Transcriptome profiling of Chlamydomonas shows that major waves of transcript accumulation in partially light/dark-synchronized cell cultures are under differential regulation by the kinases CDKA and CDKB.
Abstract
We analyzed global transcriptome changes during synchronized cell division in the green alga Chlamydomonas reinhardtii. The Chlamydomonas cell cycle consists of a long G1 phase, followed by an S/M phase with multiple rapid, alternating rounds of DNA replication and segregation. We found that the S/M period is associated with strong induction of ∼2300 genes, many with conserved roles in DNA replication or cell division. Other genes, including many involved in photosynthesis, are reciprocally downregulated in S/M, suggesting a gene expression split correlating with the temporal separation between G1 and S/M. The Chlamydomonas cell cycle is synchronized by light-dark cycles, so in principle, these transcriptional changes could be directly responsive to light or to metabolic cues. Alternatively, cell-cycle-periodic transcription may be directly regulated by cyclin-dependent kinases. To distinguish between these possibilities, we analyzed transcriptional profiles of mutants in the kinases CDKA and CDKB, as well as other mutants with distinct cell cycle blocks. Initial cell-cycle-periodic expression changes are largely CDK independent, but later regulation (induction and repression) is under differential control by CDKA and CDKB. Deviation from the wild-type transcriptional program in diverse cell cycle mutants will be an informative phenotype for further characterization of the Chlamydomonas cell cycle.
INTRODUCTION
Oscillating levels of activity of cyclin-dependent kinases (CDKs) are central to regulation of the eukaryotic cell cycle (Morgan, 2007). Additionally, regulated transcription is an important aspect of cell cycle control in many organisms (Menges et al., 2002; Bertoli et al., 2013). Many genes encoding components of the central oscillator, including CDKs and cyclins themselves, are transcriptionally regulated through the cell cycle. Specific transcription factors may be targets of CDK-dependent control, closing the regulatory circuit (Wittenberg and Reed, 2005).
Examples of the connection between CDK activity and regulated transcription in budding yeast include cyclin-dependent positive feedback at both the G1-S (Skotheim et al., 2008) and S-M (Reynolds et al., 2003) transitions, and blockage of the mitotic exit transcriptional program by CDK-dependent inhibition of transcription factor nuclear import (Moll et al., 1991). In animals, the G1-S transition is regulated by CDK-dependent phosphorylation of the transcriptional repressor Rb (Henley and Dick, 2012). Plants encode homologous Rb-related (RBR) proteins, which play diverse roles during growth and development (Gutzat et al., 2012), probably involving transcriptional regulation (Uemukai et al., 2005; Sabelli et al., 2013). The regulatory circuits controlling expression of cell-cycle-regulatory genes simultaneously control many other genes, so that broad transcriptional programs come under CDK control.
In contrast to cell-cycle-intrinsic regulation, cell cycle control genes are frequently regulated exogenously. For example, cyclin D transcription in animals and plants is induced by growth factors (Sherr, 1993; Riou-Khamlichi et al., 1999); transcription of the CDK-activating phosphatase string/CDC25 is tightly regulated by developmental signals in Drosophila melanogaster embryos (Edgar et al., 1994), and many environmental cell-cycle-inhibitory stimuli induce cyclin-dependent kinase inhibitors such as p27 (Sherr and Roberts, 1999). Furthermore, it has been suggested that much of the broad cell-cycle-periodic gene expression program in budding yeast will cycle in the absence of CDK activity (Orlando et al., 2008), perhaps due to cyclical activation of a transcription factor cascade.
Photosynthetic organisms exhibit very strong transcriptional responses to light (Jiao et al., 2007) and a strong diurnal/circadian transcriptional program (Schaffer et al., 2001; Michael and McClung, 2003). In many organisms, cell division is also entrained to the diurnal cycle (Goto and Johnson, 1995; Moulager et al., 2007). This provides an interesting context in which to examine the interrelationship between exogenous controls and CDK activity in periodic gene expression.
In the green alga Chlamydomonas reinhardtii, many cell-cycle-regulatory genes are tightly regulated through a diurnal cycle, and cell division itself is suppressed during the day, when cells grow many-fold, with rapid division cycles occurring at night (Bisova et al., 2005).
Previously, we isolated temperature-sensitive mutants in the two cell-cycle-regulatory cyclin-dependent kinases in Chlamydomonas (Tulin and Cross, 2014): CDKA, the closest sequelog to yeast and animal CDK1/CDC2, and CDKB, a CDK found in all members of the plant superkingdom but absent elsewhere (Vandepoele et al., 2002). Analysis of these mutants suggested strong regulatory divergence between them, with CDKA primarily acting early to promote cell cycle initiation. In contrast, CDKB acts later, specifically promoting nuclear division and mitosis (Tulin and Cross, 2014). Here, we characterize the diurnal and cell cycle transcriptional regulatory program in Chlamydomonas and its regulation by CDKA and CDKB.
RESULTS
Growth and Sampling of Liquid Cultures
The mitotic cell cycle in Chlamydomonas, grown under favorable conditions, is characterized by a long G1 phase, during which cell size can increase greatly. This period of growth is followed by rapidly alternating rounds of S phase and mitosis (S/M), generating up to 16 daughter cells. In light-dark synchronized populations, G1 occupies most of the light phase and the S/M program is restricted to the dark (Figure 1A). See Cross and Umen (2015) for a detailed review of the Chlamydomonas cell cycle.
Figure 1.
Culturing and Sampling Wild-Type and cdk Mutant Cells.
Parallel cultures of wild type, cdka-1, and cdkb-1 were transferred to 33°C (restrictive temperature) at time zero.
(A) Schematic of the Chlamydomonas mitotic cell cycle in a light/dark-synchronized culture.
(B) Cell number and density by Coulter counter.
(C) DNA content by FACS. Wild-type cells replicate DNA between 14 and 16 h, as indicated by an increase in ploidy. Release of daughter cells is indicated by a return to 1C DNA content (arrow, newborn). cdka-1 cells have not initiated DNA replication by 18 h. cdkb-1 cells initiate replication at 16 h and arrest after one round of DNA replication (2C).
(D) Correlations of independent wild-type cDNA libraries from the same or different time points. R2 = variation explained by linear fit through the origin.
We analyzed global transcriptomes through a single diurnal cycle in the wild type compared with parallel cdka-1 and cdkb-1 cultures. Cultures were maintained on a 14:10 light/dark cycle at 21°C and shifted to 33°C (permissive and restrictive temperatures, respectively, for the cdk mutants) at the beginning of the 14-h light period for the experiment (wild-type cultures received the same temperature treatments). Samples were collected for determination of cell density and volume (Coulter counter), DNA content (flow cytometry; fluorescence-activated cell sorting [FACS]), and RNA isolation. We also purified CDK kinase and measured enzymatic activity (Cks1-purified kinase) as described previously (Tulin and Cross, 2014).
Wild-type cells grew from an initial size of 100 to 200 fl to a predivision size of ∼600 fl at 14 h (Figure 1B). Cks1-purified kinase activity began to rise at ∼10 h and was declining by 16 h (Supplemental Figure 1). Between 14 and 16 h, DNA replication is evident from accumulation of high-ploidy cells by FACS (Figure 1C; Supplemental Figure 1), representing S/M cells that are undergoing multiple rounds of division before release of daughter cells (Figure 1A). Daughter cell release (hatching) occurs between 16 and 18 h, indicated by a sharp increase in cell number and a coincident drop in median cell volume (Figure 1B) with a return to a G1 DNA content (newborn, Figure 1C; note that the FACS peak for newborn cells is shifted slightly to the left compared with G1 cells just prior to replication, probably due to accumulation of chloroplast DNA in G1 cells). Synchrony is imperfect, as reflected by co-occurrence of cells in different stages in later time points.
Both cdka-1 and cdkb-1 mutant cells showed a near-wild-type increase in cell volume during the first 14 h at the restrictive temperature (Figure 1C). They failed to undergo cell division, as shown by constant cell number and sustained large cell volume after 14 h. cdka-1 mutant cells failed to replicate DNA during the course of the experiment, whereas cdkb-1 cells initiated DNA replication with a slight delay compared with the wild type, but arrested after one replication (2C). The multimodal FACS profile at early time points in cdka-1 (Figure 1C) reflects delayed completion of the preceding mitosis, caused by partial penetrance of the cdka-1 mutation at the permissive temperature. As reported previously (Tulin and Cross, 2014), Cks1 kinase activity rose with a moderate delay in cdkb-1 cells, but failed to decline significantly by the end of the experiment (Supplemental Figure 1B). Accumulation of kinase activity in cdka-1 cells was strongly reduced.
We performed three experiments using wild-type culture with somewhat different time points chosen; in two experiments, cdka-1 was analyzed in parallel, and in one of these, cdkb-1 was analyzed. We prepared RNA samples and generated Illumina cDNA libraries from polyadenylated mRNA. Using TopHat (Kim et al., 2013), we mapped ∼90% of the reads (6 to 12 million per library) to unique Chlamydomonas gene models (Phytozome v.5.5 gene models). Raw read counts were obtained by HTSeq (Anders et al., 2014; Supplemental Data Set 1). Normalized read counts (fragments per kilobase of exon per million fragments mapped) from independent biological replicates (same or adjacent time points and same genotype, whether from the same or different experiments) were highly correlated (R2 > 90% in all cases). Lower correlations were observed between libraries prepared from mid-G1 compared with S/M (Figure 1D), whether from the same or different experiments, indicating global changes in transcriptome between these times. A high correlation of adjacent time points means that there are effectively multiple biological replicates for all samples. Conclusions below are all supported by multiple time points and samples.
Differential Gene Expression between Mid-G1 and S/M
To focus on genes regulated specifically in relation to the cell cycle, we initially tested for differentially expressed genes, comparing mid-G1 (5 to 7 h) and the peak of S/M (14 to 16 h). The S/M period should be enriched for genes associated with the division phase; we chose the mid-G1 time to reduce the contribution of genes specifically responding to the dark-light shift or the temperature shift. The mid-G1 point is 3 to 5 h before detectable DNA replication and a similar time before induction of Cdk kinase activity (Supplemental Figure 1). More than 90% of mid-G1 cells were elongated with two beating cilia. The 14- to 16-h S/M population marks the peak time of cell division. Cells were morphologically heterogeneous, including large predivision cells, cells undergoing cleavage and septation, and recently divided daughter cells (Supplemental Figure 1C). Cellular DNA content was also heterogeneous, but DNA replication was clearly ongoing in many of these cells.
Between mid-G1 and S/M, 3834 genes were differentially expressed: 2342 upregulated in S/M and 1492 downregulated (Supplemental Data Set 2) (false discovery rate of 0.001 using the edgeR package; Robinson et al., 2010) (see Methods). On average, upregulation in S/M was associated with much greater fold changes (Figure 2).
Figure 2.
Differentially Expressed Genes.
Wild-type cells were samples in mid-G1 (t = 5 or 7 h) and during the S/M phase (t = 16 h) in three independent experiments.
(A) Differentially expressed genes were calculated by edgeR and plotted as log(fold-change) versus log(counts per million). Positive log(fold-change) indicates differential upregulation during S/M, and negative log(fold-change) indicates differential upregulation in mid-G1. Horizontal black bars represent 4-fold up- or downregulation.
(B) Distribution of functional classes among differentially expressed genes, nondifferential genes, and all genes (17,741 gene models). Genes implicated in photosynthesis and metabolism are overrepresented in mid-G1. Genes implicated in cell cycle, tubulin/cilia, and cell wall are overrepresented in S/M.
Gene annotation (Supplemental Data Set 3) showed a clear distinction between genes implicated in photosynthesis and translation (high in mid-G1) and genes annotated in the cell cycle, basal-body/cilia/microtubule (BB-cilia-MT), and cell wall functional classes (upregulated during S/M) (Figure 2B). This striking stratification of functional classes likely reflects the different physiological requirements associated with photosynthesis-dependent cell growth (G1) versus cell division (S/M).
Global Differences between the Wild Type and cdk Mutants during the S/M Phase
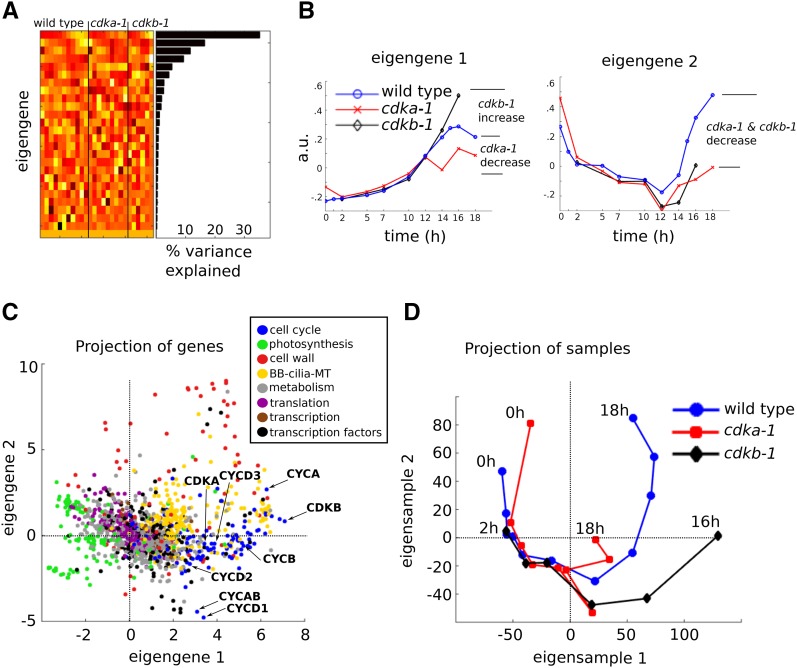
We used principal component analysis (PCA; Methods) to explore the global differences between wild-type and cdk mutant transcript accumulation patterns (Alter et al., 2000). PCA performed on the full data set, containing normalized read count data from both wild-type and cdk mutant samples, generated 26 principal components: uncorrelated vectors that independently explain variation among the genes (eigengenes). The first two eigengenes together explained ∼50% of the gene expression variability in the data set (Figure 3), and both described CDK-dependent effects at late time points (after 12 h), coinciding with S/M. Eigengene 1 indicated opposing effects of the cdka-1 and cdkb-1 mutations relative to the wild type during S/M, whereas eigengene 2 indicated similar effects of both cdk mutants (Figure 3B). Notably, both eigengenes indicated similar expression in all three strains (wild type, cdka-1, and cdkb-1) at early time points (0 to 10 h) after lights on, coinciding with the G1 growth phase of the cell cycle.
Figure 3.
Principal Component Analysis.
Normalized read counts from wild-type, cdka-1, and cdkb-1 cDNA libraries were analyzed by PCA.
(A) Heat map display of the principal components, eigengenes, arranged as rows, and the corresponding percentage of the variance explained by each eigengene.
(B) Line plots of the first two eigengenes.
(C) Projection of the genes on the 2D space spanned by eigengenes 1 and 2. Only genes that were assigned to one of eight functional categories are shown. Cyclins and CDKs of particular interest are marked.
(D) Projections of the samples on the 2D space spanned by eigensamples 1 and 2.
A projection of functionally classified genes onto the subspace spanned by the first two eigengenes (Figure 3C) revealed a gene expression split between photosynthesis genes (large negative component along eigengene 1) versus cell cycle and BB-cilia-MT genes (large positive component along eigengene 1). It also highlighted genes with a putative role in cell wall biosynthesis with large positive components along eigengene 2.
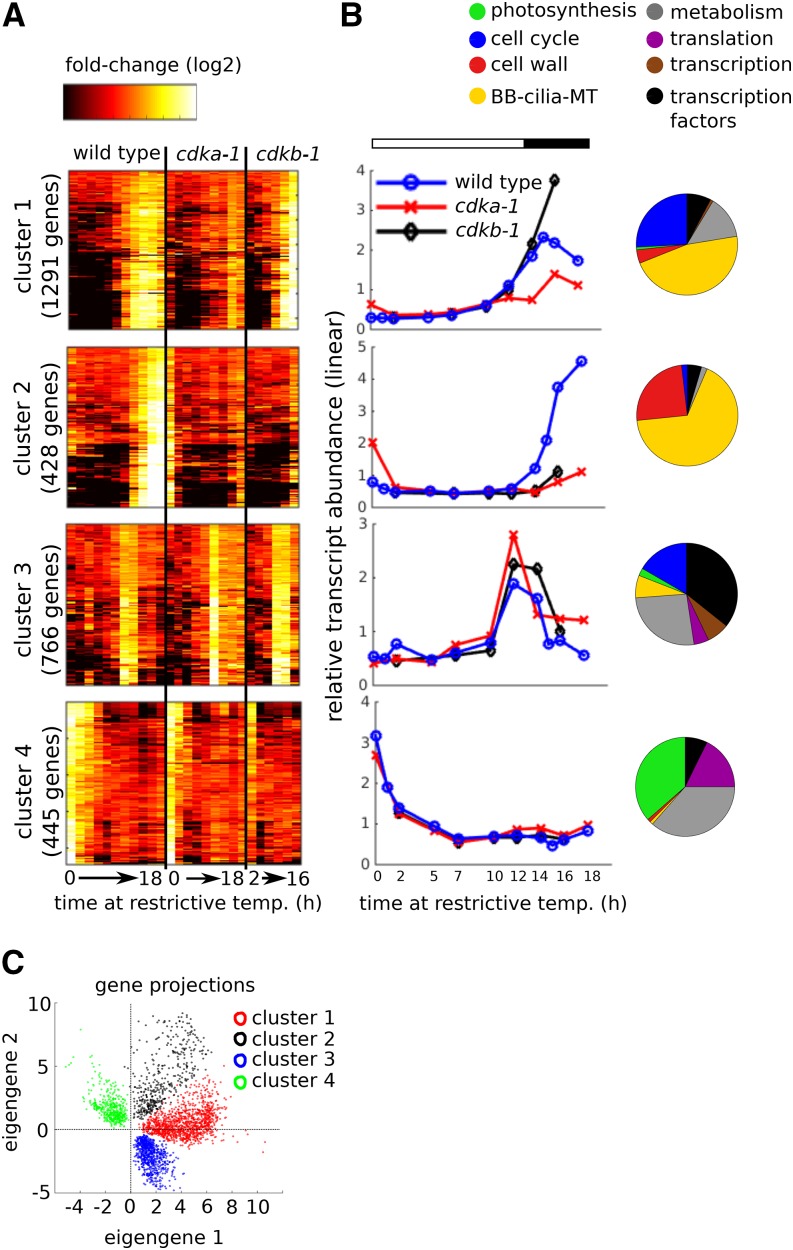
The gene projection plot suggested that the magnitudes of eigengenes 1 and 2 contribution may be used as a starting point to organize the gene expression data. Indeed, inspection of the genes with the largest positive and negative components along eigengenes 1 and 2 suggested four distinct gene expression patterns that characterized most of the variably expressed genes (Supplemental Figure 2). We chose a representative gene from each pattern as a query to search the data set for similar genes using a 0.8 correlation coefficient as a somewhat arbitrary threshold to score similarity. This procedure generated four clusters with distinct transcript accumulation patterns (Figure 4), which occupied distinct regions in the eigengene 1/2 space (Figure 4B).
Figure 4.
CDK-Dependent and CDK-Independent Transcriptional Programs.
(A) Clusters 1 through 4 represent major patterns of transcript accumulation during one cell cycle. Rows = genes; columns = samples. Samples are arranged from early to late for each genotype (wild type: 0, 1, 2, 5, 7, 10, 12, 14, 15, 16, and 18 h; cdka-1: 0, 2, 5, 7, 10, 12, 14, 16, and 18 h; cdkb-1: 2, 7, 10, 12, 14, and 16 h). The genes were clustered hierarchically within each cluster for improved visualization.
(B) Average relative transcript abundance for each cluster plotted on a linear scale. Pie charts indicate distribution of functional classes within each cluster.
(C) Projection of the genes in the four clusters onto the eigengene 1/2 subspace.
Cluster 1 (1291 genes) contained genes that increased from a low level in wild-type and cdkb-1 mutant cells around the 12-h time point, with a clearly reduced accumulation in cdka-1. Although there is no guarantee that eigengenes will directly resemble any actual genes in the data set, the cluster 1 pattern of expression in wild type and cdk mutants was strikingly similar to eigengene 1. Cluster 2 (428 genes) contained genes that increased more sharply and a few hours later in wild-type cells. Cluster 2 transcripts remained low both cdka-1 and cdkb-1 mutant cells. Cluster 3 (766 genes) showed a transient peak in transcript abundance at 12 to 14 h, and cluster 4 (445 genes) showed an initial decrease during the first 5 h of the light period. Clusters 3 and 4 were essentially CDK independent.
A majority of genes in clusters 1 (64%) and 2 (67%) were detected by differential expression analysis as upregulated during S/M, relative to mid-G1 in wild-type cells (Figure 2A). Only 11% of genes in cluster 3 were scored as differentially upregulated due to the specific time points used for differential expression analysis. See Supplemental Data Set 4 for a complete listing of genes in clusters 1 through 4.
As well as eigengenes, PCA also computes eigensamples, which are uncorrelated vectors explaining variation between samples. Plotting the contribution of eigensamples 1 and 2 to successive time points in the data set (Figure 3D) revealed a nearly circular orbit for the wild-type time course, consistent with nearly complete return of the cells to the starting condition by the end of the experiment. Initially, the cdka-1 and cdkb-1 time courses on the same plot were very similar to the wild type, but diverged strongly from the wild type and from each other starting at ∼10 h, consistent with the analysis of eigengenes 1 and 2 above. These divergences are highly statistically significant (Supplemental Figure 3), implying a strong differential contribution of CDKA and CDKB to global transcriptional regulation, starting in mid-diurnal cycle.
Substructure and Differential CDK Regulation of the Mitotic Transcriptional Program
Clusters 1 and 2 (∼1300 and ∼400 genes, respectively) together contain ∼10% of the total genes in the genome. Therefore, these clusters represent a major transcriptional program specific to cell division. Cluster 1 turns on approximately concomitant with DNA replication (Figure 1) and CDK activation (Supplemental Figure 1) and is enriched for genes involved in DNA replication and mitotic processes such as spindle assembly. CDKA is required for full activation of cluster 1. In contrast, CDKB is not required for cluster 1 induction but may be required for its later shutoff.
Cluster 2 turns on a few hours after cluster 1 and shows strong dependence on both CDKA and CDKB. This cluster is heavily enriched in genes with a putative role in cell wall biosynthesis (annotated as pherophorin or hydroxyproline-rich on phytozome.org).
BB-cilia-MT genes were enriched in both clusters 1 and 2. However, further analysis demonstrated a functional split in these genes between the two clusters. A large majority of BB-cilia-MT genes in cluster 2 (61/73, 83%) were previously shown to encode components of the flagellar proteome (Pazour et al., 2005), compared with 9/54 genes (17%) in cluster 1. Instead, the BB-cilia-MT genes in cluster 1 mostly coded for tubulins, microtubule-directed motors (kinesins), and structural components of the basal body/centriole complex (POC genes; Keller et. al., 2005). Indeed, several of these proteins, such as POC1 (Keller et al., 2009), VFL1 (Silflow et al., 2001), and VFL3 (Esparza et al., 2013), are known to be required for basal body assembly and integrity.
We tested for association between upregulation following experimental deflagellation (Albee et al., 2013) and cluster membership of BB-cilia-MT genes. Of the BB-cilia-MT genes in cluster 2, 67/73 (92%) were upregulated after deflagellation, compared with 16/54 (30%) for cluster 1 (P < 0.001).
Thus, transcript accumulation during S/M may be separated into two programs: early and late. The early program requires CDKA, but not CDKB, for full induction, whereas the late program depends on both CDKA and CDKB. Biologically, the early program is likely associated with S phase and mitosis. Indeed, 14/38 DIV genes, with confirmed essential functions in DNA replication and nuclear division (Tulin and Cross, 2014) were in cluster 1, and none were in cluster 2. In contrast, cluster 2 may be associated with later events, such as cell wall assembly and cilia regrowth following nuclear division.
Our results are consistent with published results (Wood et al., 2012; Panchy et al., 2014) showing late expression of ciliogenesis genes compared with other cell-division-related genes. In our data, sorting by time of expression in the wild-type cell cycle allows some separation of these two classes, but with substantial overlap such that many genes cannot be unambiguously associated with early versus late expression. The cluster-based classification derived from PCA analysis of the complete data set, incorporating the cdka and cdkb mutants, greatly sharpens the split because CDKB is required for expression of late, but not early, genes, resulting in a near-digital classification. This observation indicates the value of incorporating expression in mutant backgrounds for understanding gene expression programs, especially since independent functional information supports the vector-based classifications.
CDK-Independent Transcript Accumulation
Genes in cluster 3 (766 genes) exhibited a transient increase in transcript abundance between 12 and 14 h that was similar in wild-type, cdka-1, and cdkb-1 mutant cells. Thus, cluster 3 apparently represents a CDK-independent transcriptional program that is activated nearly coincident with the G1-to-S/M transition. This indicates that the cdka-1 and cdkb-1 mutants are transcriptionally competent at the time when these mutations prevent or delay robust activation of cluster 1. CDK-independent regulation of cluster 3 genes was further supported by the cdka-1 cdkb-1 double mutant. This mutant has undetectable Cks1-purified kinase activity at the restrictive temperature and arrests permanently in G1 (Tulin and Cross, 2014). Nevertheless, the cdka-1 cdkb-1 double mutant accumulated cluster 3 transcripts with near-wild-type timing (Supplemental Figure 4).
Cluster 4 contained transcripts that decreased from an initially high level over the first 5 h. We found a strong enrichment of genes implicated in plastid ribosome function in this cluster, consistent with a previous study reporting timing of expression of plastid ribosomal genes (Kucho et al., 2005).
Confirmation with Plate Synchronization
Previously, we reported that cell cycle synchrony in wild-type cells could be markedly increased by growing cells on agar plates rather than in liquid cultures (Tulin and Cross, 2014). This procedure was critical for analyzing anaphase-promoting complex mutants, which had nonspecific growth defects in liquid culture that obscured their specific cell cycle phenotypes (Tulin and Cross, 2014). In this protocol, the cells are precultured on plates for 2 d with 14/10 h light/dark, with transfer to high temperature and continuous light at the beginning of the third light period. Overall cell cycle progression was highly synchronous and temporally advanced in this protocol, with wild-type cells completing cell division at ∼11 h after the temperature shift (compared with 14 to 16 h for liquid-grown cells).
We used this protocol to confirm the above conclusions regarding cdka-1 and cdkb-1. We also included the div44-1/bsl1-1 mutant for comparison. BSL1 is a Kelch-domain-containing phosphatase that is universal in the green plant lineage. The BSL family has key roles in brassinosteroid signaling and an unidentified essential role in higher plants (Maselli et al., 2014). We found that BSL1 was essential for the mitotic cell cycle in Chlamydomonas (Tulin and Cross, 2014), and the bsl1-1 cellular phenotype was similar to that of cdkb-1. Because we hoped that differential transcriptional response to absence of CDKB or BSL1 might clarify the extent to which they act in the same cellular pathways, we addressed this issue next.
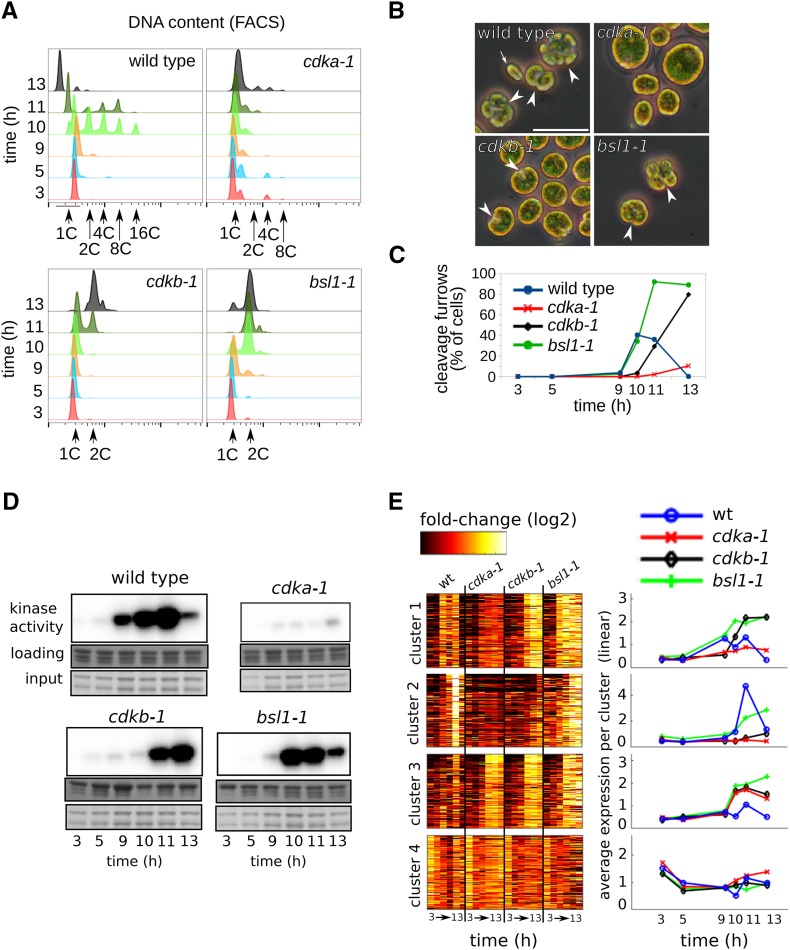
The wild type, cdka-1, cdkb-1, and bsl1-1 were grown to low density on agar plates in 14:10 light-dark, at 21°C, and shifted to 33°C at time zero (lights on). Samples were collected at 3, 5, 9, 10, 11, and 13 h following illumination. Most wild-type cells completed S/M between 9 and 11 h (accumulation of high-ploidy [2-16C] cells) and had released daughters by 13 h, as evidenced by the disappearance of high-ploidy cells and appearance of a left-shifted ‘newborn’ peak in DNA flow cytometry (Figure 5). Progression through S/M was accompanied by the accumulation of cells with clearly visible cleavage structures (Figure 5B), and a sharp rise and fall in Cks1-purified kinase activity (Figure 5D). At the 10 h time point ∼80% of the cells were in S/M (with 2C or higher DNA content), indicating a high degree of cell cycle synchrony.
Figure 5.
Cell Cycle Analysis of Plate-Grown Wild-Type and Mutant Cells.
Wild-type, cdka-1, cdkb-1, and bsl1-1 mutant cells were grown in parallel on agar plates and shifted to restrictive temperature (33°C) at time zero.
(A) DNA content as determined by FACS. Wild-type cells go through S/M phase at 10 to 11 h, as determined by accumulation of cells with 2, 4, 8, and 16C DNA content. Newborn cells emerge at 13 h with 1C DNA. cdka-1 fails to replicate during the course of the experiment. cdkb-1 initiate DNA replication ∼1 h after the wild type and arrest with once-replicated 2C DNA. The bsl1-1 mutant initiates DNA replication with normal timing and arrests with 2C DNA.
(B) Representative micrographs of cells at the 11-h time point. White arrowheads, cleavage furrow; white arrow, newborn wild-type cell. Bar = 25 μm.
(C) Quantification of cleavage furrow development.
(D) Cks1-associated kinase activity. The sample for cdka-1 at 3 h was lost.
(E) Transcript abundance (relative to mean for each gene) is shown on a log2 scale for clusters 1 through 4 (left panel). Average transcript abundance for each cluster and genotype is shown on a linear scale (right panel).
cdka-1 cells were drastically delayed in entry into cell division, indicated by failure to replicate DNA, turn on Cks1-purified kinase activity, or develop cleavage furrows. In contrast, cdkb-1 cells developed a single notch structure (unlike the many cleavage planes in wild-type cells); they completed only a single round of DNA replication, arresting with 2C DNA content, and activated Cks1-purified kinase activity to wild-type levels or above. The sharp decline in Cks1-purified kinase observed in the wild type did not occur in cdkb-1 cells.
These results with cdka-1 and cdkb-1 cells confirm previous findings that showed distinct regulatory roles of CDKA and CDKB (Tulin and Cross, 2014; see above). The improved plate synchrony allowed clear detection of a moderate delay in cdkb-1 cells both for induction of Cks1-purified kinase and for initiation of DNA replication (Figure 5): A majority of wild-type cells have completed at least one round of DNA replication between 9 and 10 h, but cdkb-1 cells show significant DNA replication between 10 and 11 h. This difference, while modest relative to the resolution of the time points we took, is reproducible in multiple experiments.
Analysis of transcriptome data from this protocol (Supplemental Data Set 5), comparing the wild type and the cdk mutants, confirmed the conclusions reached from liquid synchrony experiments (Figure 5E). In particular, CDKA was required for efficient activation of both clusters 1 and 2. In contrast, CDKB was only required for activation of cluster 2. Also confirming results using liquid cultures, the cdkb-1 mutant arrested with high levels of cluster 1 transcripts, indicating that CDKB may be needed for turning off expression of cluster 1 genes.
Correlated with the moderate delay in activation of DNA replication and Cks1-purified kinase in cdkb-1 in the plate protocol noted above, cluster 1 transcriptional activation appeared to be delayed as well, although the time resolution before 9 h is limited. We could reproduce this delay between the wild type and cdkb-1 by RT-qPCR in an independent plate growth time course, by looking at two genes strongly induced on S/M entry: CYCB and CDKB (Supplemental Figure 5). These two genes reproduced the general behavior of cluster 1: delayed induction in cdkb-1 relative to the wild type and sustained high levels through the end of the time course. Furthermore, the cell wall gene GAS28 (Hoffmann and Beck, 2005; representing cluster 2) failed to turn on in cdkb-1 when assayed by RT-qPCR (Supplemental Figure 5). Thus, the overall picture of CDK-dependent and -independent transcriptional control is very similar with this new data set for clusters 1 and 2.
In contrast, cluster 3 genes showed a qualitatively different behavior when assayed in the plate protocol. Although strong activation of cluster 3 genes occurred in both cdk mutants and in bsl1-1, we detected only a small increase in abundance in the wild type. The reason for this discrepancy is currently unknown.
Analysis of div and gex Mutants
We previously isolated mutants with temperature-sensitive mutations in genes required for cell division in Chlamydomonas. Characterization of these mutants indicated that one class (the G1 exit GEX genes) are likely required during G1 to effect the transition to S/M, whereas the DIV genes play diverse and mutant-specific roles once the S/M program is underway. We were interested to learn if transcriptional profiles of arrested mutants had any relationship to their point of cytological cell cycle arrest.
We sequenced cDNA from two gex and eight div mutants at their arrest points, after 14 to 16 h (Supplemental Data Set 1) growth in liquid cultures at the restrictive temperature (a time by which parallel wild-type cultures were in the last stages of the division cycle) and compared the average relative transcript abundances of the clusters established above. Quantification of DNA content (Supplemental Figure 6) indicated that the gex mutants arrested with unreplicated DNA as described previously. The div mutants were heterogeneous with respect to DNA replication: some arrested with essentially unreplicated DNA (div20-1 and div17-1), some with extensively rereplicated DNA (div24-1 and div43-1), and some with ∼2C DNA content (div13-1 and bsl1-1).
The broad picture from transcriptome analysis of these mutants suggests that in the gex mutants, which fail to enter S/M, clusters 1 and 2 did not accumulate to above wild-type pre-S/M levels (Figure 6). In contrast, the div mutants displayed uniformly high induction (up to or above wild-type levels) of cluster 1 genes. The div mutants represent several distinct S/M defects, including DNA replication (div17-1) or spindle formation (div24-1) failure. This result suggests that successful completion of neither DNA replication nor nuclear division is required for cluster 1 activation once CDKA-dependent cell cycle entry has occurred. Cluster 2 showed strong activation to wild-type levels only in a subset of div mutants (div13-1/pol-alpha, div24-1/tfce, div42-1/sgt1, and div43-1/auroraB).
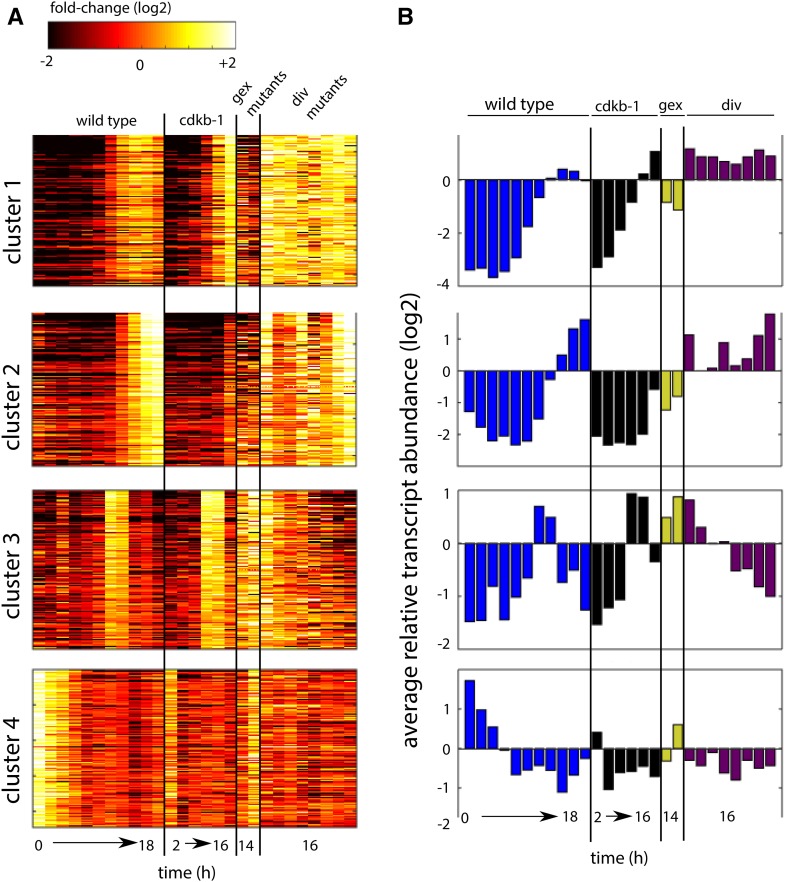
Figure 6.
Transcript Accumulation in div and gex Mutants.
(A) Transcript accumulation in the wild type and ckdb-1 (same time points as in Figure 4), the gex mutants (gex1-3 and gex26-1), and div mutants (div13-1, div20-1, div17-1, div24-1, div44-1, div39-1, div42-1, and div43-1). Genes (rows) in each cluster same as in Figure 4.
(B) Average transcript abundance for each strain and time point.
Interpretation of cluster 3 is complicated by the fact that it is only transiently activated in the wild type and that the mutants are each represented by a single (end point) sample. Nevertheless, we observed high levels of cluster 3 transcripts in both gex mutants, similar to peak wild-type levels. This result is consistent with the idea that cluster 3 is regulated independently of CDKA-dependent cell cycle entry, which is blocked by the gex1-3 and gex26-1 mutations. Finally, we noted low levels of cluster 4 transcripts in all arrested div and gex mutants except gex26-1, which carries a mutation in the rRNA processing factor NOP52. Cluster 4 contains many cytoplasmic and plastid translation-related genes, and we speculate that loss of NOP52 function may lead to reduced translational capacity, perhaps preventing efficient inhibition of genes involved in translation through a feedback mechanism.
The analysis above does not address the question of whether individual mutants additionally have specific transcriptional fingerprints. We think it is likely that such fingerprints do exist; however, these mutants were examined with only one or two time points each and that is not enough to clearly analyze a mutant-specific pattern. We address this question with div44-1/bsl1-1 below.
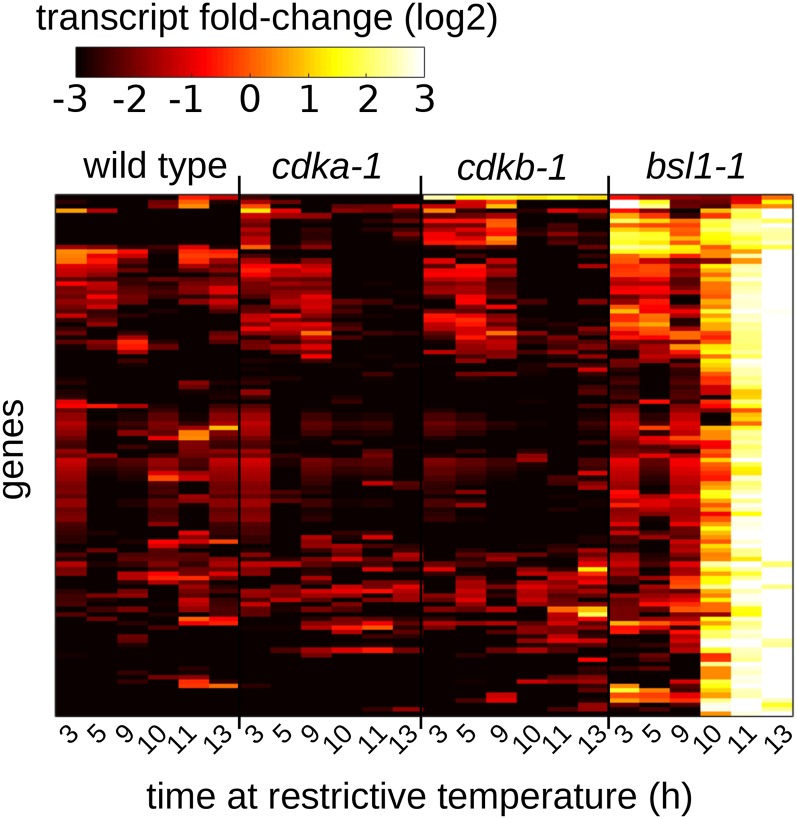
Strong Transcriptional Responses following Loss of BSL1
DIV44 is clearly homologous to the kelch-domain-containing phosphatase BSL1 found in land plants (Tulin and Cross, 2014). In Arabidopsis thaliana, BSL phosphatases are critical intermediates in the brassinosteroid pathway (Mora-García et al., 2004), and they also carry out an unidentified essential role (Maselli et al., 2014). In Chlamydomonas, DIV44 is required for mitosis but not for the first round of DNA replication (Tulin and Cross, 2014). We confirm this finding here (Figure 7) and further find that, unlike cdkb-1 cells, bsl1-1 cells initiate replication with no detectable delay. In bsl1-1 cells, the appearance of multiple cleavage planes, suggestive of cytokinetic progression, and the rise and fall of Cks1-purified kinase activity both occurred with timing similar to wild-type cells. Thus, BSL1 appears to be specifically required for the nuclear division cycle after the first round of replication, whereas CDKB is required more broadly for mitotic progression after the first S phase.
Figure 7.
Loss of BSL1 Is Associated with a Strong Transcriptional Response.
Transcript accumulation in wild-type, cdka-1, cdkb-1, and bsl1-1 cells at the indicated times following a shift to restrictive temperature. The cell cycle arrest in bsl1-1 at 11 to 13 h (see Figure 5) is associated with strong induction of 115 genes.
In bsl1-1 cells, early mitotic genes were induced to a high level (cluster 1, Figure 5). In addition, we identified a cluster of 115 genes that were strongly upregulated specifically in the bsl1-1 mutant concomitant with the cell cycle arrest (10 to 13 h), and these genes were not induced in a parallel cdkb-1 mutant culture. Annotations for these genes (Supplemental Data Set 6) did not suggest a clear unifying biological theme. However, this expression pattern suggests that different mutants will have unique transcriptional fingerprints that should help identify the function of the mutated gene.
It is striking that cdkb-1 and bsl1-1, which both arrest after a single cycle of DNA replication with similar cytology, have such sharply different underlying gene expression patterns.
Expression Patterns of Cyclin Genes and CDKs
Cyclins are key regulators of the eukaryotic cell cycle, and their transcription is frequently cell cycle regulated. Chlamydomonas encodes six cyclins homologous to members of the main cell-cycle-regulatory cyclin families in plants: three CYCDs, one each of CYCA and CYCB, and one hybrid CYCAB. Cyclin transcripts (except CYCD1) were previously shown to accumulate strongly during S/M (Bisova et al., 2005).
We found that all six cyclin genes, as well as CDKA and CDKB, were differentially upregulated in S/M relative to mid-G1 (Supplemental Data Set 2). In addition, we were able to distinguish between CDK-dependent and CDK-independent cyclin gene activation. Hierarchical clustering of the CDK and cyclin genes indicated similar pattern of expression for (1) CDKA, CYCD2, and CYCD3; (2) CYCAB and CYCD1; and (3) CDKB, CYCA, and CYCB (Figure 8A). The CDKA-containing cluster was characterized by relatively low fold-change induction between mid-G1 and S/M and little to no effect of the cdka-1 mutation. In contrast, the CDKB-containing cluster showed a much greater fold-change induction in wild-type cells and a clear delay in transcript accumulation in the cdka-1 mutant. Interestingly, the two cyclins CYCAB and CYCD1 displayed a transient CDK-independent peak in transcript accumulation at the 12- to 14-h time point. There was good qualitative agreement between these results and data obtained by the plate synchronization method (Supplemental Figure 7).
Figure 8.
Transcript Accumulation from Core Cell Cycle Genes.
(A) Hierarchical clustering (Euclidean distance) of transcript abundance patterns of CDKA, CDKB, and six cyclins.
(B) Individual traces of relative transcript abundances for each gene in wild-type, cdka-1, and cdkb-1 cells.
DISCUSSION
In eukarotyes, cell cycle decisions are regulated by a central CDK oscillator. Many components of the oscillator are encoded by periodically expressed genes; furthermore, CDK activity is known to regulate many transcription factors required for broad periodic transcriptional programs (Bertoli et al., 2013; Haase and Wittenberg, 2014). This suggests mutual regulation and a closed regulatory loop.
On the other hand, external factors such as light (Shepherd et al., 1983; López-Juez et al., 2008) could be key drivers of genome-wide transcriptional changes independently of CDKs. This distinction may be particularly relevant to photosynthetic organisms such as Chlamydomonas, in which the cell cycle is normally tightly coupled to diurnal light-dark changes.
Our results suggest a logical combination of these two views. Early changes in transcription, including initial activation of the S/M-related cell cycle cluster, are largely or entirely CDK independent, providing a likely dependence of CDK activation on environmental and/or cell growth signals. However, full transcriptional induction and subsequent repression are under differential regulation by CDKA and CDKB, suggesting a regulatory switch from extrinsic to intrinsic controls.
Functional Integration of Transcription with Cell Cycle Progression
The cell cycle in Chlamydomonas can be divided into two parts. The G1 phase is a long period of photosynthesis-dependent cell growth. This is followed by the S phase/mitosis period (S/M), during which the cell divides several times in rapid succession. The cell cycle is normally (though not obligatorily) tightly coupled to diurnal light-dark changes, such that G1 occupies the day and the divisions occur in the dark.
The G1-to-S/M transition entails a major shift in cell structure and function, including retraction of cilia, initiation of DNA replication, and subsequent chromosome partitioning in mitosis. There is also a sharp increase in Cks1-purified CDK kinase activity at this point (Bisova et al., 2005; Figure 6). Once all divisions have been completed, the resulting daughter cells hatch from the mother cell to start a new period of G1 growth. Completion of S/M is temporally associated with regrowth of cilia and assembly of new cell walls in the daughters (Cross and Umen, 2015).
We found that the G1-S/M transition is associated with activation of a transcriptional program containing ∼4000 genes, many of which may be linked to cell division, basal body/cilia-, and cell wall-related processes. Genes implicated in photosynthesis were coordinately downregulated at this time, reflecting the shift from cell growth to cell division (Figure 2).
Within S/M, we detected two temporally separated waves of transcript accumulation: an early wave (cluster 1, Figure 4) enriched in genes implicated in S phase and mitosis (cell cycle functional category) and a later wave (cluster 2, Figure 4) containing many genes that may be involved in cell wall biosynthesis, as well as genes that are upregulated during cilia regeneration. These two waves are under differential control by CDKA and CDKB.
Our results confirm previous findings that, in highly synchronized wild-type populations (Wood et al., 2012), transcript accumulation for IFT27 (a gene involved in intraflagellar transport and cilium growth) occurred after CYCB. This relative ordering of transcripts implicated in cell division and cilia-related processes was also found in a recent investigation of genome-wide transcript patterns in Chlamydomonas over one light-dark cycle (Panchy et al., 2014). The differential regulation by CDKA and CDKB implies a mechanistic separation between these two waves and provides a sharp annotation-independent means of sorting genes into early or late.
The PCA-based structuring of the transcriptional program in the wild type and mutants is unbiased and independent of gene annotations and functional categories. Therefore, the fact that this analysis segregates genes along borders that subsequently turn out to separate functional categories provides clear independent evidence that this structuring is biologically meaningful. The clusters contain many additional genes without such functional assignments, and it is likely that their presence in these clusters will signify related function.
In the green alga Ostreococcus tauri, cell division and accumulation of many cell-cycle-related genes is governed, at least in part, by an endogenous circadian clock. Differences in cyclin gene expression suggested distinct regulatory mechanisms with respect to circadian control versus other signals such as light or cell size (Moulager et al., 2010). Some of the diurnal transcriptional responses we report here could be under control of the Chlamydomonas circadian clock (Kucho et al., 2005; Matsuo et al., 2008; Matsuo and Ishiura, 2010; Niwa et al., 2013). We saw little or no CDK dependence for expression of circadian-regulated genes (Supplemental Figure 8; Kucho et al., 2005; Matsuo et al., 2008), suggesting some independence of the circadian and cell cycle clocks. However, we caution that comparisons to circadian-controlled transcripts from other studies should be considered preliminary because the presence of a robust circadian clock has not been established in our strain background (Matsuo et al., 2008).
The abundance of a given protein may be only partially explained by the abundance of its transcript, depending on the degree of posttranscriptional control (Vogel and Marcotte, 2012). During yeast meiosis, transcriptionally coregulated genes can differ in the time of their translation (Brar et al., 2012). Translational control has been demonstrated in Chlamydomonas for the expression of light-harvesting complex genes in response to light stress (Wobbe et al., 2008). Such multilayered controls of gene expression are likely to be found in the Chlamydomonas cell cycle as well.
Even in the absence of translational control, the abundance of highly stable proteins will not change rapidly in response to changes in transcript levels, although the rate of change of abundance will respond. For short-lived proteins (either due to constitutive turnover with half-lives significantly shorter than the cell cycle time or to cell-cycle-regulated turnover), transcript abundance is a much better indicator of protein abundance. Most cell cycle control proteins are well known to be unstable due to either constitutive degradation (e.g., G1 cyclins in yeast; Schneider et al., 1998) or to regulated degradation at specific points in the cycle (e.g., KIP/CIP inhibitors and mitotic cyclins) (Morgan, 2007). A-type, B-type, and D-type cyclins, KRP inhibitors, and CDKB are known to be unstable in plant systems (Inagaki and Umeda, 2011).
In plants and in the green alga O. tauri, CDKB is nearly absent in G1 cells (Corellou et al., 2005; Adachi et al., 2006), so mitotic accumulation of CDKB transcripts is essential for timely appearance of CDKB protein. The close correspondence between accumulation of mitotic transcripts (including many cyclins and CDKB) and the sharp induction of Cks1-purified kinase activity [almost surely due to CDKA and CDKB in complex with cyclin(s); Harashima and Schnittger, 2012] suggests that transcriptional control is a primary regulator of protein abundance and activity, although further work is required to establish this.
Thus, knowing patterns of cell-cycle-regulated transcript accumulation is only a first step in understanding control of protein abundance, which integrates transcription, mRNA degradation, translation, and protein degradation.
Differential Regulation of Transcription of Cell Division Genes by CDKA and CDKB
Our previous analysis based on cytological events suggested that CDKA is an upstream initiator of the S/M program because a number of cell cycle events were strongly delayed by the cdka-1 mutation. The CDKB gene acts later; it is not required for initiation of the S/M program, but it is essential for entry into the first mitosis. Wild-type cells undergo several alternating S/M rounds, accumulating up to 16C DNA content, and CDKB is required for reinitiating DNA replication after the first round. Arrested cdkb-1 cells contain very high levels of Cks1-purified kinase activity, likely due to failure of inactivation of CDKA.
Mirroring its effects on overall cell cycle progression, the cdka-1 mutation resulted in delayed accumulation of most S/M transcripts (clusters 1 and 2, Figure 4). This effect was strongest for cluster 2, which contains many genes in the BB-cilia-MT and cell wall functional classes.
CDKB has a more complex transcriptional phenotype. Cluster 1 genes turned on and transcripts accumulated to high levels (Figures 4 and 5), which remained high through the end of the experiment. This result indicated that CDKB is not required for induction of early S/M transcription, but needed for its eventual shutoff. Transcripts that normally turn on late in S/M (cluster 2) did not turn on in cdkb-1.
There is a notable parallel between transcription of cluster 1 and activation of Cks1-purified kinase in the wild type, cdka-1, and cdkb-1 revealed in the high-synchrony time course (Figure 5; including specific aspects such as a delay in activation and failure of later shutoff in cdkb-1). Causality is unclear: Kinase activity might directly regulate cluster 1 transcription, or cluster 1 transcription might lead to kinase activity. Finding several cyclins and CDKB itself in cluster 1 is consistent with the latter idea. These ideas are not mutually exclusive, and mutual dependence of CDK kinase activity and cyclin transcription (positive feedback) has been characterized in budding yeast at two different cell cycle transitions (Amon et al., 1993; Skotheim et al., 2008).
Many genes required for DNA replication are found in cluster 1, so cluster 1 induction may be required for replication. This could explain the delay both in activation of transcription of cluster 1, and in initiation of replication in the cdkb-1 mutant. However, better time resolution before the 9-h time point will be necessary to substantiate this idea. Notably, the bsl1-1 mutant activates cluster 1 genes and initiates DNA replication with wild-type timing.
Negative Feedback Control in Chlamydomonas Cell Cycle
We previously proposed a negative feedback model to account for the cdka-1 and cdkb-1 cell cycle phenotypes described above (Tulin and Cross, 2014). This simple model has two components: activation of CDKB by CDKA and inactivation of CDKA by CDKB (negative feedback). The model was designed to account for cytology and DNA replication status as well as Cks1-purified kinase activity in the arrested mutants.
The transcriptome results presented here extend this model. Since CDKB is itself a cluster 1 gene, as is CYCA, it is possible that CDKA-dependent transcriptional activation provides at least part of a mechanism by which CDKA might activate CDKB. CDKB is essentially inactive without a cyclin partner (Harashima and Schnittger, 2012).
Such a transcriptional relay mechanism may be analogous to the situation in Arabidopsis where CDKA-dependent transcriptional activation of CDKB is an important feature of cell cycle control (Nowack et al., 2012). In Arabidopsis, a critical link between CDKA and CDKB is the transcriptional repressor RBR. Phosphorylation of RBR by CDKA likely releases E2F family transcription factors to activate target genes, among them CDKB. In Chlamydomonas, the RBR homolog MAT3 and E2F regulate cell size, but the relationship between MAT3 and CDKA in transcriptional regulation is still unclear (Umen and Goodenough, 2001; Fang et al., 2006; Olson et al., 2010). In our preliminary experiments, cdka-1 mat3 double mutants seem to have phenotypes intermediate between the single mutants. We do not know to what extent, if at all, the transcriptional defects in cdka-1 would be rescued by simultaneous deletion of mat3. This is a technically challenging determination because it is difficult to obtain synchronous mat3 cultures (Fang et al., 2006).
Another possible link between CDK activity and transcriptional activation are the three-repeat MYB proteins (Feller et al., 2011), which have been shown to promote expression of mitosis-specific genes in both tobacco BY-2 cells (Ito et al., 2001) and Arabidopsis (Haga et al., 2007). Chlamydomonas encodes a single gene with three MYB domains (Cre12.g522400), and this gene may be homologous to the MYB3R1 and MYB3R4 genes in Arabidopsis. Cre12.g522400 was upregulated in dividing cells (cluster 1), suggesting a function at that time. In plants, triple-repeat MYB transcription factors bind to the mitosis-specific activator motif AACGG (Ito et al., 1998). We did not detect any notable enrichment of this motif in a 600-bp segment upstream of the translation start site in the cell cycle cluster (cluster 1, Figure 4) compared with all genes. Intriguingly, we detected five copies of this sequence upstream of Cre12.g522400 itself.
Candidate cis-acting sequences were described that might control diurnal expression patterns in Chlamydomonas (Panchy et al., 2014). None of these sequences had a clear resemblance to the three-repeat MYB consensus.
Coupling between Environment, Cell Cycle Execution, and Transcriptional Control
As noted above, the requirement for CDKA activity for activation of cluster 1 is only partial, since many transcripts in cluster 1 showed a substantial but delayed increase in the cdka-1 mutant. Also, a subset of cell-cycle-regulated genes showed no requirement for CDKA or CDKB for activation (cluster 3). Interestingly, this set of genes included the two cyclins CYCD1 and CYCAB, as well as several putative transcription factors (including components of the MAT3 pathway), and a gene (Cre04.g223200) with a CDK-like C-terminal kinase domain. CYCD1, but not CYCAB, contains a C-terminal LxCxE signature, a conserved motif that is known to be important for the interaction between D-type cyclins and RBR in animals (Chan et al., 2001).
These observations suggest a model in which initial accumulation of cell cycle transcripts is independent of CDK activity, but the trigger for this CDK-independent accumulation is unknown. Mutation of gex genes, which have at least partial cell growth defects, strongly limited the induction of these genes, suggesting that their induction may be dependent on rapid cell growth. The induced genes include CDKB as well as cyclins potentially required for activation of both CDKA and CDKB. The final pulse of high-level expression depends on CDKA (likely in combination with some cyclin); speed but not extent of induction depends on CDKB. Inactivation at the end of S/M also depends on CDKB.
It is interesting to consider to what extent these transcriptional controls might be direct (as is observed in CDK-dependent phosphorylation of transcriptional regulators in budding yeast) or indirect, working through dependency of transcription on CDK-mediated cell cycle events. For example, nuclear division is dependent on CDKB, so if some genes are specifically induced conditional on nuclear division, they would be indirectly dependent on CDKB. Figure 6 shows that cluster 1 (enriched for cell cycle and BB-cilia-MT genes) is expressed well in mutants blocked for the first (div20-1 and div17-1) or second (cdkb-1 and div13-1) cycle of DNA replication, as well as in a mutant blocked for spindle morphogenesis and cytokinesis (div24-1). All these mutants are defective for nuclear division and completion of cytokinesis. Therefore, neither of these clusters shows obvious dependence on completion of specific cytological cell cycle events. Cluster 2 (enriched for cell wall genes) shows a more complicated pattern (Figure 6), with strong dependence on CDKB (and CDKA; Figure 3) but variable dependence on other DIV genes. This initial survey then suggests that most of these regulated genes are not responding to actual completion of cell cycle events. The nature of the underlying regulatory network, and how this network is regulated by CDKs and other proteins such as BSL1, is a subject for future work.
METHODS
Strains and Media
CC-124 (mt−) and congenic iso10 (mt+) (here M10 and P10) were provided by Susan Dutcher and represent the wild type. Isolation and characterization of mutant strains has been described previously (Tulin and Cross, 2014). The cdka-1 and cdkb-1 mutants used in these experiments were backcrossed at least four times to CC-124 and showed stable and uniform segregation of the mutant phenotypes. Other mutant strains used were backcrossed at least twice.
TAP medium was prepared as described (Dutcher, 1995; Harris, 2008).
Culturing Conditions and Time-Course Experiments
For liquid culturing, cells were grown in liquid TAP medium in glass flasks under 14/10-h light/dark cycles illuminated with fluorescent bulbs (Ecosmart; 150 W, 2800 lumens) at ∼195 μmol photons s−1 m−2. For time-course experiments, the cultures were diluted to OD750 = 0.05 at the beginning of the light phase and shifted to a water bath at 33°C. This resulted in partial synchronization of wild-type cells, as indicated by an enrichment of dividing cells between 14 and 16 h (light microscopy) and cells with higher order DNA content (FACS).
For time-course experiments on agar plates, ∼106 cells were plated on TAP agar (1.5%) plates and grown for 2 d at 21°C under 14/10-h light/dark before being shifted to 33°C at the beginning of the light phase. The light intensity at the level of the plates was ∼50 μmol photons s−1 m−2. Cells were harvested by washing the plates with 10 mL of prewarmed TAP medium and gently detached from the surface with a glass pipette. This protocol resulted in a peak of cell division in the wild type at 10 to 11 h after the transfer to high temperature.
Genetic Analysis
Genetic crosses were performed essentially as described (Dutcher, 1995; Tulin and Cross, 2014). Briefly, gametogenesis was induced by transferring cells to nitrogen-free medium. Following gametogenesis, plus and minus gametes were mixed together to initiate mating. Mated cells were spread on TAP agar plates and matured into zygotes during a 5-d incubation in the dark. Tetrad dissection was using a Zeiss Axioskop 40 Tetrad microscope.
FACS, Coulter Counter, and Kinase Activity Measurements
Determination of DNA content (FACS), cell volume (Coulter counter), and Cks1-associated kinase activity was performed as previously described (Tulin and Cross, 2014). Briefly, roughly 106 cells were fixed in 3:1 ethanol:acetic acid, washed in PBS, treated with RNase A, and stained with 500 nM Sytox Green (Invitrogen). FACS data were recorded on a BD Accuri C6 instrument (BD Biosciences).
cDNA Isolation and Sequencing
A volume of Chlamydomonas reinhardtii liquid culture, normalized by OD to correspond to 30 mL of an OD750 = 0.05 culture, was collected for each time point and added to crushed ice. Cells were spun down (∼400g, 5 min, 4°C ) and washed once with ice-cold water. The pellet was lysed in TriZol (Life Technologies) according to the manufacturer’s instructions. RNA integrity was assessed on an Agilent 2100 Bioanalyzer (RNA integrity number was between 6 and 8), and RNA concentration was measured by a NanoDrop (Thermo Scientific). Approximately 2 μg total RNA was then used to prepare cDNA libraries using the TruSeq RNA kit (Illumina) according to the manufacturer’s instructions. The mean size of adapter-ligated cDNA was 260 bp, as determined on a 2200 TapeStation (Agilent Technologies). Twelve cDNA libraries, each with a different bar code, were pooled in equal proportions by NanoDrop estimate and sequenced in one lane on a HiSeq2000 instrument (Illumina) by Beijing Genomics Institute (Hong Kong), using 50-bp single-end reads. Alignment to the reference genome was done by TopHat (Kim et al., 2013) using the published genome (Phytozome v5.5) annotation as a guide. This generated between 10 and 14 million mapped fragments for most libraries, distributed over the 17,741 gene models. Read counts per gene model were obtained using the HTSeq software (Anders et al., 2014). Read quality control was done with the FastQC package (Babraham Bioinformatics) and generally indicated high quality (Phred score > 30) over the length of the reads.
Differential Expression Analysis
Differential gene expression was performed by edgeR (Robinson et al., 2010) on wild-type cultures only. Three samples represented mid-G1: t = 7 h (two samples) and t = 5 h (one sample). Three libraries represented the S/M phase at t = 14 h and three at t = 16 h. Tests for differential expression were done separately for mid-G1 versus S/M phase at 14 and 16 h. Any gene with a false discovery rate of <0.001 was considered differentially expressed. Genes that were upregulated at 14 or 16 h, relative to mid-G1, were filtered for duplicates and combined into a single nonredundant set. Similarly, genes that were downregulated at 14 or 16 h, relative to mid-G1, were filtered for duplicates and combined into a single set. This generated three nonoverlapping sets of genes (1) upregulated during S/M phase relative to mid-G1, (2) downregulated during S/M phase relative to mid-G1, and (3) not differentially expressed between mid-G1 and S/M phase (Supplemental Data Set 2).
Principal Component Analysis
The raw read count data was pretreated as follows. We collected all sequence libraries originating from the wild-type, cdka-1, and cdkb-1 strains (listed in columns 1 to 34 of Supplemental Data Set 1) and standardized the raw read counts by dividing by the average read count for each library. Next, all libraries from the same strain and time point were averaged. The resulting matrix was 17,741 rows × 26 columns, with the rows corresponding to the gene models, the columns to samples, and the entries in the matrix to column-averaged raw read counts. Finally, each row was normalized by its row average (across all 26 samples) and centered around zero. This generated the matrix A.
We performed PCA according to A = USVT by the Matlab (MathWorks) command svd. The principal components, here referred to as eigengenes, are in the rows of VT and correspond to the eigenvectors of the covariance matrix ATA. The eigensamples are in the columns of U and correspond to the eigenvectors of AAT. We inverted the sign of the first eigengene so that the shape of the eigengene matched the pattern of S/M phase transcript accumulation. Projections (plotted in Figures 2C and D) were calculated as UTA (for the samples) and AV (for the genes).
Gene Annotations
We assembled a list of annotations associating 1441 genes with one of eight functional classes: cell cycle, photosynthesis, cell wall, tubulin/cilia, metabolism, transcription, translation, and transcription factors. The assignment of a particular gene to a functional class was based in part on published reports and in part by keywords and Gene Ontology terms using the file “Creinhardtii_281_v5.5.annotation_info.txt,” available on Phytozome v.10. Briefly, the cell cycle category (112 genes) contains conserved genes involved in DNA replication (e.g., origin recognition, replicative enzymes, and deoxyribonucleotide synthesis) and chromosome cohesion and regulation of cell cycle progression (e.g., cyclins, CDKs and APC components). The photosynthesis category (144 genes) contains components of photosystems I and II, chlorophyll binding proteins, as well as components of plastid ribosomes and the carbon concentration mechanism. Genes in the cell wall functional class were identified by searching Phytozome v.10 with the keywords pherophorin and hydroxyproline-rich. The basal body/cilia/microtubule (BB-cilia-MT) functional class (481 genes) was created by searching the gene annotation available at phytozome.org for the Gene Ontology terms GO:0000226, GO:0007018, GO:0030286, GO:0005874, GO:0003777, and GO:0003774. We also included in the BB-cilia-MT class published genes in the flagellar (Pazour et al., 2005) and basal body proteomes (Keller et al., 2005) and genes with a likely function in basal body assembly (see Chlamydomonas Source Book, Vol. 3, Table 2.1B; Harris, 2008). The metabolism category (445 genes) was assembled by searching the annotations for the GO terms GO:0009058 and GO:0008152. The translation (197 genes) and transcription (55 genes) categories contain core components of protein translation (ribosome genes and rRNA processing) and DNA-directed RNA transcription, respectively. The complete gene annotations file used can be found in Supplemental Data Set 3.
Quantitative Real-Time PCR
Approximately 5 × 107 cells were spun down, the pellet was lysed in Trizol, and RNA was precipitated according to the manufacturer's instructions. DNA was digested and the RNA was further purified by the Qiagen RNEasy kit with DNase digestion. The concentration was ∼200 ng/μL, as determined by absorbance at 280 nm. For each sample, 400 ng RNA was used for cDNA synthesis (Roche) using random hexamer primers. Primers for qPCR were designed using the Primer3 software (Rozen and Skaletsky, 2000): PTB1-for, GCCTACTCGCCCAGCATC, and PTB1-rev, TGTTGGTGCGGTTGAGCA; CDKB-for, ACCTGCACCGCATCTTCC, and CDKB-rev, GGGTGGTTGATCGCCTCC; CYCB-for, TGCCCAGCGACTACATGAC, and CYCB-rev, CGTCTCGGGCATCAGCTT; GAS28-for, CGACCCCACGCAAAAGTG, and GAS28-rev, TTGCCGTCCACAGTCACG.
RT-qPCR was performed in 25-mL total volume of SYBR green PCR mix (Applied Biosystems) with 0.4 μM of each primer and a 1:5 dilution of cDNA. The PCR protocol was 95°C for 10 min, then 40 cycles of 95°C for 15 s (melting) and 63°C for 60 s (annealing/elongation). Amplification efficiency was determined to be nearly 100% for CDKB and PTB1 and ∼90% for CYCB and GAS28 over a 100-fold dilution range of cDNA input.
Accession Numbers
Accession numbers (Phytozome v.10) of Chlamydomonas genes in this study are as follows: Cre10.g465900 (CDKA), Cre08.g372550 (CDKB), Cre04.g220700 (DIV43), Cre15.g634701 (DIV20), Cre12.g491050 (DIV17), Cre02.g095113 (DIV24), Cre12.g513600 (DIV42), Cre01.g050850 (DIV44), Cre09.g396661 (DIV39), Cre01.g019200 (GEX26), and Cre13.g561800 (GEX1).
Supplemental Data
Supplemental Figure 1. Cell cycle analysis of wild-type and cdk mutant strains.
Supplemental Figure 2. PCA-guided cluster construction.
Supplemental Figure 3. PCA significance estimation.
Supplemental Figure 4. Transcript accumulation in the cdka-1 cdkb-1 double mutant.
Supplemental Figure 5. Wild type and cdkb-1 RT-qPCR.
Supplemental Figure 6. Determination of DNA content in div and gex mutants by FACS.
Supplemental Figure 7. Transcript abundance of CDKA, CDKB, and six cyclin genes.
Supplemental Figure 8. Largely CDK-independent transcript accumulation of putatively circadian clock-controlled genes.
Supplemental Data Set 1. Raw read counts for 54 liquid-grown samples.
Supplemental Data Set 2. Result of edgeR analysis, identifying genes that are differentially expressed (upregulated or downregulated) between G1 and S/M.
Supplemental Data Set 3. Functional categories used in this study.
Supplemental Data Set 4. Cluster 1 to 4 gene IDs and annotations.
Supplemental Data Set 5. Raw read counts for 24 plate-grown samples.
Supplemental Data Set 6. Gene IDs and annotation for genes in the BSL1 cluster.
Supplementary Material
Acknowledgments
We thank S. Dutcher and J. Umen for Chlamydomonas strains, advice, and useful discussion. We also thank J.S. Rahi for critical comments and K. Pekani for managing the laboratory and assisting in carrying out experiments. This work was supported by National Institutes of Health Grant 5R01GM078153-07 and by The Rockefeller University.
AUTHOR CONTRIBUTIONS
F.T. and F.C. designed and executed the experiments and wrote the article.
Glossary
- CDK
cyclin-dependent kinase
- FACS
fluorescence-activated cell sorting
- PCA
principal component analysis
References
- Adachi S., Uchimiya H., Umeda M. (2006). Expression of B2-type cyclin-dependent kinase is controlled by protein degradation in Arabidopsis thaliana. Plant Cell Physiol. 47: 1683–1686. [DOI] [PubMed] [Google Scholar]
- Albee A.J., Kwan A.L., Lin H., Granas D., Stormo G.D., Dutcher S.K. (2013). Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3 (Bethesda) 3: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter O., Brown P.O., Botstein D. (2000). Singular value decomposition for genome-wide expression data processing and modeling. Proc. Natl. Acad. Sci. USA 97: 10101–10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon A., Tyers M., Futcher B., Nasmyth K. (1993). Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74: 993–1007. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. (2014). HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoli C., Skotheim J.M., de Bruin R.A. (2013). Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 14: 518–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisova K., Krylov D.M., Umen J.G. (2005). Genome-wide annotation and expression profiling of cell cycle regulatory genes in Chlamydomonas reinhardtii. Plant Physiol. 137: 475–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar G.A., Yassour M., Friedman N., Regev A., Ingolia N.T., Weissman J.S. (2012). High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H.M., Smith L., La Thangue N.B. (2001). Role of LXCXE motif-dependent interactions in the activity of the retinoblastoma protein. Oncogene 20: 6152–6163. [DOI] [PubMed] [Google Scholar]
- Corellou F., Camasses A., Ligat L., Peaucellier G., Bouget F.Y. (2005). Atypical regulation of a green lineage-specific B-type cyclin-dependent kinase. Plant Physiol. 138: 1627–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F.R., Umen J.G. (2015). The Chlamydomonas cell cycle. Plant J. 82: 370–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K. (1995). Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 47: 531–540. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Lehman D.A., O’Farrell P.H. (1994). Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle. Development 120: 3131–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparza J.M., O'Toole E., Li L., Giddings T.H., Kozak B., Albee A.J., Dutcher S.K. (2013). Katanin localization requires triplet microtubules in Chlamydomonas reinhardtii PLoS ONE 8: e53940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S.-C., de los Reyes C., Umen J.G. (2006). Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller A., Machemer K., Braun E.L., Grotewold E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66: 94–116. [DOI] [PubMed] [Google Scholar]
- Goto K., Johnson C.H. (1995). Is the cell division cycle gated by a circadian clock? The case of Chlamydomonas reinhardtii. J. Cell Biol. 129: 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzat R., Borghi L., Gruissem W. (2012). Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci. 17: 139–148. [DOI] [PubMed] [Google Scholar]
- Haase S.B., Wittenberg C. (2014). Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics 196: 65–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga N., Kato K., Murase M., Araki S., Kubo M., Demura T., Suzuki K., Müller I., Voss U., Jürgens G., Ito M. (2007). R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134: 1101–1110. [DOI] [PubMed] [Google Scholar]
- Harashima H., Schnittger A. (2012). Robust reconstitution of active cell-cycle control complexes from co-expressed proteins in bacteria. Plant Methods 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. (2008). The Chlamydomonas Sourcebook: Introduction into Chlamydomonas and Its Laboratory Use. (San Diego, CA: Elsevier Academic Press; ). [Google Scholar]
- Henley S.A., Dick F.A. (2012). The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 7: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann X.K., Beck C.F. (2005). Mating-induced shedding of cell walls, removal of walls from vegetative cells, and osmotic stress induce presumed cell wall genes in Chlamydomonas. Plant Physiol. 139: 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki S., Umeda M. (2011). Cell-cycle control and plant development. Int. Rev. Cell Mol. Biol. 291: 227–261. [DOI] [PubMed] [Google Scholar]
- Ito M., Araki S., Matsunaga S., Itoh T., Nishihama R., Machida Y., Doonan J.H., Watanabe A. (2001). G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors. Plant Cell 13: 1891–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Iwase M., Kodama H., Lavisse P., Komamine A., Nishihama R., Machida Y., Watanabe A. (1998). A novel cis-acting element in promoters of plant B-type cyclin genes activates M phase-specific transcription. Plant Cell 10: 331–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lau O.S., Deng X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8: 217–230. [DOI] [PubMed] [Google Scholar]
- Keller L.C., Geimer S., Romijn E., Yates J. III, Zamora I., Marshall W.F. (2009). Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell 20: 1150–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L.C., Romijn E.P., Zamora I., Yates J.R. III, Marshall W.F. (2005). Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15: 1090–1098. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K., Okamoto K., Tabata S., Fukuzawa H., Ishiura M. (2005). Identification of novel clock-controlled genes by cDNA macroarray analysis in Chlamydomonas reinhardtii. Plant Mol. Biol. 57: 889–906. [DOI] [PubMed] [Google Scholar]
- López-Juez E., Dillon E., Magyar Z., Khan S., Hazeldine S., de Jager S.M., Murray J.A., Beemster G.T., Bögre L., Shanahan H. (2008). Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20: 947–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maselli G.A., Slamovits C.H., Bianchi J.I., Vilarrasa-Blasi J., Caño-Delgado A.I., Mora-García S. (2014). Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol. 164: 1527–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Ishiura M. (2010). New insights into the circadian clock in Chlamydomonas. Int. Rev. Cell Mol. Biol. 280: 281–314. [DOI] [PubMed] [Google Scholar]
- Matsuo T., Okamoto K., Onai K., Niwa Y., Shimogawara K., Ishiura M. (2008). A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev. 22: 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M., Hennig L., Gruissem W., Murray J.A. (2002). Cell cycle-regulated gene expression in Arabidopsis. J. Biol. Chem. 277: 41987–42002. [DOI] [PubMed] [Google Scholar]
- Michael T.P., McClung C.R. (2003). Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiol. 132: 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll T., Tebb G., Surana U., Robitsch H., Nasmyth K. (1991). The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66: 743–758. [DOI] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan D. (2007). The Cell Cycle: Principles of Control. (London: New Science Press; ). [Google Scholar]
- Moulager M., Corellou F., Vergé V., Escande M.-L., Bouget F.-Y. (2010). Integration of light signals by the retinoblastoma pathway in the control of S phase entry in the picophytoplanktonic cell Ostreococcus. PLoS Genet. 6: e1000957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulager M., Monnier A., Jesson B., Bouvet R., Mosser J., Schwartz C., Garnier L., Corellou F., Bouget F.-Y. (2007). Light-dependent regulation of cell division in Ostreococcus: evidence for a major transcriptional input. Plant Physiol. 144: 1360–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Matsuo T., Onai K., Kato D., Tachikawa M., Ishiura M. (2013). Phase-resetting mechanism of the circadian clock in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 110: 13666–13671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowack M.K., Harashima H., Dissmeyer N., Zhao X., Bouyer D., Weimer A.K., De Winter F., Yang F., Schnittger A. (2012). Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev. Cell 22: 1030–1040. [DOI] [PubMed] [Google Scholar]
- Olson B.J.S.C., Oberholzer M., Li Y., Zones J.M., Kohli H.S., Bisova K., Fang S.-C., Meisenhelder J., Hunter T., Umen J.G. (2010). Regulation of the Chlamydomonas cell cycle by a stable, chromatin-associated retinoblastoma tumor suppressor complex. Plant Cell 22: 3331–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando D.A., Lin C.Y., Bernard A., Wang J.Y., Socolar J.E., Iversen E.S., Hartemink A.J., Haase S.B. (2008). Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature 453: 944–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N., Wu G., Newton L., Tsai C.-H., Chen J., Benning C., Farré E.M., Shiu S.-H. (2014). Prevalence, evolution, and cis-regulation of diel transcription in Chlamydomonas reinhardtii. G3 (Bethesda) 4: 2461–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Agrin N., Leszyk J., Witman G.B. (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170: 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds D., Shi B.J., McLean C., Katsis F., Kemp B., Dalton S. (2003). Recruitment of Thr 319-phosphorylated Ndd1p to the FHA domain of Fkh2p requires Clb kinase activity: a mechanism for CLB cluster gene activation. Genes Dev. 17: 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C., Huntley R., Jacqmard A., Murray J.A. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Sabelli P.A., Liu Y., Dante R.A., Lizarraga L.E., Nguyen H.N., Brown S.W., Klingler J.P., Yu J., LaBrant E., Layton T.M., Feldman M., Larkins B.A. (2013). Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proc. Natl. Acad. Sci. USA 110: E1827–E1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R., Landgraf J., Accerbi M., Simon V., Larson M., Wisman E. (2001). Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B.L., Patton E.E., Lanker S., Mendenhall M.D., Wittenberg C., Futcher B., Tyers M. (1998). Yeast G1 cyclins are unstable in G1 phase. Nature 395: 86–89. [DOI] [PubMed] [Google Scholar]
- Shepherd H.S., Ledoigt G., Howell S.H. (1983). Regulation of light-harvesting chlorophyll-binding protein (LHCP) mRNA accumulation during the cell cycle in Chlamydomonas reinhardi. Cell 32: 99–107. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. (1993). Mammalian G1 cyclins. Cell 73: 1059–1065. [DOI] [PubMed] [Google Scholar]
- Sherr C.J., Roberts J.M. (1999). CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13: 1501–1512. [DOI] [PubMed] [Google Scholar]
- Silflow C.D., LaVoie M., Tam L.W., Tousey S., Sanders M., Wu W., Borodovsky M., Lefebvre P.A. (2001). The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J. Cell Biol. 153: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotheim J.M., Di Talia S., Siggia E.D., Cross F.R. (2008). Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature 454: 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin F., Cross F.R. (2014). A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26: 4019–4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemukai K., Iwakawa H., Kosugi S., de Uemukai S., Kato K., Kondorosi E., Murray J.A., Ito M., Shinmyo A., Sekine M. (2005). Transcriptional activation of tobacco E2F is repressed by co-transfection with the retinoblastoma-related protein: cyclin D expression overcomes this repressor activity. Plant Mol. Biol. 57: 83–100. [DOI] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. (2001). Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15: 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Marcotte E.M. (2012). Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13: 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C., Reed S.I. (2005). Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 24: 2746–2755. [DOI] [PubMed] [Google Scholar]
- Wobbe L., Schwarz C., Nickelsen J., Kruse O. (2008). Translational control of photosynthetic gene expression in phototrophic eukaryotes. Physiol. Plant. 133: 507–515. [DOI] [PubMed] [Google Scholar]
- Wood C.R., Wang Z., Diener D., Zones J.M., Rosenbaum J., Umen J.G. (2012). IFT proteins accumulate during cell division and localize to the cleavage furrow in Chlamydomonas. PLoS One 7: e30729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.