Summary
Pluripotency represents a cell state comprising a fine-tuned pattern of transcription factor activity required for embryonic stem cell (ESC) self-renewal. TBX3 is the earliest expressed member of the T-box transcription factor family and is involved in maintenance and induction of pluripotency. Hence, TBX3 is believed to be a key member of the pluripotency circuitry, with loss of TBX3 coinciding with loss of pluripotency. We report a dynamic expression of TBX3 in vitro and in vivo using genetic reporter tools tracking TBX3 expression in mouse ESCs (mESCs). Low TBX3 levels are associated with reduced pluripotency, resembling the more mature epiblast. Notably, TBX3-low cells maintain the intrinsic capability to switch to a TBX3-high state and vice versa. Additionally, we show TBX3 to be dispensable for induction and maintenance of naive pluripotency as well as for germ cell development. These data highlight novel facets of TBX3 action in mESCs.
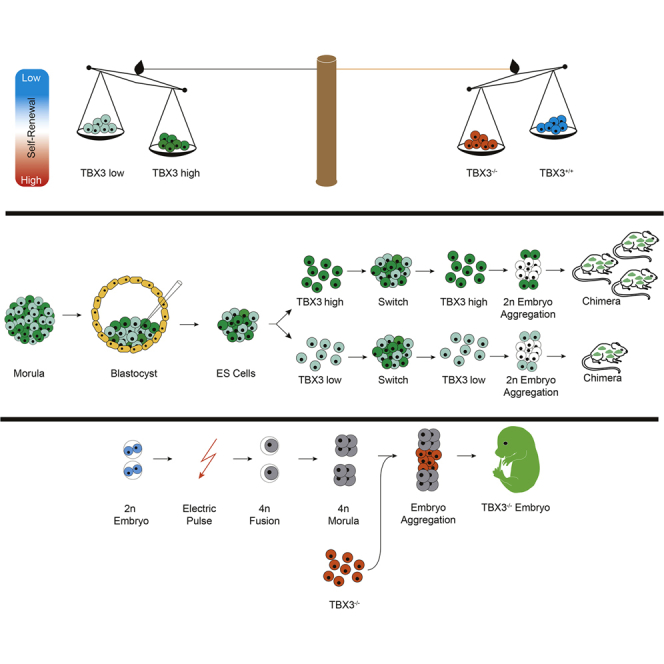
Graphical Abstract
Highlights
-
•
Tbx3 is not required for induction and maintenance of pluripotency
-
•
Tbx3 is heterogeneously expressed in pluripotent cell in vivo and in vitro
-
•
In vivo Tbx3-high cells are committed to the PE; low cells commit to the epiblast
-
•
Loss of Tbx3 leads to complex compensational processes in pluripotent stem cells
In this article, Kleger and colleagues show that Tbx3, a transcription factor of the pluripotency network is heterogeneously expressed in embryonic cells in vitro and in vivo. They show that distinct Tbx3 expression levels correlate with various embryonic fates and potency during ongoing development. Finally, they show that Tbx3 is not only dispensable for pluripotency but leads to a compensated state with a self-renewal bias.
Introduction
Pluripotent stem cells (PSCs) are characterized by continuous self-renewal while maintaining the potential to differentiate into cells of all three germ layers. Great knowledge exists about the regulatory networks that maintain pluripotency and about key players that regulate differentiation. Pluripotency exists in various states, with the ground state of naive pluripotency as the most basic state of pluripotency (Chen et al., 2013, Leitch et al., 2013, Wray et al., 2010). Here, diverse signaling pathways, in concert with a combination of key transcription factors (TFs), precisely regulate ground state conditions. Diminutive changes in their expression can either destabilize or strengthen the network (Karwacki-Neisius et al., 2013). Several network TFs are heterogeneously expressed (Chambers et al., 2007, Festuccia et al., 2012, Kalmar et al., 2009, MacArthur et al., 2012, Miyanari and Torres-Padilla, 2012, Papatsenko et al., 2015) and regulated in a highly dynamic manner to balance between stem cell self-renewal and exit from pluripotency (Faddah et al., 2013, Radzisheuskaya et al., 2013) as well as during somatic reprogramming (Takahashi and Yamanaka, 2006). Finally, even core TFs of the pluripotency network determine the exit from stemness to early cell fate determination in a competitive manner (Lu et al., 2011, Teo et al., 2011, Waghray et al., 2015, Weidgang et al., 2013).
The T-box family of TFs is involved in a variety of signaling cascades including the pluripotency network (Niwa et al., 2009). TBX3 mutually regulates the expression of key lineage TFs factors while maintaining and inducing pluripotency (Han et al., 2010a, Weidgang et al., 2013). In detail, TBX3 is directly bound by NANOG and in turn binds OCT4 and SOX2 (Han et al., 2010a). Its expression is regulated in part by the phosphatidylinositol-3-OH-kinase-Akt (PI3K) and mitogen-activated protein kinase (MAPK) pathways (Niwa et al., 2009). Moreover, TBX3 can bypass the requirement for leukemia inhibitory factor (LIF) signaling and functions upstream of NANOG in PSCs (Niwa et al., 2009). Removal of TBX3 from embryonic stem cells (ESCs) causes differentiation (Han et al., 2010a, Ivanova et al., 2006, Lee et al., 2012, Lu et al., 2011, Nishiyama et al., 2013). In contrast, TBX3 is also a crucial player in early cell fate events, driving mesendodermal and primitive endoderm (PE) specification (Kartikasari et al., 2013, Lu et al., 2011, Waghray et al., 2015, Weidgang et al., 2013). Here, we provide a comprehensive view on the definitive requirements for TBX3 to maintain and induce pluripotency and precisely characterize various TBX3-expression states in PSCs.
Results
TBX3 Is Dynamically Expressed in mESCs
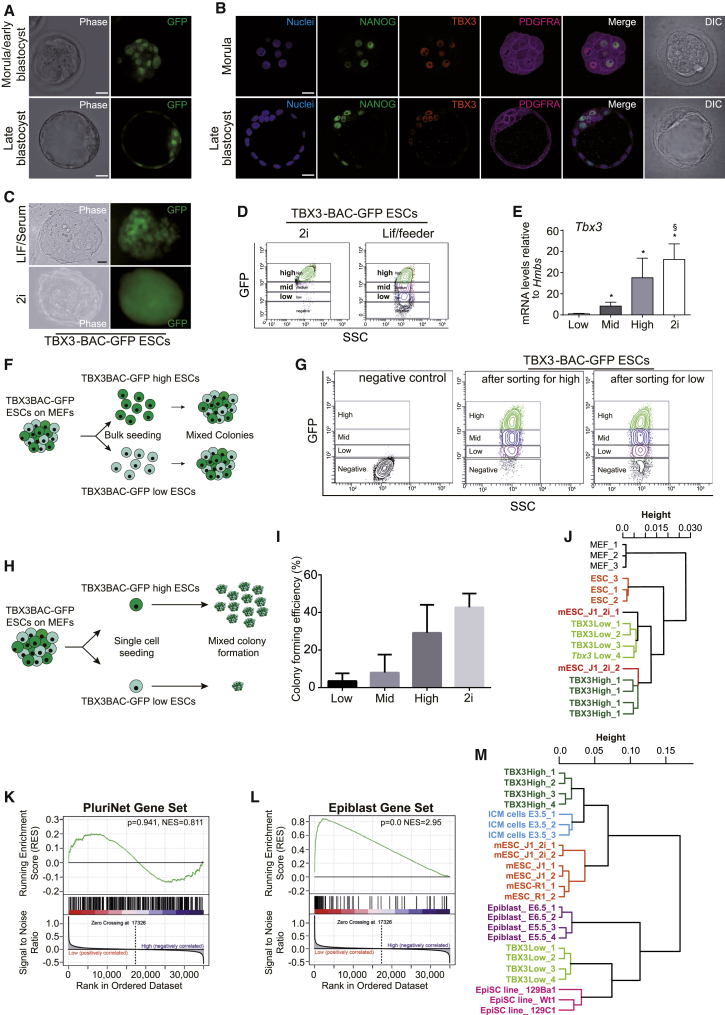
Heterogeneous expression of pluripotency TFs is present under various culture conditions, to date focused on the TF Nanog (Dietrich and Hiiragi, 2007, Xenopoulos et al., 2015). Heterogeneous Tbx3 expression has been reported in mouse ESCs (mESCs) (Niwa et al., 2009, Toyooka et al., 2008). The relevance of such heterogeneity in vitro remains divisive in vivo. To access TBX3 expression in vivo, we used a mouse strain containing a Venus-cassette (ven) to disrupt and track endogenous TBX3 locus activity (Kunasegaran et al., 2014). We observed a heterogeneous venus signal tracking TBX3 protein in both morula and blastocyst stages of murine embryos (Figure 1A). Immunohistochemistry (IHC) of wild-type embryos confirmed this observation, where NANOG-positive epiblast (EPI) cells express varying levels of TBX3 (Figure 1B). Interestingly, the inner cell mass (ICM) cells with high TBX3 expression tend to have increased PDGFRA and decreased NANOG expression, suggestive of a PE cell fate. In contrast, low TBX3 expression correlates with high NANOG expression, indicative of an EPI fate.
Figure 1.
TBX3 Is Dynamically Expressed in Mouse ESCs
(A) TBX3+/ven pre-implantation embryos at indicated time points.
(B) Wild-type pre-implantation embryos stained for NANOG (green), TBX3 (red), and PDGFRa (purple). Hoechst 33342 is shown (nuclei, blue).
(C) Representative phase contrast and GFP fluorescent images of de-novo-derived TBX3-BAC-EGFP mESCs cultured under indicated conditions. The scale bars represent 20 μm.
(D) TBX3-BAC-EGFP expression fluctuates under indicated culture conditions.
(E) qPCR for Tbx3 of TBX3-BAC-EGFP low, mid, and high sorted cells (according to D). n = 4; two individual clones from two independent experiments)
(F and G) Experimental sorting strategy and schematic results (F) of (G); n = 2, two independent clones.
(H and I) Sorting strategy and schematic results (H) of (I). Two out of four independent experiments with similar results are shown. For 2i condition, n = 2.
(J) Hierarchical clustering of gene expression profiles of TBX3GFP-high and -low ESCs with published pluripotent and somatic cells (GSE11274 and GSE58735).
(K and L) Gene set enrichment analysis shows no bias of pluripotent marker genes between TBX3GFP-high (high) and -low (low) ESCs (J) but a bias to epiblast (K) in TBX3GFP-low ESCs.
(M) Hierarchical clustering shows that TBX3GFP-low ESCs (TBX3 low) cluster close to EpiSCs and epiblast (E5.5/6.5), TBX3GFP-high ESCs (TBX3 high) are close to published ESCs (J1, R1), 2i-treated ESCs (J1_2i), and E3.5 inner cell mass (ICM cells E3.5; GSE58735 and GSE35416).
See also Figures S1 and S2.
For a global overview on Tbx3 expression in vivo at early developmental stages, we performed in silico analyses of published datasets investigating single-cell transcriptomes of morula and blastocyst stages (Blakeley et al., 2015, Deng et al., 2014, Piras et al., 2014). Violin plots depict a bimodal expression of Nanog and Tbx3, whereas Oct4 expression recapitulates a described increase of transcriptional noise with ongoing development (Piras et al., 2014) and separates just upon ICM and trophectoderm (TE) segregation (Figures S1A–S1D). Next, we applied a defined gene signature of 48 TFs capable to segregate the three lineages in pre-implantation embryos, namely PE, EPI, both defining the ICM, and TE (Guo et al., 2010). This gene signature separates two cell populations as early as in the 16-cell stage, which could be allocated to the later putative ICM and TE (Figure S1E). This separation becomes more evident with ongoing development and maximizes in the late blastocyst (Figures S1F and S1G). All clustered samples indicate heterogeneous single-embryonic-cell Tbx3 expression in vivo across both lineages (ICM and TE). Moreover, Tbx3 closely clusters with distinct TFs in the three embryonic stages regulating different lineages, supporting its role for lineage segregation (Figures S1E–S1G). We used this single-cell-based transcriptional lineage segregation to illustrate heterogeneous Tbx3 expression (Figures S1H–S1J). Box plots illustrate the greater heterogeneity of Tbx3 compared to Oct4 in the ICM, most evident in the early blastocyst (Figure S1I). Finally, for each gene in the signature, we calculated Pearson correlation for Tbx3 using all single embryonic cells at the late blastocyst stage to identify genes being positively or negatively correlated with Tbx3 (Figure S1K). We note that TBX3-high in vivo may bias toward PE (high correlation with Pdgfra), thus recapitulating our immunostainings for PDGFRA presented in Figure 1B. In pre-implantation embryos, Tbx3 expression mostly restricts to the ICM as shown by the negatively correlated TE markers (Eomes and Cdx2; Figure S1K).
To verify TBX3 heterogeneity observed in vivo in more detail, we used a transgenic TBX3GFP (TBX3-BAC-EGFP) reporter mouse, well correlating with endogenous TBX3 protein levels (Horsthuis et al., 2009). We avoided the TBX3-Venus system in that context to exclude the potential of a TBX3 haploinsufficiency phenotype and due to previous reports showing conflicting results for different Nanog knockin reporters (Frank et al., 2013, Miyanari and Torres-Padilla, 2012). Moreover, recently, a BAC-NANOG reporter was shown to be a reliable tool for tracing Nanog expression in mESCs (Xenopoulos et al., 2015). Here, we isolated mESCs from the ICM of this strain to serve as a live-time TBX3 tracking tool in vitro (Figure 1C). Under LIF/feeder conditions, we observed differential GFP expression, representing a spectrum of endogenous TBX3 expression levels. In contrast, under feeder-free 2i conditions (Frum et al., 2013, Niwa et al., 2009), GFP was homogenously expressed at high levels (Figures 1C, 1D, and S2A). Consequently, we performed FACS of LIF/feeder ESCs and isolated ESCs expressing high, mid, or low levels of TBX3GFP. As expected, reporter intensity reflects endogenous Tbx3 expression (Figures 1D and 1E), further mirrored by single-cell qPCR (Figure S2B). To investigate whether TBX3GFP-low or TBX3GFP-high ESCs can switch from one condition to the other, we sorted ESCs for respective GFP states into low- and high-expressing populations (Figure 1F). Both subpopulations regenerated bona fide ESC cultures. TBX3GFP-low cells generated colonies containing TBX3GFP-high cells and vice versa (Figure 1G). Interestingly, colony-forming efficiency was lower for single TBX3GFP-low ESCs compared to TBX3GFP-high cells. However, both gave rise to heterogeneous colonies. As expected, 2i cultures of TBX3-BAC-EGFP cells displayed highest clonal growth rates (Figures 1H and 1I).
Next, we compared transcriptional profiles of TBX3GFP-low and TBX3GFP-high mESCs (LIF/feeder) and observed a differentially regulated gene signature (Figure S2C; Tables S2, S3, and S4). Hierarchical clustering with mouse embryonic fibroblasts (MEFs) and reference ESC sets positioned TBX3-high and -low cells to the pluripotent references, excluding that TBX3-low cells simply represent differentiating ESCs (Figure 1J). PluriNet has been shown to define a common gene signature defining bona fide pluripotent cell types such as mESCs, induced pluripotent stem cells (iPSCs), and human oocytes (MacArthur et al., 2012, Müller et al., 2008). Applying this signature to TBX3GFP-low and TBX3GFP-high ESCs clearly illustrates the robust pluripotent state of both cell types (Figure 1K). Interestingly, TBX3GFP-low cells also showed enrichment for transcripts expressed in EPI stem cells (Figures 1L [Nora et al., 2012] and S2D [Kurek et al., 2015]). Next, we correlated TBX3GFP-low and TBX3GFP-high cells relative to published datasets from EpiSCs and E5.5/6.5 EPIs and from ESCs and E3.5 ICM. Consistent with the above findings, TBX3GFP-low cells clustered closer to EPI and EpiSCs than to ESCs and vice versa, whereas TBX3GFP-high cells clustered closely to the ICM, similar to reference ESCs cultured under 2i conditions (Figure 1M).
Low Levels of Tbx3 Restrict Developmental Potential In Vivo
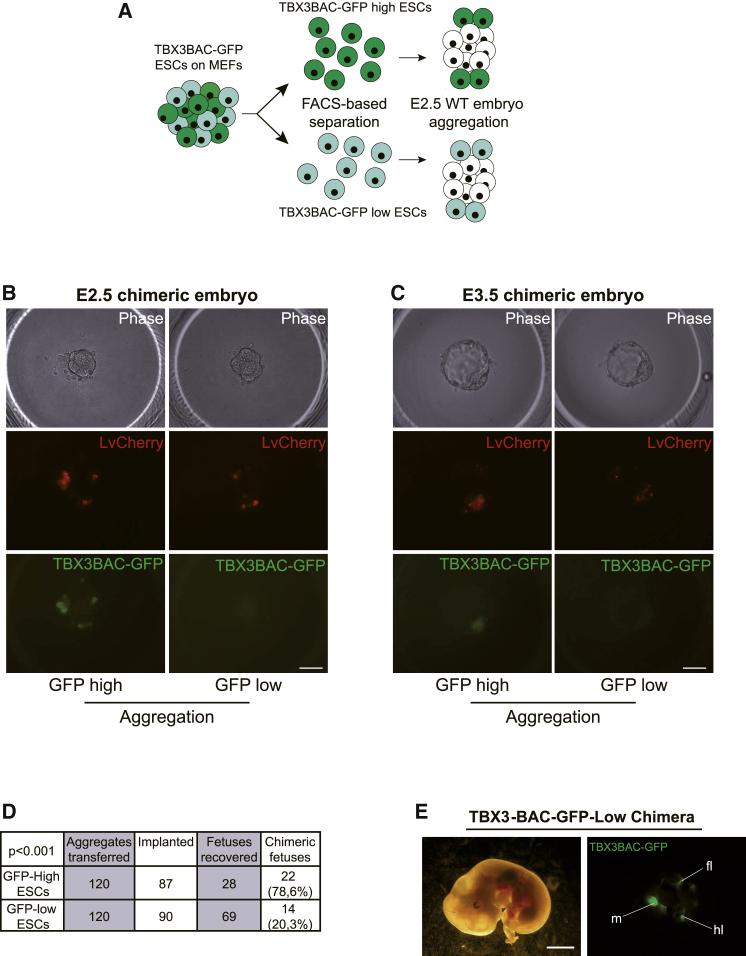
Finally, we performed embryo aggregation assays using freshly sorted LV-mCherry-labeled TBX3GFP-low or TBX3GFP-high cells and wild-type E2.5 embryos (Figure 2A). Both cell populations were capable to integrate into the ICM as observed by the constitutively expressed mCherry reporter at E3.5 (Figures 2B and 2C). 120 aggregates from each population were transferred to pseudopregnant mice for further development. Sixty-nine and 28 embryos from the TBX3GFP-low and TBX3GFP-high aggregates were recovered, of which 14 (20.3%) and 22 (78.6%) were classified to be chimeric (Figure 2D). Notably, very similar numbers of embryos implanted, although a lower number of fetuses was recovered from the TBX3GFP-high cell population. We hypothesize that the high contribution ability of TBX3GFP-high mESCs may limit the survival rate of this group as described previously (Gertsenstein et al., 2010). Chimeric embryos recapitulated previously reported Tbx3-expression patterns as seen by the BAC-EGFP signal (Figure 2E; Horsthuis et al., 2009). Interestingly, the frequency of TBX3-low chimeric embryos was overall significantly lower (Figure 2D). Unfortunately, the LV-mCherry reporter appeared to be silenced in all embryos, impeding a more detailed characterization of chimerism. This highlights that TBX3-low ESCs have a significantly reduced potential for in vivo contribution to chimeric embryos, underpinning our in vitro observations that a TBX3-low state represents a differentiation-poised but pluripotent state.
Figure 2.
Low Levels of Tbx3 Restrict Developmental Potential In Vivo
(A) Scheme of sorting strategy and 2n aggregation assay with E2.5 embryos.
(B and C) Generation of chimeric embryos by aggregating TBX3-BAC-EGFP high/low ESCs infected with LvCherry lentivirus with 2n E2.5 embryos (B) and ESCs integrated into ICM of chimeric blastocysts after 24 hr in culture (E3.5). (C) Top, phase contrast; middle, LvCherry signal for lentiviral labeling of cells to ensure visualization; bottom, GFP signal of the TBX3-BAC-EGFP high and low ESCs.
(D) Embryonic development from 2n embryo aggregation with TBX3GFP-high and -low ESCs.
(E) Phase contrast (left) and GFP fluorescence (right) of TBX3-BAC-EGFP-low chimera. fl, front limb; hl, hind limb; m, midbrain.
The scale bars represent 50 μm (B and C) and 1 mm (E).
Tbx3 Is Dispensable to Establish and Maintain Pluripotency
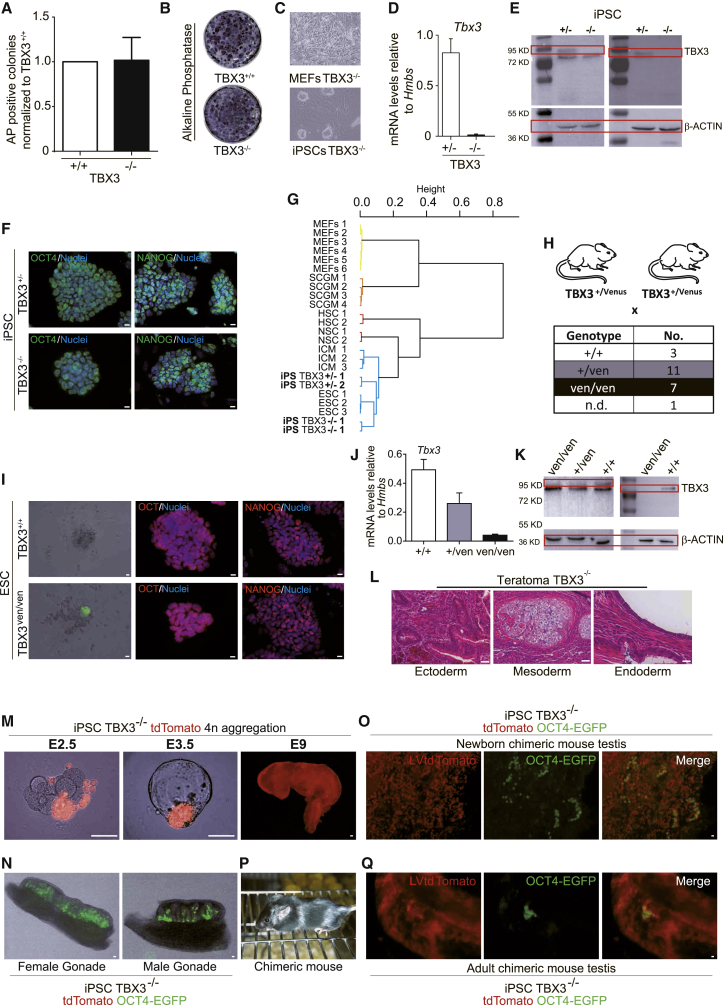
Because the spectrum of TBX3-low and -high expression is compatible with pluripotency, we investigated genetic ablation of TBX3 in induction and maintenance of pluripotency. First, low passage TBX3+/+ and TBX3−/− MEFs (Frank et al., 2013, Kumar et al., 2014) were assayed for reprogramming toward iPSCs. These MEFs did not show significant differences in their proliferative capacity over three passages (data not shown). Interestingly, alkaline phosphatase (AP)-positive iPSC colonies appeared in similar numbers in TBX3−/− MEFs compared to wild-type controls (Figures 3A–3C). We also assessed colony formation kinetics by counting iPSC-like colonies without observing any difference. Using higher passages of TBX3−/− MEFs led to fewer iPSC colonies (data not shown). This observation would be in line with previously reported data showing that CAPERα/TBX3 repressor complex is required to prevent senescence, the latter representing an established barrier for reprogramming (Banito et al., 2009, Kumar et al., 2014). Indeed, TBX3 mRNA and protein were absent in TBX3−/− iPSC lines (Figures 3D and 3E). These colonies displayed normal dome-shaped morphology and pluripotency marker expression (Figures 3F and S3A). Transcriptome cluster analysis of TBX3−/− iPSCs further confirmed pluripotency (Figure 3G). To complement these findings, we investigated again the TBX3-Venus mouse strain (see also Figure 1A). Mating heterozygous mice with subsequent embryo isolation allowed us to culture E3.5 blastocysts for de novo ESC derivation (Czechanski et al., 2014). From 24 blastocysts, 22 ESC lines were expanded, 3 TBX3+/+, 11 TBX3ven/+, 7 TBX3ven/ven, and 1 line undetermined (Figure 3H). Morphology and marker staining confirmed pluripotency, whereas the Venus reporter signal localized to the outgrowing ICM (Figures 3I, S3B, and S3C). FACS analysis for the Venus reporter as well as mRNA and protein analysis confirmed the TBX3-null status (Figures 3J, 3K, S3D, and S3E).
Figure 3.
Tbx3 Is Dispensable to Induce and Maintain Pluripotency
(A and B) Quantification (A) and representative AP+ colonies (B) day 12 after reprogramming of TBX3+/+ and TBX3−/− MEFs. Three independent experiments performed with triplicate technical replicates are shown.
(C) Morphology of OKS-infected TBX3−/− MEFs (top) picked and expanded TBX3−/− iPSCs (bottom).
(D) Tbx3 expression in TBX3+/− and (TBX3−/−) iPSCs. Two independent experiments performed with triplicate technical replicates are shown.
(E) TBX3 western blot in iPSCs (two different antibodies). Representative images of at least three independent experiments are shown.
(F) Pluripotency marker IHC in generated iPSCs of the indicated genotypes.
(G) Hierarchical clustering of gene expression profiles shows that iPSCs cluster with published ESC/ICM datasets.
(H) Crossing scheme to obtain TBX3ven/ven-null mice and overview of derivation frequency; ND, not determined.
(I) Pluripotency marker IHC in de novo ESCs of the indicated genotypes.
(J and K) TBX3 mRNA (J; three independent experiments performed with triplicate technical replicates) and protein (K) expression confirms TBX3-null phenotype in TBX3ven/ven ESCs (two TBX3 antibodies). Representative images of at least three independent experiments are shown.
(L) TBX3−/− iPSCs form teratomas with all three germ layers (ecto-, meso-, and endoderm) in NMRI mice.
(M) Generation of “all-PSC”-derived TBX3-null embryos using 4n embryo aggregation from TBX3−/− iPSCs. Developmental stage as indicated. To ensure visualization of TBX3-null progeny, TBX3−/− iPSCs were constitutively labeled with a tdTomato-expressing lentivirus (LVtdTomato).
(N) Germline contribution of TBX3−/− iPSCs carrying an OCT4-EGFP reporter and LVtdTomato after 2n aggregation assay in female (left) and male (right) gonads at E12.5.
(O) Representative fluorescent image of newborn chimeric mouse testis after 2n aggregation assay of OCT4-EGFP and LVtdTomato-infected TBX3−/− iPSCs.
(P) TBX3−/− iPSCs contribute to adult chimeric mice after 2n aggregation assay (light coat color).
(Q) Representative fluorescent image of adult chimeric mouse testis after 2n aggregation assay of OCT4-EGFP and LVtdTomato-infected TBX3−/− iPSCs.
The scale bars represent 50 μm. See also Figures S3 and S4.
Tbx3 Is Dispensable for the Formation of “All-PSC”-Derived Mice and Germ Cell Development
A hallmark of PSCs is the ability to form teratomas and to generate chimeric embryos upon blastocyst injection. Indeed, TBX3−/− iPSCs generated teratomas (Figure 3L). Surprisingly, even diploid (2n) embryo aggregation of TBX3−/− ESCs led to high-grade chimeras at E9.5 (Figure S4A). Moreover, both TBX3−/− ESCs and iPSCs were capable of generating all-PSC-derived embryos (Figures 3M and S4A) in tetraploid (4n) embryo aggregation, the most stringent test for pluripotency (Stadtfeld et al., 2010). In line with the previously reported TBX3−/− phenotype (Frank et al., 2013), all-PSC embryos (∼85%) were found developmentally arrested and dead at E11.5 (Figure S4B; Table 1).
Table 1.
Quantitative Overview of 2n and 4n Embryo Aggregation of the Indicated Cell and Genotypes
| Cell Type | Approach | Aggregation | Transferred | E9.5 |
E11.5 |
GFP Positive | ||
|---|---|---|---|---|---|---|---|---|
| Placenta Found | Fetus Found | Placenta Found | Fetus Found | |||||
| iPSC TBX3Δflox/Δflox | 4n | 50 | 47 | − | − | 41 | 6 | − |
| iPSC TBX3Δflox/Δflox+ tdTomato | 4n | 50 | 46 | 12 | 10 | 29 | 6 | − |
| iPSC TBX3Δflox/Δflox + tdTomato | 4n | 40 | 40 | 20 | 8 | − | − | − |
| iPSC TBX3Δflox/Δflox + tdTomato | 2n | 47 | 47 | 35 | 22 | − | − | − |
| ESC TBX3ven/ven Clone 4a | 4n | 20 | 20 | 19 | 14 | − | − | − |
| ESC TBX3ven/ven Clone 4a | 2n | 27 | 27 | 21 | 15 | − | − | − |
| ESC TBX3+/+ Clone 21 | 4n | 20 | 20 | 10 | 8 | − | − | − |
| ESC TBX3+/+ Clone 21 | 2n | 18 | 17 | 8 | 6 | − | − | − |
| ESC TBX3ven/ven Clone 5 + GFP | 4n | 54 | 53 | 40 | 8 | − | − | 8 |
| ESC TBX3ven/ven Clone 5 + GFP | 2n | 48 | 48 | 41 | 24 | − | − | 13 |
To assess germline contribution of TBX3−/− PSCs, we introduced an OCT4-EGFP reporter into tdTomato-labeled TBX3−/− iPSCs (Figure S4C). In a 2n aggregation assay, high germline contribution of TBX3−/− cells in five out of 12 E12.5 chimeric fetuses (presence of OCT4-EGFP-positive cells in gonads) indicated that TBX3 is not required for germ cell development (Figure 3N). Large clusters of OCT4-EGFP-positive spermatogonia stem cells (SSCs) were observed in the seminiferous tubules of a newborn mouse chimera (Figure 3O). However, the seminiferous tubules of an adult chimeric mouse showed very few and small clusters of OCT4-EGFP-positive cells, despite the presence of large amounts of tdTomato-positive TBX3−/−-iPSC-derived cells (Figures 3P and 3Q). Spermatozoa from this mouse were used for in vitro fertilization (IVF) with oocytes from B6C3F1 mice, but no germline transmission could be achieved in 44 developed blastocysts as judged by tdTomato and OCT4-EGFP fluorescence. In addition, a chimeric TBX3-null mouse gave rise to four litters with 54 F1 pups in total and none of them was RFP positive in the somatic part or OCT4-EGFP positive in gonads. We conclude that, although TBX3 is not required for germ cell development, it seems to be essential for the maintenance of SSCs after birth.
TBX3 Removal from the Pluripotency Circuitry Delays Lineage Commitment
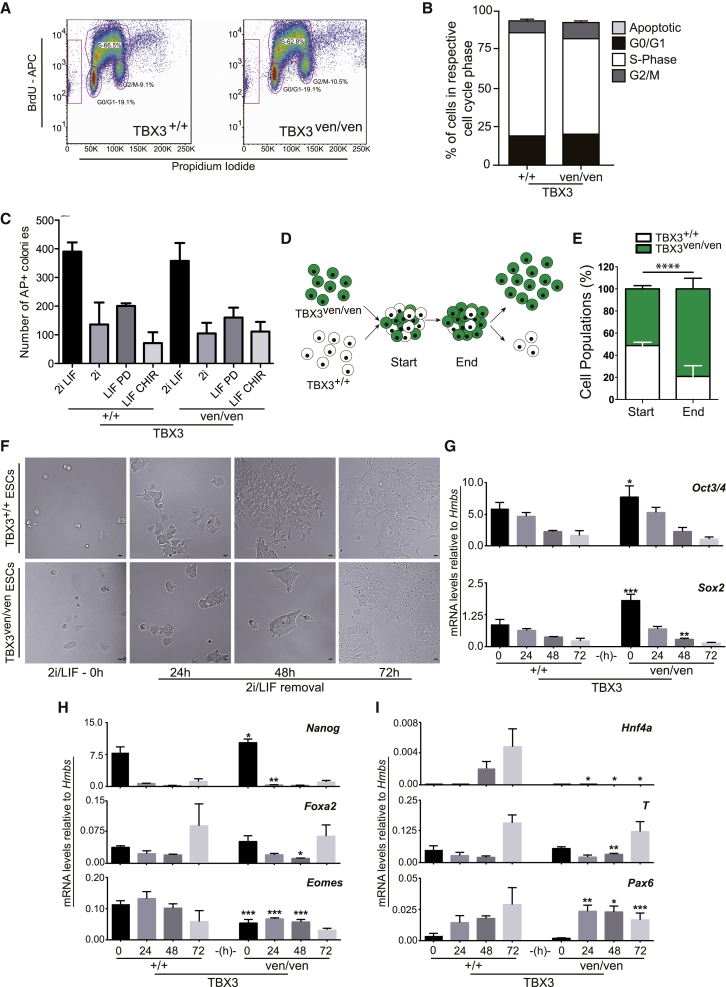
Next, we investigated the cell-cycle profile of TBX3−/− ESCs using two complementary methods without observing significant differences compared to controls (Figures 4A and 4B; data not shown). Also, there was no difference in single-cell colony formation capacity in TBX3 wild-type versus null ESCs, irrespective of removal of any LIF/2i component. This indicates that LIF/STAT3, ERK, and GSK3 signaling is unperturbed in TBX3−/− cells (Figure 4C). Interestingly, TBX3−/− cells outperformed their wild-type counterparts in a cell competition assay, indicating enhanced self-renewal in TBX3−/− ESCs (Figures 4D and 4E). It has been shown that reducing OCT4 levels directs an even more robust pluripotent state (Karwacki-Neisius et al., 2013). On a global gene expression level, only TBX3−/− iPSCs, but not ESCs, display greater enrichment of the PluriNet signature (data not shown).
Figure 4.
Characterization of TBX3-Null ESCs
(A) BrdU-FACS of TBX3ven/ven and TBX3+/+ ESC lines. Gate at y axis, apoptotic cells; lower left circle, G0/G1 population; lower right circle, G2/M population; upper big gate, S phase.
(B) Quantification of BrdU incorporation summarized from three TBX3ven/ven and a TBX3+/+ ESC line. n = 3 (three individual clones). DAPI-based cell-cycle analysis revealed no difference between TBX3ven/ven and TBX3+/+ ESCs (data not shown).
(C) Quantified AP+ colonies after removing individual LIF/2i components. N2B27 backbone medium. n = 3, for two independent clones with duplicate technical replicates.
(D) Co-culture strategy to compare self-renewal capacity of TBX3+/+ and TBX3ven/ven mESC under LIF/2i. Analysis at start (P0) and end of experiment (P3) for venus expression via FACS is shown. n = 3, for two independent clones with duplicate technical replicates.
(E) Quantification of cell populations from (D) after three passages.
(F) Phase images of TBX3ven/ven and TBX3+/+ mESC at 0 hr, 24 hr, 48 hr, and 72 hr after withdrawal of LIF and 2i. Delayed differentiation of TBX3ven/ven as evident by preserved ESC morphology. The scale bars represent 20 μm.
(G–I) mRNA levels of pluripotency markers (Oct3/4, Sox2, and Nanog), early endodermal markers (Foxa2, Eomes, and Hnf4a), T (brachyury) mesodermal, and Pax6 as ectodermal marker in TBX3+/+ and TBX3ven/ven mESC at respective time points of LIF/2i withdrawal. Two clones per genotype are shown.
Two independent experiments performed with triplicate technical replicates are shown. See also Figure S5.
To test the differentiation capacity of TBX3−/− ESCs in vitro, we removed LIF/2i in a serum-based medium to allow spontaneous differentiation. TBX3−/− ESCs displayed a delay in losing typical ESC morphology compared to controls (Figures 4F and S5). Of note, TBX3−/− ESCs showed higher mRNA baseline levels of Oct4, Sox2, and Nanog (Figures 4G and 4H). Downregulation of pluripotency markers assimilated rapidly, whereas upregulation of the endodermal differentiation markers was limited in TBX3-null cells. In contrast, brachyury (T) upregulation occurred earlier in TBX3−/− cells in line with a recent study (Waghray et al., 2015; Figures 4H and 4I). This supports previous observations showing that TBX3 fine-tunes early cell fate choices in a dose-dependent manner (Kartikasari et al., 2013, Lu et al., 2011, Weidgang et al., 2013).
Loss of Tbx3 Leads to Complex Compensational Processes
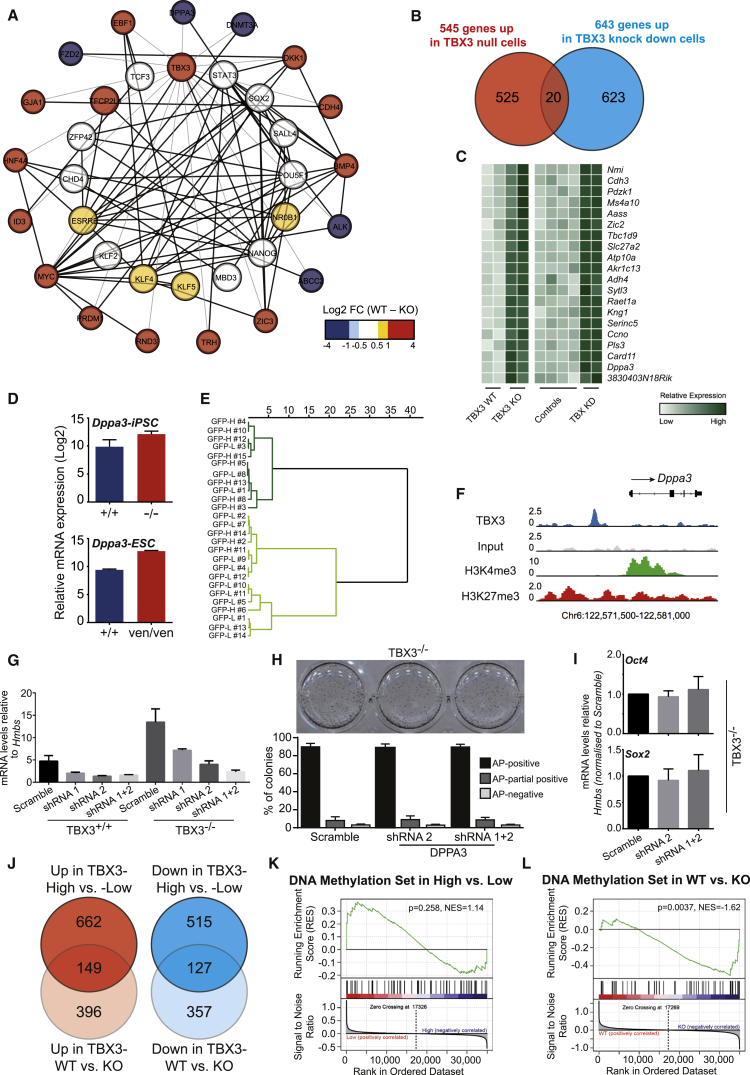
Previously, a simplified pluripotency circuitry was described (Dunn et al., 2014). We combined this network with TBX3-ChIP-sequencing data (Han et al., 2010a) and our gene expression data of wild-type versus TBX3−/− ESCs cultured under LIF/2i conditions to obtain a more global view on the molecular changes upon TBX3 removal. Figure 5A shows a network in which the inner circle describes core factors of pluripotency, whereas the outer circle depicts TBX3-bound genes that are differentially regulated in TBX3−/− ESCs. Most pluripotency markers in the inner circle did not show significant changes in their gene expression, although Klf4/5, Esrrb, Tfcp2l1, and NrOb1 were slightly downregulated in TBX3−/− ESCs compared to controls (yellow/red color in Figure 5A) and qPCR indicated upregulation of Oct4, Sox2, and Nanog (Figures 4G and 4H). TBX3 is a transcriptional repressor (Bakker et al., 2008). We hypothesize that loss of TBX3 may lead to an upregulation of specific genes impacting on pluripotency that are usually repressed by TBX3. TBX3-bound genes in the outer circle showed that Dppa3, Fzd2, Abcc2, Alk, and Dnmt3a transcripts were significantly increased in TBX3−/− ESCs (Figure 5A). To narrow down this list, we compared the upregulated genes in TBX3−/− ESCs with the TBX3 target genes identified by a Tbx3 knockdown experiment (Han et al., 2010a, Nishiyama et al., 2013), resulting in an overlap of 20 candidate genes (Figures 5B and 5C). The sole overlapping candidate between this approach and our network was Dppa3/Stella, a gene described to impact on pluripotency and germ cell development (Hayashi et al., 2008, Payer et al., 2006, Waghray et al., 2015, Xu et al., 2015). Our microarray data and single-cell qPCR confirmed the reciprocal relationship between Tbx3 and Dppa3 (Figures 5D, 5E, and 5G). Additionally, the DPPA3 promoter is occupied by TBX3 and possesses bivalent modifications as expected for genes regulating pluripotency (Figure 5F). Unexpectedly, the knockdown of Dppa3 in TBX3−/− iPSCs did not impact on pluripotency as shown by colony formation assays and qPCR (Figures 5G–5I). We conclude that compensational mechanisms allow a TBX3−/− state independent of DPPA3 and most likely involve a complex compensational network, not only involving direct (as suggested by Figure 5A) but also indirect TBX3 target genes. Still, it appears rather antithetic that downregulation of TBX3 inversely correlates with developmental potential, whereas complete loss of TBX3 leads to overcompensation and a reduced propensity to differentiate. To address this point, we looked for overlap among the differentially regulated genes in TBX3-low and -high and TBX3−/− and wild-type ESCs. Due to the developmental differences of these cell states, only a limited number of genes were commonly regulated (Figure 5J; Tables S5, S6, S7, and S8). Interestingly, a gene set associated with “DNA methylation” was enriched only in TBX3−/− cells and not in TBX3-low cells, indicating global epigenetic differences, particularly in TBX3−/− cells (Figures 5K and 5L).
Figure 5.
TBX3 Depletion Leads to Complex Compensational Processes
(A) Molecular circuitry of pluripotency markers (inner layer) and TBX3 target genes (outer layer) generated by the STRING database. The color of a node reflects the change in gene expression level between wild-type (WT) and TBX3-null (KO) ESCs. Black edge shows an evident interaction (confidence score > 0.7) between two connected genes from the STRING database. The gray edge connects TBX3 with its target genes identified by ChIP-seq data (GSE19219; Han et al., 2010a).
(B) Venn diagram of upregulated genes in TBX3-null ESCs and Tbx3 knockdown ESCs (GSE26520), compared to respective wild-type controls, shows 20 candidate genes.
(C) Heatmap of gene expression level of these candidate genes. Light green, below median expression; dark green, above median.
(D) The expression level of Dppa3 in TBX3+/+and TBX3−/− iPSCs, wild-type (+/+), and TBX3-null (ven/ven) ESCs.
(E) Clustering of single-cell Dppa3 expression (n = 26) suggests the reciprocal expression between Tbx3 and Dppa3 (hypergeometric test; p = 0.023). Dark green, group of low Dppa3 expression; light green, group of high Dppa3 expression. TBX3-low-sorted single cell is shown (GFP-L no. X). TBX3-high-sorted single cell is shown (GFP-H no. X).
(F) TBX3 occupancy (GSE19219) and H3K4me3/H3K27me3 bivalent modifications (GSE31039) of DPPA3 promoter region in ESCs.
(G) Dppa3 expression in WT or TBX3−/− iPSCs upon transduction with scramble control virus or various combinations of different shRNAs targeting Dppa3. Two independent experiments performed with triplicate technical replicates are shown.
(H) Colony formation of TBX3−/− iPSC lines with shRNAs against Dppa3 upon clonal density seeding. Top, representative AP-stained plates; bottom, quantification of three independent experiments performed with triplicate technical replicates. Black bar, AP positive; grey bar, partially AP positive; light gray bar, AP negative.
(I) qPCR of pluripotency markers in Dppa3 knockdown and scramble control TBX3−/− iPSC lines (three independent experiments performed with duplicate technical replicates).
(J) Comparisons of differentially regulated genes between TBX3-high and -low ESC and between TBX3 WT (+/+) and TBX3-null (ven/ven) ESCs.
(K and L) The GO terms of DNA methylation (GO0006306) show no bias toward TBX3-high or -low ESCs (K) but were significantly enriched in TBX3-null ESCs (L).
Discussion
A series of studies has reported that TBX3 was indispensable for maintenance of pluripotency. In line, TBX3 gain of function facilitated induction of pluripotency (Han et al., 2010a, Ivanova et al., 2006, Lee et al., 2012, Lu et al., 2011, Nishiyama et al., 2013) but interestingly also lineage commitment (Kartikasari et al., 2013, Lu et al., 2011, Weidgang et al., 2013). In line with a recent study (Waghray et al., 2015), we provide clear evidence that TBX3 is absolutely dispensable for both maintenance and induction of pluripotency. Additionally, we show that its removal alters pluripotency exit. In a wild-type context, TBX3 is heterogeneously expressed in the developing embryo in vivo but also in ESCs in vitro and various TBX3 expression levels correlate with distinct developmental potential.
Heterogeneous expression of pluripotency factors has been described and studied in detail for Nanog when cultured under LIF/FCS conditions. Single-cell resolution confirmed this observation and correlates a TF-low state with a more committed EPI-like state (Murayama et al., 2015, Papatsenko et al., 2015, Sugimoto et al., 2015). The advent of serum-free LIF/2i cultures, which virtually erases TF heterogeneity, questions this observation as a relic of outdated culture conditions, whereas others envision it as a stochastic advantage mimicking the dynamic fluctuations in the developing embryo (Martello and Smith, 2014, Torres-Padilla and Chambers, 2014). Though heterogeneity in vivo is established as early as in the four- to eight-cell stage (e.g., NANOG), reciprocal lineage patterning between NANOG/CDX2 occurs later (Dietrich and Hiiragi, 2007). These lineage segregation events have been defined into at least three rapid but sequential events in vivo (Schrode et al., 2013), whereas mESCs cultured under LIF/2i conditions are arguably more similar to the EPI cells found in diapause embryos (Abranches et al., 2014, Murayama et al., 2015, Papatsenko et al., 2015, Sugimoto et al., 2015). Therefore, heterogeneity found in mESCs cultured in LIF/FCS might be a closer reflection of the heterogeneity found in vivo in pre-implantation blastocysts. Notably, neither culture regime precisely recapitulates the in vivo situation (Macfarlan et al., 2012). In turn, most likely each cell culture system presents distinct advantages to study various facets of pluripotency and lineage commitment.
We complemented such in vitro observations with in vivo assays that allow to rigorously test the pluripotent potential of defined cell populations. In the context of wild-type mESCs, expression of Tbx3 inversely correlates with the capacity to form chimeric embryos and thereby supports the hypothesis that TBX3-low cells resemble committed EPI-like cells (Papatsenko et al., 2015). Moreover, we show that, even after cross-platform adjustment, the transcriptome of our TBX3-high cells converges on that of the ICM, whereas TBX3-low cells better resemble the E5.5/6.5 EPI.
Our data identify dynamic fluctuations in TBX3-expression levels to be linked with developmental potency of PSCs and hint that, within the heterogeneous TBX3 expression found in the late blastocyst, TBX3-high correlates with a Nanog-low state and thus PE cell fate specification (Niwa et al., 2009, Xenopoulos et al., 2015). This was independently confirmed by re-analyzing RNA sequencing data of single embryonic cells in pre-implantation embryos. Interestingly, a recent report described occasional transitions in NANOG expression in vivo from the PE back to the EPI (Xenopoulos et al., 2015). It would be of interest in the future to perform live-time imaging studies to track the fate of various TBX3 states to their progeny in pre- and postimplantation embryonic stages.
We were able to generate TBX3-null PSCs showing all hallmarks of pluripotency. This strongly contradicts several previous reports (Han et al., 2010a, Ivanova et al., 2006, Lee et al., 2012, Lu et al., 2011, Nishiyama et al., 2013) but supports a very recent study (Waghray et al., 2015). Additionally, it seems to be in conflict with our findings that TBX3-low cells represent a differentiation-poised late EPI state. A series of reports revealed strong variability between different EPI lines with various degrees of lineage bias and developmental potential (Bernemann et al., 2011, Han et al., 2010b, Murayama et al., 2015, Sugimoto et al., 2015). Nevertheless, germline transmission or even the generation of all-PSC mice upon 4n embryo aggregation has never been reported for EpiSCs (Bernemann et al., 2011, Han et al., 2010b). Intriguingly, recent studies implicated that the pluripotency circuitry can be perturbed in both directions, either toward differentiation or to a pluripotency arrested state with a hampered differentiation capacity as shown for the reduction of OCT4 expression (Karwacki-Neisius et al., 2013, Radzisheuskaya et al., 2013). We observe similar behaviors in TBX3-null cells. These cells not only keep their undifferentiated state longer than wild-type counterparts upon LIF/2i removal but also outperform wild-type cells in a competition assay. They also successfully formed all-PSC-derived mouse embryos in a 4n assay displaying the reported TBX3-null phenotypes (Davenport et al., 2003, Frank et al., 2013).
Previous studies identified TBX3 as an important driver and enhancer for iPSC formation (Han et al., 2010a). In our hands, reprogramming of TBX3-null MEFs occurred with similar efficiencies and kinetics compared to wild-type MEFs. Expanded clones retain the full ground state pluripotency. Interestingly, normal germline development occurred in TBX3-null mice, although it seems to be essential for the maintenance of SSCs after birth based on our observation using IVF with sperm from a TBX3-null iPSC chimeric mouse. The TBX2 subfamily (TBX2, 3, 4, and 5) has been previously implicated in the development of the reproductive system (Douglas et al., 2012), and overexpressing TBX3 with the Yamanaka factors generated iPSCs with higher capacity for germline contribution (Han et al., 2010a). TBX3 is only expressed in genital ducts, and deletion of TBX4, a close family member of TBX3 (Bertolessi et al., 2015), led to a reduced amount of primordial germ cells but with subsequent normal fertility (Douglas et al., 2013). Further studies need to clarify the precise role of TBX3 for germ cell development, particularly in light of the intimate connection between DPPA3/STELLA and TBX3 (Douglas et al., 2013, Waghray et al., 2015). Thus, TBX3 supports but is not absolutely required to maintain and install pluripotency and germ cell development, probably due to an alternate pluripotency network. Transcriptome analysis and downstream bioinformatical analysis of TBX3-null PSCs helped to identify putative compensators revealing DPPA3 as the most promising candidate, in line with a recent study (Waghray et al., 2015). Although we confirmed an intimate connection between TBX3 and DPPA3, self-renewal of TBX3-null cells remained stable upon depletion of DPPA3. Most likely several lines of compensation may allow ongoing self-renewal in TBX3-null cells. In fact, Waghray et al. (2015) also observed just partial compensation by DPPA3 in the TBX3-null state. Most likely, differences in the genetic background or distinct cell culture conditions further impact the observed differences between our study and Waghray et al. Nevertheless, the intimate connection between TBX3 and DPPA3 remains and needs to be explored in detail in future studies.
Our observations of an enriched DNA-methylation signature only in TBX3-null and not in the TBX3-low cells indicate additional epigenetic mechanisms that may be implicated and warrant further investigation (Figure 5L). Notably, DPPA3 and DNMT3a were found to be associated during cellular reprogramming and DNA methyltransferases are critical for both genome integrity and differentiation capacity of mESCs (Baubec et al., 2015, Xu et al., 2015). Moreover, OCT4 haploinsufficiency stabilizes the pluripotency circuitry not only at the transcriptional level but also at the epigenetic level via higher occupancy of key regulatory elements supporting pluripotency (Karwacki-Neisius et al., 2013, Radzisheuskaya et al., 2013). Taken together, we have deciphered further aspects of TBX3 activity within the pluripotency circuitry and identify variable pluripotency states, which underpin the dual role of pluripotency for both indefinite self-renewal and cell lineage specification.
Experimental Procedures
Full methods accompany this paper.
Ethics Statement
All animal experiments were performed in compliance with the guidelines for the welfare of experimental animals issued by the Federal Government of Germany, the NIH, and the Max Planck Society. The experiments in this study were approved by the review board of the Land Baden-Wuerttemberg and the ethics committee of Ulm University.
Reprogramming and PSC Culture
MEFs were isolated and reprogrammed according to standard protocols from TBX3+/− and TBX3−/− mice (Kleger et al., 2012).
Isolation of mESCs from Blastocysts
TBX3-BAC-EGFP mESCs and TBX3-KI-Venus mESCs were derived using previously reported methods by breeding TBX3-BAC-EGFP and TBX3-KI-Venus heterozygous mice, respectively (Bryja et al., 2006).
Western Blot
Western blotting was performed according to standard procedures (Linta et al., 2012). Whole-cell extracts (50–100 μg) prepared using RIPA lysis buffer containing 50 mM Tris HCl (pH 8.0), 150 mM NaCl, 0.05% Na-Deoxycholat, 1% NP40, 0.01% SDS, and 1 mM PMSF supplemented with Complete Protease Inhibitor Cocktail Tablets (Roche) were subjected to SDS-PAGE.
IHC
IHC was performed as described in Russell et al. (2015) and Stockmann et al. (2013). Cells at different time points of differentiation were fixed using 4% paraformaldehyde (PFA). Nuclei were stained with DAPI. Images were captured using an upright fluorescence Zeiss Axioimager Z1 microscope and analyzed using Axiovision software (Zeiss).
Microarray Data Analysis
The DNA microarray data were pre-processed and normalized by the limma package (Wettenhall and Smyth, 2004). The differentially expressed genes of a pairwise comparison were detected using limma t test with criteria of fold change >2 and p value < 0.05. The p value was further adjusted by the procedure of Benjamini and Hochberg (Klipper-Aurbach et al., 1995).
Author Contributions
Study concept and design, A.K., A.I., and S.L.; acquisition of the main part of experimental data, R.R., M.I., G.W., and Q.L.; experimental support, L.L., M.H., M.K., S.R., W.B., and O.S.; microarray analysis, Q.L., M.Z., and A.K.; 2n/4n embryo aggregation, G.W. and H.S.; TBX3−/− mice and MEF cultures, P.K.P. and A.M.; single-cell analysis, S.R. and M.K.; analysis and interpretation of data, A.K., A.I., R.R., Q.L., and S.L.; drafting of the manuscript, A.K., A.I., and S.L.; critical revision of the manuscript for important intellectual content, R.R., Q.L., G.W., A.L., S.R., K.A., A.M., M.Z., A.I., S.L., and A.K.; study supervision: A.I., A.K., and S.L.
Acknowledgments
The authors thank Ralf Köhntop and Sabine Conrad for excellent technical assistance and V. Christoffels for providing TBX3venus/venus and TBX3-BAC-EGFP mice. This study was funded by the Deutsche Forschungsgemeinschaft (DFG, K.L. 2544/1-1, S.L. BO1718/4-1, M.Z. ZE432/5-2), Forschungskern SyStaR to A.K., BIU (Böhringer Ingelheim Ulm to A.K.), Fortüne (Faculty of Medicine Tübingen to L.L.), and Else-Kröner-Fresenius-Stiftung (2011_A200; to A.K. and S.L.). A.K. is indebted to the Baden-Württemberg Stiftung for the financial support of this research project by the Eliteprogramme for Postdocs. A.K. is an Else-Kröner-Fresenius Memorial Fellow.
Published: December 8, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, five figures, and eight tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2015.11.003.
Accession Numbers
The accession number for the microarray data reported in this paper is GEO: GSE73862.
Supplemental Information
References
- Abranches E., Guedes A.M., Moravec M., Maamar H., Svoboda P., Raj A., Henrique D. Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency. Development. 2014;141:2770–2779. doi: 10.1242/dev.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker M.L., Boukens B.J., Mommersteeg M.T., Brons J.F., Wakker V., Moorman A.F., Christoffels V.M. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Banito A., Rashid S.T., Acosta J.C., Li S., Pereira C.F., Geti I., Pinho S., Silva J.C., Azuara V., Walsh M. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T., Colombo D.F., Wirbelauer C., Schmidt J., Burger L., Krebs A.R., Akalin A., Schübeler D. Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature. 2015;520:243–247. doi: 10.1038/nature14176. [DOI] [PubMed] [Google Scholar]
- Bernemann C., Greber B., Ko K., Sterneckert J., Han D.W., Araúzo-Bravo M.J., Schöler H.R. Distinct developmental ground states of epiblast stem cell lines determine different pluripotency features. Stem Cells. 2011;29:1496–1503. doi: 10.1002/stem.709. [DOI] [PubMed] [Google Scholar]
- Bertolessi M., Linta L., Seufferlein T., Kleger A., Liebau S. A fresh look on T-box factor action in early embryogenesis (T-box factors in early development) Stem Cells Dev. 2015;24:1833–1851. doi: 10.1089/scd.2015.0102. [DOI] [PubMed] [Google Scholar]
- Blakeley P., Fogarty N.M., Del Valle I., Wamaitha S.E., Hu T.X., Elder K., Snell P., Christie L., Robson P., Niakan K.K. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. 2015;142:3151–3165. doi: 10.1242/dev.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryja V., Bonilla S., Arenas E. Derivation of mouse embryonic stem cells. Nat. Protoc. 2006;1:2082–2087. doi: 10.1038/nprot.2006.355. [DOI] [PubMed] [Google Scholar]
- Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen Y., Blair K., Smith A. Robust self-renewal of rat embryonic stem cells requires fine-tuning of glycogen synthase kinase-3 inhibition. Stem Cell Reports. 2013;1:209–217. doi: 10.1016/j.stemcr.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechanski A., Byers C., Greenstein I., Schrode N., Donahue L.R., Hadjantonakis A.K., Reinholdt L.G. Derivation and characterization of mouse embryonic stem cells from permissive and nonpermissive strains. Nat. Protoc. 2014;9:559–574. doi: 10.1038/nprot.2014.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport T.G., Jerome-Majewska L.A., Papaioannou V.E. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- Deng Q., Ramsköld D., Reinius B., Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- Dietrich J.E., Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Douglas N.C., Heng K., Sauer M.V., Papaioannou V.E. Dynamic expression of Tbx2 subfamily genes in development of the mouse reproductive system. Dev. Dyn. 2012;241:365–375. doi: 10.1002/dvdy.23710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas N.C., Arora R., Chen C.Y., Sauer M.V., Papaioannou V.E. Investigating the role of tbx4 in the female germline in mice. Biol. Reprod. 2013;89:148. doi: 10.1095/biolreprod.113.107649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S.J., Martello G., Yordanov B., Emmott S., Smith A.G. Defining an essential transcription factor program for naïve pluripotency. Science. 2014;344:1156–1160. doi: 10.1126/science.1248882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faddah D.A., Wang H., Cheng A.W., Katz Y., Buganim Y., Jaenisch R. Single-cell analysis reveals that expression of nanog is biallelic and equally variable as that of other pluripotency factors in mouse ESCs. Cell Stem Cell. 2013;13:23–29. doi: 10.1016/j.stem.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., Wong F., Yates A., Tomlinson S.R., Chambers I. Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell. 2012;11:477–490. doi: 10.1016/j.stem.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.U., Emechebe U., Thomas K.R., Moon A.M. Mouse TBX3 mutants suggest novel molecular mechanisms for Ulnar-mammary syndrome. PLoS ONE. 2013;8:e67841. doi: 10.1371/journal.pone.0067841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum T., Halbisen M.A., Wang C., Amiri H., Robson P., Ralston A. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev. Cell. 2013;25:610–622. doi: 10.1016/j.devcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertsenstein M., Nutter L.M., Reid T., Pereira M., Stanford W.L., Rossant J., Nagy A. Efficient generation of germ line transmitting chimeras from C57BL/6N ES cells by aggregation with outbred host embryos. PLoS ONE. 2010;5:e11260. doi: 10.1371/journal.pone.0011260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Huss M., Tong G.Q., Wang C., Li Sun L., Clarke N.D., Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev. Cell. 2010;18:675–685. doi: 10.1016/j.devcel.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Han J., Yuan P., Yang H., Zhang J., Soh B.S., Li P., Lim S.L., Cao S., Tay J., Orlov Y.L. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Joo J.Y., Greber B., Araúzo-Bravo M.J., Bernemann C., Ko K., Wu G., Stehling M., Do J.T., Schöler H.R. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Lopes S.M., Tang F., Surani M.A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthuis T., Buermans H.P., Brons J.F., Verkerk A.O., Bakker M.L., Wakker V., Clout D.E., Moorman A.F., ’t Hoen P.A., Christoffels V.M. Gene expression profiling of the forming atrioventricular node using a novel tbx3-based node-specific transgenic reporter. Circ. Res. 2009;105:61–69. doi: 10.1161/CIRCRESAHA.108.192443. [DOI] [PubMed] [Google Scholar]
- Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Kalmar T., Lim C., Hayward P., Muñoz-Descalzo S., Nichols J., Garcia-Ojalvo J., Martinez Arias A. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartikasari A.E., Zhou J.X., Kanji M.S., Chan D.N., Sinha A., Grapin-Botton A., Magnuson M.A., Lowry W.E., Bhushan A. The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 2013;32:1393–1408. doi: 10.1038/emboj.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacki-Neisius V., Göke J., Osorno R., Halbritter F., Ng J.H., Weiße A.Y., Wong F.C., Gagliardi A., Mullin N.P., Festuccia N. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell. 2013;12:531–545. doi: 10.1016/j.stem.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleger A., Mahaddalkar P.U., Katz S.F., Lechel A., Joo J.Y., Loya K., Lin Q., Hartmann D., Liebau S., Kraus J.M. Increased reprogramming capacity of mouse liver progenitor cells, compared with differentiated liver cells, requires the BAF complex. Gastroenterology. 2012;142:907–917. doi: 10.1053/j.gastro.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Klipper-Aurbach Y., Wasserman M., Braunspiegel-Weintrob N., Borstein D., Peleg S., Assa S., Karp M., Benjamini Y., Hochberg Y., Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Med. Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Kumar P.P., Emechebe U., Smith R., Franklin S., Moore B., Yandell M., Lessnick S.L., Moon A.M. Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. eLife. 2014;3:e02805. doi: 10.7554/eLife.02805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunasegaran K., Ho V., Chang T.H., De Silva D., Bakker M.L., Christoffels V.M., Pietersen A.M. Transcriptional repressor Tbx3 is required for the hormone-sensing cell lineage in mammary epithelium. PLoS ONE. 2014;9:e110191. doi: 10.1371/journal.pone.0110191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek D., Neagu A., Tastemel M., Tüysüz N., Lehmann J., van de Werken H.J., Philipsen S., van der Linden R., Maas A., van IJcken W.F. Endogenous WNT signals mediate BMP-induced and spontaneous differentiation of epiblast stem cells and human embryonic stem cells. Stem Cell Reports. 2015;4:114–128. doi: 10.1016/j.stemcr.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.F., Su J., Sevilla A., Gingold J., Schaniel C., Lemischka I.R. Combining competition assays with genetic complementation strategies to dissect mouse embryonic stem cell self-renewal and pluripotency. Nat. Protoc. 2012;7:729–748. doi: 10.1038/nprot.2012.018. [DOI] [PubMed] [Google Scholar]
- Leitch H.G., Nichols J., Humphreys P., Mulas C., Martello G., Lee C., Jones K., Surani M.A., Smith A. Rebuilding pluripotency from primordial germ cells. Stem Cell Reports. 2013;1:66–78. doi: 10.1016/j.stemcr.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linta L., Stockmann M., Kleinhans K.N., Böckers A., Storch A., Zaehres H., Lin Q., Barbi G., Böckers T.M., Kleger A., Liebau S. Rat embryonic fibroblasts improve reprogramming of human keratinocytes into induced pluripotent stem cells. Stem Cells Dev. 2012;21:965–976. doi: 10.1089/scd.2011.0026. [DOI] [PubMed] [Google Scholar]
- Lu R., Yang A., Jin Y. Dual functions of T-box 3 (Tbx3) in the control of self-renewal and extraembryonic endoderm differentiation in mouse embryonic stem cells. J. Biol. Chem. 2011;286:8425–8436. doi: 10.1074/jbc.M110.202150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur B.D., Sevilla A., Lenz M., Müller F.J., Schuldt B.M., Schuppert A.A., Ridden S.J., Stumpf P.S., Fidalgo M., Ma’ayan A. Nanog-dependent feedback loops regulate murine embryonic stem cell heterogeneity. Nat. Cell Biol. 2012;14:1139–1147. doi: 10.1038/ncb2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T.S., Gifford W.D., Driscoll S., Lettieri K., Rowe H.M., Bonanomi D., Firth A., Singer O., Trono D., Pfaff S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Smith A. The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Miyanari Y., Torres-Padilla M.E. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Müller F.J., Laurent L.C., Kostka D., Ulitsky I., Williams R., Lu C., Park I.H., Rao M.S., Shamir R., Schwartz P.H. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama H., Masaki H., Sato H., Hayama T., Yamaguchi T., Nakauchi H. Successful reprogramming of epiblast stem cells by blocking nuclear localization of β-catenin. Stem Cell Reports. 2015;4:103–113. doi: 10.1016/j.stemcr.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Sharov A.A., Piao Y., Amano M., Amano T., Hoang H.G., Binder B.Y., Tapnio R., Bassey U., Malinou J.N. Systematic repression of transcription factors reveals limited patterns of gene expression changes in ES cells. Sci. Rep. 2013;3:1390. doi: 10.1038/srep01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Ogawa K., Shimosato D., Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D., Darr H., Kulakovskiy I.V., Waghray A., Makeev V.J., MacArthur B.D., Lemischka I.R. Single-cell analyses of ESCs reveal alternative pluripotent cell states and molecular mechanisms that control self-renewal. Stem Cell Reports. 2015;5:207–220. doi: 10.1016/j.stemcr.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payer B., Chuva de Sousa Lopes S.M., Barton S.C., Lee C., Saitou M., Surani M.A. Generation of stella-GFP transgenic mice: a novel tool to study germ cell development. Genesis. 2006;44:75–83. doi: 10.1002/gene.20187. [DOI] [PubMed] [Google Scholar]
- Piras V., Tomita M., Selvarajoo K. Transcriptome-wide variability in single embryonic development cells. Sci. Rep. 2014;4:7137. doi: 10.1038/srep07137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzisheuskaya A., Chia Gle.B., dos Santos R.L., Theunissen T.W., Castro L.F., Nichols J., Silva J.C. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat. Cell Biol. 2013;15:579–590. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R., Perkhofer L., Liebau S., Lin Q., Lechel A., Feld F.M., Hessmann E., Gaedcke J., Güthle M., Zenke M. Loss of ATM accelerates pancreatic cancer formation and epithelial-mesenchymal transition. Nat. Commun. 2015;6:7677. doi: 10.1038/ncomms8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrode N., Xenopoulos P., Piliszek A., Frankenberg S., Plusa B., Hadjantonakis A.K. Anatomy of a blastocyst: cell behaviors driving cell fate choice and morphogenesis in the early mouse embryo. Genesis. 2013;51:219–233. doi: 10.1002/dvg.22368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockmann M., Linta L., Föhr K.J., Boeckers A., Ludolph A.C., Kuh G.F., Udvardi P.T., Proepper C., Storch A., Kleger A. Developmental and functional nature of human iPSC derived motoneurons. Stem Cell Rev. 2013;9:475–492. doi: 10.1007/s12015-011-9329-4. [DOI] [PubMed] [Google Scholar]
- Sugimoto M., Kondo M., Koga Y., Shiura H., Ikeda R., Hirose M., Ogura A., Murakami A., Yoshiki A., Chuva de Sousa Lopes S.M., Abe K. A simple and robust method for establishing homogeneous mouse epiblast stem cell lines by wnt inhibition. Stem Cell Reports. 2015;4:744–757. doi: 10.1016/j.stemcr.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Teo A.K., Arnold S.J., Trotter M.W., Brown S., Ang L.T., Chng Z., Robertson E.J., Dunn N.R., Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Padilla M.E., Chambers I. Transcription factor heterogeneity in pluripotent stem cells: a stochastic advantage. Development. 2014;141:2173–2181. doi: 10.1242/dev.102624. [DOI] [PubMed] [Google Scholar]
- Toyooka Y., Shimosato D., Murakami K., Takahashi K., Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Waghray A., Saiz N., Jayaprakash A.D., Freire A.G., Papatsenko D., Pereira C.F., Lee D.F., Brosh R., Chang B., Darr H. Tbx3 controls Dppa3 levels and exit from pluripotency toward mesoderm. Stem Cell Reports. 2015;5:97–110. doi: 10.1016/j.stemcr.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidgang C.E., Russell R., Tata P.R., Kühl S.J., Illing A., Müller M., Lin Q., Brunner C., Boeckers T.M., Bauer K. TBX3 directs cell-fate decision toward mesendoderm. Stem Cell Reports. 2013;1:248–265. doi: 10.1016/j.stemcr.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettenhall J.M., Smyth G.K. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Smith A.G. The ground state of pluripotency. Biochem. Soc. Trans. 2010;38:1027–1032. doi: 10.1042/BST0381027. [DOI] [PubMed] [Google Scholar]
- Xenopoulos P., Kang M., Puliafito A., Di Talia S., Hadjantonakis A.K. Heterogeneities in Nanog expression drive stable commitment to pluripotency in the mouse blastocyst. Cell Rep. 2015;10:1508–1520. doi: 10.1016/j.celrep.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Smorag L., Nakamura T., Kimura T., Dressel R., Fitzner A., Tan X., Linke M., Zechner U., Engel W., Pantakani D.V. Dppa3 expression is critical for generation of fully reprogrammed iPS cells and maintenance of Dlk1-Dio3 imprinting. Nat. Commun. 2015;6:6008. doi: 10.1038/ncomms7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.