D53-like SMXLs regulate shoot branching and leaf development through the strigolactone signaling pathway by forming a complex with TPR2 and repressing transcription in Arabidopsis.
Abstract
Strigolactones (SLs) are carotenoid-derived phytohormones that control many aspects of plant development, including shoot branching, leaf shape, stem secondary thickening, and lateral root growth. In rice (Oryza sativa), SL signaling requires the degradation of DWARF53 (D53), mediated by a complex including D14 and D3, but in Arabidopsis thaliana, the components and mechanism of SL signaling involving the D3 ortholog MORE AXILLARY GROWTH2 (MAX2) are unknown. Here, we show that SL-dependent regulation of shoot branching in Arabidopsis requires three D53-like proteins, SUPPRESSOR OF MORE AXILLARY GROWTH2-LIKE6 (SMXL6), SMXL7, and SMXL8. The smxl6 smxl7 smxl8 triple mutant suppresses the highly branched phenotypes of max2 and the SL-deficient mutant max3. Overexpression of a mutant form of SMXL6 that is resistant to SL-induced ubiquitination and degradation enhances shoot branching. Exogenous application of the SL analog rac-GR24 causes ubiquitination and degradation of SMXL6, 7, and 8; this requires D14 and MAX2. D53-like SMXLs form complexes with MAX2 and TOPLESS-RELATED PROTEIN2 (TPR2) and interact with D14 in a GR24-responsive manner. Furthermore, D53-like SMXLs exhibit TPR2-dependent transcriptional repression activity and repress the expression of BRANCHED1. Our findings reveal that in Arabidopsis, D53-like SMXLs act with TPR2 to repress transcription and so allow lateral bud outgrowth but that SL-induced degradation of D53-like proteins activates transcription to inhibit outgrowth.
INTRODUCTION
Strigolactones (SLs) are a group of carotenoid-derived lactones produced by plants and initially characterized as rhizosphere signals that enable root-parasitic plants to detect their hosts (Cook et al., 1966) and then as signals for mycorrhizal fungi to form symbiotic associations (Akiyama et al., 2005). More recently, SLs have been reported to function as root-to-shoot phytohormones that suppress shoot branching by inhibiting the outgrowth of axillary buds (Gomez-Roldan et al., 2008; Umehara et al., 2008). Components of SL biosynthesis and signaling have been identified through analysis of highly branched mutants in dicotyledonous plants and of high tillering dwarf mutants in rice (Oryza sativa). These include more axillary growth (max) in Arabidopsis thaliana, tillering dwarf mutants (d mutants) in rice, ramosus in pea (Pisum sativum), and decreased apical dominance (dad) in Petunia hybrida (Ferguson and Beveridge, 2009; Xie et al., 2010; Bennett and Leyser, 2014; Smith and Li, 2014; Xiong et al., 2014; Al-Babili and Bouwmeester, 2015). These studies reveal a conserved role for SLs in regulating axillary bud outgrowth (Domagalska and Leyser, 2011; Ruyter-Spira et al., 2013). SLs also control root development, leaf morphology, leaf senescence, shoot gravitropism, and secondary growth of the stem (Ruyter-Spira et al., 2013; Sang et al., 2014).
During the first step of SL biosynthesis, all-trans-β-carotene is isomerized by DWARF27 (D27) at the C-9 position to form 9-cis-β-carotene (Lin et al., 2009; Alder et al., 2012). Subsequent cleavages by carotenoid cleavage dioxygenases 7 and 8 (CCD7 and CCD8) produce carlactone, a key endogenous precursor of SLs (Sorefan et al., 2003; Booker et al., 2004; Seto et al., 2014). Recently, the Arabidopsis cytochrome P450 MAX1 was demonstrated to catalyze the conversion of carlactone to carlactonoic acid, which is further converted to a strigolactone-like compound, methyl carlactonoate (Abe et al., 2014). However, in rice, MAX1 orthologs were found to mediate the stereo-selective conversion of (Z)-(11R)-carlactone predominately into ent-2'-epi-5-deoxystrigol (also known as 4-deoxyorobanchol [4DO]) and then to orobanchol (Zhang et al., 2014). The MAX1 proteins apparently catalyze the formation of a range of different SLs all with a common butenolide moiety, but researchers commonly employ the chemically synthesized analog GR24 (Scaffidi et al., 2014; Al-Babili and Bouwmeester, 2015), which is usually prepared and supplied as a mixture of two stereoisomers, one with the configuration of 5-deoxystrigol (5DS) and the other, its enantiomer (ent-5DS). This racemic mixture, known as rac-GR24, effectively stimulates SL signaling, but the enantiomer can stimulate the karrikin signaling pathway (Scaffidi et al., 2014). Therefore, purified GR24 stereoisomers with the configuration of 5DS or 4DO, designated as GR245DS and GR244DO, respectively, are used to stimulate SL signaling specifically (Scaffidi et al., 2014; Zhao et al., 2015).
Perception and signaling of SL in rice requires its interaction with D14, a proposed SL receptor, which interacts with D3, an F-box protein of the Skp-Cullin-F-box (SCF) E3 ubiquitin ligase complex (Smith and Li, 2014). The crystal structures of D14 proteins from rice, petunia (DAD2), and Arabidopsis ( D14) revealed an α/β-fold hydrolase with a large internal cavity capable of accommodating SLs and containing a canonical catalytic triad (Hamiaux et al., 2012; Bythell-Douglas et al., 2013; Kagiyama et al., 2013; Zhao et al., 2013, 2015). The catalytic triad of DAD2/D14 is required for hydrolysis of SLs, which can trigger interaction between DAD2 and MAX2 in petunia and interaction between D14 and D3 in rice, as part of SCF-mediated signal transduction (Hamiaux et al., 2012; Smith and Waters, 2012; Jiang et al., 2013; Zhou et al., 2013; Zhao et al., 2014, 2015). Furthermore, the crystal structure of D14 incubated with rac-GR24 revealed a derivative of the butenolide covalently attached to the active-site Ser residue (Zhao et al., 2013). It was proposed that the SL-induced D14-D3 complex could target proteins for ubiquitination and degradation, since that is the typical function of SCF-E3 complexes.
Rice D53 is a target for SL-dependent degradation by the SCFD3 ubiquitination complex (Jiang et al., 2013; Zhou et al., 2013). The d53 dominant mutant shows high tillering and dwarf phenotypes and is resistant to exogenous application of rac-GR24. Treatment of the wild type with rac-GR24 causes D53 degradation via the ubiquitin-proteasome system in a D14- and D3-dependent process. The D53 protein was further shown to interact with members of the TOPLESS-RELATED PROTEIN (TPR) family of transcriptional corepressors, which could potentially repress the activities of its downstream transcription factors (Smith and Li, 2014; Xiong et al., 2014).
A protein related to D53 was independently identified in Arabidopsis by virtue of its role in seed germination in response to karrikins (KARs), compounds in smoke from wildfires. KARs are structurally similar to SLs and also depend on MAX2 for signaling, but instead of D14, they employ a paralog known as KARRIKIN INSENSITIVE2 (KAI2) (Nelson et al., 2011; Waters et al., 2012). Selection of a suppressor of the max2 seed dormancy and long hypocotyl phenotype led to the identification of SUPPRESSOR OF MAX2-1 (SMAX1) (Stanga et al., 2013). Arabidopsis contains seven genes closely related to SMAX1, designated SMAX1-LIKE (SMXL2 to SMXL8). Three of these proteins (SMXL6, SMXL7, and SMXL8) share 36 to 41% identity with D53 (Jiang et al., 2013; Stanga et al., 2013), and we designate them D53-like SMXLs. The D53-like SMXLs are expressed in leaves and axillary branches and are induced by rac-GR24, suggesting that they may have a role in shoot development (Stanga et al., 2013). In this study, we show that D53-like SMXLs regulate shoot branching and leaf development through the SL signaling pathway. These SMXL proteins can form a complex with the TPR2 protein and function as repressors of transcription but are targeted for degradation by SL-dependent interaction of D14 with SMXLs and with MAX2, leading to repression of outgrowth of axillary buds in Arabidopsis.
RESULTS
D53-Like SMXLs Regulate Shoot Branching in Arabidopsis
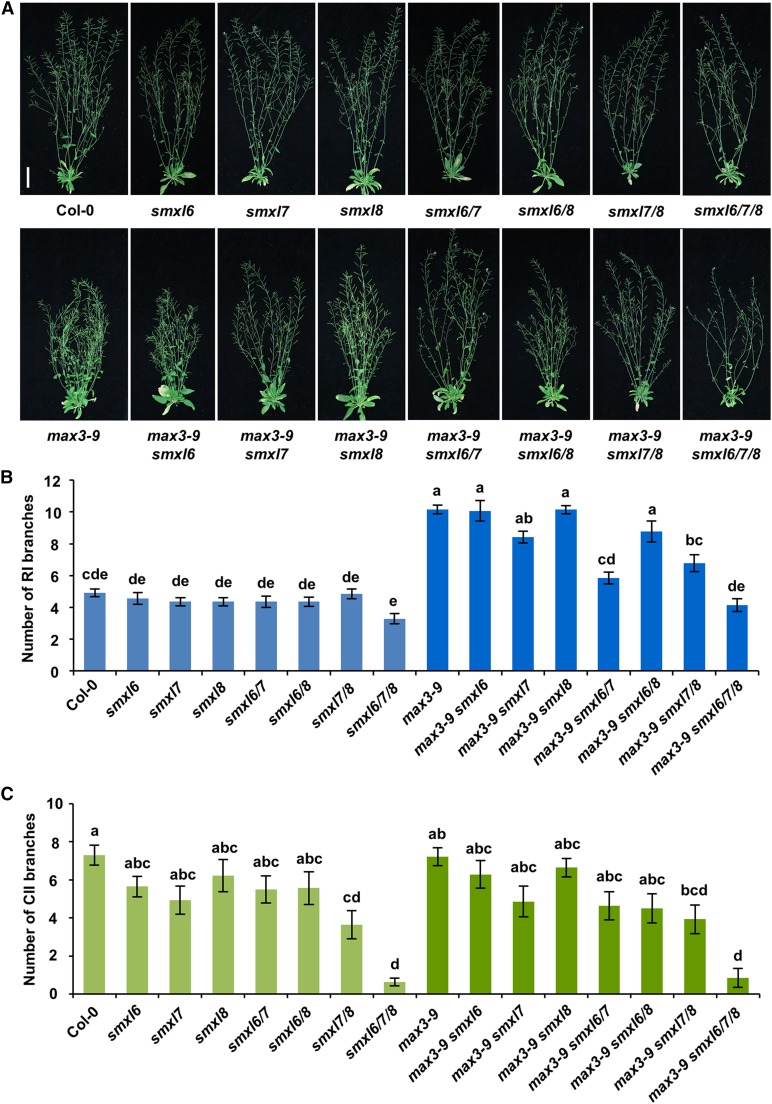
Bioinformatic analysis revealed that Arabidopsis has three orthologs of rice D53: SMXL6 (D53-like-2), SMXL7 (D53-like-1), and SMXL8 (D53-like-3) (Supplemental Figure 1), which, here, we refer to collectively as D53-like SMXLs. To investigate the roles of SMXL6, 7, and 8 in the Arabidopsis SL signaling pathway, we systematically characterized the smxl6, smxl7, and smxl8 mutants and their double and triple mutants in depth. Sequencing analyses of smxl6, smxl7, and smxl8 mutant plants confirmed that there are T-DNA insertions in the first intron of SMXL6, the third exon of SMXL7, and the first exon of SMXL8 (Supplemental Figure 2A), which disrupt the transcription of each target gene, but not the other two genes (Supplemental Figure 2B). The SMXL7 transcript was hardly detected in the smxl7 mutant, suggesting that this is effectively a null mutant, and although transcripts upstream of the T-DNA insertion were detected in smxl6 and smxl8, any protein produced would be severely truncated and lack EAR motifs, so would be expected to be nonfunctional (Supplemental Figure 2C). These smxl mutants and the double mutants of smxl6/7 and smxl6/8 displayed similar phenotypes to the wild type, while the double mutants smxl6/7 and smxl7/8 partially repressed the highly branched phenotype of max3-9, a SL biosynthesis mutant (Booker et al., 2004), and smxl7/8 formed fewer secondary cauline branches (Figures 1A to 1C). Moreover, the triple mutant smxl6/7/8 completely restored max3-9 and displayed a significant decrease in secondary cauline branch number, suggesting that SMXL6, SMXL7, and SMXL8 function redundantly in promoting the outgrowth of axillary buds. In addition, smxl6/7 and smxl7/8 partially rescued the multiple-branching phenotype of max2-1, an important SL signaling mutant (Stirnberg et al., 2002, 2007), and the smxl6/7/8 triple mutant was able to completely rescue max2-1, suggesting that MAX2 could be required for removal of D53-like SMXLs (Supplemental Figure 3).
Figure 1.
Shoot Architecture of smxl Mutants and Suppression of Shoot Branching in the max3-9 Mutant.
(A) Shoots of representative plants after 7 weeks of growth in a 16-h-light/8-h-dark photoperiod. All mutations are in the Col-0 background, and genotypes of mutants are as indicated. Bar = 5 cm.
(B) Number of primary rosette (RI) branches of at least 0.5 cm recorded in plants shown in (A). Values are means ± se (n = 14); significant differences revealed by Tukey's multiple comparison test are indicated by letters above bars (P < 0.05).
(C) Number of secondary cauline (CII) branches of at least 0.5 cm recorded in plants shown in (A). Values are means ± se (n = 14); significant differences revealed by Tukey's multiple comparison test are indicated by letters above bars (P < 0.05).
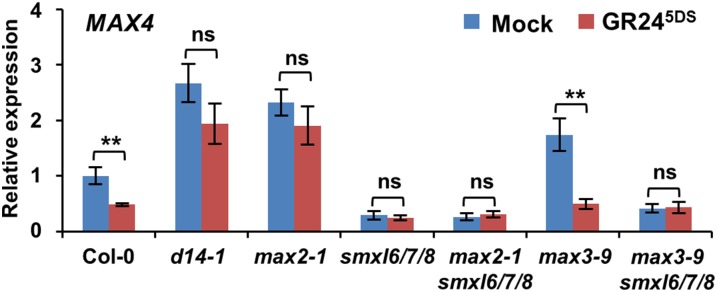
The expression of MAX4, which is involved in the SL biosynthetic pathway, is subject to feedback inhibition through the SL signaling system (Mashiguchi et al., 2009). We therefore compared the expression levels of MAX4 in the wild-type, max3-9, smxl6/7/8, max3-9 smxl6/7/8, d14-1, and max2-1 seedlings in response to GR245DS, a GR24 stereoisomer that regulates hypocotyl elongation and secondary shoot growth in a D14-dependent manner (Scaffidi et al., 2014). As expected, expression of MAX4 increased in d14-1, max2-1, and max3-9 plants, but it decreased in smxl6/7/8, max3-9 smxl6/7/8, and max2-1 smxl6/7/8 plants (Figure 2). Furthermore, expression of MAX4 was greatly downregulated by GR245DS treatment in the wild type and max3-9, whereas it remained stable in d14-1, max2-1, smxl6/7/8, max3-9 smxl6/7/8, and max2-1 smxl6/7/8 (Figure 2), suggesting that SL signaling is disrupted by mutations in SMXL6, SMXL7, and SMXL8.
Figure 2.
Expression of MAX4 Is Repressed in smxl6/7/8 and Is Unresponsive to GR245DS.
Expression of MAX4 in 10-d-old seedlings of Col-0 and the indicated mutants, each treated with 5 µM GR245DS in 0.5× MS liquid medium for 4 h. Values are means ± se (n = 3); ns, no significant difference; **P < 0.01 indicated by Student’s t test.
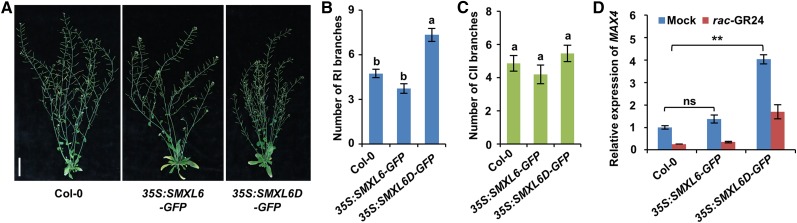
We further generated 35S:SMXL6-GFP and 35S:SMXL6D-GFP transgenic plants and investigated the effects of SMXL6D on shoot branching. The SMXL6D protein bears a deletion of four amino acid residues that were also mutated or deleted in d53 in rice (Supplemental Figure 4) and conferred resistance to degradation (Jiang et al., 2013; Zhou et al., 2013). The 35S:SMXL6-GFP plants displayed similar phenotypes to the wild type, whereas the 35S:SMXL6D-GFP plants formed more primary rosette branches and secondary cauline branches (Figures 3A to 3C; Supplemental Figure 5). Consistent with this phenomenon, compared with the wild type, 35S:SMXL6D-GFP plants had more than a 3-fold increase in the expression level of MAX4, while 35S:SMXL6-GFP plants showed no significant change (Figure 3D). Treatment of wild-type and 35S:SMXL6-GFP plants with rac-GR24 caused a strong decrease of the MAX4 expression level to ∼25% and a less decrease to ∼40% in 35S:SMXL6D-GFP plants (Figure 3D). In 35S:SMXL6D-GFP plants, MAX4 is still expected to respond partially to rac-GR24 treatment due to the contribution of endogenous SMXL6, 7, and 8 in SL signaling. Taken together, these results suggested that overexpression of SMXL6D compromises SL signaling.
Figure 3.
Overexpression of SMXL6D-GFP Enhances the Outgrowth of Axillary Buds and MAX4 Expression.
(A) Shoots of representative wild-type (Col-0) and transgenic plants expressing 35S:SMXL6-GFP or 35S:SMXL6D-GFP genes after 7 weeks of growth in a 16-h-light/8-h-dark photoperiod. Bar = 5 cm.
(B) Number of primary rosette (RI) branches of at least 0.5 cm recorded in plants shown in (A). Values are means ± se (n = 15); significant differences revealed by Tukey's multiple comparison test are indicated by different letters above bars (P < 0.05).
(C) Number of secondary cauline (CII) branches of at least 0.5 cm recorded in plants shown in (A). Values are means ± se (n = 15); Tukey's multiple comparison test revealed no significant differences (P < 0.05).
(D) RNA levels of MAX4 relative to ACTIN2 in 10-d-old seedlings of wild-type (Col-0) and transgenic plants expressing 35S:SMXL6-GFP or 35S:SMXL6D-GFP. Seedlings were treated with 5 µM rac-GR24 in 0.5× MS liquid medium for 4 h before isolation of RNA for RT-qPCR. Values are means ± se (n = 3); ns, no significant difference; asterisks indicate significant difference (**P < 0.01) revealed by Student’s t test.
D53-Like SMXLs Regulate Leaf Morphology
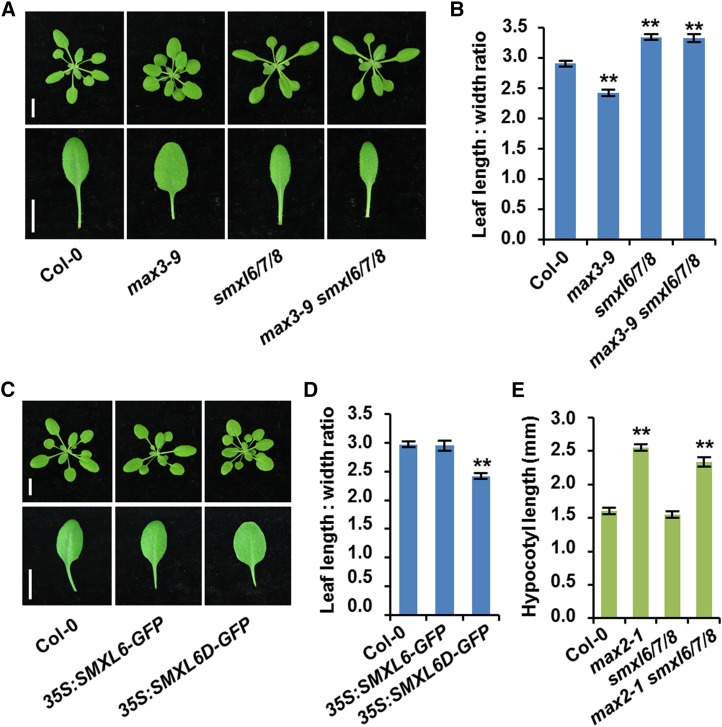
SLs influence leaf lamina shape (Stirnberg et al., 2002; Scaffidi et al., 2013). The max3-9 mutant exhibited rounder, broader laminas than wild-type leaves, whereas the smxl6/7/8 triple mutant displayed longer, narrower laminas (Figure 4A). The ratio of leaf length to width decreased in max3-9 leaves, but increased in smxl6/7/8 (Figure 4B). Moreover, the smxl6/7/8 phenotype could not be rescued by max3-9 (Figures 4A and 4B). In addition, the leaf shapes of 35S:SMXL6-GFP plants were similar to those of the wild type, while leaves of 35S:SMXL6D-GFP plants were rounder than the wild type (Figures 4C and 4D). These observations are consistent with D53-like SMXLs acting downstream of MAX3 in the regulation of leaf morphology by SL signaling. Whereas SLs repress shoot branching and promote narrower leaves, D53-like SMXLs have the opposite effects, promoting shoot branching and rounder leaves.
Figure 4.
Regulation of Leaf Shape by SMXL6, 7, and 8 in Arabidopsis.
(A) Rosettes and the fifth leaves of 3-week-old wild type (Col-0) and the indicated mutants. Bars = 1 cm.
(B) Quantitative analysis on the ratio of leaf length to leaf width for the fifth leaves shown in (A). Values are represented as mean ± se (n = 12); **P < 0.01 indicated by Student’s t test.
(C) Rosettes and the fifth leaves of 3-week-old wild-type (Col-0) and transgenic plants expressing 35S:SMXL6-GFP and 35S:SMXL6D-GFP. Bars = 1 cm.
(D) Quantitative analysis on the ratio of leaf length to leaf width for the fifth leaves shown in (C). Values are represented as mean ± se (n = 12); **P < 0.01 indicated by Student’s t test.
(E) Hypocotyl length of 7-d-old light-grown seedlings in the wild type (Col-0) and the indicated mutants. Values are represented as mean ± se (n = 16); **P < 0.01 indicated by Student’s t test.
Karrikins regulate Arabidopsis seed dormancy and seedling development through KAI2 and MAX2, and the kai2 and max2 mutants exhibit increased seed dormancy and abnormal seedling photomorphogenesis (Nelson et al., 2011; Waters et al., 2012). The SMAX1 protein acts in association with MAX2 to regulate the KAI2 signaling pathway (Stanga et al., 2013). To examine the possible effects of D53-like SMXLs on seedlings, we compared the hypocotyl length of wild-type, max2-1, smxl6/7/8, and max2-1 smxl6/7/8 seedlings after 7 d of growth. The results showed that the max2-1 seedlings formed longer hypocotyls than the wild type, while smxl6/7/8 displayed no significant difference in hypocotyl length (Figure 4E), suggesting that D53-like SMXLs are not involved in the control of seedling development.
SL-Induced SMXL6 Degradation Requires D14 and MAX2
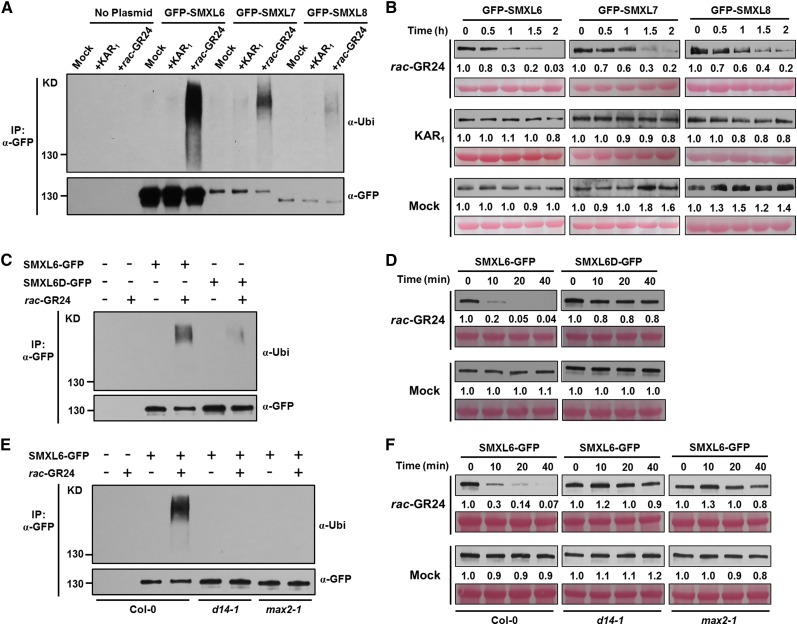
To further investigate the roles of D53-like SMXLs in SL signaling, we first examined their polyubiquitination in response to rac-GR24 and to KAR1, a specific activator of the KAI2-signaling pathway. To achieve this, protoplasts were prepared from Arabidopsis leaves and transformed with plasmids for transient expression of GFP-SMXL6, GFP-SMXL7, and GFP-SMXL8. After 12 h incubation, protoplasts were treated with proteasome inhibitor MG132 for 1 h, then with rac-GR24 or KAR1 for a further 1 h, before proteins were extracted for affinity purification using agarose-immobilized anti-GFP monoclonal antibody prior to immunoblot analysis. Upon rac-GR24 treatment, the GFP-SMXL6/7/8 recombinant proteins were polyubiquitinated within 1 h in wild-type protoplasts. In contrast, KAR1 failed to induce the ubiquitination of GFP-SMXL6, 7, or 8 (Figure 5A). Moreover, the GFP-SMXL6/7/8 recombinant proteins were degraded within 2 h in response to GR24 treatment but remained stable upon KAR1 treatment (Figure 5B). These results demonstrated that D53-like SMXLs are polyubiquitinated and degraded in response to SLs, similar to D53 in rice.
Figure 5.
GFP-SMXL6, 7, and 8 Fusion Proteins Are Polyubiquitinated and Degraded in Response to rac-GR24 Treatment in Arabidopsis.
(A) Ubiquitination of GFP-SMXL6, GFP-SMXL7, and GFP-SMXL8 in protoplasts. Arabidopsis (Col-0) protoplasts were transformed with plasmids encoding each fusion protein, incubated for a 12-h period of protein synthesis, then pretreated with 50 µM MG132 for 1 h and treated with 40 µM rac-GR24 or 40 µM KAR1 for 1 h. Proteins were isolated for immunoprecipitation with agarose-conjugated anti-GFP monoclonal antibody followed by immunoblotting with antiubiquitin antibody (upper panel) or anti-GFP (lower panel).
(B) Levels of GFP-SMXL6, GFP-SMXL7, and GFP-SMXL8 proteins in wild-type protoplasts after plasmid transformation and treatment with 40 µM rac-GR24 or 40 µM KAR1 for the times indicated. Proteins were detected by immunoblotting with anti-GFP monoclonal antibody. Relative amounts of proteins were determined by densitometry and normalized to loadings determined by Ponceau staining (red) and expressed relative to the value at zero time.
(C) Ubiquitination of SMXL6-GFP and SMXL6D-GFP in wild-type (Col-0) Arabidopsis containing 35S:SMXL6-GFP and 35S:SMXL6D-GFP transgenes. Seedlings were treated after 10 d of growth with 50 µM MG132 for 1 h, then with 2 µM rac-GR24 in 0.5× MS liquid medium for 10 min. Proteins were detected as in (A).
(D) Levels of SMXL6-GFP and SMXL6D-GFP proteins in wild-type (Col-0) Arabidopsis containing 35S:SMXL6-GFP and 35S:SMXL6D-GFP transgenes. Seedlings were treated after 10 d of growth with 2 µM rac-GR24 in 0.5× MS liquid medium for the times indicated. Proteins were detected as in (B).
(E) Ubiquitination of SMXL6-GFP in wild-type (Col-0), max2-1, and d14-1 transgenic plants each expressing 35S:SMXL6-GFP. Seedlings were treated after 10 d of growth with 50 µM MG132 for 1 h, then with 2 µM rac-GR24 in 0.5× MS liquid medium for 10 min. Proteins were detected as in (A).
(F) Level of SMXL6-GFP in wild-type (Col-0), max2-1, and d14-1 transgenic plants each expressing 35S:SMXL6-GFP. Seedlings were treated after 10 d of growth with 2 µM rac-GR24 in 0.5× MS liquid medium for the times indicated. Proteins were detected as in (B).
We further investigated whether the mutant SMXL6D is resistant to ubiquitination and degradation as expected from its ability to enhance outgrowth of axillary buds. When seedlings of 35S:SMXL6-GFP and 35S:SMXL6D-GFP transgenic plants were treated with rac-GR24 for 10 min, the ubiquitination of SMXL6-GFP was triggered, while the ubiquitination of SMXL6D-GFP was obviously attenuated (Figure 5C). Similarly, treatment with rac-GR24 for up to 40 min led to rapid degradation of SMXL6-GFP, but not of SMXL6D-GFP in stable transgenic plants, suggesting that deletion of the four amino acid residues in SMXL6 could confer its resistance to degradation (Figure 5D).
To determine if ubiquitination and degradation of SMXL6-GFP depend on MAX2 and D14, we generated transgenic plants expressing 35S:SMXL6-GFP in the max2-1 and d14-1 mutant backgrounds. After treatment with rac-GR24 for 10 min, the ubiquitination of SMXL6-GFP was detected in 35S:SMXL6-GFP seedlings but not in 35S:SMXL6-GFP/max2-1 and 35S:SMXL6-GFP/d14-1 seedlings (Figure 5E). Upon rac-GR24 treatment, degradation of SMXL6-GFP was dramatically attenuated in max2-1 and d14-1 (Figure 5F). Collectively, these data indicate that the D53-like SMXLs are subject to D14- and MAX2-dependent degradation in response to SL signaling.
D53-Like SMXLs Interact with MAX2 and D14
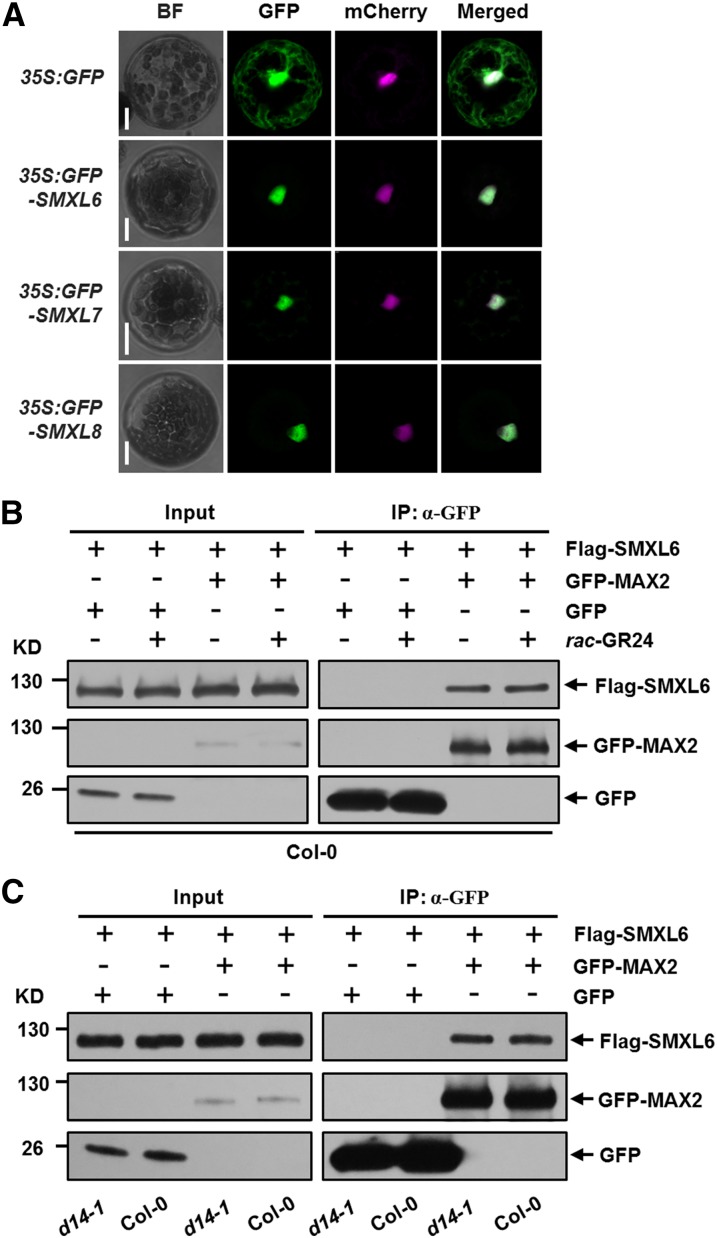
MAX2 has been reported to localize in the nucleus (Shen et al., 2007; Stirnberg et al., 2007), and D14 was detected in both nucleus and cytoplasm (Chevalier et al., 2014). To further understand the relationships between D53-like SMXLs, MAX2, and D14, we first asked whether the subcellular distributions of GFP-SMXL6, 7, and 8 overlap with MAX2 or D14 and then investigated whether individual SMXL proteins could interact with them. Using a transient expression system, we found that the GFP-SMXL6, GFP-SMXL7, and GFP-SMXL8 proteins colocalized with SV40NLS-mCherry (mCherry-tagged with a strong nuclear localization sequence), which was used to label the nucleus in protoplasts (Ye et al., 2012), suggesting that SMXL6, 7, and 8 proteins localized in the nucleus (Figure 6A). We then set up a coimmunoprecipitation (co-IP) system using Arabidopsis protoplasts to detect in vivo interaction between D53-like SMXLs and MAX2. In protoplasts prepared from the wild type, Flag-SMXL6, Flag-SMXL7, and Flag-SMXL8 were found to interact with GFP-MAX2 with or without treatment with rac-GR24, suggesting that each SMXL could form a complex with MAX2 in vivo (Figure 6B; Supplemental Figure 6). Furthermore, Flag-SMXL6 was found to interact with GFP-MAX2 in protoplasts prepared from d14-1 mutants, suggesting that D14 may be unessential for the interaction between SMXL6 and MAX2 (Figure 6C).
Figure 6.
Interactions between SMXL6 and MAX2.
(A) Subcellular localizations of GFP, GFP-SMXL6, GFP-SMXL7, and GFP-SMXL8 in Arabidopsis wild-type (Col-0) protoplasts. A plasmid construct expressing 35S:SV40NLS-mCherry was cotransformed to label the nucleus. BF, bright-field. Bar = 10 μm.
(B) In vivo interaction between Flag-SMXL6 and GFP-MAX2 revealed by co-IP assay in protoplasts prepared from the wild type (Col-0). After transformation and incubation for 11 h, protoplasts were pretreated with 40 µM rac-GR24 for 1 h, then cells were broken and immunoprecipitation (IP) with agarose-conjugated anti-GFP monoclonal antibody was performed under 40 µM rac-GR24 treatment, following which the SMXL6 recombinant protein was detected with an anti-Flag monoclonal antibody, while GFP-MAX2 fusion protein and GFP were detected with an anti-GFP monoclonal antibody. Input means total protein lysate without immunoprecipitation.
(C) In vivo interaction between Flag-SMXL6 and GFP-MAX2 revealed by the co-IP assay in protoplasts prepared from the wild type (Col-0) and d14-1. Following transformation and incubation for 12 h, cells were broken and immunoprecipitation and immunoblot were conducted as in (B).
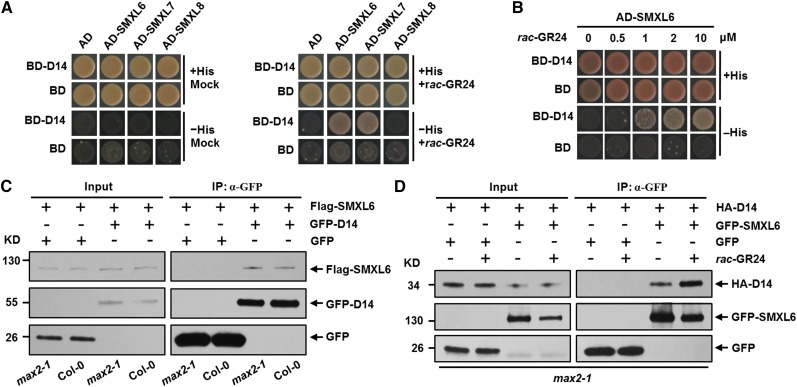
To test interactions of D53-like SMXLs with D14, we first employed a yeast two-hybrid assay. In response to rac-GR24 treatment, SMXL6 and SMXL7 directly interacted with D14 in yeast cells in a concentration-dependent manner (Figures 7A and 7B; Supplemental Figure 7). We further investigated the interaction between SMXL6 and D14 in wild-type and max2-1 protoplasts using the co-IP approach. Flag-SMXL6 was found to interact with GFP-D14 in both the wild type and max2-1 (Figure 7C), demonstrating that the SMXL6-D14 interaction does not require MAX2. However, the amount of SMXL6 detected was very low, possibly because it is continually degraded in protoplasts. This presented a challenge for investigating whether GR24 could stimulate formation of the D14-SMXL6 complex because GR24 will stimulate the degradation of SMXL6 in protoplasts. Since MAX2 is not required for formation of this complex, we therefore performed an experiment using max2-1 protoplasts, in which SMXL6 should be relatively stable under rac-GR24 treatment, and found that the interaction between GFP-SMXL6 and HA-D14 was enhanced (Figure 7D), suggesting that SLs could strengthen the SMXL6-D14 interaction. Taken together, these results indicate that the perception of SLs by D14 could potentially trigger the formation of a complex containing D14, SMXL6, 7, or 8, and MAX2, which could be required for the SL-induced degradation of the D53-like SMXLs.
Figure 7.
Interactions between D53-Like SMXLs and Arabidopsis D14.
(A) SMXL6 and SMXL7 interact with D14 in yeast cells upon rac-GR24 treatment. Yeast cells were cotransformed with constructs encoding the binding domain (BD) fused to D14 and the activation domain (AD) fused to each SMXL. Cells were plated on selective media in the absence (left panel) and presence (right panel) of 10 µM rac-GR24.
(B) The interaction between SMXL6 and D14 responds to rac-GR24 in a dose-dependent manner. Yeast cells were cotransformed with constructs encoding BD-D14 and AD-SMXL6. Cells were plated on selective media in the presence of increasing amount of rac-GR24.
(C) In vivo interaction between Flag-SMXL6 and GFP-D14 revealed by the co-IP assay in protoplasts made from the wild type and max2-1. After transformation and incubation for 12 h, cells were broken and then immunoprecipitation (IP) with agarose-conjugated anti-GFP monoclonal antibody was performed, following which the SMXL6 recombinant protein was detected with an anti-Flag monoclonal antibody, while GFP-D14 fusion protein and GFP were detected with an anti-GFP monoclonal antibody. Input represents total protein lysate without immunoprecipitation.
(D) In vivo interaction between HA-D14 and GFP-SMXL6 revealed by the co-IP assay in max2-1 protoplasts in the absence or presence of rac-GR24. After transformation and incubation for 11 h, protoplasts were pretreated with 100 µM rac-GR24 for 1 h and then cells were broken and immunoprecipitation (IP) with agarose-conjugated anti-GFP monoclonal antibody was performed under 100 µM rac-GR24 treatment, following which the HA-D14 recombinant protein was detected with an anti-HA monoclonal antibody, while GFP-SMXL6 fusion protein and GFP were detected with an anti-GFP monoclonal antibody.
D53-Like SMXLs Interact with TPR2
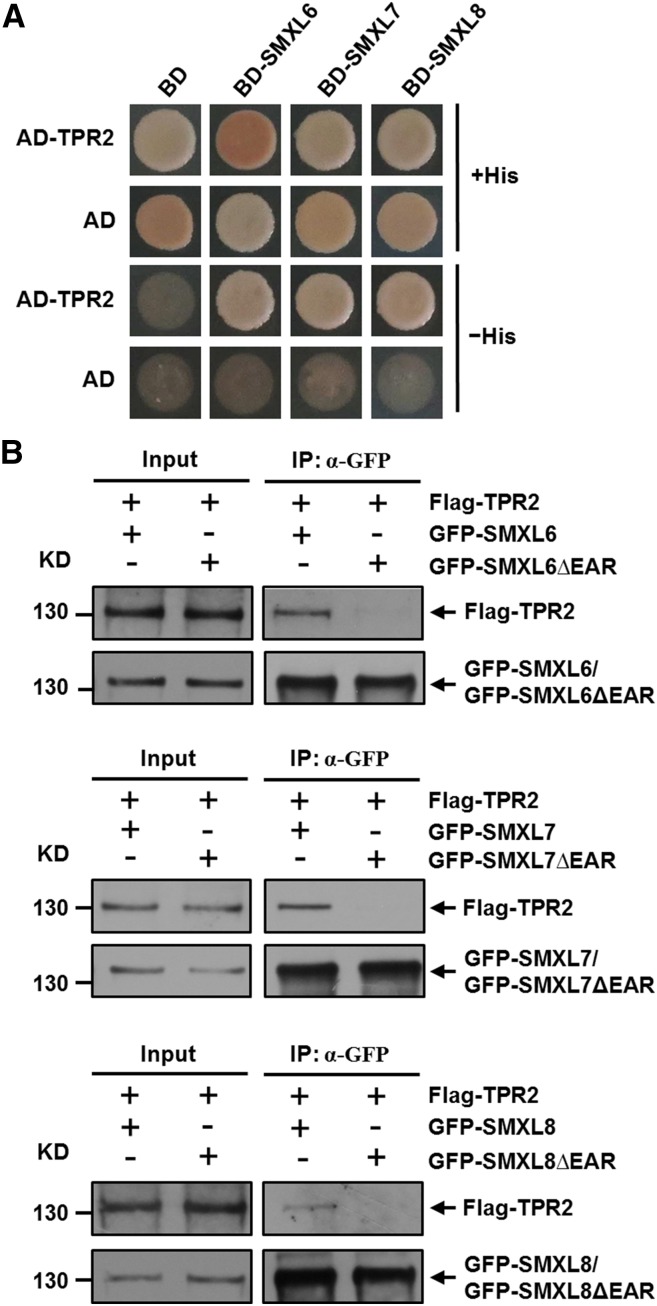
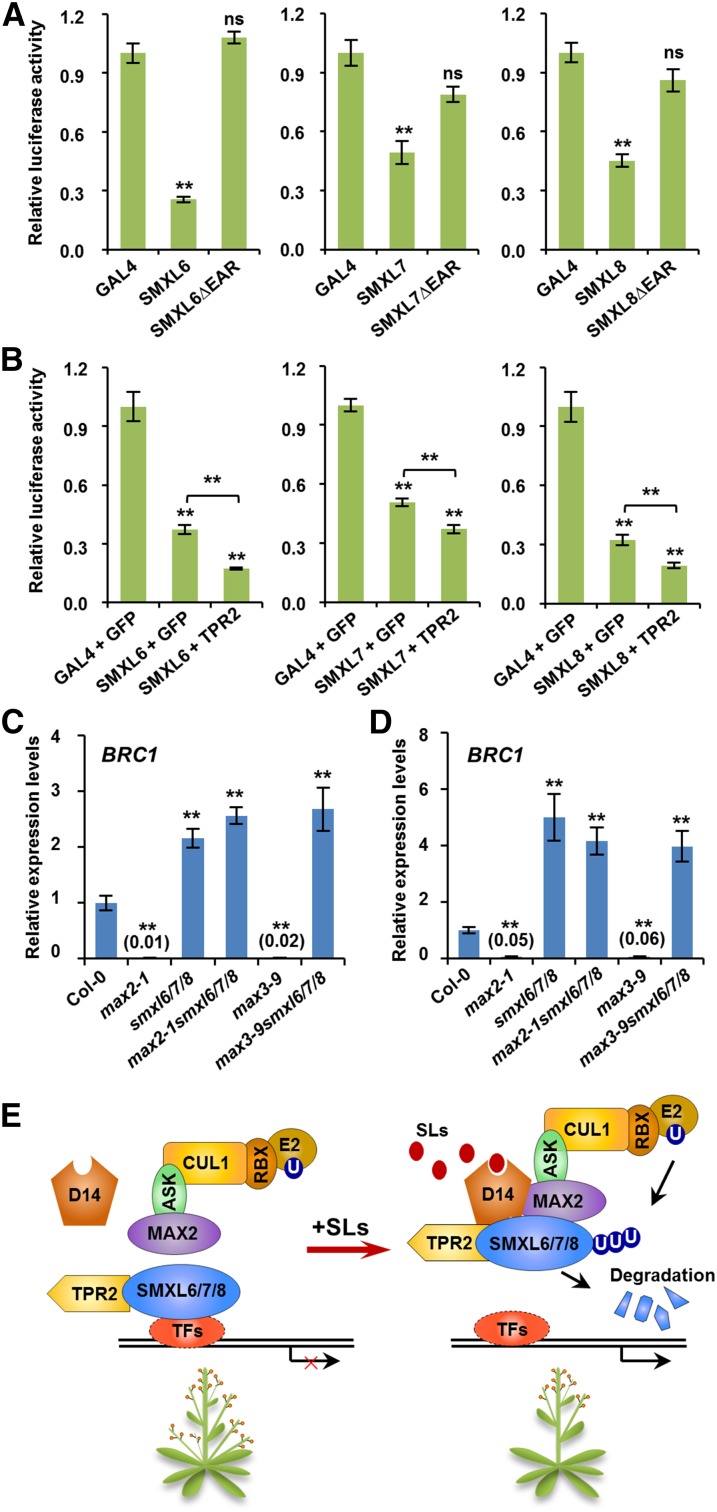
TOPLESS and TPR proteins are important corepressors that regulate multiple developmental processes and functions in plant hormone signaling pathways (Long et al., 2006; Szemenyei et al., 2008; Pauwels et al., 2010; Causier et al., 2012). Rice D53 interacts with rice TPR proteins when both are expressed in mammalian cells, and D53 expressed in rice calli interacts in a pull-down assay with bacterially produced GST-tagged TPR2 (Jiang et al., 2013). In Arabidopsis, each SMXL6/7/8 contains a conserved ethylene-responsive element binding factor-associated amphiphilic repression (EAR) motif near the C-terminal end comprising the amino acids L-D-L-N-L-P (Supplemental Figure 8). We investigated the interactions between each SMXL6/7/8 and TPR2 in the yeast two-hybrid and co-IP assays, since the EAR motif is critical for interaction with TOPLESS (TPL) and TPR proteins (Szemenyei et al., 2008; Pauwels et al., 2010). The results showed clearly that each SMXL6/7/8 was able to interact with TPR2 in yeast cells (Figure 8A). Next we conducted transient assays using Arabidopsis protoplasts to express GFP-SMXLs and Flag-TPR2 protein for co-IP analysis. We observed that SMXL6 could interact with TPR2, but SMXL6ΔEAR (SMXL6 lacking the EAR motif) could not form a complex with TPR2, indicating that interaction between SMXL6 and TPR2 depends on the EAR motif (Figure 8B). Similar results were obtained for both SMXL7 and SMXL8 (Figure 8B). Together, these results indicate that TPR2 can form a complex with SMXL6, SMXL7, and SMXL8 in vivo.
Figure 8.
SMXL6, 7, and 8 Interact with TPR2.
(A) Interactions of TPR2 with SMXL6, 7, and 8 in yeast cells. Yeast cells were cotransformed with constructs encoding the binding domain (BD) fused to each SMXL and the activation domain (AD) fused to TPR2 and plated on selective media.
(B) Interactions in Arabidopsis protoplasts of SMXL6, SMXL7, or SMXL8 with TPR2. Protoplasts were prepared from wild type (Col-0) and cotransformed with genes encoding Flag-TPR2 and either GFP-SMXL or GFP-SMXLΔEAR (both for each of SMXL6, 7, and 8). After incubation for 12 h, cells were broken and immunoprecipitation (IP) with agarose-conjugated anti-GFP monoclonal antibody was performed, following which the Flag-TPR2 was detected with an anti-Flag monoclonal antibody, while GFP-SMXL and GFP-SMXLΔEAR proteins were detected with an anti-GFP monoclonal antibody.
D53-Like SMXLs Display Transcriptional Repression Activities
The interaction between D53-like SMXLs and TPR2 raises the possibility that such complexes could regulate transcription in response to SLs. We therefore investigated the effects of SMXL6, SMXL7, and SMXL8 on transcriptional activities in Arabidopsis protoplasts. We made constructs that use the 35S promoter to express fusion proteins of the GAL4 binding domain (GAL4BD) with each SMXL, with or without the EAR motif (Supplemental Figure 8). Protoplasts were cotransformed with a reporter gene comprising the 35S promoter linked to the GAL4 upstream activation sequence driving luciferase expression. Compared with GAL4 alone, the relative luciferase activities were dramatically reduced when GAL4 was fused with SMXL6, SMXL7, or SMXL8, and this was dependent on the EAR motif (Figure 9A). Because the EAR motif is essential for interactions between TPR2 and D53-like SMXLs, we considered the possibility that TPR2 could influence their effects on transcriptional activity. Indeed, the relative luciferase activities were further reduced when TPR2 was coexpressed with SMXL6, SMXL7, or SMXL8 (Figure 9B). These results suggest that D53-like SMXLs can bring about transcriptional repression in association with TPR2 in Arabidopsis cells.
Figure 9.
SMXL6, 7, and 8 Cooperate with TPR2 to Repress the Expression of SL-Responsive Genes.
(A) SMXL6, 7, and 8 show transcriptional repression activities in transient expression assays in Arabidopsis. Protoplasts were cotransformed with two plasmids (Supplemental Figure 8), one comprising a luciferase reporter with upstream enhancer sequence and the other encoding GAL4, a GAL4-SMXL fusion, or GAL4-SMXLΔEAR fusion. After 12 h of incubation, luciferase was assayed. Values are means ± se (n = 4); ns, no significant difference; **P < 0.01 determined by Student’s t test.
(B) The effects of TPR2 on transcriptional repression activities of SMXL6, 7, and 8 in Arabidopsis. Protoplasts were cotransformed with three plasmids (Supplemental Figure 8): one comprising the luciferase reporter, another encoding either GAL4 or GAL4-SMXL fusion, and the third either GFP (as a control) or TPR2. After 12 h of incubation, luciferase was assayed. Values are means ± se (n = 4); ns, no significant difference; **P < 0.01 determined by Student’s t test.
(C) and (D) Expression of BRC1 in nonelongated axillary buds of primary rosette (RI) branches (C) and secondary cauline (CII) branches (D) of Col-0 and the mutants indicated. Values are means ± se (n = 3 or 4); **P < 0.01 determined by Student’s t test.
(E) A model of the SL signaling complex in Arabidopsis that includes SL-dependent interaction of Arabidopsis D14 with both MAX2 and SMXL proteins, although the sequence in which these interactions occur is not known. It is not known if BRC1 is a direct or indirect target of this SL signaling mechanism. ASK, CUL1, RBX, and E2 are components of the ubiquitination complex. U, Ubiquitin; TFs, transcription factors (unidentified).
We then explored whether D53-like SMXLs regulate the expression of BRC1, which encodes a TEOSINTE BRANCHED1/CYCLOIDEA/PCF transcription factor that is rapidly upregulated after SL treatment in pea and Arabidopsis (Aguilar-Martínez et al., 2007; Mashiguchi et al., 2009; Braun et al., 2012). In nonelongated axillary buds of both primary rosette (RI) and secondary cauline (CII) branches, the BRC1 gene was strongly repressed in max2-1 and max3-9 but was induced in smxl6/7/8, max2-1 smxl6/7/8, and max3-9 smxl6/7/8 mutants (Figures 9C and 9D), suggesting that absence of SMXL6, 7, and 8 releases the transcriptional repression of BRC1. Meanwhile, in nonelongated axillary buds of CII branches, the expression levels of HB53, a downstream target gene of BRC1 (González-Grandío et al., 2013), was also downregulated in max2-1 and max3-9 but upregulated in smxl6/7/8, max2-1 smxl6/7/8, and max3-9 smxl6/7/8 mutants (Supplemental Figure 9A). Similar results were obtained when detecting the expression of BRC1 and HB53 in young seedlings (Supplemental Figures 9B and 9C). In summary, these data indicate that D53-like SMXLs function as critical components of SL signaling through recruitment of TPR2 and transcriptional inhibition of key genes including BRC1.
DISCUSSION
The degree of branching, leaf shape, and leaf angle are critical features of plant architecture that affect growth and productivity (Wang and Li, 2008). Growth of a new branch leads to increased light and carbon capture, and potentially more flowers. The control of axillary bud outgrowth by SLs has profound effects on the regulation of shoot architecture in response to nutrients and light. Here, we characterized the functions of Arabidopsis SMXL6, SMXL7, and SMXL8, which are orthologs of D53 in rice, and revealed that the function of these D53-like SMXLs is to promote the outgrowth of lateral buds and that SL signaling targets these proteins for degradation, thus inhibiting bud outgrowth. We found that the smxl6/7/8 triple mutant displayed a decreased shoot-branching phenotype and completely suppressed the bushy phenotype of max3-9 and max2-1 mutants, whereas the 35S:SMXL6D-GFP transgenic plants produce more branches, indicating that SMXL6/7/8 proteins are functionally redundant important components in shoot-branching regulation in Arabidopsis. Therefore, rice D53 and the three Arabidopsis orthologs have similar functions. We have further shown that the D53-like SMXLs influence leaf shape and that this is SL regulated. It is not yet known if D53 or its ortholog in rice (D53-like) influence leaf development in rice, but this is worthy of investigation in view of the role of D53-like proteins in Arabidopsis leaf development.
SMXL6, SMXL7, SMXL8, and SMAX1 belong to an eight-gene family in Arabidopsis, but they are expressed to different extents in particular plant tissues and they respond differentially to SLs. At the transcriptional level, SMXL6, 7, and 8 could be significantly induced by rac-GR24, whereas SMAX1 showed no such response (Stanga et al., 2013). The smxl6/7/8 and smax1 mutants restore different aspects of the max2 mutant phenotype. The smxl6/7/8 triple mutant plants completely suppress the highly branched phenotypes of max2, but show little effect on seedling development. In contrast, smax1 rescues the seed germination and seedling photomorphogenesis phenotypes of max2 but does not affect the axillary shoot growth (Stanga et al., 2013). Therefore, different SMXL family members play diverse roles in plant development. The functions of other members of the SMXL family remain to be determined (Smith and Li, 2014).
The molecular mechanisms by which D53 and D53-like proteins regulate the expression of downstream target genes are still an open question (Bennett and Leyser, 2014). We found that SMXL6, 7, and 8 interacted with transcriptional corepressor TPR2 (Figure 8) and showed transcriptional repression activities in vivo (Figures 9A and 9B). Notably, the transcriptional repression activities of SMXL6, 7, and 8 are enhanced by their interaction with TPR2. In addition, SMXL6, 7, and 8 regulate the expression level of BRC1 (Figures 9C and 9D; Supplemental Figure 9B), which is an important early responsive gene of the SL signaling pathway (Aguilar-Martínez et al., 2007; Mashiguchi et al., 2009; Braun et al., 2012). BRC1 has been suggested to negatively regulate the development of rosette and cauline branches in Arabidopsis (Aguilar-Martínez et al., 2007; Chen et al., 2013). Thus, suppression of rosette and cauline branch development in smxl6/7/8, max2-1 smxl6/7/8, and max3-9 smxl6/7/8 mutants may be explained by upregulation of the expression of BRC1.
The expression of HB53, which is a target gene activated by BRC1, is also increased in the smxl6/7/8 mutant (Supplemental Figures 9A and 9C), presumably as a result of the induction of BRC1. Based on these data, we hypothesize that SMXL6, 7, and 8 may repress downstream target genes through repressing the activities of unknown transcription factors (TFs). A large-scale screen for interacting proteins using SMXL6, 7, and 8 or D53 and genetic screening for suppressors or enhancers of max or dwarf mutants will contribute to identifying those unknown TFs. However, it is important to note that SMXL6, 7, and 8 possess putative chaperonin functions, and the EAR motif has been reported to interact with the TPL proteins that are also involved in vesicle trafficking, suggesting that SMXL6, 7, and 8 may regulate plant development through SL-induced posttranscriptional events (Waldie et al., 2014). Further studies on the underlying mechanisms of how SMXL6, 7, and 8 proteins repress SL signaling will contribute to understanding how plant architecture is regulated.
The next major question centers on the mechanism by which SLs bring about degradation of SMXLs. The ubiquitination and degradation of D53-like SMXLs is triggered by rac-GR24 and is dependent on D14 and MAX2 (Figure 5). The d53 dominant mutation results in a deletion of five amino acids and an amino acid substitution, which leads to obvious attenuation of the protein to be ubiquitinated and degraded (Jiang et al., 2013; Zhou et al., 2013). Importantly, we could recapitulate this mutation in Arabidopsis by deletion of four amino acids from the same region of SMXL6 (Supplemental Figure 4). Overexpressing this mutant form of SMXL6 gene could moderately mimic the shoot branching and leaf phenotypes of max2-1 and d14-1 mutants (Figures 3 and 4). Therefore, degradation of D53-like SMXL proteins is a key part of the SL-mediated control of Arabidopsis leaf development and shoot branching.
Remarkably, we have shown that the D53-like SMXLs, MAX2, and D14 can interact in pairwise combinations, but those involving D14 were strengthened by GR24, consistent with observations that D14 proteins hydrolyzing rac-GR24 undergo a change in conformation (Hamiaux et al., 2012; Zhao et al., 2013). We do not know if SMXLs, D14, and MAX2 are recruited into a single complex by GR24, but this is deduced to be the case for D14, D53, and D3 in rice (Jiang et al., 2013; Zhou et al., 2013). It is also unknown if TPR2 remains associated with SMXL proteins when they associated with D14 and MAX2.
Based on our findings here, we proposed a model of the SL signaling pathway in Arabidopsis (Figure 9E). In the absence of SLs, SMXL6, 7, and 8 bind to TPR2 and unknown TFs to repress the expression of target genes. However, in the presence of SLs, D14 binds and hydrolyzes the SLs, triggering the formation of a D14-SMXL-SCFMAX2 complex for the ubiquitination and degradation of SMXL6, 7, and 8. This relieves the repression of unknown TFs and activates gene expression (Figure 9E). Crucially, we do not know the sequence in which different proteins interact, and this has a direct bearing on our understanding of the perception of SLs in plant cells. Several different scenarios fit the available information. Our finding that interaction of D14 and SMXL proteins is stimulated by GR24 indicates that in principle this could be the earliest event in SL perception, and the D14-SMXL proteins could represent a SL receptor complex. Alternatively, D14 could first undergo SL-dependent interaction with MAX2, following which SMXL could be recruited into the complex for degradation. However, MAX2 or the SCFMAX2 complex could be bound to SMXL proteins that are associated with their target genes and, thereafter, both could be removed as a complex as a result of SL-mediated association with D14. Identifying all the components in this mechanism and determining the sequence of interaction events are critical to understanding the perception of SLs by plant cells, but D53-like and SMXL proteins are clearly absolutely central to SL mode of action.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana materials used in this work include the wild-type ecotype Columbia (Col-0) and the following mutants: smxl6 (CS847925/SAIL_1285_H05), smxl7 (SALK_082032), smxl8 (SALK_126406), max3-9 (Booker et al., 2004), d14-1 (Waters et al., 2012), and max2-1 (Stirnberg et al., 2002). Arabidopsis seeds were surface sterilized, vernalized at 4°C for 2 to 4 d, and germinated on 0.5× Murashige and Skoog (MS) medium containing 1.0% (w/v) sucrose and 0.7% (w/v) agar. For morphological observations, 10-d-old seedlings were transferred to pots containing a 2:1 vermiculite:soil mixture saturated with 0.3× MS medium. Plants of Arabidopsis were grown under a 16-h-light/8-h-dark cycle with a light intensity of 60 to 80 μE m−2 s−1 at ∼21°C as described previously (Dai et al., 2006).
Chemicals and Reagents
Synthetic strigolactone GR24 was obtained as a racemic mixture (rac-GR24) comprising equal amounts of GR245DS and GR24ent-5DS, from Chiralix. Purified stereoisomers GR245DS were obtained from Strigolab. MG132 was obtained from Calbiochem.
Vector Construction and Plant Transformation
To construct the 35S:SMXL6-GFP plasmid, pWM101 (Ding et al., 2006) was modified by replacing the 35S poly(A) sequence with the GFP coding region and NOS terminator. The coding sequence of SMXL6 was amplified with primers SMXL6-OE-F and SMXL6-OE-R and was cloned into SalI- and KpnI-digested pWM101. To construct the 35S:SMXL6D-GFP plasmid, the mutant form of SMXL6 was derived from pDONR222-SMXL6 by site-directed mutagenesis using primers SMXL6DEL-F and SMXL6DEL-R, then amplified with primers SMXL6-OE-F and SMXL6-OE-R, and subsequently cloned into pWM101. The 35S:SMXL6-GFP and 35S:SMXL6D-GFP recombinant plasmids were introduced into Agrobacterium tumefaciens strain EHA105 and transformed into wild-type, d14-1, and max2-1 plants through the Agrobacterium-mediated floral dip method (Clough and Bent, 1998).
To construct plasmids for yeast two-hybrid assays, the coding sequence of D14 was amplified using primers D14-BK-F and D14-BK-R and was cloned into pGBKT7 (Clontech) to generate BD-D14. The coding sequence of SMXL6 was amplified with primers SMXL6-BK-F and SMXL6-BK-R and was cloned into pGBKT7 to generate BD-SMXL6. The coding sequence of SMXL6 was amplified with primers SMXL6-BK-F and SMXL6-AD-R and was cloned into pGADT7 (Clontech) to generate AD-SMXL6. The coding sequence of SMXL7 was amplified with primers SMXL7-BK-F and SMXL7-BK-R and was cloned into pGBKT7 and pGADT7 to generate BD-SMXL7 and AD-SMXL7, respectively. The coding sequence of SMXL8 was amplified with primers SMXL8-BK-F and SMXL8-BK-R and was cloned into pGBKT7 and pGADT7 to generate BD-SMXL8 and AD-SMXL8, respectively. The coding sequence of TPR2 was amplified with primers TPR2-BK-F and TPR2-BK-R and was cloned into pGBKT7 to generate BD-TPR2.
To construct tag-fused transient expression plasmids for co-IP assays, the pBeacon-EGFP, pBeacon-3×Flag, and pBeacon-3×HA vectors were generated through replacing the fragments of e35S, mRFP, and P35S in pBeacon-RFP (Bargmann and Birnbaum, 2009) with a recombinant fragment containing P35S and enhanced GFP (eGFP), a recombinant fragment containing P35S and 3×Flag, and a recombinant fragment containing P35S and 3×HA, respectively. To construct the GFP-SMXL6/7/8 and 3×Flag–SMXL6/7/8 plasmids, the full-length coding sequence of SMXL6/7/8 was amplified with primers SMXL6/7/8-GW-F and SMXL6/7/8-GW-R, then cloned into the Gateway entry vector pDONR222 (Invitrogen) by Gateway BP reaction and subsequently recombined to the transient expression vectors pBeacon-eGFP and pBeacon-3×Flag by Gateway LR reaction. To construct the GFP-SMXL6∆EAR, GFP-SMXL7∆EAR, and GFP-SMXL8∆EAR plasmids, the coding sequences of SMXL6/7/8∆EAR were derived from pDONR222-SMXL6/7/8 by site-directed mutagenesis using primers of SMXL6∆EAR-F and SMXL6∆EAR-R, SMXL7∆EAR-F and SMXL7∆EAR-R, and SMXL8∆EAR-F and SMXL8∆EAR-R, respectively, and subsequently recombined into the transient expression vectors pBeacon-eGFP by Gateway LR reaction. To construct the GFP-MAX2 plasmid, a codon-optimized MAX2 coding sequence was synthesized and inserted into vector pDONR222 and then to a transient expression vector pBeacon-eGFP. To construct the GFP-D14 plasmid, the D14 coding sequence amplified by primers D14-GW-F and D14-GW-R was inserted into vector pDONR222 and then to a transient expression vector pBeacon-eGFP. To construct the 3×HA-D14 plasmid, the D14 coding sequence amplified by primers D14-GW-F and D14-GW-R was inserted into vector pDONR222 and then to a transient expression vector pBeacon-3×Flag. To construct the 3×Flag-TPR2 plasmid, the coding sequence of TPR2 was amplified with primers TPR2-GW-F and TPR2-GW-R, then cloned into pDONR222 by Gateway BP reaction and subsequently recombined to pBeacon-3×Flag by Gateway LR reaction.
To construct recombinant plasmids used in transcriptional activity assays, the full-length SMXL6/7/8 coding sequence was amplified with the primer pairs of SMXL6-GAL4-F and SMXL6-GAL4-R, SMXL7-GAL4-F and SMXL7-GAL4-R, and SMXL8-GAL4-F and SMXL8-GAL4-R, respectively. Then, amplified DNA fragments were inserted into the SalI- and KpnI-digested GAL4-BD vector (Lu et al., 2013) to generate GAL4BD-SMXL6/7/8 constructs. To construct the GAL4BD-SMXL6∆EAR, GAL4BD-SMXL7∆EAR, and GAL4BD-SMXL8∆EAR plasmids, the coding sequences of SMXL6/7/8∆EAR were amplified from pDONR222-SMXL6/7/8∆EAR using primer pairs of SMXL6-GAL4-F and SMXL6-GAL4-R, SMXL7-GAL4-F and SMXL7-GAL4-R, and SMXL8-GAL4-F and SMXL8-GAL4-R and then cloned into GAL4-BD vector. The sequences of all the primers used in this work are shown in Supplemental Table 1.
Gene Expression Analysis
Ten-day-old seedlings of Col-0, max2-1, d14-1, max3-9, smxl6/7/8, max3-9 smxl6/7/8, 35S:SMXL6-GFP, and 35S:SMXL6D-GFP plants were collected into 0.5× MS liquid medium and treated with 5 μM GR245DS or rac-GR24 in the greenhouse for 4 h. Nonelongated axillary buds of primary rosette branches and secondary cauline branches were also collected for RNA extraction. Total RNA was prepared using a TRIzol kit (Invitrogen) according to the user manual. RNA samples (each containing 12.5 μg RNA) were treated with TUBRO DNase (Ambion). Then, first-strand cDNA was synthesized using the oligo(dT) and random primers with the SSIII first-strand synthesis system (Invitrogen). Real-time PCR experiments were performed using gene-specific primers (Supplemental Table 1) on a CFX 96 real-time PCR detection system (Bio-Rad) in a total volume of 10 μL system containing 2 μL diluted cDNA, 0.3 μM gene-specific primers, and 5 μL SsoFast EvaGreen supermix (Bio-Rad). Arabidopsis ACTIN2 (ACT2) was used as the internal control.
Leaf Morphology Analysis
Plants were grown for 3 weeks after germination, and the fifth leaves were harvested, laid flat on the surface of an agar medium plate, and photographed for further analysis. For each genotype, ∼12 plants were used for observation of leaf morphology. The leaf length (the distance between leaf tip and the base of petiole) and leaf width (the greatest distance across the leaf lamina perpendicular to the proximal/distal axis of the leaf) were measured manually using ImageJ software (http://rsbweb.nih.gov/ij/) as described previously (Scaffidi et al., 2013).
Hypocotyl Measurements
Seeds were surface sterilized, vernalized at 4°C for 3 d, and germinated on 0.5× MS medium. Seedlings were grown at 21°C for 7 d at light intensity of 60 to 80 μE m−2 s−1 under a 16-h-light/8-h-dark cycle as previously described (Stirnberg et al., 2002). Hypocotyl length was measured using ImageJ software.
Co-IP Assay
Protoplasts generated from the mesophyll cells of Arabidopsis were transformed with transient expression plasmids as described (Yoo et al., 2007). After incubation at 21°C for 12 h or incubation for 11 h followed by another 1 h with rac-GR24, protoplasts were collected in protein extraction buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) Nonidet P-40, and 1×complete protease inhibitor cocktail (Roche). The lysate was centrifuged at 20,000g for 10 min at 4°C, and the supernatant was taken for co-IP experiments. Following the supplier’s instruction, 25 μL of the agarose-conjugated anti-GFP monoclonal antibody (MBL) was added into 1 mL of total extracted protein and incubated at 4°C for 3 h in the presence or absence of rac-GR24. The beads were washed three times with extraction buffer containing 0.01% (v/v) Nonidet P-40 and then eluted with 25 μL of SDS-PAGE sample buffer for immunoblot analysis. The proteins of GFP-MAX2, GFP-D14, GFP-SMXL6, GFP-SMXL6∆EAR, GFP-SMXL7, GFP-SMXL7∆EAR, GFP-SMXL8, GFP-SMXL8∆EAR, and GFP were detected by mouse anti-GFP monoclonal antibody (Roche) at a 1:2000 dilution. The Flag-SMXL6, Flag-SMXL7, Flag-SMXL8, and Flag-TPR2 proteins were detected by mouse anti-FLAG monoclonal antibody (Abmart) at a 1:2000 dilution. The HA-D14 protein was detected by mouse anti-HA monoclonal antibody (Millipore) at a 1:2000 dilution.
In Vivo Ubiquitination Assay
Protoplasts generated from mesophyll cells were transformed with transient expression plasmids as described (Yoo et al., 2007). After incubation at 21°C for 12 h in W5 solution, the protoplasts were collected and then pretreated with 50 µM MG132 for 1 h and treated with 40 μM rac-GR24 or 40 μM KAR1 at 21°C for 1 h. Protoplasts were then collected in the extraction buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) Nonidet P-40, and 1× complete protease inhibitor cocktail (Roche). The lysates were centrifuged at 20,000g for 10 min at 4°C. The supernatant was taken for immunoprecipitation and subsequent immunoblot using antiubiquitin and anti-GFP antibodies. In detail, 25 μL of anti-GFP monoclonal antibody-conjugated agarose (MBL) was added into 1 mL total extracted proteins and incubated at 4°C for 3 h with gentle rotation. The beads were washed three times with the extraction buffer containing 0.01% (v/v) Nonidet P-40 and then eluted with 25 μL of the SDS-PAGE sample buffer for protein blotting. Mouse antiubiquitin monoclonal antibody (Cell Signaling Technology) was used at a 1:3000 dilution, and mouse anti-GFP monoclonal antibody (Roche) was used at a 1:2000 dilution. In an independent experiment system, the 10-d-old seedlings of 35S:SMXL6-GFP, 35S:SMXL6D-GFP, 35S:SMXL6-GFP/d14-1, and 35S:SMXL6-GFP/max2-1 transgenic plants were collected into 0.5× MS liquid medium and treated with 2 μM rac-GR24 or acetone in greenhouse for 10 min. Total proteins were extracted and taken for immunoprecipitation and immunoblot as mentioned above.
Yeast Two-Hybrid Assay
To detect the interactions between Arabidopsis D14 and SMXL6, 7, and 8, the fusion constructs of BD-D14, AD-SMXL6, AD-SMXL7, AD-SMXL8, and control plasmids AD and BD were transformed into Gold Yeast (Clontech) cells by the lithium acetate-mediated method. The transformed yeast strains were plated on SD/-Leu-Trp medium (Clontech) at 28°C for 2 d. Interactions in yeast were tested on SD/-Leu-Trp-His (Clontech) medium in the presence or absence of rac-GR24. To detect the interactions between TPR2 and SMXL6/7/8, the fusion constructs of AD-TPR2, BD-SMXL6, BD-SMXL7, BD-SMXL8, and control plasmids AD and BD were transformed into Gold Yeast cells and plated on SD/-Leu-Trp medium at 28°C for 2 d, then tested on SD/-Leu-Trp-His medium.
Microscopy Analyses
To observe the subcellular localizations of GFP, GFP-SMXL6, GFP-SMXL7, and GFP-SMXL8, the plasmids of pBI221-GFP (Lin et al., 2009) and GFP-SMXL6/7/8 were transformed into Arabidopsis protoplasts made form mesophyll cells as described (Yoo et al., 2007). A plasmid construct expressing SV40NLS-mCherry was cotransformed to label the nucleus (Ye et al., 2012). The GFP and mCherry signals were examined under a confocal microscope (FluoView FV1000; Olympus) at the excitation wavelengths of 488 and 559 nm, respectively. The images presented in this article were obtained by reconstruction of three to six neighboring sections using the Olympus Fluoview software.
Transcriptional Activity Assay in Protoplasts
To detect the transcriptional activities of SMXL6, 7, and 8, the plasmid combinations of GAL4BD-SMXL6/7/8, 35S:LUC, and pRTL were introduced into Arabidopsis protoplasts as described (Yoo et al., 2007); meanwhile, the combinations of GAL4BD-SMXL6/7/8∆EAR, 35S:LUC, and pRTL were introduced into protoplasts. The plasmids of GAL4BD, pRTL, and 35S:LUC were used as a control. After incubation at 21°C for 12 h, the luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. To investigate the influence of TPR2 in transcriptional activities of SMXL6/7/8, the plasmid combinations of GAL4BD-SMXL6/7/8, 35S:GFP, 35S:LUC, and pRTL were introduced into Arabidopsis protoplasts; meanwhile, the combinations of GAL4BD-SMXL6/7/8, 3×Flag-TPR2, 35S:LUC, and pRTL were introduced into protoplasts. The plasmids of GAL4BD, 35S:GFP, pRTL, and 35S:LUC were used as a control.
Genetic Analysis
Arabidopsis double, triple, and quadruple mutants were generated from the crosses of relevant homozygous single or double mutants and identified from the F2 or F3 progeny. Genotyping of the smxl6, smxl7, smxl8, max3-9, d14-1, and max2-1 mutants were performed by PCR using primers listed in Supplemental Table 1. In detail, smxl6 was genotyped using the primer pairs smxl6RP + LB1 and smxl6LP + smxl6RP. The smxl7 mutant was genotyped using the primer pairs smxl7RP + LBb1 and smxl7LP + smxl7RP. The smxl8 mutant was genotyped using the primer pairs smxl8RP + LBb1 and smxl8LP + smxl8RP. The d14-1 mutant was genotyped using the primer pairs d14RP + L4_WiscLoxHS and d14LP + d14RP. The wild type and max3-9 were amplified with primers max3-9-F and max3-9-R and formed fragments of 120 and 131 bp, respectively. The PCR products of the wild type and max2-1 amplified with primers max2-1-F and max2-1-R were digested with ApoI.
Phylogenetic Analysis
The sequence of D53, D53-like, SMAX1, and SMXL2, 3, 4, 5, 6, 7, and 8 were extracted from the Phytozome 9.0 (http://www.phytozome.net) and TAIR (http://www.arabidopsis.org) databases. Multiple sequence alignment of the protein sequences was done using Clustalw2 (see Supplemental Data Set 1 for detailed information). A maximum likelihood phylogenetic tree was constructed using MEGA 5.1 software (Tamura et al., 2011) using default parameters.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: AT1G07200 (SMXL6), AT2G29970 (SMXL7), AT2G40130 (SMXL8), AT3G03990 (D14), AT2G42620 (MAX2), AT2G44990 (MAX3), AT3G18550 (BRC1), AT5G66700 (HB53), AT3G18780 (ACT2), AT1G04130 (TPR2), AT5G57710 (SMAX1), AT4G30350 (SMXL2), AT3G52490 (SMXL3), AT4G29920 (SMXL4), AT5G57130 (SMXL5), Os11g0104300 (D53), and Os12g0104300 (D53-like).
Supplemental Data
Supplemental Figure 1. Phylogenetic tree of D53, D53-Like, and SMXL family proteins.
Supplemental Figure 2. Identification of smxl6, smxl7, and smxl8 mutants in Arabidopsis.
Supplemental Figure 3. The shoot-branching phenotypes of max2-1 are repressed by smxl6/7/8.
Supplemental Figure 4. Schematic diagrams showing the amino acid changes of d53 and SMXL6D proteins.
Supplemental Figure 5. The 35S:SMXL6D-GFP transgenic plants form more branches than the wild type.
Supplemental Figure 6. SMXL7 and SMXL8 interact with MAX2.
Supplemental Figure 7. Interaction between SMXL7 and D14 in yeast is stimulated by increasing rac-GR24 concentration.
Supplemental Figure 8. Schematic diagrams showing the constructs used in the transient gene expression assays in protoplasts.
Supplemental Figure 9. Expression of HB53 and BRC1 genes is reduced in max2 and max3 mutants but enhanced in smxl6/7/8.
Supplemental Table 1. Primers used in this study.
Supplemental Data Set 1. Text file of alignment used for phylogenetic analysis in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank the ABRC stock center for providing smxl6 (CS847925), smxl7 (SALK_082032), and smxl8 (SALK_126406) seeds, the NASC for providing d14-1 (N913109) seeds, and Ottoline Leyser at Cambridge University for providing max2-1 and max3-9 seeds. We also thank H. Eric Xu at the Shanghai Institute of Materia Medica, Chinese Academy of Sciences, for providing the chemical KAR1 and Yijun Qi at Tsinghua University for providing the SV40NLS-mCherry plasmid. This work was supported by National Natural Science Foundation of China (Grants 90917021 and 91335204) and by the award of a Chinese Academy of Sciences Senior International Scientist Visiting Professorship to S.M.S. (2013T1S0013).
AUTHOR CONTRIBUTIONS
L.W. and B.W. designed and performed experiments and analyzed data. L.J., X. Liu, X. Li, Z.L., and X.M. performed some of the experiments. J.L. designed research and conceived and supervised the project. B.W., L.W., Y.W., S.M.S., and J.L. analyzed the data and wrote the article. All authors commented on the article.
Glossary
- SL
strigolactone
- 4DO
4-deoxyorobanchol
- 5DS
5-deoxystrigol
- co-IP
coimmunoprecipitation
- TF
transcription factor
- MS
Murashige and Skoog
Footnotes
Articles can be viewed online without a subscription.
References
- Abe S., et al. (2014). Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. USA 111: 18084–18089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Martínez J.A., Poza-Carrión C., Cubas P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Matsuzaki K., Hayashi H. (2005). Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435: 824–827. [DOI] [PubMed] [Google Scholar]
- Al-Babili S., Bouwmeester H.J. (2015). Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66: 161–186. [DOI] [PubMed] [Google Scholar]
- Alder A., Jamil M., Marzorati M., Bruno M., Vermathen M., Bigler P., Ghisla S., Bouwmeester H., Beyer P., Al-Babili S. (2012). The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science 335: 1348–1351. [DOI] [PubMed] [Google Scholar]
- Bargmann B.O., Birnbaum K.D. (2009). Positive fluorescent selection permits precise, rapid, and in-depth overexpression analysis in plant protoplasts. Plant Physiol. 149: 1231–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T., Leyser O. (2014). Strigolactone signalling: standing on the shoulders of DWARFs. Curr. Opin. Plant Biol. 22: 7–13. [DOI] [PubMed] [Google Scholar]
- Booker J., Auldridge M., Wills S., McCarty D., Klee H., Leyser O. (2004). MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 14: 1232–1238. [DOI] [PubMed] [Google Scholar]
- Braun N., et al. (2012). The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 158: 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bythell-Douglas R., Waters M.T., Scaffidi A., Flematti G.R., Smith S.M., Bond C.S. (2013). The structure of the karrikin-insensitive protein (KAI2) in Arabidopsis thaliana. PLoS One 8: e54758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhou X., Xi L., Li J., Zhao R., Ma N., Zhao L. (2013). Roles of DgBRC1 in regulation of lateral branching in chrysanthemum (Dendranthema × grandiflora cv. Jinba). PLoS One 8: e61717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier F., Nieminen K., Sánchez-Ferrero J.C., Rodríguez M.L., Chagoyen M., Hardtke C.S., Cubas P. (2014). Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26: 1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cook C.E., Whichard L.P., Turner B., Wall M.E., Egley G.H. (1966). Germination of witchweed (Striga lutea Lour.): isolation and properties of a potent stimulant. Science 154: 1189–1190. [DOI] [PubMed] [Google Scholar]
- Dai Y., Wang H., Li B., Huang J., Liu X., Zhou Y., Mou Z., Li J. (2006). Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 18: 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y.H., Liu N.Y., Tang Z.S., Liu J., Yang W.C. (2006). Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18: 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domagalska M.A., Leyser O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12: 211–221. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Beveridge C.A. (2009). Roles for auxin, cytokinin, and strigolactone in regulating shoot branching. Plant Physiol. 149: 1929–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roldan V., et al. (2008). Strigolactone inhibition of shoot branching. Nature 455: 189–194. [DOI] [PubMed] [Google Scholar]
- González-Grandío E., Poza-Carrión C., Sorzano C.O., Cubas P. (2013). BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 25: 834–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiaux C., Drummond R.S., Janssen B.J., Ledger S.E., Cooney J.M., Newcomb R.D., Snowden K.C. (2012). DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22: 2032–2036. [DOI] [PubMed] [Google Scholar]
- Jiang L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504: 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M., Hirano Y., Mori T., Kim S.Y., Kyozuka J., Seto Y., Yamaguchi S., Hakoshima T. (2013). Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes Cells 18: 147–160. [DOI] [PubMed] [Google Scholar]
- Lin H., Wang R., Qian Q., Yan M., Meng X., Fu Z., Yan C., Jiang B., Su Z., Li J., Wang Y. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21: 1512–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523. [DOI] [PubMed] [Google Scholar]
- Lu Z., et al. (2013). Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25: 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K., Sasaki E., Shimada Y., Nagae M., Ueno K., Nakano T., Yoneyama K., Suzuki Y., Asami T. (2009). Feedback-regulation of strigolactone biosynthetic genes and strigolactone-regulated genes in Arabidopsis. Biosci. Biotechnol. Biochem. 73: 2460–2465. [DOI] [PubMed] [Google Scholar]
- Nelson D.C., Scaffidi A., Dun E.A., Waters M.T., Flematti G.R., Dixon K.W., Beveridge C.A., Ghisalberti E.L., Smith S.M. (2011). F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 108: 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyter-Spira C., Al-Babili S., van der Krol S., Bouwmeester H. (2013). The biology of strigolactones. Trends Plant Sci. 18: 72–83. [DOI] [PubMed] [Google Scholar]
- Sang D., et al. (2014). Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. USA 111: 11199–11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi A., Waters M.T., Ghisalberti E.L., Dixon K.W., Flematti G.R., Smith S.M. (2013). Carlactone-independent seedling morphogenesis in Arabidopsis. Plant J. 76: 1–9. [DOI] [PubMed] [Google Scholar]
- Scaffidi A., Waters M.T., Sun Y.K., Skelton B.W., Dixon K.W., Ghisalberti E.L., Flematti G.R., Smith S.M. (2014). Strigolactone hormones and their stereoisomers signal through two related receptor proteins to induce different physiological responses in Arabidopsis. Plant Physiol. 165: 1221–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y., Sado A., Asami K., Hanada A., Umehara M., Akiyama K., Yamaguchi S. (2014). Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. USA 111: 1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Luong P., Huq E. (2007). The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol. 145: 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Li J. (2014). Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 21: 23–29. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Waters M.T. (2012). Strigolactones: destruction-dependent perception? Curr. Biol. 22: R924–R927. [DOI] [PubMed] [Google Scholar]
- Sorefan K., Booker J., Haurogné K., Goussot M., Bainbridge K., Foo E., Chatfield S., Ward S., Beveridge C., Rameau C., Leyser O. (2003). MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 17: 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga J.P., Smith S.M., Briggs W.R., Nelson D.C. (2013). SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 163: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Furner I.J., Ottoline Leyser H.M. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50: 80–94. [DOI] [PubMed] [Google Scholar]
- Stirnberg P., van De Sande K., Leyser H.M. (2002). MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 129: 1131–1141. [DOI] [PubMed] [Google Scholar]
- Szemenyei H., Hannon M., Long J.A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., Yamaguchi S. (2008). Inhibition of shoot branching by new terpenoid plant hormones. Nature 455: 195–200. [DOI] [PubMed] [Google Scholar]
- Waldie T., McCulloch H., Leyser O. (2014). Strigolactones and the control of plant development: lessons from shoot branching. Plant J. 79: 607–622. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li J. (2008). Molecular basis of plant architecture. Annu. Rev. Plant Biol. 59: 253–279. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Nelson D.C., Scaffidi A., Flematti G.R., Sun Y.K., Dixon K.W., Smith S.M. (2012). Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139: 1285–1295. [DOI] [PubMed] [Google Scholar]
- Xie X., Yoneyama K., Yoneyama K. (2010). The strigolactone story. Annu. Rev. Phytopathol. 48: 93–117. [DOI] [PubMed] [Google Scholar]
- Xiong G., Wang Y., Li J. (2014). Action of strigolactones in plants. Enzymes 35: 57–84. [DOI] [PubMed] [Google Scholar]
- Ye R., Wang W., Iki T., Liu C., Wu Y., Ishikawa M., Zhou X., Qi Y. (2012). Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol. Cell 46: 859–870. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang Y., et al. (2014). Rice cytochrome P450 MAX1 homologs catalyze distinct steps in strigolactone biosynthesis. Nat. Chem. Biol. 10: 1028–1033. [DOI] [PubMed] [Google Scholar]
- Zhao J., Wang T., Wang M., Liu Y., Yuan S., Gao Y., Yin L., Sun W., Peng L., Zhang W., Wan J., Li X. (2014). DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol. 55: 1096–1109. [DOI] [PubMed] [Google Scholar]
- Zhao L.H., et al. (2013). Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23: 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.H., et al. (2015). Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25: 1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., et al. (2013). D14-SCFD3-dependent degradation of D53 regulates strigolactone signalling. Nature 504: 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.