Strigolactones regulate shoot branching through MAX2-mediated degradation of a clade of SMAX1-LIKE proteins.
Abstract
The plant hormones strigolactones and smoke-derived karrikins are butenolide signals that control distinct aspects of plant development. Perception of both molecules in Arabidopsis thaliana requires the F-box protein MORE AXILLARY GROWTH2 (MAX2). Recent studies suggest that the homologous SUPPRESSOR OF MAX2 1 (SMAX1) in Arabidopsis and DWARF53 (D53) in rice (Oryza sativa) are downstream targets of MAX2. Through an extensive analysis of loss-of-function mutants, we demonstrate that the Arabidopsis SMAX1-LIKE genes SMXL6, SMXL7, and SMXL8 are co-orthologs of rice D53 that promote shoot branching. SMXL7 is degraded rapidly after treatment with the synthetic strigolactone mixture rac-GR24. Like D53, SMXL7 degradation is MAX2- and D14-dependent and can be prevented by deletion of a putative P-loop. Loss of SMXL6,7,8 suppresses several other strigolactone-related phenotypes in max2, including increased auxin transport and PIN1 accumulation, and increased lateral root density. Although only SMAX1 regulates germination and hypocotyl elongation, SMAX1 and SMXL6,7,8 have complementary roles in the control of leaf morphology. Our data indicate that SMAX1 and SMXL6,7,8 repress karrikin and strigolactone signaling, respectively, and suggest that all MAX2-dependent growth effects are mediated by degradation of SMAX1/SMXL proteins. We propose that functional diversification within the SMXL family enabled responses to different butenolide signals through a shared regulatory mechanism.
INTRODUCTION
Strigolactones (SLs) and karrikins (KARs) are butenolide compounds that have recently been identified as regulators of plant growth and development. SLs were first found in root exudates as compounds that stimulate germination of parasitic weeds in the Orobanchaceae family (Cook et al., 1966). Although parasitic weeds exploit SLs as a host-detection signal, SL exudation into the rhizosphere is beneficial to plants. Particularly under nutrient-poor conditions, SLs promote arbuscular mycorrhizal symbioses that enable exchange of carbon for nitrogen, phosphorus, and water (Akiyama and Hayashi, 2006; reviewed in Xie et al., 2010). Recently, SLs have been recognized as plant hormones that influence multiple aspects of development, including shoot branching (tillering), root architecture, leaf senescence, and secondary growth (Gomez-Roldan et al., 2008; Umehara et al., 2008; Agusti et al., 2011; Kapulnik et al., 2011; Ruyter-Spira et al., 2011; Rasmussen et al., 2012; Yamada et al., 2014; Ueda and Kusaba, 2015). SLs are synthesized from carotenoids by the DWARF27 class carotenoid isomerase and the CCD7/MORE AXILLARY GROWTH3 (MAX3) and CCD8/MAX4 classes of carotenoid cleavage dioxygenases; the sequential action of these enzymes produces carlactone (Alder et al., 2012). Other enzymes, including MAX1, a cytochrome P450, subsequently act upon carlactone to produce a large range of biologically active strigolactones (Zhang et al., 2014; reviewed in Al-Babili and Bouwmeester, 2015).
KARs are chemical signals found in smoke that promote seed germination of a wide range of species (Flematti et al., 2004; Nelson et al., 2009; reviewed in Nelson et al., 2012). KAR application also enhances seedling responses to light in Arabidopsis thaliana and promotes seedling vigor in crop plants (Jain et al., 2006; Kulkarni et al., 2006; van Staden et al., 2006; Nelson et al., 2010). To date, there is no evidence that plants synthesize KARs, but genetic evidence suggests that KARs mimic an as yet unknown endogenous butenolide molecule (KAI2 ligand [KL]) that is not SL (Flematti et al., 2013; Waters et al., 2014).
Remarkably, SL and KAR signaling both depend upon the activity of the F-box protein MAX2, which forms part of a Skp-Cullin-F-box (SCF) complex (Stirnberg et al., 2002; Stirnberg et al., 2007; Nelson et al., 2011). SCF complexes act by ligating ubiquitin moieties to target proteins, often resulting in their degradation by the 26S proteasome (Somers and Fujiwara, 2009). The putative receptors for SLs and KARs are the closely related α/β-hydrolases DWARF14 (D14) and KARRIKIN INSENSITIVE2 (KAI2), respectively. These are ancient paralogs that are present throughout angiosperms (Waters et al., 2012, 2013). D14 and KAI2 require an intact catalytic triad for signal transduction (Hamiaux et al., 2012; Waters et al., 2014). Ligand binding or hydrolysis is thought to induce conformational changes in the receptors that alter their interactions with downstream signaling partners, including MAX2 (Hamiaux et al., 2012). The differences between SL-insensitive d14 phenotypes and KAR-insensitive kai2 phenotypes show that SL and KAR/KL regulate distinct aspects of MAX2-dependent development and that the max2 phenotype reflects a combination of d14 and kai2 effects (Waters et al., 2012). Since both signaling pathways act through SCFMAX2, it is unclear how specific developmental responses to SL and KAR/KL are mediated. Identifying the targets of MAX2 and understanding how they mediate specificity is a key objective for elucidating the mechanisms of SL and KAR/KL signaling.
To date, several candidates have been suggested as targets of SCFMAX2. Based on biochemical approaches, the DELLA family of transcriptional activators, which are growth repressors targeted for degradation by gibberellins, and the BES1 family of brassinosteroid response factors have been proposed to be MAX2 targets (Nakamura et al., 2013; Wang et al., 2013). A third class of putative targets, SMAX1-LIKE (SMXL) proteins, was identified primarily on the basis of genetic approaches. A screen for suppressors of max2 in Arabidopsis led to the identification of smax1, which suppresses the seed germination and seedling growth phenotypes of max2 but does not affect its shoot architecture, lateral root growth, or senescence (Stanga et al., 2013). D53, a homolog of SMAX1 in rice (Oryza sativa), was identified from a semidominant d53 mutation that has a SL-insensitive, high tillering phenotype similar to that of d14 and d3, the rice ortholog of MAX2 (Jiang et al., 2013; Zhou et al., 2013). SL promotes physical interaction of D14 with D53 and D3, and the D53 protein is rapidly degraded following SL treatment in a D3- and D14-dependent manner. The d53 mutant protein, however, is resistant to SL-induced degradation (Jiang et al., 2013; Zhou et al., 2013). This suggests a mechanism in which SL promotes formation of an SCFD3-D14-D53 complex. This leads to polyubiquitination and degradation of D53, which enables growth responses to SL.
SMAX1 and D53 are members of a wider, uncharacterized SMXL protein family that has weak similarity to Class 1 Hsp100/ClpB proteins (Jiang et al., 2013; Stanga et al., 2013; Zhou et al., 2013). Convergence on the same gene family through independent approaches in two species strengthens the evidence that SMXL proteins are bona fide MAX2 targets. It also furthers the parallel between SL and KAR signaling pathways seen at the receptor level. Therefore, a promising hypothesis is that different aspects of MAX2-dependent signaling are mediated by degradation of different SMXL proteins. In this study, we perform an extensive analysis of loss-of-function mutants to determine the contributions of SMAX1, SMXL6, SMXL7, and SMXL8 to growth responses downstream of MAX2. We demonstrate that SMXL6, SMXL7, and SMXL8 are redundant orthologs of D53 that regulate shoot branching and other SL-regulated processes. We therefore establish specific relationships between SMAX1 and KAI2-KAR/KL-regulated growth and between SMXL6,7,8 and D14-SL-regulated growth.
RESULTS
SMXL6, SMXL7, and SMXL8 Control Branching in Arabidopsis
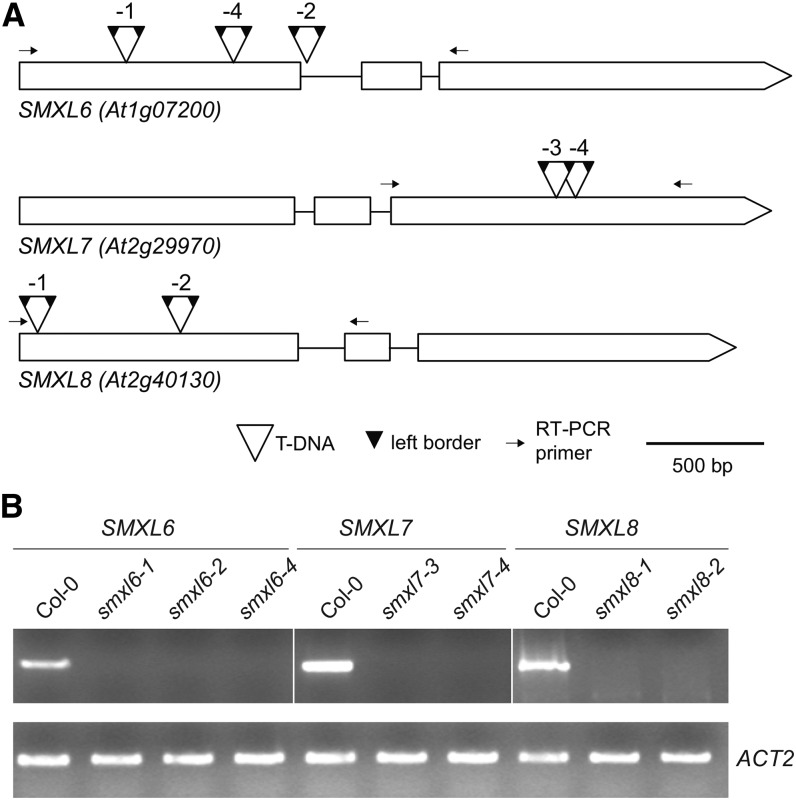
The SMXL family in Arabidopsis is composed of eight genes that can be divided into four clades present in all angiosperms: (1) SMAX1 and SMXL2, (2) SMXL3, (3) SMXL4 and SMXL5, and (4) SMXL6, SMXL7, and SMXL8 (Stanga et al., 2013; Zhou et al., 2013). To investigate whether SMXL genes control shoot branching, we constitutively expressed artificial microRNAs (amiRNAs) that target SMXL4 and SMXL5 (smxl45-ami), and SMXL6, SMXL7, and SMXL8 (smxl678-ami) in the max2-1 mutant background (Supplemental Figure 1). The increased branching phenotype of max2 was reduced by smxl678-ami in most transgenic lines, but not by smxl45-ami, implicating SMXL6, SMXL7, or SMXL8 in branching control. This approach was useful to overcome genetic redundancy in the SMXL family, but we observed variability in branching suppression among the transgenic lines (Supplemental Figure 1). To enable straightforward genetic analyses, we isolated multiple T-DNA insertion alleles for SMXL6, SMXL7, and SMXL8 from publicly available mutant collections. All T-DNA insertions disrupted the production of full-length transcripts (Figure 1).
Figure 1.
Mutant Alleles of SMXL6, SMXL7, and SMXL8.
(A) T-DNA insertion positions for smxl6-1 (SAIL_86_H04), smxl6-2 (SAIL_1285_H05), smxl6-4 (SALK_049115), smxl7-3 (WiDsLox339_C04), smxl7-4 (SALK_082032), smxl8-1 (SALK_025338C), and smxl8-2 (SALK_126406C). Boxes indicate exons, open triangles indicate the positions of T-DNAs, and small dark triangles indicate left borders. Most insertions had two outward-facing left borders, implying T-DNA concatamers. Left border insertion positions in the gene sequence were defined by Sanger sequencing and are described in Supplemental Table 2.
(B) End-point RT-PCR analysis of SMXL6 (30 cycles), SMXL7 (30 cycles), and SMXL8 (40 cycles) expression in respective T-DNA mutant lines. ACT2 (30 cycles) was used as an internal reference gene. RT-PCR primers span the T-DNA insertion sites, as indicated in (A).
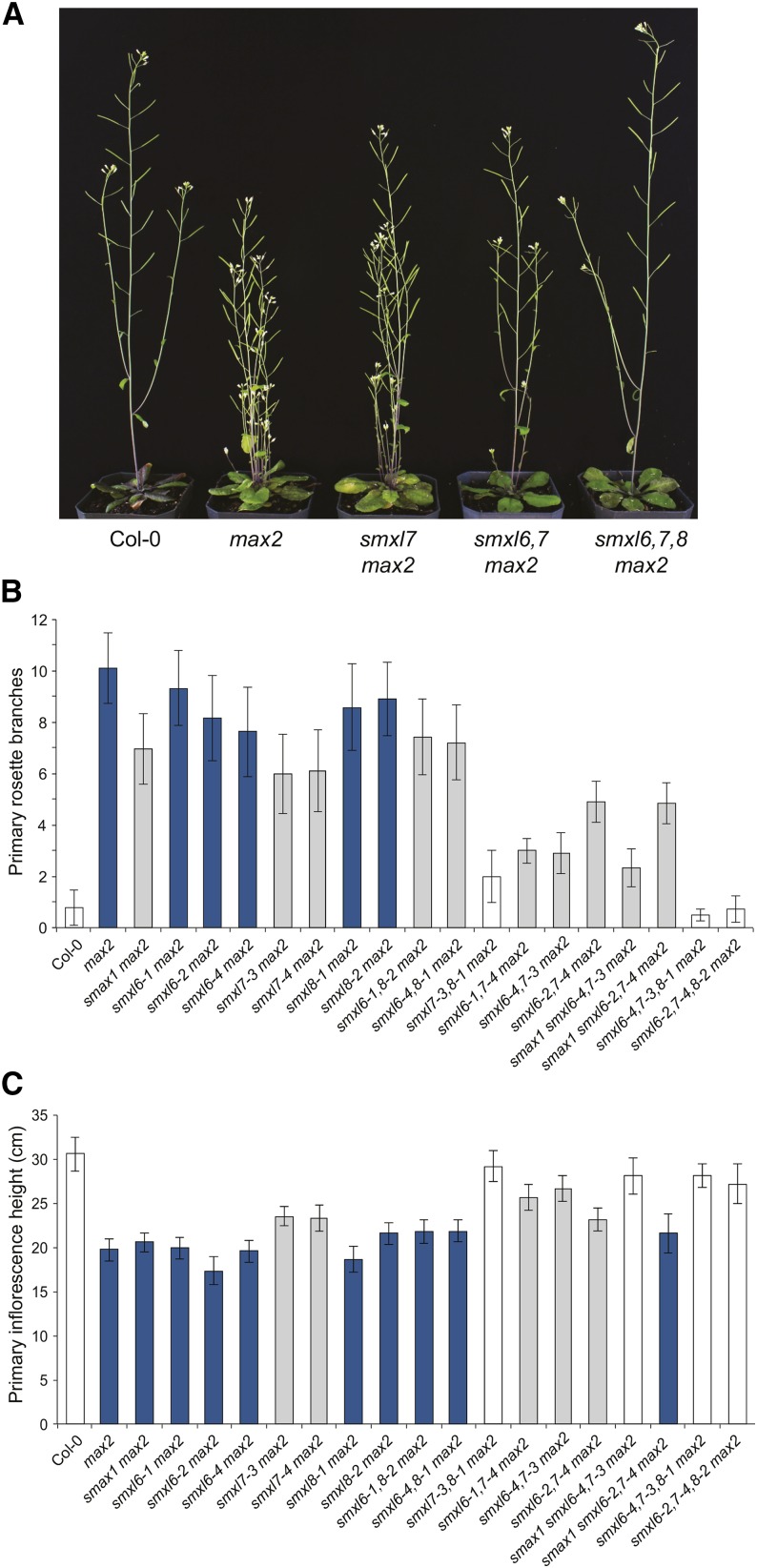
We first tested whether smxl6, smxl7, or smxl8 alleles suppress the max2-1 shoot branching phenotype. Branching and inflorescence heights of smxl6 max2 and smxl8 max2 allelic combinations were all similar to max2 (Figure 2). However, both smxl7-3 max2 and smxl7-4 max2 had significantly reduced branch numbers and increased height compared with max2 (Figure 2). To eliminate functional redundancy among these genes, we also created higher order smxl mutant combinations. Rosette branching and inflorescence height were restored to wild type levels in smxl7,8 max2 and smxl6,7,8 max2 (Figure 2; Supplemental Figure 2A). Although not quite phenotypically wild type, smxl6,7 max2 had reduced branching relative to smxl7 max2, while smxl6,8 max2 only showed a slight reduction in branching relative to max2 (Figure 2B). We conclude that SMXL7 has the predominant role in branching control in Arabidopsis but that SMXL6 and SMXL8 also contribute significantly.
Figure 2.
SMXL6, SMXL7, and SMXL8 Promote Shoot Branching Downstream of MAX2.
(A) Representative images of adult Col-0 (wild type), max2-1, smxl7-3 max2-1, smxl6-4,7-3 max2-1, and smxl6-4,7-3,8-1 max2-1 plants.
(B) Primary rosette branch number.
(C) Height of the primary inflorescence.
Plants were grown under a LD photoperiod (16 h light/8 h dark). Images and data were collected for each plant at 10 d postanthesis. For (B) and (C), graphs show mean ± 99% CI, n ≥ 17. Bar colors indicate no significant difference to Col-0 (white), no significant difference to max2 (dark blue), and significant differences to both Col-0 and max2 (gray). P < 0.001 (P = 0.02, with Bonferroni correction for 19 comparisons), Student’s t test.
The function of these genes with respect to shoot branching is most apparent in the max2 background, as rosette branching of smxl mutants alone was not significantly different from the wild type (Supplemental Figures 2B and 2C). The smxl6,7,8 mutant has the same number of rosette branches as the wild type but has a slight decrease in cauline branch number (Supplemental Figures 3A and 3C). Contrary to our prior observations (Stanga et al., 2013), we observed a mild but significant reduction in branching in smax1-2 max2 compared with max2 (Figure 2B). However, smax1 did not enhance branching suppression in either of two smxl6,7 max2 allelic combinations (Figure 2B), so if SMAX1 has a role in branching, it is relatively limited. This is supported by the observation that smxl6,7,8 mutations were sufficient to fully suppress max2 shoot phenotypes.
SMXL6,7,8 Promote Auxin Transport and PIN1 Accumulation in the Stem
Auxin transport within the inflorescence stem is increased in SL-deficient and SL-insensitive mutants (Bennett et al., 2006). This is attributed to increased accumulation of the auxin efflux carrier PIN1 at the basal plasma membrane of xylem parenchyma cells in the polar auxin transport stream (Bennett et al., 2006; Crawford et al., 2010; Shinohara et al., 2013). PIN1 levels at the plasma membrane are rapidly reduced following rac-GR24 treatment in a MAX2-dependent, translation-independent manner, and altered dynamics of PIN1 membrane localization are at least partially responsible for the increase in shoot branching observed in SL mutants (Bennett et al., 2006; Prusinkiewicz et al., 2009; Shinohara et al., 2013). PIN1 has thus been suggested as an effector of SL signaling (Shinohara et al., 2013).
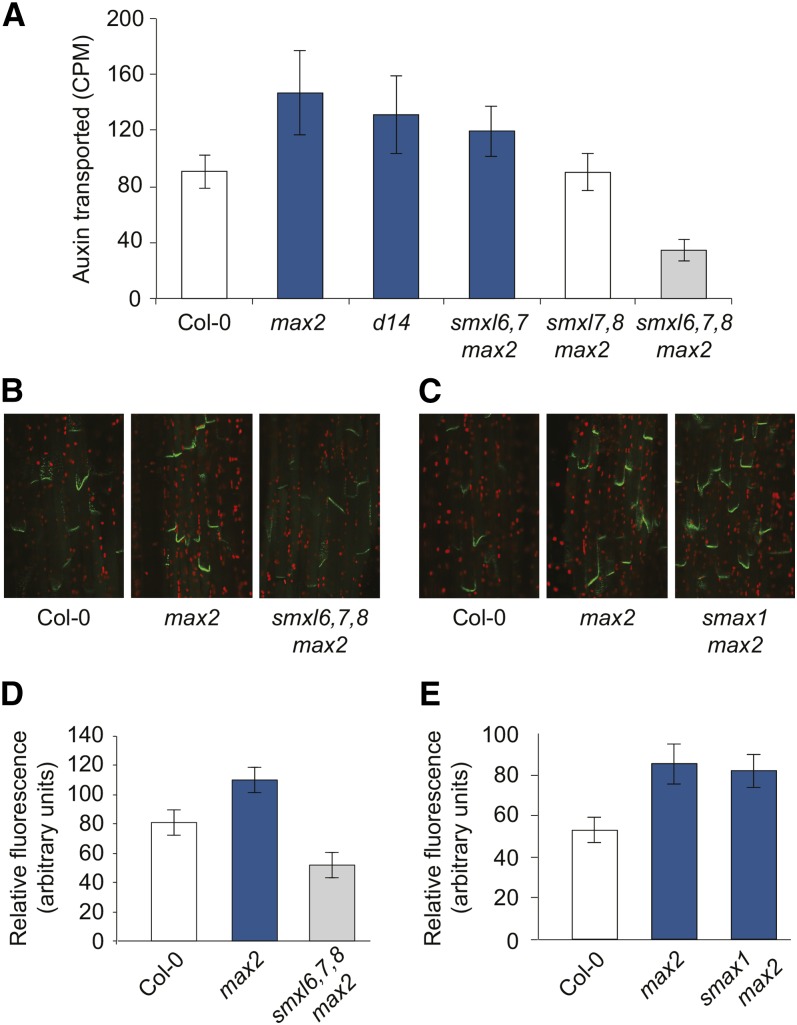
We tested whether SMXL proteins regulate PIN1-mediated auxin transport. We observed that the increased level of auxin transport in max2 was suppressed to wild-type levels in smxl7,8 max2 and even further reduced in smxl6,7,8 max2 (Figure 3A). The smxl6,7,8 mutant also has reduced auxin transport compared with the wild type (Supplemental Figure 3D). Thus, there is a correlation between auxin transport level in the stem and axillary bud outgrowth, although exceptions such as smxl6,7 max2 can be observed (Figure 3A). Consistent with these results, we observed reduced PIN1-GFP accumulation at the basal plasma membrane of xylem parenchyma cells in the basal internodes of smxl6,7,8 max2 (Figures 3B and 3D). In contrast, PIN1-GFP accumulation at the plasma membrane in max2 was not reduced by smax1 (Figures 3C and 3E). These data indicate that accumulation of PIN1 at the membrane is regulated by SMXL6,7,8 activity, as a downstream outcome of MAX2-dependent signaling.
Figure 3.
SMXL6,7,8 Promote Auxin Transport and PIN1 Accumulation.
(A) Auxin transport in stem segments from basal internodes. Mean ± 95% CI, n = 15 to 20.
(B) Confocal laser scanning fluorescence microscopy of PIN1-GFP in stem segments from basal internodes of Col-0, max2, and smxl6,7,8 max2.
(C) Confocal laser scanning fluorescence microscopy of PIN1-GFP in stem segments from basal internodes of Col-0, max2, and smax1 max2.
(D) PIN1-GFP fluorescence intensity at the basal plasma membrane in stem segments, as exemplified in (B). Mean ± 99% CI, n = 40 (five basal membranes in eight plants per genotype).
(E) PIN1-GFP fluorescence intensity at the basal plasma membrane in stem segments, as exemplified in (C). Mean ± 99% CI, n = 40 (five basal membranes in eight plants per genotype).
Bar colors indicate no significant difference to Col-0 (white), no significant difference to max2 (dark blue), and significant differences to both Col-0 and max2 (gray). Student’s t test, P < 0.05 for (A) and P < 0.01 for (D) and (E). PIN1-GFP is from Xu et al. (2006).
SMXL6,7,8 Repress BRC1/TCP18 Expression in Axillary Buds
The TCP domain (TEOSINTE BRANCHED1, CYCLOIDEA, and PCF) transcription factor BRC1/TCP18 has been implicated as another downstream effector of MAX2 signaling in the regulation of shoot branching. brc1 mutants have increased, SL-resistant shoot branching, BRC1 expression is upregulated after rac-GR24 treatment, and BRC1 transcription is reduced in max2, d14, and SL-deficient mutants (Aguilar-Martínez et al., 2007; Dun et al., 2012; Chevalier et al., 2014). We thus tested whether BRC1 might also act downstream of SMXL6,7,8. Consistent with prior reports, BRC1 expression was reduced more than 10-fold in the axillary buds of max2 rosette leaves. Conversely, BRC1 expression was upregulated more than 60-fold in smxl6,7,8 max2 relative to max2 but was not recovered in smax1 max2 (Figure 4). This observation is consistent with the suppression of branching seen in smxl6,7,8 max2 plants and suggests that repression of BRC1 transcription is one mechanism by which SMXL6,7,8 promote bud outgrowth.
Figure 4.
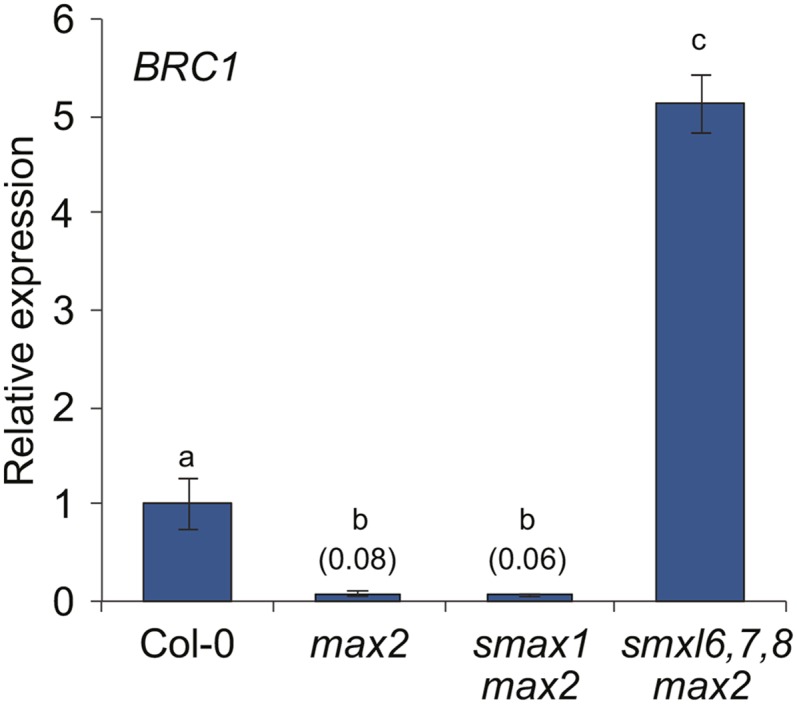
SMXL6,7,8 Repress BRC1 Expression in Axillary Buds.
RT-qPCR analysis of BRC1/TCP18 gene expression in nonelongated axillary buds of Col-0, max2-1, smax1-2 max2-1, and smxl6-4,7-3,8-1 max2-1 collected 7 d after anthesis. Expression of BRC1 is relative to CACS internal reference gene. Expression values are scaled to the Col-0 level of expression. Expression for max2-1 and smax1-2 max2-1 is indicated in parentheses. Mean ± se; n = 3 to 4 pooled tissue samples, four plants per pool. ANOVA with post-hoc Fisher’s LSD, P < 0.01.
SMXL6,7,8 Promote Lateral Root Growth
The max2 mutant has increased lateral root density, which can be attributed partially to SL insensitivity, as SL-deficient max1 and max4 mutants have an intermediate lateral root density phenotype between the wild type and max2 (Kapulnik et al., 2011; Ruyter-Spira et al., 2011). We assessed whether SMXL6,7,8 might also act downstream of MAX2 in this response. We found that smxl6,7,8 had lower lateral root density than the wild type and strongly suppressed the increased lateral root density phenotype of max2 (Figure 5; Supplemental Figure 3H). In contrast, lateral root density in smax1 max2 seedlings was not significantly different from max2 (Figure 5).
Figure 5.
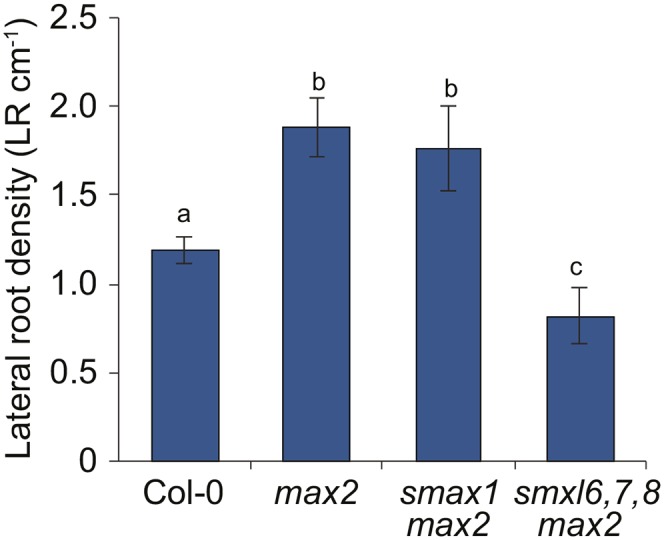
Lateral Root Formation Is Suppressed by smxl6,7,8.
Lateral root density of 10-d-old seedlings. Mean ± se; n = 6 experiments, ≥10 roots measured per genotype in each experiment. ANOVA with post-hoc Student’s paired t test, P < 0.05.
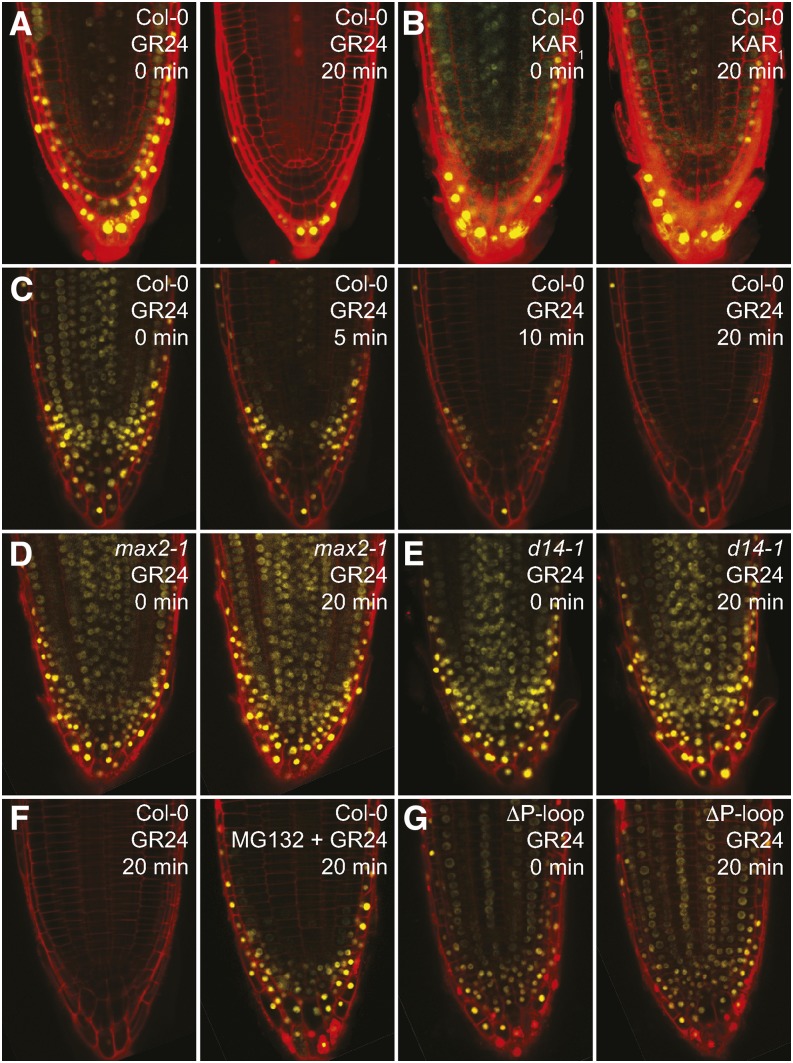
rac-GR24 Triggers Rapid Degradation of SMXL7
Our data support the hypothesis that SMXL6,7,8 are targets of SCFMAX2 in Arabidopsis. To test this, we examined the posttranslational regulation of SMXL7 by SL and KAR1. We introduced a 35Spro:SMXL7-YFP reporter into a wild-type (Col-0) background. SMXL7-YFP, which localizes to the nucleus, was degraded by 1 μM rac-GR24, but not KAR1, within 20 min (Figures 6A and 6B). At a 10 μM rac-GR24 concentration, SMXL7-YFP was noticeably reduced after 5 min of treatment and was undetectable after 20 min (Figure 6C). SMXL7-YFP was not degraded after rac-GR24 treatment in the max2 and d14 backgrounds (Figures 6D and 6E). Consistent with SL-induced proteolysis via SCFMAX2, pretreatment with the 26S proteasome inhibitor MG132 reduced rac-GR24-triggered destabilization of SMXL7-YFP (Figure 6F). These results support a role for D14 and SCFMAX2 in targeting SMXL7 for degradation after SL perception and are consistent with the regulation of D53 by SL in rice (Jiang et al., 2013; Zhou et al., 2013). Notably, KAR1 had no effect on SMXL7 and the d14 mutation was sufficient to stabilize SMXL7. This suggests that KAI2 does not regulate SMXL7.
Figure 6.
rac-GR24 Triggers Rapid Degradation of SMXL7.
(A) and (B) Col-0 35Spro:SMXL7-YFP roots after 0 and 20 min of treatment with 1 μM rac-GR24 (A) or 1 μM KAR1 (B).
(C) Col-0 35Spro:SMXL7-YFP after 0, 5, 10, and 20 min treatment with 10 μM rac-GR24.
(D) max2-1 35Spro:SMXL7-YFP after 0 and 20 min of treatment with 10 μM rac-GR24.
(E) d14-1 35Spro:SMXL7-YFP after 0 and 20 min of treatment with 10 μM rac-GR24.
(F) Col-0 35Spro:SMXL7-YFP after 20 min of treatment with 10 μM rac-GR24, without and with a 1-h pretreatment with 50 μM MG132, a 26S proteasome inhibitor.
(G) An eight-amino acid sequence (FRGKTVVD) containing a putative P-loop was removed from SMXL7-YFP. Col-0 35Spro:SMXL7ΔP-loop-YFP after 0 and 20 min of treatment with 10 μM rac-GR24.
The semidominant d53 mutation in rice is a 15-bp deletion that results in substitution of the amino acids RGKTGI with a single threonine residue. This form of d53 is resistant to rac-GR24-induced degradation (Jiang et al., 2013; Zhou et al., 2013). The RGKT component is a conserved motif in the Arabidopsis SMAX1, SMXL2, and SMXL6,7,8 proteins (and their rice homologs) but is not present in SMXL3,4,5 (Supplemental Figure 4). It is similar to the phosphate binding P-loop (or Walker A) consensus sequence Gx4GK[T/S] and therefore may bind ATP or GTP (Saraste et al., 1990). For instance, deletion of an RGKT motif from the protein phosphatase 2C ABI2, which is involved in ABA signaling, strongly reduced its phosphatase activity (Sun et al., 2011). To test whether this motif is required for SCFMAX2-dependent degradation of SMXL7-YFP, we created a ΔP-loop variant of SMXL7 (deletion of amino acids 718 to 725). Similar to d53, SMXL7ΔP-loop was resistant to degradation after rac-GR24 treatment (Figure 6G). Based on the evidence of similarity in sequence, developmental roles, and posttranslational regulation by SL, we conclude that SMXL7 and D53 are orthologs.
SMXL6,7,8 Affect Cotyledon Expansion but Not Germination or Hypocotyl Elongation
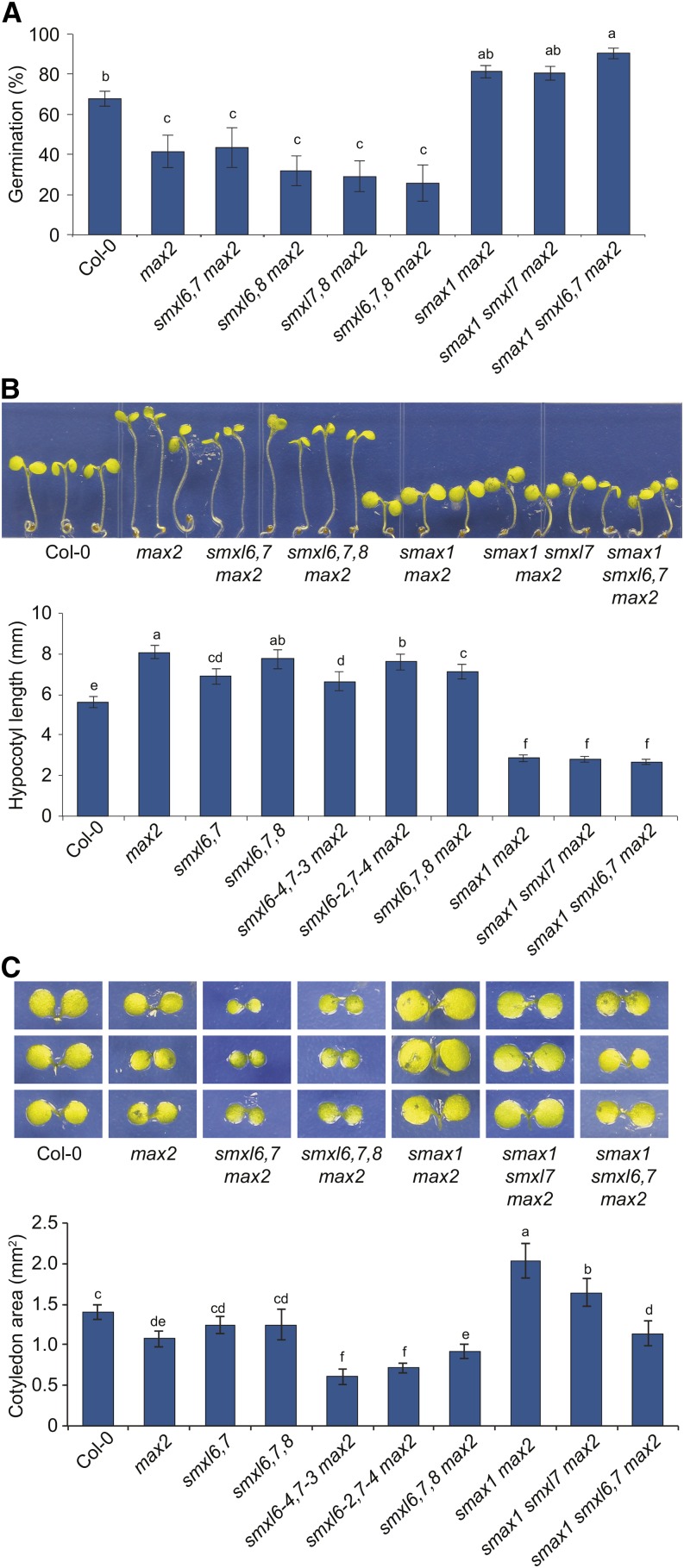
We next examined whether SMXL6,7,8 may be SCFMAX2 targets in other developmental contexts. max2 seed are insensitive to the germination-promoting effects of karrikins and have increased primary dormancy (Nelson et al., 2011). The dormancy phenotype of max2 is suppressed by smax1 (Stanga et al., 2013). Loss of SMXL6,7,8 did not significantly increase germination of max2 nor did smxl6,7 further enhance smax1 max2 germination (Figure 7A). Thus, SMXL6,7,8 do not regulate Arabidopsis seed dormancy.
Figure 7.
Distinct Roles for SMXL6,7,8 and SMAX1 during Germination and Seedling Growth.
(A) Germination of primary dormant seed after 66 h imbibition at 21°C in LD photoperiod on 0.8% (w/v) Bacto-agar media. Mean ± se; n = 6 to 7 batches of seed, 75 seed tested per batch. ANOVA with post-hoc Fisher’s LSD, P < 0.025.
(B) Hypocotyl lengths of 5-d-old seedlings grown under continuous red light (30 μE) for 4 d at 21°C. Mean ± 99% CI; n = 45 seedlings (15 seedlings from three replicate plates). ANOVA with post-hoc Fisher’s LSD, P < 0.01.
(C) Cotyledon surface area of seedlings grown as described in (B). Mean ± 99% CI; n = 36 cotyledons from 18 seedlings. ANOVA with post-hoc Fisher’s LSD, P < 0.01.
max2 seedlings grown under continuous red light have elongated hypocotyls, consistent with decreased responses to light (Stirnberg et al., 2002; Shen et al., 2007). We tested whether SMXL6,7,8 might regulate this process but did not observe consistent suppression of the max2 hypocotyl phenotype among several smxl6,7 max2 and smxl6,7,8 max2 allelic combinations (Figure 7B; Supplemental Figure 5A). The max2 hypocotyl phenotype was strongly suppressed by smax1, producing even shorter seedlings than the wild type, but the hypocotyl length of smax1 smxl6,7 max2 was not different from that of smax1 max2. We conclude that SMXL6,7,8 do not promote hypocotyl elongation.
max2 mutant seedlings also have reduced cotyledon expansion (Shen et al., 2007). Consistent with the results above, this phenotype is suppressed by smax1, producing significantly larger cotyledons than the wild type (Stanga et al., 2013). In contrast, we observed that two smxl6,7 alleles actually enhanced the max2 phenotype (Figure 7C). Thus, SMAX1 and SMXL6,7 have opposite and antagonistic effects on cotyledon expansion (Figure 7C). Neither SMAX1 or SMXL6,7 may be considered epistatic to the other, as the smax1 smxl6,7 max2 phenotype is intermediate to smax1 max2 and smxl6,7 max2. Curiously, smxl6,7,8 max2 had cotyledons that were larger than smxl6,7 max2 and not different than max2 (Figure 7C; Supplemental Figure 5B), suggesting that SMXL8 may have an opposite effect on cotyledon expansion than SMXL6 and SMXL7 or that loss of all three genes affects growth capacity differently than the loss of two genes.
These results implicate SMAX1, but not SMXL6,7,8, as the likely target of MAX2 during germination and seedling photomorphogenesis. It should be noted that the dormancy and seedling growth phenotypes of max2 are not seen in SL-deficient mutants or d14 and appear to be kai2-specific defects (Nelson et al., 2011; Shen et al., 2012; Waters et al., 2012). Moreover, (1) KAR but not natural SL (stereoisomers with a 2′R configuration) treatments promote Arabidopsis germination; (2) KAR promotes cotyledon expansion, but rac-GR24, which acts through both KAI2 and D14, inhibits cotyledon expansion; and (3) SL-deficient max mutants and d14, but not kai2, have increased axillary branching (Nelson et al., 2010; Waters et al., 2012; Scaffidi et al., 2013, 2014). Therefore smax1 suppresses max2 phenotypes associated with KAR/KL-KAI2 regulated growth, and smxl6,7,8 suppresses max2 phenotypes associated with SL-D14-regulated growth.
SMAX1 and SMXL6,7,8 Influence Different Aspects of Leaf Morphology
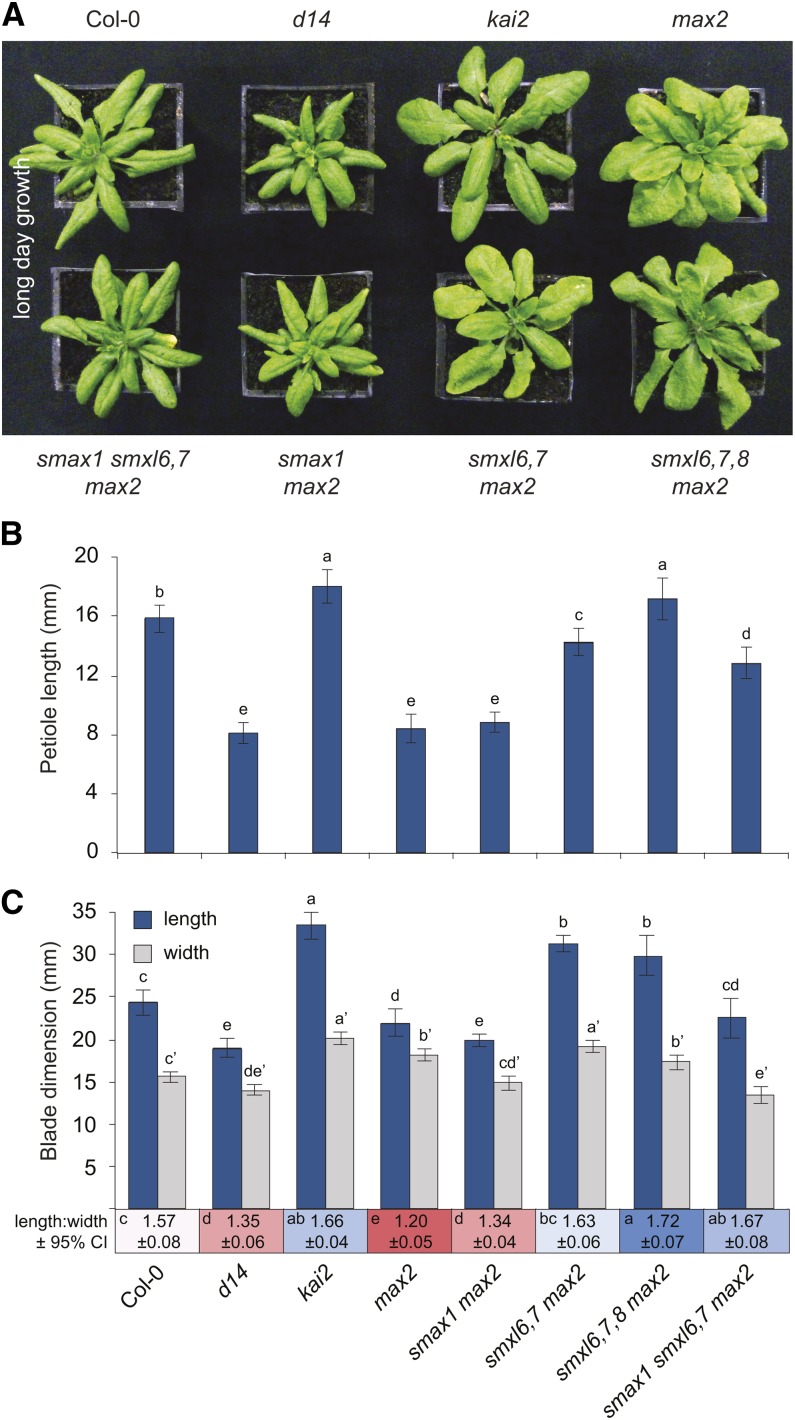
To further investigate this correlation, we examined the contributions of SMXL genes to leaf morphology, which is regulated by both KAI2 and D14. SL-deficient mutants and d14 have a distinct leaf phenotype under long-day (LD) photoperiods (16 h light/8 h dark), with reduced petiole and blade lengths (Figure 8) (Stirnberg et al., 2002; Booker et al., 2004; Scaffidi et al., 2013). In kai2 leaves, blade length and width are both increased. The max2 mutant shares the general rosette morphology of d14, including shorter petiole length, but has a rounder, wider blade than d14; this may represent a hybrid of the d14 and kai2 phenotypes (Figure 8).
Figure 8.
SMXL6,7,8 and SMAX1 Have Different Effects on Leaf Morphology.
(A) Rosettes of 28-d-old plants grown under LD photoperiod (16 h light/8 h dark). d14-1, kai2-1, max2-1, smax1-2, smxl6-4, smxl7-3, and smxl8-1 alleles were used. kai2-1, first identified in Landsberg erecta, was backcrossed twice into the Col-0 background.
(B) Petiole length of the 7th leaf of 35-d-old plants grown under LD photoperiod. Mean ± 95% CI; n = 11 to 12. ANOVA with post-hoc Fisher’s LSD, P < 0.05.
(C) Blade length (blue), not including the petiole, and width (gray) of the 7th leaf of 35-d-old plants grown under LD photoperiod. Mean ± 95% CI; n = 11 to 12. ANOVA with post-hoc Fisher’s LSD, P < 0.05. Prime symbol (e.g., a′) differentiates statistical tests of width measurements from length measurements. Mean length:width ratio ± 95% CI is shown below the x axis. Statistical groups for length:width ratios are indicated in each colored box.
We observed that petiole elongation was inhibited specifically by SMXL6,7,8. The shortened petiole phenotype of max2 was the same as d14, and max2 petiole elongation was additively restored by smxl6, smxl7, and smxl8 mutations (Figure 8B; Supplemental Figures 3F and 6A). KAI2 and SMAX1 had little or no effect on petiole elongation. However, SMAX1 does promote leaf blade growth, as smax1 caused a reduction in the blade width and length of max2 and smxl6,7 max2 (Figure 8C; Supplemental Figure 6B). In contrast, smxl6,7 mutations increased blade elongation of max2, suggesting that SMXL6 and SMXL7 repress leaf growth under these conditions. Consistent with the idea that D14 targets SMXL6,7,8 for degradation and KAI2 may target SMAX1 for degradation, d14 effects on leaf growth were opposite to smxl6,7,8 and kai2 effects were opposite to smax1. Moreover, d14 shared leaf morphology traits with smax1 max2, and kai2 had similar morphology to smxl6,7 max2 and smxl6,7,8 max2; thus, different aspects of the hybrid d14-kai2 phenotype of max2 were suppressed by removing subsets of the SMXL family. Suppression of both subsets of genes restored max2 leaves to essentially wild type dimensions, as shown by smax1 smxl6,7 max2 (Figure 8C; Supplemental Figure 6B).
Tissue-Specific Expression Patterns of SMAX1 and SMXL6,7,8
The different developmental roles of SMAX1 and SMXL6,7,8 could result from distinct expression patterns. Among the SMXL family, SMAX1 is expressed most highly in seed, seedlings, and leaves, and SMXL7 is expressed most highly in axillary branches (Stanga et al., 2013). This suggested that further correlation might be found between the expression of SMXL family members and their functions.
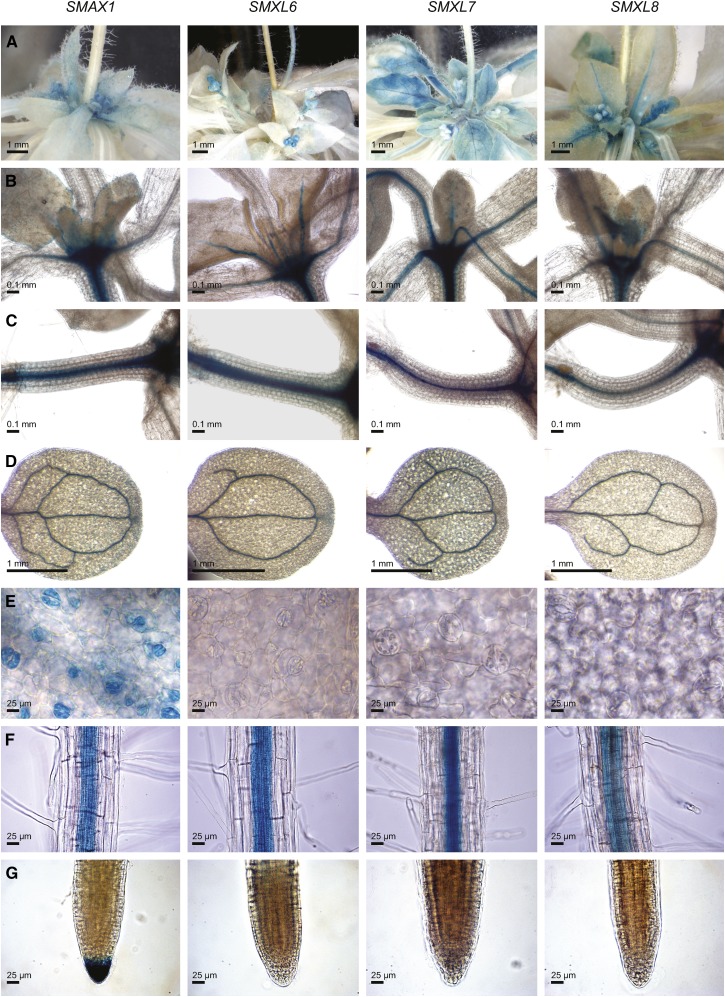
We examined the tissue-specific expression of SMAX1, SMXL6, SMXL7, and SMXL8 through promoter fusions to a GUS-GFP reporter. We observed similar patterns of expression for the SMXL6, SMXL7, and SMXL8 transcriptional reporters in the primary rosette buds and expanding leaves of adult rosettes, the vasculature of the hypocotyls, cotyledons, and mature roots, and in the midvein and petioles of young leaves (Figures 9A to 9D and 9F). The SMAX1 transcriptional reporter had a similar pattern of expression in these tissues but also produced GUS staining in the young leaf periphery, stomata, and the root cap of primary roots (Figures 9B, 9E, and 9G). SMAX1pro:GUS-GFP expression was also visible in the caps of lateral roots but not during the early stages of lateral root emergence (Supplemental Figures 7A to 7D). SMAX1pro:GUS-GFP expression in seedling hypocotyls was somewhat variable, showing strong expression in the hypocotyl vasculature in some seedlings and little or no expression in others (Figure 9C; Supplemental Figure 7E). A clear correlation between SMAX1 and SMXL6,7,8 expression and their mutant phenotypes was not apparent in these data, suggesting that tissue-specific transcriptional regulation is not sufficient to explain the different developmental roles of these genes.
Figure 9.
Tissue-Specific GUS Expression from SMAX1, SMXL6, SMXL7, and SMXL8 Promoters.
Expression of promoter:GUS-GFP fusions in the primary rosette buds and young leaves of 4.5-week-old plants (A), shoot apex and developing leaves (B), hypocotyl (C), cotyledonary veins (D), stomata on first true leaves (E), vasculature in the maturation zone of primary roots (F), and primary root cap (G). (B) to (G) are tissues of 10-d-old light-grown seedlings. For each tissue, the different SMAX1 and SMXL transcriptional reporters were stained at the same time, for the same duration.
SMXL7 and SMAX1 May Interact with TOPLESS Proteins
An alternative explanation for the differences between SMAX1 and SMXL6,7,8 functions might be that they have different protein-protein interactions with downstream signaling partners. The downstream signal transduction partners of the SMXL family are unknown, but three potential ethylene-responsive element binding factor amphiphilic repression (EAR) motifs in D53 that were identified by Jiang et al. (2013) provide clues about their identity. EAR motifs were initially identified as transcriptional repression domains in ethylene signaling (Ohta et al., 2001). Subsequent analysis has shown that EAR domains are required for interactions with the C-terminal to lissencephaly homology (CTLH) domain of the TOPLESS (TPL) and TOPLESS-RELATED (TPR) family of transcriptional corepressors (Szemenyei et al., 2008). EAR motifs are typically defined as LxLxL, DLNxxP, or the hybrid L/FDLNL/FxP (Ohta et al., 2001; Kagale et al., 2010). Only one of the candidate EAR motifs from D53 is evolutionarily conserved across angiosperm SMXL sequences (Bennett and Leyser, 2014), but it is often a noncanonical sequence that only retains strict conservation of the L/FDLN portion of the motif (Supplemental Figure 8A). Therefore, TPL and TPR corepressors are reasonable candidates for proteins that interact with SMXL family members. Indeed, D53 was previously reported to interact with rice TPL-related proteins TPR1, TPR2, and TPR3 in a mammalian two-hybrid assay, and the TPR2-D53 interaction was supported by an in vitro pull-down assay (Jiang et al., 2013). SMXL6 was also identified as a TPR3 interactor in a yeast two-hybrid whole-plant library screen with TPL/TPR baits (Causier et al., 2012).
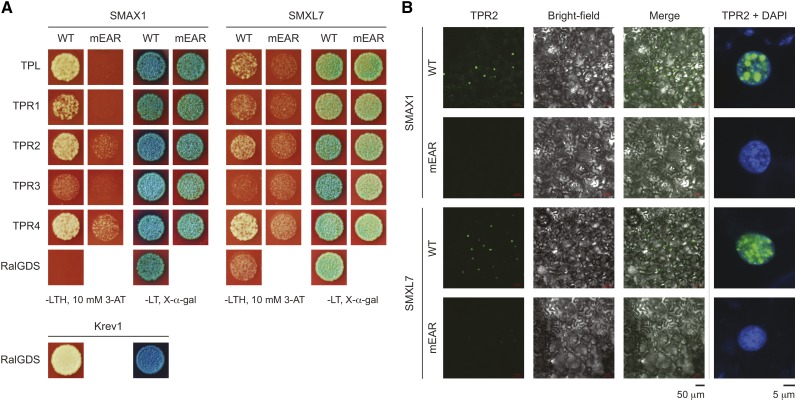
We tested whether SMAX1 and SMXL7 can interact with TPL/TPR proteins in a yeast two-hybrid assay. SMAX1 interacted with TPL, TPR2, and TPR4, and to a lesser extent with TPR1. Mutation of the putative EAR motif (mEAR) weakened or abolished growth on selective media (Figure 10A). SMXL7 had a similar, but weaker, pattern of interactions than SMAX1. SMXL7mEAR mutants did not show better growth than the negative control test with RalGDS. In comparison to the strong positive control RalGDS-Krev1 interaction, yeast two-hybrid interactions involving SMAX1 or SMXL7 were weak. β-Galactosidase activity was also weak in all interactions and did not appear substantially different between most wild-type and mEAR versions of SMAX1 and SMXL7 (Figure 10A).
Figure 10.
EAR Motif-Dependent Interaction of SMAX1 and SMXL7 with TOPLESS Family Proteins.
(A) Yeast two-hybrid interaction tests of SMAX1 and SMXL7 with TPL and TPR proteins TPR1-TPR4. SMAX1/SMXL7 proteins were bait (N-terminal GAL4 DNA binding domain fusions), and TPL/TPR proteins were prey (N-terminal GAL4 activation domain fusions). RalGDS was a negative control for interactions with SMAX1 and SMXL7. RalGDS + Krev1 was a strong positive control. SMAX1 and SMXL7 variants with a mutated EAR domain (mEAR, sequence FDLNQ or LDLNL modified to ADANA) were also tested. Growth is shown for equally diluted colonies grown for 3 d at 30°C on -Leu/Trp/His (-LTH) dropout media with 10 mM 3-amino-1,2,4-triazole (3-AT). Qualitative LacZ assays were performed by growing colonies on -Leu/Trp (-LT) double dropout media with 80 μg/mL X-α-Gal for 30 h.
(B) BiFC tests for interaction between SMAX1, SMAX1mEAR, SMXL7, and SMXL7mEAR with TPR2. The N-terminal portion of GFP was fused to the N terminus of TPR2, and the C-terminal portion of GFP was fused to the N terminus of SMAX1/SMXL7 proteins. Confocal fluorescence microscopy images of transiently transformed N. benthamiana leaves were taken with the same exposure settings across construct comparisons. A representative field of view is shown (left). Representative nuclei (right) are shown with GFP signal (green) overlaid onto 4′,6-diamidino-2-phenylindole (DAPI) signal (blue).
As a complementary approach, we tested whether SMAX1 and SMXL7 can interact with TPR2 through a transient bimolecular fluorescence complementation (BiFC) assay in Nicotiana benthamiana leaves. We observed associations between SMAX1 and SMXL7 with TPR2, which were localized to subnuclear speckles. These interactions were abolished by mutation of the EAR motif, even though the mEAR isoforms of SMAX1 and SMXL7 were present in equivalent amounts as the wild-type isoforms (Figure 10B; Supplemental Figure 8B).
These data, interaction studies by others (Jiang et al., 2013; Zhou et al., 2013), and the strong conservation of the EAR motif suggest that SMXL proteins interact with TPL/TPR in an EAR-motif dependent manner. Therefore, it is tempting to speculate that SMXL proteins function in a transcriptional corepressor complex, similar to Aux/IAAs in auxin signaling and NINJA in jasmonate signaling (Tiwari et al., 2004; Pauwels et al., 2010). However, further analysis is required to determine whether the EAR motif is in fact necessary for SMXL function, whether SMXL complexes are associated with DNA, and whether SMXL have repressive effects on expression of KAR- and SL-responsive genes. Other EAR motif-interacting proteins, such as SIN3-ASSOCIATED POLYPEPTIDE P18 and LEUNIG/LEUNIG_HOMOLOG, must also be considered as candidate interaction partners. In any case, there is currently no evidence for preferences among the SMXL family for different TPL/TPR. Therefore, discrimination among other signal transduction partners may be necessary for differential growth control by SMXL proteins.
DISCUSSION
In this article, we describe the characterization of SMXL6, SMXL7, and SMXL8 from Arabidopsis and show that they are co-orthologs of rice D53. The original d53 mutation is hypermorphic, and while such mutations are useful because they can overcome functional redundancy, they can also give misleading interpretations of genetic relationships or obscure the roles of individual genes within a family. Here, we use genetic resources in Arabidopsis to perform an extensive loss-of-function analysis of the SMXL6,7,8/D53 clade. SMXL6,7,8 have conserved roles in the regulation of shoot branching and act upstream of the auxin efflux carrier PIN1 and the TCP transcription factor BRC1 in this process. Our analysis of rac-GR24-induced SMXL7 protein degradation supports it as a proteolytic target of MAX2 and D14 activity, which corresponds well to the previously described regulation of the D53 protein (Jiang et al., 2013; Zhou et al., 2013). We show that SMXL6,7,8 regulate multiple aspects of development including leaf morphology and root architecture. We demonstrate that SMAX1 and SMXL6,7,8 have distinct roles in development but can also have overlapping, antagonistic effects within the same tissues (e.g., cotyledons and leaves). Finally, we provide support for in vivo interactions between SMAX1 and SMXL7 with TPR2, suggesting a potential function for SMXL proteins in transcriptional regulation. Several themes emerge from our analysis, which we discuss below.
SMXL6,7,8 Are the Major Targets of MAX2-Dependent SL Signaling
In this study, we observed that smxl6,7,8 max2 phenotypes mimic most wild-type responses to SL, but smax1 max2 phenotypes mimic wild-type responses to KAR. For example, KAR, but not SL, promotes germination, and we observed reduced dormancy in smax1 max2 but not smxl6,7,8 max2. rac-GR24 suppresses cotyledon expansion in seedlings, but KAR promotes expansion; correspondingly, smxl6,7,8 max2 have smaller cotyledons, but smax1 max2 have enlarged cotyledons. Finally, SL, but not KAR, represses axillary branching, and smxl6,7,8 max2 have a clear reduction in primary rosette branch numbers.
As SCFMAX2 mediates the signaling of at least two different butenolide molecules, the max2 phenotype is a combination of SL-insensitive phenotypes (as observed in d14) and KAR/KL-insensitive phenotypes (as observed in kai2). This is particularly apparent in rosettes, in which max2 is phenotypically intermediate to kai2 and d14 (Figures 8A and 11A). Interestingly, SMAX1 and SMXL6,7 have very different effects on leaf morphology in short days versus long-day conditions (Figures 8A and 11A). Nonetheless, under both photoperiods, it is clear that smxl6, smxl7, and smxl8 suppress max2 phenotypes that are shared with d14; thus, the smxl6,7 max2 rosette resembles kai2. Each SL-related phenotype in max2 that we examined was completely suppressed by the loss of SMXL6, 7, and 8, which strongly suggests that these proteins are major proteolytic targets of SCFMAX2-mediated SL signaling. Consistent with a lack of germination or seedling phenotypes in d14 and SL biosynthesis mutants, the smxl6,7,8 max2 mutants were not different from max2 in germination and hypocotyl elongation (Figure 7). In contrast, we show that smax1 suppresses max2 phenotypes that are shared with kai2, and as such the smax1 max2 mutant rosettes resemble d14 (Figures 8A and 11A). Thus, SMAX1 is likely to be the major target of SCFMAX2 during KAR/KL-KAI2 signaling. We did not examine phenotypes such as leaf senescence, drought tolerance, secondary growth, adventitious rooting, or root hair initiation in this study, but we hypothesize that these phenotypes are also regulated by SMXL proteins.
Figure 11.
SMAX1 and SMXL6,7,8 Have Complementary Roles in MAX2-Regulated Development.
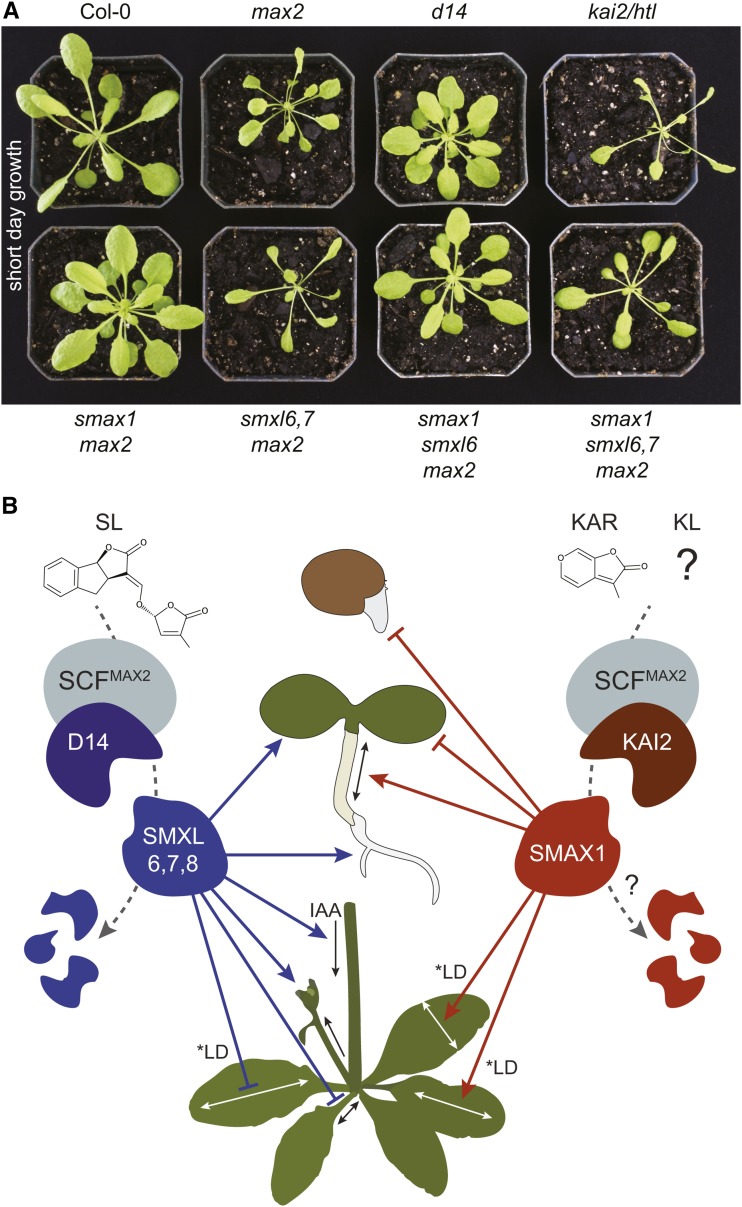
(A) Rosettes of 37-d-old plants grown under a short-day photoperiod (8 h light/16 h dark). The kai2/htl allele is htl-3 (Toh et al., 2014).
(B) Model of SL and KAR/KL signaling. SMXL6,7,8 act in processes controlled by D14, and SMAX1 acts in processes controlled by KAI2. SMAX1 represses germination and seedling responses to light (e.g., reduces cotyledon expansion and promotes hypocotyl elongation) and promotes medio-lateral blade expansion (i.e., blade width) and elongation in long days. SMXL6,7,8 promote branching, auxin transport, PIN1 accumulation at the basal plasma membrane (data not shown), and lateral root density but inhibit petiole elongation and proximo-distal blade expansion in long days (i.e., blade elongation). *LD denotes control in a long-day photoperiod; under short-day conditions, SMAX1 and SMXL6,7,8 have different effects on leaf growth but are still antagonistic (A). Loss of SMXL6,7 inhibits cotyledon expansion, although this is countered somewhat by loss of SMXL8. D53/SMXL7 interacts with D14 after SL perception and is targeted for degradation in a MAX2-dependent manner (Figure 6; Jiang et al., 2013; Zhou et al., 2013; Umehara et al., 2015). A similar mechanism for SMAX1 degradation after KAR/KL perception by KAI2 is hypothesized.
The discovery that MAX2 mediates signaling in response to the perception of two types of butenolide molecules raises questions about how specific downstream responses could occur (Nelson et al., 2011). Our results show that SMAX1 and SMXL6,7,8 expression overlaps in many tissues and furthermore that SMAX1 and SMXL6,7,8 can have antagonistic effects within the same tissues. It is thus unlikely that differential expression of these proteins is sufficient to account for the specific outcomes of MAX2-dependent signaling. Instead, our results support a model in which specific protein-protein interactions between KAI2-SMAX1 and D14-SMXL6/7/8 confer the specificity of MAX2 signaling (Figure 11B) (Bennett and Leyser, 2014). This hypothesis is supported by recent KAI2 and D14 promoter-swapping experiments, which demonstrated that the distinct roles of KAI2 and D14 in plant growth are not due to transcriptional regulation (Waters et al., 2015). Physical interactions between D14 and D53 have been demonstrated (Jiang et al., 2013; Zhou et al., 2013), and yeast two-hybrid assays also support interaction between SMXL7 and D14 in the presence of SL analogs (Umehara et al., 2015). However, KAI2 and D14 affinities for different SMXL protein partners have not been reported. It should be noted that under some experimental conditions, there might be some cross-interaction between these anticipated receptor-target pairs. For instance, exogenous SL application inhibits hypocotyl elongation through D14 [e.g., GR24 or (+)-5-deoxystrigol application is effective in the kai2 mutant, but not in kai2 d14] (Scaffidi et al., 2014; Toh et al., 2014; Umehara et al., 2015). However, smxl6,7,8 max2, which otherwise shows constitutive SL responses, has no suppression of hypocotyl elongation (Figure 7B). This suggests that SMAX1, or another SMXL family member that can regulate hypocotyl growth, is targeted by D14 following exogenous SL treatment. However, we do not believe that this response to exogenous SL is physiologically relevant, as d14 and SL-deficient mutants have normal hypocotyl elongation (Shen et al., 2007, 2012; Nelson et al., 2011; Waters et al., 2012; Umehara et al., 2015). Thus, even though D14 can mediate a response to exogenous SL, endogenous SL does not appear to control hypocotyl growth.
Evolutionary Diversification of Butenolide Signaling Pathways
Therefore, we hypothesize that the ligand-induced formation of separate receptor-SMXL complexes permits their interaction with SCFMAX2 and subsequent degradation of the SMXL protein. One of the intriguing aspects of SL signaling is that the D14 clade arose within the vascular plants, but KAI2 proteins can be traced back to charophyte algae (Waldie et al., 2014). However, SL synthesis and responses to rac-GR24 have been identified in basally diverging land plants such as the moss Physcomitrella patens (Proust et al., 2011). One possible explanation for these observations is that SL and other butenolide molecules in basally diverging land plants signal through a KAI2-SMXL pathway similar to that in angiosperms. Consistent with this, SMXL proteins are present in P. patens (Bennett and Leyser, 2014; Waldie et al., 2014). It is possible that wholesale duplication of this signaling pathway in the vascular plant lineage gave rise to the D14 and SMXL6,7,8/D53 clades, and that subsequent coevolution of the receptor-target pairs created separate signaling pathways for SL and KAR/KL (Bennett and Leyser, 2014). Interestingly, this signaling system has evolved more recently at the receptor level in parasitic plants in the Orobanchaceae family, in which SL perception through KAI2 is the basis of host-triggered germination (Conn et al., 2015; Toh et al., 2015; Tsuchiya et al., 2015).
SMXL Proteins Are Likely the Only Proteolytic Targets of SCFMAX2
All the max2 phenotypes we have examined so far can be attributed to excess SMAX1 and/or SMXL6,7,8 activity, which is strikingly illustrated by the broadly wild-type phenotype of smax1 smxl6,7 max2 mutant rosettes (Figures 8 and 11A). This suggests that SMXL proteins are likely to be the only direct proteolytic targets of SCFMAX2-dependent signaling. Among the four other unstudied SMXL proteins in Arabidopsis, SMXL3, SMXL4, and SMXL5 lack the RGKT motif that is required for MAX2-mediated protein degradation of D53/SMXL7 (Supplemental Figure 4). As such, these proteins may not be proteolytic targets of MAX2. Nevertheless, we did not examine every max2 phenotype, and it is quite possible that SMXL3,4,5 function in other MAX2-regulated processes. SMAX1 and SMXL2 are very closely related proteins that likely represent a recent duplication; as such, SMXL2 may have redundant functions with SMAX1 (Stanga et al., 2013).
Our analysis calls into question prior assertions that BES1 and DELLA proteins are targets of SCFMAX2. The identification of BES1 was primarily through biochemical means, and the conclusion that it is a MAX2 target hinges on the report that BES1-RNAi lines suppress the max2 shoot branching phenotype (Wang et al., 2013). However, the three transgenic lines described by Wang et al. had severe growth defects, including dwarfism, reduced inflorescence height, highly curled leaves, and reduced fertility, and appeared even more severely affected in the max2 background than in the wild type. It is therefore unlikely that max2 branching suppression by BES1-RNAi is a specific epistatic interaction, rather than an effect caused by a highly pleiotropic mutant phenotype. In contrast, smxl6,7,8 mutations restore max2 to an approximately wild-type shoot morphology, thus showing a highly specific genetic interaction consistent with being bona fide targets of max2. In the case of DELLA proteins, the biological relevance of the reported interaction between the DELLA protein SLR1 from rice and the strigolactone receptor D14 has not yet been demonstrated (Nakamura et al., 2013). Protein-protein interaction studies can provide powerful means to identify new components of signaling pathways or modes of crosstalk, and we do not necessarily exclude the possibility of crosstalk between strigolactone signaling and brassinosteroid or gibberellin signaling. However, cautious interpretation of these results is advised in the absence of supporting genetic evidence.
SMXL Interactions with Auxin
We demonstrated that SMXL6,7,8 promote auxin transport in the stem and accumulation of PIN1-GFP at the basal plasma membrane of cells in the polar auxin transport stream. Cycloheximide does not prevent the rapid removal of PIN1-GFP from the plasma membrane after rac-GR24 treatment, indicating that de novo protein synthesis is not required for SL effects on auxin transport capacity (Shinohara et al., 2013). This observation challenges the current hypothesis that SMXL proteins act through transcriptional regulation and suggests that transcription-independent functions should also be considered. For instance, it is possible that a heat shock protein-like function of SMXL proteins may be involved in PIN1 regulation.
It also remains to be determined whether SMXL genes influence other PIN proteins or affect auxin transport in other tissues. Auxin dose–response curves often show a bell shape, in which too little or too much auxin can cause similar, nonoptimal growth effects (Collett et al., 2000; Ruyter-Spira et al., 2011). It is tempting to speculate that unexpected observations such as elongated smxl6,7 and smxl6,7,8 hypocotyls compared with the wild type, and reduced phenotypic severity in smxl6,7,8 max2 cotyledons compared with smxl6,7 max2 may be caused by shifts in auxin distribution or availability (Figures 7B and 7C; Supplemental Figure 5).
SMXL6,7,8 Are Epistatic to MAX2
A notable feature of our data is that the suppression of max2 by smxl6,7,8 often “overshoots” the wild type, resulting in phenotypes that are quantitatively opposite to the effect of max2. For instance, lateral root density is ∼50% less than the wild type in smxl6,7,8 max2 (Figure 5), while PIN1-GFP and auxin transport levels are ∼40 and ∼60% less than the wild type, respectively (Figure 3A). Intriguingly, we found that several of these phenotypes are replicated in a MAX2 background, such that smxl6,7,8 is similar to smxl6,7,8 max2 in growth habit and branching pattern (Supplemental Figures 3A and 3C), lateral root growth (Supplemental Figure 3H), and auxin transport level (Supplemental Figure 3D). There are obvious differences in leaf shape between these genotypes, but these result from the residual KAI2-related phenotypes in the max2 background (Supplemental Figures 3B, 3E, and 3F). In a wild-type MAX2 background, without these conflicting phenotypes, smxl6,7,8 causes narrowing of the leaf blade and an increase in petiole length, emphasizing the qualitative and quantitative differences between smxl6,7,8 and the wild type (Supplemental Figure 3). Taken together, these data show that SMXL6,7,8 are epistatic to MAX2-mediated SL signaling, such that their mutation causes the same phenotypes irrespective of whether SL signaling is functional or not. This suggests that SMXLs are not simply repressors of SL signaling, but are growth regulators whose activity is repressed by SL signaling. Wild-type morphology in Arabidopsis thus represents a balance between too little SMXL activity (e.g., smxl6,7,8) and unrestrained SMXL activity (e.g., max2).
METHODS
Plant Materials
The max2-1 (Stirnberg et al., 2002), d14-1 (Waters et al., 2012), kai2-1 (Waters et al., 2012), htl-3 allele (Toh et al., 2014), smax1-2 max2-1 (Stanga et al., 2013), and PIN1pro:PIN1-GFP (Xu et al., 2006) lines have been described previously. T-DNA alleles described here are smxl6-1 (SAIL_86_H04), smxl6-2 (SAIL_1285_H05), smxl6-4 (SALK_049115), smxl7-3 (WiDsLox339_C04), smxl7-4 (SALK_082032), smxl8-1 (SALK_025338C), and smxl8-2 (SALK_126406C). T-DNA alleles were generated by Alonso et al. (2003), Sessions et al. (2002), and Woody et al. (2007). All lines are in the Col-0 background. To validate the knockout effects of the T-DNA alleles, RNA was extracted from 7-d-old seedlings with the Spectrum Total Plant RNA extraction kit (Sigma-Aldrich) and reverse transcribed with Superscript III (Invitrogen) and anchored oligo(dT) primers. PCR was performed on cDNA for 30 or 40 cycles, as noted. Primers for genotyping and RT-PCR analysis are described in Supplemental Table 1. Except where other alleles are indicated, simplified mutant notation in figures (e.g., smax1 smxl6,7 max2) refers to the max2-1, smax1-2, smxl6-4, smxl7-3, and smxl8-1 alleles.
Plant Growth Conditions
Mature plants for leaf morphology, auxin transport, and PIN1-GFP analysis were grown on Levington’s F2 compost under a LD photoperiod (16 h light/8 h dark) at 22°C/18°C day/night in controlled environment rooms with light provided by white fluorescent tubes (∼150 μmol m−2 s−1). For other assays, seeds were plated on 0.5× Murashige and Skoog (MS) basal medium with Gamborg’s vitamins (Sigma-Aldrich M0404), pH 5.7, 0.8% (w/v) Bacto agar plates, stratified 3 d in the dark at 4°C, and grown for 7 d under LD photoperiod (∼50 to 70 μmol m−2 s−1) at 21°C before transfer to soil (Fafard 3B) and further growth under LD photoperiod (∼80 to 110 μmol m−2 s−1) at 21°C. Light intensity for plants grown in short days (8 h light/16 h dark) was ∼150 to 200 μmol m−2 s−1.
Statistical Analysis
Confidence intervals were calculated with the t-distribution. ANOVA and post-hoc comparisons of means with Student’s t test (Fisher’s LSD) were performed in JMP Pro 11.
amiRNAs
amiRNAs were designed by Web MicroRNA Designer (WMD3) at http://wmd3.weigelworld.org/ (Ossowski et al., 2008). Primers for amiRNA cloning are described in Supplemental Table 1. attB adapter sequences were included during amiRNA construction for Gateway cloning. amiRNAs were cloned into pDONR221 entry vector and then transferred into a pEarlyGate100 vector (Earley et al., 2006) that drives expression from a 35S promoter. Constructs were introduced into max2-1 and max2-1 PIN1-GFP by the Agrobacterium tumefaciens floral dip method (Clough and Bent, 1998) and selected for BASTAr. Homozygous transgenic lines were acquired for T2 lines that had a 3:1 segregation ratio of BASTAr, suggesting a single T-DNA insertion locus.
Axillary Branching and Height Assay
The position of plants within flats was randomized to account for environmental variation. Primary rosette branches at least 1 cm in length were counted for each plant at 10 d postanthesis, or as noted. The primary inflorescence height was measured on the same day as branching.
Auxin Transport Assays
Auxin transport assays were modified from Crawford et al. (2010). The 18-mm stem segments from basal internodes were excised and the apical end placed in 30 μL ATS solution without sucrose, pH 5.6, containing 1 μM 14C-IAA (American Radiolabeled Chemicals). After 6 h incubation, the basal 5 mm of the stem segment was excised, placed in 200 μL scintillation liquid, and shaken overnight at 400 rpm prior to scintillation counting (Perkin-Elmer MicroBeta2).
PIN1-GFP Quantitation
Hand sections of the vascular bundles of basal internodal stem segments of 6-week-old plants were embedded in agar plates. PIN1-GFP was imaged by confocal laser scanning microscopy using a Zeiss LSM700 imaging system with 20× water immersion lenses (final magnification 200×). Excitation was performed using 488-nm (15% laser power) and 555-nm (6%) lasers. Chloroplast autofluorescence was detected above 600 nm and GFP fluorescence below 555 nm. The same settings for GFP detection were used within experiments for each line. Fluorescence intensity in the GFP channel was performed on nonsaturated images using Zeiss ZEN software.
Gene Expression Analysis
RNA was prepared with the Sigma-Aldrich Spectrum Total Plant RNA extraction kit from nonelongated axillary buds (collected 7 d after anthesis) of plants grown in LD photoperiod. Real-time quantitative RT-PCR was performed as previously described (Stanga et al., 2013). Primers for BRC1 and CACS reference genes are described in Supplemental Table 1.
SMXL7 Degradation
The SMXL7 coding sequence was cloned into a pDONR221 entry vector (Life Technologies). The SMXL7ΔP-loop variant, lacking amino acids 718 to 725 (FRGKTVVD), was made with the Q5 Site-Directed Mutagenesis Kit (NEB). Primers are described in Supplemental Table 1. SMXL7 and SMXL7ΔP-loop entry clones were subcloned into a pEarlyGate101 destination vector (Earley et al., 2006), between the 35S promoter and a C-terminal YFP tag. The resultant 35Spro:SMXL7-YFP and SMXL7ΔP-loop-YFP constructs were transformed into the wild type (Col-0) and max2-1 genetic backgrounds using the Agrobacterium floral dip method (Clough and Bent, 1998). The roots of 3- to 5-d-old T3 homozygous seedlings were treated as described, mounted on glass slides in 10 μM propidium iodide solution, and imaged by confocal laser scanning microscopy using a Zeiss LSM780 imaging system with 20× lenses. Excitation was performed using a 514-nm laser. Propidium iodide was detected above 600 nm and YFP fluorescence below 555 nm. The same settings for YFP detection were used within experiments for each line.
Germination Assay
Plants grown under LD photoperiod at 21°C were harvested when most of the siliques were brown (∼2 months of growth in soil) and dried in paper bags at room temperature for 3 d. Seeds were cleaned and stored at −80°C to preserve primary seed dormancy. Seeds were surface sterilized with a 70% (v/v) ethanol and 0.05% (v/v) Triton X-100 solution for 5 min, rinsed with 70% (v/v) ethanol, rinsed with 95% (v/v) ethanol, and dried on filter paper. Seeds were plated on 0.8% (w/v) Bacto-agar (Difco) and incubated in white light (∼50 to 70 μmol m−2 s−1) at 21°C under LD photoperiod. Germination was indicated by the emergence of the radicle. Independent seed batches grown under similar conditions were assessed.
Seedling Photomorphogenesis Assay
Surface-sterilized seeds were plated on solid 0.5× MS medium and stratified in the dark at 4°C for 3 d. Seeds were treated with 3 h of white light (∼50 to 70 μmol m−2 s−1) at 21°C, 21 h of dark at 21°C, and then grown for 4 d at 21°C in continuous red light (∼30 μmol m−2 s−1; Philips GreenPower LED Research Module HF Deep Red). Hypocotyl lengths and cotyledon areas were measured from digital images using ImageJ (NIH).
Lateral Root Assay
Seedlings were grown using ATS media (Lincoln et al., 1990) with 1% (w/v) sucrose, solidified with 0.8% (w/v) plant agar, in 10-cm square plates. Plates were oriented vertically in growth chambers with a LD photoperiod (22°C/18°C), with light provided by white fluorescent tubes (intensity ∼150 μmol m−2 s−1). Roots were analyzed at 10 d postgermination (dpg). Plates were imaged using a flatbed scanner at 600 dpi. Primary root length was then measured using ImageJ. Lateral roots and primordia that were visible in the scanned images were scored. The lateral root density along the whole root was calculated separately for each root.
Leaf Morphology Assay
The 7th leaf of each plant was marked with indelible marker at ∼4 weeks postgermination. These leaves were provisionally measured at 35 dpg and then again at 37 dpg to confirm that growth of these leaves was arrested. The maximum length and width of the leaf blade were measured, as well as the length of the petiole.
Promoter:GUS Reporter Assay
Approximately 3 kb of DNA 5′ to the start codon of SMAX1, SMXL6, SMXL7, and SMXL8 was PCR amplified from wild-type (Col-0) genomic DNA using Primestar GXL high-fidelity DNA polymerase (Clontech), cloned by BP reaction into Gateway entry vector pDONR221 (Invitrogen), and sequenced to confirm orientation and reading frame. Primers are described in Supplemental Table 1. Reverse primers included 6 or 12 of the first coding nucleotides of each gene to ensure full transcriptional fusions. Promoters were subcloned by LR reaction into pHGWFS7, a vector for GUS and GFP fusions (Karimi et al., 2002). Constructs were transformed into Col-0 plants by Agrobacterium floral dip method. T1 transformants were selected for resistance to 25 μg/mL hygromycin. GUS analysis was performed in the T2 generation and compared across multiple independent transgenic lines to ensure consistent tissue-specific expression patterns. Surface-sterilized seeds were plated on 0.5× MS medium, stratified 3 d at 4°C in dark, and grown at 21°C in white light under LD photoperiod. After 10 d of growth, seedlings were either stained for GUS expression or transferred to soil and grown under LD photoperiod. Whole plants were assayed for GUS expression after bolting (∼30 to 34 d old). GUS staining was according to Bomblies (2000). Seedlings were stained for 12 h and adult tissues for 16 h. Representative images are shown.
Yeast Two-Hybrid Assays
Yeast two-hybrid was performed with the Proquest system (Invitrogen). pDEST32 and pDEST22 constructs were generated by Gateway cloning (Invitrogen) from sequenced entry vectors. Primers for cloning are described in Supplemental Table 1. mEAR site-directed mutants were generated with InFusion (Clontech). The MaV203 strain of Saccharomyces cerevisiae was transformed with pDEST32 bait plasmid and a pDEST22 prey plasmid by the polyethylene glycol-mediated transformation method (Gietz and Schiestl, 2007). Transformants were selected on -Leu/-Trp double dropout media and confirmed by colony PCR. To compare growth, yeast were grown overnight in -Leu/-Trp media, diluted to final concentrations of 0.005 and 0.0005 OD550, spotted onto -Leu/-Trp/-His plates containing 5 or 10 mM 3-amino-1,2,4-triazole, and grown 3 d at 30°C. To compare β-galactosidase activity, diluted colonies were grown on -Leu/-Trp plates containing 80 μg/mL X-α-Gal for 30 h at 30°C.
Bimolecular Fluorescence Complementation Analysis
BiFC expression plasmids (Gehl et al., 2009) were generated by Gateway cloning from sequenced entry vectors and transformed in Agrobacterium (strain C58C1). Transient transformation of Nicotiana benthamiana was according to Voinnet et al. (2003), except the OD600 of p19 carriers and each BiFC vector carrier strain was 0.3 and 0.4, respectively, for a total OD600 of 1.1 after resuspension in the inoculant buffer (10 mM MES, pH 5.6, 10 mM MgCl2, and 150 μM acetosyringone). In addition, cells were incubated for 2 to 4 h at room temperature in the inoculation buffer. Each interaction pair was injected into two different plants aged 4 to 5 weeks old. Leaf clippings were collected 2 to 4 d after injections and viewed under a Zeiss 510 LSM confocal microscope. Images were taken at 200× and 400×. To test whether a lack of mEAR interaction with TPR2 was due to low mEAR protein levels, tissue coinfected with SMAX1, smax1mEAR, SMXL7, or smxl7mEAR with TPR2 BiFC vectors was collected 2 d after infection. Protein was extracted in 400 mM sucrose, 10 mM KCl, 1 mM MgCl2, 0.01% (v/v) Triton X-100, 200 mM Tris, pH7.2, and 3% (v/v) DTT. Protein concentration was normalized by Bradford assay (Bio-Rad). Total protein (34 μg) for each sample was separated by 10% SDS-PAGE. Protein blots on polyvinylidene fluoride membrane were probed with mouse monoclonal anti-HA HA-7 primary antibody (Sigma-Aldrich) at 1:1000 dilution and anti-mouse IgG horseradish peroxidase-linked secondary antibody (Cell Signaling) at 1:10,000 dilution. GE Amersham ECL Western Blotting Detection Reagent was used for detection.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACT2 (At3g18780), BRC1 (At3g18550), CACS (At5g46630), D14 (At3g03990), KAI2 (At4g37470), MAX2 (At2g42620), SMAX1 (At5g57710), SMXL4 (At4g29920), SMXL5 (At5g57130), SMXL6 (At1g07200), SMXL7 (At2g29970), SMXL8 (At2g40130), TPL (At1g15750), TPR1 (At1g80490), TPR2 (At3g16830), TPR3 (At5g27030), and TPR4 (At3g15880).
Supplemental Data
Supplemental Figure 1. Targeting of SMXL6,7,8 with artificial miRNAs reduces branching in max2.
Supplemental Figure 2. Shoot phenotypes of additional smxl6, smxl7, and smxl8 mutants.
Supplemental Figure 3. Shoot, leaf, and root phenotypes of smxl6,7,8.
Supplemental Figure 4. Putative P-loop motif in the SMXL family.
Supplemental Figure 5. Seedling morphology phenotypes of additional smxl6, smxl7, and smxl8 alleles.
Supplemental Figure 6. Specific contributions of SMXL6, SMXL7, and SMXL8 to leaf morphology.
Supplemental Figure 7. Additional examples of SMAX1pro:GUS-GFP expression in 10-d-old seedlings.
Supplemental Figure 8. A conserved C-terminal EAR motif in the SMXL/D53 family in Arabidopsis and rice.
Supplemental Table 1. Primer sequences.
Supplemental Table 2. T-DNA positions of smxl alleles.
Supplementary Material
Acknowledgments
We thank Shigeo Toh and Peter McCourt for the htl-3 allele of KAI2/HTL. Funding support was provided by the University of Georgia Research Foundation and the National Science Foundation (IOS-1350561) to D.C.N.; NIGMS National Institutes of Health Award T32GM007103 to N.M.; Chinese Government Scholarship PhD Program (Sichuan Agriculture University) to Y.L.; and European Research Council (294514 EnCoDe) and the Gatsby Foundation (GAT3272C) to T.B. and O.L.
AUTHOR CONTRIBUTIONS
I.S., T.B., N.M., Y.L., J.P.S., and D.C.N. designed the research. I.S., T.B., N.M., Y.L., J.P.S., A.A., and D.C.N. performed research and analyzed data. T.B., O.L., and D.C.N. wrote the article.
Glossary
- SL
strigolactone
- KAR
karrikin
- KL
KAI2 ligand
- LD
long-day
- BiFC
bimolecular fluorescence complementation
- amiRNA
artificial microRNA
- MS
Murashige and Skoog
- dpg
days postgermination
References
- Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agusti J., Herold S., Schwarz M., Sanchez P., Ljung K., Dun E.A., Brewer P.B., Beveridge C.A., Sieberer T., Sehr E.M., Greb T. (2011). Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc. Natl. Acad. Sci. USA 108: 20242–20247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Hayashi H. (2006). Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann. Bot. (Lond.) 97: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Babili S., Bouwmeester H.J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66: 161–186. [DOI] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Bennett T., Leyser O. (2014). Strigolactone signalling: standing on the shoulders of DWARFs. Curr. Opin. Plant Biol. 22: 7–13. [DOI] [PubMed] [Google Scholar]
- Bennett T., Sieberer T., Willett B., Booker J., Luschnig C., Leyser O. (2006). The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16: 553–563. [DOI] [PubMed] [Google Scholar]
- Bomblies K. (2000). Whole mount GUS staining. In Arabidopsis: A Laboratory Manual, Weigel D., Glazebrook J., eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; ), pp. 243–245. [Google Scholar]
- Booker J., Auldridge M., Wills S., McCarty D., Klee H., Leyser O. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14: 1232–1238. [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F., Nieminen K., Sánchez-Ferrero J.C., Rodríguez M.L., Chagoyen M., Hardtke C.S., Cubas P. (2014). Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26: 1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collett C.E., Harberd N.P., Leyser O. (2000). Hormonal interactions in the control of Arabidopsis hypocotyl elongation. Plant Physiol. 124: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn C.E., Bythell-Douglas R., Neumann D., Yoshida S., Whittington B., Westwood J.H., Shirasu K., Bond C.S., Dyer K.A., Nelson D.C. (2015). PLANT EVOLUTION. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349: 540–543. [DOI] [PubMed] [Google Scholar]
- Cook C.E., Whichard L.P., Turner B., Wall M.E., Egley G.H. (1966). Germination of witchweed (Striga lutea Lour.): Isolation and properties of a potent stimulant. Science 154: 1189–1190. [DOI] [PubMed] [Google Scholar]
- Crawford S., Shinohara N., Sieberer T., Williamson L., George G., Hepworth J., Müller D., Domagalska M.A., Leyser O. (2010a). Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 137: 2905–2913. [DOI] [PubMed] [Google Scholar]
- Dun E.A., de Saint Germain A., Rameau C., Beveridge C.A. (2012). Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 158: 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629. [DOI] [PubMed] [Google Scholar]
- Flematti G.R., Ghisalberti E.L., Dixon K.W., Trengove R.D. (2004). A compound from smoke that promotes seed germination. Science 305: 977. [DOI] [PubMed] [Google Scholar]
- Flematti G.R., Waters M.T., Scaffidi A., Merritt D.J., Ghisalberti E.L., Dixon K.W., Smith S.M. (2013). Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol. Plant 6: 29–37. [DOI] [PubMed] [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.R., Hänsch R. (2009). New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2: 1051–1058. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. (2007). Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2: 35–37. [DOI] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- Hamiaux C., Drummond R.S., Janssen B.J., Ledger S.E., Cooney J.M., Newcomb R.D., Snowden K.C. (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22: 2032–2036. [DOI] [PubMed] [Google Scholar]
- Jain N., Kulkarni M.G., van Staden J. (2006). A butenolide, isolated from smoke, can overcome the detrimental effects of extreme temperatures during tomato seed germination. Plant Growth Regul. 49: 263–267. [Google Scholar]
- Jiang L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S., Links M.G., Rozwadowski K. (2010). Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiol. 152: 1109–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y., et al. (2011). Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233: 209–216. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kulkarni M.G., Sparg S.G., Light M.E., van Staden J. (2006). Stimulation of rice (Oryza sativa L.) seedling vigour by smoke-water and butenolide. J. Agron. Crop Sci. 192: 395–398. [Google Scholar]
- Lincoln C., Britton J.H., Estelle M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., et al. (2013). Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4: 2613. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Flematti G.R., Ghisalberti E.L., Dixon K.W., Smith S.M. (2012). Regulation of seed germination and seedling growth by chemical signals from burning vegetation. Annu. Rev. Plant Biol. 63: 107–130. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Flematti G.R., Riseborough J.A., Ghisalberti E.L., Dixon K.W., Smith S.M. (2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 7095–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.C., Riseborough J.A., Flematti G.R., Stevens J., Ghisalberti E.L., Dixon K.W., Smith S.M. (2009). Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149: 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., Beveridge C.A., Ghisalberti E.L., Smith S.M. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Matsui K., Hiratsu K., Shinshi H., Ohme-Takagi M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schwab R., Weigel D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53: 674–690. [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust H., Hoffmann B., Xie X., Yoneyama K., Schaefer D.G., Yoneyama K., Nogué F., Rameau C. (2011). Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138: 1531–1539. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P., Crawford S., Smith R.S., Ljung K., Bennett T., Ongaro V., Leyser O. (2009). Control of bud activation by an auxin transport switch. Proc. Natl. Acad. Sci. USA 106: 17431–17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A., Mason M.G., De Cuyper C., Brewer P.B., Herold S., Agusti J., Geelen D., Greb T., Goormachtig S., Beeckman T., Beveridge C.A. (2012). Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 158: 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C., Kohlen W., Charnikhova T., van Zeijl A., van Bezouwen L., de Ruijter N., Cardoso C., Lopez-Raez J.A., Matusova R., Bours R., Verstappen F., Bouwmeester H. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: another belowground role for strigolactones? Plant Physiol. 155: 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M., Sibbald P.R., Wittinghofer A. (1990). The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15: 430–434. [DOI] [PubMed] [Google Scholar]
- Scaffidi A., Waters M.T., Ghisalberti E.L., Dixon K.W., Flematti G.R., Smith S.M. (2013). Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J. 76: 1–9. [DOI] [PubMed] [Google Scholar]
- Scaffidi A., Waters M.T., Sun Y.K., Skelton B.W., Dixon K.W., Ghisalberti E.L., Flematti G.R., Smith S.M. (2014). Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165: 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Luong P., Huq E. (2007). The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol. 145: 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Zhu L., Bu Q.Y., Huq E. (2012). MAX2 affects multiple hormones to promote photomorphogenesis. Mol. Plant 5: 750–762. [DOI] [PubMed] [Google Scholar]
- Shinohara N., Taylor C., Leyser O. (2013). Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol. 11: e1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D.E., Fujiwara S. (2009). Thinking outside the F-box: novel ligands for novel receptors. Trends Plant Sci. 14: 206–213. [DOI] [PubMed] [Google Scholar]
- Stanga J.P., Smith S.M., Briggs W.R., Nelson D.C. (2013). SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Furner I.J., Ottoline Leyser H.M. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50: 80–94. [DOI] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande K., Leyser H.M. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Sun H.L., Wang X.J., Ding W.H., Zhu S.Y., Zhao R., Zhang Y.X., Xin Q., Wang X.F., Zhang D.P. (2011). Identification of an important site for function of the type 2C protein phosphatase ABI2 in abscisic acid signalling in Arabidopsis. J. Exp. Bot. 62: 5713–5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen G., Guilfoyle T.J. (2004). Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S., Holbrook-Smith D., Stogios P.J., Onopriyenko O., Lumba S., Tsuchiya Y., Savchenko A., McCourt P. (2015). Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350: 203–207. [DOI] [PubMed] [Google Scholar]
- Toh S., Holbrook-Smith D., Stokes M.E., Tsuchiya Y., McCourt P. (2014). Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem. Biol. 21: 988–998. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y., Yoshimura M., Sato Y., Kuwata K., Toh S., Holbrook-Smith D., Zhang H., McCourt P., Itami K., Kinoshita T., Hagihara S. (2015). PARASITIC PLANTS. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349: 864–868. [DOI] [PubMed] [Google Scholar]
- Ueda H., Kusaba M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis. Plant Physiol. 169: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Cao M., Akiyama K., Akatsu T., Seto Y., Hanada A., Li W., Takeda-Kamiya N., Morimoto Y., Yamaguchi S. (2015). Structural requirements of strigolactones for shoot branching inhibition in rice and Arabidopsis. Plant Cell Physiol. 56: 1059–1072. [DOI] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- van Staden J., Sparg S.G., Kulkarni M.G., Light M.E. (2006). Post-germination effects of the smoke-derived compound 3-methyl-2H-furo[2,3-c]pyran-2-one, and its potential as a preconditioning agent. Field Crops Res. 98: 98–105. [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Waldie T., McCulloch H., Leyser O. (2014). Strigolactones and the control of plant development: lessons from shoot branching. Plant J. 79: 607–622. [DOI] [PubMed] [Google Scholar]
- Wang Y., Sun S., Zhu W., Jia K., Yang H., Wang X. (2013). Strigolactone/MAX2-induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev. Cell 27: 681–688. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., Smith S.M. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Flematti G.R., Smith S.M. (2013). The origins and mechanisms of karrikin signalling. Curr. Opin. Plant Biol. 16: 667–673. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Moulin S.L., Sun Y.K., Flematti G.R., Smith S.M. (2015). A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in Arabidopsis development but cannot mediate responses to karrikins or strigolactones. Plant Cell 27: 1925–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M.T., Scaffidi A., Sun Y.K., Flematti G.R., Smith S.M. (2014). The karrikin response system of Arabidopsis. Plant J. 79: 623–631. [DOI] [PubMed] [Google Scholar]
- Woody S.T., Austin-Phillips S., Amasino R.M., Krysan P.J. (2007). The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant Res. 120: 157–165. [DOI] [PubMed] [Google Scholar]
- Xie X., Yoneyama K., Yoneyama K. (2010). The strigolactone story. Annu. Rev. Phytopathol. 48: 93–117. [DOI] [PubMed] [Google Scholar]
- Xu J., Hofhuis H., Heidstra R., Sauer M., Friml J., Scheres B. (2006). A molecular framework for plant regeneration. Science 311: 385–388. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Furusawa S., Nagasaka S., Shimomura K., Yamaguchi S., Umehara M. (2014). Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240: 399–408. [DOI] [PubMed] [Google Scholar]
- Zhang Y., et al. (2014). Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 10: 1028–1033. [DOI] [PubMed] [Google Scholar]
- Zhou F., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.