The time-calibrated evolutionary history of Brassicaceae is characterized by polyploidization and rapid genomic stabilization indicating polyploidization as driver for future diversification.
Abstract
The Brassicaceae include several major crop plants and numerous important model species in comparative evolutionary research such as Arabidopsis, Brassica, Boechera, Thellungiella, and Arabis species. As any evolutionary hypothesis needs to be placed in a temporal context, reliably dated major splits within the evolution of Brassicaceae are essential. We present a comprehensive time-calibrated framework with important divergence time estimates based on whole-chloroplast sequence data for 29 Brassicaceae species. Diversification of the Brassicaceae crown group started at the Eocene-to-Oligocene transition. Subsequent major evolutionary splits are dated to ∼20 million years ago, coinciding with the Oligocene-to-Miocene transition, with increasing drought and aridity and transient glaciation events. The age of the Arabidopsis thaliana crown group is 6 million years ago, at the Miocene and Pliocene border. The overall species richness of the family is well explained by high levels of neopolyploidy (43% in total), but this trend is neither directly associated with an increase in genome size nor is there a general lineage-specific constraint. Our results highlight polyploidization as an important source for generating new evolutionary lineages adapted to changing environments. We conclude that species radiation, paralleled by high levels of neopolyploidization, follows genome size decrease, stabilization, and genetic diploidization.
INTRODUCTION
Whole-genome duplications (WGDs; or polyploidization) have played a major role in plant evolution, and a series of important WGDs that occurred throughout the evolutionary past of land plants have been identified (De Bodt et al., 2005; Tang et al., 2010; Vanneste et al., 2014). Consequently, all angiosperm lineages can be considered polyploid (Amborella Genome Project, 2013), and many of them have undergone multiple WGD events. However, major questions concerning polyploid evolution have been discussed since Stebbins (1938, 1950) (reviewed in Soltis et al., 2014), and there is ongoing discussion about the importance of polyploidization for plant species diversification (Mayrose et al., 2011; Arrigo and Barker, 2012; Soltis et al., 2014). Do polyploids have an advantage over their diploid ancestors, and how is this realized? What causes species richness to be positively correlated with polyploidy? Why is polyploidy followed by genetic diploidization, often associated with genome downsizing? Iterative cycles of polyploidization and subsequent genome diploidization seem to bring significant evolutionary and adaptive advantages to polyploid plants, as illustrated by many examples (reviewed in Soltis et al., 2004, 2007). Nonetheless, it remains unclear which specific processes or parameters initiate an increase in ploidy level and what triggers might be responsible for polyploidization (Flagel and Wendel, 2009).
The Brassicaceae are characterized by remarkable speciation rates, which are the highest among those reported for any land plant group (e.g., genus Draba [Jordon-Thaden and Koch, 2008] and Arabis [Karl and Koch, 2013]). Additionally, tribal and family-wide lineage-through-time plots indicate accelerated evolutionary lineage and species number increases over the past 30 million years (Couvreur et al., 2010; Karl and Koch, 2013). Some of this diversification is correlated with polyploidization. Because WGDs are assumed to have played a major role in the speciation and diversification of angiosperms (De Bodt et al., 2005; Soltis et al., 2009; Tang et al., 2010; Jiao et al., 2011), the significance of ancient polyploidy has also been investigated across the Brassicaceae family (Lysak et al., 2009; Franzke et al., 2011; Lysak and Koch, 2011; Kagale et al., 2014). Based on the elapsed time since their occurrence and the level of genome diploidization, WGDs have been classified into three categories, as neo-, meso-, and paleopolyploid events (Mandáková et al., 2010a; Lysak and Koch, 2011; Kagale et al., 2014).
The ancestral Brassicaceae-specific paleopolyploidization event is known as the α-duplication (At-α) and is shared among all Brassicaceae lineages. This WGD was first shown by Vision et al. (2000) and Bowers et al. (2003), later shown to be Brassicaceae-specific (Schranz et al., 2012; Haudry et al., 2013; Hofberger et al., 2013), and recently its maximum age was estimated to 47.1 million years ago (mya; Kagale et al., 2014), thereby also providing the maximum age of the Brassicaceae stem group. The older β-WGD (At-β) was dated to 124 mya (Kagale et al., 2014) and is assumed not to be shared with the Caricaceae (Ming et al., 2008). However, in an angiosperm-wide time-calibrated phylogeny including all reliable fossils, the Caricaceae node was set to 81.6 million years (Magallón et al., 2015), thereby corresponding to the maximum age of the β-WGD. The discrepancy might be explained by the different approaches used to calculate divergence times: via relaxed evolutionary models (Magallón et al., 2015) or extrapolating a constant synonymous substitution rate (Kagale et al., 2014). Recently, a Brassicales-wide transcriptome analysis provided phylogenetic placements of these two genome duplication events and the estimates are 40 and 88 mya for At-α and At-β, respectively (Edger et al., 2015).
Mesopolyploidization events in Brassicaceae have been defined previously (Mandáková et al., 2010a; Lysak and Koch, 2011). Although mesopolyploid WGDs are obscured by extensive chromosomal and genetic diploidization processes, such as chromosome rearrangements, chromosome number reduction, genome downsizing, and fractionation, duplicated genomic regions are still detectable by comparative genetic and cytogenomic approaches (Mandáková et al., 2010a). The majority of mesopolyploidization events, frequently followed by later neopolyploidization, are connected to the early evolution of various Brassicaceae tribes (Lysak and Koch, 2011). In neopolyploids, chromosome numbers are not reduced and most duplicated genomic blocks have not been reshuffled yet (Lysak and Koch, 2011).
To date, four tribes (Heliophileae [Mandáková et al., 2012], Microlepidieae [Joly et al., 2009; Mandáková et al., 2010a, 2010b], Brassiceae and Brassicas [Lysak et al., 2005; Wang et al., 2011; Cheng et al., 2013], and Biscutelleae [C. Geiser, T. Mandáková, N. Arrigo, M.A. Lysak, and C. Parisod, unpublished results]) and the genus Leavenworthia (Haudry et al., 2013) have been shown to be mesopolyploids, which have undergone post-At-α polyploidizations. However, the total number of mesopolyploidization events might be higher, considering isolated lineages characterized by extreme long branches in molecular phylogenetic reconstructions, as exemplified by the tribes Kernereae, Buniadeae, Heliophileae, and Cochlearieae. Some additional mesopolyploidy events have been postulated (Pringlea and Stanleya [Kagale et al., 2014] and Caulanthus [Burrell et al., 2011], all from tribe Thelypodieae; Physarieae [Lysak et al., 2009]), pending further validation.
The mustard family belongs to the order Brassicales (core eudicots and rosids) and consists of ∼3740 species (Kiefer et al., 2014). Together with two additional families, Cleomaceae and Capparaceae, the Brassicaceae are grouped into the “core Brassicales” (Beilstein et al., 2010) comprising 4440 species in total. The core Brassicales are distributed mostly in temperate regions of the world, with much fewer representatives in subtropical regions. The core Brassicales are further complemented by an additional 14 families with much lower species numbers, all belonging to the order Brassicales (245 species in total; Stevens, 2001), and a preference for tropical regions. Brassicales is part of the Malvidae, and together with Fabidae, it forms the monophyletic Rosidae (Ruhfel et al., 2014). Saxifragales and Vitales are sister to the Rosidae clade and all together form the Superrosidae (Ruhfel et al., 2014) with a crown group age of ∼122 million years (Magallón et al., 2015). The systematics, taxonomy, and evolutionary history of the Brassicaceae have long been controversial, although representatives of the family have always been the focus of systematists and taxonomists (Koch et al., 2003; Koch and Al-Shehbaz, 2009).
However, boundaries between related genera were often poorly delimited and attempts to group species diversity into higher order entities often failed in circumscribing monophyletic groups (e.g., for an overview: Arabis [Koch et al., 1999, 2000, 2001], Arabidopsis [Koch et al., 2008; Koch and German, 2013], Lepidium [Mummenhoff et al., 2009], Thlaspi [Mummenhoff et al., 1997], and family-wide [Al-Shehbaz, 2012]). Therefore, past concepts combining genera into so-called tribes resulted not only in artificial but also contradicting concepts (reviewed in Koch and Al-Shehbaz, 2009; Lysak and Koch, 2011; Al-Shehbaz, 2012). For a long time, this was also reflected by a lack of agreement on the number and boundaries of phylogenetically defined tribes and genera, giving rise to several contradictory classification systems (reviewed in Warwick et al., 2010), and often led to wrong conclusions on trait and character evolution. In the most up-to-date overview, also used in BrassiBase (http://brassibase.cos.uni-heidelberg.de/; Koch et al., 2012; Kiefer et al., 2014), the mustard family comprises 3740 species classified within 325 genera assigned to 51 tribes (Al-Shehbaz, 2012; BrassiBase, 2015). Only 15 genera with 22 species are not yet assigned to any tribe (Al-Shehbaz, 2012; Kiefer et al., 2014), representing <0.75% of the family-wide species diversity. Within the Brassicaceae, four major infrafamiliar evolutionary lineages have been repeatedly described (lineages I to III plus the sister group Aethionemeae; Bailey et al., 2006; Beilstein et al., 2006; Koch et al., 2007; Franzke et al., 2009; Lysak et al., 2009; Couvreur et al., 2010). However, it has also been concluded that this simple view of four clearly separated lineages might not reflect well the evolutionary history of more than 50 tribes (Franzke et al., 2010) because of incongruences among published studies and lineage II most likely not being a monophyletic clade. Thus, debate is ongoing and various explanations have been proposed to explain unresolved or even contradicting phylogenetic hypotheses (e.g., hard polytomies in phylogenetic reconstruction, reticulate evolution, and extensive polyploidization; Couvreur et al., 2011; Franzke et al., 2011).
Although the enormous present day abundance of both auto- and allopolyploids in Brassicaceae is obvious (Jordon-Thaden and Koch, 2008), we lack a detailed overview for the whole family. Considering that we recognize paleo- and mesopolyploidization as important driving forces of past genome evolution in crucifers, we can hypothesize that neopolyploids are a potential source of future lineage diversification. Their percentage should be very high if polyploidization was a widespread and important process during the Brassicaceae evolutionary history, particularly in facilitating adaptation to changing environmental conditions (e.g., drought and cold, but also heat and sometimes extreme wet conditions). Some first and very rough estimates suggested ∼37% (Appel and Al-Shehbaz, 2002; Warwick and Al-Shehbaz, 2006) or 50% (Koch et al., 2003) neopolyploid Brassicaceae species. Unraveling the evolution of neopolyploids in a spatio-temporal context is challenging as the time scales considered are much shorter, with the Pleistocene periods of glaciation and deglaciation cycling in 100,000-year intervals over a total period of only 2 million years (Imbrie et al., 1993).
Here, we present a time-calibrated phylogenetic tree for the Brassicaceae based on whole-chloroplast genome sequences. We analyzed not only a representative set of Brassicaceae genomes, but also included a large set of angiosperm chloroplast genomes with the aim of placing the evolution of Brassicaceae into the context of large-scale phylogenetic studies focusing either on systematics (Ruhfel et al., 2014) or on angiosperm-wide divergence time estimates (Magallón et al., 2015). We then tested the idea that neopolyploidization has driven the enormous radiation within the entire family by mining the literature for chromosome number reports from the past 100 years and putting these data into the context of genome size evolution as a potential indicator of WGD. While relating ploidy information to tribal-wide genome size estimates and base chromosome numbers, we also looked for footprints of putative mesopolyploidization events.
RESULTS
Phylogenetic Reconstructions and Divergence Time Estimates
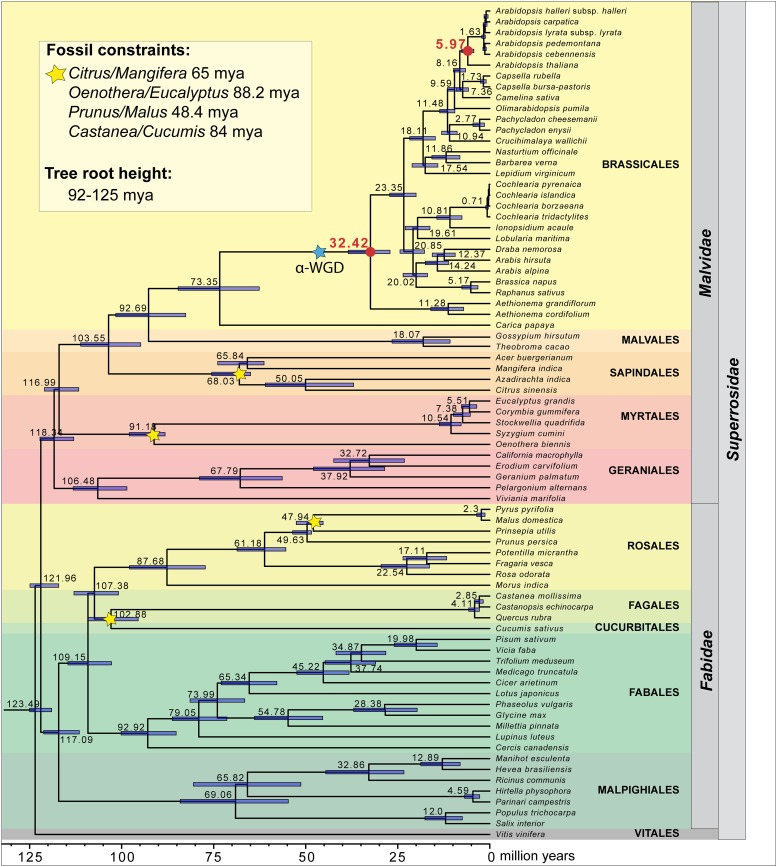
The phylogenetic relationships based on whole-chloroplast genomes among taxa from the whole Superrosidae clade were fully in congruence with a large-scale angiosperm data set also using whole-chloroplast genome sequences (Ruhfel et al., 2014). Consequently, the maximum likelihood (ML) trees for the Brassicaceae generated here are shown in Supplemental Figure 1, including bootstrap values. Importantly, the fossil-constrained BEAST analysis (Figure 1) resulted in an identical tree topology compared with the ML trees (Supplemental Figure 1). The BEAST analysis was compared with the most recent angiosperm-wide calibrated phylogenetic tree (Magallón et al., 2015); indeed, the four divergence time estimates we used for fossil calibration perfectly fit estimates by Magallón et al. (2015). These four constraints are considered the most reliable source for calibrating this particular superclade. Our chloroplast genome data indicated that the Brassicaceae crown group radiation dates back 32.4 mya. The corresponding stem group age was calculated by Magallón et al. (2015) with a maximum of ∼44.2 mya including the Capparaceae. This gives an early origin of the Brassicaceae in the Eocene, but the first radiation occurred at the transition from Eocene to Oligocene (32.4 mya) with the split of Aethionemeae from the rest of the family (Supplemental Figure 2). Much later, the split at the Eocene-Oligocene boundary was followed by a second split dated to 23.5 mya and subsequent rapid radiation of major lineages and tribes. These radiation events correspond exactly to the Oligocene-Miocene boundary (Supplemental Figure 2). The two case studies included here with increased taxon sampling, tribe Cochlearieae and the genus Arabidopsis, are also fully in agreement with what we “know” about their evolutionary history. The deep split within Cochlearieae, separating the Mediterranean Ionopsidium from the arctic-alpine and Nordic Cochlearia, occurred 10.8 mya, in agreement with earlier molecular estimates and paleoenvironmental correlations (Koch, 2012). Radiation within the polyploid complex and reticulate genus Cochlearia from within tribe Cochlearieae happened exclusively during the Pleistocene with the deepest split observed at 0.7 mya.
Figure 1.
BEAST Analysis of Whole-Chloroplast Genome Sequence Data from the Superrosidae Clade.
Fossil constraints and tree root height are indicated accordingly. The α-WGD was placed on the figure at 47 million years (maximum age estimated in Kagale et al., 2014). Divergence times are indicated with 95% confidence intervals.
Similarly, the crown group age of the genus Arabidopsis was set to 5.97 mya. The Messinian salinity crisis ended 5.33 mya (Garcia-Castellanos et al., 2009), and we could hypothesize that this period had considerable influence on the northern Mediterranean and the Balkan regions and, thereby, on the deepest split found within the Arabidopsis lineage. However, subsequent radiation among all existing perennial Arabidopsis species started 1.63 mya, close to the onset of Pleistocene glaciation and deglaciation cycles ∼2 mya.
Cytogenetic Analyses
The total number of identified neopolyploid taxa was 715 (43.3%) out of 1653 analyzed taxa with available data and their frequency varied widely among tribes. We classified a taxon as neopolyploid if it had undergone an additional polyploidization event after the α-WGD or after any documented subsequent mesopolyploidization. Consequently, the large majority of neopolyploids was identified based on chromosome number variation (ploidy level variation) within a genus or among closely related genera mostly without a change in respective base chromosome numbers.
Our survey of haploid chromosome numbers and the percentage of neopolyploids per tribe is summarized in Table 1. Note that the number of neopolyploids may vary slightly depending on taxonomic concepts (e.g., when recognizing intraspecific cytotypes as distinct species). A completely revised species checklist was used here for the chromosome number survey to minimize confusing taxonomic issues (Supplemental Data Set 1; the new Brassicaceae species checklist is also implemented in the BrassiBase knowledge database, http://brassibase.cos.uni-heidelberg.de/; Koch et al., 2012; Kiefer et al., 2014). Treating infraspecific ploidy level variations (cytotypes) as the same species results in slightly higher numbers of neopolyploids (<1% over the whole data set) than if kept as separate taxonomic entities.
Table 1. Number of Genera and Species Described within the 51 Tribes.
| Tribe | [Genera: Species] | Base Chr. Nos. (Doubtful?) | % Neopolyploids [No. of Taxa with Data] | Mean 1 Cx GS (sd) [No. of Taxa with Data] | Mean 1C GS (sd) | Mean 1 Cx GS (in Mb) | % Annuals |
|---|---|---|---|---|---|---|---|
| Aethionemeae | [1: 47] | 11,12 (7?,8) | 44 [18] | 0.37 (0.05) [7] | 0.51 (0.19) | 288 | 14 |
| Alysseae | [22: 275] | 8 (7,10,11,12,15) | 32 [143] | 1.11 (0.54) [15] | 1.22 (0.65) | 866 | 22 |
| Alyssopsideae | [4: 9] | 8 (7?) | 50 [4] | 0.18 (0.01) [3] | 0.35 (0.14) | 140 | 50 |
| Anastaticeae | [13: 67] | 9-13 (7,21,23,35) | 19 [27] | 0.60 (0.30) [7] | 0.69 (0.41) | 468 | 32 |
| Anchonieae | [9: 77] | 6-8 (5?,13) | 9 [33] | 2.11 (0.46) [10] | 2.11 (0.46) | 1645 | 28 |
| Aphragmeae | [1: 11] | 7,8 | 0 [2] | No data | No data | 0 | |
| Arabideae | [17: 508] | 8 (6,7,9–11,13,15) | 63 [211] | 0.36 (0.10) [23] | 0.44 (0.26) | 280 | 8 |
| Asteae | [1: 1] | 10 | 0 [1] | No data | No data | 0 | |
| Biscutelleae | [2: 46] | 6,8,9 | 19 [36] | 0.92 (0.17) [5] | 1.16 (0.59) | 717 | 25 |
| Bivonaeeae | [1: 1] | No data | No data | No data | No data | 100 | |
| Boechereae | [9: 128] | 7 (6,8,9,15,22) | 45 [55] | 0.24 [2] | 0.24 | 187 | 0 |
| Brassiceae | [47: 229] | 7-12, 14,15,17 (12,13) | 28 [190] | 0.85 (0.38) [73] | 1.05 (0.90) | 663 | 43 |
| Buniadeae | [1: 2] | 7 | 50 [2] | 2.37 (0.36) [2] | 2.37 (0.36) | 0 | |
| Calepineae | [3:9] | 7 | 75 [4] | 0.20 [1] | 0.30 | 156 | 100 |
| Camelineae | [8: 33] | 8 (6,7,10,12,13,19) | 50 [22] | 0.26 (0.05) [17] | 0.38 (0.17) | 203 | 54 |
| Cardamineae | [12: 349] | 8 (5–7,10,11,15) | 63 [160] | 0.31 (0.08) [16] | 0.62 (0.45) | 241 | 26 |
| Chorisporeae | [4: 54] | 7 (9) | 33 [12] | 0.57 (0.36) [3] | 0.57 (0.36) | 444 | 49 |
| Cochlearieae | [2: 29] | 6,7 (8,11) | 76 [21] | 0.37 (0.07) [11] | 0.73 (0.40) | 288 | 29 |
| Coluteocarpeae | [3: 128] | 7 | 31 [49] | 0.33 (0.12) [15] | 0.40 (0.20) | 257 | 8 |
| Conringieae | [2: 9] | 7 (8?,9?) | 20 [5] | 0.23 [1] | 0.23 | 179 | 100 |
| Cremolobeae | [3: 32] | 11 | 0 [1] | No data | No data | 100 | |
| Crucihimalayeae | [3: 13] | 8 (7) | 0 [8] | 0.38 (0.03) [4] | 0.32 (0.10) | 296 | 0 |
| Descurainieae | [6: 46] | 7 (6,8) | 44 [34] | 0.20 (0.02) [10] | 0.24 (0.05) | 156 | 53 |
| Dontostemoneae | [2: 17] | 7 | 33 [6] | 1.99 [1] | 3.97 | 1552 | 33 |
| Erysimeae | [1: 215] | 6–9 (10,11,13) | 66 [115] | 0.33 (0.09) [8] | 0.51 (0.18) | 257 | 4 |
| Euclidieae | [25: 151] | 7 | 36 [44] | 0.99 (0.57) [9] | 1.11 (0.60) | 772 | 36 |
| Eudemeae | [6: 32] | 7,9,11 | 0 [2] | 0.73 [1] | 0.73 | 569 | 0 |
| Eutremeae | [3: 35] | 7 | 78 [9] | 0.32 [1] | 0.32 | 250 | 13 |
| Halimolobeae | [5: 39] | 8 | 22 [9] | 0.17 (0.01) [4] | 0.17 (0.01) | 132 | 24 |
| Heliophileae | [1: 90] | 10,11 | 14 [7] | 0.39 (0.07) [4] | 0.46 (0.07) | 304 | 86 |
| Hesperideae | [2: 36] | 6,7 (8,10?) | 29 [21] | 4.33 (0.12) [3] | 5.71 (1.83) | 3377 | 0 |
| Iberideae | [2: 31] | 7–11 | 27 [26] | 0.66 (0.25) [8] | 0.69 (0.21) | 514 | 28 |
| Isatideae | [5: 91] | 7 (6,8) | 39 [36] | 0.35 (0.04) [11] | 0.38 (0.08) | 273 | 53 |
| Kernereae | [2: 2] | 7 (8) | 0 [2] | 0.20 [1] | 0.20 | 156 | 100 |
| Lepidieae | [3: 265] | 8 (6,7) | 73 [67] | 0.30 (0.15) [10] | 0.66 (0.39) | 234 | 44 |
| Malcolmieae | [1: 6] | 7,8 | 14 [7] | 0.26 [1] | 0.26 | 203 | 100 |
| Megacarpaeeae | [2: 11] | 7,9 | 0 [2] | No data | No data | 50 | |
| Microlepidieae | [16: 59] | 4–6 | 7 [15] | 0.50 (0.05) [3] | 0.50 (0.56) | 390 | 67 |
| Notothlaspideae | [1: 2] | 19? | 100 [1] | No data | No data | 0 | |
| Oreophytoneae | [2: 6] | 8 | 33 [3] | 0.19 [2] | 0.19 | 148 | 0 |
| Physarieae | [7: 135] | 4–10 | 46 [82] | 0.78 (0.87) [6] | 1.09 (0.94) | 608 | 20 |
| Schizopetaleae | [2: 15] | 9,10 | 0 [1] | No data | No data | 100 | |
| Scoliaxoneae | [1: 1] | No data | No data | No data | No data | 0 | |
| Shehbazieae | [1: 1] | No data | No data | No data | No data | 0 | |
| Sisymbrieae | [1: 42] | 7 (8) | 35 [26] | 0.31 (0.06) [7] | 0.34 (0.07) | 242 | 61 |
| Smelowskieae | [1: 25] | 6 | 62 [13] | 0.26 [2] | 0.38 | 202 | 0 |
| Stevenieae | [3: 11] | 8,15 | 60 [5] | 0.46 (0.10) [3] | 1.06 (0.14) | 359 | 16 |
| Thelypodieae | [28: 249] | 10,11,13,14 (9,12,17) | 9 [75] | 0.71 (0.30) [10] | 0.82 (0.51) | 553 | 53 |
| Thlaspideae | [12: 35] | 7 (5,8,9) | 18 [17] | 0.50 (0.24) [6] | 0.65 (0.42) | 390 | 35 |
| Turritideae | [1: 2] | 6 (7,8) | 50 [2] | 0.24 [1] | 0.24 | 187 | 0 |
| Yinshanieae | [1: 13] | 6,7 | 63 [8] | No data | No data | 78 | |
| Unassigned | [15: 22] | 7,13,14 | 50 [22] | Unclear [4] | 0.75 [0.55) | 50 | |
| [325: 3740] | 43.3% [1653] | 0.66 (0.06) [331] | 0.75 (0.09) | 514 | 28 |
Percentage of neopolyploids per tribe has been calculated based on data for chromosome numbers from 1653 species. For the same 1653 species, life cycle was evaluated from the literature and the percentage of annuals per tribe is provided. Mean tribal 1 Cx and 1C genome size is provided in picograms (pg).
Neopolyploidy Is Only Weakly Correlated with Species Richness
If we contrast the percentage of neopolyploids with species richness (Figure 2), several very general observations are remarkable: (1) species richness of tribes is positively but only weakly correlated with the number of polyploids (r = 0.176, P < 0.01), (2) six tribes (Arabideae, Brassiceae, Cardamineae, Erysimeae, Lepidieae, and Thelypodieae) comprise nearly 50% of the 3740 Brassicaceae species, with 50% of neopolyploids found only in these six tribes. Interestingly, all six tribes belong to lineages I and II, with none in lineage III. Another minor observation (3) is that genera not assigned to any tribe yet, because of conflicting and incongruent phylogenetic signals, also consist of at least 50% polyploids. This might be indicative of a hybrid origin and thus reflect reported difficulties in reconstructing their phylogenetic relationships (Beilstein et al., 2010; Couvreur et al., 2011).
Figure 2.
Species Number per Tribe versus Percentage of Neopolyploids.
Major Brassicaceae lineages are indicated by different colors. The six most species-rich tribes (indicated by black circled dots) comprise nearly 50% of the species diversity of the whole family with more than 200 species each. Mesopolyploidy has been reported for five tribes to date (tribe abbreviations in bold).
Tribes with 100 to 200 species (Alysseae, Boechereae, Coluteocarpeae, Euclidieae, and Physarieae) have between 30 and 50% neopolyploids and form a heterogeneous group with tribes belonging to all three lineages. The tribe Physarieae (135 spp, 46% neopolyploids) might be species and polyploidy rich because of a purported WGD (Lysak et al., 2009), whereas the high number of neopolyploids in Boechereae (128 spp, 45% neopolyploids) is due to the apomictic mode of reproduction (Kiefer et al., 2009a, 2009b; Kiefer and Koch, 2012; Mandáková et al., 2015; Mau et al., 2015). Numerous triploid apomicts in the tribe Boechereae have been shown not to be necessarily an evolutionary dead end, being mostly facultative and not obligate apomicts (Aliyu et al., 2010). Alysseae (275 spp, 32% polyploids), Coluteocarpeae (128 spp, 31% polyploids), and Euclidieae (151 spp, 36% polyploids) most likely have not undergone a mesopolyploid WGD.
Genome Size Evolution and Chromosome Number Variation
Testing the various hypotheses for how genome size (GS) might have changed during polyploidization and radiation processes, we found no significant correlation [Kendall’s tau-b coefficient TB = 0.294, Spearman’s rank order coefficient ρ (rho) = 0.382] between holoploid GS and holoploid chromosome number (Table 2). This contradicts a hypothesis of continuous GS increase over evolutionary time scales (e.g., as significantly shown for Fritillaria; Kelly et al., 2015) and, inversely, suggests rapid genome reorganization and genome downsizing after polyploidization (Leitch and Bennett, 2004; Weiss-Schneeweiss et al., 2006; Lysak et al., 2009).
Table 2. Correlation Analysis.
| Kendall’s tau-b Coefficient TB/Spearman’s Rank Order Coefficient ρ (rho) | (1) Holoploid chromosome number (n = 4–68) | (2) Monoploid Chromosome Number (x = 4–19) |
|---|---|---|
| (1) Holoploid genome size (1 C-value) | 0.294**/0.382** | – |
| (2) Monoploid genome size (1 Cx value) | – | 0.337**/0.441** |
(1) Correlation between holoploid genome size (1 C-value) and holoploid chromosome number (n) across the Brassicaceae (331 species analyzed in total). (2) Correlation between monoploid GS (1 Cx value) and monoploid (base) chromosome number (x) across the Brassicaceae. The monoploid chromosome number of each taxon was adopted from the literature or deduced under the assumption of an ancestral crucifer karyotype of n = 8 for lineages I and III (Lysak et al., 2006; Schranz et al., 2006) and n = 7 for (extended) lineage II (Mandáková and Lysak, 2008). **P < 0.01.
Monoploid GS and monoploid chromosome number also showed no significant correlation [Kendall’s tau-b coefficient TB = 0.337, Spearman’s rank order coefficient ρ (rho) = 0.441]. This finding also contradicts the idea of increasing GS per monoploid genome, thereby indicating effective shrinkage and diploidization of genomes after WGD.
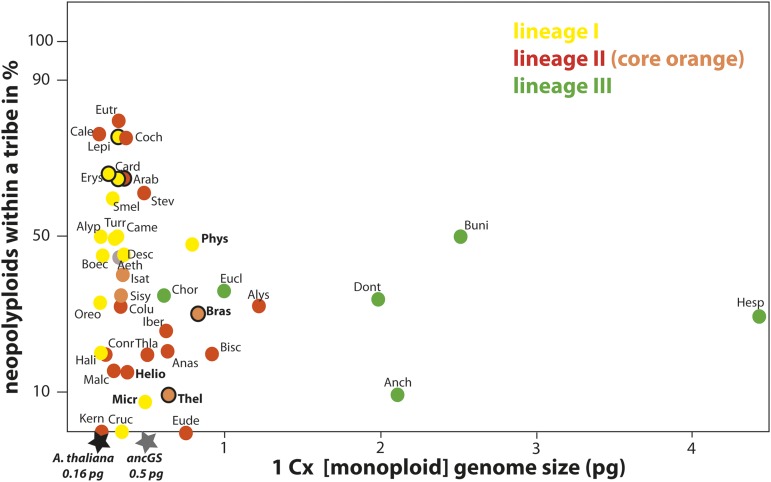
GS within the Brassicaceae family was low in general (mean 1 Cx = 0.66 ± 0.30 pg) (Table 1, Figure 3), but the average GS for lineage III tribes (mean 1 Cx = 2.06 ± 1.01 pg) was comparatively high and significantly different from the family’s mean (Table 1, Figure 3).
Figure 3.
Mean Monoploid Genome Size (1 Cx) per Tribe versus Percentage of Neopolyploids.
Major Brassicaceae lineages are indicated by different colors. The mean GS for six lineage III tribes exceeds the ancestral genome size (ancGS) of ∼0.5 pg (Lysak et al., 2009). Mesopolyploidy has been reported for five tribes to date (tribe abbreviations in bold). The six most species-rich tribes (more than 200 species each) are indicated by black circled dots.
We extrapolated GS values into genomes sizes in megabase pairs following Hohmann et al. (2014) with 1 pg DNA = 780 Mb. These data might be a valuable resource for many genomic analyses (following an Arabidopsis example from Hohmann et al. [2014]; e.g., Arabidopsis thaliana 1 Cx 0.173 pg = 135 Mb; Arabidopsis cebennensis 1 Cx 0.281 pg = 219 Mb). These values in megabase pairs are close to genome sizes ignoring (peri)centromers, repeat-rich regions, and rDNA, as these regions frequently have not been sequenced and/or assembled. Consequently, “real” genome sizes are then underestimated by ∼25% (Bennett et al., 2003).
Impact of Polyploidization on GS Variation
To test the idea that polyploidization during the evolutionary history of the family was a prerequisite for subsequent diversification and radiation of the different main lineages and tribes, we investigated the relationship between the percentage of polyploids per tribe and mean tribal GS.
The results are shown in Figure 3 and some remarkable trends can be seen: (1) All main evolutionary lineages tend to reduce their monoploid GS close to the inferred ancestral genome size (ancGS) of ∼0.5 pg, i.e., 390 Mb (Lysak et al., 2009) or even lower, close to the smallest genome size (A. thaliana, 0.16 pg); (2) all tribes that are species rich (>200 taxa, black-circled dots; see also Figure 2) and in parallel highly polyploid (>50% neopolyploids) have monoploid GS smaller than the ancGS (Arabideae, Cardamineae, Erysimeae, and Lepidieae).
Interestingly, all tribes assigned to lineage III exceed the ancGS with their tribal means, by up to 9-fold in Hesperideae. These tribes contain <50% neopolyploids and with the exception of Euclidieae are relatively species poor (Table 1, Figure 2).
To look for potential correlation at the family level between GS variation and life strategy, we screened the relevant literature and original species descriptions for lifestyles (annual/winter annual and monocarpic; biennial and monocarpic; and polycarpic) in species with known cytogenetic data (Supplemental Data Set 2). We found a weak but significant relationship (rank correlation test r = 0.165, P < 0.01) between GS and developmental lifestyle, with GS in annuals being significantly (P < 0.01) lower (0.631, sd = 0.31) than in bi/perennials (0.902, sd = 0.62). Across the whole Brassicaceae, 28% of species are annuals and a comparable proportion of annual species (27%) is found in all six tribes of lineage III (Figure 3). However, among the six lineage III tribes with largest genomes among Brassicaceae, Chorisporeae and Euclidieae have smaller genomes and 35% annual species, whereas the four tribes (Anchonieae, Buniadeae, Dontostemoneae, and Hesperideae) possessing larger GS contain only 18% annuals (Figure 3).
DISCUSSION
Here, we aimed to provide a biogeographic-paleoenvironmental concept for crucifer evolution and radiation highlighting that important environmental/climatic transitions correlate with diversification and radiation of Brassicaceae from the onset of the Oligocene. These climatic changes provided new ecological and geographical spaces into which the infra-familiar evolutionary lineages could evolve. In parallel, the increase in biodiversity was accompanied by major changes in the genomic makeup. Here, multiple and often clade-specific polyploidization events played an important role, although we acknowledge polyploidization alone cannot always explain subsequent species radiations. It seems that the ability of crucifers to diploidize and stabilize their duplicated genomes rapidly is important in maintaining the diversity created via adaptive evolution within drastically changing environments, thereby enabling them to maintain genetic diversity as a whole over long periods of time.
Brassicaceae Species Radiation and Paleoenvironment
The onset of crown group diversification of Brassicaceae started with the split of tribe Aethionemeae at the transition from Eocene to Oligocene (32.4 mya). Later evolutionary splits are found at the Oligocene-Miocene boundary followed by a rapid radiation of major lineages and tribes. The split of Aethionemeae from the rest of the family might favor the idea that the family originated in the Irano-Turanian region because this tribe shows clear affinities with this region as outlined much earlier (Hedge, 1976; Al-Shehbaz et al., 2006) and the whole Brassicaceae has its highest worldwide species richness in this region (Koch and Kiefer, 2006).
For most tribes of the Brassicaceae, we have not elaborated detailed phylo/biogeographic scenarios that enable us to link paleoenvironmental and climatic data with species evolution in a spatio-temporal context. However, a more general picture emerged from Couvreur et al. (2010) with limited DNA sequence data that included sequence information from the mitochondrial genome. A very similar scenario is depicted by the results presented here (Supplemental Figure 2). Tribes from monophyletic lineage I all evolved with stem group ages ranging from 9 to 17 mya. Representative tribes from monophyletic lineage III show consistent emergence at ∼17 mya with a maximum stem age of ∼21 mya. The only difference is that the stem ages of main lineages I and II are overestimated at 27 and 28 mya, respectively, by Couvreur et al. (2010). This discrepancy could be explained by phylogenetic uncertainties in tree reconstruction (e.g., assuming the monophyly of lineage II, which is obviously misleading; Franzke et al., 2011).
Various contributions in recent years have aimed to set the evolutionary past of crucifers into a broader biogeographical-ecological and environmental context. At the tribal level, Arias et al. (2014) estimated the stem age of tribe Brassiceae at ∼24 mya and hypothesized it originated close to the Arabian Peninsula and Saharan Africa, with its various sublineages diversifying during the Miocene when the Anatolian plate collided with Africa and Arabia and the Mediterranean was cut off from the Indian Ocean (Rögl, 1999; Arias and Pires, 2012). This age might be overestimated because it is based on calibration with a fossil presumably from tribe Thlaspideae with a minimum age of ∼30 mya (Beilstein et al., 2010).
Interestingly, subsequent radiation of various genera from tribe Brassiceae overlaps with the Messinian salinity crisis (5.96 to 5.33 mya), which caused a severe drawdown of the sea level and major aridification around the Mediterranean basin (Thompson et al., 2005; Blondel et al., 2010; Garcia-Castellanos and Villaseñor, 2011). Consequently, the Messinian salinity crisis could have promoted lineage diversification either via adaptive radiation or through geographically induced allopatric events. The most species-rich tribe, Arabideae, with more than 500 species, has been demonstrated to have a stem group age of ∼18 mya, also close to the Oligocene-Miocene boundary (Karl and Koch, 2013). Here, we can assume an Irano-Turanian origin of the tribe with subsequent radiations mostly in the Eastern Mediterranean, but also in various alpine regions in Eurasia (Karl and Koch, 2013). The Late Miocene with its generally more stable climate throughout the Tortonian, 11 to 17 mya, is considered very important for subsequent species radiation in Arabideae. The crown group age of Cochlearieae was estimated to ∼13 mya (Koch, 2012), well in agreement with a much older stem group age that might be close to 20 mya as estimated in this study (Figure 1). Cochlearieae experienced a long evolutionary stasis throughout the whole Late Miocene, and species radiation of the cold-adapted genus Cochlearia started earliest after migration to northern regions outside the Mediterranean and with the onset of glaciation and deglaciation cycles in the Pleistocene.
A. thaliana crown group divergence was previously dated to ∼5.1 to 5.4 mya using one plastid and two nuclear markers (Koch et al., 2000), which is in agreement with the mean estimate of 5.8 mya presented here based on the whole-chloroplast genome. Similar to radiation within the young polyploid complex Cochlearia, subsequent speciation within perennial Arabidopsis species also started during Pleistocene glaciation and deglaciation cycles. This is also in agreement with phylogeographic-evolutionary studies of A. thaliana’s perennial relatives (Koch et al., 2008; Hohmann et al., 2014), placing any split within this genus in the Pleistocene epoch (Jakobsson et al., 2006; Koch and Matschinger, 2007; Schmickl et al., 2010, 2012; Roux et al., 2011; Pauwels et al., 2012; Tsuchimatsu et al., 2012).
Some major confusion and incongruences among divergence time estimates within the Arabidopsis clade arose from a much older crown group divergence time estimate of ∼13 mya (Beilstein et al., 2010). Two- to threefold higher estimates are found throughout Beilstein et al. (2010), which consequently provides a higher estimate for the Brassicaceae crown group radiation at 54 mya and contradicts the recently published, major angiosperm-wide data set (Magallón et al., 2015). The most likely explanation for this major incongruence is a fossil (Thlaspi primaerum; Becker, 1961; Wing, 1987) used to constrain the stem age of tribe Thlaspideae at ∼30 mya (Beilstein et al., 2010). The argument for using this fossil to set a minimum age for Thlaspideae is weak for various reasons. The most important concern is that there is no convincing argument to believe that the specimen belongs to Thlaspi (Franzke et al., 2010), a genus that has been notoriously difficult when relying on morphological characters only (Mummenhoff et al., 1997). However, fully in accordance would be a more parsimonious perspective, also followed by Magallón et al. (2015), namely, using the T. primaerum fossil as a minimum age for Brassicaceae. Consequently, past estimates of Brassicaceae crown group age (Aethionemeae versus the rest of the family; 30 to 60 mya [Koch et al., 2000, 2001], 24 to 40 mya [Ermolaeva et al., 2003], 34 mya [Schranz and Mitchell-Olds, 2006], 24 to 40 mya [Henry et al., 2006], 40 mya [Fawcett et al., 2009], and 32 mya [Edger et al., 2015]) are all in agreement with our estimate of 34 mya. It should also be mentioned that another study (Franzke et al., 2009) produced divergence time estimates consistently 2 to 3 times younger than the values proposed here. The reasons remain unclear but it could be assumed that (1) only a single secondary constraint was used and (2) that the rate of nucleotide evolution was correlated across the tree, thereby not accounting for uncertainties in the phylogenetic hypothesis and placement of fossil age constraint in general.
Polyploidization as a Source of Past and Future Diversification in Brassicaceae
Although polyploidy has long been considered important for plant speciation and diversification (Stebbins, 1938, 1950; Ehrendorfer, 1980), the advent of -omics technologies within the last decade has demonstrated that every seed plant on Earth carries a genome that has experienced at least a single round of whole-genome duplication (Amborella Genome Project, 2013). It is also widely accepted that genome multiplications drive taxonomic, phenotypic, and ecological diversification (Soltis et al., 2009; Tank et al., 2015). Following WGD, large fractions of a duplicated genome are eliminated over several millions of years of evolution. This process is not only a simple mechanistic vehicle to stabilize and maintain the general functions of the genome, associated with genome downsizing and chromosome number decreases (Lim et al., 2007; Leitch and Leitch, 2008; Lysak et al., 2009), but also provides a playground for gene neofunctionalization (e.g., flower evolution [Eckardt, 2006], the glucosinolate pathway [Yang et al., 2014], and silencing/loss of duplicated gene copies). Naturally, these diploidization processes are subject to selection, and any outcome is a result of interactions between intrinsic and extrinsic factors, such as the environment and biotic and abiotic interactions; thus, increased species diversification and adaptive radiation often follows the initial polyploidization. A similar concept has been first introduced by Schranz et al. (2012) as “WGD radiation lag-time hypothesis,” stating that among angiosperm lineages often WGD contributed to the evolution of key defining traits, and after lineage splitting and (secondary) dispersal events in the background of major environmental changes species radiations might happen. This concept has been further tested and proven by Tank et al. (2015). However, ongoing diversification and radiation is not necessarily connected to additional polyploidization, but should be associated with rapid genome downsizing and diploidization. Consequently, polyploidization per se should be only weakly correlated with species number increases over longer periods of time.
As exemplified by Brassicaceae, this is exactly what we observe across tribes, which define monophyletic clades representing a time span of 20 to 10 million years. Despite polyploidization, the tribes on average do not show any significant genome size increases (Table 2). On the contrary, genome downsizing toward or even below the ancGS value is prevalent across the family (Figure 2).
An excellent example of neofunctionalization is a major plant-insect coevolutionary innovation: the pierid (butterfly) glucosinolate detoxification mechanism, which has been reviewed recently (Wheat et al., 2007; Beilstein et al., 2010). This mechanism is matched by new glucosinolate pathways leading to new chemical defense compounds. Here, WGD in the Brassicales and Brassicaceae played a major role (Hofberger et al., 2013) in establishing new functions. It is intriguing that Pierinae fossils, a subfamily of the Pieridae with modern relatives in the Brassicales feeding clade, appear 34 mya (Braby and Trueman, 2006; Wheat et al., 2007), thereby suggesting that the increased diversification of the Pierinae resulted from a host-plant shift onto Brassicales/Brassicaceae, which matches perfectly the onset of crown group radiation in Brassicaceae ∼32 mya. Several members of the subfamily Pierinae also feed on host plants from the families Capparidaceae, Santalaceae, and Loranthaceae, and among members of the other three subfamilies of the Pieridae there are many tropical lineages (DeVries, 2001). This coincides with the fact that the estimated stem age of the Pieridae family is between 82.5 and 111 mya and the early evolution of tropical Brassicales is in the same temporal dimensions (Wheat et al., 2007; Beilstein et al., 2010; Edger et al., 2015).
Unusual Genome Size Increase in Tribes of Lineage III
The overall tendency of genome downsizing across the Brassicaceae is not followed by lineage III. Four tribes from lineage III (Anchonieae, Buniadeae, Dontostemoneae, and Hesperideae) harbor species with the largest genomes in the family, while the remaining two tribes (Chorisporeae and Euclidieae) have smaller but still relatively large genome sizes (Figure 2, Table 1). These findings are congruent with those of Lysak et al. (2009). This group, particularly the analyzed genera Bunias, Hesperis, Matthiola, and Clausia, has paradoxical karyological characteristics: exceptionally large genomes for crucifer species, but few (n = 6 to 7) very large chromosomes. Interestingly, based on the available data, none of these tribes have undergone an extensive radiation compared with the most species-rich tribes nor are they characterized by a high percentage of polyploids (Figure 1). The larger genome sizes in most tribes of lineage III are positively correlated with the percentage of perennial and polycarpic species in these tribes. This is in accordance with the observed relationship between DNA content and lifestyle expressed as minimum generation time (reviewed in Leitch and Bennett, 2007). In lineage III genomes, the GS increase through accumulation of repetitive sequences probably limits the option of being an ephemeral or annual, in favor of the perennial polycarpic lifestyle. In Chorisporeae and Euclidieae, the higher proportion of annual species is associated with genomes being significantly smaller than in the four remaining tribes.
As discussed by Lysak et al. (2009), the unusual association of large genome sizes, large chromosomes and lower-than-ancestral chromosome numbers can be explained by several nonexclusive processes, namely, a still overlooked WGD event prior to the diversification of lineage III followed by independent diploidizations, resulting in quasidiploid genomes with ancestral-like or lower chromosome numbers. However, a species from Hesperideae (Hesperis matronalis; a neopolyploid species with 2n = 24) was investigated recently (Kagale et al., 2014), but a past mesopolyploidization was not detected in this genome. Alternative scenarios include amplification of repetitive elements only in the four tribes (Anchonieae, Buniadeae, Dontostemoneae, and Hesperideae) or the existence of an effective downsizing mechanism in Chorisporeae and Euclidieae. Clearly, a WGD followed by diploidizing descending dysploidy and amplification of retrotransposons and satellite repeats are mutually nonexclusive and can act in parallel. When looking for parallels across Brassicaceae, both alternatives show some support. Genome evolution through WGD diploidization cycles toward small diploid-like genomes with small chromosomes is characteristic of Brassiceae, Heliophileae, and Microlepidieae, but evolution in some Physarieae taxa most likely represents a pattern of WGD followed by diploidization and descending dysploidy toward large genomes with a few large chromosomes (Lysak et al., 2009).
Polyploidization and Radiation in the Brassicaceae
Although the long-term importance of polyploidy, and particularly its association with lower diversification rates, was disputed recently (Mayrose et al., 2011; Arrigo and Barker, 2012), a number of large-scale radiations were preceded by WGD events (Soltis et al., 2009; Jiao et al., 2012; Vanneste et al., 2014). In Brassicaceae, mesopolyploid WGDs postdating the α-WGD most likely accelerated diversification and species radiation in the Brassiceae (Lysak et al., 2005), the New Zealand and Australian Microlepidieae (Mandáková et al., 2010a, 2010b), Heliophileae (Mandáková et al., 2012), Biscutelleae (C. Geiser, T. Mandáková, N. Arrigo, M.A. Lysak, and C. Parisod, unpublished results), Thelypodieae (Burrell et al., 2011), Leavenworthia (Cardamineae; Haudry et al., 2013), and possibly in Physarieae (Lysak et al., 2009).
The extent of post-WGD diploidization varies among polyploid lineages according to the age of these events, ecogeographic conditions, and intrinsic factors. Whereas diploidization progressed to low quasidiploid chromosome numbers in Microlepidieae and Physarieae, the chromosome counts in Thelypodieae, Heliophileae, and Brassiceae remained relatively high (≥n = 20). We argue that genome diploidization often associated with descending dysploidy can further promote diversification through speciation and cladogenesis (origin of new genera), presumably through chromosome rearrangements creating reproductive barriers to infraspecific gene flow. Furthermore, diploidized mesopolyploid species may produce neopolyploid taxa via autopolyploidization or allopolyploidization. This would explain the coincidence between pronounced karyological (multiple monoploid chromosome numbers in genera and tribes), morphological and taxonomic diversity, eco-geographic variation, and frequently reticulate and complex phylogenetic relationships in mesopolyploid genera and tribes.
An increase in the number of polyploids coupled with increased speciation rates and species richness has been demonstrated for a few significant groups within the Brassicaceae: tribe Arabideae, and in particular for the largest genus in the family, Draba, with ∼400 species (Jordon-Thaden et al., 2013); tribe Heliophileae (Mandáková et al., 2012); and Cochlearia within tribe Cochlearieae (Koch et al., 1998). These radiations are all recent diversifications seen within the scenarios of increased aridity in the Late Pliocene and of alternating cold and warm phases during Pleistocene glaciation and deglaciation cycles (Warren and Hawkins, 2006; Mandáková et al., 2012).
In summary, we conclude that polyploidization in Brassicaceae is not an evolutionary dead end, but it is a prerequisite for subsequent diversification and radiation. Therefore, it will be exciting to see what future studies on other polyploid-rich angiosperm plant families reveal.
METHODS
Plant Material and Taxon Sampling
Chromosome number information from ∼1653 Brassicaceae species has been accumulated in recent years; we obtained the details using the Cytogenetics Tool from the BrassiBase knowledge database system for cytogenetic analysis (Warwick and Al-Shehbaz, 2006; Koch et al., 2012; Kiefer et al., 2014). The corresponding taxon list is provided in Supplemental Data Set 1. An overview of the data indicating species coverage per individual tribe is provided in Table 3. The information gathered included chromosome numbers and a survey of ploidy levels and corresponding base chromosome numbers, but also categorization of neopolyploidy and mesopolyploidy (if documented). The base chromosome numbers found within the various tribes varied from 4 to 19 (Lysak et al., 2006, 2009); these are also provided (based on Warwick and Al-Shehbaz, 2006). Genome size data were collected from the literature, but a considerable number of taxa (334 in total) were newly investigated for this study (Supplemental Data Set 3).
Table 3. Coverage of Cytogenetic Data Sorted for Taxonomic Levels.
| Taxonomic Level | 51 Tribes | 325 Genera | 3740 Species |
|---|---|---|---|
| Chromosome countsa | 47 (92%) | 230 (71%) | 1653 (44%) |
| Genome sizesb | 41 (80%) | 127 (39%) | 331 (9%) |
A summary of the numbers of species/genera and tribes and currently incorporated data in BrassiBase and its coverage of taxonomic ranks within the Brassicaceae (total number and percentage of total number) used for the analyses conducted here.
Chromosome counts are currently available from 9035 data points collected from 1421 references.
Genome size data represent 331 taxa in BrassiBase. Full data can be downloaded without any restriction from BrassiBase (http://brassibase.cos.uni-heidelberg.de/; Kiefer et al., 2014).
For all taxa with cytogenetic information available, we surveyed the relevant literature (e.g., flora and species descriptions) but also consulted original voucher material from various herbaria to collect life cycle information (annual/winter annual, biennial, perennial, and polycarpic).
Cytogenetic Analyses
It is well documented with several examples at various taxonomical levels (species complexes, genera, and tribes) that crucifer evolution is often characterized by hybridization and polyploidization. Therefore, here we aimed to provide a comprehensive overview of the occurrence of neopolyploidy across the entire family. We further aimed to compare the phenomenon of neopolyploidy tribe-wise with corresponding base chromosome numbers and average genome size across tribes and the whole family.
Nuclear DNA content was determined using flow cytometry following a simplified two-step protocol (Doležel et al., 2007). Approximately 10 mm2 of fresh leaf tissue from each plant to be analyzed was chopped together with an appropriate volume of the respective internal reference standard (Raphanus sativus cv Saxa, 1.11 pg/2C; Solanum lycopersicum cv Stupicke, 1.96 pg/2C; Glycine max cv Polanka, 2.5 pg/2C; Zea mays cv CE-777, 5.43 pg/2C; Pisum sativum cv Citrad, 9.09 pg/2C) (Doležel et al., 1992) using a sharp razor blade in a Petri dish containing 0.5 mL of ice-cold lysis buffer LB01 (Doležel et al., 1989) supplemented with 2 µg mL−1 β-mercaptoethanol. The suspension was filtered through a 30-μm CellTrics filter (Sysmec-Partec), and 1.5 mL of LB01 buffer was added as well as propidium iodide and RNase (both to a final concentration of 50 μg mL−1). After 90 min incubation on ice, the relative fluorescence intensity of 5000 particles was recorded using a flow cytometer (CyFlow Space; Sysmex-Partec) equipped with a green (532-nm) solid-state laser. We applied the following stringent criteria to get precise and stable flow cytometric results: (1) Only analyses where the coefficient of variation of the sample peak was below 5% were taken into account; (2) each sample was measured twice on different days to minimize potential random instrumental drift (Doležel and Bartoš, 2005); and (3) if the between-day variation exceeded a 5% threshold, another measurement was made and the most remote value was discarded when the sample was reanalyzed. The histograms were evaluated with the FloMax FCS 2.0 program (Sysmex-Partec). The data were deposited in BrassiBase (Koch et al., 2012; Kiefer et al., 2014).
Chromosome number reports from recent decades were critically evaluated regarding taxonomy and reliability. The complete documentation and citation list (with 1421 citations) is accessible via BrassiBase (Kiefer et al., 2014).
The cytogenetic data were first analyzed in a descriptive way to provide a comprehensive overview. We then tested two hypotheses in more detail. (1) As a result of past and recent polyploidization events, the holoploid chromosome number (n, whole set of chromosomes) increases and the genome size of the whole chromosome set (holoploid genome size, C-value) is assumed to increase as well. We tested whether this correlation was true or if rapid genome downsizing over the entire data set suggested rapid genetic stabilization and diploidization of whole genomes leading rapidly to mesopolyploids. (2) As a consequence of WGD, the base chromosome number (x, haploid chromosome number of a series of chromosome numbers within a taxon) might increase and genome size corrected for recent polyploidy (monoploid genome size, 1 Cx value) might be assumed to increase as well. Statistical rank correlation tests were performed with SPSS v19.0 (IBM)
Taxon Sampling for Phylogenetic Analyses and Divergence Time Estimates
Genomic data for phylogenetic analysis and divergence time estimates were either retrieved from the NCBI or newly generated for this study. Our aim was to include all significant lineages of the Superrosidae clade exemplified earlier in an angiosperm-wide chloroplast genome data set (Ruhfel et al., 2014), but in particular the sister relationships among lineages where reliable fossil evidence was used for angiosperm-wide divergence time estimates (Magallón et al., 2015). In addition to data available at GenBank/NCBI, we included the chloroplast genomes of Mangifera indica from Azim et al. (2014) and Malus domesticus from http://rosaceae.org (Velasco et al., 2010). Vitis vinifera was used as an outgroup to the Superrosidae clade. Finally, we added 12 newly sequenced chloroplast genomes from various Brassicaceae including five chloroplast genomes from Arabidopsis representing all major lineages of the genus (Hohmann et al., 2014), as well as their close relatives Capsella rubella and Camelina sativa. Five genomes were added from Cochlearia and Ionopsidium representing the best documented example of reticulate evolution that has generated a series of auto- and allopolyploids (Koch et al., 1998, 1999, 2003; Koch, 2002, 2012). In total, we included 29 Brassicaceae chloroplast genomes from 11 tribes and all three major evolutionary lineages. A summary of accession codes and biological resources is given in Supplemental Data Set 4.
DNA Extraction, Library Preparation, and Next-Generation Sequencing
DNA was extracted from either fresh or dried leaf material using the Invisorb Spin Plant Mini Kit (STRATEC Biomedical). Total genomic DNA libraries were prepared and sequenced at the CellNetworks Deep Sequencing Core Facility (Heidelberg) on an Illumina HiSequation 2000 in 100-bp paired-end mode with a library insert size of 200 to 400 bp.
De Novo Sequence Assembly of Chloroplast Genomes
Complete chloroplast genomes were assembled in CLC Genomics Workbench v6.0.4 (CLC Bio). Quality and adapter trimming was conducted using a minimum per-base quality of 0.001 (corresponding to a phred score of 30) and minimum sequence length of 50 bp. For the Arabidopsis, Capsella, and Camelina chloroplasts, trimmed paired reads were assembled using the legacy version of the CLC de novo assembly algorithm (length fraction 0.9, similarity 0.9, and appropriate distance settings). Chloroplast contigs were identified using BLASTn (with default parameters) and aligned manually to the closest related, published complete chloroplast sequence (Arabidopsis thaliana for Arabidopsis species, Capsella bursa-pastoris for Capsella rubella, and Camelina sativa) using PhyDE v0.9971 (Müller et al., 2010).
Gaps between nonoverlapping contigs were filled from the reference sequence, creating a preliminary pseudo-reference. To obtain the complete chloroplast genome and as a quality control, the reads were then mapped back to this sequence in CLC using the legacy version algorithm with a length fraction of 0.9, a similarity fraction of 0.9, and appropriate distance settings. Misalignments and mismatches between the reads and pseudo-reference were detected by checking the mapping manually and using the CLC variant detection (minimum coverage 1; variant probability 0.1). Subsequently, the sequence was adjusted and the variant detection steps were repeated until no further variants were found. The de novo assembly of the four Cochlearia samples and Ionopsidium acaule was performed according to the workflow described above with the following slight modifications: The length and similarity fractions of the standard CLC de novo assembly algorithm were set to 0.8 and the cp genome of Cochlearia borzaeana, made up of two overlapping de novo contigs, was used as a reference for further contig alignments.
Alignments and Phylogenetic Data Matrix
Complete chloroplast sequences were aligned using MAFFT v7.017 (Katoh et al., 2002, 2005) implemented in Geneious v7.1.7 (Biomatters). The FFT-NS-ix1000 algorithm was used, with the 200 PAM/k = 2 scoring matrix, a gap open penalty of 1.53, and an offset value of 0.123. Alignments were constructed for every order separately, or, if not possible because of major rearrangements in the order of genes, for smaller data sets. Coding sequences were then extracted from the alignments according to published annotations, and introns were excluded. Finally, the gene alignments of all orders were combined, and genes with no missing data were selected, i.e., coding sequence annotations had to be present for all sequences, except Mangifera indica and Malus domesticus (which were not annotated, so the annotation of the closest relative was used); pseudogenes were excluded. This resulted in a set of 73 genes, including 51 protein-coding genes, 19 tRNAs, and three rRNAs. All genes were realigned (settings as above) and checked manually, and for protein-coding genes start and stop codons were excluded from further analyses because of their high potential for homoplasy.
Indels were excluded from the alignments using Gblocks v0.91b (Castresana, 2000). The minimum length of a block was set to 2 bp, and because of the high sequence divergence in the data set, nonconserved blocks were also saved. The remaining data set comprised 38,622 bp of sequence information, of which 15,010 characters were variable.
To account for rate heterogeneity among genes, the data set was partitioned into subsets of genes evolving at a similar rate under the same substitution model using PartitionFinder v1.1.1 (Lanfear et al., 2012, 2014). Branch lengths were allowed to be unlinked, and only models implemented in BEAST were tested, with BIC used for model selection in a greedy search. Three subsets were found, of 20,655, 11,210, and 6757 bp (7388, 6343, and 1279 variable characters, respectively), each with GTR+Γ+I as the substitution model. Alignments are provided in Supplemental Data Set 5.
Phylogenetic Analysis
Phylogenetic trees were reconstructed with ML using RAxML v8.1.16 (Stamatakis, 2014) based on the three partitioned sequence alignments described above. A rapid bootstrap analysis and search for the best-scoring ML tree was conducted with 1000 bootstrap replicates. The partitioned data set with per-partition estimation of branch lengths was used with GTR+I+Γ as the substitution model and Vitis vinifera was chosen as an outgroup. Initial ML analyses used a larger data set. For computational feasibility of BEAST analyses, we excluded closely related species within well represented clades but aimed to keep representatives of all major lineages, major crops, and taxa with whole nuclear genome sequence information available.
Divergence Time Estimation and Fossil Calibration
Divergence time estimation was conducted in BEAST v1.7.5 (Drummond et al., 2012) using independent site and clock models, but a combined partition tree for the three partitions (GTR+I+Γ). Vitis was defined as an outgroup. The ML tree was used as a starting tree after it was made ultrametric with node ages to fit constraints using the R package APE version 3.1-4 (Paradis et al., 2004).
We used an uncorrelated lognormal relaxed clock approach with estimated rates to account for rate variation among branches (Drummond et al., 2006). We chose four fossil constraints that were also used in the most recent and comprehensive angiosperm-wide temporal analyses and were in accordance with Magallón et al. (2015). Following Njuguna et al. (2013), the minimum age for the Prunus/Malus split was set to 48.4 mya (Benedict et al., 2011) and the Castanea/Cucumis split to 84 mya (Sims et al., 1999; Moore et al., 2010). Following Bell et al. (2010), the minimum age for Mangifera/Citrus was set to 65 mya (Knobloch and Mai, 1986) and for Oenothera/Eucalyptus to 88.2 mya (Takahashi et al., 1999). The recently introduced fossil from within the Brassicaceae (Thlaspi primaerum) (Beilstein et al., 2010) was not included and we followed Magallón et al. (2015), who used this taxon to set a minimum age for the Brassicaceae crown group of ∼25 mya. However, because all phylogenetic-temporal analysis demonstrated higher ages than that for the radiation of the family, there was no need to include fossil evidence that is still under debate (Franzke et al., 2011). A uniform distribution was used for all four fossil constraints with a maximum age of 125 mya. The root was calibrated with a uniform distribution between 92 and 125 Ma (minimum age of 125 mya to crown eudicots; Magallón et al., 2015). We also tested whether exponential or log normal distributions of fossil calibrations resulted in different age estimates by running the same analyses but with 100,000,000 generations. However, no substantial differences were found, so we chose uniform distributions, which use the least assumptions and assume the same model used in the most recent contribution (Beilstein et al., 2010).
We ran two independent MCMC runs with 500,000,000 generations each and sampling parameters every 50,000 generations. LogCombiner v1.7.5 (Drummond et al., 2012) was used to combine trees from the two runs and the first 50,000,000 generations of each run were discarded as burn-in. The resulting 18,000 trees were combined to a maximum clade credibility tree in TreeAnnotator v1.7.5 (Drummond et al., 2012) and visualized in FigTree v1.4.1 (Drummond et al., 2012).
Life Cycle and Genome Size
For all taxa with cytogenetic data available, the relevant literature and material (floras, databases, original voucher specimens via digitally available databases such as JSTOR plant) were inspected to survey information on their life cycles. We categorized life cycles into (1) annual and winter-annual (both monocarpic), (2) biennial (monocarpic), and (3) perennial (polycarpic). We used rank correlation tests to compare this information with genome size data (also categorized into [1] <0.5 pg Cx values, smaller than the ancestral crucifer genome size; [2] 0.5 to 1.0 pg Cx values; and [3] >1.0 pg Cx values).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers LN866844-LN866848 and LN877380-LN877386.
Supplemental Data
Supplemental Figure 1. Maximum likelihood tree for the Superrosidae clade based on chloroplast genome sequence information.
Supplemental Figure 2. Summarizing cartoon of phylogenetic relationships, cytogenetic features and age estimates among the various tribes of Brassicaceae.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.qd0c2.
Supplemental Data Set 1. Brassicaceae species checklist for cytogenetically investigated taxa and respective chromosome numbers (data export from BrassiBase; http://brassibase.cos.uni-heidelberg.de/).
Supplemental Data Set 2. Species list with data for Brassicaceae life cycle.
Supplemental Data Set 3. Species list for genome size estimates of Brassicaceae species.
Supplemental Data Set 4. Sources of plant material and sequence data used for chloroplast genome analysis.
Supplemental Data Set 5. Alignments used for phylogenetic analysis.
Supplementary Material
Acknowledgments
We thank Dmitry German for help with the taxonomic species checklist and scoring of life cycle data and Markus Kiefer for substantial support for in-house IT infrastructure and program running. We also thank Roswitha Schmickl, Peter Sack, Susanne Ball, and Thorsten Jakob for excellent plant care and help in the lab estimating chromosome numbers and genome sizes. This work was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft; Grants KO2302/11 and KO2302/13) to M.A.K. and by the Czech Science Foundation (13-10159S) to M.A.L.
AUTHOR CONTRIBUTIONS
N.H. and E.M.W. conducted genomic analysis and contributed to writing the article. M.A.L. helped with interpretations of cytogenetic data, contributed to the discussion, and wrote the article. M.A.K. designed the research and experimental setup, evaluated and analyzed cytogenetic and biodiversity data, and wrote the article.
Glossary
- WGD
whole-genome duplication
- mya
million years ago
- ML
maximum likelihood
- GS
genome size
Footnotes
Articles can be viewed online without a subscription.
References
- Aliyu O.M., Schranz M.E., Sharbel T.F. (2010). Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am. J. Bot. 97: 1719–1731. [DOI] [PubMed] [Google Scholar]
- Al-Shehbaz I.A. (2012). A generic and tribal synopsis of the Brassicaceae (Cruciferae). Taxon 61: 931–954. [Google Scholar]
- Al-Shehbaz I.A., Beilstein M.A., Kellogg E.A. (2006). Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Syst. Evol. 259: 89–120. [Google Scholar]
- Amborella Genome Project (2013). The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Appel O., Al-Shehbaz I.A. (2002). Cruciferae. In The Families and Genera of Vascular Plants, Vol. V, Kubitzki K., ed (Heidelberg, Germany: Springer Berlin; ), pp. 75–174. [Google Scholar]
- Arias T., Pires J.C. (2012). A fully resolved chloroplast phylogeny of the brassica crops and wild relatives (Brassicaceae: Brassiceae): Novel clades and potential taxonomic implications. Taxon 61: 980–988. [Google Scholar]
- Arias T., Beilstein M.A., Tang M., McKain M.R., Pires J.C. (2014). Diversification times among Brassica (Brassicaceae) crops suggest hybrid formation after 20 million years of divergence. Am. J. Bot. 101: 86–91. [DOI] [PubMed] [Google Scholar]
- Arrigo N., Barker M.S. (2012). Rarely successful polyploids and their legacy in plant genomes. Curr. Opin. Plant Biol. 15: 140–146. [DOI] [PubMed] [Google Scholar]
- Azim M.K., Khan I.A., Zhang Y. (2014). Characterization of mango (Mangifera indica L.) transcriptome and chloroplast genome. Plant Mol. Biol. 85: 193–208. [DOI] [PubMed] [Google Scholar]
- Bailey C.D., Koch M.A., Mayer M., Mummenhoff K., O’Kane S.L. Jr., Warwick S.I., Windham M.D., Al-Shehbaz I.A. (2006). Toward a global phylogeny of the Brassicaceae. Mol. Biol. Evol. 23: 2142–2160. [DOI] [PubMed] [Google Scholar]
- Becker H.F. (1961). Oligocene plants from the upper Ruby river basin, southwestern Montana. GSA Memoirs 82: 1–122. [Google Scholar]
- Beilstein M.A., Al-Shehbaz I.A., Kellogg E.A. (2006). Brassicaceae phylogeny and trichome evolution. Am. J. Bot. 93: 607–619. [DOI] [PubMed] [Google Scholar]
- Beilstein M.A., Nagalingum N.S., Clements M.D., Manchester S.R., Mathews S. (2010). Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 18724–18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict J.C., DeVore M.L., Pigg K.B. (2011). Prunus and Oemleria (Rosaceae) flowers from the late early Eocene Republic flora of northeastern Washington state, USA. Int. J. Plant Sci. 172: 948–958. [Google Scholar]
- Bennett M.D., Leitch I.J., Price H.J., Johnston J.S. (2003). Comparisons with Caenorhbditis (∼100 Mb) and Drosophila (∼175 Mb) using flow cytometry show genome size in Arabidopsis to be ∼157 Mb and thus ∼25% larger than the Arabidopsis Genome Initiative estimates of ∼125 Mb. Ann. Bot. (Lond.) 91: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel J., Aronson J., Bodiou J.Y., Boeuf G. (2010). The Mediterranean Region: Biological Diversity through Time and Space. (Oxford, UK: Oxford University Press; ). [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438. [DOI] [PubMed] [Google Scholar]
- Braby M.F., Trueman J.W.H. (2006). Evolution of larval host plant associations and adaptive radiation in pierid butterflies. J. Evol. Biol. 19: 1677–1690. [DOI] [PubMed] [Google Scholar]
- Burrell A.M., Taylor K.G., Williams R.J., Cantrell R.T., Menz M.A., Pepper A.E. (2011). A comparative genomic map for Caulanthus amplexicaulis and related species (Brassicaceae). Mol. Ecol. 20: 784–798. [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Cheng F., Mandáková T., Wu J., Xie Q., Lysak M.A., Wang X. (2013). Deciphering the diploid ancestral genome of the Mesohexaploid Brassica rapa. Plant Cell 25: 1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur T.L., Franzke A., Al-Shehbaz I.A., Bakker F.T., Koch M.A., Mummenhoff K. (2010). Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 27: 55–71. [DOI] [PubMed] [Google Scholar]
- De Bodt S., Maere S., Van de Peer Y. (2005). Genome duplication and the origin of angiosperms. Trends Ecol. Evol. (Amst.) 20: 591–597. [DOI] [PubMed] [Google Scholar]
- DeVries P.J. (2001). Butterflies. In The Encyclopedia of Biodiversity, I–V, Levin S.A., ed (San Diego, CA: Academic Press; ), pp. 559–573. [Google Scholar]
- Doležel J., Bartoš J. (2005). Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. (Lond.) 95: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doležel J., Binarová P., Lucretti S. (1989). Analysis of nuclear DNA content in plant cells by flow cytometry. Biol. Plant. 31: 113–120. [Google Scholar]
- Doležel J., Greilhuber J., Suda J. (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2: 2233–2244. [DOI] [PubMed] [Google Scholar]
- Doležel J., Sgorbati S., Lucretti S. (1992). Comparison of three fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 85: 625–631. [Google Scholar]
- Drummond A.J., Ho S.Y.W., Phillips M.J., Rambaut A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biol. 4: e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Suchard M.A., Xie D., Rambaut A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckardt N.A. (2006). Functional Divergence of AP3 Genes in the MAD World of Flower Development. Plant Cell 18: 1779–1781. [Google Scholar]
- Edger P.P., et al. (2015). The butterfly plant arms-race escalated by gene and genome duplications. Proc. Natl. Acad. Sci. USA 112: 8362–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrendorfer F. (1980). Polyploidy and distribution. In Polyploidy. Biological Relevance, Lewis W.H., ed (New York, London: Plenum Press; ), pp. 45–59. [Google Scholar]
- Ermolaeva M.D., Wu M., Eisen J.A., Salzberg S.L. (2003). The age of the Arabidopsis thaliana genome duplication. Plant Mol. Biol. 51: 859–866. [DOI] [PubMed] [Google Scholar]
- Fawcett J.A., Maere S., Van de Peer Y. (2009). Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 106: 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L.E., Wendel J.F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183: 557–564. [DOI] [PubMed] [Google Scholar]
- Franzke A., German D., Al-Shehbaz I.A., Mummenhoff K. (2009). Arabidopsis family ties: molecular phylogeny and age estimates in Brassicaceae. Taxon 58: 425–437. [Google Scholar]
- Franzke A., Koch M.A., Couvreur T.L.P., Lysak M.A., Mummenhoff K. (2010). On the age of the mustard family (Brassicaceae). Nature 467: 755. [Google Scholar]
- Franzke A., Lysak M.A., Al-Shehbaz I.A., Koch M.A., Mummenhoff K. (2011). Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 16: 108–116. [DOI] [PubMed] [Google Scholar]
- Garcia-Castellanos D., Estrada F., Jiménez-Munt I., Gorini C., Fernàndez M., Vergés J., De Vicente R. (2009). Catastrophic flood of the Mediterranean after the Messinian salinity crisis. Nature 462: 778–781. [DOI] [PubMed] [Google Scholar]
- Garcia-Castellanos D., Villaseñor A. (2011). Messinian salinity crisis regulated by competing tectonics and erosion at the Gibraltar arc. Nature 480: 359–363. [DOI] [PubMed] [Google Scholar]
- Haudry A., et al. (2013). An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 45: 891–898. [DOI] [PubMed] [Google Scholar]
- Hedge I.C. (1976). A systematic and geographical survey of the Old World Cruciferae. In The Biology and Chemistry of the Cruciferae, Vaughan J.G., Macleod A.J., Jones B.M.G., eds (London: Academic Press; ), pp. 1–45. [Google Scholar]
- Henry Y., Bedhomme M., Blanc G. (2006). History, protohistory and prehistory of the Arabidopsis thaliana chromosome complement. Trends Plant Sci. 11: 267–273. [DOI] [PubMed] [Google Scholar]
- Hofberger J.A., Lyons E., Edger P.P., Chris Pires J., Eric Schranz M. (2013). Whole genome and tandem duplicate retention facilitated glucosinolate pathway diversification in the mustard family. Genome Biol. Evol. 5: 2155–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann N., Schmickl R., Chiang T.-Y., Lučanová M., Kolář F., Marhold K., Koch M.A. (2014). Taming the wild: resolving the gene pools of non-model Arabidopsis lineages. BMC Evol. Biol. 14: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbrie J., et al. (1993). On the structure and origin of major glaciation cycles 2. The 100,000‐year cycle. Paleoceanography 8: 699–735. [Google Scholar]
- Jakobsson M., Hagenblad J., Tavaré S., Säll T., Halldén C., Lind-Halldén C., Nordborg M. (2006). A unique recent origin of the allotetraploid species Arabidopsis suecica: Evidence from nuclear DNA markers. Mol. Biol. Evol. 23: 1217–1231. [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Jiao Y., et al. (2012). A genome triplication associated with early diversification of the core eudicots. Genome Biol. 13: R3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly S., Heenan P.B., Lockhart P.J. (2009). A Pleistocene inter-tribal allopolyploidization event precedes the species radiation of Pachycladon (Brassicaceae) in New Zealand. Mol. Phylogenet. Evol. 51: 365–372. [DOI] [PubMed] [Google Scholar]
- Jordon-Thaden I., Koch M. (2008). Species richness and polyploid patterns in the genus Draba (Brassicaceae): a first global perspective. Plant Ecol. Divers. 1: 255–263. [Google Scholar]
- Jordon-Thaden I.E., Al-Shehbaz I.A., Koch M.A. (2013). Species richness of the globally distributed, arctic–alpine genus Draba L. (Brassicaceae). Alp. Bot. 123: 97–106. [Google Scholar]
- Kagale S., Robinson S.J., Nixon J., Xiao R., Huebert T., Condie J., Kessler D., Clarke W.E., Edger P.P., Links M.G., Sharpe A.G., Parkin I.A.P. (2014). Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26: 2777–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl R., Koch M.A. (2013). A world-wide perspective on crucifer speciation and evolution: phylogenetics, biogeography and trait evolution in tribe Arabideae. Ann. Bot. (Lond.) 112: 983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Kuma K., Toh H., Miyata T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly L.J., et al. (2015). Analysis of the giant genomes of Fritillaria (Liliaceae) indicates that a lack of DNA removal characterizes extreme expansions in genome size. New Phytol. 8: http://dx.doi.org/10.1111/nph.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer C., Dobeš C., Koch M.A. (2009a). Boechera or not? Phylogeny and phylogeography of eastern North American Boechera species (Brassicaceae). Taxon 58: 1109–1121. [Google Scholar]
- Kiefer C., Dobeš C., Sharbel T.F., Koch M.A. (2009b). Phylogeographic structure of the chloroplast DNA gene pool in North American Boechera--a genus and continental-wide perspective. Mol. Phylogenet. Evol. 52: 303–311. [DOI] [PubMed] [Google Scholar]
- Kiefer C., Koch M.A. (2012). A continental-wide perspective: the genepool of nuclear encoded ribosomal DNA and single-copy gene sequences in North American Boechera (Brassicaceae). PLoS One 7: e36491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer M., Schmickl R., German D.A., Mandáková T., Lysak M.A., Al-Shehbaz I.A., Franzke A., Mummenhoff K., Stamatakis A., Koch M.A. (2014). BrassiBase: introduction to a novel knowledge database on Brassicaceae evolution. Plant Cell Physiol. 55: e3. [DOI] [PubMed] [Google Scholar]
- Knobloch E.D., Mai D.H. (1986). Monograph of the fruits and seeds in the Cretaceous of Central Europe. Rozpravy Ústreõdního Ústavu Geologickéh 47: 1–219. [Google Scholar]
- Koch M. (2002). Genetic differentiation and speciation in prealpine Cochlearia: Allohexaploid Cochlearia bavarica Vogt (Brassicaceae) compared to its diploid ancestor Cochlearia pyrenaica DC. in Germany and Austria. Plant Syst. Evol. 232: 35–49. [Google Scholar]
- Koch M., Bishop J., Mitchell‐Olds T. (1999). Molecular systematics and evolution of Arabidopsis and Arabis. Plant Biol. 1: 529–537. [Google Scholar]
- Koch M., Haubold B., Mitchell-Olds T. (2001). Molecular systematics of the Brassicaceae: evidence from coding plastidic matK and nuclear Chs sequences. Am. J. Bot. 88: 534–544. [PubMed] [Google Scholar]
- Koch M., Huthmann M., Hurka H. (1998). Isozymes, speciation and evolution in the polyploid complex Cochlearia L. (Brassicaceae). Bot. Acta 111: 411–425. [Google Scholar]
- Koch M., Al-Shehbaz I.A., Mummenhoff K. (2003). Molecular systematics, evolution, and population biology in the mustard family (Brassicaceae). Ann. Mo. Bot. Gard. 90: 151–171. [Google Scholar]
- Koch M.A. (2012). Mid-Miocene divergence of Ionopsidium and Cochlearia and its impact on the systematics and biogeography of the tribe Cochlearieae (Brassicaceae). Taxon 61: 76–92. [Google Scholar]
- Koch M.A., Al-Shehbaz I.A. (2009). Molecular systematics and evolution of “wild” crucifers (Brassicaceae or Cruciferae). In Biology and Breeding of Crucifers, S.K. Gupta, ed (Boca Raton, FL: Taylor and Francis Group; ), pp. 1–19. [Google Scholar]
- Koch M.A., Dobeš C., Kiefer C., Schmickl R., Klimes L., Lysak M.A. (2007). Supernetwork identifies multiple events of plastid trnF(GAA) pseudogene evolution in the Brassicaceae. Mol. Biol. Evol. 24: 63–73. [DOI] [PubMed] [Google Scholar]
- Koch M.A., German D.A. (2013). Taxonomy and systematics are key to biological information: Arabidopsis, Eutrema (Thellungiella), Noccaea and Schrenkiella (Brassicaceae) as examples. Front. Plant Sci. 4: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Haubold B., Mitchell-Olds T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498. [DOI] [PubMed] [Google Scholar]
- Koch M.A., Kiefer C. (2006). Molecules and migration: biogeographical studies in cruciferous plants. Plant Syst. Evol. 259: 121–142. [Google Scholar]
- Koch M.A., Kiefer M., German D.A., Al-Shehbaz I.A., Franzke A., Mummenhoff K., Schmickl R. (2012). BrassiBase: Tools and biological resources to study characters and traits in the Brassicaceae—version 1.1. Taxon 61: 1001–1009. [Google Scholar]
- Koch M.A., Matschinger M. (2007). Evolution and genetic differentiation among relatives of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 6272–6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch M.A., Wernisch M., Schmickl R. (2008). Arabidopsis thaliana’s wild relatives: an updated overview on systematics, taxonomy and evolution. Taxon 57: 933. [Google Scholar]
- Lanfear R., Calcott B., Kainer D., Mayer C., Stamatakis A. (2014). Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 14: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R., Calcott B., Ho S.Y., Guindon S. (2012). Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Leitch I.J., Bennett M.D. (2007). Genome size and its uses: the impact of flow cytometry. In Flow Cytometry with Plant Cells, J. Doležel, J. Greilhuber, and J. Suda, eds (Weinheim, Germany: Wiley-VCH Verlag), pp. 153–176. [Google Scholar]
- Leitch I.J., Bennett M.D. (2004). Genome downsizing in polyploid plants. Biol. J. Linn. Soc. Lond. 82: 651–663. [Google Scholar]
- Leitch A.R., Leitch I.J. (2008). Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. [DOI] [PubMed] [Google Scholar]
- Lim K.Y., Kovarik A., Matyasek R., Chase M.W., Clarkson J.J., Grandbastien M.A., Leitch A.R. (2007). Sequence of events leading to near-complete genome turnover in allopolyploid Nicotiana within five million years. New Phytol. 175: 756–763. [DOI] [PubMed] [Google Scholar]
- Lysak M.A., Berr A., Pecinka A., Schmidt R., McBreen K., Schubert I. (2006). Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103: 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Koch M.A. (2011). Phylogeny, genome, and karyotype evolution of crucifers (Brassicaceae). In Genetics and Genomics of the Brassicaceae, Schmidt R., Bancroft I., eds (New York: Springer; ), pp. 1–31. [Google Scholar]
- Lysak M.A., Koch M.A., Beaulieu J.M., Meister A., Leitch I.J. (2009). The dynamic ups and downs of genome size evolution in Brassicaceae. Mol. Biol. Evol. 26: 85–98. [DOI] [PubMed] [Google Scholar]
- Lysak M.A., Koch M.A., Pecinka A., Schubert I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S., Gómez‐Acevedo S., Sánchez‐Reyes L.L., Hernández‐Hernández T. (2015). A metacalibrated time‐tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207: 437–453. [DOI] [PubMed] [Google Scholar]
- Mandáková T., Heenan P.B., Lysak M.A. (2010b). Island species radiation and karyotypic stasis in Pachycladon allopolyploids. BMC Evol. Biol. 10: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Joly S., Krzywinski M., Mummenhoff K., Lysak M.A. (2010a). Fast diploidization in close mesopolyploid relatives of Arabidopsis. Plant Cell 22: 2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Lysak M.A. (2008). Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae). Plant Cell 20: 2559–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandáková T., Mummenhoff K., Al-Shehbaz I.A., Mucina L., Mühlhausen A., Lysak M.A. (2012). Whole-genome triplication and species radiation in the southern African tribe Heliophileae (Brassicaceae). Taxon 61: 989–1000. [Google Scholar]
- Mandáková T., Schranz M.E., Sharbel T.F., de Jong H., Lysak M.A. (2015). Karyotype evolution in apomictic Boechera and the origin of the aberrant chromosomes. Plant J. 82: 785–793. [DOI] [PubMed] [Google Scholar]
- Mau M., Lovell J.T., Corral J.M., Kiefer C., Koch M.A., Aliyu O.M., McKay J., Sharbel T.F. (2015). Hybrid apomicts trapped in the ecological niches of their sexual ancestors. Proc. Natl. Acad. Sci. USA 112: E2357–E2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrose I., Zhan S.H., Rothfels C.J., Magnuson-Ford K., Barker M.S., Rieseberg L.H., Otto S.P. (2011). Recently formed polyploid plants diversify at lower rates. Science 333: 1257. [DOI] [PubMed] [Google Scholar]
- Ming R., et al. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452: 991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Soltis P.S., Bell C.D., Burleigh J.G., Soltis D.E. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 107: 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummenhoff K., Franzke A., Koch M. (1997). Molecular data reveal convergence in fruit characters used in the classification of Thlaspi s.l. (Brassicaceae). Bot. J. Linn. Soc. 125: 183–199. [Google Scholar]
- Mummenhoff K., Polster A., Mühlhausen A., Theissen G. (2009). Lepidium as a model system for studying the evolution of fruit development in Brassicaceae. J. Exp. Bot. 60: 1503–1513. [DOI] [PubMed] [Google Scholar]
- Müller K., Quandt D., Müller J., Neinhuis C. (2010). PhyDE, Version 0.9971: Phylogenetic Data Editor. Available at http://www.phyde.de.
- Njuguna W., Liston A., Cronn R., Ashman T.-L., Bassil N. (2013). Insights into phylogeny, sex function and age of Fragaria based on whole chloroplast genome sequencing. Mol. Phylogenet. Evol. 66: 17–29. [DOI] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Pauwels M., Vekemans X., Godé C., Frérot H., Castric V., Saumitou-Laprade P. (2012). Nuclear and chloroplast DNA phylogeography reveals vicariance among European populations of the model species for the study of metal tolerance, Arabidopsis halleri (Brassicaceae). New Phytol. 193: 916–928. [DOI] [PubMed] [Google Scholar]
- Roux C., Castric V., Pauwels M., Wright S.I., Saumitou-Laprade P., Vekemans X. (2011). Does speciation between Arabidopsis halleri and Arabidopsis lyrata coincide with major changes in a molecular target of adaptation? PLoS One 6: e26872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rögl F. (1999). Mediterranean and Paratethys. Facts and hypotheses of an Oligocene to Miocene paleogeography (short overview). Geologica Carpathica 50: 339–349. [Google Scholar]
- Ruhfel B.R., Gitzendanner M.A., Soltis P.S., Soltis D.E., Burleigh J.G. (2014). From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 14: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl R., Jørgensen M.H., Brysting A.K., Koch M.A. (2010). The evolutionary history of the Arabidopsis lyrata complex: a hybrid in the amphi-Beringian area closes a large distribution gap and builds up a genetic barrier. BMC Evol. Biol. 10: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl R., Paule J., Klein J., Marhold K., Koch M.A. (2012). The evolutionary history of the Arabidopsis arenosa complex: diverse tetraploids mask the Western Carpathian center of species and genetic diversity. PLoS One 7: e42691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M.E., Lysak M.A., Mitchell-Olds T. (2006). The ABC’s of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11: 535–542. [DOI] [PubMed] [Google Scholar]
- Schranz M.E., Mitchell-Olds T. (2006). Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. Plant Cell 18: 1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M.E., Mohammadin S., Edger P.P. (2012). Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 15: 147–153. [DOI] [PubMed] [Google Scholar]
- Sims H.J., Herendeen P.S., Lupia R., Christopher R.A., Crane P.R. (1999). Fossil flowers with Normapolles pollen from the Upper Cretaceous of southeastern North America. Rev. Palaeobot. Palynol. 106: 131–151. [Google Scholar]
- Soltis D.E., Albert V.A., Leebens-Mack J., Bell C.D., Paterson A.H., Zheng C., Sankoff D., Depamphilis C.W., Wall P.K., Soltis P.S. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96: 336–348. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Liu X., Marchant D.B., Visger C.J., Soltis D.E. (2014). Polyploidy and novelty: Gottlieb’s legacy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Schemske D.W., Hancock J.F., Thompson J.N., Husband B.C., Judd W.S. (2007). Autopolyploidy in angiosperms: have we grossly underestimated the number of species? Taxon 56: 13–30. [Google Scholar]
- Soltis D.E., Soltis P.S., Tate J.A. (2004). Advances in the study of polyploidy since plant speciation. New Phytol. 161: 173–191. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G.L., Jr. (1938). Cytological characteristics associated with the different growth habits in the dicotyledons. Am. J. Bot. 25: 189–198. [Google Scholar]
- Stebbins G.L., Jr. (1950). Variation and Evolution in Plants. (London, UK: Oxford University Press; ). [Google Scholar]
- Stevens P.F. (2001). Angiosperm Phylogeny Website 2001, onwards, Version 13. http://www.mobot.org/MOBOT/research/APweb/.
- Takahashi M., Crane P.R., Ando H. (1999). Esgueiria futabensis sp. nov., a new angiosperm flower from the Upper Cretaceous (Lower Coniacian) of northeastern Honshu, Japan. Paleontological Research 3: 81–87. [Google Scholar]
- Tang H., Bowers J.E., Wang X., Paterson A.H. (2010). Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl. Acad. Sci. USA 107: 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank D.C., Eastman J.M., Pennell M.W., Soltis P.S., Soltis D.E., Hinchliff C.E., Brown J.W., Sessa E.B., Harmon L.J. (2015). Nested radiations and the pulse of angiosperm diversification: increased diversification rates often follow whole genome duplications. New Phytol. 207: 454–467. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Lavergne S., Affre L., Gaudeul M., Debussche M. (2005). Ecological differentiation of Mediterranean endemic plants. Taxon 54: 967–976. [Google Scholar]
- Tsuchimatsu T., Kaiser P., Yew C.-L., Bachelier J.B., Shimizu K.K. (2012). Recent loss of self-incompatibility by degradation of the male component in allotetraploid Arabidopsis kamchatica. PLoS Genet. 8: e1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste K., Maere S., Van de Peer Y. (2014). Tangled up in two: a burst of genome duplications at the end of the Cretaceous and the consequences for plant evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369: 20130353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco R., et al. (2010). The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 42: 833–839. [DOI] [PubMed] [Google Scholar]
- Vision T.J., Brown D.G., Tanksley S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117. [DOI] [PubMed] [Google Scholar]
- Wang X., et al. ; Brassica rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Warren B.H., Hawkins J.A. (2006). The distribution of species diversity across a flora’s component lineages: dating the Cape’s ‘relicts’. Proc. Biol. Sci. 273: 2149–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwick S.I., Al-Shehbaz I.A. (2006). Brassicaceae: Chromosome number index and database on CD-Rom. Plant Syst. Evol. 259: 237–248. [Google Scholar]
- Warwick S.I., Mummenhoff K., Sauder C., Koch M.A., Al-Shehbaz I.A. (2010). Closing the gaps: Phylogenetic relationships in the Brassicaceae based on DNA sequence data of nuclear ribosomal ITS region. Plant Syst. Evol. 285: 209–232. [Google Scholar]
- Weiss-Schneeweiss H., Greilhuber J., Schneeweiss G.M. (2006). Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am. J. Bot. 93: 148–156. [DOI] [PubMed] [Google Scholar]
- Wheat C.W., Vogel H., Wittstock U., Braby M.F., Underwood D., Mitchell-Olds T. (2007). The genetic basis of a plant-insect coevolutionary key innovation. Proc. Natl. Acad. Sci. USA 104: 20427–20431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing S.L. (1987). Eocene and Oligocene floras and vegetation of the Rocky Mountains. Ann. Mo. Bot. Gard. 74: 748–784. [Google Scholar]
- Yang J., Song N., Zhao X., Qi X., Hu Z., Zhang M. (2014). Genome survey sequencing provides clues into glucosinolate biosynthesis and flowering pathway evolution in allotetraploid Brassica juncea. BMC Genomics 15: e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.