Global identification of RNA modifications that affect the Watson-Crick base-pairing edge across the Arabidopsis transcriptome provides insights into the many functions of these important additions.
Abstract
Posttranscriptional chemical modification of RNA bases is a widespread and physiologically relevant regulator of RNA maturation, stability, and function. While modifications are best characterized in short, noncoding RNAs such as tRNAs, growing evidence indicates that mRNAs and long noncoding RNAs (lncRNAs) are likewise modified. Here, we apply our high-throughput annotation of modified ribonucleotides (HAMR) pipeline to identify and classify modifications that affect Watson-Crick base pairing at three different levels of the Arabidopsis thaliana transcriptome (polyadenylated, small, and degrading RNAs). We find this type of modifications primarily within uncapped, degrading mRNAs and lncRNAs, suggesting they are the cause or consequence of RNA turnover. Additionally, modifications within stable mRNAs tend to occur in alternatively spliced introns, suggesting they regulate splicing. Furthermore, these modifications target mRNAs with coherent functions, including stress responses. Thus, our comprehensive analysis across multiple RNA classes yields insights into the functions of covalent RNA modifications in plant transcriptomes.
INTRODUCTION
Across prokaryotes and eukaryotes, RNA chemical modification is both widespread and physiologically relevant. While modifications are best characterized in noncoding tRNAs and rRNAs, mRNAs have also been found to contain N6-methyladenosine (m6A) (Horowitz et al., 1984; Dominissini et al., 2012; Meyer et al., 2012), 5-methylcytosine (m5C) (Squires et al., 2012), inosine (I) (Li et al., 2009; Wulff et al., 2011), and pseudouridine (Y) (Carlile et al., 2014; Schwartz et al., 2014b). Additionally, there is a growing body of evidence to support the functional significance of RNA modifications within mRNAs. For instance, in mouse (Mus musculus), spliceosome assembly disruption and changes in mRNA localization were observed upon knockdown of the oxidative demethylase ALKBH5, which removes methyl groups from RNA (Zheng et al., 2013). Furthermore, the presence of certain methylated bases in human cell lines anticorrelates with mRNA stability (Schwartz et al., 2014a). However, coding and noncoding RNAs likely share the same modifying enzymes (Lee, Kim, and Kim, 2014), and specifically testing the function of mRNA modification through genetic ablation of these proteins is difficult. Thus, the functional consequences of most mRNA modifications are still unclear.
The best characterized mRNA modification to date is m6A, which is enriched around the stop codon, suggesting interplay with the translation and degradation machinery (Meyer et al., 2012). This mark is also enriched at alternatively spliced introns and over long exons (Dominissini et al., 2012), suggesting a role in modulating splicing. Moreover, Y modifications in tRNAs stabilize secondary structures (Sundaram et al., 2000; Kierzek et al., 2014) and may do the same in mRNAs in which they are incorporated (Carlile et al., 2014; Schwartz et al., 2014b). Similarly, as tRNA modifications are known to direct cleavage of internally transcribed spacers, mRNA modifications could likewise direct transcript cleavage and subsequent turnover (Hughes and Ares, 1991; Kiss-László et al., 1996). Thus, chemical modifications likely have widespread and varied effects across the eukaryotic transcriptome. However, our knowledge of the mRNA modification sites and their functional consequences is currently limited.
Here, we comprehensively identify mRNA modifications using high-throughput annotation of modified ribonucleotides (HAMR) (Ryvkin et al., 2013). HAMR exploits the tendency of certain covalent RNA modifications, including those known to be common in tRNAs, to interfere with Watson-Crick base pairing and cause reverse transcriptase (RT) to stall and/or misincorporate nucleotides during reverse transcription. This in turn produces a characteristic pattern of RT mistakes, which present in deep sequencing as mismatches from the reference genome. Working on this premise, HAMR tabulates high confidence (quality score >30, error probability <1/1000) mismatches and tests for significance by (1) ruling out that the changes are merely sequencing error and (2) excluding single nucleotide polymorphisms (SNPs) or editing sites (Figure 1). To this end, we focus on modification-induced errors that have a trinucleotide substitution pattern and do not have a clear bias toward any single base misincorporation in order to avoid SNPs and sites of RNA editing (Ryvkin et al., 2013). These stringent filtering steps require high read coverage, and as a result, HAMR is designed to minimize false positives at the expense of likely missing a portion of the modified transcriptome. Moreover, modifications such as m6A, which do not significantly affect the Watson-Crick base-pairing edge, will not be detected by HAMR. Nonetheless, this algorithm provides a high-throughput, robust, and generalized in silico method to detect RNA modifications that affect Watson-Crick base pairing in eukaryotic transcriptomes. Such HAMR-predicted modifications include but are not limited to 3-methyl cytosine (m3C), 1-methyl guanosine (m1G), and 1-methyl adenosine (m1A) (Ryvkin et al., 2013). This algorithm also incorporates a validated (Ryvkin et al., 2013) machine learning step into the analysis that allows prediction of modification identity (e.g., m3C) based on the specific trinucleotide substitution pattern that we observe at every HAMR-predicted modification site. This analytical approach is based on our previous observation that each type of covalent RNA modification directs a distinct trinucleotide RT incorporation pattern based on their differential base-pairing properties (Ryvkin et al., 2013).
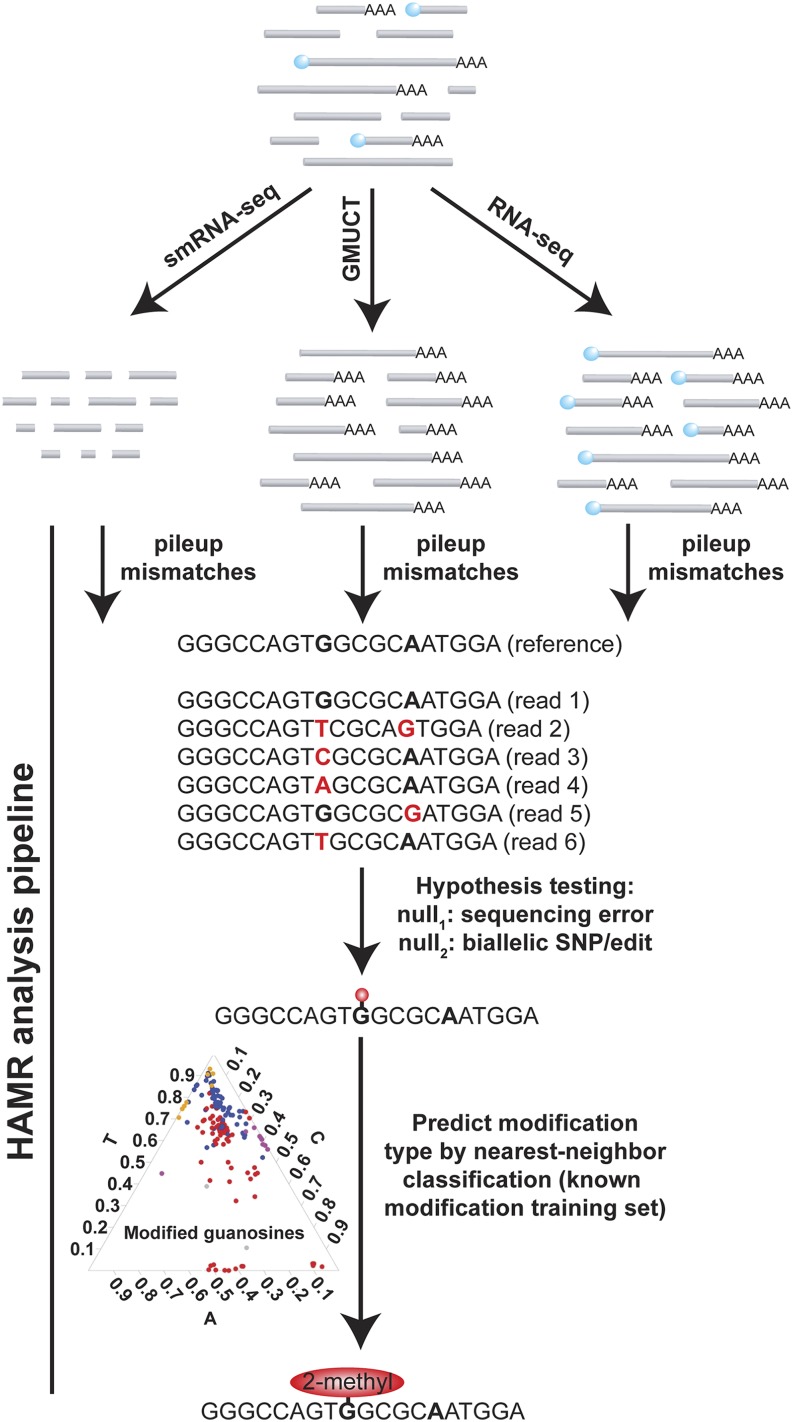
Figure 1.
Study Design to Comprehensively Identify Covalent, HAMR-Predicted Modifications in the Arabidopsis Transcriptome.
smRNA, poly(A)+-selected RNA, and poly(A)+-selected GMUCT (Gregory et al., 2008; Willmann et al., 2014) libraries were constructed in parallel. GMUCT specifically captures transcripts without a 7-methylguanosine cap (light-blue circles). The HAMR analysis pipeline was then run on the resulting data sets. Specifically, reads are mapped to their reference genome, and mismatches (red bases) for each base (bolded bases) are tabulated. After two rounds of hypothesis testing, predicted modifications are then classified, based on a training set of known tRNA modifications from S. cerevisiae.
Here, we apply the HAMR analysis pipeline to RNA sequencing data for the poly(A)+ and small portions of the transcriptome (RNA-seq and smRNA-seq, respectively), as well as uncapped and degrading RNAs via global mapping of uncapped and cleaved transcripts (GMUCT) (Gregory et al., 2008; Willmann et al., 2014). We identify, classify, and functionally characterize RNA modifications in Arabidopsis thaliana and then test whether the results generalize to human RNAs (Figure 1). In total, our results provide a global view of HAMR-predicted modifications across eukaryotic transcriptomes, allowing us to begin teasing apart their functional significance in posttranscriptional regulation.
RESULTS AND DISCUSSION
Using HAMR to Predict RNA Modification Sites That Affect the Watson-Crick Base-Pairing Edge throughout the Arabidopsis Transcriptome
In general, uncapped fragments derived from mRNAs in eukaryotic transcriptomes are generated by decapping or endonucleolytic cleavage, and these RNA fragments are then rapidly recognized and degraded by 5′ to 3′ (e.g., XRN4) (Gazzani et al., 2004) and 3′ to 5′ (e.g., exosome) (Chekanova et al., 2007) exonucleases. Thus, they represent the degrading fraction of the transcriptome. Through GMUCT (Gregory et al., 2008; Willmann et al., 2014), we surveyed the polyadenylated, uncapped, degrading transcriptome of unopened Arabidopsis flower buds. We then paired these data with data from small RNA sequencing (smRNA-seq) and poly(A)+-selected RNA sequencing (RNA-seq) of this same tissue to identify HAMR-predicted modifications at multiple levels of the plant transcriptome (Figure 1).
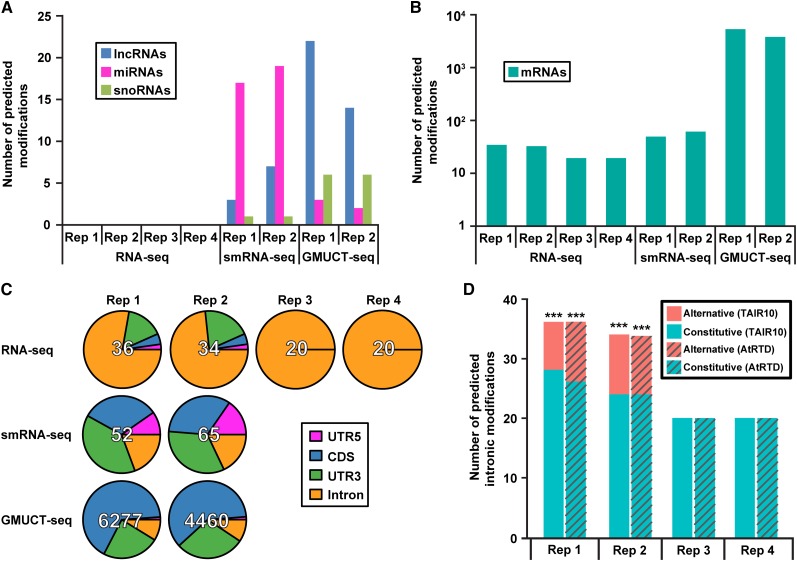
To do this, we ran the HAMR pipeline on the set of uniquely mapping reads from these three RNA-seq approaches (see Methods). From this analysis, we observed differing numbers of HAMR-predicted modifications for different classes of RNA at the three different levels of the transcriptome. For instance, we found that long noncoding RNAs (lncRNAs) and small nucleolar RNAs (snoRNAs) contained the most HAMR-predicted modifications within the GMUCT data set, while a few and none were identified when analyzing the smRNA- and RNA-seq data sets, respectively (Figure 2A). These results suggest that there may be a link between HAMR-predicted modifications and degradation for lncRNAs and snoRNAs. In contrast, HAMR-predicted modifications in microRNAs (miRNAs) were most abundant within smRNA-seq compared with GMUCT and RNA-seq data sets (Figure 2A). Among mRNAs, we observed an average of 5368 HAMR-predicted modifications in two replicates of GMUCT data. In contrast, an average of only 58 modifications was observed in two replicates of smRNA-seq and 27 in four replicates of RNA-seq data (Figure 2B). Thus, we observed a strong enrichment of HAMR-predicted modifications within degrading mRNAs compared with stable, poly(A)+ mRNAs (hereafter stable mRNAs) and mRNA-derived smRNAs (Figure 2B). Interestingly, this strong enrichment of modifications within uncapped, degrading mRNAs compared with stable mRNAs or mRNA-derived smRNAs was also seen using the same three RNA sequencing data types from two human cell lines (ENCODE Project Consortium, 2012; Huelga et al., 2012; Willmann, Berkowitz, and Gregory, 2014) (Supplemental Figures 1A and 1B), suggesting that our observations generalize to other eukaryotic organisms.
Figure 2.
HAMR-Predicted Modifications in Arabidopsis Mark Uncapped and Alternative Spliced Transcripts.
(A) and (B) Total number of modifications predicted in (A) noncoding RNAs (lncRNAs [blue bars], miRNAs [magenta bars], and snoRNAs [green bars]) and coding mRNAs are plotted for each data set (B).
(C) Relative transcript location of predicted modifications in mRNAs. Modifications that lie outside of mRNAs are excluded from this analysis.
(D) Localization of modifications to alternative versus constitutive introns. Enrichment was calculated with a Fisher’s exact test. Asterisks denote P value <1 × 10−12. Analysis was performed using transcriptome annotations from TAIR10 (solid bars) or AtRTD (hatched bars) (Zhang et al., 2015).
Since the statistical power of HAMR depends upon sequencing depth (Ryvkin et al., 2013), we took several approaches to ensure that our observed differences in HAMR-predicted modifications were not artifacts of varying sequencing coverage of transcriptome nucleotides, spurious read mapping, or differential processing of sequencing reads that are a consequence of the differential library preparations necessary for each sequencing technique. To first test that potential differences in sequencing coverage of transcriptome nucleotides between libraries was not leading to the differential identification of HAMR-predicted modifications, we downsampled all libraries to equal numbers of uniquely mapping reads. We then looked at total sequencing read coverage of each nucleotide of the Arabidopsis transcriptome. From this analysis, we found that different libraries displayed varying distributions of read coverage, notably with GMUCT and RNA-seq skewed toward higher read coverage, with GMUCT having a few nucleotides that had extremely high read depth, while smRNA-seq showed lower overall coverage (Supplemental Figure 2A). This suggests that GMUCT could have more RNA bases with sufficient read coverage for HAMR to call a modification site (HAMR-accessible bases) than smRNA and to a lesser extent RNA-seq. From this analysis, we also found that for all three sequencing approaches, the minimum coverage at a HAMR-predicted modification site was 50 reads covering that base (Supplemental Figure 2A, black dashed line), so we defined HAMR-accessible bases as those with at least this level of depth. We then normalized total modification number to total HAMR-accessible bases for the data sets from all three sequencing approaches and found that mRNAs still have an average of 1207 HAMR-predicted modifications per million accessible bases in GMUCT, compared with 602 in smRNA-seq and 15 in RNA-seq (Supplemental Figure 2B). This jump in the number of smRNA-seq-predicted modifications suggests that mRNA-derived smRNAs may have more modifications that are simply not called by the HAMR pipeline due to the generally low levels of small RNA processing from mRNAs (Supplemental Figure 2A). Since this normalization might not fully control for the proportion of nucleotides that have very high read depth in GMUCT experiments compared with both RNA- and smRNA-seq (Supplemental Figure 2A, right-hand side of the graph), we also defined a set of different coverage thresholds (1000, 500, 250, and 100 reads), above which modifications were ignored (Supplemental Figure 2C). Again, the major trends in numbers of modifications were not altered, even when setting the upper thresholds to relatively low numbers of sequencing reads (e.g., 100 reads) (Supplemental Figure 2C). This discrepancy in HAMR-predicted modifications between the different sequencing approaches was also still observed even after combining this upper limit thresholding with normalization to HAMR-accessible bases (Supplemental Figure 2D). In total, these results indicate that the overall differences in HAMR-predicted modifications between the three RNA-seq approaches are not a consequence of differential sequencing depth at RNA nucleotides.
We had previously demonstrated that HAMR results were consistent across an array of high-throughput sequence read mapping software programs even when analyzing the highly repetitive human transcriptome (Ryvkin et al., 2013). However, certain high-throughput sequence read mapping software may produce spurious uniquely mapping reads without exhaustively searching for matches across the whole transcriptome. Therefore, although Arabidopsis mRNAs do not generally contain large amounts of repetitive sequence, we still controlled for this possibility by repeating our analysis on repeat-masked (A.F.A. Smit, R. Hubley, and P. Green, 2013; RepeatMasker Open-4.0, http://www.repeatmasker.org) data and observed no change to the number of HAMR-predicted modifications for GMUCT or RNA-seq and only a slight reduction in the number of modifications on smRNAs (Supplemental Figures 2E and 2F, repeat-masked data). Finally, the different types of RNA-seq libraries were subjected to different adaptor trimming strategies based on the relation between sequencing read size (50 nucleotide reads) and expected fragment size (see Methods). To address this, we ran the uniform strategy of concatenating all reads (reads with and without adapter trimming) for all three library types. Once again, treating all libraries the same and analyzing all reads together did not alter the observed trends in differential modification calls between the three different sequencing libraries (Supplemental Figures 2E and 2F, all concatenated data). In total, these control analyses verify that uncapped, degrading mRNAs are strongly enriched for RNA modifications that affect the Watson-Crick base-pairing edge compared with stable mRNAs or mRNA-derived smRNAs.
Validation of HAMR-Predicted Modification Sites in the Arabidopsis Transcriptome
Many of the covalent modifications within yeast (Saccharomyces cerevisiae) tRNAs have been identified and characterized through years of extensive research (Björk et al., 1987; Grosjean et al., 1997; Hopper and Phizicky, 2003; El Yacoubi et al., 2012; Machnicka et al., 2013). For this reason, the machine learning algorithm that HAMR uses to classify the type of modification occurring at each predicted site uses the substitution patterns from a yeast smRNA-seq data set at known tRNA modification sites as its training set (Ryvkin et al., 2013). Furthermore, through homology comparisons of yeast tRNAs to those from other organisms, the orthologous modification sites can be identified (Ryvkin et al., 2013). Therefore, as a positive control verifying that HAMR was detecting bona fide modification sites in the Arabidopsis transcriptome, we derived “known” Arabidopsis tRNA modification sites as those with extensive homology to known modified sites in S. cerevisiae. Specifically, the yeast data were compiled from the Modomics database (Dunin-Horkawicz et al., 2006) and aligned to Arabidopsis tRNAs. Modifications within regions of homology were mapped from yeast to Arabidopsis using a custom pipeline incorporating tRNAscan (Lowe and Eddy, 1997) and LocARNA (Will et al., 2007) (see Methods) (Supplemental Files 1 and 2). As tRNA loci are highly duplicated, we then filtered our two smRNA-seq data sets to allow multimapping reads that align exclusively to tRNAs (see Methods). Additionally, we cannot unambiguously determine modifications at specific tRNA loci, so we performed all analyses at the level of tRNA family consensus sequences. After running HAMR on two replicates of smRNA-seq, we observed that 23 of 48 (48%) and 24 of 52 (46%) of predicted modification sites correspond to these well-defined modification sites. This level of overlap between HAMR-predicted and known modification sites is significantly (P value <1 × 10−7, Fisher’s exact test) higher than random sampling alone (∼11% success rate) (Supplemental Figure 3A). To ensure these results are not specific to our library preparation, we also analyzed a species- and tissue-matched smRNA data set generated by another group (Li et al., 2015) and observed comparable levels of known modification sites identified in tRNAs (P value <1 × 10−7, Fisher’s exact test) (Supplemental Figure 3B). Finally, we tested the true positive rate versus the false positive rate at various threshold settings (receiver operating characteristic) for HAMR identification of these known tRNA modification sites (see Methods), which confirmed the ability of HAMR to identify known modification sites in Arabidopsis tRNAs (area under curve = 69.87) (Supplemental Figures 3C and 3D). Thus, HAMR identifies a significant number of tRNA modification sites in the Arabidopsis transcriptome with known homology to yeast, demonstrating its predictive power for studying these covalent additions to plant RNA.
HAMR takes advantage of the propensity of RT to misincorporate nucleotides at modification sites that affect the Watson-Crick base-pairing edge. However, another consequence of RT encountering such a modification is to stall or terminate elongation and fall off the template (Foley et al., 2015). For this reason, such blocks to RT extension have been used for previous identification of covalent modifications to tRNA molecules (Woodson et al., 1993; Talkish et al., 2014). Therefore, to further validate HAMR-predicted modification sites in Arabidopsis mRNAs, we tested whether these specific nucleotide positions coincide with RT stalls that were recently identified in the control samples for dimethyl sulfate (DMS) sequencing (Structure-seq) experiments (Ding et al., 2014). Unlike our RNA-seq data, these Structure-seq libraries are not fragmented, and they unambiguously define RT stalls as the very 5′ nucleotide of their sequencing reads (Ding et al., 2014). Importantly, these Structure-seq control data sets measure RT extension inhibition in the absence of DMS treatment, which indicates they are unrelated to the addition of exogenous DMS adducts and are specifically measuring blocks to normal RT extension by the presence of an RNA modification that affects the Watson-Crick base pairing edge. Using this approach, we found that HAMR-predicted modification sites in the degrading fraction of mRNAs identified by GMUCT significantly coincide with RT extension inhibition sites (all P values <1 × 10−20, Fisher’s exact test) (Supplemental Figure 4A) and overlap with a greater number of RT stalls per site (all P values <1 × 10−39, Wilcoxon rank sum test) (Supplemental Figure 4B), as measured in the DMS control experiments compared with a background of all mRNA bases. In total, these findings provide strong evidence that HAMR detects bona fide modification sites in Arabidopsis mRNAs and that this class of covalent additions is enriched in the degrading fraction of these molecules.
Characterization of the HAMR-Predicted Modifications in the Arabidopsis Transcriptome
To better understand the potential functions of HAMR-predicted RNA modifications, we determined whether they were enriched in any particular regions of Arabidopsis mRNA molecules. From this analysis, we found that modifications called using HAMR on Arabidopsis GMUCT data tended to localize within the coding sequence and 3′ untranslated region (UTR), whereas HAMR-predicted modifications from the RNA-seq data sets were almost exclusively localized to introns (Figure 2C). Regarding the human transcriptome, we found that these results for the GMUCT and RNA-seq data sets are entirely recapitulated in both HEK293T (human embryonic kidney cells) and HeLa cell lines (Supplemental Figure 5A). Furthermore, modifications in mRNAs called by HAMR using the HEK293T and HeLa smRNA-seq data set are mostly found in mRNA introns, where the majority of human miRNA stem-loop precursors are known to reside (Supplemental Figure 5A). In contrast, modification sites in Arabidopsis mRNAs identified by HAMR using smRNA-seq data display no real bias toward any specific mRNA region (Figure 2C), consistent with the relative paucity of miRNA precursors residing in Arabidopsis introns or other mRNA sequences.
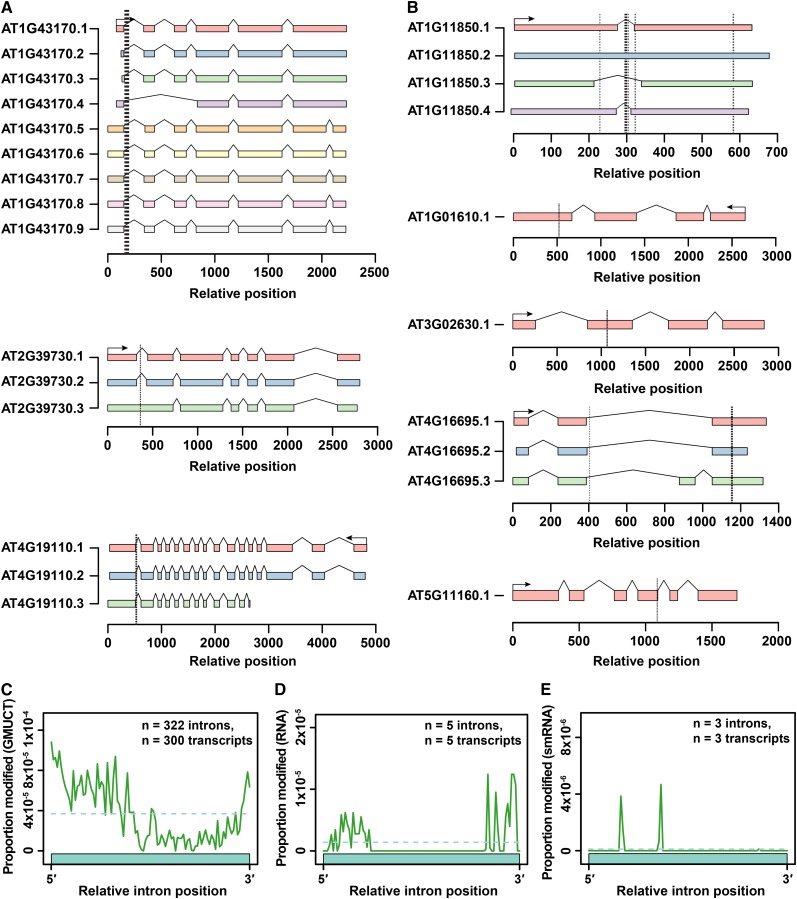
Intriguingly, a closer inspection of all of HAMR-predicted modification sites in stable mRNAs identified using the RNA-seq data sets from both Arabidopsis and human revealed that these covalent additions are significantly enriched (all P values <1 × 10−12, Fisher’s exact test) in or near introns annotated as being alternatively spliced (Figure 2D; Supplemental Figure 5B). Analysis of an expanded Arabidopsis transcriptome annotation (atRTD) (Zhang et al., 2015) yields comparable results (Figure 2D). Furthermore, seven modification sites identified with both RNA-seq replicates 1 and 2 lie within the splice donor site (first six nucleotides) of introns within AT1G43710, AT4G19110, AT4G25080, and AT4G38510 (Figure 3A). It is worth noting that even those that are currently annotated as constitutively spliced introns are most likely novel retained intron events given that they can be captured by a poly(A)+-selected RNA-seq approach. In support of this idea, over 50% of the HAMR-predicted modification sites lie within the Arabidopsis ribosomal protein L3 gene (AT1G43170), which has nine annotated isoforms and a known retained intron event within the 3′ UTR, as well as a novel retained intron in the 5′ UTR identified by our analysis here (Figure 3A). Similar examples exist for other transcripts with modifications predicted by HAMR using the RNA-seq data (Figure 3A) but are less common for transcripts with modifications predicted by analyzing data from the GMUCT approach (Figure 3B).
Figure 3.
HAMR-Predicted Modifications Mark Various Transcriptome Features.
(A) HAMR modifications predicted in three specific Arabidopsis transcripts with HAMR-predicted modifications identified by analyzing GMUCT data sets (uncapped RNAs).
(B) Five specific Arabidopsis transcripts with HAMR-predicted modifications identified by analyzing the RNA-seq data sets (stable mRNAs). For both (A) and (B), the vertical dashed, black lines indicate the relative position of each modification. The thickness of the lines indicates the number of modifications clustered at the specified positions, with thicker and thinner lines indicating more or fewer, respectively. In plus strand transcripts, relative position 0 indicates the very 5′ end. In minus strand transcripts, relative position 0 indicates the 3′ end. All known splice variants of these seven transcripts are shown in these figures.
(C) to (E) Relative position of intronic HAMR-predicted modification sites from analyzing GMUCT (C), RNA-seq (D), and smRNA-seq (E) data sets plotted across the length-normalized average of all annotated TAIR10 introns.
We also observed a significant enrichment (P value → 0, Fisher’s exact test) of HAMR-predicted modifications identified in human stable mRNAs using the human RNA-seq data within introns that were annotated to be alternatively spliced (ENCODE Project Consortium, 2012; Huelga et al., 2012). However, this bias was either much less common or was not observed for HAMR-predicted modifications identified using the smRNA-seq data from the two different cell lines for this analysis (Supplemental Figure 5B). In total, our findings for HAMR-predicted modifications identified in both Arabidopsis and human stable mRNAs using RNA-seq data suggests a role for this class of modifications in the regulation of alternative splicing. This hypothesis is further supported by the fact that most of these modification sites are proximal to the splice donor/acceptor sites of these alternatively spliced introns (Figures 3C to 3E; Supplemental Figures 5C to 5E), with some lying directly within donor site sequences. In total, these results reveal that modifications in uncapped, degrading mRNAs are prevalent in the coding sequence and 3′ UTR, while those in stable transcripts are associated with specific alternative splicing events in both plants and humans. It is noteworthy that another RNA chemical modification, m6A, has also been found to cluster near specific alternatively spliced exons and introns (Dominissini et al., 2012). Taken together, this combination of findings suggests that in general, RNA modifications in stable mRNAs may play a significant role in regulating the processes of alternative splicing in eukaryotic transcriptomes. This hypothesis will require further testing.
Uncapped and Stable mRNAs Contain Different Proportions of Specific RNA Modifications
As described above, the HAMR analysis pipeline includes a step to determine the actual modification at each predicted site based on a machine learning approach where known modification sites in yeast tRNAs are used as the training set (Ryvkin et al., 2013). As a first test that this approach could identify the actual modification at predicted sites in Arabidopsis, we tested if the classifier would call the correct identity at “known” modification sites as determined by homology with yeast tRNAs (Supplemental Figures 3A and 3B). From this analysis, we found that the HAMR modification classifier correctly predicted the exact modification type at ∼50% of these known modification sites in Arabidopsis tRNAs (Supplemental Figure 3D and Supplemental Table 1). Therefore, we were comfortable using this approach to determine the identity of the specific modifications predicted using the three different RNA-seq approaches.
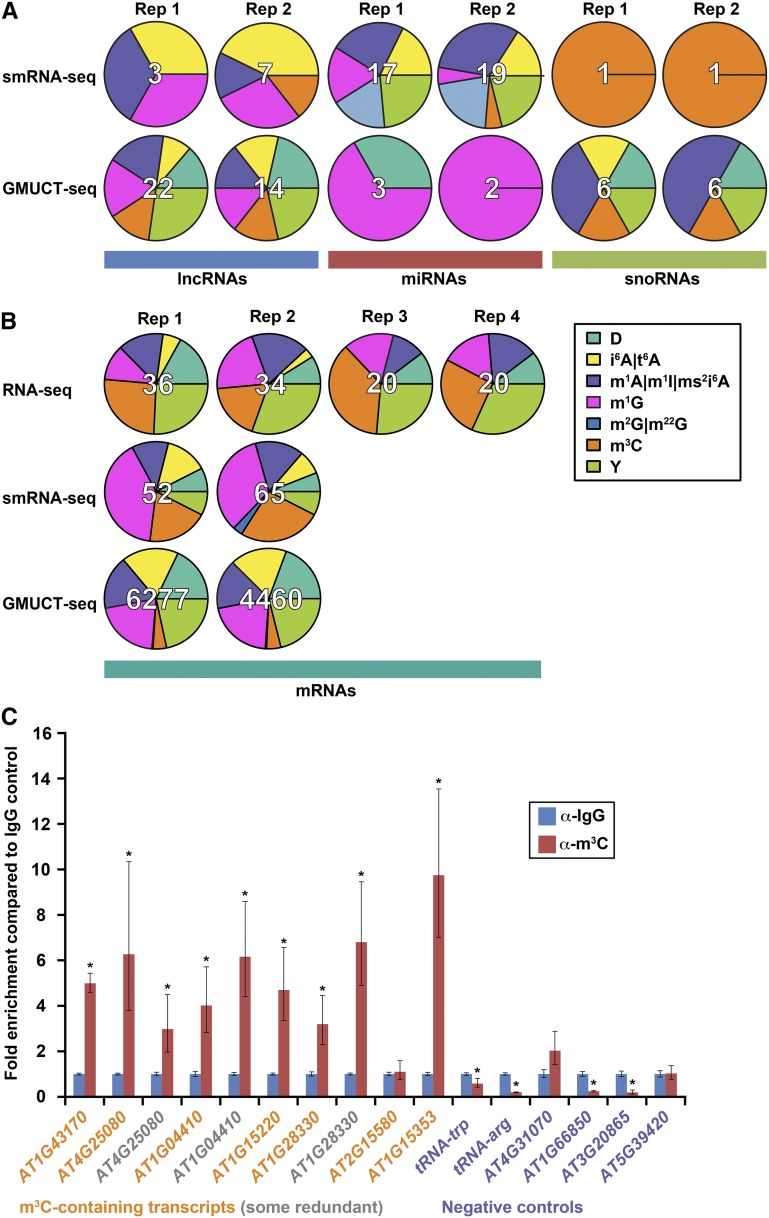
Using this machine learning-based classifier (Figure 1), we identified a wide range of modification types in both noncoding (Figure 4A) and coding RNAs (Figure 4B). Interestingly, the modification types between different classes of RNAs (lncRNAs, miRNAs, snoRNAs, and mRNAs) were quite distinct in their total quantities, but in general mostly consisted of the same few types of modifications. The most common types of modifications that HAMR could distinguish were m3C, Y, m1A, m1G, dihydrouridylation (D), N6-isopentenyladenosylation (i6A), and threonylcarbamoyladenosylation (t6A). In lncRNAs, D and Y sites were only identified for HAMR-predicted modification sites found with GMUCT data (Figure 4A), while m1G, i6A/t6A, m3C, and m1A sites were found using both GMUCT and smRNA-seq data. In miRNAs, we revealed that Y, m1A, i6A/ t6A, and m2G are only observed in smRNA-seq data, but the modification sites identified with the GMUCT data were classified mostly as m1G or D (Figure 4A). For snoRNAs, we uncovered only a single predicted m3C site in both replicates. Conversely, HAMR-predicted modification sites for the GMUCT data sets were a mix of m1A, i6A/t6A, D, Y, and m3C (Figure 3A). In total, these results reveal that different collections of modifications that affect Watson-Crick base pairing are found in noncoding RNAs, including lncRNAs, that have been processed into smRNAs compared with those that are uncapped.
Figure 4.
HAMR Predicts a Variety of Known and Novel Modification Types in the Arabidopsis Transcriptome.
(A) and (B) Distribution of the predicted identity of HAMR modifications in noncoding RNAs (A) and coding RNAs (B), as determined by nearest-neighbor classification using a training set of known tRNA modifications from S. cerevisiae.
(C) Immunoprecipitations of transcripts predicted to contain m3C modifications. RT-qPCR analysis of two transcripts (AT1G43170 and AT4G25080) predicted to contain m3C based upon RNA-seq data, five transcripts (AT1G04410, AT1G15220, AT1G28330, AT2G15580, and AT3G15353) predicted to contain m3C based upon GMUCT, and six transcripts/tRNA families (tRNA-Arg [anticodon: AGT], tRNA-Trp [anticodon: CCA], AT1G66850, AT3G20865, AT4G31070, and AT5G39420) not predicted to contain m3C. The RT-qPCR data for all transcripts was normalized to tRNA-ala (anticodon:AGC), which is well known to not contain m3C in any other organism, making it the most reliable negative control. Fold enrichment over an IgG nonspecific antibody control (y axis) is plotted for each transcript. RT-qPCRs were performed on two biological and three technical replicates. Error bars indicate ± se of the mean. P values were calculated with a Student’s t test, as previously described (Ryvkin et al., 2013). Asterisk denotes P value < 0.05.
In coding mRNAs, we found that the identified modifications included previously characterized adenosine methylation (m1A) and Y sites (Squires et al., 2012; Carlile et al., 2014; Schwartz et al., 2014b), as well as novel cytosine (m3C) and guanosine methylation (m1G), dihydrouridylation (D), N6- isopentenyladenosylation (i6A), and threonylcarbamoyladenosylation (t6A) (Figure 4B; Supplemental Figure 6). As in noncoding RNAs, the distribution of these modification types is distinct between stable RNA, smRNA, and uncapped, degrading transcripts. For instance, m3C and m1G modifications tend to be much more common in stable RNAs and mRNA-derived smRNAs, respectively, compared with the overall distribution of these covalent additions in uncapped, degrading transcripts identified by GMUCT in both Arabidopsis and human data (Figure 3B; Supplemental Figure 6). Conversely, uncapped, degrading mRNAs as identified by HAMR analysis of GMUCT data demonstrate much higher levels of D and i6A /t6A compared with stable mRNAs and mRNA-derived smRNAs in both plants and humans (Figure 4B; Supplemental Figure 6), suggesting that these modifications may be the cause or consequence of protein-coding transcript turnover in eukaryotic transcriptomes. In total, these results reveal that the different collections of transcripts in eukaryotic transcriptomes are marked by distinct distributions of covalent modifications that affect the Watson-Crick base pairing edge.
To experimentally validate both HAMR and the machine learning-based prediction of modification identity, we performed m3C RNA immunoprecipitations on RNAs predicted to contain this modification alongside negative controls with no predicted m3C. Using RT-qPCR on fractions of RNAs immunoprecipitated (IP) with either an antibody specific for m3C or an IgG control, we measured the abundance of two mRNAs predicted to contain m3C using the RNA-seq data, five mRNAs predicted using the GMUCT data, and six mRNAs that were not predicted to contain such modification sites in any of the HAMR analyses (Figure 4C). We normalized RT-qPCR measurements in the two IP fractions to tRNA-ala (anticodon:AGC), which is known to be devoid of m3C in all other eukaryotic organisms and which HAMR does not predict to contain m3C in Arabidopsis (Supplemental Files 1 and 2). Thus, this RNA serves as the most confident negative control locus for our analyses. We found that six of the seven transcripts tested (86%) were significantly (all P values < 0.01, Student’s t test) enriched in the m3C fractions compared with the nonspecific antibody control (Figure 4C). Notably, one of these transcripts (AT4G25080) contained a predicted m3C site within the splice donor sequences (Figure 3A). For the one mRNA (AT2G15580) that was predicted to contain an m3C site but that was not validated by this approach, this result could be a consequence of an incorrect modification site call (part of the 5% false discovery rate [FDR]) or misclassification by the machine learning approach of the HAMR pipeline. Regardless, 86% of the predicted m3C sites could be experimentally validated, providing evidence for the robustness of the identification and classification of modification sites by the HAMR approach (Figure 4C). For the putative negative control loci (those predicted not to contain an m3C site), we found that all of these RNAs had similar or significantly (all P values < 0.01, Student’s t test) lower levels in the m3C IP fractions compared with the IgG control (Figure 4C). These results support the HAMR prediction that these loci truly lack an m3C modification site. In total, these results indicate that, in general, HAMR identified and classified bona fide covalent modification sites that affect the Watson-Crick base-pairing edge within the Arabidopsis and human (Ryvkin et al., 2013) transcriptomes and that these modifications are enriched within degrading mRNAs.
The Proportion of Uncapped Transcripts and Number of HAMR-Predicted Modifications Positively Correlate for Arabidopsis mRNAs
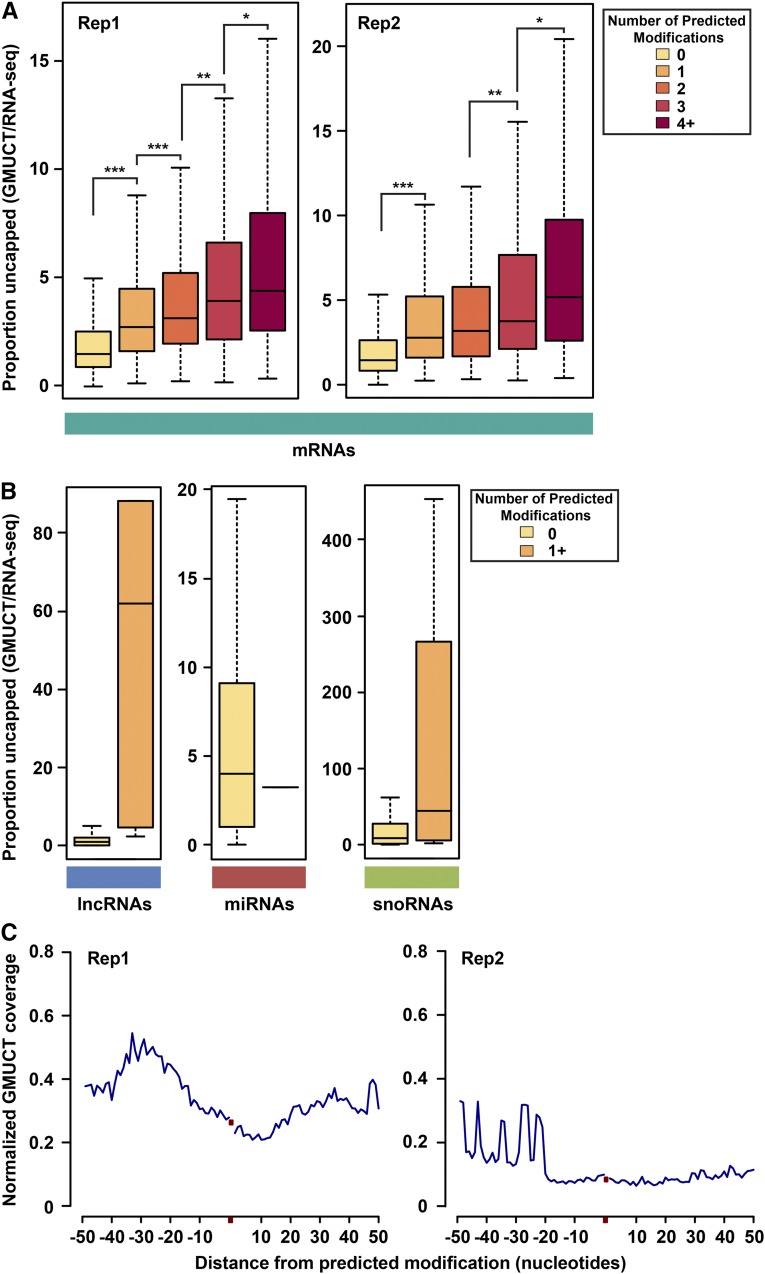
We found that uncapped, degrading transcripts as interrogated by GMUCT were the most enriched class of transcripts for HAMR-predicted covalent modifications within our analyses (Figure 2B; Supplemental Figure 1). Therefore, we wanted to test whether these Watson-Crick base-pairing edge affecting modifications correlate with the proportion of steady state transcripts in an uncapped state (proportion uncapped) (Figure 5A; Supplemental Figure 7A), as measured by GMUCT reads (steady state uncapped population) normalized to RNA-seq reads (steady state total transcript population). We previously used this measure as an approximation of the overall percentage of transcripts that are undergoing turnover (Li et al., 2012). Using this approach, we observed a monotonic increase in the total levels of transcripts that are found in the uncapped and likely degrading fraction of transcripts as the number of predicted modification sites in mRNAs increases (Figure 5A). Interestingly, the majority of these stepwise increases were significant (all P values < 0.01, Wilcoxon rank sum test), and comparison of all transcripts containing HAMR-predicted modifications to all transcripts that are not identified as containing these modifications also yields highly significant differences (P → 0, Wilcoxon rank sum test). Furthermore, we observed the same trends across two independent replicates of GMUCT and RNA-seq (Figure 5A). Similar trends were also observed in human (HEK293T and HeLa) cells, though not all stepwise comparisons reached detectable significance in our analyses (Supplemental Figure 7A).
Figure 5.
Arabidopsis RNAs with HAMR-Predicted Modifications Have Higher Levels of Uncapped Transcripts.
(A) and (B) Distribution of proportion uncapped (total GMUCT reads per transcript normalized to total RNA-seq reads) per transcript for coding mRNAs (A) and a representative replicate for noncoding RNAs (B). P values were calculated with a Wilcoxon rank sum test; one asterisk denotes P value < 0.01, two asterisks denotes P value < 0.001, and three asterisks denotes P value <1 × 10−5. Only a single miRNA was predicted to contain a modification using GMUCT data, so it is represented as a single line.
(C) Averaged GMUCT coverage profiles 50 bp up- and downstream of all predicted mRNA modification sites, normalized to RNA-seq read abundance. Red dots indicate the position of the predicted modification and are plotted within 50 bp up- and downstream flanking regions. Modifications within 50 bp of the mRNA 5′ or 3′ ends were given correspondingly shorter flanking regions.
Interestingly, modified lncRNAs and snoRNAs, but not miRNAs, likewise showed a similar trend, where transcripts with HAMR-predicted modifications had a higher proportion of their populations in the uncapped, degrading proportion of the transcriptome compared with those without these covalent additions, although not at detectable significance. However, this lack of significance is most likely a consequence of the low numbers of detected modification sites in these classes of RNAs (Figures 2B and 5B). In summary, these findings reveal that higher levels of HAMR-predicted covalent modifications in mRNAs in both plants and humans correlate with increased proportions of those transcripts in the uncapped, degrading fraction of transcripts as measured by GMUCT. In total, these findings suggest that covalent RNA modifications that affect the Watson-Crick base-pairing edge are a cause or consequence of RNA turnover in eukaryotic transcriptomes.
Since GMUCT maps the precise position of RNA cleavage events in detected transcripts, we then sought to determine whether the predicted modified positions within mRNAs were in close proximity to specific cleavage events. We tested this because such a finding would suggest that these modifications could be the signal for an RNA cleaving enzyme to initiate the degradation process. To test this idea, we examined the 50 nucleotides up- and downstream of HAMR-predicted modification sites (Figure 5C). This analysis revealed no specific peak or pattern in GMUCT cleavage signal in this 100-bp window surrounding HAMR-predicted modification sites (Figure 5C). These results suggest modification-associated uncapping and RNA turnover does not require a specific cleavage event related to the site of covalent addition but is either a consequence of the degradation process and/or induces the turnover of these transcripts by normal 5′-to-3′ and 3′-to-5′ exonucleolytic mechanisms. Intriguingly, seven transcripts containing HAMR-predicted modifications in the GMUCT data sets overlapped with the set of 33 transcripts recently found to undergo nonsense-mediated decay in an alternative splicing-dependent manner (Kalyna et al., 2012), suggesting nonsense-mediated decay might be one such turnover mechanism. In contrast, HAMR-predicted modification sites in the human (HEK293T and HeLa) cells showed a small peak in average GMUCT cleavage signal directly upstream (Supplemental Figure 7B) of HAMR-predicted modification sites, suggesting that a mechanism of modification-induced cleavage may be active in humans. Thus, HAMR-predicted modifications may function differently in plants and humans. However, this hypothesis will require future testing.
Stress-Responsive mRNAs Are Enriched for RNA Modifications That Affect the Watson-Crick Base-Pairing Edge
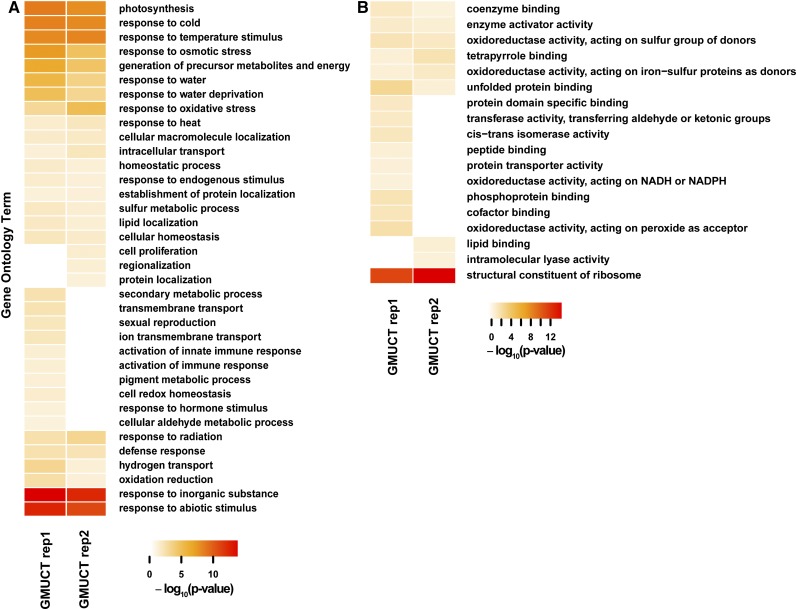
Our finding that HAMR-predicted covalent modifications were enriched in degrading mRNAs as identified by GMUCT (Figure 5) suggested the intriguing possibility that this could be a mechanism for regulating the levels of mRNAs encoding proteins with common cellular functions. To test this hypothesis, we searched for overrepresented Gene Ontology (GO) terms among the collection of modified mRNAs identified using the GMUCT data. To reduce any bias in reporting GO terms for this collection of mRNAs, we identified all GO terms within three branches of the “biological process” and “molecular function” roots, as determined by a depth first search (Vandivier et al., 2013). From this analysis, we observed a significant (FDR < 0.05) enrichment for transcripts encoding ribosomal proteins for both Arabidopsis and human uncapped transcripts identified by GMUCT (Figure 6; Supplemental Figure 8). Additionally, for Arabidopsis uncapped, degrading transcripts containing HAMR-predicted modifications, we also observed a significant (FDR < 0.05) enrichment of transcripts encoding proteins involved in photosynthesis, as well as a variety of biotic and abiotic stress response terms, including “defense response,” “response to water,” “response to cold,” “response to heat,” “response to radiation,” and “response to oxidative stress” (Figure 6A). Relatedly, for human uncapped, degrading transcripts containing HAMR-predicted modifications identified by GMUCT, we found significant (FDR < 0.05) enrichment of transcripts encoding proteins involved in “cell death” and “cell cycle” (Supplemental Figure 8A). Conversely, we did not observe any measurable enrichment for the transcripts with HAMR-predicted modifications in our smRNA-seq and RNA-seq data sets, which is likely a consequence of the low levels of these covalent additions identified by HAMR analysis of data from these approaches. In total, the overrepresentation of certain biological functions such as stress responses and cell cycle among uncapped transcripts with HAMR-predicted modifications but not in stable mRNAs or mRNA-derived smRNAs suggests that addition of modifications that affect the Watson-Crick base-pairing edge targets specific sets of transcripts for degradation to maintain their proper levels in the cell. This hypothesis will require further testing.
Figure 6.
Arabidopsis Transcripts with HAMR-Predicted Modifications Encode Proteins with Coherent Functions.
Biological process (A) and molecular function (B) GO terms are reported if they are significantly enriched (FDR < 0.05) over a background of all “HAMR-accessible transcripts” with at least 100 uniquely mapping reads. Analyses were performed using the DAVID package (Huang et al., 2009). Terms are only reported if they are separated from their ancestor term by no more than two parents, as determined by a depth first search as previously described (Vandivier et al., 2013). Lack of color denotes lack of significance.
In conclusion, we present evidence that covalent modifications of mRNA bases that affect the Watson-Crick base-pairing edge are strongly enriched in uncapped, degrading mRNAs in both Arabidopsis and two human cell lines and are usually found within exonic portions of these transcripts. In contrast, the identified modifications in stable mRNAs tend to occur in alternatively spliced introns of protein-coding transcripts and often accumulate in or near the splice donor and acceptor sites. Together, these results suggest a potential role for HAMR-predicted modifications in modulating specific alternative splicing events. Moreover, we found that specific HAMR-predicted modifications tend to occur in stable mRNAs (e.g., m3C), whereas others tend to label uncapped, degrading transcripts (e.g., i6A). These results suggest that certain classes of chemical modifications mark transcripts that are being degraded in eukaryotic transcriptomes. However, whether this is a cause or consequence of the RNA degradation process requires further investigation. Finally, we found that mRNA modifications mark transcripts that encode proteins with specific functions, many of which are involved in stress responses in both Arabidopsis and humans. These results suggest that modifications mark these classes of mRNA molecules for degradation to maintain them as mostly unstable during normal development, as was profiled in our experiments here. However, this hypothesis will require future testing during specific stress responses in both Arabidopsis and humans. In total, our study provides a resource for studying mRNA chemical modifications that affect the Watson-Crick base-pairing edge and identifies a potentially novel mechanism for initiating and/or maintaining mRNA degradation in eukaryotic transcriptomes.
METHODS
Plant Materials
Immature flower bud clusters from the Columbia (Col-0) ecotype of Arabidopsis thaliana grown under 16-h-light/8-h-dark cycles using 2800 lumen, 4100K fluorescent light bulbs at 22°C were used for all experiments and analyses described in this study.
Human Materials
HeLa and HEK293T cells were seeded in 15-cm standard Corning tissue culture dishes (Sigma-Aldrich) and grown to 90% confluence (∼18 million cells) in DMEM medium (Life Technologies) supplemented with l-glutamine, 4.5 g/L d-glucose, 10% fetal bovine serum (Atlanta Biologics), and Pen/Strep (Fisher Scientific).
RNA Extraction
For Arabidopsis, bud tissue was ground with a mortar and pestle under liquid nitrogen. For human cell lines, cells were scraped, pelleted, and homogenized. For both Arabidopsis and human cell lines, RNA was extracted using Qiazol (Qiagen) and further purified with the miRNeasy mini kit (Qiagen) per the manufacturer’s protocol.
Library Preparation and Sequencing
RNA-seq, smRNA-seq, and GMUCT libraries were constructed as previously described (Gregory et al., 2008; Li et al., 2012; Willmann et al., 2014). Both RNA-seq and GMUCT libraries were subjected to two rounds of poly(A)+ selection using oligo(dT) Dynabeads (Thermo Fisher Scientific). All libraries were ligated to TruSeq smRNA adaptors (Illumina) and were sequenced on an Illumina HiSequation 2000 (Illumina) using the 50-bp single-end sequencing approach. All sequencing was performed according to the manufacturer’s instructions.
Previously Published Data Sets
Human RNA-seq data for HeLa cells were downloaded from the ENCODE Caltech RNA-seq compendium (Gene Expression Omnibus [GEO] accession number GSM958739) (ENCODE Project Consortium, 2012). Human RNA-seq data for HEK293T cells were downloaded from GEO accession GSE34995 (Huelga et al., 2012). Human GMUCT data were downloaded from GEO accession GSE47121 (Willmann et al., 2014). Additional plant smRNA-seq data were downloaded from GEO accession GSE57215 (Li et al., 2015). RT stalling data (Structure-seq) were downloaded from SRA accession SRP027216 (Ding et al., 2014).
Genome Annotation
All analyses in plants were performed using the TAIR10 genome assembly, and all analyses in humans were performed using the UCSC hg19 RefSeq assembly. Alternative and constitutive introns were identified using the TAIR10 transcriptome annotation, as well as the AtRTD alternate transcriptome annotation (https://ics.hutton.ac.uk/atRTD/) (Zhang et al., 2015). Repeat-subtracted genomes (repeat-masked) for TAIR10 were produced with the RepeatMasker package (A.F.A. Smit, R. Hubley, and P. Green, 2013; RepeatMasker Open-4.0, http://www.repeatmasker.org).
Read Processing and Alignment
Read processing and alignment were performed as previously described (Li et al., 2012) with slight modifications. Briefly, sequencing reads were first trimmed to remove 3′ sequencing adapters. For libraries where the expected range of insert lengths are all less than the read length (i.e., smRNA-seq libraries), only trimmed reads were retained. For libraries where the expected range of insert lengths are all greater than the read length (i.e., RNA-seq libraries), only untrimmed reads were retained. For libraries where the expected range of insert lengths includes insert lengths of both classes (i.e., GMUCT libraries), trimmed and untrimmed reads were concatenated and aligned together. Reads were aligned to the Arabidopsis genome version TAIR10 or the human genome version hg19. Only uniquely mapping reads were allowed, except for tRNA analyses (see below).
tRNA Read Processing and Alignment
tRNA amino acid-anticodon families were annotated with tRNAscan (Lowe and Eddy, 1997). For each amino acid-anticodon family of tRNAs, a consensus sequence was constructed through multiple alignment of all loci with LocARNA (Will et al., 2007) and selection of the most abundant nucleotide at each aligned position. Any consensus nucleotides with biallelic SNPs were retained since HAMR will filter these in hypothesis testing, while a few rare triallelic SNPs were excluded since these could potentially lead to HAMR artifacts. smRNA reads were first aligned to the Arabidopsis genome version TAIR10, allowing multimappers. Reads that mapped exclusively to tRNAs were retained. This subset of reads was then remapped to the tRNA consensus sequence set. Downstream analyses were performed using consensus coordinates, as described previously (Ryvkin et al., 2013).
HAMR
HAMR was performed as previously described (Ryvkin et al., 2013). For each set of mapped reads, deviations from the reference sequence (mismatches) with a quality score >30 (error rate <0.001) are tabulated for each base in either the Arabidopsis genome version TAIR10, human genome version hg19, or TAIR10 tRNA consensus sequence set. Each base with mismatches was tested for significant enrichment of mismatches using a binomial distribution, with the conservative assumption that the sequencing error rate is 0.01. Bases that pass this filter are then tested against the null hypothesis that the genotype is biallelic. Each possible biallelic genotype is tested, again using a binomial distribution. Significant deviation from all possible biallelic genotypes is used as evidence of modification, as modification-induced errors should be semirandom and not have a clear bias toward any single base substitution, as would be true with SNPs or RNA editing (Ryvkin et al., 2013). Each predicted modified base was then classified using nearest-neighbor machine learning, as described previously (Ryvkin et al., 2013). Known tRNA modifications in Saccharomyces cerevisiae (from the MODOMICS database) (Dunin-Horkawicz et al., 2006) were used previously (Ryvkin et al., 2013) to construct the training set.
Definition of HAMR-Accessible Bases and Transcripts
In Arabidopsis, the minimum base coverage at an observed modification in either GMUCT, smRNA-seq, or RNA-seq was always 50 reads per base (50×). Thus, any base with at least 50× coverage was designated as HAMR-accessible. For comparison, the minimum coverage for humans, though not included in any analyses, was 10×. The minimum number of uniquely mapping reads to call a transcript as modified was 100 for Arabidopsis and 10 for humans. Thus, transcripts with at least 100 or 10 uniquely mapping reads were designated as HAMR-accessible in Arabidopsis and humans, respectively.
RNA Immunoprecipitation
Total RNA was immunoprecipitated with 10 μg of an undiluted IgG nonspecific control antibody (Cell Signaling) or an anti-3-methylcytosine (m3C) antibody (Active Motif). Forty microliters of Dynabeads Protein A (Thermo Fisher Scientific) were washed with 1× Dulbecco’s phosphate-buffered saline (DPBS; Thermo Fisher Scientific) and coupled to the 10 μg of antibody in DPBS by rocking at room temperature for 1 h. Beads were washed again twice with DPBS. Five micrograms of RNA was denatured at 70°C for 5 min, placed on ice for 3 min, and then incubated with the bead-linked antibodies in IP buffer (140 mM NaCl, 0.05% [v/v] Triton X-100, and 10 mM Tris, all from ultrapure, RNase-free stocks dissolved in DEPC-treated water and filter sterilized at 0.22 μM). Bead/RNA mix was rocked at 4°C for 2 h. Bound RNA was washed three times in IP buffer and then eluted in Trizol (Thermo Fisher Scientific), precipitated, and washed.
RT-qPCR
Primers were designed using PrimerBlast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). tRNA primers were designed against tRNA family consensus sequences. Primer sequences are listed in Supplemental Table 2. RNA was reverse transcribed using random hexamers and then preamplified with SsoAdvanced PreAmp Supermix (Bio-Rad Laboratories) per the manufacturer’s protocol using a mix of all primers listed above. Quantitative PCR was performed using SYBR Green 2X master mix (Thermo Fisher Scientific) in a StepOne machine (Thermo Fisher Scientific) on two biological and three technical replicates.
GO Enrichment
GO enrichment analyses of transcripts with predicted modifications was performed using the DAVID online tool (Huang et al., 2009) as previously described (Vandivier et al., 2013). All HAMR-accessible transcripts (i.e., those with comparable coverage to modified transcripts) were used as the background set for this analysis.
Statistical Analyses
All statistical analyses were performed using the R software package (http://www.r-project.org/), including P values for all hypothesis testing. See figure legends for specific statistical tests used to assess significance.
Accession Numbers
All smRNA-seq, RNA-seq, and GMUCT data generated for this study were deposited in the GEO under accession number GSE66224. Additionally, HAMR-predicted modifications are available under the same GEO accession or at http://gregorylab.bio.upenn.edu/HAMR_degradome/. Sequence data for genes mentioned in this article can be found in the GenBank/EMBL libraries under accession numbers NM_100321 for AT1G04410; NM_202105 and NM_101390 for AT1G15220; NM_179390, NM_102599, NM_001160906, and NM_179389 for AT1G28330; NM_202237, NM_103469, NM_001036069, NM_001084202, and NM_103496 for AT1G43170; NM_105356 for AT1G66850; NM_127119 for AT2G15580; NM_112401 for AT3G15353; NM_112978 for AT3G20865; NM_118030, NM_179076, and NM_001084939 for AT4G19110; NM_179108, NM_118640, NM_179107, and NM_001125580 for AT4G25080; NM_119257 for AT4G31070; NM_120012, NM_001036731, NM_001036730, and NM_202978 for AT4G38510; and NM_123304 for AT5G39420.
Supplemental Data
Supplemental Figure 1. HAMR-predicted modifications in two human cell lines.
Supplemental Figure 2. Differences in the number of HAMR-predicted modifications are not artifacts of differences in library preparation, overall size, or transcriptome coverage.
Supplemental Figure 3. HAMR captures a large proportion of known tRNA modification sites in the Arabidopsis transcriptome.
Supplemental Figure 4. Sites of HAMR-predicted modifications are enriched in reverse transcriptase stalls.
Supplemental Figure 5. HAMR-predicted modifications in two human cell lines mark uncapped and alternatively spliced transcripts.
Supplemental Figure 6. HAMR predicts a variety of known and novel modification types in the human transcriptome.
Supplemental Figure 7. Human RNAs with HAMR-predicted modifications have higher levels of uncapped transcripts.
Supplemental Figure 8. Human transcripts with HAMR-predicted modifications encode proteins with coherent functions.
Supplemental Table 1. HAMR correctly classifies a portion of homology-based predicted tRNA locus modification sites.
Supplemental Table 2. Primer sequences used for RT-qPCR.
Supplemental File 1. Homology-based prediction of Arabidopsis tRNA family modification sites.
Supplemental File 2. Homology-based prediction of Arabidopsis tRNA locus modification sites.
Supplementary Material
Acknowledgments
We thank members of the Wang and Gregory labs for their helpful discussions and comments on the article. This work was funded by the National Science Foundation (Career Award MCB-1053846, MCB-1243947, and IOS-1444490 to B.D.G.) and the National Institute of General Medical Sciences (R01-GM099962 to L.-S.W. and B.D.G. and 5T32GM007229-37 to L.E.V.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
AUTHOR CONTRIBUTIONS
B.D.G. designed the study. L.E.V., R.C., P.P.K., and I.M.S. compiled and performed the RNA-seq and validation experiments with input from B.D.G. L.E.V., R.C., and P.P.K. compiled all annotation data and carried out the computational analysis with input from B.D.G. and L.-S.W. L.E.V. and B.D.G. wrote the article. All authors read and approved the article.
Glossary
- HAMR
high-throughput annotation of modified ribonucleotides
- RT
reverse transcriptase
- SNP
single nucleotide polymorphism
- GMUCT
global mapping of uncapped and cleaved transcripts
- smRNA-seq
small RNA sequencing
- RNA-seq
RNA sequencing
- lncRNA
long noncoding RNA
- snoRNA
small nucleolar RNA
- miRNA
microRNA
- DMS
dimethyl sulfate
- UTR
untranslated region
- IP
immunoprecipitated
- GO
Gene Ontology
- FDR
false discovery rate
- DPBS
Dulbecco’s phosphate-buffered saline
- GEO
Gene Expression Omnibus
Footnotes
Articles can be viewed online without a subscription.
References
- Björk G.R., Ericson J.U., Gustafsson C.E.D., Hagervall T.G., Jönsson Y.H., Wikström P.M. (1987). Transfer RNA modification. Annu. Rev. Biochem. 56: 263–287. [DOI] [PubMed] [Google Scholar]
- Carlile T.M., Rojas-Duran M.F., Zinshteyn B., Shin H., Bartoli K.M., Gilbert W.V. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova J.A., et al. (2007). Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131: 1340–1353. [DOI] [PubMed] [Google Scholar]
- Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M. (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505: 696–700. [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. [DOI] [PubMed] [Google Scholar]
- Dunin-Horkawicz S., Czerwoniec A., Gajda M.J., Feder M., Grosjean H., Bujnicki J.M. (2006). MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 34: D145–D149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B., Bailly M., de Crécy-Lagard V. (2012). Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 46: 69–95. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley S.W., Vandivier L.E., Kuksa P.P., Gregory B.D. (2015). Transcriptome-wide measurement of plant RNA secondary structure. Curr. Opin. Plant Biol. 27: 36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani S., Lawrenson T., Woodward C., Headon D., Sablowski R. (2004). A link between mRNA turnover and RNA interference in Arabidopsis. Science 306: 1046–1048. [DOI] [PubMed] [Google Scholar]
- Gregory B.D., O’Malley R.C., Lister R., Urich M.A., Tonti-Filippini J., Chen H., Millar A.H., Ecker J.R. (2008). A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell 14: 854–866. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Szweykowska-Kulinska Z., Motorin Y., Fasiolo F., Simos G. (1997). Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: a review. Biochimie 79: 293–302. [DOI] [PubMed] [Google Scholar]
- Hopper A.K., Phizicky E.M. (2003). tRNA transfers to the limelight. Genes Dev. 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Horowitz S., Horowitz A., Nilsen T.W., Munns T.W., Rottman F.M. (1984). Mapping of N6-methyladenosine residues in bovine prolactin mRNA. Proc. Natl. Acad. Sci. USA 81: 5667–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huelga S.C., Vu A.Q., Arnold J.D., Liang T.Y., Liu P.P., Yan B.Y., Donohue J.P., Shiue L., Hoon S., Brenner S., Ares M. Jr., Yeo G.W. (2012). Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Reports 1: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.M., Ares M. Jr. (1991). Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 10: 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40: 2454–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R. (2014). The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 42: 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-László Z., Henry Y., Bachellerie J.-P., Caizergues-Ferrer M., Kiss T. (1996). Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Lee M., Kim B., Kim V.N. (2014). Emerging roles of RNA modification: m(6)A and U-tail. Cell 158: 980–987. [DOI] [PubMed] [Google Scholar]
- Li F., Zheng Q., Vandivier L.E., Willmann M.R., Chen Y., Gregory B.D. (2012). Regulatory impact of RNA secondary structure across the Arabidopsis transcriptome. Plant Cell 24: 4346–4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.B., Levanon E.Y., Yoon J.-K., Aach J., Xie B., Leproust E., Zhang K., Gao Y., Church G.M. (2009). Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213. [DOI] [PubMed] [Google Scholar]
- Li S., Vandivier L.E., Tu B., Gao L., Won S.Y., Li S., Zheng B., Gregory B.D., Chen X. (2015). Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 25: 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T.M., Eddy S.R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka M.A., et al. (2013). MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 41: D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin P., Leung Y.Y., Silverman I.M., Childress M., Valladares O., Dragomir I., Gregory B.D., Wang L.S. (2013). HAMR: high-throughput annotation of modified ribonucleotides. RNA 19: 1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., et al. (2014a). Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Reports 8: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Bernstein D.A., Mumbach M.R., Jovanovic M., Herbst R.H., León-Ricardo B.X., Engreitz J.M., Guttman M., Satija R., Lander E.S., Fink G., Regev A. (2014b). Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T. (2012). Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40: 5023–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M., Durant P.C., Davis D.R. (2000). Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 39: 12575–12584. [DOI] [PubMed] [Google Scholar]
- Talkish J., May G., Lin Y., Woolford J.L. Jr., McManus C.J. (2014). Mod-seq: high-throughput sequencing for chemical probing of RNA structure. RNA 20: 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandivier L., Li F., Zheng Q., Willmann M., Chen Y., Gregory B. (2013). Arabidopsis mRNA secondary structure correlates with protein function and domains. Plant Signal. Behav. 8: e24301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann M.R., Berkowitz N.D., Gregory B.D. (2014). Improved genome-wide mapping of uncapped and cleaved transcripts in eukaryotes--GMUCT 2.0. Methods 67: 64–73. [DOI] [PubMed] [Google Scholar]
- Will S., Reiche K., Hofacker I.L., Stadler P.F., Backofen R. (2007). Inferring noncoding RNA families and classes by means of genome-scale structure-based clustering. PLOS Comput. Biol. 3: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson S.A., Muller J.G., Burrows C.J., Rokita S.E. (1993). A primer extension assay for modification of guanine by Ni(II) complexes. Nucleic Acids Res. 21: 5524–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff B.-E., Sakurai M., Nishikura K. (2011). Elucidating the inosinome: global approaches to adenosine-to-inosine RNA editing. Nat. Rev. Genet. 12: 81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., et al. (2015). AtRTD - a comprehensive reference transcript dataset resource for accurate quantification of transcript-specific expression in Arabidopsis thaliana. New Phytol. 208: 96–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.