Chemical compounds in plant root exudates influence the growth of neighboring plants by interfering with their chromatin configuration and gene expression.
Abstract
To secure their access to water, light, and nutrients, many plant species have developed allelopathic strategies to suppress competitors. To this end, they release into the rhizosphere phytotoxic substances that inhibit the germination and growth of neighbors. Despite the importance of allelopathy in shaping natural plant communities and for agricultural production, the underlying molecular mechanisms are largely unknown. Here, we report that allelochemicals derived from the common class of cyclic hydroxamic acid root exudates directly affect the chromatin-modifying machinery in Arabidopsis thaliana. These allelochemicals inhibit histone deacetylases both in vitro and in vivo and exert their activity through locus-specific alterations of histone acetylation and associated gene expression. Our multilevel analysis collectively shows how plant-plant interactions interfere with a fundamental cellular process, histone acetylation, by targeting an evolutionarily highly conserved class of enzymes.
INTRODUCTION
Many plant species produce and release allelochemicals, bioactive secondary metabolites that can inhibit germination or growth of neighboring plants (Belz, 2007; Macías et al., 2007). Such chemical clashes are generally referred to as “allelopathy” (Molisch, 1937). They can occur between individuals of the same or of different species and strongly influence both natural and managed ecosystems (Inderjit et al., 2011).
A major route of allelochemical release into the environment is exudation from the roots of living plants into their immediate surroundings, i.e., the soil rhizosphere (Bais et al., 2006; Belz, 2007). Grasses in particular have the ability to interfere with and to outcompete neighboring plants via the biosynthesis of phytotoxic root exudates. Cyclic hydroxamic acids (e.g., benzoxazinoids or benzoxazinones), with their main representatives DIBOA [4-dihydroxy-2H-1,4-benzoxazin-3(4H)-one] and its methoxylated analog DIMBOA [2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one], were among the first active allelochemicals identified and are present in root exudates of several major cereal crops (Belz and Hurle, 2005; Macías et al., 2006; Belz, 2007). In nature, these substances typically undergo rapid degradation via the intermediates benzoxazolin-2(3H)-one (BOA) and 6-methoxybenzoxazolin-2(3H)-one (MBOA). The resulting benzoxazolinone metabolites are much more stable, and in particular, the aminophenoxazinone compounds 2-amino-3H-phenoxazin-3-one (APO) and 2-amino-7-methoxy-3H-phenoxazin-3-one (AMPO) are highly potent phytotoxins (Macías et al., 2006, 2009). The precursors DIBOA and DIMBOA as well as their degradation products APO and AMPO have been detected in target plants (Macías et al., 2014). As they exert physiological effects on a variety of plant species, it has been surmised that these allelochemicals act on highly conserved central cellular pathways or structures (Duke, 2007; Dayan et al., 2009). In vivo bioassays confirmed growth inhibitory activity for hydroxamic-acid-derived allelochemicals, such as APO and AMPO, that is comparable to that of commercial herbicides (Macías et al., 2006). However, the modes of action and molecular targets affected by this type of allelopathic interaction between plants have remained mostly unresolved (Duke, 2010).
Histone acetylation is a highly conserved molecular process, which like other posttranslational histone modifications, defines chromatin condensation and, thus, DNA accessibility (Ha et al., 2011). As a key component of chromatin-mediated control of gene expression, histone acetylation typically marks actively transcribed genes (Shahbazian and Grunstein, 2007; Wang et al., 2009). Two antagonistically acting enzyme families, histone acetyltransferases and histone deacetylases (HDACs; in plants also called HDAs), establish and remove histone acetylation, respectively. Histone acetylation patterns in somatic cells are dynamically balanced by histone acetyltransferase and HDAC activities in response to intrinsic or environmental signals. Moreover, as chromatin-associated marks, they potentially play a role in the maintenance or reestablishment of chromatin composition after mitosis and/or meiosis (Feinberg, 2008; Pray, 2008; Blevins et al., 2014). HDACs, which belong to a class of enzymes that is evolutionarily conserved from prokaryotes to eukaryotes (Gregoretti et al., 2004; Menegola et al., 2006), are important for both animal and plant physiology and development (Menegola et al., 2006; Hollender and Liu, 2008; Kucharski et al., 2008). In plants, histone acetylation/deacetylation plays a role in many regulatory processes, such as rapid responses to internal or external signals (Benhamed et al., 2006; Hollender and Liu, 2008) or the switch from vegetative to reproductive growth (He et al., 2003). Accordingly, suppression of Arabidopsis thaliana HDAC genes such as HD1 (HDA19) or HDA6, the latter being important for large-scale silencing of ribosomal gene sets, leads to a wide range of developmental abnormalities (Tian and Chen, 2001; Probst et al., 2004; Tian et al., 2005; Earley et al., 2006).
Here, we report that the physiological effects of naturally occurring hydroxamic acid derivatives during allelopathy result from chromatin modifications that are mediated by direct HDAC inhibition. We not only provide biochemical evidence for such activity, but also demonstrate that these root-derived allelochemicals affect functional histone acetylation and gene expression on a genome-wide scale. Our findings lead to a mechanistically informed model for the molecular mode of action of allelopathic aminophenoxazinones in the target plant and provide insights into the potent allelopathic defense and competition strategy of hydroxamic-acid-producing plants.
RESULTS
Docking Simulation of APO and AMPO to HDACs
The allelochemicals DIBOA and DIMBOA from root exudates are cyclic hydroxamic acids. Although different in their molecular geometry, they contain the same functional group as the noncyclic hydroxamic acids trichostatin-A (TSA) and suberoylanilide hydroxamic acid (SAHA), two established inhibitors of mammalian HDACs (Supplemental Figure 1). DIBOA and DIMBOA typically undergo rapid degradation in natural environments. The resulting product APO is a highly potent phytotoxin, which is also the case, although to a lesser extent, for AMPO (Macías et al., 2006, 2009). The allelochemicals APO and AMPO as well as their precursors have been detected in plants in proximity to DI(M)BOA donor plants (Macías et al., 2014).
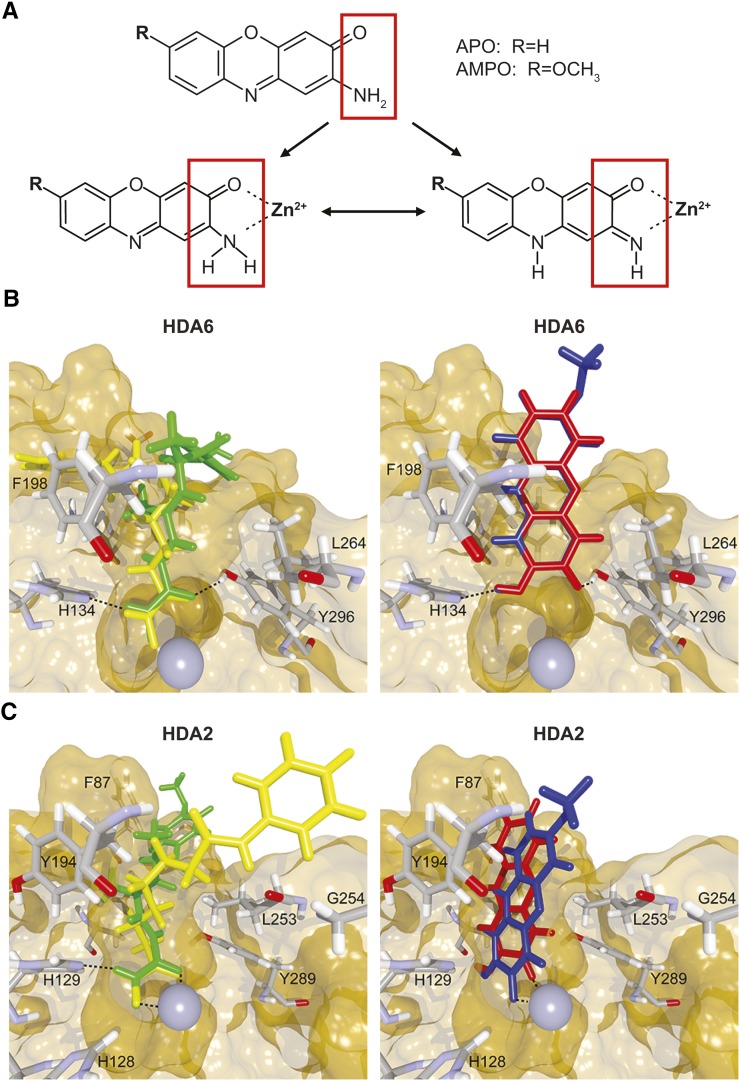
Even though APO and AMPO have different molecular geometry than their respective precursors (Figure 1A; Supplemental Figure 1), we speculated that they might have activities similar to those of noncyclic hydroxamic acids. We first assessed in silico the binding potential of hydroxamic acids to different HDAC proteins, based on the high sequence conservation of HDACs across organisms (Supplemental Figure 2). In comparative analyses of docking into the crystal structures of human class I and class II HDACs and of bacterial histone deacetylase-like protein (HDLP) (Finnin et al., 1999; Somoza et al., 2004; Bottomley et al., 2008), APO and AMPO behaved similarly to TSA and SAHA (Supplemental Figure 3). In the absence of available crystal structures for plant HDACs, we performed docking analyses for these enzymes using homology models of Arabidopsis HDA6 (class I) and HDA2 (class III) (Supplemental Figure 4). Despite differences between TSA/SAHA and APO/AMPO in forming bonds to the zinc ion of HDA6 and HDA2, and in some hydrogen bond formations with HDA2 (Figures 1B and 1C), APO and AMPO were predicted to fit into the binding pocket for both homology models of Arabidopsis HDAs with reasonable affinities (between −14.9 and −16.3 score units in all cases). Altogether, the in silico data suggest, with some caveats, potential binding of APO and AMPO to HDACs in a manner similar to interactions between known HDAC inhibitors and their targets.
Figure 1.
Binding of APO and AMPO to Arabidopsis HDACs.
(A) Chemical structure of the cyclic-hydroxamic-acid-derived allelochemicals APO and AMPO. Both substances have the capacity to coordinate a zinc ion in a bidentate fashion.
(B) and (C) Simulated docking of TSA (green), SAHA (yellow), APO (red), and AMPO (blue) to the binding pocket of Arabidopsis HDA6 (B) and HDA2 (C) (surface representation), with crystal structure reference coordinates of TSA (atom colors). Residues with predicted interactions with the ligands are shown in stick representation; the zinc ion of the HDAC is shown as a gray sphere. Figures were rendered with BALLView and POVRay (v3.6).
Inhibition of HDACs by APO and AMPO
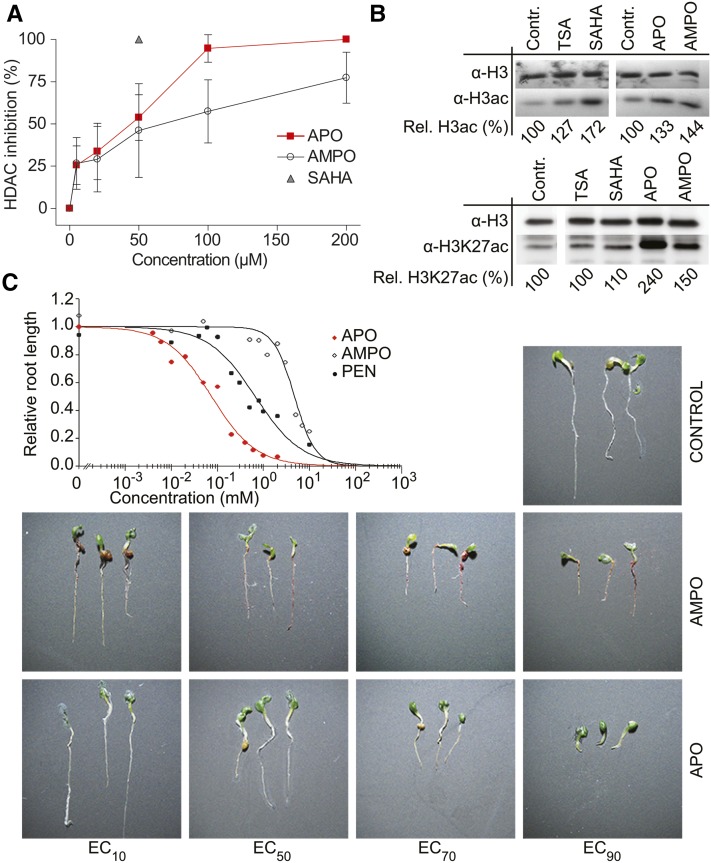
Given the discrepancies and limitations in docking analyses using homology models, we tested directly whether APO and AMPO could inhibit HDACs. To this end, we determined HDAC activity in nuclear extracts of Arabidopsis seedlings, before and after application of APO and AMPO. Both substances showed a distinct dose-dependent HDAC inhibitory effect (Figure 2A). APO was the more potent HDAC inhibitor, and at a concentration of 50 µM reduced Arabidopsis HDAC activity by more than half. When tested in the same concentration range as APO and AMPO, the root-exuded precursor substances DIBOA and DIMBOA and the intermediates BOA and MBOA did not inhibit plant HDACs (Supplemental Figure 5). These results were in line with a previous report that the half-maximal effective concentration (EC50) of BOA in a root growth inhibition assay was 540 µM (Baerson et al., 2005), i.e., a concentration 10 times higher than the effective APO and AMPO concentrations for HDAC inhibition. This suggests that the root-secreted precursor substances need to be metabolized or chemically converted to become potent HDAC inhibitors. In accordance with the high conservation of HDACs across eukaryotes (Supplemental Figure 1), both APO and AMPO dose-dependently reduced the activity of human class I, II, and IV HDACs (Supplemental Figures 6 and 7), all of which have a zinc-dependent catalytic domain, similar to Arabidopsis HDA6 and HDA2 (Schäfer and Jung, 2005; Marks and Xu, 2009).
Figure 2.
HDAC and Growth Inhibition by Allelochemicals.
(A) HDAC inhibition by APO and AMPO in Arabidopsis nuclear extracts. SAHA (50 µM) was used as reference inhibitor. Data points represent mean values ± sd of three independent experiments performed in duplicate.
(B) Immunoblot showing acetylation of histone H3 (α-H3ac) and of lysine 27 acetylation of histone H3 (α-H3K27ac). Seedlings were treated for 24 h with EC50 concentrations of APO, AMPO, SAHA, or TSA 5 d after germination; upper row shows equal protein loading (histone protein H3) on a parallel blot. Relative amounts of acetylated histone were estimated by densitometric analysis; values are relative to the respective untreated control.
(C) Concentration-response assays on root growth in seedlings. The commercially available herbicide PEN was used as a positive control. Root length was measured 6 d after sowing (≥0.1 mm minimum root length), and concentration-response curves were calculated using a logistic regression model. The quality of curve fitting was verified by F test for lack-of-fit based on ANOVAs (α = 0.05). Right panel: visualization of the concentration-dependent growth inhibitory effect of APO and AMPO on seedlings 6 d after sowing.
To assess whether APO and AMPO affected HDAC activity in vivo, we incubated seedlings with EC50 concentrations of either substance and determined global acetylation of histone H3 (H3ac) in chromatin extracts. Levels of hyperacetylated H3 increased within 24 h of treatment with APO, AMPO, TSA, and SAHA, while only APO and AMPO treatments resulted in an increase of H3 lysine 27 acetylation levels (Figure 2B), indicating that these substances have a substantial and specific effect on histone acetylation in vivo.
Similar Phytotoxicity of HDAC Inhibitors and Conventional Herbicides
To investigate the physiological effects of aminophenoxazinones APO and AMPO in vivo, we performed concentration-response assays with seedlings. Both allelochemicals effectively reduced the growth of Arabidopsis and lettuce (Lactuca sativa) seedlings (Figure 2C; Supplemental Figure 8). The EC50 concentrations of APO for Arabidopsis growth inhibition were in a similar range as for the in vitro HDAC inhibition assays (Figure 2A; Supplemental Table 1). Pendimethalin (PEN), a commercially available preemergent herbicide inhibiting cell division, showed effects that were intermediate between those of APO and AMPO in Arabidopsis (Figure 2C; Supplemental Table 1), although it outperformed both allelochemicals in lettuce (Supplemental Figure 8 and Supplemental Table 1). When germinated on APO- or AMPO-supplemented media, 6-d-old Arabidopsis seedlings showed severe root growth inhibition, whereas shoot development was little affected (Figure 2C). In conclusion, APO and AMPO can inhibit different aspects of plant growth at concentrations comparable to the effective concentration of commercial herbicides. The root growth defects were comparable to those seen in plants lacking function of HDA6 and HDA19 (RPD3/HDA1 superfamily; Supplemental Figure 9) and of HDT1 and HDT3 (plant-specific HD2 family) (Colville et al., 2011), supporting the hypothesis that this class of allelochemicals affects the germination and development of surrounding competitors by inhibiting one or several HDACs.
Effects of APO on H3 Acetylation and Gene Expression
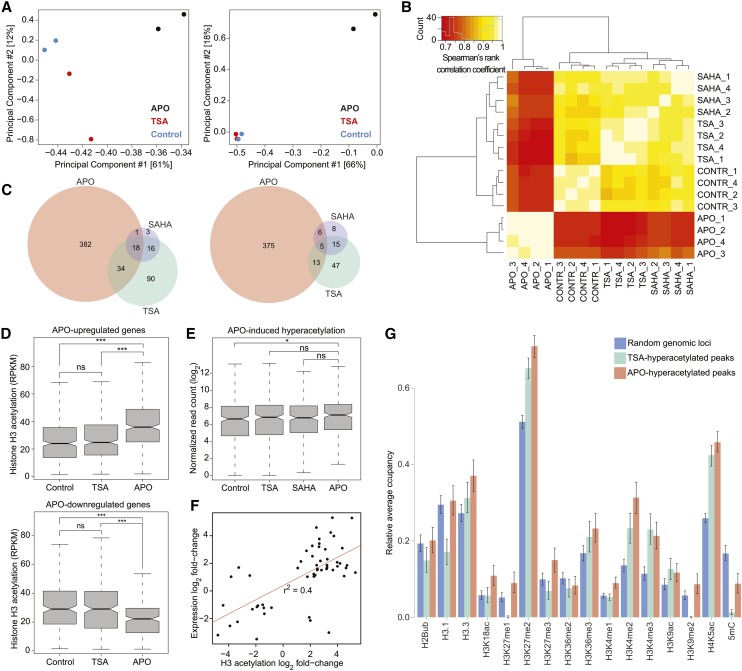
To determine the effects of APO on H3ac at specific loci, we performed chromatin immunoprecipitation of histone H3ac, followed by next-generation sequencing (ChIP-seq). We focused our analysis on APO, due to its superior allelopathic activity, its higher solubility in aqueous medium, and its lower lipophilicity compared with AMPO (Macías et al., 2006); mock- and TSA-treated seedlings served as controls. We validated successful enrichment for H3ac by quantitative real-time PCR (Supplemental Figure 10 and Supplemental Table 2). Using input, non-antibody-treated DNA to estimate background signal, we called peaks for H3ac enrichment (Supplemental Figure 11 and Supplemental Data Set 1). Principal component analysis showed that samples treated with APO for 24 h were the most divergent compared with control and TSA-treated samples, with replicates clustering together (Figure 3A; Supplemental Figure 12A). We identified 406 hyper- and 362 hypoacetylated peaks after APO treatment and 304 hyper- and 117 hypoacetylated peaks after TSA treatment; peak coordinates and proximal annotated genes are listed in Supplemental Data Set 2. Only 34 hyper- and 18 hypoacetylated peaks overlapped between the two treatments. Thus, although both substances inhibited HDACs in vitro and induced increased total H3ac (Figure 2B), their effects on histone acetylation at individual loci after 24 h varied, suggesting partial specificity of the substances and both direct and indirect effects on H3ac.
Figure 3.
Locus-Specific Effect of APO on Histone Acetylation and Gene Expression.
(A) Principal component analyses of histone acetylation on all identified acetylated genomic regions (left panel) and on peaks identified as differentially acetylated between APO- and non-APO-treated samples (right panel) using the DESeq2 implementation of the DiffBind package. Variation explained by the respective principal component is given in brackets.
(B) Bihierarchical clustering of normalized read counts at differentially expressed genes. Numbers 1 to 4 indicate biological replicates.
(C) Venn diagrams showing the overlap of upregulated (left) and downregulated (right) genes after 24-h treatment with EC50 concentrations of APO, TSA, and SAHA.
(D) H3 acetylation in control-, TSA-, and APO-treated samples at peaks overlapping with APO-upregulated (upper panel) and APO-downregulated genes (lower panel). RPKM, reads per kilobase per million; ns, not significant; asterisks, significant difference in acetylation compared with control treatment: *P < 0.05 and ***P < 0.001, unpaired two-tailed Student’s t test.
(E) Expression in control-, TSA-, and APO-treated samples of genes with hyperacetylated H3 levels in response to APO treatment (*P < 0.05, unpaired two-tailed Student’s t test; ns, not significant).
(F) Correlation between H3ac level changes and gene expression changes at loci that had both differential histone acetylation levels and were differentially expressed in a comparison of control and APO-treated samples. Red line represents linear regression.
(G) Chromatin landscape in Arabidopsis Col-0 according to Wang et al. (2015), at randomly chosen genomic loci and at loci hyperacetylated by APO or TSA. Relative occupancy was calculated as the fraction of hyperacetylated regions covered by the respective mark. Bars represent the mean; errors indicate 95% confidence intervals.
To understand how APO-induced changes in histone acetylation affected gene expression, we performed RNA-seq on the same material that we had used for ChIP-seq, including samples treated with SAHA. Of the three treatments, APO had the strongest effect on gene expression and SAHA the weakest (Figures 3B and 3C). We identified 435 up- and 399 downregulated genes (common variance >2-fold, per gene variance false discovery rate < 0.05) after 24 h exposure to EC50 concentrations of APO (Supplemental Data Set 3). One-third of the genes that were differentially expressed in TSA-treated samples were also affected by APO, which is a much greater overlap than expected by chance (P < 0.001, hypergeometric test). Almost all SAHA-upregulated genes were also induced by either APO or TSA, and, notably, there were no genes that were upregulated by one HDAC inhibitor, but downregulated by one of the other inhibitors. Altogether, our data indicate that APO partially acts on the same gene networks as known HDAC inhibitors, while also causing specific responses at the transcriptional level.
The chromatin environment of APO-upregulated genes included significantly increased H3ac levels (P < 0.001, unpaired two-tailed Student’s t test), while conversely APO-downregulated genes had lower H3ac levels compared with the control (P < 0.001) (Figure 3D). Accordingly, in APO-treated seedlings genes with hyperacetylated H3 were on average more highly expressed than in untreated seedlings (Figure 3E; P < 0.05). Of 288 genes overlapping with APO-hyperacetylated regions in APO-treated seedlings, 37 (13%) corresponded to APO-upregulated genes (Supplemental Data Set 4), with only 3 out of 346 hypoacetylated regions overlapping with APO-upregulated genes. At the 54 loci that showed significant APO effects on both acetylation and expression, changes in histone acetylation levels positively correlated with changes in expression levels (P < 0.001, Pearson’s correlation coefficient = 0.65) (Figure 3F). We conclude from these data that a substantial fraction of genes differentially expressed upon APO treatment respond to direct effects of APO on local H3ac.
In contrast to APO, TSA treatment did not lead to a significant increase in H3ac levels at TSA-upregulated genes (Supplemental Figure 13). The overall genetic context of APO-hyperacetylated regions was different from that of TSA-hyperacetylated regions (P < 0.01, χ2 test). TSA preferentially led to hyperacetylation of intergenic sequences, while transcribed regions of protein-coding genes were less affected than in APO-treated seedlings (Supplemental Figure 14). We did not observe substantial differences in the annotation of hypoacetylated regions from APO- and TSA-treated seedlings (P = 0.151, χ2 test; Supplemental Figure 14).
We compared APO- and TSA-responsive regions of the genome to the global chromatin landscape reported for untreated Arabidopsis plants (Wang et al., 2015). Although the overall chromatin configuration was similar between regions that became hyperacetylated in APO- and TSA-treated plants, those targeted by APO were markedly enriched in the transcriptionally relevant modifications H3K27, H3K9, and DNA methylation, indicating different epigenetic states between genomic loci affected by these two compounds (Figure 3G). Together, these results suggest that TSA and APO, although they both inhibit HDACs and lead to alterations in histone acetylation levels, act in different (de)acetylation pathways, maybe by binding to different plant HDACs.
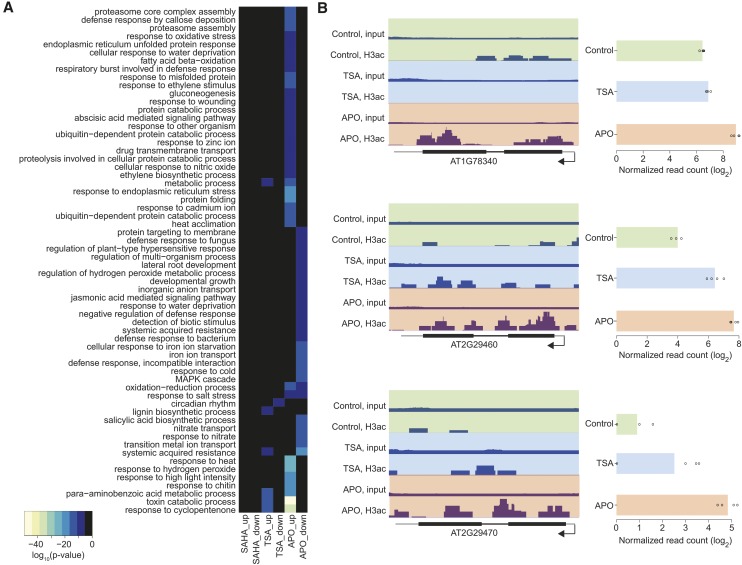
Gene Ontology (GO) analysis revealed that genes involved in detoxification and in response to abiotic and biotic stresses were overrepresented among APO- and TSA-upregulated genes (Figure 4A; Supplemental Figure 15 and Supplemental Data Set 3). This was in accordance with a previous microarray analysis of Arabidopsis seedlings treated with the APO-precursor BOA (Baerson et al., 2005). Almost half of the genes reported to be upregulated by high concentrations of BOA (540 µM) coincided with genes upregulated after APO treatment in our analysis. APO-downregulated genes were overrepresented, among other categories, for developmental growth and lateral root development (Figure 4A). A few gene families, e.g., for glutathione S-transferases (Figure 4B), were overrepresented among the genes that displayed both increased mRNA and increased H3ac levels after APO treatment.
Figure 4.
APO Affects Genes Related to Detoxification and Stress Response.
(A) Overrepresented biological functions of genes induced or repressed by HDAC inhibitor treatment. Heat map shows P values of GO terms that were significantly overrepresented (P < 0.05) in at least one treatment. Black color indicates that the respective GO term was not significantly overrepresented after the corresponding treatment.
(B) Left panel: H3 acetylation, represented by dark-blue stacks, at genomic loci corresponding to three differentially expressed glutathione S-transferase genes. Thick and thin black horizontal bars represent exons and introns, respectively, gray horizontal bars represent UTRs; arrows indicate direction of transcription. Track height was adjusted to the maximum value for each locus. Right panel shows the expression of the same genes. Bars represent the mean across four replicates; points indicate individual values.
DISCUSSION
The release of allelochemicals to influence the germination and growth of neighboring plants is an important component of plant defense and survival strategies (Belz, 2007; Macías et al., 2007). Understanding the molecular mechanisms of allelopathy is therefore of general interest, from ecology to breeding and farming. We used a combination of in silico simulations, biochemical in vitro assays, in vivo experiments, and whole-genome expression and chromatin profiling to assess the action of allelochemicals as potential inhibitors of HDACs, a class of evolutionarily highly conserved enzymes (Gregoretti et al., 2004; Menegola et al., 2006). Our analyses support the conclusion that derivatives of plant hydroxamic acid compounds can exhibit allelochemical effects by directly inhibiting the activity of target plant HDACs. They thereby affect chromatin-based gene regulation and ultimately decelerate or prevent germination and growth of competitors.
The allelochemicals APO and AMPO broadly inhibit HDAC activity in vitro, with similar effects on plant and human enzymes. Although we cannot exclude that inhibition of HDAC activity in Arabidopsis nuclear extracts (Figure 2A) is an indirect consequence of treatment with allelochemicals, the effect on pure recombinant human HDACs (Supplemental Figures 6 and 7) suggests a direct inhibition of this enzyme class by APO and AMPO, which is in line with our in silico docking results. Although structurally similar, APO is a stronger HDAC and growth inhibitor than AMPO. Published studies on phytotoxicity and effects on microbes corroborate differences between the physiological activity of APO and AMPO (Anzai et al., 1960), which can be explained through differences in lipophilicity (Macías et al., 2006). Our results suggest that part of the effect could also be explained by different efficacies in terms of solubility and HDAC inhibition, the latter notion being supported by the stronger increase of H3K27 acetylation levels after APO treatment compared with AMPO treatment (Figure 2B).
The formation of APO from root-exuded DIBOA as well as the absorption of APO by exposed plants has been confirmed (Krogh et al., 2006; Rice et al., 2012), but little is known about the rate of synthesis and the availability of APO and AMPO in the soil under natural conditions. The presence of these allelochemicals in the rhizosphere depends on factors such as species, cultivar, or plant density (Huang et al., 2003; Belz and Hurle, 2005; Macías et al., 2014). Maybe the best estimate for naturally occurring APO concentrations can be gained from studies of exudation of the APO precursor DIBOA from roots: In hydroponic cultures, DIBOA released from rye (Secale cereale) and durum (Triticum durum) roots accumulated to concentrations of 300 and 30 µM, respectively (Belz and Hurle, 2005; Macías et al., 2014). Given the equimolar conversion of DIBOA to BOA, and a conversion rate of 10:1 of BOA into APO (Gents et al., 2005; Understrup et al., 2005), this would result in APO concentrations of 30 and 3 µM, respectively. These concentrations are in the range determined to be effective in our assays, which supports HDAC inhibition by APO as a realistic model for allelopathy.
Chromatin modification provides a means for rapid cellular responses to changing conditions; by modulating chromatin composition and accessibility, histone acetylation and deacetylation constitute both on-off switches and rheostats for gene expression (Pray, 2008). Treatment of plants with APO for 24 h resulted in elevated histone H3 acetylation, which was frequently associated with transcriptional upregulation (Figure 3). This is consistent with APO (and potentially the closely related AMPO) entering plant cells, where it inhibits HDACs. That APO treatment also resulted in the reduced expression of several hundred genes and that this repression correlated with a reduction of H3 acetylation could point to secondary effects that resulted from developmental or physiological defects. Further studies will be necessary to resolve early and late target genes of this type of allelopathic inhibition.
TSA and SAHA, which are known as potent HDAC inhibitors from other systems, had a weaker effect than APO on plant gene expression. In contrast to APO, we did not observe a correlation between TSA-induced changes in gene expression and H3 acetylation, even though all tested HDAC inhibitors led to a global increase in H3ac and to growth inhibition. This discrepancy suggests that TSA and APO act on different HDACs in the plant, similar to clinically applied HDAC inhibitors, which display characteristic target profiles in humans (Bantscheff et al., 2011; West and Johnstone, 2014). Specificity for the target HDAC might also result in the deacetylation of different histones and/or histone residues, a notion supported by the observation that TSA and APO affected acetylation patterns in different genomic contexts and in different epigenetic states (Figure 3G; Supplemental Figure 14). Moreover, the increase in H3 lysine 27 acetylation was particularly pronounced in seedlings treated with APO compared with the other compounds (Figure 2B). Of note, the plant-derived aminophenoxazinone APO showed the clearest correlation between effects on H3ac and gene expression in Arabidopsis, in agreement with APO effects being physiologically most relevant. Characterized mutants in single HDAC genes show less severe slowdown of root growth than plants treated with high concentrations of APO (Supplemental Figure 9; Colville et al., 2011), which suggests that the allelochemical either acts on other HDACs or that it inhibits more than one HDAC. Elucidating the specificity of the allelochemicals APO and AMPO in inhibiting different plant HDACs, and the consequences for different histones and for modifications at different histone residues, should therefore allow a better understanding of their direct chromatin targets.
Highly conserved molecular structures have emerged as prime targets in plant-pathogen interactions (Bent and Mackey, 2007). One example is provided by fungal plant pathogens that produce peptidic HDAC inhibitors (Brosch et al., 1995). Here, we have shown that this strategy is not only used for interactions between different kingdoms of life, but also for plant-plant allelopathic interactions. The conservation of the target raises the question how the donor can avoid HDAC inhibition by its own allelochemicals. APO and AMPO are the main stable intermediates that usually develop after precursor substances have been released by plant roots into the rhizosphere; thus, these compounds are not directly produced within the donor plant itself (Belz and Hurle, 2005; Macías et al., 2006, 2009; Belz, 2007). Accordingly, neither the APO and AMPO precursors DIBOA and DIMBOA nor the BOA and MBOA intermediates had strong effects on HDAC activity (Supplemental Figure 5).
The allelochemical producers likely protect themselves either by preventing APO and AMPO uptake or by having means of sequestering or degrading them. As HDACs are highly conserved, it is improbable that resistance is established at the level of these target enzymes. Our observation that a considerable fraction of genes upregulated after exposure to APO were related to transport and chemical modification may be consistent with degradation or sequestration of these toxic compounds to the vacuole, with donors of allelochemicals having pathways that are simply more efficient than those in other plants. Alternatively, the root microbiome of the donor plant might contribute to the degradation of phytotoxins and thus prevent the emitter from damage by its own exudate. Allelochemicals might further act preferentially during particular developmental stages. In this scenario, only already established plants release the compound to prevent germination and early growth of nearby competitors, themselves remaining unaffected. The elucidation of the molecular details of emission and resistance needs further experimental analysis.
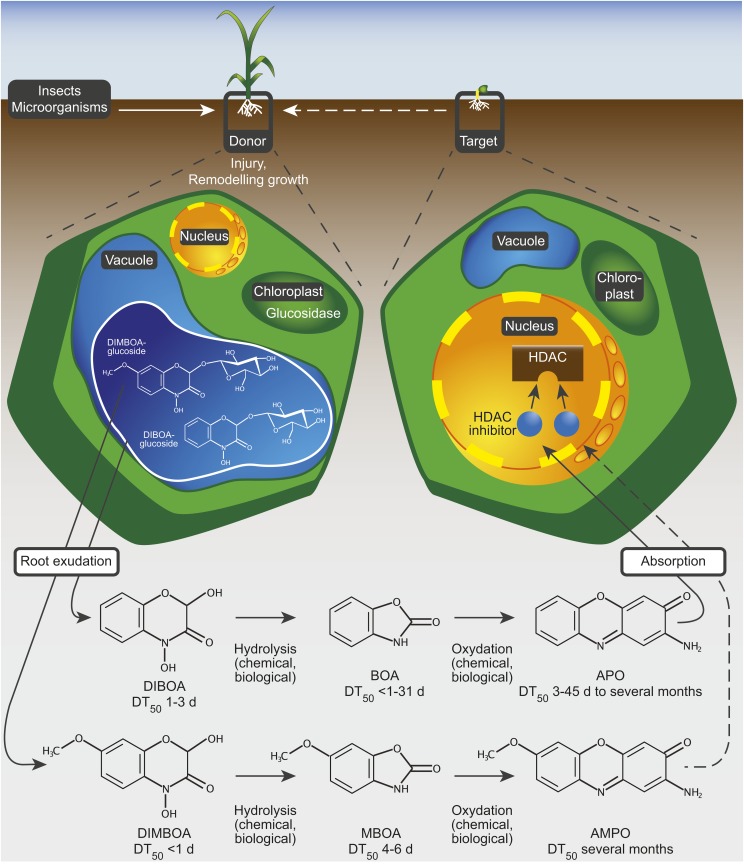
In summary, this study reports a molecular mechanism by which plants interfere with the growth or development of their competitors via a plant-derived soil-borne allelochemical. We propose a model whereby a donor plant exudes benzoxazinone compounds into the rhizosphere, where they are converted into more stable and biologically highly active products such as aminophenoxazinones (Figure 5). Subsequently, these enter the cells of target plants where they inhibit HDAC enzymes and affect the regulation of gene expression, ultimately causing a slowdown of growth and development and providing the donor plant with a competitive advantage.
Figure 5.
Model of a Chromatin-Based Mode of Action of the Allelochemicals APO and AMPO.
A donor plant exudes DIBOA and/or DIMBOA into the rhizosphere as a consequence of injury, plant age, or target plant interaction. These two unstable cyclic hydroxamic acids diffuse as parent compounds from the roots of the donor plant into the surrounding rhizosphere and are rapidly converted into the intermediates BOA and MBOA. Both intermediates are further converted into the more stable compounds APO and AMPO, which are absorbed by the target plant. APO and AMPO broadly inhibit HDAC enzymes and thereby modify the chromatin pattern of target cells, which profoundly affects plants in an early developmental stage. DT50, dissipation time 50%, according to Macías et al. (2004, 2005) and Understrup et al. (2005).
METHODS
Reagents
DIBOA, DIMBOA, APO, and AMPO were chemically synthesized by EMC (EMC Microcollections); BOA and MBOA were purchased from Sigma-Aldrich. For internal controls, APO and AMPO were generated according to the biological degradation path as follows: BOA and MBOA were dissolved separately in water up to saturation and kept at room temperature in darkness under aerated conditions for ∼14 d. After filtration, the solution was partitioned two times against 3-fold volumes of ethyl acetate. The ethyl acetate extracts were pooled, filtered over anhydrous sodium sulfate, and evaporated to complete dryness under vacuum. The residue was redissolved in a one-to-one mixture (v/v) of acetonitrile (ACN) and water and fractionated by preparative HPLC. A Phenomenex Synergi 10 Hydro-RP 80 column (250 mm by 15 mm [5 mm]) was used and eluted with a gradient of 20% ACN and 80% Na2HPO4 buffer for 0 to 16 min, 95% ACN and 5% buffer for 17 to 18 min, then reequilibrated to starting conditions (6 mL/min flow rate). Injection volume was 100 µL. APO was identified in the fraction ranging from 15.3 to 16.1 min, and AMPO was detected at 15.4 to 16.3 min. APO and AMPO were dissolved in DMSO for HDAC inhibitor screening and HDAC profiling assays, and in ethanol for the in vivo concentration-response assays. The HDAC inhibitors SAHA (IBL) and TSA (Sigma-Aldrich) were dissolved in DMSO for HDAC inhibitor screening and HDAC profiling assays, and in ethanol for in vivo concentration-response assays. The commercial herbicide Stomp SC (400 g/L PEN; BASF) served as a reference; the compound was used as formulated product and mixed in demineralized water to give various test solutions.
Plant Material
HDAC loss-of-function mutants were ordered from the European Arabidopsis Stock Centre (www.arabidopsis.info) under the following reference numbers: N66153 (hda6-6; also known as axe1-5) (Murfett et al., 2001), N66154 (hda6-7; also known as rts1-1) (Aufsatz et al., 2002), and N31355 (hda19-1; also known as athd1-t1) (Tian et al., 2003).
Sequence Alignment
Multiple sequence alignment of human HDAC8 (class I) and HDAC4 (class II), bacterial HDLP (Aquifex aeolicus), and Arabidopsis thaliana HDA1/HDA19, HDA2, and HDA6 was performed using MAFFT version 6 (Katoh et al., 2002; Katoh and Toh, 2008) and visualized with ESPript (Gouet et al., 1999). Sequences of human HDACs were obtained from PDB IDs 1T64 (Somoza et al., 2004) and 2VQM (Bottomley et al., 2008); the HDLP sequence of A. aeolicus was obtained from PDB ID 1C3R (Finnin et al., 1999); Arabidopsis sequences were retrieved from The Arabidopsis Information Resource (Rhee et al., 2003) using gene models AT4G38130.1 (HDA1/19, class I), AT5G63110.1 (HDA6, class I), and AT5G26040.2 (HDA2, class III).
Docking Simulation
Homology models for Arabidopsis HDAs were created using MODELER 9v8 (Sali and Blundell, 1993). We built models of HDA6 (class I) and HDA2 (class III). In both cases, the crystal structure of the catalytic core of HDLP (PDB ID 1C3R; Finnin et al., 1999) was used as template. The template has a sequence identity of 35% to HDA6 and 27% to HDA2, respectively. The target-template alignment was calculated using the SALIGN routine of MODELER. For each alignment, 50 models were calculated (standard automodel class), and for each model, an automated loop refinement was performed (loop model class, refinement mode slow). The zinc ions in the templates were copied to the target models and treated as rigid during both initial model building and loop refinement, using the BLK functionality of MODELER. The resulting homology models were evaluated with ProSA-web (Wiederstein and Sippl, 2007).
Predictive docking of APO and AMPO into the homology models of Arabidopsis HDA6 and HDA2, into human HDAC8 and HDAC4, and into A. aeolicus HDLP was performed using FlexX version 2.2.1 (Rarey et al., 1996). For HDAC8, the binding site of PDB ID 1T64 (Somoza et al., 2004), a human HDAC8 structure with bound inhibitor TSA, was used and preprocessed (side chain orientation, protonation state) for docking with Reduce version 3.10 (Word et al., 1999). For human HDAC4, PDB-ID 2VQM (Bottomley et al., 2008) was used, a structure with a bound hydroxamate inhibitor. For the A. aeolicus HDLP, PDB ID 1C3R (Finnin et al., 1999) with bound TSA was used. Residues within 8 Å radius around the crystal structure reference coordinates of the inhibitors were used as putative binding site. Binding sites of the homology models of HDA6 and HDA2 were selected similar to the binding sites of the crystal structures. In all cases the internal cavity (Wang et al., 2004) was not considered for binding, as it is unlikely that APO and AMPO can enter this cavity due to their rigidity. The receptor description files for FlexX were generated using RDFWizard version 1.3 (Raub et al., 2008). The torsion angle for Tyr-306 in HDAC8 was manually adjusted such that the hydroxyl group faces toward the nearest hydrogen bond acceptor of TSA. Tautomeric states of APO and AMPO were chosen that are capable of a bidentate binding to the zinc ion of the HDACs. Conformations of the ligands suitable for docking were obtained by energy minimization using the MMFF94 force field (Halgren, 1996) as implemented in BALLView (Moll et al., 2006). In the case of HDAC4, two water molecules have been reported to mediate the binding of a hydroxamic acid inhibitor (Bottomley et al., 2008). Both water molecules (A 2296 and A 2375 in PDB ID 2VQM) were retained in the binding site, and their respective hydrogen positions were optimized with the MMFF94 force field in context of the binding site with the bound hydroxamic acid inhibitor. For docking, the ring system of APO/AMPO was held rigid. FlexX was used with standard scoring function and standard parameters. Only the RMS value for triangle clustering was changed to 0.8 Å to obtain a larger number of placements of the rigid fragment of APO/AMPO.
Plant HDAC Inhibitor Assay
Extract of functional nuclear proteins as source for plant HDAC enzymes was produced using the Plant Nuclei Isolation/Extraction Kit (CelLytic PN; Sigma-Aldrich) according to the manufacturer’s protocol. As starting material, young leaves (40 g) of Arabidopsis Col-0 were collected without leaf stalks and freshly ground in liquid nitrogen to a fine powder. The extract was frozen at −80°C until further use. Determination of HDAC inhibitory activity of APO and AMPO was performed with the HDAC assay kit (Active Motif) based on fluorescence detection (excitation wavelength 340 nm and emission wavelength 450 nm). This assay enables the screening of potential inhibitor compounds in nuclear extracts of different origin. The HDAC assay was performed as described in the manual, with the following changes, introduced after consultation with the manufacturer. Per reaction, we used 30 μL of undiluted Arabidopsis nuclear extract. To increase the signal strength, the incubation time was extended to 2 h at 37°C. The treatment with the commercial HDAC inhibitor SAHA (50 µM) served as positive control for complete inhibition of all HDAC enzymes in the assay and was set accordingly to 0% HDAC activity; this value was used as reference for the calculation of the specific inhibition values of APO or AMPO.
Human HDAC Profiling Assay
The human HDAC profiling assay was based on fluorometric measurement and performed by Scottish Biomedical. Each purified human HDAC enzyme was incubated separately with a fluorogenic substrate, which comprises an acetylated lysine side chain and a 7-amino-4-methylcoumarin group, together with the substance to be analyzed (APO or AMPO). Cleavage of the substrate by the specific HDAC enzyme leads to release of a fluorescent compound, the signal of which is detected. Activation or inhibition of the HDAC enzyme is estimated from the strength of the fluorescent signal. We determined inhibition of human HDACs 1 to 11 individually by both APO and AMPO tested at increasing concentrations (5, 20, 100, and 200 μM).
Bioassays with Arabidopsis
Growth inhibition assays were conducted as concentration-response experiments in 24-well plates (Nunc) with Arabidopsis Col-0. Ten seeds were placed on one layer of filter paper (Filter Discs grade 289; Sartorius) per well and exposed to 8 to 12 concentrations of the different compounds. PEN, SAHA, TSA, and APO were tested with six replications at a concentration range of 0 to 10 mM for PEN, 0 to 0.5 mM for SAHA, 0 to 0.2 mM for TSA, and 0 to 2 mM for APO. AMPO was tested with three replications at a concentration range of 0 to 10 mM. Test concentrations were prepared from a stock solution in ethanol. Test concentrations of PEN were obtained by a dilution series from the formulated product. Plates were kept at 4°C for 3 d and subsequently cultivated in a growth chamber (24/18°C, 50/0 µE/m2s photosynthetic active radiation [12/12 h]). Root length was measured after 6 d.
Bioassays with Lettuce
Growth inhibition assays were conducted as concentration-response experiments in Petri dishes (4.5-cm diameter) with lettuce (Lactuca sativa var capitata cv Maikönig). Fifteen seeds were placed on one layer of filter paper (Rundfilter 595; Schleicher and Schuell) per Petri dish and exposed to 8 to 10 concentrations of the different compounds. PEN, SAHA, and TSA were tested with six replications at a concentration range of 0 to 0.5 mM. The aminophenoxazinones were tested with six control replicates and with three replications at a concentration range of 0.1 to 10 mM for APO and with two replications at 1 to 20 mM for AMPO. Test concentrations were prepared from a stock solution in ethanol. Test concentrations of PEN were obtained by a dilution series from the formulated product. Petri dishes were cultivated in a completely randomized design in a growth chamber (24/18°C, 50/0 µE/m2s photosynthetic active radiation [12/12 h]). Root length was measured after 5 d.
Statistical Analysis: EC50 Estimation
Effective concentrations causing a 50% inhibition in root length (y) of the test plants were calculated using the following logistic concentration-response model (Finney, 1978; Streibig, 1988): y = D/(1+e(b*ln(x/EC50)), where D denotes the expected response of the untreated control, EC50 denotes the concentration (x) causing 50% inhibition, and b denotes the rate of change around EC50. The model was fitted to the data by nonlinear regression analysis using IBM SPSS Statistics. Variance of responses was stabilized at each concentration using the inverse sd of replicates as weight. The quality of curve fitting was verified by F test for lack-of-fit based on ANOVA (α = 0.05).
Immunoblot Analysis
Chromatin samples were obtained as detailed in the ChIP procedure without cross-linking or sonication. Twenty micrograms of protein samples, as estimated by the bicinchoninic acid method, were loaded on 14% LiDs Tris-Tricine gels and blotted onto polyvinylidene fluoride membranes before immunodetection using antibodies (Merck Millipore) recognizing the histone H3 unmodified C terminus (ref 07-690), acetyl-histone H3 (ref 06-599), or anti-acetyl H3K27 (ref 07-360) antibodies. Immunoblots were done from two biological replicates.
Statistical Analysis of Annotation Spectra
We first generated a union of all H3ac peaks from the control-, APO-, and TSA-treated samples. Let n1 and n2 be the number of APO-hyperacetylated and TSA-hyperacetylated regions. From the union of peaks we drew 1000 random subsets each of size n1 and n2. For each iteration, we performed a χ2 test between a random set of size n1 and a random set of size n2. We compared the test statistic of the comparison of real APO-hyperacetylated and TSA-hyperacetylated peaks to the distribution of test statistics from the comparisons of the random subsets and calculated an empirical P value (Supplemental Figure 14). We repeated the process for APO-hypoacetylated and TSA-hypoacetylated peaks, subsampling random sets of the respective sizes.
Chromatin Immunoprecipitation for ChIP-seq
For each sample, we pooled two biological replicates from the same material that we used for RNA extraction (see below), resulting in two biological replicates per treatment. After stratification for 4 d at 4°C, seedlings were germinated and grown in 12-well plates in 5 mL water at 23°C, 16/8 h light/dark. Six days after germination, the medium was supplemented with APO and TSA and dissolved in ethanol, to a final concentration corresponding to the EC50 concentration; the control sample was supplemented with the identical volume of ethanol. After 24 h, seedlings were fixated in 1% formaldehyde for 30 min. After several washes, excess liquid was removed and the material was flash-frozen in liquid nitrogen. Chromatin extraction and immunoprecipitation were adapted from Kaufmann et al. (2010) and performed as described by Posé et al. (2013). We used α-H3 (Merck Millipore; ref 07-690) and α-H3ac antibodies (Merck Millipore; ref 06-599). Library preparation for ChIP-seq was performed using the TruSeq ChIP-seq kit (Illumina) according to the manufacturer’s instructions. Size selection from 200 to 400 bp after adapter ligation was done on a Blue Pippin instrument (Sage Science) using a 1.5% gel.
ChIP Enrichment Validation by Quantitative Real-Time PCR
Enrichment of acetylated H3 on the indicated genes in untreated or APO-/AMPO-treated samples was detected by qRT-PCR. We analyzed two loci of the Arabidopsis genome that, according to published data (Wang et al., 2015), were either devoid of (AT1G37110) or studded with (AT5G62690) H3ac. Of each ChIP eluate, 2 μL was 15-fold diluted to a total volume of 30 µL; 4.5 μL was used in qRT-PCR (10-μL setup) using the CFX384 Touch real-time PCR detection system (Bio-Rad) with Maxima SYBR Green qPCR Master Mixes (Thermo Fisher Scientific). The performed two-step PCR protocol was as follows: 95°C for 10 min, 45 cycles of 95°C for 15 s, and 55°C for 60 s. Sequences of primers targeting TUBULIN β-CHAIN2 (TUB2; AT5G62690) and transposable element AT1TE46405 (AT1G37110) are listed in Supplemental Table 2. The input sample was used as loading control to balance the deviation of genomic DNA entry to the ChIP. Fold changes were determined from Δc(t) values (input – target) for both target primer pairs, taking into account the individual primer efficiencies obtained by means of serial dilution (factor 5). Relative histone3-acetylation rates were determined by normalizing the enrichment of H3ac to the H3 control at both loci (H3-Ac IP fraction/H3 IP fraction).
ChIP-seq and Peak Calling
ChIP-seq libraries were sequenced on Illumina MiSeq and HiSequation 2000 instruments with 50- and 100-bp single-end reads, respectively. We trimmed reads for adapter sequences using the software skewer (v0.1.120) (Jiang et al., 2014). Reads were then quality-filtered using SHORE (v0.9.0) (Ossowski et al., 2008), and reads trimmed to <30 bp due to low quality were discarded. Mapping against the TAIR9 version of the Arabidopsis reference genome (www.arabidopsis.org) was performed using SHORE, and alignment files were converted to sam and bam formats using SHORE and Samtools (v1.18). We called peaks for H3ac enrichment using MACS (v2) (Zhang et al., 2008), with “broad” mode enabled and bandwidth set to 1000. We filtered duplicated reads using the “auto” mode of the keep-dup function. We followed the guidelines of the ENCODE project in order to identify confident peak calls (Landt et al., 2012; Bailey et al., 2013) (Supplemental Figure 11). First, we ran MACS2 on every individual sample and subjected the peak calls of the two biological replicates of each treatment to an IDR (irreproducibility discovery rate) analysis (Li et al., 2011). Second, we merged the alignments of respective biological replicates and performed the same MACS analysis on the combined data. Third, we split the combined alignments of the biological replicates into two pseudoreplicate alignment sets and repeated the MACS and IDR analyses (Supplemental Figure 11). Only peaks that were detected in all three approaches were retained for further analysis. Quality control summary statistics are listed in Supplemental Table 3.
Differential ChIP Analysis
Differential ChIP analysis was performed using the package DiffBind (v1.8.5) (Ross-Innes et al., 2012) in R (v3.1.0) (www.cran.r-project.org); only alignments that had been retained by MACS2 in the previous peak calling were used in the analysis. DiffBind was run with the edgeR (v3.4.1) or DESeq2 (v1.2.1) methods enabled, whereby edgeR was more conservative and detected only a subset of peaks identified by DESeq2 (Supplemental Figures 12B to 12D). Only differential peaks identified by both methods were used in the further analysis.
Peak Annotation and Overlap
We used the TAIR10 annotation for genes, exons, introns, untranslated regions (UTRs), and transposable elements. Positions and regions were hierarchically assigned to annotated elements in the order coding sequence > intron > 5′ UTR > 3′ UTR > transposon > intergenic space; as intergenic we defined coordinates that were not annotated either as coding sequence, intron, UTR, or transposon. We assigned regions to annotated elements by base pair; each position in the region was assigned in the above-mentioned order. A region can thus stretch over several annotated elements. Overlaps of different sets of peaks were tested using bedtools intersect v.2.17.
Transcriptome Analysis
For each treatment, we extracted RNA from four biological replicates (see “Chromatin Immunoprecipitation for ChIP-seq” above; samples for RNA extraction were flash-frozen before the ChIP fixation step). Total RNA was extracted from frozen material using the RNeasy Plant Mini kit (Qiagen). RNA sequencing libraries were generated using the TruSeq RNA Sample Prep kit (Illumina) according to the manufacturer’s instructions and sequenced on an Illumina HiSeq2000 instrument as 100-bp single-end reads in a 16-plex pool.
Reads were quality-filtered using SHORE (Ossowski et al., 2008); reads trimmed to <40 bp due to low quality were discarded. Reads were mapped to the TAIR10 version of Arabidopsis annotated genes (www.arabidopsis.org) using bwa (v0.7.5a). Read counts for each gene were generated using a custom perl script. We performed differential expression analysis using the DESeq package (Anders and Huber, 2010) in R (v3.1.0); dispersion was estimated with the “per-condition” method and “maximum” sharing mode. Genes were considered as differentially expressed if the expression level between samples differed by >2-fold and if the Bonferroni-corrected P value was <0.05. All differentially expressed genes are listed in Supplemental Data Set 3.
Validation of RNA-seq Data
For first-strand cDNA synthesis, we primed 0.9 µg total RNA with oligo(dT)18 (RevertAid; Thermo Scientific). The resulting product was 10-fold diluted to a final volume of 200 µL; 2 μL was applied to qPCR using the CFX384 Touch real-time PCR detection system (Bio-Rad) with Maxima SYBR Green qPCR Master Mix (Thermo Scientific). The performed protocol was 95°C for 5 min, 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 20 s, followed by 72°C for 7 min. For validation of RNA-seq data, we first generated a random subset (n = 67) from all genes that were differentially expressed after APO treatment. We selected six genes total, two for each range of basic expression (low, <100 read counts; medium, 400 to 600 read counts; high, >1000 read counts) and analyzed their expression levels by quantitative real-time PCR in three biological replicates each of control-, APO-, and TSA-treated seedlings (Supplemental Figure 16). As reference gene to estimate the deviation of RNA input to the cDNA synthesis reaction, we used TUB2 (AT5G62690), the transcription level of which was not affected by APO according to our RNA-seq data. Fold changes were determined employing relative quantification, and the required primer efficiencies were determined by serial dilution. Values for APO- and TSA-treated samples were normalized to those of the untreated control and compared with RNA-seq results. Overall correlation of fold changes between the RNA-seq and qPCR experiments was high (R2 = 88.6). Primers used for qPCR are listed in Supplemental Table 4.
GO Analysis
We used the R package “goseq” (v1.14.0) to assess differentially expressed genes for overrepresentation of GO terms after calculating a probability weighting function based on gene length. All expressed genes (read counts summed over all samples >20) were used as background set. P values were calculated for each GO identifier that was listed in the “org.At.tairGO2TAIR” package and that was represented by at least one expressed gene; only GO identifiers with a Bonferroni-corrected P value < 0.05 were considered as overrepresented. GO identifiers were mapped to the corresponding GO terms using the “GO.db” package.
Accession Numbers
Human HDACs referred to in this study are entered in the NCBI Protein Database under the reference IDs 1T64 (HDAC8) and 2VQM (HDAC4); the reference entry of bacterial HDLP is 1C3R. Arabidopsis HDAC sequences were retrieved from The Arabidopsis Information Resource using gene models AT4G38130.1 (HDA1/19), AT5G63110.1 (HDA6), and AT5G26040.2 (HDA2). The RNA- and ChIP-seq data have been deposited with the European Nucleotide Archive under accession number PRJEB8886. ChIP-seq and RNA-seq data have been uploaded to and can be displayed in the genome browser of the EPIC consortium (https://www.plant-epigenome.org/; https://genomevolution.org/wiki/index.php/EPIC-CoGe), accessible at http://genomevolution.org/r/939v.
Supplemental Data
Supplemental Figure 1. Chemical structures of hydroxamic acid HDAC inhibitors and allelochemicals.
Supplemental Figure 2. HDAC conservation across different organisms.
Supplemental Figure 3. Modeled binding of known and putative HDAC inhibitors to human and bacterial HDACs.
Supplemental Figure 4. Homology models of Arabidopsis HDACs.
Supplemental Figure 5. DIBOA, DIMBOA, BOA, and MBOA do not inhibit plant-derived HDAC enzymes.
Supplemental Figure 6. Inhibition of human class I, II, and IV HDAC enzymes by APO.
Supplemental Figure 7. Inhibition of human class I, II, and IV HDAC enzymes by AMPO.
Supplemental Figure 8. Exposure to hydroxamic acids and their derivatives inhibits plant growth in lettuce.
Supplemental Figure 9. Growth defects in HDAC loss-of-function mutants.
Supplemental Figure 10. Validation of ChIP enrichment for H3ac by quantitative real-time PCR.
Supplemental Figure 11. Schematic representation of the ChIP peak calling and filtering.
Supplemental Figure 12. Differential ChIP-seq analysis.
Supplemental Figure 13. Correlation between TSA-dependent gene expression and H3 acetylation.
Supplemental Figure 14. Annotation of acetylated and differentially acetylated regions.
Supplemental Figure 15. Summary of Gene Ontology term analysis of differentially expressed genes.
Supplemental Figure 16. Validation of RNA-seq by quantitative real-time PCR.
Supplemental Table 1. Effective concentrations for different HDAC inhibitors.
Supplemental Table 2. Primer sequences for chromatin immunoprecipitation analyses.
Supplemental Table 3. Quality control analysis of ChIP peak calling.
Supplemental Table 4. Primer sequences for RNA-seq validation by qRT-PCR.
Supplemental Data Set 1. ChIP-seq peak calls using MACS2 and peaks after IDR filtering.
Supplemental Data Set 2. Differentially acetylated regions.
Supplemental Data Set 3. Differentially expressed genes.
Supplemental Data Set 4. Differentially expressed genes overlapping with differentially acetylated loci.
Supplementary Material
Acknowledgments
We thank R. Schwab, A. Possart, S. Armeanu, C. Busch, G. Wang, and R. Johnstone for stimulating discussions and comments on the manuscript and J. Chen for providing seed material. We also thank A. Heyn, A. Schenk, J. Schoene, I. Smirnow, and C. Leischner for excellent technical assistance, M. Vötsch for help with graphic design, and C. Lanz for support with Illumina sequencing. This work was supported in part by grants from the Deutsche Forschungsgemeinschaft (SFB 685, SFB 773, and BE 4189/1-2), from the Ministry for Nutrition and Rural Territories and the Ministry for Science, Research, and Arts of Baden-Württemberg, Germany (Kap. 0802, Title 98174), from the Wissenschaftsförderung der Deutschen Brauwirtschaft (Project B103), from the European Research Council (ERC Advanced Grant 340602 ‘IMMUNEMESIS’), and by the Max Planck Society. S.V. was supported by a grant from the Jürgen Manchot Stiftung and the Innovation Grant of the University of Tübingen. F.B. was supported by the French ANR agency (ANR-11-JSV2-003-01). A.B. was supported by a grant from the Fortüne Program of the University Clinic Tübingen (1966-0-0). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
AUTHOR CONTRIBUTIONS
S.V., C.B., R.G.B., U.M.L., and M.B. designed the research. S.V., C.B., R.G.B., A.K., A.B., K.v.H., A.W., A.B., F.B., G.Z., T.L. and O.K. performed the experiments; S.V., C.B., R.G.B., A.K., A.B., K.v.H., A.W., A.B., F.B., O.K., U.M.L., and M.B. analyzed the data. C.B., S.V., R.G.B., A.K., F.B., O.K., D.W., U.M.L., and M.B. wrote the article. S.V., C.B., U.M.L., and M.B. supervised the project. All authors discussed and commented on the article.
Glossary
- DIBOA
4-dihydroxy-2H-1,4-benzoxazin-3(4H)-one
- DIMBOA
2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one
- MBOA
6-methoxybenzoxazolin-2(3H)-one
- BOA
benzoxazolin-2(3H)-one
- APO
2-amino-3H-phenoxazin-3-one
- AMPO
2-amino-7-methoxy-3H-phenoxazin-3-one
- HDAC
histone deacetylase
- SAHA
suberoylanilide hydroxamic acid
- TSA
trichostatin-A
- PEN
pendimethalin
- ChIP-seq
chromatin immunoprecipitation followed by next-generation sequencing
- GO
Gene Ontology
- ACN
acetonitrile
- UTR
untranslated region
- IDR
irreproducibility discovery rate
Footnotes
Articles can be viewed online without a subscription.
References
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai K., Isono K., Okuma K., Suzuki S. (1960). The new antibiotics, questiomycins A and B. J. Antibiot. 13: 125–132. [PubMed] [Google Scholar]
- Aufsatz W., Mette M.F., van der Winden J., Matzke M., Matzke A.J. (2002). HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 21: 6832–6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerson S.R., Sánchez-Moreiras A., Pedrol-Bonjoch N., Schulz M., Kagan I.A., Agarwal A.K., Reigosa M.J., Duke S.O. (2005). Detoxification and transcriptome response in Arabidopsis seedlings exposed to the allelochemical benzoxazolin-2(3H)-one. J. Biol. Chem. 280: 21867–21881. [DOI] [PubMed] [Google Scholar]
- Bailey T., Krajewski P., Ladunga I., Lefebvre C., Li Q., Liu T., Madrigal P., Taslim C., Zhang J. (2013). Practical guidelines for the comprehensive analysis of ChIP-seq data. PLOS Comput. Biol. 9: e1003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bais H.P., Weir T.L., Perry L.G., Gilroy S., Vivanco J.M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57: 233–266. [DOI] [PubMed] [Google Scholar]
- Bantscheff M., et al. (2011). Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 29: 255–265. [DOI] [PubMed] [Google Scholar]
- Belz R.G. (2007). Allelopathy in crop/weed interactions--an update. Pest Manag. Sci. 63: 308–326. [DOI] [PubMed] [Google Scholar]
- Belz R.G., Hurle K. (2005). Differential exudation of two benzoxazinoids--one of the determining factors for seedling allelopathy of Triticeae species. J. Agric. Food Chem. 53: 250–261. [DOI] [PubMed] [Google Scholar]
- Benhamed M., Bertrand C., Servet C., Zhou D.X. (2006). Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 18: 2893–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436. [DOI] [PubMed] [Google Scholar]
- Blevins T., Pontvianne F., Cocklin R., Podicheti R., Chandrasekhara C., Yerneni S., Braun C., Lee B., Rusch D., Mockaitis K., Tang H., Pikaard C.S. (2014). A two-step process for epigenetic inheritance in Arabidopsis. Mol. Cell 54: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley M.J., Lo Surdo P., Di Giovine P., Cirillo A., Scarpelli R., Ferrigno F., Jones P., Neddermann P., De Francesco R., Steinkühler C., Gallinari P., Carfí A. (2008). Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 283: 26694–26704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch G., Ransom R., Lechner T., Walton J.D., Loidl P. (1995). Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7: 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colville A., Alhattab R., Hu M., Labbé H., Xing T., Miki B. (2011). Role of HD2 genes in seed germination and early seedling growth in Arabidopsis. Plant Cell Rep. 30: 1969–1979. [DOI] [PubMed] [Google Scholar]
- Dayan F.E., Cantrell C.L., Duke S.O. (2009). Natural products in crop protection. Bioorg. Med. Chem. 17: 4022–4034. [DOI] [PubMed] [Google Scholar]
- Duke S.O. (2007). Weeding with allelochemicals and allelopathy--a commentary. Pest Manag. Sci. 63: 307. [DOI] [PubMed] [Google Scholar]
- Duke S.O. (2010). Allelopathy - current status of research and future of the discipline: a commentary. Allelopathy J. 25: 17–30. [Google Scholar]
- Earley K., Lawrence R.J., Pontes O., Reuther R., Enciso A.J., Silva M., Neves N., Gross M., Viegas W., Pikaard C.S. (2006). Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 20: 1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.P. (2008). Epigenetics at the epicenter of modern medicine. JAMA 299: 1345–1350. [DOI] [PubMed] [Google Scholar]
- Finney D.J. (1978). Statistical Method of Biological Assay. (London: Charles Griffin & Co. Ltd; ). [Google Scholar]
- Finnin M.S., Donigian J.R., Cohen A., Richon V.M., Rifkind R.A., Marks P.A., Breslow R., Pavletich N.P. (1999). Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401: 188–193. [DOI] [PubMed] [Google Scholar]
- Gents M.B., Nielsen S.T., Mortensen A.G., Christophersen C., Fomsgaard I.S. (2005). Transformation products of 2-benzoxazolinone (BOA) in soil. Chemosphere 61: 74–84. [DOI] [PubMed] [Google Scholar]
- Gouet P., Courcelle E., Stuart D.I., Métoz F. (1999). ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308. [DOI] [PubMed] [Google Scholar]
- Gregoretti I.V., Lee Y.M., Goodson H.V. (2004). Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338: 17–31. [DOI] [PubMed] [Google Scholar]
- Ha M., Ng D.W., Li W.H., Chen Z.J. (2011). Coordinated histone modifications are associated with gene expression variation within and between species. Genome Res. 21: 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T.A. (1996). Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 17: 490–519. [Google Scholar]
- He Y., Michaels S.D., Amasino R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754. [DOI] [PubMed] [Google Scholar]
- Hollender C., Liu Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50: 875–885. [DOI] [PubMed] [Google Scholar]
- Huang Z., Haig T., Wu H., An M., Pratley J. (2003). Correlation between phytotoxicity on annual ryegrass (Lolium rigidum) and production dynamics of allelochemicals within root exudates of an allelopathic wheat. J. Chem. Ecol. 29: 2263–2279. [DOI] [PubMed] [Google Scholar]
- Inderjit W., Wardle D.A., Karban R., Callaway R.M. (2011). The ecosystem and evolutionary contexts of allelopathy. Trends Ecol. Evol. (Amst.) 26: 655–662. [DOI] [PubMed] [Google Scholar]
- Jiang H., Lei R., Ding S.W., Zhu S. (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics 15: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Toh H. (2008). Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9: 286–298. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Krogh S.S., Mensz S.J., Nielsen S.T., Mortensen A.G., Christophersen C., Fomsgaard I.S. (2006). Fate of benzoxazinone allelochemicals in soil after incorporation of wheat and rye sprouts. J. Agric. Food Chem. 54: 1064–1074. [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka J., Foret S., Maleszka R. (2008). Nutritional control of reproductive status in honeybees via DNA methylation. Science 319: 1827–1830. [DOI] [PubMed] [Google Scholar]
- Landt S.G., et al. (2012). ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22: 1813–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Brown J., Huang H., Bickel P. (2011). Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5: 1752–1779. [Google Scholar]
- Macías F.A., Marín D., Oliveros-Bastidas A., Castellano D., Simonet A.M., Molinillo J.M. (2006). Structure-activity relationship (SAR) studies of benzoxazinones, their degradation products, and analogues. Phytotoxicity on problematic weeds Avena fatua L. and Lolium rigidum Gaud. J. Agric. Food Chem. 54: 1040–1048. [DOI] [PubMed] [Google Scholar]
- Macías F.A., Marín D., Oliveros-Bastidas A., Molinillo J.M. (2009). Rediscovering the bioactivity and ecological role of 1,4-benzoxazinones. Nat. Prod. Rep. 26: 478–489. [DOI] [PubMed] [Google Scholar]
- Macías F.A., Molinillo J.M., Varela R.M., Galindo J.C. (2007). Allelopathy--a natural alternative for weed control. Pest Manag. Sci. 63: 327–348. [DOI] [PubMed] [Google Scholar]
- Macías F.A., Oliveros-Bastidas A., Marín D., Castellano D., Simonet A.M., Molinillo J.M. (2004). Degradation studies on benzoxazinoids. Soil degradation dynamics of 2,4-dihydroxy-7-methoxy-(2H)-1,4-benzoxazin-3(4H)-one (DIMBOA) and its degradation products, phytotoxic allelochemicals from gramineae. J. Agric. Food Chem. 52: 6402–6413. [DOI] [PubMed] [Google Scholar]
- Macías F.A., Oliveros-Bastidas A., Marín D., Castellano D., Simonet A.M., Molinillo J.M. (2005). Degradation studies on benzoxazinoids. Soil degradation dynamics of (2R)-2-O-beta-D-glucopyranosyl-4-hydroxy-(2H)- 1,4-benzoxazin-3(4H)-one (DIBOA-Glc) and its degradation products, phytotoxic allelochemicals from Gramineae. J. Agric. Food Chem. 53: 554–561. [DOI] [PubMed] [Google Scholar]
- Macías F.A., Oliveros-Bastidas A., Marín D., Chinchilla N., Castellano D., Molinillo J.M. (2014). Evidence for an allelopathic interaction between rye and wild oats. J. Agric. Food Chem. 62: 9450–9457. [DOI] [PubMed] [Google Scholar]
- Marks P.A., Xu W.S. (2009). Histone deacetylase inhibitors: Potential in cancer therapy. J. Cell. Biochem. 107: 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegola E., Di Renzo F., Broccia M.L., Giavini E. (2006). Inhibition of histone deacetylase as a new mechanism of teratogenesis. Birth Defects Res. C Embryo Today 78: 345–353. [DOI] [PubMed] [Google Scholar]
- Molisch H. (1937). Der Einfluss einer Pflanze auf eine andere - Allelopathie. (Jena, Germany: Fischer; ). [Google Scholar]
- Moll A., Hildebrandt A., Lenhof H.P., Kohlbacher O. (2006). BALLView: a tool for research and education in molecular modeling. Bioinformatics 22: 365–366. [DOI] [PubMed] [Google Scholar]
- Murfett J., Wang X.J., Hagen G., Guilfoyle T.J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13: 1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D. (2008). Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posé D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G.C., Immink R.G., Schmid M. (2013). Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 503: 414–417. [DOI] [PubMed] [Google Scholar]
- Pray L. (2008). At the flick of a switch: epigenetic drugs. Chem. Biol. 15: 640–641. [DOI] [PubMed] [Google Scholar]
- Probst A.V., Fagard M., Proux F., Mourrain P., Boutet S., Earley K., Lawrence R.J., Pikaard C.S., Murfett J., Furner I., Vaucheret H., Mittelsten Scheid O. (2004). Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell 16: 1021–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rarey M., Kramer B., Lengauer T., Klebe G. (1996). A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 261: 470–489. [DOI] [PubMed] [Google Scholar]
- Raub S., Steffen A., Kämper A., Marian C.M. (2008). AIScore chemically diverse empirical scoring function employing quantum chemical binding energies of hydrogen-bonded complexes. J. Chem. Inf. Model. 48: 1492–1510. [DOI] [PubMed] [Google Scholar]
- Rhee S.Y., et al. (2003). The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 31: 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C.P., Cai G., Teasdale J.R. (2012). Concentrations and allelopathic effects of benzoxazinoid compounds in soil treated with rye (Secale cereale) cover crop. J. Agric. Food Chem. 60: 4471–4479. [DOI] [PubMed] [Google Scholar]
- Ross-Innes C.S., et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. (1993). Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815. [DOI] [PubMed] [Google Scholar]
- Schäfer S., Jung M. (2005). Chromatin modifications as targets for new anticancer drugs. Arch. Pharm. (Weinheim) 338: 347–357. [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76: 75–100. [DOI] [PubMed] [Google Scholar]
- Somoza J.R., et al. (2004). Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure 12: 1325–1334. [DOI] [PubMed] [Google Scholar]
- Streibig J.C. (1988). Herbicide bioassay. Weed Res. 28: 479–484. [Google Scholar]
- Tian L., Chen Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98: 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Fong M.P., Wang J.J., Wei N.E., Jiang H., Doerge R.W., Chen Z.J. (2005). Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169: 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Wang J., Fong M.P., Chen M., Cao H., Gelvin S.B., Chen Z.J. (2003). Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics 165: 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Understrup A.G., Ravnskov S., Hansen H.C., Fomsgaard I.S. (2005). Biotransformation of 2-benzoxazolinone to 2-amino-(3H)-phenoxazin-3-one and 2-acetylamino-(3H)-phenoxazin-3-one in soil. J. Chem. Ecol. 31: 1205–1222. [DOI] [PubMed] [Google Scholar]
- Wang C., Liu C., Roqueiro D., Grimm D., Schwab R., Becker C., Lanz C., Weigel D. (2015). Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 25: 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.F., Wiest O., Helquist P., Lan-Hargest H.Y., Wiech N.L. (2004). On the function of the 14 A long internal cavity of histone deacetylase-like protein: implications for the design of histone deacetylase inhibitors. J. Med. Chem. 47: 3409–3417. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K. (2009). Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138: 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.C., Johnstone R.W. (2014). New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124: 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M., Sippl M.J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35: W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Word J.M., Lovell S.C., Richardson J.S., Richardson D.C. (1999). Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 285: 1735–1747. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.