LEC1 interacts with PIF4 to mediate postembryonic development through coregulating expression of hypocotyl elongation-related genes via direct binding to the G-box element in the dark.
Abstract
Plants undergo postembryonic growth during the developmental transition from germinating seeds to seedlings. Recent studies suggest LEAFY COTYLEDON1 (LEC1), initially identified as a central regulator in embryogenesis and seed maturation in Arabidopsis thaliana, plays a distinct role in postembryonic development. However, the mechanism by which LEC1 regulates nonembryonic development still remains elusive. In this study, we observed etiolation-related phenotypes in early seedlings of lec1 mutants and inducible LEC1 overexpression transgenic lines. Consistent with this, LEC1 promotes the expression of hypocotyl elongation-related genes in a darkness-dependent manner in spite of the comparable LEC1 transcript levels in the light- and dark-grown seedlings. Furthermore, we show that LEC1 interacts with PHYTOCHROME-INTERACTING FACTOR4 (PIF4), a major transcription modulator in postgermination development, to interdependently regulate hypocotyl elongation-related genes via direct binding to G-box element in the dark. Moreover, loss of LEC1 function suppresses the elongated hypocotyl phenotype of PIF-overaccumulating plants; conversely, inducible overexpression of LEC1 does not rescue the short hypocotyl in pif4 mutants. Our findings reveal that LEC1 acts as a coactivator of PIFs in transcriptional regulation during postembryonic growth, providing a possible mechanism by which plants fine-tune morphological development for their survival during the transition from the embryonic phase to seedling establishment.
INTRODUCTION
Plants have evolved sophisticated sensory systems to perceive changes in light quality, intensity, direction, and periodicity (Chen et al., 2004). Seedlings grown in the light develop short hypocotyls and open cotyledons, which is referred to as photomorphogenesis. However, during the developmental transition from germinating seeds to seedlings, plants kept in darkness adopt a skotomorphogenic or etiolation program, including rapid elongation of the hypocotyl, tightly folded apical hook, and closed cotyledons, ensuring that limited seed reserves are used effectively to survive until the plant is exposed to light (Von Arnim and Deng, 1996; Josse and Halliday, 2008).
PHYTOCHROME INTERACTING FACTORs (PIFs), a subgroup of basic helix-loop-helix (bHLH) transcription factors, act as central modulators in the postgermination stage (Castillon et al., 2007; Bae and Choi, 2008; Leivar and Quail, 2011; Leivar and Monte, 2014). Mutants deficient in PIFs, such as pif3, pif4, and the pif1 pif3 pif4 pif5 (pifq) quadruple mutant, show partial or constitutive photomorphogenesis in the dark; overexpression of PIF3, PIF4, or PIF5 leads to high induction of genes involved in skotomorphogenesis and relevant etiolation phenotype, suggesting PIFs contribute to the maintenance of skotomorphogenesis state (Huq and Quail, 2002; Kim et al., 2003; Leivar et al., 2008). Previous studies demonstrated that PIFs are destabilized in light-grown seedlings, which is the result of the light-triggered nuclear relocalization of phytochromes (Khanna et al., 2004; Al-Sady et al., 2006). For example, phytochrome B (phyB) is a red/far-red photoreceptor involved in the regulation of deetiolation and its null mutant shows an etiolation-like phenotype. Red light induces the Pr form of phyB into the active Pfr form, which relocates to the nucleus and interacts with PIFs through the conserved active phytochrome binding motif. The interaction between phyB and PIFs results in phosphorylation of PIFs and their subsequent degradation by the 26S proteasome, thus relieving the repression of light responses (Leivar et al., 2008; Leivar and Quail, 2011; Leivar and Monte, 2014). Conversely, light-induced phosphorylation of PIFs is also necessary for the degradation of phyB. A recent study reported that recruitment of LRB E3 ubiquitin ligases to the PIF3-phyB complex facilitates concurrent polyubiquitination and degradation of PIF3 and phyB, which assures destruction of the receptor and its immediate signaling partner when “lighting up” in plants (Ni et al., 2014).
Besides the well-characterized phyB-PIF regulatory module in the light, several PIF cofactors, such as DELLA, BZR1, HDA15, and PKL, affect PIFs’ transcriptional activity on targets via epigenetic regulation and thus allow the integration of light and other cellular pathways (de Lucas et al., 2008; Feng et al., 2008; Oh et al., 2012; Zhang et al., 2014). Especially, in addition to functioning in hypocotyl elongation in light conditions, the chromatin-remodeling factor PKL also negatively regulates expression of LEAFY COTYLEDON1 (LEC1) and the embryonic program, implying a possible mechanistic connection between embryogenesis and light signaling (Zhang et al., 2012; Jing et al., 2013).
LEC1, one of the first nuclear factor Y (NF-Y) family members identified in plants, acts as a central regulator controlling embryogenesis and seed maturation in Arabidopsis thaliana (Meinke et al., 1994; West et al., 1994; Parcy et al., 1997; Lotan et al., 1998; Harada, 2001). Consistent with this, LEC1 is predominantly expressed during early and late stages of seed development (Le et al., 2010). Strong evidence supporting the pivotal role of LEC1 in embryo development came with the finding that ectopic LEC1 expression was sufficient to induce somatic embryogenesis from vegetative cells (Lotan et al., 1998). In contrast, the LEC1 null mutant causes a pleiotropic phenotype, including abnormal embryo and seed desiccation intolerance, defects in reserve accumulation, leafy cotyledons, and a reduced capability for hypocotyl elongation, indicating that in addition to its vital function in embryogenesis, LEC1 also plays a distinct role in postembryonic development (Lotan et al., 1998; Harada, 2001; Brocard-Gifford et al., 2003). However, the molecular mechanism of such nonembryonic functions of LEC1 still remains elusive.
In this study, we isolated a LEC1 knockdown mutant that mostly phenocopies the LEC1 null allele but with higher desiccation tolerance in mature seeds, which provides a more efficient genetic tool to examine LEC1 function in plants. Primarily, we found that LEC1 and PIF4, one of the major transcription modulators in skotomorphogenesis, interdependently regulate hypocotyl elongation of early seedlings in a darkness-dependent manner. LEC1 binds to the G-box in the promoters of hypocotyl-related genes via protein-protein interaction with PIF4 in Arabidopsis. Our findings reveal that LEC1 acts as a coactivator of PIFs in transcriptional regulation of postembryonic growth, providing a possible mechanism by which plants can fine-tune morphological development for their survival during the transition from the embryonic phase to seedling establishment.
RESULTS
Identification and Characterization of the lec1-4 Mutant
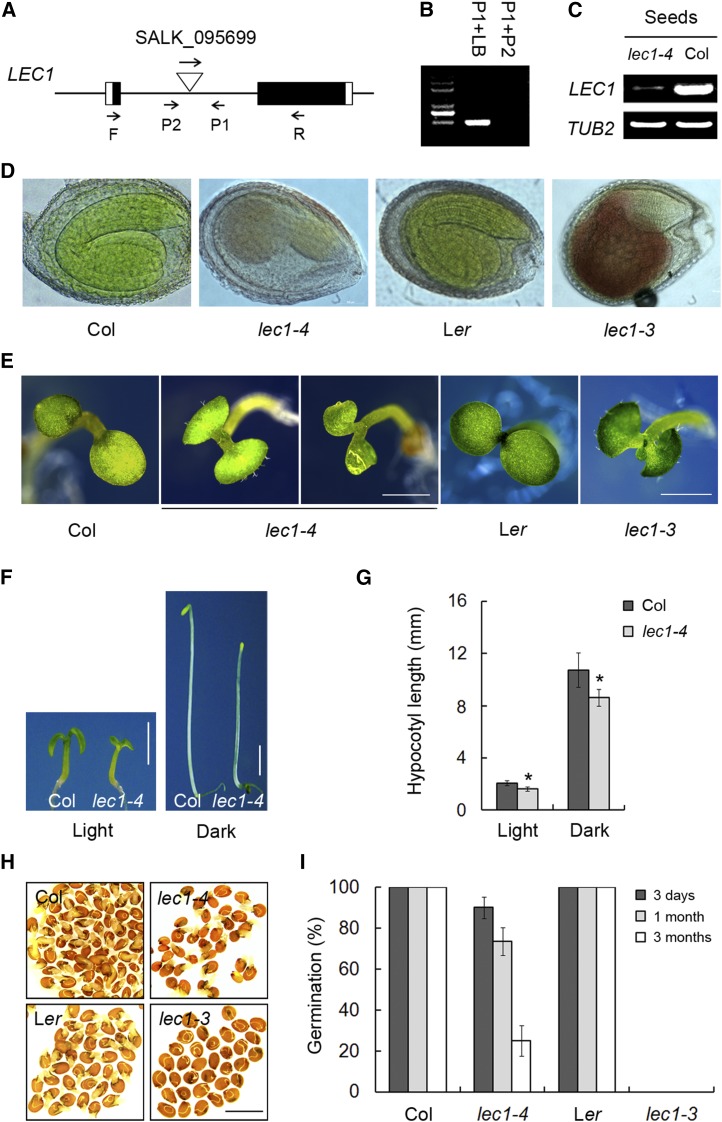
To investigate the LEC1-mediated regulatory mechanisms acting during the transition from seed to seedling establishment, we identified a mutant (SALK_095699) in the Columbia (Col) background containing a T-DNA insertion in the LEC1 intron from the ABRC (Figures 1A and 1B). We named this mutant lec1-4, based on our subsequent characterization described below. RT-PCR showed that the expression of LEC1 only remained low in abundance in developing lec1-4 mutant seeds in comparison to highly expressed LEC1 in the wild type (Figure 1C). Similar to previously reported LEC1 null mutants such as lec1-3, the typical pleiotropic phenotypes of lec1-4, including defective embryos, abnormal seeds, developed trichomes on cotyledons, short hypocotyls, and less apical hook formation in the dark (Figures 1D to 1G; Supplemental Figures 1A and 1B). However, some of the lec1-4 phenotypes are weaker than those in lec1-3 null mutants, probably due to the leaky expression of LEC1 (Figure 1C). For example, ∼90% freshly harvested mature seeds from the lec1-4 mutants are viable in spite of reduced germination rate following longer storage, whereas the seed embryo of the lec1-3 null allele is desiccation-intolerant and has to be rescued before maturation in order to continue growth (Figures 1H and 1I; West et al., 1994). Thus, it is more convenient for us to get lec1-4 homozygous recessive plants whenever needed. In addition, in lec1-4 mutants, the amount of trichomes developed on the cotyledons was greatly variable and a few of seedlings resembling the wild type can also be observed (Supplemental Figure 1C). These results indicate that the defective phenotype observed is due to knockdown of LEC1. This is further corroborated by a complementation test in which a 3.8-kb genomic fragment of LEC1 fused with a 3×FLAG tag (pLEC1:LEC1-FLAG) was able to rescue the lec1 typical phenotypes (Supplemental Figures 2A to 2F). Taking these observations together, we defined this mutant as a lec1 allele and designated it lec1-4. Because of the high viability of seeds after desiccation, lec1-4 mutant is more suitable to be used for genetic analysis.
Figure 1.
Identification of a LEC1 Knockdown Allele in Arabidopsis.
(A) Schematic diagram showing the T-DNA insertion site in lec1-4 (SALK_095699). Exons are represented by boxes and introns by lines between the boxes. Black boxes represent the LEC1 coding regions.
(B) PCR of genomic DNA confirms that lec1-4 is a homozygous insertion mutant. The primers used are shown in (A). LB indicates the T-DNA left border primer.
(C) LEC1 expression is low in lec1-4 in comparison to the wild type by RT-PCR using the primers F and R shown in (A). Total RNA was isolated from developing seeds (6 d after pollination) of lec1-4 and Col plants.
(D) The lec1 mutants exhibit abnormal embryos in comparison to the wild type. Bar = 1 mm.
(E) The lec1 mutants develop trichomes on their cotyledons or have shrunken cotyledons in comparison to the wild type. Five-day-old seedlings grown in the light were used for investigation. Bar = 1 mm.
(F) Five-day-old seedlings of lec1-4 mutants grown in the light or dark show shorter hypocotyls than the wild type. Bar = 2 mm.
(G) Hypocotyl length statistics of the wild type and lec1-4 seedlings shown in (F). Data represent mean ± sd of at least 30 seedlings. Asterisks indicate significant differences between Col and lec1-4 mutants (P < 0.05, by Student’s t test).
(H) Germination investigation of the freshly harvested dry seeds from Col, lec1-4, Landsberg erecta (Ler), and lec1-3. Germination rate of seeds was measured after transfer to 22°C for 1 d from 4°C stratification.
(I) Germination rate of the Col, lec1-4, Ler, and lec1-3 seeds with a 3-d, 1-month, or 3-month storage period after harvest.
LEC1 Functions in Postembryonic Development
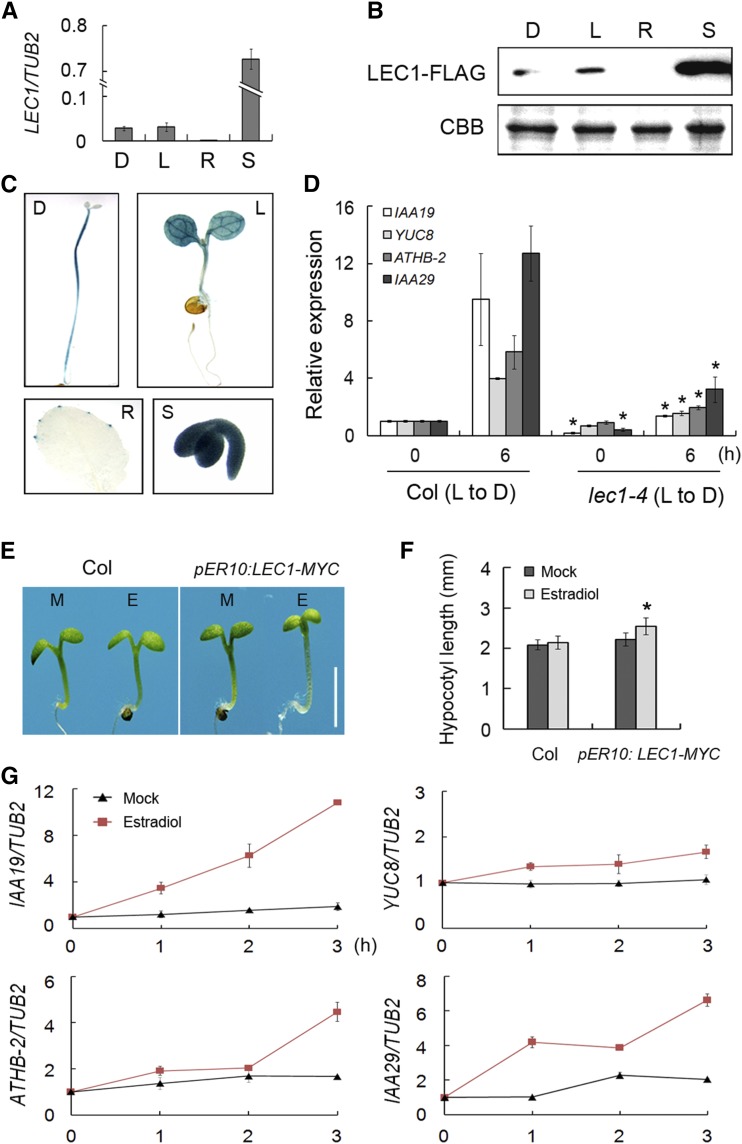
LEC1 acts as a crucial regulator of embryogenesis and seed development (Meinke et al., 1994; West et al., 1994; Parcy et al., 1997; Lotan et al., 1998; Harada, 2001). However, observation of the short hypocotyl and reduced apical hook formation in lec1 mutants suggests a definite role of LEC1 in etiolated growth during postembryonic development (Figures 1F and 1G; Supplemental Figure 1B; Brocard-Gifford et al., 2003; Junker and Bäumlein, 2012). To further confirm whether LEC1 functions in etiolation, we first examined the expression of LEC1 in 5-d-old seedlings grown under light and dark conditions. The results showed that LEC1 had comparable expression in seedlings grown in the light and dark; it also had little expression in rosette leaves and high expression in developing seeds (Figure 2A). Immunoblot assay of lec1-4 pLEC1:LEC1-FLAG transgenic seedlings demonstrated that LEC1 protein expression was consistent with its transcription pattern, and light had no effect on LEC1 abundance (Figure 2B). Next, we generated transgenic plants harboring GUS driven by an ∼2-kb fragment of the LEC1 promoter region (pLEC1:GUS) to examine the tissue-specific expression of LEC1. Consistent with the expression pattern of LEC1, GUS staining was found in seedlings grown in the light and dark and developing seeds but barely in rosette leaves (Figure 1C). These results imply the potential role of LEC1 in early seedling development.
Figure 2.
LEC1 Is Involved in Hypocotyl Elongation.
(A) Quantitative RT-PCR analysis showing LEC1 expression in 5-d-old Col wild-type seedling grown in the dark (D) or in the light (L). Tissues from rosette leaves (R) and developing seeds (S) were used as negative and positive controls, respectively. The β-tubulin gene (TUB2) was amplified as an internal control.
(B) Immunoblot analysis of LEC1-FLAG protein expressed in tissues indicated in (A). The lec1-4 pLEC1:LEC1-FLAG transgenic line was used to examine the LEC1 protein level. Coomassie blue staining (CBB) was used as a loading control.
(C) GUS staining of pLEC1:GUS transgenic plants. Tissues detected are indicated in (A).
(D) Quantitative RT-PCR analysis showing repression of hypocotyl elongation-related gene expression in lec1-4 in comparison to the wild type. Three-day-old lec1-4 and Col seedlings grown in the light (0 h) or then transferred to darkness for 6 h (6 h) were harvested. Relative gene expression was calculated by comparing the values to that of Col at 0 h. TUB2 was amplified as an internal control. Asterisks indicate significant differences between Col and lec1-4 mutant (P < 0.05, by Student’s t test). L to D, light to darkness.
(E) Hypocotyl length of the pER10:LEC1-MYC transgenic line. Five-day-old Col and pER10:LEC1-MYC seedlings grown in the light with 10 μM estradiol (E) or mock treatment (M) were used for investigation. Bar = 2 mm.
(F) Hypocotyl length statistics of the wild type and pER10:LEC1-MYC seedlings shown in (E). Data represent mean ± sd of at least 30 seedlings. Asterisk indicates significant difference between pER10:LEC1-MYC with estradiol and mock treatment (P < 0.05, by Student’s t test).
(G) Estradiol-induced LEC1 overexpression promotes the expression of hypocotyl elongation-related genes. Three-day-old pER10:LEC1-MYC seedlings were treated with 10 μM estradiol or mock-treated and immediately transferred to darkness for the indicated time. Relative gene expression was calculated by comparing the values to that at 0 h. TUB2 was amplified as an internal control.
To learn whether LEC1 regulates the expression of genes involved in etiolated growth, we measured transcript levels of IAA19, YUC8, ATHB-2, and IAA29, four typical genes involved in dark-induced hypocotyl elongation (Kunihiro et al., 2011; Sun et al., 2012; Zhang et al., 2013), in Col and lec1-4. At 6 h after the transition from light to darkness, expression of all four genes was dramatically induced in wild-type seedlings. In contrast, increase in transcript levels of these genes in darkness was significantly compromised in lec1-4 mutants, indicating that LEC1 is important for activation of hypocotyl elongation-related genes in the dark (Figure 2D).
Since constitutive expression of LEC1 could cause serious defects including embryo lethality or embryo-like structures in vegetative tissues (Lotan et al., 1998; Mu et al., 2008; Junker and Bäumlein, 2012), we employed an estradiol-inducible transgenic plant harboring pER10:LEC1-MYC to further verify LEC1 function. RT-PCR and immunoblot analyses showed that LEC1 expression was promptly induced and specific LEC1 protein accumulation was detected in estradiol-treated pER10:LEC1-MYC plants (Supplemental Figures 3A and 3B). As expected, estradiol induced longer hypocotyls in pER10:LEC1-MYC seedlings, while hypocotyl elongation was not affected in estradiol-treated wild type and mock-treated pER10:LEC1-MYC (Figures 2E and 2F). To further confirm the regulation of LEC1 on hypocotyl elongation-related genes, we measured transcript levels of IAA19, YUC8, ATHB-2, and IAA29 in 3-d-old pER10:LEC1-MYC seedlings that were transferred to the darkness or continuous light over a time course with estradiol treatment. Consistent with the phenotype of the transgenic plant, IAA19, YUC8, ATHB-2, and IAA29 transcript levels were dramatically elevated by induction of LEC1 in the dark (Figure 2G). Interestingly, under light conditions, we detected no significant promotion of expression of these genes (Supplemental Figure 3C), suggesting that LEC1 regulates hypocotyl elongation in a dark-dependent manner.
LEC1 Interacts with PIF4
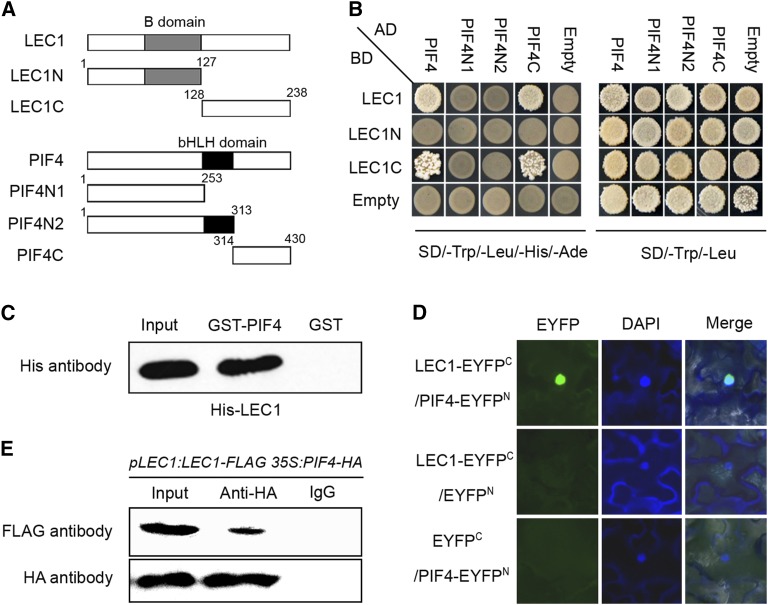
NF-Y subunits interact with other transcription factors to modulate gene expression in plant development and stress responses (Liu and Howell, 2010; Hou et al., 2014). LEC1 belongs to the NF-YB family; thus, we reasoned that LEC1 might function with other partners in the regulation of hypocotyl elongation during postembryonic development. PIF transcription factors, including PIF1, PIF3, PIF4, and PIF5, function as central modulators in skotomorphogenic growth, and pif loss-of-function mutants show reduced hypocotyl length, like lec1-4 mutants (Supplemental Figures 4A and 4B). Also, PIFs directly regulate expression of IAA19, YUC8, ATHB-2, and IAA29 (Supplemental Figure 4C; Kunihiro et al., 2011; Hornitschek et al., 2012; Oh et al., 2012; Sun et al., 2012; Zhang et al., 2013); thus, it is possible that LEC1 may coregulate hypocotyl elongation with PIFs. Since overexpression of LEC1 did not affect PIF4 transcript levels (Supplemental Figure 4D), we tested the protein interaction between LEC1 and PIFs using a yeast two-hybrid assay. Strikingly, the results demonstrated a strong interaction between LEC1 and PIF4 and a weaker interaction between LEC and PIF3, but detected no interaction between LEC1 and PIF1 or PIF5 or between L1L, the closest homolog of LEC1 in Arabidopsis, and all indicated PIFs (Supplemental Figure 5). In our observations, the L1L null mutant (l1l-1) showed less of a change in hypocotyl length, suggesting that L1L is not involved in postembryonic growth (Supplemental Figure 6). We further generated two truncated versions of LEC1 and three versions of PIF4 to identify the functional domains required for the interaction (Figure 3A). The B-domain in LEC1 is a conserved domain in the NF-YB family and probably functions in NF-Y trimerization (Calvenzani et al., 2012; Hackenberg et al., 2012). The bHLH domain in PIFs was reported to be a DNA binding domain (Toledo-Ortiz et al., 2003). We found that neither the B-domain of LEC1 nor the bHLH domain of PIF4 was involved in the protein interaction; rather, the C-terminal domains of LEC1 and PIF4 were sufficient and necessary for their interaction (Figure 3B). This indicates that the LEC1-PIF4 interaction may not influence PIF4-DNA binding and may not compete for NF-YA/LEC1/NF-YC trimerization. We next performed pull-down assays to verify the interaction between PIF4 and LEC1, using purified GST- and His-tagged proteins. Again, we found that GST-PIF4, but not GST alone, precipitated His-LEC1 in vitro (Figure 3C). In addition, bimolecular fluorescence complementation (BiFC) analysis revealed the interaction of LEC1 and PIF4 in the nuclei of living plant cells (Figure 3D). Coimmunoprecipitation of protein extracts from pLEC1:LEC1-FLAG 35S:PIF4-HA seedlings further confirmed the interaction between LEC1 and PIF4 in vivo (Figure 3E). Taken together, these results indicate that LEC1 might regulate plant postembryonic growth through its interaction with PIF4.
Figure 3.
LEC1 Physically Interacts with PIF4 in Vitro and in Vivo.
(A) Sketches showing the domain structures of LEC1 and PIF4 and various deletions.
(B) Yeast two-hybrid assays showing the interactions between LEC1, PIF4, and their derivatives. Transformed yeast cells were grown on SD/-Trp/-Leu/-His/-Ade and SD/-Trp/-Leu medium.
(C) Pull-down assay showing direct interaction between His-LEC1 and GST-PIF4 fusion proteins in vitro. His-LEC1 protein was incubated with immobilized GST or GST-PIF4 proteins, and immunoprecipitated fractions were detected by anti-His antibody.
(D) BiFC assay showing LEC1-EYFPC and PIF4-EYFPN interact to form a functional EYFP in the nucleus. DAPI, fluorescence of 4′,6-diamidino-2-phenylindole; Merge, merge of EYFP and DAPI.
(E) In vivo interaction of PIF4 and LEC1 in Arabidopsis. Plant nuclear extracts from 5-d-old pLEC1:LEC1-FLAG 35S:PIF4-HA seedling grown in the dark were immunoprecipitated by either anti-HA antibody or preimmune serum (IgG). The coimmunoprecipitated proteins were detected by anti-FLAG antibody.
LEC1 and PIF4 Coregulate Hypocotyl Elongation-Related Genes via Direct Binding to the G-Box
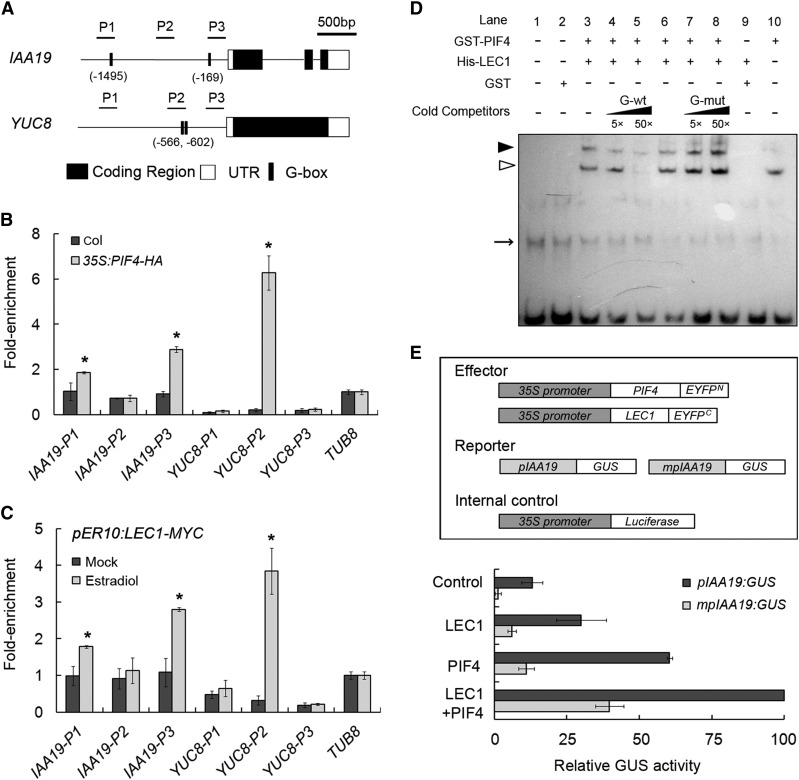
Previous studies showed that PIFs promote skotomorphogenesis in the dark via specific binding to G-box or E-box variants in downstream genes (Huq and Quail, 2002; Hornitschek et al., 2012; Oh et al., 2012; Zhang et al., 2013). To learn how the interaction between LEC1 and PIF4 regulates the expression of genes involved in hypocotyl elongation, we performed chromatin immunoprecipitation (ChIP) assays to measure the DNA enrichment of G-box in IAA19 and YUC8, downstream genes of both LEC1 and PIF4 (Figure 2G; Supplemental Figure 4C). Consistent with previous reports, the results showed that PIF4 strongly bound to the G-box-containing regions at P1 and P3 in IAA19 promoter and P2 in YUC8 promoter (Figure 4B). Furthermore, a similar DNA enrichment pattern in IAA19 or YUC8 by the inducible LEC1-MYC protein was also detected in the dark-treated pER10:LEC1-MYC seedlings, whereas no significant binding of LEC1 to these genes was observed in light-grown plants (Figure 4C; Supplemental Figure 3D). ChIP analysis for IAA29 and ATHB-2, two other downstream genes of LEC1 and PIF4 (Figure 2G; Supplemental Figure 4C), also showed similar results to those observed for IAA19 and YUC8 (Supplemental Figure 7), indicating that, in the dark, LEC1 and PIF4 cotarget the promoters of downstream genes probably via direct binding to G-box elements.
Figure 4.
LEC1 and PIF4 Coregulate the Hypocotyl Elongation-Related Genes via Directly Binding to the G-Box.
(A) Schematic diagram of the IAA19 and YUC8 genomic regions. P1 to P3 indicate fragments for ChIP-qPCR amplification. Numbers indicate the positions of G-box elements in IAA19 and YUC8 promoters relative to ATG.
(B) ChIP analysis of PIF4-HA binding to G-box containing region in IAA19 and YUC8 genes upon precipitation with anti-HA antibody. Three-day-old of 35S:PIF4-HA and Col seedlings were transferred to darkness for an additional 2 d and harvested for ChIP assay. Data represent mean ± sd of triplicates. Asterisks indicate significant changes in ChIP enrichment in 35S:PIF4-HA compared with the Col sample (P < 0.05, by Student’s t test).
(C) ChIP analysis of LEC1-MYC binding to the G-box containing region in IAA19 and YUC8 genes upon precipitation with anti-MYC antibody. Three-day-old of pER10:LEC1-MYC seedlings were treated with 10 μM estradiol or mock and immediately transferred to darkness for an additional two days and harvested for ChIP assay. Data represent mean ± sd of triplicates. Asterisks indicate significant changes in ChIP enrichment in estradiol-treated sample compared with the mock-treated sample (P < 0.05, by Student’s t test).
(D) EMSA assay of the PIF4-LEC1 complex binding to the G-box in IAA19 promoter (P3 region). The biotinylated probe containing the G-box element was incubated with GST-PIF4 (Lane 10), His-LEC1 (Lane 9), or their mixture (lanes 3 to 8), while the probe incubated with no protein (lane 1) or GST protein (lane 2) was used as negative control. Nonlabeled probes in 5- and 50-fold molar excess relative to the biotinylated probe containing G-box (G-wt, CACGTG) or mutated G-box (G-mut, CACGGG), respectively, were used as cold competitors. White and black arrowheads indicate the specific bindings of PIF4 protein and PIF4-LEC1 complex to the biotinylated probe, respectively, while arrow indicates nonspecific bands.
(E) Transient expression assays of IAA19 transcriptional activity modulated by LEC1 and PIF4 in Arabidopsis mesophyll protoplasts. Various constructs used in transient expression assays are shown in the upper panel. Either pIAA19:GUS or mpIAA19:GUS was cotransformed with effectors or empty vector (Control) into Col mesophyll protoplasts. Relative GUS activity (GUS/Luciferase) that indicates the level of IAA19 expression activated by various effectors is shown in the lower panel. Values are mean ± sd of five biological replicates.
Since the LEC1 protein contains no identified DNA binding domain (Calvenzani et al., 2012), we speculated that LEC1 may bind to IAA19 and YUC8 by associating with PIF4, which contributes to the direct DNA affinity. To test this hypothesis, electrophoretic mobility shift assays (EMSAs) were performed using GST-PIF4 and His-LEC1 recombinant proteins and a biotinylated probe containing the G-box element in the P3 region of IAA19. The EMSAs also used a nonlabeled probe with mutated G-box as cold competitor (Supplemental Figure 8A). The results showed that PIF4 bound to the G-box element, whereas no binding occurred between LEC1 or GST protein and the P3 probe (Figure 4D). Remarkably, a band of slower mobility was detected when PIF4 was incubated with LEC1, indicating that these two proteins bind to the G-box as a heterodimer (Figure 4D).
To further examine the regulation of PIF4 and LEC1 on the expression of their target genes, we performed transient expression assays using ∼2 kb of IAA19 promoter fused to the GUS gene as a reporter (pIAA19:GUS). Effector constructs for PIF4-EYFPN or LEC1-EYFPC were expressed under the control of 35S promoter and transfected together with the reporter construct into Arabidopsis mesophyll protoplasts. Addition of either LEC1 or PIF4 significantly activated the expression of IAA19, whereas IAA19 activation by LEC1 alone was abolished in pifq mutants (Figure 4E; Supplemental Figure 9), showing PIF4-dependent regulation of LEC1 on IAA19. Notably, higher GUS activity was detected when LEC1 and PIF4 proteins were coexpressed compared with that when either LEC1 or PIF4 was expressed alone (Figure 4E). However, when site-specific mutations were introduced into the two G-boxes (CACGTG to CACGgG) in mpIAA19:GUS (Supplemental Figure 8B), such disruption of G-box element dramatically reduced the expression of IAA19 despite the presence of LEC1, PIF4, or both LEC1 and PIF4 (Figure 4E), indicating that the G-box element is essential for LEC1-PIF4-mediated activation of IAA19.
Taken together, these findings strongly support the idea that PIF4 and LEC1 coregulate downstream genes to regulate hypocotyl elongation.
LEC1 and PIF4 Act Interdependently in the Regulation of Hypocotyl Elongation
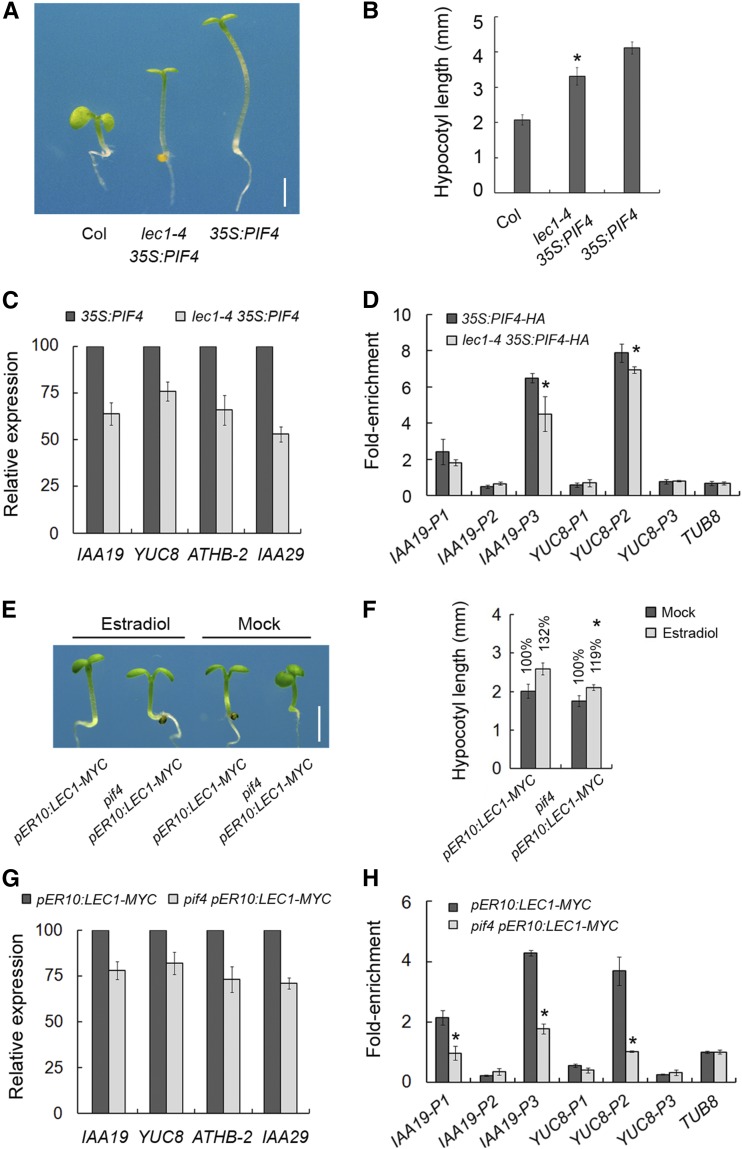
PIFs, including PIF1, PIF3, PIF4, and PIF5, function with overlapping redundancy in dark-grown seedlings to promote etiolated growth, and the photoactivated Phy-triggered degradation of PIFs induces deetiolation (Leivar et al., 2008). To further investigate the genetic role of LEC1 in postembryonic growth, we crossed lec1-4 with 35S:PIF4, 35S:PIF3-MYC, and the phyb mutant in which the deetiolation response in the light is suppressed (Reed et al., 1993; Zhang et al., 2013). Hypocotyl elongation of 35S:PIF4, 35S:PIF3-MYC, and phyb was significantly reduced in the lec1-4 background (Figures 5A and 5B; Supplemental Figures 10A and 10B). Consistent with this, expression of the hypocotyl elongation-related genes was also compromised in lec1-4 35S:PIF4 plants, compared with that in 35S:PIF4 (Figure 5C). Together with the ChIP analysis, which showed that the association of PIF4-HA with G-box in IAA19 and YUC8 was attenuated in lec1-4 mutants (Figure 5D), these results suggest that LEC1 promotes PIF-mediated hypocotyl elongation through enhancing DNA affinity of PIF4 to target genes.
Figure 5.
LEC1 Is Required for PIF4-Mediated Hypocotyl Elongation.
(A) Hypocotyl lengths of the wild-type, 35S:PIF4, and lec1-4 35S:PIF4 plants. Five-day-old seedlings grown in the light were used for investigation. Bar = 2 mm.
(B) Hypocotyl length statistics of the wild-type, 35S:PIF4, and lec1-4 35S:PIF4 seedlings shown in (A). Data represent mean ± sd of at least 30 seedlings. Asterisks indicate significant difference between 35S:PIF4 and lec1-4 35S:PIF4 (P < 0.05, by Student’s t test).
(C) Quantitative RT-PCR analysis showing hypocotyl elongation-related genes expression in 35S:PIF4 and lec1-4 35S:PIF4. Three-day-old seedlings grown in the light were transferred to darkness for 6 h and then harvested for RNA extraction and further analysis. Relative gene expression levels were normalized against the expression of TUB2 and those in 35S:PIF4 seedlings were designated as 100%.
(D) ChIP analysis of PIF4-HA binding to G-box containing region in IAA19 and YUC8 genes upon precipitation with anti-HA antibody. Three-day-old 35S:PIF4-HA and lec1-4 35S:PIF4-HA seedlings were transferred to darkness for an additional 2 d and harvested for ChIP assay. Data represent mean ± sd of triplicates. Asterisks indicate significant changes in ChIP enrichment in lec1-4 35S:PIF4-HA compared with 35S:PIF4-HA sample (P < 0.05, by Student’s t test).
(E) Hypocotyl lengths of pER10:LEC1-MYC and pif4 pER10:LEC1-MYC transgenic line. Five-day-old seedlings grown in the light with 10 μM estradiol or mock treatment were used for investigation. Bar = 2 mm.
(F) Hypocotyl length statistics of pER10:LEC1-MYC and pif4 pER10:LEC1-MYC shown in (E). The percentage indicates the relative hypocotyl length with estradiol treatment against that with mock treatment (designated as 100%). Data represent mean ± sd of at least 30 seedlings. Asterisks indicate significant difference of relative hypocotyl length in pif4 pER10:LEC1-MYC compared with pER10:LEC1-MYC seedlings (P < 0.05, by Student’s t test).
(G) Quantitative RT-PCR analysis showing expression of hypocotyl elongation-related genes in pER10:LEC1-MYC and pif4 pER10:LEC1-MYC. Three-day-old seedlings were treated with 10 μM estradiol and immediately transferred to darkness for 3 h and then harvested for RNA extraction and further analysis. Relative gene expression levels were normalized against the expression of TUB2 and those in pER10:LEC1-MYC seedlings were designated as 100%.
(H) ChIP analysis of LEC1-MYC binding to G-box-containing region in IAA19 and YUC8 genes upon precipitation with anti-HA antibody. Three-day-old pER10:LEC1-MYC and pif4 pER10:LEC1-MYC seedlings were treated with 10 μM estradiol and immediately transferred to darkness for additional 2 d and harvested for ChIP experiment. Data represent mean ± sd of triplicates. Asterisks indicate significant changes in ChIP enrichment in pif4 pER10:LEC1-MYC compared with pER10:LEC1-MYC sample (P < 0.05, by Student’s t test).
In turn, we crossed pER10:LEC1-MYC to pif4 to examine whether LEC1 function in hypocotyl elongation depends on PIF4. Loss of PIF4 function remarkably impaired the elongated hypocotyl induced by LEC1-MYC (Figures 5E and 5F), and expression of the hypocotyl elongation-related genes was consistently decreased in pif4 pER10:LEC1-MYC compared with that in pER10:LEC1-MYC when treated with estradiol in the dark (Figure 5G). Further ChIP assays showed that the DNA enrichment of IAA19 and YUC8 by LEC1-MYC was attenuated in pif4 mutants (Figure 5H).
In addition, gibberellic acid (GA) and brassinosteroids (BRs) promote hypocotyl elongation depending on PIFs activity, while PIF4 functions on hypocotyl growth at high temperature through mediating auxin biosynthesis (de Lucas et al., 2008; Feng et al., 2008; Franklin et al., 2011; Oh et al., 2012; Bernardo-García et al., 2014). Our observations demonstrated that, resembling that in pif4, the effect of GA or brassinolide on hypocotyl elongation was attenuated, and, by contrast, the synthetic auxin picloram largely rescued the short hypocotyl phenotype at high temperature in the lec1-4 mutant (Supplemental Figures 11A to 11C). Collectively, these results strongly suggest that LEC1 and PIF4 function interdependently in the regulation of hypocotyl growth.
DISCUSSION
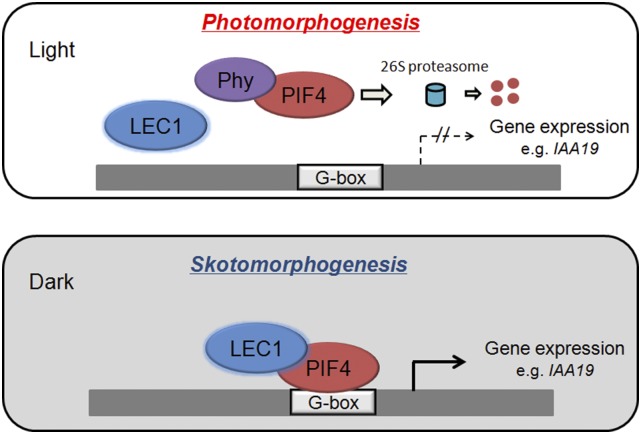
LEC1 acts as a central regulator in embryogenesis, and it also plays a distinct role in postembryonic growth in Arabidopsis (Meinke et al., 1994; West et al., 1994; Parcy et al., 1997; Lotan et al., 1998; Harada, 2001; Junker and Bäumlein, 2012). However, the molecular mechanism by which LEC1 regulates such nonembryonic development so far remains elusive. Here, we demonstrated that LEC1 promotes etiolation growth via protein-protein interaction with PIF4, one of the key transcriptional activators in etiolation. Similar to pif4, the lec1 mutant exhibits defects in hypocotyl elongation and apical hook formation in darkness, while inducible overexpression of LEC1 promotes hypocotyl elongation of seedlings (Figures 2E; Supplemental Figures 4A and 4B; Huq and Quail, 2002; Junker et al., 2012). Protein interaction analyses, transient expression assays, EMSA, and ChIP analyses showed that LEC1 and PIF4 interdependently regulate hypocotyl elongation-related genes via direct binding to G-box in a dark-dependent manner. Corresponding to this, loss of LEC1 function suppresses the elongated hypocotyl phenotype of PIF-overaccumulating plants, including 35S:PIF4, 35S:PIF3-MYC, and phyb; conversely, inducible overexpression of LEC1 does not rescue the short hypocotyl phenotype of pif4. These results support a model in which LEC1 acts as a coactivator of PIFs in transcriptional regulation of postembryonic growth. Briefly, in the light, photoactivated phytochromes (Phy) interact with PIFs and trigger their codegradation by the 26S proteasome, thus abolishing the expression of the etiolation-related genes and promoting photomorphogenesis. In the dark, LEC1 interacts with stabilized PIF4 to coregulate the etiolation-related genes via direct binding to the G-box, thus promoting skotomorphogenic growth (Figure 6). The transition from embryonic phase to seedling establishment is crucial for successful sporophyte development of plants. Because LEC1 plays indispensable roles in embryogenesis and postgerminative growth, both of which involve specification of hypocotyl and cotyledon development, this model provides new insights into the possible mechanism by which plants fine-tune morphological development for their survival during the seed-seedling switch when facing adverse conditions such as a lack of light.
Figure 6.
A Model Illustrating the Proposed Role of LEC1 and PIF4 in the Transcriptional Regulation during Postembryonic Growth.
In the light, photoactivated phytochromes (Phy) interact with PIF4 and trigger their codegradation by the 26S proteasome, thus abolishing the expression of the etiolation-related genes and promoting photomorphogenesis. In the dark, LEC1 interacts with stabilized PIF4 to coregulate the etiolation-related genes via direct binding to the G-box, thus promoting skotomorphogenic growth.
LEC1 expression in dark-grown seedlings was previously reported (Warpeha et al., 2007; Siefers et al., 2009). In this study, however, we showed that LEC1 expressed at comparable levels in seedlings grown in the light and the dark. Because lec1 mutants also have shorter hypocotyls than the wild type in light-grown seedlings, in which no PIF4 protein was detected in contrast to its significant accumulation in the dark-grown seedlings (Figure 1F; Supplemental Figure 12), combined the observation that inducible LEC1 had no effect on the expression of downstream genes of PIF4 in the light (Supplemental Figures 3C and 3D), we cannot exclude the possibility that LEC1 plays an extra role in the light via a PIF4-independent mechanism.
GA mediates hypocotyl elongation through the interaction between DELLA proteins, the key GA signaling repressors, and PIFs (de Lucas et al., 2008; Feng et al., 2008), while BR promotes hypocotyl elongation through PIF activity (Oh et al., 2012; Bernardo-García et al., 2014). Moreover, PIF4 promotes hypocotyl elongation at high temperature by mediating auxin biosynthesis (Franklin et al., 2011). Notably, a recent study showed that ectopically expressed LEC1 from an inducible 35S:LEC1-GR transgene regulates the expression of the auxin synthesis gene YUC10 and IAA accumulation in the elongation zone at hypocotyl-root junction, as well as the expression of genes involved in BR metabolism (Junker et al., 2012). We demonstrate that like PIF4, LEC1 also mediates GA and BR effects on the promotion of hypocotyl growth, while the synthetic auxin picloram significantly rescues the short hypocotyl phenotype responding to high temperature in both pif4 and lec1 (Supplemental Figure 11). Combined with the fact that LEC1 and PIF4 cotarget auxin-responsive genes, such as IAA19 and YUC8, these findings further confirm the interdependent regulation of LEC1 and PIF4 presented in this model.
Seed embryos of LEC1 null mutants are desiccation-intolerant and have to be rescued prior to desiccation for producing seedlings, causing problems in maintenance of homozygous recessive plants (West et al., 1994). In this study, we identified a lec1 mutant, designated lec1-4, in which LEC1 transcripts remain at a low level compared with that in the wild type. The lec1-4 mutants show typical pleiotropic phenotypes in LEC1 null alleles, including defective embryo, abnormal seeds, and postgerminative growth. More importantly, most lec1-4 embryos are tolerant to desiccation so that ∼90% of freshly harvested mutant seeds are viable, in spite that longer storage gradually reduces the germination rate. Because of the convenience in producing lec1 homozygous plants, this allele provides an efficient genetic tool for us to learn how LEC1 functions in plant development.
LEC1 encodes a protein related to the B-domain of NF-Y subunit, which can interact with NF-YC subunits and subsequently with the NF-YA subunit to form a heterotrimer binding to the CCAAT box (Calvenzani et al., 2012; Hackenberg et al., 2012). A de novo motif search showed that two motifs, the G-box-containing motif and the CCAAT box motif, are overrepresented in putative LEC1 target genes (Junker et al., 2012). However, disruption of the G-box but not the CCAAT box abolished the activation of seed storage-related genes by NF-YC2/bZIP67/LEC1 (or L1L) in mesophyll protoplasts (Yamamoto et al., 2009; Mendes et al., 2013). In this study, we demonstrate that the association with the G-box via PIFs is necessary for LEC1 to promote the expression of genes in response to skotomorphogenic growth. Although we observed no obvious binding of LEC1 to CCAAT in hypocotyl elongation-related genes in ChIP assays, whether the potentially functional CCAAT box is involved in LEC1-PIF regulation still needs further investigation.
To survive in the dark, seedlings undergo skotomorphogenesis by a complicated regulatory mechanism. Our findings shed light on the mechanism of LEC1-PIF module function in dark-adapted responses. However, studies have shown that PIFs also interact with several central regulators in multiple signals, which allows the integration of light and other cellular pathways to fine-tune plant growth (de Lucas et al., 2008; Feng et al., 2008; Oh et al., 2012; Liu et al., 2013; Zhang et al., 2014). The relationship between LEC1 and these regulators in early development of seedlings needs to be clarified. In addition, since LEC1 acts a key regulator in embryo development, including embryonic hypocotyl elongation, which was impaired in the lec1 mutant (West et al., 1994), and some bHLH transcriptional factors function in embryo development (Kondou et al., 2008; Chandler et al., 2009; Sun et al., 2010), these findings indicate that a LEC1-PIF-like regulatory interaction could occur between LEC1 and other bHLHs in embryogenesis. Further investigation of these issues will provide a comprehensive understanding of LEC1 functions in plant development.
METHODS
Plant Materials and Growth Conditions
The used plant materials including lec1-3 (Sugliani et al., 2009), pif4-2 (Sun et al., 2012), pifq (Zhang et al., 2013), phyb-9 (Reed et al., 1993), 35S:PIF4-HA (de Lucas et al., 2008), 35S:PIF4 (de Lucas et al., 2008), and 35S:PIF3-MYC (Zhang et al., 2013) were previously described. All Arabidopsis thaliana plants used in this study are in Col background except lec1-3, which is in the Landsberg erecta background. The lec1-4 (SALK_095699) and l1l-1 (SALK_118236) mutant seeds were obtained from the ABRC. For experiment analyses, sterilized seeds sown on half-strength Murashige and Skoog (MS) medium containing no sucrose were incubated at 4°C in darkness for 3 d and then transferred to 22°C under full-spectrum white fluorescent light (16 h light/8 h dark) or in the dark for treatments as indicated.
Plasmid Construction and Plant Transformation
For the pLEC1:LEC1-FLAG construct, the 3.8-kb genomic fragment of LEC1 without stop codon and 3′ untranslated region was amplified and cloned into the pPZPY122-FLAG binary vector (Hou et al., 2010). For pLEC1:GUS construct, ∼2 kb promoter of LEC1 was cloned into pCAMBIA 1391Z vector. The estradiol-inducible pER10:LEC1-MYC construct was previously described (Mu et al., 2008). Primers used for plasmid construction are listed in Supplemental Table 1. Transgenic plants harboring pLEC1:LEC1-FLAG, pLEC1:GUS, or pER10:LEC1-MYC were selected on half-strength MS medium supplemented with gentamycin, hygromycin, or kanamycin, respectively.
Hypocotyl Length Analysis
Seedlings grown on half-strength MS medium were photographed with a digital camera. The hypocotyl length was measured by Digimizer Image Analysis software.
Gene Expression Analysis
Growth conditions and treatment of seedlings were described in the text. Total RNA was extracted using the Plant RNA Kit (OMEGA) and reverse transcribed using the M-MLV reverse transcriptase (Promega). Quantitative PCR was performed in triplicates with the SYBR Premix ExTaq Mix (Takara Bio) following the manufacturer’s instructions. The relative expression level was normalized to that of a TUB2 internal control. Primers used for gene expression analysis are listed in Supplemental Table 1.
Yeast Two-Hybrid Assay
The coding regions of LEC1, L1L, and PIFs or truncated versions of LEC1 and PIF4 were amplified and cloned into pGBKT7 and pGADT7 (Clontech), respectively. Primers used are listed in Supplemental Table 1. Yeast two-hybrid assays were performed using the Yeastmaker Yeast Transformation System 2 (Clontech). Yeast AH109 cells were cotransformed with specific bait and prey constructs. All yeast transformants were grown on SD/-Trp/-Leu/-His/-Ade medium for selection or interaction tests.
Pull-Down and Coimmunoprecipitation
Full-length cDNAs of PIF4 and LEC1 were cloned into pGEX-4T-1 (Pharmacia) and pQE30 (Qiagen) vectors to produce GST-PIF4 and His-LEC1 proteins, respectively. Primers used for the construction are listed in Supplemental Table 1. GST and His fusion recombinant proteins were induced by isopropyl β-d-1-thiogalactopyranoside and expressed in Escherichia coli Rosetta (DE3; Novagen). The soluble GST and His fusion proteins were purified using Glutathione Sepharose Beads (Amersham Biosciences) or Ni-NTA agarose beads (Qiagen), following the manufacturers’ instructions. For pull-down assays, 2 μg His-LEC1 was incubated in the binding buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 1 mM EDTA) with immobilized GST or GST fusion protein at 4°C for 2 h. After washing with binding buffer, proteins retained on the beads were subsequently resolved by SDS-PAGE and detected with anti-His antibody (BGI). For coimmunoprecipitation, 3-d-old pLEC1:LEC1-FLAG 35S:PIF4-HA seedlings were kept in darkness for additional 2 d and then total proteins were extracted. Total proteins were extracted with extraction buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM DTT, and 1% Triton X-100). After centrifugation, the supernatants were incubated with anti-HA agarose conjugate (Sigma-Aldrich) in the coimmunoprecipitation buffer (50 mM HEPES, pH 7.5, 150 mM KCl, 10 μM ZnSO4, 5 mM MgCl2, and 1% Triton X-100) at 4°C for 2 h. After washing with coimmunoprecipitation buffer three times, the proteins bound to beads were resolved by SDS-PAGE and detected by anti-FLAG or anti-HA antibody (Santa Cruz Biotechnology).
BiFC Assay
Full-length cDNAs of LEC1 and PIF4 were cloned into the pGreen binary vector HY105 containing C- or N-terminal fusions of EYFP to generate 35S:LEC1-EYFPC and 35S:PIF4-EYFPN, then cotransformed into tobacco (Nicotiana benthamiana) leaf epidermal cells by infiltration as previously published (Hou et al., 2014). After infiltration, tobacco was grown for 2 d and transferred to darkness for 24 h before detection of the YFP signals. The YFP fluorescence was determined using a fluorescence microscope (Leica). DAPI (4',6-diamidino-2-phenylindole) staining was used to show the nucleus.
ChIP Assay
ChIP assays were performed as previously described (Hou et al., 2014). Briefly, for 35S:PIF4-HA, 3-d-old seedlings were transferred to darkness for additional 2 d and harvested for fixation. For pER10:LEC1-MYC, 3-d-old seedlings were treated with 10 μM estradiol and immediately transferred to darkness for additional 2 d and harvested for fixation. Chromatin was isolated and sonicated to generate DNA fragment with an average size of ∼500 bp. The solubilized chromatin was immunoprecipitated by Protein G PLUS agarose (Santa Cruz Biotechnology) with anti-HA (Santa Cruz Biotechnology), anti-MYC (Santa Cruz Biotechnology), and the coimmunoprecipitated DNA was recovered and analyzed by quantitative PCR with SYBR Premix ExTaq Mix (Takara Bio). Relative fold-enrichment was calculated by normalizing the amount of a target DNA fragment against that of a TUB8 genomic fragment and then against the respective input DNA samples. The primers used are listed in Supplemental Table 1.
EMSA
The EMSA assay was performed using the LightShift Chemiluminescent EMSA kit (Pierce). The 5′-end biotinylated oligonucleotide containing G-box in the P3 region of IAA19 promoter was used as a probe, while nonlabeled probe containing the native G-box (CACGTG) or mutated G-box (CACGgG) was used as cold competitor (Supplemental Figure 8A). Recombinant His-LEC1 and GST-PIF4 proteins were used for protein-DNA binding.
Transient Expression Assay
To generate the pIAA19:GUS reporter construct, ∼2 kb IAA19 promoter was cloned into HY107-containing GUS gene (Hou et al., 2014). mpIAA19:GUS carrying two mutated G-box elements (CACGTG to CACGGG) was generated by overlapping PCR reaction from pIAA19:GUS construct (Supplemental Figure 8B). 35S:LEC1-EYFPC and 35S:PIF4-EYFPN were used as effector constructs and a construct containing the firefly luciferase driven by 35S promoter in pGreen-35S was used as an internal control to evaluate the protoplast transfection efficiency. All primers used for generating constructs for transient expression assays are listed in Supplemental Table 1. Arabidopsis mesophyll protoplasts were prepared, transfected, and cultured as previously described (Yoo et al., 2007). Relative GUS activity was calculated by normalizing against the luciferase activity, and the data presented are the averages of five biological replicates.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: LEC1 (AT1G21970), L1L (AT5G47670), PIF3 (AT1g09530), PIF4 (AT2G43010), IAA19 (AT3G15540), ATHB-2 (AT4G16780), YUC8 (AT4G28720), IAA29 (AT4G32280), TUB2 (AT5G62690), and TUB8 (At5g23860).
Supplemental Data
Supplemental Figure 1. Characterization of the lec1-4 Mutant.
Supplemental Figure 2. pLEC1:LEC1-FLAG Complements Growth Defects of lec1-4.
Supplemental Figure 3. Characterization of pER10:LEC1-MYC Transgenic Line.
Supplemental Figure 4. Hypocotyl Length and Gene Expression Analysis in lec1-4, pif4, pifq, and pER10:LEC1-MYC Transgenic Plants.
Supplemental Figure 5. Yeast Two-Hybrid Assays Showing the Interactions between LEC1, L1L, and PIFs.
Supplemental Figure 6. Hypocotyl Phenotype of the l1l-1 Mutant.
Supplemental Figure 7. ChIP Analysis of LEC1-MYC Binding in IAA29 and ATHB-2 Genes in the Dark and Light.
Supplemental Figure 8. Mutated Versions of the IAA19 Sequence Used in EMSA and Transient Expression Assays.
Supplemental Figure 9. Transient Expression Assays of IAA19 Transcriptional Activity Modulated by LEC1.
Supplemental Figure 10. lec1-4 Suppresses the Long Hypocotyl Phenotype of 35S:PIF3-MYC and phyb.
Supplemental Figure 11. Effects of GA, Auxin, and BR on Hypocotyl Elongation of lec1-4 and pif4.
Supplemental Figure 12. Immunoblot Analysis of PIF4 Protein.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank C. Fankhauser for providing 35S:PIF4-HA seeds, C. Li for pif4-2 seeds, W.J.J. Soppe for lec1-3 seeds, J. Li for bri1-301 seeds, the ABRC for lec1-4 and l1l-1 seeds, J. Zuo for pER10:LEC1-MYC plasmid, and W. Xi and H. Yu for critical reading of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31370342, 31301055, and 31300239), the “Hundred Talents” program of the Chinese Academy of Sciences, and the Natural Science Foundation of Guangdong Province (S2013010014954 and S2013040013147).
AUTHOR CONTRIBUTIONS
M.H. and X.H. designed the research. M.H., Y.H., X.L., and Y.L. performed research. M.H., Y.H., and X.H. analyzed data. M.H. and X.H. wrote the article.
Glossary
- bHLH
basic helix-loop-helix
- BiFC
bimolecular fluorescence complementation
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- GA
gibberellic acid
- BR
brassinosteroid
- MS
Murashige and Skoog
Footnotes
Articles can be viewed online without a subscription.
References
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446. [DOI] [PubMed] [Google Scholar]
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311. [DOI] [PubMed] [Google Scholar]
- Bernardo-García S., de Lucas M., Martínez C., Espinosa-Ruiz A., Davière J.M., Prat S. (2014). BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28: 1681–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford I.M., Lynch T.J., Finkelstein R.R. (2003). Regulatory networks in seeds integrating developmental, abscisic acid, sugar, and light signaling. Plant Physiol. 131: 78–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvenzani V., Testoni B., Gusmaroli G., Lorenzo M., Gnesutta N., Petroni K., Mantovani R., Tonelli C. (2012). Interactions and CCAAT-binding of Arabidopsis thaliana NF-Y subunits. PLoS One 7: e42902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A., Shen H., Huq E. (2007). Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12: 514–521. [DOI] [PubMed] [Google Scholar]
- Chandler J.W., Cole M., Flier A., Werr W. (2009). BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNROSCHEN and DORNROSCHEN-LIKE. Plant Mol. Biol. 69: 57–68. [DOI] [PubMed] [Google Scholar]
- Chen M., Chory J., Fankhauser C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38: 87–117. [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S., Yu P., Breen G., Cohen J.D., Wigge P.A., Gray W.M. (2011). Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 108: 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D., Wu Y., Voigt A., Adams R., Schramm P., Grimm B. (2012). Studies on differential nuclear translocation mechanism and assembly of the three subunits of the Arabidopsis thaliana transcription factor NF-Y. Mol. Plant 5: 876–888. [DOI] [PubMed] [Google Scholar]
- Harada J.J. (2001). Role of Arabidopsis LEAFY COTYLEDON genes in seed development. J. Plant Physiol. 158: 405–409. [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Hou X., Lee L.Y.C., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 19: 884–894. [DOI] [PubMed] [Google Scholar]
- Hou X., Zhou J., Liu C., Liu L., Shen L., Yu H. (2014). Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 5: 4601. [DOI] [PubMed] [Google Scholar]
- Huq E., Quail P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21: 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Y., Zhang D., Wang X., Tang W., Wang W., Huai J., Xu G., Chen D., Li Y., Lin R. (2013). Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josse E.M., Halliday K.J. (2008). Skotomorphogenesis: the dark side of light signalling. Curr. Biol. 18: R1144–R1146. [DOI] [PubMed] [Google Scholar]
- Junker A., Bäumlein H. (2012). Multifunctionality of the LEC1 transcription factor during plant development. Plant Signal. Behav. 7: 1718–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker A., et al. (2012). Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 71: 427–442. [DOI] [PubMed] [Google Scholar]
- Khanna R., Huq E., Kikis E.A., Al-Sady B., Lanzatella C., Quail P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16: 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yi H., Choi G., Shin B., Song P.S., Choi G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15: 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondou Y., Nakazawa M., Kawashima M., Ichikawa T., Yoshizumi T., Suzuki K., Ishikawa A., Koshi T., Matsui R., Muto S., Matsui M. (2008). RETARDED GROWTH OF EMBRYO1, a new basic helix-loop-helix protein, expresses in endosperm to control embryo growth. Plant Physiol. 147: 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro A., Yamashino T., Nakamichi N., Niwa Y., Nakanishi H., Mizuno T. (2011). Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 52: 1315–1329. [DOI] [PubMed] [Google Scholar]
- Le B.H., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. USA 107: 8063–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E. (2014). PIFs: systems integrators in plant development. Plant Cell 26: 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22: 782–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Chen C.Y., Wang K.C., Luo M., Tai R., Yuan L., Zhao M., Yang S., Tian G., Cui Y., Hsieh H.L., Wu K. (2013). PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25: 1258–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205. [DOI] [PubMed] [Google Scholar]
- Meinke D.W., Franzmann L.H., Nickle T.C., Yeung E.C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes A., Kelly A.A., van Erp H., Shaw E., Powers S.J., Kurup S., Eastmond P.J. (2013). bZIP67 regulates the omega-3 fatty acid content of Arabidopsis seed oil by activating fatty acid desaturase3. Plant Cell 25: 3104–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Tan H., Zheng Q., Fu F., Liang Y., Zhang J., Yang X., Wang T., Chong K., Wang X.J., Zuo J. (2008). LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol. 148: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W., Xu S.L., Tepperman J.M., Stanley D.J., Maltby D.A., Gross J.D., Burlingame A.L., Wang Z.Y., Quail P.H. (2014). A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 344: 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F., Valon C., Kohara A., Miséra S., Giraudat J. (1997). The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9: 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W., Nagpal P., Poole D.S., Furuya M., Chory J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siefers N., Dang K.K., Kumimoto R.W., Bynum W.E. IV, Tayrose G., Holt B.F. III (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugliani M., Rajjou L., Clerkx E.J.M., Koornneef M., Soppe W.J.J. (2009). Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive3-5 (abi3-5) and leafy cotyledon1-3 (lec1-3). New Phytol. 184: 898–908. [DOI] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Shantharaj D., Kang X., Ni M. (2010). Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 13: 611–620. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G., Huq E., Quail P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Arnim A., Deng X.W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47: 215–243. [DOI] [PubMed] [Google Scholar]
- Warpeha K.M., Upadhyay S., Yeh J., Adamiak J., Hawkins S.I., Lapik Y.R., Anderson M.B., Kaufman L.S. (2007). The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 143: 1590–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M., Yee K.M., Danao J., Zimmerman J.L., Fischer R.L., Goldberg R.B., Harada J.J. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6: 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Kagaya Y., Toyoshima R., Kagaya M., Takeda S., Hattori T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58: 843–856. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang D., Jing Y., Jiang Z., Lin R. (2014). The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26: 2472–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bishop B., Ringenberg W., Muir W.M., Ogas J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159: 418–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.