Underlying the jasmonate receptor COI1-mediated delay of flowering, the APETALA2 transcription factors TOE1 and TOE2 interact with a subset of JAZ repressors to regulate the expression of FT.
Abstract
Flowering time of plants must be tightly regulated to maximize reproductive success. Plants have evolved sophisticated signaling network to coordinate the timing of flowering in response to their ever-changing environmental conditions. Besides being a key immune signal, the lipid-derived plant hormone jasmonate (JA) also regulates a wide range of developmental processes including flowering time. Here, we report that the CORONATINE INSENSITIVE1 (COI1)-dependent signaling pathway delays the flowering time of Arabidopsis thaliana by inhibiting the expression of the florigen gene FLOWERING LOCUS T (FT). We provide genetic and biochemical evidence that the APETALA2 transcription factors (TFs) TARGET OF EAT1 (TOE1) and TOE2 interact with a subset of JAZ (jasmonate-ZIM domain) proteins and repress the transcription of FT. Our results support a scenario that, when plants encounter stress conditions, bioactive JA promotes COI1-dependent degradation of JAZs. Degradation of the JAZ repressors liberates the transcriptional function of the TOEs to repress the expression of FT and thereby triggers the signaling cascades to delay flowering. Our study identified interacting pairs of TF and JAZ transcriptional regulators that underlie JA-mediated regulation of flowering, suggesting that JA signals are converted into specific context-dependent responses by matching pairs of TF and JAZ proteins.
INTRODUCTION
The strict regulation of flowering time of higher plants is essential for reproductive success, enabling completion of seed development in favorable environmental conditions. The timing of flowering is coordinately controlled by various endogenous (e.g., development and age) and environmental (e.g., photoperiod and temperature) signals. Dedicated studies in the model plant Arabidopsis thaliana have led to the definition of several genetic pathways involved in flowering time control. It is generally believed that endogenous signals regulate flowering time through the autonomous and gibberellic acid pathways, whereas environmental signals regulate flowering time through the photoperiod and vernalization pathways (Fornara et al., 2010; Srikanth and Schmid, 2011; Andrés and Coupland, 2012; Song et al., 2013). Ultimately, signaling information in all these pathways is integrated into flowering genetic networks through a small group of floral pathway integrators, such as FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CO1, and LEAFY, whose accumulation in the shoot apical meristem (SAM) promotes the identity switching of the SAM from vegetative to reproductive (Srikanth and Schmid, 2011; Song et al., 2013). Furthermore, an ever-growing body of evidence indicates that different environmental constraints, such as cold, drought, salinity, and pathogen infection, also affect the timing of flowering (Wang et al., 2011; Yaish et al., 2011; Li et al., 2012; Bolouri Moghaddam and Van den Ende, 2013; Riboni et al., 2014). The emerging picture is that these environmental constraints often exert their effects on flowering through modulating the expression of the floral integrators.
Among the best characterized floral integrators is FT, which encodes an evolutionarily conserved small globular protein that is translocated from the leaves to the SAM to initiate floral transition (Kardailsky et al., 1999). To enable flowering to be properly timed, plants have evolved diverse regulatory mechanisms to keep the FT expression in check. For example, the transcriptional expression of FT is positively regulated by CONSTANS (CO), which encodes a putative zinc finger transcription factor (TF) (Putterill et al., 1995). Under favorable conditions, light signaling and the circadian clock coordinately control the activity of CO, which directly regulates the transcription of FT to promote flowering (Suárez-López et al., 2001; An et al., 2004; Imaizumi et al., 2005; Sawa et al., 2007; Fornara et al., 2009). On the other hand, the expression of FT is also negatively regulated by several transcriptional repressors, such as FLOWERING LOCUS C (Helliwell et al., 2006; Searle et al., 2006), SHORT VEGETATIVE PHASE (Lee et al., 2007), TEMPRANILLO1 (Castillejo and Pelaz, 2008), and SCHLAFMŰTZE (SMZ) (Mathieu et al., 2009). Under unfavorable conditions for flowering or during the juvenile developmental phases, these repressors, as well as their related transcriptional regulators, bind to specific cis-elements in the FT locus and repress its expression to prevent precocious flowering (Song et al., 2013). Together, these studies highlight the central role of FT in mediating the crosstalk between stress signaling and the internal genetic flowering network.
The fatty acid-derived plant hormone jasmonate (JA) is a prime example of a small molecule that orchestrates physiological and phenotypic plasticity of plants in their ever-changing environments. Besides being a major immune hormone, JA also regulates a wide range of developmental processes, including root growth (Staswick et al., 1992; Feys et al., 1994; Pauwels et al., 2010), male and female fertility (McConn and Browse, 1996; Sanders et al., 2000; Stintzi and Browse, 2000; Cheng et al., 2009), trichome formation, and anthocyanin accumulation (Franceschi and Grimes, 1991; Shan et al., 2009). Remarkable recent progress in dissecting the JA signaling pathway has revealed that three major molecular components are involved in JA-signaled transcriptional reprogramming: the F-box protein CORONATINE INSENSITIVE1 (COI1) that forms a functional E3 ubiquitin ligase SCFCOI1 and acts as an essential component of the JA perception machinery, the jasmonate-ZIM domain (JAZ) family of transcriptional repressors that are targeted by SCFCOI1 for degradation, and TFs that regulate the expression of JA-responsive genes. Upon hormone accumulation and perception, degradation of JAZ repressors by SCFCOI1 relieves the repression of TFs, thereby initiating the transcriptional reprogramming of the cell and the activation (derepression) of JA responses (Chini et al., 2007; Thines et al., 2007; Yan et al., 2009; Pauwels et al., 2010).
The basic helix-loop-helix (bHLH) TF MYC2 and its closely related paralogs bHLH TF MYC3 and MYC4 are the most extensively studied JAZ-interacting TFs. These MYC TFs interact with most of the 12 JAZ proteins and act redundantly to regulate diverse aspects of JA responses, including root growth inhibition and defense gene expression (Chen et al., 2011; Fernández-Calvo et al., 2011; Kazan and Manners, 2013; Schweizer et al., 2013). In addition to these MYC proteins, recent studies have identified numerous TFs that interact with JAZ proteins and regulate different aspects of JA-dependent transcriptional responses (Qi et al., 2011; Song et al., 2011; Zhu et al., 2011; Hu et al., 2013; Jiang et al., 2014).
Although JA has been implicated in regulating flowering time in several plant species (Krajncic et al., 2006; Robson et al., 2010), the exact role of JA in regulating this important physiological process and the underlying molecular mechanism remains obscure. Here, we report that the COI1-dependent signaling pathway delays flowering of Arabidopsis by repressing the transcriptional expression of FT. We discovered that the APETALA2 (AP2) family TFs TARGET OF EAT1 (TOE1) and TOE2 interact with a subset of JAZ proteins and repress the transcription of FT. Our study uncovers the transcriptional mechanism of JA in regulating flowering time and highlights how the specificity of JA responses is determined.
RESULTS
COI1-Dependent Signaling Pathway Delays Flowering of Arabidopsis
As a first step to investigate the molecular mechanism underlying JA regulation of flowering time, we examined the flowering phenotype of the coi1-2 mutant, which harbors a point mutation of the JA receptor gene COI1 (Xu et al., 2002). Under long-day (LD) conditions, coi1-2 exhibited an early flowering phenotype, as measured by total rosette leaf number (RLN) and days from germination to flowering (DTF) (Figures 1A to 1C). Similarly, under short-day (SD) conditions, coi1-2 plants also flowered earlier than their wild-type counterparts (Figures 1D and 1E). These results are consistent with previous observations that the COI1-dependent signaling pathway delays flowering of Arabidopsis (Robson et al., 2010; Yang et al., 2012).
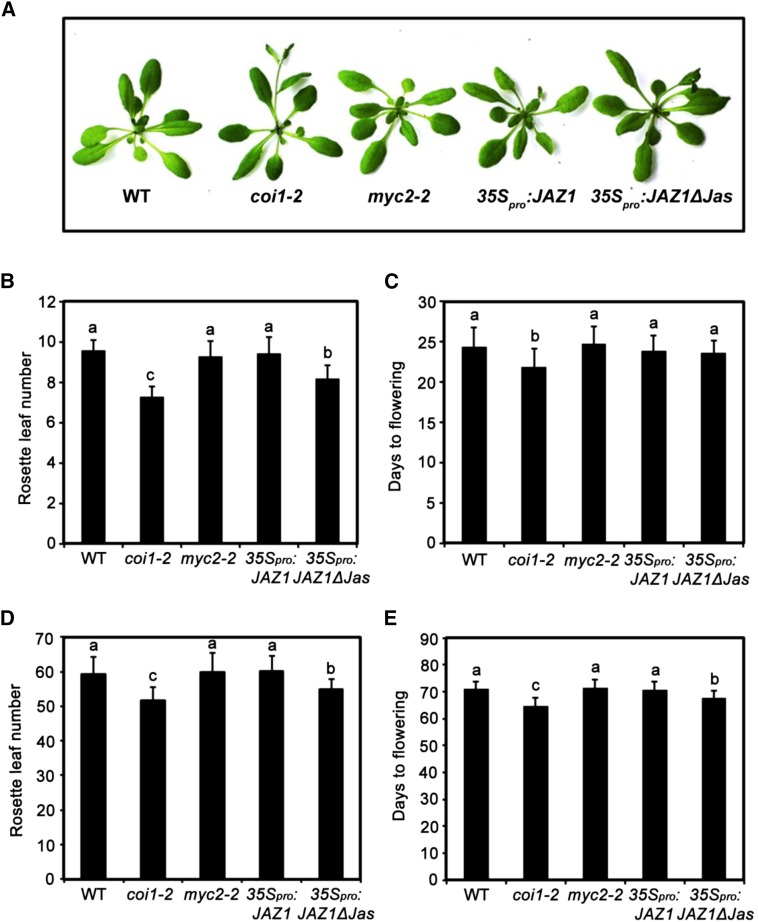
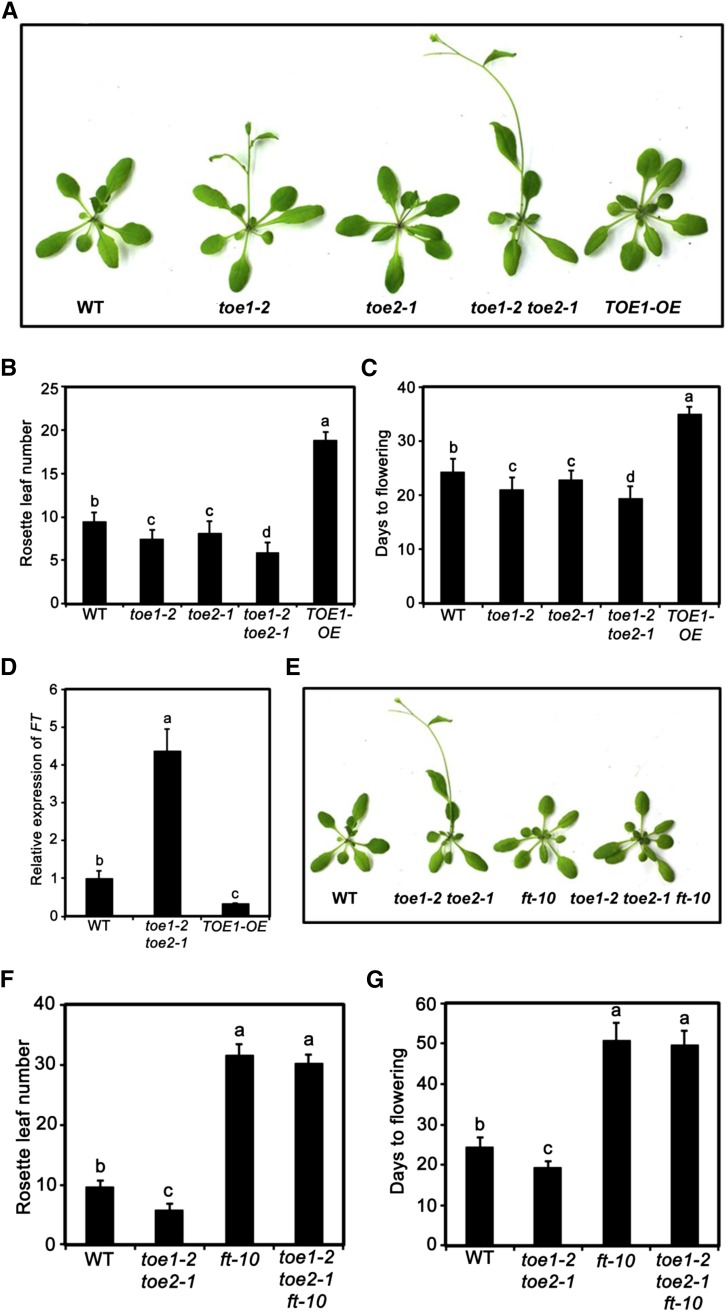
Figure 1.
Flowering Phenotype of JA-Related Mutants and Transgenic Plants.
(A) Representative images of the indicated genotypes showing their flowering phenotype under LD conditions.
(B) and (C) Flowering phenotype of the indicated genotypes assessed by RLN (B) and DTF (C) under LD conditions.
(D) and (E) Flowering phenotype of the indicated genotypes assessed by RLN (D) and DTF (E) under SD conditions.
For (B) to (E), values are means ± sd of 20 to 30 plants. The experiments were repeated at least three times with similar results. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
The finding that the COI1-dependent signaling pathway delays flowering raised a possibility that Arabidopsis mutants defective in JA biosynthesis may exhibit an early flowering phenotype like coi1-2. To test this, we examined the flowering phenotype of aos, a mutant with disrupted expression of the JA biosynthetic gene ALLENE OXIDASE SYNTHESIS (AOS) (Vick and Zimmerman, 1987; Song et al., 1993; Park et al., 2002). Interestingly, under LD or SD conditions, the flowering time phenotype of aos was similar to its wild-type counterpart (Supplemental Figure 1), indicating that AOS-dependent JA biosynthesis is dispensable for the control of flowering time.
To identify other molecular components involved in COI1-dependent regulation of flowering time, we asked whether genetic manipulation of JAZ1, which encodes a substrate of the SCFCOI1 E3 ubiquitin ligase that acts as a transcriptional repressor of JA-responsive genes, could lead to altered flowering time of plants. Toward this goal, we generated 35Spro:JAZ1 and 35Spro:JAZ1∆Jas plants (Supplemental Figure 2), which ectopically express the JAZ1 cDNA and a Jas-domain deletion version of JAZ1 cDNA, respectively. When grown under LD conditions, 35Spro:JAZ1∆Jas plants showed similar DTF as the wild type, but the RLN was significantly reduced (Figures 1B and 1C). When grown under SD conditions, 35Spro:JAZ1∆Jas plants displayed an early flowering phenotype, both measured by RLN and DTF (Figures 1D and 1E). Parallel experiments indicated that 35Spro:JAZ1 plants did not display an early flowering phenotype under LD or SD conditions (Figures 1A to 1E). These results are consistent with the observations that overexpression of a full-length JAZ1 cDNA did not lead to altered JA response, but overexpression of a dominant-negative form (Jas-domain deletion version) of JAZ1 resulted in a JA-insensitive phenotype like coi1 (Thines et al., 2007).
Considering that MYC2 regulates diverse aspects of JA responses (Boter et al., 2004; Lorenzo et al., 2004; Dombrecht et al., 2007; Zhai et al., 2013), we examined whether this master regulator is also involved in COI1-mediated regulation of flowering time. Interestingly, in both LD and SD conditions, the flowering time of the myc2-2 mutant, which harbors a T-DNA insertion that disrupts the function of MYC2 (Boter et al., 2004), was comparable to its wild-type counterpart (Figures 1A to 1E), indicating that MYC2 is not involved in COI1-dependent regulation of flowering time.
JA Delays Flowering Time through Repressing FT Expression
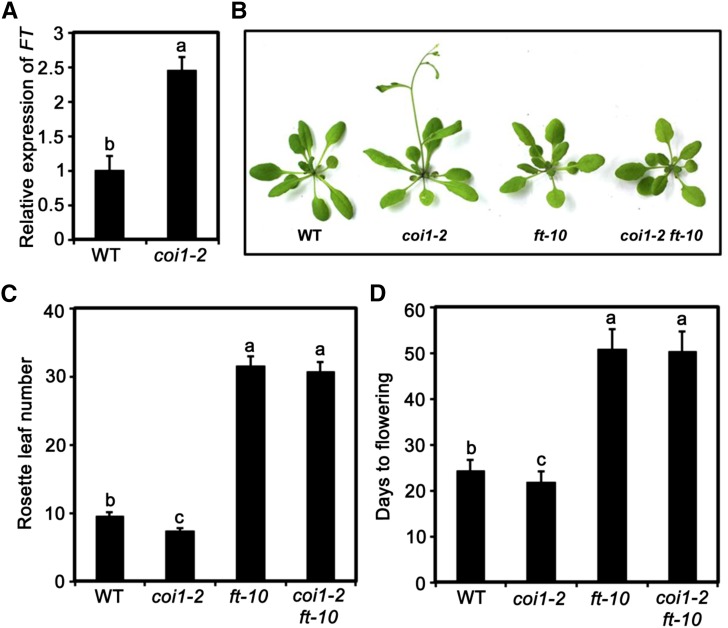
The finding that JA delays flowering suggests a paradigm in which plants are able integrate the effect of the COI1-dependent signaling pathway into the internal flowering genetic network. Considering that FT is a central node of floral integration and that many external inputs are channeled into the transcriptional regulation of FT to regulate flowering (Corbesier et al., 2007; Tamaki et al., 2007; Andrés and Coupland, 2012; Pin and Nilsson, 2012), we compared the expression levels of FT between coi1-2 and wild-type plants. As shown in Figure 2A, in the middle of LDs (at Zeitgeber time 8 [ZT8], i.e., 8 h after light on), transcript levels of FT in coi1-2 plants were substantially higher than those in wild-type plants, suggesting a possibility that the early flowering phenotype of coi1-2 is due to ectopic FT expression. To confirm this genetically, we introduced the ft-10 mutation (Yoo et al., 2005) into the coi1-2 mutant background through crossing. As shown in Figures 2B to 2D, the early flowering phenotype of coi1-2 was largely suppressed by ft-10. Together, these results demonstrate that COI1 mediates the delay of flowering through repressing the expression of FT.
Figure 2.
The ft-10 Mutation Suppresses the Early Flowering Phenotype of coi1-2.
(A) Expression of FT in the indicated genotypes. Ten-day-old plants grown under normal growth conditions (23°C, LD) were harvested at ZT8 for total RNA extraction and qRT-PCR assays. Values are means ± sd of three technical replicates. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
(B) Representative images of the indicated genotypes showing their flowering phenotypes under LD conditions.
(C) and (D) Flowering phenotype of the indicated genotypes assessed by RLN (C) and DTF (D) under LD conditions. Values are means ± sd of 20 to 30 plants. The experiment was repeated three times with similar results. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
A Subset of JAZ Proteins Interact with the AP2 TFs TOE1 and TOE2
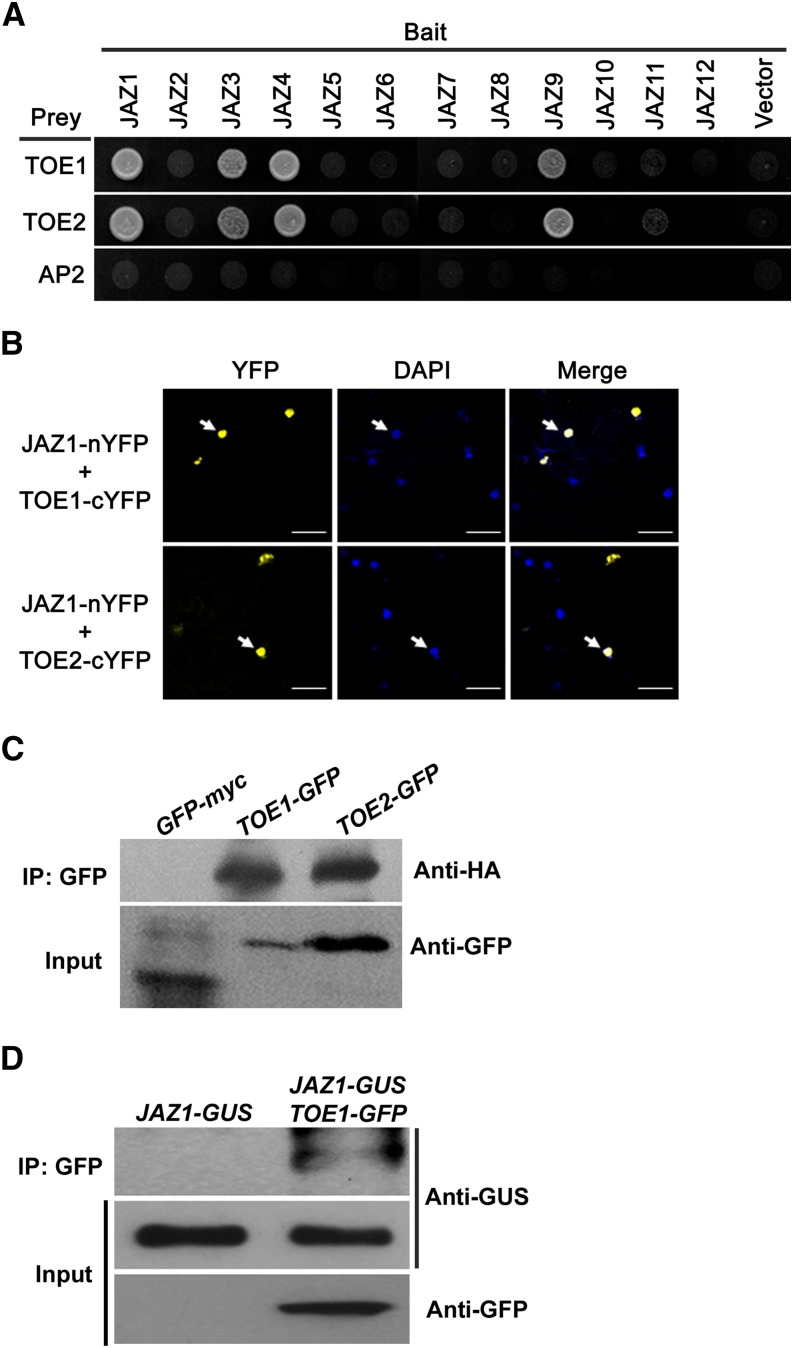
The finding that MYC2 does not play a role in JA-mediated regulation of flowering implies the existence of TFs other than MYC2 that interact with JAZ proteins to orchestrate COI1-mediated regulation of flowering. To identify these potential TFs, we conducted yeast two-hybrid (Y2H) screening using JAZ1 as a bait (BD-JAZ1). Among the identified JAZ1-interacting proteins, we focused on two AP2 family TFs TOE1 and TOE2 (Figure 3A; Supplemental Figure 3), which have been shown to play an important role in the regulation of flowering (Aukerman and Sakai, 2003; Jung et al., 2007).
Figure 3.
TOE1 and TOE2 Interact with a Subset of JAZs.
(A) Y2H assay to detect interactions of TOE1 and TOE2 with the 12 JAZ proteins. Yeast cells cotransformed with pGADT7-TOE1, pGADT7-TOE2, or pGADT7-AP2 (preys) and pGBKT7-JAZ1-12 (bait) were selected and subsequently grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as transformation control (shown in Supplemental Figure 3) or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. pGADT7-TOE1, pGADT7-TOE2, and pGADT7-AP2 cotransformed with pGBDT7 vector were included as a control.
(B) BiFC assays to detect the interaction of JAZ1 with TOE1 and TOE2. JAZ1 was fused to the N-terminal fragment of YFP (nYFP); TOE1 and TOE2 were fused to the C-terminal fragment of YFP (cYFP). The localization of the nuclei was detected by 4′,6-diamidino-2-phenylindole (DAPI) staining. Bars = 50 μm.
(C) Co-IP assays to verify the interaction of JAZ1 with TOE1 and TOE2. Protein extracts from N. benthamiana leaves coexpressing JAZ1 with TOE1, TOE2, or GFP were immunoprecipitated (IP) with or without GFP antibody-bound agarose beads, and immunoprecipitated proteins were analyzed by immunoblotting using anti-HA and anti-GFP antibodies. The experiments were repeated three times with similar results.
(D) Co-IP assays to verify the interaction of JAZ1 with TOE1 in vivo. Protein extracts from transgenic plants carrying both 35Spro:JAZ1-GUS and 35Spro:TOE1-GFP (TOE1-GFP JAZ1-GUS) or from transgenic plants harboring 35Spro:JAZ1-GUS (JAZ-GUS) were immunoprecipitated with GFP antibody, and immunoprecipitated proteins were analyzed by immunoblotting using anti-GUS and anti-GFP antibodies. The experiments were repeated three times with similar results.
We then employed a YFP bimolecular fluorescence complementation (BiFC) system (Weinthal and Tzfira, 2009) to confirm the interaction of JAZ1 with TOE1 and TOE2. For these experiments, JAZ1 was fused to the N-terminal fragment of YFP (nYFP) to produce nYFP-JAZ1, whereas TOE1 and TOE2 were fused to the C-terminal fragment of YFP (cYFP) to produce cYFP-TOE1 and cYFP-TOE2. When the nYFP-JAZ1 construct was transiently coexpressed with cYFP-TOE1 or cYFP-TOE2 in tobacco (Nicotiana benthamiana) leaf cells, reconstituted fluorescence in the nucleus can be observed (Figure 3B), indicating that JAZ1 interacts with TOE1 and TOE2 in planta.
Interaction of JAZ1 with TOE1 and TOE2 was further confirmed with coimmunoprecipitation experiments. For these assays, the JAZ1-HA construct was coexpressed with TOE1-GFP, TOE2-GFP, or GFP-myc in N. benthamiana leaves. Cell extracts were immunopurified with an anti-GFP antibody, and the resultant immunoprecipitates were fractionated via SDS-PAGE and detected with an anti-HA antibody. As shown in Figure 3C, the JAZ1-HA fusion protein can be detected in cell extracts when it was coexpressed with TOE1-GFP or TOE2-GFP. As a negative control, the JAZ1-HA fusion protein cannot be detected in cell extracts when it was coexpressed with GFP-myc (Figure 3C).
Next, we generated a transgenic line expressing 35Spro:TOE1-GFP (Supplemental Figure 4) and crossed it with the previously reported line expressing 35Spro:JAZ1-GUS (Thines et al., 2007). Cell extracts from the resulting F1 plants were immunopurified with an anti-GFP antibody and detected with an anti-GUS antibody. As shown in Figure 3D, the JAZ1-GUS fusion protein was detected in the resultant immunoprecipitates, suggesting that TOE1 interacts with JAZ1 in plant cells.
We then tested whether other members of the Arabidopsis JAZ family proteins also interact with TOE1 and TOE2. In Y2H assays, JAZ1, JAZ3, JAZ4, and JAZ9 interacted with TOE1 and TOE2, but the remaining members of the JAZ family (i.e., JAZ2, JAZ5, JAZ6, JAZ7, JAZ8, JAZ10, JAZ11, and JAZ12) did not show obvious interaction with TOE1 and TOE2 (Figure 3A; Supplemental Figure 3). As a negative control, testing the interaction of the 12 JAZ proteins with the TF AP2, which is a close homolog of TOE1 and TOE2, did not render any positive results in our parallel Y2H assays (Figure 3A; Supplemental Figure 3). Together, these results support a scenario in which TOE1 and TOE2 are targets of a subset of JAZ proteins and these TF repressor complexes orchestrate COI1-mediated regulation of flowering.
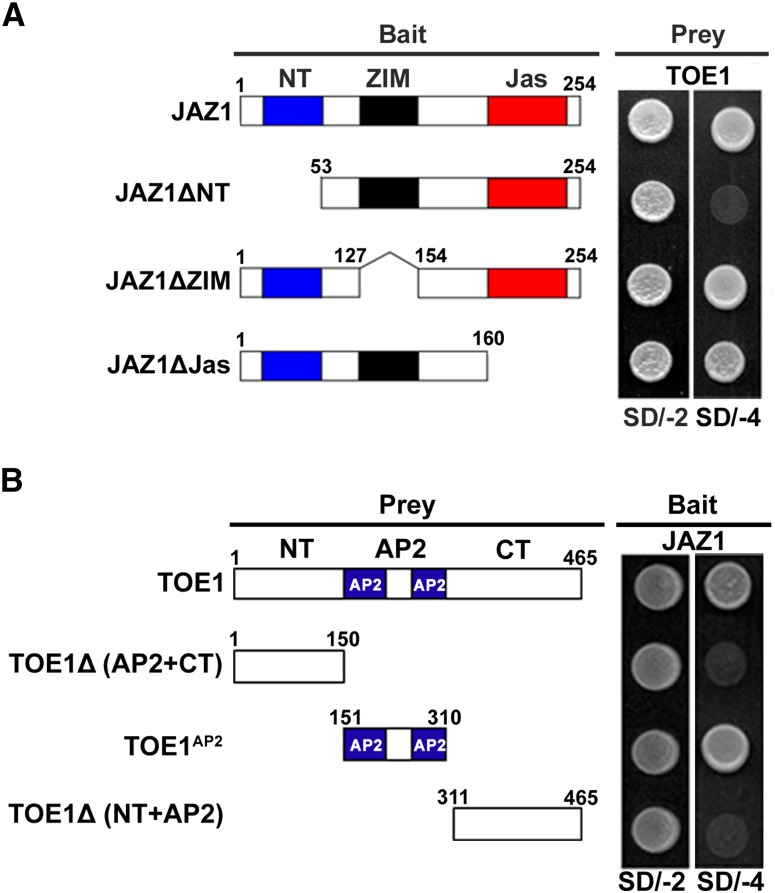
Mapping of Protein Domains Involved in JAZ1-TOE1 Interaction
To examine which protein domain of JAZ1 is critical for its interaction with TOE1, we fused the full-length or different truncations of JAZ1 (Figure 4A) to the Gal4-DNA binding domain to generate bait vectors and fused the full-length TOE1 to the GAL4 activation domain to generate the prey vector. Y2H assays indicated that full-length JAZ1 interacts with TOE1 and that deletion of the ZIM domain and Jas domain of JAZ1 does not affect this interaction (Figure 4A), whereas deletion of the N-terminal domain (NT) of JAZ1 eliminated the interaction between JAZ1 and TOE1 (Figure 4A). These results support the notion that the NT domain of JAZ1 is involved in its interaction with TOE1.
Figure 4.
Mapping of the Protein Domains Involved in the Interaction of TOE1 with JAZ1 Using Y2H Assays.
(A) Based on the schematic protein structure of JAZ1, full-length JAZ1 or its derivatives (pGBKT7-JAZ1 or pGBKT7-JAZ1 derivatives) were tested for interactions with TOE1 (pGADT7-TOE1). Yeast cells cotransformed with pGBKT7-JAZ1 or pGBKT7-JAZ1 derivatives (baits) and pGADT7-TOE1 (prey) were grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as transformation control or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. The different truncations of JAZ1 are represented.
(B) Based on the schematic protein structure of TOE1, full-length TOE1 or its derivatives (pGADT7-TOE1 or pGADT7-TOE1 derivatives) were tested for interactions with JAZ1 (pGBKT7-JAZ1). Yeast cells cotransformed with pGADT7-TOE1 or pGADT7-TOE1 derivatives (prey) and pGBKT7-JAZ1 (bait) were grown on yeast synthetic dropout lacking Leu and Trp (SD/-2) as transformation control or on selective media lacking Ade, His, Leu, and Trp (SD/-4) to test protein interactions. The different domains of TOE1 are represented.
Similarly, to determine which protein domain of TOE1 is responsible for its interaction with JAZ1, we representatively divided the TOE1 protein into the NT domain, the middle fragment containing the two AP2 domains and the C-terminal domain (Figure 4B). We then fused the full-length or different truncations of TOE1 with the GAL4 activation domain to generate prey vector and fused full-length JAZ1 with the GAL4-DNA binding domain to generate the bait vector. Y2H assays indicated that the full-length and the middle AP2 domain of TOE1 interact with JAZ1 (Figure 4B), whereas deletion of the middle AP2 domain plus the C-terminal domain of TOE1 or deletion of the NT domain plus the middle AP2 domain of TOE1 abolish its interaction with JAZ1 (Figure 4B). These results suggest that the middle AP2 domain of TOE1 is important for its interaction with JAZ1.
TOE1 Represses the Expression of FT
TOE1 and TOE2 belong to a subfamily of AP2-like transcription factors, including AP2 itself, TOE1, TOE2, TOE3, SMZ, and its paralog SCHNARCHZAPFEN (Aukerman and Sakai, 2003; Schmid et al., 2003). A wealth of evidence indicated that these six AP2 transcription factors are all the targets of microRNA 172 and they play distinct yet overlapping functions in regulating flowering as well as other developmental processes (Jung et al., 2007; Mathieu et al., 2009; Yant et al., 2010). To understand the physiological relevance of the interactions between the JAZ repressors and the TOE TFs, we examined the role of TOE1 and TOE2 in flowering time regulation. Under our growth conditions, the T-DNA insertion mutants toe1-2 and toe2-1 (Aukerman and Sakai, 2003) both displayed an early flowering phenotype (Figures 5A to 5C), and the early flowering phenotype of the toe1-2 toe2-1 double mutant was much stronger than that of either single mutant lines (Figures 5A to 5C), suggesting that TOE1 and TOE2 have overlapping functions in the regulation of flowering. By contrast, TOE1-OE plants, which express a TOE1-4myc fusion driven by the 35S promoter (Supplemental Figure 4), displayed a late flowering phenotype (Figures 5A to 5C). These results are consistent with previous observations that TOE1 and TOE2 act as repressors of flowering (Schmid et al., 2003; Jung et al., 2007). We then examined the expression levels of FT in the toe1-2 toe2-1 double mutant and TOE1-OE plants. As shown in Figure 5D, FT transcript levels in the toe1-2 toe2-1 double mutant were remarkably higher than those in the wild type. By contrast, FT transcript levels in TOE1-OE plants were substantially lower than those in wild-type plants.
Figure 5.
TOE1 and TOE2 Delay Flowering through Negatively Regulating the Expression of FT.
(A) Representative image of the indicated genotypes showing their flowering phenotype under LD conditions.
(B) and (C) Flowering phenotype of the indicated genotypes assessed by RLN (B) and DTF (C) under LD conditions. Values are means ± sd of 20 to 30 plants. The experiment was repeated three times with similar results. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
(D) Expression of FT in the indicated genotypes. Ten-day-old plants grown under normal growth conditions (23°C, LD) were harvested at ZT8 for total RNA extraction and qRT-PCR assays. Values are means ± sd of three technical replicates. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
(E) to (G) the ft-10 mutation suppresses the early flowering phenotype of the toe1-2 toe2-1 double mutant. Flowering phenotype of the indicated genotypes under LD conditions are shown by representative image (E) or assessed by RLN (F) and DTF (G). Values are means ± sd of 20 to 30 plants. The experiment was repeated three times with similar results. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
The above results point to the possibility that TOE1 and TOE2 regulate flowering by modulating the transcriptional expression of FT. To verify this genetically, we generated a toe1-2 toe2-1 ft-10 triple mutant line and found that the early flowering phenotype of the toe1-2 toe2-1 double mutant was largely suppressed by the ft-10 mutation (Figures 5E to 5G). Together, these results lead us to conclude that TOE1 and TOE2 negatively regulate flowering by repressing the transcription of FT.
TOE1 Binds the Promoter and Downstream Regulatory Region of the FT Locus
To test the hypothesis that TOE1 represses FT expression by binding to its chromatin, we performed chromatin immunoprecipitation (ChIP) assays using 35Spro:TOE1-GFP plants (Supplemental Figure 4) and an anti-GFP antibody. Coprecipitating DNAs were quantified by qRT-PCR with a series of primer sets covering the upstream and downstream regulatory regions as well as the gene body of the FT locus (Figure 6A). Using a high-throughput protein binding microarray assay (Franco-Zorrilla et al., 2014), we showed that the putative TOE binding site (TBS) is AACCTACGA. Indeed, our sequence analysis revealed the existence of a TBS-like (i.e., AACCTAAGA) motif at region C of the FT promoter region (Figure 6A). As shown in Figure 6B, TOE1 enrichment levels at region C (located in the upstream region of FT) and region R (located in the downstream region of FT) were markedly higher than those at other regions. It was recently shown that TOE1 also associates with the FT promoter at region F and region H (Figure 6A), which contain AT-rich elements (Dinh et al., 2012; Zhang et al., 2015). In our ChIP-qPCR experiments, however, TOE1 enrichment levels at region F and region H were much lower than those at region C and region R (Figure 6B). These results indicate that TOE1 preferentially binds region C and region R of the FT chromatin. Interestingly, our sequence analysis failed to identify any TBS or TBS-like motif at region R, suggesting that TOE1 may also recognize DNA sequences other than TBS or TBS-like.
Figure 6.
JAZ1 Represses the Transcriptional Function of TOE1 and TOE2.
(A) Schematic diagram of FT indicating the amplicons and probe used for the ChIP-qPCR assay and EMSA. Positions of the transcription start site (TSS) and transcription termination site (TTS) are indicated.
(B) ChIP-qPCR assays showing that TOE1 associates with the FT locus. Chromatin of transgenic plants expressing 35Spro:TOE1-GFP (TOE1-GFP) was immunoprecipitated with an anti-GFP antibody, and no antibody immunoprecipitates served as control. Immunoprecipitated chromatin was analyzed by qRT-PCR using primers corresponding to the amplicons represented by the schematic diagram of FT (A). ChIP signal was displayed as the percentage of total input DNA. Values are means ± sd of three technical replicates.
(C) Dynamic recruitment of TOE1 to the FT locus. ChIP assays were performed as in (B), except that 35Spro:TOE1-GFP plants were treated with 100 μM MeJA for 30 min before cross-linking. Values are means ± sd of three technical replicates. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
(D) EMSA showing that the TOE1-MBP fusion protein binds to the DNA probes of FT in vitro. Biotin-labeled probes were incubated with TOE1-MBP protein or TOE1-MBP and JAZ1-His proteins, and the free and bound DNAs were separated on an acrylamide gel. As indicated, unlabeled probe and unlabeled mutant probe were used as competitors. Mu, mutated probe in which the 5′-AACCTAAGA-3′ motif was replaced with 5′-TTTTTTTT-3′. A gradient concentration of JAZ1-His was applied (0.5 μg for +; 1.0 μg for ++; 1.5 μg for +++).
(E) Transient expression assays showing that JAZ1 counteracts the function of TOE1 to repress FT expression. Representative images of N. benthamiana leaves 72 h after infiltration are shown. The bottom panel indicates the infiltrated constructs.
(F) Quantitative analysis of luminescence intensity in (E). Values are means ± sd of five independent determinations. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
(G) qRT-PCR analysis of TOE1 expression in the infiltrated leaf areas shown in (E). Total RNA was extracted from leaves of N. benthamiana coinfiltrated with the indicated constructs. Values are means ± sd of five independent determinations.
We then conducted a DNA electrophoretic mobility shift assay (EMSA) to confirm that TOE1 binds the TBS-like motif in vitro. For this purpose, full-length TOE1 protein was expressed as a maltose binding protein (MBP) fusion protein in Escherichia coli and affinity purified. As shown in Figure 6D, the TOE1-MBP fusion proteins were able to bind DNA probes containing the TBS-like motif located at region C of the FT promoter. Moreover, the addition of an excess amount of unlabeled DNA probe effectively reduced the binding of TOE-MBP to the labeled DNA probe (Figure 6D). Parallel experiments indicated that a mutant form of the DNA probe failed to compete the binding of TOE1-MBP to the TBS-like motif (Figure 6D). Together, these results reveal that TOE1 regulates FT expression through direct association with its chromatin.
JAZ1 Represses the Transcriptional Function of TOE1
Our findings that TOE TFs are targets of a subset of JAZs suggest that these JAZ repressors might have a negative effect on the transcriptional function of TOE TFs. To test this, we verified the effect of JAZ1 on the transcriptional function of TOE1 using the well-established transient expression assay in N. benthamiana (Chen et al., 2011; Zhai et al., 2013). For this purpose, we generated a FTpro:LUC reporter, in which luciferase (LUC) was fused with the FT promoter. When the FTpro:LUC reporter was infiltrated into N. benthamiana leaves, a substantial amount of LUC activity could be detected (Figures 6E to 6G), indicating that endogenous factors of N. benthamiana may activate the expression of FTpro:LUC. Coexpression of FTpro:LUC with the 35Spro:TOE1 construct led to an obvious reduction of luminescence intensity (Figures 6E to 6G), suggesting that 35Spro:TOE1 represses the expression of FTpro:LUC. When the 35Spro:JAZ1 construct was added, the luminescence intensity of the reporter was dramatically higher than that of the coexpression combination of 35Spro:TOE1 and FTpro:LUC (Figures 6E to 6G), suggesting that the repression effect of 35Spro:TOE1 on FTpro:LUC expression was counteracted by 35Spro:JAZ1. Importantly, when 35Spro:JAZ1 was replaced by the 35Spro:JAZ1ΔNT construct, in which the JAZ1 NT domain that is important for its interaction with TOE1 was deleted, JAZ1-mediated repression of the transcriptional function of TOE1 was reversed (Figures 6E to 6G). Taken together, these results support the notion that JAZ1 interacts with TOE1 and thereby relieves the repression effect of TOE1 on FT.
Considering that the AP2 domain of TOE1 is involved in its interaction with JAZ1 and that the AP2 domain of TOE family TFs is important for DNA binding (Okamuro et al., 1997), we asked whether JAZ1 affects the DNA binding ability of TOE1. In the above-described EMSA experiments, we found that the addition of a gradient amount of JAZ1-His fusion protein shows a negligible effect on the ability of TOE1-MBP to bind a DNA probe containing the TBS-like motif (Figure 6D), indicating that JAZ1 does not interfere with the DNA binding ability of TOE1 in vitro. However, in our ChIP-qPCR experiments, TOE1 binding levels at regions C and region R of the FT chromatin were significantly increased upon methyl jasmonate (MeJA) treatment (Figure 6C), suggesting that JAZ1 indeed affects the DNA binding ability of TOE1 in vivo. We reasoned that, in plant cells, the ability of JAZ1 to affect the DNA binding ability of TOE1 may rely on additional cofactors (i.e., JAZ-interacting corepressors).
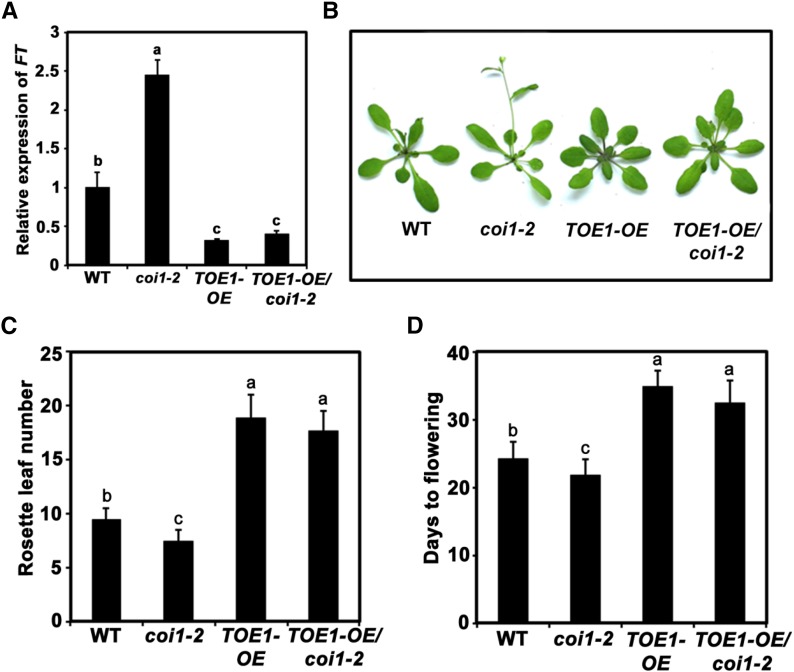
Transgenic Expression of TOE1 Selectively Rescues the Early Flowering Phenotype of the coi1-2 Mutant
In light of our current understanding of the molecular mechanisms of JA-triggered gene transcription, our findings support a scenario in which JAZ proteins are overaccumulated in coi1 mutants and hence attenuate the function of the TOE TFs to repress the expression of FT. Therefore, FT expression levels are elevated in coi1, which accounts for the early flowering phenotype of this mutant. We reasoned that ectopic expression of TOE1 could reduce the expression levels of FT and therefore restore the early flowering phenotype of coi1. To test this, the above-described TOE1-OE plants were crossed with coi1-2 to generate TOE1-OE/coi1-2 plants (Supplemental Figure 4). Examination of FT expression revealed that, in contrast with coi1-2 plants, which exhibited substantially higher levels of FT transcripts than did wild-type plants, the levels of FT transcripts in TOE1-OE/coi1-2 plants were much lower than in wild-type plants (Figure 7A). Consistently, as assessed by RLN or DTF, TOE1-OE/coi1-2 plants displayed a late flowering phenotype (Figures 7B to 7D), indicating that ectopic expression of TOE1 restored the early flowering phenotype of the coi1-2 mutant. Taken together, these results substantiated our conclusion that the TOE-JAZ complex-orchestrated regulation of FT transcription underlies COI1-dependent regulation of flowering.
Figure 7.
Transgenic Expression of TOE1 Rescues the Early Flowering of coi1-2.
(A) Expression of FT in the indicated genotypes. Ten-day-old plants grown under normal growth conditions (23°C, LD) were harvested at ZT8 for total RNA extraction. Values are means ± sd of three technical replicates. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
(B) Representative images of the indicated genotypes showing their flowering phenotype under LD conditions.
(C) and (D) Flowering phenotype of the indicated genotypes assessed by RLN (C) and DTF (D). Values are means ± sd of 20 to 30 plants. The experiment was repeated three times with similar results. ANOVA was performed for statistical analysis; bars with different letters are significantly different from each other (P < 0.01).
We then asked whether ectopic expression of TOE1 could restore the JA-insensitive phenotype of coi1-2 in root growth inhibition and defense gene expression. Our standard JA response assays revealed that, in terms of JA-induced root growth inhibition and JA-induced VEGETATIVE STORAGE PROTEIN1 (VSP1) (Berger et al., 1995) or PDF 1.2 (Penninckx et al., 1996) expression, the JA responses of TOE1-OE/coi1-2 plants were essentially similar to those of coi1-2 plants (Supplemental Figures 5A to 5C), indicating that ectopic expression of TOE1 failed to restore the JA response deficiency of coi1-2 in root growth inhibition and defense gene expression. These results are consistent with the fact that, in terms of JA-induced root growth inhibition and defense gene expression, the JA responses of the toe1-2 toe2-1 double mutant were comparable to those of its wild type (Supplemental Figures 6A to 6C). Together, these results support the notion that the TOE-JAZ interaction complexes are specifically involved in COI1-mediated regulation of flowering in Arabidopsis.
DISCUSSION
The striking capacity of plants to adapt their growth and development to their ever-changing environments is mediated by an array of small-molecule hormones that regulate virtually every biological process during the life cycle. Here, we provide evidence that, in addition to regulating the immune response, the JA receptor COI1 also plays an important role in regulating the plasticity of flowering time. First, the coi1-2 mutant displays an early flowering phenotype (Figure 1), which can be largely suppressed by the ft-10 mutation (Figure 2), suggesting that COI1 negatively regulates flowering through repressing the expression of the florigen gene FT. Second, the AP2 TFs TOE1 and TOE2 repress FT transcription by binding to its chromatin (Figures 5 and 6). Third, a subset of JAZ proteins physically interacts with TOE1 and TOE2 and JAZ1 is able to relieve the repression effect of TOE1 on FT transcription (Figure 3, 4, and 6). Finally, overexpression of TOE1 rescues the early flowering phenotype of the coi1-2 mutant (Figure 7). Collectively, these results not only ascribe an important physiological role to the COI1-dependent signaling pathway in regulating flowering, but also illustrate the underlying transcriptional mechanism. Our findings are consistent with the long-standing observations that, in addition to promoting the immune response, JA generally represses vegetative growth while promoting reproductive development (Wasternack, 2007; Browse, 2009; Wasternack and Hause, 2013; Huot et al., 2014). The differential effects of JA on vegetative growth and reproductive development have already indicated that this hormone has an important role in ensuring successful seed set and propagation.
Given that adaptive variations in the timing to flowering are important for maximizing plant survival and thereby enabling reproductive success, the finding that the COI1-dependent signaling pathway delays flowering reflects that, when encountering stress conditions that trigger elevated JA levels, plants employ the TOE/JAZ interaction complex (which is a JA signaling module-mediated transcriptional regulator of FT) to integrate COI1-dependent signaling into the internal flowering pathway to delay flowering, thereby ensuring that seeds set under the most favorable conditions.
Intriguingly, however, a knockout mutation of AOS does not affect flowering time, indicating that even though AOS-dependent JA biosynthesis plays an essential role in the wound response and male fertility (Park et al., 2002), this gene is dispensable for the control of flowering time. Arabidopsis AOS (At5g42650) encodes the cytochrome P450 enzyme CYP74A. The primary sequences of CYP74s are significantly different from those of other P450s (Bak et al., 2011). Based on the primary sequence, it is believed that only one copy of AOS is present in the Arabidopsis genome (Park et al., 2002; Bak et al., 2011). However, we cannot rule out the possibility that other members of the CYP74 family (i.e., CYP74B2, At4g15440) play a role in JA biosynthesis (Bak et al., 2011). These results are consistent with the long-standing observations that JA biosynthesis contributing to different aspects of the JA response (i.e., wound response, development, and other stress responses) are differentially regulated. For example, DEFECTIVE IN ANTHER DEHISCENCE1 (DAD1), which encodes a chloroplast-targeted lipase, mainly affects male fertility and anther dehiscence, but shows minor effects on the wound response (Ishiguro et al., 2001), whereas DONGLE, a homolog of DAD1, mainly contributes to the rapid JA burst after wounding (Hyun et al., 2008). Other examples in which different members of a gene family contribute to different aspects of the JA response come from the JA synthesis gene family 13-LIPOXYGENASE. Among the four LOX genes in the Arabidopsis genome (i.e., LOX2, LOX3, LOX4, and LOX6), it is already known that LOX2 is mainly involved in wound-induced JA biosynthesis but has a negligible effect on fertility (Glauser et al., 2009; Seltmann et al., 2010), whereas LOX3 and LOX4 act redundantly to regulate fertility (Caldelari et al., 2011). It is of great interest to identify the JA biosynthesis genes that are specifically involved in the control of flowering.
The finding that the COI1-dependent signaling pathway delays flowering but JA biosynthesis mutants lack the delayed flowering time phenotype raised the possibility that COI1 regulates flowering in response to factors other than JA itself. Among the candidates of these factors are coronatine (COR), a phytotoxin produced by several pathovars of the bacterial pathogen Pseudomonas syringae (Feys et al., 1994). COR is a structural and functional mimic of JA-Ile, the bioactive form of JA (Fonseca et al., 2009; Yan et al., 2009; Sheard et al., 2010). It is well established that, like JA-Ile, COR directly targets the COI1-JAZ coreceptor complex to promote the degradation of the JAZ repressors and thereby activates JA responses (Fonseca et al., 2009; Yan et al., 2009; Sheard et al., 2010). It is reasonable to speculate that, in response to COR produced by the attacking pathogens, plants activate COI1-dependent signaling to regulate flowering time as an adaptation strategy.
As a major immune hormone, JA mainly regulates plant defense responses to mechanical wounding, insect attack, and pathogen infection. In addition, this hormone also regulates diverse aspects of plant growth and development. An interesting question in the field of JA studies concerns how the JA signals are converted into specific context-dependent responses. Our results that TOE1 and TOE2 together with their interacting JAZ repressors specifically regulate COI1-dependent flowering, but not other COI1-dependent JA responses (i.e., defense gene expression and root growth inhibition), suggest that the specificity of the JA response is determined by matching pairs of TFs and their interacting JAZ repressors. In line with this scenario, recent studies have identified a growing number of interacting pairs between TFs and JAZs that regulate different aspects of JA-dependent transcriptional responses. For example, the WD-repeat/bHLH/MYB transcriptional complexes interact with a subset of JAZs to regulate JA-mediated anthocyanin accumulation and trichome formation (Qi et al., 2011). The R2R3-MYB TFs MYB21 and MYB24 interact with JAZ1, JAZ8, and JAZ11 to regulate JA-dependent male fertility (Song et al., 2011). The bHLH-type TFs INDUCER OF CBF EXPRESSION1 (ICE1) and ICE2 interact with JAZ4 and JAZ9 to regulate JA-dependent freezing tolerance (Hu et al., 2013). The WRKY family TF WRKY57 interacts with JAZ4 and JAZ8 to regulate JA-induced leaf senescence (Jiang et al., 2014). It is reasonable to speculate that future studies will identify more interacting pairs of TFs and JAZs that regulate different aspects of JA-dependent transcriptional responses, which will deepen our understanding of the molecular mechanisms of JA actions.
Interestingly, in contrast to most of the known JAZ-TF interaction pairs, in which the Jas domain of JAZ proteins and the transcription activation domain of TFs are involved in JAZ-TF interaction (reviewed in Pauwels and Goossens, 2011), we found that the NT domain of JAZ1 and the AP2 domain of TOE1 are involved in the JAZ1-TOE1 interaction. This might reflect that JAZ proteins can adopt diverse mechanisms to attenuate the function of their interacting TFs in a context-specific manner. Considering that the AP2 domain of TOE1 is likely to be important for DNA binding (Okamuro et al., 1997), our results support that, in addition to repressing the transcriptional activity of TOE1 and TOE2, JAZ proteins also affect the DNA binding ability of these TFs. In line with this hypothesis, our ChIP-qPCR assays revealed that JA treatment indeed enhanced the binding of TOE1 to the FT locus. Together, these results support a scenario in which, in the steady state, the activity of TOE TFs is repressed by their interacting JAZ repressors. Upon activation of the COI1-dependent signaling pathway, JAZ repressors are degraded and TOE TFs repress the expression of FT.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as the wild type. Some of the plant materials used in this study were previously described: coi1-2 (Xu et al., 2002); myc2-2 (Boter et al., 2004); toe1-2, toe2-1, and the toe1-2 toe2-1 double mutant (Aukerman and Sakai, 2003); and 35Spro:JAZ1-GUS (Thines et al., 2007). Seeds of ft-10 (CS9869) were obtained from the ABRC (Alonso et al., 2003).
The coi1-2 ft-10 double mutant and the toe1-2 toe2-1 ft-10 triple mutant were generated by crossing parental single or double homozygous lines. The resulting F2 segregating progenies were genotyped to identify plants homozygous for each locus. The toe1-2 (Salk_069677) and toe2-1 (Salk_065370) mutations were identified with PCR amplification to confirm the presence of T-DNA insertions in both genes.
Plants were grown in plastic pots containing a mixture of substrate and vermiculite (3:1) or in Murashige and Skoog medium supplemented with 1% (w/v) sucrose and 0.8% (w/v) agar for in vitro culture. Controlled environmental conditions were provided in growth chambers at 22°C and 70% relative humidity. Plants were illuminated with cool-white fluorescent lights (∼120 µmol m–2 s–1). LD conditions consisted of 16 h of light followed by 8 h of darkness; SD conditions consisted of 8 h of light followed by 16 h of darkness. Nicotiana benthamiana was grown under a 16-h-light (28°C)/8-h-dark (22°C) photoperiod.
Measurement of Flowering Time
Measurement of flowering time was performed as previously described (Chen et al., 2012). Flowering time was scored as DTF (the number of days from germination to the first appearance of buds at the apex) and RLN (the total number of rosette leaves after the main stem has bolted 1 cm). For each replicate, flowering time was recorded from at least 20 plants per genotype. Data are the averages of three replicates. Statistical significance was determined using ANOVA.
Plant Treatment
JA-mediated root growth inhibition assays were performed as previously described (Chen et al., 2011). For MeJA-induced defense gene expression, 10-d-old seedlings were treated as recently described (Zhai et al., 2013).
DNA Constructs and Plant Transformation
DNA constructs for plant transformation were generated following standard molecular biology protocols and Gateway (Invitrogen) technology. The full-length coding sequences of TOE1 and TOE2 were amplified with Gateway-compatible primers. The PCR product was cloned using pENTR Directional TOPO cloning kits (Invitrogen) and then recombined with the binary vector pGWB6 (35S promoter, N-GFP) to generate the 35Spro:TOE1-GFP and 35Spro:TOE2-GFP constructs, respectively. The full-length coding sequence of TOE1 was also cloned into the pGWB18 vector (35S promoter, N-4myc) to generate the 35Spro:TOE1-4myc (TOE1-OE) construct. A similar approach was used to generate the 35Spro:JAZ1 (35S promoter, C-GFP), 35Spro:JAZ1∆Jas (35S promoter, C-GFP), and 35Spro:JAZ1∆NT (35S promoter, C-GFP) constructs. All primers used for DNA construct generation are listed in Supplemental Table 1.
The above constructs were then transformed into Agrobacterium tumefaciens strain GV3101 (pMP90), which was used for transformation of Arabidopsis plants via the floral dip method. Transformants were selected based on their resistance to hygromycin. Homozygous T3 or T4 transgenic seedlings were used for phenotype and molecular characterization.
Y2H Assays
The full-length coding sequence of JAZ1 was amplified with the listed primers (Supplemental Table 1). Enzyme-digested PCR products were cloned into the pGBKT7 vector for the Y2H screening of the pGADT7-based Arabidopsis cDNA library, which was constructed with mRNAs isolated from Arabidopsis seedlings. Y2H assays were based on Matchmaker GAL4 two-hybrid systems (Clontech). Yeast transformants were exhaustively selected on SD/-Ade/-His/-Leu/-Trp/X-α-Gal (4 mg/mL) medium. Putative JAZ1 interacting clones were characterized and sequenced. TOE1 and TOE2 were identified through the screening.
To verify the interactions of JAZs and their domains with TOE1 and TOE2, 11 other JAZs (JAZ2, JAZ3, JAZ4, JAZ5, JAZ6, JAZ7, JAZ8, JAZ9, JAZ10, JAZ11, and JAZ12) and the related domains were fused with the BD domain in the pGBKT7 vector individually. TOE1, TOE2, and the derivatives of TOE1 were fused with the AD domain in the pGADT7 vector. Primers used for the constructs are listed in Supplemental Table 1. Constructs to test interaction were cotransformed into the yeast strain Saccharomyces cerevisiae AH109. The presence of the transgenes was confirmed by growth on an SD/-Leu/-Trp plate. To assess protein interactions, the transformed yeasts were suspended in liquid SD/-Leu/-Trp to OD = 1.0. Five microliters of suspended yeast was spread on the plates containing SD/-Ade/-His/-Leu/-Trp medium. The interactions were observed after 3 d of incubation at 30°C.
BiFC Assays
Full-length coding sequences of TOE1, TOE2, and JAZ1 were cloned into the binary N-terminal fragment of YFP or the C-terminal fragment of YFP vector through the Gateway reaction with the pENTR vector system (Invitrogen) and sequences were verified. Primers for the construction are listed in Supplemental Table 1. The resulting constructs were then introduced into Agrobacterium strain GV3101(pMP90). N. benthamiana infiltration was performed as described (Song et al., 2011). After infiltration, plants were incubated for at least 50 h before observation. The YFP fluorescence was imaged under a Leica confocal laser scanning microscope (Leica Microsystems). Leaves were infiltrated with 2 μg/mL 4′,6-diamidino-2-phenylindole for nuclei staining 2 h before observation.
Co-IP Assays
Co-IP assays were performed according to a published procedure (Liu et al., 2010) with minor modifications. Briefly, agrobacterial strains carrying constructs of TOE1-GFP or TOE2-GFP, JAZ1-HA, as well as p19 genes were coinfiltrated into N. benthamiana leaves. Agrobacterial strains carrying constructs of GFP-myc were used as a control. The infiltrated parts of N. benthamiana leaves were harvested and then the leaf tissue was ground in liquid nitrogen and resuspended in extraction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.2% Nonidet P-40, 0.6 mM PMSF, and 20 μΜ MG132 with 1× Roche protease inhibitor cocktail). After protein extraction, 20 μL protein G plus agarose (Santa Cruz) was added to the 2-mg extracts to reduce nonspecific immunoglobulin binding. After 1 h of incubation, the supernatant was transferred to a new tube. GFP antibody-bound agarose beads (MBL) were then added to each reaction for 1 h at 4°C. The precipitated samples were washed at least four times with the lysis buffer and then eluted by adding 1× SDS protein loading buffer with boiling for 5 min. To test the interaction of JAZ1 with TOE1 in plants, 10-d-old transgenic plants carrying both 35Spro:JAZ1-GUS and 35Spro:TOE1-GFP (JAZ1-GUS TOE1-GFP) or 35Spro:JAZ1-GUS only (JAZ1-GUS) were used in co-IP assays, and 3 mg of protein extracts were precleared with the protein G plus agarose beads and incubated with GFP antibody (Abcam) and the protein G plus agarose beads at 4°C for 4 h.
Transient Expression Assay in N. benthamiana Leaves
The transient expression assays were performed in N. benthamiana leaves as previously described (Chen et al., 2011). The FT promoter was amplified with Gateway-compatible primers. The PCR product was cloned using pENTR Directional TOPO cloning kits (Invitrogen) and then recombined with the binary vector pGWB35 to generate the reporter construct FTpro:LUC. The TOE1, JAZ1, and JAZ1ΔNT effector constructs were the above-described 35Spro:TOE1-GFP (35Spro:TOE1), 35Spro:JAZ1-GFP (35Spro:JAZ1), and 35Spro:JAZ1ΔNT -GFP (35Spro:JAZ1ΔNT). We used a low-light cooled CCD imaging apparatus (NightOWL II LB983 with Indigo software) to capture the LUC image and to count luminescence intensity. The leaves were sprayed with 0.5 mM luciferin and were placed in darkness for 3 min before luminescence detection.
RNA Extraction and Gene Expression Analyses
For qRT-PCR analysis, total RNA was extracted from 10-d-old seedlings using Trizol (Invitrogen) reagent. cDNA was prepared from 2 µg of total RNA with Superscript III reverse transcriptase (Invitrogen) and quantified with a cycler apparatus (Roche 480) with the SYBR Green kit (Takara) according to the manufacturer’s instructions. Expression levels of target genes were normalized to ACTIN7. Primers used for qRT-PCR are listed in Supplemental Table 1.
ChIP-qPCR Assay
Ten-day-old seedlings of 35Spro:TOE1-GFP (TOE1-GFP) plants were treated with or without 100 μM MeJA for 30 min, and 1.5 g of the samples was harvested and cross-linked in 1% formaldehyde at room temperature for 10 min and neutralized with 0.125 M glycine. Chromatin was sonicated with a Bioruptor 20 times (30 s on/30 s off cycles and high-power output) in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% SDS, 1% Triton X-100, 0.1% sodium deoxycholate, and 1 mM PMSF with 1× Roche protease inhibitor cocktail) and then incubated at 4°C for 4 h with GFP antibody (Roche). Immunoprecipitated complexes were collected using protein A beads (Millipore) and washed with LiCl washing buffer (0.25 M LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.0). Reverse protein DNA cross-linking was performed by incubating the immunoprecipitated complexes at 95°C for 10 min. DNA was recovered with a QIAquick PCR purification kit (Qiagen) and analyzed by quantitative real-time PCR (qRT-PCR) using primers as described (Supplemental Table 1). Chromatin precipitated without antibody was used as a negative control, while the chromatin isolated before precipitation was used as an input control. Three independent biological repeats were performed.
EMSA
To construct plasmids for the expression of recombinant TOE1 and JAZ1 protein in Escherichia coli, the full-length coding sequences of TOE1 and JAZ1 were amplified and cloned into pMAL-c2 and pET28a vectors, respectively. Oligonucleotide probes were synthesized and labeled with biotin at the 3′ end (Invitrogen). EMSA was performed using a LightShift Chemiluminescent EMSA kit (Thermo Scientific). Briefly, biotin-labeled probes were incubated in 1× binding buffer, 2.5% glycerol, 50 mM KCl, 5 mM MgCl2, and 10 mM EDTA with or without proteins at room temperature for 20 min. For unlabeled probe competition, unlabeled probes were added to the reactions. The probe sequence was as follows: 5′-GGTTACCAAATATTTTGGATTTCAACCTAAGATGAAGTTAACCTATGGACGT-3′; the mutated probe (Mu) sequence was as follows: 5′-GGTTACCAAATATTTTGGATTTCTTTTTTTTTGAAGTTAACCTATGGACGT-3′.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: TOE1 (At2g28550), TOE2 (At5g60120), AP2 (At4g36920), FT (At1g65480), MYC2 (At1g32640), COI1 (At2g39940), JAZ1 (At1g19180), JAZ2 (At1g74950), JAZ3 (At3g17860), JAZ4 (At1g48500), JAZ5 (At1g17380), JAZ6 (At1g72450), JAZ7 (At2g34600), JAZ8 (At1g30135), JAZ9 (At1g70700), JAZ10 (At5g13220), JAZ11 (At3g43440), JAZ12 (At5g20900), VSP1 (At5g24780), PDF1.2 (At5g44420), and ACTIN7 (At5g09810).
Supplemental Data
Supplemental Figure 1. Flowering Phenotype of JA Biosynthesis Mutant aos.
Supplemental Figure 2. Generation of 35Spro:JAZ1 and 35Spro:JAZ1∆Jas Plants.
Supplemental Figure 3. Control Growth of Yeast Cells Transformed with TOE1, TOE2, or AP2 with JAZs in Y2H Assays.
Supplemental Figure 4. Generation of Transgenic Plants Containing 35Spro:TOE1 in the Genetic Background of Col-0 and coi1-2.
Supplemental Figure 5. Transgenic Expression of TOE1 Failed to Restore the Defective JA Response of coi1-2 in Root Growth Inhibition and Defense Gene Expression.
Supplemental Figure 6. JA-Induced Root Growth Inhibition and Defense Gene Expression in the Indicated Genotypes.
Supplemental Table 1. List of the DNA Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank John Browse (Washington State University Pullman, WA) for providing materials. This work was supported by the National Basic Research Program of China (2015CB942900), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB11030200), the National Natural Science Foundation of China (31030006, 91317039, and 31371223), and the State Key Laboratory of Plant Genomics of China (2015B0129-04).
AUTHOR CONTRIBUTIONS
C.L. and Q.W. designed the research. C.L. conceived and supervised the project. Q.Z., X.Z., and F.W. performed most of the experiments. H.F., L.D., L.X., and M.Z. generated the constructs and the transgenic plants. Q.Z. and C.L. analyzed the data. Q.Z., Q.W., and C.L. wrote the article.
Glossary
- SAM
shoot apical meristem
- TF
transcription factor
- JA
jasmonate
- bHLH
basic helix-loop-helix
- LD
long-day
- RLN
rosette leaf number
- SD
short-day
- DTF
days from germination to flowering
- ZT8
Zeitgeber time 8
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- NT
N-terminal
- ChIP
chromatin immunoprecipitation
- TBS
TOE binding site
- EMSA
electrophoretic mobility shift assay
- MeJA
methyl jasmonate
- COR
coronatine
Footnotes
Articles can be viewed online without a subscription.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- An H., Roussot C., Suárez-López P., Corbesier L., Vincent C., Piñeiro M., Hepworth S., Mouradov A., Justin S., Turnbull C., Coupland G. (2004). CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626. [DOI] [PubMed] [Google Scholar]
- Andrés F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13: 627–639. [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S., Beisson F., Bishop G., Hamberger B., Höfer R., Paquette S., Werck-Reichhart D. (2011). Cytochromes p450. Arabidopsis Book 9: e0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S., Bell E., Sadka A., Mullet J.E. (1995). Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol. Biol. 27: 933–942. [DOI] [PubMed] [Google Scholar]
- Bolouri Moghaddam M.R., Van den Ende W. (2013). Sugars, the clock and transition to flowering. Front. Plant Sci. 4: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M., Ruíz-Rivero O., Abdeen A., Prat S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18: 1577–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009). Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60: 183–205. [DOI] [PubMed] [Google Scholar]
- Caldelari D., Wang G., Farmer E.E., Dong X. (2011). Arabidopsis lox3 lox4 double mutants are male sterile and defective in global proliferative arrest. Plant Mol. Biol. 75: 25–33. [DOI] [PubMed] [Google Scholar]
- Castillejo C., Pelaz S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18: 1338–1343. [DOI] [PubMed] [Google Scholar]
- Chen Q., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23: 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Jiang H., Li L., Zhai Q., Qi L., Zhou W., Liu X., Li H., Zheng W., Sun J., Li C. (2012). The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. [DOI] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033. [DOI] [PubMed] [Google Scholar]
- Dombrecht B., Xue G.P., Sprague S.J., Kirkegaard J.A., Ross J.J., Reid J.B., Fitt G.P., Sewelam N., Schenk P.M., Manners J.M., Kazan K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh T.T., Girke T., Liu X., Yant L., Schmid M., Chen X. (2012). The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139: 1978–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B., Benedetti C.E., Penfold C.N., Turner J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., Miersch O., Wasternack C., Solano R. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: Control of flowering in Arabidopsis. Cell 141: 550–, 550.e1–550.e2.. [DOI] [PubMed] [Google Scholar]
- Fornara F., Panigrahi K.C., Gissot L., Sauerbrunn N., Rühl M., Jarillo J.A., Coupland G. (2009). Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17: 75–86. [DOI] [PubMed] [Google Scholar]
- Franceschi V.R., Grimes H.D. (1991). Induction of soybean vegetative storage proteins and anthocyanins by low-level atmospheric methyl jasmonate. Proc. Natl. Acad. Sci. USA 88: 6745–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., López-Vidriero I., Carrasco J.L., Godoy M., Vera P., Solano R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauser G., Dubugnon L., Mousavi S.A., Rudaz S., Wolfender J.L., Farmer E.E. (2009). Velocity estimates for signal propagation leading to systemic jasmonic acid accumulation in wounded Arabidopsis. J. Biol. Chem. 284: 34506–34513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell C.A., Wood C.C., Robertson M., James Peacock W., Dennis E.S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46: 183–192. [DOI] [PubMed] [Google Scholar]
- Hu Y., Jiang L., Wang F., Yu D. (2013). Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B., Yao J., Montgomery B.L., He S.Y. (2014). Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7: 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y., et al. (2008). Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev. Cell 14: 183–192. [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Kawai-Oda A., Ueda J., Nishida I., Okada K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13: 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid- and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell 26: 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kazan K., Manners J.M. (2013). MYC2: the master in action. Mol. Plant 6: 686–703. [DOI] [PubMed] [Google Scholar]
- Krajncic B., Kristl J., Janzekovic I. (2006). Possible role of jasmonic acid in the regulation of floral induction, evocation and floral differentiation in Lemna minor L. Plant Physiol. Biochem. 44: 752–758. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21: 397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., et al. (2012). The U-Box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol. 159: 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhang Y., Tang S., Zhao Q., Zhang Z., Zhang H., Dong L., Guo H., Xie Q. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61: 893–903. [DOI] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Mürdter F., Küttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M., Browse J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8: 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro J.K., Caster B., Villarroel R., Van Montagu M., Jofuku K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12. [DOI] [PubMed] [Google Scholar]
- Pauwels L., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., Goossens A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23: 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx I.A., Eggermont K., Terras F.R., Thomma B.P., De Samblanx G.W., Buchala A., Métraux J.P., Manners J.M., Broekaert W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin P.A., Nilsson O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35: 1742–1755. [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Qi T., Song S., Ren Q., Wu D., Huang H., Chen Y., Fan M., Peng W., Ren C., Xie D. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni M., Robustelli Test A., Galbiati M., Tonelli C., Conti L. (2014). Environmental stress and flowering time: The photoperiodic connection. Plant Signal. Behav. 9: e29036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Okamoto H., Patrick E., Harris S.R., Wasternack C., Brearley C., Turner J.G. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders P.M., Lee P.Y., Biesgen C., Boone J.D., Beals T.P., Weiler E.W., Goldberg R.B. (2000). The arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa M., Nusinow D.A., Kay S.A., Imaizumi T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schweizer F., Fernández-Calvo P., Zander M., Diez-Diaz M., Fonseca S., Glauser G., Lewsey M.G., Ecker J.R., Solano R., Reymond P. (2013). Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell 25: 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20: 898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann M.A., Stingl N.E., Lautenschlaeger J.K., Krischke M., Mueller M.J., Berger S. (2010). Differential impact of lipoxygenase 2 and jasmonates on natural and stress-induced senescence in Arabidopsis. Plant Physiol. 152: 1940–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Zhang Y., Peng W., Wang Z., Xie D. (2009). Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 60: 3849–3860. [DOI] [PubMed] [Google Scholar]
- Sheard L.B., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Ito S., Imaizumi T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18: 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.C., Funk C.D., Brash A.R. (1993). Molecular cloning of an allene oxide synthase: a cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc. Natl. Acad. Sci. USA 90: 8519–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68: 2013–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick P.E., Su W., Howell S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89: 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Browse J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97: 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036. [DOI] [PubMed] [Google Scholar]
- Thines B., Katsir L., Melotto M., Niu Y., Mandaokar A., Liu G., Nomura K., He S.Y., Howe G.A., Browse J. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665. [DOI] [PubMed] [Google Scholar]
- Vick B.A., Zimmerman D.C. (1987). Pathways of fatty acid hydroperoxide metabolism in spinach leaf chloroplasts. Plant Physiol. 85: 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.F., Seabolt S., Hamdoun S., Ng G., Park J., Lu H. (2011). Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol. 156: 1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100: 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C., Hause B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. (Lond.) 111: 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinthal D., Tzfira T. (2009). Imaging protein-protein interactions in plant cells by bimolecular fluorescence complementation assay. Trends Plant Sci. 14: 59–63. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14: 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish M.W., Colasanti J., Rothstein S.J. (2011). The role of epigenetic processes in controlling flowering time in plants exposed to stress. J. Exp. Bot. 62: 3727–3735. [DOI] [PubMed] [Google Scholar]
- Yan J., Zhang C., Gu M., Bai Z., Zhang W., Qi T., Cheng Z., Peng W., Luo H., Nan F., Wang Z., Xie D. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21: 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.L., et al. (2012). Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl. Acad. Sci. USA 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T.T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.K., Chung K.S., Kim J., Lee J.H., Hong S.M., Yoo S.J., Yoo S.Y., Lee J.S., Ahn J.H. (2005). CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiol. 139: 770–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q., Yan L., Tan D., Chen R., Sun J., Gao L., Dong M.Q., Wang Y., Li C. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9: e1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Wang L., Zeng L., Zhang C., Ma H. (2015). Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 29: 975–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., et al. (2011). Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 12539–12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.