conjunction with RGS2S405A. Double (RGS2S405A, S269A), triple (RGS2S405A, S269A, S277A), quadruple (RGS2quadA; RGS2S405A, S269A, S277A, T267A), quintuple (RGSpentaA; RGS2S405A, S269A, S277A, T267A, T428A) and sextuple (RGShexaA; RGS2S405A, S269A, S277A, T267A, T428A, S437A) recombinant proteins were purified and used as substrates in phosphorylation assays in the presence of C-terminal NFR1α protein. In comparison to RGS2S405A alone, each additional mutation lead to lower phosphorylation, with almost no phosphorylation detected in the RGS2hexaA protein (Figure 9D).
To evaluate whether the phosphorylation of RGS proteins by NFR1α has any influence on the regulation of the G-protein cycle, we generated phospho-mimic versions of RGS by replacing the respective serine and threonine residues to aspartic acid: RGS2S405D and RGS2hexaD (RGS2S405D, S269D, S277D, T267D, T428D, S437D). Native and phospho-mimic versions of RGS2 were evaluated for their GAP activity in the presence of GTP-loaded Gα. Recombinant purified RGS proteins were incubated with the Gα protein and an increase in the rate of GTP hydrolysis was measured using Pi release assay (Figure 10) and a real-time BODIPY fluorescence-based assay (Supplemental Figure 15). RGS2hexaD exhibited significantly higher rate of Pi release from the Gα protein compared with the native RGS2. The amount of Pi released in the presence of the GAP activity-dead RGS2E319K was used as a control in these assays (Figure 10). To test whether RGS phosphorylation has any effect on its interaction with Gα or NFR1, we evaluated the interaction between mutant versions of proteins with Gα and NFR1α in a yeast-based split ubiquitin system. No obvious change in interaction ability or strength was observed (Supplemental Figures 16A and 16B). Additionally, we also evaluated the interaction between the inactive version of NFR1α (NFR1αT473A) with Gα1 and C-terminal region of RGS2 in yeast-based and co-IP assays. No difference in interaction strength was observed (Supplemental Figures 16C, 16D, and 17).
Constitutive Expression of Phospho-Mimic RGS Proteins Results in Significant Increases in Nodule Formation
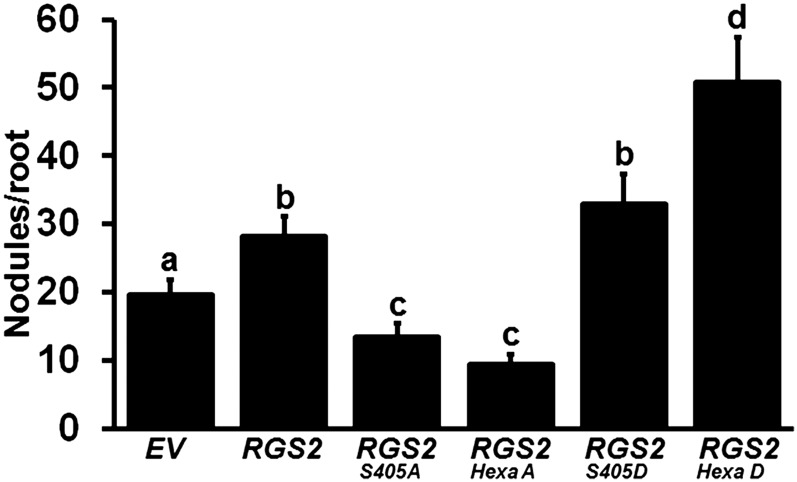
To determine the in planta effect of RGS phosphorylation on nodule formation in soybean, the phospho-mimic versions of RGS were cloned into overexpression vector (driven by the CvMV promoter) and transformed into soybean hairy roots. All transgenic roots were tested for increased transcript accumulation (Supplemental Figure 18). Clear differences in nodule numbers were observed when roots were transformed with native versus phospho amino acid variant constructs. Overexpression of phospho-deficient mutant RGS2S405A or RGS2hexaA resulted in a decrease in nodule number compared with the native RGS2 overexpression. Importantly, nodule number significantly increased in the presence of phospho-mimic mutants, RGS2S405D and RGS2hexaD, compared with what was observed with the overexpression of native RGS2 (Figure 11). Similar results were obtained when using only the C-terminal regions of native and mutant RGS proteins driven by CvMV or Enod40 promoters (Supplemental Figure 19). These data suggest that phosphorylation of RGS proteins by NFR1 receptors positively regulates nodule formation in soybean.
Figure 11.
Nodule Formation on Transgenic Soybean Hairy Roots Overexpressing Phospho-Dead and Phospho-Mimic Versions of RGS Protein.
Native RGS2 gene, single mutant phospho-dead and phospho-mimic versions (RGS2S405A and RGS2S405D), and hexa mutant phospho-dead and phospho-mimic versions (RGS2HexaA and RGS2HexaD) of RGS2 genes driven by the CvMV promoter were used for hairy root transformation. Nodules developed on overexpression roots were counted at 32 dpi with B. japonicum and compared with the EV-containing hairy roots. The data represent average of three biological replicates (40 to 50 individual plants/biological replicate) containing transgenic nodulated roots. Different letters indicate significant differences (Dunn’s multiple comparisons test, P < 0.05) between samples.
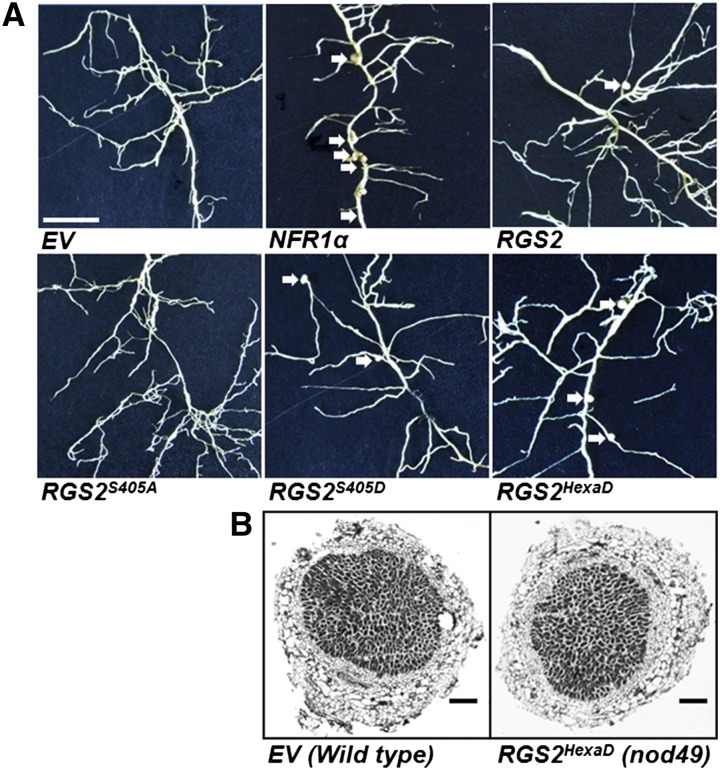
We further assessed the ability of RGS proteins to restore nodule formation in the non-nodulating nod49 mutant of soybean, which lacks a functional NFR1α. Expression of native NFR1α gene (used as a positive control) partially restored nodulation (Figure 12, Table 1). Overexpression of RGS2 and its phospho-mimic versions RGS2S405D and RGS2HexaD also resulted in development of few nodules on nod49 mutant hairy roots (Figure 12 A), whereas no nodules were observed on roots transformed with EV or phospho-dead version of RGS2 (RGS2S405A and RGS2hexaA). The nodules that developed on the RGS2HexaD transformed nod49 roots exhibited normal morphology, similar to the nodules formed on wild-type roots (Figure 12B). However, the nodule numbers in all these cases were significantly lower than the nodules formed on a normal hairy root (Figure 12, Table 1).
Figure 12.
Partial Restoration of Nodule Formation on nod49 Mutant Hairy Roots by Overexpressing NFR1α and Phospho-Mimic RGS Proteins.
(A) Images of nod49 mutant (non-nodulating mutant lacking NFR1α) hairy roots expressing EV, native NFR1α gene, or different versions of RGS2 gene. Expression of CvMV promoter-driven NFR1α is able to partially restore nodule formation on nod49 mutants, whereas plants transformed with EV or RGS2S405A never formed any nodules. Overexpression of native or phospho-mimic versions of RGS2 (RGS2S405D and RGS2HexaD) also resulted in nodule development on some hairy roots (quantitative data in Table 1). The data represent average of two biological replicates containing 20 to 25 transgenic nodulated roots per construct per experiment. The sample size for the constructs containing single phospho-mimic or phospho-dead version of the proteins was 10 to 12 transgenic roots. Bar = 1 mm.
(B) Light micrographs (4×) of sections of nodules formed on EV-transformed wild-type roots and RGS2HexaD transformed nod49 roots. Semithin (5 mm thickness) wax sections, obtained using a microtome, were observed under a light microscope. Bars = 200 μm.
Table 1. The Effect of Overexpression of GmNFR1α and Different Phospho-Mimic and Phospho-Deficient Version of GmRGS2 Gene on Soybean Nodulation.
| Construct | No. of Plants | Total No. of Nodulated Plants | Nodules/Nodulated Plants |
|---|---|---|---|
| EV | 40 | 0 | 0 |
| GmNFR1α | 45 | 34 | 5.46 ± 1.25 |
| GmRGS2 | 47 | 13 | 1.10 ± 0.14 |
| GmRGS2S405A | 20 | 0 | 0 |
| GmRGS2S405D | 24 | 18 | 2.17 ± 0.26 |
| GmRGS2HexaA | 40 | 0 | 0 |
| GmRGS2HexaD | 57 | 42 | 3.14 ± 0.20 |
DISCUSSION
The Role of G-Protein Complex during Nodule Formation in Soybean
Heterotrimeric G-proteins are involved in regulation of a range of physiological processes in plants, similar to what has been discovered in yeast and metazoans. The simplicity of G-protein complex system in Arabidopsis, together with the ease of performing direct genetic analysis of loss-of-function mutants, has enabled the identification of a multitude of processes that involve G-proteins. Phenotypic analysis combined with large-scale transcriptomic analyses have also identified several modes of G-protein function, some similar to what exists in other systems and others specific to plants (Pandey et al., 2010; Urano and Jones, 2014).
The inherent nature of the G-protein cycle predicts different regulatory modes. Activation of the G-protein cycle results in generation of Gα and freed Gβγ subunits and depending on the involvement of both these entities or only one, the modes are defined as classical modes I and II, respectively. In classical mode Ia, both Gα and Gβγ interact with downstream effectors. The phenotypes of mutants lacking either of these subunits are similar, as has been demonstrated during G-protein-regulated leaf shape and abscisic acid responses in Arabidopsis (Perfus-Barbeoch et al., 2004). In classical mode Ib, only Gα interacts with the downstream effectors, but the Gβγ subunits are required for its correct localization. In this case too, the phenotypes of mutants lacking one or both subunits are similar (Pandey et al., 2010). In classical II mode, only the Gβγ dimer interacts with the downstream effectors. Pathways controlled by this regulatory mode exhibit opposite phenotypes due to lack of Gα or Gβγ, as has been seen during the development of lateral roots or stomata. In such cases, the lack of Gβγ results in abrogation of signal transduction, whereas the lack of Gα results in constitutive signaling by Gβγ since it can no longer be sequestered in its inactive conformation (Chen et al., 2006; Pandey et al., 2010). Our previous results with G-protein regulation of nodule development suggested the involvement of Gβγ subunits, but the effect of Gα subunits remained inconclusive (Choudhury and Pandey, 2013). Furthermore, additional regulatory steps such as the involvement of RGS protein and its effects on the activation/deactivation of G-protein cycle during nodule formation were also not known.
The data presented here clearly show a distinct role of Gα proteins and regulation of their activity by RGS proteins during nodule formation in soybean. Effective silencing of Gα proteins resulted in higher nodule numbers compared with the EV-containing hairy roots, which is opposite of the effect of Gβ and Gγ silencing (Figures 1A and 1B). Moreover, overexpression of individual proteins, using two different promoters, resulted in lower nodule numbers, corroborating the data obtained by RNAi approaches (Figure 1C). Furthermore, there were differences between the group I Gα proteins (Gα1 and Gα4) versus group II Gα proteins (Gα2 and Gα3), with group II proteins showing stronger phenotypes (Figure 1C). These two subgroups also exhibit different expression levels during nodule development, with group I proteins showing relatively high expression (Choudhury and Pandey, 2013). It may be that the effective increase in group II Gα proteins is much higher due to their lower basal expression in hairy roots and nodules, thereby resulting in stronger suppression of nodule formation. Alternatively, there might be inherent differences between these proteins, as we have shown previously during the complementation of yeast Gpa1 mutants in pheromone response pathways (Roy Choudhury et al., 2014). However, it is clear that Gα proteins are negative regulators of nodule formation in soybean (Figure 1). Whether the Gβγ proteins are involved only in maintaining Gα protein in trimeric combination or also have additional independent roles during regulation of nodule formation cannot be resolved at this point.
RGS proteins are central to the regulation of the G-protein cycle, especially in plants (Urano and Jones, 2014). It has been proposed that GTP hydrolysis by Gα protein is the rate-limiting step of the G-protein cycle in plants, in contrast to mammalian systems, where GDP to GTP exchange is the rate-limiting step (Johnston et al., 2007). RGS genes displayed a nodule development-dependent increase in their expression and the B. japonicum-infected roots had a significantly higher level of RGS proteins compared with the noninfected roots (Figure 2A). Furthermore, altering the level of RGS proteins by RNAi-mediated silencing or by overexpression resulted in lower and higher nodule number per root, respectively (Figures 2 and 5). Both the early events during nodulation such as expression of marker genes, root hair deformation, and formation of nodule primordia (Figure 3), and the later stages of nodule development (Figures 2F and 4) were affected by altered expression of G-protein complex genes. A higher percentage of nodules formed on the RGS-RNAi hairy roots were not fully developed (Figures 2F and 4).
Also, the role of RGS proteins during regulation of nodule formation is not independent of the G-protein cycle per se. Higher expression of the C-terminal RGS domain-containing region has the same effects as the full-length proteins, and an inactive RGS protein, RGS2E319K, which has no effect on the GTPase activity of Gα protein (Figure 10), does not affect nodule formation (Figure 5C). Gene expression analysis suggests that at least a subset of nodulation marker genes are oppositely regulated in Gα-RNAi lines versus RGS RNAi lines (Figures 3A, 3B, and 6). These genes could be direct downstream targets of regulation via G-protein-mediated pathways. Future large-scale analysis of gene expression changes in different G-protein mutants will shed light on such targets. However, these data do suggest that the role of RGS proteins during nodule development is to keep Gα in its inactive, trimeric conformation. Overexpression of RGS proteins leads to lower levels of free Gα, resulting in more nodules, whereas the reverse is true for lower RGS levels.
Phosphorylation of RGS Proteins by NFR1
The paucity of classical G-protein coupled receptors has posed interesting questions about the activation of G-protein cycle in plants. Very few proteins which may act upstream of G-proteins have been identified. These include several receptor-like kinases (RLKs) that have been shown to directly interact with Gα proteins (Bommert et al., 2013; Choudhury and Pandey, 2013). Similarly, many established pathways that are controlled by well-defined RLKs have been shown to involve members of G-protein complex (Llorente et al., 2005; Bommert et al., 2013; Liu et al., 2013; Ishida et al., 2014). However, in most cases, the mechanism by which the receptors regulate the G-protein cycle remains unknown. We have previously shown that the soybean Gα proteins interact with the NFR1 receptors, but there was no effect of this interaction on Gα activity (Choudhury and Pandey, 2013). Our current work shows that the Gα proteins are not phosphorylated by NFR1. The precise effect of such an interaction remains unknown; however it is possible that interaction with NFR1 changes the availability of Gα proteins for additional effectors or interacting partners.
Interestingly, RGS proteins can also interact with the NFR1 receptors (Figure 7), and the three proteins likely form a tripartite complex in vivo (Figure 8). This is exciting as plant RGS proteins are plasma membrane localized due to the presence of the seven-transmembrane domain and are likely present in close proximity to the plasma membrane-localized NFR1 receptors and Gα proteins. However, in contrast to Gα, the RGS proteins are efficiently phosphorylated by NFR1 receptors (Figure 9A). NFR1 proteins are well-established kinases and are known to phosphorylate the NFR5 coreceptors, but the extent to which they phosphorylate additional proteins in cells was not known (Madsen et al., 2011). Similarly, RGS proteins are well-known phosphorylation substrates in metazoan signaling systems (Garrison et al., 1999; Hollinger et al., 2003; Sokal et al., 2003; Huang et al., 2007; Moroi et al., 2007; Xie et al., 2010; Kach et al., 2012) and during sugar signaling in Arabidopsis, but their phosphorylation by a receptor-like kinase may suggest a novel regulatory mode of G-protein signaling pathways in plants. Phosphorylation of RGS proteins affects their biochemical activity (Figure 10), and this effect is clearly biologically relevant because overexpression of the phospho-mimic versions of the proteins results in significantly higher nodule numbers in wild-type soybean hairy roots (Figure 11) and partial restoration of the nodulation phenotype in nod49 mutant roots (Figure 12, Table 1). Incidentally, during sugar signaling pathway in Arabidopsis, RGS1 is phosphorylated by a WNK family kinase, and this phosphorylation alters its plasma membrane localization, thereby making it unavailable to accelerate G-protein hydrolysis (Urano et al., 2012b). In comparison, RGS phosphorylation by NFR1, which occurs at different sites than that by WNK family kinases, affects its activity, but not localization (Supplemental Figure 20). It may be that during the regulation of signaling pathways that require an active Gα, RGS protein is removed from the vicinity of G-proteins to maintain them in an active conformation. On the contrary, pathways that are negatively regulated by an active Gα protein may require active RGS at the plasma membrane to maintain the G-proteins in their trimeric conformation. These data can possibly be expanded to other signaling systems where the involvement of G-proteins has been shown in conjunction with various RLKs. It is likely that other RLKs can also phosphorylate RGS, thereby regulating G-protein-mediated responses. Further analyses of additional RLKs in the context of their effect on RGS activity and/or localization may shed light on these mechanistic details and possibly identify novel, plant-specific regulatory mechanisms.
Mechanism of G-Protein Regulation of Nodule Formation
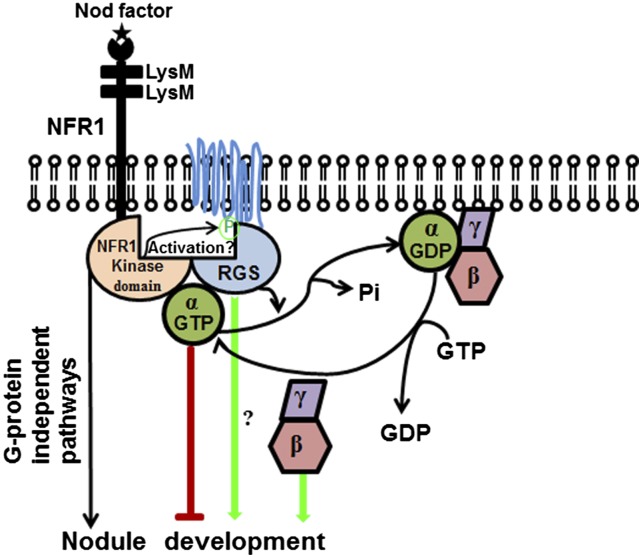
Based on these data, we propose the following model for G-protein complex-dependent regulation of nodulation in soybean (Figure 13). During nodule development, free (active) Gα proteins act as negative regulators of signaling. Nod factor binding activates NFR1 receptors, which phosphorylate RGS, thereby activating them. Active, phosphorylated RGS maintains Gα proteins in their trimeric, inactive conformation, thus allowing for nodulation. Higher expression of Gβγ proteins sequesters the available pool of Gα in trimeric conformation, resulting in higher nodule numbers. Additionally, an independent role of RGS proteins or freed Gβγ proteins in directly interacting with downstream effectors to regulate nodulation cannot be ruled out at this stage (Figure 13). These data, together with previously identified roles of RGS in the Arabidopsis sugar signaling pathway, suggest that plants possibly use the RGS-based regulation of G-protein deactivation as a predominant control mechanism, compared with metazoans, which essentially use a GPCR-based regulation of G-protein activation as a key mechanism. However, the prevalence of such a regulatory mechanism is notable also in the context of plants such as rice (Oryza sativa), Brachypodium distachyon, and many other grasses that do not have a canonical RGS homolog in their genomes. Whether novel, yet unidentified proteins fulfill such a regulatory role, or the G-protein cycle is regulated differently in these plants remains to be investigated.
Figure 13.
Proposed Model for G-Protein-Regulated Nodule Formation in Soybean.
Both G-protein-dependent and -independent pathways are shown.
Our model also opens up several questions, most important being: What is downstream of the G-protein cycle during nodulation? It is well established that the nuclear-localized CCaMK proteins are central to nodule development (Gleason et al., 2006; Tirichine et al., 2006; Takeda et al., 2012). How might the chain of events started at the level of plasma membrane-localized G-proteins be continued to the nuclear proteins? As we have mentioned previously, multiple downstream components proposed to be important for successful nodulation have been linked to G-protein components in other organisms. Future studies targeted toward dissecting the individual signaling components will certainly unravel many of these pathways. Another important question is whether there are proteins or signaling complex(es) in addition to G-proteins, which are involved in transducing the signal from plasma membrane to intracellular components. Nodulation is an important, high energy-demanding event and plants must have evolved multiple layers of regulation to intricately balance it under any given environment. What these additional components might be and how they might interact with the G-protein-regulated processes will be an active area of future research.
METHODS
Plant Material and Construction of Vectors
Soybean (Glycine max) wild-type (‘Williams 82’ and ‘Bragg’) and mutant (super-nodulating nitrate tolerant symbiotic382 ‘nts382’ and a non-nodulating ‘nod49’) seeds were grown on Pro-Mix BX soil (Premier Horticulture) in the greenhouse (16 h light/8 h dark) at 25°C.
Mutant versions of Gα1 and RGS2 proteins were prepared by site-directed mutagenesis using the QuikChange PCR method (Agilent). For overexpression constructs, native and site-directed mutant versions of genes were cloned into the pCR8/GW vector (Invitrogen) and confirmed by sequencing. Each overexpression construct was transferred by Gateway-based cloning into CvMV promoter and Enod40 promoter containing binary vectors (pCAMGFP-CvMV-GWi and pCAMGFP-GmEnod40-2p:GW) using LR clonase (Choudhury and Pandey, 2013). The RNAi constructs were expressed under the control of the FMV promoter in CGT11017A vector (Govindarajulu et al., 2008). After sequence verification, all constructs including empty vectors (used as controls), were transformed into Agrobacterium rhizogenes strain K599.
Hairy Root Transformation and Evaluation of Nodulation Phenotypes
Shoot apices with a single fully expanded trifoliate leaf from 2-week-old soybean plants were used for hairy root transformation. The hairy root transformation of soybean was performed essentially as described previously (Govindarajulu et al., 2009; Choudhury and Pandey, 2013). After transformation, the plants were maintained in nitrogen-free media. To assay the deformed root hairs and the nodule primordia, plants were infected with Bradyrhizobium japonicum (strain USDA110). Root segments 3 cm below the root-hypocotyl junction were cut and harvested at 4 and 6 dpi, respectively. Roots were fixed with ethanol:glacial acetic acid (3:1) for 2 h and stained with 0.01% methylene blue for 15 min (Wang et al., 2014b). The stained transgenic roots were observed by light microscopy to detect the deformed root hairs and nodule primordia in three different biological replicates each with n = 10 to 12 roots. Root hairs with curvy/wavy growth direction, bulged tips, or branching at 4 dpi with B. japonicum were considered as deformed root hairs (Supplemental Figure 6A). Nodule primordia were counted at 6 dpi with B. japonicum. Early nodule primordia were differentiated from the lateral root primordia by their cell division patterns. Nodule primordia typically have a higher frequency of cell division in the cortical layer, whereas the lateral root primordia have a higher frequency of cell division in the pericycle and the endodermis. The later stages were obvious due to the positioning of vascular bundles (central in lateral root primordia and peripheral in nodule primordia), and their shapes (elongated lateral root primordia and round nodule primordia) (Supplemental Figure 6B). Nodule number was counted at 8, 16, and 32 dpi with B. japonicum. Three biological replicates were used for each construct and nodule number was quantified from at least 40 to 50 transgenic hairy roots in each individual experiment (total 120 to 150 transgenic hairy root/per construct). The data were averaged, and statistically significant values were determined by Mann-Whitney U test.
RNA Isolation and Real-Time Quantitative PCR
The soybean hairy roots were collected at different time points after B. japonicum infection. Total RNA was isolated from uninoculated and inoculated hairy roots, using Trizol reagent (Invitrogen). RNA samples were treated with DNase I to remove the genomic DNA. First-strand cDNA was synthesized using the Superscript III RT kit (Invitrogen). Real-time quantitative PCRs were performed as previously described (Bisht et al., 2011) with a StepOne Plus real-time PCR system (Applied Biosciences) using the SYBR advantage qPCR premix (Clontech). The oligonucleotide primers used for PCR are listed in Supplemental Table 1. Two different biological replicates with three technical replicates each were performed for each experiment. The data were averaged and statistically significant values were determined by Student’s t test.
Microscopy
For analysis of the nodule cross sections, mature nodules from RNAi- and empty vector-containing transgenic roots were fixed in 2% glutaraldehyde containing 0.1 M PIPES buffer (pH 6.8) for 2 h. Sample preparation and microscopic observation were performed as described previously (Choudhury and Pandey, 2013). For phase contrast microscopy, nodules from RGS-RNAi and EV-transformed lines were fixed by high-pressure freezing and packed in 0.1 M PIPES buffer (pH 6.8) plus 300 mM sucrose. Samples were freeze substituted for 5 d at −80°C in 2% osmium tetroxide in acetone, then slowly thawed to room temperature, rinsed in acetone, and embedded in Spurr’s resin. Nodule sections (0.5 μm thick) were imaged using Nikon Eclipse 800 wide-field microscope.
For the reporter gene expression analysis, putative promoter regions of Gα1 (∼2 kb), RGS2 (∼1.5 kb), and NFR1α (∼1.5 kb) were amplified from cv Williams 82 genomic DNA and cloned in pcr8GW vector (Invitrogen). Sequence confirmed clones were then introduced into pYXT1 or pYXT2 destination vectors carrying the GUS and GFP reporter genes, respectively (Xiao et al., 2005). These constructs were transformed using the hairy root transformation system and transgenic roots were inoculated with B. japonicum. The detection of GFP fluorescence and GUS assay were performed 10 d after B. japonicum infection in hairy roots and different days after infection in nodules. To observe the cellular level expression, 28-d-old mature nodules were sectioned followed by GUS staining.
Protein-Protein Interaction Assays and Immunoblotting
Interaction assays between Gα, RGS, and NFR proteins using split ubiquitin-based and BiFC-based assays were performed essentially as previously described (Bisht et al., 2011; Choudhury et al., 2012). At least two independent transformations for the split ubiquitin-based assays and four independent transformations for BiFC-based assays were performed. For co-IP assays, 3- to 4-week-old tobacco (Nicotiana tabacum) plants were used for Agrobacterium tumefaciens-mediated transient expression of soybean RGS2 fused to HA tag, soybean NFR1α fused to Myc tag, and soybean Gα1 protein fused to the Flag tag. After 3 to 4 d, proteins were extracted from inoculated leaves in the extraction buffer containing 250 mM sucrose, 25 mM HEPES-KOH, pH 7.5, 10 mM MgCl2, 1 mM PMSF, 1 mM DTT, and 1% Triton X-100 with protease inhibitor cocktail (Sigma-Aldrich). The homogenate was centrifuged at 8000g for 10 min to remove debris. For each immunoprecipitation experiment, ∼200 μg of protein extracts was incubated with anti-Myc antibody in the presence of binding buffer (25 mM HEPES-KOH, pH 7.5, 100 mM NaCl, 3 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 1 mM PMSF, and protease inhibitor cocktail). The immunocomplex was adsorbed into binding buffer equilibrated with protein A agarose according to the manufacturer’s protocol (Sigma-Aldrich). The mixture was incubated overnight at 4°C with gentle rotation. Protein A agarose beads were washed three times with 1 mL of binding buffer by gentle shaking followed by centrifugation. Lastly, pulled-down proteins were separated on SDS-PAGE and analyzed by immunoblotting with anti-HA, anti-Myc, and anti-Flag antibodies (Sigma-Aldrich). To detect RGS protein levels, microsomal protein fractions isolated from B. japonicum inoculated or noninoculated hairy roots were used for immunoblotting with RGS antibodies, essentially according to Bisht et al. (2011).
Recombinant Protein Purification and in Vitro GTPase Activity Assay
Native and mutant versions of Gα, C-terminal region of RGS, and C-terminal region of NFR1α proteins were cloned into pET28a vector (Novagen), and the proteins were expressed and purified as described previously (Bisht et al., 2011). GAP activity of native and mutant versions RGS proteins was assayed using the ENZchek phosphate assay kit (Invitrogen) essentially according to Choudhury et al. (2012). Real-time fluorescence-based GTP binding and GTP hydrolysis assays were performed using BODIPY-GTP FL (Choudhury et al., 2013).
Accession Numbers
Sequence data from this article can be found in the SoyKB (http://soykb.org/) under the following accession numbers: Gα1 (Glyma04g05960.1), Gα2 (Glyma17g34450.1), Gα3 (Glyma14g11140.1), Gα4 (Glyma06g05960.1), RGS1 (Glyma18g01490.1), RGS2 (Glyma11g37540.1), Enod40 (Glyma01g03470), Nodulin35 (Glyma10g23790), Apyrase GS52 (Glyma16g04750), and Calmodulin (CaM)-like protein (Glyma02g06680).
Supplemental Data
Supplemental Figure 1. Constructs used for RNAi-mediated silencing of soybean Gα and RGS driven by FMV promoter and for overexpression of Gα and RGS driven by constitutive (CvMV) and nodule-specific (Enod40) promoter.
Supplemental Figure 2. Transcript levels of soybean Gα genes in Gα-RNAi hairy roots and nodulation phenotypes.
Supplemental Figure 3. Transcript levels of soybean Gα genes in Gα-overexpressing hairy roots and nodulation phenotypes.
Supplemental Figure 4. B. japonicum-induced expression of soybean RGS1 and RGS2 in wild-type, non-nodulating, and supernodulating soybean hairy roots at different time points.
Supplemental Figure 5. Study of expression levels of RGS genes and root hair deformation in RGS-RNAi silenced transgenic hairy roots.
Supplemental Figure 6. Phenotypes of root hairs, lateral root primordia, nodule primordia, and roots in Gα-RNAi and RGS-RNAi transgenic lines.
Supplemental Figure 7. Expression levels of RGS genes in RGS-overexpressing transgenic hairy roots.
Supplemental Figure 8. Domain architecture of soybean NFR1α.
Supplemental Figure 9. Interaction between soybean RGS1 and RGS2 with NFR1α and NFR1β using split ubiquitin-based interaction assay.
Supplemental Figure 10. Interaction between RGS (in 77-nEYFP-N1) and NFR1 (in 78-cEYFP-N1) using bimolecular fluorescence complementation assay.
Supplemental Figure 11. Analysis of promoter of Gα1, RGS2, and NFR1α.
Supplemental Figure 12. Detection of phosphorylated amino acid residues in NFR1α C-terminal kinase domain by LC-MS/MS after in vitro phosphorylation assay.
Supplemental Figure 13. Detection of phosphorylated amino acid residues in RGS2 C-terminal region by LC-MS/MS after in vitro phosphorylation assay.
Supplemental Figure 14. Detection of phosphorylation sites within soybean RGS2 C-terminal region by LC-MS/MS after in vitro phosphorylation assay.
Supplemental Figure 15. Changes of GTPase activity of Gα1 in the presence of different mutant versions of RGS2.
Supplemental Figure 16. Interaction between Gα1 with wild-type and mutant RGS2 and NFR1α using split ubiquitin-based interaction assay.
Supplemental Figure 17. Myc-tagged wild-type NFR1α and mutant NFR1αT473A associate with soybean RGS2 in vivo.
Supplemental Figure 18. Transcript level of soybean RGS2 in native and mutant RGS-overexpressing hairy roots.
Supplemental Figure 19. Nodule formation on transgenic soybean hairy roots overexpressing phospho-dead and phospho-mimic versions of C-terminal RGS2.
Supplemental Figure 20. Localization of native and mutant RGS2.
Supplemental Table 1. Primers used in experiments described in the article.
Supplementary Material
Acknowledgments
This research was supported by USDA/AFRI (2010-65116-20454) and USDA/AFRI (2014-04107) grants to S.P. We thank the very patient and diligent undergraduate students and technicians in the lab, including Emma Longworth-Mills, Caitlin Brenner, Henry Hellmuth, and Jenny Wilson for counting thousands of nodules on soybean roots. Jenny Wilson was supported by an NSF-REU grant (DBI-0156581) to S.P. We also acknowledge the help from R. Howard Berg (Danforth Center Microscopy Facility), Sophie Alvarez, Mike Naldrett (Danforth Center Proteomics and Mass Spectrometry Facility), and Chris Taylor (Ohio State University) for help during different stages of experiments and Alan M. Jones (University of North Carolina, Chapel Hill) for Arabidopsis RGS1 antibodies.
AUTHOR CONTRIBUTIONS
S.P. conceived, directed, and supervised the study. S.R.C. conducted all of the experimental work. Both authors contributed to designing experiments, interpreting results, and writing the article.
Glossary
- CCaMK
calcium/calmodulin-dependent protein kinase
- GPCR
G-protein-coupled receptor
- FMV
Figwort mosaic virus
- dpi
days postinoculation
- EV
empty vector
- CvMV
Cassava vein mosaic virus
- BiFC
bimolecular fluorescence complementation
- co-IP
coimmunoprecipitation
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
Footnotes
Articles can be viewed online without a subscription.
References
- Bergmann H., Preddie E., Verma D.P. (1983). Nodulin-35: a subunit of specific uricase (uricase II) induced and localized in the uninfected cells of soybean nodules. EMBO J. 2: 2333–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisht N.C., Jez J.M., Pandey S. (2011). An elaborate heterotrimeric G-protein family from soybean expands the diversity of plant G-protein networks. New Phytol. 190: 35–48. [DOI] [PubMed] [Google Scholar]
- Blanco F.A., Meschini E.P., Zanetti M.E., Aguilar O.M. (2009). A small GTPase of the Rab family is required for root hair formation and preinfection stages of the common bean-Rhizobium symbiotic association. Plant Cell 21: 2797–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommert P., Je B.I., Goldshmidt A., Jackson D. (2013). The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558. [DOI] [PubMed] [Google Scholar]
- Broghammer A., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 109: 13859–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Vera T.M., Vanhauwe J., Thomas T.O., Medkova M., Preininger A., Mazzoni M.R., Hamm H.E. (2003). Insights into G protein structure, function, and regulation. Endocr. Rev. 24: 765–781. [DOI] [PubMed] [Google Scholar]
- Chakravorty D., Trusov Y., Zhang W., Acharya B.R., Sheahan M.B., McCurdy D.W., Assmann S.M., Botella J.R. (2011). An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant J. 67: 840–851. [DOI] [PubMed] [Google Scholar]
- Chen J.G., Gao Y., Jones A.M. (2006). Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol. 141: 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.G., Willard F.S., Huang J., Liang J., Chasse S.A., Jones A.M., Siderovski D.P. (2003). A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301: 1728–1731. [DOI] [PubMed] [Google Scholar]
- Chen T., Zhu H., Ke D., Cai K., Wang C., Gou H., Hong Z., Zhang Z. (2012). A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell 24: 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S.R., Bisht N.C., Thompson R., Todorov O., Pandey S. (2011). Conventional and novel Gγ protein families constitute the heterotrimeric G-protein signaling network in soybean. PLoS One 6: e23361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S.R., Pandey S. (2013). Specific subunits of heterotrimeric G proteins play important roles during nodulation in soybean. Plant Physiol. 162: 522–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S.R., Westfall C.S., Hackenberg D., Pandey S. (2013). Measurement of GTP-binding and GTPase activity of heterotrimeric Gα proteins. Methods Mol. Biol. 1043: 13–20. [DOI] [PubMed] [Google Scholar]
- Choudhury S.R., Westfall C.S., Laborde J.P., Bisht N.C., Jez J.M., Pandey S. (2012). Two chimeric regulators of G-protein signaling (RGS) proteins differentially modulate soybean heterotrimeric G-protein cycle. J. Biol. Chem. 287: 17870–17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullimore J.V., Ranjeva R., Bono J.J. (2001). Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci. 6: 24–30. [DOI] [PubMed] [Google Scholar]
- Currie K.P. (2010). G protein modulation of CaV2 voltage-gated calcium channels. Channels (Austin) 4: 497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas P., Coste B., Gamper N., Shapiro M.S. (2005). Phosphoinositide lipid second messengers: new paradigms for calcium channel modulation. Neuron 47: 179–182. [DOI] [PubMed] [Google Scholar]
- den Hartog M., Musgrave A., Munnik T. (2001). Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: a role for phospholipase C and D in root hair deformation. Plant J. 25: 55–65. [DOI] [PubMed] [Google Scholar]
- Desbrosses G.J., Stougaard J. (2011). Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 10: 348–358. [DOI] [PubMed] [Google Scholar]
- Downie J.A. (2014). Legume nodulation. Curr. Biol. 24: R184–R190. [DOI] [PubMed] [Google Scholar]
- Fernandez-Pascual M., Lucas M.M., de Felipe M.R., Boscá L., Hirt H., Golvano M.P. (2006). Involvement of mitogen-activated protein kinases in the symbiosis Bradyrhizobium-Lupinus. J. Exp. Bot. 57: 2735–2742. [DOI] [PubMed] [Google Scholar]
- Garrison T.R., Zhang Y., Pausch M., Apanovitch D., Aebersold R., Dohlman H.G. (1999). Feedback phosphorylation of an RGS protein by MAP kinase in yeast. J. Biol. Chem. 274: 36387–36391. [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B.W., Oldroyd G.E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Govindarajulu M., Elmore J.M., Fester T., Taylor C.G. (2008). Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol. Plant Microbe Interact. 21: 1027–1035. [DOI] [PubMed] [Google Scholar]
- Govindarajulu M., Kim S.Y., Libault M., Berg R.H., Tanaka K., Stacey G., Taylor C.G. (2009). GS52 ecto-apyrase plays a critical role during soybean nodulation. Plant Physiol. 149: 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Banba M., Shimoda Y., Kouchi H., Hayashi M., Imaizumi-Anraku H. (2010). A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J. 63: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Muñoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollinger S., Ramineni S., Hepler J.R. (2003). Phosphorylation of RGS14 by protein kinase A potentiates its activity toward G alpha i. Biochemistry 42: 811–819. [DOI] [PubMed] [Google Scholar]
- Huang J., Zhou H., Mahavadi S., Sriwai W., Murthy K.S. (2007). Inhibition of Galphaq-dependent PLC-beta1 activity by PKG and PKA is mediated by phosphorylation of RGS4 and GRK2. Am. J. Physiol. Cell Physiol. 292: C200–C208. [DOI] [PubMed] [Google Scholar]
- Ishida T., et al. (2014). Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep. 15: 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston C.A., Taylor J.P., Gao Y., Kimple A.J., Grigston J.C., Chen J.G., Siderovski D.P., Jones A.M., Willard F.S. (2007). GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc. Natl. Acad. Sci. USA 104: 17317–17322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kach J., Sethakorn N., Dulin N.O. (2012). A finer tuning of G-protein signaling through regulated control of RGS proteins. Am. J. Physiol. Heart Circ. Physiol. 303: H19–H35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke D., Fang Q., Chen C., Zhu H., Chen T., Chang X., Yuan S., Kang H., Ma L., Hong Z., Zhang Z. (2012). The small GTPase ROP6 interacts with NFR5 and is involved in nodule formation in Lotus japonicus. Plant Physiol. 159: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z., et al. (2007). 3-Hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell 19: 3974–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H., et al. (2004). Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res. 11: 263–274. [DOI] [PubMed] [Google Scholar]
- Lambert N.A., Johnston C.A., Cappell S.D., Kuravi S., Kimple A.J., Willard F.S., Siderovski D.P. (2010). Regulators of G-protein signaling accelerate GPCR signaling kinetics and govern sensitivity solely by accelerating GTPase activity. Proc. Natl. Acad. Sci. USA 107: 7066–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Singh S., Hossain M.S., Andersen S.U., Ross L., Bonetta D., Zhou Y., Sato S., Tabata S., Stougaard J., Szczyglowski K., Parniske M. (2012). Negative regulation of CCaMK is essential for symbiotic infection. Plant J. 72: 572–584. [DOI] [PubMed] [Google Scholar]
- Libault M., Joshi T., Takahashi K., Hurley-Sommer A., Puricelli K., Blake S., Finger R.E., Taylor C.G., Xu D., Nguyen H.T., Stacey G. (2009). Large-scale analysis of putative soybean regulatory gene expression identifies a Myb gene involved in soybean nodule development. Plant Physiol. 151: 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libault M., Zhang X.C., Govindarajulu M., Qiu J., Ong Y.T., Brechenmacher L., Berg R.H., Hurley-Sommer A., Taylor C.G., Stacey G. (2010). A member of the highly conserved FWL (tomato FW2.2-like) gene family is essential for soybean nodule organogenesis. Plant J. 62: 852–864. [DOI] [PubMed] [Google Scholar]
- Liu J., Ding P., Sun T., Nitta Y., Dong O., Huang X., Yang W., Li X., Botella J.R., Zhang Y. (2013). Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol. 161: 2146–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F., Alonso-Blanco C., Sánchez-Rodriguez C., Jorda L., Molina A. (2005). ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 43: 165–180. [DOI] [PubMed] [Google Scholar]
- Lopez-Ilasaca M. (1998). Signaling from G-protein-coupled receptors to mitogen-activated protein (MAP)-kinase cascades. Biochem. Pharmacol. 56: 269–277. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Antolín-Llovera M., Grossmann C., Ye J., Vieweg S., Broghammer A., Krusell L., Radutoiu S., Jensen O.N., Stougaard J., Parniske M. (2011). Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 65: 404–417. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCudden C.R., Hains M.D., Kimple R.J., Siderovski D.P., Willard F.S. (2005). G-protein signaling: back to the future. Cell. Mol. Life Sci. 62: 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Shaw S.L., Long S.R. (2004). Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 101: 10217–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi K., Nishiyama M., Kawabata S., Ichiba H., Yajima T., Kimura S. (2007). Phosphorylation of Ser166 in RGS5 by protein kinase C causes loss of RGS function. Life Sci. 81: 40–50. [DOI] [PubMed] [Google Scholar]
- Munnik T. (2001). Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 6: 227–233. [DOI] [PubMed] [Google Scholar]
- Murray J.D., et al. (2011). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65: 244–252. [DOI] [PubMed] [Google Scholar]
- Offermanns S. (2003). G-proteins as transducers in transmembrane signalling. Prog. Biophys. Mol. Biol. 83: 101–130. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2006). Nuclear calcium changes at the core of symbiosis signalling. Curr. Opin. Plant Biol. 9: 351–357. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Murray J.D., Poole P.S., Downie J.A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45: 119–144. [DOI] [PubMed] [Google Scholar]
- Pandey S., Wang R.S., Wilson L., Li S., Zhao Z., Gookin T.E., Assmann S.M., Albert R. (2010). Boolean modeling of transcriptome data reveals novel modes of heterotrimeric G-protein action. Mol. Syst. Biol. 6: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Jhon D.Y., Lee C.W., Lee K.H., Rhee S.G. (1993). Activation of phospholipase C isozymes by G protein beta gamma subunits. J. Biol. Chem. 268: 4573–4576. [PubMed] [Google Scholar]
- Perfus-Barbeoch L., Jones A.M., Assmann S.M. (2004). Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr. Opin. Plant Biol. 7: 719–731. [DOI] [PubMed] [Google Scholar]
- Popp C., Ott T. (2011). Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr. Opin. Plant Biol. 14: 458–467. [DOI] [PubMed] [Google Scholar]
- Qi M., Elion E.A. (2005). MAP kinase pathways. J. Cell Sci. 118: 3569–3572. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Jurkiewicz A., Fukai E., Quistgaard E.M., Albrektsen A.S., James E.K., Thirup S., Stougaard J. (2007). LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26: 3923–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan O., Wu X., Govindarajulu M., Libault M., Neece D.J., Oh M.H., Berg R.H., Stacey G., Taylor C.G., Huber S.C., Clough S.J. (2012). 14-3-3 proteins SGF14c and SGF14l play critical roles during soybean nodulation. Plant Physiol. 160: 2125–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routray P., Miller J.B., Du L., Oldroyd G., Poovaiah B.W. (2013). Phosphorylation of S344 in the calmodulin-binding domain negatively affects CCaMK function during bacterial and fungal symbioses. Plant J. 76: 287–296. [DOI] [PubMed] [Google Scholar]
- Roy Choudhury S., Wang Y., Pandey S. (2014). Soya bean Gα proteins with distinct biochemical properties exhibit differential ability to complement Saccharomyces cerevisiae gpa1 mutant. Biochem. J. 461: 75–85. [DOI] [PubMed] [Google Scholar]
- Siderovski D.P., Willard F.S. (2005). The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int. J. Biol. Sci. 1: 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152. [DOI] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Sokal I., Hu G., Liang Y., Mao M., Wensel T.G., Palczewski K. (2003). Identification of protein kinase C isozymes responsible for the phosphorylation of photoreceptor-specific RGS9-1 at Ser475. J. Biol. Chem. 278: 8316–8325. [DOI] [PubMed] [Google Scholar]
- Takeda N., Maekawa T., Hayashi M. (2012). Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell 24: 810–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156. [DOI] [PubMed] [Google Scholar]
- Udvardi M.K., Scheible W.R. (2005). Plant science. GRAS genes and the symbiotic green revolution. Science 308: 1749–1750. [DOI] [PubMed] [Google Scholar]
- Um J.H., Kim S., Kim Y.K., Song S.B., Lee S.H., Verma D.P., Cheon C.I. (2013). RNA interference-mediated repression of S6 kinase 1 impairs root nodule development in soybean. Mol. Cells 35: 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Jones A.M. (2014). Heterotrimeric G protein-coupled signaling in plants. Annu. Rev. Plant Biol. 65: 365–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Jones J.C., Wang H., Matthews M., Bradford W., Bennetzen J.L., Jones A.M. (2012a). G protein activation without a GEF in the plant kingdom. PLoS Genet. 8: e1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D., Phan N., Jones J.C., Yang J., Huang J., Grigston J., Taylor J.P., Jones A.M. (2012b). Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat. Cell Biol. 14: 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xie Z.P., Staehelin C. (2014a). Functional analysis of chimeric lysin motif domain receptors mediating Nod factor-induced defense signaling in Arabidopsis thaliana and chitin-induced nodulation signaling in Lotus japonicus. Plant J. 78: 56–69. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang L., Zou Y., Chen L., Cai Z., Zhang S., Zhao F., Tian Y., Jiang Q., Ferguson B.J., Gresshoff P.M., Li X. (2014b). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26: 4782–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.L., Smith S.R., Ishmael N., Redman J.C., Kumar N., Monaghan E.L., Ayele M., Haas B.J., Wu H.C., Town C.D. (2005). Analysis of the cDNAs of hypothetical genes on Arabidopsis chromosome 2 reveals numerous transcript variants. Plant Physiol. 139: 1323–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Yang Z., Druey K.M. (2010). Phosphorylation of RGS13 by the cyclic AMP-dependent protein kinase inhibits RGS13 degradation. J. Mol. Cell Biol. 2: 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.C., Katinakis P., Hendriks P., Smolders A., de Vries F., Spee J., van Kammen A., Bisseling T., Franssen H. (1993). Characterization of GmENOD40, a gene showing novel patterns of cell-specific expression during soybean nodule development. Plant J. 3: 573–585. [DOI] [PubMed] [Google Scholar]
- Zhang L., Hu G., Cheng Y., Huang J. (2008). Heterotrimeric G protein alpha and beta subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Dev. Biol. 324: 68–75. [DOI] [PubMed] [Google Scholar]
- Zhu H., Chen T., Zhu M., Fang Q., Kang H., Hong Z., Zhang Z. (2008). A novel ARID DNA-binding protein interacts with SymRK and is expressed during early nodule development in Lotus japonicus. Plant Physiol. 148: 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Birnbaumer L. (1996). G protein subunits and the stimulation of phospholipase C by Gs-and Gi-coupled receptors: Lack of receptor selectivity of Galpha(16) and evidence for a synergic interaction between Gbeta gamma and the alpha subunit of a receptor activated G protein. Proc. Natl. Acad. Sci. USA 93: 2827–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.