The maize PI/GLO ortholog Zmm16/sts1 is a B class gene that interacts with the zygomorphy and sex determination pathways of maize.
Abstract
In monocots and eudicots, B class function specifies second and third whorl floral organ identity as described in the classic ABCE model. Grass B class APETALA3/DEFICIENS orthologs have been functionally characterized; here, we describe the positional cloning and characterization of a maize (Zea mays) PISTILLATA/GLOBOSA ortholog Zea mays mads16 (Zmm16)/sterile tassel silky ear1 (sts1). We show that, similar to many eudicots, all the maize B class proteins bind DNA as obligate heterodimers and positively regulate their own expression. However, sts1 mutants have novel phenotypes that provide insight into two derived aspects of maize flower development: carpel abortion and floral asymmetry. Specifically, we show that carpel abortion acts downstream of organ identity and requires the growth-promoting factor grassy tillers1 and that the maize B class genes are expressed asymmetrically, likely in response to zygomorphy of grass floral primordia. Further investigation reveals that floral phyllotactic patterning is also zygomorphic, suggesting significant mechanistic differences with the well-characterized models of floral polarity. These unexpected results show that despite extensive study of B class gene functions in diverse flowering plants, novel insights can be gained from careful investigation of homeotic mutants outside the core eudicot model species.
INTRODUCTION
Most flowers possess a stereotypical ground plan, composed of radially symmetric, concentric whorls of distinct floral organs (from outside to inside: sepals, petals, stamens, and carpels). The flowers of grasses, including maize (Zea mays), are derived in several respects compared with the stereotypical pattern characteristic of both eudicots and monocots. Developmental genetic studies contrasting floral development between species that maintain stereotypical floral morphology (e.g., Arabidopsis thaliana) with species like maize, with more derived floral morphology, can reveal both what is conserved across angiosperms, as well as developmental pathways that have evolved to produce novel morphologies.
While the stamen and carpel whorls of maize are similar in both position and morphology with reproductive organs in other angiosperms, the outer sterile floral whorls of maize (and other grasses) are not so readily homologized (Clifford, 1987; Whipple and Schmidt, 2006). Consequently, the outer floral whorls have been given grass-specific names. The outermost sterile organ is called the lemma, while opposite and internal to the lemma is a bikeeled structure called the palea. Internal to the lemma and palea is a whorl of two (three or more in some grasses) short bulbous structures called lodicules that engorge in male flowers at anthesis and force them open. In addition to the presence of grass-specific organs, maize flowers are derived in two other notable respects. First, as is typical of many grasses, the lodicule whorl initiates only the two lateral members, while the medial lodicule fails to form, breaking the typical trimerous floral symmetry of monocots and making the maize flower zygomorphic (Rudall and Bateman, 2004). Additional asymmetry is present in the wide spacing of lateral stamens and in the fusion of two lateral carpels to form the silk (style), while the medial carpel contains the single ovule and does not contribute to the silk (Irish et al., 2003). Second, maize is monoecious, bearing unisexual flowers on distinct inflorescences. Male flowers are formed in a terminal apical inflorescence (tassel) by abortion of the carpel whorl, while female flowers are formed in axillary inflorescences (ears) by arrest of stamen growth (Cheng et al., 1983).
The ABCE model of floral development explains floral patterning by the combinatorial activity of four different gene functions (Coen and Meyerowitz, 1991; Pelaz et al., 2000). While the model is not strictly conserved across all angiosperms (Litt, 2007; Litt and Kramer, 2010), it provides a useful baseline against which to gauge morphological divergence, as is present in maize. According to the model, A function alone specifies sepal identity in the first (outermost) floral whorl. A and B function together confer petal identity in the second whorl. Combined B and C function confers stamen identity in the third whorl, and C function alone confers carpel identity in the fourth whorl (Bowman et al., 1991; Coen and Meyerowitz, 1991; Honma and Goto, 2001). E function is required throughout all four whorls to establish floral organ identity (Pelaz et al., 2000; Honma and Goto, 2001). The genetic factors controlling A, B, C, and E function have been identified and, with the exception of the A class gene APETALA2, they all belong to the MADS box family of transcription factor genes. The products of these MADS box genes regulate transcription by binding to specific DNA sequences known as CArG-boxes (Riechmann and Meyerowitz, 1997). Despite the unusual morphology of grass flowers, studies in both rice (Oryza sativa; Nagasawa et al., 2003; Yamaguchi et al., 2006; Dreni et al., 2011) and maize (Mena et al., 1996; Ambrose et al., 2000; Whipple et al., 2004) indicate that B and C functions are largely conserved between grasses and eudicot species, where these functions were originally defined. That C function is conserved across these species is perhaps not surprising, considering that their reproductive organs are clearly homologous. Given the unclear homology of the outer floral whorls in grasses, analyses of B class function have been more informative in the grasses, providing strong evidence that lodicules are modified second whorl petals (Ambrose et al., 2000; Whipple et al., 2007), while the palea and/or lemma may be homologous to sepals although that is less certain (Lombardo and Yoshida, 2015).
Limited evidence suggests that another critical aspect of B class function described in the eudicots is also conserved. Eudicot B class proteins are involved in unusual protein and regulatory interactions with important evolutionary implications. Unlike the other floral MADS box proteins, which bind DNA as homodimers (Huang et al., 1993; Riechmann et al., 1996a, 1996b; West et al., 1998; Egea-Cortines et al., 1999), many eudicot B class proteins bind DNA exclusively as heterodimers composed of an APETALA3/DEFICIENS (AP3/DEF) ortholog bound to a PISTILLATA/GLOBOSA (PI/GLO) ortholog (Schwarz-Sommer et al., 1992; Riechmann et al., 1996a, 1996b; Egea-Cortines et al., 1999). In addition, the B class obligate heterodimer either directly or indirectly positively autoregulates B class gene expression in tissues where both genes have been activated (Schwarz-Sommer et al., 1992; Goto and Meyerowitz, 1994; Jack et al., 1994). Although floral MADS box complexes are involved in a number of cross-regulatory relationships (Kaufmann et al., 2009), the B class combination of obligate heterodimerization and autoregulation is rare and may have played an important role in the evolution of a robust B class expression domain (Winter et al., 2002; Lenser et al., 2009). While some maize B class proteins form obligate heterodimers (Whipple et al., 2004), not all have been assayed and it remains unclear if they are involved in autoregulatory feedback loops.
Monoecy and zygomorphy are both derived aspects of maize floral morphology with unclear links to floral patterning. While sex determination and carpel abortion in tassel florets have received considerable attention (DeLong et al., 1993; Dellaporta and Calderon-Urrea, 1994; Calderon-Urrea and Dellaporta, 1999; Acosta et al., 2009), it is not yet clear if the abortion of reproductive organs is dependent on whorl position or organ identity. Significantly less attention has been given to the molecular underpinnings of zygomorphy in maize or any other grass. In contrast to radially symmetric flowers, most grasses have a single plane of symmetry (zygomorphy), as evidenced in maize by the suppression of the medial-adaxial lodicule, wide spacing of the lateral stamens, and modification of the medial adaxial carpel (Irish et al., 2003). In Antirrhinum, where floral zygomorphy is best understood, asymmetry appears to be established independently of both ABC MADS box gene floral patterning (Carpenter and Coen, 1990; Coen and Meyerowitz, 1991), as well as the initial phyllotactic patterning of the floral organs. However, there is no a priori reason that the zygomorphy exhibited by grass flowers would employ the same mechanism, especially considering that zygomorphy in the grass family arose independently from the order that contains Antirrhinum.
Here, our characterization of a B class mutant, sterile tassel silky ear1 (sts1), helps shed light on both conserved and divergent aspects of maize flower development. We show that sts1 is caused by loss of function in the maize PI/GLO-like gene Zea mays mads16 (Zmm16). We demonstrate that ZMM16/STS1, as well as its paralogs ZMM18 and ZMM29, form obligate heterodimers with the maize AP3/DEF ortholog SILKY1 and that Zmm16/sts1 and silky1 (si1) are involved in positive regulation of all four of the maize B class genes. Our investigation of ZMM16/STS1 protein localization, as well as of Zmm16/sts1 and si1 expression, reveals that the maize B class expression domain is asymmetric. Closer examinations of organ initiation inferred from auxin dynamics indicate that phyllotactic patterning of the medial lodicule is also directly affected by maize zygomorphy. These differences point to intriguing differences in the maize zygomorphy program. Finally, we demonstrate through double mutant and gene expression analyses that carpel arrest/abortion in tassel florets occurs downstream of organ identity and requires the growth promoting factor grassy tillers1 (gt1).
RESULTS
The sts1 Mutant Phenotype Is Consistent with Loss of B Function
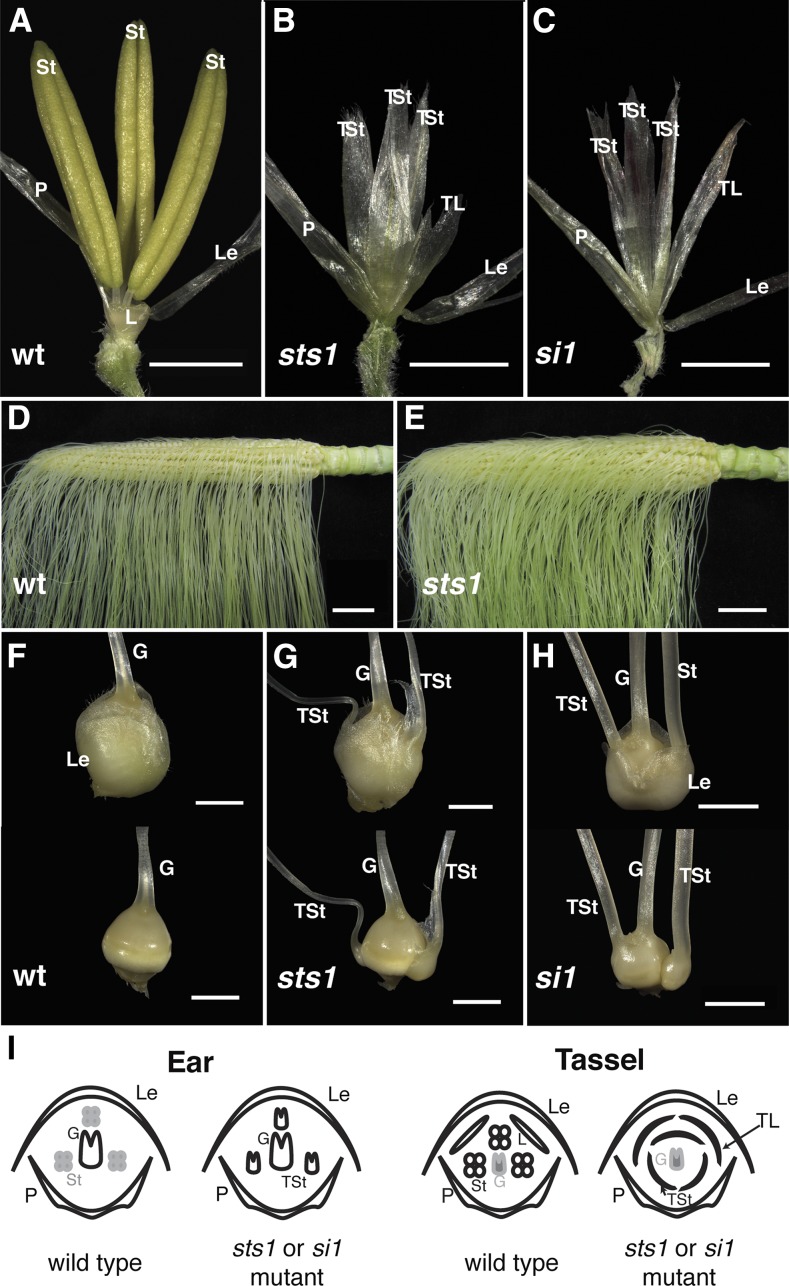
The sts1 mutant was discovered in a forward genetic screen for mutants affecting tassel floret development. sts1 mutants had no discernible vegetative phenotype, but floral development was dramatically altered, particularly in the tassel florets. While the lemma and palea of sts1 mutant tassel florets were indistinguishable from the wild type (Figure 1A), the second whorl lodicules were replaced by lemma/palea-like structures. Similar lemma/palea-like organs were also found in the third whorl normally composed of stamens (Figure 1B). Thus, sts1 mutants had a homeotic conversion of both second whorl lodicules and third whorl stamens into lemmas/paleae in tassel florets.
Figure 1.
Phenotypes of the sts1 and si1 Mutants.
(A) Wild-type tassel floret.
(B) and (C) sts1-1 (B) and si1-mum2 (C) tassel florets showing conversion of both stamens and lodicules into lemma/palea-like organs.
(D) Wild-type ear.
(E) sts1-1 mutant ear.
(F) Wild-type ear spikelet (above) and floret (below) with glumes removed.
(G) and (H) sts1-1 (G) and si-mum2 (H) mutant ear spikelets (above) and florets (below) with glumes removed showing transformation of aborted stamens to carpels
(I) Floral diagrams of wild-type and B class mutant ear and tassel florets.
G, gynoecium; St, stamen; TSt, transformed stamen; L, lodicule; TL, transformed lodicule; P, palea; Le, lemma. Bars = 2 cm in (D) and (E) and 2 mm in all other panels.
The ears of sts1 mutants had a higher density of silks compared with wild-type siblings (cf. Figures 1D and 1E). Closer inspection revealed that this higher silk density was due to the formation of extra carpels within the florets of sts1 ears. The number of extra carpels varied from floret to floret, with some florets having one extra carpel and others having two or three. Regardless of number, these additional carpels were always found in the third whorl normally occupied by arrested stamens (Figure 1G), indicating a homeotic conversion of stamens to carpels in sts1 ears.
The homeotic transformation of second and third whorl organs in sts1 resembled B class mutants in the ear florets; however, in tassel florets, stamens were transformed into lemma/palea-like organs rather than into carpels as would be predicted for a B class mutant. Unlike sts1, tassel florets of the B class mutant si1 were reported to have the expected stamen to carpel transformation (Ambrose et al., 2000), although only the si1-R and si1-5 alleles were described, neither of which has been molecularly characterized. To determine if this is the case for all si1 alleles, we analyzed the phenotype of si1-mum2, an RNA null mutant (see expression analysis below), which harbors a MuDR transposon in the 4th intron. Unlike si1-R and si1-5, the si1-mum2 tassel floret phenotype was identical to that of sts1, with both second and third whorls homeotically transformed into lemma/palea-like organs (Figure 1C). The ear floret phenotype of si1-mum2 resembled sts1, except that extra carpels were frequently fused to the central gynoecium (Figures 1E and 1H). Thus, sts1 strongly resembled the B class mutant si1, as both had an unusual stamen to lemma/palea transformation only in tassel florets.
Floral Development in sts1
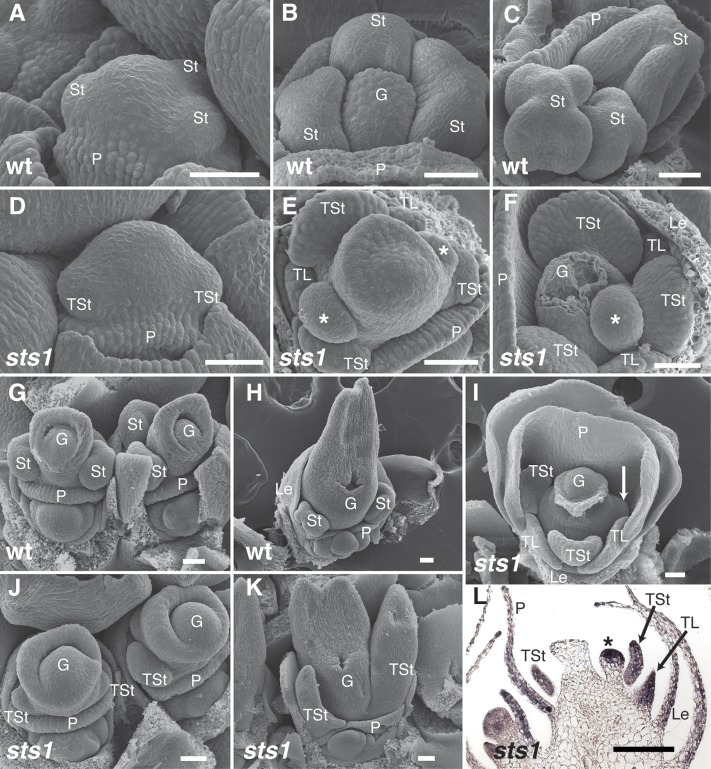
To gain a clearer understanding of the developmental basis of the unusual transformation in sts1 male florets, early developmental stages were analyzed using scanning electron microscopy. Shortly after initiation of the palea ridge, sts1 and wild-type male floral meristems were indistinguishable (Figures 2A and 2D). Following the initiation of palea primordia, wild-type florets formed three bulbous stamen primordia (Figures 2B and 2C). This was not the case for organs of the same position in sts1 florets, which were developmentally delayed and became ab/adaxially flattened (Figures 2E and 2F). Occasionally, third whorl organs failed to initiate in sts1 mutants (Figure 2I). Interestingly, we also noticed that ectopic primordia often initiated in the axils of the transformed third whorl organs (asterisk in Figures 2E and 2F). The number and position of these ectopic primordia varied: A few florets lacked ectopic primordia, many contained a single ectopic primordium in the axil of the medial transformed stamen, and others contained two ectopic primordia in the axils of both of the lateral transformed stamens. Second whorl primordia in sts1 were also ab/adaxially flattened when compared with the bulbous lodicule primordia of wild-type male florets (Figures 2E and 2F).
Figure 2.
Early Floral Development in sts1-1 Mutants.
(A) to (C) Scanning electron micrographs of wild-type tassel florets at successively older stages.
(D) to (F) Scanning electron micrographs of sts1-1 tassel florets at similar developmental stages as in (A) to (C). No clear differences compared with the wild type are observable in the earliest stage ([A] and [D]), but after floral organs emerge, sts1-1 mutants show transformation of lodicules and stamens into flattened lemma/palea-like organs, as well as ectopic floral organs (asterisk) that appear directly above the transformed stamens.
(G) and (H) Ear florets of the wild type.
(I) sts1-1 tassel floret showing failure of one lateral stamen to grow (arrow).
(J) and (K) Ear florets of sts1-1 mutants have transformation of stamens into carpels.
(L) In situ hybridization of lateral organ marker Zyb15 on sts1-1 tassel floret reveals that the ectopic primordium has a lateral organ identity.
G, gynoecium; St, stamen; P, palea; Le, lemma; TSt, transformed stamen; TL, transformed lodicule; Le, lemma. Asterisk indicates ectopic organ. Bar = 50 µm in (A) to (K) and 100 µm in (L).
At early stages of development, sts1 and wild-type ear florets resembled wild-type tassel counterparts (data not shown). Unlike tassel florets, as the third whorl organs developed in sts1 ears, they assumed a clear carpel morphology and developed a distinct silk (stigma) (Figures 2G, 2H, 2J, and 2K). We detected no ectopic primordia in sts1 ear florets.
The apparent axillary position and rounded shape of the ectopic organs of sts1 tassel florets suggested that they could be meristematic. Alternatively, they could be additional lateral organs formed in a more proliferative third whorl. To better determine the identity of these ectopic primordia, we examined expression of Zea mays yabby15 (Zyb15) in young sts1-1 tassel florets. Zyb15, like other YABBY orthologs, is a robust marker of lateral organs that is absent from meristems (Juarez et al., 2004; Goldshmidt et al., 2008; Sarojam et al., 2010; Whipple et al., 2010), and its expression was observed in the palea, lemma, transformed lodicules, transformed stamens, and young carpels as expected. Zyb15 was also expressed in the ectopic primordia, supporting the latter interpretation of an abnormally proliferating third whorl and suggesting that sts1 is necessary (at least in part) for limiting stamens number to three (Figure 2L).
Positional Cloning of sts1
To identify the genetic lesion responsible for the sts1 phenotype, we mapped the original sts1 mutant (sts1-1) using bulked-segregant analysis (Michelmore et al., 1991). Our mapping localized sts1-1 to the long arm of chromosome 3 (complete linkage, 0/44 recombinant chromosomes, with the simple sequence repeat marker umc1266). Because the sts1 phenotype is so similar to other reported loss of B function phenotypes, we hypothesized that it would map to a B class gene. While SI1 is not located on chromosome 3 (Ambrose et al., 2000), the PI/GLO ortholog Zmm16 (GRMZM2G110153) is only 3.39 Mb distal to umc1266 and was a likely sts1 candidate (Supplemental Figure 1). Sequencing of Zmm16 revealed that sts1-1 contained a G > A transition in the putative splice donor site of the third Zmm16 intron. A second sts1 allele was identified via a noncomplementation screen with sts1-1. This allele, referred to as sts1-2, has a G > A transition in the putative splice acceptor site of the fourth intron of Zmm16.
Additional confirmation that mutations in Zmm16 cause the sts1 phenotype was provided by a phenotypic rescue experiment using a Zmm16-YFP transgene expressed from its native promoter. Analysis of YFP signal in plants containing this transgene showed that Zmm16-YFP largely recapitulated the known expression pattern of endogenous Zmm16 (see below). Zmm16-YFP transgenic plants were crossed to sts1-1 mutants and the resulting F1 selfed. F2 progeny confirmed that Zmm16-YFP fully rescues the floret phenotype of sts1 (Supplemental Figure 2) and will henceforth be referred to as sts1-YFP.
STS1 Protein Localization during Ear and Tassel Floret Development
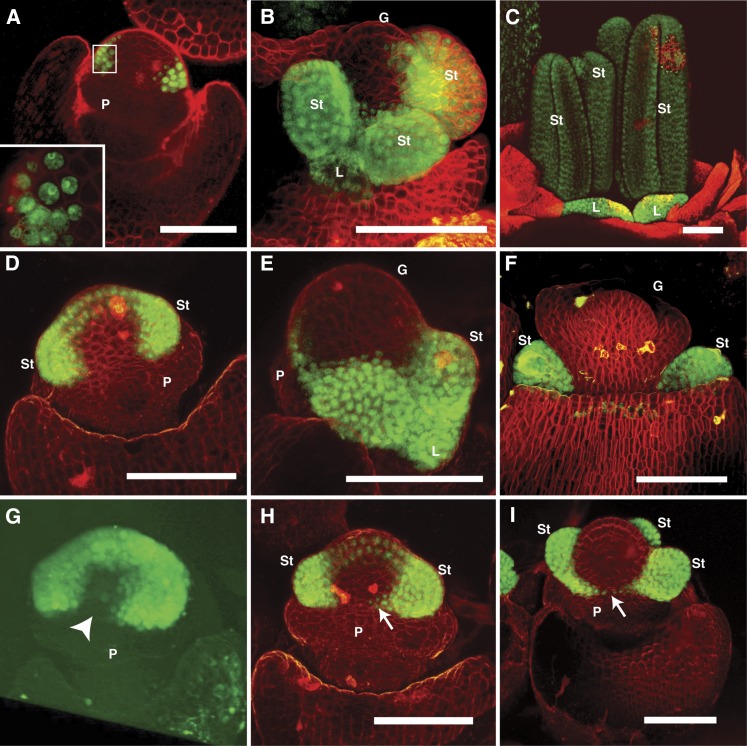
As a further investigation of the role of sts1 in floral development, we characterized STS1-YFP localization in developing tassel and ear florets. At early floral stages in both ears and tassels, STS1-YFP was observed in a broad, asymmetric, horseshoe shape, partially encircling the dome of the floral meristem (Figures 3A, 3D, and 3G). STS1-YFP signal was never detected in the adaxial region of the floral meristem, where the medial lodicule would arise if the second floral whorl were trimerous and actinomorphic (radially symmetric). In tassel florets, STS1-YFP was discernable before the stamen primordia began to bulge out from the meristem (Figure 3A). Thus, a zygomorphic STS1-YFP localization pattern is established before any morphological indication of zygomorphy. At these early stages, STS1-YFP was cytoplasmic in cells at the periphery of its expression domain, becoming nuclear in cells toward the interior of the developing floret (Figure 3A, inset). In all later stages examined, STS1-YFP was consistently nuclear. The cytoplasmic localization in early stages may indicate that SI1 is not coexpressed in these cells, as it has been demonstrated that coexpression of AP3 and PI is necessary for nuclear localization in Arabidopsis (McGonigle et al., 1996).
Figure 3.
STS1-YFP Localization.
(A) to (C) STS1-YFP localization in early (A), mid (B), and late (C) stages of tassel florets. Expression is detected throughout second and third whorl organs from early to late stages. Inset in (A) shows that in early stages, STS1-YFP is detected in the nucleus in cells proximal to the meristem, while cells more distal from the meristem also have cytoplasmic STS1-YFP.
(D) to (F) STS1-YFP localization in early (D), mid (E), and late (F) stages of ear florets. STS1-YFP is present throughout the development of second and third whorl organs, but absent from the gynoecium.
(G) to (I) Asymmetric localization of STS1-YFP during early stages of floret development. Three-dimensional reconstruction of STS1-YFP domain in an early tassel floret (G); note the absence of signal in an adaxial medial domain (arrowhead) that corresponds to the position of the missing medial lodicule. As stamens begin to initiate (H), STS1-YFP signal begins to appear in cells at the margins of the adaxial medial domain (arrow in [H]), eventually forming a narrow continuous stripe across the domain (arrow in [I]).
P, palea; St, stamen; L, lodicule; G, gynoecium. Bar = 100 µm in all panels.
At later stages, as stamens began to initiate, STS1-YFP localized to a narrow band in the adaxial medial domain in ear florets (Figures 3H and 3I). The structure and placement of tassel florets made imaging this region of the floral meristem extremely challenging, so we could not determine if a similar connecting band forms in tassel florets. Although STS1-YFP expression was maintained in stamens and lodicules until late in development (Figures 2C and 2F), no signal was detected in the gynoecium at any stage of floral development in either the ear or the tassel (Figures 3A, 3B, and 3D to 3F). This is in keeping with the results of earlier in situ hybridization experiments that showed no STS1 expression in the developing gynoecium of tassel florets (Whipple et al., 2004). However, Münster et al. (2001) report expression of sts1, as well as Zmm18/29, in the gynoecia of female florets, as assessed by RNA in situ hybridization. In addition, in maize expression browsers (Sekhon et al., 2011; qTELLER.com), sts1 expression is found in silks. We never observed STS1-YFP signal in developing silks (data not shown). This distinction from our results might be due to posttranscriptional regulation of STS1 in the gynoecium of ear florets, STS1-YFP protein levels in the gynoecium below the detection limits of our microscope, incomplete recapitulation of the native sts1 expression pattern by the transgene, or inability of the previous expression studies to distinguish among the maize PI/GLO paralogs.
Early Asymmetry in Maize Floret Development
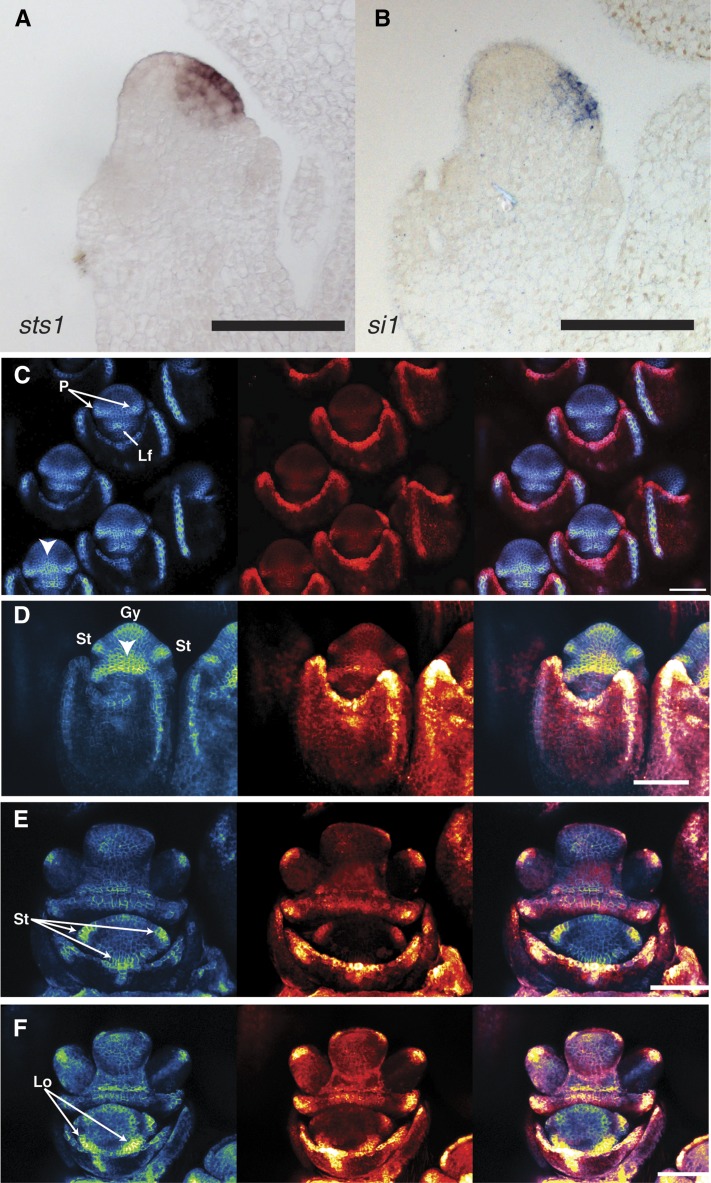
The asymmetric localization of the STS1-YFP construct was unexpected. In order to determine if B class gene transcripts show a similar pattern, we performed in situ hybridization of sts1 and si1. Both medial sections (Figures 4A and 4B) and serial sections through the flower (Supplemental Figures 3 and 4) demonstrated that the maize B class transcripts are absent from the adaxial region of the flower consistent with the STS1-YFP localization. These results confirm that B class genes are asymmetrically expressed at an early stage of flower development.
Figure 4.
Zygomorphy in B Class Expression and Phyllotaxy of Maize Florets.
(A) Medial section of male floret hybridized with anti-sts1 probe. Expression is observed in the abaxial domain, but absent from the adaxial domain.
(B) Section similar to (A) hybridized with anti-si1 probe shows similar expression pattern to sts1.
(C) to (F) Confocal z-stacks of Zm-PIN1-YFP (left), DR5-RFP (center), and merge (left). Early stages of the upper floret ([C], top) show two distinct PIN1 foci corresponding to the palea anlagen. As the florets mature, PIN1 and DR5 both increase in the center of the palea (arrowhead in [C], bottom). Later floral stages (D), when stamen primordia begin to emerge, show a broad area of PIN1 activity associated with the palea that spreads throughout the adaxial region of the flower primordium (arrowhead). Three distinct stamen PIN1 DR5 anlagen can be detected at early stages of the lower floret (E); however, only two lodicule anlagen are marked by PIN1 and DR5 (F).
Gy, gynoecium; Lo, lodicule; Lf, lower floret; P, palea; St, stamen. Bar = 100 µm.
Lack of B class gene expression in the region of the medial-adaxial lodicule points to a distinct mechanism establishing floral zygomorphy in maize compared with other better-characterized systems with zygomorphic flowers. In Antirrhinum majus, and indeed in many zygomorphic flowers, all expected organs initiate, but growth of some is inhibited or altered by the zygomorphy program (Carpenter and Coen, 1990; Rudall and Bateman, 2004; Wang et al., 2008; Hardy et al., 2009). In contrast, there is no morphological evidence for the initiation of the medial-adaxial lodicule in maize. This suggests that in maize, the zygomorphy mechanism acts at a very early stage of primordium establishment, as opposed to inhibiting outgrowth of primordia with typical phyllotactic patterning. The earliest known marker for phyllotactic patterning of lateral organs (including floral primordia) is an auxin maximum, which can be seen by PIN1/DR5 upregulation at incipient primordia (Benková et al., 2003; Heisler et al., 2005; Petrásek et al., 2006; Wisniewska et al., 2006). We looked at PIN1a-YFP and the synthetic auxin response promoter DR5rev:RFP (Gallavotti et al., 2008) in maize wild-type male florets to determine if the medial-adaxial lodicule is initiated as are the organs affected by the zygomorphy program in other systems.
We could find no evidence for an individualized auxin maximum to mark the site of the medial-adaxial lodicule. After the lemma forms, the next floral primordium to initiate is the palea. Two distinct PIN1/DR5 loci initiate at early stages of palea development, with a reduced but still detectable upregulation in a narrow band between these foci (Figure 4C). Later, when the medial region of the palea begins to grow out, this same region has a strong PIN1/DR5 signal that expands both adaxially and laterally into the future morphologically undifferentiated region between the palea and the rest of the flower (Figures 4C and 4D). We observed clear PIN1/DR5 foci for all stamen primordia (Figures 4D and 4E) and the two lateral lodicules (Figure 4F). Unlike the stamens and two lateral lodicules, we observed no distinct PIN1/DR5 focus for the medial-adaxial lodicule. However, we did see a broad region of PIN1/DR5 associated with the palea that expanded into the expected region of the medial-adaxial lodicule. Although the palea-associated auxin maximum expanded into the expected region of the medial lodicule, there was never a distinct focus of PIN1/DR5 upregulation as seen for the lateral lodicules. That early stages of both floral primordium initiation and B class gene expression are adaxially repressed underscores significant differences with taxa like A. majus in the mechanism by which the maize zygomorphy program is performed.
RNAi Knockdown of sts1 Paralogs Has No Effect on Floret Phenotype
Phylogenetic reconstructions indicate that grasses have two paralogous PI/GLO lineages (Kim et al., 2004; Whipple et al., 2007; Wei and Ge, 2011). Zmm16/sts1 belongs to the PI-2 clade, while maize has two representatives in the PI-1 clade (Zmm18 and Zmm29) that resulted from a recent tandem duplication (Münster et al., 2001). ZMM18 and ZMM29 protein products are 97.2% similar to each other, and they are also very similar to STS1 (∼84% similar) (Münster et al., 2001). In addition, sts1, Zmm18, and Zmm29 have very similar expression patterns (Münster et al., 2001; Whipple et al., 2004). Given these similarities, it is surprising that mutation of sts1 alone is sufficient to produce a strong loss of B function phenotype and suggests that Zmm18 and Zmm29 are not fully functionally redundant with Zmm16/sts1. To better understand the functions of ZMM18 and ZMM29, we knocked down their expression using an RNAi construct designed to recognize both Zmm18 and Zmm29, but not sts1. Ten independent T0 transgenic RNAi lines were obtained, and none of these had an obvious floral phenotype. We assayed each of the 10 T0 lines by RT-PCR for reduced Zmm18/29 levels and identified two lines with reduced expression (data not shown). These two lines were crossed to the wild type, and from the resulting progeny, transgene (+) and transgene (−) individuals were again assayed by RT-qPCR for Zmm18/29 levels (Supplemental Figure 5). Both lines showed significant reduction in Zmm18/29, and one line had strong downregulation (∼17-fold), yet still the florets had no detectable mutant phenotype (data not shown). These results suggest sts1 can cover for reduction in Zmm18/29, but this redundancy is not mutual, as Zmm18/29 cannot cover for reduced sts1. However, we have no direct evidence that Zmm18/29 is redundant with sts1, even though it would be expected based on sequence and expression similarity.
Obligate Heterodimerization and Autoregulation of Maize B Class Genes
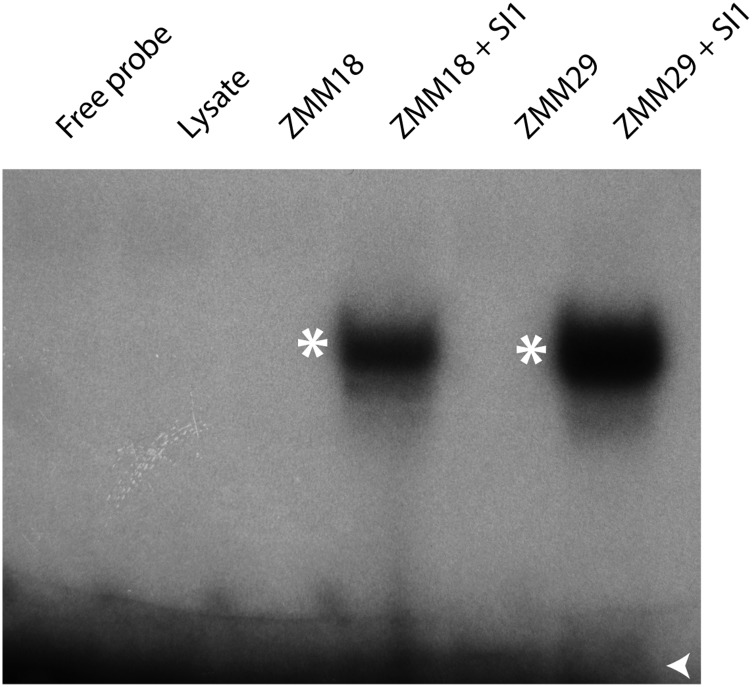
Previous analysis of maize B class function showed that SI1 and ZMM16/STS1 interact as obligate heterodimers (Whipple et al., 2004). However, it was not known if ZMM18 or ZMM29 also formed obligate heterodimers with SI1. To test this, we performed electromobility shift assays (EMSAs) using a radiolabeled CArG-box probe derived from the AP3 promoter (Whipple et al., 2004). Neither ZMM18 nor ZMM29 were capable of binding the probe alone, but both bound DNA in combination with SI1 (Figure 5). Thus, all maize B class proteins bind DNA as obligate heterodimers similar to the core eudicots.
Figure 5.
Maize PI/GLO Orthologs Bind DNA as an Obligate Heterodimer.
EMSA using in vitro-transcribed and translated ZMM18, ZMM29, and SI1 proteins and P32-labeled CArG-box probe. Neither ZMM18 nor ZMM29 on its own is capable of binding the CArG-box probe, but both bind DNA with the maize AP3/DEF ortholog SI1. Free probe indicated with an open arrowhead and shifted bands by an asterisk.
B class gene expression in Arabidopsis is initiated in part by the activity of LEAFY and UNUSUAL FLORAL ORGANS (Parcy et al., 1998) but then maintained by positive autoregulation (Schwarz-Sommer et al., 1992; Goto and Meyerowitz, 1994; Jack et al., 1994). After initiation of maize B class genes, it is not clear whether they also maintain their own expression as is seen in other eudicots. To determine whether the maize SI1/STS1 heterodimer positively regulates B class genes, we compared the expression of sts1, si1, and Zmm18/Zmm29 in sts1-1, si1-mum2, and wild-type florets. Because of extremely high coding sequence similarity and probable functional redundancy between Zmm18 and Zmm29, we chose to assay expression of these two genes together using primers that do not distinguish between them. Expression of all of the B class genes was analyzed by RT-qPCR using cDNA isolated from three pools of successively older developmental stages.
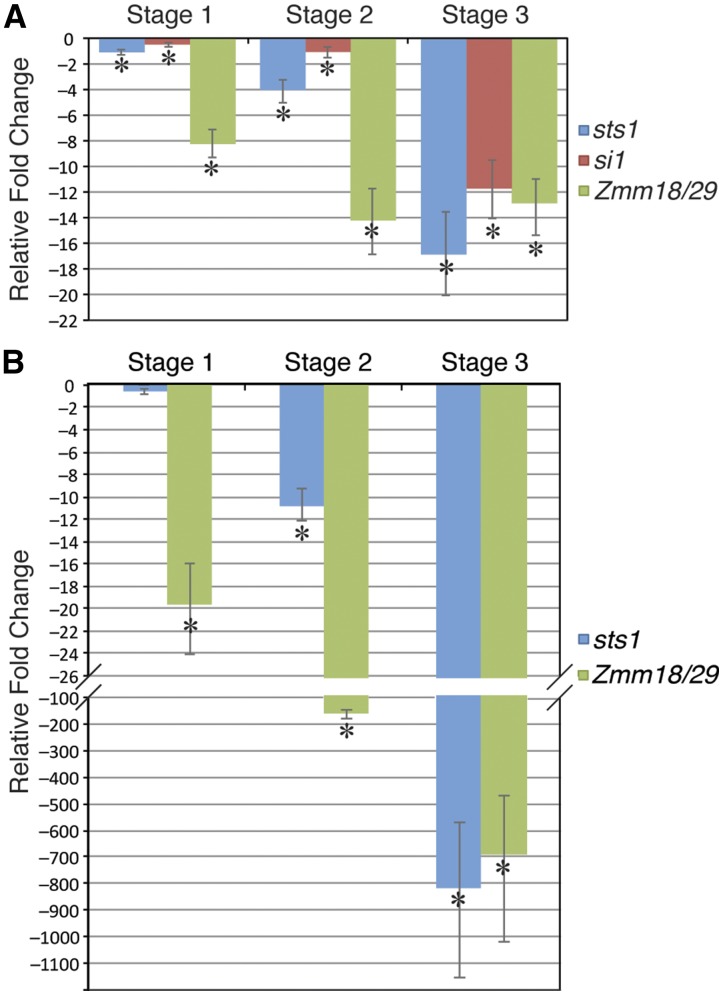
At early stages of sts1 florets (stage 1: see Methods for stage descriptions), there was a slight but significant decrease in the expression of si1 (1.5-fold), which became much more pronounced at later stages of development (2.1-fold decrease at stage 2 and 12.8-fold decrease at stage 3) (Figure 6A). Zmm18/Zmm29 expression was strongly decreased in sts1-1 inflorescences at all stages (9.3- to 14-fold decrease). sts1 transcript levels were also decreased in sts1-1 mutants relative to the wild type. However, this decrease was more pronounced at later stages similar to what was observed for si1. In si1-mum2 mutants, si1 transcripts were indistinguishable from background. sts1 transcript levels were not significantly affected at the earliest stage in si1 mutants, but decreased at later stages (11- to 800-fold) as was seen for si1 transcript levels in sts1 mutants. Zmm18/Zmm29 expression in si1-mum2 tassels was strongly decreased at all stages similar to what was observed in sts1 mutants (Figure 6B).
Figure 6.
Positive Autoregulation of Maize B Class Genes.
(A) RT-qPCR expression analysis of B class genes in successively older stages of sts1-1 tassel florets. Expression of all B class genes show significant downregulation at all stages, but the downregulation is more dramatic at later stages.
(B) RT-qPCR expression analysis of B class genes in si1-mum2 tassel florets.
In both (A) and (B), fold change is calculated relative to wild-type controls at the same stage. Error bars show sd from the mean, while asterisk indicates downregulation that is significant at P < 0.05 as determined by a t test.
Taken together, these expression data are consistent with a positive autoregulatory feedback loop involving the SI1/STS1 heterodimer. At the earliest stages, sts1 and si1 transcript levels are only mildly affected, consistent with SI1/STS1 heterodimer-independent initial activation in the young florets of these stages. However, in later floral stages, sts1 and si1 clearly fail to maintain their own expression as would be predicted by positive autoregulation. The lack of Zmm18/29 expression at even the earliest stages of development in sts1 or si1 mutants was unexpected and suggests that the SI1/STS1 heterodimer is required to initiate and/or amplify Zmm18/29 expression. That Zmm18/29 are transcriptionally downstream of sts1 and si1 helps explain the lack of redundancy of sts1 with these closely related paralogs and the consequent strong homeotic phenotype of the sts1 mutant. Since no phenotype was observed in the Zmm18/29 RNAi lines, it is possible that sts1, Zmm18, and Zmm29 do indeed act redundantly but only after SI1/STS1-dependent initiation of Zmm18/29.
Unexpected Transformation of Stamens in Male Florets Is Likely a Consequence of Carpel Abortion, Which Functions Independently of Whorl Position and Requires gt1 Activity
The distinct transformation of third whorl organs in sts1 ear versus tassel florets was not readily explained by known B class gene functions. The ABCE model predicts that third whorl organs of B class mutants will express C class genes alone and thus assume a carpel identity. Since sts1 mutants produce lemma/palea-like organs in the third whorl, one possibility is that C class gene expression is reduced in the third whorl of sts1 mutants, possibly as a result of a positive regulation of C class expression by B class genes, as has been seen in several ranunculids (Gonçalves et al., 2013; Lange et al., 2013). Under this scenario, we would expect to see a reduction in C class gene expression in the third whorl of sts1 male florets. To test this, we examined expression of two maize C class genes Zea agamous1 (Zag1) and Zmm2.
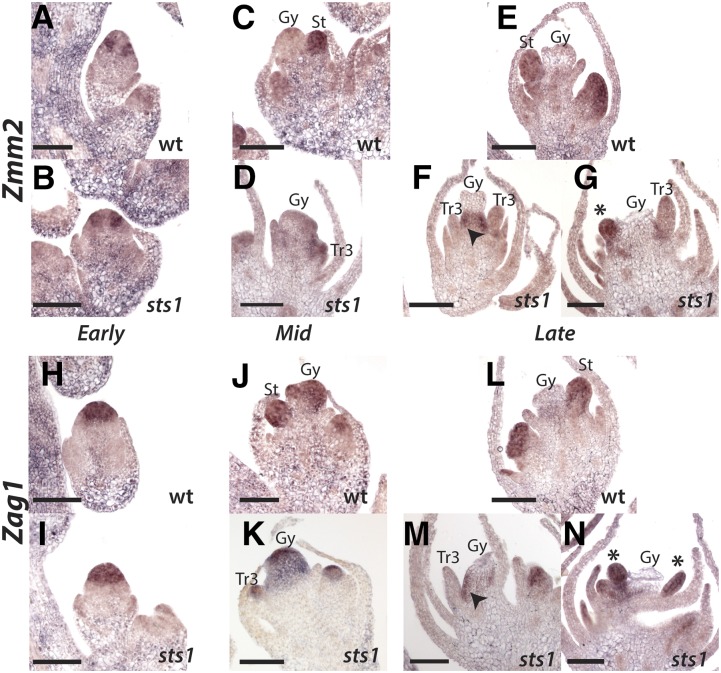
At an early stage, before the emergence of floral primordia, Zmm2 is expressed in a ring of cells surrounding the center of the meristem of both wild-type and sts1 mutant florets (Figures 7A and 7B). At a later stage, when stamen primordia have initiated in the wild type, Zmm2 was strongly expressed in emerging stamens but absent from the central gynoecium (Figure 7C). The same stage in sts1 showed similar localization of Zmm2, although expression was weak in the transformed third whorl organs compared with stamens in wild-type flowers (Figure 7D). At a later stage after emergence of the gynoecium, Zmm2 expression was maintained in wild-type stamens (Figure 7E). In sts1 mutants at this stage (and slightly later, after carpel abortion has begun), Zmm2 was either absent or weakly expressed in transformed third whorl organs of sts1 (Figures 7F and 7G). Interestingly, Zmm2 was strongly expressed at both the base of gynoecium (Figure 7F) and in young ectopic primordia that emerge from this region (Figure 7G). Zag1 showed a partially overlapping, but distinct expression pattern, compared with Zmm2. At an early stage, Zag1 was expressed throughout the center of the meristem, with no apparent difference between wild-type and sts1 florets (Figures 7H and 7I). After emergence of stamens and the gynoecial ridge, strong Zag1 expression was maintained in the stamens and gynoecium (Figure 7J). In sts1 mutants, Zag1 was similarly maintained in the gynoecium and transformed third whorl organs (Figure 7K). At a later stage, after carpel abortion had begun, Zag1 was downregulated in the gynoecium but maintained in the stamens of the wild type (Figure 7L). At a similar stage in sts1 mutants, Zag1 was downregulated in the aborting tissues of the gynoecium and transformed third whorl organs (Figure 7M). Similar to Zmm2, we also observed Zag1 in the region at the base of the gynoecium (Figure 7M) and in ectopic primordia that emerge from this region (Figure 7N). In sum, we found maize Zmm2 and Zag1 to be expressed largely as expected for C class genes, with the exception that Zmm2 was consistently absent from carpel primordia, and both C class genes were downregulated in the aborting gynoecium. Expression of these C class genes in sts1 mutants was largely consistent with the pattern in the wild type, with expression in third whorl organs expected to have a carpel identity and decreasing as these organs matured, consistent with the fate of carpels in male florets. The main difference in sts1 mutants was that both C class genes were expressed in a novel domain proximal to aborting tissue of the gynoecium, which we never observed in the wild type. In sum, C class genes were eventually downregulated in transformed third whorl organs, although downregulation occurred later in organ development and is likely a consequence of carpel abortion rather than a loss of C class positive regulation in the third whorl of sts1 mutants.
Figure 7.
C Class Gene Expression in Male Florets of the Wild Type and sts1.
(A) to (G) Anti-Zmm2 probe hybridized to male florets of wild-type (top) and sts1 mutants (bottom) at early, middle, and late stages. In early stages, both the wild type (A) and sts1 (B) have expression in the presumptive future stamen whorl, which is absent from the central domain of the meristem that will give rise to carpels. In middle stages, when stamen primordia emerge, Zmm2 is expressed in stamens of the wild type (C) and weakly in transformed third whorl organs of sts1 (D). At later stages, Zmm2 expression is maintained strongly in growing stamens (E) but absent from transformed third whorl organs ([F] and [G]). In late sts1 florets, Zmm2 is expressed at the base of the aborting gynoecium (arrowhead in [F]) and in ectopic third whorl organs (asterisk in [G]).
(H) to (N) Anti-Zag1 probe hybridized as in (A) to (G). Early Zag1 expression is throughout the center of the floral meristem in both the wild type and sts1 ([H] and [I]). Expression continues in the stamens and gynoecium of middle staged florets of the wild type (J), as well as the gynoecium and transformed third whorl of sts1. At later stages, Zag1 is absent from the aborting gynoecium of the wild type but maintained in the stamen. In late sts1 florets, Zag1 is absent from the aborting gynoecium as well as the transformed third whorl (M). Similar to Zmm2, there is some Zag1 expression at the base of the gynoecium (arrowhead in [M]) and in ectopic primordia (asterisk in [N]) of sts1 mutants. Gy, gynoecium; St, stamen; Tr3, transformed third whorl organs. Asterisk indicates ectopic organs.
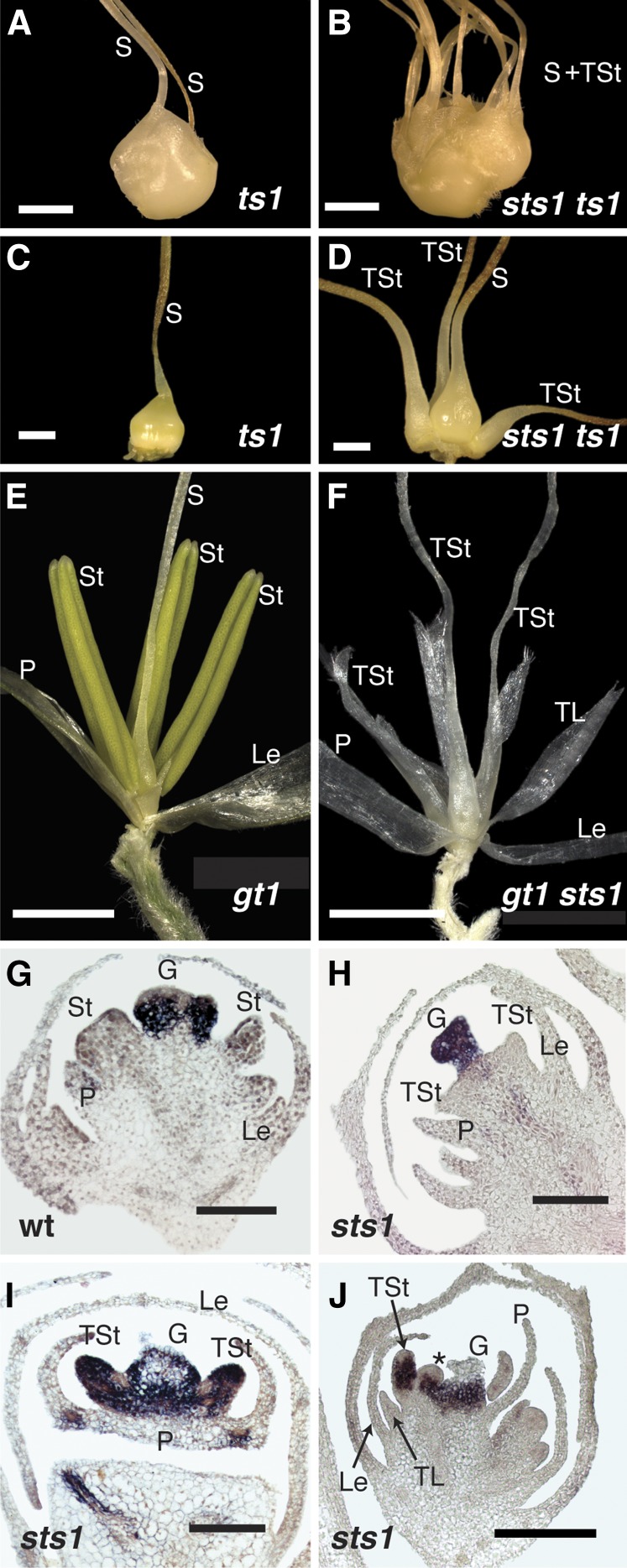
Given that C class gene expression in transformed third whorl organs of sts1 mutants largely parallels C class expression in the aborting carpel, and the fact that transformation of the stamen whorl to lemma/palea was limited to male florets in sts1, we reasoned that the maize male sex determination pathway could be involved. Specifically, we hypothesized that third whorl organs in sts1 mutants with homeotic carpel identity activate the carpel abortion pathway. To test this, we generated double mutants between sts1-1 and tassel seed1 (ts1) (Acosta et al., 2009). ts1 mutants show a nearly complete feminization of tassels, including growth of a fertile gynoecium in the central whorl, arrest of third whorl stamen growth, glume reduction, and feminization of other secondary sexual traits (Figures 8A and 8C). Tassel florets of the ts1 sts1-1 double mutants were feminized like the ts1 mutant but in addition the third whorl was fully transformed into carpels similar to ear florets of sts1 mutants (Figures 8B and 8D), suggesting that the unusual third whorl phenotype of sts1 is indeed the result of the carpel abortion pathway of male florets.
Figure 8.
Carpel Abortion Occurs Downstream of Organ Identity, Not Position.
(A) Tassel spikelet of a ts1 mutant showing complete feminization of both primary and secondary sex characters. A silk emerges from both the upper and lower floret.
(B) Tassel spikelet of sts1 ts1 double mutant, showing the emergence of silks and transformed stamens.
(C) Upper floret dissected from ts1 spikelet shown in (A).
(D) Upper floret dissected from sts1 ts1 spikelet shown in (B).
(E) Tassel floret of gt1 showing normal masculine secondary sex characteristics but failure to abort the carpel in the center of the floret.
(F) Tassel floret of gt1 sts1 double mutant, with stamens transformed into organs with partial carpel identity evidenced by a distinct silk at their apex.
(G) to (J) In situ hybridization with gt1 probe on wild-type (G) and sts1 tassel florets ([H] to [J]). There is strong gt1 expression in the central gynoecium of both the wild-type and sts1. During early stages of third whorl organ initiation in sts1 florets (H), gt1 is not detected in transformed stamens. In later stages ([I] and [J]), gt1 is strongly express in third whorl transformed stamens, as well as in ectopic organs (J).
G, gynoecium; Le, lemma; P, palea; S, silk; St, stamen; TSt, transformed stamen; TL, transformed lodicule. Asterisk indicates ectopic organ. Bar = 2.0 mm in (A) to (F) and 100 µm in (G) to (J).
While the ts1 sts1 phenotype was consistent with our hypothesis, given that ts1 mutants are feminized in both primary and secondary sex characters (essentially turning the tassel into an ear), it is perhaps not surprising that the ts1 sts1 tassel would resemble the sts1 ear. To control for secondary sexual characters affected by ts1, we also made a double mutant with grassy tillers1 (gt1), which fails to inhibit growth of carpels in the central whorl of the male floret but does not regulate any other sex-specific morphology (Whipple et al., 2011) (Figure 8E). gt1-1 sts1-1 tassel florets showed the expected B class lodicule transformation, but third whorl organs had varying degrees of carpeloidy. We did not see a complete conversion of third whorl organs into carpels, but rather mosaic organs that usually had carpel tissue at the base and frequently a short silk (stigma) at the apex, with varying degrees of lemma/palea-like tissue at the margins (Figure 8F). Occasionally these mosaic organs contained anther tissue (Supplemental Figure 6). The partial carpel identity of these mosaic organs confirm that gt1 is in part responsible for the lack of carpel transformation of sts1 male florets, but that other factors likely contribute. Thus, the growth of lemma/palea-like organs in the third whorl of sts1 mutants can be largely explained by activation of the carpel abortion pathway caused by homeotic transformation of stamens to carpels. As was seen in the gynoecium of male florets, C class expression in the third whorl of sts1 was eventually lost, leaving a “default” lemma/palea identity for these organs. For reasons that are not immediately clear, male floret third whorl organs are not fully aborted in sts1. Our results also suggest that in the gt1 sts1 double mutant background other factors (likely Zmm18/29) are sufficient to provide some stamen identity in a stochastic manner.
gt1 is strongly expressed in the carpel of male florets, consistent with its role in carpel repression (Whipple et al., 2011). Based on the gt1 sts1 phenotype, we expected to observe ectopic gt1 expression in third whorl organs of sts1 mutants. Consequently, we performed in situ hybridization of gt1 in sts1 tassels. After formation of the gynoecial ridge, gt1 expression was observed exclusively in central carpel primordia in both sts1-1 and the wild type (Figures 8G and 8H). While gt1 expression was not detected in third whorl organs of sts1-1 in these early stages, third whorl organs of later stages clearly expressed gt1, consistent with its growth repression role in carpeloid tissue of third whorl organs in sts1 male florets (Figures 8I and 8J). Interestingly, gt1 expression was also observed in the ectopic lateral primordia of the sts1 third whorl, further supporting that these organs have a carpel identity (Figure 8J).
DISCUSSION
sts1 Is Zmm16, a Maize PI/GLO Ortholog Required for B Class Function
Here, we demonstrate that sts1 is the maize B class MADS box gene Zmm16. We show that sts1 specifies organ identity in floral whorls 2 and 3, similar to the AP3/DEF ortholog si1. Interestingly, sts1 exhibits no redundancy with paralogous Zmm18/29 nor is there evidence for organ subfunctionalization of the maize PI/GLO genes as reported in rice (Prasad and Vijayraghavan, 2003; Yadav et al., 2007). The lack of redundancy appears to result from a requirement for the SI1/STS1 heterodimer to initiate Zmm18/29 expression. We could detect no phenotype in a strongly silenced Zmm18/29 RNAi line, suggesting that after initiation of Zmm18/29, the maize PI/GLO orthologs function redundantly. Silencing studies in rice indicate that the sts1 ortholog Os-MADS2 is required for lodicule identity, while Os-MADS2 acts redundantly with Os-MADS4 (the Zmm18/29 ortholog) in specifying stamen identity (Prasad and Vijayraghavan, 2003; Yadav et al., 2007). The apparent incongruity between maize and rice PI/GLO paralog functions could either result from incomplete silencing in rice or a lack of organ-specific expression subfunctionalization in maize. Resolution of this awaits characterization of complete loss-of-function alleles for Os-MADS2 and Os-MADS4.
Our results confirm previous studies showing that many aspects of maize B class function are conserved among monocots and eudicots. Specifically, we show that monocot B function is required for the identity of second and third whorl organs and that this activity requires the coexpression of an AP3/DEF ortholog with a PI/GLO ortholog, which then heterodimerize in an obligate manner. However, our analysis of the maize B class function also lends insight into divergent aspects of maize development. Namely, we show that early B class expression and early phyllotactic patterning of lodicules is asymmetric, that carpel abortion acts downstream of organ identity, and that maize B function regulates determinacy of the third whorl of developing male florets. Below, we consider implications of our results to the evolution of B class function in the angiosperms as well as unexpected connections to monoecy, zygomorphy, and determinacy in maize florets.
Obligate Heterodimerization and Autoregulation of the Maize B Class Proteins
Protein-protein interactions are crucial for the activity of floral homeotic MADS box proteins. For example, dimerization is necessary for binding of the MADS domain to DNA (Pellegrini et al., 1995; Riechmann et al., 1996a). In addition, higher-order MADS complexes form and are thought to provide specific identity to each whorl as described in the quartet model (Honma and Goto, 2001; Pelaz et al., 2001; Theissen and Saedler, 2001). Unlike all other floral MADS box proteins, which bind DNA as homodimers, B class AP3 and PI orthologs from the eudicots bind DNA as obligate AP3/PI heterodimers (Riechmann et al., 1996a). In addition, AP3/PI positively autoregulate their expression. Since both AP3 and PI must be present in order to function, any shift in the B class expression domain must involve a tandem shift in both AP3 and PI in order to have functional consequences. Thus, heterodimerization may provide a canalization mechanism restricting the domain of B class function (Krizek and Meyerowitz, 1996). Computational simulations show that the combination of obligate heterodimerization and autoregulation provides a more robust ON or OFF switch in response to an initial activating signal than does homodimerization and autoregulation (Lenser et al., 2009). Although careful functional tests are still lacking, B class heterodimerization and autoregulation have been proposed to have important evolutionary consequences. Specifically, the angiosperm AP3 and PI lineages are the result of a gene duplication event that occurred early after angiosperm divergence from the gymnosperms (Kramer et al., 1998; Kramer and Irish, 2000; Kim et al., 2004). Analyses of obligate heterodimerization among B class proteins in gymnosperms and monocots suggest that eudicot obligate heterodimerization evolved from an ancestral homodimerizing state (Winter et al., 2002). After the AP3/PI duplication, the paralogs were likely capable of both homo- and heterodimerization, and over time obligate heterodimerization evolved by successive loss of homodimerization first in the AP3 lineage and later in the PI lineage (Winter et al., 2002; Melzer et al., 2014). Reports of PI-like homodimers in the monocots (Winter et al., 2002; Kanno et al., 2003; Tsai et al., 2005; Whipple and Schmidt, 2006) may help explain the variability in petaloidy seen in the first two whorls of many monocot flowers.
In maize, the B class proteins SI1 and STS1 (ZMM16) were previously shown to bind DNA as obligate heterodimers (Whipple et al., 2004). Here, we show that the maize PI co-orthologs ZMM18 and ZMM29 also bind DNA as obligate heterodimers. In addition, we show that the SI1/STS1 heterodimer is required to maintain si1 and sts1 expression and to initiate robust Zmm18/29 expression. Thus, maize B class proteins show the unusual obligate heterodimerization/autoregulation combination described in eudicot species. While this seems to support the view that obligate heterodimerization/autoregulation of maize B class proteins is conserved with eudicots, the occurrence of homodimerizing PI orthologs in non-grass monocots complicates the issue. Indeed, the PI ortholog of Joinvillea, a close outgroup to the grass family, can also homodimerize (Whipple and Schmidt, 2006), suggesting that the obligate heterodimerization characteristic of maize and some rice B class proteins (Moon et al., 1999; Yao et al., 2008) evolved independently. Further sampling and functional analysis of hetero- versus homodimerizing monocot B class proteins may help resolve the contribution, if any, of obligate heterodimerization to the evolution of floral development.
Maize Floral Zygomorphy and B Class Function
Floral zygomorphy is a derived condition, characteristic of a few large and important families (e.g., Orchidaceae, Poaceae, Asteraceae, and Fabaceae), although dispersed throughout the angiosperms (Endress, 2012). In the eudicots, there is evidence that the CYCLOIDEA (CYC)-like TCP transcription factors have been repeatedly recruited to control the development of zygomorphy (Luo et al., 1996, 1999; Feng et al., 2006; Yang et al., 2012). In the monocots, the role of the CYC-like genes in floral development (if any) remains undefined. In Commelina (Commelinales) and the Zingiberales, there is some evidence for a correlation between CYC-like gene expression and the development of floral zygomorphy (Bartlett and Specht, 2011; Preston and Hileman, 2012). Functional studies of the CYC-like genes in the grasses implicate them in the control of shoot branching (Doebley et al., 1995; Kebrom et al., 2006), inflorescence branch angle and leaf morphology (Bai et al., 2012), and lemma versus palea identity in rice (Yuan et al., 2009), not structural floral zygomorphy. In a number of orchids, the development of distinct second and third whorl tepals associated with zygomorphy is associated with variability in AP3/DEF homolog expression and variability in floral MADS box complex formation (Mondragón-Palomino and Theissen, 2008, 2011; Hsu et al., 2015). Aside from these sparse examples, the genetic underpinnings of floral zygomorphy in the broader monocots remain largely mysterious.
A fully symmetric flower in the monocots would be expected to have alternating trimerous whorls of floral organs, with each organ equally spaced and similar in final shape. Within the order that contains the grasses, the Poales, zygomorphy is rare, although it is pervasive in the grass family (Rudall and Bateman, 2004; Endress, 2012). Although most often discussed in terms of the second and third floral whorls (petal and stamen homologs), zygomorphy can affect all four whorls of a flower, and symmetry patterns can differ within a flower (Endress, 2012). Several aspects of the maize flower demonstrate asymmetry. Leaving aside the palea and lemma, which are of uncertain homology, the lodicule, stamen, and gynoecium (second, third, and fourth whorls) each have some degree of zygomorphy. The lodicule whorl of maize, and indeed many grasses, is structurally zygomorphic due to the suppression of the adaxial-medial lodicule. The third whorl produces all three expected stamen primordia, but their orientation is not equidistant (120°), rather the two adaxial-lateral stamens are displaced abaxially, offset in a straight line (180°) with the gynoecium. Of the three carpels of the maize gynoecium, only the abaxial-lateral two grow and fuse to form the silk (style), while the adaxial-medial carpel does not form a silk and is ovulate making the fourth whorl zygomorphic as well. As noted by Irish et al. (2003), the concentration of zygomorphy effects on the adaxial side of the maize flower is similar to the situation in A. majus, where CYC and its homologs inhibit growth adaxially/dorsally. However, beyond this similarity, we demonstrate here that several aspects of zygomorphy establishment in maize are distinct from what is seen in A. majus. In particular, the inhibited lodicule shows no evidence of phyllotactic patterning. In many zygomorphic flowers, like those of A. majus, inhibited primordia are initiated but their subsequent growth repressed (Carpenter and Coen, 1990; Rudall and Bateman, 2004; Wang et al., 2008; Hardy et al., 2009). Furthermore, in maize, B class gene expression is asymmetrically absent from the adaxial domain. Although later stages were examined, a similar B class asymmetry was reported in other monocots (Mondragón-Palomino and Theissen, 2008, 2011; Preston and Hileman, 2012; Hsu et al., 2015). This contrasts with A. majus where the zygomorphy program acts independently (i.e., in parallel) of ABC floral organ patterning (Carpenter and Coen, 1990). Thus, maize has an unusual zygomorphy program in that both floral phyllotaxis and early expression of B class genes are directly influenced. While our data do not point to a direct role for B class genes in establishing asymmetry, they do reveal interesting differences between maize and A. majus in how the zygomorphy program is implemented.
How the zygomorphy program is established in maize remains an open question. Eudicot systems, where polar expression of TCP genes plays a key role in zygomorphy establishment, represent a paradigm case of evolutionary parallelism (Specht and Howarth, 2015). Thus, it is possible that TCP genes were also recruited in the origin of asymmetry characteristic of many grasses. Although, as noted above, no characterized grass TCP gene has a floral zygomorphy phenotype. In maize, the wandering carpel mutant occasionally has symmetrical gynoecia, but the mutation is weakly expressive and has no reported effect on the symmetry of lodicules or stamens (Irish et al., 2003). Indeed, no bona fide peloric (i.e., symmetric) mutant has been described in the grasses. This is somewhat surprising, given the common occurrence of peloric mutants outside the grass family and the extensive mutant screens performed in rice and maize. The lack of symmetry mutants is not sufficient to rule out the possibility that a distinct genetic program for zygomorphy exists in maize, possibly involving TCP genes as is so common in eudicots. Nevertheless, the lack of mutants combined with distinct features of zygomorphy in maize underscores the need to seriously consider other mechanistic explanations. One intriguing possibility was originally proposed by Cocucci and Anton (1988). After a comparative morphological study of grass flowers, they hypothesized that the placement and shape of the palea produces an inhibitive zone that represses initiation and growth in neighboring organs of other whorls. Such inhibitive zones, created by prominent or precocious development of particular floral organs, have been frequently noted in comparative morphological studies in other systems as well (Hofmeister, 1868; Buzgo, 2001; Remizowa et al., 2013). At the molecular level, such a palea inhibition zone could be explained by auxin dynamics in the floral meristem. Specifically, models of phyllotaxy involving formation of auxin maxima by dynamic localization of the auxin efflux protein PIN1 suggest that a large auxin sink would inhibit formation of any nearby auxin maxima (Reinhardt et al., 2003; Smith et al., 2006). Interestingly, our examination of maize PIN1a and DR5 in the maize floral meristem indicates that the palea indeed forms a large, persistent auxin maximum. This could explain the failure to initiate a medial lodicule close to the palea, as well as the wide spacing of the stamens. Such a model would be less consistent with a distinct genetic determinant of floral polarity and more consistent with indirect effects of physical and geometric constraints. Whether the palea auxin maximum is responsible for inhibition of the lodicule warrants further study. Even if the palea inhibition zone is real and causative, it still raises the question of how the palea itself is asymmetrically patterned at the adaxial side of the floret.
Carpel Abortion Is a Complex Process Involving gt1 and Acting Downstream of Organ Identity
As a monoecious plant, maize produces male and female flowers on separate inflorescences, the apical tassel and lateral ear, respectively. The maize sex determination pathway is not limited to the sex organs, but also influences the development of sterile organ and rachis morphology (Dellaporta and Calderon-Urrea, 1994; Irish, 1996). The classic feminizing mutants in maize, ts1 and ts2, affect both primary and secondary sex characteristics with tassels resembling ears in many respects, including failure to abort carpels and modification of the outer sterile organs of the spikelet and floret. ts1 and ts2 both encode biosynthetic enzymes (DeLong et al., 1993; Acosta et al., 2009) that may act in the jasmonic acid biosynthesis pathway (Acosta et al., 2009), although there is some evidence that ts2 produces a signal directly involved in the programmed cell death of the carpel (Calderon-Urrea and Dellaporta, 1999). Compared with ts1 and ts2, the role of gt1 in sex determination is much more limited. Tassel florets of gt1 mutants grow stunted or deformed carpels but maintain the proper secondary sex characteristics. gt1 encodes a homeodomain leucine-zipper (HD-Zip) transcription factor that inhibits growth in dormant tiller buds and the carpel of male florets (Whipple et al., 2011). Taken together, a more detailed pathway is emerging for male sex determination. ts1 and ts2 appear to be regulating hormonal control of male identity, which is upstream of both carpel abortion and modification of secondary sex morphology of the sterile organs. gt1, on the other hand, acts significantly downstream of this signal, and only to inhibit carpel growth. Notably, the function of gt1 does not suggest a direct role in programmed cell death, but rather growth inhibition and dormancy. Thus, carpel abortion in maize is a complex process that involves genes inhibiting growth (gt1) and other unidentified factors necessary for programmed cell death.
Our results make it clear that B class-dependent establishment of floral organ identity acts upstream of the maize carpel abortion pathway and that tassel floret carpel abortion requires the correct organ identity, not position. This is in contrast to cucumber (Cucumis sativus), where organ abortion occurs in a whorl-specific manner regardless of organ identity (Kater et al., 2001). This is not altogether surprising, however, considering that floral unisexuality has evolved numerous times by recruitment of a diverse array of developmental processes (Diggle et al., 2011). Reduced expression of C class genes in the gynoecium of the wild type and in the transformed third whorl organs of sts1 indicate that the maize carpel abortion pathway eventually abolishes C class-directed organ identity, which helps explain the palea/lemma identity in the sts1 whorl. It is interesting that the carpel abortion pathway activated ectopically in the third whorl of sts1 tassel florets is frequently incomplete, although occasionally third whorl organs of sts1 are fully suppressed (Figure 2I). The incomplete abortion could be a result of incomplete transformation to carpel identity of the sts1 third whorl. Consistent with this explanation is the observation in rice that the CRABS CLAW ortholog DROOPING LEAF (DL) establishes carpel identity in parallel with C class activity (Yamaguchi et al., 2004). If the maize DL ortholog also promotes carpel identity in parallel to C class function, this activity could be lacking in the third whorl of sts1 mutants, resulting in a partial carpel identity for this whorl, although this seems unlikely considering that the rice B class mutant, superwoman, expresses DL ectopically in the third whorl (Yamaguchi et al., 2004). Alternatively, third whorl organs of sts1 mutants have a complete carpel identity, but the carpel abortion pathway is not as robust as in the fourth whorl. A more careful examination of si1 alleles, si1-R and si1-5, or the modifier loci associated with the genetic background of those alleles that allows for carpel growth in the third whorl could help resolve this question.
B Class Function and Floral Determinacy
Another unexpected result of our phenotypic characterization of sts1 was that tassel florets produce ectopic third whorl organs with an apparent carpel identity, suggesting that in maize B function is required to limit proliferation of the third whorl. However, B class mutants in both maize and rice enhance the indeterminacy of C class mutants (Ambrose et al., 2000; Yun et al., 2013), indicating that in grasses, B class activity positively contributes more broadly to floral meristem determinacy, and not just determinacy of the third whorl. It is interesting to note that the region where ectopic primordia form in the male florets of sts1 is also marked by ectopic C class gene expression, while wild-type male florets downregulate C class genes in this domain. This could be explained by a late reactivation of meristematic activity (and associated C class expression) at the base of the aborting carpel. This indeterminacy in sts1 contrasts with Antirrhinum, where B function negatively regulates floral meristem determinacy (Tröbner et al., 1992; Davies et al., 1999). Nevertheless, ectopic third whorl organs were only observed in sts1 male florets and not in female florets, perhaps implicating an interaction with the sex determination pathway. In light of this sexual dimorphism of sts1 expressivity, it is interesting to note that the cucumber temperature sensitive gp mutant is also indeterminate when grown under low temperature, but only in the male flowers (Kater et al., 2001). It is not immediately clear why the determinacy function of B class genes would depend on the sex determination pathway in maize and cucumber, but it suggests again that careful examination of ABCE mutants in divergent systems can reveal unexpected phenotypes that provide a window into divergent aspects of floral development.
METHODS
Genetic Stocks
sts1-1 was isolated from a forward genetic screen for floral mutants. The M2 plants used in this screen were generated via ethyl methanesulfonate (EMS) treatment of A619 maize (Zea mays) inbreds and subsequent selfing of the resultant M1 plants (Neuffer, 1994). sts1-2 was isolated from a screen for noncomplementation with sts1-1. The 4000 M1 plants used in this screen were generated via pollination of sts1-1 ears with EMS-mutagenized pollen (Neuffer, 1994) from a different maize inbred, Mo17. gt1-1 was also isolated from a screen of EMS-treated A619 M2s (Whipple et al., 2011). si1-mum2 was provided by Robert Schmidt (Ambrose et al., 2000), and ts1 was provided by Jane Dorweiler, who obtained it from the Maize GDB stock center.
The STS1-YFP transgene was created by in-frame fusion of YFP to the C terminus of STS1 (details are below) and transformed into Hi Type II hybrid embryos was performed by the Iowa State University plant transformation facility via Agrobacterium tumefaciens-mediated transformation (Frame et al., 2002).
RNAi constructs were prepared using nearly identical DNA sequences from the 5′ and 3′ untranslated regions (UTRs) of Zmm18 and Zmm29. Zmm18/Zmm29-specific 5′ UTR sequence was amplified via PCR using Zmm18/Zmm29-5′SpeI-For and Zmm18/Zmm29-5′-Rev as primers (all primer sequences are in Supplemental Table 1). 3′ UTR sequence specific to these genes was also PCR amplified (using Zmm18/Zmm29-3′-For and Zmm18/Zmm29-3′XmaI-Rev as primers). The desired products of these two reactions were fused via an additional PCR using Zmm18/Zmm29-5′SpeI-For and Zmm18/ZMM29-3′XmaI-Rev on a mixture of the 5′ and 3′ fragments. The resulting 321 bp 5′/3′ fusion and a second copy (created by amplification with Zmm18/Zmm29-5′AscI-For and Zmm18/Zmm29-3′AvrII-Rev as primers) were cloned in an inverted-repeat orientation into pMCG1005. These two inverted repeats were separated by a waxy-a intron in order to promote in vivo formation of an RNA hairpin loop. The completed RNAi construct was transformed via Agrobacterium-mediated transformation (Frame et al., 2002) into Hi Type II hybrid embryos by the Iowa State University plant transformation facility.
Double Mutants
ts1 sts1 double mutants were generated by crossing ts1 plants to sts1-1 homozygotes. F1s from these crosses were grown to maturity and selfed. The F2 plants were also grown to maturity and then screened for feminization of the tassel, indicating ts1 homozygosity, and these were genotyped for sts1-1. gt1 sts1 double mutants were created by crossing gt1-1 plants to sts1-1 homozygotes. Resultant F1s were grown to maturity and selfed. F2 plants were grown to maturity and screened for the homozygous gt1-1 tillering phenotype. These tillering mutants were then genotyped for sts1-1.
Genotyping
The sts1 sequence spanning the site of the EMS-induced sts1-1 mutation was amplified via PCR using sts1-CAPS-For and sts1-CAPS-Rev as primers. Amplicons from these reactions were digested using BPU10I and separated on a 1% agarose gel. Wild-type sts1 amplicons are cut once by BPU10I. The resulting fragments are 224 and 556 bp in length. sts1-1 amplicons are not cut by BPU10I and are 780 bp long. For plants containing sts1-YFP transgenes, endogenous sts1 sequence was amplified using sts1-CAPS-For and sts1-CAPS-Rev1 as primers. sts1-CAPS-Rev1 spans sequence that is interrupted in sts1-YFP transgenes by insertion of YFP. Therefore, transgenic sts1-YFP is not amplified in PCRs that use these two primers. Following PCR, the amplicons of endogenous sts1 were digested using BPU10I and separated on a 1% agarose gel. Wild-type sts1 amplicons from these PCRs are cut twice by BPU10I. Resulting fragments are 180, 557, and 787 bp long. sts1-1 amplicons from these PCRs are cut once by BPU10I and are 180 bp and 1344 bp long.
sts1-YFP transgenic plants, which carry the bar selectable marker, were scored via leaf treatment with Finale (5.78% glufosinate-ammonium) diluted in water (1:90 Finale:water). This dilution was applied to a 4-cm region on the ad- and abaxial surfaces of a mature maize leaf using a Q-tip. Treated leaves were scored 1 week after treatment for signs of chlorosis and desiccation. Plants showing no significant affect from the Finale treatment were scored as carrying the transgene. PCR using sts1-and YFP-specific primers on genomic DNA from some of these confirmed the presence of transgene.
Wild-type si1 sequence was amplified by PCR using primers si1-For and si1-R. Because these primers anneal to DNA sequence on either side of the si1-mum2 Mutator transposon insertion, they do not amplify si1-mum2 sequence. In a separate reaction, si1-mum2 sequence was amplified by PCR using SI1-For and Mu-TIR6. The SI1-For and Mu-TIR6 primer pair does not amplify wild-type sequence because the Mu-TIR6 primer is specific to the Mutator transposon.
The gt1-1 mutation was genotyped with a CAPS marker as described by Whipple et al. (2011).
Scanning Electron and Confocal Microscopy
Inflorescences were fixed in freshly prepared FAA (3.7% formaldehyde, 5% acetic acid, and 50% ethanol) for 16 h or more. Following fixation they were washed with 35% ethanol and 15% ethanol. They were then washed three times with 0.1 M sodium cacodylate buffer and postfixed for 4 h in 1% osmium tetroxide in 0.1 M sodium cacodylate. Following postfixation, the samples were rinsed six times with deionized water, then dehydrated through a graded ethanol series (starting with 10% ethanol, and then progressing through 30% ethanol, 50% ethanol, 70% ethanol, 95% ethanol, and 100% ethanol) and dried with a critical point drier. These dried samples were mounted, dissected, sputter-coated with gold palladium, and then viewed at 10 kV accelerating voltage with a FEI XL30 environmental scanning electron microscope. STS1-YFP fluorescence was imaged by confocal and two-photon microscopy as described (Bommert et al., 2013). PIN1-YFP and DR5-RFP were imaged with a Zeiss 710, using freshly dissected live tissue. YFP and RFP were excited with a 488- and 561-nm laser, respectively, in two separate channels. Raw images were edited and Z-stacks created with ImageJ.
RNA in Situ Hybridization
Wild-type and sts1-1 inflorescences were fixed overnight at 4°C in 4% in paraformaldehyde in 1× PBS. These fixed samples were then dehydrated in ethanol, transitioned into Histo-Clear (National Diagnostics), embedded in Paraplast, sectioned, and hybridized according to the protocol previously described by Jackson et al. (1994). Hybridizations were performed using the gt1 or Zyb15 antisense digoxygenin-labeled RNA probe described by Whipple et al. (2011). Antisense probes for Zag1 and Zmm2 were created by PCR amplification of two small fragments (150 to 250 bp) from the 3′ region of the cDNA that spanned exon 7-8 or exon 9, respectively. These fragments were subcloned into pGEM T-easy (Invitrogen), linearized with SalI, and T7 antisense digoxygenin-UTP labeled RNA generated by the MegaScript kit (Life Technologies). A similar approach was taken to generate sts1 and si1 probes, but these were made to Exon1 and Exon7. In all cases, equal amounts of the two probes were mixed prior to hybridization. Primers for the probe constructs are in Supplemental Table 1.
RT-qPCR Expression Analysis
For Zmm18/Zmm29 RNAi and wild-type control lines, total RNA was extracted from unopened spikelets of recently emerged tassels. For sts1-1 and si1-mum2 mutants and wild-type siblings, male inflorescences were dissected and divided into three stages and frozen in liquid nitrogen. Stage 1 included inflorescence branch primordia ∼1 cm long, with florets of stages A-early G as described (Cheng et al., 1983). Stage 2 comprised 2.5-cm-long central spikes, containing florets of stages G-I. Stage 3 contained mature, but unopened spikelets from recently emerged tassels. RNA was extracted from these tissues using TRI reagent (Sigma-Aldrich) and the Qiagen RNeasy Plant Mini Kit (Qiagen) as follows: Frozen tissue was ground in liquid nitrogen using mortar and pestle, mixed with 1 mL of TRI reagent and transferred to a clean tube. Chloroform (200 μL) was added, and the mixture was vortexed and centrifuged at 15,000g for 10 min. The resulting supernatant (200 μL) was added to 700 μL of Qiagen RLT buffer, and 500 μL 100% ethanol was then added to the mixture and subsequently mixed by vortexing. This mixture was added to an RNeasy Mini Spin Column and purified according to the manufacturer’s protocol with the following modifications: (1) replacement of the RW1 buffer wash step with two washes using 750 μL of 80% ethanol, and (2) final elution with 11 μL of RNase-free water. Resulting RNA samples were treated with DNase (Promega) following the manufacturer’s protocol and reverse transcribed using an oligo(dT) primer and SuperScript III RT (Invitrogen) according to the manufacturer’s protocol.
sts1, si1, and Zmm18/29 expression was analyzed by qPCR of cDNA using TaqMan chemistry. qPCR reactions were performed on a StepOnePlus Real-Time PCR machine using the TaqMan Gene Expression Master Mix (Applied Biosystems). Reaction volumes were 20 μL and consisted of 10 μL of TaqMan Gene Expression Master Mix, 2 μL of 6 µM forward primer, 2 μL of 6 µM reverse primer, 2 μL of 2.5 µM TaqMan probe, 2 μL of molecular grade water, and 2 μL of 50 ng/μL cDNA. Each reaction was performed in triplicate under the following cycling parameters: 50°C for 2 min, 95°C for 10 min, and 40 cycles of two-step amplification (95°C for 15 s followed by 60°C for 1 min). Each reaction plate assayed the expression of B class genes and the endogenous αTubulin control gene in three wild-type biological replicates and three mutant biological replicates. No-RT controls were also included in these plates. ΔCt, ΔΔCt, and standard deviations were calculated by the StepOne software v2.2.2. Significance of ΔΔCt values was determined by a two-tailed Student’s t test. Average fold change was determined using the 2ΔΔCt method (Livak and Schmittgen, 2001), and fold change range was determined via 2ΔΔCt + SD and 2ΔΔCt − SD.
EMSAs
Full-length cDNAs for Zmm18, Zmm29, and silky1 were cloned into the pSPUTK expression vector (Stratagene). Protein was synthesized using the TnT Quick Coupled Transcription/Translation system (Promega). EMSAs were performed as previously described (Whipple et al., 2004).
Creation of sts1-YFP Transgene
A C-terminal, in-frame fusion of YFP to the genomic sequence of sts1 including ∼4.2 kb of promoter 5′ to the start codon, complete sts1 coding sequence including the introns, and ∼1.2 kb of sequence 3′ of the stop codon, was created using the MultiSite Gateway Three Fragment System (Invitrogen) by a modification of the methods described by Mohanty et al. (2009). All fragments were amplified using Phusion Taq polymerase (New England Biolabs). The 5′ promoter and coding sequence up to the stop codon was amplified with primers sts1_attB4 and sts1_attB1(R) and cloned into the pDONR P4-P1R vector using BP recombinase (Invitrogen). The coding 3′ UTR was amplified with primers sts1_attB2 and sts1_attB3(R) and cloned into the pDONR P2R-P3 vector. The citrineYFP fragment was amplified with primers YFP_attB1 and YFP_attB2(R) and cloned into pDONR221 vector using BP recombinase (Invitrogen). The pDONR P4-P1R, pDONR P2R-P3, and pDONR221 vector fragments were combined and transferred to the pTF101 Gateway-compatible maize transformation vector by a multisite LR recombination reaction (Invitrogen). Confirmed clones were transferred to Agrobacterium and transformed into maize, as described (Mohanty et al., 2009).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: gt1, GRMZM2G005624; si1, GRMZM2G139073; sts1/Zmm16, GRMZM2G110153; tubulin, AC195340.3_FG001; zag1, GRMZM2G052890; Zmm2, GRMZM2G359952; Zmm18, GRMZM5G805387; Zmm29, GRMZM2G152862; and Zyb15, GRMZM2G079293.
Supplemental Data
Supplemental Figure 1. Positional cloning of sts1.
Supplemental Figure 2. Complementation of sts1-1 by Zmm16-YFP.
Supplemental Figure 3. sts1 expression in male florets.
Supplemental Figure 4. si1 expression in male florets.
Supplemental Figure 5. Expression analysis of Zmm18/29 RNAi knockdown lines.
Supplemental Figure 6. Partial stamen identity in gt1 sts1 double mutants.
Supplemental Table 1. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Jane Dorweiler for sharing ts1 seed. Anonymous reviewers provided helpful comments to improve an earlier version of this manuscript. Funding for this work was provided by grants from the National Science Foundation (IOS-1025121 to C.J.W.; MCB-1027445 and IOS-1238202 to D.P.J.).
AUTHOR CONTRIBUTIONS
C.J.W., M.E.B., R.J.S., and D.P.J. designed the research. S.K.W., M.E.B., C.J.W., Z.T., D.H.H., A.G., and S.D. performed the research. S.K.W., M.E.B., and C.J.W. analyzed the data. C.J.W., M.E.B., and S.K.W. wrote the article.
Glossary
- EMSA
electromobility shift assay
- EMS
ethyl methanesulfonate
- UTR
untranslated region
Footnotes
Articles can be viewed online without a subscription.
References
- Acosta I.F., Laparra H., Romero S.P., Schmelz E., Hamberg M., Mottinger J.P., Moreno M.A., Dellaporta S.L. (2009). tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323: 262–265. [DOI] [PubMed] [Google Scholar]
- Ambrose B.A., Lerner D.R., Ciceri P., Padilla C.M., Yanofsky M.F., Schmidt R.J. (2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5: 569–579. [DOI] [PubMed] [Google Scholar]
- Bai F., Reinheimer R., Durantini D., Kellogg E.A., Schmidt R.J. (2012). TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc. Natl. Acad. Sci. USA 109: 12225–12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett M.E., Specht C.D. (2011). Changes in expression pattern of the teosinte branched1-like genes in the Zingiberales provide a mechanism for evolutionary shifts in symmetry across the order. Am. J. Bot. 98: 227–243. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bommert P., Je B.I., Goldshmidt A., Jackson D. (2013). The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502: 555–558. [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20. [DOI] [PubMed] [Google Scholar]
- Buzgo M. (2001). Flower structure and development of Araceae compared with alismatids and Acoraceae. Bot. J. Linn. Soc. 136: 393–425. [Google Scholar]
- Calderon-Urrea A., Dellaporta S.L. (1999). Cell death and cell protection genes determine the fate of pistils in maize. Development 126: 435–441. [DOI] [PubMed] [Google Scholar]
- Carpenter R., Coen E.S. (1990). Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev. 4: 1483–1493. [DOI] [PubMed] [Google Scholar]
- Cheng P.C., Greyson R.I., Walden D.B. (1983). Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am. J. Bot. 70: 450–462. [Google Scholar]
- Clifford H.T. (1987). Spikelet and floral morphology. In Grass Systematics and Evolution, Soderstrom T.R., Hilu K.W., Campbell C.S., Barkworth M.E., eds (Washington, D.C.: Smithsonian Institution Press; ), pp. 21–30. [Google Scholar]
- Cocucci A.E., Anton A.M. (1988). The grass flower - Suggestions on its origin and evolution. Flora 181: 353–362. [Google Scholar]
- Coen E.S., Meyerowitz E.M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- Davies B., Motte P., Keck E., Saedler H., Sommer H., Schwarz-Sommer Z. (1999). PLENA and FARINELLI: redundancy and regulatory interactions between two Antirrhinum MADS-box factors controlling flower development. EMBO J. 18: 4023–4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta S.L., Calderon-Urrea A. (1994). The sex determination process in maize. Science 266: 1501–1505. [DOI] [PubMed] [Google Scholar]
- DeLong A., Calderon-Urrea A., Dellaporta S.L. (1993). Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74: 757–768. [DOI] [PubMed] [Google Scholar]
- Diggle P.K., Di Stilio V.S., Gschwend A.R., Golenberg E.M., Moore R.C., Russell J.R.W., Sinclair J.P. (2011). Multiple developmental processes underlie sex differentiation in angiosperms. Trends Genet. 27: 368–376. [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A., Gustus C. (1995). teosinte branched1 and the origin of maize: evidence for epistasis and the evolution of dominance. Genetics 141: 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L., Pilatone A., Yun D., Erreni S., Pajoro A., Caporali E., Zhang D., Kater M.M. (2011). Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18: 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress P.K. (2012). The immense diversity of floral monosymmetry and asymmetry across angiosperms. Bot. Rev. 78: 345–397. [Google Scholar]
- Feng X., et al. (2006). Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. USA 103: 4970–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame B.R., Shou H., Chikwamba R.K., Zhang Z., Xiang C., Fonger T.M., Pegg S.E.K., Li B., Nettleton D.S., Pei D., Wang K. (2002). Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol. 129: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A., Yang Y., Schmidt R.J., Jackson D. (2008). The relationship between auxin transport and maize branching. Plant Physiol. 147: 1913–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldshmidt A., Alvarez J.P., Bowman J.L., Eshed Y. (2008). Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell 20: 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves B., Nougué O., Jabbour F., Ridel C., Morin H., Laufs P., Manicacci D., Damerval C. (2013). An APETALA3 homolog controls both petal identity and floral meristem patterning in Nigella damascena L. (Ranunculaceae). Plant J. 76: 223–235. [DOI] [PubMed] [Google Scholar]
- Goto K., Meyerowitz E.M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8: 1548–1560. [DOI] [PubMed] [Google Scholar]
- Hardy C.R., Sloat L.L., Faden R.B. (2009). Floral organogenesis and the developmental basis for pollinator deception in the asiatic dayflower, Commelina communis (Commelinaceae). Am. J. Bot. 96: 1236–1244. [DOI] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hofmeister W. (1868). Allgemeine morphologie der gewachse. In Handbuch der Physiologischen Botanik, de Barry A., Irmisch T., and Sachs J., eds (Leipzig, Germany: W. Engelman; ), pp. 405–664. [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529. [DOI] [PubMed] [Google Scholar]
- Hsu H.-F., Hsu W.-H., Lee Y.-I., Mao W.-T., Yang J.-Y., Li J.-Y., Yang C.-H. (2015). Model for perianth formation in orchids. Nat. Plants 1: 15046. [Google Scholar]
- Huang H., Mizukami Y., Hu Y., Ma H. (1993). Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 21: 4769–4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish E.E. (1996). Regulation of sex determination in maize. BioEssays 18: 363–369. [Google Scholar]
- Irish E.E., Szymkowiak E.J., Garrels K. (2003). The wandering carpel mutation of Zea mays (Gramineae) causes misorientation and loss of zygomorphy in flowers and two-seeded kernels. Am. J. Bot. 90: 551–560. [DOI] [PubMed] [Google Scholar]
- Jack T., Fox G.L., Meyerowitz E.M. (1994). Arabidopsis homeotic gene APETALA3 ectopic expression: transcriptional and posttranscriptional regulation determine floral organ identity. Cell 76: 703–716. [DOI] [PubMed] [Google Scholar]
- Jackson D., Veit B., Hake S. (1994). Expression of maize Knotted1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413. [Google Scholar]
- Juarez M.T., Twigg R.W., Timmermans M.C. (2004). Specification of adaxial cell fate during maize leaf development. Development 131: 4533–4544. [DOI] [PubMed] [Google Scholar]
- Kanno A., Saeki H., Kameya T., Saedler H., Theissen G. (2003). Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol. Biol. 52: 831–841. [DOI] [PubMed] [Google Scholar]
- Kater M.M., Franken J., Carney K.J., Colombo L., Angenent G.C. (2001). Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell 13: 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom T.H., Burson B.L., Finlayson S.A. (2006). Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 140: 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Yoo M.J., Albert V.A., Farris J.S., Soltis P.S., Soltis D.E. (2004). Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 91: 2102–2118. [DOI] [PubMed] [Google Scholar]
- Kramer E.M., Dorit R.L., Irish V.F. (1998). Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149: 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.M., Irish V.F. (2000). Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. Int. J. Plant Sci. 161: S29–S40. [Google Scholar]
- Krizek B.A., Meyerowitz E.M. (1996). The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development 122: 11–22. [DOI] [PubMed] [Google Scholar]
- Lange M., Orashakova S., Lange S., Melzer R., Theißen G., Smyth D.R., Becker A. (2013). The seirena B class floral homeotic mutant of California poppy (Eschscholzia californica) reveals a function of the enigmatic PI motif in the formation of specific multimeric MADS domain protein complexes. Plant Cell 25: 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenser T., Theissen G., Dittrich P. (2009). Developmental robustness by obligate interaction of class B floral homeotic genes and proteins. PLOS Comput. Biol. 5: e1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt A. (2007). An evaluation of A-function: Evidence from the APETALA1 and APETALA2 gene lineages. Int. J. Plant Sci. 168: 73–91. [Google Scholar]
- Litt A., Kramer E.M. (2010). The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 21: 129–137. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lombardo F., Yoshida H. (2015). Interpreting lemma and palea homologies: a point of view from rice floral mutants. Front. Plant Sci. 6: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Copsey L., Vincent C., Clark J., Coen E. (1999). Control of organ asymmetry in flowers of Antirrhinum. Cell 99: 367–376. [DOI] [PubMed] [Google Scholar]
- Luo D., Carpenter R., Vincent C., Copsey L., Coen E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383: 794–799. [DOI] [PubMed] [Google Scholar]
- McGonigle B., Bouhidel K., Irish V.F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10: 1812–1821. [DOI] [PubMed] [Google Scholar]
- Melzer R., Härter A., Rümpler F., Kim S., Soltis P.S., Soltis D.E., Theißen G. (2014). DEF- and GLO-like proteins may have lost most of their interaction partners during angiosperm evolution. Ann. Bot. (Lond.) 114: 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M., Ambrose B.A., Meeley R.B., Briggs S.P., Yanofsky M.F., Schmidt R.J. (1996). Diversification of C-function activity in maize flower development. Science 274: 1537–1540. [DOI] [PubMed] [Google Scholar]
- Michelmore R.W., Paran I., Kesseli R.V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88: 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty A., Luo A., DeBlasio S., Ling X., Yang Y., Tuthill D.E., Williams K.E., Hill D., Zadrozny T., Chan A., Sylvester A.W., Jackson D. (2009). Advancing cell biology and functional genomics in maize using fluorescent protein-tagged lines. Plant Physiol. 149: 601–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Theissen G. (2008). MADS about the evolution of orchid flowers. Trends Plant Sci. 13: 51–59. [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Theissen G. (2011). Conserved differential expression of paralogous DEFICIENS- and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: refining the ‘orchid code’. Plant J. 66: 1008–1019. [DOI] [PubMed] [Google Scholar]
- Moon Y.H., Jung J.Y., Kang H.G., An G. (1999). Identification of a rice APETALA3 homologue by yeast two-hybrid screening. Plant Mol. Biol. 40: 167–177. [DOI] [PubMed] [Google Scholar]
- Münster T., Wingen L.U., Faigl W., Werth S., Saedler H., Theissen G. (2001). Characterization of three GLOBOSA-like MADS-box genes from maize: evidence for ancient paralogy in one class of floral homeotic B-function genes of grasses. Gene 262: 1–13. [DOI] [PubMed] [Google Scholar]
- Nagasawa N., Miyoshi M., Sano Y., Satoh H., Hirano H., Sakai H., Nagato Y. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718. [DOI] [PubMed] [Google Scholar]
- Neuffer M.G. (1994). Mutagenesis. In The Maize Handbook, Freeling M., Walbot V., eds (New York: Springer-Verlag; ), pp. 212–218. [Google Scholar]
- Parcy F., Nilsson O., Busch M.A., Lee I., Weigel D. (1998). A genetic framework for floral patterning. Nature 395: 561–566. [DOI] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203. [DOI] [PubMed] [Google Scholar]
- Pelaz S., Tapia-López R., Alvarez-Buylla E.R., Yanofsky M.F. (2001). Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11: 182–184. [DOI] [PubMed] [Google Scholar]
- Pellegrini L., Tan S., Richmond T.J. (1995). Structure of serum response factor core bound to DNA. Nature 376: 490–498. [DOI] [PubMed] [Google Scholar]
- Petrásek J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918. [DOI] [PubMed] [Google Scholar]
- Prasad K., Vijayraghavan U. (2003). Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ patterning. Genetics 165: 2301–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J.C., Hileman L.C. (2012). Parallel evolution of TCP and B-class genes in Commelinaceae flower bilateral symmetry. Evodevo 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Remizowa M.V., Rudall P.J., Choob V.V., Sokoloff D.D. (2013). Racemose inflorescences of monocots: structural and morphogenetic interaction at the flower/inflorescence level. Ann. Bot. (Lond.) 112: 1553–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Krizek B.A., Meyerowitz E.M. (1996a). Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA 93: 4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L., Meyerowitz E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378: 1079–1101. [PubMed] [Google Scholar]
- Riechmann J.L., Wang M., Meyerowitz E.M. (1996b). DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 24: 3134–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall P.J., Bateman R.M. (2004). Evolution of zygomorphy in monocot flowers: iterative patterns and developmental constraints. New Phytol. 162: 25–44. [Google Scholar]
- Sarojam R., Sappl P.G., Goldshmidt A., Efroni I., Floyd S.K., Eshed Y., Bowman J.L. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22: 2113–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Hue I., Huijser P., Flor P.J., Hansen R., Tetens F., Lönnig W.E., Saedler H., Sommer H. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11: 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhon R.S., Lin H., Childs K.L., Hansey C.N., Buell C.R., de Leon N., Kaeppler S.M. (2011). Genome-wide atlas of transcription during maize development. Plant J. 66: 553–563. [DOI] [PubMed] [Google Scholar]
- Smith R.S., Guyomarc’h S., Mandel T., Reinhardt D., Kuhlemeier C., Prusinkiewicz P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103: 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht C.D., Howarth D.G. (2015). Adaptation in flower form: a comparative evodevo approach. New Phytol. 206: 74–90. [DOI] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409: 469–471. [DOI] [PubMed] [Google Scholar]
- Tröbner W., Ramirez L., Motte P., Hue I., Huijser P., Lönnig W.E., Saedler H., Sommer H., Schwarz-Sommer Z. (1992). GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11: 4693–4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W.C., Lee P.F., Chen H.I., Hsiao Y.Y., Wei W.J., Pan Z.J., Chuang M.H., Kuoh C.S., Chen W.H., Chen H.H. (2005). PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol. 46: 1125–1139. [DOI] [PubMed] [Google Scholar]
- Wang Z., et al. (2008). Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc. Natl. Acad. Sci. USA 105: 10414–10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R.X., Ge S. (2011). Evolutionary history and complementary selective relaxation of the duplicated PI genes in grasses. J. Integr. Plant Biol. 53: 682–693. [DOI] [PubMed] [Google Scholar]
- West A.G., Causier B.E., Davies B., Sharrocks A.D. (1998). DNA binding and dimerisation determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res. 26: 5277–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C.J., Ciceri P., Padilla C.M., Ambrose B.A., Bandong S.L., Schmidt R.J. (2004). Conservation of B-class floral homeotic gene function between maize and Arabidopsis. Development 131: 6083–6091. [DOI] [PubMed] [Google Scholar]
- Whipple C.J., Hall D.H., DeBlasio S., Taguchi-Shiobara F., Schmidt R.J., Jackson D.P. (2010). A conserved mechanism of bract suppression in the grass family. Plant Cell 22: 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C.J., Kebrom T.H., Weber A.L., Yang F., Hall D., Meeley R., Schmidt R., Doebley J., Brutnell T.P., Jackson D.P. (2011). grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 108: E506–E512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple C.J., Schmidt R.J. (2006). Genetics of grass flower development. Adv. Bot. Res. 44: 385–424. [Google Scholar]
- Whipple C.J., Zanis M.J., Kellogg E.A., Schmidt R.J. (2007). Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc. Natl. Acad. Sci. USA 104: 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K.U., Weiser C., Kaufmann K., Bohne A., Kirchner C., Kanno A., Saedler H., Theissen G. (2002). Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Mol. Biol. Evol. 19: 587–596. [DOI] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertová D., Brewer P.B., Ruzicka K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Yadav S.R., Prasad K., Vijayraghavan U. (2007). Divergent regulatory OsMADS2 functions control size, shape and differentiation of the highly derived rice floret second-whorl organ. Genetics 176: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Lee D.Y., Miyao A., Hirochika H., An G., Hirano H.Y. (2006). Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Pang H.B., Liu B.L., Qiu Z.J., Gao Q., Wei L., Dong Y., Wang Y.Z. (2012). Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24: 1834–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S.G., Ohmori S., Kimizu M., Yoshida H. (2008). Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol. 49: 853–857. [DOI] [PubMed] [Google Scholar]
- Yuan Z., Gao S., Xue D.W., Luo D., Li L.T., Ding S.Y., Yao X., Wilson Z.A., Qian Q., Zhang D.B. (2009). RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 149: 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun D., Liang W., Dreni L., Yin C., Zhou Z., Kater M.M., Zhang D. (2013). OsMADS16 genetically interacts with OsMADS3 and OsMADS58 in specifying floral patterning in rice. Mol. Plant 6: 743–756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.