Summary
Histone methylation is involved in the regulation of many cellular processes. In the past two years, several histone demethylases including BHC110/LSD1 have been characterized. BHC110, the first known histone lysine demethylase, removes methyl groups from methylated histone H3 lysine 4 and has been found in many multi-protein complexes. Using one-step affinity purification, we have isolated enzymatically active BHC110-containing complexes. Here, we detail the methods used for the isolation and characterization of these histone demethylase complexes from a human stable cell line.
Keywords: histone methylation, histone demethylase, protein complex, BHC110/LSD1
Introduction
Histone methylation is a form of posttranslational histone modification that plays an important role in chromatin organization, genomic imprinting and transcriptional regulation (Kouzarides, 2002; Margueron et al., 2005; Martin and Zhang, 2005). This modification occurs at lysine (K) and arginine residues. While arginine methylation generally correlates with gene activation (Kouzarides, 2002), lysine methylation is linked to gene activation or repression, depending on the lysine methylated. Generally, methylation at H3K4, H3K36 and H3K79 residues denotes gene activation, whereas methylation at H3K9, H3K27 and H4K20 residues signifies repression (Martin and Zhang, 2005; Sims et al., 2003). In addition, these lysine residues can be mono-, di- or tri-methylated.
Though histone methylation was originally considered irreversible, recent studies have demonstrated that it can be reversed by several histone demethylases including PAD4/PADI4, BHC110/LSD1 and a new family of JmjC domain-containing histone demethylases (Cloos et al., 2006; Cuthbert et al., 2004; Fodor et al., 2006; Klose et al., 2006; Shi et al., 2004; Tsukada et al., 2006; Wang et al., 2004; Whetstine et al., 2006; Yamane et al., 2006). In particular, BHC110/LSD1 belongs to a family of FAD-dependent polyamine oxidases and removes methyl groups from mono- or di-methyl H3K4 by an oxidation-based mechanism (Shi et al., 2004). It should be noted that dimethyl H3K4 is associated with actively transcribed genes while monomethyl H3K4 is enriched at the end of genes in yeast.
BHC110 was found to be a component of several multiprotein complexes (Ballas et al., 2001; Eimer et al., 2002; Hakimi et al., 2002; Humphrey et al., 2001; Jarriault and Greenwald, 2002; Shi et al., 2003; Tong et al., 1998; You et al., 2001) including BHC (BRAF-HDAC Complex). BHC was isolated as a multiprotein complex containing histone deacetylase 1 and 2 (Hakimi et al., 2002; Marmorstein et al., 2001), and has been shown to mediate silencing of neuron-specific genes by REST (RE1-silencing transcription factor) in differentiated non-neuronal cells (Hakimi et al., 2002). Subsequently, we isolated this complex using a two-step purification involving P11 column fractionation followed by an affinity chromatography using antibodies to BHC110 (Hakimi et al., 2003). More recently, we developed an epitope-tagged stable cell line expressing Flag-BHC110 that allowed the isolation of BHC110-containing complexes through a single-step affinity-purification (Lee et al., 2005). Importantly, we have demonstrated that this anti-FLAG affinity-purified complex is enzymatically active and that the subunit composition of the complex is nearly identical to that of the complexes isolated by other chromatographic methods. In this chapter, we describe protocols used for the purification of the histone demethylase complexes and the analysis of their enzymatic activities.
Materials and Reagents
Plasmid: pFLAG-CMV2 (Sigma) vector containing BHC110 cDNA.
Media: Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS, antibiotics/antimycotics, 2 mM glutamine and 2.5 μg/ml puromycin.
Affinity purification: Anti-FLAG M2-agarose affinity resin (Sigma A-2220), FLAG peptide (5 μg/μl, 10 × stock solution, Sigma), 10 ml poly-prep chromatography column (BioRad)
Wash and assay buffers: BC250 or BC500 (20 mM Tris (pH 8.0), 250 or 500 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10% Glycerol, 0.2 mM PMSF, 1 mM DTT, 1 μg/ml Aprotinin, 2.5 μg/ml Leupeptin, and 1 μg/ml Pepstatin (Roche)), histone demethylase (HDM) assay buffer A (50 mM Tris pH 8.5, 50 mM KCl, 5 mM MgCl2, 5% glycerol, 0.2 mM PMSF and 1 mM DTT).
TCA precipitation: TCA (100% w/v), ice-cold acetone
Enzyme substrates: dimethyl H3K4 peptide (Upstate), bulk histones (Sigma H9250), nucleosomes purified from HeLa nuclear pellet as previously described (Barak et al., 2003).
Electrophoresis: 1× gel loading buffer (62.5 mM Tris (pH 6.8), 5% β-mercaptoethanol, 2% SDS, 10% glycerol, 0.01% Bromophenol blue), 1× gel running buffer (25 mM Tris (pH 8.3), 190 mM glycine, and 0.1% SDS), precast 4–20% Tris-glycine SDS-polyacrylamide gel (Invitrogen).
Western transfer and blotting: 1× Western transfer buffer (25 mM Tris (pH 8.3), 190 mM glycine, and 20% methanol), Polyvinylidene fluoride membrane (PVDF, Immobilon-P, Millipore), 1× TTBS (25 mM Tris (pH 7.4), 150 mM NaCl, and 0.05% Tween 20).
Antibodies: Anti-dimethyl H3K4 (Upstate 12–460), anti-monomethyl H3K4 (Upstate 07–436), anti-acetyl H3 K9/K14 (Upstate 06–599), anti-H3 antibody (Abcam ab1791), and anti-FLAG M2 antibody (Sigma F3165).
Western developing solution: 10 ml alkaline phosphatase (AP) buffer (100 mM Tris-HCl (9.5), 100 mM NaCl, 5 mM MgCl2) + 33 μl of BCIP (50 mg/ml 5-Bromo-4-Chloro-3-Indolyl phosphate p-Toluidine salt in 100% N, N-dimethylformamide, final concentration = 165 μg/ml buffer) + 66 μl of NBT (50 mg/ml Nitro-Blue Tetrazolium Chloride in 70% N, N-dimethylformamide, final concentration =330 μg/ml buffer)
Affinity Purification of BHC
As the first step for anti-FLAG affinity purification of BHC, we generated a stable cell line expressing FLAG-tagged BHC110. In brief, we cotransfected HEK293 cells with two plasmids; one encodes FLAG-BHC110 while the other contains a puromycin resistant gene. We used puromycin selection rather than the commonly used neomycin selection because puromycin is more toxic to parent cells and less expensive. Each stable cell line should be generated from a monoclonal population. After expansion of stable clones, we screened more than 20 clones by small-scale immunoprecipitation of whole cell extracts from 10 million cells and then by checking eluates by Western blot analysis. We selected a positive clone expressing the highest level of BHC110. A more detailed protocol for generating stable cell lines has been previously described (Barak and Shiekhattar, 2004).
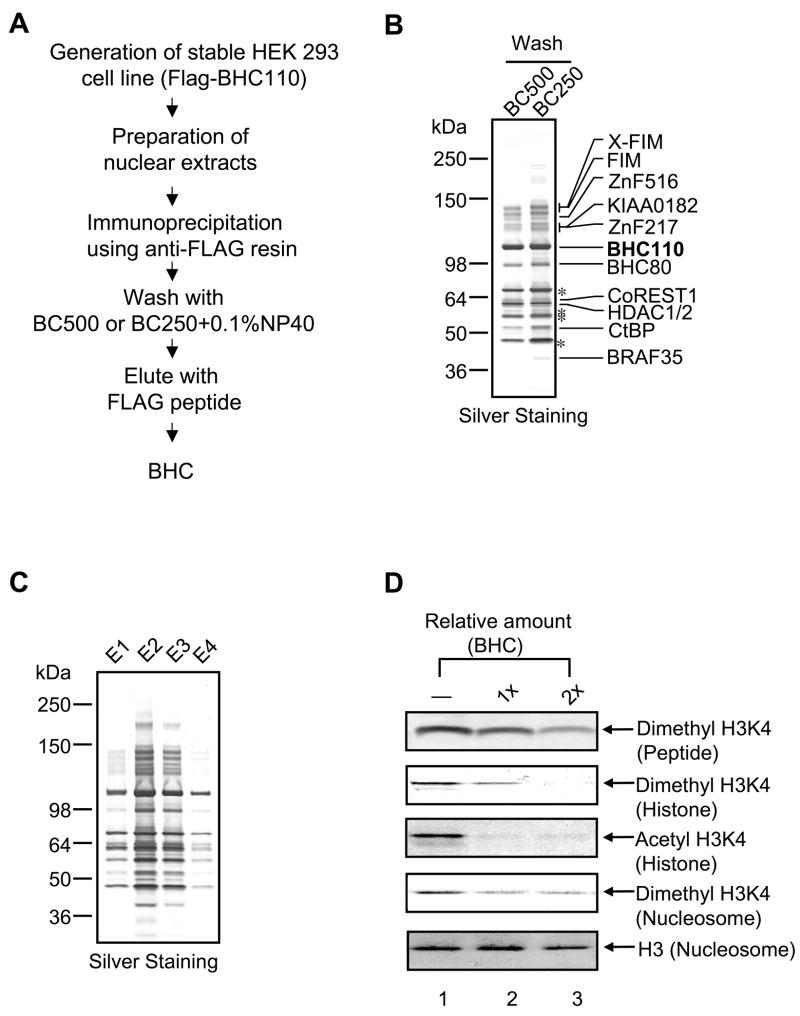
Nuclear extracts (150–200 mg) were isolated from the stable cell line. In brief, the stable cell line expressing FLAG-BHC110 was expanded to 100 × 15cm tissue culture dishes. Cells were harvested at confluence, collected by centrifugation, and rinsed using PBS. Nuclear extracts were prepared and dialyzed against BC50 containing 50 mM KCl. The resulting nuclear extracts were mixed with anti-FLAG M2 affinity resin. After anti-FLAG beads were washed extensively, BHC was eluted from the beads and analyzed by silver staining and Western blotting. Here we used two wash methods involving extensive washes with BC250 or BC500 containing 0.1% NP40. We have found that although BHC isolated by both wash methods shared almost identical subunits, extensive washes with BC250 resulted in better enrichment of BHC110-associated polypeptides while maximizing complex recovery and minimizing contaminant polypeptides (Fig. 1B). BHC110-associated polypeptides were identified by liquid chromatography–tandem mass spectroscopy as previously described (Lee et al., 2005) (Fig. 1B and 1C). The amount of BHC110 in the complex was deduced by comparing bands on a silver-stained gel with known amounts of BSA.
Fig. 1. Isolation of BHC by FLAG-affinity purification and analysis of the complex activity.
(A) Schematic representation of BHC isolation. (B) Silver staining of BHC following extensive washes with BC500 or BC250 containing 0.1% NP40. (C) A typical elution profile of BHCE. 1, the first eluate; E2, the second eluate; E3, the third eluate; E4, the fourth eluate. (D) Evaluation of enzymatic activity of BHC. Peptide, histones or nucleosomes were used as substrates, and reaction mixtures were analyzed by SDS-PAGE, followed by Western blotting. H3 bands serve as internal controls to monitor variation in preparation and gel analysis of reaction mixtures. “1×” corresponds to 100–125 ng of BHC110 in the complex. Acetyl H3 represents acetyl H3 K9/K14.
Protocol
Add anti-FLAG M2 affinity resin to the nuclear extracts. Use 0.5 ml of 50% slurry per 100 mg of nuclear extracts. Beads (0.5 ml) should be washed at least once with 20–40 resin bed volumes (10 ml) of PBS prior to immunoprecipitation.
Incubate for 2 – 5 h with rotation. Longer incubation time such as overnight may increase nonspecific background without increasing protein yield.
Pellet the beads by centrifugation at 200g for 5 min in a refrigerated centrifuge. Save the unbound fraction by transferring the supernatant to a new tube and store it at − 80°C.
Add 20–40 resin bed volumes of BC250 or BC500 containing 0.1% NP40 to beads and mix gently by inverting by hand.
Pellet the beads by centrifugation at 200g for 5 min in a refrigerated centrifuge and aspirate the supernatant.
Repeat steps 4 and 5 four times.
Add 20–40 resin bed volumes of HDM assay buffer A to beads and mix gently by inverting with hand.
Pellet the beads by centrifugation at 200g for 5 min in a refrigerated centrifuge and aspirate the supernatant.
Transfer beads onto a 3 ml poly-prep chromatography column (BioRad) and equilibrate beads by passing with 10 resin bed volume of HDM assay buffer A at gravimetric flow rate.
FLAG-BHC110-containing complex can now be eluted using FLAG peptide. Add one resin bed volume of HDM assay buffer A supplemented with 0.4 μg/μl FLAG peptide and collect the first eluate
Add one resin bed volume of HDM assay buffer A supplemented with 0.4 μg/μl FLAG peptide and cap the column. Mix beads, incubate at 4 °C for 10 min, and collect the second eluate.
For the third and fourth eluate, add one resin bed volume of HDM assay buffer A supplemented with 0.4 μg/μl FLAG peptide and collect eluates.
Eluted fractions should be analyzed for purity by 4–12% gradient PAGE followed by silver staining and Western blotting.
Demethylation Assay
In the assay, the complex was incubated with various substrates such as peptide, bulk histones, mono- or poly- nucleosomes. Since we eluted BHC in the assay buffer, the complex could be directly used for the assay without dialysis. Moreover, we prepared all substrates in the assay buffer to directly use for the assay. Importantly, we were able to assess not only histone demethylation activity but also HDAC activity under the same assay conditions (Lee et al., 2005; Lee et al., 2006). It should be noted that this complex contains two enzymatic subunits: the histone demethylase BHC110 and the histone deacetylases HDAC1/2 (Hakimi et al., 2003). The reaction mixture was analyzed by SDS-PAGE, followed by Western blotting. Antibodies against di- (or mono-) methyl H3K4 and acetyl H3-K9/K14 were used to detect methylation and acetylation levels, respectively. Demethylation and deacetylation activities were monitored by observing decreases of band intensities compared to control bands (Fig. 1D).
Protocol
Prepare 2 μg/μl bulk histones in HDM assay buffer A. Dialyze nucleosomes against HDM assay buffer A. Prepare mononucleosomes by treating nucleosomes with micrococcal nuclease according to manufacturer’s instructions.
Mix peptide (0.1 μg) or bulk histones (4 μg) with 0 – 200 ng of BHC110 in the complex in the histone demethylase (HDM) assay buffer A in a final volume of 10 μl. For polynucleosomes (0.3 μg) or mononucleosomes (0.3 μg), supplement NP40 at a final concentration of 0.1%.
Incubate for 4 –16 h at 37°C.
Add 10 μl of 2 × gel loading buffer and 16 μl of 1 × gel loading buffer, mix by vortexing, and heat-denature at 95°C for 3 min.
Load 4 μl mixture for peptide and bulk histone (8 μl mixture for mono- and poly-nucleosomes) onto 4–20 % SDS-PAGE gel and run gel until the bromophenol blue dye front reaches two third of the total gel length.
Electrotransfer the proteins onto a PVDF membrane at 4°C for 2 h at 100 volts. After transfer, block membranes with 5% (w/v) nonfat dry milk for 30 – 60 min at room temperature (RT) and rinse in TTBS.
Incubate with a 1:500 dilution of anti- di-(or mono-) methyl K4-H3 or acetyl K9/K14-H3 antibodies for 1 h at RT and wash with TTBS for 5 min four times.
Incubate with a 1:5,000 dilution of calf intestinal AP-conjugated, anti-rabbit IgG secondary antibody for 1 h at RT and wash with TTBS for 5 min four times.
Visualize the protein bands (purple bands) by incubating membranes in AP buffer containing 165 μg/ml BCIP and 330 μg/ml NBT. Stop the developing reaction by rinsing with water.
Acknowledgments
R.S. was supported by a grant from NIH (GM61204).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, Rose DW, Rosenfeld MG, Brehm P, Mandel G. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak OG, Shiekhattar R. Preparation and assays for mammalian ISWI complexes. Methods Enzymol. 2004;377:389–401. doi: 10.1016/S0076-6879(03)77025-6. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T, Hansen KH, Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Eimer S, Lakowski B, Donhauser R, Baumeister R. Loss of spr-5 bypasses the requirement for the C. elegans presenilin sel-12 by derepressing hop-1. EMBO J. 2002;21:5787–5796. doi: 10.1093/emboj/cdf561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J Biol Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, Howard BH. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. 2001;276:6817–6824. doi: 10.1074/jbc.M007372200. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Greenwald I. Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans. Genes Dev. 2002;16:2713–2728. doi: 10.1101/gad.1022402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Marmorstein LY, Kinev AV, Chan GK, Bochar DA, Beniya H, Epstein JA, Yen TJ, Shiekhattar R. A human BRCA2 complex containing a structural DNA binding component influences cell cycle progression. Cell. 2001;104:247–257. doi: 10.1016/s0092-8674(01)00209-4. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Tong JK, Hassig CA, Schnitzler GR, Kingston RE, Schreiber SL. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998;395:917–921. doi: 10.1038/27699. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of Histone Lysine Trimethylation by the JMJD2 Family of Histone Demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST- human histone deacetylase complex. Proc Natl Acad Sci U S A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]