Fibrillin 5 is required for plastoquinone-9 (PQ-9) biosynthesis through its interaction with solanesyl diphosphate synthases 1 and 2, which biosynthesize the solanesyl moiety of PQ-9.
Abstract
Fibrillins are lipid-associated proteins in plastids and are ubiquitous in plants. They accumulate in chromoplasts and sequester carotenoids during the development of flowers and fruits. However, little is known about the functions of fibrillins in leaf tissues. Here, we identified fibrillin 5 (FBN5), which is essential for plastoquinone-9 (PQ-9) biosynthesis in Arabidopsis thaliana. Homozygous fbn5-1 mutations were seedling-lethal, and XVE:FBN5-B transgenic plants expressing low levels of FBN5-B had a slower growth rate and were smaller than wild-type plants. In chloroplasts, FBN5-B specifically interacted with solanesyl diphosphate synthases (SPSs) 1 and 2, which biosynthesize the solanesyl moiety of PQ-9. Plants containing defective FBN5-B accumulated less PQ-9 and its cyclized product, plastochromanol-8, but the levels of tocopherols were not affected. The reduced PQ-9 content of XVE:FBN5-B transgenic plants was consistent with their lower photosynthetic performance and higher levels of hydrogen peroxide under cold stress. These results indicate that FBN5-B is required for PQ-9 biosynthesis through its interaction with SPS. Our study adds FBN5 as a structural component involved in the biosynthesis of PQ-9. FBN5 binding to the hydrophobic solanesyl moiety, which is generated by SPS1 and SPS2, in FBN5-B/SPS homodimeric complexes stimulates the enzyme activity of SPS1 and SPS2.
INTRODUCTION
Fibrillins are lipid-associated proteins found in all photosynthetic organisms, from cyanobacteria to plants (Pozueta-Romero et al., 1997; Kessler et al., 1999; Ytterberg et al., 2006; Simkin et al., 2007; Cunningham et al., 2010). They were first identified in fibrils, the thread-like structures involved in carotenoid storage in the chromoplasts of bell pepper (Capsicum annuum) fruit (Newman et al., 1989; Deruère et al., 1994), and were called fibrillins because of their high abundance in fibrils. Members of the fibrillin protein family have been given many different names over the years, including plastid-lipid-associated protein (Pozueta-Romero et al., 1997; Ting et al., 1998), chromoplast-specific carotenoid-associated protein (Vishnevetsky et al., 1996), and plastoglobulins (Kessler et al., 1999). Recently, Singh and McNellis (2011) proposed that this type of proteins be called fibrillins, abbreviated FBN.
Fibrillins can be divided into 12 subfamilies, ranging from algae to plants (Singh and McNellis, 2011). The fibrillin proteins appear quite diverse, with molecular masses in the range of 21 to 42 kD, pI values of 4 to 9, hydrophobic profiles, and plastid localization. Fibrillins are distributed throughout the stroma or are associated with the thylakoid membranes and with lipid globules (Vidi et al., 2006; Lundquist et al., 2012). This diversity suggests that each fibrillin family member has specific biological functions (Laizet et al., 2004; Singh and McNellis, 2011; Lundquist et al., 2012). Arabidopsis thaliana contains 14 genes with sequence similarity to fibrillin genes. All the fibrillins of Arabidopsis share three blocks of similarity in the mature proteins (Laizet et al., 2004) and contain hydrophobic domains that associate with lipids. Singh and McNellis (2011) showed that the 14 fibrillins expressed in Arabidopsis contain a predicted lipocalin motif 1 within conserved block 1 and suggested that the fibrillin family is involved in binding to and transporting a range of small hydrophobic molecules. The residues near the fibrillin C terminus, including aspartic acid, are well conserved in the fibrillins (Singh and McNellis, 2011).
Fibrillins play a structural role in the formation of lipoprotein structures in plastids. Deruère et al. (1994) demonstrated that a 32-kD protein, designated fibrillin, purified from the chromoplasts of a ripening bell pepper fruit, reconstituted the fibril structure in vitro when it was combined with carotenoids and polar lipids. Furthermore, the overexpression of a pepper fibrillin in tobacco (Nicotiana tabacum) increased the numbers of plastoglobules (Rey et al., 2000). In addition to their structural roles, fibrillins are involved in photosynthesis during plant development, tolerance of photooxidative stress, and resistance to biotic stress (Rey et al., 2000; Leitner-Dagan et al., 2006; Simkin et al., 2007; Cunningham et al., 2010; Singh et al., 2010; Singh and McNellis, 2011). Recently, fibrillin mutants of Arabidopsis have extended our understanding of the functions of the fibrillin proteins. The Arabidopsis FIBRILLIN1 (FBN1) gene family is involved in abscisic acid-mediated protection from photoinhibition (Yang et al., 2006), and the FBN1 and FBN2 gene families modulate jasmonate biosynthesis during light and cold stress (Youssef et al., 2010). Apple tree (Malus domestica) and Arabidopsis lacking FBN4 are susceptible to abiotic and biotic stresses, which are accompanied by lower levels of plastoquinone-9 (2,3-dimethyl-6-solanesyl-1,4-benzoquinone; PQ-9) in their plastoglobules. It has been suggested that FBN4, with its lipocalin domain, is involved in the transport of PQ-9 and other molecules from the thylakoids to the plastoglobules (Singh et al., 2010, 2012). However, the precise biological functions of the fibrillins require further investigation.
In plants, the lipid-soluble plastid-localized electron carrier PQ-9 is essential for oxygenic photoautotrophs. It is involved in photosynthetic electron transport and acts as a mobile redox carrier (Trebst, 1978) and a cofactor in the desaturation of phytoene. Consequently, PQ-9 functions in carotenoid biosynthesis (Norris et al., 1995). Reduced PQ-9 also has antioxidant activity in plants under stress conditions (Kruk and Trebst, 2008; Szymańska and Kruk, 2010; Nowicka and Kruk, 2012). Furthermore, the redox state of PQ-9 mediates a number of photosynthetic responses, such as state transitions, photosystem stoichiometry, and the biosynthesis of the proteins of the photosynthetic apparatus (Allen, 1995; Maxwell et al., 1995; Melis et al., 1996; Pfannschmidt et al., 2001). In chloroplasts, PQ-9 is located in the thylakoids, where it occurs in the free form or is associated with the QA and QB sites of photosystem II (PSII). These forms of plastoquinone (PQ) are likely part of the photoactive PQ pool and participate in both electron transport and antioxidant functions (Kruk and Karpinski, 2006). PQ-9 is also found in plastoglobules and the inner membrane of plastids (Tevini and Steinmüller, 1985; Eugeni Piller et al., 2012; Singh et al., 2012). These PQ-9 molecules are thought to be nonphotoactive, storage forms that are used to replenish the photoactive PQ pool (Zbierzak et al., 2010; Singh et al., 2012). Previous studies have shown that PQ-9 biosynthesis occurs in the membranes of the plastid envelope (Hutson and Threlfall, 1980; Soll et al., 1980, 1985; Joyard et al., 2009).
The biosynthetic pathways and enzymes involved in the production of PQ-9 and tocopherols in plants have been clarified with genetic, genomic, and biochemical studies in Arabidopsis (DellaPenna and Pogson, 2006; Sadre et al., 2006; Block et al., 2013). Homogentisate (HGA) is the common head group of PQ-9 and tocopherols in plants. PQ-9 biosynthesis shares the formation of the aromatic HGA ring with the tocopherol pathway. Whereas HGA is condensed with phytyl diphosphate by homogentisate phytyltransferase (VTE2) in tocopherol biosynthesis, the first committed step in PQ-9 biosynthesis is the condensation of homogentisate and solanesyl diphosphate (SPP), which is catalyzed by homogentisate solanesyltransferase (HST). HST uses HGA (derived from the shikimate pathway) and SPP (biosynthesized in the plastid nonmevolonate isoprenoid pathway) as substrates and catalyzes their decarboxylation and prenylation to yield 2-methyl-6-prenyl-1,4-benzoquinol (MSBQ). MSBQ is then methylated by MSBQ/2-methyl-6-phytyl-1,4-benzoquinone methyltransferase (VTE3) to yield PQ-9, which is cyclized into plastochromanol-8 (PC-8) by tocopherol cyclase (VTE1). PC-8 and the tocopherols are lipid-soluble antioxidants essential for seed desiccation and quiescence in Arabidopsis (Mène-Saffrané et al., 2010). The solanesyl moiety is generated by the trans-type consecutive condensation of isopentenyl diphosphate (IPP; C5) with farnesyl diphosphate (C15) or geranylgeranyl diphosphate (GGPP; C20) and is catalyzed by solanesyl diphosphate synthases (SPSs) (Hirooka et al., 2003). The SPSs of Arabidopsis (SPS1 and SPS2), tomato (Solanum lycopersicum) (SlSPS), and rice (Oryza sativa) (OsSPS2) produce SPP in vitro, and the enzymes are targeted to the plastids in planta (Jun et al., 2004; Ohara et al., 2010; Block et al., 2013; Jones et al., 2013). The corresponding single or double knockout mutants of Arabidopsis display either reduced or absent production of PQ-9 and PC-8 (Block et al., 2013). All three SPS enzymes of Arabidopsis function as homodimers (Hirooka et al., 2003; Jun et al., 2004; Hsieh et al., 2011).
In this study, we identified and characterized an fbn5-1 T-DNA insertion mutant of Arabidopsis with a seedling-lethal phenotype. FBN5 is a phylogenetically distant relative of other fibrillins in Arabidopsis (Singh and McNellis, 2011), and it localizes to the stroma, with a lower pI and lower hydrophobicity than the other fibrillins (Lundquist et al., 2012). Therefore, FBN5 may play different roles from those previously reported for the fibrillins that localize primarily to the plastoglobules (Trebst, 1978; Yang et al., 2006; Singh et al., 2010, 2012; Youssef et al., 2010). Surprisingly, we found that FBN5 specifically interacts with two solanesyl diphosphate synthases, SPS1 and SPS2, in the chloroplasts. A deficiency of FBN5 results in reduced levels of PQ-9 and PC-8 in the leaves, suggesting that FBN5 plays a critical role in PQ-9 biosynthesis and is therefore essential for plant development and growth. We propose a model for PQ-9 biosynthesis in plants in which FBN5 functions in concert with SPS.
RESULTS
Mutation of FBN5 Results in Seedling-Lethal Phenotype
In this study, we investigated the function of the FBN proteins by identifying homozygous mutants among T-DNA insertion lines from the TAIR database (http://www.arabidopsis.org). Among 11 T-DNA insertion mutants of FBN genes, seedlings homozygous for mutations in FBN5 were seedling-lethal, which was an unexpected phenotype for a mutated structural protein. Plants of the T-DNA insertion line SALK_064597, which has an insertion in exon 1 of FBN5 (Figure 1A), were grown in soil and genotyped by PCR analysis using primers specific for the T-DNA and FBN5 (Figure 1B). Of 12 plants tested, three were wild type and nine were heterozygous for the T-DNA insertion. No homozygous mutant plants were identified. These results suggest that the homozygous mutant is embryo- or seedling-lethal. When progeny seeds from one of the heterozygous plants, designated FBN5/fbn5-1, were germinated on agar medium in the absence of sucrose, approximately one-quarter of the seedlings quickly turned white and stopped growing (Figure 1C). Murashige and Skoog (MS) medium containing 1% sucrose allowed some growth of the seedlings, but they eventually died.
Figure 1.
Identification and Characterization of an fbn5-1 Homozygous Plant.
(A) Structure of the FBN5 locus showing the location of the T-DNA insertion in the fbn5-1 line (SALK_064597). Exons are shown with boxes; introns are shown with lines. The locations of the oligonucleotide primers used to genotype the fbn5-1 mutant and for RT-PCR are indicated.
(B) Genotyping of wild-type and fbn5-1 T-DNA insertion mutant plants by PCR. A 1.2-kb PCR product was detected using the LP + RP primers, but not with the LBa1 + RP primers in wild-type segregants. In fbn5-1 segregants, a 1-kb band was detected using the Lba1 + RP but not the LP + RP primers.
(C) The fbn5-1 mutation is seedling-lethal. fbn5-1 seedlings germinated on agar medium lacking sucrose, but stopped growing and became chlorotic. Limited growth of fbn5-1 plants on medium containing 1% sucrose. Bars = 1 cm.
(D) RT-PCR indicates that the fbn5-1 plant is a null mutant. Full-length FBN5-A and FBN5-B transcripts, amplified with the P1 + P3 primers and P1 + P2 primers, respectively, were detected in wild-type segregants but not in fbn5-1 plants. Amplification of the ACTIN gene (ACT7, At5g09810) was a control. Molecular mass markers are shown in the left lane of each panel.
To test whether the seedling-lethal phenotype cosegregated with the fbn5-1 mutation, 120 progeny plants obtained from one heterozygote on medium containing 1% sucrose segregated in a green:white phenotypic ratio of 97:23. Among these plants, 10 randomly selected white seedlings were homozygous for the fbn5-1 mutation and 12 randomly selected green seedlings were either wild type or heterozygous at the FBN5 locus. Taken together, these results indicate that the seedling-lethal phenotype segregates as a single, recessive mutation (23 of 120; χ2 = 2.17; P > 0.95) and cosegregates with the fbn5-1 T-DNA insertion.
The FBN5 locus expresses two differentially spliced transcripts according to the TAIR database. We designated the shorter variant FBN5-A and the longer variant FBN5-B (Supplemental Figure 1A). FBN5-A differs from FBN5-B in the deletion of 14 amino acids at the COOH terminus and the substitution of 10 amino acids in the remaining part of the COOH terminus by alternative splicing (Supplemental Figures 1B and 1C). The fibrillin domain occurs at residues 85 to 267 of the FBN5-B polypeptide (Supplemental Figures 1B and 1C). PROSITE (http://expasy.org) did not identify a lipocalin motif in FBN5, whereas FBN4 was predicted to contain the first motif of lipocalin, which is the most highly conserved in the lipocalin proteins (Singh and McNellis, 2011). FBN5 contains a region similar (53.8% identity; 84.6 similarity) to the lipocalin motif 1 in FBN4 (Supplemental Figure 1C). This possible lipocalin motif 1 in FBN5 has an invariant glycine, followed by an invariant tryptophan and an aromatic residue (tyrosine), which are the characteristically conserved residues of lipocalin motif 1 (Flower et al., 1993).
RT-PCR analysis revealed that the FBN5-A and FBN5-B transcripts were undetectable in the RNA from fbn5-1 plants, whereas they were detected in the wild type (Figure 1D). The two transcripts showed similar expression patterns in most tissues, and the transcript levels were highest in the photosynthetic tissues, including in seedlings, developing leaves, stems, and unopened flowers (Supplemental Figure 2A). To further investigate the expression of FBN5, we cloned a 2.7-kb fragment upstream from the FBN5 initiation codon, fused it to the GUS reporter gene, and used the construct to transform wild-type Arabidopsis plants. GUS activity was strong in the young seedlings, developing roots, developing leaves, and young stems and sepals of the flowers, but was not detectable in siliques or petals (Supplemental Figure 2B).
To confirm that the fbn5-1 null mutation is directly responsible for the seedling-lethal phenotype, FBN5-A and FBN5-B cDNAs under the control of the CaMV 35S promoter were constructed and used to transform heterozygous FBN5/fbn5-1 plants. Thirty-two BASTA-resistant transformants showing a healthy, green phenotype were selected from each group (FBN5-A and FBN5-B). Among the FBN5-A transgenic plants, 11 and 21 plants were genotyped as wild type and fbn5-1/FBN5, respectively; no fbn5-1 plants contained the FBN5-A cDNA. Among the 32 FBN5-B healthy green transformants, 18 were wild-type plants, eight were fbn5/FBN5 plants, and six were genotyped as fbn5-1 plants. The FBN5-B-complemented homozygous fbn5-1 plants showed healthy green growth, similar to that of wild-type plants in soil (Supplemental Figures 3A and 3B). This indicates that the FBN5-B cDNA complemented the seedling-lethal phenotype but the FBN5-A cDNA did not. Genomic DNA PCR using the P1 and P2 primers generated products of ∼1.7 and 0.8 kb from the wild-type and fbn5-1+35S:FBN5-B plants, respectively, indicating that the latter were true fbn5-1 plants transformed with FBN5-B cDNA. The expression of FBN5-B was higher in both the cauline and rosette leaves of the fbn5-1+35S:FBN5-B plants than in those of the wild-type plants (Supplemental Figures 3C and 3D). Collectively, these results indicate that the fbn5-1 mutation causes the seedling-lethal phenotype and that the FBN5-B gene product is required for autotrophic growth. The maximum photosynthetic performance of PSII was lower in the leaves of the fbn5-1 plants (Fv/Fm = 0.65) than in those of the wild-type plants (Fv/Fm = 0.8) grown on medium containing 1% sucrose. The fbn5-1 plants also accumulated more H2O2 than the wild-type plants (Supplemental Figure 4).
FBN5 Localizes in the Chloroplasts
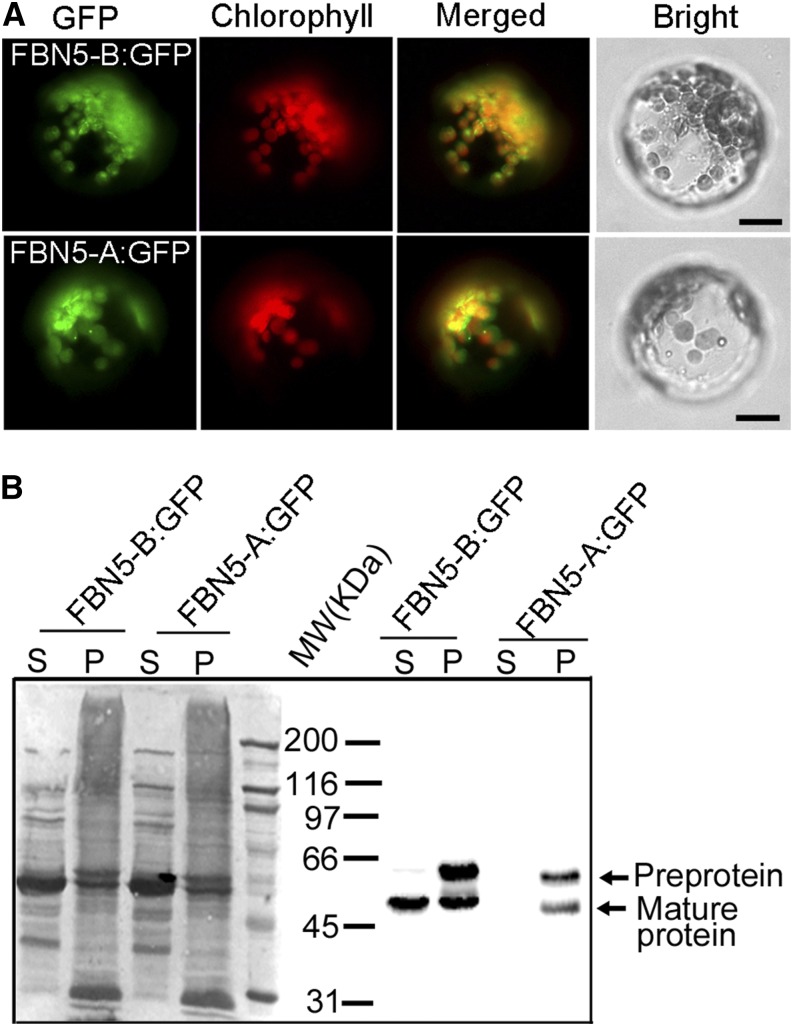
It was previously reported that FBN5 is present in the stroma of chloroplasts (Lundquist et al., 2012). To confirm that FBN5 is targeted to the chloroplasts, we constructed GFP fusions with FBN5-A and FBN5-B cDNA under the control of the CaMV 35S promoter for transient expression in protoplasts derived from Arabidopsis leaves. The GFP signal in the transformed protoplasts colocalized with the autofluorescence of the chlorophylls in the chloroplasts (Figure 2A). The soluble and pellet fractions were collected from the transformed protoplasts, and the FBN5 fusion proteins were detected with an anti-GFP antibody. Both the FBN5-B:GFP and FBN5-A:GFP isoforms were detected, with approximately equal amounts of preprotein and mature protein in the pellet fractions of the protoplasts. It seems that the transient overexpression of FBN5 produced high levels of preproteins, which were probably improperly targeted to the plastid. Interestingly, FBN5-B:GFP was the only mature form detected in the soluble fraction (Figure 2B). These results indicate that the functional FBN5-B protein is targeted to the chloroplast and processed into the mature protein in the soluble portion of the organelle.
Figure 2.
Localization of FBN5 in Chloroplasts.
(A) Subcellular localization of the FBN5-A:GFP and FBN5-B:GFP fusion proteins. A sequence encoding GFP was fused to the 3′ ends of the full-length FBN5-A and FBN5-B cDNAs, which were transcribed from the CaMV 35S promoter. Transient expression of these constructs in protoplasts derived from Arabidopsis mesophyll cells resulted in colocalization of GFP fluorescence patterns with the autofluorescence of chlorophyll. Bars = 20 µm.
(B) SDS-PAGE of proteins in the soluble (S) and pellet (P) fractions of protoplasts expressing FBN5-B:GFP or FBN5-A:GFP fusion proteins and immunoblots showing the localization of FBN5 proteins in the soluble and pellet fractions of protoplasts.
FBN5 Mutation Reduces the Levels of PQ-9 and PQ-8
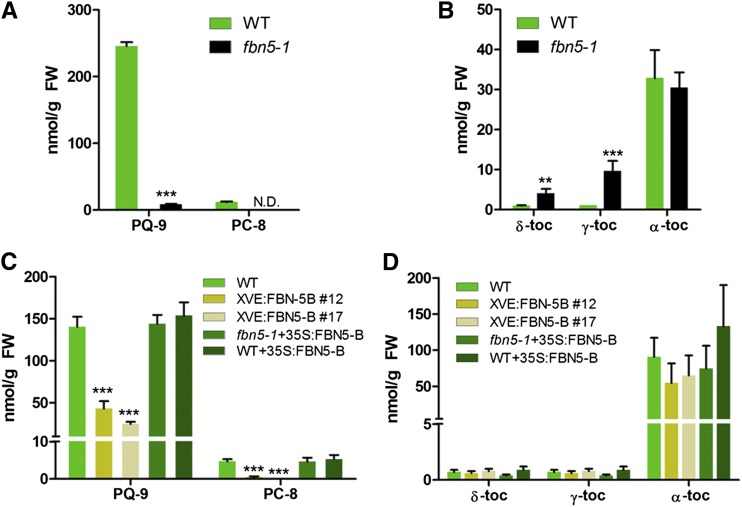
We used HPLC to determine the amounts of PQ-9, tocochromanols, carotenoids, and chlorophylls in the mutant and wild-type plants because the photosynthetic capacity of the fbn5-1 plants was reduced and FBN4 is involved in plastoquinone accumulation in the plastoglobules (Singh et al., 2012). Although fbn5-1 is seedling-lethal when plants are grown in soil, seedlings cultivated on agar medium supplemented with 1% sucrose were pale green, survived for several weeks, and produced sufficient tissue for biochemical analyses. The amount of PQ-9 was reduced ∼28-fold in the leaves of the fbn5-1 plants compared with that in the wild-type plants of a similar age (Figure 3A). We did not detect any PC-8, which is the product of PC-9 cyclization, in the leaves of the fbn5-1 plants, unlike in the wild-type plants (Figure 3A). The tocopherol (TC) profile in the leaves of the wild-type plants consisted of α-TC, γ-TC, and δ-TC in an ∼94:3:3 ratio. In the fbn5-1 leaves, the levels of δ-TC and γ-TC were elevated relative to those in the wild-type plants, whereas α-TC and the total tocopherols were not significantly different (Figure 3B). Thus, it seems that the incompletely developed fbn5-1 plants accumulated higher levels of δ-TC and γ-TC. The levels of lutein and β-carotene in the fbn5-1 plants were reduced by ∼14 and 46%, respectively, compared with those in the wild-type plants, whereas neoxanthin and violaxanthin were only slightly reduced (Supplemental Figure 5A). The level of antheraxanthin was increased in the fbn5-1 plants, indicating that they were more photostressed than the wild-type plants (Supplemental Figure 5A). Both chlorophylls were significantly reduced in the fbn5-1 plants compared with the wild-type plants (Supplemental Figure 5B).
Figure 3.
Quantification of PQ-9 and Tocochromanols in Arabidopsis Leaves.
Reverse-phase HPLC was used to analyze total lipids in the leaves of 3-week-old wild-type (WT) and fbn5-1 plants grown on MS medium (+1% sucrose) ([A] and [B]) and in 6-week-old wild-type and transgenic plants grown in soil ([C] and [D]). The quantities of individual tocopherols, PQ-9, and PC-8 were determined relative to standards. Asterisks represent significance relative to the wild type by Student’s t test: **P < 0.01 and ***P < 0.001. Data are means ± sd (n = 3 to 4). N.D., not detected; FW, fresh weight.
To investigate whether the lack of PQ-9 and PC-8 is FBN5 mutation specific or is just one of the pleiotropic defects occurring in any pale-green mutant, we assessed the levels of PQ-9, tocochromanols, carotenoids, and chlorophylls in four known pale-green mutants. The tested mutants were defective in nuclear genes encoding essential chloroplast proteins, namely, PDS2 (encoding homogentisate solanesyltransferase), GDC1 (encoding grana-deficient chloroplast 1), SBP (encoding the Calvin-Benson cycle enzyme sedoheptulose-1,7-bisphosphatase), and AAE14 (encoding acyl-activating enzyme 14) (Tian et al., 2007; Kim et al., 2008; Cui et al., 2011; Liu et al., 2012; Savage et al., 2013). The gdc1-2, sbp-1, and aae14-1 pale-green homozygous mutants were null, whereas the pds2-1 homozygous mutant was leaky because the T-DNA insertion disrupted its promoter region (Supplemental Figure 6). Consistent with previous findings, all of these pale-green mutants exhibited pale-green cotyledons and true leaves, with slow growth on medium containing sucrose (Supplemental Figure 6). The total tocopherols in these pale-green mutants did not differ from those in the wild-type plants, although the compositions of their tocopherols varied (Supplemental Table 1). However, all of the pale-green mutants showed significantly reduced levels of PQ-9 and PC-8: in order of severity, pds2-1 > sbp-1 > aae14-1 > gdc1-2 plants. The total carotenoids and chlorophylls were also significantly reduced in the plants (aae14-1 > gdc1-2 > sbp-1 > pds2-1 plants) (Supplemental Table 2). Therefore, it is likely that lower levels of PQ-9, PC-8, carotenoids, and chlorophylls in the pale-green mutants are pleiotropic defects, possibly attributable to the limited accumulation of the photosynthetic machinery. However, the extent of the reductions in these molecules seems to be related to the specific mutation. Moreover, the reductions in PQ-9 and PC-8 did not correlate with the reductions in the carotenoids and chlorophylls in the pale-green mutants. For example, gdc1-2 plants seemed to be more affected in the accumulation of carotenoids and chlorophylls than in the accumulation of PQ-9, whereas the converse was true of pds2-1 plants. The aae14-1 plants showed the lowest levels of carotenoids and chlorophylls, but accumulated more PQ-9 and PC-8 than the sbp-1, pds2-1, or fbn5-1 plants. In particular, the pds2-1 plants, which are directly defective in PQ-9 biosynthesis (Norris et al., 1995; Tian et al., 2007), showed similar levels of PQ-9 and PC-8, as well as total carotenoids and chlorophylls, to those of the fbn5-1 plants. These results suggest that the lack of PQ-9 and PC-8 in the fbn5-1 plants is specific for the FBN5 mutation.
Because the fbn5-1 mutation was seedling-lethal, we studied the function of FBN5 by generating transgenic fbn5-1 plants with an FBN5-leaky phenotype. A β-estradiol-inducible XVE:FBN5-B construct was used to transform FBN5/fbn5-1 plants, and the fbn5-1 lines were selected with PCR genotyping. These transgenic plants survived when germinated in soil because of the leaky expression of XVE:FBN5-B, despite the absence of β-estradiol (Supplemental Figure 7A). Two independent T4 homozygous lines, XVE:FBN5-B#12 and XVE:FBN5-B#17, were characterized and used for HPLC analysis. The XVE:FBN5-B#12 and #17 transgenic plants were clearly smaller, with slower growth rates, than the wild-type plants (Supplemental Figures 7A to 7C). The growth of the complemented T4 homozygous line (fbn5-1+35S:FBN5-B) and a T4-overexpressing line of wild-type plants (WT+35S:FBN5-B) was similar to that of the wild-type plants (Supplemental Figures 7A to 7C). The rosette size of the plants is determined during the course of plant development, and we measured the fresh weight of the plants that grew to maturity (33 d old). The fresh weights of the fbn5-1+35S:FBN5-B and WT+35S:FBN5-B transgenic plants did not differ significantly from those of the wild-type plants, whereas the fresh weights of the XVE:FBN5-B#12 and #17 transgenic plants were reduced by 68 and 83%, respectively (Supplemental Figures 7B and 7C). The contents of PQ-9, tocochromanols, carotenoids, and chlorophylls were measured in the leaves of 6-week-old plants grown in soil (Figure 3; Supplemental Figure 5). The levels of PQ-9 were reduced ∼3- and 5-fold, and PC-8 was reduced nearly 9- and 38-fold in the XVE:FBN5-B#12 and #17 transgenic plants, respectively (Figure 3C). The tocopherol contents of the XVE:FBN5-B#12 and #17 transgenic plants did not differ significantly from those of the wild-type plants (Figure 3D). However, the levels of PQ-9 and PC-8 in the fbn5-1+35S:FBN5-B and WT+35S:FBN5-B transgenic plants were similar to those in the wild-type plants (Figures 3C). Notably, in the XVE:FBN5-B#12 and #17 transgenic plants, the reduced contents of PQ-9 and PC-8 coincided with reduced levels of the FBN5 transcript (Figure 3C; Supplemental Figure 7D). However, the increased levels of FBN5 transcripts, by ∼12-fold and 6-fold in the fbn5-1+35S:FBN5-B and WT+35S:FBN5-B transgenic plants, respectively, did not affect the accumulation of PQ-9 or PC-8 (Figure 3; Supplemental Figure 7D). The total carotenoid contents did not differ between the plants. However, the XVE:FBN5-B#12 and #17 transgenic plants had more antheraxanthin compared with the wild-type plants and fbn5-1+35S:FBN5-B and WT+35S:FBN5-B transgenic plants, and so must have been photostressed under these growth conditions (Supplemental Figure 5C). Taken together, our results for the knockout and knockdown mutants of FBN5 suggest that FBN5 is involved in the biosynthesis of PQ-9 and PC-8 in Arabidopsis leaves.
FBN5-B Interacts with SPSs Involved in PQ-9 Biosynthesis
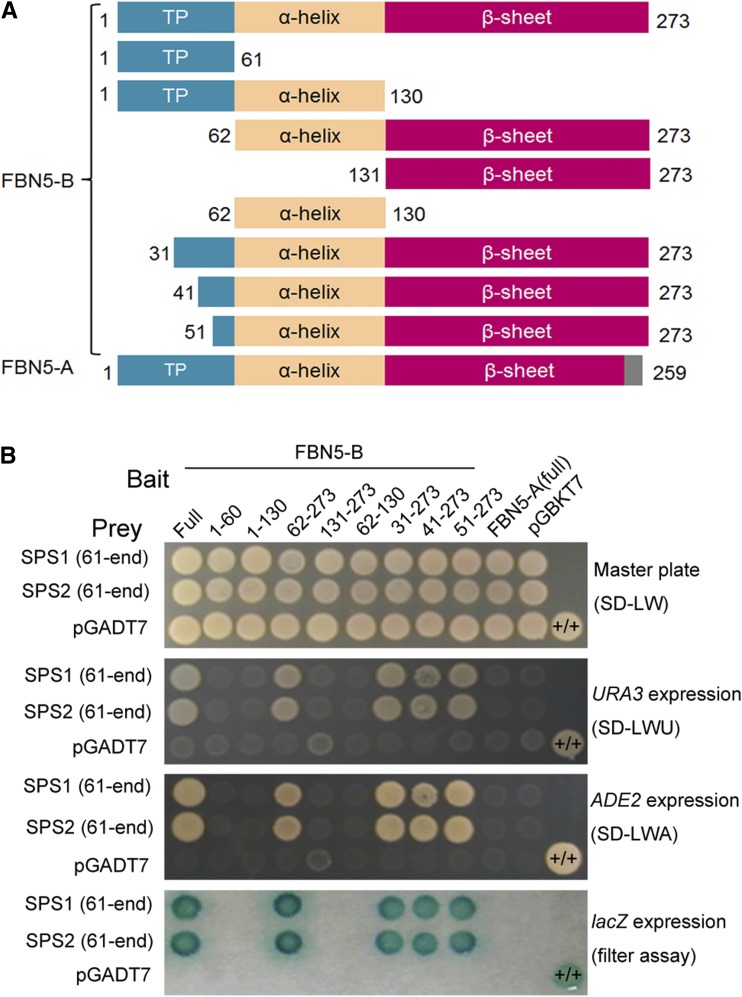
FBN5 lacks obvious enzyme-associated protein domains, suggesting that it plays a structural role in PQ-9 biosynthesis. Accordingly, we tested whether FBN5 interacts with the proteins involved in the PQ-9 biosynthetic pathway using the yeast two-hybrid (Y2H) system. After confirming that the fusion of the full-length FBN5-B cDNA to the GAL4 DNA binding domain (BD fusion) did not cause self-transcription, this fusion protein was used as the bait to screen Arabidopsis cDNA prey libraries. From a total of 1.24 × 107 yeast transformants, 20 independent colonies were positive for lacZ activation and URA3 and ADE2 autotrophy. DNA sequence analysis revealed that all 20 cDNA fusions encoded the mature portion of SPS1 (At1g78510), lacking the first 60 N-terminal amino acid residues of the polypeptide. Arabidopsis has two paralogous solanesyl diphosphate synthases, SPS1 and SPS2 (At1g17050), and these proteins assemble the prenyl chain of PQ-9 in plastids (Jun et al., 2004; Block et al., 2013). Because SPS1 and SPS2 share 80% identical residues (Hirooka et al., 2005), the interaction between the preprotein of FBN5-B (full-length peptide) and SPS2 was also examined. Although mature SPS2 showed a strong interaction with the preprotein of FBN5-B, the preprotein of SPS2 barely interacted with the FBN5-B preprotein (Supplemental Figure 8). The SPS1 preprotein also did not interact with the FBN5-B preprotein (Supplemental Figure 8). These results indicate that the transit peptides of SPS1 and SPS2 abolish their interactions with FBN5-B.
To further characterize the specificity and structural requirements for the interaction between FBN5-B and SPS, the interactions between different variants of FBN5-B and mature SPS1 or mature SPS2 were analyzed. The construction of the FBN5-B variants was based on the transit peptide, α-helix domain, and β-sheet domain of the mature protein using a structural algorithm (http://www.compbio.dundee.ac.uk/www-jpred/) (Figure 4A). The results clearly indicated that the transit peptide, α-helix domain, and β-sheet domain of FBN5-B alone did not interact with any SPS, whereas the FBN5-B preprotein and mature FBN5-B did interact with SPS. However, the FBN5-A preprotein, a product of alternative splicing, did not interact with either mature SPS1 or mature SPS2. These results suggest that the overall integrity and the C-terminal region of FBN5-B are important for its interaction with the SPS proteins (Figure 4B). The transit peptide of FBN5-B did not abrogate the interaction with the SPS proteins. The failure of overexpressed FBN5-A to interact with the SPS proteins in the Y2H analysis or to complement the seedling-lethal fbn5-1 plants supports the conclusion that the interaction between FBN5-B and the SPS proteins is essential for plant growth.
Figure 4.
Interaction Domain Mapping of FBN5 with SPS1 and SPS2.
(A) Schematic diagrams of the bait constructs of FBN5-B variants and full FBN5-A fused to the GAL4 DNA binding domain. The numbers represent amino acid positions at the start and stop positions of the constructs. Full-length FBN5 contains chloroplast-targeting sequences and α-helix and β-sheet structures.
(B) Specificity of the FBN5 interaction with either mature SPS1 or mature SPS2 in the Y2H system. Interactions of FBN5 variants with either mature SPS1 or mature SPS2 fused to the GAL4 DNA-activation domain were examined in each yeast transformant (PBN204) on growth-selective medium lacking Leu and Trp (SD-LW) or on selective medium also lacking Ura (SD-LWU) or Ade (SD-LWA). β-Galactosidase activity was tested in each colony on SD-LW medium. Positive control was yeast transformed with the PTB (polypyrimidine tract binding protein) bait plasmid and PTB prey plasmid (+/+). PTB is a homodimeric protein. Negative control was a cell transformed with the parental bait vector (pGBKT7) and prey vector (pGADT7).
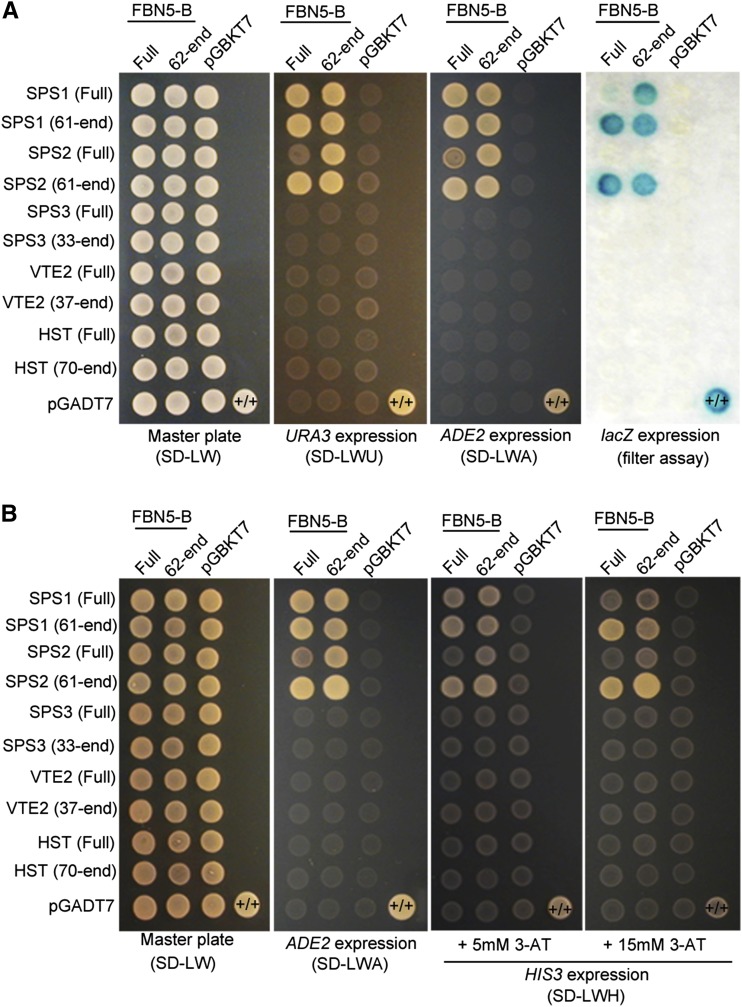
Because it is possible that the enzymes involved in the biosynthesis of PQ-9 and the tocopherols form a multienzyme system that functions in metabolic channeling, we examined the potential interactions of FBN5-B with eight other proteins: SPS3, fanesyl diphosphate synthase 1 (FPS1), FPS2, geranylgeranly diphosphate synthase 1 (GGPS1), VTE2, VTE3, HST, and geranylgeranly reductase (GGR), which are metabolically connected (Supplemental Figure 9A). However, none of them interacted with the FBN5-B preprotein (Supplemental Figure 9B), which may have been because the preprotein form of each enzyme was used as the prey. Therefore, we tested the binding of FBN5-B to mature SPS3, VTE2, and HST because either their substrates or products are highly hydrophobic (Figure 5). Both the Y2H analyses in yeast strains PBN204 (Figure 5A) and AH109 (Figure 5B) clearly showed that neither the FBN5-B preprotein nor mature FBN5-B interacted with either the preprotein or mature forms of SPS3, VTE2, or HST. A test of the intensity of the interaction between the proteins in AH109 cells on medium containing 3-amino-1,2,3-triazole, a competitive inhibitor of the yeast HIS3 protein, revealed that the interactions between FBN5-B and the mature forms of SPS1 and SPS2 were stronger than its interactions with the positive control proteins (Figure 5B). It is noteworthy that FBN5-B did not interact with SPS3, another SPS, responsible for the solanesyl moiety of ubiquinone-9 in Arabidopsis (Ducluzeau et al., 2012). In summary, both the preprotein and mature form of FBN5-B specifically interacts with mature SPS1 and mature SPS2, but not with other enzymes involved in the biosynthesis of hydrophobic compounds, suggesting that FBN5-B plays a role associated with SPP biosynthesis.
Figure 5.
Y2H Experiments to Test Interactions between FBN5-B and SPS1, SPS2, SPS3, VTE2, and HST Enzymes.
Either the FBN5-B preprotein (full) or mature FBN5-B (62-end) was fused to the GAL4 DNA binding domain (pGBKT7 vector). Either the preprotein (full) or mature form of SPS1, SPS2, SPS3, VTE2, or HST was fused to GAL4 DNA-activation domain (pGADT7 vector).
(A) Transformed yeasts (PBN204) were dropped onto selective medium lacking Leu and Trp (SD-LW) or selective medium also lacking Ura (SD-LWU) or Ade (SD-LWA). β-Galactosidase activity in each colony was tested on SD-LWA medium. Positive control was yeast transformed with the PTB bait plasmid and PTB prey plasmid (+/+). PTB is a homodimeric protein. Negative control was a cell transformed with the parental bait vector (pGBKT7) and prey vector (pGADT7).
(B) The interaction strength test of FBN5-B with SPS1, SPS2, SPS3, VTE2 and HST. The transformants (AH109) were dropped onto selective medium also lacking His (SD-LWH) but containing 5 or 15 mM 3-amino-1,2,3-triazole (3-AT), a competitive inhibitor of the yeast HIS3 protein.
FBN5-B Interacts with SPS in Chloroplasts
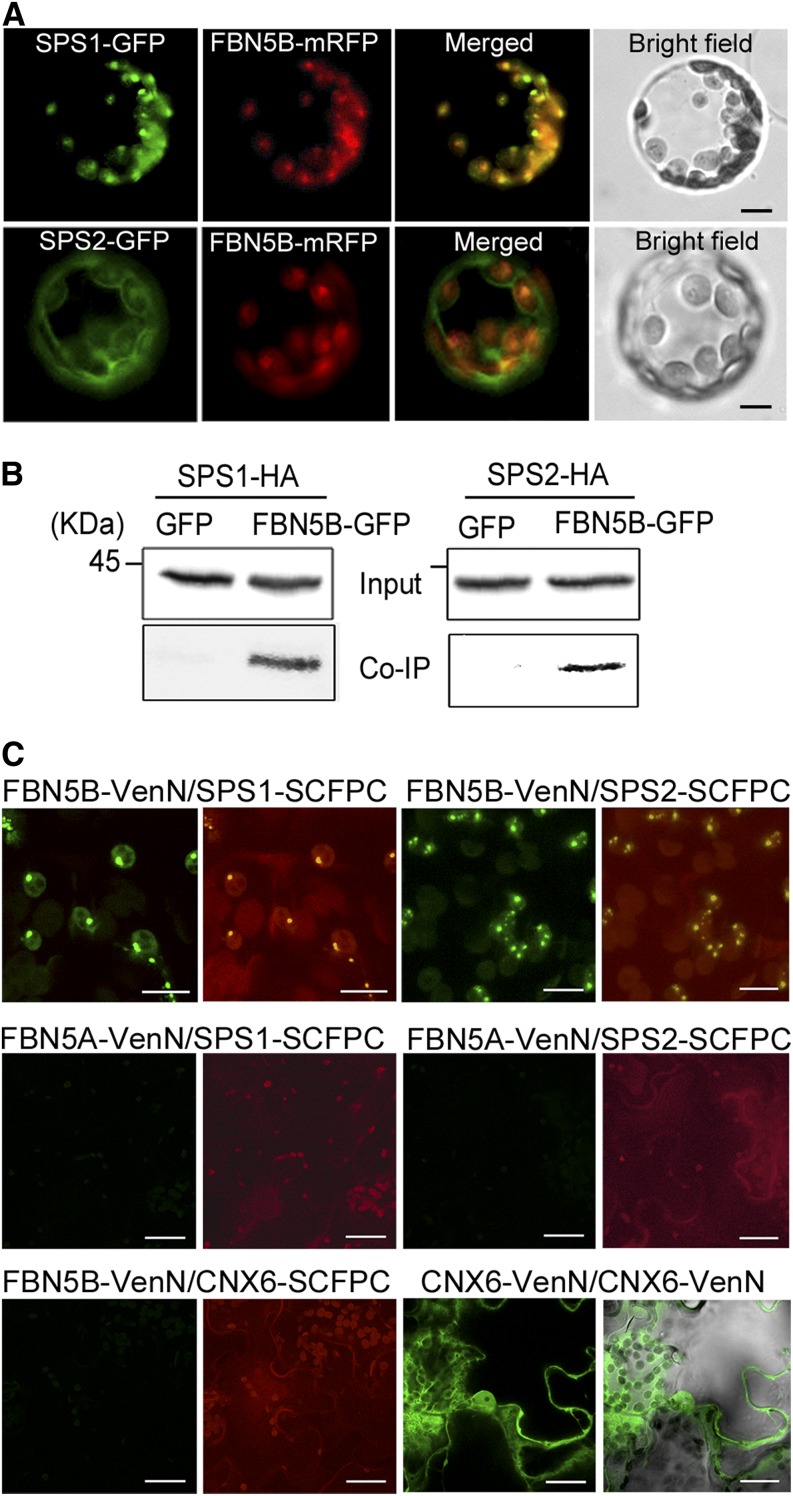
We demonstrated above that FBN5-B localizes to the chloroplasts (Figure 2). Our data for the transient expression of SPS1-GFP and SPS2-GFP in protoplasts (Supplemental Figure 10), together with previous studies (Jun et al., 2004; Block et al., 2013), indicate that both SPS1 and SPS2 are targeted to the chloroplasts. To further examine the colocalization of FBN5-B with SPS1 or SPS2, their full-length cDNAs were cloned in frame with modified RFP (mRFP) under the control of the CaMV 35S promoter. The resulting construct, 35S:FBN5B-mRFP, also containing either 35S:SPS1-GFP or 35S:SPS2-GFP, was transiently coexpressed in Arabidopsis protoplasts. SPS1-GFP colocalized with FBN5B-mRFP in the chloroplasts, evident as overlapping green and red fluorescence (Figure 6A). Although some green fluorescence associated with SPS2-GFP appeared in the envelope membrane and cytosol, it was primarily located in the chloroplasts and colocalized with FBN5B-mRFP (Figure 6A). The signal for SPS2-GFP in the envelope membrane and in the cytosol could be attributable to its overexpression.
Figure 6.
Interaction of FBN5-B with SPS1 and SPS2 in Chloroplasts.
(A) Colocalization of FBN5B with SPS1 or SPS2. Representative confocal images are shown of the GFP and mRFP signals in FBN5B-mRFP Arabidopsis protoplasts transformed with either SPS1-GFP or SPS2-GFP.
(B) Immunoblot analysis of SPS1 and SPS2 using anti-HA antibody (upper panel, input). Arabidopsis protoplasts transiently expressing FBN5B-GFP with either SPS1-HA or SPS2-HA. Coimmunoprecipitated SPS1 and SPS2 were detected with an anti-HA antibody (lower panel, Co-IP).
(C) BiFC analysis of epidermal cells from N. benthamiana. FBN5B-VenusN interacted with SPS1-SCFPC and SPS2-SCFPC (upper panels), but FBN5A-VenusN did not interact with the fusion proteins (middle panels). Negative and positive controls are shown in the lower left and right panels, respectively. Chloroplasts were visualized with the autofluorescence of chlorophyll. Bars = 20 µm.
We next tested whether FBN5-B interacts with SPS1 or SPS2 in planta using coimmunoprecipitation (Co-IP) and bimolecular fluorescence complementation (BiFC) assays. In the Co-IP assay, total protein extracts from Arabidopsis protoplasts coexpressing GFP-tagged FBN5-B and either hemagglutinin (HA)-tagged SPS1 or HA-tagged SPS2 were incubated with anti-GFP affinity agarose. The immunoprecipitated proteins were detected with immunoblotting using an anti-HA antibody. As shown in Figure 6B, FBN5-B coimmunoprecipitated with SPS1 and SPS2, which suggests that these proteins interact in planta.
The BiFC analysis confirmed the in planta interactions between FBN5-B and SPS1 or SPS2 in the chloroplasts (Figure 6C). In this experiment, epidermal cells from Nicotiana benthamiana leaves were transiently cotransformed with six different binary constructs expressed under the CaMV 35S promoter. Two days after infiltration, green granular signals were detected in the chloroplasts when FBN5-B-VenusN was paired with either SPS1-SCFPC or SPS2-SCFPC, indicating their physical interaction in vivo (Figure 6C; Supplemental Movies 1 and 2). However, no green signal was observed when FBN5-A-VenusN was paired with SPS1-SCFPC or SPS2-SCFPC (Figure 6C), indicating that SPS1 and SPS2 interact specifically with FBN5-B, but not with FBN5-A. Based on our results, we conclude that FBN5-B interacts physically with SPS1 and SPS2 in chloroplasts. However, we note that to confirm these findings, the proteins must be stably expressed in Arabidopsis under their endogenous promoters.
Expression of Tocochromanols and PQ-9 Biosynthesis Genes Is Not Altered in fbn5-1
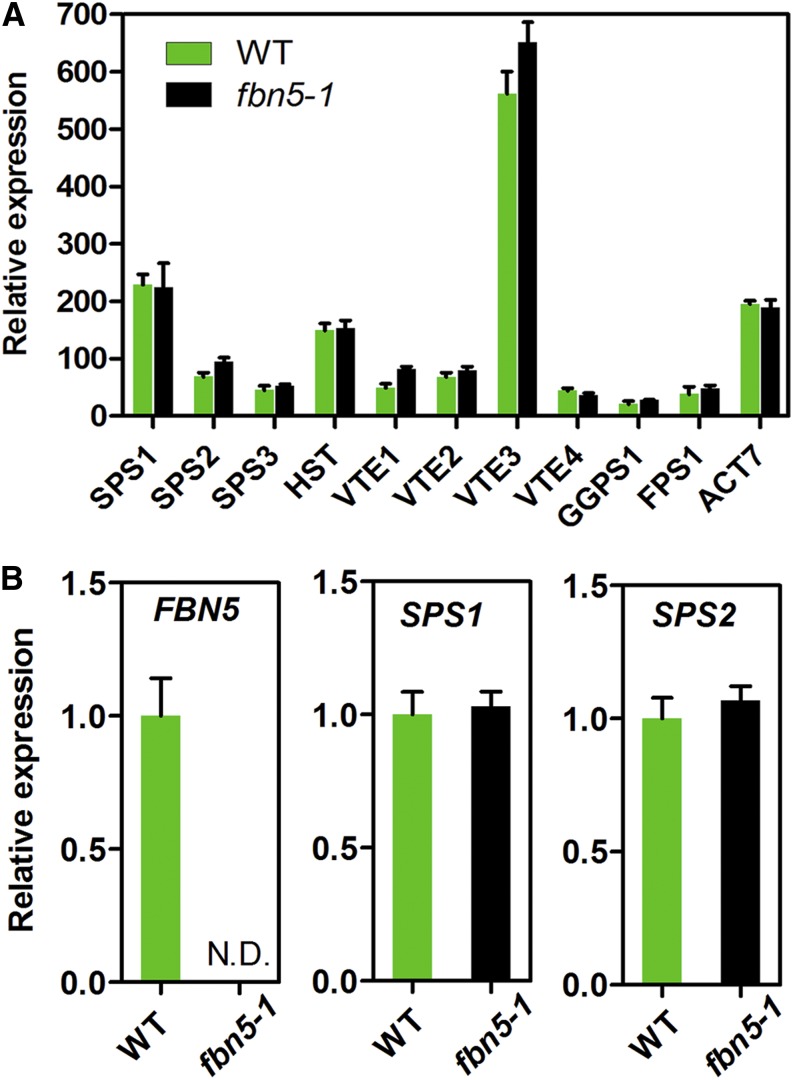
To determine whether the PQ-9 deficiency in the fbn5-1 plants is attributable to perturbation in the transcript abundance of the genes encoding the enzymes for tocochromanols and PQ-9 biosynthesis, we monitored the expression levels of GGPS1, HST, VTE2, VTE1, VTE3, VTE4 (γ-tocopherol methyltransferase), SPS1, and SPS2 with microarray data in wild-type and fbn5-1 plants. The FPS1 and SPS3 genes, which are involved in the biosynthesis of the solanesyl moiety of ubiquinone-9 in Arabidopsis, were also included. As shown in Figure 7A, the transcript abundances of these genes did not differ significantly in the wild-type and fbn5-1 plants. Using real-time PCR, we also confirmed that there were no differences in the transcription levels of SPS1 and SPS2 in the wild-type and fbn5-1 plants, except for the knocked-out FBN5 gene (Figure 7B). These results indicate that the deficiency of PQ-9 in fbn5-1 plants is specifically associated with FBN5.
Figure 7.
Transcript Levels of Genes Involved in Prenylquinone Biosynthesis in Wild-Type and fbn5-1 Plants.
(A) Comparative expression of 10 genes involved in prenylquinone biosynthesis, with ACT7 control. Expression was measured via microarray analysis.
(B) Determination of relative expression of FBN5, SPS1, and SPS2 in wild-type and fbn5-1 plants with real-time RT-PCR. RNA samples were extracted from 3-week-old wild-type and fbn5-1 homozygous mutant seedlings grown on MS medium + 1% sucrose. ACT7 expression was used for normalization. Significant differences in gene expression in the wild-type and fbn5-1 plants were determined with Student’s t test. Data are means ± se (n = 4).
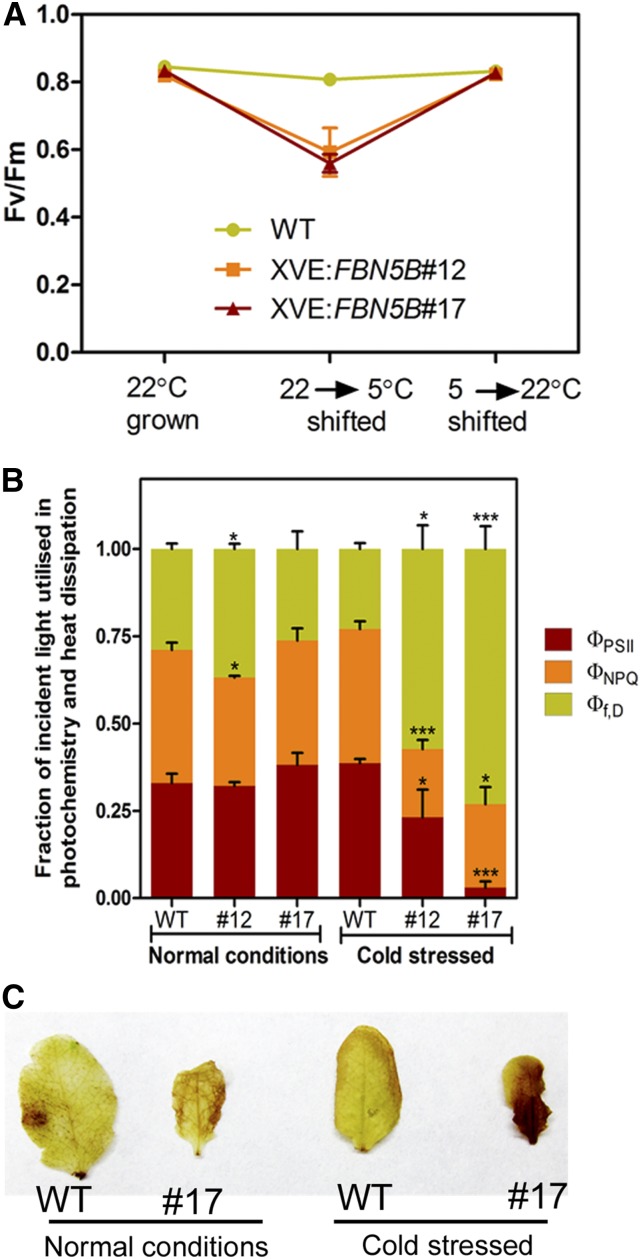
FBN5-B-Deficient Plants Are Sensitive to Cold Stress
To determine the effects of FBN5 deficiency on photosynthesis, various photosynthetic parameters were measured in wild-type and XVE:FBN5-B transgenic plants before their exposure to cold stress, after a 5-d period of cold stress, and after their transfer to normal temperatures. The maximum quantum efficiency of PSII photochemistry (Fv/Fm) was unchanged in the wild-type plants subjected to cold stress. However, Fv/Fm decreased significantly in the XVE:FBN5-B transgenic plants under cold stress, but was fully restored after their transfer to normal conditions (Figure 8A). During cold stress, the defect in FBN5 primarily affected the photochemical efficiency (ФPSII) and slightly reduced the light-dependent thermal dissipation component of nonphotochemical quenching (ФNPQ) (Figure 8B). H2O2 was assayed by visualizing the amount of precipitate formed after incubation with diaminobenzidine (DAB) solution. There were no differences in DAB staining before and after cold stress in the wild-type plants. However, cold stress treatment significantly increased DAB staining in the XVE:FBN5B#17 leaves (Figure 8C).
Figure 8.
Effects of Short-Term Cold Stress on Photosynthetic Properties of PSII and Hydrogen Peroxide Production in Wild-Type and XVE:FBN5B Leaves.
Plants grown for 4 weeks at 22°C, with a 16/8-h light/dark photoperiod at 100 μmol m–2 s–1 illumination were considered to be cultivated under normal conditions. Plants were transferred to 5 to 7°C with a 16/8-h light/dark photoperiod under 60 μmol m–2 s–1 illumination for 5 d and then transferred to normal conditions to recover.
(A) Fv/Fm measurements after each treatment. Data are means ± sd (n = 3).
(B) Fraction of absorbed light consumed by photochemistry (ФPSII), ∆pH- and xanthophyll-regulated thermal dissipation (ФNPQ), and the sum of fluorescence and constitutive thermal dissipation (ФF,D). Asterisks denote significant differences between wild-type and XVE:FBN5B transgenic plants by Student’s t test: *P < 0.05 and ***P < 0.001. Data are means ± sd (n = 3).
(C) H2O2 detected in wild-type and XVE:FBN5B#17 leaves from normal and cold-stressed plants, determined with DAB solution staining.
DISCUSSION
We characterized an fbn5-1 T-DNA insertion mutant to determine how a defect in FBN5, a structural lipid-associated protein, produces a seedling-lethal phenotype. Our study is not the first to report a seedling-lethal phenotype for the fbn5-1 T-DNA insertion mutant. Savage et al. (2013) found that FBN5 is an essential nuclear gene encoding a protein that targets the chloroplast, although they did not study the gene in depth. Consistent with our results (Figure 1), segregating seeds from parent plants heterozygous for fbn5-1 yielded no homozygotes that could survive in soil. Furthermore, the seedlings homozygous for the fbn5-1 allele were small and pale green, with or without sucrose supplementation (Savage et al., 2013). In our study, we confirmed that the FBN5 mutation is responsible for the lethal phenotype by complementing fbn5-1 plants with 35S:FBN5-B (Supplemental Figure 3).
The fbn5-1 plants and previous findings with the pds2 mutant (Norris et al., 1995; Tian et al., 2007) and sps1 sps2 double mutant (Block et al., 2013) demonstrated the absolute requirement for PQ-9 during plant development. Phytoene desaturase requires quinones and molecular oxygen as electron acceptors to pass electrons from phytoene (Mayer et al., 1990). An Arabidopsis pds2 (HST) mutant accumulated phytoene but not downstream carotenoids, indicating that PQ-9 is an essential component of phytoene desaturation (Norris et al., 1995). However, in our study, fbn5-1 and pds2-1 (a leaky mutant of pds2) plants biosynthesized downstream carotenoids without accumulating phytoene, although the total carotenoid levels were reduced relative to that in wild-type plants (Supplemental Figure 5A and Supplemental Table 2). It is likely that even the extremely low levels of PQ-9 in the fbn5-1 and pds2-1 plants were sufficient for phytoene desaturation, but resulted in reduced carotenoid biosynthesis (Figure 3A; Supplemental Tables 1 and 2). The similar carotenoid levels in the XVE:FBN5-B transgenic and wild-type plants implies that a certain level of PQ-9 is sufficient for phytoene desaturase activity (Supplemental Figure 5C). The lower photosynthetic efficiency and higher levels of H2O2 observed in the fbn5-1 plants (Supplemental Figure 4) and the enhanced sensitivity to cold in the XVE:FBN5-B transgenic plants (Figure 8) are probably attributable to the deficiency in PQ-9 biosynthesis. A limited PQ pool reduces the forward steps in electron transport, resulting in a reduced state of QA. This generates reactive oxygen species and highly oxidized radicals, which damage PSII (Napiwotzki et al., 1997; Vass, 2012). Under cold stress, the PSII excitation pressure is higher than under normal growth conditions (Wise, 1995; Gray et al., 2003; Ensminger et al., 2006). Therefore, the lower levels of PQ-9 in the XVE:FBN5-B transgenic plants might increase the proportion of “closed” PSII reaction centers under cold stress relative to that under normal growth conditions, resulting in lower Fv/Fm and the greater accumulation of H2O2 in the leaves.
As shown in Figure 3, the amounts of PQ-9 and PC-8 were significantly reduced in both the FBN5 knockout and knockdown plants, whereas the levels of total tocopherols were not affected (Figure 3). PQ-9 and tocopherols both have homogentisate head groups. These results indicate that the homogentisate synthesis in the fbn5-1 plants was normal. Therefore, we conclude that FBN5 is specifically involved in the biosynthesis of the solanesyl moiety and/or the transfer of the solanesyl moiety to homogentisate. Block et al. (2013) demonstrated that SPS1 and SPS2 are functionally redundant in PQ-9 biosynthesis. Consistent with that finding, we have shown that FBN-5B interacts with both SPS1 and SPS2 (Figures 4 and 6), thus explaining the similar phenotypes of the fbn5-1 plants and the sps1 sps2 double mutant plants (Figures 1 and 3). It is unlikely that FBN5-B forms a complex with HST that functions in PQ-9 biosynthesis because FBN5-B did not interact with HST (Figure 5). However, we cannot exclude the possibility that biosynthesis of PQ-9 occurs by metabolic channeling because the interaction between SPS1 or SPS2 and HST was not examined. It must be noted that FBN5-B did not interact with other enzymes involved in the biosynthesis of hydrophobic compounds, such as tocochromanols or benzoprenylquinones (Figure 5; Supplemental Figure 9). Furthermore, the fbn5-1 mutation did not alter the transcript levels of the genes involved in the synthesis of tocochromanols or benzoprenylquinones. Specifically, the expression of SPS1, SPS2, and HST, which are directly responsible for PQ-9 biosynthesis, did not fluctuate in the fbn5-1 plants (Figure 7). These results confirm that the binding of FBN5-B to SPS1 and SPS2 is essential for PQ-9 biosynthesis.
What role does the physical binding of FBN5-B to SPS play in PQ-9 biosynthesis? It has been reported that the long-chain polyprenyl pyrophosphate synthase often requires detergent or another factor for its optimal activity because its product release is slow. Several in vitro studies have demonstrated that long-chain trans-polyprenyl diphosphate synthases from several organisms require the addition of phospholipids or detergent to the assay buffer to enhance their product release and maintain efficient turnover (Pan et al., 2013). The activity of SPS isolated from Micrococcus luteus was enhanced in the presence of BSA or a high molecular mass fraction (HMF) separated from cell-free extracts of M. luteus (Ohnuma et al., 1991). This suggests that HMF contains a factor that binds to the polyprenyl products and removes them from the active site of the enzyme to facilitate and maintain the turnover of catalysis. The steady state rate of octaprenyl pyrophosphate synthase purified from Escherichia coli also accelerated 3-fold in the presence of Triton X and HMF (Pan et al., 2002). Recently, mitochondrial coenzyme Q9 was reported to be a lipid binding protein that associates physically with coenzyme Q7 (Lohman et al., 2014). The authors suggested that COQ9 binds immature COQ species and presents them to COQ7 to allow coenzyme biosynthesis. Singh and McNellis (2011) showed that lipocalin motif 1 is conserved among the Arabidopsis fibrillins and suggested that the fibrillin family is involved in binding to and transporting small, hydrophobic molecules. Consistent with the lipocalin activity of the fibrillins, it has been suggested that FBN4 is involved in trafficking PQ-9 and other small hydrophobic molecules from the thylakoids to the plastoglobules (Singh et al., 2012). FBN5 contains a possible lipocalin motif 1, which encompasses amino acid residues 131 to 142 (Supplemental Figure 1) (Singh and McNellis, 2011). Based on these studies, we conclude that FBN-5B is involved in the biosynthesis of the solanesyl moiety, insofar as the lipid binding domain of FBN5-B binds to the hydrophobic solanesyl moieties that are generated by SPS1 and SPS2. Thus, FBN5-B stimulates the enzymatic activity of SPS1 and SPS2. When FBN5-B is deficient, the release of the solanesyl moiety from the enzyme and the enzyme turnover become slower, delaying PQ-9 biosynthesis. However, to support our hypothesis, in vitro experiments are required to determine whether the presence of FBN5-B enhances the reaction efficiency of SPS1 or SPS2.
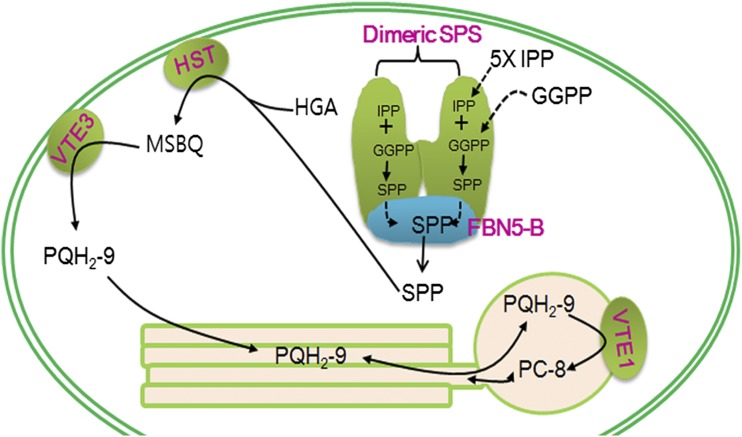
Our work has shown that a plastid-lipid-associated protein, FBN5, is required for PQ-9 biosynthesis through its physical and functional interactions with SPS1 and SPS2 in Arabidopsis. The PQ-9 biosynthetic enzymes are well characterized, and our work presents FBN5 as a new component of the PQ-9 biosynthetic process. We have also shown that at least one member of the fibrillin family is physically and functionally associated with other enzymes. Therefore, given the plastid-lipid binding domain in the fibrillins, future studies that clarify the relationships between fibrillins and other enzymatic reactions, especially those producing hydrophobic molecules, will be exciting. We propose a model for PQ-9 biosynthesis in Arabidopsis based on our results and data in the literature (Figure 9). In our model, SPS of the FBN5-B/dimeric SPS complex catalyzes the sequential condensation of five molecules of IPP with GGPP to yield the all-trans form of SPP. FBN5-B of the FBN5-B/dimeric SPS complex then removes SPP from the catalytic site of SPS. The SPP released from FBN5-B or the SPP bound to FBN5-B is condensed with HGA by HST to produce MSBQ. Finally, MSBQ is converted to PQH2-9 by VTE3, followed by its translocation to the thylakoids and plastoglobules, and some PQH2-9 is cyclized to PC-8 by VTE1 in the plastoglobules.
Figure 9.
Proposed Model for the Biosynthesis of PQ-9 and PC-8 in Chloroplasts.
SPS of the FBN5-B/homodimeric SPS complex binds to GGPP and IPP and then condenses five molecules of IPP to GGPP, yielding SPP. Synthesized SPP is removed from SPS when it binds to FBN5-B. SPP released from FBN5-B or SPP bound to FBN5-B is condensed with HGA by HST, generating MSBQ. MSBQ is converted to PQH2-9 by VTE3. VTE1 cyclizes PQH2-9 to PC-8 in plastoglobules.
METHODS
Plant Materials and Growth Conditions
Mutants fbn5-1 (SALK_064597), pds2-1 (SALK_024357), aae14-1 (SALK_060226), gdc1-2 (SALK_151530C), and sbp-1 (SALK_090549C) were acquired from the ABRC. Arabidopsis thaliana ecotype Columbia-0 was grown in soil or on agar plates containing 0.5× MS medium supplemented with/without sucrose under a 16-h-light/8-h-dark photoperiod with 100 µmol m–2 s–1 fluorescent light at 22°C. Nicotiana benthamiana (tobacco) plants were grown under a 16-h-light/8-h-dark photoperiod at 23°C under 100 µmol m–2 s–1 fluorescent light. Arabidopsis leaf tissues were genotyped with PCR using genomic DNA as the template. The primers used in this study are listed in Supplemental Table 3.
RT-PCR Analysis
Total RNA was extracted from the leaf tissues using TRIzol reagent (Sigma-Aldrich) according to the manufacturer’s instructions. cDNA was synthesized from the total RNA using the PrimeScript 1st Strand cDNA synthesis kit (Takara), as recommended by the manufacturer. The PCR reactions were performed with Takara Ex Taq DNA Polymerase and the appropriate primer pairs. The PCR cycling parameters were (1) denaturation, 30 s at 94°C; (2) annealing, 30 s at 58°C; and (3) extension, 60 s at 72°C. The reactions were run for 32 cycles.
Complementation of the Arabidopsis fbn5-1 Mutation
The full-length FBN5 coding sequences (FBN5-A and FBN5-B) were amplified from cDNA obtained from Arabidopsis leaves using KOD Hot Start DNA Polymerase (Novagen). The amplified products were cloned into the Gateway transformation vector pENTR/D-TOPO (Invitrogen) with the TOPO reaction, according to the manufacturer’s instructions. pENTR-FBN5-A and pENTR-FBN5-B were recombined with LR reactions into the plant transformation vector, pB2GW7.0, where FBN5-A and FBN5-B were controlled by the CaMV 35S promoter (Karimi et al., 2002). Competent Agrobacterium tumefaciens cells (GV3101) were transformed with the constructs pB2GW7.0-FBN5-A and pB2GW7.0-FBN5-B with freeze-thaw methods. Heterozygous FBN5/fbn5-1 Arabidopsis plants were transformed with the Agrobacterium described above, using the floral dip transformation method (Clough and Bent, 1998), and selected by their resistance to BASTA (Bayer). Genomic DNA was extracted from the leaves of the BASTA-resistant plants and used to identify the homozygous fbn5-1 plants. pENTR-FBN5-B was recombined with the LR reaction into the pMDC7 destination vector (Zuo et al., 2000) to be expressed under the control of the XVE β-estradiol-inducible promoter and was then used to transform heterozygous FBN5/fbn5-1 Arabidopsis plants, which were screened as described above.
Histochemical Analysis of GUS Expression
A 2.7-kb region of the FBN5 genomic DNA including the 5′-untranslated region was amplified and cloned into pENTR/D-TOPO and then transferred into the pMDC163 destination vector with the LR reaction (Curtis and Grossniklaus, 2003). Arabidopsis was transformed with the construct, as described above, and the transgenic plants were selected by their hygromycin resistance. The plant tissues were vacuum infiltrated with GUS staining buffer, as described previously (Kim et al., 2008). The GUS staining buffer contained 50 mM sodium phosphate buffer (pH 7.0), 0.1% Triton X-100, 2 mM potassium ferrocyanide, 10 mM EDTA, 100 µg mL–1 chloramphenicol, and 1 mg mL–1 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid. The infiltrated tissues were incubated at 37°C overnight. The nonspecific stains and colored pigments were then removed with 70% ethanol.
Microarray and qRT-PCR Analyses
Affymetrix GeneAtlas Arabidopsis Gene 1.1 ST Array Strips were used to comparatively analyze the genes expressed in the wild-type and fbn5-1 seedlings. Four biological replicates of total RNA were prepared from 3-week-old wild-type and fbn5-1 seedlings. Probe preparation, hybridization, and the signal normalization process were as described by Lee et al. (2008). The raw microarray data were deposited in the Gene Expression Omnibus under accession number GSE68275 (http://www.ncbi.nlm.nih.gov/geo).
Real-time PCR was performed with SYBR Green Premix Ex Taq II (Takara) and the CFX96 Touch Real-Time PCR detection system (Bio-Rad Laboratories), as specified by the manufacturer. The primers for real-time qRT-PCR are listed in Supplemental Table 3.
Y2H Screen and Interaction Assay
The FBN5-B coding region was amplified by PCR with primers containing restriction sites and was cloned in frame between the EcoRI and BamHI sites of the bait vector (pGBKT7) to form pGBKT7-FBN5-B. Y2H screening of FBN5-B was performed in yeast strain PBN204 containing three reporter genes (URA3, lacZ, and ADE2) under the control of different GAL promoters. Yeast transformants containing the FBN5-B bait and an Arabidopsis cDNA AD library were plated on selection medium (SD-leucine, tryptophan, uracil [SD-LWU]) that supported the growth of yeast cells containing both bait and prey plasmids, thus yielding proteins that interacted with each other. After the yeast colonies were selected on uracil-deficient medium, we monitored the activity of β-galactosidase. The URA+ and lacZ+ colonies were also examined on adenosine-deficient medium. To construct the Y2H systems, the full-length coding sequences of the following Arabidopsis genes were amplified from the cDNA from Arabidopsis leaves: SPS1 (At1g78510), SPS2 (At1g17050), SPS3 (At2g34630), FPS1 (At5g47770), FPS2 (At4g17190), GGPS1 (At4g36810), VTE2 (At2g18950), VTE3 (At3g63410), HST (At3g11945), and GGR (At4g38450). The coding sequences for SPS1, SPS2, SPS3, VTE2, and HST, without the chloroplast transit peptide sequences, were also amplified. The amplified products were cloned into the pGADT7 vector (prey). The bait (pGBKT7-FBN5-B) and prey plasmids were introduced into two yeast strains: PBN204 (containing URA3, ADE2, and lacZ as reporters) and AH109 (containing HIS3, ADE2, and lacZ). The transformants were spotted onto SD-LW medium. After incubation for 3 d at 30°C, the colonies were replica-plated onto several selective media. To test the interactions between the FBN5 variants and either mature SPS1 or mature SPS2, the corresponding sequences of the FBN5 variants were cloned into pGBKT7. The coding sequences for SPS1 and SPS2, without their transit peptide sequences, were cloned into pGADT7. The Y2H experiment was performed as described above.
Subcellular Localization
The FBN5-A, FBN5-B, SPS1, and SPS2 full-length coding regions, each containing a BamHI site, were amplified from the cDNA of Arabidopsis leaves using KOD Hot Start DNA Polymerase (Novagen). The amplified FBN5-A sequence was fused in frame to the superfolder GFP (sGFP) gene in vector 326sGFP and FBN5-B sequence was to the mRFP gene in the vector 326mRFP and to the sGFP gene in vector 326sGFP. The amplified SPS1 and SPS2 sequences were fused in frame to the sGFP gene in vector 326sGFP. For the transient expression of the proteins, mesophyll protoplasts were prepared from 3-week-old Arabidopsis plants and transformed with the above-mentioned plasmids, as described previously (Jin et al., 2001). About 1.5 × 106 protoplasts were transfected with 20 µg plasmid DNA, and the transfected protoplasts were then incubated in the dark. Fluorescence images of the protoplasts were taken with a fluorescence microscope (Axioplan 2; Carl Zeiss) equipped with a 40×/0.75 objective (Plan-Neofluar) and a cooled CCD camera (Senicam; PCO Imaging). The filter sets used were XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23; Omega) for sGFP and XF33/E (exciter, 535DF35; dichroic, 570DRLP; emitter, 605DF50; Omega) for mRFP. The protoplasts were harvested 24 h after transformation and resuspended in sonication buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1 mM EDTA) and complete protease inhibitor cocktail (Roche). The protoplasts were lysed with brief sonication and centrifuged at 10,000g for 10 min to remove the debris. The proteins were separated with SDS-PAGE and analyzed with immunoblotting with a monoclonal anti-GFP antibody (Clontech). The protein blots were developed with protein gel blot detection solution (Supex; Neuronex) and visualized with the LAS3000 image capture system (Fuji Film).
BiFC
The entire coding regions of CNX6, FBN5-A, FBN5-B, SPS1, and SPS2 were cloned into the destination vectors pDEST-GWVYNE (Venus yellow fluorescent protein amino acids 1 to 173) (CNX6, FBN5-A, and FBN5-B) and pDEST-GWSCYCE (SCFP3A super cyan fluorescent protein amino acids 156 to 239) (CNX6, SPS1, and SPS2). FBN5-B-VenusN and CNX6-VenusC were used as a negative control; CNX6 proteins form a dimer in the heterotetramer molybdopterin synthase complex (Gehl et al., 2009). CNX6-VenusN and CNX6-VenusC were used as a positive control; a CNX6 homodimer is formed by the reconstitution of green signals from CNX6-VenusN and CNX6-VenusC. We tested the following combinations for interaction: FBN5-A-VenusN and SPS1-SCFPC, FBN5-A-VenusN and SPS2-SCFPC, FBN5-B-VenusN and SPS1-SCFPC, and FBN5-B-VenusN and SPS2-SCFPC (Gehl et al., 2009). The constructs were transiently introduced into the leaves of N. benthamiana by Agrobacterium GV3103 infiltration (Gehl et al., 2009). The plants were maintained in the dark for 2 d and then analyzed with confocal microscopy under CLSM (Nikon C2 confocal microscope).
Co-IP
The FBN5-B full-length coding region, containing XbaI and BamHI sites, was fused in frame to the HA gene cloned in vector 326HA vector (Min et al., 2007). FBN5-B-HA was transiently expressed with either SPS1-sGFP or SPS2-sGFP in mesophyll protoplasts of Arabidopsis, and proteins were extracted from protoplasts as described above. Protein extracts in immunoprecipitation buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% Triton X-100, and complete protease inhibitor cocktail [Roche]) were incubated with an anti-GFP antibody (Clontech) for 3 h at 4°C and then incubated with protein A-Sepharose beads (Amersham) for 1 h at 4°C. The beads were washed three times with immunoprecipitation buffer. After washing, the proteins were released by incubation for 10 min in SDS sample buffer at 98°C and analyzed with immunoblotting using an anti-HA (Roche) or anti-GFP antibody (Clontech).
Measurement of Photosynthetic Parameters
The maximal photochemical activity of PSII (Fv/Fm) was measured with a portable chlorophyll fluorimeter (Walz) under atmospheric conditions. Fv/Fm was measured in leaves that were dark adapted for 10 min and was calculated as (Fm – Fo)/Fm, where Fo is the initial chlorophyll fluorescence level and Fm is the maximal fluorescence level, determined with an intense pulse of white light. The stress-dependent reduction in Fv/Fm values was interpreted as the photoinhibition of PSII. The utilization of absorbed light energy was measured with a pulse amplitude modulation fluorimeter (PAM 101; Walz) on 4-week-old plants, as described previously (Hendrickson et al., 2004). After dark adaptation for 10 min, Fo and Fm were measured, and then the steady state chlorophyll fluorescence, Fs, was determined under illumination with 700 µmol m–2 s–1 actinic light.
Reactive Oxygen Species Determination
Three individual plants of each genotype were submerged in 20 mL DAB (Sigma-Aldrich) staining solution (0.1% [w/v] DAB, pH 3.0, and 10 mM NaH2PO4), introduced by vacuum infiltration. The DAB-infiltrated tissues were then incubated in the dark at ambient temperature for 24 h (Daudi and O’Brien, 2012).
HPLC Analysis
For the analysis of carotenoids, frozen tissues were ground in liquid nitrogen, and the pigments were extracted and analyzed with HPLC using a Shimadzu LC-20AD chromatograph, as previously described (Tian and DellaPenna, 2001). The HPLC peak areas at 440 nm were integrated. Chromatography was conducted at 30°C on a C18 reverse-phase column (5 µM Supelco Discovery C18 column, 150 × 4.6 mm). The chromatographic conditions were a flow rate of 1.0 mL/min with solvent A (acetonitrile:water = 9:1 v/v, plus 0.1% triethylamine) and solvent B (ethyl acetate), using the following gradient: 0 to 5 min, 0 to 33.3% B; 5 to 13 min, 33.3 to 66.7% B; 13 to 13.2 min, 100% B; 13.2 to 15.9 min, 0% B. To analyze PQ-9 and the tocochromanols, the total lipids were extracted from frozen tissues, as described previously (Bligh and Dyer, 1959). The samples in ethanol were analyzed by HPLC on a C18 column at 30°C and separated in isocratic mode with methanol at a flow rate of 1.0 mL/min. PQ-9 was detected spectrophotometrically at 255 nm. The tocopherols and PC-8 were detected fluorimetrically (290 nm excitation; 330 nm emission). The compounds were quantified with their corresponding external calibration standards, and the data were corrected by comparison with the recovery of rac-Tocol (Matreya) as the internal standard.
Accession Numbers
Sequence data from this study can be found in the GenBank/EMBL libraries under the following accession numbers: AAE14 (At1g30520), ACT2 (At3g18780), ACT7 (At5g09810), CNX6 (At2g43760), FBN1 (At4g04020/At22240), FBN2 (At2g35490), FBN4 (At3g23400), FBN5 (At5g09820), FPS1 (At5g47770), FPS2 (At4g17190), GDC1 (At1g50900), GGPS1 (At4g36810), GGR (At4g38450), HST/PDS2 (At3g11945), OsSPS2 (AK066579), PTB (CAA43973), SBP (At3g55800), SlSPS (DQ889204), SPS1 (At1g78510), SPS2 (At1g17050), SPS3 (At2g34630), VTE1 (At4g32770), VTE2 (At2g18950), VTE3 (At3g63410), and VTE4 (At1g64970).
Supplemental Data
Supplemental Figure 1. Gene Structure of FBN5.
Supplemental Figure 2. Expression Patterns of FBN5.
Supplemental Figure 3. Genetic Complementation of the Seedling-Lethal Phenotype of fbn5-1 Plants with FBN5-B cDNA.
Supplemental Figure 4. H2O2 Detection in Wild-Type and fbn5-1 Plants by Staining with DAB.
Supplemental Figure 5. Quantification of Carotenoids and Chlorophylls in Arabidopsis Leaves with Reverse-Phase HPLC.
Supplemental Figure 6. Phenotypes of Pale-Green Mutants and Wild-Type Plants on 1% Sucrose-Supplemented MS Medium.
Supplemental Figure 7. Growth and Fresh Weight of FBN5 Transgenic Plants.
Supplemental Figure 8. Interactions of Preprotein and Mature SPS with Full-Length FBN5-B.
Supplemental Figure 9. Enzymes Tested for Their Interaction with FBN5-B Using Yeast Two-Hybrid Analysis.
Supplemental Figure 10. Subcellular Localization of SPS1 and SP2.
Supplemental Table 1. Quantification of Tocochromanols and Prenylquinones in the Leaves of Each Pale-Green Mutant.
Supplemental Table 2. Quantification of Carotenoids and Chlorophylls in the Leaves of Each Pale-Green Mutant.
Supplemental Table 3. Oligonucleotides Used in This Study.
Supplemental Movie 1. Interaction of FBN5-B and SPS1 in Chloroplasts.
Supplemental Movie 2. Interaction of FBN5-B and SPS2 in Chloroplasts.
Supplementary Material
Acknowledgments
This work was supported by the National Academy of Agricultural Science (Project PJ01007505), the Cooperative Research Program for Agricultural Science and Technology Development (SSAC Grant PJ01108101), and the Rural Development Administration, Republic of Korea.
AUTHOR CONTRIBUTIONS
E.-H.K. and H.U.K. designed the study, performed most of the experiments, and wrote the article. Y.L. performed the imaging analysis with fluorescence microscopy, the SDS-PAGE and immunoblotting for protein localization, and the Co-IP experiments. All authors discussed the results and commented on the article.
Glossary
- PSII
photosystem II
- PQ
plastoquinone
- HGA
homogentisate
- SPP
solanesyl diphosphate
- MSBQ
2-methyl-6-prenyl-1,4-benzoquinol
- GGPP
geranylgeranyl diphosphate
- IPP
isopentenyl diphosphate
- SPS
solanesyl diphosphate synthase
- MS
Murashige and Skoog
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation
- Co-IP
coimmunoprecipitation
- HA
hemagglutinin
- DAB
diaminobenzidine
- HMF
high molecular mass fraction
References
- Allen J.F. (1995). Thylakoid protein phosphorylation, state 1–state 2 transitions and photosystem stoichiometry adjustment: redox control at multiple level of gene expression. Physiol. Plant. 93: 196–205. [Google Scholar]
- Bligh E.G., Dyer W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- Block A., Fristedt R., Rogers S., Kumar J., Barnes B., Barnes J., Elowsky C.G., Wamboldt Y., Mackenzie S.A., Redding K., Merchant S.S., Basset G.J. (2013). Functional modeling identifies paralogous solanesyl-diphosphate synthases that assemble the side chain of plastoquinone-9 in plastids. J. Biol. Chem. 288: 27594–27606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cui Y.-L., Jia Q.-S., Yin Q.-Q., Lin G.-N., Kong M.-M., Yang Z.-N. (2011). The GDC1 gene encodes a novel ankyrin domain-containing protein that is essential for grana formation in Arabidopsis. Plant Physiol. 155: 130–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F.X. Jr., Tice A.B., Pham C., Gantt E. (2010). Inactivation of genes encoding plastoglobuli-like proteins in Synechocystis sp. PCC 6803 leads to a light-sensitive phenotype. J. Bacteriol. 192: 1700–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A., O’Brien J. (2012). Detection of hydrogen peroxide by DAB staining in Arabidopsis leaves. Bio Protoc. 2: e263. [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D., Pogson B.J. (2006). Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 57: 711–738. [DOI] [PubMed] [Google Scholar]
- Deruère J., Römer S., d’Harlingue A., Backhaus R.A., Kuntz M., Camara B. (1994). Fibril assembly and carotenoid overaccumulation in chromoplasts: a model for supramolecular lipoprotein structures. Plant Cell 6: 119–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducluzeau A.-L., Wamboldt Y., Elowsky C.G., Mackenzie S.A., Schuurink R.C., Basset G.J.C. (2012). Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J. 69: 366–375. [DOI] [PubMed] [Google Scholar]
- Ensminger I., Busch F., Huner N.P.A. (2006). Photostasis and cold acclimation: sensing low temperature through photosynthesis. Physiol. Plant. 126: 28–44. [Google Scholar]
- Eugeni Piller L., Abraham M., Dörmann P., Kessler F., Besagni C. (2012). Plastid lipid droplets at the crossroads of prenylquinone metabolism. J. Exp. Bot. 63: 1609–1618. [DOI] [PubMed] [Google Scholar]
- Flower D.R., North A.C.T., Attwood T.K. (1993). Structure and sequence relationships in the lipocalins and related proteins. Protein Sci. 2: 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehl C., Waadt R., Kudla J., Mendel R.-R., Hänsch R. (2009). New Gateway vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol. Plant 2: 1051–1058. [DOI] [PubMed] [Google Scholar]
- Gray G.R., Hope B.J., Qin X., Taylor B.G., Whitehead C. (2003). The characterization of photoinhibition and recovery during cold acclimation in Arabidopsis thaliana using chlorophyll fluorescence imaging. Physiol. Plant. 119: 365–375. [Google Scholar]
- Hendrickson L., Furbank R.T., Chow W.S. (2004). A simple alternative approach to assessing the fate of absorbed light energy using chlorophyll fluorescence. Photosynth. Res. 82: 73–81. [DOI] [PubMed] [Google Scholar]
- Hirooka K., Bamba T., Fukusaki E., Kobayashi A. (2003). Cloning and kinetic characterization of Arabidopsis thaliana solanesyl diphosphate synthase. Biochem. J. 370: 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirooka K., Izumi Y., An C.-I., Nakazawa Y., Fukusaki E., Kobayashi A. (2005). Functional analysis of two solanesyl diphosphate synthases from Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 69: 592–601. [DOI] [PubMed] [Google Scholar]
- Hsieh F.L., Chang T.-H., Ko T.-P., Wang A.H.-J. (2011). Structure and mechanism of an Arabidopsis medium/long-chain-length prenyl pyrophosphate synthase. Plant Physiol. 155: 1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson K.G., Threlfall D.R. (1980). Synthesis of plastoquinone-9 and phytylplastoquinone from homogentisate in lettuce chloroplasts. Biochim. Biophys. Acta 632: 630–648. [DOI] [PubMed] [Google Scholar]
- Jin J.B., Kim Y.A., Kim S.J., Lee S.H., Kim D.H., Cheong G.W., Hwang I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.O., Perez-Fons L., Robertson F.P., Bramley P.M., Fraser P.D. (2013). Functional characterization of long-chain prenyl diphosphate synthases from tomato. Biochem. J. 449: 729–740. [DOI] [PubMed] [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., Rolland N. (2009). Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2: 1154–1180. [DOI] [PubMed] [Google Scholar]
- Jun L., Saiki R., Tatsumi K., Nakagawa T., Kawamukai M. (2004). Identification and subcellular localization of two solanesyl diphosphate synthases from Arabidopsis thaliana. Plant Cell Physiol. 45: 1882–1888. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kessler F., Schnell D., Blobel G. (1999). Identification of proteins associated with plastoglobules isolated from pea (Pisum sativum L.) chloroplasts. Planta 208: 107–113. [DOI] [PubMed] [Google Scholar]
- Kim H.U., van Oostende C., Basset G.J.C., Browse J. (2008). The AAE14 gene encodes the Arabidopsis o-succinylbenzoyl-CoA ligase that is essential for phylloquinone synthesis and photosystem-I function. Plant J. 54: 272–283. [DOI] [PubMed] [Google Scholar]
- Kruk J., Karpinski S. (2006). An HPLC-based method of estimation of the total redox state of plastoquinone in chloroplasts, the size of the photochemically active plastoquinone-pool and its redox state in thylakoids of Arabidopsis. Biochim. Biophys. Acta 1757: 1669–1675. [DOI] [PubMed] [Google Scholar]
- Kruk J., Trebst A. (2008). Plastoquinol as a singlet oxygen scavenger in photosystem II. Biochim. Biophys. Acta 1777: 154–162. [DOI] [PubMed] [Google Scholar]
- Laizet Y., Pontier D., Mache R., Kuntz M. (2004). Subfamily organization and phylogenetic origin of genes encoding plastid lipid-associated proteins of the fibrillin type. J. Genome Sci. Technol. 3: 19–28. [Google Scholar]
- Lee S.C., Lim M.H., Kim J.A., Lee S.I., Kim J.S., Jin M., Kwon S.J., Mun J.H., Kim Y.K., Kim H.U., Hur Y., Park B.S. (2008). Transcriptome analysis in Brassica rapa under the abiotic stresses using Brassica 24K oligo microarray. Mol. Cells 26: 595–605. [PubMed] [Google Scholar]
- Leitner-Dagan Y., Ovadis M., Shklarman E., Elad Y., Rav David D., Vainstein A. (2006). Expression and functional analyses of the plastid lipid-associated protein CHRC suggest its role in chromoplastogenesis and stress. Plant Physiol. 142: 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.-L., Yu H.-D., Guan Y., Li J.-K., Guo F.-Q. (2012). Carbonylation and loss-of-function analyses of SBPase reveal its metabolic interface role in oxidative stress, carbon assimilation, and multiple aspects of growth and development in Arabidopsis. Mol. Plant 5: 1082–1099. [DOI] [PubMed] [Google Scholar]
- Lohman D.C., et al. (2014). Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. USA 111: E4697–E4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P.K., Poliakov A., Bhuiyan N.H., Zybailov B., Sun Q., van Wijk K.J. (2012). The functional network of the Arabidopsis plastoglobule proteome based on quantitative proteomics and genome-wide coexpression analysis. Plant Physiol. 158: 1172–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell D.P., Laudenbach D.E., Huner N. (1995). Redox regulation of light-harvesting complex II and cab mRNA abundance in Dunaliella salina. Plant Physiol. 109: 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P., Beyer P., Kleinig H. (1990). Quinone compounds are able to replace molecular oxygen as terminal electron acceptor in phytoene desaturation in chromoplasts of Narcissus pseudonarcissus L. Eur. J. Biochem. 191: 359–363. [DOI] [PubMed] [Google Scholar]
- Melis A., Murakami A., Nemson J.A., Aizawa K., Ohki K., Fujita Y. (1996). Chromatic regulation inChlamydomonas reinhardtii alters photosystem stoichiometry and improves the quantum efficiency of photosynthesis. Photosynth. Res. 47: 253–265. [DOI] [PubMed] [Google Scholar]
- Mène-Saffrané L., Jones A.D., DellaPenna D. (2010). Plastochromanol-8 and tocopherols are essential lipid-soluble antioxidants during seed desiccation and quiescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 107: 17815–17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min M.K., Kim S.J., Miao Y., Shin J., Jiang L., Hwang I. (2007). Overexpression of Arabidopsis AGD7 causes relocation of Golgi-localized proteins to the endoplasmic reticulum and inhibits protein trafficking in plant cells. Plant Physiol. 143: 1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napiwotzki A., Bergmann A., Decker K., Legall H., Eckert H.-J., Eichler H.-J., Renger G. (1997). Acceptor side photoinhibition in PSII: On the possible effects of the functional integrity of the PSII donor side on photoinhibition of stable charge separation. Photosynth. Res. 52: 199–213. [Google Scholar]
- Newman L.A., Hadjeb N., Price C.A. (1989). Synthesis of two chromoplast-specific proteins during fruit development in Capsicum annuum. Plant Physiol. 91: 455–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris S.R., Barrette T.R., DellaPenna D. (1995). Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka B., Kruk J. (2012). Plastoquinol is more active than α-tocopherol in singlet oxygen scavenging during high light stress of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 1817: 389–394. [DOI] [PubMed] [Google Scholar]
- Ohara K., Sasaki K., Yazaki K. (2010). Two solanesyl diphosphate synthases with different subcellular localizations and their respective physiological roles in Oryza sativa. J. Exp. Bot. 61: 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuma S., Koyama T., Ogura K. (1991). Purification of solanesyl-diphosphate synthase from Micrococcus luteus. A new class of prenyltransferase. J. Biol. Chem. 266: 23706–23713. [PubMed] [Google Scholar]
- Pan J.-J., Kuo T.-H., Chen Y.-K., Yang L.-W., Liang P.-H. (2002). Insight into the activation mechanism of Escherichia coli octaprenyl pyrophosphate synthase derived from pre-steady-state kinetic analysis. Biochim. Biophys. Acta 1594: 64–73. [DOI] [PubMed] [Google Scholar]
- Pan J.-J., Ramamoorthy G., Poulter C.D. (2013). Dependence of the product chain-length on detergents for long-chain E-polyprenyl diphosphate synthases. Biochemistry 52: 5002–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T., Schütze K., Brost M., Oelmüller R. (2001). A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J. Biol. Chem. 276: 36125–36130. [DOI] [PubMed] [Google Scholar]
- Pozueta-Romero J., Rafia F., Houlné G., Cheniclet C., Carde J.P., Schantz M.L., Schantz R. (1997). A ubiquitous plant housekeeping gene, PAP, encodes a major protein component of bell pepper chromoplasts. Plant Physiol. 115: 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P., Gillet B., Römer S., Eymery F., Massimino J., Peltier G., Kuntz M. (2000). Over-expression of a pepper plastid lipid-associated protein in tobacco leads to changes in plastid ultrastructure and plant development upon stress. Plant J. 21: 483–494. [DOI] [PubMed] [Google Scholar]
- Sadre R., Gruber J., Frentzen M. (2006). Characterization of homogentisate prenyltransferases involved in plastoquinone-9 and tocochromanol biosynthesis. FEBS Lett. 580: 5357–5362. [DOI] [PubMed] [Google Scholar]
- Savage L.J., Imre K.M., Hall D.A., Last R.L. (2013). Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins. PLoS One 8: e73291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A.J., Gaffé J., Alcaraz J.P., Carde J.P., Bramley P.M., Fraser P.D., Kuntz M. (2007). Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 68: 1545–1556. [DOI] [PubMed] [Google Scholar]
- Singh D.K., Laremore T.N., Smith P.B., Maximova S.N., McNellis T.W. (2012). Knockdown of FIBRILLIN4 gene expression in apple decreases plastoglobule plastoquinone content. PLoS One 7: e47547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.K., Maximova S.N., Jensen P.J., Lehman B.L., Ngugi H.K., McNellis T.W. (2010). FIBRILLIN4 is required for plastoglobule development and stress resistance in apple and Arabidopsis. Plant Physiol. 154: 1281–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D.K., McNellis T.W. (2011). Fibrillin protein function: the tip of the iceberg? Trends Plant Sci. 16: 432–441. [DOI] [PubMed] [Google Scholar]
- Soll J., Kemmerling M., Schultz G. (1980). Tocopherol and plastoquinone synthesis in spinach chloroplasts subfractions. Arch. Biochem. Biophys. 204: 544–550. [DOI] [PubMed] [Google Scholar]
- Soll J., Schultz G., Joyard J., Douce R., Block M.A. (1985). Localization and synthesis of prenylquinones in isolated outer and inner envelope membranes from spinach chloroplasts. Arch. Biochem. Biophys. 238: 290–299. [DOI] [PubMed] [Google Scholar]
- Szymańska R., Kruk J. (2010). Plastoquinol is the main prenyllipid synthesized during acclimation to high light conditions in Arabidopsis and is converted to plastochromanol by tocopherol cyclase. Plant Cell Physiol. 51: 537–545. [DOI] [PubMed] [Google Scholar]
- Tevini M., Steinmüller D. (1985). Composition and function of plastoglobuli: II. Lipid composition of leaves and plastoglobuli during beech leaf senescence. Planta 163: 91–96. [DOI] [PubMed] [Google Scholar]
- Tian L., DellaPenna D. (2001). Characterization of a second carotenoid β-hydroxylase gene from Arabidopsis and its relationship to the LUT1 locus. Plant Mol. Biol. 47: 379–388. [DOI] [PubMed] [Google Scholar]
- Tian L., DellaPenna D., Dixon R.A. (2007). The pds2 mutation is a lesion in the Arabidopsis homogentisate solanesyltransferase gene involved in plastoquinone biosynthesis. Planta 226: 1067–1073. [DOI] [PubMed] [Google Scholar]
- Ting J.T.L., Wu S.S., Ratnayake C., Huang A.H. (1998). Constituents of the tapetosomes and elaioplasts in Brassica campestris tapetum and their degradation and retention during microsporogenesis. Plant J. 16: 541–551. [DOI] [PubMed] [Google Scholar]
- Trebst A. (1978). Plastoquinones in photosynthesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 284: 591–599. [Google Scholar]
- Vass I. (2012). Molecular mechanisms of photodamage in the Photosystem II complex. Biochim. Biophys. Acta 1817: 209–217. [DOI] [PubMed] [Google Scholar]
- Vidi P.A., Kanwischer M., Baginsky S., Austin J.R., Csucs G., Dörmann P., Kessler F., Bréhélin C. (2006). Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J. Biol. Chem. 281: 11225–11234. [DOI] [PubMed] [Google Scholar]
- Vishnevetsky M., Ovadis M., Itzhaki H., Levy M., Libal-Weksler Y., Adam Z., Vainstein A. (1996). Molecular cloning of a carotenoid-associated protein from Cucumis sativus corollas: homologous genes involved in carotenoid sequestration in chromoplasts. Plant J. 10: 1111–1118. [DOI] [PubMed] [Google Scholar]
- Wise R.R. (1995). Chilling-enhanced photooxidation: The production, action and study of reactive oxygen species produced during chilling in the light. Photosynth. Res. 45: 79–97. [DOI] [PubMed] [Google Scholar]
- Yang Y., Sulpice R., Himmelbach A., Meinhard M., Christmann A., Grill E. (2006). Fibrillin expression is regulated by abscisic acid response regulators and is involved in abscisic acid-mediated photoprotection. Proc. Natl. Acad. Sci. USA 103: 6061–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ytterberg A.J., Peltier J.B., van Wijk K.J. (2006). Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 140: 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef A., Laizet Y., Block M.A., Maréchal E., Alcaraz J.-P., Larson T.R., Pontier D., Gaffé J., Kuntz M. (2010). Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J. 61: 436–445. [DOI] [PubMed] [Google Scholar]
- Zbierzak A.M., Kanwischer M., Wille C., Vidi P.-A., Giavalisco P., Lohmann A., Briesen I., Porfirova S., Bréhélin C., Kessler F., Dörmann P. (2010). Intersection of the tocopherol and plastoquinol metabolic pathways at the plastoglobule. Biochem. J. 425: 389–399. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chau N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in plants. Plant J. 24: 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.