A novel regulator of transcription initiation in the central cell mediates pollen tube attraction in a non-cell-autonomous manner.
Abstract
In flowering plants, sperm cells are delivered to the embryo sac by a pollen tube guided by female signals. Both the gametic and synergid cells contribute to pollen tube attraction. Synergids secrete peptide signals that lure the tube, while the role of the gametic cells is unknown. Previously, we showed that CENTRAL CELL GUIDANCE (CCG) is essential for pollen tube attraction in Arabidopsis thaliana, but the molecular mechanism is unclear. Here, we identified CCG BINDING PROTEIN1 (CBP1) and demonstrated that it interacts with CCG, Mediator subunits, RNA polymerase II (Pol II), and central cell-specific AGAMOUS-like transcription factors. In addition, CCG interacts with TATA-box Binding Protein 1 and Pol II as a TFIIB-like transcription factor. CBP1-knockdown ovules are defective in pollen tube attraction. Expression profiling revealed that cysteine-rich peptide (CRP) transcripts were downregulated in ccg ovules. CCG and CBP1 coregulate a subset of CRPs in the central cell and the synergids, including the attractant LURE1. CBP1 is extensively expressed in multiple vegetative tissues and specifically in the central cell in reproductive growth. We propose that CBP1, via interaction with CCG and the Mediator complex, connects transcription factors and the Pol II machinery to regulate pollen tube attraction.
INTRODUCTION
Chemotactic guidance is a common mechanism for male-female recognition. Flowering plants have evolved unique multicellular gametophytes and a double fertilization mechanism. The female gametophyte (embryo sac) is composed of seven cells, of which the egg and the central cell are fertilized by two sperm cells from the same male gametophyte (pollen grain) to form the embryo and endosperm of a seed, respectively. The pollen grain consists of two sperm cells and a large vegetative cell, which germinates a tubular structure—the pollen tube—to deliver the immobile sperms to the embryo sac, a phenomenon called siphonogamy. The pollen tube grows to the embryo sac in a polar fashion and is guided by signals from the female, a process called pollen tube guidance, that requires intimate interaction between the embryo sac and the tube. Pollen tube guidance is regulated by the sporophytic tissues when growing in the style and transmitting tract, then by the female gametophyte when approaching the ovule, and finally it enters the embryo sac through the micropylar opening (Palanivelu et al., 2003; Palanivelu and Tsukamoto, 2012). During the gametophytic guidance process, two short-range signals are defined: the funicular and the micropylar signals (Higashiyama et al., 2003). The funicular signal attracts the pollen tube from the surface of the septum to the ovule, and the micropylar signal guides the pollen tube from the funicular surface to the receptive synergid cells at the micropyle of the embryo sac (Shimizu and Okada, 2000; Takeuchi and Higashiyama, 2011).
The embryo sac has been shown to actively regulate the micropylar pollen tube guidance process by laser ablation and genetic studies in Torenia fournieri and Arabidopsis thaliana (Shimizu and Okada, 2000; Higashiyama et al., 2001; Shimizu et al., 2008). Mutation of the synergid-specific transcription factor MYB98 abolishes micropylar guidance (Kasahara et al., 2005). The LURE family is defensin-like polypeptides derived from the synergids and has been shown to be the chemotactic guidance signals for the pollen tube (Okuda et al., 2009; Takeuchi and Higashiyama, 2012). In maize (Zea mays), the small diffusible peptide EGG APPARATUS1 specifically expressed in the egg apparatus functions as the attractant (Márton et al., 2005, 2012). GEX3, an egg-expressed gene, is also required for pollen tube guidance (Alandete-Saez et al., 2008). magatama3 (maa3), a mutant defective in central cell maturation, exhibits pollen tube attraction defect as well (Shimizu et al., 2008). In the central cell guidance (ccg) mutant, embryo sac development is normal, but micropylar pollen tube guidance is abolished (Chen et al., 2007). CCG is expressed specifically in the central cell, and the central cell-specific expression of CCG driven by the FERTILIZATION-INDEPENDENT SEED2 promoter rescues the guidance defect indicating the indispensable role of the central cell in pollen tube guidance. Immunostaining analysis showed that Arabidopsis LURE1 peptides are not detected in myb98, maa3, and ccg ovules (Takeuchi and Higashiyama, 2012), suggesting that there might be intercellular interactions between the central cell and the synergids to coordinate LURE1 production. CCG encodes a nuclear protein with a conserved N-terminal zinc ribbon domain that is typical for the transcription factor TFIIB family (Knutson, 2013). It has been proposed that CCG plays a role in transcription initiation, but the underlying mechanism remains unclear.
Transcription initiation of eukaryotic protein-coding genes is mediated by the basal transcription machinery, which includes RNA polymerase II (Pol II) and general transcription factors (TFs). TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH form a complex known as the preinitiation complex (PIC). A relatively conserved transcription initiation complex is employed from archaea to humans and plants. Transcription initiation requires interaction between Pol II and TFIIB, which was proposed to be a selector for transcription initiation and serves as a target of transcription activator (Li et al., 1994). The assembly of the PIC initiates with the binding of TATA-box Binding Protein (TBP) and TFIIB to the starting site, and then the Pol II and TFIIF are recruited. Here, the zinc ribbon domain of TFIIB mediates its association with TBP, Pol II, and TFIIF (Bushnell et al., 2004; Wang and Roberts, 2010).
Transcription initiation also requires the Mediator complex, which is a conserved central coactivator of transcription. The Mediator interacts with the transcription activator at specific DNA sites and recruits the PIC. It functions as a large conserved complex to transmit the effects of activator on the general transcription machinery and Pol II in yeast and metazoan organisms (Conaway and Conaway, 2011). The underlying mechanism of PIC formation through the Mediator remains poorly understood. In plants, Mediator regulates diverse cell signaling processes (Chen et al., 2012). A range of Mediator subunits have been shown to specifically activate signaling pathways in plant development or the environmental response (Kidd et al., 2011). MED5 is involved in cell wall lignification and the darkness response (Bonawitz et al., 2014; Hemsley et al., 2014). MED14/SWP (STRUWWELPETER), MED15, MED16/SFR6/IEN1, and MED19 regulate the plant immune response (Caillaud et al., 2013; Zhang et al., 2013). MED25/PFT1 modulates organ size (Xu and Li, 2011), light (Klose et al., 2012), hormone (Chen et al., 2012), and flowering signaling (Bäckström et al., 2007). MED12 and MED13 have been shown to regulate pattern formation during embryogenesis (Gillmor et al., 2010). However, no Mediator or related regulators have been found to regulate male-female interaction.
Here, we report the identification and functional analysis of Arabidopsis CCG BINDING PROTEIN1 (CBP1), a CCG binding protein. We demonstrated that CBP1 interacts with CCG, the C-terminal domain (CTD) of the NRPB1 subunit of RNA Pol II (NRPB1_CTD), the Mediator subunits MED7 and MED9, and a series of AGAMOUS-like (AGL) transcription factors. In addition, CCG interacts with TBP1, TFIIF, and NRPB1_CTD, and CBP1 can form a tetramer in vitro. CBP1 is conserved in plants and shows no homology to animal or yeast proteins. Our biochemical data imply that CBP1 is a functional counterpart of the Mediator subunit or a plant-specific regulator of transcription initiation. Expression and genetic analysis showed that CBP1 is expressed in the central cell and plays a role in pollen tube attraction. Furthermore, expression profiling revealed an enrichment of CYSTEINE-RICH PEPTIDEs (CRPs) regulated by CCG. We confirmed the downregulation of several central cell- and/or synergid-expressed CRPs, including LURE1 in ccg and cbp1. Thus, our findings define a transcriptional initiation role of CBP1 in the central cell to regulate the function of synergid cells in pollen tube guidance.
RESULTS
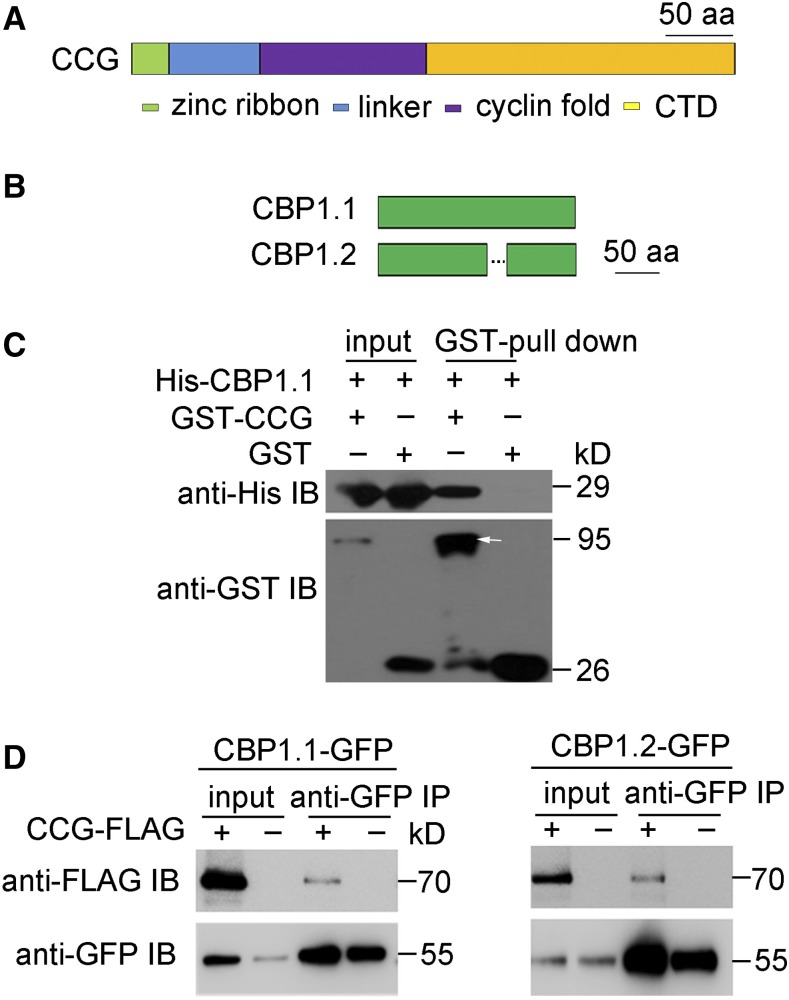
CBP1 Interacts with CCG
To investigate the molecular role of CCG in pollen tube guidance, yeast two-hybrid screening with CCG (Figure 1A) as bait was performed. Several potential candidates, designated CBP, were isolated. CBP1 corresponds to AT2G15890, which encodes two proteins, CBP1.1 and CBP1.2, respectively, via alternative splicing (Supplemental Figures 1A and 1B). Compared with CBP1.1, CBP1.2 contains a 28-amino acid deletion in the middle portion (Figure 1B). Both CBP1.1 and CBP1.2 are expressed in the mature ovules, and CBP1.1 expression is much higher than CBP1.2, suggesting that CBP1.1 is the major splicing form (Supplemental Figure 1C).
Figure 1.
CBP1 Is a CCG Binding Protein.
(A) Protein structure of CCG. aa, amino acids.
(B) Protein structure of CBP1.1 and CBP1.2.
(C) In vitro binding assay with the indicated recombinant proteins showed direct interaction between CBP1 and CCG. Arrow indicates the target protein.
(D) Immunoblotting (IB) of GFP immunoprecipitates from lysates of protoplasts transiently transformed with CBP1-GFP and CCG-Flag.
To further confirm the interaction between CCG and CBP1, pull-down and coimmunoprecipitation (Co-IP) assays were performed. Glutathione S-transferase (GST)-tagged CCG (GST-CCG) and histidine-tagged CBP1 (His-CBP1) proteins expressed in Escherichia coli were mixed and then subjected to a GST pull-down assay. The results showed that GST-CCG indeed pulled down His-CBP1 (Figure 1C). Similarly, CCG-Flag was coexpressed in protoplasts with either CBP1.1-GFP or CBP1.2-GFP. As controls, CBP1.1-GFP or CBP1.2-GFP alone was also transformed into protoplast cells. A Co-IP assay with GFP antibody showed that both CBP1.1-GFP and CBP1.2-GFP were able to precipitate CCG-Flag (Figure 1D). Together, these data indicate that CBP1 interacts with CCG both in vitro and in vivo.
CBP1 is a novel protein without known function; its homologs are found in higher plants and Physcomitrella patens, suggesting that these proteins are evolutionarily conserved across plant species. BLUSTP searches for homologs of CBP1 using the database at the National Center for Biotechnology Information, the Joint Genome Institute, and Pfam database reveal no functional information. Alignment of CBP1 homologs shows two conserved domains designated as the CI and CII domains and two highly variable regions, designated as VI and VII. The deleted region in AtCBP1.2 spans the CI and VII domains (Supplemental Figure 2).
CBP1 Expression Pattern and Subcellular Localization
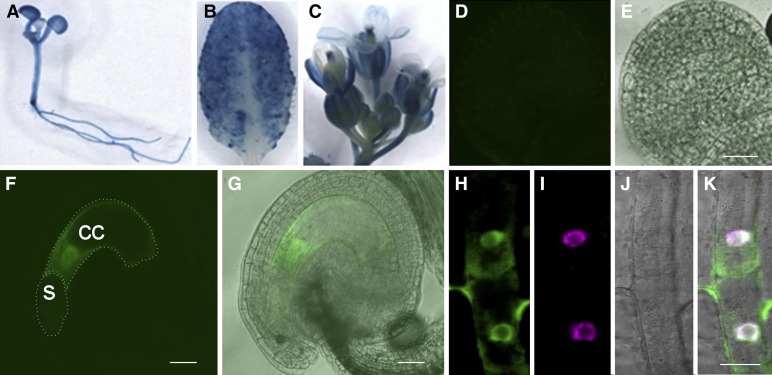
To investigate the tissue-specific expression pattern of CBP1, the β-glucuronidase (GUS) reporter was inserted into the CBP1 gene and introduced into Arabidopsis. In the transgenic plants harboring the CBP1-GUS genomic fusion construct driven by the 2.474-kb native promoter, GUS activity was detected in seedlings, leaves, inflorescences, and flowers (Figures 2A to 2C). To characterize the subcellular localization of CBP1, we replaced the GUS coding sequence with three tandem GFP repeats (3×GFP). In the three independent transgenic lines analyzed, GFP was detected in the mature embryo sac, but not in the immature embryo sac or integuments (Figures 2D to 2G). The GFP signal is predominantly localized in the nucleus of the central cell, with extremely faint GFP signal in the synergids (Supplemental Figures 3A to 3C). These findings indicated that CBP1 is ubiquitously expressed in vegetative tissues and predominately in the central cell of the embryo sac.
Figure 2.
Expression Pattern and Subcellular Localization of CBP1.
(A) to (C) The genomic fusion of CBP1-GUS under the native promoter showed expression in the seedling, leaf, and inflorescence.
(D) to (G) Confocal laser scanning microscopy images of the ovules from ProCBP1:CBP1-3×GFP plants.
(D) and (E) No GFP fluorescence was detected in the immature ovules.
(F) and (G) The fusion protein CBP1-3×GFP is localized in the nucleus and cytoplasm of the central cell. cc, central cell, s, synergids.
(H) to (K) Confocal laser scanning microscopy images showing the nuclear and cytoplasmic localization of CBP1 in the root of Pro35S:GFP-CBP1.1 transgenic plants (H).
(I) Fluorescence of DNA by 4′,6-diamidino-2-phenylindole staining of the nucleus.
(J) Bright-field image.
(K) Merged image.
Bars = 20 µm.
To confirm the expression of CBP1 in the central cell, we monitored the promoter activity by a ccg complementation assay. The ProCBP1:CCG construct was introduced into the ccg/CCG mutant plant. The ovule abortion ratio of three ProCBP1:CCG transgenic lines decreases significantly from 42% (which is the abortion ratio of the ccg/CCG mutant) to 13.4% in the T1 generation, and full seed set was recovered in the T2 selfed ProCBP1:CCG plants (Supplemental Table 1). This indicated that the CBP1 promoter is indeed active in the central cell of the ovule.
To further confirm the subcellular localization, GFP-CBP1 fusion driven by the constitutive cauliflower mosaic virus 35S promoter was introduced into Arabidopsis. The GFP signal was detected in the nucleus and cytoplasm in root cells of the transgenic lines (Figures 2H to 2J). Similar results were observed in the root of the plants expressing CBP1-3×GFP fusion (Supplemental Figures 4A to 4D). These results indicate that both the N-terminal and C-terminal GFP fusions of CBP1 localized in the nucleus and cytoplasm. In addition, CBP1.1-GFP and CBP1.2-GFP show the same subcellular localization in tobacco (Nicotiana tabacum) epidermal cells (Supplemental Figures 4E to 4J).
CCG and CBP1 Interact with the Transcription Initiation Machinery
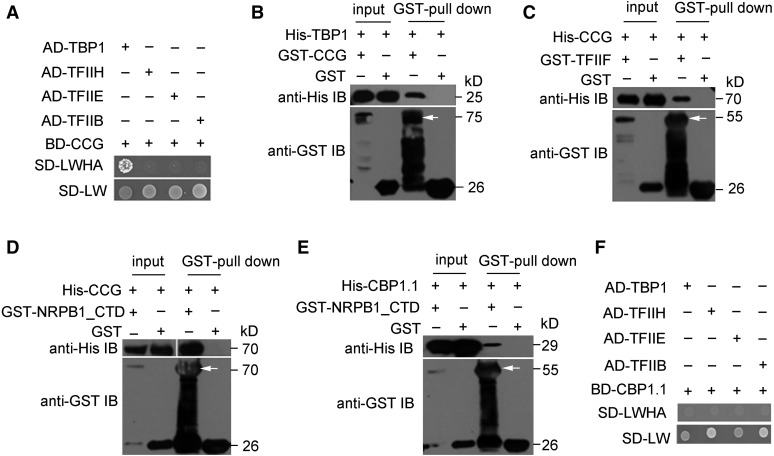
Previously, CCG was shown to interact with the general transcription factor TFIIF in yeast and the N-terminal zinc finger domain is interchangeable with that of TFIIB, indicating a possible role of CCG as a TFIIB protein (Chen et al., 2007). Here, we show that CCG interacts with the general transcription factor TBP1 and the CTD of NRPB1 (AT4G35800), a subunit of RNA polymerase II, in yeast and pull-down assays (Figured 3A to 3D). Furthermore, we verified the interaction between CCG and TFIIF by a pull-down assay. Similarly, CBP1 interacts with NRPB1_CTD, while not with TFIIB, TFIIE, TFIIH, and TBP1 (Figures 3E and 3F). The interaction with TBP1, TFIIF, and Pol II supports the notion that CCG functions as a TFIIB-like transcription factor in transcription initiation.
Figure 3.
Interaction of CCG and CBP1 with Components of the Transcription Preinitiation Complex.
(A) CCG interacted with TBP1 in a yeast two-hybrid assay.
(B) CCG interacted with TBP1 in a GST pull-down assay.
(C) CCG interacted with TFIIF in a GST pull-down assay.
(D) and (E) CCG and CBP1.1 interacted with the CTD of RNA POLYMERASE II LARGE SUBUNIT NRPB1 (NRPB1_CTD) in a GST pull-down assay.
(F) CBP1.1 showed no direct interaction with TFII-B, -E, and -H and TBP1 in a yeast two-hybrid assay.
Arrows indicate the target proteins.
CBP1 Interacts with Mediator and AGAMOUS-Like Transcription Factors
The interactions between CCG, CBP1, and Pol II suggest that CBP1 most likely serves as a coregulator for transcription initiation. It has been well established that the Mediator complex functions as a bridge between transcription activator and the Pol II complex in basal and regulatory transcription from yeast to human (Bäckström et al., 2007). In plants, almost all conserved and several plant-specific Mediator subunits are biochemically purified or found by in silico studies except MED1 (Bäckström et al., 2007; Mathur et al., 2011). MED1 interacts directly with MED7 and MED9 in yeast and human (Kang et al., 2001; Guglielmi et al., 2004; Tsai et al., 2014). These reports led us to explore whether CBP1 could interact with the Mediator complex or if it functions as a MED1 directly. We tested the possible interaction of CBP1 with MED7 and MED9 by the yeast two-hybrid assay, although CBP1 displays almost no homology to the human MED1 (Supplemental Figure 5). We found that CBP1.2 interacts directly with MED9 and two MED7 homologs (AT5G03220 named MED7a and AT5G03500 named MED7b) in yeast (Figure 4A). An in vivo Co-IP assay further confirmed the interactions between CBP1.1 and CBP1.2 with MED7a, MED7b, and MED9, respectively (Figures 4B and 4C).
Figure 4.
CBP1 Interacts with MEDs, AGLs, and Itself.
(A) CBP1.2 interacted with MED7a, MED7b, and MED9 in a yeast two-hybrid assay.
(B) and (C) CBP1.1 and CBP1.2 interacted with Mediator subunits in plants.
(D) and (E) CBP1.1 interacted with itself in a yeast two-hybrid assay (D) and in a GST pull-down assay (E).
(F) CBP1.1 interacted with itself in planta.
(G) Interaction of CBP1 with AGL transcription factors expressed in the central cell detected by a yeast two-hybrid assay.
+, Positive interaction; –, no interaction. S, self-activation of the cells expressing the BD-fused AGLs. Arrow in (E) indicates the fusion protein.
In contrast to the more than 1000 amino acids of MED1 from human and yeast, CBP1 is much shorter, containing 203 amino acids for CBP1.1 and 175 amino acids for CBP1.2. This raises the possibility that CBP1 functions as an oligomer. To check whether CBP1 forms oligomers, we performed yeast two-hybrid, in vitro pull-down, and Co-IP assays. Yeast cells transformed with both AD-CBP1 and BD-CBP1 constructs grew well on the selective growth medium (Figure 4D). His-CBP1 and GST-CBP1 fusion proteins interact in vitro, as shown by the pull-down assay (Figure 4E). Furthermore, the Co-IP assay with transiently cotransformed protoplasts with CBP1-HA and CBP1-Flag constructs shows that CBP1 indeed interacts with itself (Figure 4F). Blue-native gel-based electrophoresis shows that CBP1 forms a tetramer in vitro (Supplemental Figure 6).
AGL transcription activators belong to the plant type I MADS domain subfamily and have been demonstrated to regulate reproductive development (Portereiko et al., 2006; Colombo et al., 2008; Kang et al., 2008; Steffen et al., 2008). A number of AGL transcription factors have been reported to be expressed specifically in the central cell and endosperm (Bemer et al., 2010). We examined whether CBP1 interacts with the 16 AGLs expressed in the central cell by yeast two-hybrid assay and detected eight interactions (Figure 4G). We found that CBP1 interacts with the Mβ-type MADS box proteins AGL49, AGL53, AGL75, AGL81, AGL82, and AGL103 and Mγ-type AGL80, but not with the Mα-type or AGL61, which form a heterodimer with AGL80 (Steffen et al., 2008). These CBP1-interacting AGLs may function as direct transcription factors maintaining the proper function of the central cell in pollen tube attraction.
CBP1 Functions in Micropylar Pollen Tube Attraction
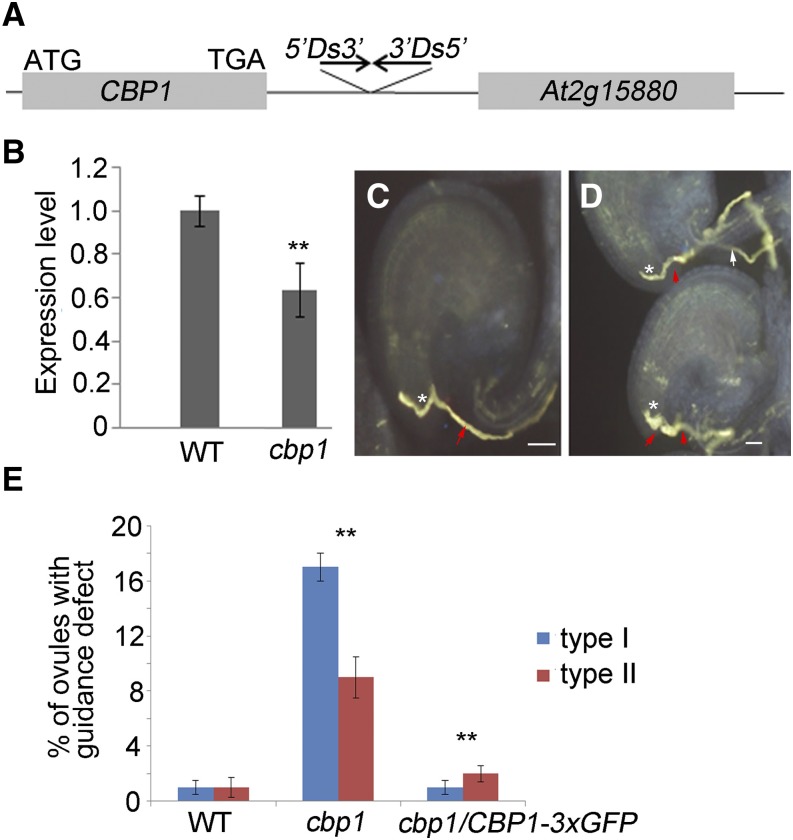
To analyze the function of CBP1 in development and reproductive processes, we obtained a homozygous Ds insertion mutant line CS852557 from the Arabidopsis Biological Resource Center. Thermal asymmetric interlaced-PCR results show that two Ds elements are inserted in reverse direction in the 3′ untranslated region, 578 bp downstream of the stop codon of CBP1 (Figure 5A). Transcript level analysis shows that CBP1 transcript is knocked down to about half of the wild-type level (Figure 5B). cbp1 contains a full seed set and exhibits no apparent growth defect under normal growth conditions. In contrast to the wild-type ovules, which are approached by only one pollen tube and rarely with two (2% ± 0.7%, n = 100), the cbp1 ovules are often approached by two (26%, n = 127). It was observed that when one pollen tube approaches but fails to enter the micropyle, the second enters (17% ± 1%, n = 70, P < 0.01) or two pollen tubes enter the micropyle simultaneously (9% ± 1.5%, n = 57, P < 0.01) (Figures 5C and 5D). The ProCBP1:CBP1-3×GFP-TerCBP1 construct can completely rescue the guidance defect in the T2 generation (Figure 5E; 3% ± 0.3%, n = 136), indicating that the phenotype is caused by the decreased transcription of CBP1.
Figure 5.
The Knockdown Ds Insertion Mutant cbp1 Showing Pollen Tube Attraction Defect and Polytubey.
(A) Schematic diagram of insertion of two reverse Ds elements in the 3′ untranslated region of CBP1.
(B) Real-time quantitative RT-PCR showing knockdown of CBP1 transcript in the cbp1 ovules. Each expression level was normalized to that of eIF1α. The data are the means ± sd of three independent experiments.
(C) The wild-type ovule attracted the wild-type pollen tube.
(D) The cbp1 ovules show two types of attraction defect to the wild-type pollen tubes: type I, one pollen tube failed to enter the micropyle (white arrow), and the second entered the micropyle (red arrow on the top); type II, two pollen tubes entering the micropyle (red arrows at the bottom).
(E) The average ratio (means ± sd) of two types of pollen tube attraction defects of cbp1.
Asterisks in (C) and (D) denote the micropyle; asterisks in (B) and (E) denote statistically significant differences to the wild type (Student’s t test, **P < 0.01). Bars = 20 µm.
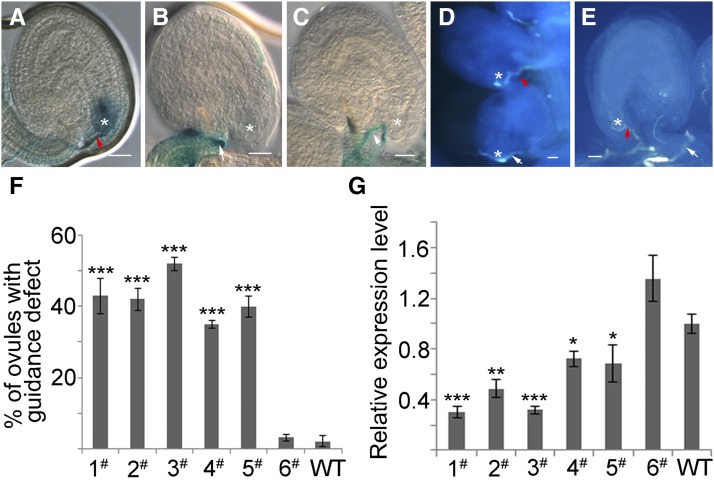
Due to the lack of null mutants of CBP1, an artificial microRNA approach was employed to further verify the function of CBP1. amiRCBP1-a and amiRCBP1-b constructs were introduced into the wild-type plants. Six randomly selected T1 lines were subjected to further analysis. Pistils hand-pollinated by the wild-type pollen or pollen carrying GUS protein driven by the LAT52 promoter were subjected to aniline blue or GUS staining 24 h after pollination. For each line with reduced CBP1 transcript, 35 to 52% ovules showed defective pollen tube attraction (Figure 6). About half the seeds were aborted in lines with lower CBP1 transcript (1# to 3#) due to defects to attract the pollen tubes (Figures 6A to 6D). However, the lines with medium transcript reduction (4# and 5#) exhibited no seed abortion, although they did have defects in pollen tube attraction (Figure 6E). The line with a comparable CBP1 level to the wild type (6#) exhibited no pollen tube attraction defect. Furthermore, no defect in embryo sac development was observed in these six lines (n = 100 for each line; Supplemental Figure 3D). Together, these data suggest that CBP1 regulates pollen tube attraction.
Figure 6.
Knockdown of CBP1 by Artificial MicroRNA Causes Pollen Tube Attraction Defect.
(A) to (E) Wild-type ovule attracted the pollen tube into the micropyle normally (A). About half of the ovules in amiRCBP1 plants were defective in pollen tube attraction ([B] to [E]). Red arrows indicate the normally attracted pollen tubes and white arrows indicate the misguided pollen tubes. Asterisks indicate the micropyle. Bars = 20 µm.
(A) to (C) GUS activity staining of wild-type pollen tubes carrying LAT52:GUS grown for 24 h in the amiRCBP1 pistil.
(D) and (E) Aniline blue staining of wild-type pollen tubes grown for 24 h in the pistil.
(F) The percentage of ovules with pollen tube attraction defect in different transgenic lines (1# to 6#).
(G) The relative expression level of CBP1 in the transgenic lines indicated in (F). 1# to 5# showed significantly severe guidance defect in the T1 generation; 6# with normal transcript level showed no obvious difference to the wild type (n = 230 ovules for each line). Data are presented as means ± sd for three replicates (Student’s t test, ***P < 0.001, **P < 0.01, and *P < 0.05).
CRPs Are Overrepresented in the CCG-Regulated Genes
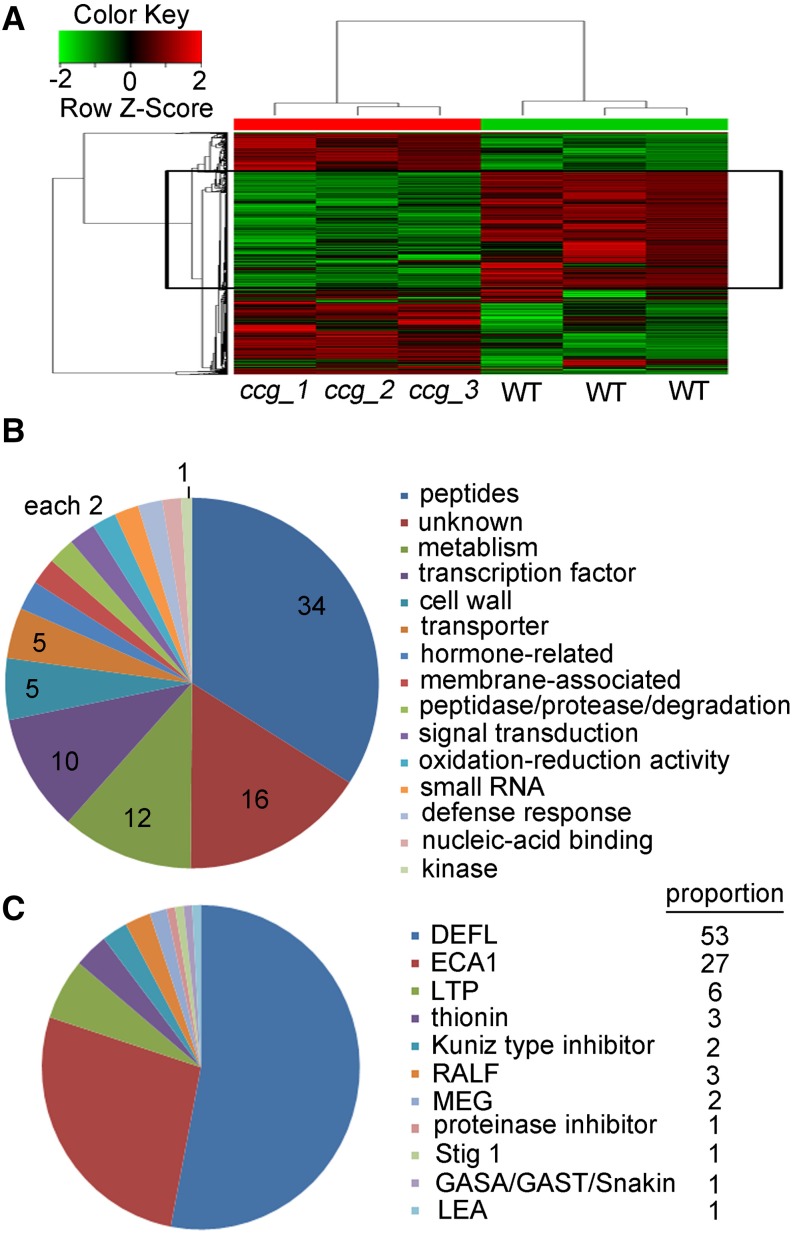
To identify genes regulated by CCG in the embryo sacs, we conducted an RNA profiling assay using the wild-type and ccg ovules. The ccg/CCG mutant and wild-type plant were grown for 12 h after emasculation. The ccg/CCG pistils were hand-pollinated with the wild-type pollen, and the wild-type pistils were left unpollinated. After 1 d, the wild-type unfertilized ovules and smaller unfertilized ccg ovules were picked up manually, using a stereoscope, and subjected to ATH1 microarray analysis. Cluster analysis indicated that the repeatability of these three microarray replications is of good quality (Figure 7A). Compared with the wild-type ovules, 428 genes are downregulated and 1067 genes are upregulated by at least 3-folds in ccg ovules. Considering the unavoidable contamination of mistargeted pollen tubes on the funiculus or integument of ccg ovules, we focused on the downregulated genes for further analysis. Gene Ontology analysis showed that among the 428 downregulated genes, 147 genes (34%) encode secreted proteins with signal peptides among which 115 genes encode CRPs (Figure 7B; Supplemental Data Set 1). Other genes are associated with metabolism (12%), transcription (10%), cell wall (5%), transporter (5%), and 16% with unknown function. The CRPs are divided into 11 classes according to previous classification (Silverstein et al., 2007). Among the downregulated CRP genes, the DEFENSIN-LIKE and ECA1 (EARLY CULTURE ABUNDANT1) genes account for 57 and 23%, respectively (Figure 7C). These findings suggest that CCG likely regulates the transcription of CRP genes in the embryo sac.
Figure 7.
Distribution and Classification of the Genes Downregulated in ccg.
(A) Microarray analysis showing genes that are differentially expressed between ccg and wild-type ovules. Black box: genes primarily downregulated in ccg.
(B) Gene Ontology classification of genes downregulated more than 3-fold in ccg ovules. Numbers on the diagram indicate the proportion of each Gene Ontology term.
(C) Categories of CRPs downregulated more than 3-fold in ccg ovules.
CRPs in the Central Cell Are Downregulated in ccg and cbp1 Ovules
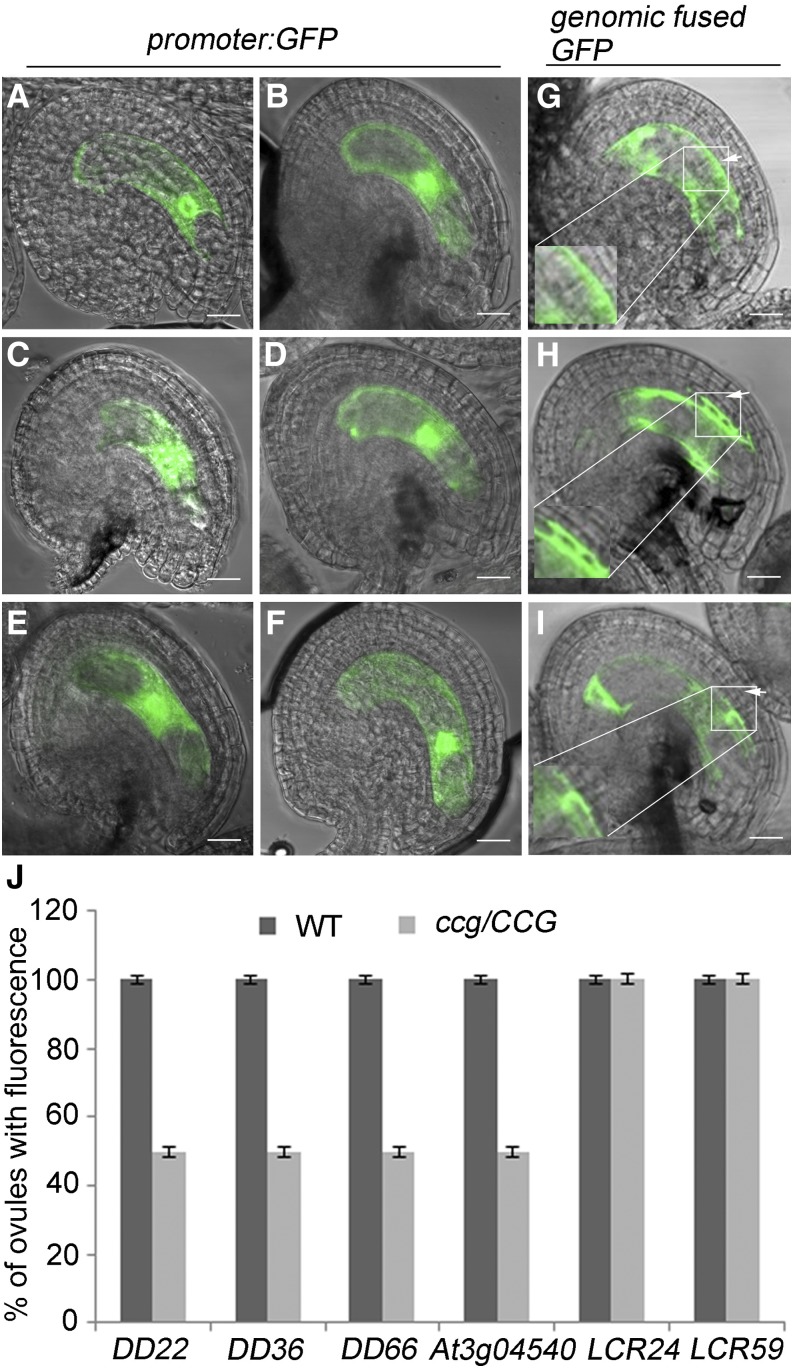
CRPs are small signaling peptides enriched in the embryo sac and play critical roles in pollen tube guidance, fertilization, and early embryogenesis (Jones-Rhoades et al., 2007; Okuda et al., 2009; Sprunck et al., 2012; Takeuchi and Higashiyama, 2012; Costa et al., 2014). Six small cysteine-rich peptides, DOWNREGULATED IN dif1 22 (DD22), DD36, DD66, low molecular weight cysteine-Rich 24 (LCR24), LCR59, and AT3G04540, were reported to be expressed specifically in the central cell (Wuest et al., 2010). In our array data, DD22 and DD66 were downregulated more than 16- and 4-fold, respectively. To test the expression of these genes in ccg ovules, promoter-GFP reporter constructs were introduced into ccg/CCG and wild-type plants. In homozygous transgenic wild-type plants, DD22, DD36, DD66, LCR24, LCR59, and AT3G04540 are indeed specifically expressed in the central cells of the ovules (Figures 8A to 8F). In ccg/CCG plants with homozygous transgenes, only 50% of the ovules express DD22-, DD36-, DD66-, and AT3G04540 promoter-driven GFP (Figures 8J). This suggests that expression of these genes in ccg ovules is impaired. By contrast, the number of ovules expressing LCR24 and LCR59 was ∼100% in the ccg/CCG plant, indicating that both LCR24 and LCR29 are not affected by CCG mutation. In addition, DD36-GFP and At3g04540-GFP fusion proteins were secreted to the surrounding integument layers but not DD22-GFP (Figures 8G to 8I). qRT-PCR analysis also showed that the expression of DD22, DD36, DD66, and AT3G04540 was significantly reduced in ccg ovules, while the expression of LCR24 and LCR59 was not affected (Supplemental Figure 7). Only the expression of DD22 was significantly reduced in the cbp1 pistil, possibly because of the residual CBP1 transcript. Together, these data suggest that the expression of the central cell-expressed CRPs genes is differentially regulated by the CCG-CBP1 complex.
Figure 8.
Central Cell-Expressed CRPs Are Downregulated in ccg Ovules.
(A) to (I) Ovules of the wild-type transgenic plants transformed with free GFP under the native promoter ([A] to [F]) and genomic fusion GFP ([G] to [I]). (A) ProDD22:GFP; (B) ProDD36:GFP; (C) ProDD66:GFP; (D) ProLCR24:GFP; (E) ProLCR59:GFP; (F) ProAt3g04540:GFP; (G) ProDD22:DD22-GFP; (H) ProDD36:DD36-GFP; (I) ProAt3g04540:At3g04540-GFP. Arrows: cell layers of the integument. Bars = 20 µm.
(J) The ratio of ovules expressing CRP genes to the total number (n = 100 for each experiment) of ovules in the wild type and ccg/CCG mutant. The plants analyzed are homozygotes for the transgenes.
CRP Genes in Synergid Cells Are Also Downregulated in ccg and cbp1 Ovules
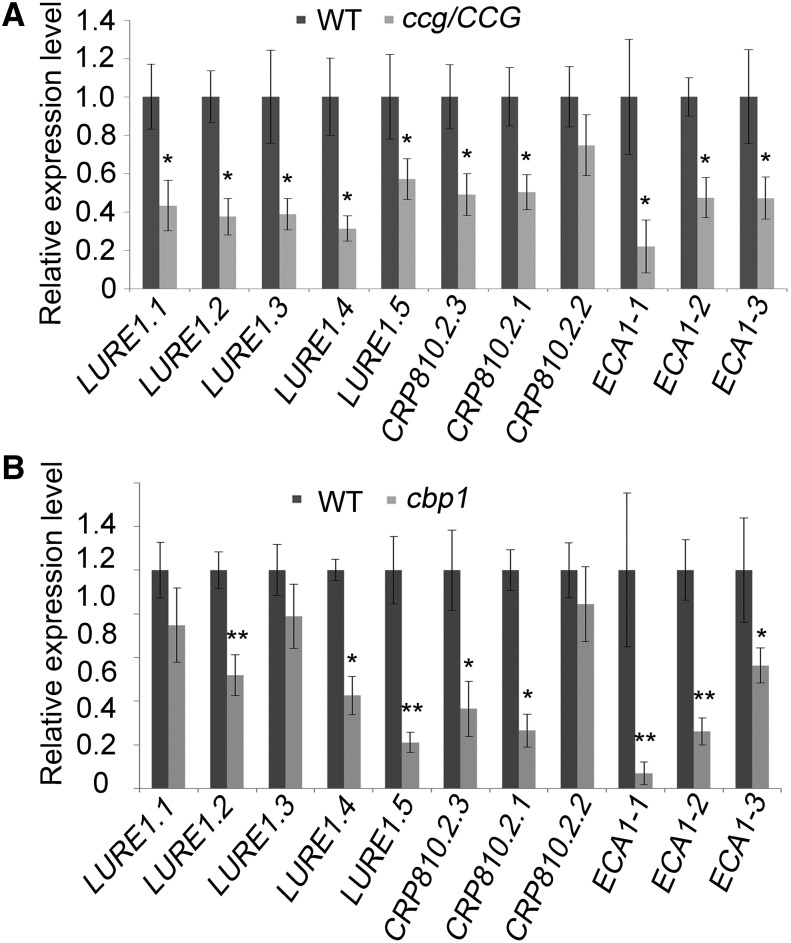
Among the 43 ECA1 genes down-regulated in ccg, four genes (AT5G35405, AT2G21727, AT2G14378, AT5G42895) have been shown to be expressed specifically in the synergids (Jones-Rhoades et al., 2007). This indicates that the central cell-expressed CCG is required for the proper expression of genes in other cells of the embryo sac. MYB98 is a synergid-specific transcription factor that regulates the expression of the female attractant LURE1 (Takeuchi and Higashiyama, 2012). qRT-PCR analysis showed that 22 DD-type genes regulated by MYB98 and MYB98 (DD53) are downregulated in ccg ovules (Supplemental Figure 8). The expression of the five LURE1 genes, three LURE1 homologs (CRP810.2.1, CRP810.2.2, and CRP810.2.3), and three synergid ECA1 genes (ECA1-1, ECA1-2, and ECA1-3) regulated by MYB98 were examined in the ccg and cbp1 mutants. The expression level of these genes is decreased in ccg and cbp1 ovules (Figure 9). The three CRP810.2 genes are expressed in synergid cells and secreted to the filiform apparatus, indicating a possible role in pollen tube attraction (Supplemental Figure 9). The three ECA1 genes belong to the DUF784 family and are regulated by MYB98, but their molecular functions are unknown (Jones-Rhoades et al., 2007). These findings indicate that MYB98-regulated transcription in the synergids is impaired by loss of CCG or CBP1 function in the central cell.
Figure 9.
LURE1 and Synergid Cell-Expressed CRPs Are Downregulated in ccg and cbp1.
(A) LURE1, CRP810.2, and ECA1 genes are downregulated in ccg/CCG. Real-time qRT-PCR was performed with cDNA synthesized from total RNA extracted from ccg ovules.
(B) LURE1, CRP810.2, and ECA1 genes are downregulated in cbp1 ovules. Real-time qRT-PCR was performed with cDNA synthesized from total RNA extracted from pistils from cbp1 homozygotes. Data are presented as means ± sd for three replicates. Student’s t test, **P < 0.01, *P < 0.05.
DISCUSSION
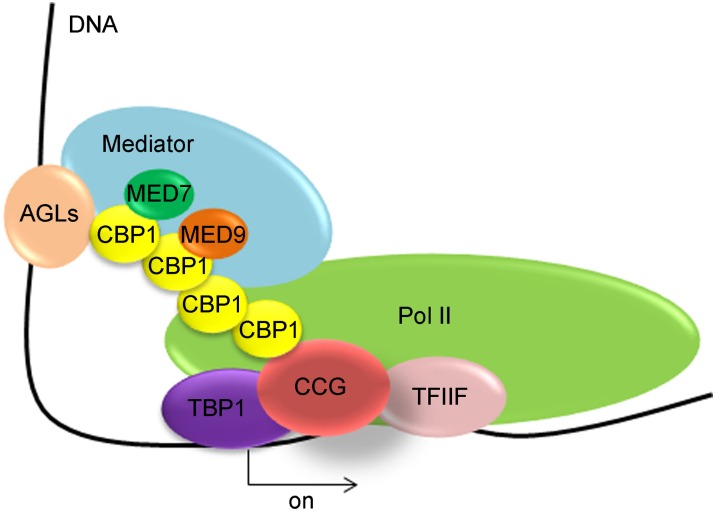
Here, we report the identification and functional analysis of a CCG-interacting protein CBP1 that plays a transcriptional role in pollen tube attraction mediated by the central cell. Our data show that CCG interacts with Pol II, TFIIF, and TBP1, suggesting a TFIIB-like function of CCG. CBP1, on the other hand, interacts with MED7, MED9, Pol II, and several AGL transcription factors. These findings strongly suggest that the CCG-CBP1 complex acts as a transcription regulator within the central cell. Further expression profiling showed that many CRPs, including both central cell- and synergid-expressed, are regulated by CCG. Based on these findings, we propose that CBP1, together with CCG, recruits the Mediator and transcription machinery to AGL transcription factors to modulate the expression of genes that regulate pollen tube guidance within the central cell in flowering plants (Figure 10). The possibility that CBP1 functions in pollen tube guidance directly via the synergid, as implied by its faint expression in the synergid, cannot be excluded.
Figure 10.
Proposed Model for CCG-CBP1 Function in Mediator-Mediated Transcription Initiation in the Central Cell.
CBP1 recruits CCG, Mediator subunits, and Pol II to AGL transcription factors to promote the transcription of target genes.
CBP1 Functions in Transcription Initiation
Mediator functions in activator-dependent and basal transcription in yeast as shown by genetic and biochemical analysis. The modular structure and components of Mediator are conserved from single-cell to multicellular organisms, but the sequence homology of subunits is low among yeast, human, and plants. Due to the lack of sequence similarity, it is hard to determine the evolutionary relationship of CBP1 with MED1 in other organisms or the origin of CBP1 after the divergence of plants. The oligomerization makes CBP1 potentially function as a platform or a bridge for both the Mediator and Pol II transcription machinery and possibly for the transcriptional activator. Oligomerization capacity might confer more flexibility of the mediator complex and is reconciled with the hinge function of the middle module. MED1 in mammals interacts with multiple nuclear receptors in a ligand-dependent fashion (Ito and Roeder, 2001). This interaction triggers recruitment of the MED complex to nuclear receptor-targeted genes. Then, the head and middle of Mediator modules recruit Pol II and general transcription factors to the target DNA to initiate transcription. Similarly, CBP1 interacts with AGL transcription factors. In animals, MED1 is essential for the development of the placenta and embryo, which is similar to the essential function of CBP1 in sexual reproduction (Ito et al., 2000; Landles et al., 2003). The sequence diversification of Mediator subunits may adapt to the highly evolved transcription factors in plants (Bäckström et al., 2007). The nearly total loss of sequence conservation of CBP1 between plants, yeast, and metazoans indicates that its adapted role in plant sexual reproduction was highly specified during evolution. The distinct functions of Mediator subunits are evidenced by their differential roles in development and the stress response (Wang and Chen, 2004; Gillmor et al., 2010). The finding that AGLs interact with CBP1 will be helpful to decipher how the MED complex regulates specific processes and how these signaling pathways are integrated.
The identification of CBP1 as a Mediator subunit or regulator in Arabidopsis is significant in several aspects. First, CBP1 has not been isolated in the Mediator complex by biochemical purification, possibly because of low protein abundance or cell type-specific expression. Second, in silico prediction using yeast or metazoan MED1 failed to detect any sequence similarity in plants. Third, the essential role of CBP1 in reproduction possibly causes a lethal mutation and the subtle phenotype of the available mutant from the public seed stock make the functional characterization more difficult. Expression of CBP1 has been shown to be induced by pathogen stress and repressed by abscisic acid (Leonhardt et al., 2004; Huibers et al., 2009), indicating a possible role in the environmental response. AGL80 regulates the central cell and endosperm development, but not pollen tube attraction, and AGL81 is also expressed in the endosperm (Portereiko et al., 2006; Bemer et al., 2010). The interaction of CBP1 with AGL80 and AGL81 implies that CBP1 may also be involved in endosperm development. MED7 and MED9 have not been functionally studied in Arabidopsis. The extensive expression of these two genes in the carpel and sporophytic tissues implies functionality in multiple signaling processes (Winter et al., 2007). The functional characterization of Mediator components in sexual reproduction will improve our understanding of the transcription regulation mechanism in this highly specific process.
CBP1 may mediate PIC assembly through direct interaction with CCG and Pol II. Early studies showed the direct interaction of TFIIH with MED4, MED11, and MED15, interaction of TBP with MED8, and interaction of TFIID with MED26 in yeast and animals (Sakurai and Fukasawa, 2000; Giot et al., 2003; Esnault et al., 2008). The interaction of mediators with general transcription factors functions in the recruitment of Pol II (Esnault et al., 2008). The direct interaction of the Mediator subunit with general transcription factors has not been found, to our knowledge, in plants, and our finding provides clues into the formation of the PIC.
CCG Functions as a General Transcription Factor
The TFIIB family consists of 14 members in Arabidopsis (Knutson, 2013). In contrast to TFIIB1 and TFIIB2 containing an N-terminal zinc ribbon domain, a linker domain, and two cyclin fold repeats, CCG contains an extended C-terminal domain (Figure 1A). The tfiib1 mutant has been shown to be male sterile and bear lethal homozygotes (Zhou et al., 2013). Another TFIIB-like protein pBRP2 is Brassicacea specific and expressed specifically in the reproductive organs and seeds. Loss of pBRP2 function affects endosperm development (Cavel et al., 2011). The phenotypic differences of ccg, tfiib1, and pbrp2 indicate different DNA target specificity of TFIIB proteins. It has been reported that differential use of TBP and TFIIB renders altered transcriptional specificity (Tansey and Herr, 1997).
Phylogenic analysis classifies CCG as the RNA polymerase I (Pol I) TFIIB-like transcription factor by sequence homology with the yeast and human counterpart Rrn7 and TAF1B (Knutson and Hahn, 2011; Naidu et al., 2011; Knutson, 2013). Pol I transcribes the ribosomal DNA into rRNA precursor, which is essential for cell viability. The specificity of downstream genes and the specific expression pattern of CCG make it unlikely that CCG functions as an obligatory TFIIB-like protein of Pol I. In another scenario, mutants of ribosome processing components YAOZHE and SLOW WALKER1 show slower embryo sac development, but not specifically a guidance defect (Shi et al., 2005; Li et al., 2010). All findings imply that CCG functions in the selective expression of a specific subset of genes mediated by the Pol II machinery.
Intercellular Communication in the Embryo Sac
Intercellular communication mediated by small molecules is a vital mechanism for coordinating plant development and survival (Sager and Lee, 2014; Yadav et al., 2014; Grienenberger and Fletcher, 2015) and for reproduction, including pollen tube attraction and reception, gamete fusion, and embryo sac and embryo development (Dresselhaus and Franklin-Tong, 2013). Our knowledge of the intercellular communication within the embryo sac is extremely limited. In maize, the egg cell-secreted peptide EAL1 restricts the antipodal cell from acquiring central cell fate (Krohn et al., 2012). Mutation of the mitochondrial protein FIONA in the central cell has been shown to extend the antipodal lifespan (Kägi et al., 2010). Similarly, the synergid-specific Arabidopsis LURE1 peptide encoding genes are selectively downregulated in ccg ovules. In addition, the MYB98 expression pattern in the synergid cells is not changed in ccg, as we previously showed (Chen et al., 2007), but the expression level is substantially decreased in the ccg ovules. Arabidopsis LURE1 is under the indirect transcriptional regulation of MYB98 (Punwani et al., 2008). It is possible that the guidance defect of ccg ovules is partially due to the downregulation of MYB98, though the direct role of the central cell cannot be excluded. These results demonstrate that the central cell transcriptionally regulates the function of the synergid. The central cell has been recently suggested to be required for the maturation of the synergid in T. fournieri (Susaki et al., 2015). This gamete-regulated cell fate or function of accessory cells may be a general mechanism in flowering plants to ensure coordination of gamete maturation and pollen tube attraction.
One interesting question is how CCG in the central cell affects the expression of MYB98 in the synergids. Apoplastic peptides, RNAs, hormones, and mobile transcription factors all contribute to the diversity of the intercellular signaling network (Han et al., 2014). Small secreted peptides have been shown to function as intercellular signaling molecules, such as EPIDERMAL PATTERNING FACTOR1 in stomatal patterning, EMBRYO SURROUNDING FACTOR1 in embryo patterning, EGG CELL1 in sperm cell activation, LURE1 in pollen tube attraction, and ES4 in maize pollen tube reception (Amien et al., 2010; Sprunck et al., 2012; Costa et al., 2014; Grienenberger and Fletcher, 2015). Secreted peptides accounted for a large proportion of the downstream genes of CCG. These peptides may activate cell surface receptors to regulate the intracellular signals. To verify this hypothesis, both functional peptides from the central cell and the receptors in the synergid cells must be identified. Microinjection experiments in T. fournieri suggested that small molecules of up to 10 kD undergo symplastic transport from the central cell to the egg cell and synergid (Han et al., 2000). Plasmodesmata have been observed in the embryo sac (Huang and Russell, 1992; Russell, 1992). Mobile transcription factors and microRNAs have been reported to specify cell fate (Nakajima et al., 2001; Muraro et al., 2014) and are candidates for entering the synergids through plasmodesmata from the central cell. Transcription factors and small proteins with unknown function were indeed identified in our profiling of ccg. In addition, the lack of a cell wall between the egg cell, central cell, and synergids exposes the plasma membranes of these cells to each other (Huang and Russell, 1992). This feature might also contribute to the cell-cell communication. Identifying the intercellular signal will provide valuable insight into the mechanism underlying the non-cell-autonomous regulation of pollen tube attraction.
METHODS
Plant Materials and Growth Conditions
ccg mutant seeds were described previously (Chen et al., 2007), and Col-0 and cbp1 (CS852557) seeds were obtained from the ABRC. Arabidopsis thaliana seeds were sterilized for 10 min in 20% bleach, washed five times in sterile water, and germinated on Murashige and Skoog (MS) medium containing 1% sucrose and 0.8% agar. After about a week of growth in the greenhouse (16 h of light/8 h of dark, 22°C), seedlings were transferred to soil and grown under the same conditions.
Yeast Two-Hybrid Screen
Bait was produced by CCG fused with the GAL4 DNA binding domain (BD) from the yeast expression plasmid pGBKT7 (Clontech). Prey was expressed as proteins fused with the GAL4 activation domain from pGADT7 (Clontech). An Arabidopsis cDNA library of inflorescences was screened with the strain AH109 expressing BD-CCG. Full-length coding sequences of TBP1, TFIIB, TFIIE, TFIIH, CBP1.1, MED7a, MED7b, MED9, and all AGLs mentioned in the text were cloned to pGBKT7 or pGADT7 to generate the fusion constructs. All the primers used in this article were listed in Supplemental Data Set 2. Positive interaction was confirmed by the ability of growth on selective medium lacking amino acids Trp, Leu, His, and Ade.
In Vitro Pull-Down Assay
Coding sequences of all genes were cloned to pGEX4T-2 or pET28a to generate the fusion constructs with GST or His tag. The fusion constructs were transformed into Escherichia coli strain BL21. Transformants were grown to a concentration of OD600 = 0.6 in the 37°C shaker and then induced to express the fusion protein by incubation in growth culture supplemented with 0.5 mM isopropyl β-d-1-thiogalactopyranoside for 5 to 6 h at 22°C. The cells were lysed in a buffer containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 1 mM DTT. Equal amounts of supernatant of two interacting proteins were mixed and the supernatant was incubated with glutathione agarose beads (GE) overnight at 4°C. The beads were collected by centrifugation and then washed five times with buffer containing 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM DTT, and 1% Triton X-100. Finally, the proteins bound on the beads were boiled with 1× SDS loading buffer in a 95 to 100°C water bath and then subjected to SDS-PAGE and immunoblot with anti-GST (GE) and anti-His (Santa Cruz) antibody.
Blue-Native PAGE
Two hundred milliliters of E. coli cell culture expressing His-CBP1 (0.5 mM isopropyl β-d-1-thiogalactopyranoside, 16°C, overnight) was centrifuged at 8000 rpm at 4°C for 2 min, and the pellet was resuspended in 8 mL cold lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, and 1× proteinase inhibitor cocktail [Roche], pH 8.0) followed by sonication on ice (3-s bursts with 3-s cooling periods, 15 min in total). The lysate was centrifuged at 10,000g at 4°C for 20 min and the supernatant was transferred into a new 15-mL Eppendorf tube. The supernatant was incubated with the 200 μL prewashed Ni-NTA agarose beads (Qiagen) at 15 rpm, 4°C, overnight. The Ni-NTA agarose beads were collected at 500g, 4°C for 2 min, washed five times with lysis buffer, and then incubated with 1 mL elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, and 1× proteinase inhibitor cocktail, pH 8.0) for 6 h, 4°C, for 15rpm. After centrifugation at 500g, 4°C, for 2 min, the supernatant was transferred into a new 1.5-mL Eppendorf tube. The prepared sample was separated on a 4 to 16% NativePAGE Bis-Tris Gel (BN1002BOX; Novex) according to standard procedures.
Real-Time Quantitative RT-PCR
Total RNA was extracted from different tissues with TRIzol reagent (Invitrogen) and then treated with DNase I (RNase-free DNase kit; Qiagen) to remove any contaminating DNA. SuperScript III Reverse Transcriptase (Invitrogen) was used for reverse transcription reactions. Quantitative PCR was performed with Power SYBR Green PCR Master Mix on the Bio-Rad C1000 Thermal Cycler using eIF1α as the internal control for quantitative normalization. The specificity of all RT-PCR products was examined on 2.5% agarose gels.
GUS and Aniline Blue Staining
GUS activity was assayed histochemically with GUS staining solution (100 mM sodium phosphate, pH 6.8, 2 mM potassium ferrocyanide, 2 mM ferricyanide, 10 mM EDTA, 0.1% Triton X-100, 20% methanol, and 1 mM X-Gluc [Biosynth]) for 8 to 12 h at 37°C. The stained materials were decolored by 70% ethanol at room temperature and observed with a Zeiss Stemi 2000-C microscope. For pollen tube observation, a Zeiss Axioskop2 Plus microscope was used. Aniline blue (0.1% in KPO4, pH 8.0) was used to stain the callose of pollen tubes.
Protein Expression and Localization in Planta
For GUS and GFP reporters, the genomic DNA fragments were amplified and fused to the GUS and eGFP sequence in the pCAMBIA1300 backbone, and the terminator sequence of CBP1 was inserted downstream to generate PCBP1:CBP1-GUS-TerCBP1 and PCBP1:CBP1-3×GFP-TerCBP1. Plasmid pCAMBIA1300-Pro35S:eGFP-CBP1.1-TerNOS was generated by insertion of the CBP1 coding sequence into the pCAMBIA1300-Pro35S:eGFP-TerNOS backbone. All the plasmids were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. The Arabidopsis plants were transformed according to the floral dip method (Clough and Bent, 1998). Transgenic plants were selected on MS agar medium containing 50 mg/L hygromycin. To characterize the expression pattern of CRPs, ovules from five hygromycin-resistant plants were observed with a Zeiss confocal laser scanning microscope (Carl Zeiss Meta 510). For the localization in roots, transgenic seedlings grown vertically on MS medium for 1 week were mounted in water and analyzed on a laser scanning microscope. The microscope settings for the excitation wavelengths were as follows: GFP, 488 nm; 4′,6-diamidino-2-phenylindole, 405 nm; and mCherry, 543 nm. For tobacco (Nicotiana tabacum) leaf transient transformation, GV3101 containing pCAMBIA1300-Pro35S:CBP1.1-GFP-TerNOS and pCAMBIA1300-Pro35S:CBP1.2-GFP-TerNOS was used following standard procedures. For the peptide expression in planta, the following length of promoter upstream of the ATG start codon and terminator downstream of the stop codon, respectively, were used: DD22 (935 and 620 bp), DD36 (1997 and 589 bp), At3g04540 (2476 and 586 bp), CRP810.2.1 (2440 and 720 bp), CRP810.2.2 (1915 and 912 bp), CRP810.2.3 (1787 and 705 bp), DD66 (2428 bp, TerNOS), LCR59 (2385 bp, TerNOS), and LCR24 (2426 bp, TerNOS).
Transient Expression and Co-IP
The full-length coding sequences of genes were cloned to fuse with C-terminal GFP, 3×FLAG, or 3×HA in the pBSKII backbone under the 35S promoter and terminated by the poly(A) terminator to generate the fusion constructs. The transient expression constructs were cotransformed into Arabidopsis protoplasts by the polyethylene glycol method (Yoo et al., 2007). The protoplasts were harvested 12 h after transformation and lysed in lysis buffer (0.05 M HEPES-KOH, pH 7.5, 150 mM KCl, 1 mM EDTA, 0.1% Triton-X 100, and 1 mM DTT with freshly added 1× proteinase inhibitor cocktail). The lysis was centrifuged at 10,000g at 4°C for 10 min, and the supernatant was subjected to Co-IP with anti-GFP agarose beads (ChromoTek) for 3 h with 360° shaking at 4°C. The beads were washed with lysis buffer 6 times, diluted in 1× loading buffer, and boiled for 5 min before SDS-PAGE. The following immunoblot reactions were performed according to standard procedures with anti-Flag (Sigma-Aldrich), anti-Myc (Sigma-Aldrich), anti-HA (Santa Cruz), and anti-GFP-HRP (Miltenyi Biotec) antibody.
Artificial MicroRNA Construction
Primers were designed using the online tool http://wmd3.weigelworld.org/cgi-bin/webapp.cgi. CBP1a-1 (GATGTACTATAATAGGCACGCAGTCTCTCTTTTGTATTCC) pairing to CBP1a-2 (GACTGCGTGCCTATTATAGTACATCAAAGAGAATCAATGA), CBP1a-3 (GACTACGTGCCTATTTTAGTACTTCACAGGTCGTGATATG) pairing to CBP1a-4 (GAAGTACTAAAATAGGCACGTAGTCTACATATATATTCCT), CBP1b-1 (GATTAGTACTATAATAGGCACACTCTCTCTTTTGTATTCC) pairing to CBP1b-2 (GAGTGTGCCTATTATAGTACTAATCAAAGAGAATCAATGA), and CBP1b-3 (GAGTATGCCTATTATTGTACTATTCACAGGTCGTGATATG) pairing to CBP1b-4 (GAATAGTACAATAATAGGCATACTCTACATATATATTCCT) were used to amplify the target sequence and construct the hairpin structure. The fragments were finally cloned into the pCAMBIA1300-ProCBP1-TerNOS backbone and transformed into the wild-type plants.
Accession Numbers
Arabidopsis sequence data from this article can be found in the Arabidopsis Genome Initiative database or the GenBank/EMBL data library under the following accession numbers: CCG (At2g02955), AtCBP1 (AT2G15890), DD22 (At5g38330), DD36 (At3g24510), DD66 (At1g60985), LCR24 (At4g29285), LCR59 (At4g30070), LURE1.1 (At5g43285), LURE1.2 (At5g43510), LURE1.3 (At5g43513), LURE1.4 (At5g43518), LURE1.5 (At5g43525), CRP810.2.1 (AT5G48515), CRP810.2.2 (AT5G48595), CRP810.2.3 (AT5G48605), ECA1-1 (AT1G57775), ECA1-2 (AT3G30383), ECA1-3 (AT5G42567), TFIIE (AT1G03280), TFIIB1 (AT2G41630), TFIIF (AT1G75510), TFIIH (AT1G05055), TBP1 (AT3G13445), NRPB1 (AT4G35800), MED7a (AT5G03220), MED7b (AT5G03500), MED9 (AT1G55080), Al-CBP1 (XP_002885987.1), Bn-CBP1 (CDX89682.1), Vv-CBP1 (XP_002284884.1), Gm-CBP1 (NP_001237958.1), Mt-CBP1 (XP_003608052.1), Zm-CBP1 (NP_001158997.1), Sb-CBP1 (XP_002463831.1), Oj-CBP1 (NP_001173652.1), and Pp-CBP1 (XP_001770736.1).
Supplemental Data
Supplemental Figure 1. Two splicing forms of CBP1.
Supplemental Figure 2. Sequence alignment of CBP1 homologs from different species.
Supplemental Figure 3. CBP1-3×GFP fluorescence is predominantly localized in the central cell.
Supplemental Figure 4. CBP1 localization in the nucleus and cytoplasm.
Supplemental Figure 5. Protein alignment of Homo sapiens MED1 and AtCBP1.
Supplemental Figure 6. CBP1 forms a tetramer in vitro.
Supplemental Figure 7. Central cell-expressed CRPs are downregulated in ccg and cbp1.
Supplemental Figure 8. MYB98 and the DD-type genes downstream of MYB98 are downregulated in ccg ovules.
Supplemental Figure 9. CRP810.2 genes are expressed in the synergid cells.
Supplemental Table 1. Genetic complementation data of ccg by expression of CCG under the CBP1 promoter.
Supplemental Data Set 1. The genes encoding secreted peptides down-regulated in ccg ovules.
Supplemental Data Set 2. Primers used in this article.
Supplementary Material
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2013CB945103) and the National Natural Science Foundation of China (30830063).
AUTHOR CONTRIBUTIONS
W.-C.Y., H.-J.L., and S.-S.Z. conceived and designed the experiments. S.-S.Z., M.-X.Z., and H.-J.L. performed the experiments. T.W., L.L., Y.X., D.-Q.W., and J.L. provided technical assistance. H.-J.L., W.-C.Y., and S.-S.Z. analyzed the data and wrote the article.
Glossary
- PIC
preinitiation complex
- Co-IP
coimmunoprecipitation
- MS
Murashige and Skoog
- CTD
C-terminal domain
Footnotes
Articles can be viewed online without a subscription.
References
- Alandete-Saez M., Ron M., McCormick S. (2008). GEX3, expressed in the male gametophyte and in the egg cell of Arabidopsis thaliana, is essential for micropylar pollen tube guidance and plays a role during early embryogenesis. Mol. Plant 1: 586–598. [DOI] [PubMed] [Google Scholar]
- Amien S., Kliwer I., Márton M.L., Debener T., Geiger D., Becker D., Dresselhaus T. (2010). Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel KZM1. PLoS Biol. 8: e1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007). Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26: 717–729. [DOI] [PubMed] [Google Scholar]
- Bemer M., Heijmans K., Airoldi C., Davies B., Angenent G.C. (2010). An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol. 154: 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N.D., Kim J.I., Tobimatsu Y., Ciesielski P.N., Anderson N.A., Ximenes E., Maeda J., Ralph J., Donohoe B.S., Ladisch M., Chapple C. (2014). Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509: 376–380. [DOI] [PubMed] [Google Scholar]
- Bushnell D.A., Westover K.D., Davis R.E., Kornberg R.D. (2004). Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science 303: 983–988. [DOI] [PubMed] [Google Scholar]
- Caillaud M.C., Asai S., Rallapalli G., Piquerez S., Fabro G., Jones J.D. (2013). A downy mildew effector attenuates salicylic acid-triggered immunity in Arabidopsis by interacting with the host mediator complex. PLoS Biol. 11: e1001732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavel E., Pillot M., Pontier D., Lahmy S., Bies-Etheve N., Vega D., Grimanelli D., Lagrange T. (2011). A plant-specific transcription factor IIB-related protein, pBRP2, is involved in endosperm growth control. PLoS One 6: e17216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., Li C. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.H., Li H.J., Shi D.Q., Yuan L., Liu J., Sreenivasan R., Baskar R., Grossniklaus U., Yang W.C. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell 19: 3563–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colombo M., Masiero S., Vanzulli S., Lardelli P., Kater M.M., Colombo L. (2008). AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J. 54: 1037–1048. [DOI] [PubMed] [Google Scholar]
- Conaway R.C., Conaway J.W. (2011). Origins and activity of the Mediator complex. Semin. Cell Dev. Biol. 22: 729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L.M., et al. (2014). Central cell-derived peptides regulate early embryo patterning in flowering plants. Science 344: 168–172. [DOI] [PubMed] [Google Scholar]
- Dresselhaus T., Franklin-Tong N. (2013). Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6: 1018–1036. [DOI] [PubMed] [Google Scholar]
- Esnault C., Ghavi-Helm Y., Brun S., Soutourina J., Van Berkum N., Boschiero C., Holstege F., Werner M. (2008). Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31: 337–346. [DOI] [PubMed] [Google Scholar]
- Gillmor C.S., Park M.Y., Smith M.R., Pepitone R., Kerstetter R.A., Poethig R.S. (2010). The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137: 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giot L., et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736. [DOI] [PubMed] [Google Scholar]
- Grienenberger E., Fletcher J.C. (2015). Polypeptide signaling molecules in plant development. Curr. Opin. Plant Biol. 23: 8–14. [DOI] [PubMed] [Google Scholar]
- Guglielmi B., van Berkum N.L., Klapholz B., Bijma T., Boube M., Boschiero C., Bourbon H.M., Holstege F.C., Werner M. (2004). A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32: 5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Kumar D., Chen H., Wu S., Kim J.Y. (2014). Transcription factor-mediated cell-to-cell signalling in plants. J. Exp. Bot. 65: 1737–1749. [DOI] [PubMed] [Google Scholar]
- Han Y.-Z., Huang B.-Q., Zee S.-Y., Yuan M. (2000). Symplastic communication between the central cell and the egg apparatus cells in the embryo sac of Torenia fournieri Lind. before and during fertilization. Planta 211: 158–162. [DOI] [PubMed] [Google Scholar]
- Hemsley P.A., Hurst C.H., Kaliyadasa E., Lamb R., Knight M.R., De Cothi E.A., Steele J.F., Knight H. (2014). The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26: 465–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama T., Kuroiwa H., Kuroiwa T. (2003). Pollen-tube guidance: beacons from the female gametophyte. Curr. Opin. Plant Biol. 6: 36–41. [DOI] [PubMed] [Google Scholar]
- Higashiyama T., Yabe S., Sasaki N., Nishimura Y., Miyagishima S, Kuroiwa H., Kuroiwa T. (2001). Pollen tube attraction by the synergid cell. Science 293: 1480–1483. [DOI] [PubMed] [Google Scholar]
- Huang B.-Q., Russell S.D. (1992). Female germ unit: organization, isolation, and function. Int. Rev. Cytol. 140: 233–293. [Google Scholar]
- Huibers R.P., de Jong M., Dekter R.W., Van den Ackerveken G. (2009). Disease-specific expression of host genes during downy mildew infection of Arabidopsis. Mol. Plant Microbe Interact. 22: 1104–1115. [DOI] [PubMed] [Google Scholar]
- Ito M., Roeder R.G. (2001). The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol. Metab. 12: 127–134. [DOI] [PubMed] [Google Scholar]
- Ito M., Yuan C.X., Okano H.J., Darnell R.B., Roeder R.G. (2000). Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5: 683–693. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades M.W., Borevitz J.O., Preuss D. (2007). Genome-wide expression profiling of the Arabidopsis female gametophyte identifies families of small, secreted proteins. PLoS Genet. 3: 1848–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kägi C., Baumann N., Nielsen N., Stierhof Y.D., Gross-Hardt R. (2010). The gametic central cell of Arabidopsis determines the lifespan of adjacent accessory cells. Proc. Natl. Acad. Sci. USA 107: 22350–22355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang I.H., Steffen J.G., Portereiko M.F., Lloyd A., Drews G.N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.S., Kim S.H., Hwang M.S., Han S.J., Lee Y.C., Kim Y.J. (2001). The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276: 42003–42010. [DOI] [PubMed] [Google Scholar]
- Kasahara R.D., Portereiko M.F., Sandaklie-Nikolova L., Rabiger D.S., Drews G.N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17: 2981–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B.N., Cahill D.M., Manners J.M., Schenk P.M., Kazan K. (2011). Diverse roles of the Mediator complex in plants. Semin. Cell Dev. Biol. 22: 741–748. [DOI] [PubMed] [Google Scholar]
- Klose C., Büche C., Fernandez A.P., Schäfer E., Zwick E., Kretsch T. (2012). The mediator complex subunit PFT1 interferes with COP1 and HY5 in the regulation of Arabidopsis light signaling. Plant Physiol. 160: 289–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson B.A. (2013). Emergence and expansion of TFIIB-like factors in the plant kingdom. Gene 526: 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson B.A., Hahn S. (2011). Yeast Rrn7 and human TAF1B are TFIIB-related RNA polymerase I general transcription factors. Science 333: 1637–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn N.G., Lausser A., Juranić M., Dresselhaus T. (2012). Egg cell signaling by the secreted peptide ZmEAL1 controls antipodal cell fate. Dev. Cell 23: 219–225. [DOI] [PubMed] [Google Scholar]
- Landles C., Chalk S., Steel J.H., Rosewell I., Spencer-Dene B., Lalani N., Parker M.G. (2003). The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Mol. Endocrinol. 17: 2418–2435. [DOI] [PubMed] [Google Scholar]
- Leonhardt N., Kwak J.M., Robert N., Waner D., Leonhardt G., Schroeder J.I. (2004). Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.J., Liu N.Y., Shi D.Q., Liu J., Yang W.C. (2010). YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol. 10: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Flanagan P.M., Tschochner H., Kornberg R.D. (1994). RNA polymerase II initiation factor interactions and transcription start site selection. Science 263: 805–807. [DOI] [PubMed] [Google Scholar]
- Márton M.L., Cordts S., Broadhvest J., Dresselhaus T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576. [DOI] [PubMed] [Google Scholar]
- Márton M.L., Fastner A., Uebler S., Dresselhaus T. (2012). Overcoming hybridization barriers by the secretion of the maize pollen tube attractant ZmEA1 from Arabidopsis ovules. Curr. Biol. 22: 1194–1198. [DOI] [PubMed] [Google Scholar]
- Mathur S., Vyas S., Kapoor S., Tyagi A.K. (2011). The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 157: 1609–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraro D., et al. (2014). Integration of hormonal signaling networks and mobile microRNAs is required for vascular patterning in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 111: 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidu S., Friedrich J.K., Russell J., Zomerdijk J.C. (2011). TAF1B is a TFIIB-like component of the basal transcription machinery for RNA polymerase I. Science 333: 1640–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Sena G., Nawy T., Benfey P.N. (2001). Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311. [DOI] [PubMed] [Google Scholar]
- Okuda S., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458: 357–361. [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Tsukamoto T. (2012). Pathfinding in angiosperm reproduction: pollen tube guidance by pistils ensures successful double fertilization. Wiley Interdiscip. Rev. Dev. Biol. 1: 96–113. [DOI] [PubMed] [Google Scholar]
- Palanivelu R., Brass L., Edlund A.F., Preuss D. (2003). Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59. [DOI] [PubMed] [Google Scholar]
- Portereiko M.F., Lloyd A., Steffen J.G., Punwani J.A., Otsuga D., Drews G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punwani J.A., Rabiger D.S., Lloyd A., Drews G.N. (2008). The MYB98 subcircuit of the synergid gene regulatory network includes genes directly and indirectly regulated by MYB98. Plant J. 55: 406–414. [DOI] [PubMed] [Google Scholar]
- Russell S.D. (1992). Double fertilization. Int. Rev. Cytol. 140: 357–388. [Google Scholar]
- Sager R., Lee J.Y. (2014). Plasmodesmata in integrated cell signalling: insights from development and environmental signals and stresses. J. Exp. Bot. 65: 6337–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai H., Fukasawa T. (2000). Functional connections between mediator components and general transcription factors of Saccharomyces cerevisiae. J. Biol. Chem. 275: 37251–37256. [DOI] [PubMed] [Google Scholar]
- Shi D.Q., Liu J., Xiang Y.H., Ye D., Sundaresan V., Yang W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17: 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K.K., Okada K. (2000). Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127: 4511–4518. [DOI] [PubMed] [Google Scholar]
- Shimizu K.K., Ito T., Ishiguro S., Okada K. (2008). MAA3 (MAGATAMA3) helicase gene is required for female gametophyte development and pollen tube guidance in Arabidopsis thaliana. Plant Cell Physiol. 49: 1478–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein K.A., Moskal W.A. Jr., Wu H.C., Underwood B.A., Graham M.A., Town C.D., VandenBosch K.A. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51: 262–280. [DOI] [PubMed] [Google Scholar]
- Sprunck S., Rademacher S., Vogler F., Gheyselinck J., Grossniklaus U., Dresselhaus T. (2012). Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 338: 1093–1097. [DOI] [PubMed] [Google Scholar]
- Steffen J.G., Kang I.H., Portereiko M.F., Lloyd A., Drews G.N. (2008). AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol. 148: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki D., Takeuchi H., Tsutsui H., Kurihara D., Higashiyama T. (2015). Live imaging and laser disruption reveal the dynamics and cell-cell communication during Torenia fournieri female gametophyte development. Plant Cell Physiol. 56: 1031–1041. [DOI] [PubMed] [Google Scholar]
- Takeuchi H., Higashiyama T. (2011). Attraction of tip-growing pollen tubes by the female gametophyte. Curr. Opin. Plant Biol. 14: 614–621. [DOI] [PubMed] [Google Scholar]
- Takeuchi H., Higashiyama T. (2012). A species-specific cluster of defensin-like genes encodes diffusible pollen tube attractants in Arabidopsis. PLoS Biol. 10: e1001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey W.P., Herr W. (1997). Selective use of TBP and TFIIB revealed by a TATA-TBP-TFIIB array with altered specificity. Science 275: 829–831. [DOI] [PubMed] [Google Scholar]
- Tsai K.L., Tomomori-Sato C., Sato S., Conaway R.C., Conaway J.W., Asturias F.J. (2014). Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157: 1430–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Chen X. (2004). HUA ENHANCER3 reveals a role for a cyclin-dependent protein kinase in the specification of floral organ identity in Arabidopsis. Development 131: 3147–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Roberts S.G.E. (2010). New insights into the role of TFIIB in transcription initiation. Transcription 1: 126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuest S.E., Vijverberg K., Schmidt A., Weiss M., Gheyselinck J., Lohr M., Wellmer F., Rahnenführer J., von Mering C., Grossniklaus U. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20: 506–512. [DOI] [PubMed] [Google Scholar]
- Xu R., Li Y. (2011). Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138: 4545–4554. [DOI] [PubMed] [Google Scholar]
- Yadav S.R., Yan D., Sevilem I., Helariutta Y. (2014). Plasmodesmata-mediated intercellular signaling during plant growth and development. Front. Plant Sci. 5: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yao J., Zhang Y., Sun Y., Mou Z. (2013). The Arabidopsis Mediator complex subunits MED14/SWP and MED16/SFR6/IEN1 differentially regulate defense gene expression in plant immune responses. Plant J. 75: 484–497. [DOI] [PubMed] [Google Scholar]
- Zhou J.J., Liang Y., Niu Q.K., Chen L.Q., Zhang X.Q., Ye D. (2013). The Arabidopsis general transcription factor TFIIB1 (AtTFIIB1) is required for pollen tube growth and endosperm development. J. Exp. Bot. 64: 2205–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.