EXB1 plays pivotal roles in shoot branching by positively regulating the key branch genes RAX1, RAX2, and RAX3 and by affecting auxin pathways.
Abstract
Plant shoot branching is pivotal for developmental plasticity and crop yield. The formation of branch meristems is regulated by several key transcription factors including REGULATOR OF AXILLARY MERISTEMS1 (RAX1), RAX2, and RAX3. However, the regulatory network of shoot branching is still largely unknown. Here, we report the identification of EXCESSIVE BRANCHES1 (EXB1), which affects axillary meristem (AM) initiation and bud activity. Overexpression of EXB1 in the gain-of-function mutant exb1-D leads to severe bushy and dwarf phenotypes, which result from excessive AM initiation and elevated bud activities. EXB1 encodes the WRKY transcription factor WRKY71, which has demonstrated transactivation activities. Disruption of WRKY71/EXB1 by chimeric repressor silencing technology leads to fewer branches, indicating that EXB1 plays important roles in the control of shoot branching. We demonstrate that EXB1 controls AM initiation by positively regulating the transcription of RAX1, RAX2, and RAX3. Disruption of the RAX genes partially rescues the branching phenotype caused by EXB1 overexpression. We further show that EXB1 also regulates auxin homeostasis in control of shoot branching. Our data demonstrate that EXB1 plays pivotal roles in shoot branching by regulating both transcription of RAX genes and auxin pathways.
INTRODUCTION
Plant shoot branching affects plant architecture and thus is an important trait for plant breeding and domestication (Doebley et al., 1997; Teichmann and Muhr, 2015). The ideal plant architecture favors crop production and management (Jiao et al., 2010). Modification of shoot branching can improve plant architecture for economic benefits (Teichmann and Muhr, 2015). Alterations of shoot branching can also help plants to morphologically adapt to environmental changes (Domagalska and Leyser, 2011). Shoot branches develop from axillary meristems (AMs) initiated in the boundary zone that separates leaf primordia from the shoot apex. AMs first form axillary buds with a few leaves and then the buds can either stay dormant or develop into shoot branches (McSteen and Leyser, 2005). Consequently, AM initiation is a prerequisite for shoot branching.
AM initiation starts from a group of small and slowly dividing cells in the axils of leaf primordia. Phytohormone homeostasis and signaling have been reported to be essential for AM initiation (Q. Wang et al., 2014; Y. Wang et al., 2014). First, the auxin minimum mediated by the auxin transporter PIN-FORMED1 (PIN1) in the boundary zone is a prerequisite for AM initiation. The polar localization of PIN1 proteins in cell membranes directs the auxin flows and the kinase PINOID regulates the polar localization of PIN1 (Reinhardt et al., 2003; Cheng et al., 2007; Huang et al., 2010). During the initiation of a leaf primordium on the flanks of shoot apical meristems (SAMs), PIN1 protein is oriented toward a convergence point; this leads to an auxin maximum for leaf primordium formation (Benková et al., 2003). As the leaf develops, the auxin in the boundary region between the SAM and the leaf primordium is depleted by the reversed orientation of PIN1 toward the SAM to form an auxin minimum for AM initiation (Q. Wang et al., 2014; Y. Wang et al., 2014). Accordingly, the compromise of the auxin minimum in pin1 or pid mutants or caused by ectopic expression of the auxin biosynthesis gene iaaM in the boundary zone leads to deficiencies of AM initiation, whereas disruption of auxin signaling by expressing the stable AUX/IAA repressor BODENLOS in the leaf axils largely rescues the AM initiation defects in pid-9 mutants and causes the formation of AMs in the axils of cotyledons (Q. Wang et al., 2014; Y. Wang et al., 2014). Second, after the establishment of an auxin minimum in the boundary zone, the activation of cytokinin (CK) perception and signaling is required for AM initiation (Y. Wang et al., 2014). Three CK receptors, i.e., ARABIDOPSIS HISTIDINE KINASE2 (AHK2), AHK3, and AHK4/CRE1/WOL, perceive the CK signal to activate the type-B ARABIDOPSIS RESPONSE REGULATOR transcription factors (Hwang et al., 2012). The AM initiation is significantly reduced in the multiple mutants with compromised CK perception and signaling, i.e., ahk2-5 ahk3-7, ahk2-5 ahk3-7/+ cre1-2, arr1-3 arr10-5, or arr1-3 arr10-5 arr12-1, indicating that CK plays important roles during AM initiation (Y. Wang et al., 2014). Finally, the reduced local brassinosteroid (BR) accumulation is also pivotal for the establishment of the boundary zone (Arnaud and Laufs, 2013). BR is an essential plant steroid hormone controlling cell division and cell expansion (Gendron et al., 2012). In the leaf axils, BR accumulation is negatively regulated by LATERAL ORGAN BOUNDARIES (LOB), an important boundary-specific transcription factor. LOB directly upregulates PHYB ACTIVATION TAGGED SUPPRESSOR1 (BAS1). BAS1 is a cytochrome P450 enzyme that inactivates BRs by C-26 hydroxylation, thus forming a low BR concentration to reduce cell division and expansion in the boundary zone (Bell et al., 2012; Gendron et al., 2012).
Two hypotheses have been proposed to explain the origins of AMs. One is the de novo meristem formation hypothesis suggesting that the original cells forming AMs in the boundary zone adopt AM fate from the beginning (McConnell and Barton, 1998), and the other is the detached meristem hypothesis describing that the original cells of AMs detach early from SAM during the initiation of the leaf primordium and reserve AM fate from SAM (Steeves and Sussex, 1989). The fact that the meristematic identity gene SHOOT MERISTEMLESS (STM) continues to be expressed in the boundary zone supports the detached meristem hypothesis (Long and Barton, 2000; Greb et al., 2003). It is suggested that the niche of the auxin minimum maintains the expression of STM in the boundary zone, and STM subsequently promotes CK biosynthesis in the leaf axils. CK then activates WUSCHEL, a stem cell marker gene, to further establish functional AMs (Y. Wang et al., 2014). The expression of STM in leaf axils is also regulated by other regulators important for AM initiation. For example, REVOLUTA (REV) likely acts upstream of STM (Talbert et al., 1995). Because AMs are only initiated on the adaxial side of leaf petioles, the roles of REV in AM formation likely resulted from its specification of leaf adaxial fate (Otsuga et al., 2001). STM is also controlled by LATERAL SUPPRESSOR (LAS). LAS is a member of the GRAS family of putative transcription factors and is essential for AM initiation (Greb et al., 2003). Disruption of LAS in Arabidopsis thaliana or its orthologous genes, i.e., Lateral suppressor (Ls) in tomato (Solanum lycopersicum) and MONOCULM1 (MOC1) in rice (Oryza sativa), leads to a lack of AMs and, therefore, branches or tillers, indicating that the function of these genes are highly conserved. Consistent with this, both LAS and MOC1 are specifically expressed in AM initiation zone (Schumacher et al., 1999; Greb et al., 2003; Li et al., 2003). LAS is negatively regulated by the microRNA miR164 (Raman et al., 2008), which directly targets the transcripts of CUP SHAPED COTYLEDON1 (CUC1) and CUC2 for degradation (Raman et al., 2008). The mir164a, b, c triple mutant produced accessory buds in leaf axils, whereas overexpression of miR164 combined with the cuc3-2 mutant led to the abolishment of AMs, indicating that miR164, CUC1, CUC2, and CUC3 also play pivotal roles in AM initiation (Raman et al., 2008). CUC genes encode NAM-ATAF1/2-CUC2 transcription factors and are negatively regulated by BRs. Thus, the low BRs in the boundary zones facilitate the specific expression of CUC genes and AM initiation (Gendron et al., 2012).
The specific expression of CUC2 in the boundary zone is positively controlled by another important AM initiation factor, i.e., REGULATOR OF AXILLARY MERISTEMS1 (RAX1). RAX1 functions redundantly with RAX2 and RAX3 in the control of AM initiation (Keller et al., 2006). RAX genes encode proteins belonging to the R2R3 MYB family of transcription factors. The rax1-3 mutants produce fewer AMs during vegetative development, and the rax1-3 rax2-1 rax3-1 triple mutant lost nearly all AMs, suggesting that RAX genes play central roles during AM initiation (Keller et al., 2006; Müller et al., 2006). The function of RAX genes in AM formation is also conserved. BLIND (BL) is the ortholog of RAX genes in tomato and pepper (Capsicum annuum). AM initiation is strongly suppressed in bl mutants in the two species (Schmitz et al., 2002; Jeifetz et al., 2011). Overproduction of cytokinin by expressing a cytokinin synthase gene IPT8 in the leaf axil region largely rescues the deficiency of AM initiation in rax mutants, suggesting that both CUC2 and cytokinin signaling may be positively regulated by RAX proteins (Y. Wang et al., 2014). However, very little is known about the upstream regulators of RAX genes.
Here, we identify the EXB1 (EXCESSIVE BRANCHES1) gene, which encodes a WRKY transcription factor previously designated WRKY71 (Eulgem et al., 2000). The gain-of-function mutant exb1-D displays dramatically increased branches, which resulted from a combination of excessive AM initiation and elevated bud activities. Disruption of EXB1 by chimeric repressor technology significantly reduces the number of AMs. We show that EXB1 positively regulates RAX genes at the transcriptional level. Our data demonstrate that WRKY71/EXB1 is an important positive regulator of RAX genes and plays important roles in AM initiation.
RESULTS
The Arabidopsis exb1-D Mutant Exhibits Excessive Branches
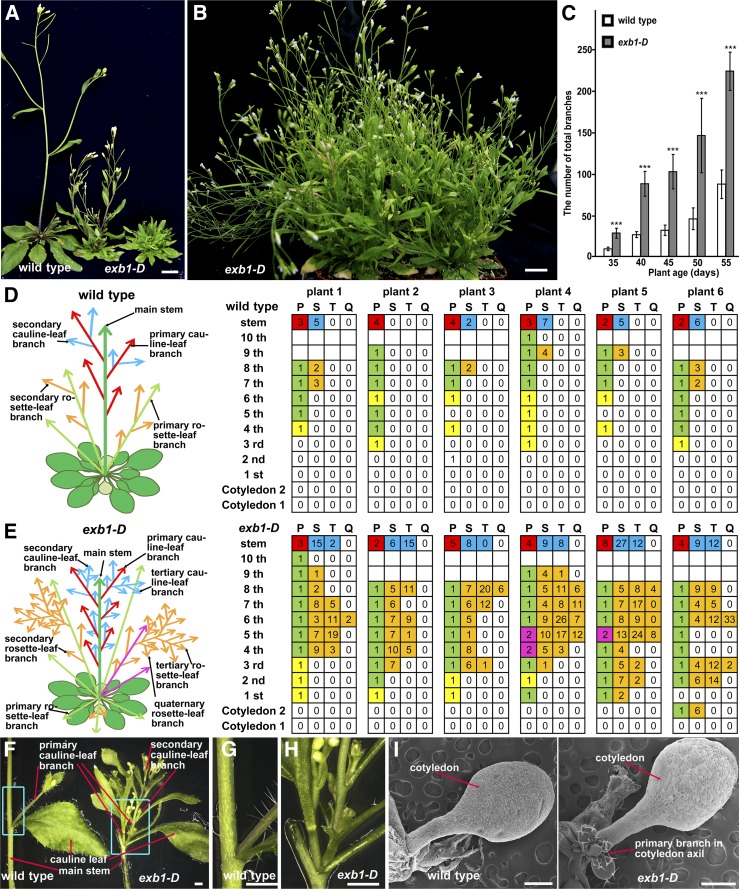
In order to study the genetic networks involved in regulation of plant architecture, we systematically screened an activation-tagging mutant collection for mutants with abnormal plant architecture (Qin et al., 2003; Li et al., 2015). One of the mutants, designated exb1-D (excessive branches 1-Dominant), produced more branches than wild-type plants (Figure 1A). At the seedling stage, no obvious differences were observed between exb1-D and wild-type plants. However, as the plants grew older, exb1-D produced many more branches than wild-type plants (Figures 1B and 1C). We quantified the number of buds and branches developed in the leaf axils. As shown in Figures 1D and 1E, the axillary buds were rarely formed in the first three rosette leaf axils in the wild type, whereas nearly all rosette leaves carried buds or branches in the axils in exb1-D. We noticed that two branches formed in a leaf axil and more high-order branches were produced in exb1-D (Figures 1E to 1H). Interestingly, in some cases, branches were even produced in the axils of cotyledons in exb1-D, which is not observed in wild-type plants (Figure 1I; Supplemental Figure 1A). These results indicated that the initiation of AMs was promoted in exb1-D. The fact that nearly all buds of exb1-D grew out to form branches and the exb1-D branches grew faster than the wild type (Figures 1D to 1H) indicated that the bud activities were highly increased in exb1-D. These analyses show that the observed excessive branching in exb1-D resulted from both increased AM initiation and elevated bud activities. The exb1-D mutant also displayed some other phenotypes, including dwarfism, deformed leaves, defective flowers with carpeloid sepals, and abnormal siliques with longer styles compared with wild-type plants (Figure 1A; Supplemental Figures 1B to 1E).
Figure 1.
The Phenotypes of exb1-D.
(A) From left to right, 40-d-old wild-type plants, heterozygous exb1-D, and homozygous exb1-D. The exb1-D mutants produced many more branches than wild-type plants.
(B) The 55-d-old homozygous exb1-D. exb1-D displayed excessive branches and bushy phenotype.
(C) Number of total branches of wild-type plants and homozygous exb1-D at different growth stages. Total branches were the average of 10 plants. Two-tailed t test was used to test the significance. Three asterisks represent P < 0.001.
(D) and (E) Schematic representation and calculation of axillary branches or buds in leaf axils in 45-d-old wild-type plants and homozygous exb1-D (n = 6). Left, schematic representation of branches in the wild type and exb1-D. Right, the statistical analysis of branches or buds in each axil of the wild type and exb1-D. Each column represents the order of branch, and each row presents the position of the axils. P, primary branch; S, secondary branch; T, tertiary branch; Q, quaternary branch. The white grids represent the bud absence, the yellow ones represent the inactive buds, the green ones indicate the outgrown branches, the orange ones indicate high order branches, the magenta ones indicate that multiple branches are formed in one axil, the red ones represent the branch in the axils of cauline leaves from main stem, and the blue ones represent the high order branches of cauline leaves. The number in the grids represents the number of buds or branches, and the blank grids means that no leaves are formed at the positions.
(F) to (H) Cauline leaf branches in the wild type or in exb1-D. exb1-D produced two branches in one axil.
(G) and (H) Close-up views of insets in (F).
(I) exb1-D produced the branch in the cotyledon axil.
Bars = 1 cm in (A) and (B), 1 mm in (F) to (H), and 0.5 mm in (I).
The exb1-D Phenotypes Are Caused by the Overexpression of At1g29860
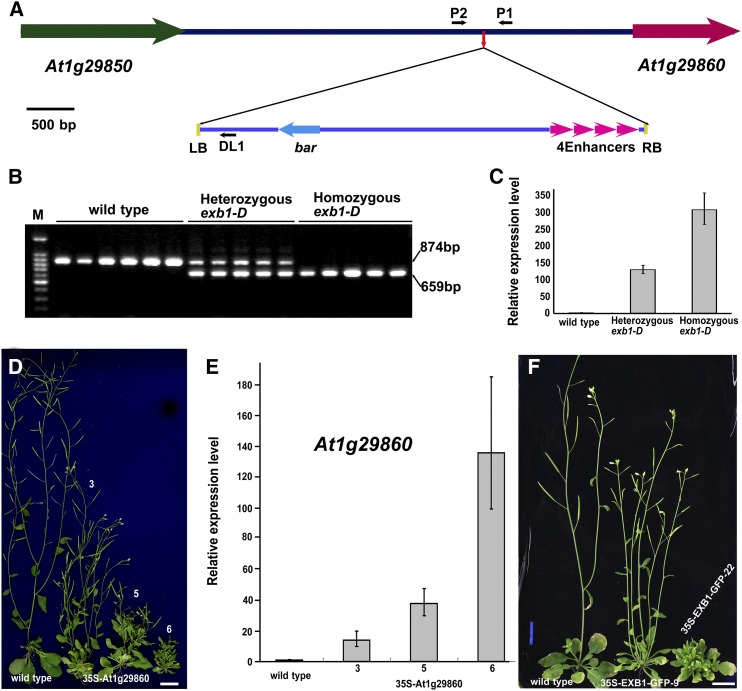
The phenotypes of exb1-D cosegregated with the resistance to herbicide phosphinothricin conferred by the bar gene in the T-DNA of the activation-tagging vector pSKI015 (Weigel et al., 2000), suggesting that the defects of exb1-D are likely caused by a T-DNA insertion. Using thermal asymmetric interlaced PCR (TAIL-PCR) (Liu and Chen, 2007), we identified one T-DNA insertion located in the intergenic region between At1g29850 and At1g29860 (Figure 2A). We further showed that the T-DNA insertion cosegregated with the phenotypes of exb1-D (Figure 2B) and led to overexpression of At1g29860 (Figure 2C). To determine whether overexpression of At1g29860 is responsible for the excessive branching in exb1-D or not, we generated transgenic At1g29860 overexpression plants using a CaMV 35S promoter. Analysis of 36 independent lines showed that 35S-At1g29860 transgenic lines produced more branches than the wild type, recapitulating the phenotypes of exb1-D (Figures 2D). RT-qPCR analysis showed that the expression levels of At1g29860 increased with increasing severity of the phenotypes (Figures 2E). These results indicate that overexpression of At1g29860 leads to the increased number of branches in exb1-D. Thus, At1g29860 was designated as EXB1. We also generated a construct of 35S-EXB1-GFP in which EXB1 was fused with GFP at its C terminus under the control of the CaMV 35S promoter. Eleven independent 35S-EXB1-GFP transgenic plants produced more branches than the wild type, further confirming that overexpression of EXB1 causes excessive branching in exb1-D (Figure 2F).
Figure 2.
The Phenotypes of exb1-D Are Caused by Overexpression of At1g29860.
(A) Schematic representation of the T-DNA insertion site in exb1-D. The colored arrows represent genes, and lines indicate the intergenic region. The small red arrow in the intergenic region indicates the T-DNA insertion site. The four magenta arrowheads represent the four CaMV 35S enhancers in pSKI015. The small black arrows represent DL1, P1, and P2 primers used in cosegregation analysis. LB, T-DNA left border; RB, T-DNA right border; 4Enhancers, four CaMV 35S enhancers; bar, Basta resistance gene.
(B) Cosegregation analysis of T-DNA insertion with the increased branches. The 874-bp DNA bands were amplified from the wild type and the 659-bp bands from exb1-D.
(C) The relative expression level of At1g29860 in the wild type, heterozygous exb1-D, and homozygous exb1-D. The expression level of EXB1 in the wild type was set to 1.0. The error bars represent the sd of three biological replicates.
(D) The exb1-D phenotypes were recapitulated by overexpressing At1g29860 using a CaMV 35S promoter. From left to right, 40-d-old wild type and three independent 35S-At1g29860 transgenic lines. Bars = 1 cm.
(E) The relative expression level of At1g29860 in the wild type and the three 35S-At1g29860 transgenic lines in (D). The expression level of At1g29860 in the wild type was set to 1.0. The error bars represent the sd of three biological replicates.
(F) The overexpression of EXB1 fused with GFP also recapitulated the phenotypes of exb1-D. From left to right, 40-d-old wild type and two independent 35S-EXB1-GFP transgenic lines. Bars = 1 cm.
EXB1 Encodes a WRKY Transcription Factor with Transactivation Activity
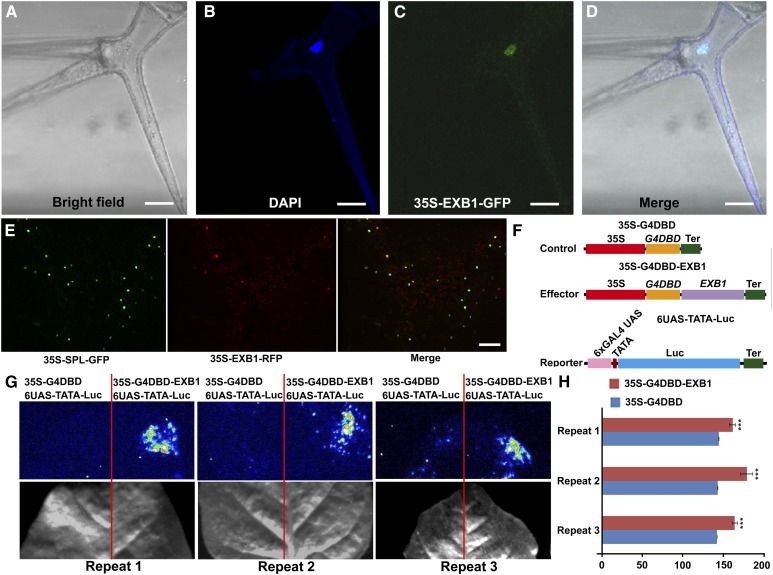
EXB1 encodes WRKY71, a protein of 282 amino acid residues containing a typical WRKY domain that consists of a conserved WRKYGQK motif and a C2H2 zinc finger-like motif (Supplemental Figure 2A). However, the function of EXB1/WRKY71 has not been characterized. In the 35S-EXB1-GFP transgenic plants, clear green fluorescence was observed in the nuclei of trichome cells, demonstrating that EXB1 may be a nuclear protein (Figures 3A to 3D). To further confirm that EXB1 is located in the nucleus, we generated 35-EXB1-RFP and 35S-SPL-GFP in which the known nuclear protein SPOROCYTELESS (SPL) was fused to GFP (Yang et al., 1999; Wei et al., 2015). Coexpression of the two fusing proteins in tobacco (Nicotiana benthamiana) leaves showed that they were colocalized in the nuclei (Figure 3E). This is consistent with the roles of EXB1 as a transcription factor.
Figure 3.
EXB1 Is a Transcriptional Activator Localized in the Nuclei.
(A) to (D) EXB1 is localized in the nucleus. The leaf trichome cell of 35S-EXB1-GFP-5 transgenic line was observed. From left to right, the trichome under bright field, DAPI staining of the nucleus, GFP fluorescence, and merge of DAPI staining and GFP fluorescence. Bars = 25 µm.
(E) Coexpression of 35-EXB1-RFP and 35S-GFP-SPL in tobacco leaves indicated that EXB1 were colocalized with SPL, a known nuclear transcriptional repressor (Yang et al., 1999; Wei et al., 2015). Bar = 100 µm.
(F) to (H) EXB1 is a transcription activator.
(F) Schematic representation of constructs using in transcriptional activity assay.
(G) and (H) Transcription activity of EXB1 was tested in tobacco leaves using a GAL4/UAS-based system. The quantitative analysis of fluorescence intensity in (G) is shown in (H). Two-tailed t test was used to test the significance. Three asterisks represent P < 0.001. 35S, CaMV 35S promoter; G4DBD, GAL4 DNA binding domain; Ter, terminator of nopaline synthase gene; 6XGAL4 UAS, six copies of GAL4 binding site UAS; TATA, TATA box of CaMV 35S promoter.
To test whether EXB1 has transactivation activity, we generated a reporter 6×UAS-TATA-Luc construct in which firefly luciferase (Luc) gene was placed under the control of six copies of GAL4 binding site (UAS) (Figure 3F). We then fused EXB1 to the GAL4 DNA binding domain (DBD) and put it under the control of the CaMV 35S promoter to generate 35S-G4DBD-EXB1. We cotransformed the 6UAS-TATA-Luc reporter with 35S-G4DBD or 35S-G4DBD-EXB1 into tobacco leaves to examine the transcriptional activity of EXB1. The firefly luciferase imaging assay showed that 35S-G4DBD-EXB1 significantly activated the expression of the Luc reporter gene (Figures 3G and 3H), indicating that EXB1 has transactivation activity.
EXB1 Is Expressed in the Leaf Axils
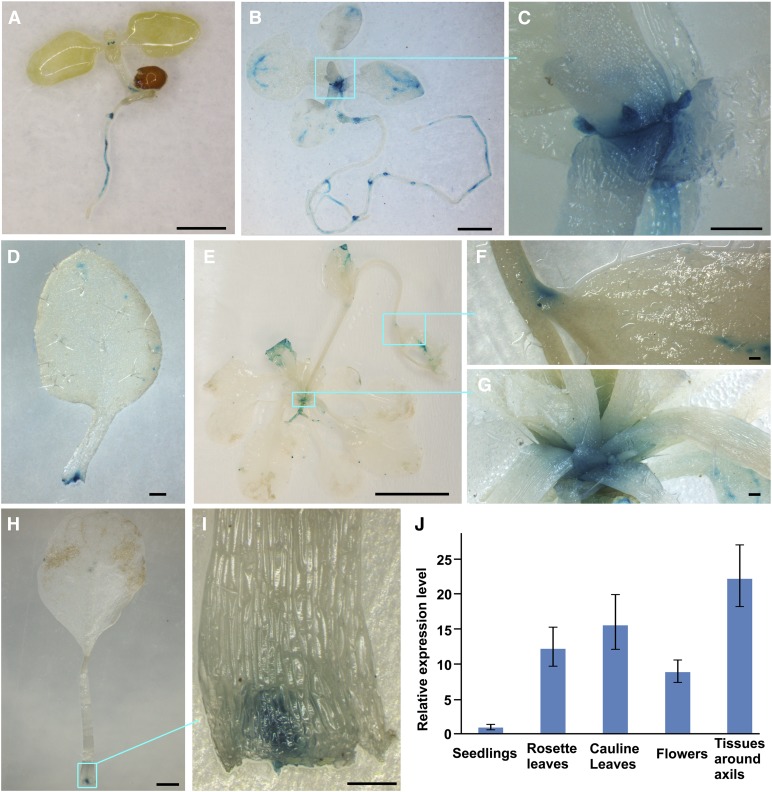
To investigate the spatial and temporal expression pattern of EXB1, we generated pEXB1-GUS in which the GUS reporter was driven by a 2700-bp promoter of EXB1. Seventeen pEXB1-GUS transgenic lines displayed a similar staining pattern. GUS activity was detected at the base of young leaves at the seedling stage (Figure 4A). As the plants grew older, GUS activity became stronger and was specifically observed at the bases of rosette leaves, in the leaf axils and in the stipules (Figures 4B to 4I). In mature plants, GUS activity became weaker, but was still clearly detectable in the axils of leaves (Figures 4E to 4I). We also observed GUS staining in hypocotyls, roots, flowers, and hydathodes (Figures 4A to 4E). Quantitative RT-PCR analysis showed that EXB1 was predominantly expressed in the tissues around AM initiation sites (Figure 4J), consistent with the expression data from GUS staining. The specific expression of EXB1 in the leaf axils was consistent with the roles of EXB1 in control of shoot branching.
Figure 4.
EXB1 Is Expressed in Leaf Axils.
(A) The 7-d-old pEXB1-GUS-11 seedling. EXB1 was strongly expressed at the base of small true leaves.
(B) The 15-d-old pEXB1-GUS-11 plant. EXB1 was strongly expressed around the axils of rosette leaves.
(C) Close-up view of the leaf axils in the cyan square in (B).
(D) One leaf dissected from the plant in (B) showed that clear and specific expression of EXB1 at the leaf base.
(E) The 35-d-old pEXB1-GUS-11 plant. EXB1 was expressed in the axils of rosette leaves and cauline leaves.
(F) Close-up view of a cauline leaf axil in the cyan square in (E).
(G) Close-up view of the rosette leaf axils in the cyan square in (E).
(H) One leaf dissected from the plant in (E) showed a specific GUS staining at the leaf base.
(I) Close-up view of the leaf base in the cyan square in (H). EXB1 was specifically expressed at the position of AM initiation site.
(J) The relative expression of EXB1 in different tissues. EXB1 was expressed at the highest level in the tissues around axils. The expression level in seedlings was set to 1.0. The error bars represent the sd of three biological replicates.
Bars = 1 mm in (A), (B), and (H), 200 µm in (C), (D), (F), (G), and (I), and 1 cm in (E).
Disruption of EXB1 by Chimeric Repressor Silencing Technology Led to Defects of Shoot Branching
To further elucidate the roles of EXB1 in shoot branching, we identified one T-DNA insertion mutant SALK_050011 for EXB1, named as exb1-1, from the publically available mutant collections, in which the EXB1 transcripts were highly reduced (Supplemental Figures 2B and 2C). We could not observe obvious phenotypes in exb1-1. Because EXB1 belongs to the WRKY superfamily of transcription factors, and there are four closely related WRKY members, i.e., WRKY8, WRKY28, WRKY48, and WRKY57, in the same clade with WRKY71/EXB1 (Supplemental Figures 2D and 2E), these WRKY members likely have functional redundancy. To demonstrate whether these four WRKYs have a redundant function with EXB1, we first overexpressed WRKY8, WRKY28, WRKY48, and WRKY57 using four CaMV 35S enhancers and the 2700-bp EXB1 promoter (4EnhEXB1p), respectively (Supplemental Figure 3A). Transgenic analysis showed that overexpression of WRKY8, WRKY28, WRKY48, and WRKY57 all caused excessive branching similar to that observed in exb1-D (Supplemental Figures 3B to 3F), indicating that the four WRKYs had similar biochemical functions to EXB1 during shoot branching. Thus, we renamed WRKY8 as EXB2, WRKY28 as EXB3, WRKY48 as EXB4, and WRKY57 as EXB5. We then cloned the promoters of EXB2, EXB3, EXB4, and EXB5. We put the GUS reporter gene under the control of the promoters to generate pEXB2-GUS, pEXB3-GUS, pEXB4-GUS, and pEXB5-GUS, respectively. GUS staining showed that the EXBs had overlapping expression with EXB1 in the leaf axils (Supplemental Figures 3G to 3J), further indicating that the EXBs play redundant roles in shoot branching.
We then identified SALK_107668 (exb2-1) for EXB2, SALK_066438 (exb4-1) for EXB4, and SALK_006260 (exb5-1) for EXB5 (Supplemental Figure 4A). A previous report and real-time PCR showed that EXB2, EXB4, and EXB5 were knocked out or knocked down in the mutants (Supplemental Figure 4B) (Xing et al., 2008). No mutants for EXB3 were available in the public databases. We generated the exb1-1 exb2-1, exb1-1 exb4-1, and exb1-1 exb5-1 double mutants, but observed no obvious branching phenotypes in these double mutants. We then generated the triple mutants exb1-1 exb2-1 exb5-1 and exb1-1 exb4-1 exb5-1. The triple mutants also did not exhibit any branching phenotypes. We crossed exb1-1 exb2-1 exb5-1 with exb1-1 exb4-1 exb5-1 and screened an F2 population consisting of more than 1000 lines for the quadruple mutant of exb1-1 exb2-1 exb4-1 exb5-1. We failed to obtain the quadruple mutant, likely because of the close linkage between EXB2 and EXB4. These results further suggested that EXB1 was highly redundant with other WRKY family proteins during shoot branching.
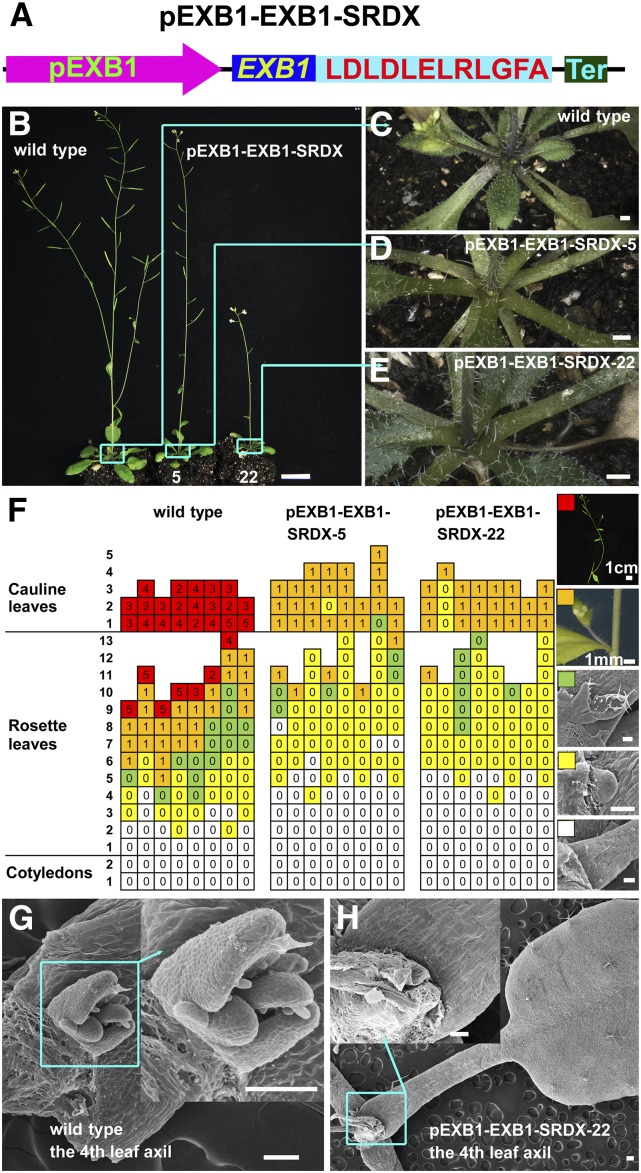
To overcome the difficulties caused by genetic redundancy, we employed chimeric repressor silencing technology (CRES-T), an efficient technology widely used to identify the functions of transcription activators (Hiratsu et al., 2003; Koyama et al., 2007; Heyl et al., 2008). In CRES-T, a transcription factor is fused to the transcriptional repression domain of SUPERMAN (SRDX), producing a chimeric protein that can dominantly repress the targets of the transcription factor (Hiratsu et al., 2003). We generated EXB1-SRDX by fusing EXB1 to an EAR motif, the active transcriptional repression domain SRDX, and expressed EXB1-SRDX in wild-type Arabidopsis under the control of the EXB1 promoter (Figure 5A). Out of 96 pEXB1-EXB1-SRDX transgenic plants, 43 plants displayed fewer branches (Figures 5B to 5E).
Figure 5.
Disruption of EXB by Localized Expression of Chimeric EXB1 Repressor Led to Compromise of Shoot Branching.
(A) Schematic representation of the pEXB1-EXB1-SRDX construct.
(B) Phenotypes of the wild type and two independent lines of pEXB1-EXB1-SRDX transgenic plants.
(C) to (E) Close-up views of rosette leaf branches in the cyan squares of the corresponding plants in (B). Bars = 1 cm in (B) and 1 mm in (C) to (E).
(F) Statistical analysis of AMs or branches in each leaf axil of 45-d-old wild-type or pEXB1-EXB1-SRDX transgenic plants. Each column represents one independent plant and each row presents the position of the axils. The typical leaf axil morphologies represented by the different color grids are shown at the right. The white grids represent the absence of AMs, the yellow ones represent the AMs with leaf primordia, the green ones indicate the buds with leaves, the orange ones indicate branches with flower buds, and the red ones represent branches with high order branches. The number in the grids represents the number of branches.
(G) and (H) Representative leaf axils of the wild-type plant and pEXB1-EXB1-SRDX-22.
Bars = 100 μm for (F) to (H) except that indicated near the bars.
We took one line with moderate phenotypes (i.e., pEXB1-EXB1-SRDX-5) and one line with strong phenotypes (i.e., pEXB1-EXB1-SRDX-22) to analyze the branching abnormalities in detail. Compared with wild-type plants, the number of primary branches, secondary branches, and tertiary branches from either pEXB1-EXB1-SRDX-5 or pEXB1-EXB1-SRDX-22 significantly decreased (Supplemental Figure 5A). We then investigated the detailed branching phenotype by analyzing the AMs or branches in every leaf axils. The results showed that AMs were frequently absent in the axils of the early rosette leaves (Figures 5F to 5H). We further grew plants under short-day conditions to observe the phenotypes. We found that the defects of AM initiation in pEXB1-EXB1-SRDX-22 were highly enhanced and AMs were deficient in more than half of the rosette leaves (Supplemental Figures 5B to 5D). These results indicate that EXB1 plays important roles in the regulation of AM initiation.
EXB1-Regulated Genes Controlling Shoot Branching
To investigate the downstream genes of EXB1, we generated the construct 4EnhpEXB1-EXB1GR in which EXB1 was fused to the sequence encoding the steroid binding domain of the rat glucocorticoid receptor (GR) (Aoyama and Chua, 1997) and was driven by four CaMV 35S enhancers and EXB1 promoter. Out of 21 4EnhpEXB1-EXB1GR transgenic lines, six lines (including 4EnhpEXB1-EXB1GR-13) were found to produce more branches than the control plants when treated with dexamethasone (DEX) (Supplemental Figure 6A), indicating that EXB1GR is functional. We then performed RNA-sequencing (RNA-seq) analysis on the tissues around leaf axils of 14-d-old 4EnhpEXB1-EXB1GR-13 transgenic lines treated with or without DEX. Our results showed that 3114 genes were differentially expressed, with 1015 upregulated and 2099 downregulated (fold change ≥ 2, P < 0.05; Supplemental Data Set 1). Because EXB1 is a transcription activator, we hypothesized that the direct targets of EXB1 could be among the genes upregulated by DEX treatment. Gene Ontology enrichment analysis of the 1015 upregulated genes with the DAVID gene functional annotation tool (Huang et al., 2009a, 2009b) showed that the genes involved in transcription regulation, plant hormone pathway, oxidative stress, oxidation reduction, and defense response were enriched (Supplemental Data Set 1). Among the overrepresented genes related to transcription regulation, three MYB family transcription factor genes, i.e., RAX1, RAX2, and RAX3, known to regulate axillary meristem initiation (Keller et al., 2006; Müller et al., 2006), were identified (Supplemental Data Set 1). Overexpression of RAX1 and RAX2 led to increased number of shoot branches (Keller et al., 2006; Müller et al., 2006), consistent with the observed exb1-D branching phenotype, suggesting that RAX1, RAX2, and RAX3 are probably downstream genes of EXB1.
RAX Genes Were Likely Regulated by EXB1 Directly
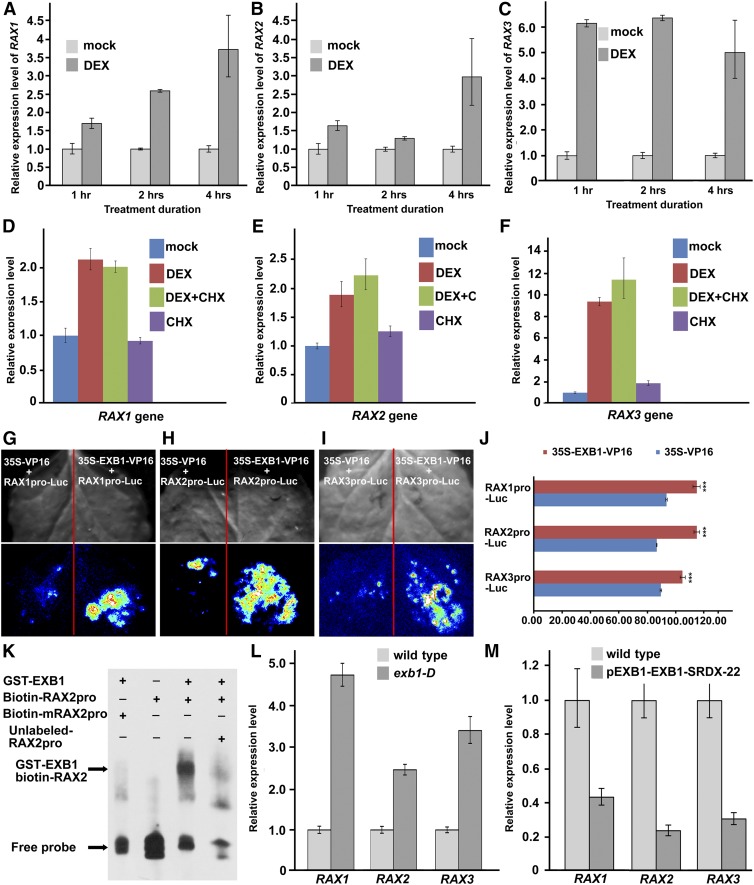
To clarify the relationship between EXB1 and RAX genes, we first examined the transcription induction of RAX genes by EXB1 in a time course. We harvested the tissues around leaf axils from the plants treated with or without DEX for 1, 2, or 4 h. The result showed that RAX1, RAX2, and RAX3 were all upregulated by DEX treatment (Figures 6A to 6C), consistent with the RNA-seq data. Interestingly, the three RAX genes were regulated at different time points after EXB1 induction. RAX1 was significantly upregulated after 2 h induction (Figure 6A). RAX2 was not upregulated until 4 h induction (Figure 6B), but RAX3 was induced as early as 1 h after EXB1 induction (Figure 6C).
Figure 6.
RAX Genes Are Likely Regulated by EXB1 Directly.
(A) to (C) Relative expression level of RAX1, RAX2, and RAX3 of 21-d-old 4EnhpEXB1-EXB1GR transgenic plants treated with DEX or mock for 1, 2, or 4 h. The expression level of corresponding genes in mock-treated plants was set to 1.0. The error bars represent the sd of three replicates.
(D) to (F) Relative expression level of RAX1, RAX2, and RAX3 in 21-d-old 4EnhpEXB1-EXB1GR transgenic plants treated with DEX, CHX, DEX plus CHX, or mock. The gene expression level in mock-treated plants was set to 1.0. The error bars represent the sd of three replicates.
(G) to (J) Interaction assays between EXB1 and the promoters of RAX1, RAX2, or RAX3 by transient expression in tobacco leaves. EXB1 was likely bind to the promoters of RAX1, RAX2, and RAX3 and activated the expression of Luc driven by the RAX1, RAX2, or RAX3 promoters.
(G) to (I) Top, bright field; bottom, fluorescence. Similar results were obtained by three biological repeats.
(J) Quantitative analysis of fluorescence intensity in (G) to (I). Two-tailed t test was used to test the significance. Three asterisks indicate P < 0.001.
(K) EMSA showed that EXB1 could bind to the W-box cis-elements in the RAX2 promoter in vitro. The RAX2 promoter fragments containing the W-box cis-elements were incubated with GST-EXB1 protein in vitro. Unlabeled RAX2 promoter fragments were used to compete for EXB1 binding, and the fragments with mutated W-box cis-elements were used in the control.
(L) Expression level of RAX1, RAX2, and RAX3 in the wild type and exb1-D.
(M) Expression level of RAX1, RAX2, and RAX3 in the wild type and pEXB1-EXB1-SRDX-22. The expression level of wild-type plants was set to 1.0. The error bars represent the sd of three replicates in (L) and (M).
We then conducted the DEX treatment experiment with or without cycloheximide (CHX), a chemical reagent, to inhibit protein synthesis (Sablowski and Meyerowitz, 1998; Sakai et al., 2001). As shown in Figures 6D to 6F, even with the CHX treatment, RAX1, RAX2, and RAX3 were still rapidly induced by DEX treatment, suggesting that induction of RAX genes by EXB1 is independent of new protein synthesis. The results suggest that these three RAX genes may be direct targets of EXB1.
To test whether EXB1 can bind to the promoter regions of the three RAX genes, we generated the construct 35S-EXB1-VP16 in which EXB1 was fused to the sequence encoding transactivator protein VP16 from herpes simplex virus and driven by a CaMV 35S promoter. We fused the promoter regions of RAX1, RAX2, or RAX3 to the firefly luciferase gene Luc and generated the reporters RAX1pro-Luc, RAX2pro-Luc, and RAX3pro-Luc. We cotransformed 35S-EXB1-VP16 or the control 35S-VP16 with RAX1pro-Luc, RAX2pro-Luc, or RAX3pro-Luc into tobacco leaves, respectively. The firefly luciferase imaging assay showed that the fluorescence was stronger in the 35S-EXB1-VP16 cotransformed with RAX1pro-Luc, RAX2pro-Luc, or RAX3pro-Luc combinations than in controls (Figures 6G to 6J), suggesting that EXB1 directly binds to the promoter regions of RAX genes in vivo.
In order to further verify the direct binding of EXB1 to RAX gene promoters, we first analyzed the possible W-box cis-element that is previously identified as the binding site for WRKY transcription factors in the promoter regions of RAX1, RAX2, or RAX3 using the signal scan program (Prestridge, 1991; Higo et al., 1999; Bakshi and Oelmüller, 2014). The results showed that numerous W-box cis-elements were found in RAX gene promoters (Supplemental Figure 7), consistent with the above results that EXB1 binds to RAX promoter regions in vivo. We then expressed the EXB1 protein and synthesized the RAX2 promoter region containing two typical W-box cis-elements for conducting electrophoretic mobility shift assay (EMSA). The sequence containing the mutated W-box (TTGAAA) was synthesized simultaneously as control. Our results showed that, when adding EXB1, the migration of biotin-labeled probes was shifted dramatically, but not the control probe containing the mutated W-box (Figure 6K). This band shift was reversed by adding competitive unlabeled probe (Figure 6K). These results further indicate that EXB1 directly binds to the promoter region of RAX2.
We further investigated the expression levels of RAX genes in the exb1-D mutant or EXB1 disruption line pEXB1-EXB1-SRDX-22. We found that all three RAX genes were significantly upregulated in exb1-D and downregulated in pEXB1-EXB1-SRDX-22 plants (Figures 6L and 6M). We then examined the known RAX downstream genes, i.e., ROX and CUC2 (Keller et al., 2006; Yang et al., 2012). Although the two genes were not rapidly upregulated after EXB1 induction (Supplemental Data Set 1), the expression of the two genes increased significantly in exb1-D (Supplemental Figure 6B). These results demonstrate that EXB1 may directly regulate RAX genes to control AM initiation during shoot branching.
The Defects of AM Initiation by EXB1 Overexpression Were Partially Rescued in rax Mutants
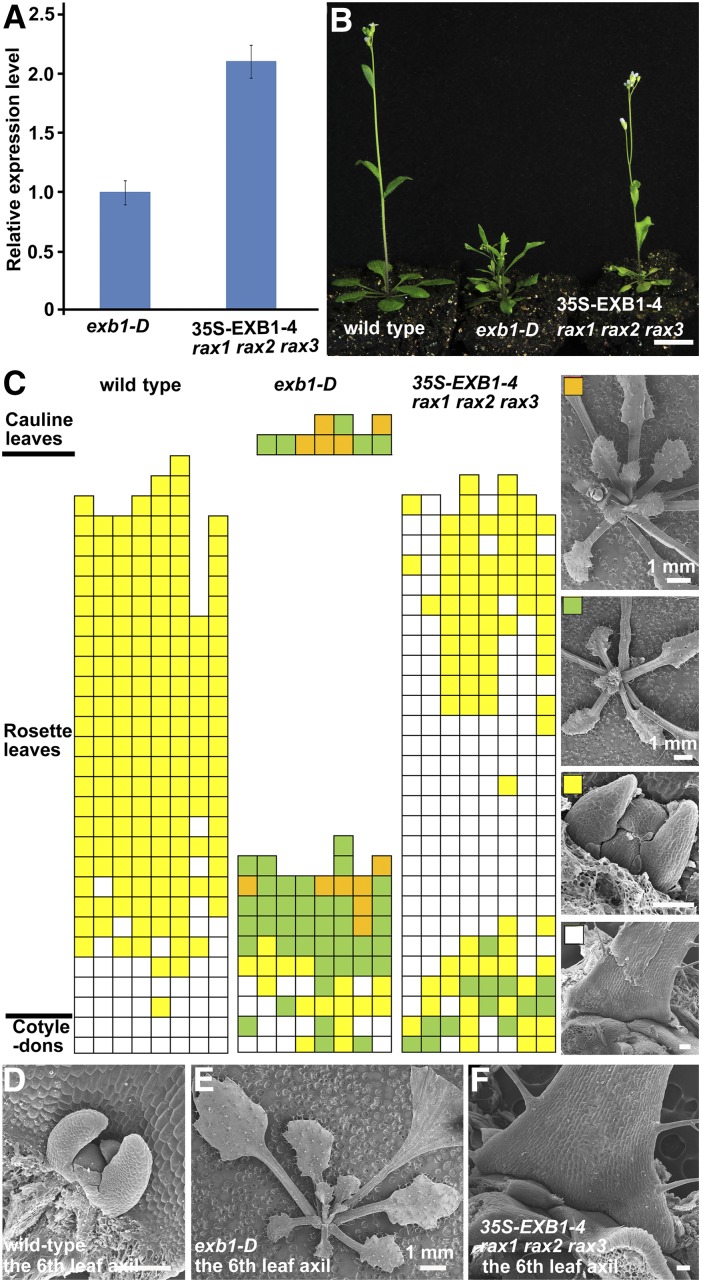
To further confirm that RAX genes act downstream of EXB1, we transformed the 35S-EXB1 construct in which EXB1 was driven by the CaMV 35S promoter into rax1 rax2 rax3 triple mutants. We obtained T2 seeds of three independent 35S-EXB1 rax1 rax2 rax3 transgenic lines and tested the expression levels of EXB1. We found that the expression level of EXB1 was significantly higher in 35S-EXB1-4 rax1 rax2 rax3 than in exb1-D (Figure 7A). Although the expression levels of EXB1 increased with increasing severities of the phenotypes of EXB1 overexpression lines (Figures 2D and 2E), our observation showed that the branches of 35S-EXB1-4 rax1 rax2 rax3 were obviously less than those of exb1-D (Figure 7B), suggesting that the defects of branching caused by EXB1 overexpression were mitigated in rax mutants. We further analyzed the detailed branching phenotype of wild-type plants, exb1-D, and 35S-EXB1-4 rax1 rax2 rax3 under a short-day condition. The results showed that AM initiation was strongly reduced in the early rosette leaves of 35S-EXB1-4 rax1 rax2 rax3 when compared with that in exb1-D (Figures 7C to 7F). The AM initiation in the later rosette leaves was even completely absent in 35S-EXB1-4 rax1 rax2 rax3 when compared with that in wild-type plants (Figures 7C to 7F), suggesting that the excessive branches caused by EXB1 overexpression could be partially rescued by disruption of RAX genes. These results suggest that RAX genes are required for the regulation of AM initiation by EXB1.
Figure 7.
RAX Genes Are Required for the Excessive Shoot Branching Caused by EXB1 Overexpression.
(A) Relative expression level of EXB1 in 35S-EXB1-4 rax1 rax2 rax3. EXB1 expression level in 35S-EXB1-4 rax1 rax2 rax3 was more than 2-fold of that in exb1-D.
(B) Shoot branches in 35S-EXB1-4 rax1 rax2 rax3 are obviously fewer than those in exb1-D in a long-day condition. Bar = 1 cm.
(C) Statistical analysis of AMs or branches in each leaf axil of 50-d-old wild type, exb1-D, or 35S-EXB1-4 rax1 rax2 rax3 in a short-day condition. Each column represents one independent plant and each row presents the position of the axils. The typical leaf axil morphologies represented by the different color grids were shown in the right of (C). The white grids represent the absence of AMs, the yellow ones represent the AMs with leaf primordia, the green ones indicate the buds with leaves, and the orange ones indicate branches with flower buds.
(D) to (F) Representative leaf axils of the wild type, exb1-D, or 35S-EXB1-4 rax1 rax2 rax3.
Bars = 100 μm for (C) to (F) except that indicated near the bars.
Auxin Homeostasis Was Disrupted in exb1-D Mutants
The exb1-D mutant displayed pleiotropic phenotypes including dwarfism and deformed leaves and flowers (Figure 1A; Supplemental Figure 1). This led us to hypothesize that some hormonal pathways might also be regulated by EXB1. Auxin is a well-known hormone affecting diverse aspects of plant development including AM initiation and bud outgrowth (Stirnberg et al., 1999; Domagalska and Leyser, 2011). To test whether the auxin pathway was affected by EXB1, we treated exb1-D at 29°C and measured hypocotyl elongation (Gray et al., 1998). The results showed that the hypocotyls of exb1-D mutants were significantly shorter than those of the wild type at high temperature (Supplemental Figure 8A), indicating that auxin homeostasis was disturbed in exb1-D. We further crossed an auxin reporter DR5-GUS to exb1-D. GUS activities were lower in exb1-D than in the wild-type control, especially at the sites of leaf axils, suggesting that auxin pathway was affected by EXB1 overexpression (Supplemental Figure 8B). We then searched our RNA-seq data and found that auxin biosynthesis genes such as TAA1, auxin transport genes like PIN5, and auxin-signaling genes including ARF11, ARF18, and IAA14 were repressed by EXB1 induction, while YUC8 and IAA7 were induced (Supplemental Table 1).
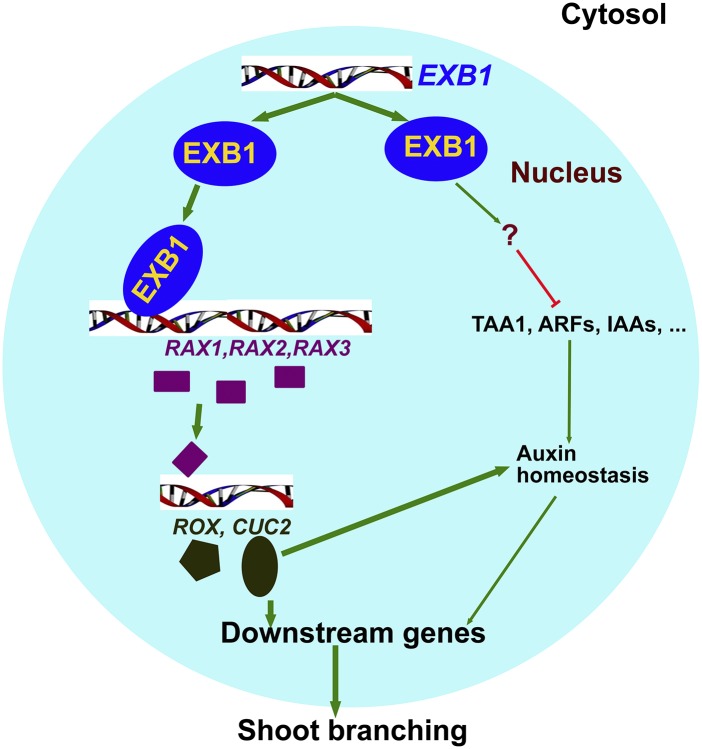
Taking these results together, we suggest that EXB1 plays very important roles in shoot branching by regulating RAX genes and auxin pathways. We propose a model that accounts for EXB1 action (Figure 8). EXB1 affects both AM initiation and bud activities during shoot branching. On one hand, EXB1 positively regulates RAX1, RAX2, and RAX3 to affect AM initiation; on the other hand, EXB1 may also regulate auxin pathways to control AM formation and bud outgrowth.
Figure 8.
Working Model for EXB1 Function.
EXB1 controls plant branching by affecting both AM initiation and bud activities. EXB1 positively regulates RAX1, RAX2, and RAX3 and thus upregulates the downstream genes CUC2 and ROX to control AM formation. EXB1 also regulates auxin pathways to affect both AM initiation and bud outgrowth.
DISCUSSION
Plant shoot branching is an important trait affecting crop production. The number of branches is mainly determined by the number of AMs and outgrown buds. In this study, we discovered that the gain-of-function mutant exb1-D produces many more branches, while disruption of EXB1 leads to fewer branches. We further demonstrate that EXB1 encodes the WRKY transcription factor WRKY71, which controls shoot branching by affecting both AM initiation and bud outgrowth. EXB1 is specifically expressed in leaf axils where AMs initiate. We further identified a molecular mechanism by which WRKY71/EXB1 regulates shoot branching by controlling RAX genes and by affecting the auxin pathway.
The Arabidopsis genome contains ∼74 WRKY transcription factors (Bakshi and Oelmüller, 2014). Extensive studies reveal that WRKY genes are involved in diverse biological processes including plant development, senescence, hormone signaling, and responses to biotic or abiotic stresses (Wei et al., 2013; Bakshi and Oelmüller, 2014). Our data demonstrate that EXB1/WRKY71 plays a central role in shoot branching. EXB1/WRKY71 contains one typical WRKY domain and is grouped into a small clade with WRKY8, WRKY28, WRKY48, WRKY57, WRKY23, and WRKY68 (Wu et al., 2005). WRKY8, WRKY28, WRKY48, and WRKY57 have overlapping expression pattern with EXB1 in the leaf axils. Ectopic expression of WRKY8, WRKY28, WRKY48, and WRKY57 by EXB1 promoter and four copies of CaMV 35S enhancers all caused excessive branches similar to those observed in exb1-D. These results suggest that the WRKY proteins of the EXB1 clade have redundant functions in shoot branching. WRKY8, WRKY28, and WRKY48 also participate in plant responses to biotic and abiotic stress (Xing et al., 2008; Wu et al., 2011; Chen et al., 2013; Hu et al., 2013), suggesting that the WRKY genes are important for both morphological adaptation and physiological adaptation to environments in plants.
It is proposed that AM formation is redundantly controlled by at least three groups of transcription factors, i.e., RAX1, ROX, and LAS (Yang et al., 2012). RAX1 has redundant function with its homologs RAX2 and RAX3 (Müller et al., 2006). Overexpression of RAX1 or RAX2 leads to short stature and produces accessory buds in cauline leaf axils, while loss of RAX gene function causes defects in AM formation (Keller et al., 2006; Müller et al., 2006). RAX1 and LAS function in two independent genetic pathways; however, RAX1 and LAS genetically interact with each other and positively regulate ROX (Yang et al., 2012). Our data indicate that EXB1 positively regulates RAX genes, indicating that EXB1 is a key player functioning upstream of RAX genes. Interestingly, EXB1 regulates RAX1, RAX2, and RAX3 temporally as well. RAX3 was upregulated as early as 1 h after EXB1 induction, and then RAX1 and RAX2 were induced at 2 and 4 h, respectively. Up to now, few transcription factors have been reported to directly regulate RAX genes. LEAFY was reported to bind to the promoter region of RAX1 directly (Chahtane et al., 2013). Our data suggested that RAX genes are very likely regulated by EXB1 directly for the following reasons. First, RAX genes were induced rapidly upon EXB1 induction. Second, induction of RAX genes was independent of new protein synthesis but was dependent on EXB1 induction. Third, EXB1 could bind to the promoter region of RAX genes in vivo and in vitro. Fourth, genetic analysis shows that disruption of RAX genes could partially rescue the phenotype of excessive branching caused by EXB1 overexpression, indicating that RAX genes act downstream of EXB1. Finally, the exb1-D mutant produced more AMs, like the plants overexpressing RAX1 or RAX2 (Keller et al., 2006; Müller et al., 2006). These findings not only demonstrate the important roles of EXB1 in AM formation, but also increase our understanding of the complex regulation of the key branch genes.
The WRKY proteins in the EXB1 clade of phylogenetic tree regulate the auxin pathway. WRKY57 affects auxin signaling by interacting with AUX/IAA protein IAA29 in the regulation of jasmonate-induced leaf senescence (Jiang et al., 2014). WRKY23 acts downstream of IAA14, AUXIN RESPONSE FACTOR7 (ARF7), and ARF19 (Grunewald et al., 2008, 2012). WRKY23 also controls auxin distribution by regulating flavonol biosynthesis during plant root and embryo development (Grunewald et al., 2012, 2013). In this article, we found that the auxin pathway was compromised in the exb1-D mutant, as shown by auxin reporter DR5-GUS analysis and by the reduced rates of hypocotyl elongation of exb1-D in high temperature treatment. This is consistent with the fact that overexpression of WRKY23 disrupts auxin transport and response (Grunewald et al., 2012). Auxin is a well-known inhibitor of bud outgrowth and an auxin minimum is required for AM initiation (Hayward et al., 2009; Stirnberg et al., 2012; Y. Wang et al., 2014). The negative regulation of auxin signaling by EXB1 may be an important reason for the phenotype of excessive branches and other pleiotropic phenotypes in exb1-D. Interestingly, overexpression of the rice WRKY72 in Arabidopsis also increases shoot branches (Yu et al., 2010), suggesting that the roles of EXB1/WRKY71 in shoot branching may be evolutionarily conserved between monocots and dicots.
METHODS
Plant Materials and Growth Conditions
All the Arabidopsis thaliana materials used for this study were Col-0 ecotype. exb1-D was a gain-of-function mutant obtained from a T-DNA insertion mutant collection (Qin et al., 2003). Half-strength Murashige and Skoog medium with or without 20 µg/mL dl-phosphinothricin or 50 µg/mL kanamycin were prepared for growing or screening the seeds of the wild type, mutants, transgenic plants, and crossed plants. The seeds were synchronized at 4°C for 2 d and were then placed at 22°C ± 2°C under long-day conditions (16 h light and 8 h dark) with light intensity of ∼170 μmol/m2/s. The seedlings were grown in soil under long-day conditions as described above. Nicotiana benthamiana plants were grown in soil under the same conditions in the greenhouse for transient expression assay and the luciferase assay.
PCR Analysis and Gene Expression Assays
Primers used in this study are listed in Supplemental Table 2. TAIL-PCR was used to identify the flanking sequences of the T-DNA insertion in exb1-D. The procedures of TAIL-PCR and the arbitrary primers (AD) and specific primers were described previously (Qin et al., 2003). Primers DL1, P1, and P2 were designed for cosegregation analysis of exb1-D. PCR was performed for 26 to 40 cycles under conditions of 94°C for 30 s, 56 to 60°C for 30 s, and 72°C for 2 min.
Total RNAs of all samples were prepared using TRIzol reagent (Invitrogen) and were reverse transcribed using the SuperScript III kit (Invitrogen) according to the user’s manual. RT-PCR or RT-qPCR was performed using the diluted cDNA as the template. RT-qPCR was performed with three biological repeats using SYBR Green Master Mix (Toyobo) as described previously (Wang et al., 2012). The cycling conditions of RT-qPCR were 94°C for 20 s, 56°C for 20 s, and 72°C for 30 s using Applied Biosystems 7500 Fast Real-time PCR system (ABI). The 2−ΔΔCT (cycle threshold) method was used to evaluate the relative expression level of each gene (Livak and Schmittgen, 2001). ACT8 was used as an internal control.
Generation of Binary Constructs and Transformation
The coding sequence of At1g29860/EXB1 was amplified from the exb1-D cDNA using primers EXB1-1 and EXB1-2. The DNA fragments were cloned into the EcoRV site of pBluescript SK+ to generate pBS-EXB1 (with ATG near the M13F primer). Alternatively, the coding sequence of EXB1 was amplified from the exb1-D cDNA by RT-PCR using primers EXB1-3 and EXB1-2 and cloned into pENTR/D-TOPO (Invitrogen) to generate pENTR/D-EXB1. pQG111 was generated by introducing the CaMV 35S fragment flanked by HindIII-EcoRI restriction sites isolated from pWM101 into pPZP111 to construct the binary vectors (Hajdukiewicz et al., 1994). The KpnI-PstI-digested fragment of pBS-EXB1 was ligated with pQG111 digested with the same restriction enzymes to generate overexpression construct 35S-At1g29860.
To generate pEXB1-EXB1-SRDX, primers EXB1-3 and EXB1-SRDX were used to amplify EXB1-SRDX using pBS-EXB1 as template and EXB1-SRDX fragment was cloned into pENTR/D-TOPO to generate pENTR/D-EXB1-SRDX. The 2700 bp of promoter before the start codon ATG of the EXB1 coding region was also isolated using EXB1P-1 and EXB1P-2 and was cloned into pDONRP4P1r (Invitrogen) to generate pEN-L4-pEXB1-R1. pEXB1-EXB1-SRDX was generated by LR reactions among pENTR/D-EXB1-SRDX, pEN-L4-pEXB1-R1, and pK7m24GW (Ghent University).
To generate 35S-EXB1-GFP, the coding region of EXB1 without stop codon was amplified from pBS-EXB1 using primers EXB1-3 and EXB1-2N and was cloned into pENTR/D-TOPO to generate pENTR/D-EXB1N. pENTR/D-EXB1N was cloned into pB7FWG2 (Ghent University) to generate 35S-EXB1-GFP using LR reactions. To study the subcellular localization of EXB1 in tobacco, 35S-RFP-EXB1 was generated through LR reaction between pENTR/D-EXB1 and pK7RWG2 (Ghent University). The coding sequence of SPL was amplified using primers SPL-1 and SPL-2 and then cloned into pENTR/D-TOPO to generate pENTR-SPL. 35S-GFP-SPL was generated through LR reaction between pENTR-SPL and pK7WGF2 (Ghent University).
To test the transcriptional activity of EXB1, GAL4UAS, and TATA box of 35S promoter was fused before the coding sequence of firefly Luc gene sequentially to generate the reporter 6UAS-TATA-Luc. To generate 35S-G4DBD-EXB1, G4DBD-EXB1 was first obtained by LR reaction between pDEST32 (Invitrogen) and pENTR/D-EXB1. The sequences of G4DBD-EXB1 and G4DBD were then amplified with the primer pairs of G4DBD-1, EXB1-2 using G4DBD-EXB1 plasmid as template and G4DBD-1, G4DBD-2 using pDEST32 plasmid as template. The fragments were cloned into pENTR/D-TOPO to generate pENTR/D-G4DBD-EXB1 and pENTR/D-G4DBD. The effector 35S-G4DBD-EXB1 and the control 35S-G4DBD were generated by LR reactions between pK2GW7 (Ghent University) with pENTR/D-G4DBD-EXB1 or pENTR/D-G4DBD.
To examine temporal and spatial expression pattern of EXB1, the 2700-bp promoter region of EXB1 was amplified using the primers EXB1P-3 and EXB1P-4 and was cloned into pENTR/D-TOPO to generate pENTR/D-pEXB1. pEXB1-GUS was generated by LR reaction between pENTR/D-pEXB1 and pKGWFS7 (Ghent University).
To generate 4EnhpEXB1-EXB1GR, the CaMV 35S enhancer tetrad was amplified using pSKI015 as template with the primers Enh-1 and Enh-2 and cloned into pQDL4R1 to generate pQDL4R1-4Enh. The GR domain was cloned from pTA7002 using primers GR-1 and GR-2 and the coding region of EXB1 was cloned from pBS-EXB1 using primers GR-3 and EXB1-2. Both fragments were fused together by PCR and cloned into pQDR2L3 vector to generate pQDR2L3-GR-EXB1. 4EnhEXB1p-EXB1GR was generated by LR reaction among pQDL4R1-4Enh, pENTR/D-pEXB1, pQDR2L3-GR-EXB1, and pB7m34GW (Ghent University).
The constructs were transformed into Agrobacterium tumefaciens GV3101/pMP90 by electroporation method and transformed into Arabidopsis as described previously by floral dip method (Clough and Bent, 1998).
Staining and Microscopy
The histochemical GUS staining analysis was described previously (Tao et al., 2013). Briefly, tissues of pEXB1-GUS transgenic lines or DR5-GUS plants were immerged in 90% acetone for 30 min on ice and washed by phosphate buffer twice. Then the samples were immerged in 0.5 mg/mL of 5-bromo-4-chloro-3-indolyl glucuronide staining buffer and vacuumed for 30 min before incubation at 37°C for ∼12 h. Then the staining buffer was replaced by 70% ethanol before observation.
For 4′,6-diamidino-2-phenylindole (DAPI) staining, leaves of 35S-EXB1-GFP transgenic plants were dissected and soaked in 1 µg/mL DAPI solution for ∼30 min. Nuclear localization of EXB1 fusion protein was observed in the trichome using a confocal laser scanning microscope (Leica TCS SPE confocal microscope).
Transient Expression Assay in Leaves of N. benthamiana
For generation of RAX1pro-Luc, RAX2pro-Luc, and RAX3pro-Luc reporter, promoters of RAX1, RAX2, and RAX3 were amplified from the total DNA of wild-type Arabidopsis using primer pairs of RAX1P-1 and RAX1P-2, RAX2P-1 and RAX2P-2, or RAX3P-1 and RAX3P-2. Promoters of RAX1, RAX2, and RAX3 were cloned into the SmaI or EcoRV site of pBluescript SK+ to generate pBS-RAX1pro, pBS-RAX2pro, and pBS-RAX3pro. The entry vectors of pQDL4R1-RAX1pro, pQDL4R1-RAX2pro, or pQDL4R1-RAX3pro were generated by ligation of the KpnI-PstI-digested fragments from pBS-RAX1pro, pBS-RAX2pro, or pBS-RAX3pro with corresponding restriction enzyme-digested pQDL4R1 fragment, respectively. The coding region of firefly Luc gene was amplified using primers FLUC-1 and FLUC-2 and cloned into pENTR/D-TOPO to generate pENTR/D-FLUC. The three reporter constructs were generated by LR reaction of the three plasmids including pH7m24GW (Ghent University), pENTR/D-FLUC and pQDL4R1-RAX1pro, pQDL4R1-RAX2pro, or pQDL4R1-RAX3pro.
To generate 35S-EXB1-VP16 or the control 35S-VP16, 35S promoter was amplified from pWM101 and then cloned into pQDL4R1 to generate pQDL4R1-35S using primers 35S-1 and 35S-2. The coding region of VP16 was amplified from pTA7002 and cloned into pQDR2L3 using primers VP16-1 and VP16-2 to generate pQDR2L3-VP16. Alternatively, VP16 was amplified using primers VP16-3 and VP16-2 and was cloned into pENTR/D-TOPO to generate pENTR/D-VP16. 35S-EXB1-VP16 was generated by LR reactions among plasmids pQDL4R1-35S, pENTR/D-EXB1N, pQDR2L3-VP16, and pK7m34GW (Ghent University). The control 35S-VP16 was generated by LR reactions between pK2GW7 and pENTR/D-VP16.
The reporter and the effector or the control were transformed into Agrobacterium GV3101/pMP90 and then were coinfiltrated with pCam-P19 into leaves of N. benthamiana as described previously (Voinnet et al., 2003). After incubation in the dark for 24 h and then in the light for 48 h, the leaves were observed using a low-light cooled CCD imaging apparatus (Lumazone 1300B; Roper Bioscience). To quantitatively analyze the fluorescence intensity, the representative region of each sample was divided into a number of small patches of 100 pixels. The average fluorescence intensity of each patch was read out by the WinView32 Software. Then the mean value and sd of readouts of all the patches for one sample were calculated. Two-tailed t test was used to test the statistical significance of the experiments.
EMSA
The GST-EXB1 construct was generated by LR reaction between pENTR/D-EXB1 and pGEX-4T1-Gateway modified from pGEX-4T1, and the resulting construct was introduced into Escherichia coli Rosetta (DE3) cell line. The induction of the recombinant GST-EXB1 proteins was performed by adding 0.2 mM isopropyl β-d-1-thiogalactopyranoside for 16 h of incubation at 18°C. The recombinant proteins were then purified using Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) according to the manufacturer’s instructions and were later used for EMSA.
The biotin-labeled and unlabeled single-strand promoter fragment of RAX2 (783 to 745 bp plus 177 to 139 bp upstream of the start codon) containing W-box were synthesized and purified by Invitrogen. The control fragment with mutated W-box was synthesized by replacing TTGACT with TTGAAA. The double-strand DNA fragments were acquired by annealing equal molar concentration of both complementary oligos in annealing buffer (10 mM Tris, pH 7.5, 1 mM EDTA, and 50 mM NaCl). The EMSA was performed using the LightShift Chemiluminescent EMSA kit (Pierce) according to the manufacturer’s instructions. The reactions were performed with only minimal reaction components [ultrapure water, 10× binding buffer, 1 μg/μL poly(dI⋅dC), together with the protein extract, biotin-labeled target DNA, and unlabeled DNA]. The reaction products were analyzed on 5% nondenaturing polyacrylamide gels.
DEX Induction Assay and RNA-seq
The 21-d-old homozygous 4EnhpEXB1-EXB1GR-13 transgenic plants were treated with 30 μM DEX, 30 μM DEX plus 100 μM CHX, or DMSO (mock). The tissues around leaf axils were collected at 1, 2, and 4 h after treatments and put into liquid nitrogen immediately. The expression levels of RAX genes were determined using RT-qPCR as described above. RNA-seq was performed on the HiSeq Illumina Hisequation 2000 platform in the Biodynamic Optical Imaging Center (BIOPIC) of Peking University. TopHat version 2.0.6 (http://tophat.cbcb.umd.edu/) was used to map each library against the Arabidopsis genome sequence index (Ensembl, TAIR10 version). Cuffdiff version 2.0.1 was run using the reference transcriptome along with the BAM files resulting from TopHat for each sample to analyze the differential expression between the two samples at gene level (Trapnell et al., 2012). Gene Ontology enrichment analysis was conducted using the DAVID gene functional annotation tool (http://david.abcc.ncifcrf.gov/) (Huang et al., 2009a, 2009b).
Scanning Electron Microscopy
The tissues from exb1-D, wild-type, and the transgenic plants were dissected and put into 30% FAA (30% ethanol [v/v], 5% glacial acetic acid [v/v], and 10% formaldehyde [v/v]) as soon as possible followed by vacuum treatment for ∼6 h at room temperature. The tissues were then fixed in 50% FAA (50% ethanol [v/v], 5% glacial acetic acid [v/v], and 10% formaldehyde [v/v]) overnight at 4°C. Then, serial ethanol dehydration (50, 70, 90, and 100% three times for 30 min each time) was conducted. The tissues were subsequently dried through CO2 critical point drying method (Leica EM CPD300). Dried tissues were observed through a scanning electron microscope (Hitachi 4700).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: EXB1 (At1g29860), EXB2 (AT5G46350), EXB3 (AT4G18170), EXB4 (AT5G49520), EXB5 (AT1G69310), RAX1 (AT5G23000), RAX2 (AT2G36890), RAX3 (AT3G49690), ROX (At5g01310), and CUC2 (AT5G53950).
Supplemental Data
Supplemental Figure 1. The Mutant exb1-D Displayed Pleiotropic Phenotypes and Produced Branches in the Axils of Cotyledons.
Supplemental Figure 2. EXB1 Encoding WRKY71, Which Shares High Similarity with WRKY8, WRKY28, WRKY48, and WRKY57 in Amino Acid Sequence.
Supplemental Figure 3. EXB1 Had Highly Redundant Function with EXB2, EXB3, EXB4, and EXB5.
Supplemental Figure 4. The Identification of exb2-1, exb4-1, and exb5-1 Mutants.
Supplemental Figure 5. Disruption of EXB1 Caused Fewer Branches in pEXB1-EXB1-SRDX Transgenic Plants.
Supplemental Figure 6. 4EnhpEXB1-EXB1GR Transgenic Plants Produced More Branches after DEX Treatment and the RAX Downstream Genes ROX and CUC2 Were Upregulated Significantly in exb1-D.
Supplemental Figure 7. The Possible W-box cis-Elements in the Promoters of RAX Genes.
Supplemental Figure 8. The Auxin Homeostasis Was Compromised by EXB1 Overexpression.
Supplemental Table 1. The Transcript Alteration of Genes in Auxin Pathway by EXB1 Induction.
Supplemental Table 2. The Primer List Used in This Work.
Supplemental Data Set 1. List of Genes Upregulated and Downregulated by DEX Treatment in 4EnhpEXB1-EXB1GR-13 Plants (Fold Change ≥2; P < 0.05) and Their Gene Ontology Enrichment Analysis.
Supplemental Data Set 2. Text File of Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 2E.
Supplementary Material
Acknowledgments
We thank Yunde Zhao (University of California at San Diego) for his valuable suggestions. We also thank Klaus Theres (Max Planck Institute for Plant Breeding Research, Germany) for kindly providing the seeds of rax1 rax2 rax3 triple mutants. This work was supported by the National Natural Science Foundation of China (Grant 31270321), by the National Transformation Science and Technology Program (Grant 2014ZX08009003-003), and by the National Key Basic Research Program of People’s Republic of China (Grant 973-2012CB944801). This work was also partially supported by the 111 Project (B06001).
AUTHOR CONTRIBUTIONS
G.Q. conceived the project. G.Q., D.G., J.Z., and X.W. designed the experiments. G.Q., D.G., J.Z., X.W., X.H., B.W., J.W., B.L., H.Y., and G.Q. performed the experiments. G.Q., D.G., Q.H., H.G., and G.Q. analyzed the data. G.Q., D.G., and L.-J.Q. wrote the article.
Glossary
- AM
axillary meristem
- SAM
shoot apical meristem
- CK
cytokinin
- BR
brassinosteroid
- TAIL-PCR
thermal asymmetric interlaced PCR
- DEX
dexamethasone
- CHX
cycloheximide
- EMSA
electrophoretic mobility shift assay
- DAPI
4′,6-diamidino-2-phenylindole
References
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. [DOI] [PubMed] [Google Scholar]
- Arnaud N., Laufs P. (2013). Plant development: brassinosteroids go out of bounds. Curr. Biol. 23: R152–R154. [DOI] [PubMed] [Google Scholar]
- Bakshi M., Oelmüller R. (2014). WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 9: e27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E.M., Lin W.C., Husbands A.Y., Yu L., Jaganatha V., Jablonska B., Mangeon A., Neff M.M., Girke T., Springer P.S. (2012). Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 109: 21146–21151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Chahtane H., et al. (2013). A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 74: 678–689. [DOI] [PubMed] [Google Scholar]
- Chen L., Zhang L., Li D., Wang F., Yu D. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 110: E1963–E1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Qin G., Dai X., Zhao Y. (2007). NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18825–18829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Doebley J., Stec A., Hubbard L. (1997). The evolution of apical dominance in maize. Nature 386: 485–488. [DOI] [PubMed] [Google Scholar]
- Domagalska M.A., Leyser O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12: 211–221. [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P.J., Robatzek S., Somssich I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Gendron J.M., Liu J.-S., Fan M., Bai M.-Y., Wenkel S., Springer P.S., Barton M.K., Wang Z.-Y. (2012). Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 21152–21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Ostin A., Sandberg G., Romano C.P., Estelle M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., Theres K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17: 1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., De Smet I., De Rybel B., Robert H.S., van de Cotte B., Willemsen V., Gheysen G., Weijers D., Friml J., Beeckman T. (2013). Tightly controlled WRKY23 expression mediates Arabidopsis embryo development. EMBO Rep. 14: 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., Karimi M., Wieczorek K., Van de Cappelle E., Wischnitzki E., Grundler F., Inzé D., Beeckman T., Gheysen G. (2008). A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 148: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W., et al. (2012). Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. USA 109: 1554–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Hayward A., Stirnberg P., Beveridge C., Leyser O. (2009). Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 151: 400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A., Ramireddy E., Brenner W.G., Riefler M., Allemeersch J., Schmülling T. (2008). The transcriptional repressor ARR1-SRDX suppresses pleiotropic cytokinin activities in Arabidopsis. Plant Physiol. 147: 1380–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34: 733–739. [DOI] [PubMed] [Google Scholar]
- Hu Y., Chen L., Wang H., Zhang L., Wang F., Yu D. (2013). Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74: 730–745. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I., Sheen J., Müller B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63: 353–380. [DOI] [PubMed] [Google Scholar]
- Jeifetz D., David-Schwartz R., Borovsky Y., Paran I. (2011). CaBLIND regulates axillary meristem initiation and transition to flowering in pepper. Planta 234: 1227–1236. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., Dong G., Zeng D., Lu Z., Zhu X., Qian Q., Li J. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42: 541–544. [DOI] [PubMed] [Google Scholar]
- Keller T., Abbott J., Moritz T., Doerner P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18: 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19: 473–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li R., Jiang Z., Gu H., Qu L.-J. (2015). ADP1 affects abundance and endocytosis of PIN-FORMED proteins in Arabidopsis. Plant Signal. Behav. 10: e973811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. (2003). Control of tillering in rice. Nature 422: 618–621. [DOI] [PubMed] [Google Scholar]
- Liu Y.G., Chen Y. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–650, 652, 654 passim. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Long J., Barton M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218: 341–353. [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Barton M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942. [DOI] [PubMed] [Google Scholar]
- McSteen P., Leyser O. (2005). Shoot branching. Annu. Rev. Plant Biol. 56: 353–374. [DOI] [PubMed] [Google Scholar]
- Müller D., Schmitz G., Theres K. (2006). Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18: 586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M.J., Drews G.N., Clark S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25: 223–236. [DOI] [PubMed] [Google Scholar]
- Prestridge D.S. (1991). SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput. Appl. Biosci. 7: 203–206. [DOI] [PubMed] [Google Scholar]
- Qin G., et al. (2003). Obtaining and analysis of flanking sequences from T-DNA transformants of Arabidopsis. Plant Sci. 165: 941–949. [Google Scholar]
- Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55: 65–76. [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. [DOI] [PubMed] [Google Scholar]
- Sablowski R.W.M., Meyerowitz E.M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92: 93–103. [DOI] [PubMed] [Google Scholar]
- Sakai H., Honma T., Aoyama T., Sato S., Kato T., Tabata S., Oka A. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521. [DOI] [PubMed] [Google Scholar]
- Schmitz G., Tillmann E., Carriero F., Fiore C., Cellini F., Theres K. (2002). The tomato Blind gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. USA 99: 1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. USA 96: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves T.A., Sussex I.M. (1989). Patterns in Plant Development, 2nd ed. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Stirnberg P., Chatfield S.P., Leyser H.M. (1999). AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 121: 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnberg P., Zhao S., Williamson L., Ward S., Leyser O. (2012). FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 71: 907–920. [DOI] [PubMed] [Google Scholar]
- Talbert P.B., Adler H.T., Parks D.W., Comai L. (1995). The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121: 2723–2735. [DOI] [PubMed] [Google Scholar]
- Tao Q., Guo D., Wei B., Zhang F., Pang C., Jiang H., Zhang J., Wei T., Gu H., Qu L.-J., Qin G. (2013). The TIE1 transcriptional repressor links TCP transcription factors with TOPLESS/TOPLESS-RELATED corepressors and modulates leaf development in Arabidopsis. Plant Cell 25: 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann T., Muhr M. (2015). Shaping plant architecture. Front. Plant Sci. 6: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956. [DOI] [PubMed] [Google Scholar]
- Wang Q., Kohlen W., Rossmann S., Vernoux T., Theres K. (2014). Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26: 2068–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.Y., Zhang L., Xing S., Ma Z., Liu J., Gu H., Qin G., Qu L.-J. (2012). Arabidopsis AtVPS15 plays essential roles in pollen germination possibly by interacting with AtVPS34. J. Genet. Genomics 39: 81–92. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang J., Shi B., Yu T., Qi J., Meyerowitz E.M., Jiao Y. (2014). The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26: 2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei B., Zhang J., Pang C., Yu H., Guo D., Jiang H., Ding M., Chen Z., Tao Q., Gu H., Qu L.-J., Qin G. (2015). The molecular mechanism of sporocyteless/nozzle in controlling Arabidopsis ovule development. Cell Res. 25: 121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Ou B., Li J., Zhao Y., Guo D., Zhu Y., Chen Z., Gu H., Li C., Qin G., Qu L.-J. (2013). Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance. PLoS One 8: e59720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.L., Guo Z.J., Wang H.H., Li J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12: 9–26. [DOI] [PubMed] [Google Scholar]
- Wu L.T., Zhong G.M., Wang J.M., Li X.F., Song X., Yang Y. (2011). Arabidopsis WRKY28 transcription factor is required for resistance to necrotrophic pathogen, Botrytis cinerea. Afr. J. Microbiol. Res. 5: 5481–5488. [Google Scholar]
- Xing D.H., Lai Z.B., Zheng Z.Y., Vinod K.M., Fan B.F., Chen Z.X. (2008). Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 1: 459–470. [DOI] [PubMed] [Google Scholar]
- Yang F., Wang Q., Schmitz G., Müller D., Theres K. (2012). The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 71: 61–70. [DOI] [PubMed] [Google Scholar]
- Yang W.-C., Ye D., Xu J., Sundaresan V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13: 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Ligang C., Liping Z., Diqiu Y. (2010). Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 35: 459–471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.