Tocopherol synthesis and growth of Arabidopsis require the activity of the phytyl-phosphate kinase VTE6 in the phosphorylation cascade of the phytol salvage pathway.
Abstract
Phytol from chlorophyll degradation can be phosphorylated to phytyl-phosphate and phytyl-diphosphate, the substrate for tocopherol (vitamin E) synthesis. A candidate for the phytyl-phosphate kinase from Arabidopsis thaliana (At1g78620) was identified via a phylogeny-based approach. This gene was designated VITAMIN E DEFICIENT6 (VTE6) because the leaves of the Arabidopsis vte6 mutants are tocopherol deficient. The vte6 mutant plants are incapable of photoautotrophic growth. Phytol and phytyl-phosphate accumulate, and the phytyl-diphosphate content is strongly decreased in vte6 leaves. Phytol feeding and enzyme assays with Arabidopsis and recombinant Escherichia coli cells demonstrated that VTE6 has phytyl-P kinase activity. Overexpression of VTE6 resulted in increased phytyl-diphosphate and tocopherol contents in seeds, indicating that VTE6 encodes phytyl-phosphate kinase. The severe growth retardation of vte6 mutants was partially rescued by introducing the phytol kinase mutation vte5. Double mutant plants (vte5 vte6) are tocopherol deficient and contain more chlorophyll, but reduced amounts of phytol and phytyl-phosphate compared with vte6 mutants, suggesting that phytol or phytyl-phosphate are detrimental to plant growth. Therefore, VTE6 represents the missing phytyl-phosphate kinase, linking phytol release from chlorophyll with tocopherol synthesis. Moreover, tocopherol synthesis in leaves depends on phytol derived from chlorophyll, not on de novo synthesis of phytyl-diphosphate from geranylgeranyl-diphosphate.
INTRODUCTION
Tocochromanols (vitamin E), a group of prenyl quinol lipids accumulating in cyanobacteria and chloroplasts of plants and green algae, are involved in the protection against oxidative stress and in adaptation to low-temperature conditions (Sattler et al., 2004; Havaux et al., 2005; Maeda et al., 2006). Depending on the side chain, three classes of tocochromanols can be distinguished, with tocopherols carrying a phytyl chain, tocotrienols harboring a geranylgeranyl chain, and plastochromanol-8 (PC-8) with a solanesyl chain. The biosynthesis of tocopherols includes the condensation of homogentisate with phytyl-diphosphate (phytyl-PP) catalyzed by homogentisate phytyl transferase (VTE2) (Collakova and DellaPenna, 2001; Savidge et al., 2002). In some plants, in particular monocotyledons, geranylgeranyl-diphosphate (GG-PP) is used for condensation with homogentisate by an alternative enzyme, homogentisate geranylgeranyl transferase, leading to the synthesis of tocotrienols (Cahoon et al., 2003; Yang et al., 2011). Subsequent methylation reactions and closure of the second ring by tocopherol cyclase (VTE1) result in the production of the four forms of tocopherol (α, β, γ, and δ) differing by the numbers and positions of the methyl groups on the chromanol ring (Dörmann, 2007; Hunter and Cahoon, 2007; Maeda and DellaPenna, 2007). PC-8 is produced from plastoquinol-9 via cyclization by VTE1 (Kleinig and Liedvogel, 1978; Mène-Saffrané et al., 2010; Zbierzak et al., 2010).
While the biosynthesis of the tocopherol head group has been studied in detail, less is known about the origin of the phytol moiety for tocopherol synthesis. Phytyl-PP is believed to largely originate from isoprenoid de novo synthesis via reduction of three double bonds in GG-PP. The corresponding enzyme, geranylgeranyl reductase (GGR), from plants has been characterized (Soll et al., 1983; Keller et al., 1998). Later, an alternative pathway for phytyl-PP production via phosphorylation of free phytol to phytyl-monophosphate (phytyl-P) and phytyl-PP was described (Ischebeck et al., 2006; Valentin et al., 2006). Free phytol can be derived from chlorophyll turnover and breakdown, in particular during senescence or chlorotic stress. Chlorophyll degradation starts with the removal of the magnesium cation, yielding pheophytin (Schelbert et al., 2009). Subsequently, phytol is cleaved from pheophytin by pheophytin pheophorbide hydrolase (PPH). Interestingly, GGR from Arabidopsis thaliana is capable of reducing the geranylgeranyl moiety in both GG-PP and the geranylgeranylated form of chlorophyll (Keller et al., 1998). Chlorophyll synthase can utilize phytyl-PP or GG-PP for prenylation of chlorophyllide (Soll et al., 1983). As the pool sizes and turnover rates of the isoprenyl-diphosphates in plants are unknown, the exact routes for chlorophyll and tocopherol synthesis are unclear. Increasing attention has been paid to the link of chlorophyll degradation and tocopherol accumulation. Overexpression of PPH in seeds resulted in a modest increase in tocopherol (Zhang et al., 2014). These results suggested that manipulation of expression of genes involved in chlorophyll breakdown reveals minor effects on seed tocopherol levels and that a PPH-independent pathway for chlorophyll dephytylation might exist in seeds.
The first enzyme of phytol phosphorylation, phytol kinase, was isolated from Arabidopsis (VTE5, At5g04490) and Synechocystis (slr1652) (Valentin et al., 2006). The Arabidopsis vte5 mutant contains 65 and 85% of tocopherol in leaves and seeds, respectively, compared with the wild type. However, it remained unclear to what extent phytol phosphorylation or the GG-PP-based de novo pathway contributes to overall phytyl-PP synthesis. Furthermore, the identity of the second enzyme of the phosphorylation pathway, phytyl-P kinase, remained unknown.
In prokaryotes, the genes of a common biosynthetic pathway are often localized in close proximity in the genome, e.g., in operons. Therefore, the sequences of enzymes of one pathway in plants can be identified by analyzing the prokaryotic genome structure, provided that the pathway of interest is present in both plants and bacteria. The phytol kinase is predicted to be present in all plants, algae, and bacteria containing chlorophyll and carrying out oxygenic photosynthesis (Ischebeck et al., 2006; Valentin et al., 2006). In agreement with this prediction, genes encoding phytol kinase were identified in Arabidopsis and Synechocystis, and related sequences were found in many other plants and photosynthetic bacteria (Valentin et al., 2006). The SEED database, harboring sequences of a large number of bacterial and plant genomes, provides the means to search for genes in subsystems (groups of genes with a related function or participating in a common pathway) (Overbeek et al., 2005; Seaver et al., 2014). Analysis of the subsystem encompassing functions involved in the metabolism of long-chain isoprenoids, including phytol kinase VTE5, undecaprenyl pyrophosphate synthetase (UppS), GGR, and chlorophyll synthase, revealed the presence of genes encoding a protein of unknown function (COG1836, pfam01940) that localize close to other isoprenoid genes in many bacterial genomes. Among Archaea, COG1836 sequences cluster with different genes of isoprenoid synthesis. In some cyanobacteria (Anabaena and Nostoc), orthologs of COG1836 are located in the genome close to phytol kinase, and in certain bacteria (Pelodyction, Symbiobacterium, and Thermoplasma) they are C-terminally fused to an ortholog of phytol kinase. Sequence orthologs of COG1836 are also present in Synechocystis (sll0875) and in Arabidopsis (At1g78620) (Seaver et al., 2014).
Here, we describe the characterization of the Arabidopsis At1g78620 gene and the corresponding insertional mutant plants. Biochemical and physiological analyses reveal that At1g78620 encodes phytyl-P kinase and that the phytol phosphorylation pathway represents the most relevant route of phytyl-PP synthesis for tocopherol production in leaves. Furthermore, we show that the phytol phosphorylation pathway is essential for plant growth and development.
RESULTS
Tocopherol Accumulation in Leaves Depends on Phytol Released from Chlorophyll
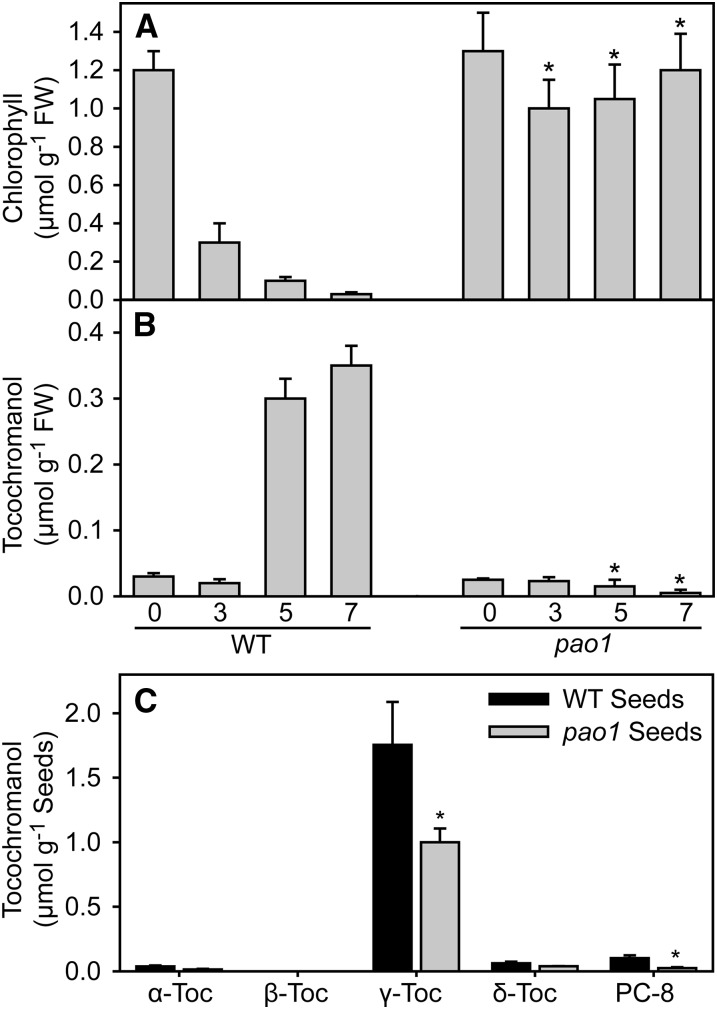
Tocopherol accumulates in leaves during stress or senescence (Collakova and DellaPenna, 2003). Phytyl-PP required for tocopherol synthesis can be derived from GG-PP from plastidial isoprenoid de novo synthesis or by phosphorylation of free phytol from chlorophyll hydrolysis. To determine the relative contribution of the phytol phosphorylation pathway for phytyl-PP synthesis destined for tocopherol production in leaves, the tocopherol contents in Arabidopsis wild type and the pheophorbide a oxygenase1 (pao1) mutant were determined during senescence. Chlorophyll degradation in the pao1 mutant is significantly retarded, due to a mutation in pheophorbide a oxygenase (PAO1) (Pruzinská et al., 2003, 2005) (Figure 1A). While total tocochromanol strongly accumulates during dark-induced senescence of detached leaves in the wild type, it remains at a very low amount in pao1 leaves (Figure 1B), indicating that all tocopherol synthesis during leaf senescence is based on phytol derived from chlorophyll degradation. In pao1 mutant seeds, the tocopherol content is reduced by 60% (Figure 1C). This result suggests an important role for phytyl-PP derived from chlorophyll degradation in the seeds, in accordance with previous findings (Valentin et al., 2006).
Figure 1.
Tocopherol Accumulation in Leaves and Seeds of the Wild Type and pao1 Mutant.
(A) Chlorophyll contents in leaves of the wild type and pao1 mutant. Detached leaves were incubated on wet filter paper in the darkness for 3, 5, and 7 d.
(B) Tocochromanol (tocopherol and plastochromanol-8) contents in leaves of the wild type and pao1.
(C) Tocochromanol (tocopherol and plastochromanol-8) contents in seeds of the wild type and pao1.
Data represent mean and sd of three to four measurements. Values significantly different from the wild type; *P < 0.05; Student’s t test. Toc, tocopherol.
The COG1836 Sequence At1g78620 Shows Similarity with Cytidylyltransferases
At1g78620 from Arabidopsis shows high similarity with COG1836 sequences from the genomes of cyanobacteria (Anabaena and Nostoc) and other photosynthetic prokaryotes (Archaeoglobus) where it clusters in close proximity with genes of isoprenoid metabolism (Seaver et al., 2014). COG1836 amino acid sequences from bacteria and plants can be organized into different groups. The first group includes sequences from Streptophyta (including At1g78620), Chlorophyta, and Cyanobacteria. A second group contains a second Arabidopsis sequence (At5g19930) and COG1836 sequences that form translational fusions with VTE5 proteins from Firmicutes and Chlorobi (Supplemental Figure 1). The two archaeal COG1836 sequences from Archaeoglobus and Thermoplasma can be placed between the At1g78620- and At5g19930-related COG1836 sequences. The At1g78620 protein sequence is more closely related to Synechocystis sll0875 (43.8% identity) than to At5g19930, a gene encoding a protein of unknown function (30.5%). At1g78620 belongs to the DUF92 family of proteins of unknown function harboring several transmembrane α-helices (Seaver et al., 2014). BLAST searches in the GenBank protein database revealed that At1g78620 shows low similarity to cytidylyltransferases, including At3g45040 (24.3%), a putative dolichol kinase (Valentin et al., 2006) (Supplemental Figure 1). At1g78620 is weakly related to phytol kinases from different organisms, including Arabidopsis (At5g04490/VTE5, 17.5% identity). The phytol kinase/VTE5 sequences cluster into a cyanobacterial/plant group containing At5g04490 (VTE5) and a related sequence (At5g58560, VTE5-like) (Fitzpatrick et al., 2011) and the second branch containing the phytol kinase domains that form fusion proteins with COG1836 sequences from Chlorobi and Firmicutes (Supplemental Figure 1) (Valentin et al., 2006).
The At1g78620 Protein Localizes to Chloroplast Envelopes
The At1g78620 sequence harbors an ∼70-amino-acid-long N-terminal extension absent from other COG1836 proteins (e.g., Synechocystis sll0875) and At5g19930 (Supplemental Figure 1). Sequence analysis with ChloroP1.1 (http://www.cbs.dtu.dk/) (Emanuelsson et al., 1999) revealed the presence of a putative N-terminal transit peptide of 65 amino acids predicted to target the protein to the chloroplast. To test the subcellular localization experimentally, the full-length At1g78620 sequence was C-terminally fused to YFP, and this construct introduced into Agrobacterium tumefaciens. After infiltration into Nicotiana benthamiana leaves, subcellular localization was observed in protoplasts by confocal laser scanning microscopy. Yellow fluorescence was observed as a ring-like structure surrounding the chlorophyll autofluorescence, indicating that At1g78620 localizes to the chloroplasts envelopes (Supplemental Figure 2). This result is in agreement with the identification of the spinach (Spinacia oleracea) ortholog of At1g78620 in isolated chloroplast envelopes by a proteomics approach (Ferro et al., 2002).
Arabidopsis At1g78620 Insertional Mutant Plants Are Incapable of Photoautotrophic Growth
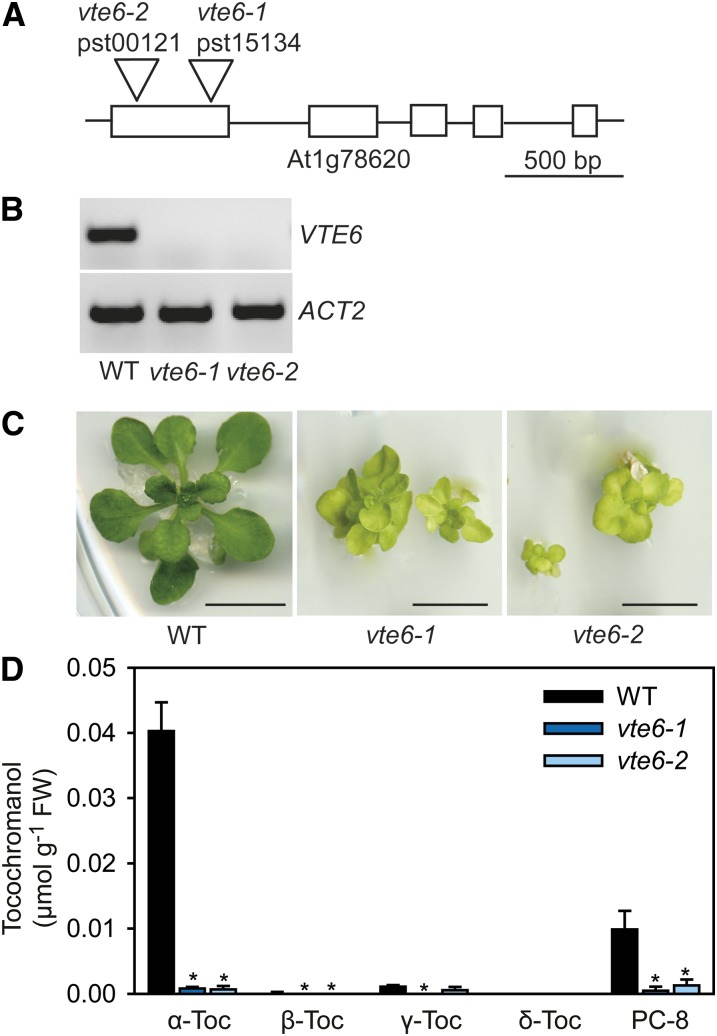
To study the role of At1g78620 in growth and phytol lipid synthesis, two Arabidopsis insertional mutant lines were obtained. The mutant plants (pst15134 and pst00121) carry transposons in the first exon of the At1g78620 gene (Figure 2A). Expression analysis by RT-PCR revealed that the At1g78620 mRNA was not detectable in the two mutant lines, suggesting that they represent null alleles (Figure 2B). The heterozygous plants of the two mutant lines grow similarly to the wild type. Homozygous pst15134 and pst00121 mutant plants are only viable on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with sucrose (Figure 2C). The plants are characterized by a stunted growth and short petioles and show a pale-green leaf color. They have a weakly developed or missing root system. Later, the plants produce a bushy rosette with small leaves but do not bolt and flower and therefore do not produce seeds. The plants die shortly after transfer to soil.
Figure 2.
Isolation and Tocochromanol Content of the vte6 Mutant from Arabidopsis.
(A) Map of the VTE6 (At1g78620) locus of Arabidopsis with exons (boxes) showing the positions of transposon insertions in the vte6 mutant alleles. Bar = 500 bp.
(B) Expression analysis of VTE6 in Arabidopsis vte6 mutant plants. The RT-PCR products were separated by agarose gel electrophoresis and stained with ethidium bromide. Actin (ACT2) was used as control.
(C) Growth of Arabidopsis vte6-1 and vte6-2 mutant plants. Plants of the wild type and the two vte6 mutant alleles were grown on MS medium with sucrose for 6 weeks. The vte6 mutant plants are homozygous as revealed by PCR of genomic DNA.
(D) Different forms of tocopherol (α-Toc, β-Toc, γ-Toc, and δ-Toc) and PC-8 were measured by fluorescence HPLC in leaves of the wild type, vte6-1, and vte6-2 grown on MS medium with sucrose for 6 weeks. Data show mean and sd of three measurements. Values significantly different from the wild type; *P < 0.05; Student’s t test.
At1g78620 Mutant Plants Are Tocopherol-Deficient
To study the role of At1g78620 in tocopherol metabolism, the amounts of the different forms of tocopherol and of PC-8 were measured by fluorescence HPLC in leaves of homozygous At1g78620 mutant plants grown on MS medium with sucrose for 6 weeks. Arabidopsis wild-type leaves mainly contain α-tocopherol and low amounts of the other tocopherol forms and of PC-8 (Figure 2D). In contrast, tocopherol and PC-8 levels were significantly reduced in the leaves of the two At1g78620 mutant lines, mostly due to the reduction in α-tocopherol by ∼98 to 100% (Figures 2D and 4A, respectively). Therefore, the mutation in the gene At1g78620 causes severe tocochromanol deficiency, and the corresponding mutant lines were renamed vte6-1 (pst15134) and vte6-2 (pst00121) for vitamin E deficient6.
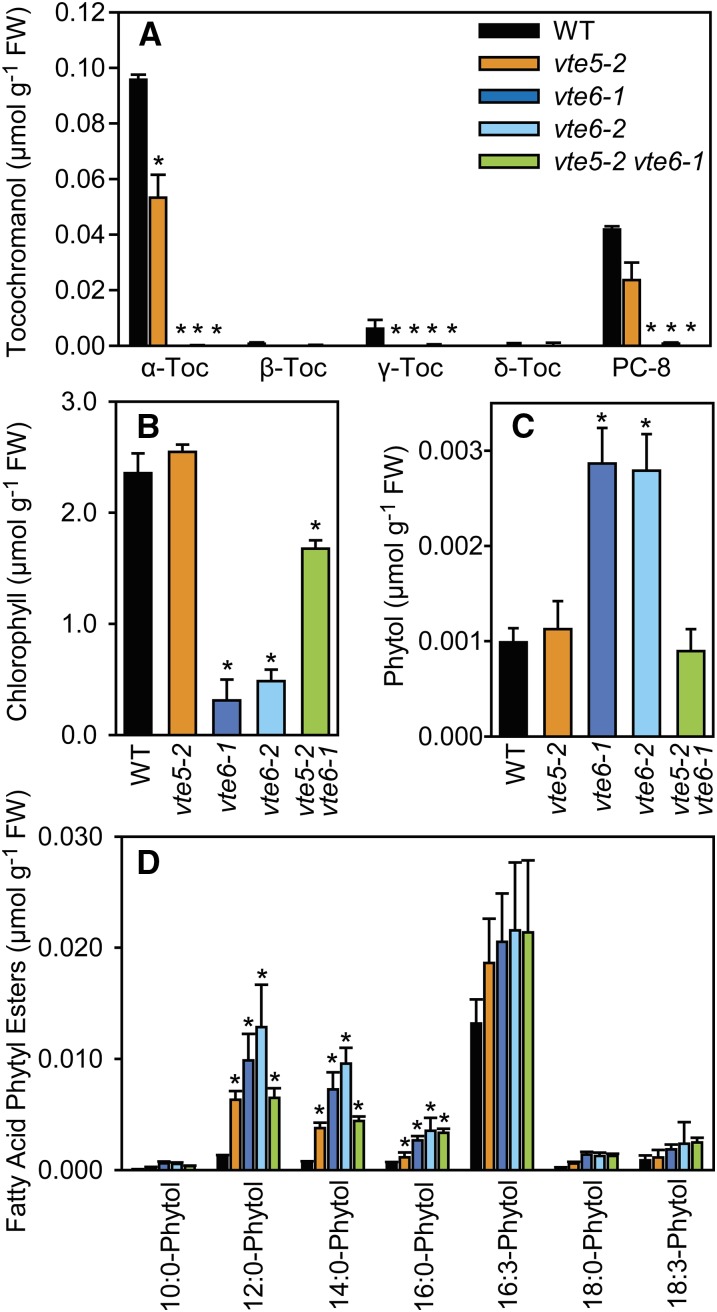
Figure 4.
Tocopherol, Chlorophyll, Phytol, and Fatty Acid Phytyl Esters in the Wild Type, vte5-2, vte6-1, vte6-2, and vte5-2 vte6-1.
(A) Tocopherol (α-Toc, β-Toc, γ-Toc, and δ-Toc) and PC-8 were measured by fluorescence HPLC.
(B) Chlorophyll was measured photometrically.
(C) The amount of free phytol was quantified by gas chromatography-mass spectrometry.
(D) Fatty acid phytyl esters were measured by direct infusion quadrupole time-of-flight tandem mass spectrometry.
Plants (wild type, vte5-2, vte6-1, vte6-2, and vte5-2 vte6-1) were grown on MS medium containing sucrose for 6 weeks. Leaves were harvested and analyzed for their lipid content. Mean and sd of four measurements are shown. Significantly different from the wild type; *P < 0.05. Chlorophyll content in vte5-2 vte6-1 was also significantly different from vte6-1 (Figure 4B); Student’s t test.
Longevity of Homozygous vte6 Seeds Is Compromised
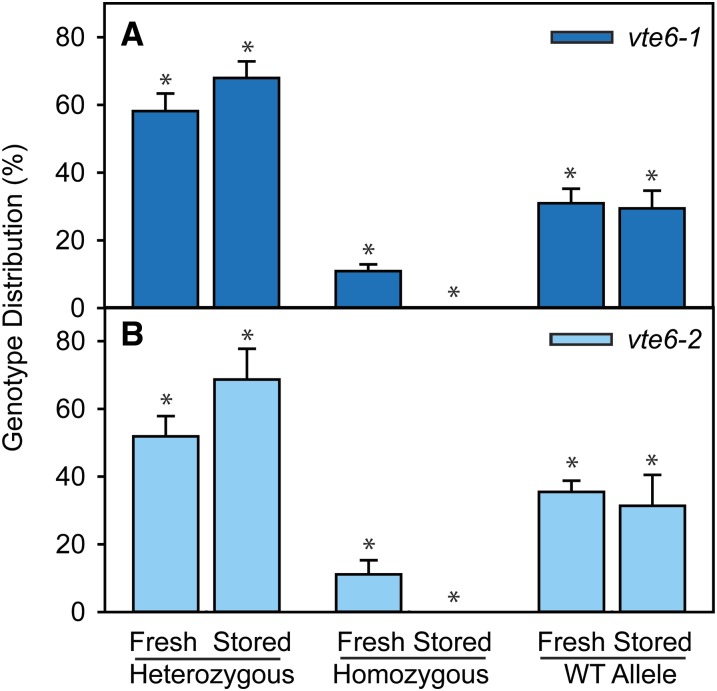
Previous studies showed that seed longevity of the tocopherol-deficient mutants vte1 and vte2 was strongly compromised (Sattler et al., 2004). This defect was associated with increased nonenzymatic oxidation of storage lipids in the tocopherol-deficient seeds of vte2. To study the seed longevity of the vte6 mutants, ∼250 seeds each from heterozygous vte6-1 and vte6-2 plants were either freshly harvested or stored for more than 3 months at room temperature. The seeds were germinated on sucrose-supplemented MS medium. The average germination rate was calculated and the genotype of the germinated plants was determined by PCR of genomic DNA. The total germination rate was 96% ± 3%, 76% ± 6%, and 91% ± 3% for freshly harvested seeds from wild-type and heterozygous vte6-1 and vte6-2 plants, respectively. It was significantly reduced in segregating vte6-2 seeds compared with the wild type after storage, when 67% ± 8% (vte6-2) and 89% ± 5% (wild type) of seeds germinated. The germination rate in freshly harvested segregating vte6-1 seeds was significantly lower (76% ± 6%) than that of wild-type seeds, yet was not altered after storage (76% ± 7%).
A segregation of 50% heterozygous plants, 25% wild-type, and 25% homozygous mutant plants would be expected for the seeds harvested from heterozygous vte6-1 or vte6-2 plants. However, only 11% of homozygous seedlings was retrieved from freshly harvested segregating vte6-1 or vte6-2 seeds, indicating that homozygous mutant seeds show significantly reduced viability (Figure 3). When seeds of heterozygous vte6-1 or vte6-2 plants were stored for 3 months at room temperature, germination of homozygous seeds was abolished. Only heterozygous or wild-type segregant seeds were able to germinate after storage. Therefore, similar to vte1 and vte2, homozygous vte6 seeds reveal a strong reduction in seed longevity, indicating that the VTE6 gene is essential for maintenance of seed viability.
Figure 3.
Decreased Germination Capacity of vte6 Seeds after Storage.
(A) Germination of seeds from a heterozygous vte6-1 plant.
(B) Germination of seeds from a heterozygous vte6-2 plant.
The seeds were germinated on MS medium with sucrose. Freshly harvested seeds stored for 3 months at room temperature were used. The genotype of the seedlings was determined by PCR of genomic DNA. The P values (asterisks) calculated from χ2 tests for the deviations of the genotype distributions of fresh or stored vte6-1 or vte6-2 seeds from the expected segregation (50:25:25) were <0.01.
The vte6 Mutation Affects Chlorophyll Content and Results in the Accumulation of Phytol and Fatty Acid Phytyl Esters
The strong reduction in tocopherol in leaves of the vte6-1 and vte6-2 mutant plants (Figures 2D and 4A) was expected to affect the amounts of other phytol-derived lipids, including chlorophyll. In agreement with the yellowish leaf color, the chlorophyll contents in vte6-1 and vte6-2 were significantly reduced as compared with the wild type (Figure 4B). The measurement of free phytol revealed a significant increase (∼3-fold) in the vte6-1 and vte6-2 leaves (Figure 4C). Previous results showed that free phytol can be used for the synthesis of fatty acid phytyl esters in chloroplasts of Arabidopsis. Quantification of fatty acid phytyl esters showed a significant increase in different molecular species, in particular 12:0-phytol, 14:0-phytol, and 16:0-phytol in vte6-1 and vte6-2 (Figure 4D).
VTE6 Harbors Phytyl-P Kinase Activity
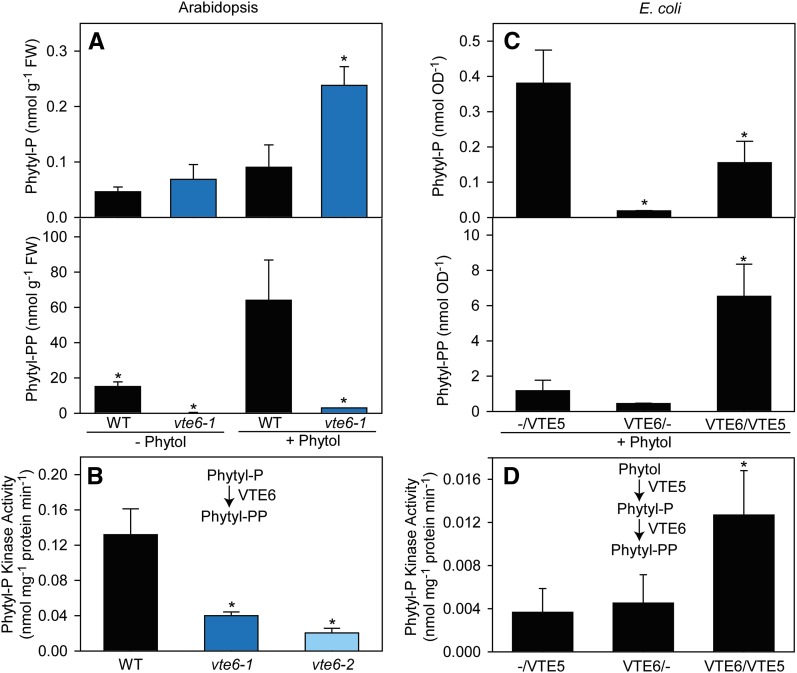
The in vivo activity of VTE6 was investigated by feeding phytol to seedlings of Arabidopsis wild type and vte6-1. Seedlings were incubated in buffer (control) or in buffer containing phytol, then seedlings were harvested and phytyl-P and phytyl-PP quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Figure 5A). Control plants incubated in buffer contained low amounts of phytyl-P. The amounts of phytyl-PP were high in the wild type, but extremely low in vte6-1 plants. Phytol feeding resulted in a strong increase of phytyl-P in vte6-1, but not in the wild type. In contrast, the phytyl-PP content in the wild type strongly increased (>4-fold) after phytol feeding, but not in vte6-1, where phytol feeding had almost no effect on the amount of phytyl-PP. In fact, the phytyl-PP content in vte6-1 was only 5% compared with the wild type.
Figure 5.
VTE6 Harbors Phytyl-P Kinase Activity.
(A) Phytol feeding experiments with Arabidopsis plants. Wild-type and vte6-1 plants were incubated in buffer without phytol or with 0.1% (v/v) phytol for 48 h, and isoprenyl-phosphates were isolated.
(B) Phytyl-P kinase assay with protein extracts from Arabidopsis wild-type, vte6-1, and vte6-2 leaves employing phytyl-P and CTP as substrates.
(C) Phytol feeding experiment with recombinant E. coli cells. Protein expression in E. coli cells harboring plasmids with either VTE5 (-/VTE5), VTE6 (VTE6/-) or both cDNAs (VTE6/VTE5) was induced and the cells incubated in the presence of phytol before extraction of isoprenyl-phosphates.
(D) Coupled phytol kinase/phytyl-P kinase assay with protein extracts from recombinant E. coli cells. Total protein was isolated from E. coli expressing VTE5 (-/VTE5), VTE6 (VTE6/-), or both proteins (VTE6/VTE5) and used for a coupled phytyl kinase/phytyl-P kinase assay employing phytol and CTP as substrates.
Isoprenyl-phosphates (phytyl-P and phytyl-PP) were measured by LC-MS/MS. Data represent mean and sd of three to four measurements. Significantly different from the wild type ([A] and [B]) or to E. coli expressing only VTE5 ([C] and [D]); *P < 0.05; Student’s t test.
Next, total protein was isolated from leaves of Arabidopsis wild-type, vte6-1, and vte6-2 plants. Phytyl-P kinase assays were done employing phytyl-P and CTP as substrates, and phytyl-PP was quantified by LC-MS/MS. The phytyl-P kinase activity with wild-type protein was significantly (3 to 6 times) higher compared with vte6-1 and vte6-2 (Figure 5B).
Phytol can be taken up and converted into phytyl-P by Escherichia coli cells expressing VTE5 (Valentin et al., 2006); this system was employed to demonstrate the activity of recombinant VTE6. To this end, the two cDNAs for VTE5 and VTE6 were introduced into E. coli as single constructs or in combination. After induction, the expression of VTE5 and VTE6 proteins was detected by immunoblot analysis (Supplemental Figure 3). Phytol was added to the E. coli cultures, and after 3 h, phytyl-P and phytyl-PP were extracted and quantified by LC-MS/MS. The amount of phytyl-P in VTE6-expressing cells was very low, suggesting that VTE6 might not harbor phytol kinase activity (Figure 5C). Cells expressing both VTE5 and VTE6 produced phytyl-P, albeit less than cells expressing only VTE5. The amounts of phytyl-PP in cells expressing either VTE5 or VTE6 were very low. However, when the two proteins were coexpressed in E. coli, large amounts of phytyl-PP accumulated (Figure 5C).
Coupled enzyme assays with protein extracts from E. coli expressing either VTE5 or VTE6 or expressing both proteins were performed employing phytol and CTP as substrates. Only minor amounts of phytyl-PP were produced with protein extracts from VTE5- or VTE6-expressing cells, indicating the presence of low phytol kinase and phytyl-P kinase activity in E. coli. Protein from cells coexpressing VTE5 and VTE6 showed significantly (∼3-fold) higher activity (Figure 5D). Taken together, phytol feeding experiments and enzyme assays with Arabidopsis wild-type and vte6 mutant plants, and with recombinant E. coli cells expressing VTE5/VTE6, demonstrate that VTE6 harbors phytyl-P kinase activity.
The Growth Defect of vte6-1 Is Alleviated in the vte5-2 vte6-1 Double Mutant
The strong growth retardation of the vte6-1 and vte6-2 mutant plants is in contrast with other tocopherol-deficient mutants (vte1, vte2, vte4, and vte5) because growth of these mutants under normal conditions is very similar to the wild type (Porfirova et al., 2002; Bergmüller et al., 2003; Maeda et al., 2006; Valentin et al., 2006). As an exception, the vte3-2 mutant shows reduction of both tocopherol and plastoquinol-9 due to a mutation in the methyltransferase VTE3; therefore, vte3-2 plants are incapable of photoautotrophic growth (Cheng et al., 2003). The tocopherol-deficient mutants vte1, vte2, vte4, and vte5 can grow on soil and produce fertile seeds, while vte6-1 and vte6-2 cannot grow photoautotrophically and are infertile. Therefore, the strong growth reduction of vte6 mutant plants cannot be explained by tocopherol deficiency per se, but must be caused by other factors. For example, it is possible that due to the block in phytyl-P phosphorylation, the accumulation of phytyl-P in vte6 plants might have negative effects on chloroplast development and growth, in contrast to other tocopherol-deficient plants, where such accumulation would not be expected. To investigate this potential scenario, and to suppress the accumulation of phytyl-P in vte6 plants, the vte5 mutation (phytol kinase) was introduced into the vte6-1 background. The Arabidopsis vte5-1 mutant carrying a premature stop codon in the VTE5 gene was previously isolated from a chemically mutagenized population (Valentin et al., 2006). An independent mutant allele (pst12490, vte5-2) was obtained that carries a transposon insertion in the first exon behind the start codon of the VTE5 gene (Figure 6A). Analysis of gene expression by RT-PCR revealed the absence of VTE5 mRNA in vte5-2 (Figure 6B), indicating that the vte5-2 mutation represents a null allele. After crossing of homozygous vte5-2 and heterozygous vte6-1 plants, double homozygous plants were identified by PCR in the F2 population. When grown on MS medium with sucrose, vte5-2 vte6-1 plants produce larger and greener leaves in contrast with vte6-1 plants (Figure 6C). The double mutant plants survive after transfer to soil. Figures 6D and 6E show 6-, 9-, and 12-week-old vte5-2 vte6-1 plants grown on soil. The morphology of the plants is stunted and bushy with many leaves. Double mutant plants can grow on soil for longer than 4 months. At this stage, wild-type and vte5-2 plants have died. Therefore, vte5-2 vte6-1 plants show a stay-green phenotype with strongly delayed senescence and extended lifetime. Old leaves do not turn yellow, but remain green and eventually wither and die (Figure 6E). These results indicate that the strong growth retardation and the incapability to grow on soil of the vte6-1 single mutant can be partially complemented by introducing the vte5-2 mutation into the genetic background of vte6-1.
Figure 6.
The vte5-2 vte6-1 Double Mutant.
(A) The vte5-2 mutant carries a transposon insertion in the first exon of VTE5 (At5g04490, phytol kinase). Exons are indicated by boxes. Bar = 500 bp.
(B) Expression analysis of the vte5-2 mutant. The RT-PCR products for VTE5 and ACT2 (control) were separated by agarose gel electrophoresis and stained with ethidium bromide.
(C) Growth of wild-type, vte5-2, vte6-1, and vte5-2 vte6-1 plants on MS medium with sucrose. Plants are 6 weeks old. Bar = 1 cm.
(D) Growth of wild-type, vte5-2, and vte5-2 vte6-1 plants on soil for 6 weeks. Bar = 2 cm.
(E) Wild-type and vte5-2 vte6-1 plants grown on soil for 9 or 12 weeks. Bar = 2 cm
Tocopherol, Phytol, and Fatty Acid Phytyl Esters in the vte5-2 vte6-1 Double Mutant
Wild-type, vte5-2, vte6-1 vte6-2, and vte5-2 vte6-1 plants were grown on MS medium with sucrose for 6 weeks, and leaves were harvested for the measurement of tocochromanols, chlorophyll, phytol, and fatty acid phytyl esters. Leaves of vte5-2 contain ∼50% less tocopherol than wild-type leaves under these conditions (Figure 4A). Tocopherol levels in leaves of vte6-1, vte6-2, and vte5-2 vte6-1 are below the detection limit. The reduced amount of tocopherol in vte6-1, vte6-2, and in the double mutant vte6-1 vte5-2 was accompanied by a significant decrease in PC-8 levels compared with the wild type (Figures 2D and 4A). The chlorophyll content is only slightly decreased in vte5-2 vte6-1 compared with the wild type and vte5-2, but it much is higher than in vte6-1 and vte6-2 (Figure 4B). Free phytol accumulates in leaves of vte6-1 and vte6-2 to ∼3 nmol g−1 fresh weight (FW), while the amounts of phytol are unchanged in vte5-2 and vte5-2 vte6-1 compared with the wild type (Figure 4C). In addition, the different forms of fatty acid phytyl esters are increased in leaves of vte5-2, vte6-1, vte6-2, and vte5-2 vte6-1 compared with the wild type, especially 12:0-phytol, 14:0-phytol, and 16:0-phytol (Figure 4D).
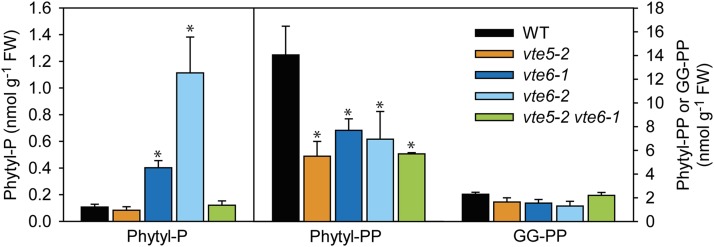
Phytyl-P Accumulates in the vte6-1 Mutant, but Not in the vte5-2 vte6-1 Double Mutant Plants
The severe growth retardation of vte6-1 was partially rescued in the vte5-2 vte6-1 double mutant, suggesting that alterations in phytol, phytyl-P, or phytyl-PP metabolism might be causal for the growth reduction of vte6-1. Phytyl-P, phytyl-PP, and GG-PP were quantified by LC-MS/MS in the leaves of plants grown on MS medium with sucrose. Arabidopsis leaves of wild-type plants contain around 15 nmol g−1 FW of phytyl-PP. The phytyl-P content in the wild type is extremely low (0.1 nmol g−1 FW). The phytyl-PP contents are significantly decreased in vte5-2, vte6-1, vte6-2, and vte5-2 vte6-1 mutant leaves to 6 to 8 nmol g−1 FW (Figure 7). The phytyl-P contents in vte6-1 and vte6-2 are significantly increased by ∼4- and 10-fold, respectively, compared with the wild type, vte5-2, and vte5-2 vte6-1. These results show that the increase in the amount of phytyl-P in vte6-1 is suppressed in the vte5-2 vte6-1 double mutant, suggesting a negative impact of phytyl-P accumulation on plant growth. The amounts of GG-PP were ∼2 nmol g−1 FW for all lines, indicating that the content of GG-PP is much lower than that of phytyl-PP and that it is not affected by changes in phytyl-P or phytyl-PP metabolism. Furthermore, these results are consistent with the presumed function of VTE6 as phytyl-P kinase because a block in phytyl-P kinase activity in vte6-1 and vte6-2 is expected to cause an accumulation of phytyl-P associated with a decrease in phytyl-PP.
Figure 7.
Quantification of Isoprenyl-Phosphates in Wild-Type, vte5-2, vte6-1, vte6-2, and vte5-2 vte6-1 Plants.
Plants were grown on MS medium with sucrose and leaves were harvested after 6 weeks. Isoprenyl-phosphates were measured by LC-MS/MS. Data show mean and sd of four to five measurements. Significantly different from the wild type; *P < 0.05; Student’s t test.
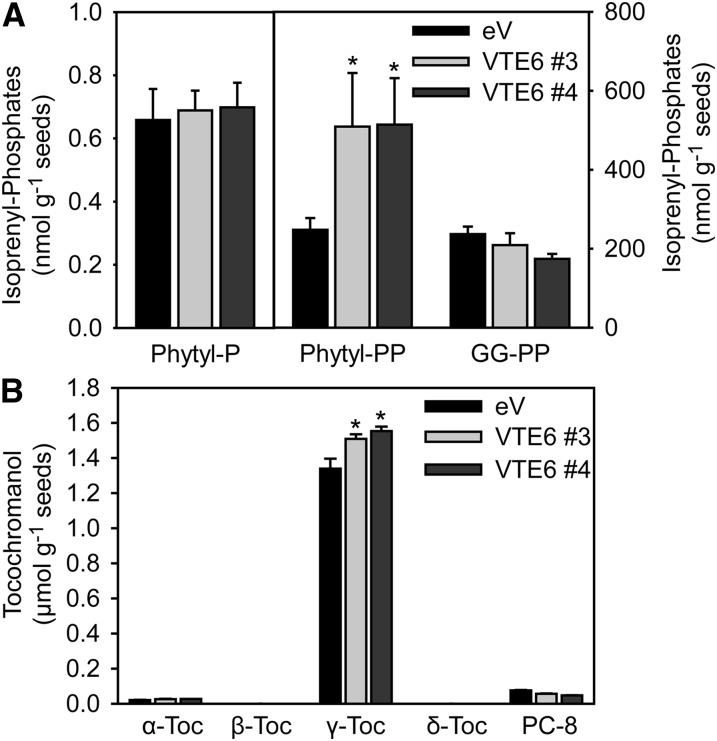
Overexpression of VTE6 Results in Increased Phytyl-PP and Tocopherol Levels in Seeds
VTE6 was overexpressed under the control of the CaMV 35S promoter in Arabidopsis wild-type plants, and the seeds of these plants were analyzed for isoprenyl-phosphates and tocochromanols. In the VTE6-overexpressing lines VTE6#3 and VTE6#4, phytyl-PP levels were significantly increased (509 and 514 nmol g−1 seeds) compared with the empty vector control line (247 nmol g−1 seeds) (Figure 8A). Phytyl-P and GG-PP levels were unaffected by overexpression of VTE6 (Figure 8A). Interestingly, the γ-tocopherol content was significantly increased in the VTE6 overexpression lines (1.5 and 1.6 µmol g−1 seeds) compared with the empty vector line (1.3 µmol g−1 seeds) (Figure 8B). Therefore, overexpression of VTE6 in seeds results in the accumulation of phytyl-PP, which can be used for prenylation of homogentisate by VTE2, finally causing an increase in tocopherol biosynthesis.
Figure 8.
Quantification of Isoprenyl-Phosphates and Tocopherol in VTE6 Overexpression Lines.
Seeds of Arabidopsis wild-type plants, empty vector (eV) control, and two lines overexpressing VTE6 (VTE6#3 and VTE6#4) were harvested from plants simultaneously grown on soil. Data show mean and sd of three to four measurements. Significantly different from the wild type; *P < 0.05; Student’s t test.
(A) Isoprenyl-phosphates were isolated from 20 mg of seeds and quantified by LC-MS/MS.
(B) Tocopherol (α-Toc, β-Toc, γ-Toc, and δ-Toc) and PC-8 were measured by fluorescence HPLC.
DISCUSSION
Tocopherol (vitamin E) is an important antioxidant in plants and animals and an essential component of the human diet. Work in recent years has established the pathways for tocopherol and tocotrienol synthesis, including the prenylation of homogentisate with a phytyl or geranylgeranyl group, respectively, with subsequent methylation and cyclization reactions (Hunter and Cahoon, 2007; Mène-Saffrané and DellaPenna, 2010). For a long time it was believed that the phytyl moiety for tocopherol synthesis exclusively originates from GG-PP via reduction by GGR. However, in the past years it became clear that tocopherol synthesis also depends on phytyl-PP derived from phytol released during chlorophyll breakdown (Figure 9). Two kinase reactions are required for the conversion of phytol into phytyl-PP (Ischebeck et al., 2006). While the gene for phytol kinase (VTE5) was characterized (Valentin et al., 2006), the identity of the second kinase remained unknown. Here, we present the characterization of a novel protein as candidate for phytyl-P kinase (VTE6). The sequence for VTE6 was identified based on its colocalization with orthologs of VTE5 and other genes of isoprenoid lipid metabolism in genomes of prokaryotic photosynthetic organisms, its coexpression pattern with genes of tocopherol metabolism, and its predicted localization in the chloroplast (Seaver et al., 2014). Furthermore, the VTE6 sequence displays weak similarity to cytidylyltransferases and to isoprenoid lipid kinases (Supplemental Figure 1).
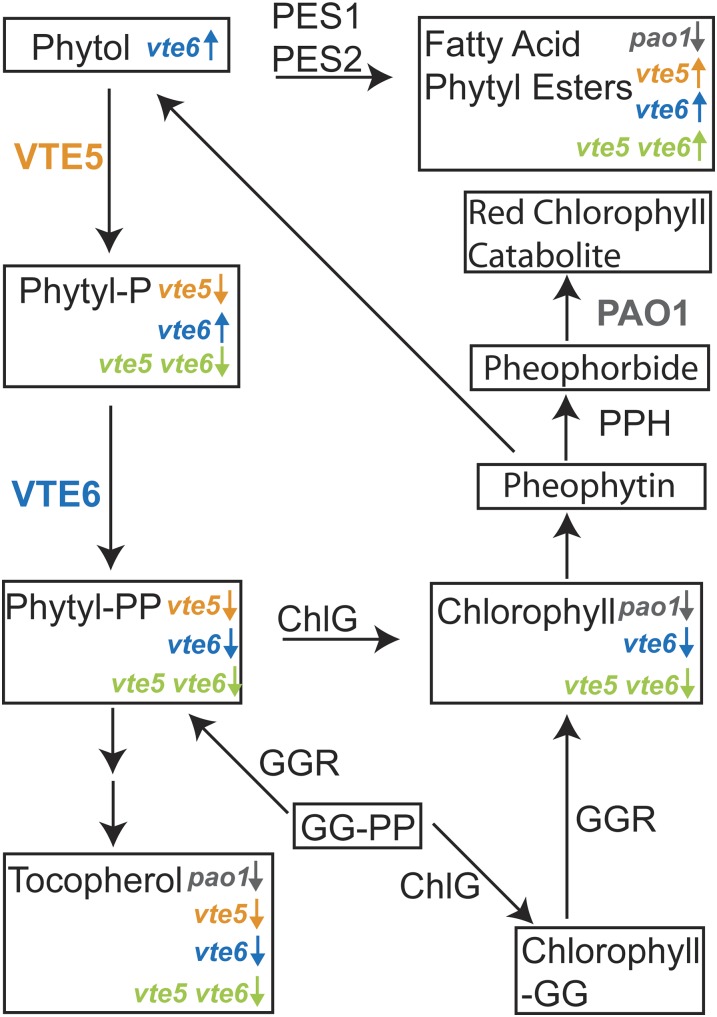
Figure 9.
Phytol Metabolism in Arabidopsis.
Phytol is derived from chlorophyll degradation via PPH or by an unknown dephytylase. PAO1 is involved in further pheophorbide catabolism. Phosphorylation by VTE5 and VTE6 results in the production of phytyl-PP, which serves as precursor for tocopherol synthesis. Phytol is also used for the synthesis of fatty acid phytyl esters by phytyl ester synthases (PES1 and PES2) (Lippold et al., 2012). GG-PP or phytyl-PP can be used to synthesize geranylgeranyl-chlorophyll (chlorophyll-GG) or chlorophyll, respectively, by chlorophyll synthase (ChlG). Chlorophyll-GG is reduced to chlorophyll by GGR. The contribution of de novo synthesis via direct reduction of GG-PP to phytyl-PP by GGR is low. Alterations in metabolite or lipid contents in the respective mutants are indicated by arrows (upregulation or downregulation).
Phytyl-P Kinase Activity
Phytyl-P kinase catalyzes the conversion of phytyl-P into phytyl-PP. Feeding experiments with Arabidopsis wild-type and vte6-1 seedlings showed that phytol is taken up by the plants and converted into phytyl-P by VTE5. Phytyl-PP levels were strongly increased in wild-type seedlings, while phytyl-PP levels were extremely low in vte6-1 plants after phytol feeding (Figure 5). On the other hand, phytyl-P, the precursor of phytyl-PP, increased in vte6-1, but not in the wild type. Furthermore, enzyme assays with Arabidopsis leaf protein showed that phytyl-P kinase activity in vte6-1 is ∼3- to 4-fold lower compared with the wild type.
Feeding experiments and enzyme assays with recombinant E. coli cells confirmed the results obtained with Arabidopsis plants. After phytol feeding, phytyl-P but not phytyl-PP accumulated in VTE5-expressing cells. In VTE5/VTE6 coexpressing cells, the accumulation of phytyl-P was attenuated, while phytyl-PP accumulated, indicating that a certain proportion of phytyl-P was converted into phytyl-PP by VTE6. In cells expressing only VTE6, neither phytyl-P nor phytyl-PP accumulated, indicating that VTE6 does not harbor phytol kinase activity, but is dependent on the presence of phytyl-P, which can only be provided by VTE5 but not by other E. coli proteins in this system.
A coupled enzyme assay with total protein extracts from E. coli and phytol and CTP as substrates showed that phytyl-PP synthesis depends on the presence of VTE6. Taken together, these experiments conclusively demonstrate that VTE6 converts phytyl-P into phytyl-PP, in agreement with the hypothesis that VTE6 encodes phytyl-P kinase.
Phytyl-PP for Tocopherol Biosynthesis in Leaves Is Almost Exclusively Provided by the Phytol Phosphorylation Pathway
Two mutant alleles, vte6-1 and vte6-2, were isolated which both showed reduced growth and changes in phytol lipid contents. Furthermore, the vte6-2 mutant was backcrossed to the wild type (see Methods), and a homozygous F2 plant was obtained that showed the same growth and lipid phenotypes as vte6-1. These experiments demonstrate that the strong growth retardation and the phytol lipid changes are genetically linked with the vte6 mutations.
Phytyl-phosphates and geranylgeranyl-phosphates are important intermediates for the synthesis of chlorophyll, tocopherol, phylloquinol, and carotenoids in plant leaves, but their measurement was hindered by their low abundance. Employing an LC-MS/MS-based strategy, the amounts of phytyl-P, phytyl-PP, and GG-PP were measured in leaves and seeds of Arabidopsis. Phytyl-P levels in wild-type leaves are extremely low (∼0.1 nmol g−1 FW), while phytyl-PP and GG-PP amount to 15 or 2 nmol g−1 FW, respectively (Figure 7). Phytyl-P accumulates in the vte6-1 mutant, in accordance with the role of VTE6 as phytyl-P kinase, while the phytyl-PP level is reduced to 50% compared with the wild type. As vte6-1 represents a null mutation (Figure 2), the residual amount of phytyl-PP presumably is derived from GG-PP reduction by GGR. Interestingly, the reduction of phytyl-PP to 50% of wild-type levels is associated with a severe decrease in tocopherol contents in leaves (by ∼98 to 100% compared with the wild type), highlighting the relevance of VTE6 for tocopherol synthesis. Therefore, the residual content of phytyl-PP in vte6 cannot be used for tocopherol synthesis. Maybe different pools of phytyl-PP exist in the chloroplast, derived from phytol phosphorylation or from GGR, and only the phytyl-PP derived from phytol phosphorylation might be accessible for tocopherol synthesis. The amount of GG-PP was not decreased in leaves of vte6-1 but was much lower than that of phytyl-PP, indicating that GG-PP reduction to phytyl-PP cannot compensate for the loss of phytyl-P phosphorylation activity to provide phytyl-PP as substrate for tocopherol synthesis. Therefore, GG-PP produced via isoprenoid de novo synthesis plays a negligible role in providing the phytyl-moiety for tocopherol synthesis in leaves.
Phytyl-PP Synthesized by VTE6 Is Used for Tocopherol Biosynthesis in Seeds
Seeds of the Arabidopsis phytol kinase mutant vte5-1 contain only 20% of wild-type tocopherol levels (Valentin et al., 2006). Therefore, it was considered that the bulk of tocopherol is synthesized from phytyl-PP that originates from the phytol phosphorylation pathway. Intriguingly, overexpression of VTE6 in Arabidopsis results in a 2-fold increase in phytyl-PP and in increased tocopherol levels in seeds. This finding is in agreement with the hypothesis that VTE6 encodes phytyl-P kinase and that it provides phytyl-PP for seed tocopherol synthesis. Furthermore, this result suggests that phytyl-PP might be limiting for tocopherol synthesis in seeds.
Longevity in vte6 Mutant Seeds Is Impaired
Seeds of the vte2 mutant are completely devoid of tocopherol and exhibit a severe reduction in longevity. Accelerated ageing at high temperature and high humidity causes a strong defect in germination rate of vte2 seeds, which was associated with nonenzymatic peroxidation of seed lipids, caused by the lack of antioxidant capacity (Sattler et al., 2004). However, apart from a delay in seedling development and rare cotyledon defects, vte2 plants develop normally, grow to full size, and produce seeds. The vte5-1 mutant contains 20 and 35% of tocopherol in the seeds and leaves, respectively, compared with the wild type and also exhibits normal plant development and growth (Valentin et al., 2006). Similarly, germination of vte5-2 seeds was not affected. In contrast, vte6-1 and vte6-2 plants can only grow on sucrose-supplemented MS medium, are pale-green and dwarfed, and do not produce seeds. Homozygous vte6 seeds do not germinate after storage for more than 3 months. These plants are infertile, and homozygous vte6 mutant seeds cannot be selected from seeds of a heterozygous plant because the seeds cannot be distinguished. Therefore, it was not possible to analyze homozygous vte6 seeds for lipid contents. Attempts to measure tocopherol, phytol, or isoprenyl-phosphates in single, segregating seeds of a heterozygous vte6 plant were not successful. However, the fact that longevity is affected in vte6 seeds, similar to vte2 seeds, suggests that vte6 seeds also suffer from a severe reduction in tocopherol content.
The Deficiency in Growth and Development of vte6 Plants Is Unrelated to the Degree of Membrane Lipid Unsaturation
Tocopherol plays a crucial role during the adaptation of growth under low-temperature conditions (Maeda et al., 2006). Tocopherol-deficient mutants including vte2 show increased sugar and carbohydrate contents in the leaves, accompanied by callose deposition at plasmodesmata in phloem parenchyma transfer cells, when grown at low temperature. Biochemical and genetic evidence showed that this effect is unrelated to oxidative stress, but it is associated with reduced desaturation of linoleic acid (18:2) to α-linolenic acid (18:3), particularly in phosphatidylcholine (PC). Therefore, when grown under normal or low temperature, PC in vte2 contains a reduced amount of 18:3 compared with the wild type. The mechanism of how tocopherol deficiency affects lipid desaturation remains unclear. It was possible that the severe growth retardation of the vte6 mutant was related to the low temperature phenotype of vte2 plants. Measurements of membrane lipids revealed that the proportion of monogalactosyldiacylglycerol is reduced in vte6-1 and vte6-2, while the relative amount of PC is increased (Supplemental Figure 4). This result indicates a general decrease in the amounts of thylakoid lipids, albeit much less severe as the decrease in chlorophyll. The molecular species of 36:6 (18:3/18:3) and 36:5 (18:3/18:2) are the predominant PC molecules carrying two C18 fatty acids. The vte6-1 and vte6-2 plants contain higher amounts of 36:6 PC than the wild type, while the amounts of 36:5 PC remain similar. Therefore, vte6 plants contain a higher degree of unsaturated C18 fatty acids in PC than the wild type, in contrast to vte2, which contains lower amounts of 36:6 compared with the wild type (Maeda et al., 2006). This result indicates that the severe growth retardation of vte6 is unrelated to the low-temperature phenotype of the vte2 mutant.
Phytyl Lipids in the vte5 and vte6 Mutants
The vte5-1 mutant of Arabidopsis was described previously (Valentin et al., 2006). We used a second mutant allele (vte5-2), which grows like the wild type and shows normal chlorophyll contents. The amount of tocopherol is reduced, as was shown for vte5-1, indicating that the insertion in vte5-2 is causal for the tocopherol deficiency, in analogy to vte5-1. The amount of tocopherol is strongly reduced in leaves of vte6 (<5%), while vte5 leaves contain considerable more tocopherol (vte5-1, 35% of the wild type, Valentin et al., 2006; vte5-2, 50% of the wild type, Figures 4 and 6). As vte5-2 represents a null allele, a second phytol kinase activity might exist in the chloroplasts, which could explain the residual capacity for the conversion of phytol into phytyl-P and the absence of phytol accumulation in vte5-2. On the other hand, VTE6 presumably is the only phytyl-P kinase in chloroplasts. Therefore, the complete block of the phytol phosphorylation pathway in vte6 might lead to the accumulation of phytyl-P and at the same time of phytol because phytol phosphorylation by VTE5 (and additional phytol kinase activities) might be downregulated, possibly by a feedback mechanism, to avoid the uncontrolled accumulation of phytyl-P.
The Severe Impairment of Growth and Development in vte6 Mutants Is Alleviated in vte5 vte6 Double Mutants
To test the hypothesis whether the block in sequestration of phytol to phytyl-P and subsequently to phytyl-PP is related to the strong growth retardation of the vte6-1 mutant, a genetic test was conducted by generating a double mutant of vte5-2 and vte6-1. Interestingly, growth of homozygous vte5-2 vte6-1 plants is strongly improved compared with vte6-1 single mutant plants. Similar to vte6-1, vte5-2 vte6-1 double mutant plants show strongly reduced amounts of tocopherol (Figure 4A).
The amount of phytyl-P is increased in vte6-1 leaves (0.4 nmol g−1 FW) but is reduced in vte5-2 vte6-1 to levels similar to the wild type and vte5-2 (around 0.1 nmol g−1 FW) (Figure 7). The amounts of free phytol follow a similar pattern, because phytol is unaffected in vte5-2 and vte5-2 vte6-1, while it is strongly accumulated in vte6-1 compared with the wild type (Figure 4C). Therefore, the introduction of the vte5-2 mutation into the vte6-1 mutant background strongly improves growth, associated with the suppression of the accumulation of phytol and phytyl-P. This pattern of changes is in accordance with the expected result of combining mutations in phytol kinase and phytyl-P kinase activities in a single plant.
The fact that phytyl-PP in vte6-1 and vte5-2 vte6-1 is reduced compared with the wild type, but that chlorophyll content in vte5-2 vte6-1 is strongly increased, indicates that the reduction in phytyl-PP content per se does not become limiting for chlorophyll synthesis in vte5-2 vte6-1, as well as in vte6-1. It is possible that phytol or phytyl-P levels, which are increased in vte6-1, affect chlorophyll synthesis activity, e.g., by competing with phytyl-PP for binding to chlorophyll synthase or by interfering with other important chloroplast pathways.
The relative amounts of galactolipids in vte6 are reduced to only a minor extent (monogalactosyldiacylglycerol drops from 42 to 33 to 36%, while digalactosyldiacylglycerol and sulfoquinovosyldiacylglycerol are not changed; Supplemental Figure 4), indicating that the overall effect on chloroplast membrane lipids is much less severe than the decrease in chlorophyll content. Furthermore, not all chloroplast lipids are decreased. Some lipids increased in vte6, in particular phytol, phytyl-P, and fatty acid phytyl esters. Therefore, the block in vte6 preferentially affects phytol-containing lipids. On the other hand, the amounts of PC-8 in vte6-1 and vte6-2 were reduced. PC-8 synthesis is not directly linked with phytyl-PP metabolism. It is possible that the block in phytol phosphorylation might affect tocochromanol synthesis (head group and/or prenyl side chain production) by feedback inhibition (see above), which would explain the general decrease in the levels of all tocochromanols in vte6. In line with this scenario, PC-8 also decreased in the vte5-2 mutant, which has normal chlorophyll levels (Figure 4), and in the seeds of the pao1 mutant (Figure 1).
The vte5-2 vte6-1 plants contain significantly higher chlorophyll contents than vte6-1 (Figure 4B). Therefore, the strong reduction in vte6 growth is primarily not caused by tocopherol deficiency or reduction in chlorophyll synthesis capacity, but might rather be associated with the accumulation of free phytol or phytyl-P. It has previously been shown that the accumulation of free alcohols is detrimental to membrane integrity (Löbbecke and Cevc, 1995). Therefore, the accumulation of phytol might destabilize chloroplast membranes affecting chlorophyll and galactolipid synthesis in vte6-1. In addition, it is possible that phytyl-P is toxic per se or that the sequestration of phytol into the phytyl-P pool is harmful for plant metabolism. In this regard, it is interesting to note that phytyl-P represents an alcohol phosphate analogous to sphingobase-1-phosphates (e.g., sphingosine-1-phosphate), which are known signaling molecules in plants and animals (Lynch et al., 2009). It is also possible that phytyl-P-derived compounds accumulate in vte6 with detrimental consequences for plant growth.
Synechocystis harbors a protein sequence (sll0875) highly related to Arabidopsis VTE6 (Supplemental Figure 1). A gene knockout strategy was employed to mutagenize sll0875 in Synechocystis by homologous recombination. However, screening of transformed Synechocystis cells after repeated rounds of selection on antibiotic medium revealed the absence of fully segregated homozygous mutant cells, indicating that this gene is essential. Therefore, the phosphorylation pathway of phytol is crucial for Arabidopsis and Synechocystis, and this essential function is independent of tocopherol deficiency.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana transposon insertion lines vte6-1 (pst15134, 13-0604-1) and vte6-2 (pst00121, 54-0912-1) were obtained from the RIKEN Bioresource Center (Tsukuba, Japan). Seeds of heterozygous vte6-1 and vte6-2 plants were germinated on MS medium containing 2% (w/v) sucrose (Murashige and Skoog, 1962). Homozygous seedlings were selected by PCR using the primers bn233 and bn234 (genomic VTE6 locus, 1255 bp); bn232 and bn234 (vte6-1, 970 bp); and bn130 and bn234 (vte6-2, 1230 bp). The phenotype of the line vte6-2 (pst00121) was complicated by the fact that heterozygous plants were severely stunted, originating from a second site mutation. After backcrossing this plant with the RIKEN transposon donor line Ds13 (N8521), a vte6-2 plant devoid of the secondary mutation was obtained.
The vte5-2 (At5g04490; pst12490, 11-6074-1) transposon mutant allelic to vte5-1 deficient in phytol kinase (Valentin et al., 2006) was obtained from the RIKEN Bioresource Center. Homozygous vte5-2 plants were isolated by PCR screening (genomic locus, bn771, bn772, 1352 bp; insertion, bn771, bn232, 1080 bp). Heterozygous vte6-1 and homozygous vte5-2 plants were crossed, and plants heterozygous for vte6-1 and homozygous for vte5-2 were selected in the F2 progeny, and double homozygous vte5-2 vte6-1 plants in the F3, by PCR screening.
The pao1 mutant (At3g44880, SALK_111333) was obtained from Stefan Hörtensteiner, University of Zürich, Switzerland (Pruzinská et al., 2005). Plants were grown at 150 µmol m−2 s−1 light at 16 h light per day, 20°C, and 55% relative humidity.
RT-PCR
RNA was extracted from leaves, and cDNA synthesis and RT-PCR were performed as described previously (Lippold et al., 2012). Oligonucleotides used for RT-PCR of VTE6, VTE5, and ACT2 are described in Supplemental Table 1.
Generation of Overexpression Lines
To generate transgenic Arabidopsis lines overexpressing VTE6, the full-length coding sequence of At1g78620/VTE6 was amplified from VTE6 cDNA (pda00492, RIKEN) by PCR adding restriction sites using the oligonucleotides bn327 (XmaJI) and bn328 (SalI) (Supplemental Table 1). The PCR products were ligated into the cloning vector pGEM-T Easy (Promega). The plasmid was digested with XmaJI and SalI (VTE6) and the insert ligated into the SpeI and SalI sites of the binary vector pLH9000 (DNA Cloning Service), yielding pLH9000-VTE6 for expression in plants under the control of the CaMV 35S promoter. The plasmid was transferred into Agrobacterium tumefaciens (GV3101-pMP90) by electroporation. Arabidopsis wild-type Col-0 plants were transformed by floral dipping (Clough and Bent, 1998). Transgenic seeds were selected by fluorescence microscopy based on the expression of the DsRed marker gene. Overexpression of VTE6 was confirmed by RT-PCR.
Measurement of Chlorophyll, Tocopherol, Phytol, and Fatty Acid Phytyl Esters
Chlorophyll was quantified photometrically (Porra et al., 1989). For the extraction of tocopherol, fatty acid phytyl esters, and free phytol, 50 mg of leaf tissue was frozen in liquid nitrogen and ground to a fine powder (Precellys homogenizer). The ground tissue was resuspended in 500 μL diethylether and phase separation achieved by centrifugation after adding 250 μL of 0.2 M H3PO4/1 M KCl. The organic phase was harvested and dried under air flow. Tocopherol was measured by fluorescence HPLC on a diol column (Zbierzak et al., 2010). Free phytol was quantified by gas chromatography-mass spectrometry after derivatization with N-methyl-N-(trimethylsilyl) trifluoroacetamide using oleyl alcohol (Sigma-Aldrich) as internal standard (Ischebeck et al., 2006).
Fatty acid phytyl esters were quantified using 17:0-phytol, synthesized from heptadecanoic acid (17:0) and phytol (Ischebeck et al., 2006). Fatty acid phytyl esters were detected by scanning for the neutral loss of the mass of 278.2974 (C20H38, [phytol-H20]) by direct infusion-tandem mass spectrometry experiments (Agilent 6530 Accurate Mass Q-TOF). The samples were infused via an HPLC-ChipCube nanospray ion source in chloroform/methanol/300 mM ammonium acetate (300:665:35, v/v/v) at 1 μL min−1 (Welti et al., 2002). The Q-TOF mass spectrometer was operated in positive ion mode with a fragmentor voltage of 270 V. Membrane glycerolipids were isolated and quantified as described (Gasulla et al., 2013).
Quantification of Isoprenyl-Phosphates by Liquid Chromatography-Mass Spectrometry
Isoprenyl-phosphates were extracted from leaves (50 mg) using isopropanol/50 mM KH2PO4/acetic acid (1:1:0.025, v/v/v) (Larson and Graham, 2001). Internal standards (0.1 nmol each of decanyl-phosphate and decanyl-diphosphate) were synthesized and purified (Joo et al., 1973) and added to the lipid extracts. Isoprenyl-phosphates were separated on a Nucleodur C8 column (50 mm × 4.6 mm, 1.8 µm; Macherey and Nagel), using a gradient of acetonitril and 5 mM aqueous ammonium acetate. The concentration of acetonitril was increased from 20 to 100% in 25 min. Mass spectrometry was performed on an Agilent 6530 Accurate Mass Q-TOF instrument with JetStream ESI source. Isoprenyl-phosphates were analyzed in negative ion mode after collision-induced dissociation, by precursor ion scanning for the phosphate group (HPO3−, m/z 78.9591). The parental ions of the isoprenyl-phosphates [M-H]− were phytyl-P (m/z 375.2664), phytyl-PP (m/z 455.2328), and GG-PP (m/z 449.1858) as previously described (Valentin et al., 2006).
Expression of VTE6 and VTE5 Proteins in Escherichia coli
The coding sequences of VTE6 and VTE5 were amplified from Arabidopsis cDNA, and specific restriction sites were added using the primer pairs CB1/CB2 and CB3/CB4 (Supplemental Table 1), respectively. The fragments were subcloned into pJet2.1 (ThermoFisher Scientific) and verified by sequencing. After digestion with BamHI/NotI (VTE6 fragment) or NdeI/FseI (VTE5 fragment), the fragments were ligated into the multiple cloning sites 1 or 2 of the expression vector pETDuet-1 (Novagen, Merck Millipore), which is designed for coexpression of two genes. Thus, VTE6 was N-terminally fused to a 6x His-tag sequence, while VTE5 was C-terminally fused to an S-tag. In addition to the pETDuet-1 plasmid containing both the VTE6 and VTE5 cDNAs, plasmids carrying either VTE6 or VTE5 were also generated. The plasmids were transformed into E. coli Rosetta (DE3) cells, and ampicillin-resistant transformants were selected. Cells were grown in Luria-Bertani medium at 37°C and 200 rpm until an OD600 of 0.7 was reached. Protein expression was induced with 1 mM isopropylthio-β-galactoside. The fusion proteins were detected by immunodetection after blotting using horseradish peroxidase-conjugated antibodies specific for His-tag (Miltenyi Biotec) and S-tag (Merck Millipore) for VTE6 and VTE5, respectively.
In Vivo Phytol Feeding
Arabidopsis seedlings (the wild-type, vte6-1, and vte6-2) were grown on MS medium with 2% (w/v) sucrose for 6 weeks and then transferred to 20 mL of 20 mM MES-KOH, pH 6.5, containing 0.1% (v/v) phytol (Sigma-Aldrich) in an Erlenmeyer flask. The flasks were incubated under continuous light (100 µmol photons m−2 s−1) for 48 h while shaking before extraction of isoprenyl-phosphates.
Feeding of E. coli cells with synthetic phytol was done as described by Valentin et al. (2006) with some modifications. Briefly, protein expression was induced using 1 mM isopropylthio-β-galactoside and the cells grown overnight at 16°C. Cells were harvested, washed, and transferred to 5 mL of Luria-Bertani medium containing carbenicillin (50 µg/mL), 5 mM phytol, and 0.2% toluene. The cells were incubated for 3 h at 30°C and harvested by centrifugation and the cell pellet used for isoprenyl-phosphate extraction.
In Vitro Phytol Kinase and Phytyl-P Kinase Assays
Proteins were isolated from Arabidopsis leaves and E. coli cell pellets as previously described (Ischebeck et al., 2006). For the phytyl-P kinase assay, 10 nmol of phytyl-P (Isoprenoids) in ethanol were transferred to a 1.5-mL microfuge tube and dried under air flow. Afterwards, 300 µg crude protein extract was mixed with 10 nmol CTP and 8 μL assay buffer (20 mM MgCl2, 50 mM Na-orthovanadate, and 0.25% CHAPS) to a final volume of 100 µL. For the coupled phytol kinase/phytyl-P kinase assay, 1 nmol of phytol instead of phytyl-P was used in the reaction. The reaction was incubated for 30 min at 30°C and terminated by freezing in liquid N2. Isoprenyl-phosphates were isolated and analyzed using LC-MS/MS as described above.
Subcellular Localization Studies of YFP-Tagged VTE6 Protein
For subcellular localization studies, the coding sequence (without stop codon) of VTE6 was PCR amplified from Arabidopsis cDNA with gene-specific primers (CB157/CB158; Supplemental Table 1) carrying Gateway recombination sites and recombined into the pUBC-YFP binary vector (Grefen et al., 2010) for C-terminal fusion of VTE6 to YFP, driven by the Ubiquitin10 promoter. The fusion construct YFP:VTE6 was verified by sequencing. Agrobacterium strain GV3101 was transformed with the plasmid and infiltrated into Nicotiana benthamiana leaves for transient expression of YFP:VTE6 fusion proteins. After 48 h, protoplasts were isolated for localization studies by a confocal laser scanning microscope (Zeiss LSM 510) as described in detail earlier (Breuers et al., 2012).
Phylogenetic and Statistical Analysis
Amino acid sequences of VTE5- and VTE6-related proteins from Arabidopsis, Synechocystis, Synechococcus, Anabaena, Nostoc, Chlorobium, Pelodyction, Symbiobacterium thermophilum, Thermoplasma acidophilum, and Archaeoglobus fulgidus were obtained from GenBank. Sequence alignments were done using ClustalW, and phylogenetic trees were constructed based on the neighbor-joining method with the bootstrap test with MEGA 6 (Tamura et al., 2013). A text file of the alignment is available as Supplemental Data Set 1.
Statistical analyses were performed using Microsoft Excel 2010. Values are significantly different according to Student’s t test (P < 0.05). The significance of differences in genotypes between the segregating population and the expected distribution (Figure 3) was calculated according to the χ2 test (P < 0.01). At least three biological replicates were used for the biochemical analyses.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under Arabidopsis locus identifiers At5g04490 and At1g78620, for VTE5 and VTE6, respectively.
Supplemental Data
Supplemental Figure 1. Phylogenetic Relationship of COG1836-Like and Phytol Kinase Sequences.
Supplemental Figure 2. Subcellular Localization of VTE6 to the Chloroplast Envelope Membranes.
Supplemental Figure 3. Expression of VTE5 and VTE6 in Escherichia coli.
Supplemental Figure 4. Membrane Glycerolipids in Leaves of the vte6 Mutant.
Supplemental Table 1. Oligonucleotides Used in This Study.
Supplemental Data Set 1. Text File of the Alignment Corresponding to the Phylogenetic Analysis in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Stefan Hörtensteiner (University of Zürich, Switzerland) for seeds of the pao1 mutant of Arabidopsis. We acknowledge the help of Brigitte Dresen-Scholz, Anne Bünder, and Omnia El Said (University of Bonn) during plant growth, genotyping, and cloning. This research was in part supported by the Deutsche Forschungsgemeinschaft (SFB 645, Project Z4 to P.D., WE 2231/8-2 to A.P.M.W. and M.E., and EXC 1028 to A.P.M.W.).
AUTHOR CONTRIBUTIONS
P.D., G.H. M.E., A.P.M.W., and A.D.H. designed the experiments. K.v.D., G.H., C.P., M.E., and M.A. performed the research. A.D.H. contributed new analytic and computational tools. All authors analyzed data. P.D., K.v.D., M.E., A.P.M.W., and A.D.H. wrote the article.
Glossary
- phytyl-PP
phytyl-diphosphate
- GG-PP
geranylgeranyl-diphosphate
- GGR
geranylgeranyl reductase
- PPH
pheophytin pheophorbide hydrolase
- MS
Murashige and Skoog
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- FW
fresh weight
- PC
phosphatidylcholine
References
- Bergmüller E., Porfirova S., Dörmann P. (2003). Characterization of an Arabidopsis mutant deficient in γ-tocopherol methyltransferase. Plant Mol. Biol. 52: 1181–1190. [DOI] [PubMed] [Google Scholar]
- Breuers F.K., Bräutigam A., Geimer S., Welzel U.Y., Stefano G., Renna L., Brandizzi F., Weber A.P. (2012). Dynamic remodeling of the plastid envelope membranes - A tool for chloroplast envelope in vivo localizations. Front. Plant Sci. 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon E.B., Hall S.E., Ripp K.G., Ganzke T.S., Hitz W.D., Coughlan S.J. (2003). Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat. Biotechnol. 21: 1082–1087. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Sattler S., Maeda H., Sakuragi Y., Bryant D.A., DellaPenna D. (2003). Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15: 2343–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Collakova E., DellaPenna D. (2001). Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 127: 1113–1124. [PMC free article] [PubMed] [Google Scholar]
- Collakova E., DellaPenna D. (2003). The role of homogentisate phytyltransferase and other tocopherol pathway enzymes in the regulation of tocopherol synthesis during abiotic stress. Plant Physiol. 133: 930–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P. (2007). Functional diversity of tocochromanols in plants. Planta 225: 269–276. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8: 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Salvi D., Riviere-Rolland H., Vermat T., Seigneurin-Berny D., Grunwald D., Garin J., Joyard J., Rolland N. (2002). Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc. Natl. Acad. Sci. USA 99: 11487–11492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick A.H., Bhandari J., Crowell D.N. (2011). Farnesol kinase is involved in farnesol metabolism, ABA signaling and flower development in Arabidopsis. Plant J. 66: 1078–1088. [DOI] [PubMed] [Google Scholar]
- Gasulla F., Vom Dorp K., Dombrink I., Zähringer U., Gisch N., Dörmann P., Bartels D. (2013). The role of lipid metabolism in the acquisition of desiccation tolerance in Craterostigma plantagineum: a comparative approach. Plant J. 75: 726–741. [DOI] [PubMed] [Google Scholar]
- Grefen C., Donald N., Hashimoto K., Kudla J., Schumacher K., Blatt M.R. (2010). A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 64: 355–365. [DOI] [PubMed] [Google Scholar]
- Havaux M., Eymery F., Porfirova S., Rey P., Dörmann P. (2005). Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17: 3451–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S.C., Cahoon E.B. (2007). Enhancing vitamin E in oilseeds: unraveling tocopherol and tocotrienol biosynthesis. Lipids 42: 97–108. [DOI] [PubMed] [Google Scholar]
- Ischebeck T., Zbierzak A.M., Kanwischer M., Dörmann P. (2006). A salvage pathway for phytol metabolism in Arabidopsis. J. Biol. Chem. 281: 2470–2477. [DOI] [PubMed] [Google Scholar]
- Joo C.N., Park C.E., Kramer J.K., Kates M. (1973). Synthesis and acid hydrolysis of monophosphate and pyrophosphate esters of phytanol and phytol. Can. J. Biochem. 51: 1527–1536. [DOI] [PubMed] [Google Scholar]
- Keller Y., Bouvier F., d’Harlingue A., Camara B. (1998). Metabolic compartmentation of plastid prenyllipid biosynthesis--evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur. J. Biochem. 251: 413–417. [DOI] [PubMed] [Google Scholar]
- Kleinig H., Liedvogel B. (1978). Fatty acid synthesis by isolated chromoplasts from the daffodil. [14C]Acetate incorporation and distribution of labelled acids. Eur. J. Biochem. 83: 499–505. [DOI] [PubMed] [Google Scholar]
- Larson T.R., Graham I.A. (2001). A novel technique for the sensitive quantifcation of acyl CoA esters from plant tissues. Plant J. 25: 115–125. [DOI] [PubMed] [Google Scholar]
- Löbbecke L., Cevc G. (1995). Effects of short-chain alcohols on the phase behavior and interdigitation of phosphatidylcholine bilayer membranes. Biochim. Biophys. Acta 1237: 59–69. [DOI] [PubMed] [Google Scholar]
- Lippold F., vom Dorp K., Abraham M., Hölzl G., Wewer V., Yilmaz J.L., Lager I., Montandon C., Besagni C., Kessler F., Stymne S., Dörmann P. (2012). Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D.V., Chen M., Cahoon E.B. (2009). Lipid signaling in Arabidopsis: no sphingosine? No problem! Trends Plant Sci. 14: 463–466. [DOI] [PubMed] [Google Scholar]
- Maeda H., DellaPenna D. (2007). Tocopherol functions in photosynthetic organisms. Curr. Opin. Plant Biol. 10: 260–265. [DOI] [PubMed] [Google Scholar]
- Maeda H., Song W., Sage T.L., DellaPenna D. (2006). Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell 18: 2710–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mène-Saffrané L., DellaPenna D. (2010). Biosynthesis, regulation and functions of tocochromanols in plants. Plant Physiol. Biochem. 48: 301–309. [DOI] [PubMed] [Google Scholar]
- Mène-Saffrané L., Jones A.D., DellaPenna D. (2010). Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed dessication and quiescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 17815–17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497. [Google Scholar]
- Overbeek R., et al. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33: 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porfirova S., Bergmüller E., Tropf S., Lemke R., Dörmann P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl. Acad. Sci. USA 99: 12495–12500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Thompson W.A., Kriedemann P.E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975: 384–394. [Google Scholar]
- Pruzinská A., Tanner G., Anders I., Roca M., Hörtensteiner S. (2003). Chlorophyll breakdown: pheophorbide a oxygenase is a Rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 100: 15259–15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A., Tanner G., Aubry S., Anders I., Moser S., Müller T., Ongania K.-H., Kräutler B., Youn J.-Y., Liljegren S.J., Hörtensteiner S. (2005). Chlorophyll breakdown in senescent Arabidopsis leaves. Characterization of chlorophyll catabolites and of chlorophyll catabolic enzymes involved in the degreening reaction. Plant Physiol. 139: 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler S.E., Gilliland L.U., Magallanes-Lundback M., Pollard M., DellaPenna D. (2004). Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16: 1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge B., Weiss J.D., Wong Y.-H.H., Lassner M.W., Mitsky T.A., Shewmaker C.K., Post-Beittenmiller D., Valentin H.E. (2002). Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol. 129: 321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S., Aubry S., Burla B., Agne B., Kessler F., Krupinska K., Hörtensteiner S. (2009). Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver S.M., et al. (2014). High-throughput comparison, functional annotation, and metabolic modeling of plant genomes using the PlantSEED resource. Proc. Natl. Acad. Sci. USA 111: 9645–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll J., Schultz G., Rüdiger W., Benz J. (1983). Hydrogenation of geranylgeraniol : two pathways exist in spinach chloroplasts. Plant Physiol. 71: 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin H.E., Lincoln K., Moshiri F., Jensen P.K., Qi Q., Venkatesh T.V., Karunanandaa B., Baszis S.R., Norris S.R., Savidge B., Gruys K.J., Last R.L. (2006). The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 18: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti R., Li W., Li M., Sang Y., Biesiada H., Zhou H.-E., Rajashekar C.B., Williams T.D., Wang X. (2002). Profiling membrane lipids in plant stress responses. Role of phospholipase D alpha in freezing-induced lipid changes in Arabidopsis. J. Biol. Chem. 277: 31994–32002. [DOI] [PubMed] [Google Scholar]
- Yang W., Cahoon R.E., Hunter S.C., Zhang C., Han J., Borgschulte T., Cahoon E.B. (2011). Vitamin E biosynthesis: functional characterization of the monocot homogentisate geranylgeranyl transferase. Plant J. 65: 206–217. [DOI] [PubMed] [Google Scholar]
- Zbierzak A.M., Kanwischer M., Wille C., Vidi P.A., Giavalisco P., Lohmann A., Briesen I., Porfirova S., Bréhélin C., Kessler F., Dörmann P. (2010). Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem. J. 425: 389–399. [DOI] [PubMed] [Google Scholar]
- Zhang W., Liu T., Ren G., Hörtensteiner S., Zhou Y., Cahoon E.B., Zhang C. (2014). Chlorophyll degradation: the tocopherol biosynthesis-related phytol hydrolase in Arabidopsis seeds is still missing. Plant Physiol. 166: 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.