A rice VILLIN2 mutant, with altered microfilament dynamics, displays malformed organs associated with weakened membrane localization of the PIN2 protein and reduced polar auxin transport.
Abstract
As a fundamental and dynamic cytoskeleton network, microfilaments (MFs) are regulated by diverse actin binding proteins (ABPs). Villins are one type of ABPs belonging to the villin/gelsolin superfamily, and their function is poorly understood in monocotyledonous plants. Here, we report the isolation and characterization of a rice (Oryza sativa) mutant defective in VILLIN2 (VLN2), which exhibits malformed organs, including twisted roots and shoots at the seedling stage. Cellular examination revealed that the twisted phenotype of the vln2 mutant is mainly caused by asymmetrical expansion of cells on the opposite sides of an organ. VLN2 is preferentially expressed in growing tissues, consistent with a role in regulating cell expansion in developing organs. Biochemically, VLN2 exhibits conserved actin filament bundling, severing and capping activities in vitro, with bundling and stabilizing activity being confirmed in vivo. In line with these findings, the vln2 mutant plants exhibit a more dynamic actin cytoskeleton network than the wild type. We show that vln2 mutant plants exhibit a hypersensitive gravitropic response, faster recycling of PIN2 (an auxin efflux carrier), and altered auxin distribution. Together, our results demonstrate that VLN2 plays an important role in regulating plant architecture by modulating MF dynamics, recycling of PIN2, and polar auxin transport.
INTRODUCTION
Rice (Oryza sativa) is a staple food for nearly half of the world’s population and a model monocotyledonous crop (Huang et al., 2010). Rice yield is directly influenced by plant architecture, such as tilling number, plant height, leaf and panicle erectness, and root morphology (Miura et al., 2010; Springer, 2010). At the cellular level, plant architecture is coordinately regulated by cell division and cell morphogenesis. Cell division provides new cells to support organ growth, whereas cell morphogenesis, a process where a cell with a predetermined fate matures into its final size and shape, contributes greatly to the formation of organ size and shape (Green, 1980). Defects either in cell division or in cell growth patterns cause abnormal organogenesis, such as abnormal shape or altered organ size and tissue composition (Buschmann et al., 2009). Although extensive studies have provided much insight into the regulation of cell division (Harashima and Schnittger, 2010), the molecular genetic control of cell morphogenesis is much less characterized in plants, especially in monocotyledonous crops.
As in other eukaryotes, the plant cytoskeleton is mainly composed of microtubules (MTs), assembled from tubulin heterodimers (α- and β-tubulin; Lloyd and Chan, 2004), and microfilaments (MFs), assembled from globular-actin (G-actin; Pollard and Cooper, 2009; Dominguez and Holmes, 2011). Molecular genetic and cytological studies of the model dicotyledonous plant Arabidopsis thaliana have generated ample evidence supporting a critical role of the cytoskeleton in regulating nearly all cellular activities, such as intracellular transport of organelles and vesicles, mitotic and meiotic cell division, and cell growth (Wasteneys and Yang, 2004; Pastuglia and Bouchez, 2007). Plant MTs are generally believed to participate in cell division by regulating spindle formation and directional cell expansion by directing cell wall deposition (Wolf et al., 2012). Mutations in genes encoding either tubulins or microtubule-associated proteins often lead to twisted organogenesis (Buschmann et al., 2004; Nakajima et al., 2004; Ishida et al., 2007; Perrin et al., 2007), which is most likely caused by abnormal deposition of the cellulose microfibrils (Buschmann et al., 2009). In rice, the formin protein RMD/BUI1 modulates directional organ growth by regulating MTs and MF array formation and stability (Yang et al., 2011; Z. Zhang et al., 2011; Li et al., 2014). However, the role of MFs in organogenesis and its underlying mechanism remain largely elusive, especially in crop plants.
In plant cells, MFs are regulated by several classes of actin binding proteins (ABPs), including the actin depolymerizing factor (ADF/cofilin), formins, LIM domain-containing proteins, capping proteins, fimbrins, and the villin (VLN)/gelsolin superfamily proteins (Hussey et al., 2006; Papuga et al., 2010; van Gisbergen and Bezanilla, 2013). These ABPs directly bind to G-actin and/or MFs and regulate MF dynamics via nucleating, severing, bundling, or capping activities (Hussey et al., 2006; Dominguez and Holmes, 2011). In addition, these ABPs themselves are regulated at the transcriptional or posttranscriptional level and their activity is affected by pH, calcium-calmodulin, phosphorylation, or phosphoinositides, thus making MFs a highly dynamic and signal responsive system (Xiang et al., 2007; Staiger et al., 2009; Papuga et al., 2010).
As an essential regulatory element to MFs, villin belongs to the villin/gelsolin superfamily, typically possessing six highly conserved gelsolin domains and a villin head piece domain (Friederich et al., 1999; Klahre et al., 2000). There are five villin genes in Arabidopsis, and their translational products share high sequence identity with each other (Khurana et al., 2010). Biochemical studies have revealed that Arabidopsis VLN1 has only Ca2+ (calcium)-independent bundling activity (Huang et al., 2005), whereas the other four Arabidopsis VLNs retain conserved bundling, capping, and severing activities (Khurana et al., 2010; Zhang et al., 2010; Y. Zhang et al., 2011b; Bao et al., 2012). Genetically, Arabidopsis villins have been implicated in polarized cell growth. Loss of Arabidopsis VLN4 and VLN5 retards root hair and pollen tube growth, respectively, but has no visible effect on the establishment of plant architecture (Zhang et al., 2010; Y. Zhang et al., 2011b). Similarly, mutation either in Arabidopsis VLN2 or VLN3 does not result in any apparent morphological changes (Bao et al., 2012; van der Honing et al., 2012), whereas the vln2 vln3 double mutant displays significantly altered morphology, including curly roots, petioles, rosette leaves, siliques, pendent stems, and completely turned inflorescences (Bao et al., 2012; van der Honing et al., 2012), suggesting functional redundancy between VLN2 and VLN3. However, the molecular mechanisms linking the functions of plant villins to plant morphogenesis remain largely unknown. The rice genome also contains five VLN genes (Khurana et al., 2010), but there has been no report on functional studies of those VLNs.
The phytohormone auxin plays an important role in regulating cell morphogenesis, directional organ growth, and response to environmental signals, which requires polar auxin transport (Zhao, 2010). Polar auxin transport is primarily determined by polar localization of the PIN-FORMED (PIN) auxin efflux carriers (Benková et al., 2003; Friml, 2003; Petrášek et al., 2006). Recent drug and cytological studies have shown that MTs play an important role in regulating clathrin-mediated endocytosis of PIN proteins and their recycling back to the plasma membrane (PM; Kleine-Vehn et al., 2008; Ambrose et al., 2013). MF dynamics have also been implicated in auxin transport and PIN endocytosis (Geldner et al., 2001; Dhonukshe et al., 2008; Nagawa et al., 2012). However, the molecular and cellular bases by which MFs regulate plant organogenesis and plant architecture remain largely unclear.
In this study, we identified a rice mutant defective in VLN2, which displays organ malformations, such as twisted roots and shoots. Microscopy analysis revealed that the twisted mutant phenotype is caused by asymmetrical cell expansion on the opposite sides of a given organ. We demonstrate that VLN2 is capable of bundling, capping, and severing actin filaments and plays an important role in bundling actin filaments in vivo. We also present evidence that VLN2 regulates cell expansion, PIN2 recycling, and polar auxin transport in rice by modulating microfilament dynamics.
RESULTS
Identification and Phenotypic Characterization of the vln2 Mutant
To investigate the molecular mechanisms governing the assembly of plant architecture in rice, we constructed a T-DNA insertion library by transforming the japonica variety Kitaake and screened for mutants with altered organ morphology. One of the identified mutants, designated vln2 (see below), displayed various morphological defects in almost every organ during vegetative and reproductive growth. The most apparent morphological changes in vln2 include twisted or bent leaf sheaths and twisted or undulated roots, reduced root growth, and fewer lateral roots (Figure 1A), floppy and wrinkled leaves (Figures 1B and 1C), floppy panicle and wavy panicle branches (Figures 1D and 1E), and caved-in grains (Figure 1F). A typical twisted phenotype was often seen in vln2 seedlings germinated on a hormone-free medium, and undulated roots were visible even in field-grown vln2 plants (Supplemental Figures 1A to 1D). A detailed analysis of seedlings germinated in water revealed that vln2 shoots displayed either left-handed (18.60% ± 0.04%) or right-handed (20.15% ± 0.02%) twisting or just a bent (61.24% ± 0.06%) phenotype (Supplemental Figures 1E and 1F). The vln2 roots showed right-handed (28.29% ± 0.05%) or left-handed (22.48% ± 0.05%) twisting or had an undulated (48.06% ± 0.01%) phenotype (Supplemental Figures 1G and 1H). These results indicate that the handedness of the twisted phenotype is rather random, which is different from that caused by the mutation in the microtubule system (Buschmann et al., 2004; Ishida et al., 2007). In addition, vln2 was semidwarfed, mainly due to reduction in length of the first and second internodes from the top (Figure 1B; Supplemental Table 1). The seed setting rate, 1000-grain weight, grain width, and thickness were all significantly decreased in field-grown vln2 plants compared with the wild type; however, the grain length was increased in vln2 (Supplemental Table 2). Thus, vln2 displayed a pleiotropic phenotype, affecting the formation of all major plant organs and most agronomic traits in rice.
Figure 1.
Phenotypes of vln2.
(A) vln2 and wild-type (WT) seedlings grown in hydroponic culture 7 DAG, showing shorter and twisted roots and leaf sheaths in vln2. The reduced lateral root formation in vln2 is shown as a close-up in comparison to the wild type.
(B) Reduced plant height and floppy architecture of vln2 at the heading stage.
(C) A wrinkled leaf phenotype was often seen in vln2.
(D) Floppy and scattered panicle in vln2.
(E) Twisted panicle axes and branches in vln2.
(F) Wrinkled and caved-in seeds in vln2.
Bars = 2 cm in (A), (D), and (E), 0.5 cm in (C), and 2 mm in (F).
Asymmetric Cell Expansion Causes Twisted Organs in vln2
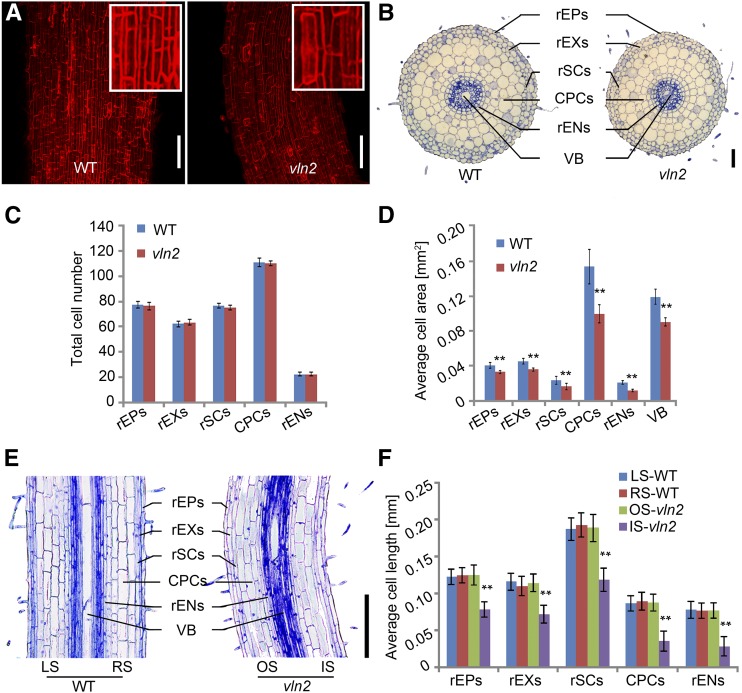
To understand the cellular basis of the twisting phenotype, we performed histological analyses on the twisted roots and leaf sheaths of the vln2 mutant. A correlation between twisted cells caused by abnormal arrangement of MTs and twisted organs has been reported previously (Nakajima et al., 2004; Buschmann et al., 2009). However, we found that both the root and sheath cells were shaped similarly in the wild type and vln2 (Figure 2A; Supplemental Figure 2A), indicating that the twisting morphology in vln2 is not related to cell shape change. Consistent with this, there were no detectable alternations in the arrangement of MTs in vln2 (Supplemental Figure 3). Microscopy examination of cross sections showed that the root and leaf sheath diameters were significantly reduced in vln2 (Figure 2B; Supplemental Figure 2B and Supplemental Table 3). Quantification analysis revealed that the decreased size was mainly due to a reduction in the size of all cell types (including root epidermal cells, root exodermis cells, root sclerenchyma cells, cortex parenchyma cells, root endodermis cells, sheath parenchyma cells, sheath epidermal cells, and sheath sclerenchyma cells), while the cell number was not obviously affected in either the roots or leaf sheaths (Figures 2C and 2D; Supplemental Figures 2C and 2D). In addition, the results from root and leaf sheath longitudinal sections showed that the cell length on the inner (concave) side of both organs was significantly shorter than that on the outer (convex) side in vln2, whereas there was no difference in cell length on either side of the two wild-type organs (Figures 2E and 2F; Supplemental Figures 2E and 2F). These observations suggest that asymmetric cell expansion on the opposite sides of the roots and leaf sheaths is mainly responsible for the twisted growth of roots and leaf sheaths. The location of the asymmetric cell expansion is dynamic during organ development, thus leading to a twisted phenotype of a growing organ.
Figure 2.
Asymmetrical Cell Expansion in the Twisted Region in vln2.
(A) Propidium iodide staining of roots, showing similar cell shape in the wild type (WT) and vln2. A close-up view of the cells was shown in the enlarged boxes.
(B) Cross sections of the wild type and vln2 at 7 DAG showing reduced root diameter in the mature zone of vln2. VB, vascular bundle; rEPs, root epidermis cells; rEXs, root exodermis cells; rSCs, root sclerenchyma cells; CPCs, cortex parenchyma cells; rENs, root endodermis cells.
(C) No significant change was found in cell numbers of root epidermis cells, root exodermis cells, root sclerenchyma cells, cortex parenchyma cells, or root endodermis cells in vln2 compared with the wild type.
(D) The size of root epidermis cells, root exodermis cells, root sclerenchyma cells, cortex parenchyma cells, root endodermis cells, and vascular bundle was significantly reduced in vln2 compared with the wild type.
(E) Longitudinal section of a bent region in the mature zone of the vln2 root and the corresponding region of the wild type (7 DAG seedlings). LS and RS, left and right side of the wild-type section, respectively; OS and IS, outer and inner side of the vln2 bent region, respectively.
(F) Differential cell lengths of root epidermis cells, root exodermis cells, root sclerenchyma cells, cortex parenchyma cells, and root endodermis cells on the outer and inner sides of the bent root region, showing asymmetrical cell expansion in vln2.
Number of cells was counted and their size was measured from ∼60 cross sections of the primary roots in (C) and (D). Length of at least 23 cells on each of 15 longitudinal sections from a primary root was measured and averaged in (F). Data from eight primary roots were used for statistical analysis. **Significantly different at P < 0.01, based on Student’s t test. Error bars indicate ± se in (C), (D), and (F). Bars = 100 μm in (A) and 200 μm in (B) and (E).
vln2 Is Defective in VLN2
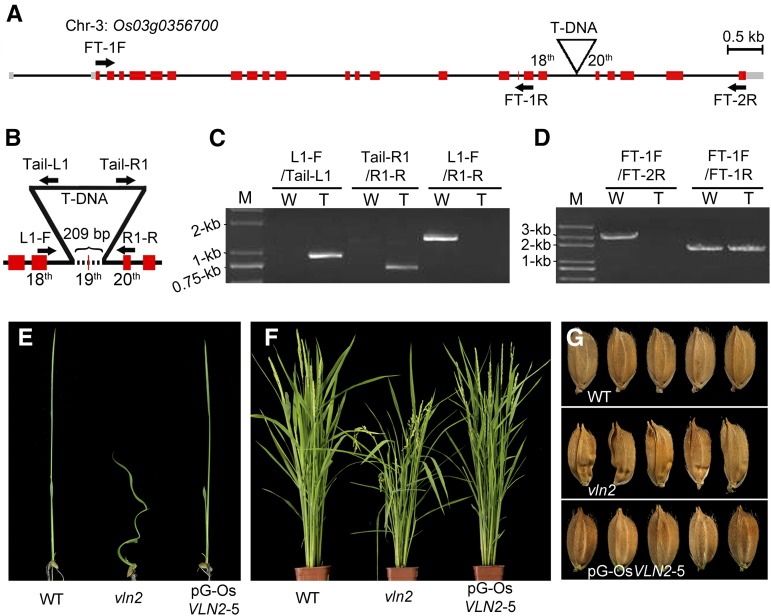
A F2 population from a cross of vln2 with the wild type displayed phenotypic segregation (242 normal and 86 malformed plants, fitting the expected 3:1 ratio: χ2 = 0.199 < χ20.05 = 3.84, df = 1), indicating that vln2 is inherited as a single nuclear recessive mutation. Our PCR analysis confirmed that all 86 malformed plants carried T-DNA, suggesting that the phenotype is most likely caused by T-DNA insertion. Using the thermal asymmetric interlaced PCR method (Liu and Huang, 1998), we obtained a flanking sequence of the T-DNA insertion site. Sequence analysis showed that the T-DNA was inserted between the 18th exon and 20th exon of Os03g0356700 in vln2 (Figures 3A and 3B). Os03g0356700 was predicted to encode a VLN-like protein, which was previously named VLN2, and there are five homologs (VLN1 to VLN5) in the rice genome (Khurana et al., 2010). The insertion also caused deletion of a 209-bp fragment spanning the 19th exon of VLN2 (Figure 3B). The integration site of the T-DNA was further confirmed by PCR using primers on T-DNA and the flanking genome (Figures 3B and 3C). To detect whether expression of VLN2 is affected in vln2, we performed RT-PCR analysis and found no full-length VLN2 cDNA, but a short transcript truncated close to the T-DNA insertion was detected in vln2 (Figures 3A and 3D). Subsequently, we fused GFP to the 5′ end of a DNA fragment including the 5′ part of the VLN2 genomic sequence and part of the T-DNA sequence to simulate T-DNA disruption in vln2 and replaced the partial T-DNA with the NOS terminator as a control construct (Supplemental Figure 4A). When transiently expressed in rice leaf protoplasts, the control but not the simulation construct produced GFP signal, although both constructs could be transcribed (Supplemental Figures 4B and 4C). Those results suggest that there is either no translation from the truncated transcript or the truncated protein is not stable if translated. To verify whether the mutant phenotype was indeed caused by a loss-of-function mutation of VLN2, a 14.8-kb genomic DNA fragment of VLN2 was cloned from the wild type and subsequently introduced into vln2. This fragment contains 23 exons and 22 introns (Figure 3A), a 3.6-kb promoter region, and a 466-bp terminator region, and our annotation analysis by ORF Finder (http://www.ncbi.nlm.nih.gov/) did not reveal any other open reading frames except VLN2. The transformants carrying the transgene showed complete complementation of the mutant phenotypes at different developmental stages, including seedling, adult plant, and seeds (Figures 3E to 3G; Supplemental Table 1). Additionally, the RNAi (RNA interference) transgenic plants of VLN2 exhibited a similar phenotype to vln2, including twisted seedlings, floppy leaves, and dwarfism (Supplemental Figures 5 and Supplemental Table 1). There was no change in expression of the other four rice VLNs (Supplemental Figure 5C). These data together suggest that the phenotype of vln2 is due to defective VLN2.
Figure 3.
Cloning and Genetic Complementation of vln2.
(A) Genomic structure of Os03g0356700 and the T-DNA insertion site on chromosome 3. Gray boxes, red boxes, and black lines indicate 5′ and 3′ untranslated region, exon, and intron, respectively. Arrows indicate the binding sites of primers used to detect the transcription of Os03g0356700 in the wild type and vln2 shown in (D).
(B) T-DNA insertion caused deletion of a 209-bp fragment (the dotted line), spanning the 19th exon and partial sequences of the 18th and 19th introns. Arrows indicate the binding sites of primers used to verify the T-DNA insertion site in (C).
(C) Confirmation of the T-DNA insertion site. Primers on both the genomic sequence and T-DNA were used and are shown in (B). W, wild type; T, the T-DNA insertion mutant vln2; M, molecular weight markers.
(D) The expression of Os03g0356700 is disrupted by T-DNA, leading to its incomplete transcription in vln2. The primers used are shown in (A). W, wild type; T, the T-DNA insertion mutant vln2; M, molecular weight markers.
(E) to (G) Complementation of the mutant phenotype by a 14.8-kb genomic vln2 sequence from the wild type, showing the restored seedling (E), flowering plant (F), and seeds (G). pG-OsVLN2, the construct containing a VLN2 genomic sequence including its promoter region.
The predicted VLN2 translational product retains the overall molecular structure of villins with six conserved gelsolin repeats (G1 to G6) and a villin headpiece (Supplemental Figure 6). Importantly, it retains the conserved residues presumably important for its actin-regulatory functions as well as Ca2+ binding activity (Supplemental Figure 6; Huang et al., 2005; Khurana et al., 2010). A distance-based neighbor-joining phylogenetic analysis revealed that VLN2, together with Arabidopsis VLN2 and VLN3 and lily (Lilium longiflorum) P-135-ABP, was categorized into group II villins (Supplemental Figure 7 and Supplemental Data Set 1; Khurana et al., 2010; Huang et al., 2015). Thus, VLN2 encodes a typical villin protein and may retain the conserved actin regulatory functions.
VLN2 Is Preferentially Expressed in Growing Tissues
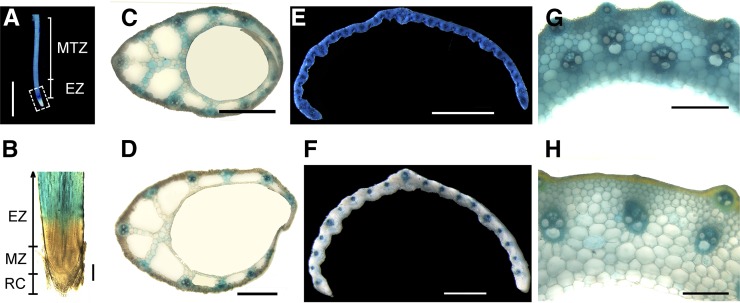
To examine the expression pattern of VLN2, qRT-PCR and promoter-GUS fusion assays were performed. The result of qRT-PCR showed that VLN2 was ubiquitously expressed in all organs examined, but much higher expression was detected in immature tissues than in mature tissues (Supplemental Figure 8). The VLN2 promoter-GUS fusion reporter gene (pOsVLN2:GUS) assay revealed a low level of expression in the meristem zone but strong expression in the elongation zone and then gradually reduced expression toward the maturation zone of roots (Figures 4A and 4B). Histochemical staining of hand-cut sections of immature leaf sheaths 10 d after germination (DAG), unexpanded flag leaves, and elongating internodes showed strong GUS activity in all cell types (Figures 4C, 4E, and 4G). By contrast, lower GUS activity was observed in cells of mature sheaths (15 DAG), expanded flag leaves, and elongated internodes (Figures 4D, 4F, and 4H). Those results together support the preferential expression of VLN2 in expanding tissues, implying an important role of VLN2 in regulating cell expansion.
Figure 4.
VLN2 Is Preferentially Expressed in Immature Tissues.
(A) GUS activity in primary root of a 4-d-old representative transgenic line carrying the pOsVLN2:GUS reporter gene. The white box highlights little activity in the root tip, in contrast to strong activity in the elongation zone (EZ) and mature zone (MTZ).
(B) Longitudinal section of a stained root tip showing little or no GUS activity in the root cap (RC) and meristem zone (MZ) but strong GUS activity in the EZ.
(C) to (H) GUS activity in immature (C) and mature (D) leaf sheath, expanding (E) and expanded (F) leaf, and elongating (G) and elongated (H) uppermost internode.
Bars = 0.8 mm in (A) and 0.2 mm in (B) to (H).
Organization and Dynamics of MFs Are Altered in the vln2 Mutant
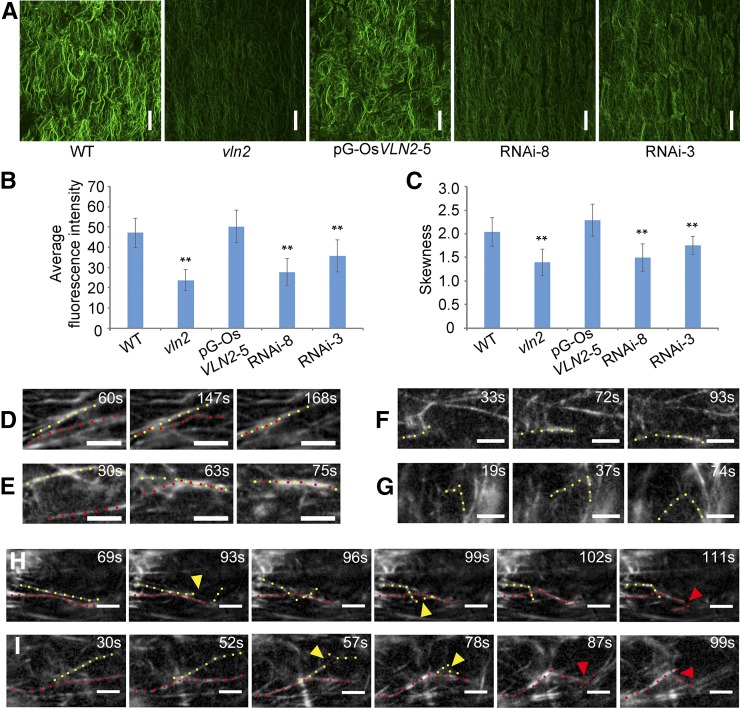
Given that VLN2 encodes a villin-like protein, we asked whether the actin cytoskeleton was affected in vln2. We selected the root elongation zone for detailed analysis since it is easily accessible to actin staining with fluorescent phalloidin. As shown in Figure 5A and consistent with the previous study (Yang et al., 2011), MFs behaved as the thick longitudinal actin cables in cells of the wild type. By contrast, the fluorescence of phalloidin staining was dimmer and the MFs were thinner in cells of vln2 and two RNAi lines (RNAi-3 and -8) under the same image acquisition conditions, and these defects were restored in vln2 complemented with a genomic sequence of VLN2 (Figure 5A). In addition, measurement of the average fluorescence intensity of MF staining revealed that the amount of MFs was significantly decreased in vln2 and the RNAi lines (Figure 5B). These results suggest that VLN2 may play a role in stabilizing MFs and/or promoting MF bundling in cells. To quantify the extent of MF bundling, we measured the parameter of skewness (a measurement of the degree of asymmetry distribution) as previously reported (Higaki et al., 2010; Khurana et al., 2010; Bao et al., 2012). Skewness was significantly decreased in vln2 and the RNAi lines, further confirming that the extent of MF bundling was decreased in these lines (Figure 5C).
Figure 5.
The Organization and Dynamics of the Actin Cytoskeleton Are Altered in vln2.
(A) Staining of MFs in the root elongation zone by Alexa-488-Phalloidin in the wild type, vln2, a complemented line (pG-OsVLN2-5), and two RNA lines (RNAi-3 and -8). Images shown are the maximum projections of all optical sections (0.4-μm interval). Bars = 20 μm.
(B) Average fluorescence intensity of MFs in the wild type, vln2, pG-OsVLN2-5, RNAi-3, and RNAi-8, analyzed by MBF-ImageJ software. Five cells were analyzed and averaged in each root and five to eight roots were used as replicates for each of the lines.
(C) Skewness was measured to determine the bundling status of MFs in the wild type, vln2, pG-OsVLN2-5, RNAi-3, and RNAi-8. Five cells were analyzed and averaged in each root and five roots were used as replicates for each of the lines.
(D) to (I) Recording of MF severing, elongating, and bundling events by VAEM, based on fABD2-GFP decoration.
(D) and (E) Actin filament bundling events in the wild type (D) and vln2 (E).
(F) and (G) Actin filament elongating events in the wild type (F) and vln2 (G).
(H) and (I) Actin filament severing events in the wild type (H) and vln2 (I).
In (B) and (C), asterisks indicate significant difference from the wild type by Student’s t test, P < 0.01, and error bars indicate ±se. In (D) to (I), bars = 5 μm; yellow and red dotted lines indicate two adjacent MFs; colored triangles indicate the place where severing events occur.
To directly visualize the effect of VLN2 on MF dynamics, we fused GFP to fABD2 (actin binding domain 2 of Arabidopsis Fimbrin1) under control of the CaMV 35S promoter (Sheahan et al., 2004). Two fABD2-GFP lines with similar expression at both the transcript and protein level (Supplemental Figures 9B and 9C) were selected for parallel comparison studies of the organization and dynamics of MFs between the wild type and vln2. Although the organization of MFs revealed by decoration with fABD2-GFP in the wild type and vln2 was slightly different compared with that revealed by fluorescent phalloidin staining (Supplemental Figure 10A), the introduction of pCaMV35S:fABD2-GFP caused neither plant development defects nor any apparent trait change of the existing phenotypes (Supplemental Figure 9A and Supplemental Table 4), indicating the suitability of this fusion protein as a visualizing marker of MFs in vivo. Consistent with the actin staining result, actin florescent intensity and skewness of the two lines were significantly reduced in epidermal cells from the root elongation zone of vln2 (Supplemental Figures 10B and 10C). We further visualized MF dynamics using variable-angle epifluorescence microscopy (VAEM) as described previously (Konopka and Bednarek, 2008; Staiger et al., 2009). Several MF dynamic events including bundling (Figures 5D and 5E; Supplemental Movies 1 and 2), elongation (Figures 5F and 5G; Supplemental Movies 3 and 4), and severing (Figures 5H and 5I; Supplemental Movies 5 and 6) could be observed in vivo, which made it possible to quantify various parameters of single MF dynamics in the two transgenic lines (Staiger et al., 2009; Henty et al., 2011; Zheng et al., 2013). First, percentage of mobile filaments (PMF) was determined as described by Lanza et al. (2012). The results showed that PMF was increased in vln2 compared with in the wild type (Table 1), indicating that MFs were more dynamic in vln2. The increase in PMF should theoretically increase the possibility for single MF to find and bundle with each other. However, the bundling frequency was significantly decreased in vln2 (Table 1), suggesting that VLN2 plays an important role in bundling MFs in vivo. This is consistent with the phalloidin staining results described above. However, the frequency of debundling was not affected in vln2 (Table 1). We also measured several other parameters of single MF dynamics, including severing frequency, maximum filament length, and maximum filament lifetime as described previously (Andrianantoandro and Pollard, 2006; Staiger et al., 2009; Henty et al., 2011). Interestingly, we found that the severing frequency was increased in vln2, which is consistent with the finding that both the maximum length and maximum lifetime of MFs were significantly decreased in vln2 (Table 1). These observations imply that other severing proteins may have increased access to MFs in vln2 due to loss of VLN2 function. Together, these data suggest that root epidermal cells in the elongation zone of vln2 have a more dynamic actin network.
Table 1. MF Dynamics Parameters in Root Epidermal Cells of the Wild Type and vln2.
| Stochastic Dynamics Parameters | Wild Type | vln2 |
|---|---|---|
| PMF (%) | 87.2 ± 3.2 | 94.4 ± 3.1** |
| Bundling frequency; events/µm2/s, × 10−5 | 0.503 ± 0.62 | 0.410 ± 0.10** |
| Debundling frequency; events/µm2/s, × 10−5 | 0.203 ± 0.05 | 0.245 ± 0.05ND |
| Max. filament length; µm | 28.76 ± 1.67 | 25.33 ± 1.33** |
| Max. filament lifetimes; s | 35.33 ± 1.97 | 20.00 ± 2.00** |
| Severing frequency; breaks/µm/s | 0.0009 ± 0.0002 | 0.0017 ± 0.0005** |
| Elongation rate; µm/s | 0.165 ± 0.04 | 0.3 ± 0.02** |
| Annealing of severed ends (%) | 2.36 ± 0.54 | 2.67 ± 0.71ND |
Measurements were taken from 5 DAG root epidermal cells (0.5 cm from the tip) of vln2 and the wild type. Values given are means ± se, with n = 231 filaments from n = 63 root epidermal cells per line. Asterisks indicate significant difference from the wild-type control value by Student’s t test, P ≤ 0.01; ND, no significant difference from wild-type control value by Student’s t test, P > 0.05.
VLN2 Is a Typical VLN-Like Protein Capable of Bundling, Capping, and Severing MFs in Vitro
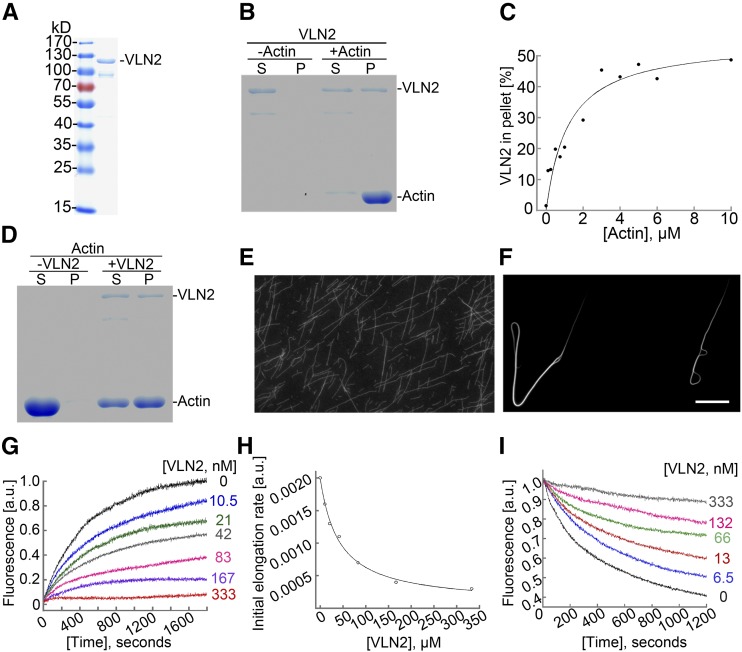
We next performed experiments to characterize the biochemical function of recombinant VLN2-6×His-tag protein (Figure 6A). We first determined the ability of recombinant VLN2 to bind MFs using a high-speed cosedimentation assay. As shown in Figure 6B, VLN2 bound to MFs. To quantify the binding affinity of VLN2 to MFs, we determined the equilibrium dissociation constant (Kd) for VLN2 binding to MFs according to Khurana et al. (2010). Data from a representative experiment are shown in Figure 6C, and the Kd was calculated to be 0.87 μM. Three independent experiments were performed in this study, and the average Kd value (mean ± sd) for VLN2 binding affinity was 1.05 ± 0.22 μM, which is similar to that of Arabidopsis VLN1 (0.6 μM) and VLN3 (0.9 μM; Khurana et al., 2010). We next used low-speed MF cosedimentation and fluorescence light microscopy assays to determine whether VLN2 could bundle MFs. Compared with actin alone, the presence of VLN2 increased the amount of sedimented actin substantially (Figure 6D; Supplemental Figure 11A). This result suggests that VLN2 was capable of organizing MFs into high-order structures, which was further confirmed by direct visualization of MFs in the absence or presence of VLN2 (Figures 6E and 6F). Since villins were shown to be a calcium-responsive protein (Yokota et al., 2005; Bao et al., 2012), we next examined whether the bundling activity of VLN2 was regulated by calcium and found that the bundling activity of VLN2 was sensitive to calcium (Supplemental Figure 11B). Thus, the data suggest that VLN2 is capable of binding to and bundling MFs.
Figure 6.
VLN2 Can Bundle and Cap MFs in Vitro.
(A) Purification of recombinant VLN2 protein in E. coli. Two micrograms of purified VLN2-6×His tag fusion protein was loaded and stained by Coomassie blue.
(B) VLN2 binds to actin filaments. A high-speed cosedimentation assay was used to determine the binding of VLN2 to filamentous actin. Three micromolars of preassembled F-actin was incubated with 0.5 μM VLN2 for 30 min at room temperature and sedimented at 100,000g for 1 h at 4°C. The proteins in the supernatant (S) and pellet (P) were resolved by SDS-PAGE.
(C) Determination of the equilibrium dissociation constant (Kd) for VLN2 binding to actin filaments. The Kd value calculated for VLN2 in this representative experiment is 0.87 μM.
(D) VLN2 bundles actin filaments. A low-speed cosedimentation assay was used to determine the bundling activity of VLN2 on MFs. Three micromolars of preassembled F-actin was incubated with 0.5 μM VLN2 for 30 min at room temperature and sedimented at 13,600g for 1 h at 4°C. The proteins in the supernatant and pellet were resolved by SDS-PAGE.
(E) and (F) Micrographs showing the actin filament status in the absence ([E]; filamentous actin) or presence ([F]; actin bundles) of VLN2. Bars = 50 μm.
(G) VLN2 inhibits seeded actin elongation. VLN2 inhibited the addition of the profilin/actin complex onto the barbed end of actin filaments in a dose-dependent manner.
(H) The initial rates of elongation were plotted for the representative experiments to determine the capping activity of VLN2. In this representative experiment, the Kd value for the binding of VLN2 to the barbed ends of MFs is 41.8 nM.
(I) VLN2 stabilizes actin filaments from dilution-mediated depolymerization in the presence of 10 nM Ca2+.
Next, we asked whether VLN2 could cap the barbed end of MFs by performing the seeded actin elongation assay and found that VLN2 inhibited actin elongation in a dosage-dependent manner (Figure 6G), suggesting that VLN2 possesses barbed end capping ability. The data were plotted and fitted to Equation 1 to yield the Kd value of 41.8 nM for VLN2 capping affinity (Figure 6H). The average Kd value (mean ± se) for VLN2 capping affinity from three independent experiments was determined to be 32.6 ± 12.5 nM. Since barbed-end capping often leads to filament stability under certain conditions, we tested whether VLN2 is able to stabilize MFs by determining its effects on dilution-mediated actin depolymerization. As shown in Figure 6I, in the presence of 10 nM free Ca2+, VLN2 protected actin from dilution-mediated depolymerization in a dosage-dependent manner, suggesting that VLN2 can stabilize MFs by capping the barbed end of MFs. Nevertheless, the possibility that side binding activity of VLN2 also contributes to MF stabilization cannot be ruled out here.
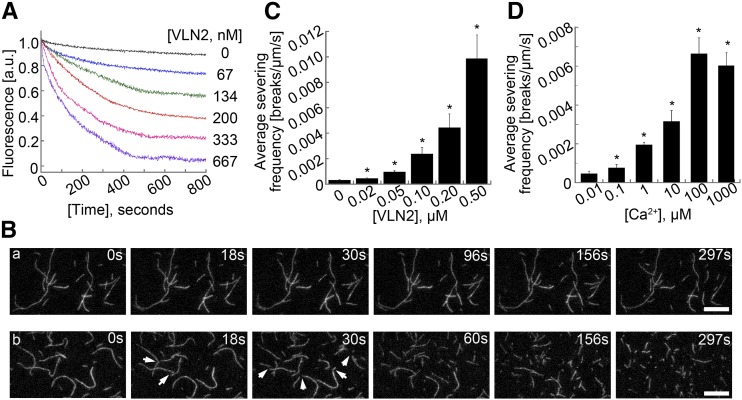
It was previously shown that Arabidopsis VLN2 and VLN5 promote dilution-mediated actin depolymerization in the presence of micromolar concentrations of calcium (Zhang et al., 2010; Bao et al., 2012). We therefore further examined the effect of VLN2 on dilution-mediated actin depolymerization in the presence of 10 μM free Ca2+ and found that VLN2 indeed promoted dilution-mediated actin depolymerization in a dosage-dependent manner (Figure 7A), differing from that in the presence of 10 nM free Ca2+ (Figure 6I). We reasoned that the altered sensitivity of VLN2 to different concentrations of Ca2+ could be due to increased filament-severing activity of VLN2 in the presence of micromolar Ca2+ concentrations. To test this, we performed time-lapse total internal reflection fluorescence microscopy (TIRFM) assays to determine the effect of VLN2 on single MF dynamics. As shown in Figure 7B, few breaks occurred along the MFs in the presence of 1 μM free Ca2+ over the observation period (Figure 7B, a; Supplemental Movie 7). However, numerous breaks were generated along the MFs after the introduction of 0.2 nM VLN2 (Figure 7B, b; Supplemental Movie 8), suggesting that VLN2 severs MFs. We next quantified the average severing frequency and found that the value increased significantly from 0.0003 ± 0.00004 breaks/μm/s (n = 3) for actin alone to 0.0044 ± 0.0012 breaks/μm/s (n = 3) in the presence of 0.2 μM VLN2 (Figure 7C). VLN2 actually possesses a dosage-dependent MF severing activity (Figure 7C). The severing activity of VLN2 is similar to that of Arabidopsis VLN2 (Bao et al., 2012) but is much more potent than that of Arabidopsis VLN3, VLN4, and VLN5 (Khurana et al., 2010; Zhang et al., 2010; Y. Zhang et al., 2011b). To examine the threshold of Ca2+ concentrations required to trigger the severing activity of VLN2, 0.2 nM VLN2 and various concentrations of Ca2+ were introduced into the reaction, and we found that 1 μM Ca2+ was sufficient to trigger a substantial amount of filament severing events (Figure 7D). Thus, these data suggest that VLN2 is able to sever MFs and that physiological Ca2+ concentrations are sufficient to trigger the severing activity.
Figure 7.
VLN2 Severs MFs in Vitro.
(A) VLN2 enhances dilution-mediated actin depolymerization in the presence of 10 μM Ca2+.
(B) VLN2 has filament severing activity. The severing activity of VLN2 was directly visualized by time-lapse TIRFM. (A) Actin alone; (B) actin + 0.2 nM VLN2. Bar = 10 μm.
(C) VLN2 severs actin filaments in a dose-dependent manner. Values represent mean ± se (n = 3).
(D) The severing activity of VLN2 is Ca2+ dependent. Values represent mean ± se (n = 3). *P < 0.05 by Student’s t test.
Taken together, the data suggest that VLN2 is a typical villin-like protein, and it can cap and bundle MFs as well as sever them in a Ca2+-dependent manner.
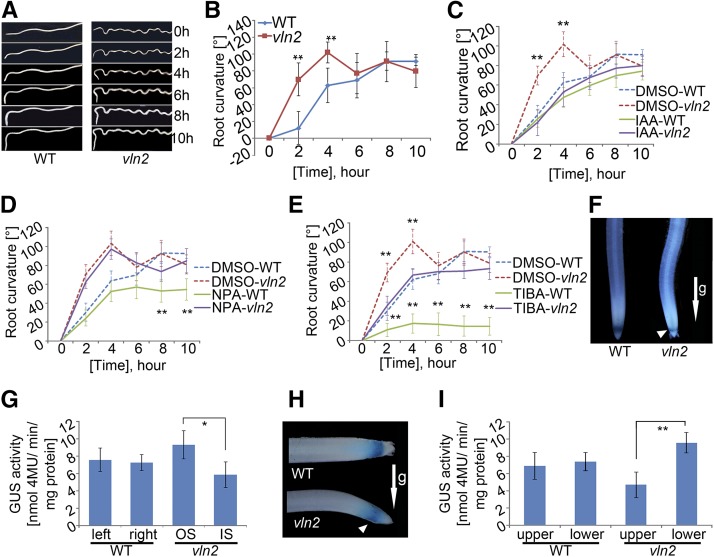
Hypergravitropism in vln2
Auxin is known to regulate root gravitropism in plants (Swarup et al., 2005; Morita, 2010). We speculated that the change of vertical to twisted growth of the root in vln2 might be associated with a change in its responsiveness to gravity. To investigate this, we compared the gravistimulated roots of the wild type and vln2 and found that vln2 curved down faster than the wild type following reorientation of the root (Figures 8A and 8B). However, treatment with exogenous indole-3-acetic acid (IAA) at a concentration as low as 10 nM abolished the hypergravitropism in vln2, whereas there was no effect on wild-type gravitropism at such a concentration (Figure 8C; Supplemental Figure 12). Furthermore, the gravitropic response was dramatically reduced in the wild type, but much less affected in vln2, when treated with the auxin efflux inhibitors TIBA (tri-iodobenzoic acid; Figure 8D) and NPA (1-N-naphthylphthalamic acid; Figure 8E). Next, we transformed an auxin response reporter, DR5:GUS, into both the wild type and vln2. Histochemical staining and GUS activity measurement of the vertically grown roots showed that in contrast to the symmetrical staining of GUS activity in the wild-type root tips, slightly stronger staining was observed on the side opposite to the newly formed curve in the elongation zone of the vln2 root tips (Figures 8F and 8G). Moreover, 20-min gravistimulation was able to induce a differential auxin distribution between the upper and lower sides of the vln2 root but not in the wild-type root (Figures 8H and 8I). Together, those results suggest that the hypergravitropism, and in turn the twisted phenotype in the vln2 roots, are related to the change in concentration and more sensitive to the gravity-induced asymmetric distribution of IAA in the root tips.
Figure 8.
The vln2 Root Has a Hypergravitropism Response.
(A) and (B) Hypersensitive gravity response of vln2 roots compared with those of the wild type during a 10-h time course after their reorientation.
(C) Suppression of hypergravitropism in vln2 roots by IAA (10 nM) treatment.
(D) and (E) vln2 roots were less sensitive to the PAT inhibitors NPA ([D]; 10 nM) and TIBA ([E]; 10 nM) with regard to gravitropic growth.
(F) and (G) Asymmetric staining (F) and quantitative analysis (G) of GUS activity in the vertically grown root of vln2 and the wild type transformed with the DR5:GUS reporter. OS and IS, outer and inner side of the curved vln2 root, respectively.
(H) and (I) Faster establishment of asymmetric auxin distribution in vln2 (H) and quantitative analysis of GUS activity (I) following 20 min gravistimulation.
Arrows indicate gravity (g) direction, and arrowheads indicate the side of higher auxin accumulation in (F) and (H). Error bars indicate sd of the means of 21 seedlings in (B), 16 seedlings in (C) and (D), and 174 seedlings in (G) and (I). Asterisks indicate significant difference at *P < 0.05 and **P < 0.01, respectively, based on Student’s t test.
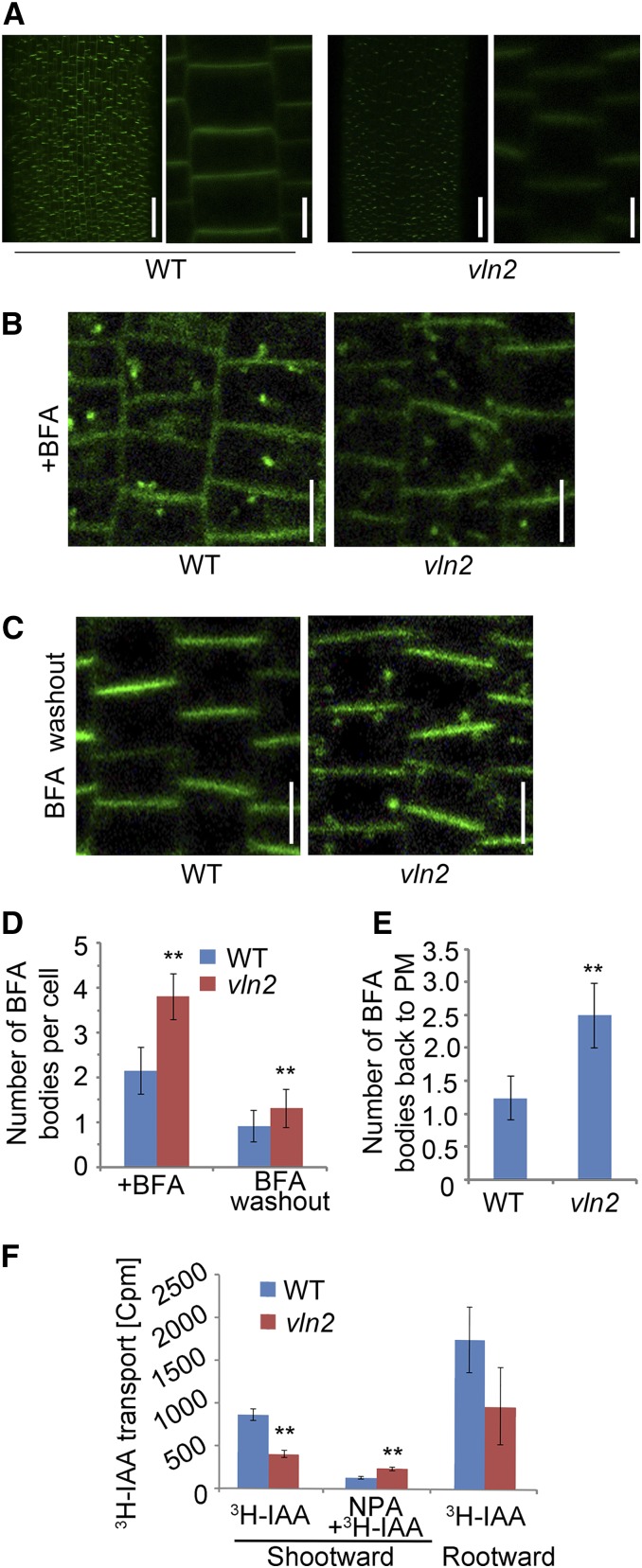
Recycling of PIN2 and Polar Auxin Transport Are Altered in vln2
Previous studies have suggested a role of MFs in regulating recycling of PIN proteins and polar auxin transport (PAT; Geldner et al., 2001). We next sought to determine whether the recycling of PIN proteins was altered in vln2. Considering that Arabidopsis PIN2 plays a key role in determining the polar distribution of auxin in the elongation zone of roots (Müller et al., 1998), we selected the closest rice PIN member, PIN2 (Wang et al., 2009), to construct a OsPIN2:GFP fusion reporter, which was transformed into the wild type. We selected a single-copy event and introduced it into vln2 by crossing to generate plants identical in term of the transgene. In general, PM-localized PIN2 was weaker in the root epidermal cells of vln2 than in those of the wild type (Figure 9A). When treated with BFA (brefeldin A), an inhibitor of vesicle budding (Baster et al., 2013), more BFA bodies were accumulated in the vln2 root cells compared with the wild type, suggesting more active internalization of PIN2 in vln2 (Figures 9B and 9D). Furthermore, PIN2 was recycled back to the PM after BFA washout, as shown by a reduced number of BFA bodies (Figures 9C and 9D). Quantification analysis indicates that more BFA bodies were recycled back to the PM after the BFA washout in vln2 than in the wild type (Figure 9E). Thus, the data suggest that the rates of secretion and recycling of PIN2 are increased in vln2. The observation that PIN2 plays a crucial role in PAT of roots (Morita, 2010) prompted us to determine whether PAT was altered in vln2. Our measurement of PAT using 3H-IAA showed that both the rate of shootward and rootward PAT was slower in vln2 than in the wild type. However, the inhibition of PAT by NPA was attenuated in vln2 compared with the wild type (Figure 9F). Thus, the data showed that the recycling and polar distribution of PIN2, as well as PAT, were altered in vln2.
Figure 9.
Increased Internalization of PIN2 and Reduced PAT in vln2.
(A) Fluorescence of PM-localized PIN2-GFP in root epidermal cells is weaker in vln2 than in the wild type. A close-up view is shown to the right of each image. Bars = 50 μm in the original images and 5 μm in the magnified ones.
(B) BFA induces internalization of PIN2-GFP in the wild type (left panel) and vln2 (right panel) 1 h after BFA treatment. Bars = 10 μm.
(C) Cycling of PIN2-GFP back to the PM 1 h after BFA treatment washout. Bars = 10 μm.
(D) Quantification of BFA bodies shown in (B) and (C).
(E) Ratio of number of BFA bodies in the BFA treatment to BFA washout samples in the wild type and vln2, respectively.
(F) Reduced PAT in vln2 root.
Thirty cells in the main root were counted and averaged in (D) and (E); n = 6 in (D) and (E) and n = 12 in (F). Error bars indicate ±sd. Asterisks indicate significant difference at P < 0.01, based on Student’s t test.
DISCUSSION
In this study, we report the isolation and characterization of a rice mutant defective in VLN2 and showed that VLN2 plays a vital role in modulating directional organ growth by regulating MF organization and dynamics. VLN2 is ubiquitously expressed in various tissues examined, with much higher expression in expanding tissues, which is consistent with its role in regulating cell expansion. VLN2 retains all typical activities of the villin family proteins, including MF bundling, severing, and capping in vitro. Moreover, direct visualization of the organization and dynamics of MFs confirmed that VLN2 is required for actin filament bundling in vivo. Furthermore, we observed a more dynamic actin cytoskeleton, hypergravitropism, faster recycling of PIN2, altered auxin distribution pattern, and impaired polar auxin transport in vln2 roots, which may explain the twisted or undulated phenotype. Thus, we have established a genetic link between VLNs, auxin transport, and directed organ growth in rice.
VLN2 Possesses Bundling, Capping, and Severing Activity
VLN2 contains all the conserved structural domains of villins and thus likely retains all biochemical functions of villins. Indeed, our biochemical assays showed that VLN2 retains capping, Ca2+-sensitive bundling, and severing activities in vitro. Consistent with the biochemical results, in vivo cellular analysis (biochemical staining and MF dynamics statistics) showed that the MF bundling status was decreased in vln2, confirming a role for VLN2 in bundling MFs in vivo. Our results, along with previous findings (Tominaga et al., 2000; Y. Zhang et al., 2011b; Bao et al., 2012; van der Honing et al., 2012; Qu et al., 2013), firmly support the notion that plant villins function as a major MF bundling factor in vivo. Our results showed that the bundling frequency was 0.503 × 10−5 events/µm2/s in root epidermal cells (Table 1), which is lower than that in Arabidopsis pollen tubes (2.3 × 10−4 events/µm2/s; Zheng et al., 2013) and hypocotyl epidermal cells (6.9 × 10−5 events/μm2/s; Hoffmann et al., 2014), suggesting a possible association between bundling frequency and a cell’s growing status. Unexpectedly, inspection of MF dynamics revealed that severing frequency is significantly increased in vln2. A possible explanation might be that debundled MFs are vulnerable to severing by other ABPs (like ADFs and VLNs; Pope et al., 1994; Dominguez, 2004). In support of this, a previous study showed that Arabidopsis VLN1-decorated MFs are resistant to the action of Arabidopsis ADF1 (Huang et al., 2005). Though our current findings do not provide direct evidence for a role of VLN2 in severing MFs in vivo, we cannot rule out the possibility that VLN2-mediated filament severing is biologically relevant. In particular, since the severing activity of VLN2 is Ca2+ dependent, it is possible that the severing activity of VLN2 is triggered under some stimulating conditions with changed Ca2+ fluxes. In addition, we found that the MF elongation rate was increased in vln2 compared with the wild type. This might be due to a more rapid production of MFs to offset the faster MF severing activity in vln2. Theoretically, loss-of-function mutation of VLN2 is unlikely to affect the translation of actin; therefore, the total amount of actin is expected to be constant in vln2 compared with that in the wild type. Our actin staining results showed that the amount of MFs decreased in vln2. It is therefore expected that the amount of G-actin might increase in vln2, which may consequently promote actin elongation. It is also possible that loss of VLN2 may upregulate the activity of some actin elongation promoting factors, like the formins (Deeks et al., 2002). The combination of these factors may lead to an increase in actin elongation to allow the cell to balance the loss of MFs in vln2. No significant differences were found in annealing of the severed ends. Whether the capping activity of VLN2 is biologically relevant remains to be examined.
Asymmetric Cell Expansion Underlies Organ Malformation in vln2 Seedlings
Previous studies reported the twisted organ growth phenotype in several MT-related mutants. For example, mutation in the genes encoding either tubulins (e.g., lefty1, lefty2, tortifolia, and tua4S178Δ) or microtubule-associated proteins (e.g., tortifolia1/spiral2, wave-dampened2, and spiral1) generally lead to twisted organogenesis (Thitamadee et al., 2002; Buschmann et al., 2004, 2009; Nakajima et al., 2004; Ishida et al., 2007; Perrin et al., 2007). The twisted organogenesis can be either left-handed (lefty1, lefty2, and tua4S178Δ) or right-handed (tor2, tortifolia1/spiral2, wave-dampened2, and spiral1). In either case, a strong correlation has been established between the organ growth direction and arrangement of the cortical microtubules, which is consistent with the proposed role of cortical microtubules in guiding the deposition of cellulose microfibrils during plant cell growth (Paredez et al., 2008; Wolf et al., 2012). According to the “Spring Model,” when cells stretch, the left-handed arrangement of MTs could lead to right-handed cell distortion, whereas right-handed organization of MTs could lead to left-handed cell distortion, thus resulting in twisted organ growth (Buschmann et al., 2009).
Our results suggest that a different mechanism may cause the twisted organ growth in vln2. First, MT arrays and amount are similar in the wild type and vln2; second, there is no detectable cell distortion observed in vln2; and third, the handedness of vln2 is random. Our in vivo analysis of vln2 showed a change in MF amount but not in arrays, supporting the earlier observation that loss-of-function mutation in the actin and associated proteins generally does not cause right- or left-handed organization of MFs (Tian et al., 2009; Yang et al., 2011). Instead, our histological analysis suggested that the asymmetric cell expansion largely explains the twisted organ growth phenotype of vln2. This is remarkably similar to the formation of the apical hook in etiolated dicotyledonous plants, which is also caused by differential cell elongation at two sides of the hypocotyl apex (Lehman et al., 1996). Earlier studies have shown that formation of an auxin maxima at the concave (inner) side of the hook is essential for hook formation and maintenance and that this process is largely determined by the AUX/LAX-mediated auxin input and PIN protein-mediated auxin efflux (Vandenbussche et al., 2010; Zádníková et al., 2010). Consistent with these findings, we found that vln2 roots display a hypergravitropic response that can be suppressed by application of exogenous IAA, but is less sensitive to the PAT inhibitors. In addition, the DR5:GUS reporter gene showed faster formation of an asymmetric auxin distribution pattern in vln2 roots when curved or placed horizontally, compared with wild-type plants. Moreover, we found reduced PM localization and more active recycling of PIN2 as well as reduced PAT in the vln2 root elongation zone. Together, our results suggest that loss-of-function mutation of VLN2 causes altered dynamics of MFs, leading to faster recycling of PIN2, altered auxin distribution and polar auxin transport, and ultimately twisted organ growth, fewer lateral roots, and reduced root length and plant height. Such a notion is consistent with the earlier reports that reduced PAT causes a shorter main root and fewer lateral root phenotype in Arabidopsis (Casimiro et al., 2001) and that a mutation in Arabidopsis Actin2 causes a higher percentage of mobile actin filaments, altered PIN2 polar distribution, and, as a result, wavy root growth (Lanza et al., 2012). Since other PINs were not investigated in vln2, we cannot rule out the possibility that recycling of other PINs is also altered in vln2, which may be partially associated with the changed PAT. A combination analysis of more rice PINs may provide a deeper understanding of the link between the actin cytoskeleton, PAT, and morphogenesis.
In further support of our proposition, a recent study showed that loss-of-function mutation in the rice RMD/BUI1 gene causes similar morphological changes as observed in the vln2 mutant, including twisting of roots and reduction of plant height (Li et al., 2014). RMD/BUI1 encodes a type II formin that plays an important role in regulating the dynamics of both MTs and MFs (Yang et al., 2011; Z. Zhang et al., 2011). Also similar to vln2, reduced PM localization of PIN2, increased BFA bodies, and altered auxin transport were documented in rmd (Li et al., 2014). Thus, both VLN2- and RMD/BUI1-mediated actin dynamics regulate organ growth presumably via regulating the recycling and localization of PIN and PAT. Thus, these studies provide direct genetic evidence establishing the connection between actin dynamics and PAT via modulating the recycling and localization of PIN proteins. It will be interesting to dissect the functional relationship between VLN2 and RMD/BUI1 in future studies.
METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) mutant vln2 was isolated from a T-DNA library (in the Kitaake background). For data collection of agronomic traits, rice plants were grown at the experimental station of the Chinese Academy of Agricultural Sciences, located in Beijing. For materials used in the lab, seedlings were germinated and grown on Murashige and Skoog (MS) medium in a growth chamber (12 h light/12 h dark at 80% relative humidity).
Histology and GUS Analysis
Technovit 7100 embedding resin (Heraeus Kulzer) was used for histological analysis according to Clément et al. (2009). Embedded tissues were sectioned (5 μm in thickness) using a Leica RM2265 microtome, stained in 0.05% toluidine blue for 15 min, and examined under a Leica CTR5000B microscope. Cell number, area, and length were measured with ImageJ software (http://rsb.info.nih.gov/ij/).
For promoter activity analysis of VLN2, hand-cut sections of homozygous T3 plants were stained in a GUS staining solution according to Jefferson et al. (1987). Younger tissues, including root, sheath, unexpanded leaf, and elongating first internode, were stained for 10 min, and mature tissues, including sheath, leaf, and elongated first internode, were stained for 2 h. For analysis of transgenic plants containing the auxin reporter DR5:GUS, 4 DAG roots vertically grown on MS medium were collected and stained for 30 min. Staining was stopped by washing with 70% ethanol. For analysis of asymmetrical GUS distribution, root tips of 0.5 cm length were divided into right and left sides for the wild type and into outer and inner sides (relative to the curve) for vln2 by cutting them in the middle; they were then collected in liquid nitrogen. GUS activity was measured according to Jefferson et al. (1987).
Cloning of vln2 and Plasmid Construction
vln2 was crossed to the wild type, and the resulting F2 plants were used for T-DNA cosegregation analysis. Thermal asymmetric interlaced PCR (Liu and Huang, 1998) was employed to clone sequences flanking the T-DNA insertion. The constructs pOsVLN2:OsVLN2, pOsVLN2:GUS, pCaMV35S:fABD2-GFP, and pOsPIN2:OsPIN2-GFP were constructed using the In-Fusion HD cloning system (Clontech), with the corresponding primers (Supplemental Table 5). For genetic complementation, a 14.8-kb VLN2 genomic fragment including a 3.2-kb promoter region and a 0.5-kb terminator region was inserted into pCAMBIA1305 using a two-step recombination procedure (step 1, primers Infu-1F/Infu1R; step 2, Infu-2F/ Infu2R), resulting in pOsVLN2:OsVLN2. For promoter activity analysis, the 3.2-kb promoter region was amplified using Infu-3F/Infu3R and constructed into pCAMBIA1305, resulting in pOsVLN2:GUS. For labeling MFs, fABD2 was cloned (Infu-4F/Infu-4R) from Arabidopsis thaliana (Columbia) and constructed into the PEGAD vector according to Wang et al. (2008), resulting in pCaMV35S:fABD2-GFP. For labeling PIN2, the promoter region together with first exon of PIN2, GFP, and the remaining part of PIN2 cDNA were amplified with Infu-5F/Infu-5R, Infu-6F/Infu-6R, and Infu-7F/Infu-7R, respectively, and then sequentially cloned into pCAMBIA1305, resulting in pOsPIN2:OsPIN2-GFP. GFP was inserted between the two hydrophobic transmembrane domains of PIN2 as described previously (Benková et al., 2003; Xu and Scheres, 2005). For the RNAi construct, two inverted repeats of a 200-bp VLN2 fragment were amplified with Infu-8F/Infu-8R or Infu-9F/Infu-9R and then cloned into pCUbi1390-ΔFAD2 (Li et al., 2013). These constructs were introduced into rice by Agrobacterium tumefaciens-mediated transformation (Hiei et al., 1994). For biochemical assays, VLN2 cDNA was amplified with Infu-10F/Infu-10R and cloned into pET30a, resulting in pT7:OsVLN2-6×His. For simulating the T-DNA interruption, GFP was inserted between a promoter region of VLN2 (amplified using primers Infu-11F/Infu-11R) and a 7.7-kb genomic sequence (spanning ATG and part of the T-DNA) amplified from vln2 (primers Infu-12F/Infu-12R) and constructed into pCAMBIA1305 using the one step recombination procedure, resulting in pOsVLN2:GFP-OsVLN2-T-DNA; as a control, a 7.4-kb genomic sequence lacking the T-DNA fragment (amplified using primers Infu-11F/Infu-13R) was instead used and linked to the NOS terminator, resulting in pOsVLN2:GFP-OsVLN2-tNOS. Sequence information of the primers can be found in Supplemental Table 5. The leaf sheath protoplast transient assay was performed as described by Y. Zhang et al. (2011a).
RT-PCR and qRT-PCR Analysis
RNA was extracted from freshly prepared samples using the Direct-zol RNA MiniPrep Kit (Zymo Research) according to the manufacturer’s instructions. The quality of extracted RNA was determined using a spectrophotometer (Nanodrop). RNA was then reverse transcribed using the QuantiTect reverse transcription kit (Qiagen). qRT-PCR was performed on the Applied Biosystems 7500 real-time PCR system (Life Technologies) using SYBR Premix Ex Taq II (TaKaRa). Rice UBIQUITIN-1 was used as an internal control. The primers used here are listed in Supplemental Table 5.
Staining and Quantification of MFs and MTs in Root Epidermal Cells
MFs were stained using the glycerol method (Yang et al., 2011). Briefly, 1-cm segments from root tips of 2 DAG seedlings were incubated in PME buffer (100 mM PIPES, 5 mM MgSO4, and 10 mM EGTA, pH 6.8) containing 300 mM m-maleimidobenzoyl-N-hydroxysuccinimide ester, 1.5% glycerol, and 0.1% Triton X-100, with gentle agitation for 30 min, followed by rinsing twice with PME. The samples were then fixed in PME supplemented with 2% paraformaldehyde for 30 min, rinsed twice with PME, and finally incubated in actin-staining buffer (PME, 1.5% glycerol, and 0.1% Triton X-100) with 0.66 mM AlexaFluor488-Phalloidin at 4°C in darkness overnight. Immunostaining of MTs was performed according to previously published methods (Yang et al., 2011). For data collection, optical sections were taken by LSM700 (Zeiss; equipped with a 40× objective), and step size was set at 0.4 μm to collect optical sections. Alexa-488 phalloidin was excited with the 488-nm line of an argon laser with the emission set to 550 to 600 nm. Images were prepared by generating projections of the optical sections through an individual epidermal cell. Average fluorescence intensity and skewness of MFs were examined according to Higaki et al. (2010).
In Vitro Biochemical Assays
To prepare recombinant VLN2-6×His, pT7:OsVLN2-6×His was expressed in Escherichia coli and purified using Ni-NTA His Bind resin (Novagen) according to the user manual. Actin from rabbit muscle was purified according to the published methods (MacLean-Fletcher and Pollard, 1980). The actin binding and bundling properties of VLN2-6×His were determined using high- and low-speed cosedimentation assays, respectively, mainly according to Khurana et al. (2010). The visualization of MFs with a wide-field fluorescence microscope was performed according to the methods reported previously (Blanchoin et al., 2010; Zhang et al., 2010). The activities of VLN2-6×His binding and capping the barbed end of MFs were examined using the seeded elongation assay as described by Huang et al. (2003). Dilution-mediated actin depolymerization assays were adopted according to a published method (Zhang et al., 2010). Direct visualization of MFs severing using VLN2-6×His was observed under a total internal reflection fluorescence microscope as described by Amann and Pollard (2001) and Michelot et al. (2005).
Recombinant Protein Preparation
The pT7:VLN2-6×His construct was transformed into E. coli strain BL21 (DE3) by a heat shock method. After induction by the addition of 0.4 mM isopropyl β-d-thiogalactopyranoside for 3 h at 37°C, the cells were collected by centrifugation (5000g for 10 min at 4°C) and resuspended in Buffer A (25 mM Tris-HCl, pH 7.9, 250 mM KCl, 5 mM imidazole, and 2 mM mercaptoethanol) containing 1 mM phenylmethylsulfonyl fluoride. The cell suspension was sonicated and the supernatant was recovered by centrifuging at 19,000g at 4°C for 1 h and then applied onto a Ni-NTA affinity column (Novagen). The column was washed with Buffer B (25 mM Tris-HCl, pH 7.9, 250 mM KCl, 20 mM imidazole, and 2 mM 2-mercaptoethanol), and the recombinant VLN2-6×His protein was eluted with Buffer C (25 mM Tris-HCl, pH 7.9, 250 mM KCl, 40 mM imidazole, and 2 mM 2-mercaptoethanol) and dialyzed against 10 mM Tris-HCl (pH 8.0). The purified VLN2-6×His protein was concentrated, aliquoted, and flash frozen in liquid nitrogen and stored at −80°C. The concentration of VLN2-6×His was determined with the Bradford assay using BSA as a standard. Actin from rabbit muscle was purified according to published methods (Spudich and Watt, 1971; MacLean-Fletcher and Pollard, 1980).
High- and Low-Speed Cosedimentation Assays
The actin binding and bundling properties of VLN2-6×His were determined by high- and low-speed cosedimentation assays, respectively, mainly according to Khurana et al. (2010) and Zhang et al. (2010). Briefly, actin was prepolymerized in 1× KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole-HCl, pH 7.0) at room temperature for 1 h, and VLN2-6×His was preclarified by centrifugation at 200,000g for 40 min at 4°C. To determine the affinity of VLN2-6×His for MFs, 0.5 μM VLN2-6×His was incubated with various amounts (0.1 to 10 μM) of MFs in 1× KMEI as above in the presence of 10 nM free Ca2+ (determined with EqCal software). After incubation at room temperature for 30 min, the mixtures were centrifuged at 186,000g for 35 min at 4°C. Equal amounts of supernatant and pellet samples were separated by 10% SDS-PAGE and then stained with Coomassie Brilliant Blue R 250. To determine the Kd values, the amount of VLN2-6×His bound to MFs was determined by densitometry analysis with ImageJ software and further plotted and fitted to a hyperbolic function using the Synergy Software Kaleidagraph (version 3.6). To analyze the MF bundling activity of VLN2-6×His, 5 μM of preassembled MFs was incubated with increasing concentrations of VLN2-6×His (0.1 to 10 μM) in 1× KMEI in a 100-μL reaction for 60 min at room temperature. After spinning the mixture at 13,600g for 40 min at 4°C, the supernatant and pellet samples were separated on 10% SDS-PAGE gels and stained with Coomassie Brilliant Blue R 250. The amount of actin was determined by densitometry with ImageJ, and the percentage of actin in the pellet was calculated and plotted against [VLN2-6×His]. The effect of [Ca2+] on the bundling activity of VLN2-6×His was determined by incubating 0.5 μM VLN2-6×His with 5 μM MFs in 1× KMEI in the presence of various amounts of free Ca2+ (0.01 to 1000 μM).
Wide-Field Fluorescence Microscopy of MFs
The visualization of MFs was performed according to the methods of Blanchoin et al. (2010) and Zhang et al. (2010). Four micromolar MFs with equal molar amounts of rhodamine-phalloidin were mixed with various amounts of VLN2-6×His in 1× KMEI and incubated at room temperature for 30 min. The mixture was diluted to 5 nM with fluorescence buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 100 mM DTT, 100 mg/mL glucoseoxidase, 15 mg/mL glucose, 20 mg/mL catalase, and 0.5% methylcellulose) and further observed under a fluorescence light microscope (Olympus) equipped with a ×60, 1.42-numerical aperture oil objective. Images were acquired with a Retiga EXi Fast 1394 CCD camera using Image-Pro Express 6.3 software.
Determination of the Binding and Capping Affinity of VLN2
The activities of VLN2-6×His binding and capping the barbed end of MFs were examined using the seeded elongation assay as described by Huang et al. (2003). Muscle actin was conjugated with pyrene iodoacetamide for the following fluorimetry assays according to Pollard and Weeds (1984). Freshly prepolymerized MF seeds (0.8 μM) were mixed with various amounts of VLN2-6×His and incubated at room temperature for 5 min. Actin elongation at the barbed end of MFs was initiated by the addition of 1 μM G-actin (5% pyrene labeled) saturated with 4 μM human profilin 1 in 1× KMEI buffer, and the elongation was further monitored by the change in pyrene fluorescence accompanying actin polymerization. The Kd value for VLN2-6×His binding to the barbed end of MFs was determined by plotting the initial elongation rate against the amount of VLN2-6×His and further fitting the data with Equation 1 (Huang et al., 2003).
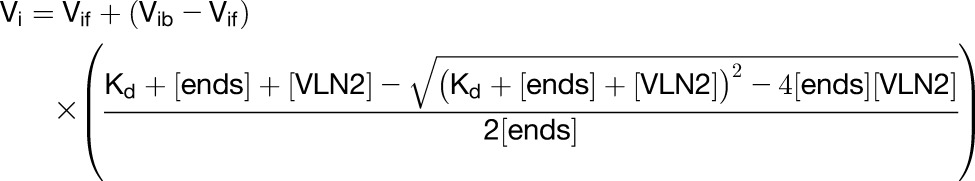 |
(1) |
In this equation, Vi is the observed rate of elongation. Vif is the rate of elongation when all the barbed ends are free. Vib is the rate of elongation when all the barbed ends are capped. [ends] is the concentration of barbed ends, and [VLN2] is the concentration of VLN2-6×His. The data were modeled with Kaleidagraph software.
Dilution-Mediated Actin Depolymerization Assays
Dilution-mediated actin depolymerization assays were performed according to a published method (Zhang et al., 2010). To determine whether VLN2-6×His can stabilize MFs, different concentrations of VLN2-6×His were incubated with 5 μM prepolymerized MFs (100% pyrene labeled) in the presence of 10 nM free Ca2+ for 5 min at room temperature, and the mixtures were diluted 25-fold into low ionic strength Buffer G with 10 nM Ca2+ to initiate the depolymerization. To determine whether VLN2-6×His can enhance actin depolymerization, various concentrations of VLN2-6×His were incubated with 5 μM prepolymerized MFs (100% pyrene labeled) for 5 min at room temperature in the presence of 10 μM free Ca2+. The mixtures were subsequently diluted 50-fold into 1× KMEI buffer with 10 μM free Ca2+.
VAEM
For microfilament dynamic statistics, seeds were first germinated on a plate containing 0.5× MS medium with 0.35% Phytogel for 2 d, and then the plate was placed vertically for another 2 d. The root top of 1 cm was cut and mounted on a variable-angle epifluorescence microscope (Olympus IX81) equipped with a 3100 oil objective. Images were collected for 180 s with an interval of 3 s using a Photometrics cascade II 512 CCD camera (Major Instruments) with microManager software. The microfilament parameters were determined and calculated as described previously (Andrianantoandro and Pollard, 2006; Staiger et al., 2009; Henty et al., 2011; Lanza et al., 2012; Li et al., 2012; Zheng et al., 2013). Severing frequency is presented as breaks/μm/s (number of breaks per unit length of MFs per second). Maximum filament length is the actin filament length between point of origin and endpoint of growth; maximum filament lifetime is the time required for MFs to reach the maximum filament length.
Gravity Response of vln2 Roots and Observation of PIN2 Cycling
Root gravitropism was observed for wild-type and vln2 seedlings vertically germinated for 4 d and then placed horizontally for a time course of 10 h, and root curvature was photographed and measured at an interval of 2 h. To determine effect of auxin on root gravity response, 4-d-old seedlings were pretreated with a series of 1, 5, 10, and 100 nM IAA for 1 h and placed horizontally for 10 h in dark. To determine effect of auxin transport inhibitors on root curvature, experiments were performed with TIBA (10 nM) and NPA (10 nM) as for IAA. For examining PIN2 recycling, roots of pOsPIN2:OsPIN2-GFP transgenic seedlings 4 DAG were incubated with 25 μM BFA for 1 h in darkness. To remove BFA, the samples were washed with water twice and then incubated in water for another hour. BFA body formation and relocalization of PIN2-GFP to PM after BFA washout in cells of the elongation zone were observed with a confocal microscope (LSM700, Zeiss; equipped with 63× oil objective) with a step size set at 1 μm. The ImageJ software was used to quantify the collected photos according to Yang et al. (2011).
PAT Assay
The PAT was performed according to the method described previously with minor modifications (Li et al., 2007). The 1.5-cm-long root tips were collected from 4-d-old seedlings germinated on a vertically placed plate containing 0.5× MS medium with 0.35% phytogel and then inserted either shootward or rootward into the same medium but with the addition of 0.1 μM 3H-labeled IAA (American Radiolabeled Chemicals) for 3 h in darkness at room temperature. As a control, 30 μM NPA was added to the medium to block active IAA transport. The air-exposed root fragment end of 0.5 cm was excised and then washed twice with 0.5× MS liquid medium. The excised segments were transferred into scintillation liquid for 18 h, and the radioactivity was then counted using a liquid scintillation counter (1450 MicroBeta TriLux; Perkin-Elmer).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Os VLN1 (Os05g0153000, NP_001054676); Os VLN2 (Os03g0356700, NP_001050140); Os VLN3 (Os06g0659300, NP_001058263); Os VLN4 (Os04g0604000, NP_001053784); Os VLN5 (Os08g0240800, NP_001061335); VLN1 (At2g29890, NP_029567); VLN2 (At2g41740, NP_565958); VLN3 (At3g57410, NP_567048); VLN4 (At4g30160, NP_194745); VLN5 (At5g57320, NP_200542); P-115-ABP (BAC77209); P-135-ABP (AAD54660); Hs-VLN (NP_009058); Hs-Gel (CAA28000); Os UBIQUITIN-1 (Os03g0234200, NP_001049479); FIM1 (At5g57090); and Os PIN2 (Os06g0660200, NP_001058268).
Supplemental Data
Supplemental Figure 1. Characterization of Malformed Shoots and Roots in vln2.
Supplemental Figure 2. Histological and Statistical Analysis of Twisted Leaf Sheath in the Wild Type and vln2.
Supplemental Figure 3. Cortical MT Array in vln2 Is Indistinguishable from That in the Wild Type.
Supplemental Figure 4. The Disrupted VLN2 Is Transcribed but No Corresponding Truncated Protein Is Produced.
Supplemental Figure 5. Phenotype of VLN2 RNAi Plants.
Supplemental Figure 6. Sequence Alignment of Os VLN2 with Other Members in the Villin/Gelsolin Family.
Supplemental Figure 7. Distance-Based Neighbor-Joining Phylogenetic Analysis of the Villin/Gelsolin Members.
Supplemental Figure 8. Quantitative RT-PCR Analysis of VLN2.
Supplemental Figure 9. Expression of the fABD2-GFP Marker Does Not Change the Existing Phenotype of Either the Wild Type or vln2.
Supplemental Figure 10. The Amount of Filamentous Actin Is Reduced and the Extent of Filament Bundling Is Decreased in vln2 Root Epidermal Cells.
Supplemental Figure 11. VLN2 Bundles MFs in a Dosage- and Ca2+-Dependent Manner.
Supplemental Figure 12. Lower IAA Concentration Treatment Does Not Alter the Root Hypergravitropism in vln2.
Supplemental Table 1. Comparison of Panicle, Internode Length, and Plant Height.
Supplemental Table 2. Grain Phenotype Comparison.
Supplemental Table 3. Diameter of Leaf Sheath and Root in the Wild Type and vln2.
Supplemental Table 4. Phenotypic Characterization of fABD2-GFP Transgenic lines.
Supplemental Table 5. Primers Used in This Study.
Supplemental Data Set. 1. Alignments Used to Generate the Phylogeny Presented in Supplemental Figure 7.
Supplemental Movie 1. Time-Lapse VAEM Visualization of Actin Bundling Event in Vivo (Wild Type).
Supplemental Movie 2. Time-Lapse VAEM Visualization of Actin Bundling Event in Vivo (vln2).
Supplemental Movie 3. Time-Lapse VAEM Visualization of Actin Elongation Event in Vivo (Wild Type).
Supplemental Movie 4. Time-Lapse VAEM Visualization of Actin Elongation Event in Vivo (vln2).
Supplemental Movie 5. Time-Lapse VAEM Visualization of Actin Severing Event in Vivo (Wild Type).
Supplemental Movie 6. Time-Lapse VAEM Visualization of Actin Severing Event in Vivo (vln2).
Supplemental Movie 7. Time-Lapse TIRFM Showing Much Reduced MF Severing Activity in the Absence of VLN2 in Vitro.
Supplemental Movie 8. Time-Lapse TIRFM Showing the MF Severing Activity of VLN2 in the Presence of VLN2 in Vitro.
Supplementary Material
Acknowledgments
This research was supported by grants from National Natural Science Foundation (31371601 and 31125004), the National Transformation Science and Technology Program (2014ZX08009-003 and 2014ZX08010-004), the 863 Program (2011AA101101), the National Science and Technology Support Program (2011BAD35B02-02 and 2013BAD01B02-16), the Jiangsu Science and Technology Development Program (BE2012303), and the Jiangsu Province 333 Talents Project (BRA2012126).
AUTHOR CONTRIBUTIONS
J. Wan, S.H., H.W., C.W., and S.W. designed the research. S.W., Y.X., J.Z., F.W., P.S., and Juan Wang performed experiments. Y.R., Jiulin Wang, X.Z., and X.G. provided technical support. J. Wan, S.H., H.W., C.W., S.W., and Y.X. analyzed the data and wrote the article.
Glossary
- MT
microtubule
- MF
microfilament
- PM
plasma membrane
- RNAi
RNA interference
- DAG
days after germination
- VAEM
variable-angle epifluorescence microscopy
- PMF
percentage of mobile filament
- TIRFM
total internal reflection fluorescence microscopy
- TIBA
tri-iodobenzoic acid
- NPA
1-N-naphthylphthalamic acid
- PAT
polar auxin transport
- BFA
brefeldin A
- IAA
indole-3-acetic acid
- MS
Murashige and Skoog
Footnotes
Articles can be viewed online without a subscription.
References
- Amann K.J., Pollard T.D. (2001). Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. USA 98: 15009–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C., Ruan Y., Gardiner J., Tamblyn L.M., Catching A., Kirik V., Marc J., Overall R., Wasteneys G.O. (2013). CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell 24: 649–659. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T.D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24: 13–23. [DOI] [PubMed] [Google Scholar]
- Bao C., Wang J., Zhang R., Zhang B., Zhang H., Zhou Y., Huang S. (2012). Arabidopsis VILLIN2 and VILLIN3 act redundantly in sclerenchyma development via bundling of actin filaments. Plant J. 71: 962–975. [DOI] [PubMed] [Google Scholar]
- Baster P., Robert S., Kleine-Vehn J., Vanneste S., Kania U., Grunewald W., De Rybel B., Beeckman T., Friml J. (2013). SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 32: 260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Boujemaa-Paterski R., Henty J.L., Khurana P., Staiger C.J. (2010). Actin dynamics in plant cells: a team effort from multiple proteins orchestrates this very fast-paced game. Curr. Opin. Plant Biol. 13: 714–723. [DOI] [PubMed] [Google Scholar]
- Buschmann H., Fabri C.O., Hauptmann M., Hutzler P., Laux T., Lloyd C.W., Schäffner A.R. (2004). Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr. Biol. 14: 1515–1521. [DOI] [PubMed] [Google Scholar]
- Buschmann H., Hauptmann M., Niessing D., Lloyd C.W., Schäffner A.R. (2009). Helical growth of the Arabidopsis mutant tortifolia2 does not depend on cell division patterns but involves handed twisting of isolated cells. Plant Cell 21: 2090–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I., Marchant A., Bhalerao R.P., Beeckman T., Dhooge S., Swarup R., Graham N., Inzé D., Sandberg G., Casero P.J., Bennett M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément M., Ketelaar T., Rodiuc N., Banora M.Y., Smertenko A., Engler G., Abad P., Hussey P.J., de Almeida Engler J. (2009). Actin-depolymerizing factor2-mediated actin dynamics are essential for root-knot nematode infection of Arabidopsis. Plant Cell 21: 2963–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks M.J., Hussey P.J., Davies B. (2002). Formins: intermediates in signal-transduction cascades that affect cytoskeletal reorganization. Trends Plant Sci. 7: 492–498. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P., et al. (2008). Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc. Natl. Acad. Sci. USA 105: 4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. (2004). Actin-binding proteins--a unifying hypothesis. Trends Biochem. Sci. 29: 572–578. [DOI] [PubMed] [Google Scholar]
- Dominguez R., Holmes K.C. (2011). Actin structure and function. Annu. Rev. Biophys. 40: 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friederich E., Vancompernolle K., Louvard D., Vandekerckhove J. (1999). Villin function in the organization of the actin cytoskeleton. Correlation of in vivo effects to its biochemical activities in vitro. J. Biol. Chem. 274: 26751–26760. [DOI] [PubMed] [Google Scholar]
- Friml J. (2003). Auxin transport - shaping the plant. Curr. Opin. Plant Biol. 6: 7–12. [DOI] [PubMed] [Google Scholar]
- Geldner N., Friml J., Stierhof Y.D., Jürgens G., Palme K. (2001). Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413: 425–428. [DOI] [PubMed] [Google Scholar]
- Green P.B. (1980). Organogenesis-a biophysical view. Annu. Rev. Plant Physiol. 31: 51–82. [Google Scholar]
- Harashima H., Schnittger A. (2010). The integration of cell division, growth and differentiation. Curr. Opin. Plant Biol. 13: 66–74. [DOI] [PubMed] [Google Scholar]
- Henty J.L., Bledsoe S.W., Khurana P., Meagher R.B., Day B., Blanchoin L., Staiger C.J. (2011). Arabidopsis actin depolymerizing factor4 modulates the stochastic dynamic behavior of actin filaments in the cortical array of epidermal cells. Plant Cell 23: 3711–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Higaki T., Kutsuna N., Sano T., Kondo N., Hasezawa S. (2010). Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J. 61: 156–165. [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Moes D., Dieterle M., Neumann K., Moreau F., Tavares Furtado A., Dumas D., Steinmetz A., Thomas C. (2014). Live cell imaging reveals actin-cytoskeleton-induced self-association of the actin-bundling protein WLIM1. J. Cell Sci. 127: 583–598. [DOI] [PubMed] [Google Scholar]
- Huang S., Blanchoin L., Kovar D.R., Staiger C.J. (2003). Arabidopsis capping protein (AtCP) is a heterodimer that regulates assembly at the barbed ends of actin filaments. J. Biol. Chem. 278: 44832–44842. [DOI] [PubMed] [Google Scholar]
- Huang S., Robinson R.C., Gao L.Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C.J. (2005). Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. Plant Cell 17: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Qu X., Zhang R. (2015). Plant villins: versatile actin regulatory proteins. J. Integr. Plant Biol. 57: 40–49. [DOI] [PubMed] [Google Scholar]
- Huang X., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42: 961–967. [DOI] [PubMed] [Google Scholar]
- Hussey P.J., Ketelaar T., Deeks M.J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57: 109–125. [DOI] [PubMed] [Google Scholar]
- Ishida T., Kaneko Y., Iwano M., Hashimoto T. (2007). Helical microtubule arrays in a collection of twisting tubulin mutants of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 8544–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana P., Henty J.L., Huang S., Staiger A.M., Blanchoin L., Staiger C.J. (2010). Arabidopsis VILLIN1 and VILLIN3 have overlapping and distinct activities in actin bundle formation and turnover. Plant Cell 22: 2727–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Friederich E., Kost B., Louvard D., Chua N.H. (2000). Villin-like actin-binding proteins are expressed ubiquitously in Arabidopsis. Plant Physiol. 122: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka C.A., Bednarek S.Y. (2008). Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J. 53: 186–196. [DOI] [PubMed] [Google Scholar]
- Lanza M., et al. (2012). Role of actin cytoskeleton in brassinosteroid signaling and in its integration with the auxin response in plants. Dev. Cell 22: 1275–1285. [DOI] [PubMed] [Google Scholar]
- Lehman A., Black R., Ecker J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85: 183–194. [DOI] [PubMed] [Google Scholar]
- Li G., Liang W., Zhang X., Ren H., Hu J., Bennett M.J., Zhang D. (2014). Rice actin-binding protein RMD is a key link in the auxin-actin regulatory loop that controls cell growth. Proc. Natl. Acad. Sci. USA 111: 10377–10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. (2013). A comprehensive genetic study reveals a crucial role of CYP90D2/D2 in regulating plant architecture in rice (Oryza sativa). New Phytol. 200: 1076–1088. [DOI] [PubMed] [Google Scholar]
- Li J., Henty-Ridilla J.L., Huang S., Wang X., Blanchoin L., Staiger C.J. (2012). Capping protein modulates the dynamic behavior of actin filaments in response to phosphatidic acid in Arabidopsis. Plant Cell 24: 3742–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Wang Y., Qian Q., Fu Z., Wang M., Zeng D., Li B., Wang X., Li J. (2007). LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 17: 402–410. [DOI] [PubMed] [Google Scholar]
- Liu Y., Huang N. (1998). Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol. Biol. Rep. 16: 175. [Google Scholar]
- Lloyd C., Chan J. (2004). Microtubules and the shape of plants to come. Nat. Rev. Mol. Cell Biol. 5: 13–22. [DOI] [PubMed] [Google Scholar]
- MacLean-Fletcher S., Pollard T.D. (1980). Identification of a factor in conventional muscle actin preparations which inhibits actin filament self-association. Biochem. Biophys. Res. Commun. 96: 18–27. [DOI] [PubMed] [Google Scholar]
- Michelot A., Guerin C., Huang S., Ingouff M., Richard S., Rodiuc N., Staiger C.J., Blanchoin L. (2005). The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. Plant Cell 17: 2296–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X.J., Ito M., Asano K., Matsuoka M., Kitano H., Ashikari M. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42: 545–549. [DOI] [PubMed] [Google Scholar]
- Morita M.T. (2010). Directional gravity sensing in gravitropism. Annu. Rev. Plant Biol. 61: 705–720. [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagawa S., Xu T., Lin D., Dhonukshe P., Zhang X., Friml J., Scheres B., Fu Y., Yang Z. (2012). ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol. 10: e1001299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K., Furutani I., Tachimoto H., Matsubara H., Hashimoto T. (2004). SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. Plant Cell 16: 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papuga J., Hoffmann C., Dieterle M., Moes D., Moreau F., Tholl S., Steinmetz A., Thomas C. (2010). Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22: 3034–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Persson S., Ehrhardt D.W., Somerville C.R. (2008). Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 147: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J., et al. (2006). PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918. [DOI] [PubMed] [Google Scholar]
- Pastuglia M., Bouchez D. (2007). Molecular encounters at microtubule ends in the plant cell cortex. Curr. Opin. Plant Biol. 10: 557–563. [DOI] [PubMed] [Google Scholar]
- Perrin R.M., Wang Y., Yuen C.Y., Will J., Masson P.H. (2007). WVD2 is a novel microtubule-associated protein in Arabidopsis thaliana. Plant J. 49: 961–971. [DOI] [PubMed] [Google Scholar]
- Pollard T.D., Cooper J.A. (2009). Actin, a central player in cell shape and movement. Science 326: 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Weeds A.G. (1984). The rate constant for ATP hydrolysis by polymerized actin. FEBS Lett. 170: 94–98. [DOI] [PubMed] [Google Scholar]
- Pope B., Way M., Matsudaira P.T., Weeds A. (1994). Characterisation of the F-actin binding domains of villin: classification of F-actin binding proteins into two groups according to their binding sites on actin. FEBS Lett. 338: 58–62. [DOI] [PubMed] [Google Scholar]
- Qu X., Zhang H., Xie Y., Wang J., Chen N., Huang S. (2013). Arabidopsis villins promote actin turnover at pollen tube tips and facilitate the construction of actin collars. Plant Cell 25: 1803–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan M.B., Staiger C.J., Rose R.J., McCurdy D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136: 3968–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N. (2010). Shaping a better rice plant. Nat. Genet. 42: 475–476. [DOI] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246: 4866–4871. [PubMed] [Google Scholar]
- Staiger C.J., Sheahan M.B., Khurana P., Wang X., McCurdy D.W., Blanchoin L. (2009). Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. J. Cell Biol. 184: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Kramer E.M., Perry P., Knox K., Leyser H.M., Haseloff J., Beemster G.T., Bhalerao R., Bennett M.J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7: 1057–1065. [DOI] [PubMed] [Google Scholar]
- Thitamadee S., Tuchihara K., Hashimoto T. (2002). Microtubule basis for left-handed helical growth in Arabidopsis. Nature 417: 193–196. [DOI] [PubMed] [Google Scholar]
- Tian M., Chaudhry F., Ruzicka D.R., Meagher R.B., Staiger C.J., Day B. (2009). Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiol. 150: 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M., Yokota E., Vidali L., Sonobe S., Hepler P.K., Shimmen T. (2000). The role of plant villin in the organization of the actin cytoskeleton, cytoplasmic streaming and the architecture of the transvacuolar strand in root hair cells of Hydrocharis. Planta 210: 836–843. [DOI] [PubMed] [Google Scholar]
- Vandenbussche F., Petrásek J., Zádníková P., Hoyerová K., Pesek B., Raz V., Swarup R., Bennett M., Zazímalová E., Benková E., Van Der Straeten D. (2010). The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development 137: 597–606. [DOI] [PubMed] [Google Scholar]
- van der Honing H.S., Kieft H., Emons A.M., Ketelaar T. (2012). Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiol. 158: 1426–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen P.A., Bezanilla M. (2013). Plant formins: membrane anchors for actin polymerization. Trends Cell Biol. 23: 227–233. [DOI] [PubMed] [Google Scholar]
- Wang J.R., Hu H., Wang G.H., Li J., Chen J.Y., Wu P. (2009). Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol. Plant 2: 823–831. [DOI] [PubMed] [Google Scholar]
- Wang Y.S., Yoo C.M., Blancaflor E.B. (2008). Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C- and N-termini of the fimbrin actin-binding domain 2. New Phytol. 177: 525–536. [DOI] [PubMed] [Google Scholar]
- Wasteneys G.O., Yang Z. (2004). New views on the plant cytoskeleton. Plant Physiol. 136: 3884–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S., Hématy K., Höfte H. (2012). Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 63: 381–407. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Huang X., Wang T., Zhang Y., Liu Q., Hussey P.J., Ren H. (2007). ACTIN BINDING PROTEIN 29 from Lilium pollen plays an important role in dynamic actin remodeling. Plant Cell 19: 1930–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Ren S., Zhang X., Gao M., Ye S., Qi Y., Zheng Y., Wang J., Zeng L., Li Q., Huang S., He Z. (2011). BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23: 661–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota E., Tominaga M., Mabuchi I., Tsuji Y., Staiger C.J., Oiwa K., Shimmen T. (2005). Plant villin, lily P-135-ABP, possesses G-actin binding activity and accelerates the polymerization and depolymerization of actin in a Ca2+-sensitive manner. Plant Cell Physiol. 46: 1690–1703. [DOI] [PubMed] [Google Scholar]
- Zádníková P., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617. [DOI] [PubMed] [Google Scholar]
- Zhang H., Qu X., Bao C., Khurana P., Wang Q., Xie Y., Zheng Y., Chen N., Blanchoin L., Staiger C.J., Huang S. (2010). Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. Plant Cell 22: 2749–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Su J., Duan S., Ao Y., Dai J., Liu J., Wang P., Li Y., Liu B., Feng D., Wang J., Wang H. (2011a). A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiao Y., Du F., Cao L., Dong H., Ren H. (2011b). Arabidopsis VILLIN4 is involved in root hair growth through regulating actin organization in a Ca2+-dependent manner. New Phytol. 190: 667–682. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Tan H., Wang Y., Li G., Liang W., Yuan Z., Hu J., Ren H., Zhang D. (2011). RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23: 681–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Xie Y., Jiang Y., Qu X., Huang S. (2013). Arabidopsis actin-depolymerizing factor7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 25: 3405–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61: 49–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.